Published online Sep 7, 2023. doi: 10.3748/wjg.v29.i33.4975
Peer-review started: May 25, 2023
First decision: July 8, 2023
Revised: July 27, 2023
Accepted: August 15, 2023
Article in press: August 15, 2023
Published online: September 7, 2023
Processing time: 98 Days and 17.8 Hours
Liver fibrosis is the common pathological process associated with the occurrence and development of various chronic liver diseases. At present, there is still a lack of effective prevention and treatment methods in clinical practice. Hepatic stellate cell (HSC) plays a key role in liver fibrogenesis. In recent years, the study of liver fibrosis targeting HSC autophagy has become a hot spot in this research field. Angiotensin-converting enzyme 2 (ACE2) is a key negative regulator of renin-angiotensin system, and its specific molecular mechanism on autophagy and liver fibrosis needs to be further explored.
To investigate the effect of ACE2 on hepatic fibrosis in mice by regulating HSC autophagy through the Adenosine monophosphate activates protein kinases (AMPK)/mammalian target of rapamycin (mTOR) pathway.
Overexpression of ACE2 in a mouse liver fibrosis model was induced by injection of liver-specific recombinant adeno-associated virus ACE2 vector (rAAV2/8-ACE2). The degree of liver fibrosis was assessed by histopathological staining and the biomarkers in mouse serum were measured by Luminex multifactor analysis. The number of apoptotic HSCs was assessed by terminal deoxynucleoitidyl transferase-mediated dUTP nick-end labeling (TUNEL) and immunofluorescence staining. Transmission electron microscopy was used to identify the changes in the number of HSC autophagosomes. The effect of ACE2 overexpression on autophagy-related proteins was evaluated by multicolor immunofluorescence staining. The expression of autophagy-related indicators and AMPK pathway-related proteins was measured by western blotting.
A mouse model of liver fibrosis was successfully established after 8 wk of intraperitoneal injection of carbon tetrachloride (CCl4). rAAV2/8-ACE2 administration reduced collagen deposition and alleviated the degree of liver fibrosis in mice. The serum levels of platelet-derived growth factor, angiopoietin-2, vascular endothelial growth factor and angiotensin II were decreased, while the levels of interleukin (IL)-10 and angiotensin- (1-7) were increased in the rAAV2/8-ACE2 group. In addition, the expression of alpha-smooth muscle actin, fibronectin, and CD31 was down-regulated in the rAAV2/8-ACE2 group. TUNEL and immunofluorescence staining showed that rAAV2/8-ACE2 injection increased HSC apoptosis. Moreover, rAAV2/8-ACE2 injection notably decreased the number of autophagosomes and the expression of autophagy-related proteins (LC3I, LC3II, Beclin-1), and affected the expression of AMPK pathway-related proteins (AMPK, p-AMPK, p-mTOR).
ACE2 overexpression can inhibit HSC activation and promote cell apoptosis by regulating HSC autophagy through the AMPK/mTOR pathway, thereby alleviating liver fibrosis and hepatic sinusoidal remodeling.
Core Tip: Liver fibrosis and cirrhosis are the common outcomes of most chronic liver diseases, and there is a lack of effective treatment at present. Angiotensin-converting enzyme 2 (ACE2), as the main target receptor for the coronavirus disease virus invasion into the human body, is one of the research hotspots. The involvement of autophagy in the activation mechanism of hepatic stellate cell (HSC) during liver fibrosis has attracted increasing attention. Our study found that ACE2 can inhibit the activation and proliferation of HSCs by regulating autophagy, and promote apoptosis of HSCs, providing new ideas for the treatment of liver fibrosis and hepatic sinusoidal remodeling.
- Citation: Wu Y, Yin AH, Sun JT, Xu WH, Zhang CQ. Angiotensin-converting enzyme 2 improves liver fibrosis in mice by regulating autophagy of hepatic stellate cells. World J Gastroenterol 2023; 29(33): 4975-4990
- URL: https://www.wjgnet.com/1007-9327/full/v29/i33/4975.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i33.4975
Liver fibrosis and cirrhosis are the common pathological processes associated with the occurrence and development of various chronic liver diseases, and there is still a lack of effective prevention and treatment methods for these conditions. In liver fibrosis and cirrhosis, excessive deposition of extracellular matrix (ECM) in the liver and regenerating nodules compress blood vessels, resulting in structural changes. In addition, hepatic sinusoidal vasoconstriction and vascular remodeling cause functional changes that ultimately lead to increased intrahepatic vascular resistance and portal pressure[1]. Hepatic stellate cells (HSCs) are located in the perisinusoidal Disse space between liver sinusoidal endothelial cells (LSECs) and hepatocytes[2]. The vasomotion of hepatic sinusoids greatly affects intrahepatic blood flow and portal venous resistance, and HSCs and LSECs play key roles in increasing intrahepatic vascular resistance and portal venous pressure. Hepatic sinusoidal vascular remodeling occurs in hepatic fibrosis and is characterized by capillarization of the hepatic sinusoids and surrounded by more contractile HSCs[3]. HSC activation is a complex and coordinated process. After activation, HSCs begin to proliferate and release excess collagen, proteoglycan and other ECM components, which in turn cause changes in the intrahepatic structure; furthermore, HSCs acquire contractility, reducing the diameter of the hepatic sinusoids and increasing resistance, leading to liver fibrosis and portal hypertension[4-6].
Autophagy is a metabolic process in which eukaryotic cells eliminate disposable or potentially dangerous cytoplasmic material. It plays a critical role in cell development, differentiation, and homeostasis. In this process, some damaged proteins or organelles are wrapped by autophagic vesicles with a double membrane structure and sent to lysosomes (animals) or vacuoles (yeast and plants) for degradation and recycling[7]. Autophagy, as a cellular housekeeper, can eliminate defective proteins and organelles, clear intracellular pathogens, and prevent the accumulation of abnormal proteins. Therefore, autophagy plays an active role in the pathology of many diseases. Growing evidence suggests that an adequate autophagic response in hepatocytes and nonparenchymal cells (HSCs, LSECs, Kupffer cells) is critical for the physiological function of the liver[8]. During hepatic fibrogenesis, the study of the mechanism of autophagy involved in HSC activation has attracted increasing attention. Autophagy increases the degradation of lipid droplets in HSCs, providing energy for HSC activation[9,10]. A study showed that after reducing HSC autophagy in mice, HSC activation was inhibited, and the degree of liver fibrosis was alleviated[11]. In recent years, the study of liver fibrosis targeting HSC autophagy has become a hot spot in this research field.
The renin-angiotensin system (RAS) is an important endocrine system that regulates vascular tone and water and electrolyte metabolism in the body. Our previous studies have confirmed that HSCs have local RAS, and activated HSCs increase the synthesis of angiotensin II (Ang II) in liver cirrhosis[12]. Under the action of angiotensin-converting enzyme 2 (ACE2), Ang II is converted to Ang- (1-7), which stimulates the Mas receptor to cause vasodilation. ACE2 is a key negative regulator of RAS, and studies have shown that it can inhibit liver fibrosis by degrading Ang II[13,14], but its specific molecular mechanism needs to be further explored. We confirmed that carvedilol could inhibit Ang II-induced HSC proliferation and contraction and improve liver fibrosis in mice[12]. The study also indicated that HSCs are the main cells expressing ACE2 in the liver. In addition, our study demonstrated that carvedilol could notably reduce HSC autophagy and inhibit HSC activation and proliferation[15]. It has been reported that ACE2 alleviates the severity of acute lung injury by inhibiting autophagy[16]. Therefore, we hypothesized that ACE2 could inhibit HSC activation and proliferation by regulating autophagy, thus improving hepatic sinusoidal remodeling and ultimately alleviating liver fibrosis and portal hypertension.
The Adenosine monophosphate activates protein kinases (AMPK)/mammalian target of rapamycin (mTOR) signaling pathway is not only an important node in the intracellular energy metabolism monitoring system but also an important upstream pathway regulating autophagy. Studies have reported that ACE2 can improve vascular endothelial dysfunction in type 2 diabetic rats with insulin resistance by regulating the AMPK/mTOR pathway[17]. In addition, ACE2 was shown to effectively modulate the AMPK/mTOR signaling pathway in a mouse model of acute lung injury[16]. Our previous study confirmed that metformin could inhibit HSC proliferation, migration and angiogenesis through the Akt/mTOR and mTOR/hypoxia inducible factor-1α (HIF-1α) pathways[18]. In this study, we evaluated the effect of ACE2 on liver fibrosis in mice and demonstrated the molecular mechanism by which ACE2 regulates HSC autophagy through the AMPK/mTOR pathway to improve liver fibrosis and hepatic sinusoidal remodeling.
The aim of this study was to determine the effect of ACE2 on HSC activation, proliferation, apoptosis and liver fibrosis by regulating autophagy. This study will provide a new direction for the prevention and targeted treatment of liver fibrosis and portal hypertension.
Forty adult male C57BL/6J mice (6-8 wk, 18-20 g) were purchased from the Experimental Animal Center of Shandong University (Jinan, China). The mice were housed in an air-conditioned room at a defined temperature (23-25 °C) for one week prior to the initiation of the experiments. All experimental protocols were approved by the Animal Care Committee of the Second Hospital, Cheeloo College of Medicine, Shandong University.
The liver fibrosis mouse model was established by intraperitoneal injection of carbon tetrachloride (CCl4, 20%, 0.5 mL/100 g) twice a week for 8 wk. To evaluate the effect of ACE2 on liver fibrosis, the liver-specific recombinant adeno-associated viral vector rAAV-ACE2 (rAAV2/8-ACE2) was injected into the tail vein 4 wk after CCl4 administration. Mice were randomly assigned to four groups (10 in each): Group 1, normal control (olive oil); Group 2, CCl4-induced liver fibrosis (CCl4); Group 3, rAAV2/8-ACE2 + CCl4; and Group 4, rAAV2/8-ACE2 + CCl4 + rapamycin (mTOR inhibitor). Rapamycin (2 mg/kg) was administered at the 6th week after the intraperitoneal injection of CCl4.
The mice were dissected after anesthesia administration, and liver tissues were removed and partially stored at -80 °C. Another section was fixed in 4% paraformaldehyde and embedded in paraffin.
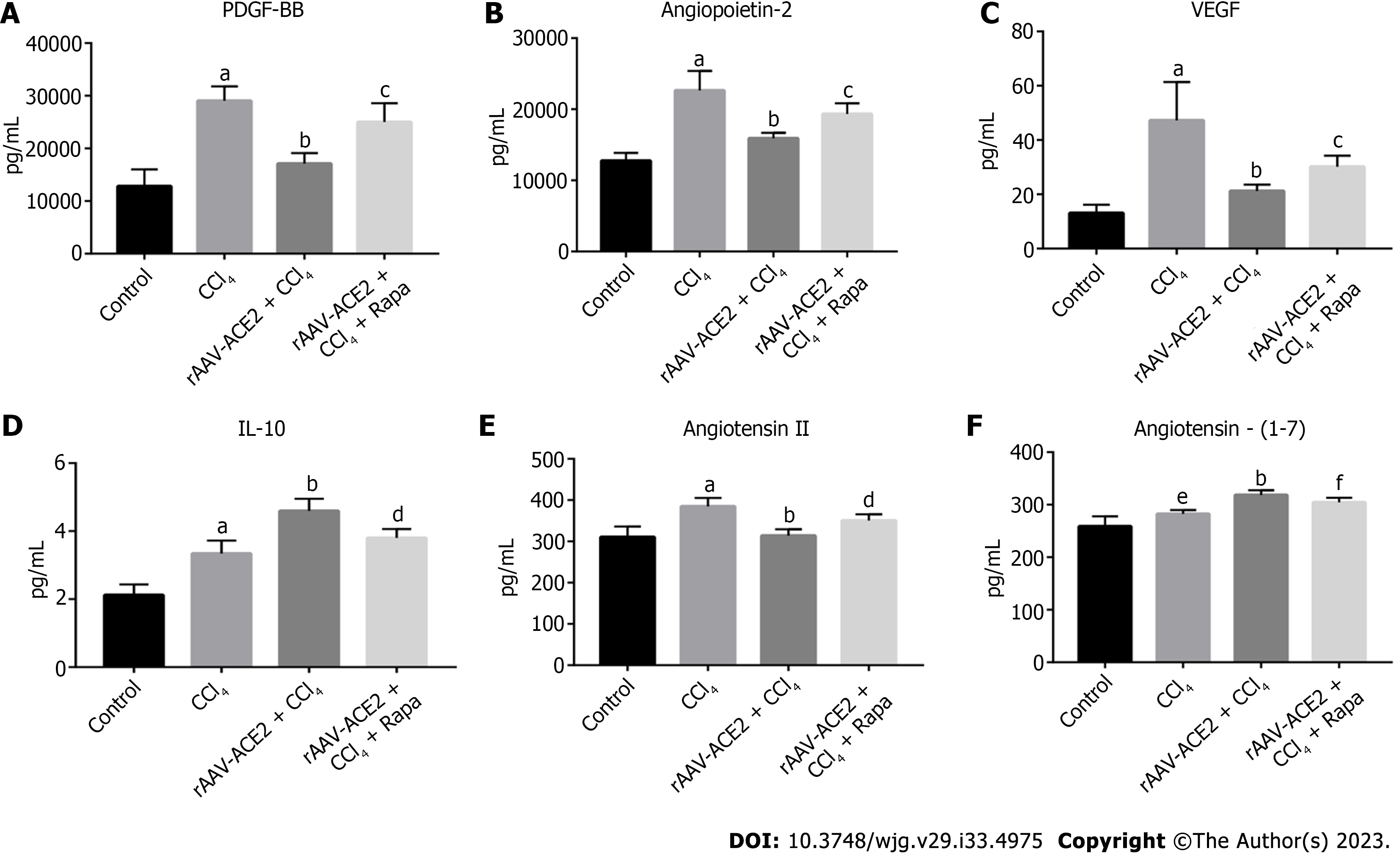
Mouse blood samples were centrifuged at 4 °C (3000 rpm) for 10 min, and the supernatant was collected. According to the manufacturer's instructions, the serum levels of platelet-derived growth factor BB (PDGF-BB), angiopoietin-2, vascular endothelial growth factor (VEGF), interleukin (IL)-10, Ang II and Ang- (1-7) were measured using Luminex multifactor assay kits and Enzyme Linked Immunosorbent Assay kits. The data were analyzed using Graph Pad Prism 8.0.
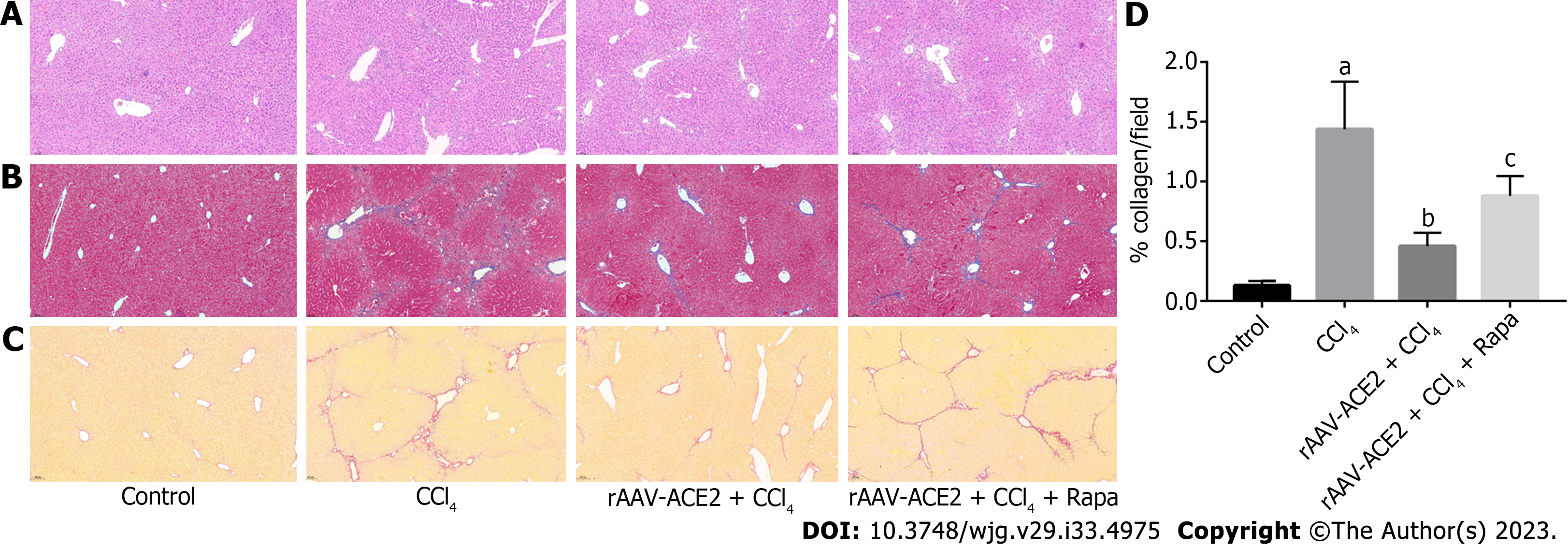
The paraffin-embedded liver tissue sections were morphologically evaluated based on hematoxylin and eosin (H&E) staining. The degree of liver fibrosis in mice was measured by Masson trichrome and Sirius red staining. According to the METAVIR scale, the degree of liver fibrosis was divided into four stages from 0 to 4 (0 - No fibrosis; 1 - Portal fibrosis; 2 - Periportal fibrosis; 3 - Bridging fibrosis; 4 - Cirrhosis). The quantity of collagen production in each group after Sirius red staining was analyzed using Image-Pro Plus 6.0 software.
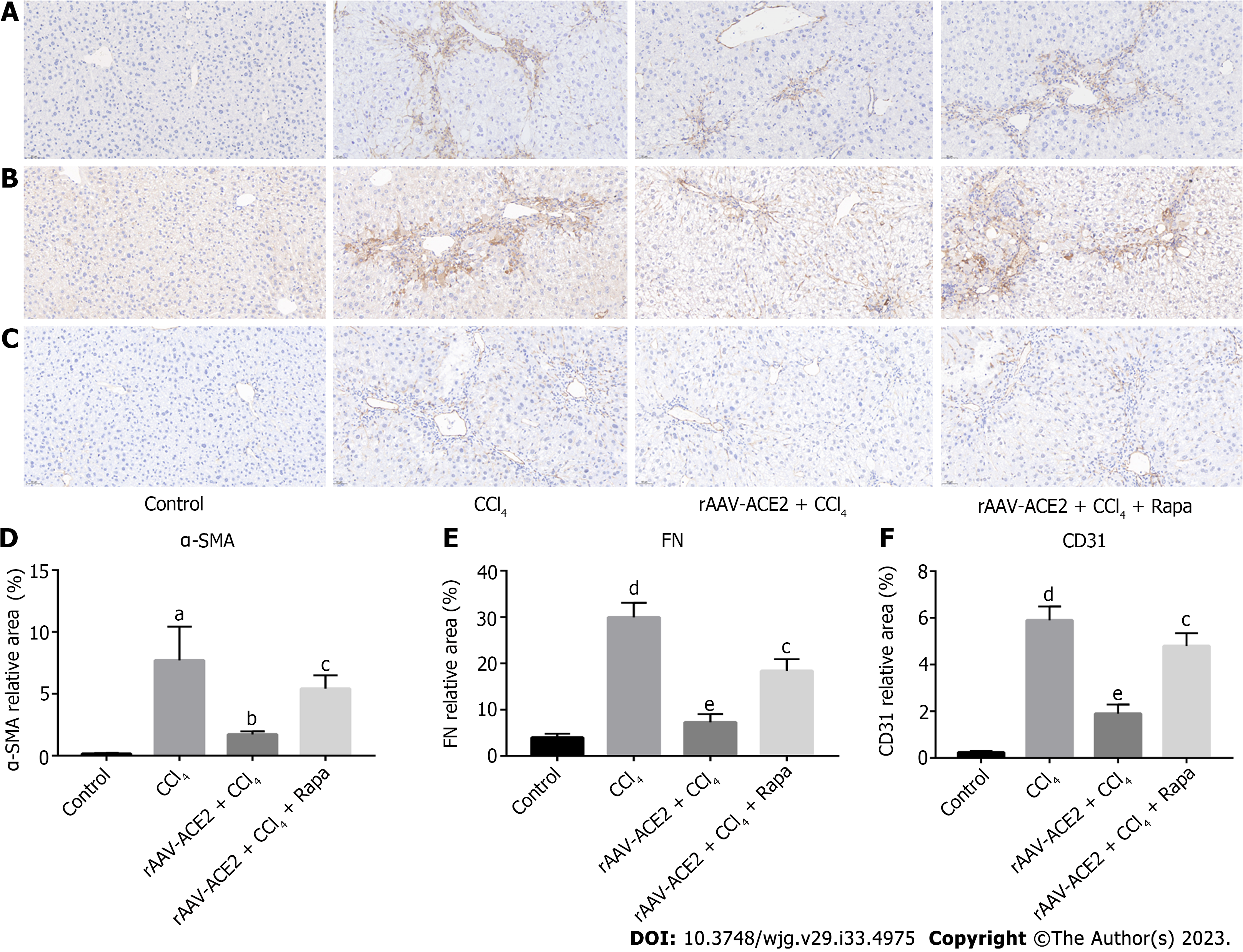
Liver tissue sections were deparaffinized, serially dehydrated in ethanol, and then incubated overnight with primary antibody at 4 °C after antigen retrieval. The primary antibodies used in the experiment included anti-alpha-smooth muscle actin (α-SMA) antibody (1:400, Abcam, United States), anti-fibronectin (FN) antibody (1:2000, Abcam, United States), and anti-CD31 antibody (1:2000, Abcam, United States). After incubation with the appropriate biotinylated secondary antibody for 30 min, the liver sections were stained with diaminobenzidine and hematoxylin. The positive staining areas appeared brownish yellow. The sections were observed under a light microscope, photographed, and then analyzed with Image-Pro Plus 6.0 software.
Fresh liver tissue sections were immobilized in electron microscopy fixative (Servicebio, Wuhan, China) for 2 h. The specimens were then immobilized in osmic acid buffer and dehydrated in ethanol. Finally, the ultrathin sections were photographed using Transmission electron microscopy (TEM) (HT7800/HT7700, Hitachi, Tokyo, Japan) after staining with 2% uranium acetate in alcohol solution. The structure of autophagosomes in each group was observed by TEM.
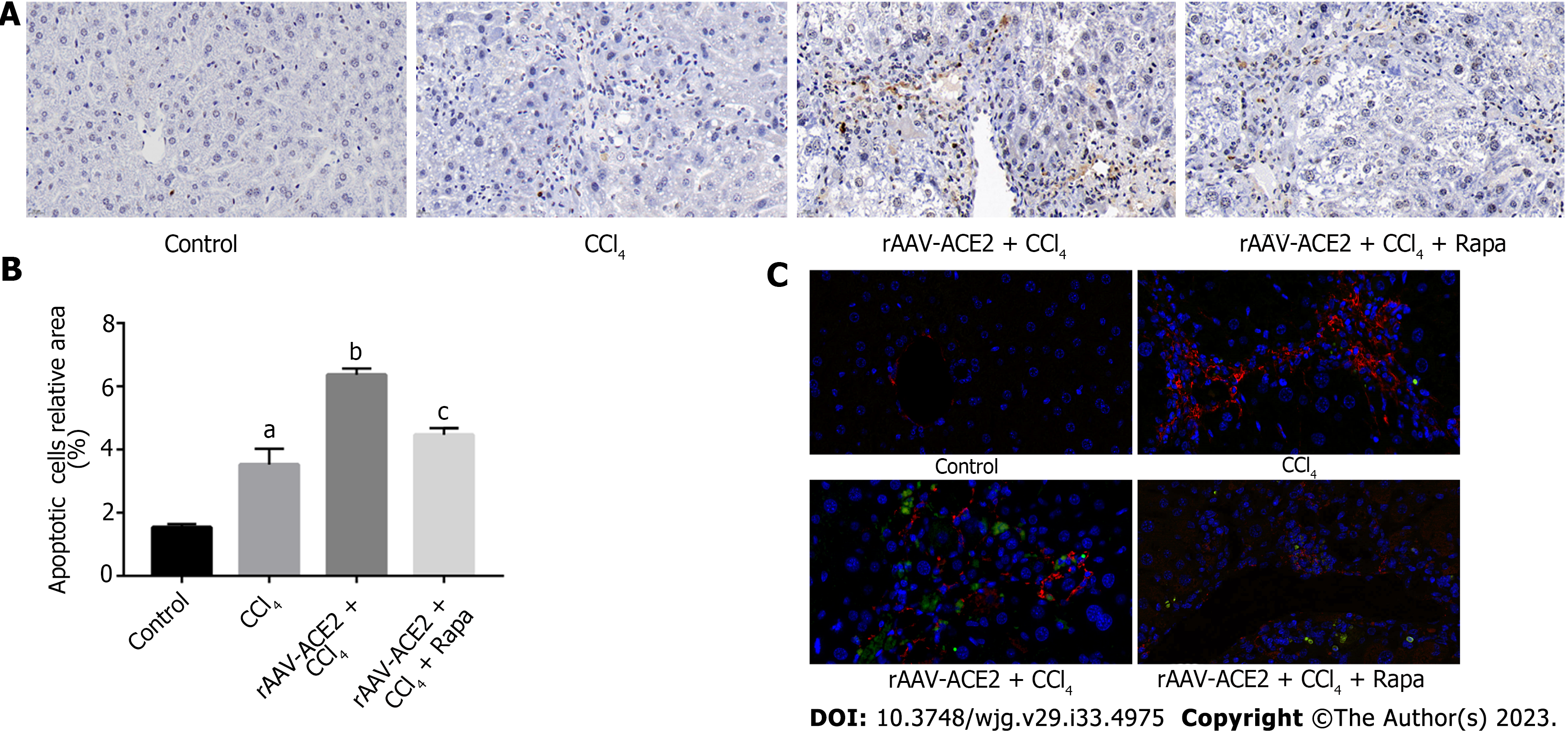
Apoptotic HSCs were localized with labeled nucleotides in TUNEL staining. The mouse liver sections were stained according to the in situ cell death detection kit (Roche, Germany) protocol. The sections were then incubated with an α-SMA primary antibody (1:500, Abcam, United States) and a CY3 goat anti-rabbit fluorescence secondary antibody (1:300, Servicebio, Wuhan, China). The nuclei were counterstained with 4',6-diamidino-2-phenylindole (DAPI) and photographed under a fluorescence microscope. The relative number of apoptotic cells in each group was analyzed using Image-Pro Plus 6.0 software.
Paraffin sections of mouse liver tissue were deparaffinized, subjected to antigen retrieval, and blocked with hydrogen peroxide and serum. The primary antibody, corresponding HRP-labeled secondary antibody, and fluorescently labeled tyramine were successively added. After microwave repair treatment, the first round of primary and secondary antibodies were eluted, and the fluorescently labeled tyramine was still attached to the target. When the second and third targets were detected, the previous steps were repeated for a new round of labeling and microwave repair processing. The fourth primary antibody and 594-labeled fluorescent secondary antibody were added, and the nuclei were then counterstained with DAPI. The slides were covered with anti-fade mounting medium. Finally, images were detected and collected with a slice scanner (pannoramic, 3Dhistech, Hungary). DAPI emits blue light; Fluorescein isothiocyanate (FITC-ACE2) emits green light; 647 (Desmin) is set to pink light; 594 (LC3) is set to purplish red light. The number of positive cells for each index was analyzed using Image-Pro Plus 6.0 software.
Mouse liver tissue proteins were extracted, and the concentration of each protein was determined. Equal amounts of protein samples were subjected to electrophoresis on 8%-12% sodium dodecylsulphate polyacrylamide gel electrophoresis gels and transferred to polyvinylidene fluoride membranes. The membranes were blocked in 5% nonfat dry milk for 1 h to block nonspecific sites and then incubated with the appropriate primary antibodies at 4 °C overnight. After incubation with the secondary antibody and membrane washing, the antibody-bound proteins were detected by chemiluminescence staining using an enhanced chemiluminescence assay kit (Millipore, United States). The density of each band was analyzed with ImageJ software.
The data are expressed as mean ± SD. Statistics were analyzed using GraphPad Prism 8.0 and SPSS 19.0 software. Statistical significance was determined by one-way ANOVA followed by LSD-t test. For all experiments, P < 0.05 was considered statistically significant.
The effect of ACE2 on CCl4-induced liver fibrosis was evaluated by H&E (Figure 1A), Masson trichrome (Figure 1B) and Sirius red staining (Figure 1C and D). Compared with those in the control group, inflammatory cell infiltration and fibrous tissue hyperplasia in the liver tissues of mice were increased after 8 wk of CCl4 injection (METAVIR > F2) (Figure 1A). In the CCl4 group, the liver architecture was widely disorganized, and the hepatic sinusoids could not be distinguished. In addition, notable collagen deposition and the formation of fibrous septa bridging the portal regions were observed in the CCl4 group. However, fibrotic tissue and inflammatory cells in the rAAV2/8-ACE2 + CCl4 group were markedly reduced compared with those in the CCl4 and rapamycin groups (METAVIR ≤ F2) (Figure 1B-D). Masson trichrome and Sirius red staining showed that collagen deposition was significantly increased in mice treated with CCl4 alone (P < 0.05). After rAAV2/8-ACE2 treatment, the degree of collagen deposition in the perisinusoidal spaces, interlobular septum and periportal zones was reduced (P < 0.05) (Figure 1B-D). The results indicated that rAAV2/8-ACE2 treatment could improve liver injury and fibrosis in mice, while rapamycin treatment increased the degree of liver fibrosis compared with that in the ACE2 overexpression group. This finding suggested that mTOR inhibitor could attenuate the antifibrotic effect of rAAV2/8-ACE2.
The levels of PDGF-BB, VEGF, angiopoietin-2, IL-10, Ang II and Ang- (1-7) in mouse serum were measured (Figure 2). PDGF signaling plays a vital role in HSC activation and angiogenesis[19]. VEGF and angiopoietin-2 are the most important regulators in the process of angiogenesis[20,21]. As a potential anti-inflammatory factor, IL-10 has been reported to inhibit the expression of many proinflammatory mediators[22]. The results showed that the levels of PDGF-BB, angiopoietin-2, VEGF, and Ang II in the CCl4 group were notably higher than those in the normal control group (P < 0.001), and rAAV2/8-ACE2 injection reduced the expression of these cytokines (P < 0.001) (Figure 2A-C and E). In addition, the results demonstrated that the levels of IL-10 and Ang- (1-7) were higher in the rAAV2/8-ACE2 group than in the CCl4 group (P < 0.001, P < 0.001), while rapamycin decreased the expression levels of these two cytokines (P < 0.01, P < 0.05) (Figure 2D and F). The results indicated that rAAV2/8-ACE2 treatment could inhibit HSC activation and angiogenesis in mice with liver fibrosis.
Effect of rAAV-ACE2 treatment on the expression of α-SMA, FN and CD31 in CCl4-induced fibrotic mice was evaluated by immunohistochemistry staining (Figure 3A-C). α-SMA is a typical marker of HSC activation and proliferation. In the immunohistochemistry staining, α-SMA positive cells were distributed along the endothelium of hepatic sinusoids in the liver tissues of CCl4-induced fibrotic mice. The number of α-SMA positive cells in the rAAV2/8-ACE2 treatment group was notably lower than that in the CCl4 and rapamycin groups (P < 0.05, P < 0.01) (Figure 3A and D).
FN is the primary protein constituting the basement membrane, and CD31 is commonly used as a vascular endothelial marker. These proteins are rarely expressed in normal liver tissues. Immunohistochemical staining revealed that the expression of these proteins was increased in the CCl4-induced liver fibrosis group (P < 0.001, P < 0.001) and decreased in the rAAV2/8-ACE2 treatment group (P < 0.001, P < 0.001). However, rapamycin increased the protein expression of FN and CD31 (P < 0.01, P < 0.01) (Figure 3B, C, E and F).
TUNEL and immunofluorescence staining were used to detect the number of apoptotic HSCs. Our results demonstrated that there were more apoptotic HSCs in the rAAV2/8-ACE2 treatment group than in the CCl4 and rapamycin groups (P < 0.05, P < 0.05) (Figure 4).
The results showed that ACE2 overexpression inhibited HSC activation and induced HSC apoptosis in fibrotic mouse liver tissues, while the mTOR inhibitor attenuated the effect of rAAV2/8-ACE2 on HSCs.
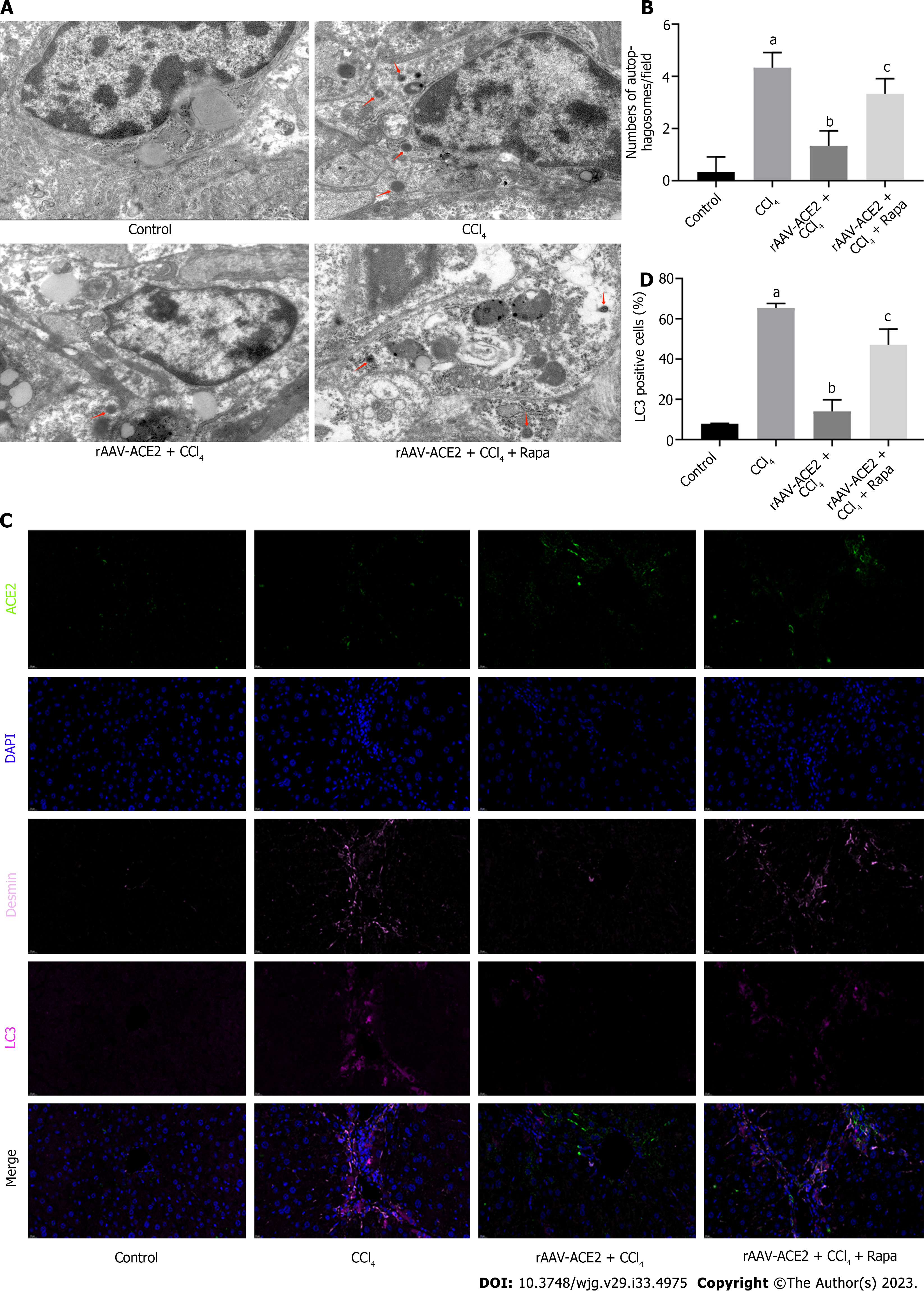
To further verify the effect of ACE2 on autophagy in liver fibrosis, a large number of autophagosomes were detected by ultrastructural analysis in the HSCs of mice in the CCl4 group. TEM analysis showed that rAAV2/8-ACE2 injection decreased the number of autophagosomes in HSCs compared with that in the CCl4 group (P < 0.05). However, autophagosomes were increased in the rAAV2/8-ACE2 + CCl4 + rapamycin group (P < 0.01) (Figure 5A and B). The results of multicolor immunofluorescence staining demonstrated that the expression of the ACE2 protein was increased after rAAV2/8-ACE2 injection, and the expression of the autophagy protein LC3 was decreased compared with that in the CCl4 group (P < 0.01). Treatment with rapamycin attenuated the inhibitory effect of ACE2 on LC3 protein expression (P < 0.05) (Figure 5C and D). These results suggested that ACE2 overexpression could reduce HSC activation and liver fibrosis by inhibiting HSC autophagy.
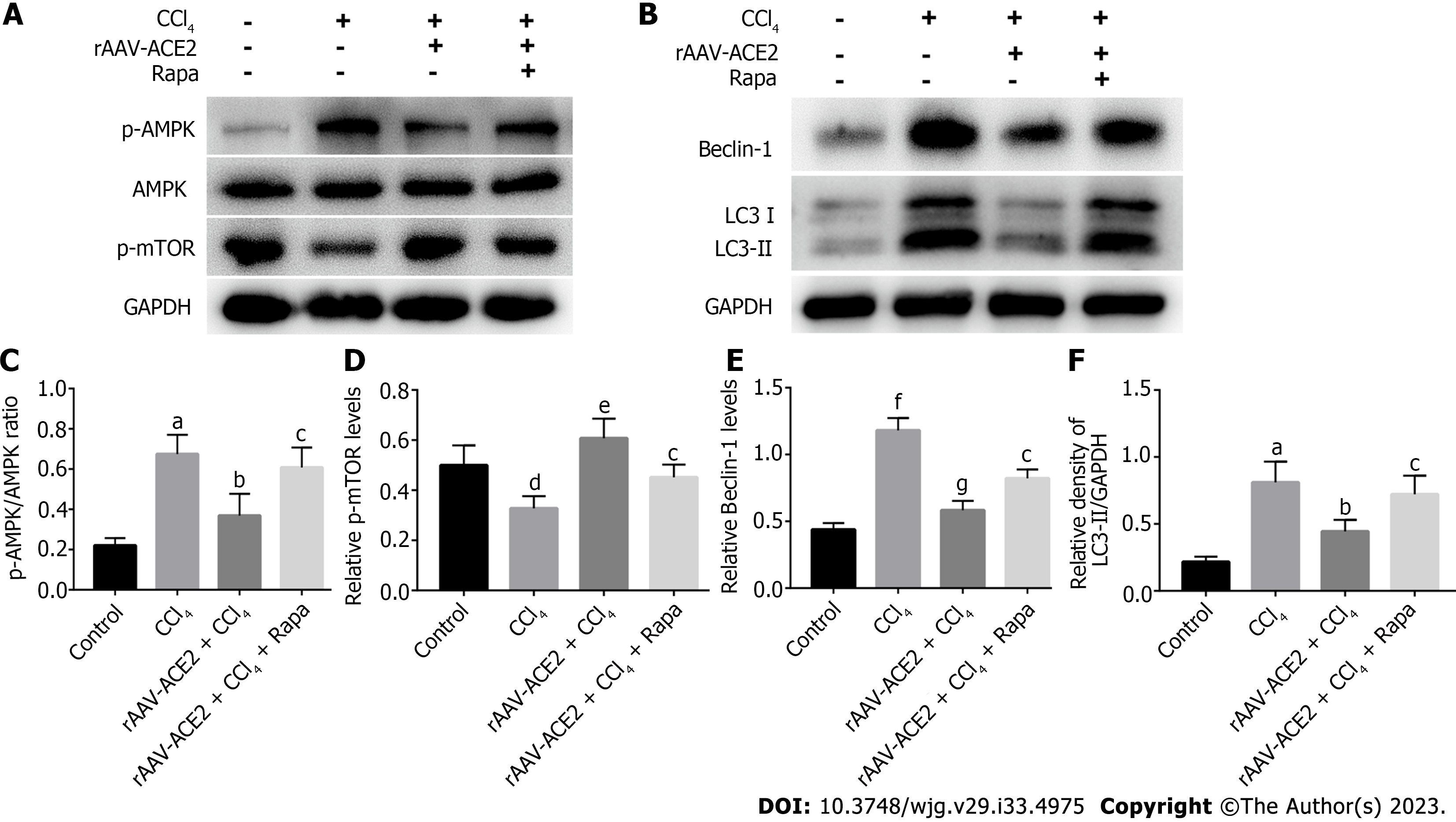
Autophagy is regulated by numerous autophagy-related genes, such as LC3 and Beclin-1. LC3II is a marker protein on the autophagosome membrane and is often considered an indicator of autophagy formation. As an autophagy-specific substrate, p62 interacts with LC3 to infiltrate into autophagosomes and is efficiently degraded by autophagolysosomes[23]. To determine the effect of ACE2 on HSC autophagy, we detected the expression of HSC autophagy-related indicators (LC3I, LC3II, Beclin-1) in the liver tissues of mice in each group by western blotting. Moreover, we verified the correlation of ACE2 with autophagy and the AMPK pathway by assessing the expression of AMPK pathway-related proteins (AMPK, p-AMPK, p-mTOR) and autophagy-related proteins (LC3I, LC3II, Beclin-1) in mouse liver tissues (Figure 6A and B). Compared with that in the control group, the p-AMPK/AMPK ratio was higher in the CCl4 group (P < 0.01). However, the ratio of p-AMPK/AMPK in the rAAV2/8-ACE2-treated group was dramatically lower than that in the CCl4 group (P < 0.05) (Figure 6A and C). In contrast, p-mTOR levels in mice in the rAAV2/8-ACE2-treated group were significantly higher than those in the CCl4 group (P < 0.01) (Figure 6A and D). In addition, the results indicated that the protein levels of Beclin-1 and LC3II in the rAAV2/8-ACE2 + CCl4 group were markedly reduced compared to those in the CCl4 alone group (P < 0.001, P < 0.05) (Figure 6B, E and F). The m-TOR inhibitor (rapamycin) affected mTOR phosphorylation and the level of autophagy proteins in liver tissues. The present study showed that rapamycin abolished the effect of rAAV2/8-ACE2 on the expression of the autophagy proteins LC3I, LC3II and Beclin-1. Compared with those in the rAAV2/8-ACE2 group, the relative Beclin-1 and LC3II levels were increased by rapamycin treatment (P < 0.05, P < 0.05) (Figure 6B, E and F). The western blot results showed that ACE2 overexpression could inhibit the expression of autophagy-related proteins in mouse liver tissues through the AMPK/mTOR pathway.
Liver fibrosis has high morbidity and mortality worldwide, and it is a compensatory response to liver inflammation and injury caused by multiple pathogenic factors[24]. In liver fibrosis, excess fibrous ECM proteins, such as collagens I and III, are deposited in the Disse space of the hepatic sinusoids[25]. Changes in ECM composition induce LSECs to lose their fenestrae and form a basement membrane, a process known as hepatic sinusoidal capillarization[26]. The activation of HSCs plays a crucial role in the process of liver fibrosis. Upon activation due to liver injury, quiescent HSCs lose their retinoid droplets, exhibit increased α-SMA expression, and release large amounts of ECM, ultimately resulting in liver fibrosis[27].
ACE2 is expressed in human alveolar epithelial cells, esophageal epithelial cells, small intestinal epithelial cells, and vascular endothelial cells[28]. Our present study found that ACE2 was also expressed in liver HSCs. In recent years, the coronavirus disease 2019 (COVID-19) virus [severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)] that caused the outbreak has been proven to invade human alveolar epithelial cells mainly through ACE2[29]. SARS-CoV-2 infection can reduce ACE2 activity, leading to an imbalance in Ang II/ACE2 regulation[30]. ACE2, which is the main target receptor for SARS-CoV-2 invasion into the human body, is currently a research hotspot. A global registry study suggested that patients with chronic liver disease and cirrhosis had higher mortality after being infected with COVID-19[31]. The baseline liver disease severity of patients with chronic liver disease and cirrhosis is closely related to the COVID-19-related incidence rates and mortality. Therefore, SARS-CoV-2 infection may exacerbate the degree of cirrhosis and portal hypertension in patients with chronic liver disease by reducing the activity of ACE2 in the liver.
ACE2 is an endogenous negative regulator that acts as a RAS "brake" to limit fibrogenesis through Ang II degradation and Ang- (1-7) formation. It was reported that the degree of liver fibrosis in ACE2 knockout mice increased after 21 d of bile duct ligation or chronic CCl4 treatment[13]. In addition to its effect on the RAS, whether ACE2 can affect liver fibrosis through other mechanisms remains unclear. In our study, a liver fibrosis model was induced by the intraperitoneal injection of CCl4 to investigate the effect of ACE2 on liver fibrosis by inhibiting autophagy. In addition, the liver-specific recombinant adeno-associated viral vector rAAV2/8-ACE2 was used in this study. ACE2 is specifically overexpressed in the liver with minimal systemic effects. Moreover, enhanced expression and activity of liver tissue-specific ACE2 can reduce local Ang II levels, increase local Ang- (1-7) levels, and minimize off-target effects[32].
Autophagy is the process of degrading defective proteins, damaged organelles, excess lipids and other harmful components in cells to maintain cellular components and homeostasis[33]. Autophagy levels are elevated in conditions of inflammation and oxidative stress, and excessive autophagy is involved in inflammatory and liver diseases[24]. Studies have demonstrated that inhibiting autophagy in HSCs reduces lipid droplet degradation, thereby preventing cell activation[9]. Autophagosome is composed of a small portion of the cytoplasm surrounded by double membranes. The digested substances are various components contained in the cytoplasm, such as mitochondria and fragments of endoplasmic reticulum, and the contents are degraded by fusion with lysosomes. Autophagy generally refers to macroautophagy, which includes two consecutive stages of autophagosome formation and degradation[15]. It has been reported that IL-10 inhibits oxidative stress-induced HSC autophagosome formation, HSC activation and liver fibrosis through the mTOR/STAT3 signaling pathway[34]. The TEM results indicated that the number of autophagosomes in the rAAV2/8-ACE2-treated group was decreased. To explore the relationship between ACE2 and autophagy, we detected the expression of the autophagy proteins LC3I, LC3II and Beclin-1 in the liver tissues of mice in each group. The results indicated that ACE2 overexpression effectively inhibited autophagy during mouse liver fibrosis.
Autophagy regulation is intricately associated with signaling pathways such as the AMPK/mTOR pathway. AMPK can inhibit mTORC1 activity by activating the TSC1/TSC2 protein heterodimer[35,36]. mTORC1 negatively regulates the initiation of autophagy through phosphorylation at Ser757 of ULK1 upon activation[36]. Compared with that in the CCl4 group, the p-AMPK/AMPK ratio was decreased (P < 0.05), while the relative expression of p-mTOR was increased in the rAAV2/8-ACE2 group (P < 0.01). The results showed that ACE2 overexpression could influence the AMPK/mTOR signaling pathway. We treated mice with an m-TOR inhibitor (rapamycin), which effectively inhibited m-TOR phosphorylation. The findings of the study indicated that ACE2 overexpression could inhibit HSC autophagy in mouse liver tissues through the AMPK/mTOR pathway. The results suggested that the AMPK/mTOR signaling pathway was an important node for ACE2 to regulate HSC autophagy.
Pathological staining showed the successful establishment of a mouse model of liver fibrosis after 8 wk of intraperitoneal injection of CCl4. rAAV2/8-ACE2 injection alleviated collagen deposition and fibrosis in the liver tissues of mice. We further investigated the mechanism by which ACE2 overexpression alleviated liver fibrosis. When liver injury occurs, HSCs are activated and proliferate, and the demand for intracellular energy increases. At this time, blocking autophagy can impair HSC activation and fibrotic activity[10]. α-SMA is an important indicator for evaluating HSC activation and proliferation. In the present study, rAAV2/8-ACE2 injection inhibited α-SMA expression and HSC activation. In addition, apoptosis plays a vital role in the proliferation, differentiation and death of HSCs, and HSC apoptosis is the key to reversing liver fibrosis[37]. TUNEL and immunofluorescence staining showed that rAAV2/8-ACE2 injection increased HSC apoptosis. Our previous study demonstrated a complex relationship between autophagy and apoptosis, and the inhibition of autophagy could induce HSC apoptosis[15]. Therefore, our findings indicated that ACE2 overexpression could alleviate liver fibrosis by regulating autophagy to inhibit HSC activation and promote apoptosis.
Intrahepatic angiogenesis and sinusoidal remodeling play an important role in the development of hepatic fibrosis and portal hypertension[38]. The inhibition of pathological angiogenesis can alleviate liver fibrosis. LSEC capillarization is associated with the accumulation of interstitial collagen in the Disse space of hepatic sinusoids and is the main pathological change in liver fibrosis[26]. The reversal of LSEC capillarization has been reported to promote HSC quiescence[39]. During cirrhosis, angiogenesis-related cytokines and receptors expressed in HSCs, such as VEGF, PDGF, and angiopoietin, can induce HSC migration, angiogenesis, and collagen production[40]. Our study demonstrated that the levels of VEGF, angiopoietin-2 and PDGF-BB were elevated in liver fibrosis, resulting in increased angiogenesis. rAAV2/8-ACE2 injection inhibited the expression levels of these angiogenesis-related factors. Therefore, the results indicated that ACE2 overexpression could effectively attenuate intrahepatic angiogenesis, thus alleviating hepatic sinusoidal resistance.
In the present study, adeno-associated viral vector technology, pathological staining, multifactor analysis, multicolor immunofluorescence staining, TEM and other advanced techniques were used to comprehensively explore the relationship and mechanism among ACE2, autophagy and liver fibrosis. However, there are still some limitations of this study. Whether ACE2 affects HSC autophagy and liver fibrosis through other pathways needs to be further explored. This study provides a new theoretical basis for the targeted treatment of liver fibrosis and portal hypertension, and its clinical application needs further research.
In summary, the study indicates that autophagy plays a crucial role in HSC activation and liver fibrosis. ACE2 overexpression can inhibit HSC activation and promote apoptosis by regulating HSC autophagy, thereby alleviating liver fibrosis and hepatic sinusoidal remodeling. Our study also demonstrates that the AMPK/mTOR pathway is involved in the effect of ACE2 on autophagy. This study may provide new ideas for exploring the molecular mechanism by which ACE2 inhibits liver fibrosis and hepatic sinusoidal remodeling.
Liver cirrhosis is a hallmark of end-stage chronic liver disease, which leads to millions of deaths each year. At present, the treatment options for liver fibrosis and cirrhosis are limited and often ineffective. Angiotensin-converting enzyme 2 (ACE2)-driven protective renin-angiotensin system (RAS) provides an effective therapeutic target for liver fibrosis. In addition, the study of liver fibrosis targeting hepatic stellate cell (HSC) autophagy has attracted more and more attention.
In addition to its effect on the RAS, whether ACE2 can affect liver fibrosis through other mechanisms remains unclear. Moreover, how to enhance the expression and activity of tissue-specific ACE2 to avoid its potential off-target effect is a problem to be solved. Using a suitable and efficient gene delivery system to achieve tissue-specific overexpression of ACE2 has pointed out a new direction for the targeted treatment of liver fibrosis.
The aim of this study is to determine the effect of ACE2 on HSC activation, proliferation, apoptosis and liver fibrosis by regulating autophagy. This study provides new ideas for exploring the molecular mechanism by which ACE2 inhibits liver fibrosis and hepatic sinusoidal remodeling.
In this study, a mouse model of liver fibrosis was constructed, and adeno-associated viral vector technology, pathological staining, multifactor analysis, multicolor immunofluorescence staining, transmission electron microscopy, TUNEL apoptosis assays, western blot analysis and other experimental methods were used to comprehensively explore the relationship and mechanism among ACE2, autophagy and liver fibrosis.
In vivo experiments showed that rAAV2/8-ACE2 treatment could inhibit HSC activation and angiogenesis, induce HSC apoptosis, and alleviate HSC proliferation and liver fibrosis by inhibiting HSC autophagy. This study also demonstrated that ACE2 overexpression could inhibit HSC autophagy in mouse liver tissues through the Adenosine monophosphate activates protein kinases (AMPK)/mammalian target of rapamycin (mTOR) pathway. The completion of this study provides new ideas for the prevention and targeted treatment of liver fibrosis and portal hypertension.
The study demonstrates that autophagy plays a crucial role in HSC activation and liver fibrosis. ACE2 overexpression can inhibit HSC activation and promote apoptosis by regulating HSC autophagy through the AMPK/mTOR pathway, thereby alleviating liver fibrosis and hepatic sinusoidal remodeling.
The pathogenesis of liver fibrosis and cirrhosis is a complex process involving the interaction of various growth factors, cytokines, and vasoactive substances. We need further clinical research to improve patient treatment outcomes through advanced technologies such as drug carrier-targeted HSC-specific therapies.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Durairajan SSK, India; Kreisel W, Germany S-Editor: Li L L-Editor: A P-Editor: Cai YX
| 1. | García-Pagán JC, Gracia-Sancho J, Bosch J. Functional aspects on the pathophysiology of portal hypertension in cirrhosis. J Hepatol. 2012;57:458-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 207] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 2. | Deleve LD, Wang X, Guo Y. Sinusoidal endothelial cells prevent rat stellate cell activation and promote reversion to quiescence. Hepatology. 2008;48:920-930. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 321] [Cited by in RCA: 295] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 3. | Coulon S, Heindryckx F, Geerts A, Van Steenkiste C, Colle I, Van Vlierberghe H. Angiogenesis in chronic liver disease and its complications. Liver Int. 2011;31:146-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 222] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 4. | Kisseleva T. The origin of fibrogenic myofibroblasts in fibrotic liver. Hepatology. 2017;65:1039-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 5. | Mimche PN, Lee CM, Mimche SM, Thapa M, Grakoui A, Henkemeyer M, Lamb TJ. EphB2 receptor tyrosine kinase promotes hepatic fibrogenesis in mice via activation of hepatic stellate cells. Sci Rep. 2018;8:2532. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 6. | Thabut D, Shah V. Intrahepatic angiogenesis and sinusoidal remodeling in chronic liver disease: new targets for the treatment of portal hypertension? J Hepatol. 2010;53:976-980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 218] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 7. | Hazari Y, Bravo-San Pedro JM, Hetz C, Galluzzi L, Kroemer G. Autophagy in hepatic adaptation to stress. J Hepatol. 2020;72:183-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 125] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 8. | Ueno T, Komatsu M. Autophagy in the liver: functions in health and disease. Nat Rev Gastroenterol Hepatol. 2017;14:170-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 398] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 9. | Hernández-Gea V, Friedman SL. Autophagy fuels tissue fibrogenesis. Autophagy. 2012;8:849-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 91] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 10. | Hernández-Gea V, Ghiassi-Nejad Z, Rozenfeld R, Gordon R, Fiel MI, Yue Z, Czaja MJ, Friedman SL. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142:938-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 520] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 11. | Seki E, Brenner DA. Recent advancement of molecular mechanisms of liver fibrosis. J Hepatobiliary Pancreat Sci. 2015;22:512-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 249] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 12. | Wu Y, Li Z, Wang S, Xiu A, Zhang C. Carvedilol Inhibits Angiotensin II-Induced Proliferation and Contraction in Hepatic Stellate Cells through the RhoA/Rho-Kinase Pathway. Biomed Res Int. 2019;2019:7932046. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 13. | Osterreicher CH, Taura K, De Minicis S, Seki E, Penz-Osterreicher M, Kodama Y, Kluwe J, Schuster M, Oudit GY, Penninger JM, Brenner DA. Angiotensin-converting-enzyme 2 inhibits liver fibrosis in mice. Hepatology. 2009;50:929-938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 112] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 14. | Mak KY, Chin R, Cunningham SC, Habib MR, Torresi J, Sharland AF, Alexander IE, Angus PW, Herath CB. ACE2 Therapy Using Adeno-associated Viral Vector Inhibits Liver Fibrosis in Mice. Mol Ther. 2015;23:1434-1443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 15. | Meng D, Li Z, Wang G, Ling L, Wu Y, Zhang C. Carvedilol attenuates liver fibrosis by suppressing autophagy and promoting apoptosis in hepatic stellate cells. Biomed Pharmacother. 2018;108:1617-1627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 16. | Zhang X, Zheng J, Yan Y, Ruan Z, Su Y, Wang J, Huang H, Zhang Y, Wang W, Gao J, Chi Y, Lu X, Liu Z. Angiotensin-converting enzyme 2 regulates autophagy in acute lung injury through AMPK/mTOR signaling. Arch Biochem Biophys. 2019;672:108061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 17. | Wang Y, Rijal B, Xu M, Li Z, An Y, Zhang F, Lu C. Renal denervation improves vascular endothelial dysfunction by inducing autophagy via AMPK/mTOR signaling activation in a rat model of type 2 diabetes mellitus with insulin resistance. Acta Diabetol. 2020;57:1227-1243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 18. | Li Z, Ding Q, Ling LP, Wu Y, Meng DX, Li X, Zhang CQ. Metformin attenuates motility, contraction, and fibrogenic response of hepatic stellate cells in vivo and in vitro by activating AMP-activated protein kinase. World J Gastroenterol. 2018;24:819-832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 31] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 19. | Greuter T, Shah VH. Hepatic sinusoids in liver injury, inflammation, and fibrosis: new pathophysiological insights. J Gastroenterol. 2016;51:511-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 102] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 20. | Fernandez M. Molecular pathophysiology of portal hypertension. Hepatology. 2015;61:1406-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 93] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 21. | An YA, Sun K, Joffin N, Zhang F, Deng Y, Donzé O, Kusminski CM, Scherer PE. Angiopoietin-2 in white adipose tissue improves metabolic homeostasis through enhanced angiogenesis. Elife. 2017;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 22. | Choi JS, Jeong IS, Park YJ, Kim SW. HGF and IL-10 expressing ALB::GFP reporter cells generated from iPSCs show robust anti-fibrotic property in acute fibrotic liver model. Stem Cell Res Ther. 2020;11:332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 23. | Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, Abdellatif M, Abdoli A, Abel S, Abeliovich H, Abildgaard MH, Abudu YP, Acevedo-Arozena A, Adamopoulos IE, Adeli K, Adolph TE, Adornetto A, Aflaki E, Agam G, Agarwal A, Aggarwal BB, Agnello M, Agostinis P, Agrewala JN, Agrotis A, Aguilar PV, Ahmad ST, Ahmed ZM, Ahumada-Castro U, Aits S, Aizawa S, Akkoc Y, Akoumianaki T, Akpinar HA, Al-Abd AM, Al-Akra L, Al-Gharaibeh A, Alaoui-Jamali MA, Alberti S, Alcocer-Gómez E, Alessandri C, Ali M, Alim Al-Bari MA, Aliwaini S, Alizadeh J, Almacellas E, Almasan A, Alonso A, Alonso GD, Altan-Bonnet N, Altieri DC, Álvarez ÉMC, Alves S, Alves da Costa C, Alzaharna MM, Amadio M, Amantini C, Amaral C, Ambrosio S, Amer AO, Ammanathan V, An Z, Andersen SU, Andrabi SA, Andrade-Silva M, Andres AM, Angelini S, Ann D, Anozie UC, Ansari MY, Antas P, Antebi A, Antón Z, Anwar T, Apetoh L, Apostolova N, Araki T, Araki Y, Arasaki K, Araújo WL, Araya J, Arden C, Arévalo MA, Arguelles S, Arias E, Arikkath J, Arimoto H, Ariosa AR, Armstrong-James D, Arnauné-Pelloquin L, Aroca A, Arroyo DS, Arsov I, Artero R, Asaro DML, Aschner M, Ashrafizadeh M, Ashur-Fabian O, Atanasov AG, Au AK, Auberger P, Auner HW, Aurelian L, Autelli R, Avagliano L, Ávalos Y, Aveic S, Aveleira CA, Avin-Wittenberg T, Aydin Y, Ayton S, Ayyadevara S, Azzopardi M, Baba M, Backer JM, Backues SK, Bae DH, Bae ON, Bae SH, Baehrecke EH, Baek A, Baek SH, Bagetta G, Bagniewska-Zadworna A, Bai H, Bai J, Bai X, Bai Y, Bairagi N, Baksi S, Balbi T, Baldari CT, Balduini W, Ballabio A, Ballester M, Balazadeh S, Balzan R, Bandopadhyay R, Banerjee S, Bánréti Á, Bao Y, Baptista MS, Baracca A, Barbati C, Bargiela A, Barilà D, Barlow PG, Barmada SJ, Barreiro E, Barreto GE, Bartek J, Bartel B, Bartolome A, Barve GR, Basagoudanavar SH, Bassham DC, Bast RC Jr, Basu A, Batoko H, Batten I, Baulieu EE, Baumgarner BL, Bayry J, Beale R, Beau I, Beaumatin F, Bechara LRG, Beck GR Jr, Beers MF, Begun J, Behrends C, Behrens GMN, Bei R, Bejarano E, Bel S, Behl C, Belaid A, Belgareh-Touzé N, Bellarosa C, Belleudi F, Belló Pérez M, Bello-Morales R, Beltran JSO, Beltran S, Benbrook DM, Bendorius M, Benitez BA, Benito-Cuesta I, Bensalem J, Berchtold MW, Berezowska S, Bergamaschi D, Bergami M, Bergmann A, Berliocchi L, Berlioz-Torrent C, Bernard A, Berthoux L, Besirli CG, Besteiro S, Betin VM, Beyaert R, Bezbradica JS, Bhaskar K, Bhatia-Kissova I, Bhattacharya R, Bhattacharya S, Bhattacharyya S, Bhuiyan MS, Bhutia SK, Bi L, Bi X, Biden TJ, Bijian K, Billes VA, Binart N, Bincoletto C, Birgisdottir AB, Bjorkoy G, Blanco G, Blas-Garcia A, Blasiak J, Blomgran R, Blomgren K, Blum JS, Boada-Romero E, Boban M, Boesze-Battaglia K, Boeuf P, Boland B, Bomont P, Bonaldo P, Bonam SR, Bonfili L, Bonifacino JS, Boone BA, Bootman MD, Bordi M, Borner C, Bornhauser BC, Borthakur G, Bosch J, Bose S, Botana LM, Botas J, Boulanger CM, Boulton ME, Bourdenx M, Bourgeois B, Bourke NM, Bousquet G, Boya P, Bozhkov PV, Bozi LHM, Bozkurt TO, Brackney DE, Brandts CH, Braun RJ, Braus GH, Bravo-Sagua R, Bravo-San Pedro JM, Brest P, Bringer MA, Briones-Herrera A, Broaddus VC, Brodersen P, Brodsky JL, Brody SL, Bronson PG, Bronstein JM, Brown CN, Brown RE, Brum PC, Brumell JH, Brunetti-Pierri N, Bruno D, Bryson-Richardson RJ, Bucci C, Buchrieser C, Bueno M, Buitrago-Molina LE, Buraschi S, Buch S, Buchan JR, Buckingham EM, Budak H, Budini M, Bultynck G, Burada F, Burgoyne JR, Burón MI, Bustos V, Büttner S, Butturini E, Byrd A, Cabas I, Cabrera-Benitez S, Cadwell K, Cai J, Cai L, Cai Q, Cairó M, Calbet JA, Caldwell GA, Caldwell KA, Call JA, Calvani R, Calvo AC, Calvo-Rubio Barrera M, Camara NO, Camonis JH, Camougrand N; Campanella M; Campbell EM, Campbell-Valois FX, Campello S, Campesi I, Campos JC, Camuzard O, Cancino J, Candido de Almeida D, Canesi L, Caniggia I, Canonico B, Cantí C, Cao B, Caraglia M, Caramés B, Carchman EH, Cardenal-Muñoz E, Cardenas C, Cardenas L, Cardoso SM, Carew JS, Carle GF, Carleton G, Carloni S, Carmona-Gutierrez D, Carneiro LA, Carnevali O, Carosi JM, Carra S, Carrier A, Carrier L, Carroll B, Carter AB, Carvalho AN, Casanova M, Casas C, Casas J, Cassioli C, Castillo EF, Castillo K, Castillo-Lluva S, Castoldi F, Castori M, Castro AF, Castro-Caldas M, Castro-Hernandez J, Castro-Obregon S, Catz SD, Cavadas C, Cavaliere F, Cavallini G, Cavinato M, Cayuela ML, Cebollada Rica P, Cecarini V, Cecconi F, Cechowska-Pasko M, Cenci S, Ceperuelo-Mallafré V, Cerqueira JJ, Cerutti JM, Cervia D, Cetintas VB, Cetrullo S, Chae HJ, Chagin AS, Chai CY, Chakrabarti G, Chakrabarti O, Chakraborty T, Chami M, Chamilos G, Chan DW, Chan EYW, Chan ED, Chan HYE, Chan HH, Chan H, Chan MTV, Chan YS, Chandra PK, Chang CP, Chang C, Chang HC, Chang K, Chao J, Chapman T, Charlet-Berguerand N, Chatterjee S, Chaube SK, Chaudhary A, Chauhan S, Chaum E, Checler F, Cheetham ME, Chen CS, Chen GC, Chen JF, Chen LL, Chen L, Chen M, Chen MK, Chen N, Chen Q, Chen RH, Chen S, Chen W, Chen XM, Chen XW, Chen X, Chen Y, Chen YG, Chen YJ, Chen YQ, Chen ZS, Chen Z, Chen ZH, Chen ZJ, Cheng H, Cheng J, Cheng SY, Cheng W, Cheng X, Cheng XT, Cheng Y, Cheng Z, Cheong H, Cheong JK, Chernyak BV, Cherry S, Cheung CFR, Cheung CHA, Cheung KH, Chevet E, Chi RJ, Chiang AKS, Chiaradonna F, Chiarelli R, Chiariello M, Chica N, Chiocca S, Chiong M, Chiou SH, Chiramel AI, Chiurchiù V, Cho DH, Choe SK, Choi AMK, Choi ME, Choudhury KR, Chow NS, Chu CT, Chua JP, Chua JJE, Chung H, Chung KP, Chung S, Chung SH, Chung YL, Cianfanelli V, Ciechomska IA, Cifuentes M, Cinque L, Cirak S, Cirone M, Clague MJ, Clarke R, Clementi E, Coccia EM, Codogno P, Cohen E, Cohen MM, Colasanti T, Colasuonno F, Colbert RA, Colell A, Čolić M, Coll NS, Collins MO, Colombo MI, Colón-Ramos DA, Combaret L, Comincini S, Cominetti MR, Consiglio A, Conte A, Conti F, Contu VR, Cookson MR, Coombs KM, Coppens I, Corasaniti MT, Corkery DP, Cordes N, Cortese K, Costa MDC, Costantino S, Costelli P, Coto-Montes A, Crack PJ, Crespo JL, Criollo A, Crippa V, Cristofani R, Csizmadia T, Cuadrado A, Cui B, Cui J, Cui Y, Culetto E, Cumino AC, Cybulsky AV, Czaja MJ, Czuczwar SJ, D'Adamo S, D'Amelio M, D'Arcangelo D, D'Lugos AC, D'Orazi G, da Silva JA, Dafsari HS, Dagda RK, Dagdas Y, Daglia M, Dai X, Dai Y, Dal Col J, Dalhaimer P, Dalla Valle L, Dallenga T, Dalmasso G, Damme M, Dando I, Dantuma NP, Darling AL, Das H, Dasarathy S, Dasari SK, Dash S, Daumke O, Dauphinee AN, Davies JS, Dávila VA, Davis RJ, Davis T, Dayalan Naidu S, De Amicis F, De Bosscher K, De Felice F, De Franceschi L, De Leonibus C, de Mattos Barbosa MG, De Meyer GRY, De Milito A, De Nunzio C, De Palma C, De Santi M, De Virgilio C, De Zio D, Debnath J, DeBosch BJ, Decuypere JP, Deehan MA, Deflorian G, DeGregori J, Dehay B, Del Rio G, Delaney JR, Delbridge LMD, Delorme-Axford E, Delpino MV, Demarchi F, Dembitz V, Demers ND, Deng H, Deng Z, Dengjel J, Dent P, Denton D, DePamphilis ML, Der CJ, Deretic V, Descoteaux A, Devis L, Devkota S, Devuyst O, Dewson G, Dharmasivam M, Dhiman R, di Bernardo D, Di Cristina M, Di Domenico F, Di Fazio P, Di Fonzo A, Di Guardo G, Di Guglielmo GM, Di Leo L, Di Malta C, Di Nardo A, Di Rienzo M, Di Sano F, Diallinas G, Diao J, Diaz-Araya G, Díaz-Laviada I, Dickinson JM, Diederich M, Dieudé M, Dikic I, Ding S, Ding WX, Dini L, Dinić J, Dinic M, Dinkova-Kostova AT, Dionne MS, Distler JHW, Diwan A, Dixon IMC, Djavaheri-Mergny M, Dobrinski I, Dobrovinskaya O, Dobrowolski R, Dobson RCJ, Đokić J, Dokmeci Emre S, Donadelli M, Dong B, Dong X, Dong Z, Dorn Ii GW, Dotsch V, Dou H, Dou J, Dowaidar M, Dridi S, Drucker L, Du A, Du C, Du G, Du HN, Du LL, du Toit A, Duan SB, Duan X, Duarte SP, Dubrovska A, Dunlop EA, Dupont N, Durán RV, Dwarakanath BS, Dyshlovoy SA, Ebrahimi-Fakhari D, Eckhart L, Edelstein CL, Efferth T, Eftekharpour E, Eichinger L, Eid N, Eisenberg T, Eissa NT, Eissa S, Ejarque M, El Andaloussi A, El-Hage N, El-Naggar S, Eleuteri AM, El-Shafey ES, Elgendy M, Eliopoulos AG, Elizalde MM, Elks PM, Elsasser HP, Elsherbiny ES, Emerling BM, Emre NCT, Eng CH, Engedal N, Engelbrecht AM, Engelsen AST, Enserink JM, Escalante R, Esclatine A, Escobar-Henriques M, Eskelinen EL, Espert L, Eusebio MO, Fabrias G, Fabrizi C, Facchiano A, Facchiano F, Fadeel B, Fader C, Faesen AC, Fairlie WD, Falcó A, Falkenburger BH, Fan D, Fan J, Fan Y, Fang EF, Fang Y, Fanto M, Farfel-Becker T, Faure M, Fazeli G, Fedele AO, Feldman AM, Feng D, Feng J, Feng L, Feng Y, Feng W, Fenz Araujo T, Ferguson TA, Fernández ÁF, Fernandez-Checa JC, Fernández-Veledo S, Fernie AR, Ferrante AW Jr, Ferraresi A, Ferrari MF, Ferreira JCB, Ferro-Novick S, Figueras A, Filadi R, Filigheddu N, Filippi-Chiela E, Filomeni G, Fimia GM, Fineschi V, Finetti F, Finkbeiner S, Fisher EA, Fisher PB, Flamigni F, Fliesler SJ, Flo TH, Florance I, Florey O, Florio T, Fodor E, Follo C, Fon EA, Forlino A, Fornai F, Fortini P, Fracassi A, Fraldi A, Franco B, Franco R, Franconi F, Frankel LB, Friedman SL, Fröhlich LF, Frühbeck G, Fuentes JM, Fujiki Y, Fujita N, Fujiwara Y, Fukuda M, Fulda S, Furic L, Furuya N, Fusco C, Gack MU, Gaffke L, Galadari S, Galasso A, Galindo MF, Gallolu Kankanamalage S, Galluzzi L, Galy V, Gammoh N, Gan B, Ganley IG, Gao F, Gao H, Gao M, Gao P, Gao SJ, Gao W, Gao X, Garcera A, Garcia MN, Garcia VE, García-Del Portillo F, Garcia-Escudero V, Garcia-Garcia A, Garcia-Macia M, García-Moreno D, Garcia-Ruiz C, García-Sanz P, Garg AD, Gargini R, Garofalo T, Garry RF, Gassen NC, Gatica D, Ge L, Ge W, Geiss-Friedlander R, Gelfi C, Genschik P, Gentle IE, Gerbino V, Gerhardt C, Germain K, Germain M, Gewirtz DA, Ghasemipour Afshar E, Ghavami S, Ghigo A, Ghosh M, Giamas G, Giampietri C, Giatromanolaki A, Gibson GE, Gibson SB, Ginet V, Giniger E, Giorgi C, Girao H, Girardin SE, Giridharan M, Giuliano S, Giulivi C, Giuriato S, Giustiniani J, Gluschko A, Goder V, Goginashvili A, Golab J, Goldstone DC, Golebiewska A, Gomes LR, Gomez R, Gómez-Sánchez R, Gomez-Puerto MC, Gomez-Sintes R, Gong Q, Goni FM, González-Gallego J, Gonzalez-Hernandez T, Gonzalez-Polo RA, Gonzalez-Reyes JA, González-Rodríguez P, Goping IS, Gorbatyuk MS, Gorbunov NV, Görgülü K, Gorojod RM, Gorski SM, Goruppi S, Gotor C, Gottlieb RA, Gozes I, Gozuacik D, Graef M, Gräler MH, Granatiero V, Grasso D, Gray JP, Green DR, Greenhough A, Gregory SL, Griffin EF, Grinstaff MW, Gros F, Grose C, Gross AS, Gruber F, Grumati P, Grune T, Gu X, Guan JL, Guardia CM, Guda K, Guerra F, Guerri C, Guha P, Guillén C, Gujar S, Gukovskaya A, Gukovsky I, Gunst J, Günther A, Guntur AR, Guo C, Guo H, Guo LW, Guo M, Gupta P, Gupta SK, Gupta S, Gupta VB, Gupta V, Gustafsson AB, Gutterman DD, H B R, Haapasalo A, Haber JE, Hać A, Hadano S, Hafrén AJ, Haidar M, Hall BS, Halldén G, Hamacher-Brady A, Hamann A, Hamasaki M, Han W, Hansen M, Hanson PI, Hao Z, Harada M, Harhaji-Trajkovic L, Hariharan N, Haroon N, Harris J, Hasegawa T, Hasima Nagoor N, Haspel JA, Haucke V, Hawkins WD, Hay BA, Haynes CM, Hayrabedyan SB, Hays TS, He C, He Q, He RR, He YW, He YY, Heakal Y, Heberle AM, Hejtmancik JF, Helgason GV, Henkel V, Herb M, Hergovich A, Herman-Antosiewicz A, Hernández A, Hernandez C, Hernandez-Diaz S, Hernandez-Gea V, Herpin A, Herreros J, Hervás JH, Hesselson D, Hetz C, Heussler VT, Higuchi Y, Hilfiker S, Hill JA, Hlavacek WS, Ho EA, Ho IHT, Ho PW, Ho SL, Ho WY, Hobbs GA, Hochstrasser M, Hoet PHM, Hofius D, Hofman P, Höhn A, Holmberg CI, Hombrebueno JR, Yi-Ren Hong CH, Hooper LV, Hoppe T, Horos R, Hoshida Y, Hsin IL, Hsu HY, Hu B, Hu D, Hu LF, Hu MC, Hu R, Hu W, Hu YC, Hu ZW, Hua F, Hua J, Hua Y, Huan C, Huang C, Huang H, Huang K, Huang MLH, Huang R, Huang S, Huang T, Huang X, Huang YJ, Huber TB, Hubert V, Hubner CA, Hughes SM, Hughes WE, Humbert M, Hummer G, Hurley JH, Hussain S, Hussey PJ, Hutabarat M, Hwang HY, Hwang S, Ieni A, Ikeda F, Imagawa Y, Imai Y, Imbriano C, Imoto M, Inman DM, Inoki K, Iovanna J, Iozzo RV, Ippolito G, Irazoqui JE, Iribarren P, Ishaq M, Ishikawa M, Ishimwe N, Isidoro C, Ismail N, Issazadeh-Navikas S, Itakura E, Ito D, Ivankovic D, Ivanova S, Iyer AKV, Izquierdo JM, Izumi M, Jäättelä M, Jabir MS, Jackson WT, Jacobo-Herrera N, Jacomin AC, Jacquin E, Jadiya P, Jaeschke H, Jagannath C, Jakobi AJ, Jakobsson J, Janji B, Jansen-Dürr P, Jansson PJ, Jantsch J, Januszewski S, Jassey A, Jean S, Jeltsch-David H, Jendelova P, Jenny A, Jensen TE, Jessen N, Jewell JL, Ji J, Jia L, Jia R, Jiang L, Jiang Q, Jiang R, Jiang T, Jiang X, Jiang Y, Jimenez-Sanchez M, Jin EJ, Jin F, Jin H, Jin L, Jin M, Jin S, Jo EK, Joffre C, Johansen T, Johnson GVW, Johnston SA, Jokitalo E, Jolly MK, Joosten LAB, Jordan J, Joseph B, Ju D, Ju JS, Ju J, Juárez E, Judith D, Juhász G, Jun Y, Jung CH, Jung SC, Jung YK, Jungbluth H, Jungverdorben J, Just S, Kaarniranta K, Kaasik A, Kabuta T, Kaganovich D, Kahana A, Kain R, Kajimura S, Kalamvoki M, Kalia M, Kalinowski DS, Kaludercic N, Kalvari I, Kaminska J, Kaminskyy VO, Kanamori H, Kanasaki K, Kang C, Kang R, Kang SS, Kaniyappan S, Kanki T, Kanneganti TD, Kanthasamy AG, Kanthasamy A, Kantorow M, Kapuy O, Karamouzis MV, Karim MR, Karmakar P, Katare RG, Kato M, Kaufmann SHE, Kauppinen A, Kaushal GP, Kaushik S, Kawasaki K, Kazan K, Ke PY, Keating DJ, Keber U, Kehrl JH, Keller KE, Keller CW, Kemper JK, Kenific CM, Kepp O, Kermorgant S, Kern A, Ketteler R, Keulers TG, Khalfin B, Khalil H, Khambu B, Khan SY, Khandelwal VKM, Khandia R, Kho W, Khobrekar NV, Khuansuwan S, Khundadze M, Killackey SA, Kim D, Kim DR, Kim DH, Kim DE, Kim EY, Kim EK, Kim HR, Kim HS, Hyung-Ryong Kim, Kim JH, Kim JK, Kim J, Kim KI, Kim PK, Kim SJ, Kimball SR, Kimchi A, Kimmelman AC, Kimura T, King MA, Kinghorn KJ, Kinsey CG, Kirkin V, Kirshenbaum LA, Kiselev SL, Kishi S, Kitamoto K, Kitaoka Y, Kitazato K, Kitsis RN, Kittler JT, Kjaerulff O, Klein PS, Klopstock T, Klucken J, Knævelsrud H, Knorr RL, Ko BCB, Ko F, Ko JL, Kobayashi H, Kobayashi S, Koch I, Koch JC, Koenig U, Kögel D, Koh YH, Koike M, Kohlwein SD, Kocaturk NM, Komatsu M, König J, Kono T, Kopp BT, Korcsmaros T, Korkmaz G, Korolchuk VI, Korsnes MS, Koskela A, Kota J, Kotake Y, Kotler ML, Kou Y, Koukourakis MI, Koustas E, Kovacs AL, Kovács T, Koya D, Kozako T, Kraft C, Krainc D, Krämer H, Krasnodembskaya AD, Kretz-Remy C, Kroemer G, Ktistakis NT, Kuchitsu K, Kuenen S, Kuerschner L, Kukar T, Kumar A, Kumar D, Kumar S, Kume S, Kumsta C, Kundu CN, Kundu M, Kunnumakkara AB, Kurgan L, Kutateladze TG, Kutlu O, Kwak S, Kwon HJ, Kwon TK, Kwon YT, Kyrmizi I, La Spada A, Labonté P, Ladoire S, Laface I, Lafont F, Lagace DC, Lahiri V, Lai Z, Laird AS, Lakkaraju A, Lamark T, Lan SH, Landajuela A, Lane DJR, Lane JD, Lang CH, Lange C, Langel Ü, Langer R, Lapaquette P, Laporte J, LaRusso NF, Lastres-Becker I, Lau WCY, Laurie GW, Lavandero S, Law BYK, Law HK, Layfield R, Le W, Le Stunff H, Leary AY, Lebrun JJ, Leck LYW, Leduc-Gaudet JP, Lee C, Lee CP, Lee DH, Lee EB, Lee EF, Lee GM, Lee HJ, Lee HK, Lee JM, Lee JS, Lee JA, Lee JY, Lee JH, Lee M, Lee MG, Lee MJ, Lee MS, Lee SY, Lee SJ, Lee SB, Lee WH, Lee YR, Lee YH, Lee Y, Lefebvre C, Legouis R, Lei YL, Lei Y, Leikin S, Leitinger G, Lemus L, Leng S, Lenoir O, Lenz G, Lenz HJ, Lenzi P, León Y, Leopoldino AM, Leschczyk C, Leskelä S, Letellier E, Leung CT, Leung PS, Leventhal JS, Levine B, Lewis PA, Ley K, Li B, Li DQ, Li J, Li K, Li L, Li M, Li PL, Li MQ, Li Q, Li S, Li T, Li W, Li X, Li YP, Li Y, Li Z, Lian J, Liang C, Liang Q, Liang W, Liang Y, Liao G, Liao L, Liao M, Liao YF, Librizzi M, Lie PPY, Lilly MA, Lim HJ, Lima TRR, Limana F, Lin C, Lin CW, Lin DS, Lin FC, Lin JD, Lin KM, Lin KH, Lin LT, Lin PH, Lin Q, Lin S, Lin SJ, Lin W, Lin X, Lin YX, Lin YS, Linden R, Lindner P, Ling SC, Lingor P, Linnemann AK, Liou YC, Lipinski MM, Lipovšek S, Lira VA, Lisiak N, Liton PB, Liu C, Liu CH, Liu CF, Liu F, Liu H, Liu HS, Liu HF, Liu J, Liu L, Liu M, Liu Q, Liu W, Liu XH, Liu X, Liu Y, Livingston JA, Lizard G, Lizcano JM, Ljubojevic-Holzer S, LLeonart ME, Llobet-Navàs D, Llorente A, Lo CH, Lobato-Márquez D, Long Q, Long YC, Loos B, Loos JA, López MG, López-Doménech G, López-Guerrero JA, López-Jiménez AT, López-Pérez Ó, López-Valero I, Lorenowicz MJ, Lorente M, Lorincz P, Lossi L, Lotersztajn S, Lovat PE, Lovell JF, Lovy A, Lőw P, Lu G, Lu H, Lu JH, Lu JJ, Lu M, Lu S, Luciani A, Lucocq JM, Ludovico P, Luftig MA, Luhr M, Luis-Ravelo D, Lum JJ, Luna-Dulcey L, Lund AH, Lund VK, Lünemann JD, Lüningschrör P, Luo H, Luo R, Luo S, Luo Z, Luparello C, Lüscher B, Luu L, Lyakhovich A, Lyamzaev KG, Lystad AH, Lytvynchuk L, Ma AC, Ma C, Ma M, Ma NF, Ma QH, Ma X, Ma Y, Ma Z, MacDougald OA, Macian F, MacIntosh GC, MacKeigan JP, Macleod KF, Maday S, Madeo F, Madesh M, Madl T, Madrigal-Matute J, Maeda A, Maejima Y, Magarinos M, Mahavadi P, Maiani E, Maiese K, Maiti P, Maiuri MC, Majello B, Major MB, Makareeva E, Malik F, Mallilankaraman K, Malorni W, Maloyan A, Mammadova N, Man GCW, Manai F, Mancias JD, Mandelkow EM, Mandell MA, Manfredi AA, Manjili MH, Manjithaya R, Manque P, Manshian BB, Manzano R, Manzoni C, Mao K, Marchese C, Marchetti S, Marconi AM, Marcucci F, Mardente S, Mareninova OA, Margeta M, Mari M, Marinelli S, Marinelli O, Mariño G, Mariotto S, Marshall RS, Marten MR, Martens S, Martin APJ, Martin KR, Martin S, Martín-Segura A, Martín-Acebes MA, Martin-Burriel I, Martin-Rincon M, Martin-Sanz P, Martina JA, Martinet W, Martinez A, Martinez J, Martinez Velazquez M, Martinez-Lopez N, Martinez-Vicente M, Martins DO, Martins JO, Martins WK, Martins-Marques T, Marzetti E, Masaldan S, Masclaux-Daubresse C, Mashek DG, Massa V, Massieu L, Masson GR, Masuelli L, Masyuk AI, Masyuk TV, Matarrese P, Matheu A, Matoba S, Matsuzaki S, Mattar P, Matte A, Mattoscio D, Mauriz JL, Mauthe M, Mauvezin C, Maverakis E, Maycotte P, Mayer J, Mazzoccoli G, Mazzoni C, Mazzulli JR, McCarty N, McDonald C, McGill MR, McKenna SL, McLaughlin B, McLoughlin F, McNiven MA, McWilliams TG, Mechta-Grigoriou F, Medeiros TC, Medina DL, Megeney LA, Megyeri K, Mehrpour M, Mehta JL, Meijer AJ, Meijer AH, Mejlvang J, Meléndez A, Melk A, Memisoglu G, Mendes AF, Meng D, Meng F, Meng T, Menna-Barreto R, Menon MB, Mercer C, Mercier AE, Mergny JL, Merighi A, Merkley SD, Merla G, Meske V, Mestre AC, Metur SP, Meyer C, Meyer H, Mi W, Mialet-Perez J, Miao J, Micale L, Miki Y, Milan E, Milczarek M, Miller DL, Miller SI, Miller S, Millward SW, Milosevic I, Minina EA, Mirzaei H, Mirzaei HR, Mirzaei M, Mishra A, Mishra N, Mishra PK, Misirkic Marjanovic M, Misasi R, Misra A, Misso G, Mitchell C, Mitou G, Miura T, Miyamoto S, Miyazaki M, Miyazaki T, Miyazawa K, Mizushima N, Mogensen TH, Mograbi B, Mohammadinejad R, Mohamud Y, Mohanty A, Mohapatra S, Möhlmann T, Mohmmed A, Moles A, Moley KH, Molinari M, Mollace V, Møller AB, Mollereau B, Mollinedo F, Montagna C, Monteiro MJ, Montella A, Montes LR, Montico B, Mony VK, Monzio Compagnoni G, Moore MN, Moosavi MA, Mora AL, Mora M, Morales-Alamo D, Moratalla R, Moreira PI, Morelli E, Moreno S, Moreno-Blas D, Moresi V, Morga B, Morgan AH, Morin F, Morishita H, Moritz OL, Moriyama M, Moriyasu Y, Morleo M, Morselli E, Moruno-Manchon JF, Moscat J, Mostowy S, Motori E, Moura AF, Moustaid-Moussa N, Mrakovcic M, Muciño-Hernández G, Mukherjee A, Mukhopadhyay S, Mulcahy Levy JM, Mulero V, Muller S, Münch C, Munjal A, Munoz-Canoves P, Muñoz-Galdeano T, Münz C, Murakawa T, Muratori C, Murphy BM, Murphy JP, Murthy A, Myöhänen TT, Mysorekar IU, Mytych J, Nabavi SM, Nabissi M, Nagy P, Nah J, Nahimana A, Nakagawa I, Nakamura K, Nakatogawa H, Nandi SS, Nanjundan M, Nanni M, Napolitano G, Nardacci R, Narita M, Nassif M, Nathan I, Natsumeda M, Naude RJ, Naumann C, Naveiras O, Navid F, Nawrocki ST, Nazarko TY, Nazio F, Negoita F, Neill T, Neisch AL, Neri LM, Netea MG, Neubert P, Neufeld TP, Neumann D, Neutzner A, Newton PT, Ney PA, Nezis IP, Ng CCW, Ng TB, Nguyen HTT, Nguyen LT, Ni HM, Ní Cheallaigh C, Ni Z, Nicolao MC, Nicoli F, Nieto-Diaz M, Nilsson P, Ning S, Niranjan R, Nishimune H, Niso-Santano M, Nixon RA, Nobili A, Nobrega C, Noda T, Nogueira-Recalde U, Nolan TM, Nombela I, Novak I, Novoa B, Nozawa T, Nukina N, Nussbaum-Krammer C, Nylandsted J, O'Donovan TR, O'Leary SM, O'Rourke EJ, O'Sullivan MP, O'Sullivan TE, Oddo S, Oehme I, Ogawa M, Ogier-Denis E, Ogmundsdottir MH, Ogretmen B, Oh GT, Oh SH, Oh YJ, Ohama T, Ohashi Y, Ohmuraya M, Oikonomou V, Ojha R, Okamoto K, Okazawa H, Oku M, Oliván S, Oliveira JMA, Ollmann M, Olzmann JA, Omari S, Omary MB, Önal G, Ondrej M, Ong SB, Ong SG, Onnis A, Orellana JA, Orellana-Muñoz S, Ortega-Villaizan MDM, Ortiz-Gonzalez XR, Ortona E, Osiewacz HD, Osman AK, Osta R, Otegui MS, Otsu K, Ott C, Ottobrini L, Ou JJ, Outeiro TF, Oynebraten I, Ozturk M, Pagès G, Pahari S, Pajares M, Pajvani UB, Pal R, Paladino S, Pallet N, Palmieri M, Palmisano G, Palumbo C, Pampaloni F, Pan L, Pan Q, Pan W, Pan X, Panasyuk G, Pandey R, Pandey UB, Pandya V, Paneni F, Pang SY, Panzarini E, Papademetrio DL, Papaleo E, Papinski D, Papp D, Park EC, Park HT, Park JM, Park JI, Park JT, Park J, Park SC, Park SY, Parola AH, Parys JB, Pasquier A, Pasquier B, Passos JF, Pastore N, Patel HH, Patschan D, Pattingre S, Pedraza-Alva G, Pedraza-Chaverri J, Pedrozo Z, Pei G, Pei J, Peled-Zehavi H, Pellegrini JM, Pelletier J, Peñalva MA, Peng D, Peng Y, Penna F, Pennuto M, Pentimalli F, Pereira CM, Pereira GJS, Pereira LC, Pereira de Almeida L, Perera ND, Pérez-Lara Á, Perez-Oliva AB, Pérez-Pérez ME, Periyasamy P, Perl A, Perrotta C, Perrotta I, Pestell RG, Petersen M, Petrache I, Petrovski G, Pfirrmann T, Pfister AS, Philips JA, Pi H, Picca A, Pickrell AM, Picot S, Pierantoni GM, Pierdominici M, Pierre P, Pierrefite-Carle V, Pierzynowska K, Pietrocola F, Pietruczuk M, Pignata C, Pimentel-Muiños FX, Pinar M, Pinheiro RO, Pinkas-Kramarski R, Pinton P, Pircs K, Piya S, Pizzo P, Plantinga TS, Platta HW, Plaza-Zabala A, Plomann M, Plotnikov EY, Plun-Favreau H, Pluta R, Pocock R, Pöggeler S, Pohl C, Poirot M, Poletti A, Ponpuak M, Popelka H, Popova B, Porta H, Porte Alcon S, Portilla-Fernandez E, Post M, Potts MB, Poulton J, Powers T, Prahlad V, Prajsnar TK, Praticò D, Prencipe R, Priault M, Proikas-Cezanne T, Promponas VJ, Proud CG, Puertollano R, Puglielli L, Pulinilkunnil T, Puri D, Puri R, Puyal J, Qi X, Qi Y, Qian W, Qiang L, Qiu Y, Quadrilatero J, Quarleri J, Raben N, Rabinowich H, Ragona D, Ragusa MJ, Rahimi N, Rahmati M, Raia V, Raimundo N, Rajasekaran NS, Ramachandra Rao S, Rami A, Ramírez-Pardo I, Ramsden DB, Randow F, Rangarajan PN, Ranieri D, Rao H, Rao L, Rao R, Rathore S, Ratnayaka JA, Ratovitski EA, Ravanan P, Ravegnini G, Ray SK, Razani B, Rebecca V, Reggiori F, Régnier-Vigouroux A, Reichert AS, Reigada D, Reiling JH, Rein T, Reipert S, Rekha RS, Ren H, Ren J, Ren W, Renault T, Renga G, Reue K, Rewitz K, Ribeiro de Andrade Ramos B, Riazuddin SA, Ribeiro-Rodrigues TM, Ricci JE, Ricci R, Riccio V, Richardson DR, Rikihisa Y, Risbud MV, Risueño RM, Ritis K, Rizza S, Rizzuto R, Roberts HC, Roberts LD, Robinson KJ, Roccheri MC, Rocchi S, Rodney GG, Rodrigues T, Rodrigues Silva VR, Rodriguez A, Rodriguez-Barrueco R, Rodriguez-Henche N, Rodriguez-Rocha H, Roelofs J, Rogers RS, Rogov VV, Rojo AI, Rolka K, Romanello V, Romani L, Romano A, Romano PS, Romeo-Guitart D, Romero LC, Romero M, Roney JC, Rongo C, Roperto S, Rosenfeldt MT, Rosenstiel P, Rosenwald AG, Roth KA, Roth L, Roth S, Rouschop KMA, Roussel BD, Roux S, Rovere-Querini P, Roy A, Rozieres A, Ruano D, Rubinsztein DC, Rubtsova MP, Ruckdeschel K, Ruckenstuhl C, Rudolf E, Rudolf R, Ruggieri A, Ruparelia AA, Rusmini P, Russell RR, Russo GL, Russo M, Russo R, Ryabaya OO, Ryan KM, Ryu KY, Sabater-Arcis M, Sachdev U, Sacher M, Sachse C, Sadhu A, Sadoshima J, Safren N, Saftig P, Sagona AP, Sahay G, Sahebkar A, Sahin M, Sahin O, Sahni S, Saito N, Saito S, Saito T, Sakai R, Sakai Y, Sakamaki JI, Saksela K, Salazar G, Salazar-Degracia A, Salekdeh GH, Saluja AK, Sampaio-Marques B, Sanchez MC, Sanchez-Alcazar JA, Sanchez-Vera V, Sancho-Shimizu V, Sanderson JT, Sandri M, Santaguida S, Santambrogio L, Santana MM, Santoni G, Sanz A, Sanz P, Saran S, Sardiello M, Sargeant TJ, Sarin A, Sarkar C, Sarkar S, Sarrias MR, Sarmah DT, Sarparanta J, Sathyanarayan A, Sathyanarayanan R, Scaglione KM, Scatozza F, Schaefer L, Schafer ZT, Schaible UE, Schapira AHV, Scharl M, Schatzl HM, Schein CH, Scheper W, Scheuring D, Schiaffino MV, Schiappacassi M, Schindl R, Schlattner U, Schmidt O, Schmitt R, Schmidt SD, Schmitz I, Schmukler E, Schneider A, Schneider BE, Schober R, Schoijet AC, Schott MB, Schramm M, Schröder B, Schuh K, Schüller C, Schulze RJ, Schürmanns L, Schwamborn JC, Schwarten M, Scialo F, Sciarretta S, Scott MJ, Scotto KW, Scovassi AI, Scrima A, Scrivo A, Sebastian D, Sebti S, Sedej S, Segatori L, Segev N, Seglen PO, Seiliez I, Seki E, Selleck SB, Sellke FW, Selsby JT, Sendtner M, Senturk S, Seranova E, Sergi C, Serra-Moreno R, Sesaki H, Settembre C, Setty SRG, Sgarbi G, Sha O, Shacka JJ, Shah JA, Shang D, Shao C, Shao F, Sharbati S, Sharkey LM, Sharma D, Sharma G, Sharma K, Sharma P, Sharma S, Shen HM, Shen H, Shen J, Shen M, Shen W, Shen Z, Sheng R, Sheng Z, Sheng ZH, Shi J, Shi X, Shi YH, Shiba-Fukushima K, Shieh JJ, Shimada Y, Shimizu S, Shimozawa M, Shintani T, Shoemaker CJ, Shojaei S, Shoji I, Shravage BV, Shridhar V, Shu CW, Shu HB, Shui K, Shukla AK, Shutt TE, Sica V, Siddiqui A, Sierra A, Sierra-Torre V, Signorelli S, Sil P, Silva BJA, Silva JD, Silva-Pavez E, Silvente-Poirot S, Simmonds RE, Simon AK, Simon HU, Simons M, Singh A, Singh LP, Singh R, Singh SV, Singh SK, Singh SB, Singh S, Singh SP, Sinha D, Sinha RA, Sinha S, Sirko A, Sirohi K, Sivridis EL, Skendros P, Skirycz A, Slaninová I, Smaili SS, Smertenko A, Smith MD, Soenen SJ, Sohn EJ, Sok SPM, Solaini G, Soldati T, Soleimanpour SA, Soler RM, Solovchenko A, Somarelli JA, Sonawane A, Song F, Song HK, Song JX, Song K, Song Z, Soria LR, Sorice M, Soukas AA, Soukup SF, Sousa D, Sousa N, Spagnuolo PA, Spector SA, Srinivas Bharath MM, St Clair D, Stagni V, Staiano L, Stalnecker CA, Stankov MV, Stathopulos PB, Stefan K, Stefan SM, Stefanis L, Steffan JS, Steinkasserer A, Stenmark H, Sterneckert J, Stevens C, Stoka V, Storch S, Stork B, Strappazzon F, Strohecker AM, Stupack DG, Su H, Su LY, Su L, Suarez-Fontes AM, Subauste CS, Subbian S, Subirada PV, Sudhandiran G, Sue CM, Sui X, Summers C, Sun G, Sun J, Sun K, Sun MX, Sun Q, Sun Y, Sun Z, Sunahara KKS, Sundberg E, Susztak K, Sutovsky P, Suzuki H, Sweeney G, Symons JD, Sze SCW, Szewczyk NJ, Tabęcka-Łonczynska A, Tabolacci C, Tacke F, Taegtmeyer H, Tafani M, Tagaya M, Tai H, Tait SWG, Takahashi Y, Takats S, Talwar P, Tam C, Tam SY, Tampellini D, Tamura A, Tan CT, Tan EK, Tan YQ, Tanaka M, Tang D, Tang J, Tang TS, Tanida I, Tao Z, Taouis M, Tatenhorst L, Tavernarakis N, Taylor A, Taylor GA, Taylor JM, Tchetina E, Tee AR, Tegeder I, Teis D, Teixeira N, Teixeira-Clerc F, Tekirdag KA, Tencomnao T, Tenreiro S, Tepikin AV, Testillano PS, Tettamanti G, Tharaux PL, Thedieck K, Thekkinghat AA, Thellung S, Thinwa JW, Thirumalaikumar VP, Thomas SM, Thomes PG, Thorburn A, Thukral L, Thum T, Thumm M, Tian L, Tichy A, Till A, Timmerman V, Titorenko VI, Todi SV, Todorova K, Toivonen JM, Tomaipitinca L, Tomar D, Tomas-Zapico C, Tomić S, Tong BC, Tong C, Tong X, Tooze SA, Torgersen ML, Torii S, Torres-López L, Torriglia A, Towers CG, Towns R, Toyokuni S, Trajkovic V, Tramontano D, Tran QG, Travassos LH, Trelford CB, Tremel S, Trougakos IP, Tsao BP, Tschan MP, Tse HF, Tse TF, Tsugawa H, Tsvetkov AS, Tumbarello DA, Tumtas Y, Tuñón MJ, Turcotte S, Turk B, Turk V, Turner BJ, Tuxworth RI, Tyler JK, Tyutereva EV, Uchiyama Y, Ugun-Klusek A, Uhlig HH, Ułamek-Kozioł M, Ulasov IV, Umekawa M, Ungermann C, Unno R, Urbe S, Uribe-Carretero E, Üstün S, Uversky VN, Vaccari T, Vaccaro MI, Vahsen BF, Vakifahmetoglu-Norberg H, Valdor R, Valente MJ, Valko A, Vallee RB, Valverde AM, Van den Berghe G, van der Veen S, Van Kaer L, van Loosdregt J, van Wijk SJL, Vandenberghe W, Vanhorebeek I, Vannier-Santos MA, Vannini N, Vanrell MC, Vantaggiato C, Varano G, Varela-Nieto I, Varga M, Vasconcelos MH, Vats S, Vavvas DG, Vega-Naredo I, Vega-Rubin-de-Celis S, Velasco G, Velázquez AP, Vellai T, Vellenga E, Velotti F, Verdier M, Verginis P, Vergne I, Verkade P, Verma M, Verstreken P, Vervliet T, Vervoorts J, Vessoni AT, Victor VM, Vidal M, Vidoni C, Vieira OV, Vierstra RD, Viganó S, Vihinen H, Vijayan V, Vila M, Vilar M, Villalba JM, Villalobo A, Villarejo-Zori B, Villarroya F, Villarroya J, Vincent O, Vindis C, Viret C, Viscomi MT, Visnjic D, Vitale I, Vocadlo DJ, Voitsekhovskaja OV, Volonté C, Volta M, Vomero M, Von Haefen C, Vooijs MA, Voos W, Vucicevic L, Wade-Martins R, Waguri S, Waite KA, Wakatsuki S, Walker DW, Walker MJ, Walker SA, Walter J, Wandosell FG, Wang B, Wang CY, Wang C, Wang D, Wang F, Wang G, Wang H, Wang HG, Wang J, Wang K, Wang L, Wang MH, Wang M, Wang N, Wang P, Wang QJ, Wang Q, Wang QK, Wang QA, Wang WT, Wang W, Wang X, Wang Y, Wang YY, Wang Z, Warnes G, Warnsmann V, Watada H, Watanabe E, Watchon M, Wawrzyńska A, Weaver TE, Wegrzyn G, Wehman AM, Wei H, Wei L, Wei T, Wei Y, Weiergräber OH, Weihl CC, Weindl G, Weiskirchen R, Wells A, Wen RH, Wen X, Werner A, Weykopf B, Wheatley SP, Whitton JL, Whitworth AJ, Wiktorska K, Wildenberg ME, Wileman T, Wilkinson S, Willbold D, Williams B, Williams RSB, Williams RL, Williamson PR, Wilson RA, Winner B, Winsor NJ, Witkin SS, Wodrich H, Woehlbier U, Wollert T, Wong E, Wong JH, Wong RW, Wong VKW, Wong WW, Wu AG, Wu C, Wu J, Wu KK, Wu M, Wu SY, Wu S, Wu WKK, Wu X, Wu YW, Wu Y, Xavier RJ, Xia H, Xia L, Xia Z, Xiang G, Xiang J, Xiang M, Xiang W, Xiao B, Xiao G, Xiao H, Xiao HT, Xiao J, Xiao L, Xiao S, Xiao Y, Xie B, Xie CM, Xie M, Xie Y, Xie Z, Xilouri M, Xu C, Xu E, Xu H, Xu J, Xu L, Xu WW, Xu X, Xue Y, Yakhine-Diop SMS, Yamaguchi M, Yamaguchi O, Yamamoto A, Yamashina S, Yan S, Yan SJ, Yan Z, Yanagi Y, Yang C, Yang DS, Yang H, Yang HT, Yang JM, Yang J, Yang L, Yang M, Yang PM, Yang Q, Yang S, Yang SF, Yang W, Yang WY, Yang X, Yang Y, Yao H, Yao S, Yao X, Yao YG, Yao YM, Yasui T, Yazdankhah M, Yen PM, Yi C, Yin XM, Yin Y, Yin Z, Ying M, Ying Z, Yip CK, Yiu SPT, Yoo YH, Yoshida K, Yoshii SR, Yoshimori T, Yousefi B, Yu B, Yu H, Yu J, Yu L, Yu ML, Yu SW, Yu VC, Yu WH, Yu Z, Yuan J, Yuan LQ, Yuan S, Yuan SF, Yuan Y, Yuan Z, Yue J, Yue Z, Yun J, Yung RL, Zacks DN, Zaffagnini G, Zambelli VO, Zanella I, Zang QS, Zanivan S, Zappavigna S, Zaragoza P, Zarbalis KS, Zarebkohan A, Zarrouk A, Zeitlin SO, Zeng J, Zeng JD, Žerovnik E, Zhan L, Zhang B, Zhang DD, Zhang H, Zhang HL, Zhang J, Zhang JP, Zhang KYB, Zhang LW, Zhang L, Zhang M, Zhang P, Zhang S, Zhang W, Zhang X, Zhang XW, Zhang XD, Zhang Y, Zhang YD, Zhang YY, Zhang Z, Zhao H, Zhao L, Zhao S, Zhao T, Zhao XF, Zhao Y, Zheng G, Zheng K, Zheng L, Zheng S, Zheng XL, Zheng Y, Zheng ZG, Zhivotovsky B, Zhong Q, Zhou A, Zhou B, Zhou C, Zhou G, Zhou H, Zhou J, Zhou K, Zhou R, Zhou XJ, Zhou Y, Zhou ZY, Zhou Z, Zhu B, Zhu C, Zhu GQ, Zhu H, Zhu WG, Zhu Y, Zhuang H, Zhuang X, Zientara-Rytter K, Zimmermann CM, Ziviani E, Zoladek T, Zong WX, Zorov DB, Zorzano A, Zou W, Zou Z, Zuryn S, Zwerschke W, Brand-Saberi B, Dong XC, Kenchappa CS, Lin Y, Oshima S, Rong Y, Sluimer JC, Stallings CL, Tong CK. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)(1). Autophagy. 2021;17:1-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 887] [Cited by in RCA: 856] [Article Influence: 214.0] [Reference Citation Analysis (0)] |
| 24. | Tao Y, Wang N, Qiu T, Sun X. The Role of Autophagy and NLRP3 Inflammasome in Liver Fibrosis. Biomed Res Int. 2020;2020:7269150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 25. | Dewidar B, Meyer C, Dooley S, Meindl-Beinker AN. TGF-β in Hepatic Stellate Cell Activation and Liver Fibrogenesis-Updated 2019. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 543] [Article Influence: 90.5] [Reference Citation Analysis (0)] |
| 26. | Ni Y, Li JM, Liu MK, Zhang TT, Wang DP, Zhou WH, Hu LZ, Lv WL. Pathological process of liver sinusoidal endothelial cells in liver diseases. World J Gastroenterol. 2017;23:7666-7677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 49] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 27. | Sui M, Jiang X, Chen J, Yang H, Zhu Y. Magnesium isoglycyrrhizinate ameliorates liver fibrosis and hepatic stellate cell activation by regulating ferroptosis signaling pathway. Biomed Pharmacother. 2018;106:125-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 119] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 28. | Durairajan SSK, Singh AK, Saravanan UB, Namachivayam M, Radhakrishnan M, Huang JD, Dhodapkar R, Zhang H. Gastrointestinal Manifestations of SARS-CoV-2: Transmission, Pathogenesis, Immunomodulation, Microflora Dysbiosis, and Clinical Implications. Viruses. 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 29. | Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15248] [Cited by in RCA: 14131] [Article Influence: 2826.2] [Reference Citation Analysis (1)] |
| 30. | Cheng H, Wang Y, Wang GQ. Organ-protective effect of angiotensin-converting enzyme 2 and its effect on the prognosis of COVID-19. J Med Virol. 2020;92:726-730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 341] [Cited by in RCA: 332] [Article Influence: 66.4] [Reference Citation Analysis (0)] |
| 31. | Moon AM, Webb GJ, Aloman C, Armstrong MJ, Cargill T, Dhanasekaran R, Genescà J, Gill US, James TW, Jones PD, Marshall A, Mells G, Perumalswami PV, Qi X, Su F, Ufere NN, Barnes E, Barritt AS, Marjot T. High mortality rates for SARS-CoV-2 infection in patients with pre-existing chronic liver disease and cirrhosis: Preliminary results from an international registry. J Hepatol. 2020;73:705-708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 203] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 32. | Warner FJ, Rajapaksha H, Shackel N, Herath CB. ACE2: from protection of liver disease to propagation of COVID-19. Clin Sci (Lond). 2020;134:3137-3158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 33. | Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069-1075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5421] [Cited by in RCA: 5293] [Article Influence: 311.4] [Reference Citation Analysis (0)] |
| 34. | Chen J, Guo Q, Chen Q, Chen Y, Chen D, Chen Z, Wang X, Huang Y. Interleukin 10 inhibits oxidative stress-induced autophagosome formation in hepatic stellate cells by activating the mTOR-STAT3 pathway. Exp Cell Res. 2022;411:113001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 35. | Cao Y, Luo Y, Zou J, Ouyang J, Cai Z, Zeng X, Ling H, Zeng T. Autophagy and its role in gastric cancer. Clin Chim Acta. 2019;489:10-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 123] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 36. | Zheng Q, Zhao K, Han X, Huff AF, Cui Q, Babcock SA, Yu S, Zhang Y. Inhibition of AMPK accentuates prolonged caloric restriction-induced change in cardiac contractile function through disruption of compensatory autophagy. Biochim Biophys Acta. 2015;1852:332-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 37. | Jin S, Li H, Han M, Ruan M, Liu Z, Zhang F, Zhang C, Choi Y, Liu B. Mesenchymal Stem Cells with Enhanced Bcl-2 Expression Promote Liver Recovery in a Rat Model of Hepatic Cirrhosis. Cell Physiol Biochem. 2016;40:1117-1128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 38. | Kong LJ, Li H, Du YJ, Pei FH, Hu Y, Zhao LL, Chen J. Vatalanib, a tyrosine kinase inhibitor, decreases hepatic fibrosis and sinusoidal capillarization in CCl4-induced fibrotic mice. Mol Med Rep. 2017;15:2604-2610. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 39. | DeLeve LD. Liver sinusoidal endothelial cells in hepatic fibrosis. Hepatology. 2015;61:1740-1746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 345] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 40. | Novo E, Cannito S, Zamara E, Valfrè di Bonzo L, Caligiuri A, Cravanzola C, Compagnone A, Colombatto S, Marra F, Pinzani M, Parola M. Proangiogenic cytokines as hypoxia-dependent factors stimulating migration of human hepatic stellate cells. Am J Pathol. 2007;170:1942-1953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 167] [Article Influence: 9.3] [Reference Citation Analysis (0)] |