Published online Aug 7, 2023. doi: 10.3748/wjg.v29.i29.4528
Peer-review started: April 5, 2023
First decision: May 23, 2023
Revised: June 11, 2023
Accepted: July 4, 2023
Article in press: July 4, 2023
Published online: August 7, 2023
Processing time: 118 Days and 21.8 Hours
Obesity plays a vital role in the occurrence and development of non-alcoholic steatohepatitis (NASH). However, the underlining mechanism is still unclear, where adipose tissue (AT) derived exosomes may actively participate. MicroRNAs (miRNAs) are commonly secreted from exosomes for cell commu
To determine the specific role of AT-derived exosomes miR-103 in developing NASH through various methods.
The expression levels of miR-103 in the AT-derived exosomes and livers were detected and compared between NASH mice and control. The effect of miR-103 on NASH progression was also explored by antagonizing miR-103, including steatosis and inflammation degree changes. The interaction between miR-103 and the autophagy-related gene phosphatase and tensin homolog (PTEN) was confirmed by dual-luciferase reporter assay. The role of the interaction between miR-103 and PTEN on autophagy was verified in NASH-like cells. Finally, the effects of miR-103 from adipose-derived exosomes on NASH and autophagy were analyzed through animal experiments.
The expression of miR-103 was increased in NASH mice, compared to the control, and inhibition of miR-103 could alleviate NASH. The results of the dual-luciferase reporter assay showed miR-103 could interact with PTEN. MiR-103-anta decreased p-AMPKa, p-mammalian target of rapamycin (mTOR), and p62 but increased the protein levels of PTEN and LC3-II/I and the number of autophagosomes in NASH mice. Similar results were also observed in NASH-like cells, and further experiments showed PTEN silencing inhibited the effect of miR-103-anta. AT derived-exosome miR-103 aggravated NASH and increased the expressions of p-AMPKa, p-mTOR, and p62 but decreased the protein levels of PTEN and LC3-II/I and the number of autophagosomes in mice.
AT derived-exosome increased the levels of miR-103 in the liver, and miR-103 aggravated NASH. Mechanically, miR-103 could interact with PTEN and inhibit autophagy.
Core Tip: Our study confirms the important role of miR-103-phosphatase and tensin homolog-autophagy axis in the pathogenesis of non-alcoholic steatohepatitis (NASH). More importantly, the elevation of miR-103 in the liver of NASH mice is partly due to adipose tissue exosome secretion and integration, which also partially explains the mechanism of obesity leading to NASH.
- Citation: Lu MM, Ren Y, Zhou YW, Xu LL, Zhang MM, Ding LP, Cheng WX, Jin X. Antagonizing adipose tissue-derived exosome miR-103-hepatocyte phosphatase and tensin homolog pathway alleviates autophagy in non-alcoholic steatohepatitis: A trans-cellular crosstalk. World J Gastroenterol 2023; 29(29): 4528-4541
- URL: https://www.wjgnet.com/1007-9327/full/v29/i29/4528.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i29.4528
Non-alcoholic fatty liver disease (NAFLD) is recognized as the hepatocellular manifestation of metabolic syndrome, characterized by hepatic lipid accumulation and inflammation, and precluded with secondary causes, such as chronic viral hepatitis, significant alcohol consumption, long-term use of steatogenic medication, and other chronic liver diseases including autoimmune hepatitis, hemochromatosis, Wilson’s disease[1]. NAFLD is categorized into NAFL, non-alcoholic steatohepatitis (NASH), fibrosis, and even cirrhosis according to histological changes in different disease stages. Among them, NASH is considered the watershed in NAFLD and is defined as the presence of 5% hepatic steatosis and ensuing hepatocyte injury[1]. According to a previous meta-analysis, the pooled overall global prevalence of NAFLD was estimated to be 25.24% [95% confidence interval (CI): 22.10-28.65], while the pooled overall NASH prevalence among biopsied NAFLD patients was 59.10% (95%CI: 47.55-69.73)[2]. Moreover, fibrosis, which is closely related to liver cirrhosis, liver cancer, and other end-stage liver diseases, is more likely to occur in NASH patients than NAFL patients[3]. Those findings support the importance of NASH and suggest that the burden of disease caused by NASH needs to be paid adequate attention. NASH has become one of the leading causes of cirrhosis and the second leading cause of liver transplantation in the United States[4,5].
NAFLD is commonly associated with metabolic comorbidities such as obesity, diabetes mellitus, and dyslipidemia[2,6]. The prevalence of obesity is as high as 51.34% (95%CI: 41.38-61.20) and 81.83% (95%CI: 55.16-94.28) among NAFLD and NASH patients, respectively[2]. The effect of obesity on NAFLD has been intensively explored. On the one hand, the expansion of adipose tissue (AT) in obese people leads to increased circulating free fatty acids (FFAs) and leptin and decreased adiponectin, which leads to intrahepatic fat accumulation. On the other hand, the chronic inflammatory state caused by obesity will further lead to the infiltration of inflammatory cells in the liver, resulting in the progression of NAFLD[7]. Due to the important role of obesity in the occurrence and development of NAFLD, in-depth research on the mechanism of obesity leading to NASH may provide new therapeutic targets.
Exosomes are extracellular vesicles secreted by various cells and serve as an essential means of intercellular communication by delivering microRNAs (miRNAs), bioactive lipids, and regulatory proteins from one cell to another[8]. Previous studies have shown that AT-derived exosomes are essential in regulating insulin sensitivity[9,10], a common manifestation of metabolic syndrome in patients with NAFLD. Previous studies have also shown that AT-derived exosome miRNAs are involved in the occurrence and development of various metabolic-related diseases[11]. Among them, miR-103 attracted our attention since previous studies have shown that miR-103 is involved in regulating insulin sensitivity[12]. The specific role of AT-derived exosomes in the development of NASH also deserves further study. Therefore, in this study, we focused on the specific role of AT-derived exosomes miR-103 in developing NASH through various methods.
This study followed the guidelines for the Care and Use of Laboratory Animals of the National Institute of Health. The animal protocol was approved by the institutional review board of the Tab of Animal Experimental Ethical Inspection of the First Affiliated Hospital of Zhejiang University. The Reference Number is 2020-1407.
C57BL/6 mice were routinely fed a high-fat diet for 12 wk to establish the NASH animal model. According to different treatments, they were initially divided into the control group (12% kcal fat, 66% kcal carbohydrate, 22% kcal protein) and the model group (60% kcal fat, 20% kcal carbohydrate, 20% kcal protein)[13]. Starting from the 13th wk, 40 mg/kg miR-negative control (NC)-anta and miR-103-anta were injected into the mice from the model group (dissolved in 0.2 mL normal saline) through the tail vein every 2 d thrice to construct miR-NC-anta model group and miR-103-anta model group. The control and model groups were injected with blank normal saline thrice (n = 10 in each group). Finally, the mice were sacrificed, where liver tissue, abdominal AT, and serum were collected and stored in a cryostorage tube at -80 °C for further analysis.
NASH-like cell model was constructed by conventional oleic acid (OA)-palmitic acid (PA) mixture culture (OA:PA = 2:1)[14]. Firstly, 128.2 mg PA (molecular weight: 256.42) was sequentially retrieved on a precision balance, 1 mL 1 M OA was added, vortex dissolved, and mixed in a small whirlpool, and then completely dissolved in a water bath at 55 °C-65 °C to obtain 1.5 M FFA mixture. After that, 1.5 M FFA with DMSO was dissolved into 0.1 M working solution. In the model group, Alpha mouse liver 12 (AML-12) cells were added with 400 μM FFA and cultured for 24 h, followed by transfection with miR-103-anta and its sh-phosphatase and tensin homolog (PTEN) and their relative controls. All further in vitro experiments were performed on those cells.
Exosomes were extracted from the filtrate according to the manufacturer’s instructions[15]. The diluted exosomes were subjected to NanoFCM (China) for transmission electron microscopy (TEM) and size distribution analysis for further confirmation.
Total RNA was isolated using Trizol (Invitrogen, United States) and reverse-transcribed into cDNA using the First Strand cDNA Synthesis Kit (TransGen, China) following the manufacturer’s instructions. Quantitative real-time polymerase chain reaction (qRT-PCR) was routinely performed using the SYBR Premix Ex Taq qPCR kit (TaKaRa, Japan). The alanine aminotransferase (ALT), aspartate aminotransferase (AST), total triglycerides (TG), total cholesterol (CHOL), superoxide dismutase (SOD), malondialdehyde (MDA), and H2O2 concentrations were detected using test kits according to the manufacturer’s instructions. Total protein was isolated using radioimmune precipitation assay buffer (TaKaRa, Japan) supplemented with a protease inhibitor (Roche, Switzerland). After quantification using the BCA Protein Assay Kit (Thermo, United States), the proteins were separated by sodium-dodecyl sulfate gel electrophoresis and transferred onto polyvinylidene fluoride membranes. The membranes were blocked and incubated overnight with antibodies against PTEN (9188T, CST), p-AMPK (ab32047, Abcam), p-mammalian target of rapamycin (mTOR) (CSB-PA271384, Cusabio), LC3 (12741T, CST), p62 (ab91526, Abcam), and GAPDH (ab245355, Abcam) at 4 °C. The membranes were incubated with HRP-conjugated secondary antibodies (ab205718, Abcam). Finally, the protein bands were detected using enhanced chemiluminescence (ECL) kits (Thermo, United States).
The hematoxylin and eosin (HE) staining was performed using the HE staining kit (C0105S, Beyotime) according to the manufacturer’s instructions. Oil red staining was performed using an oil red staining solution (G 1262, Solarbio; C0157S, Beyotime). Briefly, 5-10 μm thick fresh frozen tissue was placed on the slide and dried at room temperature for 30-60 min. The sections were fixed with 10% paraformaldehyde for 10 min, washed thrice with distilled water, and dried for several minutes. After that, the oil red was diluted with deionized water in a 3:2 ratio, with impurities removed by filter paper, and left for 10 min at room temperature. Preheated oil red was used for tissue dye in a 6 °C temperature box for 8-10 min. After the 85% propylene glycol solution was differentiated for 2-5 min, it was washed twice with distilled water and restained with hematoxylin for 30 s. After rinsing with running water for 3 min, the tablets can be sealed with glycerine gelatin.
The luciferase reporter assay was performed according to the manufacturer’s instructions of Pierce™ Cypridina-Firefly Luciferase Dual Assay Kit (16184, Thermo). Briefly, AML-12 cells were co-transfected with a 10 nM miR-103 or NC control, a 2 ng pRL-CMV, and a 20 ng firefly luciferase reporter plasmid containing PTEN of the wild-type or mutant 30-untranslated region. Then, 48 h after transfection, the cell lysates were determined by luciferase to observe the interaction between miR-103 and PTEN. The liver tissues with the size of 1 mm × 1 mm × 1 mm were fixed, dehydrated, impregnated, and embedded to make ultrathin sections (50-70 nm) and then stained with uranium acetate and lead citrate successively and dried for observation under TEM.
The frozen slices of 5-10 μm thick liver tissue were dried at room temperature for 30-60 min. They were sequentially fixed with 10% paraformaldehyde for 10 min, rinsed thrice with distilled water, and dried for several minutes. After that, the antigen was repaired by microwave at 92 °C-96 °C for 10-15 min, cooled to room temperature naturally, and sealed with 5% BSA at 37 °C for 60 min. After pouring the excess serum, LC3-II/I antibody was diluted at 1:100, added into samples, and incubated at 4 °C overnight. Rinsed with phosphate buffered saline (PBS) the next day, samples were added to the mixture of the fluorescent secondary antibody and 4’,6-diamidino-2-phenylindole at a ratio of 1:200 and incubated for 60 min at room temperature. After washing with PBS, laser confocal scanning for immunofluorescence was performed.
3T3-L1 cells were first routinely induced to differentiate into adipocytes to identify the transfer of miR-103 from adipocytes to AML-12 cells. After that, Cy3-labeled miR-103 was transfected and then co-cultured with the underlying AML-12 cells through a transwell chamber. AML-12 cells were then isolated, and immunofluorescence determined the red fluorescence value.
Cells for transfection were incubated into a 6-well plate. 5-10 μL miR-NC-anta and miR-103-anta were absorbed and diluted into 250 μL Opti-MEMⅠ reduced serum medium, mixed gently, and incubated at room temperature for 5 min. Then, 3-6 μL Lipofectamine® 2000 Reagent was diluted to 250 μL of Opti-MempI reduced-serum medium, lightly mixed, and incubated at room temperature for 5 min. Diluted miR-NC-anta, miR-103-anta, and diluted Lipofectamine® 2000 Reagent were carefully mixed, gently blended, and incubated at room temperature for 20 min to form the reagent complex. After that, cells were washed with 2 mL serum-free medium, added with 2 mL of Opti-MEMI low serum medium to each well, and then added to 500 μL of miR-NC-anta and miR-103-anta-Lipofectamine® 2000 Reagent complex. These reagents were gently mixed and prepared for use.
All data are presented as mean ± SD. Differences between the two groups were analyzed using the student’s t-test for categorical data and the chi-square method for numerical data. All statistical analyses were performed using GraphPad 9.0.2 software. Statistical significance between groups was set at P < 0.05.
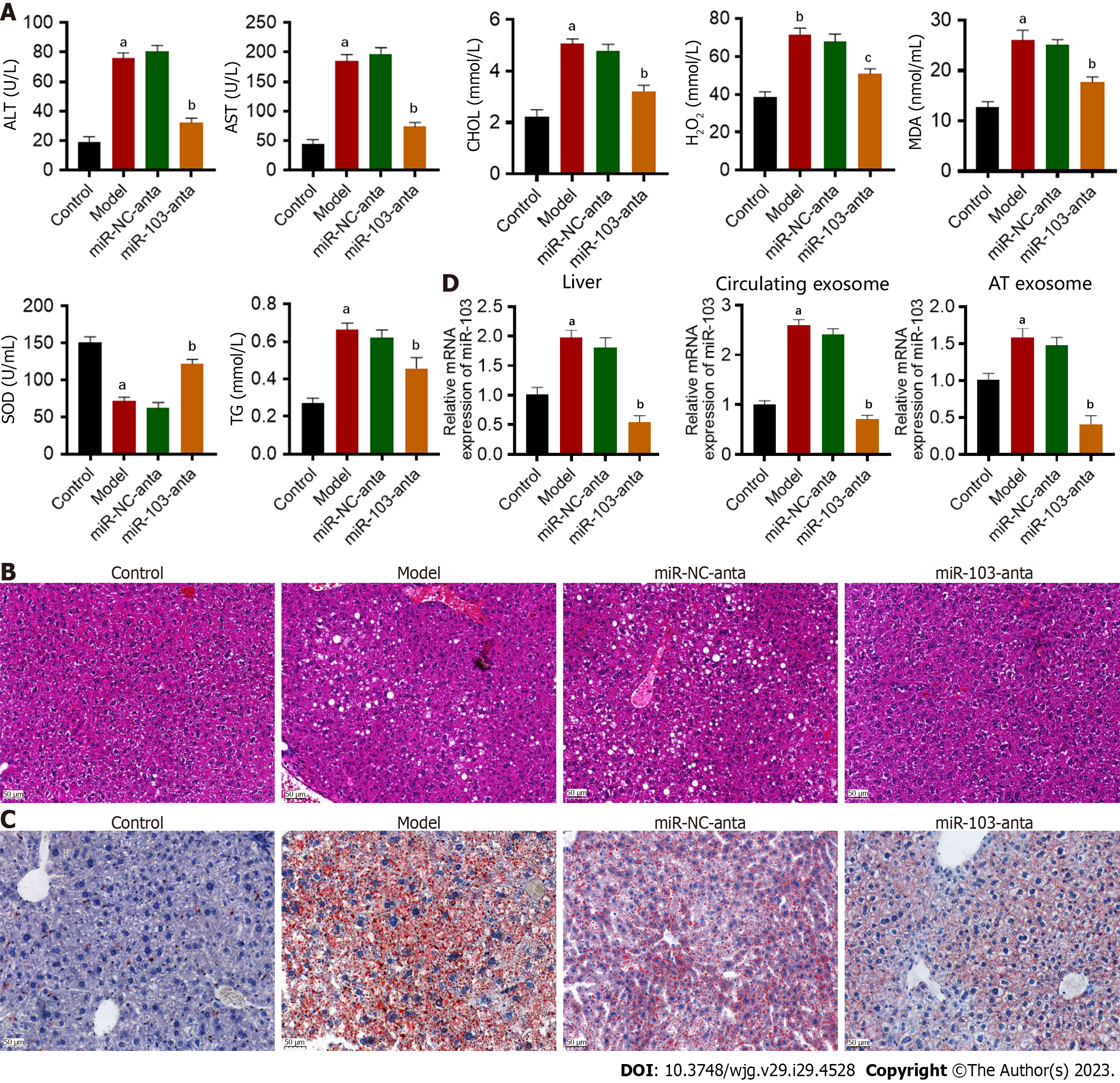
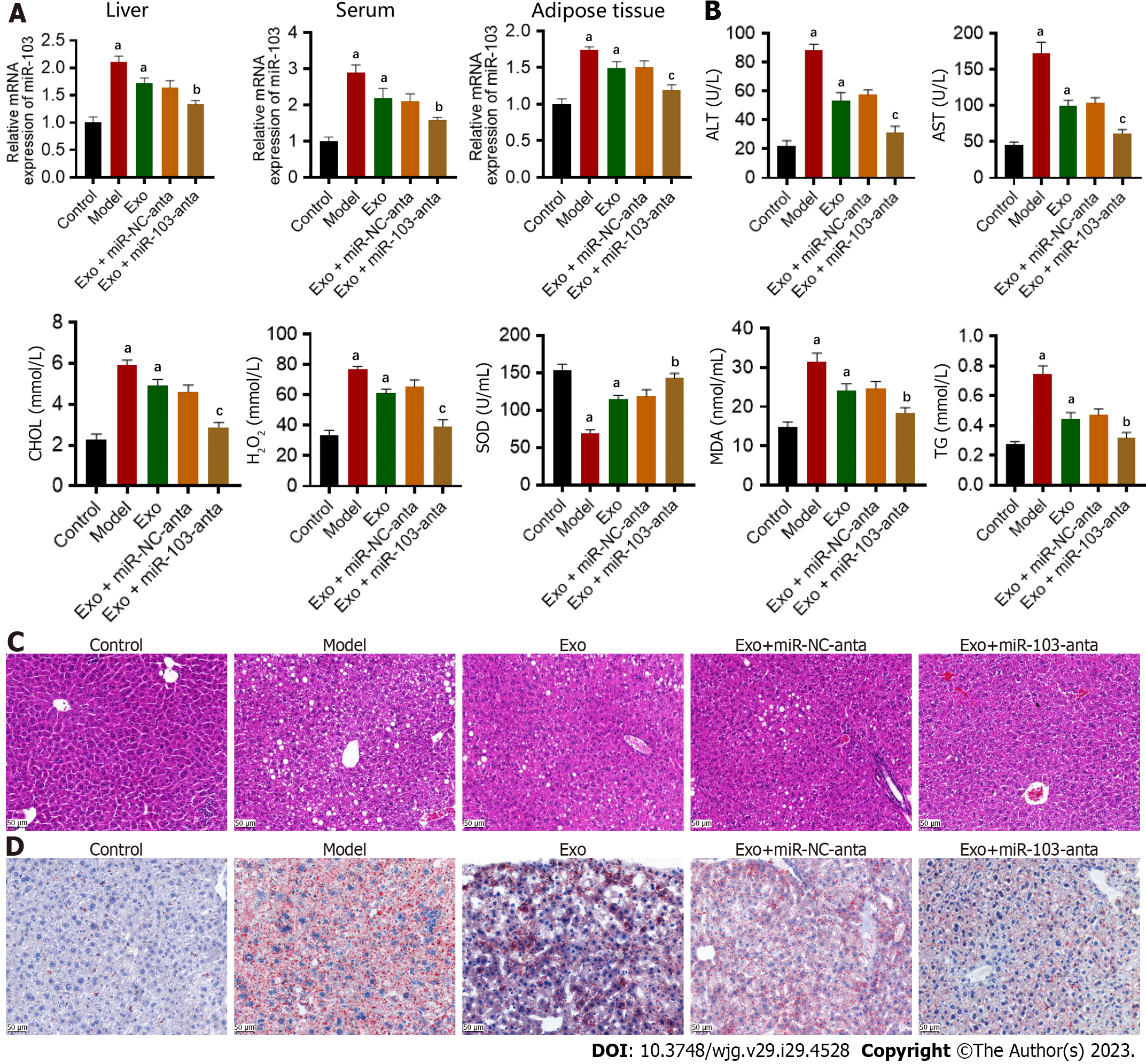
First, we successfully constructed animal models of NASH. Compared with the control, ALT, AST, TG, and CHOL were increased in NASH mice (Figure 1A). Hepatocyte ballooning, inflammatory cell infiltration, and hepatic lipid accumulation were observed in the livers of NASH mice (Figures 1B and C). Furthermore, we successfully extracted and confirmed AT-derived and circulating exosomes (Supplementary Figure 1). Subsequently, the expression of miR-103 in the livers, AT-derived exosomes, and circulating exosomes was detected by qRT-PCR. MiR-103 expression levels in the livers, AT-derived exosomes, and circulating exosomes were significantly increased in the NASH model group compared with the control group. Antagonizing miR-103 decreased miR-103 expression in NASH mice, but miR-NC-anta had no significant effect in NASH mice (Figure 1D). In addition, compared with miR-NC-anta and the model group, miR-103-anta treatment significantly reduced serum ALT and AST, decreased serum CHOL and TG, and alleviated oxidative stress (Figure 1A). Histologically, HE and oil red staining of the liver also indicated that inhibition of miR-103 alleviated hepatocyte ballooning, inflammatory cell infiltration, and hepatic lipid accumulation. (Figures 1B and C). Our above results indicate that the miR-103 level is elevated in NASH model mice, and reducing the expression of miR-103 can alleviate NASH, suggesting the potential involvement of miR-103.
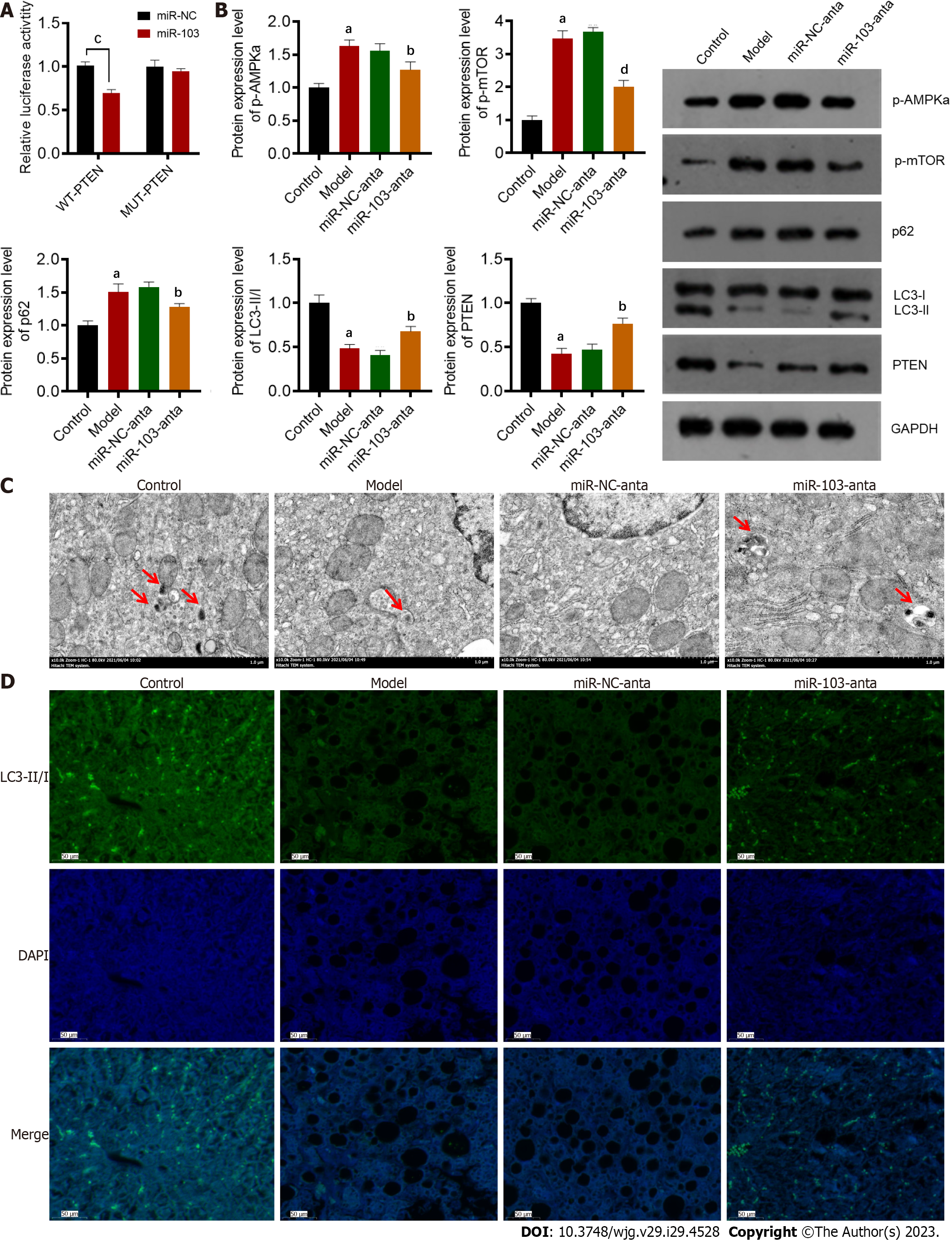
To further investigate the regulatory mechanism of miR-103 on NASH, we used TargetScan to predict its downstream targets[16]. The predicted results showed that PTEN, a gene that plays an essential role in autophagy, might interact with miR-103. Therefore, we used a dual-luciferase reporting experiment to further confirm their interaction (Figure 2A). The wild-type PTEN could interact with miR-103, while the mutant PTEN could not. Next, the protein content of PTEN and autophagy-related proteins such as p-AMPKa, p-mTOR, LC3-II/I, and p62 were determined by western blot. The results showed that compared with the control group, the expressions of p-AMPKa, p-mTOR, and p62 were significantly increased in the model group, while the protein levels of PTEN and LC3-II/I were decreased (Figure 2B). In addition, TEM and immunofluorescence staining showed that the number of autophagosomes in the liver of NASH model mice was significantly reduced compared with control mice (Figures 2C and D). However, compared with the model and miR-NC-anta groups, p-AMPKa, p-mTOR, p62, and the number of autophagosomes were significantly decreased in the miR-103-anta group. Similarly, the protein levels of PTEN and LC3-II/I was increased (Figures 2C and D). All these results indicate that autophagy is inhibited in the development of NASH, while miR-103-anta treatment could antagonize those changes. These results indicate that miR-103 interacts with PTEN and interferes with the downstream autophagy process, suggesting that the inhibition of autophagy in NASH may be attributed to the increased expression of miR-103.
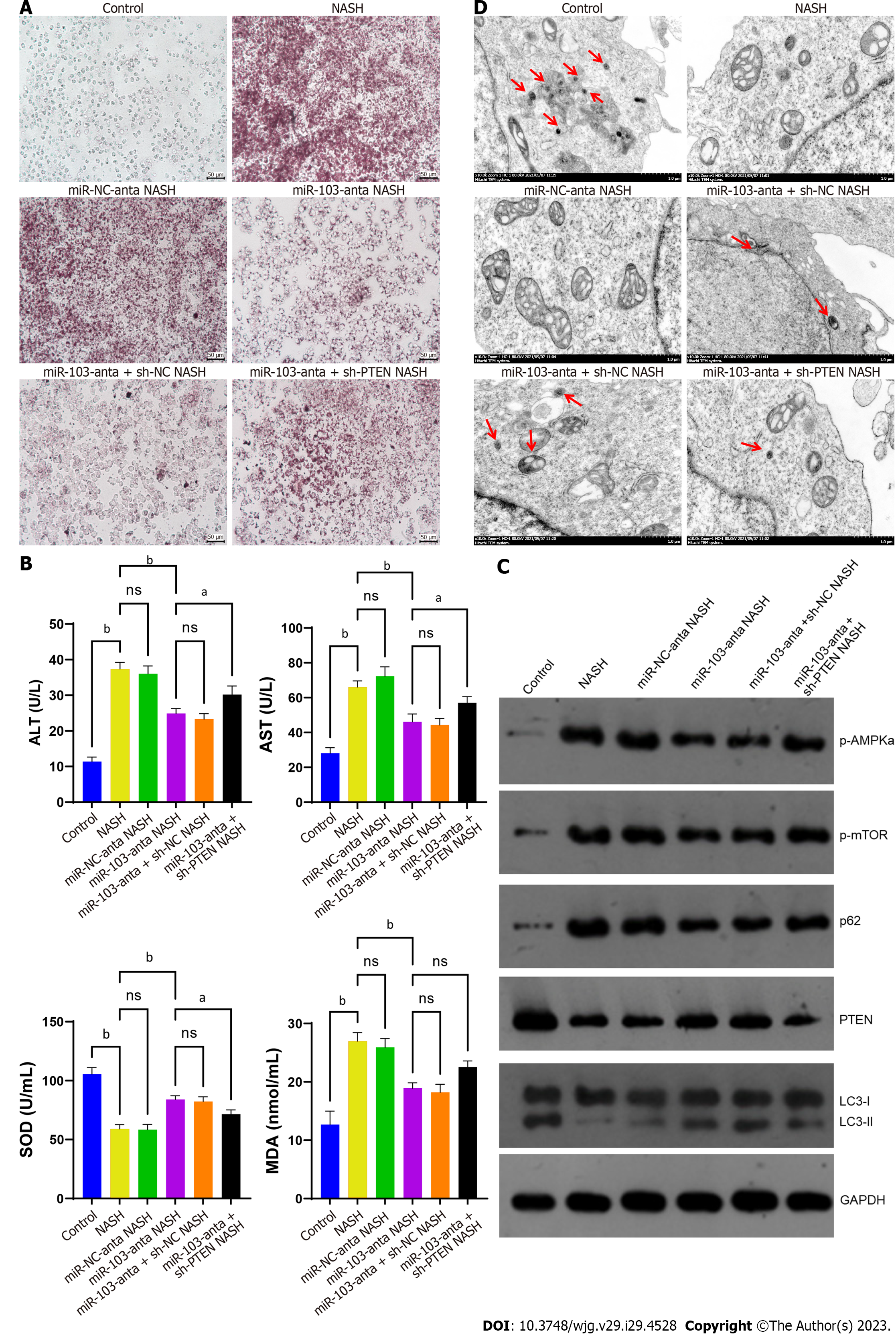
We then conducted in vitro experiments to further confirm our hypothesis. The results showed that inhibition of miR-103 expression could reduce the accumulation of lipids in NASH model cells, decrease the release of ALT and AST, and relieve oxidative stress. However, the effect of miR-103-anta was partially eliminated by silencing PTEN (Figures 3A and B). The above results indicate that miR-103 is involved in NASH formation partially through its interaction with PTEN. In addition, we detected the expression of autophagy-related proteins. We found that the expression of p-AMPKa, p-mTOR, and p62 was increased in NASH cells, while the expression of PTEN and LC3-II/I was decreased. Treatment with miR-103-anta elevated the expression of PTEN and LC3-II/I and reduced the expression of p-AMPKa, p-mTOR, and p62 in NASH cells, while PTEN silencing inhibited the effect of miR-103-anta (Figure 3C). Finally, we also found that the number of autophagosomes decreased in NASH cells, and such declination was partially antagonized after the inhibition of miR-103. Similarly, PTEN silencing inhibited the effect of miR-103-anta (Figure 3D). To sum up, we confirmed the role of the miR-103-PTEN-autophagy axis in NASH through in vitro experiments.
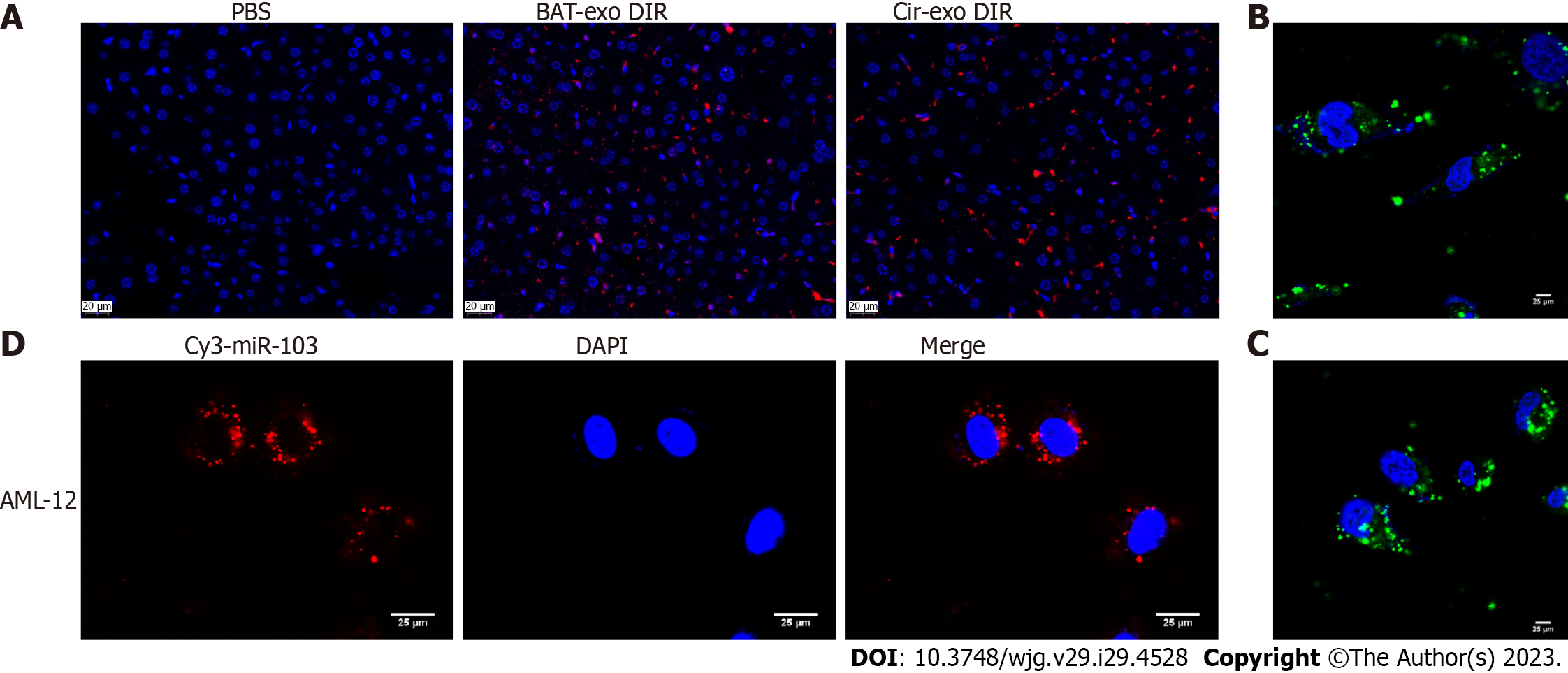
To confirm that AT-derived exosomes can encapsulate miR-103 and target the liver, we conducted in vivo and in vitro experiments. In the in vivo experiment, AT and circulating exosomes of mice were extracted. The exo-DIR complex collected by centrifugation was injected into the tail vein of mice, and the PBS group was used as a control. The fluorescence of liver tissue was observed after mice scarification. There was no fluorescence in the PBS group but observed in AT-exo-DIR and cir-exo-DIR groups, with similar fluorescence intensity (Figure 4A). In the part of the in vitro experiment, the AT exosomes and circulating exosomes were detected by the FISH probe to migrate into AML-12 cells (Figures 4B and C). Further transewell assay showed that Cy3-labeled miR-103 could be transferred from adipocytes to AML-12 cells (Figure 4D).
To verify the role of AT-derived exosome miR-103 in NASH through in vivo experiments, we extracted exosomes from mouse AT. We injected the extracted exosomes without other treatments and with miR-103-anta or miR-NC-anta into mice through the tail vein. Firstly, we found that injection of AT-derived exosomes increased the level of miR-103 in the liver, serum, and AT compared with the control group (Figure 5A). In addition, compared with the control group, treating AT-derived exosomes can increase serum ALT, AST, CHOL, and TG levels and aggravate oxidative stress (Figure 5B). Histologically, HE and oil red staining of the liver showed that treating AT-derived exosomes could aggravate hepatocyte ballooning, inflammatory cell infiltration, and intrahepatic lipid accumulation (Figures 5C and D). However, miR-103-anta treatment could partially eliminate such an effect while miR-NC-anta treatment could not, suggesting that AT-derived exosome aggravates NASH, and this effect is partly dependent on its encapsulated miR-103.
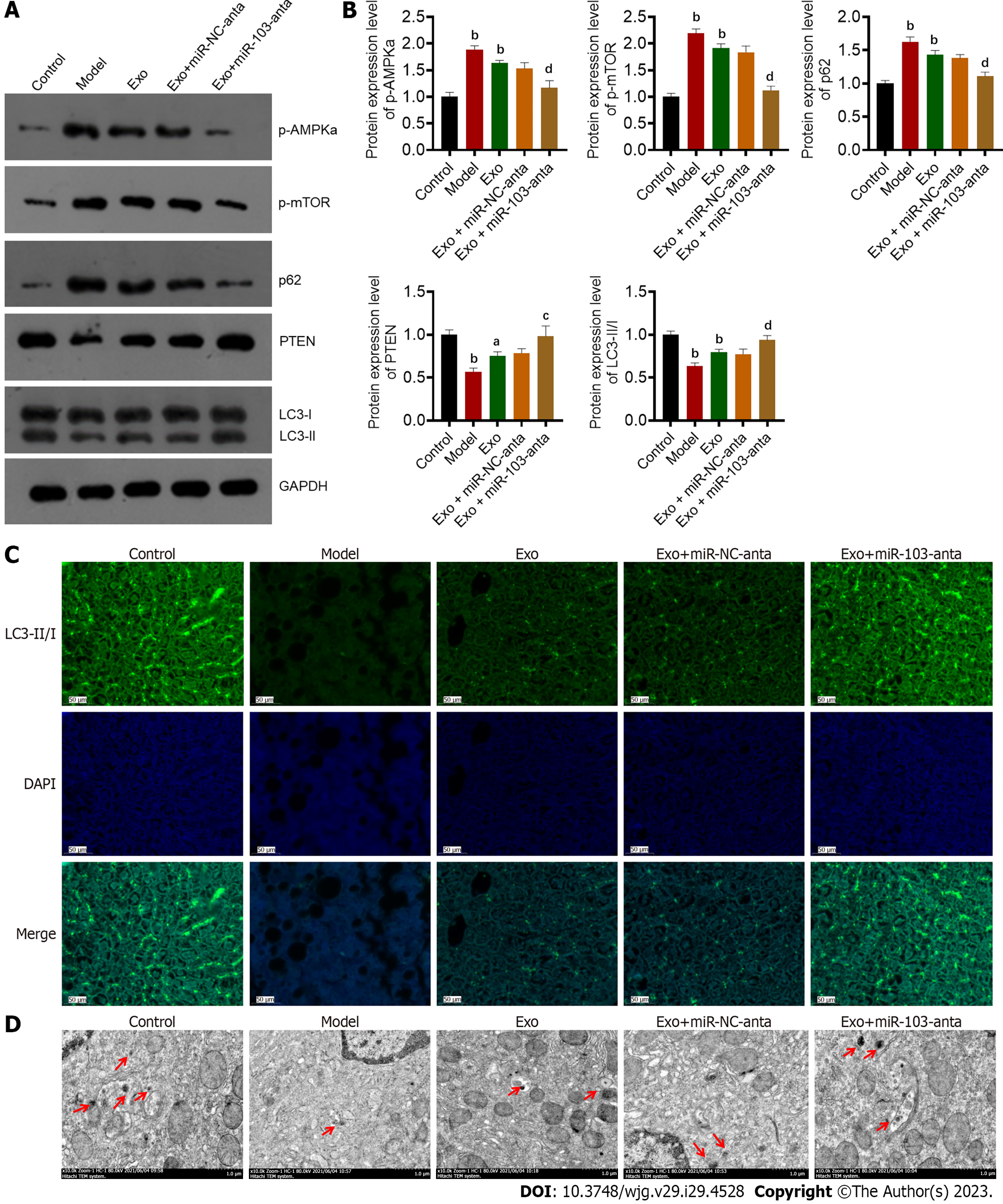
To further verify in vivo that miR-103 in AT-derived exosomes is involved in NASH formation by influencing autophagy, we detected the expression of autophagy-related proteins in the liver of mice in each group. We found that compared with the control group, the expression of p-AMPKa, p-mTOR, and p62 in the livers of AT exosome-treated group was increased, while the expression of PTEN and LC3-II/I was decreased. Further miR-103-anta treatment increased the expression level of PTEN and LC3-II/I and reduced the expression level of p-AMPKa, p-mTOR, and p62, while miR-NC-anta had no similar effect (Figures 6A and B). Finally, through TEM and immunofluorescence, we found that the number of autophagosomes in the livers of mice treated with AT-derived exosomes was decreased, while miR-103-anta could partially antagonize such effect (Figures 6C and D).
NASH is considered the watershed in the progress of NAFLD, which is more closely related to the occurrence of liver cirrhosis and other complications. The prevalence of NASH has been increasing in recent years, resulting in a big challenge in disease burden and patient suffering. For instance, the lifetime cost of care for patients with NASH was around US $222 billion in 2017 in the United States[17]. More intriguingly, many NASH patients are obese and tend to have higher healthcare costs than non-obese NASH patients[18], but the etiology is still vague. Therefore, it is necessary to study the role and underlying mechanisms of obesity on NASH and further provide possible therapeutic targets. In this study, we confirmed the important role of AT-derived exosomes miR-103 in NASH and preliminary revealed its regulation on hepatocyte autophagy through targeting PTEN, which might partially provide the mechanisms by which obesity affects NASH.
MiRNA belongs to the family of non-coding RNA, which generally consists of 22 nucleotides[19] and regulates the mRNA levels of target genes[20]. Previous studies have identified a variety of miRNAs involved in metabolism-related diseases[21]. For example, miR-200 and miR-29 families play an important role in maintaining the balance between the proliferation and differentiation of pancreatic β cells[22,23]. MiR-33a and miR-33b are involved in cholesterol and lipid metabolism[24,25]. Furthermore, several miRNAs are also targeting the liver to regulate metabolic processes. For instance, miR-122, one of the most abundant miRNAs in the liver, is involved in hepatic cholesterol and lipid metabolism[26,27]. Besides, miR-103, the focus of our study, has also been confirmed to be closely related to hepatic insulin sensitivity and the regulation of glucose homeostasis in previous studies[12]. Since NAFLD is the hepatic manifestation of metabolic syndrome, it is theoretically possible that miR-103 participates in NAFLD, but related research is still lacking. NASH was linked to menopause[28], and miR-103 was found to be linked to G protein-coupled estrogen receptor 1[29]. Therefore, the estrogen signaling pathway is the potential mechanism where miR-103 promotes NASH. However, in this study, for the first time, we showed that the miR-103 level in the liver of NASH mice was significantly increased while inhibiting miR-103 expression could alleviate NASH, suggesting that miR-103 is one of the potential targets for NASH treatment.
We further identified the AT-derived exosomes as the source of miR-103 upregulation in the NASH mouse model. There have been many studies on the role of AT-derived exosomes in the development of NAFLD in obese people[30]. For instance, Fuchs et al[31] showed that the concentration of free exosomes is significantly higher in obese with NAFLD (OB-NAFLD) patients compared with lean with normal intrahepatic triglyceride content (LEAN) and obese with normal intrahepatic triglyceride content (OB-NL) populations and that these exosomes are at least partially derived from AT. Compared with exosomes derived from the LEAN and OB-NL groups, plasma and AT-derived exosomes from the OB-NAFLD group caused insulin resistance in both myotubes and hepatocytes, demonstrated by impaired insulin signaling. However, the underlying mechanism of the above effects of AT-derived exosomes in NAFLD patients has not been further explored. Our results complement this by revealing the effect of AT-derived exosomes through the miR-103-PTEN pathway, which broadens our understanding of NASH pathogenesis from the angle of trans-cellular crosstalk.
We also confirmed that autophagy is the downstream of the action of miR-103 through in vivo and in vitro experiments. Autophagy is an evolutionarily conserved cellular degradation process that delivers some intracellular components to lysosomes for degradation[32]. Current studies suggest that autophagy includes three subtypes: Macroautophagy, microautophagy, and chaperon-mediated autophagy[33]. Autophagy plays a vital role in the liver. It involves many basic liver functions, such as glycogenolysis, gluconeogenesis, and β-oxidation[34]. Previous studies have also shown that autophagy is hampered in NAFLD patients. Our previous study also revealed that autophagy inhibition plays an important role in NASH development[35]. Furthermore, restoring autophagy through certain drugs (trehalose, rapamycin, carbamazepine, or other pharmaceutical agents) or gene targets (overexpression of Atg7 or TFEB) can also alleviate NAFLD[36]. In addition, thyroxine[37] and caffeine[38] were also identified to reduce NAFLD by regulating liver autophagy. Therefore, miR-103 is expected to be one of the therapeutic targets for its autophagy regulation capacity and needs further clinical investigation in the future.
Some limitations in this study should be acknowledged. Firstly, AT-derived exosomes contain many non-coding RNAs, and we did not detect changes in the expression of other non-coding RNAs. Moreover, inhibition of miR-103 only partially inhibited the effect of AT-derived exosomes. Therefore, the above results suggest that AT-derived exosomes promote the development of NASH in multiple ways, and miR-103 is only one of them. Further research is needed on other reasons why AT-derived exosomes promote the development of NASH. Secondly, we did not design in vivo experiments to verify whether inhibition of autophagy could abolish the role of miR-103 in NASH. Therefore, in future studies, we may improve this part of the experiment and further explore the mechanism of miR-103 affecting autophagy. Finally, the preliminary data on the hepatocyte absorption of AT-derived exosome miR-103 needs further verification.
To sum up, our study confirms the important role of miR-103-PTEN - autophagy axis in NASH, and the elevation of miR-103 in the liver of the NASH model is partly due to hepatocyte absorption of AT derived-exosomes, which also partially explains the underlining mechanism of obesity leading to NASH (Figure 7).
Non-alcoholic steatohepatitis (NASH) has become one of the leading causes of cirrhosis and the second leading cause of liver transplantation. miR-103 is involved in regulating insulin sensitivity, a common manifestation of metabolic syndrome in patients with NASH.
The specific role of miR-103 in the development of NASH also deserves further study.
To explore the specific role of miR-103 in the development of NASH and provide new therapeutic targets for NASH.
The expression levels of miR-103 were detected and compared between NASH mice and control. The effect of miR-103 on NASH progression was explored by miR-103 antagonizing, including both changes of steatosis and inflammation degree. The interaction between miR-103 and the autophagy-related gene phosphatase and tensin homolog (PTEN) was confirmed by dual-luciferase reporter assay. The role of the interaction between miR-103 and PTEN on autophagy was verified in NASH cells. Finally, the effects of miR-103 from adipose tissue (AT)-derived exosomes on NASH and autophagy were analyzed through animal experiments.
The expression of miR-103 was increased in NASH mice, compared with the control, and inhibition of miR-103 could alleviate NASH. MiR-103 could interact with PTEN. MiR-103-anta inhibited autophagy in NASH mice. Further experiments showed PTEN silencing inhibited the effect of miR-103-anta. AT-derived exosome miR-103 aggravated NASH and inhibited autophagy in mice.
AT derived-exosome increased the levels of miR-103 in the liver, and miR-103 aggravated NASH. Mechanically speaking, miR-103 could interact with PTEN and inhibit autophagy.
MiR-103 may be a potential target for NASH treatment in the future.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Herrera B, Spain; Sukocheva OA, Australia S-Editor: Wang JJ L-Editor: A P-Editor: Zhao S
| 1. | Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, Harrison SA, Brunt EM, Sanyal AJ. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67:328-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3544] [Cited by in RCA: 4949] [Article Influence: 707.0] [Reference Citation Analysis (9)] |
| 2. | Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5322] [Cited by in RCA: 7539] [Article Influence: 837.7] [Reference Citation Analysis (0)] |
| 3. | Wong VW, Wong GL, Choi PC, Chan AW, Li MK, Chan HY, Chim AM, Yu J, Sung JJ, Chan HL. Disease progression of non-alcoholic fatty liver disease: a prospective study with paired liver biopsies at 3 years. Gut. 2010;59:969-974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 491] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 4. | Wong RJ, Aguilar M, Cheung R, Perumpail RB, Harrison SA, Younossi ZM, Ahmed A. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology. 2015;148:547-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1211] [Cited by in RCA: 1382] [Article Influence: 138.2] [Reference Citation Analysis (1)] |
| 5. | Charlton MR, Burns JM, Pedersen RA, Watt KD, Heimbach JK, Dierkhising RA. Frequency and outcomes of liver transplantation for nonalcoholic steatohepatitis in the United States. Gastroenterology. 2011;141:1249-1253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 835] [Cited by in RCA: 855] [Article Influence: 61.1] [Reference Citation Analysis (0)] |
| 6. | Byrne CD, Targher G. NAFLD: a multisystem disease. J Hepatol. 2015;62:S47-S64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1516] [Cited by in RCA: 2140] [Article Influence: 214.0] [Reference Citation Analysis (0)] |
| 7. | Polyzos SA, Kountouras J, Mantzoros CS. Obesity and nonalcoholic fatty liver disease: From pathophysiology to therapeutics. Metabolism. 2019;92:82-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 453] [Cited by in RCA: 809] [Article Influence: 134.8] [Reference Citation Analysis (0)] |
| 8. | Crewe C, Scherer PE. Intercellular and interorgan crosstalk through adipocyte extracellular vesicles. Rev Endocr Metab Disord. 2022;23:61-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 39] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 9. | Ying W, Riopel M, Bandyopadhyay G, Dong Y, Birmingham A, Seo JB, Ofrecio JM, Wollam J, Hernandez-Carretero A, Fu W, Li P, Olefsky JM. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell. 2017;171:372-384.e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 591] [Cited by in RCA: 877] [Article Influence: 109.6] [Reference Citation Analysis (0)] |
| 10. | Liu T, Sun YC, Cheng P, Shao HG. Adipose tissue macrophage-derived exosomal miR-29a regulates obesity-associated insulin resistance. Biochem Biophys Res Commun. 2019;515:352-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 114] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 11. | Rottiers V, Näär AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 808] [Cited by in RCA: 894] [Article Influence: 68.8] [Reference Citation Analysis (0)] |
| 12. | Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 707] [Cited by in RCA: 796] [Article Influence: 56.9] [Reference Citation Analysis (0)] |
| 13. | Recena Aydos L, Aparecida do Amaral L, Serafim de Souza R, Jacobowski AC, Freitas Dos Santos E, Rodrigues Macedo ML. Nonalcoholic Fatty Liver Disease Induced by High-Fat Diet in C57bl/6 Models. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 101] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 14. | Wang L, Zhang X, Lin ZB, Yang PJ, Xu H, Duan JL, Ruan B, Song P, Liu JJ, Yue ZS, Fang ZQ, Hu H, Liu Z, Huang XL, Yang L, Tian S, Tao KS, Han H, Dou KF. Tripartite motif 16 ameliorates nonalcoholic steatohepatitis by promoting the degradation of phospho-TAK1. Cell Metab. 2021;33:1372-1388.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 69] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 15. | Gao J, Li X, Wang Y, Cao Y, Yao D, Sun L, Qin L, Qiu H, Zhan X. Adipocyte-derived extracellular vesicles modulate appetite and weight through mTOR signalling in the hypothalamus. Acta Physiol (Oxf). 2020;228:e13339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 59] [Article Influence: 11.8] [Reference Citation Analysis (1)] |
| 16. | Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4013] [Cited by in RCA: 5416] [Article Influence: 541.6] [Reference Citation Analysis (0)] |
| 17. | Younossi ZM, Tampi R, Priyadarshini M, Nader F, Younossi IM, Racila A. Burden of Illness and Economic Model for Patients With Nonalcoholic Steatohepatitis in the United States. Hepatology. 2019;69:564-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 166] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 18. | Allen AM, Lazarus JV, Younossi ZM. Healthcare and socioeconomic costs of NAFLD: A global framework to navigate the uncertainties. J Hepatol. 2023;79:209-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 79] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 19. | Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853-858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3501] [Cited by in RCA: 3556] [Article Influence: 148.2] [Reference Citation Analysis (0)] |
| 20. | Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1645] [Cited by in RCA: 1506] [Article Influence: 94.1] [Reference Citation Analysis (0)] |
| 21. | Vienberg S, Geiger J, Madsen S, Dalgaard LT. MicroRNAs in metabolism. Acta Physiol (Oxf). 2017;219:346-361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 283] [Cited by in RCA: 306] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 22. | Pullen TJ, da Silva Xavier G, Kelsey G, Rutter GA. miR-29a and miR-29b contribute to pancreatic beta-cell-specific silencing of monocarboxylate transporter 1 (Mct1). Mol Cell Biol. 2011;31:3182-3194. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 241] [Cited by in RCA: 221] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 23. | Belgardt BF, Ahmed K, Spranger M, Latreille M, Denzler R, Kondratiuk N, von Meyenn F, Villena FN, Herrmanns K, Bosco D, Kerr-Conte J, Pattou F, Rülicke T, Stoffel M. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat Med. 2015;21:619-627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 226] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 24. | Dávalos A, Goedeke L, Smibert P, Ramírez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, Esplugues E, Fisher EA, Penalva LO, Moore KJ, Suárez Y, Lai EC, Fernández-Hernando C. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108:9232-9237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 488] [Cited by in RCA: 536] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 25. | Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, Khatsenko OG, Kaimal V, Lees CJ, Fernandez-Hernando C, Fisher EA, Temel RE, Moore KJ. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404-407. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 550] [Cited by in RCA: 588] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 26. | Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3005] [Cited by in RCA: 3068] [Article Influence: 153.4] [Reference Citation Analysis (0)] |
| 27. | Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1612] [Cited by in RCA: 1638] [Article Influence: 86.2] [Reference Citation Analysis (0)] |
| 28. | DiStefano JK. NAFLD and NASH in Postmenopausal Women: Implications for Diagnosis and Treatment. Endocrinology. 2020;161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 147] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 29. | Fang T, Li J, Wu X. Shenmai injection improves the postoperative immune function of papillary thyroid carcinoma patients by inhibiting differentiation into Treg cells via miR-103/GPER1 axis. Drug Dev Res. 2018;79:324-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 30. | Thietart S, Rautou PE. Extracellular vesicles as biomarkers in liver diseases: A clinician's point of view. J Hepatol. 2020;73:1507-1525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 127] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 31. | Fuchs A, Samovski D, Smith GI, Cifarelli V, Farabi SS, Yoshino J, Pietka T, Chang SW, Ghosh S, Myckatyn TM, Klein S. Associations Among Adipose Tissue Immunology, Inflammation, Exosomes and Insulin Sensitivity in People With Obesity and Nonalcoholic Fatty Liver Disease. Gastroenterology. 2021;161:968-981.e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 114] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 32. | Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717-1721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2779] [Cited by in RCA: 2863] [Article Influence: 114.5] [Reference Citation Analysis (0)] |
| 33. | Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069-1075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5421] [Cited by in RCA: 5293] [Article Influence: 311.4] [Reference Citation Analysis (0)] |
| 34. | Ueno T, Komatsu M. Autophagy in the liver: functions in health and disease. Nat Rev Gastroenterol Hepatol. 2017;14:170-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 398] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 35. | Jin X, Gao J, Zheng R, Yu M, Ren Y, Yan T, Huang Y, Li Y. Antagonizing circRNA_002581-miR-122-CPEB1 axis alleviates NASH through restoring PTEN-AMPK-mTOR pathway regulated autophagy. Cell Death Dis. 2020;11:123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 36. | Qian H, Chao X, Williams J, Fulte S, Li T, Yang L, Ding WX. Autophagy in liver diseases: A review. Mol Aspects Med. 2021;82:100973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 219] [Article Influence: 54.8] [Reference Citation Analysis (0)] |
| 37. | Zhou J, Tripathi M, Ho JP, Widjaja AA, Shekeran SG, Camat MD, James A, Wu Y, Ching J, Kovalik JP, Lim KH, Cook SA, Bay BH, Singh BK, Yen PM. Thyroid Hormone Decreases Hepatic Steatosis, Inflammation, and Fibrosis in a Dietary Mouse Model of Nonalcoholic Steatohepatitis. Thyroid. 2022;32:725-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 53] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 38. | Sinha RA, Farah BL, Singh BK, Siddique MM, Li Y, Wu Y, Ilkayeva OR, Gooding J, Ching J, Zhou J, Martinez L, Xie S, Bay BH, Summers SA, Newgard CB, Yen PM. Caffeine stimulates hepatic lipid metabolism by the autophagy-lysosomal pathway in mice. Hepatology. 2014;59:1366-1380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 266] [Article Influence: 24.2] [Reference Citation Analysis (0)] |