Published online Sep 21, 2023. doi: 10.3748/wjg.v29.i35.5104
Peer-review started: June 25, 2023
First decision: August 8, 2023
Revised: August 10, 2023
Accepted: August 25, 2023
Article in press: August 25, 2023
Published online: September 21, 2023
Processing time: 81 Days and 9 Hours
Regenerating gene 4 (REG4) has been proved to be carcinogenic in some cancers, but its manifestation and possible carcinogenic mechanisms in colorectal cancer (CRC) have not yet been elucidated. Our previous study found that the drug resistance of CRC cells may be closely linked to their fat metabolism.
To explore the role of REG4 in CRC and its association with lipid droplet forma
We conducted a meta-analysis and bioinformatics and pathological analyses of REG4 expression in CRC. The effects of REG4 on the phenotypes and related protein expression were also investigated in CRC cells. We detected the impacts of REG4 on the chemoresistance and lipid droplet formation in CRC cells. Finally, we analyzed how REG4 regulated the transcription and proteasomal degradation of lipogenic enzymes in CRC cells.
Compared to normal mucosa, REG4 mRNA expression was high in CRC (P < 0.05) but protein expression was low. An inverse correlation existed between lymph node and distant metastases, tumor-node-metastasis staging or short overall survival and REG4 mRNA overexpression (P < 0.05), but vice versa for REG4 protein expression. REG4-related genes included: Chemokine activity; taste receptors; protein-DNA and DNA packing complexes; nucleosomes and chromatin; generation of second messenger molecules; programmed cell death signals; epigenetic regulation and DNA methylation; transcription repression and activation by DNA binding; insulin signaling pathway; sugar metabolism and transfer; and neurotransmitter receptors (P < 0.05). REG4 exposure or overexpression promoted proliferation, antiapoptosis, migration, and invasion of DLD-1 cells in an autocrine or paracrine manner by activating the epidermal growth factor receptor-phosphoinositide 3-kinase-Akt-nuclear factor-κB pathway. REG4 was involved in chemoresistance not through de novo lipogenesis, but lipid droplet assembly. REG4 inhibited the transcription of acetyl-CoA carboxylase 1 (ACC1) and ATP-citrate lyase (ACLY) by disassociating the complex formation of anti–acetyl (AC)-acetyl-histone 3-AC-histone 4-inhibitor of growth protein-5-si histone deacetylase;-sterol-regulatory element binding protein 1 in their promoters and induced proteasomal degradation of ACC1 or ACLY.
REG4 may be involved in chemoresistance through lipid droplet assembly. REG4 reduces expression of de novo lipid synthesis key enzymes by inhibiting transcription and promoting ubiquitination-mediated proteasomal degradation.
Core Tip: The roles of regenerating gene 4 (REG4) in colorectal cancer (CRC) and the related molecular mechanisms are still unknown. REG4 may be involved in tumorigenesis and aggressiveness of CRC via the epidermal growth factor receptor-phosphoinositide 3-kinase-Akt-nuclear factor-κB pathway, and chemoresistance through lipid droplet assembly. REG4 might be used as a useful marker for colorectal carcinogenesis and subsequent progression, as well as a potential gene therapy target.
- Citation: Zhang CY, Zhang R, Zhang L, Wang ZM, Sun HZ, Cui ZG, Zheng HC. Regenerating gene 4 promotes chemoresistance of colorectal cancer by affecting lipid droplet synthesis and assembly. World J Gastroenterol 2023; 29(35): 5104-5124
- URL: https://www.wjgnet.com/1007-9327/full/v29/i35/5104.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i35.5104
The regenerating gene 4 (REG4) gene family encodes secretory calcium-dependent lectins, which act as acute phase reactants, antiapoptotic factors, and growth agents, and contributes to cellular proliferation and differentiation, inflammation, diabetes, and cancer. To date, human REG genes include REG I (Iα and Iβ), Reg III (III and HIP/PAP), and REG4[1-3]. Of these, REG4 is transcriptionally activated by several transcription factors (e.g., Sp1 transcription factor, GATA binding protein 6 (GATA6) and caudal type homeobox 2), and the REG4 protein targets, epidermal growth factor receptor (EGF) (EGFR)/Akt/activator protein-1 and Akt/glycogen synthase kinase-3β/β-catenin/T cell factor-4 signaling pathways as a mitogen[4-8]. In pancreatic cancer cells, REG4 also enhanced invasion by upregulating matrix metalloproteinase expression and macrophage polarization through the EGFR–Akt–c-AMP response element binding protein pathway[9,10]. In gastric cancer cells, the forced expression of REG4 induced EGFR phosphorylation and inhibited 5-fluorouracil (5-FU)-induced apoptosis[11]. In ovarian cancer cells, REG4 overexpression as well as recombinant (rh) REG4 exposure prevented apoptosis, and enhanced proliferation, migration, and invasion with the hyperexpression of Wnt5a, p70s6k, survivin and vascular endothelial growth factor, and reduced expression of Bax[12].
In colorectal tissues, REG4 mRNA-positive cells are mostly enteroendocrine and goblet cells. Adenomatous and cancer cells positive for REG4 mRNA exhibited enterocyte-like, mucus-secreting, or undifferentiated features[13]. At the protein level, REG4 expression was observed in the middle and outer parts of crypts and superficial epithelium, especially goblets[14]. Oue et al[15] found that the preoperative serum REG4 concentration was not elevated in patients with colorectal cancer (CRC) at stages 0–III, but was significantly elevated in those at stage IV. Additionally, REG4 expression was significantly linked to a worse prognosis in patients with CRC as an independent predictor[15-17]. REG4 promoted migration and invasion of CRC cells via its carbohydrate-recognition domain in both autocrine and paracrine manners, which was significantly decreased by anti-REG4 antibody[18,19]. Nanakin et al[20] found that REG4 expression was stimulated by tumor necrosis factor (TNF) α, EGF, basic fibroblast growth factor, and hepatocyte growth factor in colon cancer cells, and then promoted cell proliferation and resistance to H2O2-induced apoptosis. As for therapy resistance, REG4 was markedly related to chemoresistance. Violette et al[13] discovered that REG4 protein was strongly expressed in chemoresistant rectal cancer cells, but expressed weakly in drug-sensitive rectal cancer cells. REG4 expression was found to correlate with γ-radiation sensitivity in rectal cancer patients receiving radiotherapy[21]. In radiochemotherapy-sensitive CRC cells, REG4 expression was downregulated, while it was increased in radiochemoresistant cells[22].
Leukocytes have been found to play an important role in shaping the immune microenvironment of CRC[23-25]. Among them, the interleukin (IL)-22-mediated positive effect depended on its ability to induce neutrophil chemokines into the tumor microenvironment[24]. The proinflammatory cytokine group factors (e.g. TNF-α, IL-6, IL-12 and IL-23) promoted tumor cell survival, induced angiogenesis, and facilitated cell migration by exerting antiapoptotic activity[25]. Immune checkpoint molecules, including programmed death ligand ½, B7-1/2, B7-acetyl-histone 3 (H3), B7x, V-domain Ig suppressor of T cell activation and galectin-9, were found to be effective regulators of immune activation, which play a key role in immune escape of CRC[26]. Jang[27] found that T regulatory (Treg) cells gradually increased, while CD8+ T cells and CD8+ T cell/Treg cell ratio decreased during progression of CRC. Infiltration of T cells differed between tumor regions, and the proportion in the central region of CRC was the lowest. Unfortunately, there is no report on the relationship between REG4 and the microenvironment in CRC.
In our previous study, REG4 expression gradually decreased from gastric intestinal metaplasia, adenoma, and cancer to gastritis at either the mRNA or protein level[28]. Serum REG4 levels were elevated in patients with gastric cancer, compared to those in healthy individuals. Our group subsequently developed a mouse anti-REG4 monoclonal antibody for immunohistochemistry, and found that REG4 was downregulated in CRC[3]. To understand the roles of REG4 in tumorigenesis and subsequent progression of CRC, we undertook meta-analysis, and bioinformatics, pathological and serological analyses to explore the clinicopathological and prognostic significances of REG4 in CRC. The effects of REG4 on the phenotypes of CRC cells were also studied with the detailed mechanisms clarified. We also investigated the impacts of REG4 on lipid droplet forma
A CRC cell line (DLD-1) was generously donated by Prof. Toshiro Sugiyama, Department of Gastroenterology, University of Toyama, Japan. This line was maintained in RPMI 1640 growth medium supplemented with 11% fetal bovine serum (FBS), 90 U/mL penicillin, and 90 μg/mL streptomycin in a humidified atmosphere of 5% CO2 at 37 ℃. The cells were transfected at 75% confluence 20-24 h after seeding on culture dishes (Qiagen, Hilden, Germany). The cells were transfected with pcDNA3.1-REG4 [full-length (FL)-REG4], pcDNA3.1-REG4 [nonsignal peptide (NSP)-REG4] or siRNA against histone deacetylase [si histone deacetylase (HDAC); Santa Cruz Biotechnology, Dallas, TX, United States] using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, United States). The cells were treated with high glucose (4.5 mg/L), recombinant human REG4 protein (rhREG4; R and D Systems, Minneapolis, MN, United States) or anti-REG4 antibody (R and D Systems). The cells were also exposed to 5-FU (thymidylate synthetase inhibitor); cisplatin (DDP), DNA crosslinker; suberoylanilide hydroxamic acid (SAHA); HDAC inhibitor; cycloheximide (CHX), selective inhibitor of protein synthesis to study the degradation and stability of proteins; MG132 (proteasome inhibitor to suppress protea
As a measure of cell proliferation, MTT was reduced to formazan in a mitochondrial-dependent manner. We seeded cells in 96-well plates. After culture for 24 or 48 h in a 37 ℃ incubator, 50 μL MTT reagent (5 mg/mL) was added to each well. After incubation for 4 h, 150 μL dimethyl sulfoxide was added to each well for coloration. After vigorous shaking of the plates for 30-min to achieve complete solubilization, the optical density was measured on a microplate reader (Model 680; Bio-Rad, Hercules, CA, United States) at a wavelength of 490 nm. We normalized the OD 490 values to the viability of control cells that were not treated with reagents or siRNA. Normalized viability (%) = (OD490 of treated cells/OD490 of untreated control cells) × 100. The normalized viability values were plotted to show the growth inhibition effects of treatments.
We detected phosphatidylserine externalization (on the cell membrane) using fluorescein isothiocyanate (FITC)-labeled Annexin V (Immunotech, Marseille, France) and propidium iodide (PI) to identify early apoptosis. After treatment, any remaining intact cells were collected, washed with cooled PBS at 4 ℃ and centrifuged at 700 × g for 3 min. The cell suspension (490 μL) was gently mixed with FITC-labeled Annexin-V (5 μL) and PI (5 μL). After incubation at 4 ℃ for 10 min in the dark, the cell suspension was detected by flow cytometry.
Cells (5.5 × 105 per well) were plated in six-well culture plates. When the cells reached 75% confluence, they were scraped with a tip, washed four times in PBS to reduce the number of broken cells, and cultured in FBS-free RPMI 1640 medium. We measured the area of the scratch immediately after wounding (0 h) and 24 and 48 h later using Image J. The percentage wound closure was calculated as: (Area of scratch at 0 h-area of scratch at 24 and 48 h)/area of scratch at 0 h) × 100. The values were plotted to show differences in migration between conditions.
Assays for migration were conducted by suspending 1.3 × 105 cells per 200 μL in FBS-free RPMI 1640 and seeding the upper chamber of each Transwell (BD Biosciences, Franklin Lakes, NJ, United States). As a chemoattractant, 11% FBS was added to each lower compartment. After incubation for 24 h, we scrubbed the upper surface of the Transwell membrane, washed the Transwell chamber with PBS three times and fixed cells in 100% cold methanol. The membrane was stained with crystal violet for several minutes. For the invasive assay, the same process as above was used except that each Transwell insert was coated with Matrigel (BD Biosciences).
Cells were cultured on coverslips for 12 h, fixed in 4% paraformaldehyde for 28 min and stained with Nile red (Invitrogen, Carlsbad, CA, United States; 1: 1000) for 13 min. Finally, we stained slides with 4’,6-diamidino-2-phenylindole (DAPI) and covered them with SlowFade® Gold anti-fade reagent (Thermo Fisher Scientific). The software programs, Image J and Icy, were used to acquire and analyze images.
Cells were cultured on glass slides, fixed for 10 min with 4% formaldehyde, and permeabilized for 10 min with 0.5% Triton X-100. Cells were rinsed with PBS and treated with anti-goat REG4 (R and D Systems; 1: 100) antibody for 3 h. The cells were incubated with Alexa Fluor 488 (green) anti-goat immunoglobulin G (IgG) (Invitrogen) for 50 min. Slides were mounted using SlowFade® Gold anti-fade reagent after being stained with DAPI.
A Magna ChIPTM G kit (Millipore, Burlington, MA, United States) was used to conduct ChIP. After IP of anti-acetyl (AC) H3, anti-AC-histone 4 (H4), anti-HDAC, anti–inhibitor of growth protein (ING) 5 or anti-sterol-regulatory element binding protein 1 (SREBP1) antibody (Supplementary Table 1), we used ACLY (5’-AATCGCGGGGCCGTTCTC-3’, melting temperature (TM): 57.9 ℃; 5’-CGACGAACCCCGCAAAATC-3’, TM: 55.4 ℃, -43 bp to + 81 bp) primers or ACC1 (5’-GCCCCGAATGGCAGATCC-3’, TM: 55.7 ℃; 5’-GCTCAGCGGCAGCCAATG-3’, TM: 57.4 ℃; -33 bp to + 54 bp) primers for polymerase chain reaction (PCR). IgG was used as a negative control and anti-polymerase II as a positive control. In 20 μL of mixture, DNA was amplified and separated on a 2% agarose gel.
Seven micrograms of primary antibody (Supplementary Table 1) was added to > 1 mg protein and subjected to rotation at 4 ℃ overnight. One hundred microliters of agarose A beads were added, and the mixture was rotated at 4 ℃ overnight. To exclude nonspecific binding proteins, the beads were centrifuged and washed with 1% NP40 Lysis buffer four times. The pellet was mixed using 50 μL 2 × SDS sample buffer, and heated at 100 ℃ for 10 min. The sample supernatant was used for western blotting.
Colorectal primary cancers (n = 796), adenomas (n = 62), non-neoplastic mucosa [(NNM), n = 667] and metastatic cancers in lymph nodes (n = 179) were sampled at the First Affiliated Hospital of Jinzhou Medical University between 2013 and 2022. One hundred cases of CRC and adjacent NNM were obtained from Liaoning Cancer Hospital and stored at -80 ℃ until RNA and protein extraction. Eighty patients with CRC and 50 healthy volunteers were enrolled to determinate the serum REG4 level at Liaoning Cancer Hospital and The Affiliated Hospital of Chengde Medical University. Before surgery, none of the patients had undergone radiotherapy or chemotherapy. The ethics committee of our institution authorized the research plan after giving unanimous approval to utilize tumor tissues and patient serum for clinical research.
All tissues were preserved in 10% neutral formalin, embedded in paraffin, and sectioned into 4-mm pieces. Hematoxylin and eosin (HE) staining was used to validate the histological diagnosis and other microscopic features of these sections. In HE-stained sections, representative portions of solid tumors were selected under a microscope. Using a tissue microarray, a 2-mm tissue core from each donor block was punched out and transferred to a recipient block with a maximum of 48 cores (Azumaya kin-1, Nagoya, Japan). From the recipient block, 4-μm–thick sections were sequentially cut and placed on glass slides coated with poly-lysine.
One hundred and seventy microgram samples of denatured protein were separated on 12% SDS-PAGE and then transferred to a Hybond membrane (Amersham, Chicago, IL, United States). The membrane was blocked overnight in 4.5% skimmed milk in Tris-buffered saline with Tween 20 (TBST). The membrane was treated with primary antibody for immunoblotting for 1 h (Supplementary Table 1). Following a TBST rinse, it was incubated for 1 h with anti-goat, anti-mouse or anti-rabbit IgG conjugated to [horseradish peroxidase(HRP); Dako, Glostrup, Denmark]. Bands were observed using C300 imaging system (AZURE, Peking, China) and ECL-Plus detection reagents (Santa Cruz Biotechnology).
CRC cells or tissues were used to extract total RNA with a Qiagen RNase Mini Kit. avian myeloblastosis virus transcriptase and a random primer were used to create cDNA from 2 mg total RNA (Takara, Kusatsu, Japan). The primers for REG4 were forward: 5’-CCTTTCCACAGTATCCTTCTTCCCT-3’, TM: 58.4 ℃ and reverse: 5’-TATGGCCAAAGACCCAGCTGTT-3’, TM: 58.2 ℃ (104 bp). The primers for ACLY were forward: 5’-AAACTGTGGGTCCTTTACTCG-3’, TM: 53.8 ℃ and reverse: 5’-GGATGACGATACAGCCCCTG-3’, TM: 55.6 ℃ (147 bp). The primers for ACC1 were forward: 5’-GCTGGTCCACATGAACAGG-3’, TM: 53.8 ℃ and reverse: 5’-GCCTTCTGGATATTCAGGAC
After deparaffinization with xylene and dehydration with alcohol, successive slices in a target retrieval solution were microwaved for 17 min (Dako). H2O2 at 3% in methanol was used to block endogenous peroxidase activity. To stop nonspecific binding, 4% bovine serum albumin was administered for 5 min. The sections were treated with anti-goat conjugated to horseradish peroxidase (Dako, 1: 100) antibodies for 18 min after being incubated with goat anti-human REG4 antibody (R and D Systems, 1: 50) for 17 min. To enable previously reported intermittent irradiation, all incubations were carried out in a microwave oven[28]. REG4 immunostaining was localized in the cytoplasm. For immunohistochemistry, according to the degree of color development of cell positive markers, it was divided into: Blue, negative; pale yellow, weakly positive; brown, moderately positive; dark brown, strongly positive. From five typical fields in each region, 100 cells were randomly chosen and counted by two independent observers blinded to the samples (Zhang CY and Zheng HC). Semiquantitative two-tier grading was used to determine the positive proportion of counted cells: Positive, 6%-100% and negative, 0%-5%.
We performed an ELISA (Gelatins, Shanghai, China) to determine serum REG4 concentration. One hundred microliters of standard or serum sample was incubated at 4 ℃ overnight on polystyrene microtiter plates coated with anti-REG4 antibody. Following the removal of the liquid, we added 100 μL biotin-antibody working solution to each well, which was then incubated at 37 ℃ for 2 h. After aspirating the liquid, 100 μL HRP-avidin working solution was added to each well, followed by washing three times with 350 μL wash buffer, and incubation at 37 ℃ for 1 h. The plates were washed again three times with wash buffer, followed by incubation with 90 μL tetramethylbenzidine substrate at 37 ℃ for 30 min. We dispensed 50 μL stopping solution to each well and measured the absorbance at 405 nm. Recombinant REG4 (0.312-20 ng/mL) was used as the reference standard.
PubMed, Web of Science, BIOSIS, and SciFinder were used to search the literature up to March 14, 2022. The following search phrases were entered: (Colorectal or rectal or colon or rectum) and (REG4 or REG IV) and (cancer or carcinoma or adenocarcinoma). No limitations on language or publication year were placed on the search. Inclusion criteria for studies were: (1) Studies that used immunohistochemistry to detect changes in REG4 expression in CRC; and (2) studies that used immunohistochemistry to relate REG4 expression to pathobiological behavior and CRC prognosis. Exclusion criteria included: (1) Abstracts, comments, reviews and meeting reports; (2) duplication of previous publications; (3) western blotting, RT–PCR, cDNA microarray, or transcriptomic sequencing for REG4 expression; and (4) lack of sufficient information. Two reviewers (Zhang CY and Zheng HC) independently gathered data from all relevant articles and evaluated the quality of the included studies using the Newcastle-Ottawa Scale (http://www.ohri.ca/programs/clinicalepidemiology/oxford.htm). For survival analysis, we extracted data from Kaplan-Meier curves using an Engauge Digitizer program. We calculated the hazard ratios (HRs) and accompanying 95% confidence intervals (CIs). Twelve papers regarding the correlation between REG4 expression and cancer risk, and clinicopathological or prognostic factors of CRC were found in PubMed, Web of Science, BIOSIS Citation Index, SciFinder, and CNKI (Supplementary Table 2). Samples of normal colorectal mucosa were only included in four papers[3,29-31]. In 12 investigations, a comparison was made between REG4 expression and the clinicopathological features of CRC[3,15,17,29-37]. Finally, we covered the importance of REG4 expression for prognosis in five papers [15,17,30,35,36].
We used Oncomine (www.oncomine.org, keywords: REG4, Skrzypczak, Sabates-Bellver) to analyze the REG4 expression level. The Cancer Genome Atlas (TCGA)-assembler in R program retrieved the REG4 expression and clinicopathological data for a total of 362 patients with CRC from the TCGA database (keywords, REG4). Using the Xiantao platform (https://www.xiantaozi.com/, keywords: REG4) and The University of Alabama at Birmingham Cancer Data Analysis Portal (UALCAN) database (http://ualcan.path.uab.edu, keywords: REG4), we analyzed the expression, methylation, relevant genes, and signaling pathways of the REG4 gene. A Kaplan-Meier plotter (http://kmplot.com/, keywords: 223447_at) was used to evaluate the prognostic value of REG4. Additionally, we discovered the genes that were differentially expressed by Xiantao and subjected these to protein-protein interaction (PPI) network analysis and a search of critical hub genes. Subsequently, gene ontology, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Set Enrichment Analysis (GSEA) analyses were conducted on these genes.
Hardy-Weinberg equilibrium was assessed in each study’s control group using the χ2 test. A Z test was used to establish the statistical significance of the pooled odds ratios. A fixed effect model was applied if there was no discernible heterogeneity. Instead, a random effect model was used to analyze prognostic analysis. An I2 test was used to quantify the heterogeneity impact. To measure the asymmetry of the funnel plot and evaluate publication bias, Begg’s and Egger’s tests were used to quantify this. Meta-analyses were carried out using Revman software 5.3, and Student t tests were used with SPSS 10.0 to handle data from the TCGA database. A log-rank statistic was used to compare survival curves and create Kaplan-Meier survival charts. Cox’s hazard proportional analysis was used for multivariate survival analysis. Statistical significance was defined as two-sided P < 0.05.
The expression of REG4 mRNA was higher in colorectal adenoma or CRC than in normal tissues (P < 0.05) using quantitative RT-PCR (Supplementary Figure 1A), Xiantao (Supplementary Figure 1B) and Oncomine (Supplemen
We identified distinct genes in the low and high expression groups of REG4 mRNA in CRC using a xiantao platform, and constructed a volcanic map (Supplementary Figure 3A). KEGG analysis showed that the top signaling pathway mainly included chemokine activity, taste receptor, protein-DNA and a DNA-packing complex, and nucleosome and chromatin (P < 0.05) (Supplementary Figure 3B). GSEA showed that the top signaling pathways were principally composed of a generation of second messenger molecules: programmed cell death protein 1 signaling, HDAC and histone acetyl transferase, and epigenetic regulation and DNA methylation (P < 0.05) (Supplementary Figure 3C). The upregulated genes were CCL19, CCL25, CIDEA, CMA1 and PLA2G2D, and downregulated genes were ANGPTL3, CRP, H2BC10, H4C3 and H4C6 (P < 0.05) (Supplementary Figure 3D). Cytoscape software was utilized to determine the top 10 nodes by degree and STRING software to identify the PPI pairings (Supplementary Figures 4A and B). According to a Xiantao database, CD3D, 3D3G, CD4, HLA-DRA, ZAP70, ITK, CD3E, CD247, CD28 and HLA-DRB1 were less expressed in CRC than normal tissues (P < 0.05) (Supplementary Figure 4C).
According to the Xiantao database, genes positively correlated with REG4 in CRC are shown in Supplemen
According to xiantao, the infiltration of mast cells, T cells, CD8 T cells, cytotoxic T cells, T helper (Th)1, Th2 and Th17 cells, T follicular helper (TFH) cells, TReg cells, natural killer (NK) CD56bright cells, B cells, interstitial dendritic cells (iDCs), and activated DCs (aDCs) in CRC were positively correlated with REG4 mRNA expression (P < 0.05) (Supplemen
Compared to normal mucosa, REG4 protein expression was low in CRC (P = 0.03) (Supplementary Table 3) and positively correlated with lymph node metastasis, TNM staging, and dedifferentiation of CRC (P < 0.05) (Supplementary Tables 4-
According to UALCAN (Supplementary Figure 7A), REG4 protein expression was higher in mucinous than nonmucinous adenocarcinoma (P < 0.05), and negatively correlated with chromatin modifier, p53/Rb, and the HIPPO signaling pathway (P < 0.05). Expression of REG4 protein was higher in CRC than in NNM, according to western blotting (P < 0.05) (Supplementary Figure 7B). Serum REG4 level was higher in patients with CRC than in healthy volunteers after modification by body surface area (P < 0.05) (Supplementary Figure 7C). Immunohistochemically, REG4 expression was higher in colorectal NNM than adenoma and primary cancer, and in primary than metastatic cancer (P < 0.001) (Supplementary Figure 7D and E). Mucinous adenocarcinoma showed higher REG4 expression than well-, moderately and poorly differentiated adenocarcinomas (P < 0.05) (Supplementary Table 8).
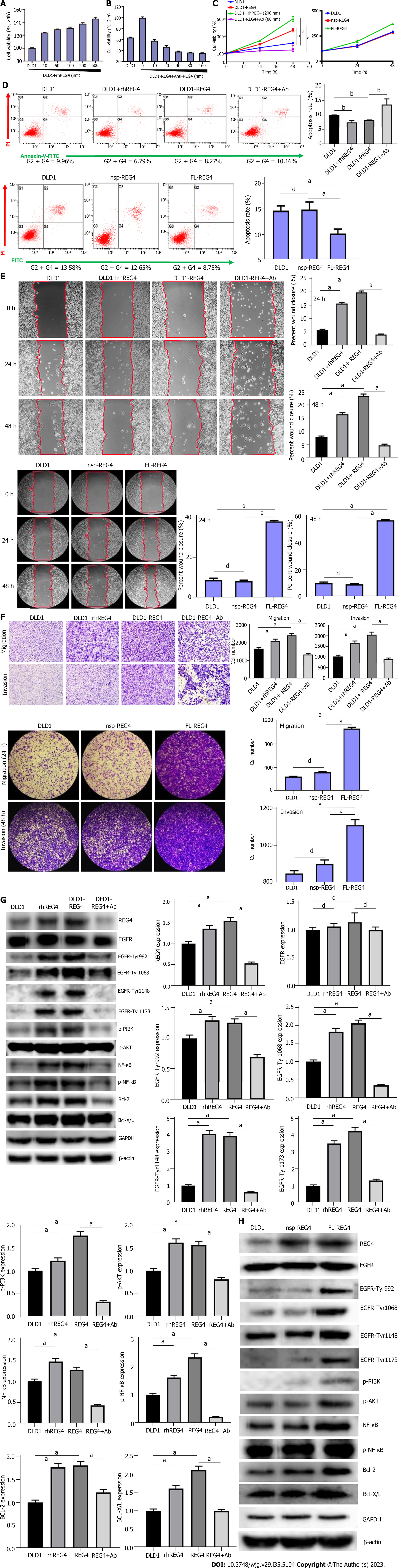
According to qPCR (P < 0.05) (Supplementary Figure 8A), western blotting (Supplementary Figure 8B), immunofluorescence (Supplementary Figure 8C), and ELISA (P < 0.05) (Supplementary Figure 8D), DLD-1 cells were successfully transfected with FL-REG4 and NSP-REG4 plasmids, as shown by higher REG4 mRNA and protein expression in transfectants than parental cells. After treatment with rhREG4, the viability of DLD-1 cells was increased in a dose-dependent manner (P < 0.05) (Figure 1A). Exposure to anti-REG4 antibody decreased the cell viability of FL-REG4-overexpressing DLD-1 cells in a dose-dependent manner (P < 0.05) (Figure 1B). Treatment with rhREG4 or FL-REG4 overexpression resulted in high viability (P < 0.05) (Figure 1C), antiapoptosis (P < 0.05) (Figure 1D), migration and invasion (P < 0.05) (Figures 1E and F) in comparison with untreated DLD-1 cells (P < 0.05). Additionally, FL-REG4 transfection and rhREG4 treatment enhanced the expression of REG4, EGFR-Tyr992, -Tyr1068, -Tyr1148, -Tyr1173, phosphorylated phosphoinositide 3-kinase (p-PI3K), p-Akt, nuclear factor (NF)-κB, p-NF-κB, Bcl-2, and Bcl-X/L compared with untreated DLD-1 cells by western blotting (P < 0.05) (Figure 1G). However, anti-REG4 antibody blocked the effects of FL-REG4 overexpression (Figure 1B-G). The effects of NSP-REG4 on aggressive phenotypes and their related protein expression in DLD-1 cells were not detectable, which differed from the effects of FL-REG4 expression and were similar to those of REG4-nontransfected CRC cells (Figure 1C-F and H).
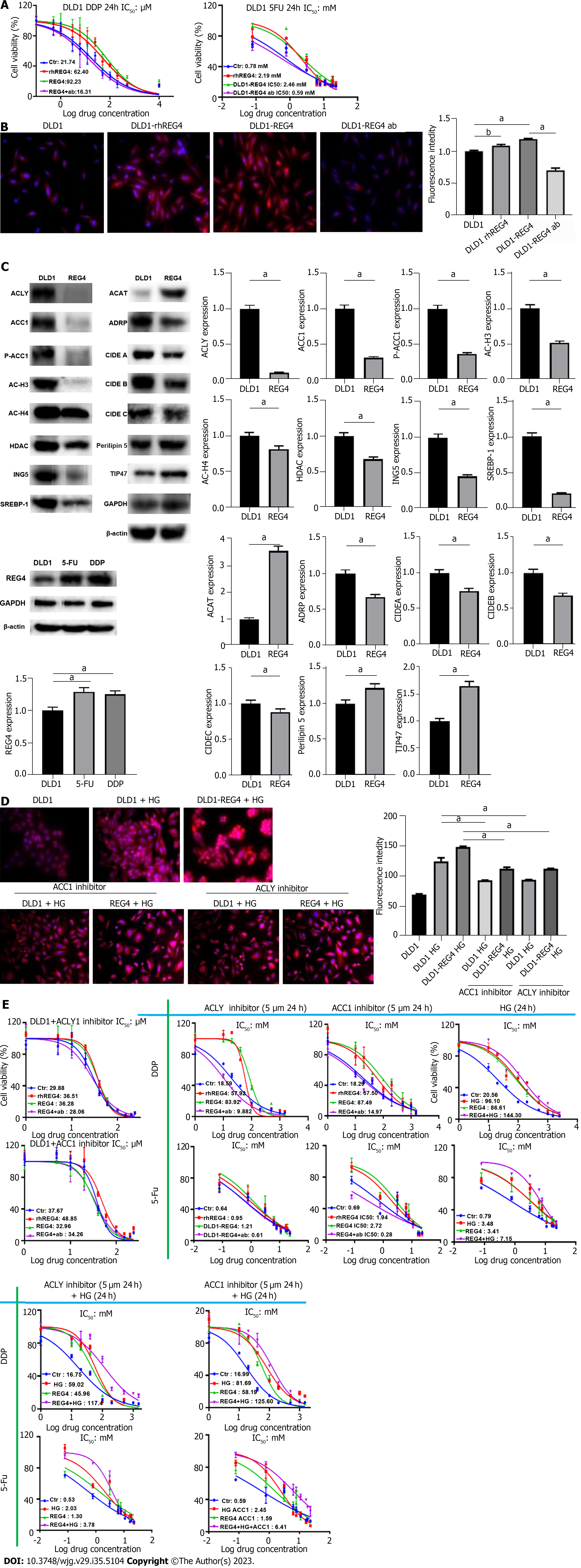
RhREG4 treatment and FL-REG4 overexpression caused DLD-1 cells to become resistant to DDP and 5-FU (P < 0.05) (Figure 2A), while anti-REG4 antibody reversed the chemoresistance of REG4-overexpressing DLD-1 cells (P < 0.05) (Figure 2A). rhREG4 treatment and FL-REG4 increased the formation of intracellular lipid droplets, as shown by Nile red staining (P < 0.05) (Figure 2B). REG4 overexpression was observed in chemoresistant DLD-1 cells. FL-REG4 overexpression enhanced the expression of acyl coenzyme A-cholesterol acyltransferase (ACAT), perilipin 5, and tail-interacting protein (TIP) 47, but weakened expression of ACLY, ACC1, p-ACC1, HDAC, adipose differentiation-related protein (ADRP), cell-death-inducing DFF45-like effector (CIDE) A, B and C, AC-H3, AC-H4, ING5, and SREBP-1 in DLD-1 cells (Figure 2C). High-glucose treatment significantly increased lipid droplet formation in DLD-1 cells, which was markedly suppressed by ACLY or ACC1 inhibitor (P < 0.05) (Figure 2D). DLD-1 cells treated with rhREG4, FL-REG4 transfection, or high glucose had decreased chemosensitivity to 5-FU and DDP, but DLD-1 cells treated with REG4 antibody, ACC1 inhibitor or ACLY inhibitor had increased chemosensitivity to 5-FU and DDP (Figure 2E). ACC1 or ACLY inhibitor reversed the insensitivity of cells to 5-FU and DDP in the high-glucose treatment group (P < 0.05).
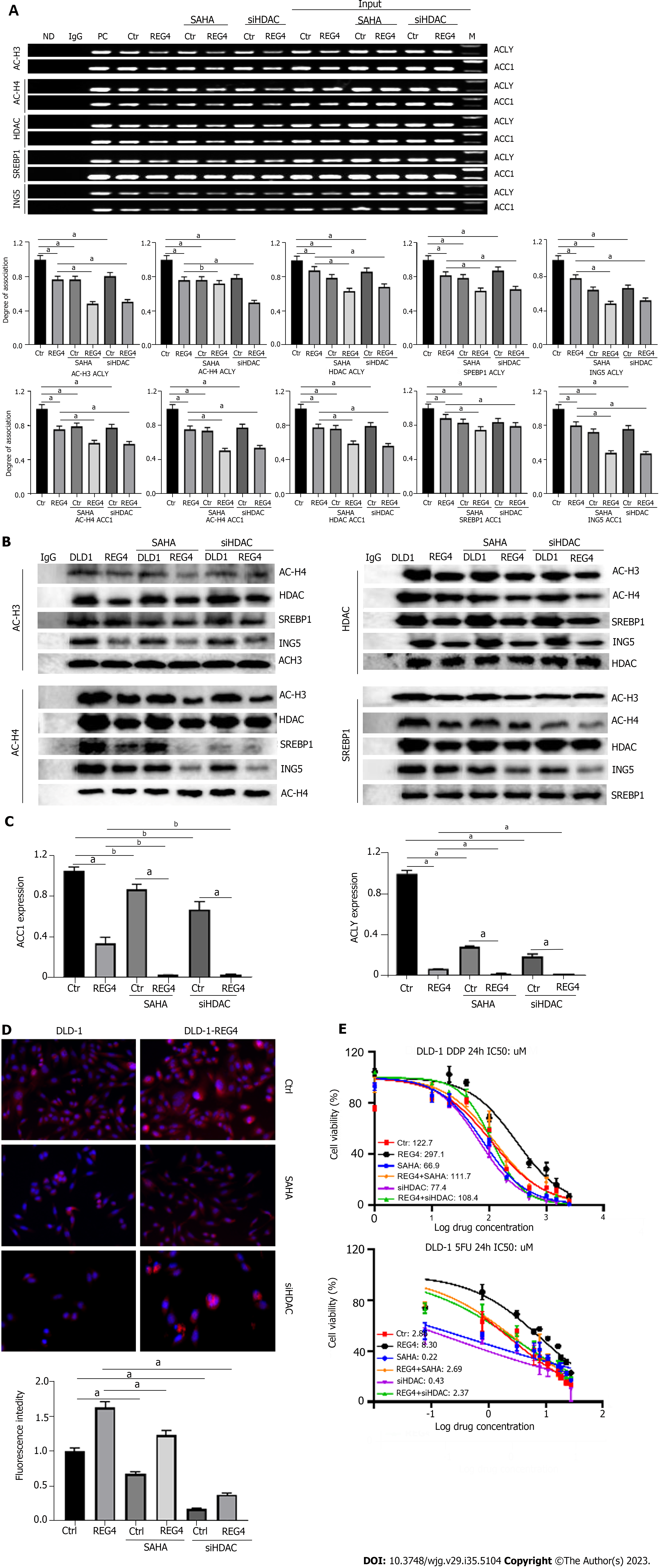
In DLD-1 cells, FL-REG4 overexpression weakened the interaction of AC-H3, AC-H4, HDAC, SREBP1 and ING5 proteins with the ACLY or ACC1 promoter, according to ChIP (P < 0.05) (Figure 3A). FL-REG4 overexpression also weakened the interaction of the five proteins (P < 0.05) (Figure 3B). Overexpression of FL-REG4 decreased ACLY and ACC1 mRNA expression (P < 0.05) (Figure 3C), and increased lipid droplet formation (P < 0.05) (Figure 3D), and increased the half maximal inhibitory concentration (IC50) of cells for 5-FU and DDP (P < 0.05) (Figure 3E). After SAHA (siHDAC inhibitor) treatment (2 μM, 24 h) and siHDAC transfection, the interaction between AC-H3, AC-H4, SREBP 1, ING5, HDAC and ACC1, ACLY promoters was weakened compared with the control group (P < 0.05) (Figure 3A), and the mRNA level of ACC1 and ACLY decreased (P < 0.05) (Figure 3B). After SAHA treatment and siHDAC transfection, the fluorescence intensity of Nile red staining decreased compared with that in the control group (P < 0.05) (Figure 3D), and the chemical sensitivity to 5-FU and DDP increased compared with that in the control group (P < 0.05) (Figure 3E).
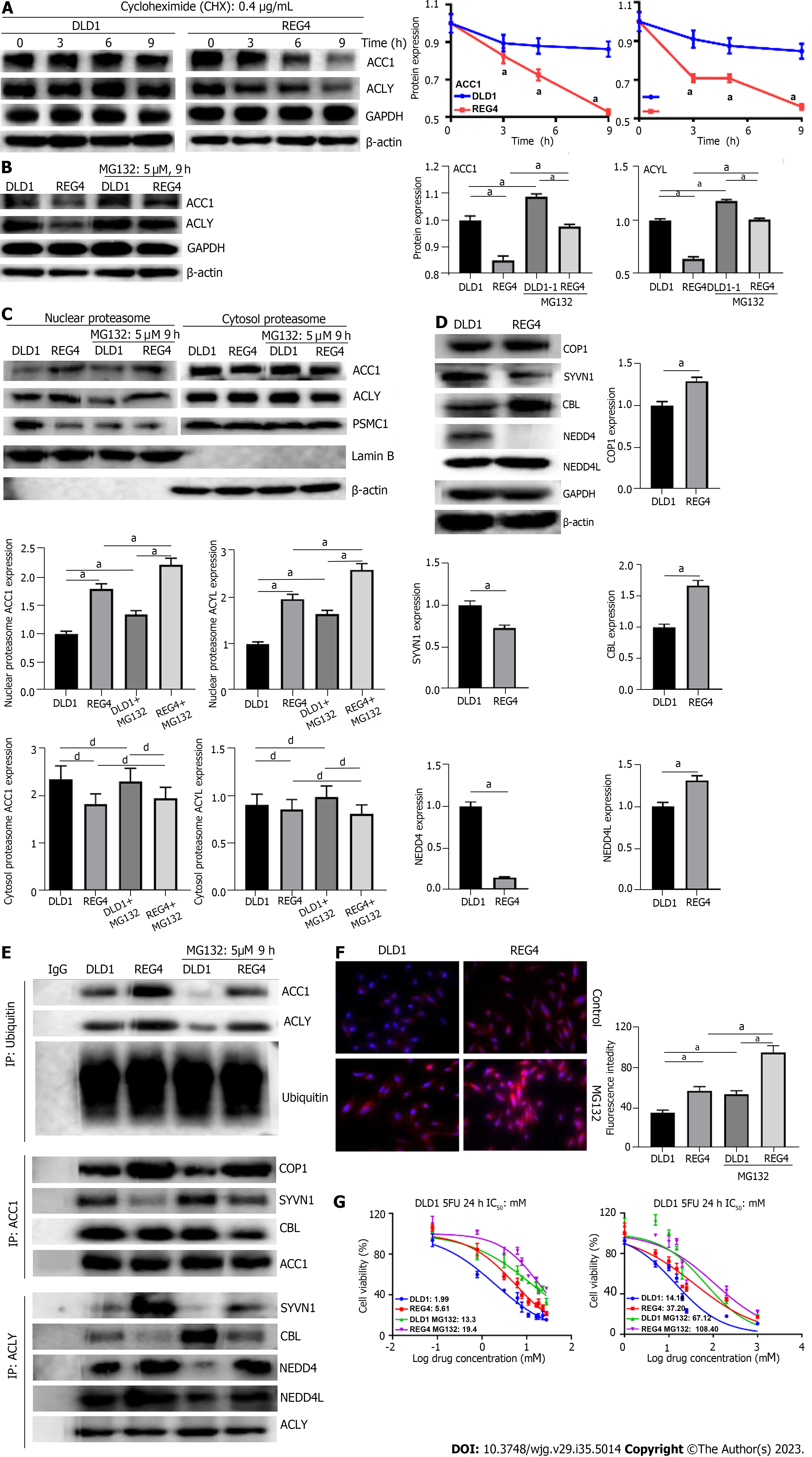
After treatment with CHX (used to inhibit the synthesis of new proteins), ACC1 and ACLY protein expression was lower in FL-REG4 transfectants than in DLD-1 cells (P < 0.05) (Figure 4A). MG132 (proteasome inhibitor) increased protein expression of ACC1 and ACLY in DLD-1 cells and FL-REG4 transfectants, which was higher in DLD-1 cells than in FL-REG4 transfectants (P < 0.05) (Figure 4B). Higher levels of ACC1 and ACLY proteins were noted in the nuclear proteasome of FL-REG4 transfectants than in DLD-1 cells, but this translocation phenomenon was not enhanced by MG132 (Figure 4C). Similar levels of ACC1 and ACLY expression were noted in the cytosolic proteasome of transfectants and parental cells, even after treatment with MG132 (Figure 4C). Regarding ubiquitin ligases, expression of COP1 E3 ubiquitin ligase (COP1), Cbl proto-oncogene (CBL) and NEDD4 like E3 ubiquitin protein ligase (NEDD4L) was higher in FL-REG4 transfectants than DLD-1 cells, but synoviolin 1 (SYVN1) and NEDD4 were lower in FL-REG4 transfectants than DLD-1 cells (Figure 4D). Co-immunoprecipitation showed that expression of ubiquitylated ACC1 and ACLY was higher in FL-REG4 transfectants than in DLD-1 cells, and pretreatment with a small dose of proteasome inhibitor MG132 (5 μM, 9 h) reduced ubiquitination of ACC1 and ACLY (Figure 4E). ACLY bound more to SYVN1, NEDD4 and NEDD4L in FL-REG4 transfectants than in DLD-1 cells, but bound less to CBL. ACC1 interacted more with COP1 in REG4 transfectants than in DLD-1 cells, but interacted less with CBL and SYVN1 (Figure 4E). MG132 pretreatment (5 μM, 24 h) aggravated lipid droplet formation (P < 0.05) (Figure 4F) and chemoresistance against 5-FU and DDP (P < 0.05) (Figure 4G) in DLD-1 cells and FL-REG4 transfectants.
The silencing of REG4 reduced cellular proliferation, but rhREG4 had the opposite effect since REG4 downregulated p21 (Cip1/WAF1) expression and upregulated cyclin D1 expression, which might promote G1/S transition[38,39]. Anti-REG4 antibody dramatically reduced the autocrine and paracrine effects of secretory REG4 on the ability of colon cancer cells to invade, migrate and proliferate[19]. REG4 overexpression may cause resistance to the irradiation-induced apoptosis of colon cancer cells. Animal studies have shown that rhREG4 increased the expression of the antiapoptotic genes, Bcl-2 and Bcl-xL, and survivin to shield normal intestinal crypt cells against irradiation-induced apoptosis[40].
We conducted bioinformatics analysis, meta-analysis, and pathological and serological studies to evaluate the clinicopathological and prognostic significance of REG4 expression in CRC. In bioinformatics analysis, we used data of different sizes and from different sources to ensure data heterogeneity, and we used qRT-PCR to verify the reliability of bioinformatics analysis. We discovered that REG4 mRNA expression was upregulated in CRC, but REG4 protein expression was downregulated, suggesting that aberrant REG4 expression could be used as a potential biomarker for colorectal tumorigenesis. In addition, we discovered a negative correlation between REG4 promoter methylation and mRNA expression in CRC. The correlation of REG4 methylation with mucinous subtypes or clinicopathological staging was opposite to that of REG4 mRNA expression. Upregulated REG4 mRNA expression might be due to promoter hypomethylation in CRC. High REG4 mRNA expression was discovered in colorectal, pancreatic, hepatic, and prostate malignancies as well as in inflammatory epithelium, dysplasia, and malignant lesions of ulcerative colon tissues[20,41-45], suggesting that upregulated REG4 mRNA was involved in the early malignant transformation of epithelial cells. Although upregulated REG4 protein expression occurred in gastric cancer, ovarian cancer, glioma, pancreatic cancer, gallbladder carcinoma, and prostate cancer[12,28,46-49], lower REG4 expression was found in CRC. In subsequent research, we will use more databases and updated data sets to analyze the expression of REG4 in CRC to verify the consistency between our studies and strengthen our conclusions.
The tumor microenvironment of CRC is regulated by many factors, such as CTLA-4, and the altered expression of these factors can lead to changes in immune responses[50]. So far, there has been no systematic study on the relationship between REG4 and immune microenvironment in CRC. Here, we studied the correlation between the expression of REG4 mRNA and the infiltration of immune cells in CRC, and found that high expression of REG4 was positively correlated with infiltration of mast cells, T cells, CD8 T cells, cytotoxic T cells, Th1, Th2 and Th17 cells, TFH cells, TReg cells, NK CD56bright cells, B cells, iDCs, and aDCs. High expression of REG4 was negatively correlated with Tcm cell infiltration. In future research, we will focus on the impact of REG4 expression on the immune environments of primary and metastatic CRC, which is also important for immunotherapy of CRC.
Previously, we discovered that expression of REG4 was substantially linked with that of mucin-2 and mucin-5AC, and that it was greater in mucinous carcinoma, signet ring cell carcinoma, and intestinal metaplasia that produced mucins[28]. REG4 was identified as a potential biomarker of mucinous ovarian cancer at both mRNA and protein levels[12,51]. Here, we also found that expression of REG4 in mucinous adenocarcinoma was greater than that in other histological subtypes at both at the mRNA and protein levels. These findings could account for REG4 protein overexpression in colorectal mucosa and poorly differentiated, signet ring cell carcinoma and undifferentiated carcinoma. REG4 expression was positively correlated with lymph node metastasis, TNM staging, poorly differentiated CRC, and had a worse prognosis, in agreement with other studies[12,28,46-49]. The opposite was the case for REG4 mRNA, suggesting that aberrant REG4 protein expression might indicate aggressiveness and prognosis of CRC. In CRCs with reduced stroma compared to those with high stroma, REG4 protein expression was considerably greater[52]. Previously, we performed REG4 immunostaining on the same tissue microarrays of CRC (Supplementary Table 1), and found similar results. Therefore, the discrepancies about REG4 mRNA and protein expression might be largely attributable to a complex process from transcription to translation and different methodologies. Serologically, the preoperative REG4 protein level was higher in CRC than in a healthy population and postoperative patients with cancer, in agreement with the finding that serum REG4 Levels were greater in pancreatic ductal adenocarcinoma than in chronic pancreatitis[53].
Bishnupuri et al[54] found that the transcriptional activator of D-type cyclins, CD44 intracytoplasmic domain, was released after REG4 connected with transmembrane CD44 and activated γ-secretase to promote proliferation and stemness of colorectal and pancreatic cancer cells. We also found that REG4 overexpression and rhREG4 treatment promoted proliferation, antiapoptosis, and migration and invasion of CRC cells. Exposure to anti-REG4 antibody inhibited the effects of REG4 overexpression on the phenotypes of CRC cells, which is consistent with our earlier result in ovarian cancer cells[12]. We also found that REG4 overexpression resulted in resistance of ovarian cancer cells to DDP or taxol by activating the PI3K-Akt-mTOR signaling pathway[55]. As a mutant KRAS-induced factor, REG4 increased cancer stem cell characteristics via Wnt/β-catenin signaling[56]. Jin et al[57] showed that REG4 increased the resistance of gastric cancer cells to 5-FU by stimulating the mitogen-activated protein kinase-ERK-Bim signaling pathway. Here, we found that expression of REG4, EGFR-Tyr992, -Tyr1068, -Tyr1148 and -Tyr1173, p-PI3K, p-Akt, NF-κB, p-NF-κB, Bcl-2, and Bcl-x/L was higher in DLD-1 cells treated with rhREG4 or transfected with REG4-overexpressing plasmid than in parental cells. Anti-REG4 antibody blocked the effects of REG4 overexpression, indicating that REG4 promoted the aggressive phenotypes by an EGFR-PI3K-Akt-NF-κB signaling pathway, in line with other studies[7,8,40]. However, the lack of effect of NSP-REG4 on aggressive phenotypes and related signaling proteins suggested that REG4 only functioned in CRC cells in an autocrine or paracrine manner. These findings demonstrated that REG4 might be a potential molecular target for gene therapy of CRC.
DLD-1 cells developed resistance to 5-FU and DDP as a result of REG4 overexpression. 5-FU- or DDP-resistant DLD-1 cells showed REG4 overexpression, suggesting a role for REG4 in chemoresistance, in line with previous studies[22,55]. The chemoresistance of CRC cells was produced by lysophosphatidylcholine acyltransferase 2-mediated lipid droplet formation[58], which was also aided by prothymosin α[59], and metastasis-associated in colon cancer[60] through SREBP-1- and fatty acid synthase-mediated and lipogenesis, respectively. Crucial enzymes for de novo fatty acid synthesis are ACC1 and ACLY, which are closely linked to chemoresistance[61]. In the liver and peritoneal tissues, lipid droplet assembly is mediated byadipocyte differentiation-related protein, CIDE, ACAT1, perilipin 5 and TIP47[62-66]. REG4-mediated lipid droplet formation might be closely linked to the upregulated expression of ACAT1, perilipin 5 and TIP47 in REG4 transfectants, but not with de novo lipogenesis, as demonstrated by the downregulated expression of ACC1 and ACLY. REG4-mediated lipid droplet formation might account for REG4-induced resistance to 5-FU and DDP, which can be reversed by ACC1 or ACLY inhibitor, but deteriorated by high glucose exposure. In combination with these discoveries, we hypothesized that REG4 may play an important role in chemoresistance, not through de novo lipogenesis, but by lipid droplet assembly, and might be used as a potential target for reversal of chemoresistance in CRC. According to the increased IC50 of DLD-1 cells after high glucose treatment, we speculate that high glucose treatment provides more glucose and increases the de novo synthesis of lipid droplets, thereby mediating the drug resistance of CRC cells.
We found that REG4-related signaling pathways included chemokine activity, taste receptors, protein-DNA and DNA packing complexes for transcription repression and activation, nucleosomes and chromatin, generation of second messenger molecules, HDAC and histone acetyltransferase for epigenetic regulation, and sugar metabolism. Therefore, we investigated the regulatory effects of REG4 on the transcription of ACC1 and ACLY. REG4 decreased expression of AC-H3, AC-H4, ING5, HDAC and SREBP1. REG4 also weakened the interaction of the five proteins with the promoters of ACC1 or ACLY, or complex formation of the five proteins, and mRNA expression of ACC1 or ACLY. The inhibitory effect of FL-REG4 transcription can be enhanced by low concentration SAHA (HDAC inhibitor) treatment or transfection with siHDAC. We speculated that HDAC hypoexpression or inactivation might increase the chemosensitivity of CRC cells by inhibiting the de novo lipogenesis, which suggests a potential clinical application for the antitumor drug SAHA. The decrease in lipid droplet formation and increase in chemotherapy sensitivity of DLD-1 cells after SAHA or siHDAC treatment indirectly confirm this view. We hypothesized that REG4 inhibited the transcription of ACC1 and ACLY by disassociating the complex formation of AC-H3-AC-H4-ING5-HDAC-SREBP1 in their promoters and releasing the combination of ACC1 and ACLY promoters and complex via HDAC-mediated deacetylation.
To date, there has been no research about the effects of REG4 on metabolic reprogramming, histone modification, and microenvironmental stress, and the interaction between REG4 and metabolic or epigenetic pathways in CRC. With regard to drug resistance, Ying et al[67] found that upregulation of REG4 mRNA was closely linked to the intrinsic drug resistance of gastric cancer cells to 5-FU. All 14 REG4-positive patients with gastric cancer showed no change or disease progression when treated with a combination of low-dose 5-FU and DDP[1]. In gastric cancer cells, REG4 enhanced the resistance to 5-FU through the MAPK–ERK–Bim pathway[1]. Anti-REG4 antibody significantly inhibited proliferation and chemosensitivity of gastric cancer cells to 5-FU, and REG4 silencing caused the loss of stemness properties[68,69]. In ovarian cancer cells, REG4 overexpression or rhREG4 treatment promoted proliferation, G2/S progression, antiapoptosis, migration, invasion, and DDP and paclitaxel[12,57]. In future research, we will also explore the potential interaction between REG4 and metabolic or epigenetic pathways in CRC, which provides a new method for the early diagnosis and targeted therapy of CRC.
Finally, we also analyzed the modulatory effects of REG4 on the protein stability of ACC1 and ACLY. Firstly, we treated DLD-1 cells and REG4 transfectants with CHX, and found that REG4 destabilized ACC1 and ACLY. REG4 promoted the recruitment of ACC1 and ACLY to the nuclear proteasome for ubiquitylation-mediated degradation, during which ACLY bound to NEDD4, SYVN1 and NEDD4L, and ACC1 bound to COP1. Overexpression of REG4 mediated lipid droplet formation and chemoresistance in DLD-1 cells. These results suggested that REG4 facilitated proteasomal degradation of ACC1 and ACLY to suppress de novo lipogenesis. We also speculate that pretreatment of proteasome inhibitor MG132 in DLD-1 cells can increase expression of ACC1 and ACLY by reducing ubiquitination of ACC1 and ACLY, thus increasing formation of lipid droplets and leading to chemoresistance. In the future, we will explore the potential role of REG4 from metabolic reprogramming and epigenetic regulation.
REG4 protein expression was decreased in CRC and positively linked with the degree of invasion, TNM stage, and dedifferentiation, but the converse was the case for REG4 mRNA expression. REG4 aggravated aggressive phenotypes in an autocrine or paracrine manner via the EGFR-PI3K-Akt-NF-κB signaling pathway. REG4 may be involved in chemoresistance, not through de novo lipogenesis, but by lipid droplet assembly. REG4 inhibited the transcription of ACC1 or ACLY by disassociating the complex formation of AC-H3-AC-H4-ING5-histone deacetylase-SREBP1 in their promoters and induced the proteasomal degradation of ACC1 and ACLY proteins. Pretreatment with high glucose might induce chemoresistance of CRC cells, which should be emphasized in clinical practice. SAHA can reverse the chemoresistance of CRC and provide a potential direction for research of DDP and 5-FU resistance of CRC. Finally, REG4 may be used as a reliable diagnostic marker for the prognosis, aggressiveness and carcinogenesis of CRC and is a potential molecular target.
Regulating gene 4 (REG4) has been proved to be carcinogenic in some cancers, but its manifestation and possible carcinogenic mechanism in colorectal cancer (CRC) have not yet been elucidated. Our previous study found that the drug resistance characteristics of CRC cells may be related to their fat metabolism.
With the aging of the world population, the incidence of CRC is increasing. For the treatment of CRC, chemoresistance has always been an urgent problem to be solved.
This study aimed to explore the role of REG4 in CRC and its association with lipid droplet formation, and the molecular mechanisms involved.
We conducted a meta-analysis and bioinformatics and pathological analysis of REG4 expression in CRC. The effects of REG4 on the phenotypes and related proteins were also investigated in CRC cells.
Compared to normal mucosa, REG4 mRNA expression was high in CRC, but protein expression was opposite. REG4-related genes included epigenetic regulation, transcription repression, sugar metabolism and transfer. REG4 exposure or overexpression promoted proliferation, antiapoptosis, migration and invasion of DLD-1 cells in an autocrine or paracrine manner by activating the epidermal growth factor receptor-phosphoinositide 3-kinase-Akt-nuclear factor-κB pathway. REG4 was involved in chemoresistance not through de novo lipogenesis, but lipid droplet assembly, which was strengthened by high glucose treatment. REG4 inhibited the transcription of acetyl-coA carboxylase 1 (ACC1) and ATP-citrate lyase (ACLY) by disassociating the complex formation of anti–acetyl (AC)-acetyl-histone 3-AC-histone 4-inhibitor of growth protein-5-si histone deacetylase-sterol-regulatory element binding protein 1 in their promoters and induced proteasomal degradation of ACC1 or ACLY.
REG4 may be an indicator of drug resistance and metabolism of tumor cells. REG4 might be a useful marker for colorectal carcinogenesis, as well as a potential gene therapy target.
This study provides new insights into a better understanding of the pathogenesis of CRC. REG4 may be used as a novel therapeutic target. However, the regulatory mechanism needs to be further explored.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Batyrbekov K, Kazakhstan; Osera S, Japan S-Editor: Qu XL L-Editor: A P-Editor: Cai YX
| 1. | Zheng HC, Xue H, Zhang CY. REG4 promotes the proliferation and anti-apoptosis of cancer. Front Cell Dev Biol. 2022;10:1012193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 2. | Hu Y, Pan C, Hu J, Zhang S. The role of Reg IV in colorectal cancer, as a potential therapeutic target. Contemp Oncol (Pozn). 2015;19:261-264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 3. | Zheng HC, Sugawara A, Okamoto H, Takasawa S, Takahashi H, Masuda S, Takano Y. Expression profile of the REG gene family in colorectal carcinoma. J Histochem Cytochem. 2011;59:106-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 4. | Wang H, Hu L, Zang M, Zhang B, Duan Y, Fan Z, Li J, Su L, Yan M, Zhu Z, Liu B, Yang Q. REG4 promotes peritoneal metastasis of gastric cancer through GPR37. Oncotarget. 2016;7:27874-27888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 51] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 5. | Kawasaki Y, Matsumura K, Miyamoto M, Tsuji S, Okuno M, Suda S, Hiyoshi M, Kitayama J, Akiyama T. REG4 is a transcriptional target of GATA6 and is essential for colorectal tumorigenesis. Sci Rep. 2015;5:14291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 6. | Naito Y, Oue N, Hinoi T, Sakamoto N, Sentani K, Ohdan H, Yanagihara K, Sasaki H, Yasui W. Reg IV is a direct target of intestinal transcriptional factor CDX2 in gastric cancer. PLoS One. 2012;7:e47545. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 7. | Bishnupuri KS, Sainathan SK, Bishnupuri K, Leahy DR, Luo Q, Anant S, Houchen CW, Dieckgraefe BK. Reg4-induced mitogenesis involves Akt-GSK3β-β-Catenin-TCF-4 signaling in human colorectal cancer. Mol Carcinog. 2014;53 Suppl 1:E169-E180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 8. | Bishnupuri KS, Luo Q, Murmu N, Houchen CW, Anant S, Dieckgraefe BK. Reg IV activates the epidermal growth factor receptor/Akt/AP-1 signaling pathway in colon adenocarcinomas. Gastroenterology. 2006;130:137-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 100] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 9. | He XJ, Jiang XT, Ma YY, Xia YJ, Wang HJ, Guan TP, Shao QS, Tao HQ. REG4 contributes to the invasiveness of pancreatic cancer by upregulating MMP-7 and MMP-9. Cancer Sci. 2012;103:2082-2091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 10. | Ma X, Wu D, Zhou S, Wan F, Liu H, Xu X, Zhao Y, Tang M. The pancreatic cancer secreted REG4 promotes macrophage polarization to M2 through EGFR/AKT/CREB pathway. Oncol Rep. 2016;35:189-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 11. | Mitani Y, Oue N, Matsumura S, Yoshida K, Noguchi T, Ito M, Tanaka S, Kuniyasu H, Kamata N, Yasui W. Reg IV is a serum biomarker for gastric cancer patients and predicts response to 5-fluorouracil-based chemotherapy. Oncogene. 2007;26:4383-4393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 96] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 12. | Chen S, Gou WF, Zhao S, Niu ZF, Zhao Y, Takano Y, Zheng HC. The role of the REG4 gene and its encoding product in ovarian epithelial carcinoma. BMC Cancer. 2015;15:471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 13. | Violette S, Festor E, Pandrea-Vasile I, Mitchell V, Adida C, Dussaulx E, Lacorte JM, Chambaz J, Lacasa M, Lesuffleur T. Reg IV, a new member of the regenerating gene family, is overexpressed in colorectal carcinomas. Int J Cancer. 2003;103:185-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 86] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 14. | Granlund Av, Beisvag V, Torp SH, Flatberg A, Kleveland PM, Ostvik AE, Waldum HL, Sandvik AK. Activation of REG family proteins in colitis. Scand J Gastroenterol. 2011;46:1316-1323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 15. | Oue N, Kuniyasu H, Noguchi T, Sentani K, Ito M, Tanaka S, Setoyama T, Sakakura C, Natsugoe S, Yasui W. Serum concentration of Reg IV in patients with colorectal cancer: overexpression and high serum levels of Reg IV are associated with liver metastasis. Oncology. 2007;72:371-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 69] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 16. | Numata M, Oshima T, Yoshihara K, Watanabe T, Tsuchida K, Tamagawa H, Yamamoto N, Shiozawa M, Morinaga S, Akaike M, Kunisaki C, Rino Y, Tanaka K, Masuda M, Imada T. Relationship between RegIV gene expression to outcomes in colorectal cancer. J Surg Oncol. 2011;104:205-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 17. | He HL, Lee YE, Shiue YL, Lee SW, Lin LC, Chen TJ, Wu TF, Hsing CH, Huang HY, Wang JY, Li CF. Overexpression of REG4 confers an independent negative prognosticator in rectal cancers receiving concurrent chemoradiotherapy. J Surg Oncol. 2014;110:1002-1010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 18. | Guo Y, Xu J, Li N, Gao F, Huang P. RegIV potentiates colorectal carcinoma cell migration and invasion via its CRD domain. Cancer Genet Cytogenet. 2010;199:38-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 19. | Rafa L, Dessein AF, Devisme L, Buob D, Truant S, Porchet N, Huet G, Buisine MP, Lesuffleur T. REG4 acts as a mitogenic, motility and pro-invasive factor for colon cancer cells. Int J Oncol. 2010;36:689-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 20. | Nanakin A, Fukui H, Fujii S, Sekikawa A, Kanda N, Hisatsune H, Seno H, Konda Y, Fujimori T, Chiba T. Expression of the REG IV gene in ulcerative colitis. Lab Invest. 2007;87:304-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 59] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 21. | Kobunai T, Watanabe T, Fukusato T. REG4, NEIL2, and BIRC5 gene expression correlates with gamma-radiation sensitivity in patients with rectal cancer receiving radiotherapy. Anticancer Res. 2011;31:4147-4153. [PubMed] |
| 22. | Gao L, Wu X, Zhang L, Dai Y, Zhu Z, Zhi Y, Wang K. REG4 is a Potential Biomarker for Radiochemotherapy Sensitivity in Colorectal Cancer. Onco Targets Ther. 2021;14:1605-1611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 23. | Jou E, Rodriguez-Rodriguez N, McKenzie ANJ. Emerging roles for IL-25 and IL-33 in colorectal cancer tumorigenesis. Front Immunol. 2022;13:981479. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 24. | Droeser RA, Iezzi G. IL-22-mediates Cross-talk between Tumor Cells and Immune Cells Associated with Favorable Prognosis in Human Colorectal Cancer. J Cell Immunol. 2021;3:118-121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 25. | Djaldetti M, Bessler H. Modulators affecting the immune dialogue between human immune and colon cancer cells. World J Gastrointest Oncol. 2014;6:129-138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 27] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 26. | Li Y, Liu Y, Zhao N, Yang X, Li Y, Zhai F, Zang X, Cui W. Checkpoint regulator B7x is epigenetically regulated by HDAC3 and mediates resistance to HDAC inhibitors by reprogramming the tumor immune environment in colorectal cancer. Cell Death Dis. 2020;11:753. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 27. | Jang TJ. Progressive Increase of Regulatory T Cells and Decrease of CD8+ T Cells and CD8+ T Cells/Regulatory T Cells Ratio during Colorectal Cancer Development. Korean J Pathol. 2013;47:443-451. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 28. | Zheng HC, Xu XY, Yu M, Takahashi H, Masuda S, Takano Y. The role of Reg IV gene and its encoding product in gastric carcinogenesis. Hum Pathol. 2010;41:59-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 29. | Zheng SQ, He J, Huang PL, Wang JH. Clinical significance of Reg IV protein expression in human colorectal cancer. Shijie Huaren Xiaohua Zazhi. 2010;18:3594-3598. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 30. | Li XH, Zheng Y, Zheng HC, Takahashi H, Yang XH, Masuda S, Takano Y. REG IV overexpression in an early stage of colorectal carcinogenesis: an immunohistochemical study. Histol Histopathol. 2010;25:473-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 31. | Li FY, Ren XB, Xu EP, Huang Q, Sheng HQ, Lv BJ, Lai MD. RegIV expression showing specificity to gastrointestinal tract and its potential role in diagnosing digestive tract neuroendocrine tumor. J Zhejiang Univ Sci B. 2010;11:258-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 32. | Wang LN, Hou SP, Liu HB, Sun XL, Zhang XJ, Zhang QQ. Expression of REG4 and surviving and clinical significance in colorectal cancer. Shiyong Yixue Zazhi. 2015;31:3347-3350. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 33. | Wang GM, Wang CX, Nie MN, Zhang LH, Diao WY. Expression of Reg IV in colon carcinoma and its relationship with angiogenesis. J Med Forum. 2016;37:7-9. [RCA] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 34. | Oue N, Mitani Y, Aung PP, Sakakura C, Takeshima Y, Kaneko M, Noguchi T, Nakayama H, Yasui W. Expression and localization of Reg IV in human neoplastic and non-neoplastic tissues: Reg IV expression is associated with intestinal and neuroendocrine differentiation in gastric adenocarcinoma. J Pathol. 2005;207:185-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 97] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 35. | Sawada T, Yashiro M, Sentani K, Oue N, Yasui W, Miyazaki K, Kai K, Fujita H, Nakamura K, Maeda K, Kakeji Y, Natsugoe S, Shirabe K, Nomura S, Shimada Y, Tomita N, Hirakawa K, Maehara Y. New molecular staging with G-factors (VEGF-C and Reg IV) by supplementing TNM classification in colorectal cancers. Oncol Rep. 2013;30:2609-2616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 36. | Zhu X, Han Y, Yuan C, Tu W, Qiu G, Lu S, Lu H, Peng Z, Zhou C. Overexpression of Reg4, alone or combined with MMP-7 overexpression, is predictive of poor prognosis in colorectal cancer. Oncol Rep. 2015;33:320-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 37. | Kaprio T, Hagström J, Mustonen H, Koskensalo S, Andersson LC, Haglund C. REG4 independently predicts better prognosis in non-mucinous colorectal cancer. PLoS One. 2014;9:e109600. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 38. | Takehara A, Eguchi H, Ohigashi H, Ishikawa O, Kasugai T, Hosokawa M, Katagiri T, Nakamura Y, Nakagawa H. Novel tumor marker REG4 detected in serum of patients with resectable pancreatic cancer and feasibility for antibody therapy targeting REG4. Cancer Sci. 2006;97:1191-1197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 64] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 39. | Liu CM, Hsieh CL, He YC, Lo SJ, Liang JA, Hsieh TF, Josson S, Chung LW, Hung MC, Sung SY. In vivo targeting of ADAM9 gene expression using lentivirus-delivered shRNA suppresses prostate cancer growth by regulating REG4 dependent cell cycle progression. PLoS One. 2013;8:e53795. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 40. | Bishnupuri KS, Luo Q, Sainathan SK, Kikuchi K, Sureban SM, Sabarinathan M, Gross JH, Aden K, May R, Houchen CW, Anant S, Dieckgraefe BK. Reg IV regulates normal intestinal and colorectal cancer cell susceptibility to radiation-induced apoptosis. Gastroenterology. 2010;138:616-626, 626.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 41. | Zhang Y, Lai M, Lv B, Gu X, Wang H, Zhu Y, Shao L, Wang G. Overexpression of Reg IV in colorectal adenoma. Cancer Lett. 2003;200:69-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 42. | Lasserre C, Colnot C, Bréchot C, Poirier F. HIP/PAP gene, encoding a C-type lectin overexpressed in primary liver cancer, is expressed in nervous system as well as in intestine and pancreas of the postimplantation mouse embryo. Am J Pathol. 1999;154:1601-1610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 43. | Lasserre C, Christa L, Simon MT, Vernier P, Bréchot C. A novel gene (HIP) activated in human primary liver cancer. Cancer Res. 1992;52:5089-5095. [PubMed] |
| 44. | Kimura N, Yonekura H, Okamoto H, Nagura H. Expression of human regenerating gene mRNA and its product in normal and neoplastic human pancreas. Cancer. 1992;70:1857-1863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 45. | Ohara S, Oue N, Matsubara A, Mita K, Hasegawa Y, Hayashi T, Usui T, Amatya VJ, Takeshima Y, Kuniyasu H, Yasui W. Reg IV is an independent prognostic factor for relapse in patients with clinically localized prostate cancer. Cancer Sci. 2008;99:1570-1577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 46. | Takayama R, Nakagawa H, Sawaki A, Mizuno N, Kawai H, Tajika M, Yatabe Y, Matsuo K, Uehara R, Ono K, Nakamura Y, Yamao K. Serum tumor antigen REG4 as a diagnostic biomarker in pancreatic ductal adenocarcinoma. J Gastroenterol. 2010;45:52-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 47. | Wang Q, Deng J, Yuan J, Wang L, Zhao Z, He S, Zhang Y, Tu Y. Oncogenic reg IV is a novel prognostic marker for glioma patient survival. Diagn Pathol. 2012;7:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 48. | Tamura H, Ohtsuka M, Washiro M, Kimura F, Shimizu H, Yoshidome H, Kato A, Seki N, Miyazaki M. Reg IV expression and clinicopathologic features of gallbladder carcinoma. Hum Pathol. 2009;40:1686-1692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 49. | Hayashi T, Matsubara A, Ohara S, Mita K, Hasegawa Y, Usui T, Arihiro K, Norimura S, Sentani K, Oue N, Yasui W. Immunohistochemical analysis of Reg IV in urogenital organs: Frequent expression of Reg IV in prostate cancer and potential utility as serum tumor marker. Oncol Rep. 2009;21:95-100. [PubMed] |
| 50. | Derakhshani A, Hashemzadeh S, Asadzadeh Z, Shadbad MA, Rasibonab F, Safarpour H, Jafarlou V, Solimando AG, Racanelli V, Singh PK, Najafi S, Javadrashid D, Brunetti O, Silvestris N, Baradaran B. Cytotoxic T-Lymphocyte Antigen-4 in Colorectal Cancer: Another Therapeutic Side of Capecitabine. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 64] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 51. | Lehtinen L, Vesterkvist P, Roering P, Korpela T, Hattara L, Kaipio K, Mpindi JP, Hynninen J, Auranen A, Davidson B, Haglund C, Iljin K, Grenman S, Siitari H, Carpen O. REG4 Is Highly Expressed in Mucinous Ovarian Cancer: A Potential Novel Serum Biomarker. PLoS One. 2016;11:e0151590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 52. | Kang G, Oh I, Pyo J, Kang D, Son B. Clinicopathological Significance and Prognostic Implications of REG4 Immunohistochemical Expression in Colorectal Cancer. Medicina (Kaunas). 2021;57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 53. | Saukkonen K, Hagström J, Mustonen H, Lehtinen L, Carpen O, Andersson LC, Seppänen H, Haglund C. Prognostic and diagnostic value of REG4 serum and tissue expression in pancreatic ductal adenocarcinoma. Tumour Biol. 2018;40:1010428318761494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 54. | Bishnupuri KS, Sainathan SK, Ciorba MA, Houchen CW, Dieckgraefe BK. Reg4 Interacts with CD44 to Regulate Proliferation and Stemness of Colorectal and Pancreatic Cancer Cells. Mol Cancer Res. 2022;20:387-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 55. | Xiang LW, Xue H, Ha MW, Yu DY, Xiao LJ, Zheng HC. The effects of REG4 expression on chemoresistance of ovarian cancer. J Obstet Gynaecol. 2022;42:3149-3157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 56. | Hwang JH, Yoon J, Cho YH, Cha PH, Park JC, Choi KY. A mutant KRAS-induced factor REG4 promotes cancer stem cell properties via Wnt/β-catenin signaling. Int J Cancer. 2020;146:2877-2890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 57. | Jin J, Lv H, Wu J, Li D, Chen K, Zhang F, Han J, Feng J, Zhang N, Yu H, Su D, Ying L. Regenerating Family Member 4 (Reg4) Enhances 5-Fluorouracil Resistance of Gastric Cancer Through Activating MAPK/Erk/Bim Signaling Pathway. Med Sci Monit. 2017;23:3715-3721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 58. | Cotte AK, Aires V, Fredon M, Limagne E, Derangère V, Thibaudin M, Humblin E, Scagliarini A, de Barros JP, Hillon P, Ghiringhelli F, Delmas D. Lysophosphatidylcholine acyltransferase 2-mediated lipid droplet production supports colorectal cancer chemoresistance. Nat Commun. 2018;9:322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 248] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 59. | Jin L, Zhu LY, Pan YL, Fu HQ, Zhang J. Prothymosin α promotes colorectal carcinoma chemoresistance through inducing lipid droplet accumulation. Mitochondrion. 2021;59:123-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 60. | Duan J, Chen L, Zhou M, Zhang J, Sun L, Huang N, Bin J, Liao Y, Liao W. MACC1 decreases the chemosensitivity of gastric cancer cells to oxaliplatin by regulating FASN expression. Oncol Rep. 2017;37:2583-2592. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 61. | Sur S, Nakanishi H, Flaveny C, Ippolito JE, McHowat J, Ford DA, Ray RB. Inhibition of the key metabolic pathways, glycolysis and lipogenesis, of oral cancer by bitter melon extract. Cell Commun Signal. 2019;17:131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 62. | Fan H, Diao H, Lu Y, Xie J, Cheng X. The relation between serum adipose differentiation-related protein and non-alcoholic fatty liver disease in type 2 diabetes mellitus. Ther Adv Endocrinol Metab. 2020;11:2042018820969025. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 63. | Kasano-Camones CI, Takizawa M, Iwasaki W, Sasaki S, Hamada M, Morimoto A, Sakaguchi M, Gonzalez FJ, Inoue Y. Synergistic regulation of hepatic Fsp27b expression by HNF4α and CREBH. Biochem Biophys Res Commun. 2020;530:432-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 64. | Ayyagari V, Li M, Pasman Z, Wang X, Louis S, Diaz-Sylvester P, Groesch K, Wilson T, Brard L. Assessment of the diagnostic and prognostic relevance of ACAT1 and CE levels in plasma, peritoneal fluid and tumor tissue of epithelial ovarian cancer patients - a pilot study. BMC Cancer. 2022;22:387. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 65. | Ethem İ, Hacıoğlu C. Effects of perilipin-5 on lipid metabolism and high-sensitivity cardiac troponin I. Rev Assoc Med Bras (1992). 2022;68:1011-1016. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 66. | Nose F, Yamaguchi T, Kato R, Aiuchi T, Obama T, Hara S, Yamamoto M, Itabe H. Crucial role of perilipin-3 (TIP47) in formation of lipid droplets and PGE2 production in HL-60-derived neutrophils. PLoS One. 2013;8:e71542. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 67. | Ying LS, Yu JL, Lu XX, Ling ZQ. Enhanced RegIV expression predicts the intrinsic 5-fluorouracil (5-FU) resistance in advanced gastric cancer. Dig Dis Sci. 2013;58:414-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 68. | Zhang XQ, Yu LT, Du P, Yin TQ, Zhang ZY, Xu Y, Li X, Li YJ, Wang M, Luo C. Single-chain Antibody Against Reg4 Suppresses Gastric Cancer Cell Growth and Enhances 5-FU-induced Cell Death in vitro. Anticancer Agents Med Chem. 2019;19:610-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 69. | Zhou W, Sun M, Wang DL, Wang Y, Jin F, Zhang YY, Yang L, Wu XL, Wu YZ. Silencing of RegIV by shRNA causes the loss of stemness properties of cancer stem cells in MKN45 gastric cancer cells. Oncol Rep. 2013;30:2685-2690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |