Published online Nov 28, 2023. doi: 10.3748/wjg.v29.i44.5919
Peer-review started: July 20, 2023
First decision: October 9, 2023
Revised: October 23, 2023
Accepted: November 14, 2023
Article in press: November 14, 2023
Published online: November 28, 2023
Processing time: 130 Days and 2.8 Hours
The role of Tousled-like kinase 1 (TLK1) in in gastric cancer (GC) remains unclear.
To investigate the expression, biological function, and underlying mechanisms of TLK1 in GC.
We measured TLK1 protein expression levels and localized TLK1 in GC cells and tissues by western blot and immunofluorescence, respectively. We transfected various GC cells with lentiviruses to create TLK1 overexpression and knockdown lines and established the functional roles of TLK1 through in vitro colony formation, 5-ethynyl-2`-deoxyuridine, and Transwell assays as well as flow cytometry. We applied bioinformatics to elucidate the signaling pathways associated with TLK1. We performed in vivo validation of TLK1 functions by inducing subcutaneous xenograft tumors in nude mice.
TLK1 was significantly upregulated in GC cells and tissues compared to their normal counterparts and was localized mainly to the nucleus. TLK1 knockdown significantly decreased colony formation, proliferation, invasion, and migration but increased apoptosis in GC cells. TLK1 overexpression had the opposite effects. Bioinformatics revealed, and subsequent experiments verified, that the tumor growth factor-beta signaling pathway was implicated in TLK1-mediated GC progression. The in vivo assays confirmed that TLK1 promotes tumorigenesis in GC.
The findings of the present study indicated that TLK1 plays a crucial role in GC progression and is, therefore, promising as a therapeutic target against this disease.
Core Tip: We demonstrated that Tousled-like kinase 1 (TLK1) is highly expressed in gastric cancer (GC), localized mainly to the nucleus, significantly promotes GC cell proliferation, invasion, and migration, and inhibits apoptosis. TLK1 may facilitate GC progression by modulating tumor growth factor-beta expression. We believe that TLK1 could be a crucial therapeutic target for GC, and propose that future investigations evaluate the feasibility and practicality of targeting TLK1 in GC treatment.
- Citation: Sun RC, Li J, Li YX, Wang HZ, Dal E, Wang ML, Li YX. Tousled-like kinase 1 promotes gastric cancer progression by regulating the tumor growth factor-beta signaling pathway. World J Gastroenterol 2023; 29(44): 5919-5934
- URL: https://www.wjgnet.com/1007-9327/full/v29/i44/5919.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i44.5919
Gastric cancer (GC) is the fifth most prevalent cancer and the third leading cause of cancer-related mortality worldwide[1]. Endoscopy is the mainstay of early-stage GC treatment whereas advanced GC must be managed through surgery and other interventions including chemotherapy and targeted therapy[2]. Despite significant progress in GC control, however, its overall survival remains unsatisfactory. Hence, potential therapeutic targets against this disease are urgently required.
Tousled kinase and Tousled-like kinase (TLK) are serine-threonine enzymes implicated in DNA replication[3], transcription[4,5], and chromatin assembly[6]. TLK promotes glioma progression[7] and modulates latent viral activation[8]. Thus, it plays a critical role in various cellular processes. The two known TLK genes are TLK1 and TLK2. However, the former has received more research attention than the latter, and prior investigations focused primarily on the mechanisms by which TLK1 regulates DNA replication and repair. It interacts with Aurora kinase and chromatin assembly factors, and together they precisely control spindle assembly and S-phase progression[9]. The ataxia-telangiectasia-mutated-checkpoint kinase (Chk1)-TLK pathway uses TLK1 as a target to direct chromatin assembly[10,11]. TLK1 collaborates with Chk1 to regulate RAD9 checkpoint clamp component A (Rad9A) phosphorylation and, by extension, modulate the DNA damage response[12,13]. It also confers resistance to ultraviolet irradiation and, therefore, helps maintain cellular integrity and survival[14-17].
Recent research efforts have aimed to clarify the roles of TLK1 in cancers. TLK1 mediates prostate cancer progression via the TLK1-MAPK-activated protein kinase 5 and TLK1-NIMA-related kinase 1 axes[18-21]. The phenothiazine analog J54 is a potent TLK1 inhibitor and a possible therapeutic agent against prostate cancer[22]. TLK1 may promote the progression of glioma[23-25] and oral cancer[26]. However, the expression patterns and functional relevance of TLK1 in GC remain to be determined. Hence, the present study aimed to elucidate the functional significance of TLK1 in GC cells and potentially identify a novel therapeutic target against this disease.
MATERIALS AND METHODS
The RNA-sequencing data obtained from The Cancer Genome Atlas (TCGA) website (https://portal.gdc.cancer.gov/) was subjected to a transcriptome analysis in R v. 4.1.2 (R Core Team, Vienna, Austria) to determine the functional relevance of TLK1 to GC. Differential gene expression between the high- and low-TLK1 expression groups was evaluated. The differentially expressed genes were then subjected to a Kyoto Encyclopedia of Genes and Genomes analysis (http://www.kegg.jp/) to identify and characterize the enrichment pathways with which they were associated. A gene set enrichment analysis was then used to analyze specific pathways and identify the interactions between them and TLK1 in GC. This multifaceted approach established novel molecular mechanisms and regulatory pathways underlying TLK1-mediated GC progression and elucidated GC biology.
The normal gastric epithelial cell line GES-1 as well as the AGS, SGC7901, MGC803, BGC823, and HGC27 GC cell lines were sourced from GeneChem (Shanghai, China) in December 2021. Short tandem repeat analyses verified the authenticity of each cell line and the final test was conducted on March 30, 2022. The cells were cultured in PMIS-1640 medium (Corning Inc., Corning, NY, United States) supplemented with 10% fetal bovine serum (FBS; Clark Bioscience, Richmond, VA, United States), 1% penicillin, and 1% streptomycin (HyClone Laboratories, Logan, UT, United States) and incubated in a Thermo Fisher incubator (Thermo Fisher Scientific, Waltham, MA, United States) under a 5% CO2 atmosphere at 37°C. They were then subcultured after they reached 80% confluence and maintained in the log phase. Strict quality control measures including regular mycoplasma testing were implemented to mitigate the risk of contamination.
Total protein was extracted with M-PER Mammalian Protein Extraction Reagent (Thermo Fisher Scientific) supplemented with phosphatase and protease inhibitors (BBI Life Sciences Corporation, Shanghai, China). Denatured sodium dodecyl sulfate-polyacrylamide gel electrophoresis was used to separate the proteins and they were then blotted onto polyvinylidene fluoride (PVDF) membranes. The latter were blocked with 5% skim milk for 1 h and incubated at 4°C overnight with primary antibodies including anti-GAPDH (1:2,500; No. 7074T; Cell Signaling Technology, Danvers, MA, United States), anti-TLK1 (1:1,000; No. 13564-1-AP; Proteintech Group, Rosemont, IL, United States), and anti-tumor growth factor-beta (TGF-β) (1:1,000; No. 346599; ZenBio, Chengdu, China) to detect the protein expression levels. The PVDF membranes were then washed with Tris-buffered saline with Tween-20 (TBST), incubated with secondary antibody at room temperature for 1 h, rinsed again with TBST, and visualized by enhanced chemiluminescence (ECL; Bridgen, Beijing, China). The ECL signals were captured on a Tanon-5200 multi-platform (Tanon Science & Technology Co. Ltd., Shanghai, China). The protein band gray values were then quantified with ImageJ software (National Institutes of Health (NIH), Bethesda, MD, United States).
Sterile slides were placed in a 24-well plate, drops of cell suspension were dispensed onto them, and they were incubated overnight. The following day, the cells were fixed with 4% formaldehyde, blocked with 5% bovine serum albumin (BSA) for 1 h, and incubated at 4°C overnight with anti-TLK1 antibody (1:1,000; No. 13564-1-AP; Proteintech Group). The next day, the slides were subjected to fluorescent secondary antibody (1:250; No. A11012; Thermo Fisher Scientific) at room temperature for 1 h. Then 4`,6-diamidino-2-phenylindole (DAPI; 1:100; No. D9542; Sigma-Aldrich Corp., Roedermark, Germany) nuclear stain was applied to them for 15 min and they were observed and photographed under a confocal laser scanning microscope (CLSM; No. LSM800; Carl Zeiss AG, Jena, Germany).
Three distinct small hairpin RNAs (shRNAs) targeting human TLK1 and a lentiviral TLK1 overexpression construct were procured from GeneChem (Shanghai, China). Cells were seeded in 12-well plates and the lentiviral particles were added to them at a multiplicity of infection = 10 in the presence of a transfection aid per the manufacturer’s instructions. The medium was replaced and the cells were passaged after 24 h and 48 h, respectively. Successful lentiviral infection was confirmed by screening the cells with a medium containing puromycin. Transfection efficiency was assessed via protein extraction. The shRNA sequences used in the experiment were as follows: sh1: 5`-GAUACAGAUACGUUUUGUACA-dTdT-3`; sh2: 5`-CUCGUAGGGUAGAAACCAAUAdTdT-3`; and sh3: 5`-GCAGGCACUUACUGGUAUUUAdTdT-3`.
Cells were seeded in six-well plates at a density of 800/well and incubated with gentle agitation under optimal conditions. The culture medium was renewed every 3 d and the experiment was terminated when cell colonies emerged. The cells were then fixed with 4% formaldehyde for 20 min, washed thrice with phosphate-buffered saline (PBS), stained with 0.1% crystal violet for 15 min, air-dried, and imaged.
An 5-ethynyl-2`-deoxyuridine (EdU) Kit (No. C0078S; Beyotime Biotechnology, Shanghai, China) was utilized for this assay. Sterile slides were aseptically placed in 12-well plates. Cells were seeded onto them and incubated overnight until the optimal cell density was attained. The next day, a 2 EdU solution was prepared according to the manufacturer’s instructions and mixed in equal proportions with the culture medium. The mixture was then added to the 12-well plates and incubated at 37°C for 2 h. The culture medium was removed and the cells were fixed with 4% paraformaldehyde for 15 min and washed with PBS containing 3% BSA (Beyotime Biotechnology). Then 0.3% Triton X-100 was added and the cells were incubated at room temperature for 15 min. The cells were rinsed and a pre-configured Click reaction solution was added to the 12-well plates. The cells were incubated for 30 min and their nuclei were stained with Hoechst33342 for 10 min. An anti-quenching agent was added and the cells were viewed under a microscope (Leica Microsystems GmbH, Wetzlar, Germany).
Cells in the log phase were seeded at optimal density and subjected to serum deprivation for 24 h. Matrigel Basement Membrane Matrix (BD Biosciences, Shanghai, China) was diluted to a working concentration and uniformly spread onto the upper layer of a Transwell chamber (Corning Inc.). The latter was incubated at 37°C for 5 h. The cell concentration was adjusted and maintained at 8 105/mL. One hundred microliters cell suspension was added to the upper chamber, 650 µL high-serum medium was added to the lower chamber, and the Transwell was incubated in a suitable environment. After a predetermined incubation period, the Transwell chambers were extracted. Their contents were then fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, and observed and photographed under a microscope (Leica Microsystems GmbH).
An Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) Apoptosis Kit (Yeasen Biotechnology, Shanghai, China) was used for this assay. Cell supernatant and digested cells were collected in a centrifuge tube, washed with PBS, and resuspended in 100 µL of 1 × binding buffer. Then 5 µL Annexin V-FITC and 10 µL PI staining solution were added and the suspension was incubated in the dark at room temperature for 15 min. Then a CytoFLEX Flow Cytometry Platform (Beckman Coulter, Brea, CA, United States) was used to detect apoptosis.
Four-Week-old male BALB/C nude mice were obtained from GemPharmatech, Jiangsu, China, and maintained under specific pathogen-free conditions. The control and experimental groups each included six mice. SGC7901 cells were digested and resuspended in PBS and 5 x 106 were subcutaneously injected into the left underarm of each mouse. During the experiment, the mice were provided with sufficient food and water, and their body weight was periodically measured. The volumes of any subcutaneous tumors that had formed were determined every 4 d. Humane euthanasia was performed when the body weight decreased by ≥ 20% and/or the tumor diameter was > 1.5 cm.
Immunohistochemical (IHC) staining was conducted per established methods[27]. Protein expression levels were independently evaluated by two pathologists blinded to the clinical information of the patients. The IHC score was calculated based on the staining intensity and area to assess the protein expression levels.
All data were analyzed with SPSS v. 22.0 (SPSS Inc., Chicago, IL, United States), GraphPad Prism v. 7.0 (GraphPad Software, La Jolla, CA, United States), and R v. 4.1.2 (R Core Team, Vienna, Austria). Student’s t-test or one-way ANOVA was used to detect significant differences between treatments. P < 0.05 was considered statistically significant (aP < 0.05).
Studies involving human participants were reviewed and approved (No. 20180323) by the Ethics Committee of Anhui Medical University, Anhui, China. All patients and participants provided written informed consent. All animal experiments were approved (No. 20180345) by the Institutional Animal Care and Use Committee of Anhui Medical University.
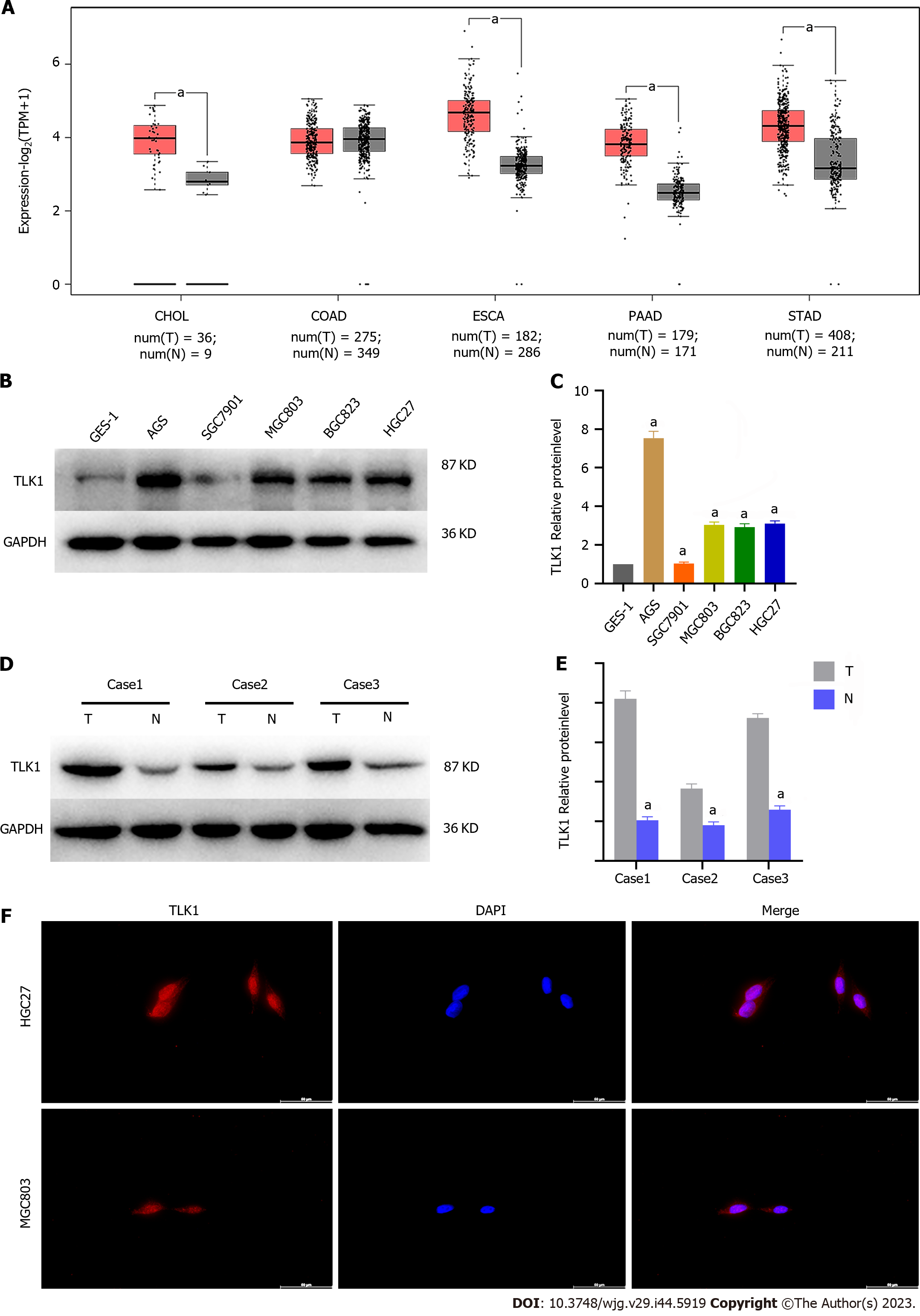
We performed a bioinformatics analysis using TCGA to determine TLK1 expression and localization in GC. We found that TLK1 was upregulated in GC and other tumors of the digestive tract (Figure 1A). We then conducted a western blot analysis to determine the TLK1 expression levels in the GC cells and tissues. We measured TLK1 protein expression in normal gastric epithelial cells (GES-1) and the GC cells AGS, SGC7901, MGC803, BGC823, and HGC27 to clarify the association between the expression and localization of TLK1 in GC. TLK1 expression was significantly higher in GC than in GES-1 cells (Figure 1B and C). We then measured TLK1 expression in GC and their adjacent normal tissues and found that it was significantly higher in the former than in the latter (Figure 1D and E). We then subjected HGC27 and MGC803 GC cells to immunofluorescence staining and observed that TLK1 was localized mainly to their nuclei (Figure 1F). The foregoing findings suggest that TLK1 is upregulated in GC cells and tissues and is localized to the nucleus.
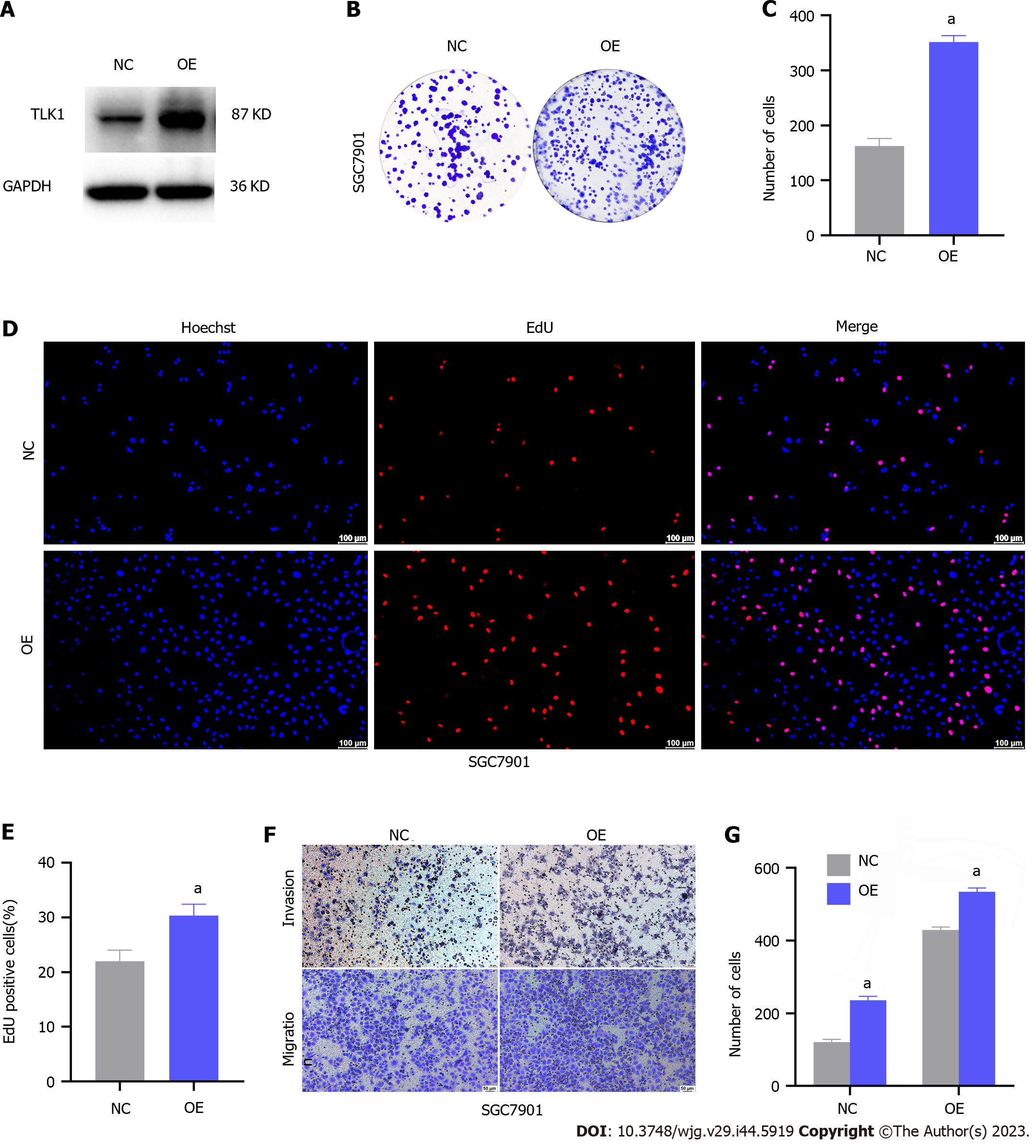
We increased TLK1 expression in SGC7901 cells via lentiviral transfection and verified its upregulation via western blot to elucidate its role in GC cells (Figure 2A). We then used a colony formation assay to assess the impact of TLK1 overexpression on GC cell proliferation. TLK overexpression substantially increased the number of SGC7901 cell colonies compared to the control (Figure 2B and C). The EdU assay demonstrated a dramatic increase in the proportion of proliferative SGC7901 cells in response to TLK1 overexpression (Figure 2D and E). The preceding experiments showed that TLK1 overexpression augments SGC7901 cell proliferation.
A Transwell assay showed that TLK1 overexpression significantly increased SGC7901 GC cell migration and invasion relative to the control (Figure 2F and G). Overall, TLK1 overexpression promoted clonal expansion, proliferation, invasion, and migration in SGC7901 GC cells.
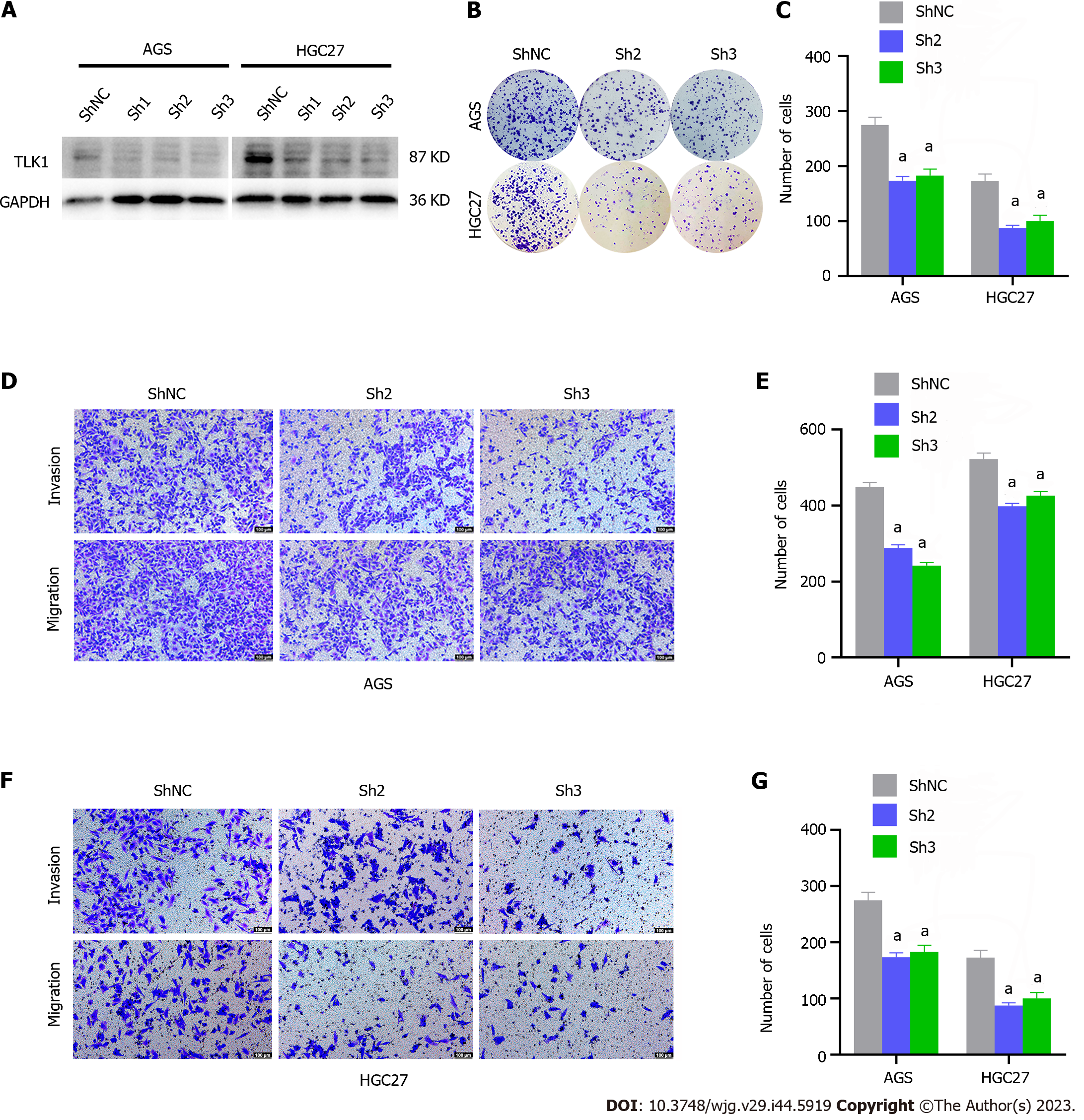
We transfected AGS and HGC27 cells with three lentiviral sequences designed to knock down/silence TLK1 and used western blot analysis to assess transfection efficiency. We selected sh2 and sh3 for the subsequent experiments as they exhibited superior knockdown efficacy (Figure 3A). A colony formation assay demonstrated that TLK1 knockdown markedly reduced the number of AGS and HGC27 cell colonies compared with the control (Figure 3B and C).
A Transwell assay revealed that TLK1 knockdown significantly diminished AGS GC cell invasion and migration relative to the control (Figure 3D and E). Similar results were obtained for HGC27 cells subjected to TLK1 knockdown (Figure 3F and G). The foregoing findings suggest that TLK1 knockdown effectively suppresses clonal formation, invasion, and migration in GC cells.
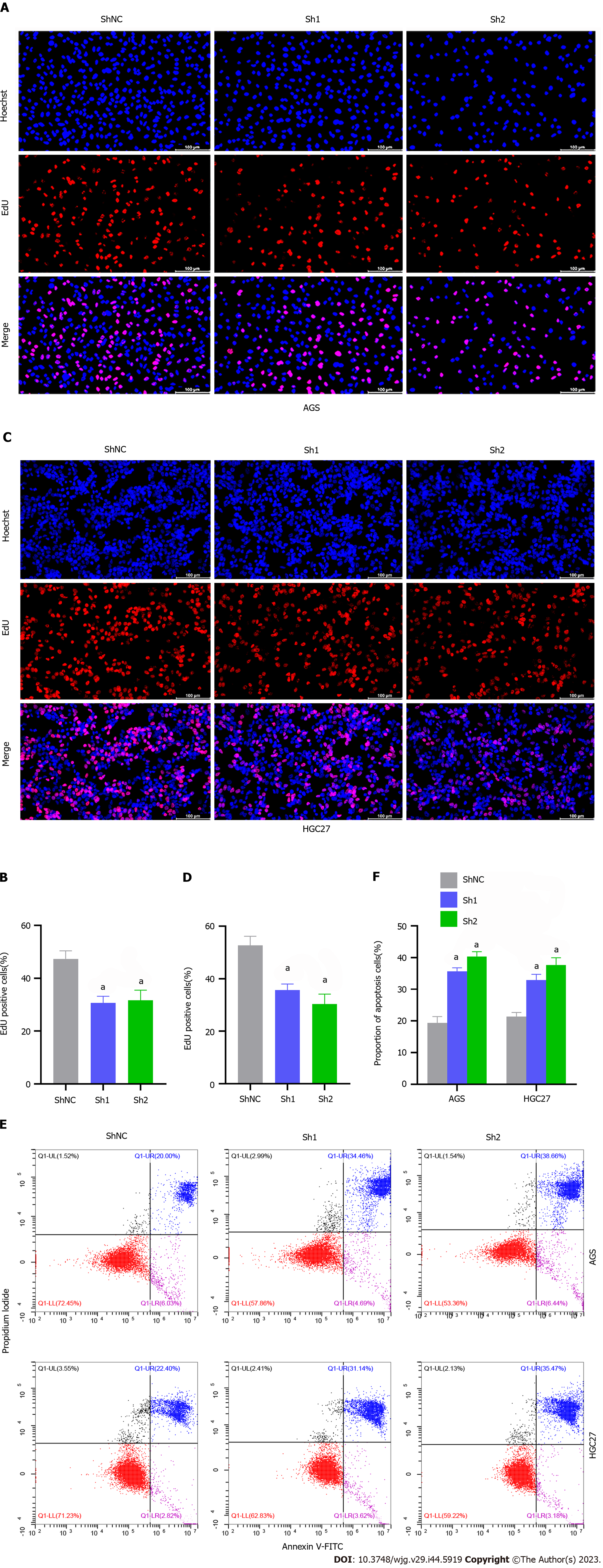
An EdU assay disclosed that TLK1 knockdown substantially inhibited AGS cell proliferation compared to the control (Figure 4A and B). Similar results were obtained for HGC27 cells (Figure 4C and D). Flow cytometry also showed that TLK1 knockdown considerably increased the apoptosis ratios in AGS and HGC27 relative to the control (Figure 4E and F).
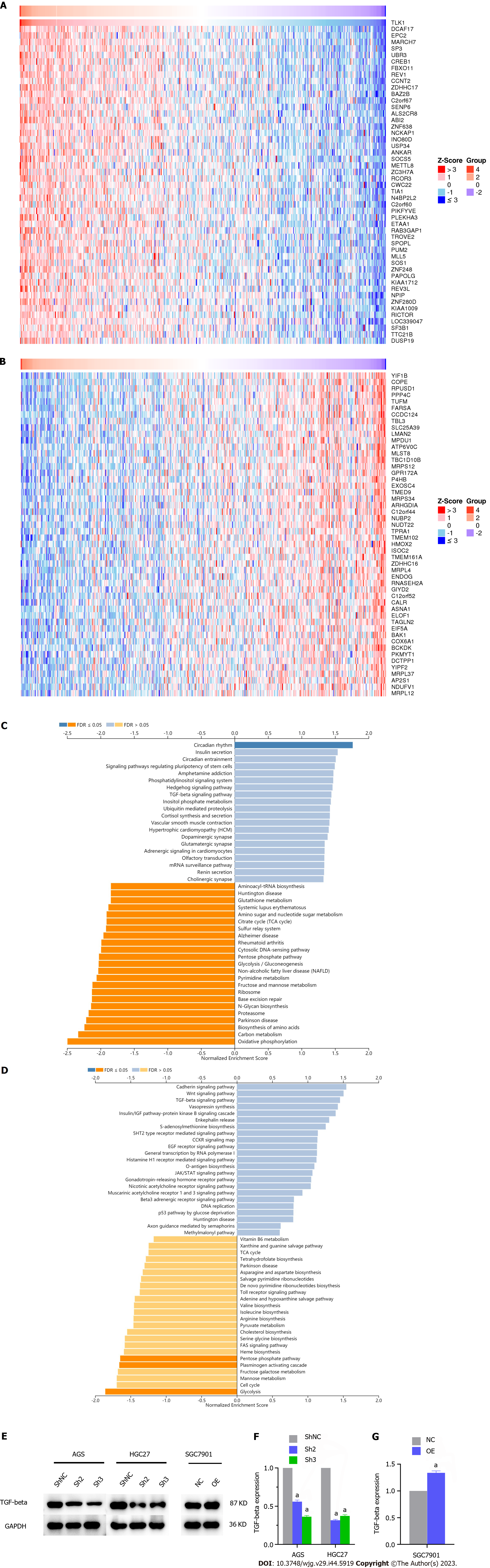
We applied bioinformatics to identify the genes associated with TLK1 and elucidate the mechanisms by which TLK1 affects GC cell clonal formation, proliferation, invasion, and migration. The expression levels of DDB1- and CUL4-associated factor 17 (DCAF17), enhancer of polycomb homolog 2 (EPC2), and membrane-associated ring-CH-type finger 7 (MARCH7) were positively correlated with that of TLK1 (Figure 5A) whereas those of COPI coat complex subunit epsilon (COPE), RNA pseudouridine synthase domain containing 1 (RPUSD1), and protein phosphatase 4, catalytic subunit (PPP4C) were negatively correlated with it (Figure 5B).
We then discovered that the TGF-β signaling pathway might mediate the impact of TLK1 on the clonal formation, proliferation, invasion, and migration of GC cells (Figure 5C and D). We then used western blot to measure TGF-β protein expression in GC cells subjected to TLK1 knockdown and overexpression. We observed that the former downregulated TGF-β in AGS and HGC27 cells whereas the latter upregulated TGF-β in SGC7901 cells. Hence, TGF-β signaling determines the influence of TLK1 on GC progression (Figure 5E-G).
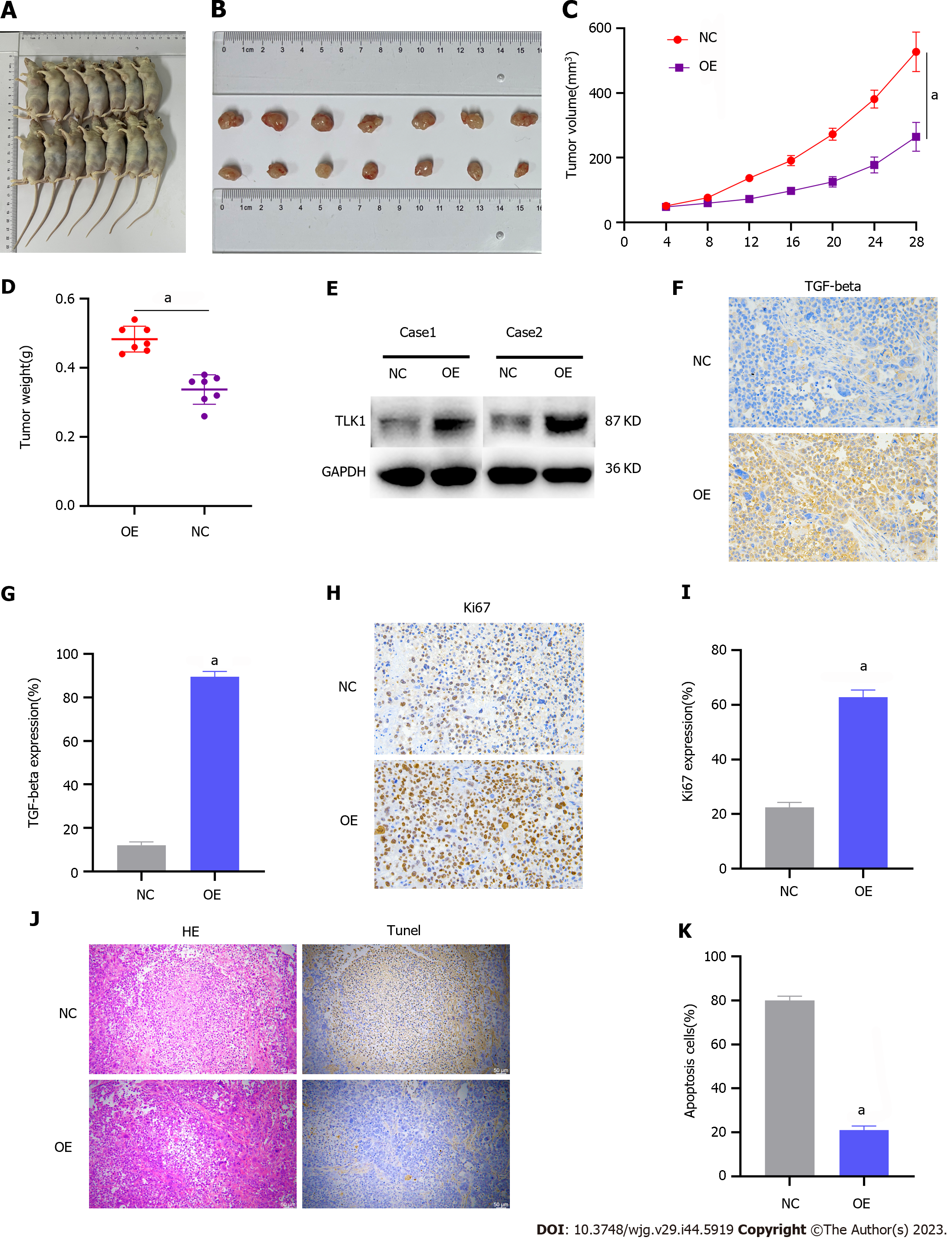
We then validated the impact of TLK1 on GC progression through subcutaneous xenograft tumor induction in nude mice. The study included control and TLK1 overexpression groups (Figure 6A). Compared to the former, the latter presented significantly larger tumor size, volume, and mass (Figure 6B–D). Western blot and IHC verified that the TLK1 and TGF-β expression levels were considerably higher in the treatment group presenting large tumors than in the control group exhibiting small tumors (Figure 6E-G).
Antigen Kiel 67 staining of the tumors revealed that cancer proliferation was markedly higher in the TLK1 overexpression group than in the control group (Figure 6H and I). Hematoxylin and eosin and terminal deoxynucleotidyl transferase dUTP nick end labeling staining exposed substantially greater apoptotic necrosis in the control group than in the TLK1 overexpression group (Figure 6J and K).
The mortality rate of advanced GC remains high despite the progress that has been made in the therapeutic approaches used against it[28]. Hence, novel treatments for GC are urgently needed. TLK promotes the progression of various malignancies. Therefore, research on TLK in the context of cancer therapy should be prioritized[18-21,23-26]. Here, we examined TLK1 expression in GC cells, investigated its effects on their functions, and used in vivo experiments to clarify how it modulates GC progression.
Previous studies reported that TLK1 was upregulated in gliomas[23,25]. In the present work, we discovered that TLK1 was significantly overexpressed in GC cells and tissues (Figure 1B–E). Immunofluorescence staining also revealed that TLK1 was localized mainly to GC cell nuclei (Figure 1F).
We then assessed the impact of TLK1 on GC cell function. TLK1 was overexpressed in the SGC7901 cell line (Figure 2A). Colony formation, EdU, and Transwell assays disclosed that TLK1 overexpression promoted SGC7901 cell proliferation, invasion, and migration (Figure 2B–G). TLK1 knockdown had the opposite effects on AGS and HGC27 cell lines (Figures 3 and 4). An earlier study reported similar findings for the roles of TLK1 in other cancer types[25]. Taken together, these results suggest that TLK1 contributes to GC progression. Our bioinformatics analysis revealed that the mechanism of TLK1 was associated with the TGF-β signaling pathway in GC (Figure 5C and D). The TGF-β signaling pathway comprises TGF-β itself, activins, nodal, bone morphogenetic proteins, growth and differentiation factors, and other factors[29,30] and plays vital roles in human embryonic development and homeostasis[31]. A recent study showed that alterations in TGF-β signaling may result in immunocompromise, fibrosis, and carcinogenesis[32]. The TGF-β signaling pathway may either inhibit or promote tumorigenesis depending upon the tumor microenvironment or cancer stage[29,33,34]. We used western blot to measure TGF-β expression in response to TLK1 overexpression or knockdown and found a positive correlation between TLK1 and TGF-β (Figure 5E and F). Thus, TLK1 may promote GC progression by upregulating TGF-β. We validated this mechanism in vivo by inducing subcutaneous xenograft tumor formation in nude mice (Figure 6) and substantiated the critical role of TLK1 in GC progression. To the best of our knowledge, the present work is one of the first to delineate the expression, localization, and functional impact of TLK1 in GC.
We demonstrated that TLK1 is highly expressed in GC, localized mainly to the nucleus, significantly promotes GC cell proliferation, invasion, and migration, and inhibits apoptosis. TLK1 may facilitate GC progression by modulating TGF-β expression. We believe that TLK1 could be a crucial therapeutic target for GC, and propose that future investigations evaluate the feasibility and practicality of targeting TLK1 in GC treatment.
Gastric cancer (GC) is the fifth most prevalent cancer and the third leading cause of cancer-related mortality worldwide. Endoscopy is the mainstay of early-stage GC treatment whereas advanced GC must be managed through surgery and other interventions including chemotherapy and targeted therapy. Despite significant progress in GC control, however, its overall survival remains unsatisfactory.
Potential therapeutic targets against GC are urgently required.
The present study aimed to elucidate the functional significance of Tousled-like kinase 1 (TLK1) in GC cells and potentially identify a novel therapeutic target against this disease.
We measured TLK1 protein expression levels and localized TLK1 in GC cells and tissues by western blot and immunofluorescence, respectively. We transfected various GC cells with lentiviruses to create TLK1 overexpression and knockdown lines and established the functional roles of TLK1 through in vitro colony formation, 5-ethynyl-2`-deoxyuridine, and Transwell assays as well as flow cytometry. We applied bioinformatics to elucidate the signaling pathways associated with TLK1. We performed in vivo validation of TLK1 functions by inducing subcutaneous xenograft tumors in nude mice.
TLK1 was significantly upregulated in GC cells and tissues compared to their normal counterparts and was localized mainly to the nucleus. TLK1 knockdown significantly decreased colony formation, proliferation, invasion, and migration but increased apoptosis in GC cells. TLK1 overexpression had the opposite effects. Bioinformatics revealed, and subsequent experiments verified, that the tumor growth factor-beta (TGF-β) signaling pathway was implicated in TLK1-mediated GC progression. The in vivo assays confirmed that TLK1 promotes tumorigenesis in GC.
We demonstrated that TLK1 is highly expressed in GC, localized mainly to the nucleus, significantly promotes GC cell proliferation, invasion, and migration, and inhibits apoptosis. TLK1 may facilitate GC progression by modulating TGF-β expression. We believe that TLK1 could be a crucial therapeutic target for GC.
Future investigations evaluate the feasibility and practicality of targeting TLK1 in GC treatment.
The authors thank the personnel at the Center for Scientific Research of the First Affiliated Hospital of Anhui Medical University for their valuable assistance with our experiments.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Buechler C, Germany; Cucu D, Romania; Ocker M, Germany S-Editor: Lin C L-Editor: A P-Editor: Chen YX
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55774] [Article Influence: 7967.7] [Reference Citation Analysis (132)] |
| 2. | Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1150] [Cited by in RCA: 2838] [Article Influence: 567.6] [Reference Citation Analysis (5)] |
| 3. | Silljé HH, Takahashi K, Tanaka K, Van Houwe G, Nigg EA. Mammalian homologues of the plant Tousled gene code for cell-cycle-regulated kinases with maximal activities linked to ongoing DNA replication. EMBO J. 1999;18:5691-5702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 118] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 4. | Han Z, Saam JR, Adams HP, Mango SE, Schumacher JM. The C. elegans Tousled-like kinase (TLK-1) has an essential role in transcription. Curr Biol. 2003;13:1921-1929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 52] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 5. | Giménez AC, Tamajón LG. Analysis of the third-order structuring of Shalom Schwartz's theory of basic human values. Heliyon. 2019;5:e01797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 6. | Silljé HH, Nigg EA. Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Curr Biol. 2001;11:1068-1073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 150] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 7. | Han Z, Riefler GM, Saam JR, Mango SE, Schumacher JM. The C. elegans Tousled-like kinase contributes to chromosome segregation as a substrate and regulator of the Aurora B kinase. Curr Biol. 2005;15:894-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 58] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 8. | Dillon PJ, Gregory SM, Tamburro K, Sanders MK, Johnson GL, Raab-Traub N, Dittmer DP, Damania B. Tousled-like kinases modulate reactivation of gammaherpesviruses from latency. Cell Host Microbe. 2013;13:204-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 9. | Li Z, Gourguechon S, Wang CC. Tousled-like kinase in a microbial eukaryote regulates spindle assembly and S-phase progression by interacting with Aurora kinase and chromatin assembly factors. J Cell Sci. 2007;120:3883-3894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 10. | Krause DR, Jonnalagadda JC, Gatei MH, Sillje HH, Zhou BB, Nigg EA, Khanna K. Suppression of Tousled-like kinase activity after DNA damage or replication block requires ATM, NBS1 and Chk1. Oncogene. 2003;22:5927-5937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 70] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 11. | Groth A, Lukas J, Nigg EA, Silljé HH, Wernstedt C, Bartek J, Hansen K. Human Tousled like kinases are targeted by an ATM- and Chk1-dependent DNA damage checkpoint. EMBO J. 2003;22:1676-1687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 131] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 12. | Sunavala-Dossabhoy G, De Benedetti A. Tousled homolog, TLK1, binds and phosphorylates Rad9; TLK1 acts as a molecular chaperone in DNA repair. DNA Repair (Amst). 2009;8:87-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 62] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 13. | Kelly R, Davey SK. Tousled-like kinase-dependent phosphorylation of Rad9 plays a role in cell cycle progression and G2/M checkpoint exit. PLoS One. 2013;8:e85859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Sen SP, De Benedetti A. TLK1B promotes repair of UV-damaged DNA through chromatin remodeling by Asf1. BMC Mol Biol. 2006;7:37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 15. | Sunavala-Dossabhoy G, Balakrishnan SK, Sen S, Nuthalapaty S, De Benedetti A. The radioresistance kinase TLK1B protects the cells by promoting repair of double strand breaks. BMC Mol Biol. 2005;6:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 40] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 16. | Canfield C, Rains J, De Benedetti A. TLK1B promotes repair of DSBs via its interaction with Rad9 and Asf1. BMC Mol Biol. 2009;10:110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 17. | Sunavala-Dossabhoy G, Fowler M, De Benedetti A. Translation of the radioresistance kinase TLK1B is induced by gamma-irradiation through activation of mTOR and phosphorylation of 4E-BP1. BMC Mol Biol. 2004;5:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 50] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 18. | Khalil MI, De Benedetti A. The TLK1-MK5 Axis Regulates Motility, Invasion, and Metastasis of Prostate Cancer Cells. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 19. | Khalil MI, Singh V, King J, De Benedetti A. TLK1-mediated MK5-S354 phosphorylation drives prostate cancer cell motility and may signify distinct pathologies. Mol Oncol. 2022;16:2537-2557. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 20. | Singh V, Khalil MI, De Benedetti A. The TLK1/Nek1 axis contributes to mitochondrial integrity and apoptosis prevention via phosphorylation of VDAC1. Cell Cycle. 2020;19:363-375. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 21. | Singh V, Jaiswal PK, Ghosh I, Koul HK, Yu X, De Benedetti A. The TLK1-Nek1 axis promotes prostate cancer progression. Cancer Lett. 2019;453:131-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 22. | Singh V, Bhoir S, Chikhale RV, Hussain J, Dwyer D, Bryce RA, Kirubakaran S, De Benedetti A. Generation of Phenothiazine with Potent Anti-TLK1 Activity for Prostate Cancer Therapy. iScience. 2020;23:101474. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Wang Z, Chen X, Liang Q, An Y, Wei M, Shi W. Inhibiting of circ-TLK1 inhibits the progression of glioma through down-regulating PANX1 via targeting miR-17-5p. J Mol Histol. 2021;52:1007-1020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 24. | Zhang Z, Liu S. The interaction between ASF1B and TLK1 promotes the malignant progression of low-grade glioma. Ann Med. 2023;55:1111-1122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 25. | Ibrahim K, Abdul Murad NA, Harun R, Jamal R. Knockdown of Tousledlike kinase 1 inhibits survival of glioblastoma multiforme cells. Int J Mol Med. 2020;46:685-699. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 26. | Hu S, Wang H, Yan D, Lu W, Gao P, Lou W, Kong X. Loss of miR-16 contributes to tumor progression by activation of tousled-like kinase 1 in oral squamous cell carcinoma. Cell Cycle. 2018;17:2284-2295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 27. | Sun R, Wu J, Chen Y, Lu M, Zhang S, Lu D, Li Y. Down regulation of Thrombospondin2 predicts poor prognosis in patients with gastric cancer. Mol Cancer. 2014;13:225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 28. | Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA Cancer J Clin. 2021;71:264-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 382] [Cited by in RCA: 1077] [Article Influence: 269.3] [Reference Citation Analysis (0)] |
| 29. | David CJ, Massagué J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat Rev Mol Cell Biol. 2018;19:419-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 615] [Article Influence: 87.9] [Reference Citation Analysis (0)] |
| 30. | Saito A, Horie M, Nagase T. TGF-β Signaling in Lung Health and Disease. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 359] [Article Influence: 51.3] [Reference Citation Analysis (0)] |
| 31. | Xu X, Zheng L, Yuan Q, Zhen G, Crane JL, Zhou X, Cao X. Transforming growth factor-β in stem cells and tissue homeostasis. Bone Res. 2018;6:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 265] [Cited by in RCA: 293] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 32. | Liu S, Ren J, Ten Dijke P. Targeting TGFβ signal transduction for cancer therapy. Signal Transduct Target Ther. 2021;6:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 239] [Article Influence: 59.8] [Reference Citation Analysis (0)] |
| 33. | Neuzillet C, Tijeras-Raballand A, Cohen R, Cros J, Faivre S, Raymond E, de Gramont A. Targeting the TGFβ pathway for cancer therapy. Pharmacol Ther. 2015;147:22-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 492] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 34. | Colak S, Ten Dijke P. Targeting TGF-β Signaling in Cancer. Trends Cancer. 2017;3:56-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 472] [Cited by in RCA: 750] [Article Influence: 93.8] [Reference Citation Analysis (0)] |