Published online Aug 21, 2023. doi: 10.3748/wjg.v29.i31.4706
Peer-review started: March 28, 2023
First decision: June 17, 2023
Revised: June 29, 2023
Accepted: August 1, 2023
Article in press: August 1, 2023
Published online: August 21, 2023
Processing time: 142 Days and 22.3 Hours
Hepatocellular carcinoma (HCC) is a malignancy with a high incidence and fatality rate worldwide. Hepatitis B virus (HBV) infection is one of the most important risk factors for its occurrence and development. Early detection of HBV-associated HCC (HBV-HCC) can improve clinical decision-making and patient outcomes. Biomarkers are extremely helpful, not only for early diagnosis, but also for the development of therapeutics. MicroRNAs (miRNAs), a subset of non-coding RNAs approximately 22 nucleotides in length, have increasingly attracted scientists’ attention due to their potential utility as biomarkers for cancer detection and therapy. HBV profoundly impacts the expression of miRNAs potentially involved in the development of hepatocarcinogenesis. In this review, we summarize the current progress on the role of miRNAs in the diagnosis and treatment of HBV-HCC. From a molecular standpoint, we discuss the mechanism by which HBV regulates miRNAs and investigate the exact effect of miRNAs on the promotion of HCC. In the near future, miRNA-based diagnostic, prognostic, and therapeutic applications will make their way into the clinical routine.
Core Tip: Hepatocellular carcinoma (HCC) is one of the most common malignancies worldwide. Hepatitis B virus (HBV) infection is one of the predominant risk factors for developing HCC. Early diagnosis and prognosis prediction are pivotal for patients with HBV-associated HCC (HBV-HCC) in their clinical management. MicroRNAs (miRNAs), a subset of non-coding RNAs, play an essential role in human diseases including HBV-HCC. Here, we summarize the role of miRNAs in the diagnosis and prognosis prediction of patients with HBV-HCC. Furthermore, we discuss the underlying mechanism by which HBV dysregulates miRNAs, and the potential role of dysregulated miRNAs in promoting hepatocarcinogenesis, laying the foundation for applying potential therapeutic targets.
- Citation: Zhang MH, Yuan YF, Liu LJ, Wei YX, Yin WY, Zheng LZY, Tang YY, Lv Z, Zhu F. Dysregulated microRNAs as a biomarker for diagnosis and prognosis of hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol 2023; 29(31): 4706-4735
- URL: https://www.wjgnet.com/1007-9327/full/v29/i31/4706.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i31.4706
According to World Health Organization (WHO) reports, in 2020, primary liver cancer is the sixth most frequently occurring cancer worldwide, with mortality ranking third among all cancers. Hepatocellular carcinoma (HCC) accounted for about 75%-85% of cases. Chronic hepatitis B virus (HBV) infection is one of the major known risk factors[1]. Although the HBV-vaccination program has greatly reduced the incidence of HBV infection, it is estimated that nearly 292 million people are living with chronic hepatitis B (CHB) globally, and approximately 900000 people die annually because of HBV-induced liver cirrhosis and HCC, according to WHO estimates[2]. Liver surgery, including liver resection and liver transplantation, has become an established therapy for HCC and cirrhosis. Early diagnosis is a key factor for liver resection. Biomarkers are widely used for the early diagnosis of several cancers. But HCC biomarkers cannot be clinically useful for early HCC diagnosis due to their low sensitivity[3]. Similarly essential is the construction of prognosis of HBV-associated HCC (HBV-HCC), which can help make treatment decisions[3]. At present, circulating nucleic acid biomarkers, including microRNAs (miRNA), are identified as possible biomarkers for the diagnosis, prognosis, and therapeutics of liver diseases, especially HBV-HCC[4].
miRNA, a subset of non-coding RNAs, possess 19-25 nucleotides in length and play crucial biological roles in the process of gene silencing. Given that up to 60% of human protein-coding genes have conserved miRNA target sites, it is not surprising that dysregulated miRNAs can disrupt homeostasis and cause diseases including cancer[5]. miRNAs play a vital role in different stages of the HBV-HCC continuum, including early HBV infection, chronic inflammation, fibrosis/cirrhosis, and the emergence of HCC[6].
Although most miRNAs are located within cells, circulating miRNAs are present in body fluids and may reflect the pathophysiology of tissues. Several desirable characteristics of circulating miRNAs, such as their high stability in biological samples, non-invasive methods for sampling, and high sensitivity and accuracy, make them suitable as biomarker candidates for cancer diagnosis, prognosis, and therapeutic response prediction[7]. In this review, we summarize the application of miRNAs as diagnostic, prognostic, and therapeutic markers in HBV-HCC. We further discuss the mechanism by which HBV affects miRNA transcription and stability. We also try to understand the mechanisms by which miRNAs participate in the development of HCC. This will provide an in-depth understanding to identify promising biomarkers in HBV-HCC diagnosis, prognosis, and treatment.
miRNAs are expressed with high tissue and cell selectivity. For example, some miRNAs, such as miR-122-5p, are particularly abundant in the liver, suggesting that certain miRNAs participate in HCC carcinogenesis and progression[8]. After tumor resection in HCC patients, some serum circulating miRNAs exhibit significant changes[9-13], indicating that circulating miRNAs may be specific non-invasive and diagnostic markers for HBV-HCC.
In clinical diagnostic tests, area under the receiver operating characteristic curve (AUC) is widely used to judge the diagnostic accuracy. Generally, an AUC between 0.7 and 0.8 is considered clinically useful, between 0.8 and 0.9 is deemed excellent, and greater than 0.9 is considered outstanding discrimination[14]. Numerous individual miRNAs have shown good diagnostic efficacy, with miR-93-5p[15], miR-122[16], miR-125b[17], miR-150[13], miR-487b[18,19], miR-768-3p[20], and miR-5193[21] achieving AUC > 0.9 in discriminating HBV-HCC patients from healthy controls (HC) (Table 1), miR-122[22], miR-125b[23], and miR-192[22] in differentiating HBV-HCC from CHB patients (Table 2), and miR-101[24] and miR-125b[17,23] in discriminating HBV-HCC from HBV related liver cirrhosis (HBV-LC) patients (Table 2). Additionally, some HBV dysregulated miRNAs show different expression profiles in the serum or plasma of HBV-HCC patients compared to control populations[25-29], suggesting a potential value of these miRNAs in diagnosing HBV-HCC. In addition, one study reveals that urine miR-93-5p demonstrates diagnostic performance comparable to plasma miR-93-5p for diagnosing early HBV-HCC. Urine sample is non-invasive and simple to perform on humans. Therefore, urine miRNAs may have more clinical application potential than plasma miRNAs[15]. However, individual miRNAs may have limitations in sensitivity and specificity due to HBV-associated chronic liver disease. In addition, despite the fact that numerous miRNAs were highly effective at distinguishing HBV-HCC patients from healthy populations or all control populations, the efficacy used to differentiate HBV-HCC from CHB or HBV-LC is nonspecific. Hence, new methods are required to improve the diagnostic efficacy of miRNAs in diagnosing HBV-HCC patients, particularly in distinguishing them from CHB and LC populations.
| miRNA | AUC | Sensitivity | Specificity | Ref. |
| miR-18a | 0.881 | 0.861 | 0.750 | [228] |
| miR-26a | 0.711 | 0.876 | 0.600 | [30] |
| miR-26a1 | 0.685 | 0.907 | 0.600 | [30] |
| miR-26a-5p | 0.762 | 0.689 | 0.744 | [38] |
| miR-27a | 0.859 | 0.933 | 0.733 | [30] |
| miR-27a1 | 0.809 | 0.926 | 0.733 | [30] |
| miR-34a | 0.736 | 0.920 | 0.600 | [21] |
| miR-93-5p | 0.906 | 0.859 | 0.954 | [15] |
| miR-93-5p | 0.903 | 0.879 | 0.938 | [15] |
| miR-93-5p | 0.905 | 0.862 | 0.954 | [15] |
| miR-101 | 0.788 | 0.761 | 0.700 | [24] |
| miR-122 | 0.984 | 0.960 | 0.940 | [16] |
| miR-122 | 0.869 | 0.816 | 0.833 | [10] |
| miR-122-5p | 0.697 | 0.489 | 0.822 | [38] |
| miR-125b | 0.835 | 0.798 | 0.867 | [30] |
| miR-125b | 0.891 | 0.859 | 0.786 | [23] |
| miR-125b | 0.940 | 0.830 | 0.960 | [17] |
| miR-125b1 | 0.822 | 0.815 | 0.867 | [30] |
| miR-141-3p | 0.758 | 0.681 | 0.833 | [38] |
| miR-143 | 0.813 | 0.776 | 0.860 | [229] |
| miR-145 | 0.852 | 0.882 | 0.780 | [229] |
| miR-150 | 0.931 | 0.825 | 0.837 | [13] |
| miR-192-5p | 0.695 | 0.719 | 0.756 | [38] |
| miR-199a-5p | 0.638 | 0.593 | 0.667 | [38] |
| miR-205 | 0.885 | 0.969 | 0.679 | [36] |
| miR-206 | 0.615 | 0.481 | 0.788 | [38] |
| miR-212 | 0.886 | 0.696 | 0.950 | [230] |
| miR-214 | 0.747 | 0.760 | 0.740 | [21] |
| miR-223 | 0.736 | 0.921 | 0.633 | [30] |
| miR-2231 | 0.822 | 0.907 | 0.633 | [30] |
| miR-433-5p | 0.736 | 0.793 | 0.644 | [38] |
| miR-487b | 0.946 | 0.888 | 0.909 | [18] |
| miR-487b | 0.929 | 0.839 | 0.928 | [19] |
| miR-768-3p | 0.908 | 0.873 | 0.800 | [20] |
| miR-1228-5p | 0.552 | 0.793 | 0.278 | [38] |
| miR-5193 | 0.993 | 0.960 | 1.000 | [21] |
| miR-6510 | 0.839 | 0.720 | 0.910 | [21] |
| Comparison | miRNA | AUC | Sensitivity | Specificity | Ref. |
| HBV-carriers | miR-20a-5p | 0.770 | 0.866 | 0.573 | [231] |
| HBV-carriers | miR-25-3p | 0.718 | 0.553 | 0.793 | [231] |
| HBV-carriers | miR-30a-5p | 0.681 | 0.642 | 0.683 | [231] |
| HBV-carriers | miR-92a-3p | 0.765 | 0.761 | 0.683 | [231] |
| HBV-carriers | miR-132-3p | 0.722 | 0.910 | 0.366 | [231] |
| HBV-carriers | miR-185-5p | 0.788 | 0.910 | 0.390 | [231] |
| HBV-carriers | miR-320a | 0.678 | 0.388 | 0.878 | [231] |
| HBV-carriers | miR-324-3p | 0.656 | 0.746 | 0.500 | [231] |
| CHB | miR-26a | 0.650 | 0.533 | 0.833 | [30] |
| CHB | miR-26a1 | 0.411 | 0.582 | 0.500 | [30] |
| CHB | miR-27a | 0.761 | 0.677 | 0.833 | [30] |
| CHB | miR-27a1 | 0.690 | 0.527 | 0.833 | [30] |
| CHB | miR-34a | 0.619 | 0.400 | 0.870 | [21] |
| CHB | miR-96 | 0.803 | 0.779 | 0.753 | [34] |
| CHB | miR-99a2 | 0.694 | 0.844 | 0.567 | [232] |
| CHB | miR-101 | 0.777 | 0.881 | 0.620 | [24] |
| CHB | miR-122 | 0.190 | 0.250 | 0.250 | [16] |
| CHB | miR-122 | 0.630 | 0.776 | 0.578 | [10] |
| CHB | miR-125b | 0.675 | 0.522 | 0.867 | [30] |
| CHB | miR-125b | 0.958 | 0.938 | 0.857 | [23] |
| CHB | miR-125b | 0.792 | 0.906 | 0.567 | [232] |
| CHB | miR-125b | 0.800 | 0.810 | 0.870 | [17] |
| CHB | miR-125b1 | 0.631 | 0.400 | 0.870 | [30] |
| CHB | miR-126 | 0.670 | 0.630 | 0.580 | [32] |
| CHB | miR-142-3p | 0.550 | 0.320 | 0.910 | [32] |
| CHB | miR-150 | 0.881 | 0.791 | 0.765 | [13] |
| CHB | miR-214 | 0.520 | 0.850 | 0.430 | [21] |
| CHB | miR-223 | 0.737 | 0.544 | 0.933 | [30] |
| CHB | miR-2231 | 0.656 | 0.782 | 0.533 | [30] |
| CHB | miR-224 | 0.846 | 0.865 | 0.745 | [11] |
| CHB | miR-487b | 0.815 | 0.836 | 0.667 | [18] |
| CHB | miR-487b | 0.856 | 0.759 | 0.897 | [19] |
| CHB | miR-768-3p | 0.819 | 0.850 | 0.727 | [20] |
| CHB | miR-5193 | 0.817 | 0.798 | 0.820 | [21] |
| CHB | miR-6510 | 0.531 | 0.810 | 0.390 | [21] |
| HBV-DN | let-7b | 0.633 | 0.825 | 0.467 | [233] |
| HBV-DN | miR-122 | 0.648 | 0.667 | 0.567 | [233] |
| HBV-LC | miR-26a-5p | 0.744 | 0.607 | 0.909 | [38] |
| HBV-LC | miR-99a2 | 0.696 | 0.967 | 0.563 | [232] |
| HBV-LC | miR-101 | 0.976 | 0.955 | 0.902 | [24] |
| HBV-LC | miR-122 | 0.675 | 0.610 | 0.760 | [37] |
| HBV-LC | miR-122-5p | 0.751 | 0.489 | 0.902 | [38] |
| HBV-LC | miR-125b | 0.958 | 0.891 | 0.881 | [23] |
| HBV-LC | miR-125b | 0.910 | 0.780 | 0.960 | [17] |
| HBV-LC | miR-126 | 0.578 | 0.550 | 0.580 | [32] |
| HBV-LC | miR-141-3p | 0.663 | 0.607 | 0.727 | [38] |
| HBV-LC | miR-142-3p | 0.566 | 0.550 | 0.630 | [32] |
| HBV-LC | miR-192-5p | 0.687 | 0.548 | 0.833 | [38] |
| HBV-LC | miR-199a-5p | 0.589 | 0.593 | 0.576 | [38] |
| HBV-LC | miR-205 | 0.781 | 0.969 | 0.542 | [36] |
| HBV-LC | miR-206 | 0.693 | 0.778 | 0.689 | [38] |
| HBV-LC | miR-224 | 0.832 | 0.865 | 0.667 | [11] |
| HBV-LC | miR-433-5p | 0.644 | 0.564 | 0.674 | [38] |
| HBV-LC | miR-1228-5p | 0.542 | 0.667 | 0.470 | [38] |
| CHB + HBV-LC | miR-18a | 0.775 | 0.772 | 0.700 | [228] |
| CHB + HBV-LC | miR-224 | 0.840 | 0.865 | 0.711 | [11] |
| CHB + HBV-LC | miR-375 | 0.768 | 0.938 | 0.639 | [234] |
Individual miRNAs are altered in different infectious diseases, nonspecific inflammation, and acute lesions in addition to cancer, revealing their lack of specificity. Forming a miRNA panel may help serve as diagnostic biomarkers for HBV-HCC (Table 3). Several miRNA panels, such as miR-21 + miR-122 + miR-192[22], miR-125b + miR-223 + miR-27a + miR-26a[30], and miR-23b + miR-423 + miR-375 + miR-23a + miR-342-3p[31] reach a high value in differentiating HBV-HCC from HC, CHB and HBV-LC patients. However, the good diagnostic efficiency of miRNA profiles does not necessitate the combination of as many miRNAs as feasible to improve diagnostic accuracy, due to the complexity of the method necessary to detect miRNAs and the lengthy timeframes involved. In some cases, the combination of multiple biomarkers showed no additive effect on HBV-HCC diagnosis[32]. Several studies demonstrate that panels with only two miRNAs can also reach a high AUC as panels with more miRNAs, such as miR-10a + miR-125b[31] and miR-15b + miR-130b[9]. Consequently, additional research is required to determine the optimal combination of the fewest possible number of miRNAs and to reduce the cost of diagnosis as much as possible, all while attaining a good diagnostic capacity.
| Comparison | miRNA | AUC | Sensitivity | Specificity | Ref. |
| HC | miR-125b + miR-2231 | 0.881 | 0.891 | 0.833 | [30] |
| HC | miR-125b + miR-26a1 | 0.884 | 0.873 | 0.867 | [30] |
| HC | miR-223 + miR-27a1 | 0.892 | 0.909 | 0.833 | [30] |
| HC | miR-223 + miR-26a1 | 0.828 | 0.927 | 0.667 | [30] |
| HC | miR-27a + miR-26a1 | 0.895 | 0.945 | 0.833 | [30] |
| HC | miR-125b + miR-223 + miR-27a + miR-26a | 0.932 | 0.865 | 0.933 | [30] |
| HC | miR-125b + miR-223 + miR-27a + miR-26a1 | 0.910 | 0.852 | 0.933 | [30] |
| HC | miR-375 + miR-25 + and let-7f | 0.997 | 0.979 | 0.991 | [31] |
| HC | miR-23b + miR-423 + miR-375 + miR-23a + miR-342-3p | 0.999 | 0.969 | 0.994 | [31] |
| HC | miR-122 + miR-192 + miR-21 + miR-223 + miR-26a + miR-27a + miR-801 | 0.941 | 0.832 | 0.939 | [44] |
| HC | miR-27b-3p + miR-192-5p | 0.823 | 0.685 | 0.952 | [235] |
| HC | miR-206 + miR-141-3p + miR-433-5p + miR-1228-5p + miR-199a-5p + miR-122-5p + miR-192-5p + miR-26a-5p | 0.893 | 0.828 | 0.833 | [38] |
| HBV-carriers | miR-20a-5p + miR-25-3p + miR-30a-5p + miR-92a-3p + miR-132-3p + miR-185-5p + miR-320a + miR-324-3p | 0.802 | 0.866 | 0.646 | [231] |
| HBV-carriers | miR-20a-5p + miR-320a + miR-324-3p + miR-375 | 0.768 | 0.650 | 0.775 | [231] |
| HBV-carriers | miR-20a-5p + miR-320a + miR-324-3p + miR-375 | 0.706 | 0.560 | 0.838 | [231] |
| CHB | miR-125b + miR-2231 | 0.680 | 0.473 | 0.867 | [30] |
| CHB | miR-125b + miR-26a1 | 0.668 | 0.509 | 0.833 | [30] |
| CHB | miR-223 + miR-27a1 | 0.714 | 0.582 | 0.833 | [30] |
| CHB | miR-223 + miR-26a1 | 0.708 | 0.509 | 0.900 | [30] |
| CHB | miR-27a + miR-26a1 | 0.741 | 0.873 | 0.533 | [30] |
| CHB | miR-125b + miR-223 + miR-27a + miR-26a | 0.761 | 0.622 | 0.867 | [30] |
| CHB | miR-125b + miR-223 + miR-27a + miR-26a1 | 0.687 | 0.818 | 0.533 | [30] |
| CHB | miR-10a + miR-125b | 0.992 | 0.985 | 0.985 | [31] |
| CHB | miR-122 + miR-192 + miR-21 + miR-223 + miR-26a + miR-27a + miR-801 | 0.842 | 0.791 | 0.764 | [44] |
| HBV-LC | miR-122 + miR-192 + miR-21 + miR-223 + miR-26a + miR-27a + miR-801 | 0.884 | 0.750 | 0.911 | [44] |
| HBV-LC | miR-27b-3p + miR-192-5p | 0.859 | 0.785 | 0.793 | [235] |
| HBV-LC | miR-206 + miR-141-3p + miR-433-5p + miR-1228-5p + miR-199a-5p + miR-122-5p + miR-192-5p + miR-26a-5p | 0.892 | 0.816 | 0.846 | [38] |
It will be better to combine miRNAs with other biomarkers in HBV-HCC diagnosis (Table 4). The most prevalent combination biomarker is alpha fetoprotein (AFP), a traditional biomarker in HCC. Obviously, the combination of most miRNAs with AFP demonstrates a high AUC and diagnostic accuracy for discriminating HBV-HCC from HC or patients with CHB, HBV-CLD, and HBV-LC, such as miR-24-3p[33], miR-96[34], miR-101[24], miR-122[35], miR-126[32], miR-142-3p[32], miR-205[36] and miR-224[11]. Another study combines miR-122 with AFP and TERT gene promoter mutations in cfDNA. The results show that it reaches a 0.98 and 0.88 AUC in discriminating HBV-HCC patients from CHB and HBV-LC patients, respectively, demonstrating a high diagnostic value in HBV-HCC[35]. Similarly, the combination of miR-122 with AFP and prothrombin induced by vitamin K deficiency or antagonist- II (PIVKA-II) reaches a 0.918 AUC in separating HBV-HCC from HBV-LC patients[37]. Consequently, the combination of single miRNA with other biomarkers may improve the diagnostic accuracy for HBV-HCC from other HBV-related diseases. Moreover, some miRNAs panels show a better diagnostic value than AFP[38], and combination of these miRNA panels with AFP further increased the efficacy[22,30,32]. Therefore, the use of miRNA profiles paired with AFP may be the optimum modality for the diagnosis of HBV-HCC.
| Comparison | miRNA | AUC | Sensitivity | Specificity | Ref. |
| HC | miR-125b + miR-27a + AFP1 | 0.937 | 0.909 | 0.933 | [30] |
| HC | miR-125b + miR-223 + miR-27a + miR-26a + AFP | 0.945 | 0.910 | 0.933 | [30] |
| HC | miR-125b + miR-223 + miR-27a + miR-26a + AFP2 | 0.972 | 0.944 | 0.900 | [30] |
| HC | miR-125b + miR-223 + miR-27a + miR-26a + AFP1 | 0.936 | 0.907 | 0.933 | [30] |
| HC | miR-125b + miR-223 + miR-27a + miR-26a + AFP1,2 | 0.956 | 0.800 | 1.000 | [30] |
| HBV-carriers | miR-20a-5p + miR-320a + miR-324-3p + miR-375 + AFP | 0.789 | 0.700 | 0.775 | [231] |
| HBV-carriers | miR-20a-5p + miR-320a + miR-324-3p + miR-375 + AFP | 0.767 | 0.640 | 0.838 | [231] |
| CHB | miR-96 + AFP | 0.889 | 0.836 | 0.824 | [34] |
| CHB | miR-126 + AFP | 0.920 | 0.840 | 0.920 | [32] |
| CHB | miR-142-3p + AFP | 0.910 | 0.860 | 0.940 | [32] |
| CHB | miR-224 + AFP | 0.867 | 0.875 | 0.765 | [11] |
| CHB | miR-125b + miR-27a + AFP1 | 0.722 | 0.600 | 0.833 | [30] |
| CHB | miR-125b + miR-223 + miR-27a + miR-26a + AFP | 0.790 | 0.689 | 0.867 | [30] |
| CHB | miR-125b + miR-223 + miR-27a + miR-26a + AFP2 | 0.833 | 0.820 | 0.767 | [30] |
| CHB | miR-125b + miR-223 + miR-27a + miR-26a + AFP1 | 0.728 | 0.582 | 0.867 | [30] |
| CHB | miR-125b + miR-223 + miR-27a + miR-26a + AFP1,2 | 0.812 | 0.704 | 0.833 | [30] |
| CHB | miR-126 + miR-142-3p + AFP | 0.930 | 0.880 | 0.970 | [32] |
| HBV-DN | let-7b + AFP | 0.706 | 0.508 | 0.767 | [233] |
| HBV-DN | miR-122 + AFP | 0.714 | 0.792 | 0.533 | [233] |
| HBV-LC | miR-99a + AFP | 0.780 | 0.719 | 0.828 | [232] |
| HBV-LC | miR-101 + AFP | 0.973 | 0.966 | 0.879 | [24] |
| HBV-LC | miR-122 + AFP + PIVKA-II | 0.918 | 0.910 | 0.880 | [37] |
| HBV-LC | miR-126 + AFP | 0.897 | 0.800 | 0.790 | [32] |
| HBV-LC | miR-142-3p + AFP | 0.899 | 0.850 | 1.000 | [32] |
| HBV-LC | miR-205 + AFP | 0.893 | 0.750 | 0.860 | [36] |
| HBV-LC | miR-224 + AFP | 0.844 | 0.969 | 0.641 | [11] |
| HBV-LC | miR-126 + miR-142-3p + AFP | 0.939 | 0.850 | 0.840 | [32] |
| CHB + HBV-LC | miR-224 + AFP | 0.857 | 0.875 | 0.722 | [11] |
In addition, miRNAs may be of particular value in the diagnosis of HBV-HCC with low AFP levels. Despite the fact that AFP is the most often utilized biomarker for HCC worldwide, serum AFP levels stay normal in 15%-30% of advanced HCC. Meanwhile, approximately 30% of early-stage HCC cannot be diagnosed via AFP measurement, which delays therapy. Therefore, it is crucial to establish biomarkers capable of identifying HCC patients with negative AFP levels[39]. Different miRNAs and miRNA panels have shown good capacity for separating HBV-HCC patients with negative AFP expression from HC and CHB patients (Table 5), such as miR-125b[23], miR-15b + miR-130b[9] miR-21 + miR-122 + miR-192[22], and miR-125b + miR-223 + miR-27a + miR-26a[30]. A meta-analysis suggests that circulating miRNAs have a relatively high diagnostic accuracy in distinguishing HBV-HCC patients with low AFP levels from non-HCC controls[40]. Therefore, circulating miRNAs may be an ideal potential diagnostic biomarker for HBV-HCC patients with low AFP levels. Notably, these miRNAs or miRNAs panels only show ordinary effects when differentiating HBV-HCC with HBV-LC, and thus, more research is needed to improve the value of miRNA in differentiating HBV-LC and HBV-HCC patients with negative AFP expression.
| AFP level in HBV-HCC patients | Comparison | miRNA | AUC | Sensitivity | Specificity | Ref. |
| < 15 ng/mL | HBV-DN | let-7b | 0.645 | 0.848 | 0.500 | [233] |
| < 15 ng/mL | HBV-DN | miR-122 | 0.629 | 0.712 | 0.577 | [233] |
| < 15 ng/mL | HBV-DN | miR-122 + let-7b | 0.646 | 0.848 | 0.500 | [233] |
| ≤ 20 ng/mL | CHB + HC | miR-26a | 0.733 | 0.868 | 0.574 | [30] |
| ≤ 20 ng/mL | CHB + HC | miR-26a1 | 0.701 | 0.880 | 0.574 | [30] |
| ≤ 20 ng/mL | CHB + HC | miR-27a | 0.832 | 0.838 | 0.723 | [30] |
| ≤ 20 ng/mL | CHB + HC | miR-27a1 | 0.771 | 0.800 | 0.723 | [30] |
| ≤ 20 ng/mL | CHB + HC | miR-125b | 0.778 | 0.760 | 0.790 | [30] |
| ≤ 20 ng/mL | CHB + HC | miR-125b1 | 0.775 | 0.800 | 0.787 | [30] |
| ≤ 20 ng/mL | CHB + HC | miR-223 | 0.759 | 0.789 | 0.702 | [30] |
| ≤ 20 ng/mL | CHB + HC | miR-2231 | 0.715 | 0.720 | 0.723 | [30] |
| < 20 ng/mL | CHB + HC | miR-15b + miR-130b | 0.980 | 0.967 | 0.915 | [9] |
| ≤ 20 ng/mL | CHB + HC | miR-125b + miR-223 + miR-27a + and miR-26a | 0.874 | 0.842 | 0.851 | [30] |
| ≤ 20 ng/mL | CHB + HC | miR-125b + miR-223 + miR-27a + miR-26a1 | 0.849 | 0.800 | 0.894 | [30] |
| < 200 ng/mL | CHB | miR-125b | 0.943 | 1.000 | 0.755 | [23] |
| < 250 ng/mL | CHB | miR-126 | 0.765 | 0.610 | 0.690 | [32] |
| < 250 ng/mL | HBV-LC | miR-126 | 0.643 | 0.610 | 0.580 | [32] |
| < 400 ng/mL | HBV-LC | miR-205 | 0.815 | 1.000 | 0.560 | [36] |
| 20-400 ng/mL | CHB + HC | miR-15b + miR-130b | 0.976 | 1.000 | 0.915 | [9] |
| < 400 ng/mL | CHB + HBV-LC + HC | miR-122 + miR-192 + miR-21 + miR-223 + miR-26a + miR-27a + miR-801 | 0.879 | 0.777 | 0.845 | [44] |
Overall, miRNAs have great potential for use in the diagnosis of HBV-HCC. Two meta-analyses also show that miRNAs attain a level between moderate and high in terms of diagnostic evaluation criteria, and also demonstrate superior diagnostic performance than AFP[41,42]. According to both studies, miR-125b demonstrates a stronger diagnostic value for HBV-related HCC than other single miRNAs[41,42]. The subgroup analysis further concludes that downregulated miRNAs, miRNA panels, and serum-type miRNAs provide the most accurate diagnostic function for HBV-HCC[42]. Notably, the majority of patients with HCC are frequently diagnosed at an advanced stage, with a 1-year survival rate of less than 50% and a 5-year survival rate of only 10%[43]. This is due to the lack of accurate early diagnostic biomarkers. Several other studies also evaluate the ability of miRNAs to discriminate between early-stage HBV-HCC and controls. In one study, the combination of AFP and miR-125b, miR-223, miR-27a, and miR-26a has the highest diagnostic accuracy for early-stage HBV-HCC[30]. Another study finds that one miRNA panel consisting of seven miRNAs shows significant diagnostic accuracy for HBV-HCC, particularly in patients with early Barcelona Clinic Liver Cancer (BCLC) stages (0/A)[44]. Consequently, these miRNA profiles may serve as possible early diagnostic markers for HBV-HCC.
Notably, in addition to circulating miRNAs, exosome miRNAs may also have diagnostic and prognostic value in HBV-HCC. Exosome miRNAs are miRNAs contained within exosomes and released by various cells. Exosome miRNAs are frequently more stable in bodily fluids than other circulating miRNAs because the exosome membrane protects them from degradation, indicating a higher value as cancer biomarkers[45]. As HBV can impact the production of exosomes and their cargos to promote HBV replication and diseases progression[46], exosomes derived from HBV-infected cells may be useful biomarkers for HBV-related diseases[47,48]. Several studies have validated and reviewed exosome-encapsulated miRNAs as circulating diagnostic markers for HCC[49,50], which may be beneficial for monitoring CHB progression and for detecting HBV-HCC at an early stage[51,52].
miRNAs, whose expression level is correlated with disease severity and survival rate in HCC patients, have shown good prognostic value for HCC[53]. For HBV-HCC patients, several miRNAs from HCC tissues or blood are found to be significantly correlated with overall survival, diseases-free survival (DFS) and progression-free survival (Table 6). For tissue miRNAs, higher expression of miR-122[54,55], miR-143[55], miR-145[56], miR-193b[57], miR-203a[58], miR-216b[59], miR-375[55,60], and miR-384[61] is associated with a better prognosis, and higher expression of miR-9-3[62], miR-10b[62], miR-21[56], miR-29a-5p[63], miR-31[62], miR-106b[64], miR-224[55], miR-371a-5p[27], miR-519c[62], miR-522[62,65], miR-523[65], miR-3188[28], miR-3682-3p[66], miR-3660[62], miR-4784[62], miR-5188[67], and miR-6883[62] is associated with a significantly poorer long-term prognosis. For circulating miRNAs, higher expression of miR-150[13], miR-223-3p[68], and miR-768-3p[20] is associated with a better prognosis, higher expression of miR-24-3p[33], miR-29a-3p[69], miR-96[34], miR-155[70], miR-192-5p[69], and miR-487b[18,19] is associated with a significantly poorer long-term prognosis. Therefore, tissues or circulating miRNAs can be a promising tool in predicting the prognosis of HBV-HCC patients. Specifically, due to the fact that miRNAs are abundant in serum exosomes, serum exosomal miRNAs can be used to predict the outcome of HCC patients. In addition, tissue miR-21[56], miR-203a[58], miR-375[60], and miR-5188[67], and serum miR-26a[30], miR-27a[30], miR-29a-3p[69], miR-125b[30], miR-150[13], miR-192-5p[69], miR-223[30,68], miR-487b[18,19], and miR-768-3p[20] are independent prognostic factors of HBV-HCC patients.
| Tissues/serum | miRNA panels | Risk/protective factors | Outcome | HR | CI | Ref. |
| Tissues | miR-9-3a | Risk | OS | - | - | [62] |
| Tissues | miR-10ba | Risk | OS | - | - | [62] |
| Tissues | miR-21b | Risk | DFS | 3.019 | 0.219-6.939 | [56] |
| Tissues | miR-29a-5pb | Risk | TTR | 0.5 | 0.3-0.8 | [63] |
| Tissues | miR-29a-5pb | Risk | OS | - | - | [63] |
| Tissues | miR-31a | Risk | OS | - | - | [62] |
| Tissues | miR-106ba | Risk | OS, DFS | - | - | [64] |
| Tissues | miR-122a | Protect | OS | - | - | [54] |
| Tissues | miR-122b | Protect | OS | - | - | [55] |
| Tissues | miR-143b | Protect | OS | - | - | [55] |
| Tissues | miR-145a | Protect | DFS | 1.12 | 0.293-2.958 | [56] |
| Tissues | miR-193bc | Protect | OS | - | - | [57] |
| Tissues | miR-203a | Protect | OS | 0.63 | 0.41-0.97 | [58] |
| Tissues | miR-216bc | Protect | OS, DFS | - | - | [59] |
| Tissues | miR-224b | Risk | OS | - | - | [55] |
| Tissues | miR-371a-5pb | Risk | OS | - | - | [27] |
| Tissues | miR-375 | Protect | DFS | - | - | [60] |
| Tissues | miR-375b | Protect | OS | - | - | [55] |
| Tissues | miR-384a | Protect | OS | - | - | [61] |
| Tissues | miR-519ca | Risk | OS | - | - | [62] |
| Tissues | miR-522a | Risk | OS | - | - | [62] |
| Tissues | miR-522b | Risk | OS | 2.19 | 1.33-3.6 | [65] |
| Tissues | miR-523b | Risk | OS | 1.5 | 1-2.44 | [65] |
| Tissues | miR-3188c | Risk | OS, DFS | - | - | [28] |
| Tissues | miR-3660a | Risk | OS | - | - | [62] |
| Tissues | miR-3682-3pc | Risk | OS | - | - | [66] |
| Tissues | miR-4784a | Risk | OS | - | - | [62] |
| Tissues | miR-5188b | Risk | OS | - | - | [67] |
| Tissues | miR-6883a | Risk | OS | - | - | [62] |
| Serum | miR-24-3pb | Risk | OS | 2.141 | 1.158-3.960 | [33] |
| Serum | miR-24-3pb | Risk | DFS | 2.055 | 1.114-3.792 | [33] |
| Serum | miR-29a-3pb | Risk | OS | 4.0 | 1.2-13.9 | [69] |
| Serum | miR-29a-3pa | Risk | PFS | - | - | [69] |
| Serum | miR-96a | Risk | OS | - | - | [34] |
| Serum | miR-150c | Protect | OS | 0.446 | 0.233-0.854 | [13] |
| Serum | miR-192-5pb | Risk | OS | - | - | [69] |
| Serum | miR-192-5pb | Risk | PFS | 2.2 | 1.1-4.2 | [69] |
| Serum | miR-487ba | Risk | OS | 2.846 | 1.139-7.114 | [19] |
| Serum | miR-487bc | Risk | OS | 2.115 | 1.083-4.132 | [18] |
| Serum | miR-768-3pb | Protect | OS | 3.057 | 1.136-8.225 | [20] |
| Plasma | miR-155 | Risk | OS | - | - | [70] |
Researchers also develop different models to predict HBV-HCC survival. One study constructs a multivariate risk model that incorporates BCLC stage, miR-192-5p, and miR-29-3p. This risk model is significantly correlated with patient survival and has a good prognostic value. Consequently, the serum miRNA signature can offer predictive value for the BCLC stage classification. In addition, one random forests model made with miRNAs can predict HBV-HCC survival well[62].
It is worthy of note that miRNAs are also correlated with the probability of HBV-HCC recurrence. Several miRNAs may serve as a potential predictor for early tumor recurrence after HCC resection. For instance, the amount of miR-29a-5p in HCC tissues is strongly linked with early HCC recurrence following surgery, including in early-stage HCCs. Stages 0 and A of BCLC are regarded as the early stages, it implies that these HCC patients may have a better prognosis. However, in clinical practice, some patients still have a bad prognosis. Predicting the prognosis of these individuals is a huge challenge for clinicians. As miR-29a-5p sensitivity and specificity may reach approximately 70% for HCC patients with BCLC 0/A stage, their miR-29a-5p expression level may provide a visual aid to distinguish them from other early-stage patients[63]. In addition, it has been discovered that the recurrence of HBV-HCC is closely associated with dysregulation of miR-21 and miR-145[56].
In addition, one study reveals that miRNAs are related with the development risk of HCC in CHB. In CHB patients who do not develop HCC, nucleos(t)ide analogue (NA) treatment restores expression of these miRNAs to near-normal levels, whereas the expression profile is not fully restored in individuals who ultimately develop HCC. Therefore, in CHB patients treated with NA, the changes in miRNAs expression may help identify HCC development risks[71].
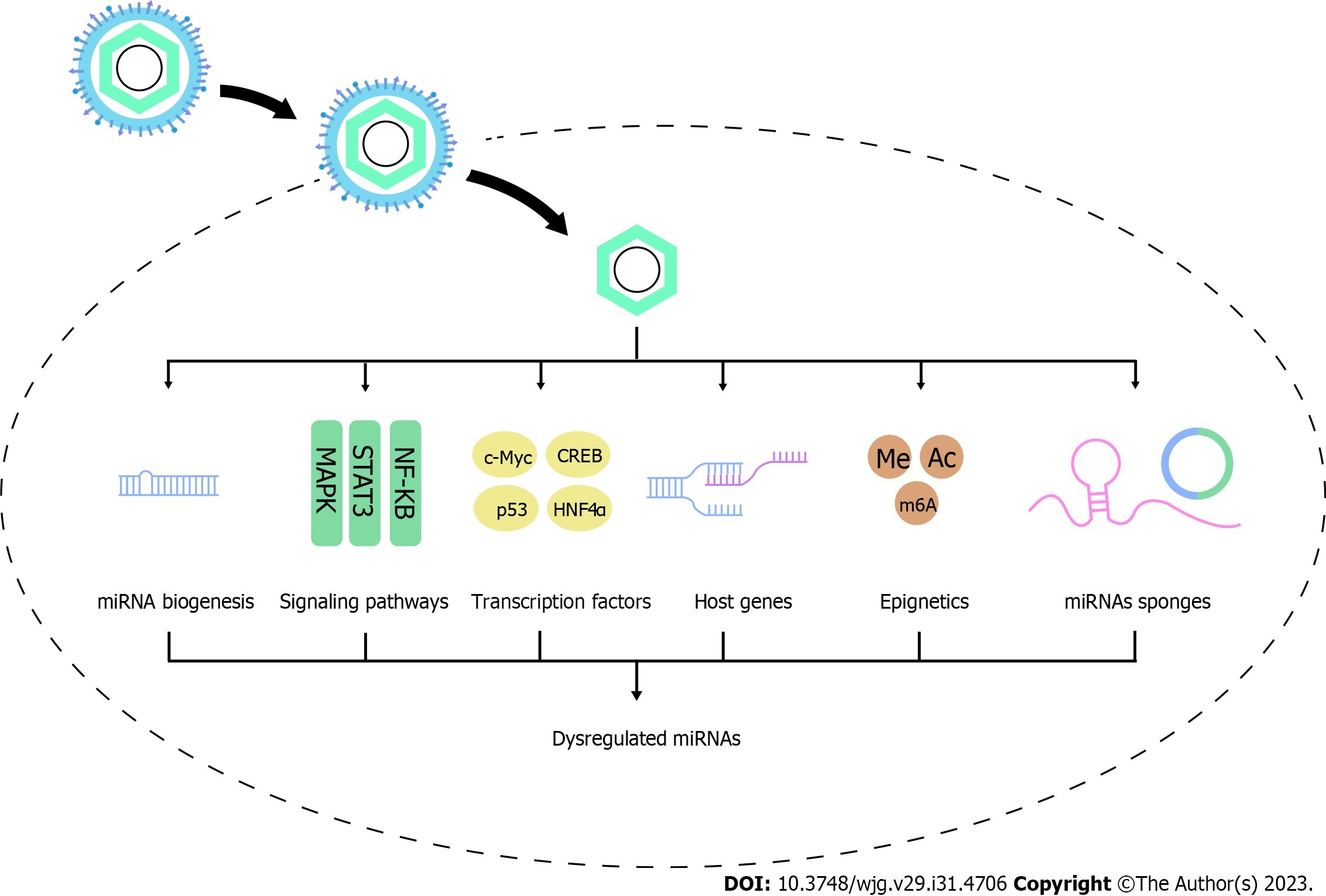
In HCC, HBV and its proteins [HBV surface antigen (HBs), HBV core antigen (HBc), HBV envelope antigen (HBe), HBV x protein (HBx), and HBV polymerase protein (HBp)] dysregulate miRNAs to promote hepatocarcinogenesis. In this part, we summarize the available studies deciphering the way in which HBV regulates the expression profiles of miRNAs through modulating miRNA processing genes and proteins and influencing transcriptional, posttranscriptional, and epigenetic mechanisms, as well as the factors affecting the regulation process, which may help to identify the novel therapeutic pathways (Figure 1).
It takes a complex process to form mature miRNAs. Firstly, miRNA genes, encoded by introns of noncoding or coding regions, are transcribed to pri-miRNAs by RNA polymerase II (Pol II) in the nucleus. Then, pri-miRNAs are spliced to pre-miRNAs by the Microprocessor complex (composed by the nuclear RNase III Drosha and DGCR8). Depending on the protein exportin 5 (EXP5), pre-miRNAs are exported to the cytosol, and are cleaved to a small RNA duplex by Dicer. The duplex is subsequently sorted and loaded onto Ago proteins, of which the guide strand is selected and preserved to form the RNA-induced silencing complex and silence gene expression[72].
There is evidence that HBV contributes to pre-miRNA production. Rather than pri-miR-18a levels, pre-miR-18a levels are associated with miR-18a elevation in HBV-HCC cases, indicating increased processing of pri- to pre-miR-18a[73]. Another study demonstrates that ectopic expression of HBx stimulates the transcription of pri-miR-1269b and hence induces the expression of pre-miR-1269b in HCC cell lines[74]. Therefore, HBV may affect miRNA expression through modulating its synthesis process.
Studies have verified a strong correlation between HBV and miRNA processing proteins. CHB patients with high HBV loads have lower mRNA and protein levels of Drosha, Dicer and Ago2 compared with patients with low viral loads[75]. In HBV-positive HCC patients, Drosha, DGCR8, Ago1, and Ago2 are significantly overexpressed[76], whereas Dicer and Ago3 are significantly downregulated in HCC tissues than that in adjacent nontumor tissues[76,77].
For Drosha protein, researchers find that HBV inhibits Drosha promoter activity to downregulate its expression. HBx are inferred to interact with SP1 and AP-2a to downregulate Drosha expression[78]. HBV can downregulate DGCR8 expression via suppressing its promoter activity through upregulating transcription factor YY-1, in which HBs and HBx may play a role[79]. No study has provided direct evidence that the EXP5 protein is controlled by HBV. PIN1 decreases mature miRNA expression by catalyzing EXP5's conformational change and reducing its ability to export pre-miRNAs from the nucleus to the cytoplasm[80]. It is reported that PIN1 can bind specifically to the HBx to synergistically increase cell proliferation[81], indicating that HBx interacts with PIN1 and may affect pre-miRNAs export. Upon export to the cytoplasm, pre-miRNA is cleaved by Dicer in Drosophila. MiRNAs and siRNAs share a similar step in splicing and partitioning[82]. Considering that HBx can inhibit the Dicer-mediated processing of dsRNAs into siRNAs[75], HBV may also modulate miRNA through inhibiting Dicer and Dicer-mediated splicing of pre-miRNAs. Researchers discover that Ago2 mRNA is repressed by miR-99a in Huh7 and Hep3B cells[83], whereas miR-99a is found to be up-regulated in serum of HBV patients[84], indicating that there may be an HBV/miR-99a/Ago2 regulatory axis.
Overall, there is still no evidence directly indicating that HBV affects miRNA expression by altering miRNA processing proteins. But due to the crucial role of these proteins in miRNA biogenesis, it is reasonable to infer that this is possible. It’s worth noting that in a study with non-viral-associated HCC samples, DGCR8, Dicer, Ago3 and Ago4 are also significantly downregulated, in which epigenetic regulation may be implicated[77]. Therefore, the regulatory role and mechanisms of HBV on miRNA machinery components still need further investigation (Figure 2).
Although miRNAs share a common synthesis machinery, specific miRNA is regulated by different transcriptional and posttranscriptional mechanisms. HBV generally leads to a range of aberration of signaling pathways, Sartorius et al[6] summarize the miRNAs that are dysregulated by HBV and are involved in regulating these signaling pathways. However, miRNAs are also regulated by these signaling pathways induced by HBV infection (Table 7).
| Upstream signaling pathway | miRNA | HBV protein | Expression | Ref. |
| ERK1/2/CREB | miR-212-3p | HBe | Up | [85] |
| MAPK/Ap1 | miR-21 | HBx | Up | [87,88] |
| MAPK/YY1 | miR-129-2 | HBV | Down | [89] |
| MAPK/YY1 | miR-203 | HBV | Down | [89] |
| MAPK/YY1 | miR-335 | HBV | Down | [89] |
| IL-6/STAT3 | miR-21 | HBx | Up | [91] |
| STAT3 | miR-328-3p | HBV, HBx, HBc | Up | [90] |
| STAT3 | miR-34a | HBx | Down | [95] |
| STAT3/SALL4 | miR-200c | HBV | Down | [94] |
| STAT3 | miR-204 | HBV | Down | [93] |
| STAT3 | miR-539 | HBx | Up | [92] |
| NF-κB | miR-23a | HBV | Up | [103] |
| NF-κB | miR-143 | HBx | Up | [98,99] |
| NF-κB | miR-146a | HBx | Up | [100] |
| NF-κB | miR-146a-5p | HBx, HBc | Up | [97] |
| NF-κB | miR-1269b | HBx | Up | [74] |
| PI3K, NF-κB | miR-155 | HBe | Up | [101] |
| IKKα/NF-κB | miR-7 | HBx | Up | [104] |
| IKKα/NF-κB | miR-21 | HBx | Up | [104] |
| IKKα/NF-κB | miR-103 | HBx | Up | [104] |
| IKKα/NF-κB | miR-107 | HBx | Up | [104] |
| Androgen pathway | miR-216a | HBx | Up | [111] |
| TLR7/NF-κB | miR-155 | HBV | Down | [106] |
| LEF-1 | miR-371a-5p | HBV | Up | [27] |
| PPARγ/NF-κB/p65 | miR-130a | HBV | Down | [105] |
MAPK pathway plays a crucial role during the innate immune response. HBeAg is able to activate ERK, one of the MAP kinases, to induce the expression of phosphorylated CREB, which is able to bind to the promoter of miR-212-3p and subsequently enhance miR-212-3p expression[85]. In addition, several AP-1 components including Fra-1, c-Jun, and JunB are found to be recruited on a miR-21 50-flanking region, thus promoting miR-21 transcription. HBx has been previously shown to activate Ap-1, which is activated predominately by MAPK signaling cascade[86]. Therefore, there is a potential HBx/MAPK/Ap1/miR-21 regulatory pathway[87,88]. Meanwhile, HBV also induces YY1 expression through MAPK signaling, and negatively regulates the expression of miR-335, miR-129-2, and miR-203[89].
STAT3 is crucial for transducing signals and regulating the expression of a wide range of genes to promote tumor progression. Studies have found that HBV, HBc and HBx but not HBs and HBp increase STAT3 phosphorylation[90], suggesting that HBV and its viral proteins underline a role in STAT3 activation. STAT3 has been proved to directly bind to several miRNAs’ promotors, increase the promotors activity, and subsequently activate miRNAs transcription. STAT3 mediates upregulation of miR-21[91], miR-328-3p[90] and miR-539[92], which are dysregulated by HBV infection. However, Huang et al[93] found that STAT3 mediates HBV-induced miR-204 suppression. HBV also activates STAT3 to induce SALL4 expression, while SALL4 suppresses miR-200c expression through directly binding to miR-200c promoter[94]. In addition, STAT3 may mediate the suppression of miR-34a caused by HBx[95].
NF-κB is a transcription factor with broad roles in gene induction in a variety of cellular responses, particularly throughout the immune system. HBV and its proteins have been shown to increase NF-κB content and facilitate its translocation from the cytoplasm to the nucleus[74,96]. Researches show that multiple miRNAs are dysregulated by HBV through modulating NF-κB signaling. For example, HBx and HBc upregulate miR-146a-5p through activating NF-κB signaling[97]. HBx upregulates miR-143[98,99], miR-146a[100] and miR-1269b[74] by activating NF-κB binding to the miRNAs promotor. Meanwhile, inhibitors of NF-κB and PI3K decrease miR-155 in HBeAg-stimulated macrophages, suggesting NF-κB and PI3K mediate HBeAg-induced miR-155 upregulation in macrophages[101]. In addition, NF-κB subunit p50 but not p65 mediates upregulation of miR-942 by LPS through binding to miR-942 promotor, and miR-942 expression is increased with progression of HBV-mediated liver fibrosis, implying a putative regulation of HBV on miR-942[102]. As a consensus p65-binding sequence (AGGGATTTCC) is located in the miR-23a promoter region, p65 dominantly represses miR-23a promoter activity, and suppresses miR-23a transcription[103]. Therefore, HBV has the potential to upregulate miR-23a by suppressing p65 expression. In addition, nuclear IKKα coordinates the transcriptional activity of NF-κB to mediate the expression of miR-7, miR-21, miR-103, and miR-107 caused by HBx[104]. Intriguingly, HBV induced PPARγ to negatively control NF-κB/p65 protein via ubiquitination and degradation. Repressed NF-κB/p65 then reduces the endogenous miR-130a expression[105]. The discrepancy in the NF-κB results may be due to the different HBV viral proteins.
Toll-like receptors (TLRs), the main cellular innate immune cell receptor, play crucial roles in immune responses against viral infections, including HBV. Sarkar et al[106] find that TLR7 expression is reduced by HBV infection, while TLR7 is able to induce miR-155 through the NF-κB pathway. Considering that HBV suppresses TLR7 and its subsequent signaling pathway including MAPK/Ap-1, NF-κB, IRF3 and IRF7[107], HBV may impact miRNAs expression through regulating TLR proteins and their signaling pathways. In addition, one study finds that HBV upregulates LEF-1, a key component of the Wnt signaling pathway, to induce miR-371a-5p expression through binding with miR-371a-5p promoter[27].
Membrane-initiated estrogen receptor (ER) and androgen receptor (AR) signaling participate significantly in physiology and disease[108]. HBx enhances AR-responsive gene expression[109], and represses ERα responsive gene transcription[110]. Therefore, HBV has the potential to regulate the miRNAs that act as AR or ERα responsive elements. HBx amplified the transcription of pri-miR-216a which is activated as a result of ligand-stimulated AR binding to the ARE site at the 5′ promoter region. However, when applied to AR-negative cells, HBx failed to stimulate an increase in pri-miR-216a[111].
HBV protein and its RNA can modulate the expression of some transcription factors or other proteins, which in turn regulate miRNAs expression (Table 8).
| Transcription factors or upstream regulatory elements | miRNA | HBV protein | Expression | Ref. |
| c-Myc | let-7 | HBx | Down | [115] |
| c-Myc | miR-15a/16 | HBx | Down | [114] |
| c-Myc | miR-17-92 | HBV | Up | [116] |
| c-Myc | miR-192 | HBx | Down | [113] |
| c-Myc | miR-3682-3p | HBx | Up | [66] |
| CREB1 | miR-520c-3p | HBx | Up | [117] |
| CREB | miR-3188 | HBx | Up | [28] |
| Survivin, Sp1 | miR-520b | HBx | Up | [118] |
| FOXO3 | miR-30b-5p | HBp | Up | [119] |
| URG11 | miR-148a | HBx | Up | [126] |
| JNK/c-Jun | miR-199a-3p | HBx | Up | [120] |
| p53 | miR-216b | HBx | Down | [59] |
| p53 | miR-148a | HBx | Down | [122] |
| Hnf4α | miR-122 | HBV | Down | [123] |
| Hnf4α | miR-548p | HBx | Down | [124] |
| DDX3 | miR-34 | HBx | Down | [127] |
HBV promotes the expression of oncogenic proteins to regulate miRNAs. c-Myc oncoprotein is a transcription factor that regulates numerous physiological processes. Chang et al[112] identify 13 miRNAs which are prominently repressed by c-Myc through binding to miRNA promotors. HBV can directly interact with c-Myc and stimulate its expression, thereby affecting the expression of miRNAs. For example, HBx induces c-Myc, and then c-Myc is recruited to a region of miR-192 promotor, which leads to decreased promotor activity, and subsequently downregulates miR-192-3p expression[113]. Similarly, HBx also downregulates miR-15a/16 expression[114], and suppresses let-7 family through c-Myc[115]. In addition, Jung et al[116] find that c-Myc mediates HBV-induced miR-17-92 overexpression. c-Myc also binds to the miR-3682-3p promoter, thus HBx may also induce miR-3682-3p expression via c-Myc[66]. CREB is an essential subset of phosphorylation-dependent transcription factors. HBx promotes CREB-mediated activation of miR-3188[28]. Similarly, HBx promotes miR-520c transcription through CREB1[117]. Meanwhile, HBx, survivin and Sp1 form a complex in the promotor of miR-520b, and the interaction between HBx and survivin or Sp1 is indispensable for the regulation of miR-520b[118]. FOXO3 is a transcriptional factor that promotes oncogenesis, HBp can promote the expression of miR-30b-5p through its interaction with FOXO3[119]. Moreover, HBx and TGF-1 induce JNK-dependent activation of c-Jun, which is then recruited to the miR-199a-3p promoter to stimulate its transcription[120].
HBV inhibits the expression of tumor suppressor proteins to regulate miRNAs. Acting as an important tumor suppressor gene, TP53 is the most frequently mutated gene in HBV-related HCC[121]. HBx can decrease the recruitment of p53 to the miR-216b promoter, and then inhibit miR-216b transcription[59]. Another research indicates that HBx can also repress miR-148a via suppressing p53-mediated activation[122]. Hepatocyte nuclear factor 4α (Hnf4α), a liver-enriched transcription factor that activates miR-122 gene transcription by binding to its promotor, is found to be repressed by HBV infection in both mRNA and protein levels[123]. Similarly, Hnf4α mediates HBx induced downregulation of miR-548p, possibly through direct binding to the miR-548p promoter[124].
In addition to these transcription factors, HBV also regulates some other proteins to affect miRNAs expression. URG4/URGCP, up-regulated by HBx, can up-regulate 77 miRNAs and down-regulate 9 miRNAs in HepG2 cells[125]. Yuan et al[126] find that HBx-induced miR-148a is dependent on oncogenic URG11. HBx also downregulates DDX3, which upregulates miR-34 expression[127]. Meanwhile, one study also identifies 75 miRNAs by ChIP-Seq whose promotor regions are putatively targeted by HBx protein, some of which have been implicated in hepatocarcinogenesis[128].
As the majority of miRNA genes are encoded in the introns of either noncoding or coding regions, multiple studies have demonstrated that only one-third of intronic miRNAs are transcribed from their own promoters. The coregulation of intronic miRNAs with their host genes can be further illustrated by their tissue- or disease-specific co-expression patterns[129].
Some miRNAs are derived from lncRNA precursors, which have the potential to be affected by HBV. LncRNA H19 has been proved to harbor a miRNA containing hairpin in its exon 1, which serves as the precursor for miRNA-675[130]. HBx upregulates H19 expression, leading to a corresponding increase of miR-675[131,132]. Therefore, HBV may affect the LncRNA-derived miRNAs. However, HBx can positively regulate miR-545/374a cluster in the Ftx lncRNA, but fails to regulate miR-421/374b cluster which is also encoded in Ftx introns. Even though miR-374a and miR-545 are transcribed off the same promoter, their abundances are not always correlated[12].
In addition, intronic miRNAs may be coupled with their host genes. miR-26b gene resides in an intron of CTDSP1. They share the same transcription start sites (TSS). miR-26b is therefore transcribed as part of its host transcription unit[133,134]. HBV downregulates miR-26b expression, partly because of the suppression of CTDSP1 mRNA transcription. Notably, the extent of the decrease in miR-26b level was greater than that of CTDSP1 mRNA, implying the other putative regulatory pathway[135]. Similarly, HBx promotes miR-106b, miR-93, and miR-25 transcription in HCC cells, whose host gene MCM7 is also co-transcribed and upregulated, suggesting that MCM7 activation may be involved in the regulation of these miRNAs by HBV[64].
Notably, despite the fact that some miRNA genes share the promoter of their host gene, the vast majority of miRNA genes have multiple TSS, and the promoters of intronic miRNAs are sometimes distinct from the promoters of their host genes.
Epigenetic mechanisms mainly include DNA methylation, posttranslational histone modifications, chromatin remodeling, ncRNA interactions and RNA modification. Despite the significant involvement of miRNAs in epigenetic regulation, miRNAs are also regulated by epigenetic modifications and involved in diverse human diseases[136]. HBV can regulate epigenetics of miRNAs, leading to the functional disruption of miRNAs and consequently promoting HCC.
HBx has been found to increase the DNA methyltransferase (DNMT) activities and promote regional hypermethylation of specific tumor suppressor genes (TSG)[137]. It has been elucidated that HBx induces DNA hypermethylation of CpG islands in miR-18b[138], miR-30e[139], miR-132[140], and miR-205 promoter[141] to affect their expression. Shang et al[89] identify that miR-335, miR-129-2, and miR-203 are repressed by HBV, but are significantly activated by 5-azacytidine, the DNMT inhibitor, indicating that HBV regulates these miRNAs through DNMT-mediated methylation. Meanwhile, Tsang et al[142] find that knockdown of HBV-upregulated YY1[89] significantly decreases DNA methylation levels in the miR-9 Loci, leading to an increased miR-9 in HCC cells. Thus, YY1 may mediate the suppression of miRNAs caused by HBV through inducing DNA methylation. Conversely, although HBx leads to overall hypomethylation, HBx highly interferes methylation levels of -550 CpG site in the miR-125a promoter, and therefore triggers miR-125 expression[143]. Further study is needed to elaborate this phenomenon.
As for the histone modification, Guerrieri et al[128] verify that HBx decreases H4 acetylation level in the promoter regions of miR-138-2, miR-224, miR-302e, miR-576-3p and miR-596, which may explain their downregulation. Conversely, HBx increases H4 acetylation at the miR-26b promoter. These imply HBx ability to regulate miRNAs by modulating the histone modification of miRNAs promotors[128]. In addition, H3K27me3 is an epigenetic modification to the DNA packaging protein Histone H3[144]. The genomic regions enriched for H3K27me3 can function as silencers to repress gene expression via chromatin interactions[145]. Knockdown of HBV-upregulated YY1 reduced not only global H3K27me3 levels, but also EZH2 and H3K27me3 promoter occupancy, leading to the increased miR-9 in HCC cells. It is also found that HBV-upregulated YY1 Leads to EZH2 recruitment for H3K27me3-mediated silencing of tumor-suppressing miRNAs[142], supporting the idea that HBV may indirectly regulate miRNAs through impacting H3K27me3 Levels mediated by EZH2 and YY1.
PPARγ, a ligand-activated transcription factor, is able to form a heterodimer with RXRα. The complex binds to DR1 and DR2 motifs in the miR-122 gene promoter to enhance miR-122 gene transcription, which will be amplified by 5-Aza-CdR (DNA methylation inhibitor) and PBA (histone deacetylation inhibitor). However, this positive regulation is abrogated by HBx which suppresses PPARγ-mediated transactivation through binding to the PPARγ DNA binding domain[146], indicating HBx may affect miR-122 epigenetics through binding and inhibiting PPARγ.
N6methyladenosine (m6A) modification is the most widespread post-transcriptional modification in mammalian mRNAs. MiRNAs can control the expression of m6A regulator, but they are also frequently modified with m6A[147]. HBV infection enhances the expression of METTL3, promotes miR-146a-5p maturation in an m6A-dependent manner[148]. In addition, Gld2 is a cytoplasmic non-canonical poly(A) RNA polymerase that adds successive AMP monomers to the 3'-end of specific RNAs. It can directly monoadenylate specific miRNAs, including miR-122, to stabilize and prolong the activity of miRNAs[149]. HBx also downregulates Gld2 expression, decreasing miR-122 3’ monoadenylation and ultimately suppressing mature miR-122 expression[150].
A variety of endogenous RNAs are able to bind to miRNAs to reduce the number of free miRNAs. These competitive endogenous RNAs (ceRNAs) mainly include lncRNAs and circRNAs, showing an increasing significance in multiple diseases[151]. HBV has been shown to regulate miRNAs through lncRNAs or circRNAs (Table 9). HBV-induced lncRNA-Unigene56159 directly targets miR-140-5p and suppresses its expression[152]. HBV infection also elevates lncRNA PCNAP1 to target miR-154[153]. HBV also enhances LncRNA n335586 to competitively bind with miR-924[154]. Meanwhile, HBx is found to stimulate lncRNA H19 to directly target to miR-138[131] and miR-22[155] through endoge
| miRNAs sponges | miRNA | Expression | Ref. | |
| LncRNA | LncRNA Unigene56159 | miR-140-5p | Down | [152] |
| LncRNA PCNAP1 | miR-154 | Down | [153] | |
| LncRNA n335586 | miR-924 | Down | [154] | |
| LncRNA H19 | miR-138 | Down | [131] | |
| LncRNA H19 | miR-22 | Down | [155] | |
| LncRNA TRERNA1 | miR-22-3p | Down | [156] | |
| LncRNA MALAT1 | miR-124 | Down | [157] | |
| LncRNA HMMR-AS1 | miR-627-3p | Down | [158] | |
| LncRNA LINC01352 | miR-135b | Up | [159] | |
| LncRNA F11-AS1 | miR-211-5p | Up | [160] | |
| LncRNA XIST | miR-192 | Down | [161] | |
| LncRNA LINC01232 | miR-708-5p | Down | [162] | |
| LncRNA TFAP2A-AS1 | miR-933 | Up | [163] | |
| LncRNA LINC00924 | miR-6755-5p | Up | [164] | |
| LncRNA HMGB1 | miR-200 | Down | [165,166] | |
| CircRNA | CircRNA ARL3 | miR-1305 | Down | [167] |
| CircRNA RNF13 | miR-424-5p | Down | [168] | |
| CircRNA BACH1 | miR-200a-3p | Down | [169] | |
| CircRNA ATP5H | miR-138-5p | Down | [170] | |
| CircRNA 0027089 | miR-136-5p | Down | [171] | |
| HBV mRNAs | HBx mRNA | miR-15a/miR-16-1 | Down | [172] |
| HBV mRNAs | miR-15a/miR-16 | Down | [173] | |
| HBV mRNAs | miR-122 | Down | [174] | |
| HBV mRNAs | let-7a | Down | [175] | |
| HBs mRNA | let-7g | Down | [176] | |
| HBx mRNA | miR-129-5p | Down | [177] | |
| HBx-LINE1 | miR-122 | Down | [179] |
HBV also regulates circRNAs to affect miRNAs (Table 9). HBx upregulates METTL3 expression to increase the m6A modification of circ-ARL3, and further favors circ-ARL3 reverse splicing and biogenesis. circ-ARL3 binds to miR-1305, antagonizing the inhibitory effects of miR-1305 on target oncogenes[167]. HBV also upregulated Circ-RNF13, as a sponge for miR-424-5p[168]. HBV upregulates Circ-BACH1, which sponges miR-200a-3p to reduce its expression[169]. Meanwhile, HBV upregulates Circ-ATP5H expression, while Circ-ATP5H directly targets miR-138-5p[170]. In HBV-positive HCC cells compared to HBV-negative HCC cells, circ_0027089 is markedly increased and specifically binds to miR-136-5p[171].
In addition to the lncRNAs and circRNAs, ceRNAs also include viral RNAs and host mRNAs[151]. HBV RNA could function as sponges to directly bind with miRNAs. Studies have implicated that HBV RNA may dysregulate miRNAs by binding to the complementary binding sites of miRNAs and depletion of miRNAs (Table 9).
HBV mRNAs, including pre-C/C (pgRNA), pre-S, S 3’-UTR, and X mRNAs, act as sponges to bind and sequester miR-15a/16-1, subsequently resulting in a depletion of miR-15a/16-1[172,173]. Li et al[174] validate that HBV mRNAs (pre-C/C (or pgRNA), pre-S, S 3’-UTR, and X mRNAs) can sponge miR-122 to inhibit its expression and function. Deng et al[175] identify that there is a let-7a complementary region in the HBV genome in HBV pre-C/C, pre-S, and S mRNAs. Notably, HBV regulates downstream targets of let-7a in a sequence-dependent manner. In addition, HBV transcripts harboring the preS2 region, such as HBV large S mRNA, can almost entirely interact with let-7g and subsequently promote HCC[176]. Ochi et al[177] find that HBx mRNA has complementary sequences with the central region of miR-129-5p, HBx mRNA interacts with the responsive element in the 3’ UTR of miR-129-5p and sequesters it from forming a complex with Ago2. It is noted that the abundances of viral RNAs may affect their regulation on miRNAs. Although HCV 5’UTR may be able to bind miR-122, it fails to change miR-122 expression like HBV mRNAs do. This discrepancy may be due to the fact that HCV mRNAs copy numbers are much less than miR-122, while HBV mRNAs copy numbers are more than miR-122[174]. Besides, HBV RNA copies per cell are much higher than those of let-7a[175], which may be essential for HBV RNA sequestration.
Of note, HBV genome gene is frequently inserted to host genes, which may lead to the transcription of the integrated virus-human chimeric fusion. It is exemplified by the discovery of a novel chimeric HBx-LINE1 RNA, which is generated from a normally silenced region of chromosome 8p11.21 after HBV integration[178]. Functioning as a long noncoding RNA (lncRNA)-like transcript, HBx-LINE1 sequesters cellular miR-122 by directly absorbing it, which ultimately leads to the depletion of miR-122[178,179].
Autophagy is the major intracellular degradation system and plays a pivotal role in multiple physiological processes[180], some of which have been delineated to function through modulating specific miRNAs. Majority of existing works have shown that HBV is able to induce autophagy. However, Lan et al[181] find that HBx transgenesis leads to a lower autophagic level, and miR-224 is preferentially recruited and degraded through autophagic progression. In addition, the selective autophagy receptor NDP52 targets Dicer and Ago2 proteins for the degradation. Autophagy is required for miRNA homeostasis and activity. Moreover, autophagy participates in the posttranscriptional regulation of Dicer mRNA, and chronic autophagy deficits impair miRNA stability after pre-miRNA processing[182]. Therefore, HBV has the potential to affect autophagy to disrupt the homeostasis of miRNAs biogenesis.
Carboxyl-terminal truncated HBx proteins (Ct-HBx, also called HBxΔC or trHBx) are variants transcribed from the mutant HBV X gene whose 3′-end are deleted during HBV genome integration into the host cells. Ct-HBx plays a pivotal role in hepatocarcinogenesis[183,184]. Ct-HBx regulates specific miRNAs more effectively than full-length HBx (HBx-FL). For instance, HBx-D35 enhances miR-21 promoter occupancy and upregulates miR-21 expression compared to HBx-FL[91]. HBxD127 also remarkably increases miR-215 expression relative to HBx[185]. A possible explanation is that C-terminal truncation may affect the binding of HBx to cellular proteins, resulting in altered miRNA gene expression patterns in cells[184]. Notably, Ct-HBx directly binds to some miRNAs promotors, such as miR-26a and miR-29c, resulting in direct transcriptional suppression which HBx-FL is unable to induce. The reason for this discrepancy may be that HBx-FL and Ct-HBx bind to different chromatin binding regions of miRNAs[186]. However, not all miRNAs are under this regulation. miR-23a and miR-27a are concordantly regulated by both HBx-FL and Ct-HBx, and their binding regions are similar[186]. For miR-146a, Ct-HBx does not lead to the same elevation as HBx-FL does[187].
HBV pre-S2 mutant protein may also play a role in the dysregulation of miRNA. HBV pre-S2 mutant induces endoplasmic reticulum stress and the mTOR signal cascade in transgenic livers and HCC tissues[188,189]. Meanwhile, Mdm2-dependent ubiquitinoylation of Drosha by mTOR activates miRNA synthesis and controls many cancer-related miRNAs[190]. Since PreS/S proteins initiate a cascade of events that lead to malignancy[189], it's worth to investigate whether PreS mutant dysregulates miRNAs.
Considering HBV integration severely disrupts host cellular gene expression, genomic loci containing miRNA sequences inserted by HBV may impact miRNA expression. It has been found that HBV DNA integration into fragile sites may alter the expression of a couple of miRNAs which are located in or near fragile sites, including miR-200a near FRA1A, miR-143, miR-145 and miR-224 near FRA5C, miRNA-17–92 cluster near FRA13D, miR-195 near FRA17A, miR-99b, miR-125a and let-7e near FRA19A, and miR-199a-1 near FRA19B[191-193]. These miRNAs have been documented in HCC[191], and there are still a great many miRNAs that are potentially dysregulated by HBV integration[192].
In addition, Yang et al[194] also find miR-602 is upregulated by HBV or HBx, and they speculate that the chromosome 9q34.3 containing miR-602 sequence is commonly integrated by HBV, which may lead to increased miR-602 expression. Similarly, Guo et al[195] speculate that HBV-induced chromosome instability caused by HBV integration may play a role in promoting the miRNAs-371-3 gene cluster expression. Further study is needed to support this hypothesis. Therefore, HBV DNA integration may alter miRNA expression, but the underlying mechanism requires additional study.
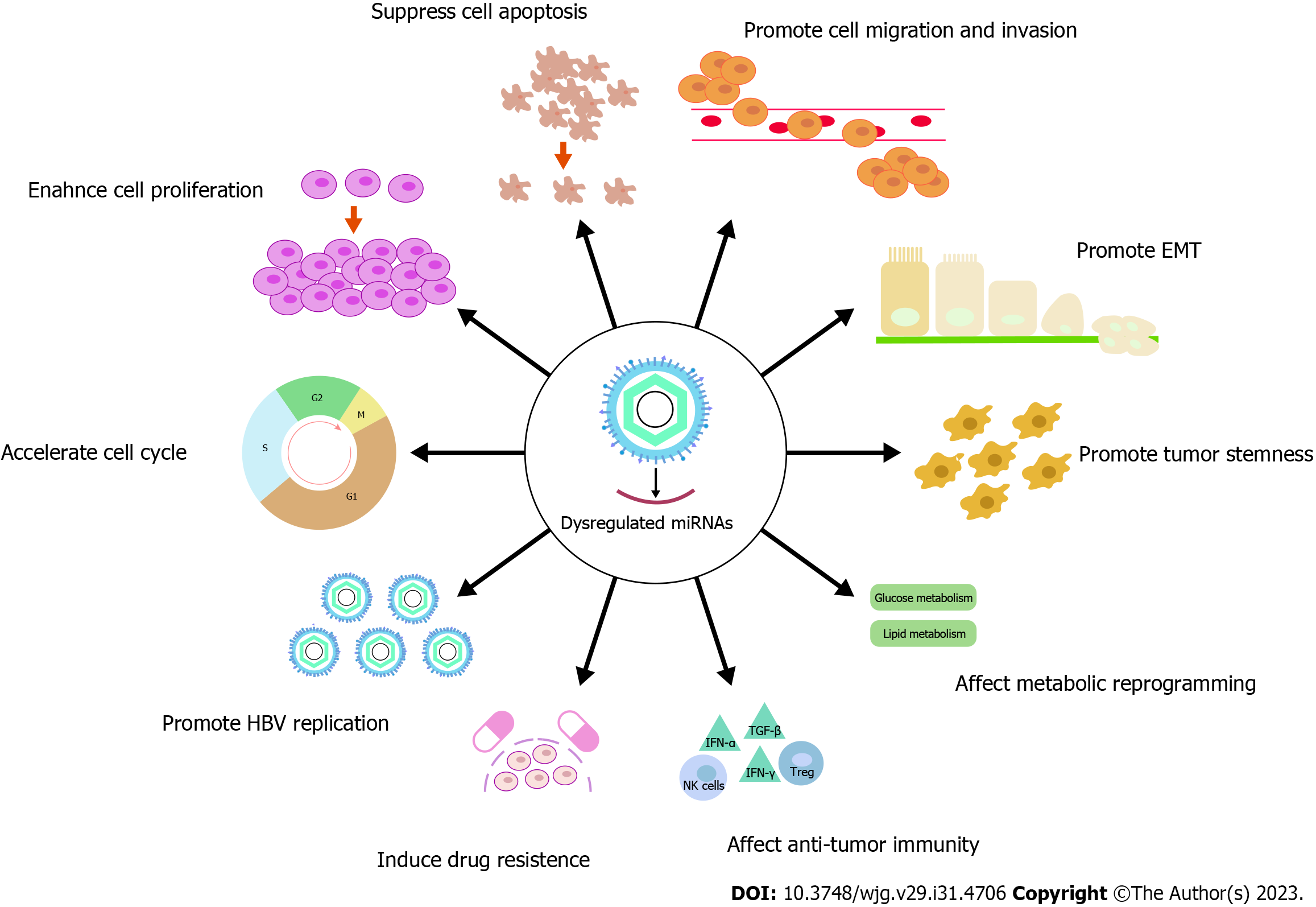
By dysregulating miRNAs, HBV exacerbates its function in the oncogenesis of HCC. Currently, multiple reviews have summarized the essential role of HBV-dysregulated miRNAs in affecting tumor cell cycle, cell proliferation, cell apoptosis, cell migration and invasion, and epithelial-mesenchymal transition (EMT)[6,196-199]. Therefore, we provided an updated supplementary list of miRNAs dysregulated by HBV and involved in these processes (Supplemen
| miRNA | HBV protein | Expression | Target genes | Abnormal function in HBV-HCC | Ref. |
| miR-7 | HBV, HBx | Up | Mapsin | Conferring HBx-mediated anoikis resistance and doxorubicin resistance | [213] |
| miR-15a/16 | HBV mRNA | Down | - | Inducing etoposide-induced apoptosis | [173] |
| miR-21 | HBV, HBx | Up | Mapsin | Conferring HBx-mediated anoikis resistance and doxorubicin resistance | [213] |
| miR-23a | HBV | Down | CCL22 | Inhibiting Tregs recruitment | [103] |
| miR-30b-5p | HBp | Up | MINPP1 | Promoting tumor growth, enhancing cell proliferation, promoting cell migration and invasion, regulating glycolytic bypass metabolism | [119] |
| miR-34a | HBV | Down | CCL22 | Inhibiting Tregs recruitment | [209] |
| miR-103 | HBV, HBx | Up | Mapsin | Conferring HBx-mediated anoikis resistance and doxorubicin resistance | [213] |
| miR-107 | HBV, HBx | Up | Mapsin | Conferring HBx-mediated anoikis resistance and doxorubicin resistance | [213] |
| miR-124 | HBx | Down | PI3K/Akt | Suppressing CSC differentiation | [157] |
| miR-135a-5p | HBc | Up | VAMP2 | Preventing Doxorubicin hydrochloride-induced apoptosis | [212] |
| miR-138 | HBV | Down | PD-1 | Regulating cytokine secretion of T cells and improving T-cell immune responses | [208] |
| miR-146a | HBV | Up | STAT1 | Suppressing IFN-induced anti-HBV effect | [187] |
| miR-152 | HBV | Down | HLA-G | Enhanced NK cytolysis against hepatoma cells | [210] |
| miR-193b | HBV | Down | Mcl-1 | Sensitizing sorafenib-induced apoptosis | [57] |
| miR-200a/200b/429 | HBx | Down | RICTOR | Impairing HCC stem cell properties, regulating glutamine metabolism, sensitizing the response to anti-PD-L1 immunotherapy | [165,166] |
| miR-203a | HBs | Down | BMI1 | Sensitizing 5-FU-induced apoptosis, impairing HCC stem cell properties | [58] |
| miR-205 | HBx | Down | ACSL1 | May promote lipogenesis | [141] [206] |
| miR-325-3p | HBV | Down | DPAGT1 | Sensitizing the response to Doxorubicin chemotherapy | [201,202] |
| miR-329 | HBV, HBx | Down | AFP | Sensitizing chemotherapy induced apoptosis | [214] |
| miR-384 | HBV, HBx | Down | PTN/PI3K/AKT/mTORC1 | Inhibiting high glucose-induced lipogenesis | [61] |
| miR-429 | HBx | Down | Rab18 | Inhibiting dysregulation of lipogenesis | [205] |
| miR-1236 | HBV, HBx | Down | AFP | Sensitizing chemotherapy induced apoptosis | [214] |
| miR-3682-3p | HBx | Up | FOXO3/PI3K/AKT1/β-catenin/c-Myc | Promoting HCC stemness | [66] |
| miR-5188 | HBx | Up | FOXO1/β-catenin | Resisting the effects of chemotherapy 5-FU, CDDP and EPI, promoting HCC stemness | [67] |
Liver cancer stem cells (CSCs) are a distinct population of HCC cells with stem cell characteristics, defining a hierarchical structure and contributing to treatment resistance and tumor recurrence. HBV is one of the most prominent players in liver CSCs. miRNAs partially mediate the stemness progression[200] (Table 11).
| Process | HBV-dysregulated miRNAs |
| Tumor stemness | miR-124[157], miR-200a/200b/429[165], miR-203[58], miR-325-3p[201], miR-3682-3p[66], miR-5188[67] |
| Metabolic reprogramming | miR-30b-5p[119], miR-200[165], miR-205[141,206], miR-384[61], miR-429[205] |
| Anti-tumor immunity | miR-23a[103], miR-34a[209], miR-138[208], miR-146a[187], miR-152[210], miR-200[165,166] |
| Drug resistance | miR-7[213], miR-21[213], miR-103[213], miR-107[213], miR-135a-5p[212], miR-5188[67], miR-15a/16[173], miR-193b[57], miR-203a[58], miR-325-3p[201,202], miR-329[214], miR-1236[214] |
In HCC cells, the expression of CD44, CD133, and EpCAM is markedly reduced by miR-124, indicating the pivotal effects of miR-124 in suppressing CSCs differentiation. HBx downregulates miR-124, and may therefore interfere CSCs differentiation[157]. In one research, HBx supports the progression of HCC via translocation and secretion of HMGB1, which regulates RICTOR expression in HCC by competitively binding to the miR-200 family[166]. Both HMGB1 and RICTOR mRNAs can augment HCC stemness characteristics in HCC[165]. HBsAg inhibits the expression of miR-203a in HCC cells. miR-203a decreases the proportion of CD133-positive HCC cells but not CD90, and it also significantly lowers the average percentage of ALDH-positive malignant stem cells. Therefore, HBV infection may promote the stemness of HCC via regulating miR-203a[58]. HBV also inhibits miR-325-3p[201], which suppresses the expression of critical stemness markers, including SOX-2, Nestin, Notch-1, OCT4, and Nanog. miR-325-3p/DPAGT1 may presumably have a role in HBV-induced HCC stemness[202]. In addition, miR-3682-3p mediates the oncogenic consequences of HBx-induced PI3K/AKT/c-Myc signaling. HBx increases stemness by elevating miR-3682-3p expression[66]. Meanwhile, miR-5188 directly targets FOXO1, which inhibits the nuclear translocation of β-catenin and promotes Wnt signaling activation and downstream tumor stemness. HBx modulates the miR-5188/FOXO1/β-catenin/c-Jun feedback loop to drive Wnt/β-catenin activation, subsequently promoting HCC stemness[67].
Metabolic reprogramming plays a crucial role in the initiation and progression of cancer. A few studies have revealed that HBV affects the process of HCC by regulating metabolism (Table 11). Aerobic glycolysis is a distinguishing feature of HCC and is responsible for regulating proliferation, immune evasion, invasion, metastasis, and drug resistance in HCC[203]. The miR-30b-5p/MINPP1 axis is capable of accelerating the conversion of glucose to lactate and 2,3-bisphosphoglycerate (2,3-BPG), as well as regulating the glycolytic bypass to generate more 2-PG for energy supplementation. HBV protein P (HBp) regulates the miR-30b-5p/MINPP1 axis, contributing to the development of HBV-positive HCC cells via glycolytic bypass[119]. RICTOR regulates glutamine metabolism via mTOR signaling[165]. HBx stimulates the translocation and secretion of HMGB1[166], which regulates RICTOR expression in HCC by binding competitively to the miR-200 family. Therefore, HBx may affect miR-200 to dysregulate glutamine metabolism. As for lipid metabolism, changes in fatty acid synthesis, β-oxidation, and cellular lipidic composition contribute to hepatocarcinogenesis[204]. HBx inhibits miR-384 and upregulates its target PTN expression, while PTN promotes hepatoma cell lipogenesis[61]. Knockdown of Rab18b decreases the lipogenesis. HBx activates Rab18 through downregulating miR-429. Therefore, HBx could enhance hepatocarcinogenesis by leading to the dysregulation of lipogenesis via the miR-429/Rab18 axis[205]. Meanwhile, HBx inhibits miR-205 expression[141], and miR-205 inhibits lipogenesis in hepatoma cells dependent on ACSL1, suggesting that HBx inhibits miR-205 to promote lipogenesis[206].
It is widely acknowledged that HBV causes chronic liver damage through aberrant immunological reactions. During chronic HBV infection in humans, adaptive immunity transitions may be immune pathogenic factors for the development of HCC[207]. A number of research have revealed the function of miRNAs in HBV-induced immunological dysregulation (Table 11). HBV infection increases the expression of miR-146a, which impairs the IFN-induced anti-HBV immune response. Additionally, inhibition of miR-146a improves IFN-α-mediated anti-HBV efficacy[187]. In HBV-HCC patients, miR-138 is significantly higher than in asymptomatic carriers. By targeting the 3'-UTR region of PD-1, miR-138 alters its expression directly. miR-138 exerts its regulatory effects on T-cell cytokine production by suppressing PD-1 expression[208].
HBV also represses some miRNAs to affect anti-tumor immunity. One study finds that HBV-elevated CCL22 induction is mediated by transcriptionally repressing miR-23a. It is hypothesized that the axis of p65/miR-23a/CCL22 is present in the HCC cells and may drive tumor progression by recruiting Tregs, particularly when HBV infection was involved[103]. In HBV-expressing HepG2.215 cells, miR-34a is downregulated, while suppressed miR-34a leads to enhanced production of chemokine CCL22, which recruits Tregs to facilitate immune escape[209]. In addition, HLA-G, which inhibits different kinds of immune cells directly, such as NK, is downregulated by miR-152 in hepatoma cells. HBV inhibits miR-152 and increases the expression of its target HLA-G, which may further suppress NK against cancer cells[210]. Additionally, mRNAs of HMGB1 regulated by HBV and RICTOR regulated by HMGB1 mediated by miR-200[166] inhibit the response to anti-PD-L1 immunotherapy in HCC by elevating PD-L1+ exosomes[165].
Chemoresistance, resulting in cancer relapse and spread, is frequently mentioned as the largest cause of cancer therapeutic failure. In HCC, HBV commonly drives chemoresistance[211]. Accumulating evidence implicates the role of miRNAs in HBV-driven chemoresistance of HCC (Table 11).
HBc upregulates miR-135a-5p to suppress VAMP2 expression, blocking doxorubicin hydrochloride-induced apoptosis in HCC[212]. Similarly, HBV-upregulated miR-5188 Leads to an increase in resistance to the chemotherapy drugs 5-FU, cisplatin, and pharmorubicin[67]. Meanwhile, it is inferred that HBx elevates miR-7, -103, -107, and -21 expression to downregulate their target mapsin. Silencing maspin boosts HCC resistance to doxorubicin and other chemotherapeutic drugs[213]. These miRNAs may contribute to HBV-induced resistance to chemotherapy.
For some anti-tumor miRNAs, HBV suppresses their expression to promote HCC drug resistance. HBV mRNA can directly sponge miR-15a/16 and inhibit the subsequent cascade of etoposide-induced apoptosis in hepatoma cells[173]. miR-193b is downregulated in HBV-positive cells and tissues. Recent research shows that it increases the sensitivity of HCC cells to sorafenib by suppressing the expression of the anti-apoptotic protein Mcl-1[57]. miR-203a reduces HCC cell viability after 5-fluorouracil (5-FU) treatment and also increases the apoptosis rate of HCC cells in response to 5-FU[58]. HBV suppresses miR-203a expression, and subsequently renders HCC cells resistant to chemotherapy drug-induced apoptosis. HBV inhibits miR-325-3p[201], which remarkably increases chemosensitivity to doxorubicin in HCC cells[202]. Similarly, HBV negatively regulates miR-329 and miR-1236 to elevate their target AFP expression, while AFP further attenuates the proapoptotic effect of chemotherapy agents cisplatinum[214].
At present, the research on HBV-dysregulated miRNAs to enhance drug resistance in HCC is in its infancy. It has been discovered that miRNA promotes tumor treatment resistance through targeted regulation of multiple drug-related genes and DNA damage repair-related genes[215]. Therefore, more in-depth studies are needed.
The majority of HBV-infected patients have strong viral replication. By promoting self-replication, HBV maintains a high titer and promotes hepatocarcinogenesis. The complex relationship between HBV replication and miRNA has been described in a number of reviews[53,199,216]. We have enumerated the currently known miRNAs dysregulated by HBV that regulate HBV replication in the Supplementary Tables 1 and 2. Intriguingly, HBx upregulates miR-125a-5p expre
DNA hypermethylation is responsible for suppressing TSGs in hepatocarcinogenesis. The inhibition of miR-101 by HBx leads to an increase in DNMT3A expression, while miR-101 inhibition or overexpression drastically affects the mRNA expression of different TSGs, demonstrating that miR-101 operates upstream to enhance TSG expression[219].
During the process of metastasis, cancer cells detaching from extracellular matrix (ECM) acquire the ability to persist in circulation by evading anoikis-induced cell death[220]. It is found that HBx induces miR-7, -103, -107, and -21 to suppress maspin expression, while maspin downregulation conferes HBx-mediated anoikis resistance in HCC cells[213]. Therefore, it can be speculated that these miRNAs may similarly confer HBx-mediated anoikis resistance.
In addition to directly affecting tumor cells, miRNA can indirectly accelerate the development of HCC via acting on other liver cells. Exosomal miR-142-3p from HBV-positive cells induces ferroptosis in HBV-infected M1-type macrophages via SLC3A2[221]. Similarly, exosomal miR-222 from HBV-infected hepatic cells boosts LX-2 cell activation by suppressing TFRC-induced ferroptosis, which ultimately exacerbates liver fibrosis[222]. Besides that, HBx-elevated P4HA2 enhances the collagen deposition in the liver in vivo and in vitro by inhibiting miR-30e, leading to liver fibrosis and liver cancer progression[139]. Moreover, HBx and TGF-β1 exposure induces the upregulation of miR-199a-3p, which contributes to the malignant transformation of hepatic progenitor cells (HPCs)[120]. As HPCs have the capacity to generate HCC with the cooperation of HBx and AFB1 in the liver microenvironment, this may provide new insight of HBV promoting HCC[223].
Due to the significant changes of miRNA in bodily fluid and tissues of HBV-HCC, its utility as a biomarker for the diagnosis of HCC incidence and prognostic risk has been extensively evaluated. However, since the PLR of miRNAs diagnosing HBV-HCC is less than 10 and the NLR is greater than 0.1[41,42], the clinical use of miRNAs for detecting HBV-HCC may still be limited. Traditional techniques for detecting miRNAs include Northern blotting, quantitative reverse transcription polymerase chain reaction (qRT-PCR), next-generation sequencing, and microarray-based hybridization[7,224]. However, quantifying miRNA in a dependable and robust manner can be challenging, and these methods may involve significant trade-offs between cost, complexity, and efficacy[7,224]. Therefore, using standardized measurements with unified standards will facilitate the collection of trustworthy miRNA data that can be compared across institutions[7]. It is crucial to minimize the influence of confounding factors, such as measurement technical characteristics, when detecting miRNA. Additionally, novel miRNA detection assays, such as miRacles which utilize conformationally responsive DNA nanoswitches, have been proved to be a simple, inexpensive, and accurate method for detecting miRNAs[224]. With the continuous development of new materials, it is anticipated that the miRNAs detection technology will increase in precision and sensitivity while decreasing in cost and operational complexity.
In addition to their use as diagnostics, miRNAs have significant promise for prognostication. Current relevant research has focused on miRNAs to predict the risk of recurrence, OS, and DFS in patients with HBV-HCC. There are few studies and insufficient data on circulating miRNAs. As circulating miRNAs offer numerous advantages, such as being convenient, safe, and noninvasive, their potential as biomarkers can be exploited further. For instance, miRNAs can be used to predict or evaluate the efficacy of neoadjuvant chemotherapy[225], radiotherapy[226], and immunotherapy[227] in cancer patients. Hence, miRNAs have the potential to anticipate therapeutic efficacy in HBV-HCC, which warrants further investigation.
Since HBV-dysregulated miRNAs play a significant role in hepatocarcinogenesis, miRNAs can be used as viable alternative therapeutic targets. Despite the fact that miRNA delivery to specific locations is hampered by many challenges, several techniques, such as conjugation, virus-associated delivery, and nanoparticles, have been researched to improve the efficacy of miRNA delivery[8]. In fact, multiple miRNA-based therapeutics have entered the clinical phase of cancer therapy. The combination of miRNAs therapy with chemotherapy, radiotherapy, and immunotherapy has shown encouraging outcomes against different malignancies[215]. Unfortunately, there is no relevant clinical research on the use of miRNAs in the treatment of HCC. Given that miRNAs play a crucial part in the occurrence and progression of HBV-HCC, the approaches of combining diverse strategies, applying complementary miRNAs together, or inventing new forms of miRNAs may bring considerable clinical benefits for HBV-HCC patients. To reach the ultimate objective of enhancing patient OS and DFS, additional research is required in this area.
HBV dysregulates miRNAs in multiple ways, thereby contributing to the occurrence and progression of HCC. Consequently, miRNAs are anticipated to become HBV-HCC biomarkers for diagnosis and prognosis. miRNAs-based therapies may also improve the efficacy of HBV-HCC. More research is required for miRNA clinical transformation.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Korkmaz P, Turkey; Oura S, Japan; Quarleri J, Argentina S-Editor: Yan JP L-Editor: A P-Editor: Zhao S
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64627] [Article Influence: 16156.8] [Reference Citation Analysis (176)] |
| 2. | World Health Organization. Global hepatitis report 2017. Geneva: World Health Organization; 2017. |
| 3. | Piñero F, Dirchwolf M, Pessôa MG. Biomarkers in Hepatocellular Carcinoma: Diagnosis, Prognosis and Treatment Response Assessment. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 342] [Cited by in RCA: 330] [Article Influence: 66.0] [Reference Citation Analysis (0)] |
| 4. | Johnson P, Zhou Q, Dao DY, Lo YMD. Circulating biomarkers in the diagnosis and management of hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2022;19:670-681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 168] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 5. | Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3368] [Cited by in RCA: 4119] [Article Influence: 374.5] [Reference Citation Analysis (1)] |
| 6. | Sartorius K, Makarova J, Sartorius B, An P, Winkler C, Chuturgoon A, Kramvis A. The Regulatory Role of MicroRNA in Hepatitis-B Virus-Associated Hepatocellular Carcinoma (HBV-HCC) Pathogenesis. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 7. | Valihrach L, Androvic P, Kubista M. Circulating miRNA analysis for cancer diagnostics and therapy. Mol Aspects Med. 2020;72:100825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 126] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 8. | Ho PTB, Clark IM, Le LTT. MicroRNA-Based Diagnosis and Therapy. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 341] [Article Influence: 113.7] [Reference Citation Analysis (0)] |
| 9. | Liu AM, Yao TJ, Wang W, Wong KF, Lee NP, Fan ST, Poon RT, Gao C, Luk JM. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: a retrospective cohort study. BMJ Open. 2012;2:e000825. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 188] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 10. | Qi P, Cheng SQ, Wang H, Li N, Chen YF, Gao CF. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS One. 2011;6:e28486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 241] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 11. | Lin L, Lu B, Yu J, Liu W, Zhou A. Serum miR-224 as a biomarker for detection of hepatocellular carcinoma at early stage. Clin Res Hepatol Gastroenterol. 2016;40:397-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 12. | Zhao Q, Li T, Qi J, Liu J, Qin C. The miR-545/374a cluster encoded in the Ftx lncRNA is overexpressed in HBV-related hepatocellular carcinoma and promotes tumorigenesis and tumor progression. PLoS One. 2014;9:e109782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 13. | Yu F, Lu Z, Chen B, Dong P, Zheng J. microRNA-150: a promising novel biomarker for hepatitis B virus-related hepatocellular carcinoma. Diagn Pathol. 2015;10:129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 14. | Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol. 2010;5:1315-1316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1283] [Cited by in RCA: 2557] [Article Influence: 170.5] [Reference Citation Analysis (0)] |
| 15. | Zhou G, Zeng Y, Luo Y, Guo S, Bao L, Zhang Q. Urine miR-93-5p is a promising biomarker for early detection of HBV-related hepatocellular carcinoma. Eur J Surg Oncol. 2022;48:95-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 16. | Quoc NB, Phuong NDN, Ngan TK, Linh NTM, Cuong PH, Chau NNB. Expression of Plasma hsa-miR122 in HBV-Related Hepatocellular Carcinoma (HCC) in Vietnamese Patients. Microrna. 2018;7:92-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 17. | Xu L, Wei B, Hui H, Liu Y. Association of serum microRNA-125b and HBV-related hepatocellular carcinoma in Chinese Han patients. Int J Clin Exp Med. 2018;11:3699-3703. |
| 18. | Li X, Guo Y, Wang X, Ge A, Wang H, Fan K, Guo C. Clinical significance of serum miR-487b in HBV-related hepatocellular carcinoma and its potential mechanism. Infect Dis (Lond). 2021;53:546-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 19. | Cao X, Yang Q, Yu Q. Increased Expression of miR-487b Is Associated With Poor Prognosis and Tumor Progression of HBV-Related Hepatocellular Carcinoma. Open Forum Infect Dis. 2020;7:ofaa498. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 20. | Cao C, Wang C. Clinical significance of serum miR-768-3p in HBV-related hepatocellular carcinoma and its potential mechanism. Clin Exp Med. 2020;20:569-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 21. | Moradi N, Paryan M, Khansarinejad B, Sarmadian H, Mondanizadeh M. Plasma Level of miR-5193 As a Novel Biomarker for Diagnosis of HBV-Related Hepatocellular Carcinoma. Hepat Mon. 2019;In Press. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 22. | Tat Trung N, Duong DC, Tong HV, Hien TTT, Hoan PQ, Bang MH, Binh MT, Ky TD, Tung NL, Thinh NT, Sang VV, Thao LTP, Bock CT, Velavan TP, Meyer CG, Song LH, Toan NL. Optimisation of quantitative miRNA panels to consolidate the diagnostic surveillance of HBV-related hepatocellular carcinoma. PLoS One. 2018;13:e0196081. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 23. | Chen S, Chen H, Gao S, Qiu S, Zhou H, Yu M, Tu J. Differential expression of plasma microRNA-125b in hepatitis B virus-related liver diseases and diagnostic potential for hepatitis B virus-induced hepatocellular carcinoma. Hepatol Res. 2017;47:312-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 24. | Xie Y, Yao Q, Butt AM, Guo J, Tian Z, Bao X, Li H, Meng Q, Lu J. Expression profiling of serum microRNA-101 in HBV-associated chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Cancer Biol Ther. 2014;15:1248-1255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (1)] |
| 25. | Mjelle R, Dima SO, Bacalbasa N, Chawla K, Sorop A, Cucu D, Herlea V, Sætrom P, Popescu I. Comprehensive transcriptomic analyses of tissue, serum, and serum exosomes from hepatocellular carcinoma patients. BMC Cancer. 2019;19:1007. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 26. | Xia G, Xie Y, He Q. Hepatitis B Virus Affected Serum MicroRNA-203a Level in Hepatocellular Carcinoma. Clin Lab. 2021;67. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 27. | Bai PS, Hou P, Kong Y. Hepatitis B virus promotes proliferation and metastasis in male Chinese hepatocellular carcinoma patients through the LEF-1/miR-371a-5p/SRCIN1/pleiotrophin/Slug pathway. Exp Cell Res. 2018;370:174-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 28. | Zhou SJ, Deng YL, Liang HF, Jaoude JC, Liu FY. Hepatitis B virus X protein promotes CREB-mediated activation of miR-3188 and Notch signaling in hepatocellular carcinoma. Cell Death Differ. 2017;24:1577-1587. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 70] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 29. | Riazalhosseini B, Mohamed R, Apalasamy YD, Langmia IM, Mohamed Z. Circulating microRNA as a marker for predicting liver disease progression in patients with chronic hepatitis B. Rev Soc Bras Med Trop. 2017;50:161-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 30. | Zuo D, Chen L, Liu X, Wang X, Xi Q, Luo Y, Zhang N, Guo H. Combination of miR-125b and miR-27a enhances sensitivity and specificity of AFP-based diagnosis of hepatocellular carcinoma. Tumour Biol. 2016;37:6539-6549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 31. | Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, Shen HB, Zhang CY, Zen K. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70:9798-9807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 378] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 32. | Ghosh A, Ghosh A, Datta S, Dasgupta D, Das S, Ray S, Gupta S, Chowdhury A, Chatterjee R, Mohapatra SK, Banerjee S. Hepatic miR-126 is a potential plasma biomarker for detection of hepatitis B virus infected hepatocellular carcinoma. Int J Cancer. 2016;138:2732-2744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 33. | Meng FL, Wang W, Jia WD. Diagnostic and prognostic significance of serum miR-24-3p in HBV-related hepatocellular carcinoma. Med Oncol. 2014;31:177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 34. | Chen Y, Dong X, Yu D, Wang X. Serum miR-96 is a promising biomarker for hepatocellular carcinoma in patients with chronic hepatitis B virus infection. Int J Clin Exp Med. 2015;8:18462-18468. [PubMed] |
| 35. | Trung NT, Hoan NX, Trung PQ, Binh MT, Van Tong H, Toan NL, Bang MH, Song LH. Clinical significance of combined circulating TERT promoter mutations and miR-122 expression for screening HBV-related hepatocellular carcinoma. Sci Rep. 2020;10:8181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 36. | Chen SS, Chen H, Gao SS, Zhou H, Qiu SL, Yu MX, Tu JC. Differential expression of plasma miR-205 in HBV-related liver diseases and diagnostic potential for HBV-induced hepatocellular carcinoma. Wuhan Daxue Xuebao (Yixueban). 2016;37:445-450. |
| 37. | Caviglia GP, Abate ML, Gaia S, Petrini E, Bosco C, Olivero A, Rosso C, Ciancio A, Pellicano R, Saracco GM, Rizzetto M, Smedile A. Risk of hepatocellular carcinoma in HBV cirrhotic patients assessed by the combination of miR-122, AFP and PIVKA-II. Panminerva Med. 2017;59:283-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 38. | Tan Y, Ge G, Pan T, Wen D, Chen L, Yu X, Zhou X, Gan J. A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PLoS One. 2014;9:e107986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 134] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 39. | Luo P, Wu S, Yu Y, Ming X, Li S, Zuo X, Tu J. Current Status and Perspective Biomarkers in AFP Negative HCC: Towards Screening for and Diagnosing Hepatocellular Carcinoma at an Earlier Stage. Pathol Oncol Res. 2020;26:599-603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 156] [Article Influence: 26.0] [Reference Citation Analysis (1)] |
| 40. | Peng C, Li Z, Xie Z, Wang Z, Ye Y, Li B, Li W. The role of circulating microRNAs for the diagnosis of hepatitis B virus-associated hepatocellular carcinoma with low alpha-fetoprotein level: a systematic review and meta-analysis. BMC Gastroenterol. 2020;20:249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 41. | Zhang WT, Gil-Gómez A, Liu CH, Gao SS, Romero-Gómez M. Diagnostic accuracy of circulating microRNA in hepatitis B virus-related hepatocellular carcinoma: a meta-analysis based on Asian data. Rev Esp Enferm Dig. 2022;114:280-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 42. | Jin X, Cai C, Qiu Y. Diagnostic Value of Circulating microRNAs in Hepatitis B Virus-Related Hepatocellular Carcinoma: A Systematic Review and Meta-Analysis. J Cancer. 2019;10:4754-4764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 43. | Shi J, Liu J, Tu X, Li B, Tong Z, Wang T, Zheng Y, Shi H, Zeng X, Chen W, Yin W, Fang W. Single-cell immune signature for detecting early-stage HCC and early assessing anti-PD-1 immunotherapy efficacy. J Immunother Cancer. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 50] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 44. | Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, Wang JF, Zhang Z, Lu S, Huang X, Wang Z, Qiu S, Wang X, Yang G, Sun H, Tang Z, Wu Y, Zhu H, Fan J. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29:4781-4788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 498] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 45. | Sanz-Rubio D, Martin-Burriel I, Gil A, Cubero P, Forner M, Khalyfa A, Marin JM. Stability of Circulating Exosomal miRNAs in Healthy Subjects. Sci Rep. 2018;8:10306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 106] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 46. | Karamichali E, Foka P, Papadopoulou G, Loukaki-Gkountara D, Andresaki K, Koskinas I, Georgopoulou U. Hepatitis Viruses Control Host Immune Responses by Modifying the Exosomal Biogenesis Pathway and Cargo. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 47. | Liu Z, Li Y, Wang Y, Bai X, Zhang Y. Exosomes in HBV infection. Clin Chim Acta. 2023;538:65-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 48. | Li S, Li S, Wu S, Chen L. Exosomes Modulate the Viral Replication and Host Immune Responses in HBV Infection. Biomed Res Int. 2019;2019:2103943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 49. | Ghosh S, Bhowmik S, Majumdar S, Goswami A, Chakraborty J, Gupta S, Aggarwal S, Ray S, Chatterjee R, Bhattacharyya S, Dutta M, Datta S, Chowdhury A, Dhali GK, Banerjee S. The exosome encapsulated microRNAs as circulating diagnostic marker for hepatocellular carcinoma with low alpha-fetoprotein. Int J Cancer. 2020;147:2934-2947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 92] [Article Influence: 18.4] [Reference Citation Analysis (1)] |
| 50. | Wei XC, Liu LJ, Zhu F. Exosomes as potential diagnosis and treatment for liver cancer. World J Gastrointest Oncol. 2022;14:334-347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 15] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (2)] |
| 51. | Pandyarajan V, Govalan R, Yang JD. Risk Factors and Biomarkers for Chronic Hepatitis B Associated Hepatocellular Carcinoma. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 52. | Bandopadhyay M, Bharadwaj M. Exosomal miRNAs in hepatitis B virus related liver disease: a new hope for biomarker. Gut Pathog. 2020;12:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 53. | Xu J, An P, Winkler CA, Yu Y. Dysregulated microRNAs in Hepatitis B Virus-Related Hepatocellular Carcinoma: Potential as Biomarkers and Therapeutic Targets. Front Oncol. 2020;10:1271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 54. | Yuan LT, Lee WJ, Yang YC, Chen BR, Yang CY, Chen MW, Chen JQ, Hsiao M, Chien MH, Hua KT. Histone Methyltransferase G9a-Promoted Progression of Hepatocellular Carcinoma Is Targeted by Liver-Specific Hsa-miR-122. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 55. | Zhang Q, Xu HF, Song WY, Zhang PJ, Song YB. Potential microRNA panel for the diagnosis and prediction of overall survival of hepatocellular carcinoma with hepatitis B virus infection. World J Gastrointest Oncol. 2020;12:383-393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 1] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 56. | Dundar HZ, Aksoy F, Aksoy SA, Tasar P, Ugras N, Tunca B, Egeli U, Cecener G, Yerci O, Kaya E. Overexpression of miR-21 Is Associated With Recurrence in Patients With Hepatitis B Virus-Mediated Hepatocellular Carcinoma Undergoing Liver Transplantation. Transplant Proc. 2019;51:1157-1161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 57. | Mao K, Zhang J, He C, Xu K, Liu J, Sun J, Wu G, Tan C, Zeng Y, Wang J, Xiao Z. Restoration of miR-193b sensitizes Hepatitis B virus-associated hepatocellular carcinoma to sorafenib. Cancer Lett. 2014;352:245-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 58. | Qin YF, Zhou ZY, Fu HW, Lin HM, Xu LB, Wu WR, Liu C, Xu XL, Zhang R. Hepatitis B Virus Surface Antigen Promotes Stemness of Hepatocellular Carcinoma through Regulating MicroRNA-203a. J Clin Transl Hepatol. 2023;11:118-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 59. | Liu FY, Zhou SJ, Deng YL, Zhang ZY, Zhang EL, Wu ZB, Huang ZY, Chen XP. MiR-216b is involved in pathogenesis and progression of hepatocellular carcinoma through HBx-miR-216b-IGF2BP2 signaling pathway. Cell Death Dis. 2015;6:e1670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 100] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 60. | Zhou N, Wu J, Wang X, Sun Z, Han Q, Zhao L. Low-level expression of microRNA-375 predicts poor prognosis in hepatocellular carcinoma. Tumour Biol. 2016;37:2145-2152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 61. | Bai PS, Xia N, Sun H, Kong Y. Pleiotrophin, a target of miR-384, promotes proliferation, metastasis and lipogenesis in HBV-related hepatocellular carcinoma. J Cell Mol Med. 2017;21:3023-3043. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 62. | Zhen Y, Xinghui Z, Chao W, Yi Z, Jinwen C, Ruifang G, Chao Z, Min Z, Chunlei G, Yan F, Lingfang D, Long S, Wenzhi S, Xiaohe L, Rong X. Several microRNAs could predict survival in patients with hepatitis B-related liver cancer. Sci Rep. 2017;7:45195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 63. | Zhu HT, Dong QZ, Sheng YY, Wei JW, Wang G, Zhou HJ, Ren N, Jia HL, Ye QH, Qin LX. MicroRNA-29a-5p is a novel predictor for early recurrence of hepatitis B virus-related hepatocellular carcinoma after surgical resection. PLoS One. 2012;7:e52393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 64. | Yen CS, Su ZR, Lee YP, Liu IT, Yen CJ. miR-106b promotes cancer progression in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol. 2016;22:5183-5192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 41] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 65. | Dai X, Huang R, Hu S, Zhou Y, Sun X, Gui P, Yu Z, Zhou P. A novel miR-0308-3p revealed by miRNA-seq of HBV-positive hepatocellular carcinoma suppresses cell proliferation and promotes G1/S arrest by targeting double CDK6/Cyclin D1 genes. Cell Biosci. 2020;10:24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 66. | Chen Q, Yang SB, Zhang YW, Han SY, Jia L, Li B, Zhang Y, Zuo S. miR-3682-3p directly targets FOXO3 and stimulates tumor stemness in hepatocellular carcinoma via a positive feedback loop involving FOXO3/PI3K/AKT/c-Myc. World J Stem Cells. 2022;14:539-555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (1)] |
| 67. | Lin X, Zuo S, Luo R, Li Y, Yu G, Zou Y, Zhou Y, Liu Z, Liu Y, Hu Y, Xie Y, Fang W. HBX-induced miR-5188 impairs FOXO1 to stimulate β-catenin nuclear translocation and promotes tumor stemness in hepatocellular carcinoma. Theranostics. 2019;9:7583-7598. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 75] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 68. | Pratedrat P, Chuaypen N, Nimsamer P, Payungporn S, Pinjaroen N, Sirichindakul B, Tangkijvanich P. Diagnostic and prognostic roles of circulating miRNA-223-3p in hepatitis B virus-related hepatocellular carcinoma. PLoS One. 2020;15:e0232211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 69. | Zhu HT, Hasan AM, Liu RB, Zhang ZC, Zhang X, Wang J, Wang HY, Wang F, Shao JY. Serum microRNA profiles as prognostic biomarkers for HBV-positive hepatocellular carcinoma. Oncotarget. 2016;7:45637-45648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 70. | Niu LJ, Huang T, Wang L, Sun XF, Zhang YM. HBX suppresses PTEN to promote the malignant progression of hepatocellular carcinoma through mi-R155 activation. Ann Hepatol. 2022;27:100688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 71. | Wakasugi H, Takahashi H, Niinuma T, Kitajima H, Oikawa R, Matsumoto N, Takeba Y, Otsubo T, Takagi M, Ariizumi Y, Suzuki M, Okuse C, Iwabuchi S, Nakano M, Akutsu N, Kang JH, Matsui T, Yamada N, Sasaki H, Yamamoto E, Kai M, Sasaki Y, Sasaki S, Tanaka Y, Yotsuyanagi H, Tsutsumi T, Yamamoto H, Tokino T, Nakase H, Suzuki H, Itoh F. Dysregulation of miRNA in chronic hepatitis B is associated with hepatocellular carcinoma risk after nucleos(t)ide analogue treatment. Cancer Lett. 2018;434:91-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 72. | Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1784] [Cited by in RCA: 1823] [Article Influence: 91.2] [Reference Citation Analysis (0)] |
| 73. | Li CL, Yeh KH, Liu WH, Chen CL, Chen DS, Chen PJ, Yeh SH. Elevated p53 promotes the processing of miR-18a to decrease estrogen receptor-α in female hepatocellular carcinoma. Int J Cancer. 2015;136:761-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 74. | Kong XX, Lv YR, Shao LP, Nong XY, Zhang GL, Zhang Y, Fan HX, Liu M, Li X, Tang H. HBx-induced MiR-1269b in NF-κB dependent manner upregulates cell division cycle 40 homolog (CDC40) to promote proliferation and migration in hepatoma cells. J Transl Med. 2016;14:189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 75. | Chinnappan M, Singh AK, Kakumani PK, Kumar G, Rooge SB, Kumari A, Varshney A, Rastogi A, Sarin SK, Malhotra P, Mukherjee SK, Bhatnagar RK. Key elements of the RNAi pathway are regulated by hepatitis B virus replication and HBx acts as a viral suppressor of RNA silencing. Biochem J. 2014;462:347-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 76. | Liu AM, Zhang C, Burchard J, Fan ST, Wong KF, Dai H, Poon RT, Luk JM. Global regulation on microRNA in hepatitis B virus-associated hepatocellular carcinoma. OMICS. 2011;15:187-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 77. | Kitagawa N, Ojima H, Shirakihara T, Shimizu H, Kokubu A, Urushidate T, Totoki Y, Kosuge T, Miyagawa S, Shibata T. Downregulation of the microRNA biogenesis components and its association with poor prognosis in hepatocellular carcinoma. Cancer Sci. 2013;104:543-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 78. | Ren M, Qin D, Li K, Qu J, Wang L, Wang Z, Huang A, Tang H. Correlation between hepatitis B virus protein and microRNA processor Drosha in cells expressing HBV. Antiviral Res. 2012;94:225-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 79. | Shan X, Ren M, Chen K, Huang A, Tang H. Regulation of the microRNA processor DGCR8 by hepatitis B virus proteins via the transcription factor YY1. Arch Virol. 2015;160:795-803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 80. | Li J, Pu W, Sun HL, Zhou JK, Fan X, Zheng Y, He J, Liu X, Xia Z, Liu L, Wei YQ, Peng Y. Pin1 impairs microRNA biogenesis by mediating conformation change of XPO5 in hepatocellular carcinoma. Cell Death Differ. 2018;25:1612-1624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 81. | Pang R, Lee TK, Poon RT, Fan ST, Wong KB, Kwong YL, Tse E. Pin1 interacts with a specific serine-proline motif of hepatitis B virus X-protein to enhance hepatocarcinogenesis. Gastroenterology. 2007;132:1088-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 105] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 82. | Förstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287-297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 361] [Cited by in RCA: 336] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 83. | Zhang J, Jin H, Liu H, Lv S, Wang B, Wang R, Ding M, Yang Y, Li L, Zhang J, Fu S, Xie D, Wu M, Zhou W, Qian Q. MiRNA-99a directly regulates AGO2 through translational repression in hepatocellular carcinoma. Oncogenesis. 2014;3:e97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 84. | Hayes CN, Akamatsu S, Tsuge M, Miki D, Akiyama R, Abe H, Ochi H, Hiraga N, Imamura M, Takahashi S, Aikata H, Kawaoka T, Kawakami Y, Ohishi W, Chayama K. Hepatitis B virus-specific miRNAs and Argonaute2 play a role in the viral life cycle. PLoS One. 2012;7:e47490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 85. | Chen W, Bian H, Xie X, Yang X, Bi B, Li C, Zhang Y, Zhu Q, Song J, Qin C, Qi J. Negative feedback loop of ERK/CREB/miR-212-3p inhibits HBeAg-induced macrophage activation. J Cell Mol Med. 2020;24:10935-10945. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 86. | Atsaves V, Leventaki V, Rassidakis GZ, Claret FX. AP-1 Transcription Factors as Regulators of Immune Responses in Cancer. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 180] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 87. | Talotta F, Cimmino A, Matarazzo MR, Casalino L, De Vita G, D'Esposito M, Di Lauro R, Verde P. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene. 2009;28:73-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 196] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 88. | Tanaka Y, Kanai F, Ichimura T, Tateishi K, Asaoka Y, Guleng B, Jazag A, Ohta M, Imamura J, Ikenoue T, Ijichi H, Kawabe T, Isobe T, Omata M. The hepatitis B virus X protein enhances AP-1 activation through interaction with Jab1. Oncogene. 2006;25:633-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 79] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 89. | Shang J, Zheng Y, Guo X, Mo J, Xie X, Xiong Y, Liu Y, Wu K, Wu J. Hepatitis B virus replication and sex-determining region Y box 4 production are tightly controlled by a novel positive feedback mechanism. Sci Rep. 2015;5:10066. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 90. | Fu X, Ouyang Y, Mo J, Li R, Fu L, Peng S. Upregulation of microRNA-328-3p by hepatitis B virus contributes to THLE-2 cell injury by downregulating FOXO4. J Transl Med. 2020;18:143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 91. | Li CH, Xu F, Chow S, Feng L, Yin D, Ng TB, Chen Y. Hepatitis B virus X protein promotes hepatocellular carcinoma transformation through interleukin-6 activation of microRNA-21 expression. Eur J Cancer. 2014;50:2560-2569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 92. | Liu Y, Feng J, Sun M, Yang G, Yuan H, Wang Y, Bu Y, Zhao M, Zhang S, Zhang X. Long non-coding RNA HULC activates HBV by modulating HBx/STAT3/miR-539/APOBEC3B signaling in HBV-related hepatocellular carcinoma. Cancer Lett. 2019;454:158-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 93. | Huang JY, Chen HL, Shih C. MicroRNA miR-204 and miR-1236 inhibit hepatitis B virus replication via two different mechanisms. Sci Rep. 2016;6:34740. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 94. | Sun C, Lan P, Han Q, Huang M, Zhang Z, Xu G, Song J, Wang J, Wei H, Zhang J, Sun R, Zhang C, Tian Z. Oncofetal gene SALL4 reactivation by hepatitis B virus counteracts miR-200c in PD-L1-induced T cell exhaustion. Nat Commun. 2018;9:1241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 79] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 95. | Zhang Y, Ren H, Li J, Xue R, Liu H, Zhu Z, Pan C, Lin Y, Hu A, Gou P, Cai J, Zhou J, Zhu W, Shi X. Elevated HMGB1 expression induced by hepatitis B virus X protein promotes epithelial-mesenchymal transition and angiogenesis through STAT3/miR-34a/NF-κB in primary liver cancer. Am J Cancer Res. 2021;11:479-494. [PubMed] |
| 96. | Yi H, Zhang Y, Yang X, Li M, Hu H, Xiong J, Wang N, Jin J, Song Y, Wang X, Chen L, Lian J. Hepatitis B Core Antigen Impairs the Polarization While Promoting the Production of Inflammatory Cytokines of M2 Macrophages via the TLR2 Pathway. Front Immunol. 2020;11:535. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 97. | Fu L, Fu X, Mo J, Li X, Li R, Peng S. miR-146a-5p enhances hepatitis B virus replication through autophagy to promote aggravation of chronic hepatitis B. IUBMB Life. 2019;71:1336-1346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 98. | Zhang X, Liu S, Hu T, He Y, Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 286] [Article Influence: 17.9] [Reference Citation Analysis (1)] |
| 99. | Yin X, Sun S, Zhao J, Yang J, Lei X, Xu C, Li K. Rs4705342 polymorphism is involved in the tumorigenesis of HBV positive HCC by altering the binding affinity of HBV induced NF-kB with the promoter region of microRNA-143. J Cell Biochem. 2018;119:5233-5242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 100. | Li JF, Dai XP, Zhang W, Sun SH, Zeng Y, Zhao GY, Kou ZH, Guo Y, Yu H, Du LY, Jiang SB, Zhou YS. Upregulation of microRNA-146a by hepatitis B virus X protein contributes to hepatitis development by downregulating complement factor H. mBio. 2015;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 101. | Wang W, Bian H, Li F, Li X, Zhang D, Sun S, Song S, Zhu Q, Ren W, Qin C, Qi J. HBeAg induces the expression of macrophage miR-155 to accelerate liver injury via promoting production of inflammatory cytokines. Cell Mol Life Sci. 2018;75:2627-2641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 102. | Tao L, Xue D, Shen D, Ma W, Zhang J, Wang X, Zhang W, Wu L, Pan K, Yang Y, Nwosu ZC, Dooley S, Seki E, Liu C. MicroRNA-942 mediates hepatic stellate cell activation by regulating BAMBI expression in human liver fibrosis. Arch Toxicol. 2018;92:2935-2946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 103. | Li ZQ, Wang HY, Zeng QL, Yan JY, Hu YS, Li H, Yu ZJ. p65/miR-23a/CCL22 axis regulated regulatory T cells recruitment in hepatitis B virus positive hepatocellular carcinoma. Cancer Med. 2020;9:711-723. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 104. | Chen WS, Liu LC, Yen CJ, Chen YJ, Chen JY, Ho CY, Liu SH, Chen CC, Huang WC. Nuclear IKKα mediates microRNA-7/-103/107/21 inductions to downregulate maspin expression in response to HBx overexpression. Oncotarget. 2016;7:56309-56323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 105. | Huang JY, Chou SF, Lee JW, Chen HL, Chen CM, Tao MH, Shih C. MicroRNA-130a can inhibit hepatitis B virus replication via targeting PGC1α and PPARγ. RNA. 2015;21:385-400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 106. | Sarkar N, Panigrahi R, Pal A, Biswas A, Singh SP, Kar SK, Bandopadhyay M, Das D, Saha D, Kanda T, Sugiyama M, Chakrabarti S, Banerjee A, Chakravarty R. Expression of microRNA-155 correlates positively with the expression of Toll-like receptor 7 and modulates hepatitis B virus via C/EBP-β in hepatocytes. J Viral Hepat. 2015;22:817-827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 107. | Sepehri Z, Kiani Z, Alavian SM, Arababadi MK, Kennedy D. The link between TLR7 signaling and hepatitis B virus infection. Life Sci. 2016;158:63-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 108. | Mauvais-Jarvis F, Lange CA, Levin ER. Membrane-Initiated Estrogen, Androgen, and Progesterone Receptor Signaling in Health and Disease. Endocr Rev. 2022;43:720-742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 55] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 109. | Yang WJ, Chang CJ, Yeh SH, Lin WH, Wang SH, Tsai TF, Chen DS, Chen PJ. Hepatitis B virus X protein enhances the transcriptional activity of the androgen receptor through c-Src and glycogen synthase kinase-3beta kinase pathways. Hepatology. 2009;49:1515-1524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 90] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 110. | Han J, Ding L, Yuan B, Yang X, Wang X, Li J, Lu Q, Huang C, Ye Q. Hepatitis B virus X protein and the estrogen receptor variant lacking exon 5 inhibit estrogen receptor signaling in hepatoma cells. Nucleic Acids Res. 2006;34:3095-3106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 111. | Chen PJ, Yeh SH, Liu WH, Lin CC, Huang HC, Chen CL, Chen DS, Chen PJ. Androgen pathway stimulates microRNA-216a transcription to suppress the tumor suppressor in lung cancer-1 gene in early hepatocarcinogenesis. Hepatology. 2012;56:632-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 93] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 112. | Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43-50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1064] [Cited by in RCA: 1045] [Article Influence: 61.5] [Reference Citation Analysis (0)] |
| 113. | Wang J, Chen J, Liu Y, Zeng X, Wei M, Wu S, Xiong Q, Song F, Yuan X, Xiao Y, Cao Y, Li C, Chen L, Guo M, Shi YB, Sun G, Guo D. Hepatitis B Virus Induces Autophagy to Promote its Replication by the Axis of miR-192-3p-XIAP Through NF kappa B Signaling. Hepatology. 2019;69:974-992. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 114. | Wu G, Yu F, Xiao Z, Xu K, Xu J, Tang W, Wang J, Song E. Hepatitis B virus X protein downregulates expression of the miR-16 family in malignant hepatocytes in vitro. Br J Cancer. 2011;105:146-153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 115. | Wu G, Huang P, Ju X, Li Z, Wang Y. Lin28B over-expression mediates the repression of let-7 by hepatitis B virus X protein in hepatoma cells. Int J Clin Exp Med. 2015;8:15108-15116. [PubMed] |
| 116. | Jung YJ, Kim JW, Park SJ, Min BY, Jang ES, Kim NY, Jeong SH, Shin CM, Lee SH, Park YS, Hwang JH, Kim N, Lee DH. c-Myc-mediated overexpression of miR-17-92 suppresses replication of hepatitis B virus in human hepatoma cells. J Med Virol. 2013;85:969-978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 117. | Liu Y, Wang J, Chen J, Wu S, Zeng X, Xiong Q, Guo Y, Sun J, Song F, Xu J, Yuan S, Li C, He Y, Wang M, Chen L, Shi YB, Guo M, Guo D, Sun G. Upregulation of miR-520c-3p via hepatitis B virus drives hepatocellular migration and invasion by the PTEN/AKT/NF-κB axis. Mol Ther Nucleic Acids. 2022;29:47-63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 118. | Zhang W, Lu Z, Kong G, Gao Y, Wang T, Wang Q, Cai N, Wang H, Liu F, Ye L, Zhang X. Hepatitis B virus X protein accelerates hepatocarcinogenesis with partner survivin through modulating miR-520b and HBXIP. Mol Cancer. 2014;13:128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 119. | Chen W, Jiang J, Gong L, Shu Z, Xiang D, Zhang X, Bi K, Diao H. Hepatitis B virus P protein initiates glycolytic bypass in HBV-related hepatocellular carcinoma via a FOXO3/miRNA-30b-5p/MINPP1 axis. J Exp Clin Cancer Res. 2021;40:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 113] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 120. | Dong KS, Chen Y, Yang G, Liao ZB, Zhang HW, Liang HF, Chen XP, Dong HH. TGF-β1 accelerates the hepatitis B virus X-induced malignant transformation of hepatic progenitor cells by upregulating miR-199a-3p. Oncogene. 2020;39:1807-1820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 121. | Gao Q, Zhu H, Dong L, Shi W, Chen R, Song Z, Huang C, Li J, Dong X, Zhou Y, Liu Q, Ma L, Wang X, Zhou J, Liu Y, Boja E, Robles AI, Ma W, Wang P, Li Y, Ding L, Wen B, Zhang B, Rodriguez H, Gao D, Zhou H, Fan J. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell. 2019;179:561-577.e22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 617] [Article Influence: 123.4] [Reference Citation Analysis (0)] |
| 122. | Xu X, Fan Z, Kang L, Han J, Jiang C, Zheng X, Zhu Z, Jiao H, Lin J, Jiang K, Ding L, Zhang H, Cheng L, Fu H, Song Y, Jiang Y, Liu J, Wang R, Du N, Ye Q. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J Clin Invest. 2013;123:630-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 140] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 123. | Wu Q, Liu HO, Liu YD, Liu WS, Pan D, Zhang WJ, Yang L, Fu Q, Xu JJ, Gu JX. Decreased expression of hepatocyte nuclear factor 4α (Hnf4α)/microRNA-122 (miR-122) axis in hepatitis B virus-associated hepatocellular carcinoma enhances potential oncogenic GALNT10 protein activity. J Biol Chem. 2015;290:1170-1185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 84] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 124. | Hu XM, Yan XH, Hu YW, Huang JL, Cao SW, Ren TY, Tang YT, Lin L, Zheng L, Wang Q. miRNA-548p suppresses hepatitis B virus X protein associated hepatocellular carcinoma by downregulating oncoprotein hepatitis B x-interacting protein. Hepatol Res. 2016;46:804-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 125. | Dodurga Y, Yonguc GN, Avci CB, Bagci G, Gunduz C, Satiroglu-Tufan NL. Investigation of microRNA expression changes in HepG2 cell line in presence of URG4/URGCP and in absence of URG4/URGCP suppressed by RNA interference. Mol Biol Rep. 2012;39:11119-11124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 126. | Yuan K, Lian Z, Sun B, Clayton MM, Ng IO, Feitelson MA. Role of miR-148a in hepatitis B associated hepatocellular carcinoma. PLoS One. 2012;7:e35331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 127. | Mishra AK, Hossain MM, Umar M, Sata TN, Yadav AK, Sah AK, Ismail M, Nayak B, Shalimar, Venugopal SK. DDX3-mediated miR-34 expression inhibits autophagy and HBV replication in hepatic cells. J Viral Hepat. 2023;30:327-334. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 128. | Guerrieri F, Belloni L, D'Andrea D, Pediconi N, Le Pera L, Testoni B, Scisciani C, Floriot O, Zoulim F, Tramontano A, Levrero M. Genome-wide identification of direct HBx genomic targets. BMC Genomics. 2017;18:184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 129. | Kwok ZH, Zhang B, Chew XH, Chan JJ, Teh V, Yang H, Kappei D, Tay Y. Systematic Analysis of Intronic miRNAs Reveals Cooperativity within the Multicomponent FTX Locus to Promote Colon Cancer Development. Cancer Res. 2021;81:1308-1320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 130. | Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659-665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 581] [Cited by in RCA: 662] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 131. | Ge L, Zhang X, Hu S, Song Y, Kong J, Zhang B, Yang X. H19 suppresses the growth of hepatoblastoma cells by promoting their apoptosis via the signaling pathways of miR-675/FADD and miR-138/PTK2. J Cell Biochem. 2019;120:5218-5231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 132. | Liu Y, Xu L, Lu B, Zhao M, Li L, Sun W, Qiu Z, Zhang B. LncRNA H19/microRNA-675/PPARα axis regulates liver cell injury and energy metabolism remodelling induced by hepatitis B X protein via Akt/mTOR signalling. Mol Immunol. 2019;116:18-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 133. | Zhu Y, Lu Y, Zhang Q, Liu JJ, Li TJ, Yang JR, Zeng C, Zhuang SM. MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucleic Acids Res. 2012;40:4615-4625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 179] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 134. | Wang H, Luo J, He Q, Yao D, Wu J, Loor JJ. miR-26b promoter analysis reveals regulatory mechanisms by lipid-related transcription factors in goat mammary epithelial cells. J Dairy Sci. 2017;100:5837-5849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 135. | Zhao F, Xu G, Zhou Y, Wang L, Xie J, Ren S, Liu S, Zhu Y. MicroRNA-26b inhibits hepatitis B virus transcription and replication by targeting the host factor CHORDC1 protein. J Biol Chem. 2014;289:35029-35041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 136. | Piletič K, Kunej T. MicroRNA epigenetic signatures in human disease. Arch Toxicol. 2016;90:2405-2419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 220] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 137. | Park IY, Sohn BH, Yu E, Suh DJ, Chung YH, Lee JH, Surzycki SJ, Lee YI. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology. 2007;132:1476-1494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 246] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 138. | Yang Z, Li J, Feng G, Wang Y, Yang G, Liu Y, Zhang S, Feng J, Zhang X. Hepatitis B virus X protein enhances hepatocarcinogenesis by depressing the targeting of NUSAP1 mRNA by miR-18b. Cancer Biol Med. 2019;16:276-287. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 139. | Feng GX, Li J, Yang Z, Zhang SQ, Liu YX, Zhang WY, Ye LH, Zhang XD. Hepatitis B virus X protein promotes the development of liver fibrosis and hepatoma through downregulation of miR-30e targeting P4HA2 mRNA. Oncogene. 2017;36:6895-6905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 140. | Wei X, Tan C, Tang C, Ren G, Xiang T, Qiu Z, Liu R, Wu Z. Epigenetic repression of miR-132 expression by the hepatitis B virus x protein in hepatitis B virus-related hepatocellular carcinoma. Cell Signal. 2013;25:1037-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 94] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 141. | Zhang T, Zhang J, Cui M, Liu F, You X, Du Y, Gao Y, Zhang S, Lu Z, Ye L, Zhang X. Hepatitis B virus X protein inhibits tumor suppressor miR-205 through inducing hypermethylation of miR-205 promoter to enhance carcinogenesis. Neoplasia. 2013;15:1282-1291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 142. | Tsang DP, Wu WK, Kang W, Lee YY, Wu F, Yu Z, Xiong L, Chan AW, Tong JH, Yang W, Li MS, Lau SS, Li X, Lee SD, Yang Y, Lai PB, Yu DY, Xu G, Lo KW, Chan MT, Wang H, Lee TL, Yu J, Wong N, Yip KY, To KF, Cheng AS. Yin Yang 1-mediated epigenetic silencing of tumour-suppressive microRNAs activates nuclear factor-κB in hepatocellular carcinoma. J Pathol. 2016;238:651-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 143. | Zhang H, Huang C, Wang Y, Lu Z, Zhuang N, Zhao D, He J, Shi L. Hepatitis B Virus X Protein Sensitizes TRAIL-Induced Hepatocyte Apoptosis by Inhibiting the E3 Ubiquitin Ligase A20. PLoS One. 2015;10:e0127329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 144. | Ferrari KJ, Scelfo A, Jammula S, Cuomo A, Barozzi I, Stützer A, Fischle W, Bonaldi T, Pasini D. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell. 2014;53:49-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 364] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 145. | Cai Y, Zhang Y, Loh YP, Tng JQ, Lim MC, Cao Z, Raju A, Lieberman Aiden E, Li S, Manikandan L, Tergaonkar V, Tucker-Kellogg G, Fullwood MJ. H3K27me3-rich genomic regions can function as silencers to repress gene expression via chromatin interactions. Nat Commun. 2021;12:719. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 169] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 146. | Song K, Han C, Zhang J, Lu D, Dash S, Feitelson M, Lim K, Wu T. Epigenetic regulation of MicroRNA-122 by peroxisome proliferator activated receptor-gamma and hepatitis b virus X protein in hepatocellular carcinoma cells. Hepatology. 2013;58:1681-1692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 117] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 147. | Feng H, Yuan X, Wu S, Yuan Y, Cui L, Lin D, Peng X, Liu X, Wang F. Effects of writers, erasers and readers within miRNA-related m6A modification in cancers. Cell Prolif. 2023;56:e13340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 32] [Reference Citation Analysis (0)] |
| 148. | Cheng D, Wu C, Li Y, Liu Y, Mo J, Fu L, Peng S. METTL3 inhibition ameliorates liver damage in mouse with hepatitis B virus-associated acute-on-chronic liver failure by regulating miR-146a-5p maturation. Biochim Biophys Acta Gene Regul Mech. 2022;1865:194782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 149. | D'Ambrogio A, Gu W, Udagawa T, Mello CC, Richter JD. Specific miRNA stabilization by Gld2-catalyzed monoadenylation. Cell Rep. 2012;2:1537-1545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 150. | Peng F, Xiao X, Jiang Y, Luo K, Tian Y, Peng M, Zhang M, Xu Y, Gong G. HBx down-regulated Gld2 plays a critical role in HBV-related dysregulation of miR-122. PLoS One. 2014;9:e92998. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 151. | Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1116] [Cited by in RCA: 1590] [Article Influence: 176.7] [Reference Citation Analysis (0)] |
| 152. | Lv J, Fan HX, Zhao XP, Lv P, Fan JY, Zhang Y, Liu M, Tang H. Long non-coding RNA Unigene56159 promotes epithelial-mesenchymal transition by acting as a ceRNA of miR-140-5p in hepatocellular carcinoma cells. Cancer Lett. 2016;382:166-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 111] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 153. | Feng J, Yang G, Liu Y, Gao Y, Zhao M, Bu Y, Yuan H, Yuan Y, Yun H, Sun M, Gao H, Zhang S, Liu Z, Yin M, Song X, Miao Z, Lin Z, Zhang X. LncRNA PCNAP1 modulates hepatitis B virus replication and enhances tumor growth of liver cancer. Theranostics. 2019;9:5227-5245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 132] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 154. | Fan H, Lv P, Mu T, Zhao X, Liu Y, Feng Y, Lv J, Liu M, Tang H. LncRNA n335586/miR-924/CKMT1A axis contributes to cell migration and invasion in hepatocellular carcinoma cells. Cancer Lett. 2018;429:89-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 155. | Li L, Han T, Liu K, Lei CG, Wang ZC, Shi GJ. LncRNA H19 promotes the development of hepatitis B related hepatocellular carcinoma through regulating microRNA-22 via EMT pathway. Eur Rev Med Pharmacol Sci. 2019;23:5392-5401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 156. | Song W, Zheng C, Liu M, Xu Y, Qian Y, Zhang Z, Su H, Li X, Wu H, Gong P, Li Y, Fan H. TRERNA1 upregulation mediated by HBx promotes sorafenib resistance and cell proliferation in HCC via targeting NRAS by sponging miR-22-3p. Mol Ther. 2021;29:2601-2616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 157. | He B, Peng F, Li W, Jiang Y. Interaction of lncRNA-MALAT1 and miR-124 regulates HBx-induced cancer stem cell properties in HepG2 through PI3K/Akt signaling. J Cell Biochem. 2019;120:2908-2918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 158. | Zhuang H, Ma X, Liu X, Li C, Li X, Wu L, Wen M, Shi W, Yang X. Hyaluronan-mediated motility receptor antisense RNA 1 promotes hepatitis B virus-related hepatocellular carcinoma progression by regulating miR-627-3p/High Mobility Group AT-hook 2 axis. Bioengineered. 2022;13:8617-8630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 159. | Huang P, Xu Q, Yan Y, Lu Y, Hu Z, Ou B, Zhang H, Mao K, Zhang J, Wang J, Xiao Z. HBx/ERα complex-mediated LINC01352 downregulation promotes HBV-related hepatocellular carcinoma via the miR-135b-APC axis. Oncogene. 2020;39:3774-3789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 160. | Deng Y, Wei Z, Huang M, Xu G, Wei W, Peng B, Nong S, Qin H. Long non-coding RNA F11-AS1 inhibits HBV-related hepatocellular carcinoma progression by regulating NR1I3 via binding to microRNA-211-5p. J Cell Mol Med. 2020;24:1848-1865. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 161. | Wang J, Yin G, Bian H, Yang J, Zhou P, Yan K, Liu C, Chen P, Zhu J, Li Z, Xue T. LncRNA XIST upregulates TRIM25 via negatively regulating miR-192 in hepatitis B virus-related hepatocellular carcinoma. Mol Med. 2021;27:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 162. | Guo L, Gao S, Sun W, Wang Y, Zhao J. Elevated LINC01232 is associated with poor prognosis and HBV infection in hepatocellular carcinoma patients and contributes to tumor progression in vitro. Clin Res Hepatol Gastroenterol. 2022;46:101813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 163. | Cheng Y, Shi W, Cui X, Sun L, Nan Y, Yao H, Fan J, Zhu L, Yu L. Long Noncoding RNA TFAP2A-AS1 Suppressed Hepatitis B Virus Replication by Modulating miR-933/HDAC11. Dis Markers. 2022;2022:7733390. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 164. | Yu K, Mei Y, Wang Z, Liu B, Deng M. LncRNA LINC00924 upregulates NDRG2 to inhibit epithelial-mesenchymal transition via sponging miR-6755-5p in hepatitis B virus-related hepatocellular carcinoma. J Med Virol. 2022;94:2702-2713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 165. | Wei Y, Tang X, Ren Y, Yang Y, Song F, Fu J, Liu S, Yu M, Chen J, Wang S, Zhang K, Tan Y, Han Z, Wei L, Zhang B, Cheng Z, Li L, Wang H. An RNA-RNA crosstalk network involving HMGB1 and RICTOR facilitates hepatocellular carcinoma tumorigenesis by promoting glutamine metabolism and impedes immunotherapy by PD-L1+ exosomes activity. Signal Transduct Target Ther. 2021;6:421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 69] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 166. | Chen S, Dong Z, Yang P, Wang X, Jin G, Yu H, Chen L, Li L, Tang L, Bai S, Yan H, Shen F, Cong W, Wen W, Wang H. Hepatitis B virus X protein stimulates high mobility group box 1 secretion and enhances hepatocellular carcinoma metastasis. Cancer Lett. 2017;394:22-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 167. | Rao X, Lai L, Li X, Wang L, Li A, Yang Q. N(6) -methyladenosine modification of circular RNA circ-ARL3 facilitates Hepatitis B virus-associated hepatocellular carcinoma via sponging miR-1305. IUBMB Life. 2021;73:408-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 63] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 168. | Chen Y, Li S, Wei Y, Xu Z, Wu X. Circ-RNF13, as an oncogene, regulates malignant progression of HBV-associated hepatocellular carcinoma cells and HBV infection through ceRNA pathway of circ-RNF13/miR-424-5p/TGIF2. Bosn J Basic Med Sci. 2021;21:555-568. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 169. | Du N, Li K, Wang Y, Song B, Zhou X, Duan S. CircRNA circBACH1 facilitates hepatitis B virus replication and hepatoma development by regulating the miR-200a-3p/MAP3K2 axis. Histol Histopathol. 2022;37:863-877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 170. | Jiang W, Wang L, Zhang Y, Li H. Circ-ATP5H Induces Hepatitis B Virus Replication and Expression by Regulating miR-138-5p/TNFAIP3 Axis. Cancer Manag Res. 2020;12:11031-11040. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 171. | He W, Zhu X, Tang X, Xiang X, Yu J, Sun H. Circ_0027089 regulates NACC1 by targeting miR-136-5p to aggravate the development of hepatitis B virus-related hepatocellular carcinoma. Anticancer Drugs. 2022;33:e336-e348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 172. | Wang Y, Jiang L, Ji X, Yang B, Zhang Y, Fu XD. Hepatitis B viral RNA directly mediates down-regulation of the tumor suppressor microRNA miR-15a/miR-16-1 in hepatocytes. J Biol Chem. 2013;288:18484-18493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 173. | Liu N, Zhang J, Jiao T, Li Z, Peng J, Cui Z, Ye X. Hepatitis B virus inhibits apoptosis of hepatoma cells by sponging the MicroRNA 15a/16 cluster. J Virol. 2013;87:13370-13378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 174. | Li C, Wang Y, Wang S, Wu B, Hao J, Fan H, Ju Y, Ding Y, Chen L, Chu X, Liu W, Ye X, Meng S. Hepatitis B virus mRNA-mediated miR-122 inhibition upregulates PTTG1-binding protein, which promotes hepatocellular carcinoma tumor growth and cell invasion. J Virol. 2013;87:2193-2205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 120] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 175. | Deng M, Hou J, Hu J, Wang S, Chen M, Chen L, Ju Y, Li C, Meng S. Hepatitis B virus mRNAs functionally sequester let-7a and enhance hepatocellular carcinoma. Cancer Lett. 2016;383:62-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 176. | Takata A, Otsuka M, Ohno M, Kishikawa T, Yoshikawa T, Koike K. Mutual antagonism between hepatitis B viral mRNA and host microRNA let-7. Sci Rep. 2016;6:23237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 177. | Ochi M, Otsuka M, Maruyama R, Koike K. HBx increases EGFR expression by inhibiting miR129-5p function. Biochem Biophys Res Commun. 2020;529:198-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 178. | Lau CC, Sun T, Ching AK, He M, Li JW, Wong AM, Co NN, Chan AW, Li PS, Lung RW, Tong JH, Lai PB, Chan HL, To KF, Chan TF, Wong N. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell. 2014;25:335-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 227] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 179. | Liang HW, Wang N, Wang Y, Wang F, Fu Z, Yan X, Zhu H, Diao W, Ding Y, Chen X, Zhang CY, Zen K. Hepatitis B virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. J Hepatol. 2016;64:278-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 101] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 180. | Levine B, Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell. 2019;176:11-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1024] [Cited by in RCA: 2062] [Article Influence: 343.7] [Reference Citation Analysis (0)] |
| 181. | Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ, Tsai TF, Lin YJ, Wu CT, Liu HS. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology. 2014;59:505-517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 163] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 182. | Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P, Voinnet O. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 2012;14:1314-1321. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 205] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 183. | Ma NF, Lau SH, Hu L, Xie D, Wu J, Yang J, Wang Y, Wu MC, Fung J, Bai X, Tzang CH, Fu L, Yang M, Su YA, Guan XY. COOH-terminal truncated HBV X protein plays key role in hepatocarcinogenesis. Clin Cancer Res. 2008;14:5061-5068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 138] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 184. | Minarovits J, Niller HH. Truncated oncoproteins of retroviruses and hepatitis B virus: A lesson in contrasts. Infect Genet Evol. 2019;73:342-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 185. | Liu F, You X, Chi X, Wang T, Ye L, Niu J, Zhang X. Hepatitis B virus X protein mutant HBxΔ127 promotes proliferation of hepatoma cells through up-regulating miR-215 targeting PTPRT. Biochem Biophys Res Commun. 2014;444:128-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 186. | Yip WK, Cheng AS, Zhu R, Lung RW, Tsang DP, Lau SS, Chen Y, Sung JG, Lai PB, Ng EK, Yu J, Wong N, To KF, Wong VW, Sung JJ, Chan HL. Carboxyl-terminal truncated HBx regulates a distinct microRNA transcription program in hepatocellular carcinoma development. PLoS One. 2011;6:e22888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 187. | Hou ZH, Han QJ, Zhang C, Tian ZG, Zhang J. miR146a impairs the IFN-induced anti-HBV immune response by downregulating STAT1 in hepatocytes. Liver Int. 2014;34:58-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 188. | Su IJ, Wang LH, Hsieh WC, Wu HC, Teng CF, Tsai HW, Huang W. The emerging role of hepatitis B virus pre-S2 deletion mutant proteins in HBV tumorigenesis. J Biomed Sci. 2014;21:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 189. | Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol. 2016;64:S84-S101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 479] [Cited by in RCA: 710] [Article Influence: 78.9] [Reference Citation Analysis (0)] |
| 190. | Zhang Y, Huang B, Wang HY, Chang A, Zheng XFS. Emerging Role of MicroRNAs in mTOR Signaling. Cell Mol Life Sci. 2017;74:2613-2625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 78] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 191. | Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537-2545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 876] [Cited by in RCA: 896] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 192. | Feitelson MA, Lee J. Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett. 2007;252:157-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 179] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 193. | Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999-3004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2908] [Cited by in RCA: 3110] [Article Influence: 148.1] [Reference Citation Analysis (0)] |
| 194. | Yang L, Ma Z, Wang D, Zhao W, Chen L, Wang G. MicroRNA-602 regulating tumor suppressive gene RASSF1A is overexpressed in hepatitis B virus-infected liver and hepatocellular carcinoma. Cancer Biol Ther. 2010;9:803-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 73] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 195. | Guo H, Liu H, Mitchelson K, Rao H, Luo M, Xie L, Sun Y, Zhang L, Lu Y, Liu R, Ren A, Liu S, Zhou S, Zhu J, Zhou Y, Huang A, Wei L, Guo Y, Cheng J. MicroRNAs-372/373 promote the expression of hepatitis B virus through the targeting of nuclear factor I/B. Hepatology. 2011;54:808-819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 196. | Zhang B, Han S, Feng B, Chu X, Chen L, Wang R. Hepatitis B virus X protein-mediated non-coding RNA aberrations in the development of human hepatocellular carcinoma. Exp Mol Med. 2017;49:e293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 197. | Sartorius K, Swadling L, An P, Makarova J, Winkler C, Chuturgoon A, Kramvis A. The Multiple Roles of Hepatitis B Virus X Protein (HBx) Dysregulated MicroRNA in Hepatitis B Virus-Associated Hepatocellular Carcinoma (HBV-HCC) and Immune Pathways. Viruses. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 198. | Sartorius K, An P, Winkler C, Chuturgoon A, Li X, Makarova J, Kramvis A. The Epigenetic Modulation of Cancer and Immune Pathways in Hepatitis B Virus-Associated Hepatocellular Carcinoma: The Influence of HBx and miRNA Dysregulation. Front Immunol. 2021;12:661204. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 199. | Xie KL, Zhang YG, Liu J, Zeng Y, Wu H. MicroRNAs associated with HBV infection and HBV-related HCC. Theranostics. 2014;4:1176-1192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 200. | Lee TK, Guan XY, Ma S. Cancer stem cells in hepatocellular carcinoma - from origin to clinical implications. Nat Rev Gastroenterol Hepatol. 2022;19:26-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 289] [Article Influence: 96.3] [Reference Citation Analysis (0)] |
| 201. | Zhang Z, Han Y, Sun G, Liu X, Jia X, Yu X. MicroRNA-325-3p inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma by down-regulation of aquaporin 5. Cell Mol Biol Lett. 2019;24:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 202. | Li R, Xu T, Wang H, Wu N, Liu F, Jia X, Mi J, Lv J, Gao H. Dysregulation of the miR-325-3p/DPAGT1 axis supports HBV-positive HCC chemoresistance. Biochem Biophys Res Commun. 2019;519:358-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 203. | Feng J, Li J, Wu L, Yu Q, Ji J, Wu J, Dai W, Guo C. Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39:126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 416] [Cited by in RCA: 419] [Article Influence: 83.8] [Reference Citation Analysis (0)] |
| 204. | Sangineto M, Villani R, Cavallone F, Romano A, Loizzi D, Serviddio G. Lipid Metabolism in Development and Progression of Hepatocellular Carcinoma. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 108] [Article Influence: 21.6] [Reference Citation Analysis (1)] |
| 205. | You X, Liu F, Zhang T, Li Y, Ye L, Zhang X. Hepatitis B virus X protein upregulates oncogene Rab18 to result in the dysregulation of lipogenesis and proliferation of hepatoma cells. Carcinogenesis. 2013;34:1644-1652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 206. | Cui M, Wang Y, Sun B, Xiao Z, Ye L, Zhang X. MiR-205 modulates abnormal lipid metabolism of hepatoma cells via targeting acyl-CoA synthetase long-chain family member 1 (ACSL1) mRNA. Biochem Biophys Res Commun. 2014;444:270-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 89] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 207. | Chen Y, Tian Z. HBV-Induced Immune Imbalance in the Development of HCC. Front Immunol. 2019;10:2048. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 189] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 208. | Liu W, Zheng X, Wang J, He Q, Li J, Zhang Z, Liu H. MicroRNA-138 Regulates T-Cell Function by Targeting PD-1 in Patients with Hepatitis B Virus-Related Liver Diseases. Lab Med. 2021;52:439-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 209. | Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, Deng Y, Zhao J, Jiang S, Yuan Y, Wang HY, Cheng SQ, Xie D, Wang XF. TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22:291-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 451] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 210. | Bian X, Si Y, Zhang M, Wei R, Yang X, Ren H, Zheng G, Wang C, Zhang Y. Down-expression of miR-152 lead to impaired anti-tumor effect of NK via upregulation of HLA-G. Tumour Biol. 2016;37:3749-3756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 211. | Sadri Nahand J, Rabiei N, Fathazam R, Taghizadieh M, Ebrahimi MS, Mahjoubin-Tehran M, Bannazadeh Baghi H, Khatami A, Abbasi-Kolli M, Mirzaei HR, Rahimian N, Darvish M, Mirzaei H. Oncogenic viruses and chemoresistance: What do we know? Pharmacol Res. 2021;170:105730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 212. | Wei XC, Xia YR, Zhou P, Xue X, Ding S, Liu LJ, Zhu F. Hepatitis B core antigen modulates exosomal miR-135a to target vesicle-associated membrane protein 2 promoting chemoresistance in hepatocellular carcinoma. World J Gastroenterol. 2021;27:8302-8322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 23] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (1)] |
| 213. | Chen WS, Yen CJ, Chen YJ, Chen JY, Wang LY, Chiu SJ, Shih WL, Ho CY, Wei TT, Pan HL, Chien PH, Hung MC, Chen CC, Huang WC. miRNA-7/21/107 contribute to HBx-induced hepatocellular carcinoma progression through suppression of maspin. Oncotarget. 2015;6:25962-25974. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 214. | Zhang C, Liu P, Zhang C. Hepatitis B virus X protein upregulates alpha-fetoprotein to promote hepatocellular carcinoma by targeting miR-1236 and miR-329. J Cell Biochem. 2020;121:2489-2499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 215. | He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi S, Xie H, Peng X, Yin W, Tao Y, Wang X. miRNA-based biomarkers, therapies, and resistance in Cancer. Int J Biol Sci. 2020;16:2628-2647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 310] [Cited by in RCA: 392] [Article Influence: 78.4] [Reference Citation Analysis (0)] |
| 216. | Lamontagne J, Steel LF, Bouchard MJ. Hepatitis B virus and microRNAs: Complex interactions affecting hepatitis B virus replication and hepatitis B virus-associated diseases. World J Gastroenterol. 2015;21:7375-7399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 49] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (1)] |
| 217. | Mosca N, Castiello F, Coppola N, Trotta MC, Sagnelli C, Pisaturo M, Sagnelli E, Russo A, Potenza N. Functional interplay between hepatitis B virus X protein and human miR-125a in HBV infection. Biochem Biophys Res Commun. 2014;449:141-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 218. | Potenza N, Papa U, Mosca N, Zerbini F, Nobile V, Russo A. Human microRNA hsa-miR-125a-5p interferes with expression of hepatitis B virus surface antigen. Nucleic Acids Res. 2011;39:5157-5163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 136] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 219. | Wei X, Xiang T, Ren G, Tan C, Liu R, Xu X, Wu Z. miR-101 is down-regulated by the hepatitis B virus x protein and induces aberrant DNA methylation by targeting DNA methyltransferase 3A. Cell Signal. 2013;25:439-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 123] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 220. | Liu Y, Lou G, Wu W, Zheng M, Shi Y, Zhao D, Chen Z. Involvement of the NF-κB pathway in multidrug resistance induced by HBx in a hepatoma cell line. J Viral Hepat. 2011;18:e439-e446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 221. | Hu Z, Yin Y, Jiang J, Yan C, Wang Y, Wang D, Li L. Exosomal miR-142-3p secreted by hepatitis B virus (HBV)-hepatocellular carcinoma (HCC) cells promotes ferroptosis of M1-type macrophages through SLC3A2 and the mechanism of HCC progression. J Gastrointest Oncol. 2022;13:754-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 45] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 222. | Zhang Q, Qu Y, Zhang Q, Li F, Li B, Li Z, Dong Y, Lu L, Cai X. Exosomes derived from hepatitis B virus-infected hepatocytes promote liver fibrosis via miR-222/TFRC axis. Cell Biol Toxicol. 2023;39:467-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 72] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 223. | Li CH, Wang YJ, Dong W, Xiang S, Liang HF, Wang HY, Dong HH, Chen L, Chen XP. Hepatic oval cell lines generate hepatocellular carcinoma following transfection with HBx gene and treatment with aflatoxin B1 in vivo. Cancer Lett. 2011;311:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 224. | Chandrasekaran AR, MacIsaac M, Dey P, Levchenko O, Zhou L, Andres M, Dey BK, Halvorsen K. Cellular microRNA detection with miRacles: microRNA- activated conditional looping of engineered switches. Sci Adv. 2019;5:eaau9443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 225. | Guo T, Tang XH, Gao XY, Zhou Y, Jin B, Deng ZQ, Hu Y, Xing XF, Li ZY, Ji JF. A liquid biopsy signature of circulating exosome-derived mRNAs, miRNAs and lncRNAs predict therapeutic efficacy to neoadjuvant chemotherapy in patients with advanced gastric cancer. Mol Cancer. 2022;21:216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 43] [Reference Citation Analysis (0)] |
| 226. | To NH, Nguyen HQ, Thiolat A, Liu B, Cohen J, Radosevic-Robin N, Belkacemi Y; TransAtlantic Radiation Oncology Network (TRONE) & Association of Radiotherapy, and Oncology of the Mediterranean Area (AROME). Radiation therapy for triple-negative breast cancer: emerging role of microRNAs as biomarkers and radiosensitivity modifiers. A systematic review. Breast Cancer Res Treat. 2022;193:265-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 227. | Peng XX, Yu R, Wu X, Wu SY, Pi C, Chen ZH, Zhang XC, Gao CY, Shao YW, Liu L, Wu YL, Zhou Q. Correlation of plasma exosomal microRNAs with the efficacy of immunotherapy in EGFR/ALK wild-type advanced non-small cell lung cancer. J Immunother Cancer. 2020;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 136] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 228. | Li L, Guo Z, Wang J, Mao Y, Gao Q. Serum miR-18a: a potential marker for hepatitis B virus-related hepatocellular carcinoma screening. Dig Dis Sci. 2012;57:2910-2916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 83] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 229. | Zhao Q, Sun X, Liu C, Li T, Cui J, Qin C. Expression of the microRNA-143/145 cluster is decreased in hepatitis B virus-associated hepatocellular carcinoma and may serve as a biomarker for tumorigenesis in patients with chronic hepatitis B. Oncol Lett. 2018;15:6115-6122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 230. | Zhang Y, Xi H, Nie X, Zhang P, Lan N, Lu Y, Liu J, Yuan W. Assessment of miR-212 and Other Biomarkers in the Diagnosis and Treatment of HBV-infection-related Liver Diseases. Curr Drug Metab. 2019;20:785-798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 231. | Wen Y, Han J, Chen J, Dong J, Xia Y, Liu J, Jiang Y, Dai J, Lu J, Jin G, Wei Q, Shen H, Sun B, Hu Z. Plasma miRNAs as early biomarkers for detecting hepatocellular carcinoma. Int J Cancer. 2015;137:1679-1690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 173] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 232. | Xiong F, Ma H, Qu Y, Wen F, Bao X, Han D, Lu J. Profiles of serum miR-99a, let-7c and miR-125b in hepatitis B virus (HBV)-associated chronic hepatitis, liver cirrhosis and hepatocellular carcinoma. Int J Clin Exp Pathol. 2016;9:7087-7095. |
| 233. | Hung CH, Hu TH, Lu SN, Kuo FY, Chen CH, Wang JH, Huang CM, Lee CM, Lin CY, Yen YH, Chiu YC. Circulating microRNAs as biomarkers for diagnosis of early hepatocellular carcinoma associated with hepatitis B virus. Int J Cancer. 2016;138:714-720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 234. | Zhang W, Fu T, Guo Z, Zhang Y, Zhang L, Su H, Long Y, Ji Z, Yan Y, Shao Z. Serum miR-375 Levels Are Closely Related to Disease Progression from HBV Infection to HBV-Related Hepatocellular Carcinoma. Biomed Res Int. 2020;2020:5819385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 235. | Zhu HT, Liu RB, Liang YY, Hasan AME, Wang HY, Shao Q, Zhang ZC, Wang J, He CY, Wang F, Shao JY. Serum microRNA profiles as diagnostic biomarkers for HBV-positive hepatocellular carcinoma. Liver Int. 2017;37:888-896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |