Published online Oct 21, 2023. doi: 10.3748/wjg.v29.i39.5452
Peer-review started: July 3, 2023
First decision: August 25, 2023
Revised: September 14, 2023
Accepted: September 26, 2023
Article in press: September 26, 2023
Published online: October 21, 2023
Processing time: 108 Days and 2.8 Hours
Oxaliplatin (Oxa) is the first-line chemotherapy drug for colorectal cancer (CRC), and Oxa resistance is crucial for treatment failure. Prostaglandin F2α synthase (PG
To explore the role and related mechanisms of PGFS in mediating Oxa resistance in CRC.
The PGFS expression level was examined in 37 pairs of CRC tissues and paracancerous tissues at both the mRNA and protein levels. Overexpression or knockdown of PGFS was performed in CRC cell lines with acquired Oxa resistance (HCT116-OxR and HCT8-OxR) and their parental cell lines (HCT116 and HCT8) to assess its influence on cell proliferation, chemoresistance, apoptosis, and DNA damage. For determination of the underlying mechanisms, CRC cells were examined for platinum-DNA adducts and reactive oxygen species (ROS) levels in the presence of a PGFS inhibitor or its products.
Both the protein and mRNA levels of PGFS were increased in the 37 examined CRC tissues compared to the adjacent normal tissues. Oxa induced PGFS expression in the parental HCT116 and HCT8 cells in a dose-dependent manner. Furthermore, overexpression of PGFS in parental CRC cells significantly attenuated Oxa-induced proliferative suppression, apoptosis, and DNA damage. In contrast, knockdown of PGFS in Oxa-resistant HCT116 and HCT8 cells (HCT116-OxR and HCT8-OxR) accentuated the effect of Oxa treatment in vitro and in vivo. The addition of the PGFS inhibitor indomethacin enhanced the cytotoxicity caused by Oxa. Treatment with the PGFS-catalyzed product PGF2α reversed the effect of PGFS knockdown on Oxa sensitivity. Interestingly, PGFS inhibited the formation of platinum-DNA adducts in a PGF2α-independent manner. PGF2α exerts its protective effect against DNA damage by reducing ROS levels.
PGFS promotes resistance to Oxa in CRC via both PGF2α-dependent and PGF2α-independent mechanisms.
Core Tip: Prostaglandin F2α synthase (PGF2α) (PGFS) is an enzymatic catalyst responsible for the biosynthesis of PGF2α. Our study revealed that PGFS exerts an inhibitory effect on the generation of reactive oxygen species by means of its downstream product PGF2α, and consequently facilitates resistance to oxaliplatin in colorectal cancer. Simultaneously, PGFS suppresses the formation of platinum-DNA adducts in a manner that is not reliant on PGF2α, which is rarely reported.
- Citation: Wang YJ, Xie XL, Liu HQ, Tian H, Jiang XY, Zhang JN, Chen SX, Liu T, Wang SL, Zhou X, Jin XX, Liu SM, Jiang HQ. Prostaglandin F2α synthase promotes oxaliplatin resistance in colorectal cancer through prostaglandin F2α-dependent and F2α-independent mechanism. World J Gastroenterol 2023; 29(39): 5452-5470
- URL: https://www.wjgnet.com/1007-9327/full/v29/i39/5452.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i39.5452
Colorectal cancer (CRC) is one of the most frequently diagnosed cancers and a leading cause of cancer death worldwide[1]. In China, CRC is the third most common cancer, and the incidence rate also exceeds that of liver cancer, ranking third in cancer[2]. The current clinical treatment regimen for CRC involves surgical resection followed by chemotherapy, radiotherapy and combination therapy. For prevention of tumor recurrence, surgery alone or combined with adjuvant chemotherapy remains the cornerstone of the treatment of nonmetastatic CRC[3]. Adjuvant oxaliplatin (Oxa)-based fluorouracil, leucovorin, and Oxa (FOLFOX) or capecitabine plus Oxa chemotherapy is the standard for patients with stage II or III colon cancer following surgery, as recommended by several treatment guidelines[4,5].
Oxa, as an alternative to cisplatin, is a third-generation platinum agent. Several multicenter clinical trials have been conducted on the therapies with Oxa shown in preclinical and single-center trials to have beneficial anticancer effects. Oxa can significantly reduce the risk of recurrence and mortality of CRC and prolong disease-free survival and overall survival[6,7]. However, resistance to chemotherapy remains a major clinical issue in the treatment of CRC[8]. However, only half of CRC patients respond to FOLFOX, and this half of chemotherapy-resistant patients also develop resistance during chemotherapy, a type of resistance known as secondary resistance. That is, cancer drug resistance includes two types: Intrinsic (also called innate or primary resistance) and acquired (also called avoidant, adaptive, or secondary resistance).
Cisplatin resistance likely occurs for complex reasons, including increased drug efflux, drug breakdown, increased repair of damaged DNA, and increased activation of prosurvival pathways or inhibition of pathways that promote cell death[9]. Several studies have also shown that for Oxa resistance, the reduction in damage formation rather than the increase in damage repair is the main factor in the acquired resistance to Oxa[10]. Compared with cisplatin, Oxa has a large platinum atom ring, which can bind more DNA molecules to form platinum-DNA complexes and cause DNA damage to cells. Then, the DNA repair mechanism is activated. Once the repair is successful, Oxa will be excreted from the body, resulting in drug resistance. If DNA damage repair fails, the cell will undergo apoptosis[11]. Several studies have shown that although Oxa has greater cell lethality than cisplatin, side effects such as ototoxicity and nephrotoxicity are weaker than those of cisplatin, and this drug can treat cisplatin-resistant tumors, but Oxa more easily results in resistance than cisplatin[12,13]. Therefore, studying the resistance to Oxa has important clinical value.
Prostaglandins (PGs) are a group of biologically active lipid mediators derived from the cyclooxygenase (COX) pathway of the arachidonic acid cascade[14]. PGs are involved in growth and development, inflammation, and cancer progression by binding to their respective receptors[15]. Prostaglandin F2α (PGF2α) is one of the main PGs produced by the COX pathway. PGF2α is synthesized by PGF2α synthase (PGFS), which is a member of the aldo-keto reductase (AKR) family and a key enzyme in steroid metabolism[16]. PGFS converts prostaglandin D2 (PGD2) into 11β-PGF2α[17,18]. Recently, the cancer-promoting effects of PGFS in prostate cancer, gastric cancer, lung cancer, and other tumors have been confirmed[19,20].
Oxidative stress is related to the occurrence and development of tumors, with reactive oxygen species (ROS) playing an important role in inducing oxidative stress. Excessive ROS production is caused by the imbalance between defense and metabolic mechanisms. Low concentrations of ROS induce cell proliferation and regulate the activation of several signaling pathways[21]; however, high concentrations of ROS induce changes in gene expression and promote the accumulation of mutations and DNA damage[22]. ROS can promote the generation of PGD2 and PGF2α[3,23], but the relationship between PGF2α and ROS and CRC treatment remains unclear.
In CRC, PGFS overexpression was reported to reduce the DNA damage caused by oxidative stress and thereby induce cisplatin resistance[24]. However, studies on the drug resistance of Oxa are scarce. Moreover, the role and mechanism of PGFS in CRC drug resistance and the relationship between PGF2α and DNA damage require further study. In this study, Oxa-resistant CRC cell lines with PGFS overexpression and knockdown were evaluated, revealing its mediating effect on Oxa resistance. Thus, this study provides novel ideas for the diagnosis and treatment of CRC.
Fresh CRC tissues and adjacent nontumor tissue samples were acquired from 37 patients with CRC who underwent surgical resection at the Second Hospital of Hebei Medical University, Shijiazhuang, China. This study was approved by the Ethics Committee of the Second Hospital of Hebei Medical University (approval No. 2021-R441). Each tissue was divided into three parts: one part was directly snap-frozen in liquid nitrogen and stored at -80 ℃ for western blotting, a second part was immediately frozen with TRIzol and stored at -80 °C for RNA extraction, and the third part was immersed in 4% paraformaldehyde and stored at 4 ℃ for further paraffin embedding.
The different gene expression levels were compared between the nonresponse and response groups. The data on the expression of PGFS in CRC patients with or without response to an Oxa-containing regimen are available in the cancer treatment response gene signature database (http://ctrdb.ncpsb.org.cn/)[25]. Data were generated from the merged dataset of CTR_Microarray_35 and CTR_Microarray_90. We found that 29 out of 45 patients were nonresponsive, and the remaining 16 were responsive. The expression levels of PGFS in the two groups of patients were calculated using the website tool.
Sections were stained for hematoxylin and eosin and immunofluorescence staining. Immunohistochemistry (IHC) was performed as previously described[26]. Briefly, the slides were routinely deparaffinized, rehydrated, subjected to antigen retrieval, and incubated in 3% hydrogen peroxide to block endogenous peroxidase. Subsequently, the slides were blocked and incubated with primary antibodies against PGFS (diluted 1:1000, Proteintech, Wuhan, China) and proliferating cell nuclear antigen (PCNA) (diluted 1:500, Proteintech, Wuhan China) at 4 ℃ overnight, followed by incubation with polymer-horseradish peroxidase-conjugated anti-rabbit secondary antibody the next day. Then, the section was stained with 3,3’-diaminobenzidine and counterstained with hematoxylin, dehydrated and coverslipped.
The human CRC HCT116 cell line was obtained from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). Oxa-resistant cell lines (HCT116-OxR and HCT8-OxR) and HCT8 cells were purchased from Oulu Biotechnology (Shanghai, China). HCT116 and HCT8 cells were cultured in roswell park memorial institute (RPMI) 1640 medium (Gibco BRL, Rockville, MD, United States), which was supplemented with 10% fetal bovine serum (Gibco BRL), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 ℃ in a humidified incubator with 5% CO2. HCT116-OxR and HCT8-OxR cells were cultured in Oxa-containing RPMI 1640 medium. HCT116, HCT116-OxR, HCT8, and HCT8-OxR cells were certified by Viva Cell Biosciences Ltd. (Shanghai, China). The reagents used in this study were Oxa (Selleck, Shanghai, China), indomethacin (MCE, Shanghai, China), and PGF2α (Merck, Jiangsu Province, China).
Proteins were extracted from CRC and adjacent tissues and cells using radio immunoprecipitation assay lysis buffer with proteinase inhibitors. Protein concentrations were measured using bicinchoninic acid (Sigma-Aldrich St. Louis, MO, United States). The primary antibodies used included anti-PGFS antibody (diluted 1:500, Abways, Shanghai, China), anti-β-actin antibody (diluted 1:1000, Abways), anti-cleaved-caspase 3 antibody (diluted 1:200, Abways), anti-cleaved poly ADP-ribose polymerase (PARP) antibody (diluted 1:500, Abways), anti-PCNA (diluted 1:500, Abways), and anti-[γ-H2A histone family member X (γ-H2AX), diluted 1:500, Abways]. Briefly, protein purification was confirmed using electrophoresis on a sodium dodecyl sulfate-polyacrylamide gel, which was then transferred onto a polyvinylidene difluoride membrane. Membranes were incubated in 5% milk for 1 h before incubation with primary antibodies at 4 ℃ overnight. The following day, the membrane was washed three times with TBS with Tween-20. Secondary antibodies were incubated for 1 h at room temperature. The bands were visualized using an Odyssey infrared imaging system (LI-COR Biosciences), and protein quantification was performed using ImageJ software (National Institutes of Health, Bethesda, MD, United States).
For generation of stable reporter lines, HCT116 and HCT8 cells were infected with lentiviral particles expressing PGFS, and the empty vector was used as a control. Lentiviruses for PGFS knockdown were infected into the Oxa-resistant cell lines HCT116-OxR and HCT8-OxR following the manufacturer’s instructions. Subsequently, transfected cells were selected for 14-d incubations in 5 ng/mL puromycin. The construction of PGFS knockdown and overexpression lentiviruses was completed by GeneChem Co., Ltd. (Shanghai, China).
Total RNA was extracted from tissues using TRIzol reagent (Tiangen). Complementary DNA (cDNA) was synthesized from total RNA through reverse transcription using the PrimeScript RT reagent kit from TaKaRa Biomedical Technology Co., Ltd. (Beijing, China). cDNA was used for RT-PCR using the above PrimeScript RT reagent kit on a StepOnePlus™ RT-PCR system (Applied Biosystems, MS). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control. Specific primer sequences were designed as follows: PGFS-forward: 5’-GTC ATC CGT ATT TCA ACC GGAG-3’; PGFS-reverse: 5’-CCA CCC ATC GTT TGT CTC GTT-3’; GAPDH-forward: 5’-AGG GGG GAG CCA AAA GGG TCA-3’; GAPDH-reverse: 5’-TGG GTG GCA GTG ATG GCA TGG A-3’.
Cell viability was determined using a cell counting kit-8 (CCK-8) kit (Dojindo Laboratories, Kumamoto, Japan) following the manufacturer’s instructions. Briefly, CCK-8 solution (10 μL) was added to each well and incubated for 2 h at 37 ℃. Absorbance was measured at 450 nm using a plate reader (BioTek, Winooski, VT, United States). CCK-8 assays were used to examine the half-inhibitory concentration (IC50). The inhibition concentration (%) was calculated as [(anegative - blank) - (Aexp - blank)]/(anegative-blank) × 100%. The IC50 was calculated from the survival curves using GraphPad Prism 7 (Version X; La Jolla, CA, United States). Each assay was performed in triplicate.
Cells stably transfected with lentiviruses were reseeded (3 × 103 cells per well) in 6-well plates and cultured for two weeks. The colonies were subsequently stained with 0.05% crystal violet for visualization. After routine culture for 2 wk, colony numbers were counted under a microscope.
For analysis of early/late apoptotic or necrotic cell death, cells were measured with the Annexin V-FITC apoptosis detection kit (Beyotime, Shanghai, China) using flow cytometric analysis. We also detected total apoptosis by a terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay (Beyotime, Shanghai, China).
Stably transfected cells were seeded onto cover glasses in 24-well plates. After 24 h of intervention, cells were fixed in 4% paraformaldehyde for 30 min, permeabilized with 0.1% Triton X-100, diluted in phosphate-buffered saline (PBS) for 10 min, and blocked with 5% normal goat serum for 30 min at room temperature. The cells were incubated with rabbit anti-PGFS or rabbit anti-γ-H2AX overnight at 4 ℃. After washing with PBS, the cells were incubated with the secondary antibody (594 goat anti-rabbit immunoglobulin G) for 1 h at room temperature. After washing, the slides were counterstained with 4’6’-diamidino-2-phenylindole dihydrochloride (Sigma-Aldrich) for 5 min. Images were acquired using a laser scanning confocal microscope (Olympus, Tokyo, Japan).
Intracellular ROS levels were evaluated using a ROS assay kit (KGAF019, KeyGEN Biotech, Nanjing, China) following the manufacturer’s instructions. Briefly, cells were stained with dihydroethidium (DHE) for 30 min and analyzed for intracellular ROS levels under a fluorescence microscope (Olympus, Tokyo, Japan). The mean fluorescence intensity, as an index of the amount of ROS, was measured using ImageJ software.
The DNA concentration was determined using a nanodrop spectrophotometer (Thermo Fisher Scientific), and the DNA-bound platinum content was quantified using inductively coupled plasma-mass spectrometry.
DNA damage was detected using a comet assay. CRC cells were administered different drug concentrations and had different exposure times. The comet assay was performed according to the alkaline comet assay protocol (KeyGEN Biotech, Nanjing, China). Nuclear DNA and migrating DNA (comet tail) labeled with propidium iodide appeared red and were observed at 515-560 nm using a fluorescence microscope (Olympus, Tokyo, Japan). Comet tail length and tail moment (percentage of DNA in the tail to the total DNA) were analyzed using ImageJ software.
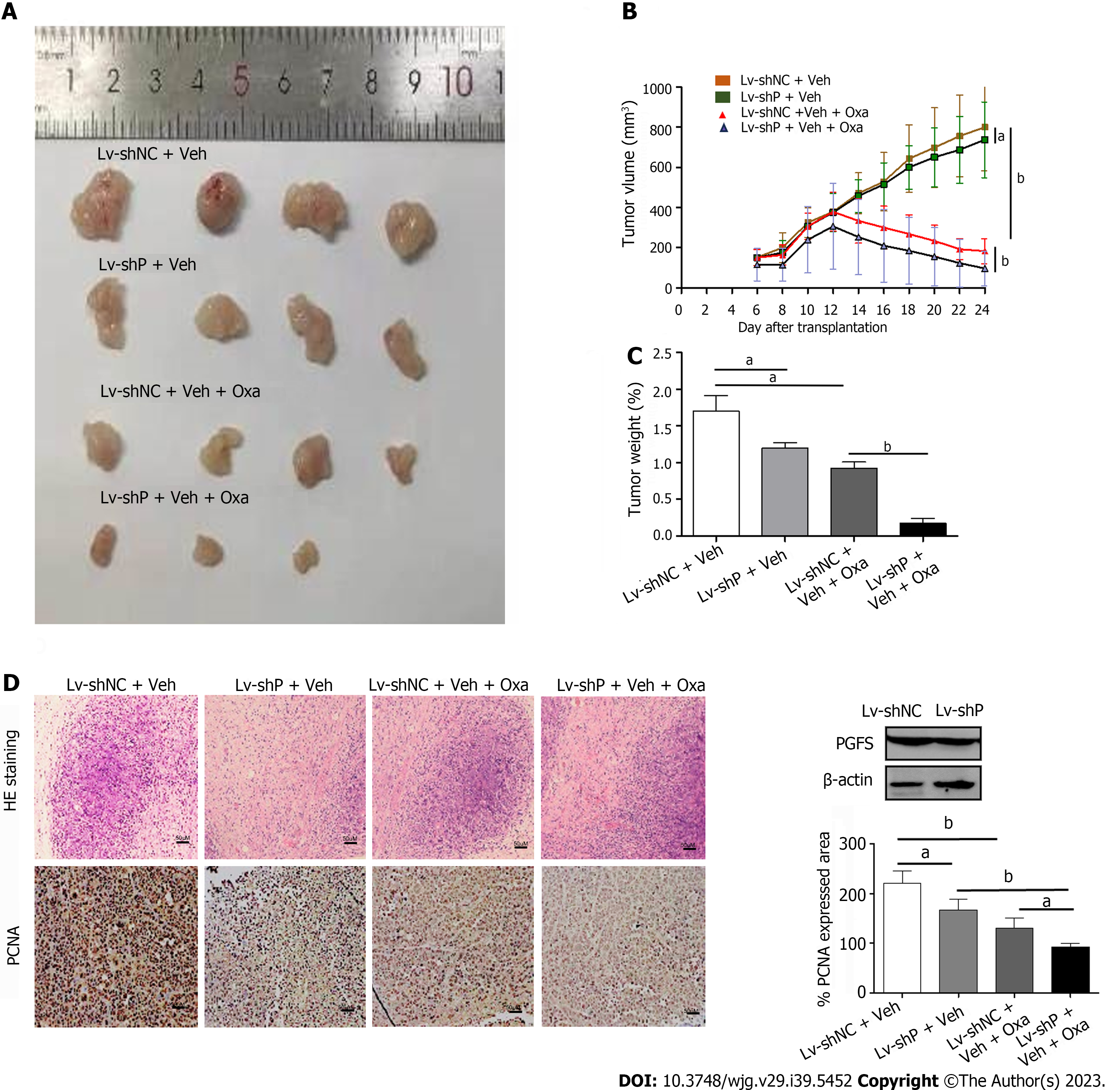
Nude mice were implanted subcutaneously with HCT8-OXR cells. When the average volume of the tumors reached approximately 100 mm3. The nude mice in the LV-shNC + Oxa and LV-shPGFS + Oxa groups were treated with Oxa by gavage at a dosage of 10 mg/kg/d for 14 d, and the volume of tumors was measured. We sacrificed the nude mice when the treatment was completed, and the tissues were fixed with 4% paraformaldehyde or immediately placed at -80 ℃. The animal protocol was designed to minimize pain and discomfort to animals. The nude mice were acclimatized to laboratory conditions for 1 wk prior to our experiment. Intragastric gavage administration was conducted by using straight gavage needles (22 gauge). All the animals in our study received humane care, and the ethics committee of the Second Hospital of Hebei Medical University approved our study (approval letter No. 2022-AE010).
Data are presented as the mean ± SD. The association between PGFS expression and clinicopathological features was evaluated using a nonparametric test. The statistical analyses included analysis of variance (ANOVA) with the Student-Newman-Keuls post hoc test and a standard two-tailed Student’s t test. Student’s t test was used for comparisons between two groups, while comparisons between three or more groups were conducted using one or two-way ANOVA with the Brown-Forsythe test for equality of group variances. Statistical analysis was performed using SPSS software version 25.0 (Chicago, IL, United States). P < 0.05 was considered statistically significant.
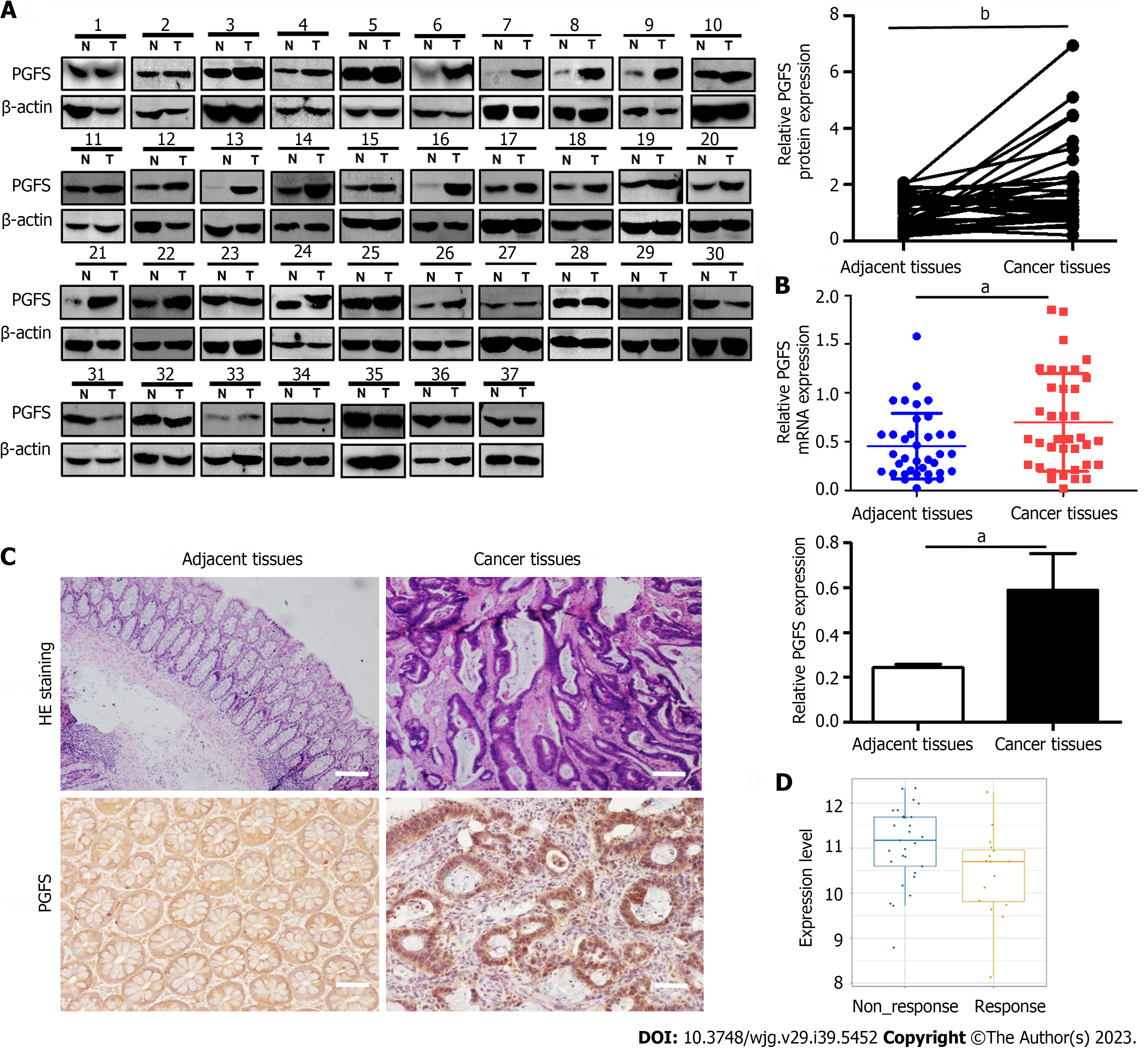
We first tested the expression of PGFS in 37 pairs of CRC tissues and matched paracarcinoma tissues. PGFS was significantly upregulated at both the protein and RNA levels in cancerous tissues compared with adjacent nontumor tissues (Figure 1A and B). IHC analysis showed that the strong positive PGFS signals were mainly detected in the nuclei of the cancerous epithelial cells. In contrast, the normal tissue samples showed low expression of PGFS homogenously distributed in the nuclei and cytoplasm (Figure 1C). Then, we investigated the relationship between PGFS and clinicopathological features. High PGFS expression was associated with advanced T stage (Tables 1 and 2). Moreover, PGFS expression was higher in the Oxa-nonresponsive group than in the Oxa-responsive group (Figure 1D).
| Variables | No. of patients | Relatives PGFS expressions (mean ± SD) | P value |
| All cases | 37 (100) | ||
| Gender | 0.376 | ||
| Male | 22 (59) | 1.84 ± 1.67 | |
| Female | 15 (41) | 1.68 ± 1.02 | |
| Age (yr) | 0.495 | ||
| ≥ 60 | 19 (51) | 1.40 ± 0.8 | |
| < 60 | 18 (49) | 2.12 ± 1.8 | |
| Tumor location | 0.066 | ||
| Colon | 15 (41) | 1.71 ± 1.2 | |
| Rectum | 22 (59) | 1.84 ± 1.65 | |
| Differentiations | 0.437 | ||
| Well and Moderately | 26 (70) | 1.65 ± 1.29 | |
| Poorly | 11 (30) | 1.81 ± 1.08 | |
| T stage | 0.023a | ||
| T1 or T2 | 25 (68) | 1.26 ± 0.52 | |
| T3 or T4 | 12 (32) | 2.47 ± 0.87 | |
| Lymph node metastasis | 0.063 | ||
| No | 25 (68) | 1.17 ± 0.56 | |
| Yes | 12 (32) | 2.09 ± 1.69 | |
| AJCC stage | 0.069 | ||
| I or II | 29 (78) | 1.44 ± 0.89 | |
| III or IV | 8 (22) | 3.09 ± 2.69 |
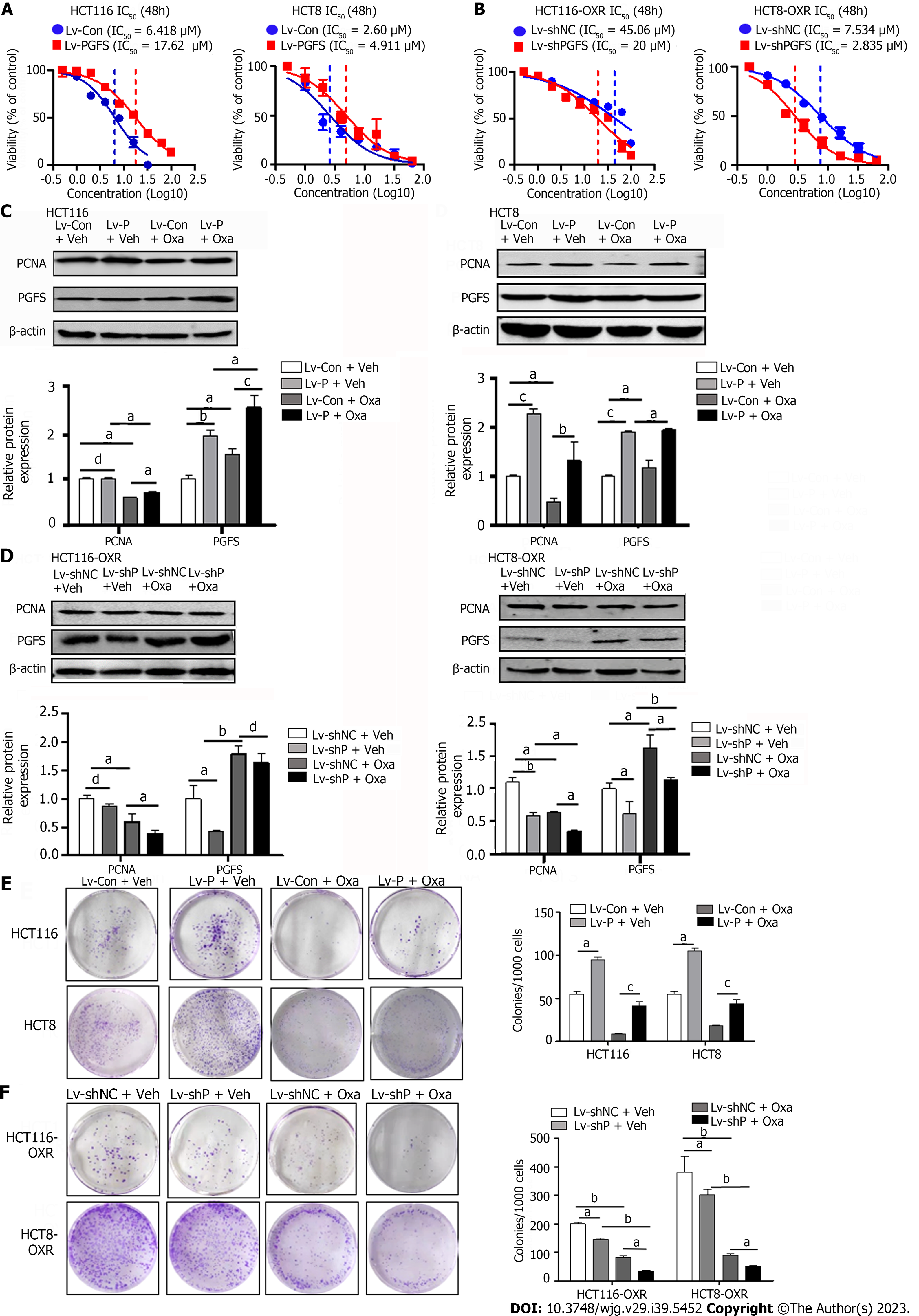
To assess the role of PGFS in Oxa resistance in CRC cells, we used two Oxa-resistant CRC cell lines derived from HCT116 and HCT8 (hereafter named HCT116-OxR and HCT8-OxR). The resistance of the two Oxa-resistant cell lines was determined by measuring the resistance index (RI), which was calculated as the IC50 ratio of drug-resistant cells to their parent cells. Both HCT116-OxR and HCT8-OxR cells showed moderate Oxa resistance, with RIs of 10.1 and 5.86, respectively (IC50-HCT116-OxR vs. IC50-HCT116: 64.2 μM vs. 6.315 μM; IC50-HCT8-OxR vs. IC50-HCT8: 6.241 μM vs. 1.064 µM; Supplementary Figure 1A). Colony formation assays revealed increased colony formation efficiency in the two Oxa-resistant cell lines (Supplementary Figure 1B). PGFS expression was significantly increased in HCT116-OxR and HCT8-OxR cells compared with their parent cells (HCT116 and HCT8) (Supplementary Figure 1C). The elevated PGFS expression in the resistant cells was confirmed using immunofluorescence staining and was largely localized in the cytoplasm and nuclei (Supplementary Figure 1D). Furthermore, Oxa induced PGFS expression in a dose-dependent manner, peaking at 24 h, in both parent and Oxa-resistant cells (Supplementary Figure 1E-H). Therefore, upregulated PGFS is associated with acquired Oxa resistance in CRC cells.
To further confirm the role of PGFS in Oxa resistance, we performed lentivirus-mediated overexpression of PGFS in HCT116 and HCT8 cells, which showed low levels of PGFS. Overexpression of PGFS was verified by western blotting (Supplementary Figure 2A). Stable overexpression of PGFS induced a significant right shift of the IC50 value for Oxa, which indicated a reduction in sensitivity to Oxa (Figure 2A). To validate this result, we designed three siRNAs to target PGFS and evaluated their knockdown efficiency in Oxa-resistant cells with high PGFS expression. The short hairpin RNA (shRNA) sequence that caused an obvious reduction in PGFS expression was cloned and inserted into a lentiviral vector for the stable knockdown of PGFS (Supplementary Figure 2B and C). Lentiviral shRNA-mediated knockdown of PGFS decreased the IC50 value for Oxa in HCT116-OxR and HCT8-OxR cells (Figure 2B). We then examined the effect of PGFS on the expression of PCNA (a marker of cell proliferation). Treatment with Oxa resulted in a decrease in PCNA expression, which was diminished by overexpression of PGFS in HCT116 and HCT8 cells (Figure 2C). In contrast, knockdown of PGFS further exacerbated the Oxa-induced reduction in PCNA expression in HCT116-OxR and HCT8-OxR cells (Figure 2D). Additionally, overexpression of PGFS restored the colony-forming capacity of CRC cells treated with Oxa. However, knockdown of PGFS caused a further decrease in clonogenic capacity (Figure 2E and F). In summary, PGFS attenuates Oxa-induced proliferation suppression.
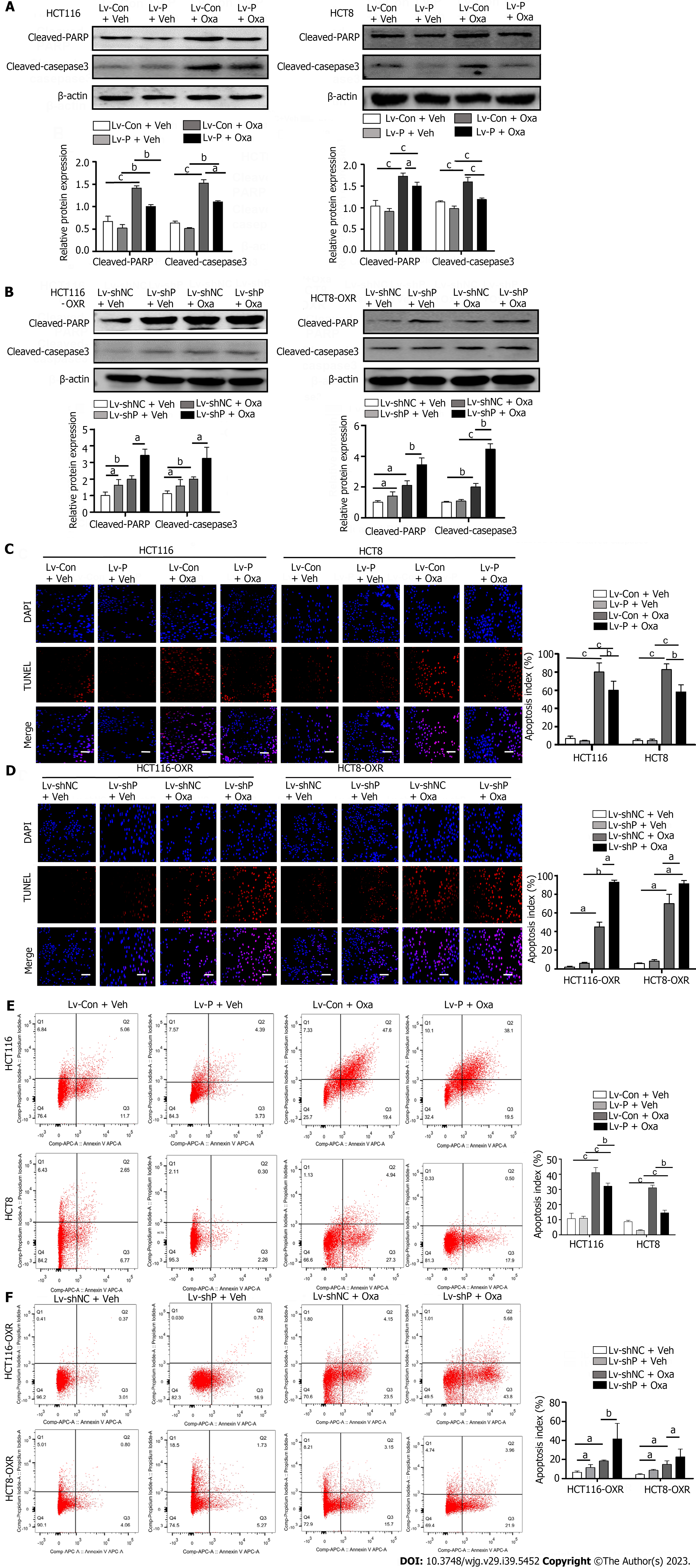
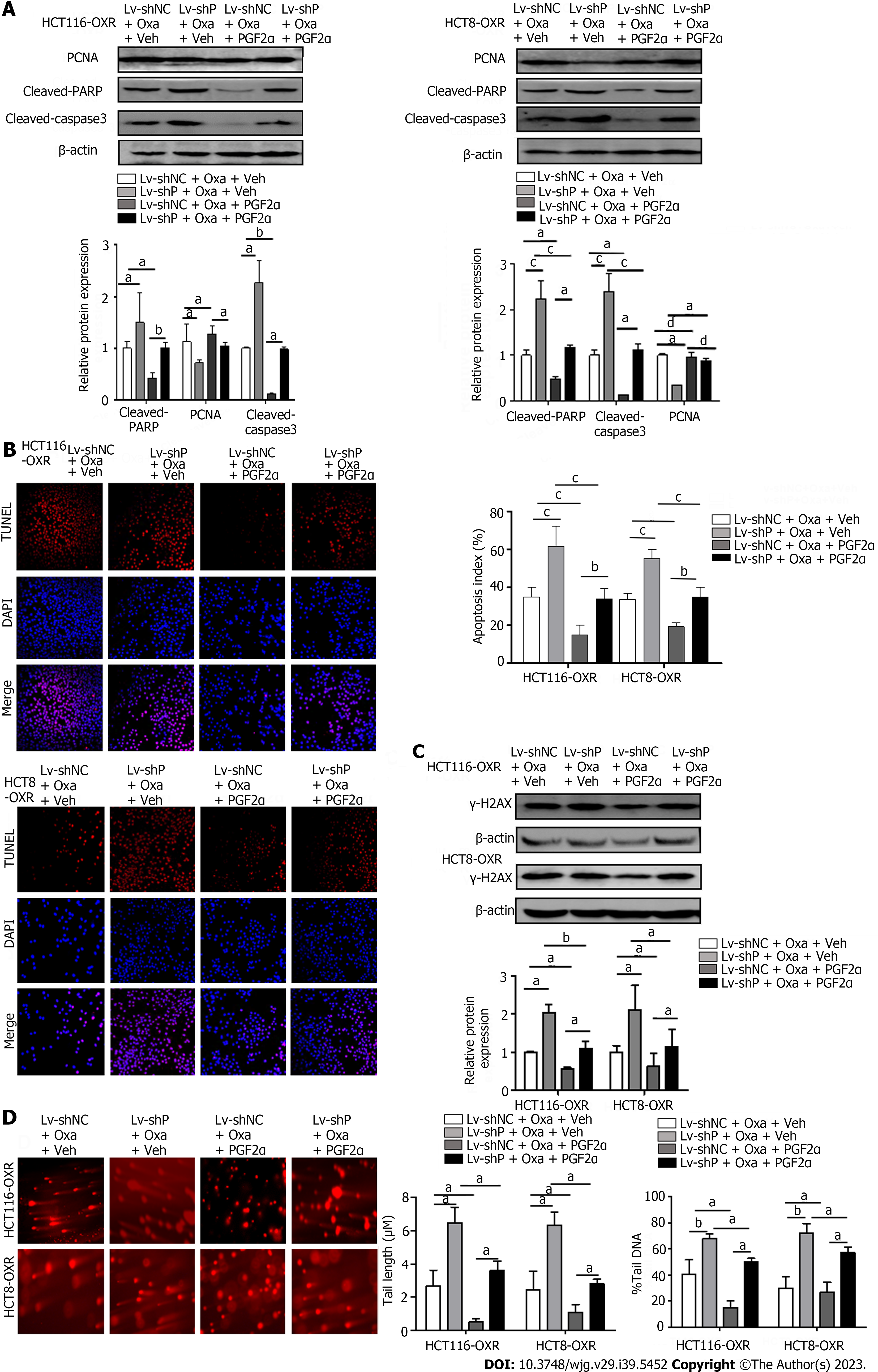
We then tested the effect of PGFS on apoptosis induced by Oxa. Western blotting showed that treatment with Oxa elevated the levels of cleaved PARP and cleaved caspase 3 in HCT116 and HCT8 cells. However, overexpression of PGFS inhibited the Oxa-induced increase in the two proteins (Figure 3A). Conversely, silencing PGFS in HCT116-OxR and HCT8-OxR cells led to a further increase in the two apoptosis markers caused by Oxa (Figure 3B). Furthermore, we examined apoptosis by TUNEL staining. We observed that PGFS overexpression reduced the increase in TUNEL-positive cells induced by Oxa treatment. In contrast, the number of apoptotic cells was further increased by infection with lentivirus expressing sh-PGFS (Figure 3C and D). In addition, we performed flow cytometry as further evidence of apoptosis. The percentage of apoptotic cells was decreased in the PGFS-overexpressing CRC cells but increased in the Oxa-resistant cells with PGFS knockdown (Figure 3E and F). These observations suggested that PGFS protects CRC cells against Oxa-induced apoptosis.
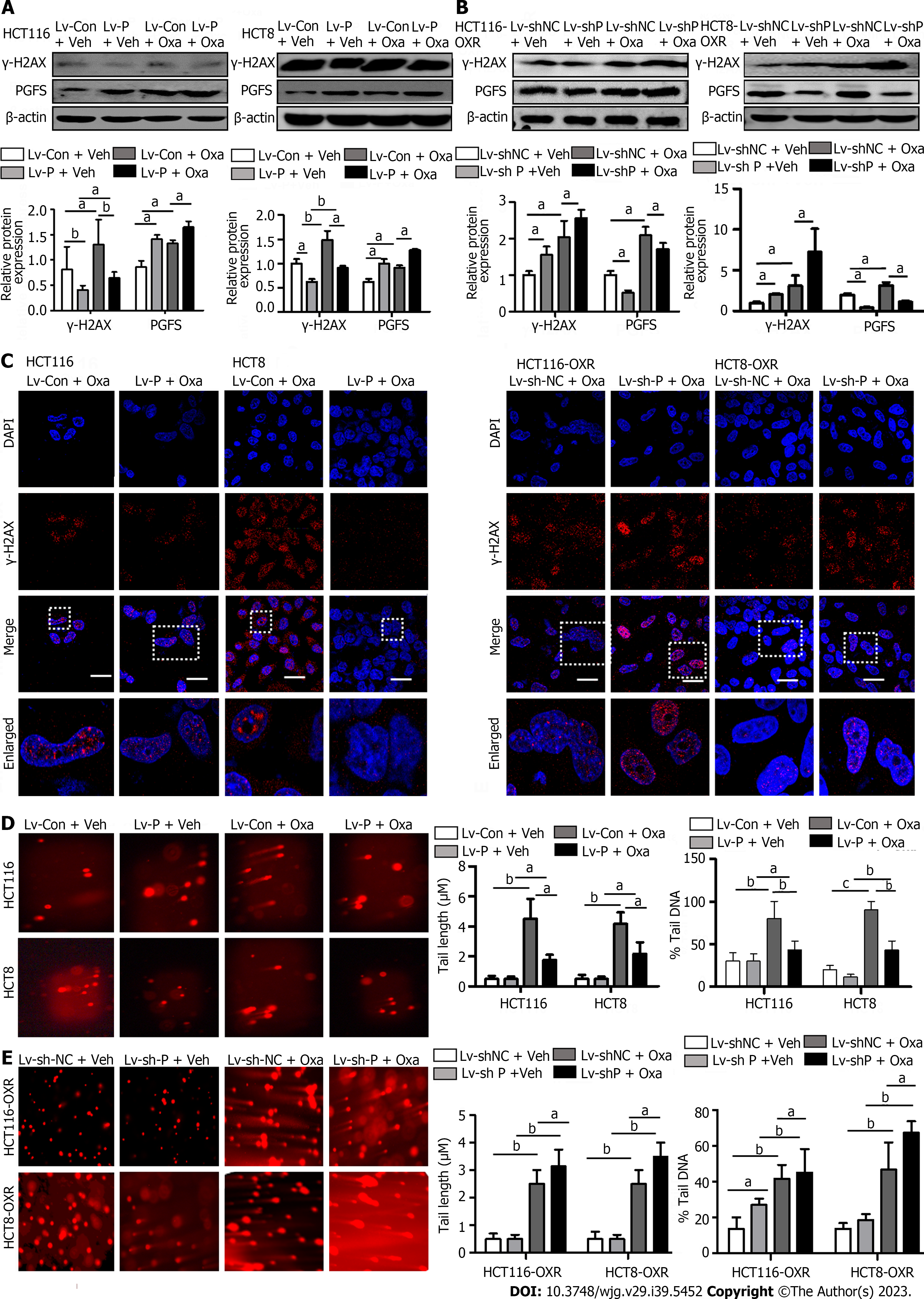
Oxa induces DNA damage by directly binding to DNA. We investigated the role of PGFS in DNA damage caused by Oxa by detecting the expression of γ-H2AX (a marker for DNA damage). The γ-H2AX level was increased by treatment with Oxa, which was diminished by PGFS overexpression in the parental CRC cells, while it was elevated by knockdown of PGFS in the Oxa-resistant CRC cells (Figure 4A and B). We then examined DNA double-strand breaks (DSBs) by γ-H2AX immunofluorescence staining. Overexpression of PGFS in HCT116 and HCT8 cells led to a reduction in the number of γ-H2AX-positive DNA damage foci induced by Oxa treatment. Conversely, the number of γ-H2AX foci was increased in the drug-resistant CRC cells upon PGFS knockdown (Figure 4C). In addition, we used a comet assay to determine DNA damage. Tail length and DNA content percentage (tail DNA%) were significantly increased after incubation with Oxa in HCT116 and HCT8 cells, and these changes were attenuated by overexpression of PGFS. Moreover, PGFS knockdown resulted in enhanced DNA damage induced by Oxa (Figure 4D and E). These results showed that PGFS is resistant to Oxa-induced DNA damage. Collectively, these findings indicated that PGFS promotes Oxa resistance in CRC cell lines.
HCT8-OxR cells with stable PGFS knockdown were inoculated subcutaneously into nude mice. Treatment with Oxa caused a decrease in tumor volume and weight, which were further decreased by knockdown of PGFS (Figure 5A-C). Immunohistochemical staining was used to evaluate PCNA expression in the xenograft tumors. Knockdown of PGFS further exacerbated the Oxa-induced reduction in PCNA expression (Figure 5D). These data indicated that PGFS knockdown enhances the sensitivity of CRC to Oxa.
PGFS catalyzed synthesis of PGF2α. To clarify whether the Oxa-resistant effect of PGFS is dependent on its enzymatic action, we blocked PGFS activity by its inhibitor indomethacin. PCNA expression was upregulated by PGFS overexpression in CRC cells treated with Oxa, and this effect was inhibited after indomethacin administration. Moreover, overexpression of PGFS attenuated the upregulation of cleaved PARP and cleaved caspase-3 caused by Oxa treatment, while indomethacin treatment reversed the effect of PGFS overexpression on the expression of apoptotic proteins (Supplementary Figure 3A). Flow cytometric analysis showed that Oxa-induced apoptosis was suppressed by infecting CRC cells with a lentivirus expressing PGFS. Treatment with indomethacin abolished the ability of PGFS to resist Oxa-induced apoptosis (Supplementary Figure 3B). Western blot analysis revealed that overexpression of PGFS inhibited the upregulation of γ-H2AX caused by Oxa, which was diminished by indomethacin incubation (Supplementary Figure 3C). A comet assay further revealed that overexpression of PGFS in CRC cells inhibited the increase in comet DNA tail length and percentage of DNA in the tail induced by Oxa, while the inhibitory function of PGFS was attenuated by indomethacin (Supplementary Figure 3D). These findings suggested that the effect of PGFS on increasing CRC resistance to Oxa depends on its PGF2α synthase activity.
PGFS catalyzes the reduction of PGD2 to PGF2α. To confirm the role of PGF2α in PGFS-mediated Oxa resistance, we performed rescue experiments by introducing PGF2α to HCT116-OxR and HCT8-OxR cells with PGFS knockdown. Western blot analysis revealed that PCNA expression was further decreased by the depletion of PGFS in HCT116 and HCT8 cells treated with Oxa, which was alleviated by the addition of PGF2α. Knockdown of PGFS further increased Oxa-induced elevation of cleaved PARP and cleaved caspase 3, which was partially rescued by PGF2α supplementation (Figure 6A). TUNEL staining results showed that the percentage of apoptotic cells was increased after the knockdown of PGFS in Oxa-treated cells, while the addition of PGF2α decreased apoptosis (Figure 6B). Moreover, knockdown of PGFS further increased the upregulation of γ-H2AX expression caused by Oxa, while supplementation with PGF2α attenuated the effect of PGFS depletion (Figure 6C). Similarly, comet assays showed that the percentage of comet tail DNA and tail length were elevated by Oxa stimulation and were further increased in HCT116-OxR and HCT8-OxR cells with PGFS knockdown, which was alleviated by the addition of PGF2α (Figure 6D). These data indicate that PGF2α, the product catalyzed by PGFS, plays an important role in PGFS-mediated Oxa resistance.
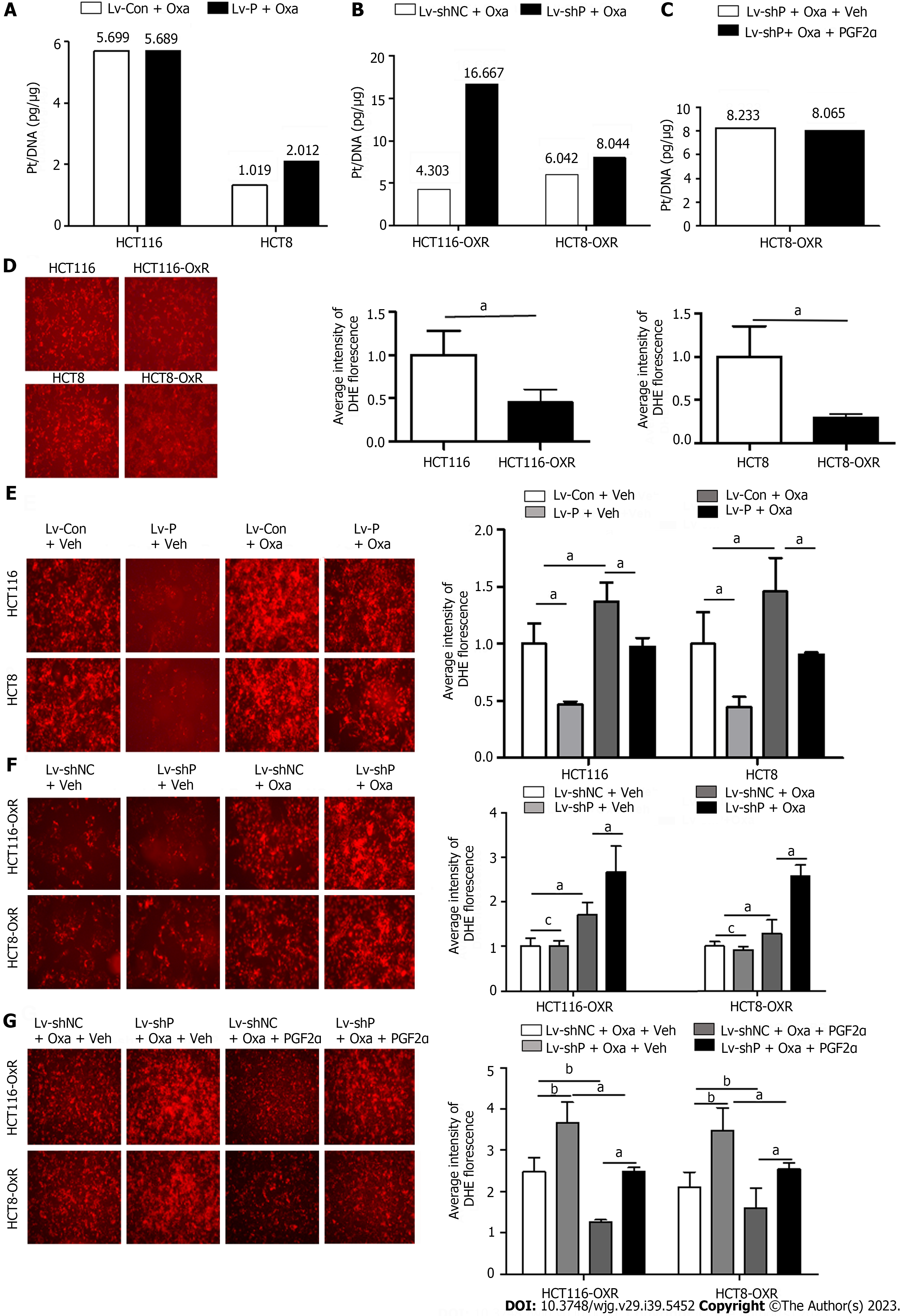
Oxa exerts its cytotoxic effect by forming platinum-DNA adducts or by generating a large amount of ROS, thereby causing DNA damage. To explore the role of PGFS in the formation of platinum-DNA adducts, we measured the amount of platinum on DNA after treatment with Oxa by inductively coupled plasma mass spectrometry. Overexpression of PGFS did not affect the platinum concentrations in HCT116 and HCT8 cells (Figure 7A). In contrast, knockdown of PGFS induced an increase in the platinum concentrations in HCT116-OxR and HCT8-OxR cells (Figure 7B). However, the addition of PGF2α did not affect the formation of platinum-DNA adducts in HCT8-OxR cells with knockdown of PGFS (Figure 7C). These findings suggested that PGFS may suppress the formation of platinum-DNA adducts in a PGF2α-independent manner.
As Oxa exerts its antitumor effect through the induction of ROS, we explored the role of PGFS in the generation of ROS by DHE staining. Compared with that of the parent CRC cells, ROS production was reduced in drug-resistant cells (Figure 7D). In addition, overexpression of PGFS led to a decrease in Oxa-induced ROS (Figure 7E). Conversely, the increased ROS induced by Oxa was further elevated by knockdown of PGFS (Figure 7F), which was alleviated by supplementation with PGF2α (Figure 7G). These data indicated that PGFS inhibits the generation of ROS in a PGF2α-dependent manner.
PGFS belongs to the AKR superfamily of NADP (H)-dependent enzymes, which play central roles in the proinflammation[20,27], proliferation[15,28] and drug resistance of tumors[29-31]. Previous studies have shown that Oxa suppresses cell proliferation and induces apoptosis in CRC cells[32]. The involvement of PGFS in resistance to cisplatin has been reported previously[24]. However, the role of PGFS in Oxa resistance in CRC and its molecular mechanism remain unclear.
Our study revealed that PGFS expression was significantly higher in human CRC tissues than in adjacent normal tissues, and PGFS may be correlated with tumor stage[28] and lymph node metastasis[33]. Therefore, how PGFS regulates CRC progression and metastasis warrants further investigation.
Induction of cell death is one of the challenges for chemotherapy. Any factors that enhance the effect of Oxa on cell proliferation and apoptosis may improve the therapeutic effect of Oxa on tumors[34,35]. PGFS-PGF2α may be a target in tumor chemotherapy. Zhou et al[33] recently found that PGFS activates nuclear factor kappaB and promotes hepatocellular carcinoma proliferation and metastasis. Other studies found that PGF2α enhances mitogen-activated protein kinase signaling and inhibits peroxisome proliferator-activated receptor-gamma, a cancer inhibitor; therefore, PGF2α promotes cancer cell proliferation and inhibits differentiation[36-38]. The present study found that PGFS was induced by Oxa in a dose-dependent manner. These findings indicated that PGFS was related to Oxa resistance in CRC. In our in vitro study, we constructed PGFS-overexpressing and PGFS knockdown cells in both parental and Oxa-resistant CRC cell lines. We proved that the effect of Oxa on apoptosis and proliferation was suppressed by PGFS overexpression and enhanced by PGFS knockdown. These results indicated that PGFS was involved in Oxa resistance in CRC.
The main mechanism of DNA damage caused by Oxa is the direct combination of platinum with DNA, ultimately destroying tumors[39]. Oxa is a platinum-based chemotherapy that induces a complex spectrum of DNA damage, including DSBs, single-strand breaks, and DNA crosslinks[40], and oxidized bases elicit DNA damage[41]. Indomethacin, a PGFS-specific inhibitor, has additive/synergistic effects on Oxa-induced proliferative inhibition, apoptosis, and DNA damage, whereas the administration of PGF2α partially alleviated the above effects[42]. In summary, our data implied that the effect of PGFS on resistance in CRC is PGF2α dependent. The knockdown of PGFS sensitizes CRC to Oxa treatment.
Several studies have demonstrated that Oxa-resistant cells can modulate the cellular resistance capacity to oxidative stress by regulating key detoxification enzymes[43,44]. The present study found that PGFS inhibited the effect of Oxa on DNA damage and thereby promoted Oxa resistance in CRC.
In the present study, PGFS was significantly increased in CRC tissues, PGFS overexpression inhibited the cytotoxic effect of Oxa on CRC cells, and downregulation of PGFS improved the efficacy of Oxa in the treatment of CRC. PGFS promotes CRC resistance to Oxa in a PGF2α-dependent manner; therefore, the PGFS-PGF2α pathway may be a potential target for the treatment of CRC patients with Oxa resistance.
PGFS knockdown effectively inhibited the resistance of CRC to Oxa. Mechanistically, PGFS knockdown increased DNA and platinum binding. Moreover, the downstream product PGF2α of PGFS was reduced. The production of ROS was increased. In summary, the above function causes reduced cell death. Hence, we surmise that downregulation of PGFS could improve the efficacy of Oxa in the treatment of CRC.
Chemoresistance is a major obstacle in colorectal cancer (CRC) therapy. Therefore, characterizing mechanisms of chemoresistance is beneficial to improve the treatment efficacy and survival rate of CRC patients. In this study, we identify the role and potential mechanism of prostaglandin F2α synthase (PGF2α) (PGFS) in drug resistance to CRC, providing a novel therapeutic target against cancer drug resistance in the treatment of CRC.
The new theory of this study is that PGFS resistance to oxaliplatin (Oxa) has two effects, one is to reduce the production of reactive oxygen species (ROS) through the generation of PGF2α products, and the other is the direct protective effect of PGFS on CRC nucleus. The new method in this study is the application of comet experiment to detect DNA damage, and the other is the application of inductively coupled plasma mass spectrometry (ICP-MS) to directly detect the platinum content of DNA in the nucleus, so as to directly detect the effect of PGFS on the binding of platinum and DNA.
This study was designed to exploit the function and mechanism of PGFS in chemoresistance in CRC. Our study reveals the different ways in which PGFS promotes chemoresistance in CRC, and provides a potential target for predicting and reversing chemoresistance of CRC.
The expression level of PGFS is assessed in 37 pairs of CRC tissues and para-cancer tissues by as detected by quantitative polymerase chain reaction and western blot. We examined the influence of PGFS overexpression or knockdown in acquired Oxa-resistant CRC cell lines (HCT116-OxR and HCT8-OxR) and their parental cell lines (HCT116 and HCT8). In order to analyze how PGFS affects colon cancer cell proliferation, A cholecystokinin octapeptide assay was utilized to determine the half-inhibitory concentration value of the cells, a plate clone formation assay was used to determine the clonogenesis ability, and an analysis of proliferating cell nuclear antigen expression was performed to determine the growth rate. Transferase dUTP nick end labeling and Annexin V/propidium iodide stainings, as well as the apoptotic markers cleaved-poly ADP-ribose polymerase and cleaved-caspase 3, were used to detect apoptosis. Western blot and cellular immunofluorescence were used to detect the expression and morphology of the DNA damage marker γ-H2AX. The DNA damage was detected by single-cell gel electrophoresis. Indomethacin, an inhibitor of prostaglandin synthase of PGFS, was used to elucidate the underlying mechanisms. Rescue experiments were conducted by introducing PGF2α, the product of PGFS, subsequent to the knockdown of PGFS. The platinum-DNA adducts were quantified using ICP-MS, and intracellular ROS levels were measured using a kit for measuring reactive oxygen species.
We found that PGFS reduced the production of ROS through its downstream product PGF2α, and thereby promotes Oxa resistance in CRC, meanwhile, it inhibited the formation of platinum-DNA adducts in a PGF2α-independent manner. The suppressive role of PGFS in the formation of platinum-DNA adducts has never been reported before. However, some questions need to be further clarified, for example, by what mechanism does PGFS suppress the formation of platinum-DNA adducts?
This study aims to explore the relationship between PGFS and the occurrence and development of CRC, and the relationship between PGFS and Oxa resistance in CRC as well as the related mechanisms. In the future, it is hoped to predict whether patients with CRC are resistant to Oxa by detecting PGFS genes.
Further work will be needed to clarify the function of PGFS in the nucleus and its mechanisms of action. Moreover, the inhibition mechanism of PGFS in the formation of platinum-DNA adducts needs to be further elucidated.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): E
P-Reviewer: Liao Z, Singapore; Liu H, United States; Sahin TT, Turkey S-Editor: Qu XL L-Editor: A P-Editor: Cai YX
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64681] [Article Influence: 16170.3] [Reference Citation Analysis (177)] |
| 2. | Yang Y, Wang HY, Chen YK, Chen JJ, Song C, Gu J. Current status of surgical treatment of rectal cancer in China. Chin Med J (Engl). 2020;133:2703-2711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 3. | Taieb J, Shi Q, Pederson L, Alberts S, Wolmark N, Van Cutsem E, de Gramont A, Kerr R, Grothey A, Lonardi S, Yoshino T, Yothers G, Sinicrope FA, Zaanan A, André T. Prognosis of microsatellite instability and/or mismatch repair deficiency stage III colon cancer patients after disease recurrence following adjuvant treatment: results of an ACCENT pooled analysis of seven studies. Ann Oncol. 2019;30:1466-1471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 109] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 4. | Glynne-Jones R, Wyrwicz L, Tiret E, Brown G, Rödel C, Cervantes A, Arnold D; ESMO Guidelines Committee. Rectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 292] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 5. | Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen YJ, Ciombor KK, Cohen S, Cooper HS, Deming D, Garrido-Laguna I, Grem JL, Gunn A, Hoffe S, Hubbard J, Hunt S, Kirilcuk N, Krishnamurthi S, Messersmith WA, Meyerhardt J, Miller ED, Mulcahy MF, Nurkin S, Overman MJ, Parikh A, Patel H, Pedersen K, Saltz L, Schneider C, Shibata D, Skibber JM, Sofocleous CT, Stoffel EM, Stotsky-Himelfarb E, Willett CG, Johnson-Chilla A, Gurski LA. NCCN Guidelines Insights: Rectal Cancer, Version 6.2020. J Natl Compr Canc Netw. 2020;18:806-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 355] [Cited by in RCA: 332] [Article Influence: 66.4] [Reference Citation Analysis (0)] |
| 6. | Tougeron D, Mouillet G, Trouilloud I, Lecomte T, Coriat R, Aparicio T, Des Guetz G, Lécaille C, Artru P, Sickersen G, Cauchin E, Sefrioui D, Boussaha T, Ferru A, Matysiak-Budnik T, Silvain C, Karayan-Tapon L, Pagès JC, Vernerey D, Bonnetain F, Michel P, Taïeb J, Zaanan A. Efficacy of Adjuvant Chemotherapy in Colon Cancer With Microsatellite Instability: A Large Multicenter AGEO Study. J Natl Cancer Inst. 2016;108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 112] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 7. | de Gramont A, Schmoll HJ, Cervantes A, Tournigand C. The evolving role of oxaliplatin in the management of colorectal cancer. Colorectal Dis. 2003;5 Suppl 3:10-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 8. | Rödel C, Graeven U, Fietkau R, Hohenberger W, Hothorn T, Arnold D, Hofheinz RD, Ghadimi M, Wolff HA, Lang-Welzenbach M, Raab HR, Wittekind C, Ströbel P, Staib L, Wilhelm M, Grabenbauer GG, Hoffmanns H, Lindemann F, Schlenska-Lange A, Folprecht G, Sauer R, Liersch T; German Rectal Cancer Study Group. Oxaliplatin added to fluorouracil-based preoperative chemoradiotherapy and postoperative chemotherapy of locally advanced rectal cancer (the German CAO/ARO/AIO-04 study): final results of the multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2015;16:979-989. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 433] [Cited by in RCA: 536] [Article Influence: 53.6] [Reference Citation Analysis (0)] |
| 9. | Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869-1883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1561] [Cited by in RCA: 2016] [Article Influence: 144.0] [Reference Citation Analysis (0)] |
| 10. | Vaughn CM, Selby CP, Yang Y, Hsu DS, Sancar A. Genome-wide single-nucleotide resolution of oxaliplatin-DNA adduct repair in drug-sensitive and -resistant colorectal cancer cell lines. J Biol Chem. 2020;295:7584-7594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 11. | Assaf KI, Nau WM. Cucurbiturils: from synthesis to high-affinity binding and catalysis. Chem Soc Rev. 2015;44:394-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 905] [Cited by in RCA: 922] [Article Influence: 92.2] [Reference Citation Analysis (0)] |
| 12. | Stordal BK, Davey MW, Davey RA. Oxaliplatin induces drug resistance more rapidly than cisplatin in H69 small cell lung cancer cells. Cancer Chemother Pharmacol. 2006;58:256-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Ekblad L, Kjellström J, Johnsson A. Reduced drug accumulation is more important in acquired resistance against oxaliplatin than against cisplatin in isogenic colon cancer cells. Anticancer Drugs. 2010;21:523-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 14. | Chu C, Wei H, Zhu W, Shen Y, Xu Q. Decreased Prostaglandin D2 Levels in Major Depressive Disorder Are Associated with Depression-Like Behaviors. Int J Neuropsychopharmacol. 2017;20:731-739. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 15. | Wang MT, Honn KV, Nie D. Cyclooxygenases, prostanoids, and tumor progression. Cancer Metastasis Rev. 2007;26:525-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 232] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 16. | Wang S, Yang Q, Fung KM, Lin HK. AKR1C2 and AKR1C3 mediated prostaglandin D2 metabolism augments the PI3K/Akt proliferative signaling pathway in human prostate cancer cells. Mol Cell Endocrinol. 2008;289:60-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 17. | Sinreih M, Anko M, Kene NH, Kocbek V, Rižner TL. Expression of AKR1B1, AKR1C3 and other genes of prostaglandin F2α biosynthesis and action in ovarian endometriosis tissue and in model cell lines. Chem Biol Interact. 2015;234:320-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 18. | Watanabe K. Recent reports about enzymes related to the synthesis of prostaglandin (PG) F(2) (PGF(2α) and 9α, 11β-PGF(2)). J Biochem. 2011;150:593-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 19. | Fung KM, Samara EN, Wong C, Metwalli A, Krlin R, Bane B, Liu CZ, Yang JT, Pitha JV, Culkin DJ, Kropp BP, Penning TM, Lin HK. Increased expression of type 2 3alpha-hydroxysteroid dehydrogenase/type 5 17beta-hydroxysteroid dehydrogenase (AKR1C3) and its relationship with androgen receptor in prostate carcinoma. Endocr Relat Cancer. 2006;13:169-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 100] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 20. | Wang HW, Lin CP, Chiu JH, Chow KC, Kuo KT, Lin CS, Wang LS. Reversal of inflammation-associated dihydrodiol dehydrogenases (AKR1C1 and AKR1C2) overexpression and drug resistance in nonsmall cell lung cancer cells by wogonin and chrysin. Int J Cancer. 2007;120:2019-2027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 92] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 21. | Baraibar MA, Liu L, Ahmed EK, Friguet B. Protein oxidative damage at the crossroads of cellular senescence, aging, and age-related diseases. Oxid Med Cell Longev. 2012;2012:919832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 22. | Srinivas US, Tan BWQ, Vellayappan BA, Jeyasekharan AD. ROS and the DNA damage response in cancer. Redox Biol. 2019;25:101084. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1147] [Cited by in RCA: 1337] [Article Influence: 222.8] [Reference Citation Analysis (0)] |
| 23. | Sugino N, Karube-Harada A, Kashida S, Takiguchi S, Kato H. Reactive oxygen species stimulate prostaglandin F2 alpha production in human endometrial stromal cells in vitro. Hum Reprod. 2001;16:1797-1801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Matsunaga T, Hojo A, Yamane Y, Endo S, El-Kabbani O, Hara A. Pathophysiological roles of aldo-keto reductases (AKR1C1 and AKR1C3) in development of cisplatin resistance in human colon cancers. Chem Biol Interact. 2013;202:234-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 25. | Liu Z, Liu J, Liu X, Wang X, Xie Q, Zhang X, Kong X, He M, Yang Y, Deng X, Yang L, Qi Y, Li J, Liu Y, Yuan L, Diao L, He F, Li D. CTR-DB, an omnibus for patient-derived gene expression signatures correlated with cancer drug response. Nucleic Acids Res. 2022;50:D1184-D1199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 26. | Tan Z, Gao L, Wang Y, Yin H, Xi Y, Wu X, Shao Y, Qiu W, Du P, Shen W, Fu L, Jia R, Zhao C, Zhang Y, Zhao Z, Sun Z, Chen H, Hu X, Xu J. PRSS contributes to cetuximab resistance in colorectal cancer. Sci Adv. 2020;6:eaax5576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 27. | Schiffer L, Bossey A, Kempegowda P, Taylor AE, Akerman I, Scheel-Toellner D, Storbeck KH, Arlt W. Peripheral blood mononuclear cells preferentially activate 11-oxygenated androgens. Eur J Endocrinol. 2021;184:353-363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | Nakarai C, Osawa K, Akiyama M, Matsubara N, Ikeuchi H, Yamano T, Hirota S, Tomita N, Usami M, Kido Y. Expression of AKR1C3 and CNN3 as markers for detection of lymph node metastases in colorectal cancer. Clin Exp Med. 2015;15:333-341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 29. | Zhao J, Xiang Y, Xiao C, Guo P, Wang D, Liu Y, Shen Y. AKR1C3 overexpression mediates methotrexate resistance in choriocarcinoma cells. Int J Med Sci. 2014;11:1089-1097. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 30. | Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev. 2004;56:185-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2875] [Cited by in RCA: 2689] [Article Influence: 128.0] [Reference Citation Analysis (0)] |
| 31. | Heibein AD, Guo B, Sprowl JA, Maclean DA, Parissenti AM. Role of aldo-keto reductases and other doxorubicin pharmacokinetic genes in doxorubicin resistance, DNA binding, and subcellular localization. BMC Cancer. 2012;12:381. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 75] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 32. | Rixe O, Ortuzar W, Alvarez M, Parker R, Reed E, Paull K, Fojo T. Oxaliplatin, tetraplatin, cisplatin, and carboplatin: spectrum of activity in drug-resistant cell lines and in the cell lines of the National Cancer Institute's Anticancer Drug Screen panel. Biochem Pharmacol. 1996;52:1855-1865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 449] [Cited by in RCA: 444] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 33. | Zhou Q, Tian W, Jiang Z, Huang T, Ge C, Liu T, Zhao F, Chen T, Cui Y, Li H, Yao M, Li J, Tian H. A Positive Feedback Loop of AKR1C3-Mediated Activation of NF-κB and STAT3 Facilitates Proliferation and Metastasis in Hepatocellular Carcinoma. Cancer Res. 2021;81:1361-1374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 64] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 34. | Ruan Y, Wang L, Lu Y. HDAC6 inhibitor, ACY1215 suppress the proliferation and induce apoptosis of gallbladder cancer cells and increased the chemotherapy effect of gemcitabine and oxaliplatin. Drug Dev Res. 2021;82:598-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 35. | Hsu HH, Chen MC, Baskaran R, Lin YM, Day CH, Lin YJ, Tu CC, Vijaya Padma V, Kuo WW, Huang CY. Oxaliplatin resistance in colorectal cancer cells is mediated via activation of ABCG2 to alleviate ER stress induced apoptosis. J Cell Physiol. 2018;233:5458-5467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 124] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 36. | Sales KJ, List T, Boddy SC, Williams AR, Anderson RA, Naor Z, Jabbour HN. A novel angiogenic role for prostaglandin F2alpha-FP receptor interaction in human endometrial adenocarcinomas. Cancer Res. 2005;65:7707-7716. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 81] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 37. | Suzuki-Yamamoto T, Nishizawa M, Fukui M, Okuda-Ashitaka E, Nakajima T, Ito S, Watanabe K. cDNA cloning, expression and characterization of human prostaglandin F synthase. FEBS Lett. 1999;462:335-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 98] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 38. | Qualtrough D, Kaidi A, Chell S, Jabbour HN, Williams AC, Paraskeva C. Prostaglandin F(2alpha) stimulates motility and invasion in colorectal tumor cells. Int J Cancer. 2007;121:734-740. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 52] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 39. | Zhang Y, Xie C, Li A, Liu X, Xing Y, Shen J, Huo Z, Zhou S, Xie Y, Cao W, Ma Y, Xu R, Cai S, Tang X, Ma D. PKI-587 enhances chemosensitivity of oxaliplatin in hepatocellular carcinoma through suppressing DNA damage repair pathway (NHEJ and HR) and PI3K/AKT/mTOR pathway. Am J Transl Res. 2019;11:5134-5149. [PubMed] |
| 40. | Liu J, Fu XQ, Zhou W, Yu HG, Yu JP, Luo HS. LY294002 potentiates the anti-cancer effect of oxaliplatin for gastric cancer via death receptor pathway. World J Gastroenterol. 2011;17:181-190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 16] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 41. | Deb S, Xu H, Tuynman J, George J, Yan Y, Li J, Ward RL, Mortensen N, Hawkins NJ, McKay MJ, Ramsay RG, Fox SB. RAD21 cohesin overexpression is a prognostic and predictive marker exacerbating poor prognosis in KRAS mutant colorectal carcinomas. Br J Cancer. 2014;110:1606-1613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 42. | Liedtke AJ, Adeniji AO, Chen M, Byrns MC, Jin Y, Christianson DW, Marnett LJ, Penning TM. Development of potent and selective indomethacin analogues for the inhibition of AKR1C3 (Type 5 17β-hydroxysteroid dehydrogenase/prostaglandin F synthase) in castrate-resistant prostate cancer. J Med Chem. 2013;56:2429-2446. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 43. | Marzano C, Gandin V, Folda A, Scutari G, Bindoli A, Rigobello MP. Inhibition of thioredoxin reductase by auranofin induces apoptosis in cisplatin-resistant human ovarian cancer cells. Free Radic Biol Med. 2007;42:872-881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 342] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 44. | Rudin CM, Yang Z, Schumaker LM, VanderWeele DJ, Newkirk K, Egorin MJ, Zuhowski EG, Cullen KJ. Inhibition of glutathione synthesis reverses Bcl-2-mediated cisplatin resistance. Cancer Res. 2003;63:312-318. [PubMed] |