Published online Jul 28, 2023. doi: 10.3748/wjg.v29.i28.4433
Peer-review started: May 24, 2023
First decision: June 14, 2023
Revised: June 19, 2023
Accepted: July 11, 2023
Article in press: July 11, 2023
Published online: July 28, 2023
Processing time: 63 Days and 10.5 Hours
Non-alcoholic fatty liver disease (NAFLD) is a major health burden with an increasing global incidence. Unfortunately, the unavailability of knowledge underlying NAFLD pathogenesis inhibits effective preventive and therapeutic measures.
To explore the molecular mechanism of NAFLD.
Whole genome sequencing (WGS) analysis was performed on liver tissues from patients with NAFLD (n = 6) and patients with normal metabolic conditions (n = 6) to identify the target genes. A NAFLD C57BL6/J mouse model induced by 16 wk of high-fat diet feeding and a hepatocyte-specific F-box only protein 2 (FBXO2) overexpression mouse model were used for in vivo studies. Plasmid transfection, co-immunoprecipitation-based mass spectrometry assays, and ubiquitination in HepG2 cells and HEK293T cells were used for in vitro studies.
A total of 30982 genes were detected in WGS analysis, with 649 up-regulated and 178 down-regulated. Expression of FBXO2, an E3 ligase, was upregulated in the liver tissues of patients with NAFLD. Hepatocyte-specific FBXO2 overexpression facilitated NAFLD-associated phenotypes in mice. Overexpression of FBXO2 aggravated odium oleate (OA)-induced lipid accumulation in HepG2 cells, resulting in an abnormal expression of genes related to lipid metabolism, such as fatty acid synthase, peroxisome proliferator-activated receptor alpha, and so on. In contrast, knocking down FBXO2 in HepG2 cells significantly alleviated the OA-induced lipid accumulation and aberrant expression of lipid metabolism genes. The hydroxyl CoA dehydrogenase alpha subunit (HADHA), a protein involved in oxidative stress, was a target of FBXO2-mediated ubiquitination. FBXO2 directly bound to HADHA and facilitated its proteasomal degradation in HepG2 and HEK293T cells. Supplementation with HADHA alleviated lipid accumulation caused by FBXO2 overexpression in HepG2 cells.
FBXO2 exacerbates lipid accumulation by targeting HADHA and is a potential therapeutic target for NAFLD.
Core Tip: This study involves an assessment of the role of F-box only protein 2 (FBXO2) in non-alcoholic fatty liver disease (NAFLD). First, based on the whole genome sequencing analysis results of liver tissues from normal controls and patients with NAFLD, the expression of FBXO2 was found to be increased during NAFLD. Hepatocyte-specific FBXO2 overexpression facilitated NAFLD-associated phenotypes in mice. In contrast, the knockdown of FBXO2 expression of HepG2 cells significantly alleviated oleate-induced lipid accumulation. Mechanistically, the hydroxyl CoA dehydrogenase alpha subunit, a protein with known role in oxidative stress reaction, is one of the protein substrates of FBXO2-mediated ubiquitination.
- Citation: Liu Z, Chen NY, Zhang Z, Zhou S, Hu SY. F-box only protein 2 exacerbates non-alcoholic fatty liver disease by targeting the hydroxyl CoA dehydrogenase alpha subunit. World J Gastroenterol 2023; 29(28): 4433-4450
- URL: https://www.wjgnet.com/1007-9327/full/v29/i28/4433.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i28.4433
Non-alcoholic fatty liver disease (NAFLD), a clinicopathologic syndrome, is characterised by an excessive deposition of fat in hepatocytes [triglyceride (TG) content exceeding 5% of the liver weight] excluding alcohol and other liver-damaging factors[1-3]. With an increase in the incidence of obesity and its associated metabolic syndrome, more than one-third of adults are affected by NAFLD globally[4-6]. NAFLD includes simple fatty liver, non-alcoholic steatohepatitis, and related cirrhosis, which can eventually develop into decompensated cirrhosis and hepatocellular carcinoma[7,8]. The key issues affecting the treatment of patients with NAFLD are atherosclerotic cardio-cerebrovascular diseases, cirrhosis, and malignant tumours associated with the metabolic syndrome[9-11]. Since the pathogenesis of NAFLD remains unclear[12], a clinically effective drug to treat NAFLD is unavailable. A healthy diet and moderate exercise remain the main measures to treat NAFLD clinically[13].
The hydroxyl CoA dehydrogenase alpha subunit (HADHA), a fatty acid β-oxidation enzyme, has been identified as a key pathogenic regulator of metabolism-related diseases, such as NAFLD, obesity, and diabetes[14-16]. Previous studies have confirmed that the expression of HADHA is decreased in L02 cells treated with free fatty acids (FFA) and in mice fed with a high-fat diet (HFD). Knocking out HADHA aggravated liver steatosis, inflammation, and oxidative stress in FFA-treated L02 cells. In addition, oxidative stress and liver damage in NAFLD mice are alleviated by the up-regulation of HADHA[17]. Meanwhile, HADHA can be affected by the ubiquitin-proteasome system (UPS), which mediates important regulatory modifications. Ubiquitination and the degradation of HADHA were reported to be mediated by ubiquitin-conjugating enzyme E2 O, thereby modulating lipid metabolism[18]. However, it remains unclear if there is another regulatory factor that could control and mediate the ubiquitination of HADHA, thus affecting the pathogenesis of HADHA-mediated NAFLD. Therefore, confirming the regulatory signaling pathway in NAFLD that inhibits or activates HADHA is necessary.
The UPS comprises an evolutionarily conserved protein degradation mechanism, including ubiquitin activation (E1), ubiquitin coupling (E2), and ubiquitin ligase (E3) enzymes[19,20]. F-box only protein 2 (FBXO2), a well-known E3 ligase, plays an indispensable role in the UPS[21] and has unique functions in various pathological processes. For example, FBXO2 regulates signal transducer and activator of transcription 3 signaling responsible for modulating the proliferation and tumorigenicity of osteosarcoma cells[22], targets glycosylated SAD1/UNC84 domain protein-2 for ubiquitination and degradation to promote the development of ovarian cancer[21] and mediates the clearance of damaged lysosomes and alters the neurodegeneration phenotype of the brain in Niemann-Pick C disease[23]. In addition, the abnormal expression of FBXO2 in obese mice disrupts glucose homeostasis through ubiquitin-mediated insulin receptor degradation[24]. However, the role of FBXO2 in NAFLD pathology remains unclear.
In this study, we found that expression of FBXO2 was upregulated in the liver tissues of patients with NAFLD when compared with those in liver tissues of normal controls via the whole genome sequencing (WGS) analysis. These findings urged us to explore whether FBXO2 can promote the progression of NAFLD and its mechanism. Our findings can clarify whether FBXO2 is a feasible therapeutic target for NAFLD and related metabolism disorders.
Liver tissue samples were collected from 10 patients with NAFLD, who underwent sleeve gastrectomy with liver biopsy at the Qilu Hospital of Shandong University, Shandong, China. Control liver tissue samples were obtained from 10 patients with normal metabolic conditions who underwent hepatic hemangioma resection at the same hospital.
The inclusion criteria were: Physical examination, laboratory investigation, ultrasound echography, and a liver biopsy consistent with the diagnosis of NAFLD, according to the Brunt criteria. Exclusion criteria were: Excess alcohol consumption (≥ 20 g/d for women and ≥ 30 g/d for men), the presence of hepatitis B virus surface antigen or hepatitis C virus antibodies in the absence of a history of vaccination, use of drugs linked to NAFLD, evidence of other specific liver diseases, such as autoimmune liver disease, and hemochromatosis[25].
A liver biopsy during surgery was performed to provide foundation to non-alcoholic steatohepatitis (NASH) score. Clinical data of the normal controls and patients with NAFLD are shown in Table 1. Written informed consent was obtained from the patients and their families. All procedures involving human liver samples were approved by the Ethics Committee of the Qilu Hospital and adhered to the principles of the Declaration of Helsinki (approval No. KYLL-2017-073).
| Normal (n = 10) | NAFLD (n = 10) | t value | P value | |
| Gender | - | - | ||
| Male, % | 4 (40) | 4 (40) | ||
| Female, % | 6 (60) | 6 (60) | ||
| Age, yr | 33.30 ± 2.83 | 32.40 ± 7.09 | 0.373 | 0.714 |
| Height, cm | 169.10 ± 7.84 | 169.80 ± 10.38 | 0.170 | 0.867 |
| Body weight, kg | 70.95 ± 16.48 | 118.04 ± 28.87 | 4.480 | < 0.001 |
| Body mass index, kg/m2 | 24.51 ± 4.02 | 40.41 ± 5.41 | 7.454 | < 0.001 |
| Waist circumference, cm | 90.80 ± 11.33 | 128.5 ± 3.98 | 9.928 | < 0.001 |
| Systolic BP, mmHg | 126.40 ± 6.33 | 134.80 ± 10.23 | 2.208 | 0.04 |
| ALT, U/L | 12.00 ± 6.77 | 69.00 ± 63.20 | 2.836 | 0.011 |
| AST, U/L | 15.40 ± 3.17 | 46.70 ± 29.74 | 3.309 | 0.004 |
| GLDH, U/L | 1.94 ± 1.22 | 7.74 ± 4.98 | 3.580 | 0.002 |
| T-Cho, mmol/L | 3.97 ± 0.52 | 4.47 ± 0.77 | 1.700 | 0.106 |
| HDL, mmol/L | 1.31 ± 0.20 | 0.97 ± 0.14 | 4.486 | < 0.001 |
| LDL, mmol/L | 2.19 ± 0.37 | 2.83 ± 0.62 | 2.824 | 0.011 |
| sdLDL, mmol/L | 0.34 ± 0.14 | 1.11 ± 0.51 | 4.580 | < 0.001 |
| TG, mmol/L | 0.62 ± 0.20 | 2.05 ± 0.67 | 6.483 | < 0.001 |
| NEFA, µmol/L | 44.41 ± 29.72 | 55.20 ± 15.05 | 1.024 | 0.319 |
| FPG, mmol/L | 4.82 ± 0.33 | 4.81 ± 1.09 | 0.014 | 0.989 |
| NASH score | 0 | 5.10 ± 1.30 | - | - |
RNA extraction and whole genome library sequencing were performed in 12 liver tissues (NAFLD, n = 6; normal, n = 6) by LC-BIO TECHNOLOGIES (Hangzhou, China) CO. Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analyses of the mRNA sequencing part were performed using the DAVID programme (https://david.ncifcrf.gov/). Bar, scatter, and Gene Set Enrichment Analysis result plots were drawn using the LC-BIO Technologies platform (https://www.omicstudio.cn/).
Eight-week-old pathogen-free C57BL/6J male mice (RRID:IMSR_JAX:000664) were purchased from SPF Biotechnology Co. (Beijing, China). Five mice were placed in each cage at a temperature of 23-26 °C with a 12 h light/dark cycle. Animals had free access to food and water. Rodent food was purchased from Xiao Shu You Tai Biotechnology (Beijing, China). The mice used in this investigation were divided randomly.
To induce NAFLD, 10 mice were fed an HFD (#D12492; Research Diets, New Brunswick, NJ, United States) for 16 wk. Control mice were fed with a standard normal chow diet (ND). After 16 wk of feeding an HFD, the weight of the mice in the HFD group exceeded that of mice in the ND group by 20%, and obvious lipid accumulation was observed in the liver. Serum intracellular TG and total cholesterol (T-Cho) levels also increased significantly in the HFD group (P < 0.001, Supplementary Table 1). Finally, eight mice met the experimental criteria. One underweight mouse and one mouse with no significant lipid deposition in the liver were excluded from any analysis.
To establish a liver-specific FBXO2 overexpression model, six mice were injected with adeno-associated virus serotype 8 (AAV8)-His-FBXO2 through the tail vein (100 μL of virus containing 5 × 1011 vg of vectors) after 6 wk of HFD feeding. An empty vector (AAV-His-vector) was injected into the corresponding control mice (n = 6). The virus-infected mice were fed with HFD for an another 16 wk. The AAV used in this study was purchased from WZ Biosciences Inc. (Shandong, China). The study was approved by the Institutional Animal Care and Use Committee of Shandong Provincial Qian Foshan Hospital (approval No. S453) in accordance with internationally accepted principles for the use of laboratory animals.
Human hepatoma cells (HepG2, ATCC; HB-8065 BCRC, #CL-0103) and human embryonic kidney 293 cells (HEK293T, ATCC; CRL-3216 DSMZ, #CL-0005) were cultured in Dulbecco’s Modified Eagle Medium, containing 10% foetal bovine serum and 1% penicillin-streptomycin, in a humidified chamber at 37 °C with 5% CO2. HepG2 cells were exposed to odium oleate (OA) dissolved in 0.5% fatty acid-free bovine serum albumin (BSA) at a final concentration of 0.3 mmol/L for 24 h to establish an in vitro cell lipid deposition model. All cells were purchased from Procell Life Science & Technology Co., Ltd. (Wuhan, China). All cell lines were authenticated via STR identification.
FBXO2 or HADHA overexpression was achieved by transfecting cells with His-FBXO2 plasmid (#NM_012168CH895916, WZ Bioscience Inc., Shandong, China) or Myc-HADHA plasmids (#BC009235HG15019-NM, Sino Biological, Beijing, China), respectively, as per the respective manufacturer’s instructions. His-vector (#66005-1-Ig, Proteintech) and Myc-vector plasmids (#16286-1-AP, Proteintech) were used as controls. FBXO2 knockdown was achieved by transfecting cells with Sh-FBXO2 plasmid (WZ Bioscience Inc). Sh-NC plasmids were used as a control. The DNA sequences of the plasmids used are shown in Supplementary material.
Briefly, liver tissues and HepG2 cells were lysed with RIPA lysis buffer containing protease inhibitors. Western blotting was performed using standard procedures[26,27]. The primary antibodies used are listed in Supplementary Table 2. Image J (RRID:SCR_003070) and GraphPad Prism software (RRID:SCR_002798) were used to analyze the western blotting results.
For the co-immunoprecipitation (co-IP) assay, HepG2 cell protein lysate was prepared. The cracking solution was then washed with 1 mL of cracking buffer (containing 100 μL of protein A/G agarose beads) and the lysate was incubated overnight with the appropriate antibodies. Next, the beads were washed thrice with cracking buffer and then re-suspended in 2 × sodium dodecyl sulfate (SDS) loading buffer. The beads were incubated at 95 °C for 5 min to elute immunoprecipitated proteins. Finally, the proteins were separated using SDS-PAGE and immunoblotted for detection. Mass spectrometry (MS)-based proteomic analysis was performed by the Jinjie Bio company (Hangzhou, China).
Total RNA was extracted from the liver tissues and HepG2 cells using TRIzol reagent (Invitrogen, #12183555). Complementary DNA (cDNA) was synthesised from the isolated RNA using a cDNA synthesis kit (Takara, #RR047A). Subsequently, SYBR Green Master Mix (Vazyme, Q111-02) and a quantitative reverse transcription polymerase chain reaction (qRT-PCR) system (LightCycle 480 II, Roche, Japan) were used to perform qRT-PCR and analyse the results. The primer sequences used in the study are listed in Supplementary Table 3.
Homeostasis model assessment of insulin resistance (HOMA-IR), oral glucose tolerance test (OGTT) and insulin tolerance test (ITT). Blood glucose levels were measured using a Glucometer Elite monitor (ACCU-CHEK, Roche, Germany) and serum insulin was measured using an insulin ELISA kit (D721159-0048, Sangon Biotech). The HOMA-IR index was calculated based on the fasting serum glucose and insulin levels. OGTT was performed via the administration of glucose (2 mg/g) after an overnight fast. ITTs were performed following the i.p. injection of 1 U/kg insulin after 4 h of fasting. OGTT and ITT were performed at 16 wk post adenovirus injection.
Blood was collected from the tail vein of mice. Serum was obtained after the blood was centrifuged at 8000 × g for 20 min at 4 °C. HepG2 cells were lysed using an ultrasonic crusher (SCINTZ-650E; Ningbo Scientz Biotechnology Co., Ningbo, China). Serum TG and T-Cho levels were measured using commercially available assay kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
Hematoxylin and eosin (H&E) staining was done to demonstrate the pattern of lipid accumulation. Oil Red-O staining was done to show the accumulation of fat droplets in the liver.
For immunohistochemistry, paraffin sections of paired human or mouse livers were deparaffinized and rehydrated. Antigens were retrieved using sodium citrate buffer. After blocking with 1% BSA for 1 h at 25 °C, the sections were sequentially incubated with anti-FBXO2 (Proteintech Cat# 14590-1-AP, RRID:AB_2104394) and the corresponding secondary antibodies. The antibodies used in the study are listed in Supplementary Table 2.
For immunofluorescence staining, cells were fixed with 4% paraformaldehyde for 25 min at 25 °C. Cells were then washed thrice with phosphate-buffered saline (PBS) and lysed with 0.1% Triton X-100 in PBS for 20 min. After washing the cells with PBS three times, the cells were incubated in 1% BSA at 37 °C for 1.5 h followed by overnight incubation at 4 °C with rabbit anti-FBXO2 and anti-HADHA antibodies (Proteintech Cat#10758-1-AP, RRID:AB_2115593). After washing thrice with PBS, the cells were incubated with the appropriate secondary antibodies for 1 h at 37 °C. Finally, the cells were stained with 4’,6-diamidino-2-phenylindole. Images were captured using a confocal microscope (IX-73, Olympus, Japan).
The HADHA ubiquitination assay was performed by transfecting HEK293T cells with FBXO2, HADHA, HA-ubiquitin, or empty vectors using Lipofectamine 2000 (#11668, Invitrogen, United States). After 24 h, the cells were washed twice with PBS and lysed in RIPA lysis buffer. The lysates were then centrifuged to obtain cytoplasmic proteins. The obtained proteins were incubated overnight with anti-HADHA antibody followed by incubation with protein A/G agarose beads at 4 °C for an additional 4 h. After washing the beads three times with lysis buffer, the beads were boiled in 2 × SDS loading buffer, separated via the SDS-PAGE, and immunoblotted. All experiments were repeated three times independently.
The t-test was performed for comparisons between two groups. Statistical significance was set at P < 0.05.
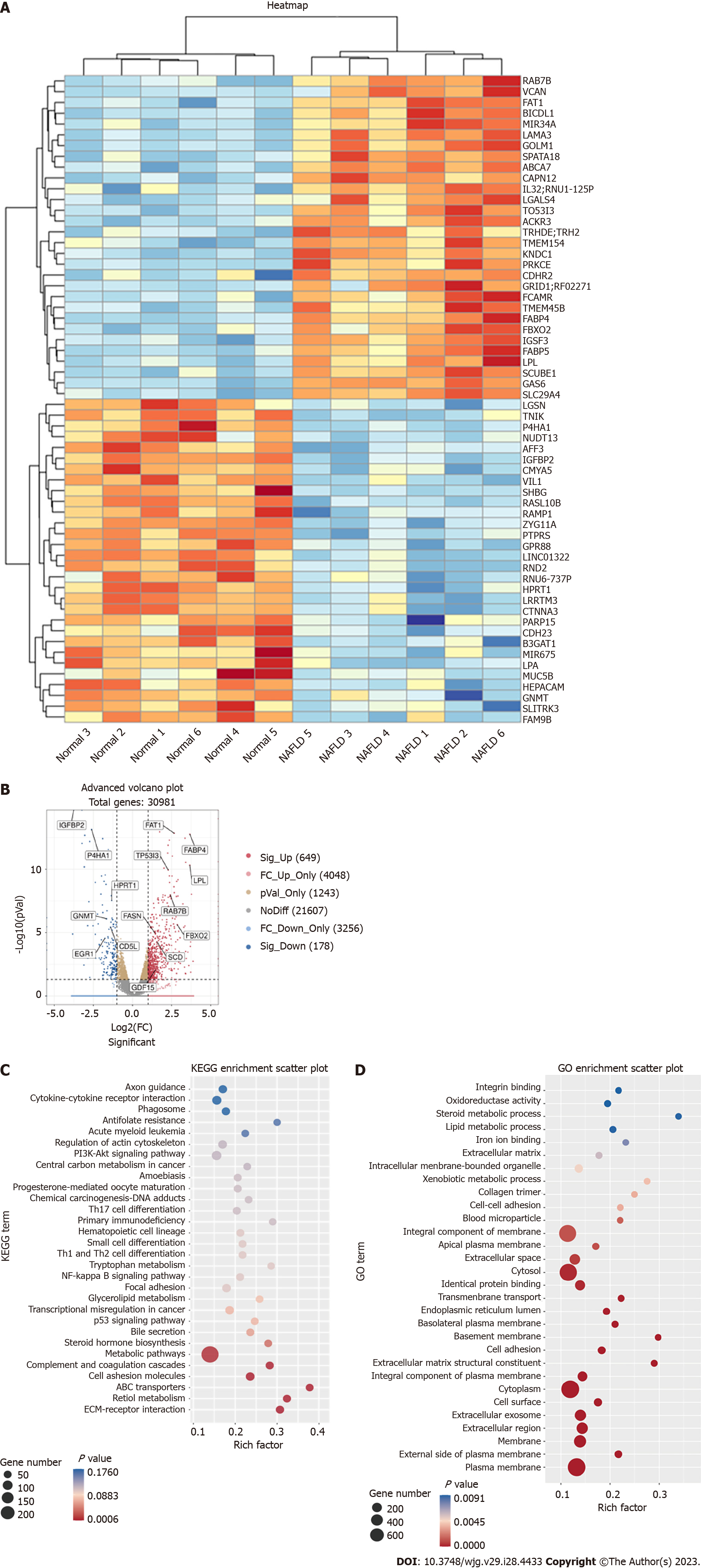
WGS was performed on the liver tissues of six patients with NAFLD and six normal controls. A total of 30982 genes were detected in the WGS analysis, of which 649 were up-regulated and 178 were down-regulated. The heat map in Figure 1A shows the 60 most significantly up- or down-regulated genes. The volcano plot in Figure 1B depicted an overview of the gene sequencing results [log2 (fold change) > 1; P < 0.05]. Some genes marked in our sequencing analysis were closely related to NAFLD[26,28-30], such as fatty acid synthase (FASN), growth differentiation factor 15 and insulin-like growth factor binding protein 2 (Figure 1B). GO and KEGG analyses were also performed (Figures 1C and D, Supple
Among the upregulated genes, the mRNA level of FBXO2 was significantly upregulated among patients with NAFLD. According to previous studies, the abnormal expression of FBXO2 in obese mice disrupts glucose homeostasis through ubiquitin-mediated insulin receptor degradation. We also found that sleeve gastrectomy could downregulate FBXO2 and activate in the phosphatidylinositol 3-kinase-protein kinase B (PI3K-AKT) pathway[31]. In our GO and KEGG analysis, some differential genes were enriched in the PI3K-AKT pathway. Hence, we speculated that FBXO2 may play a role in regulating lipid metabolism in NAFLD.
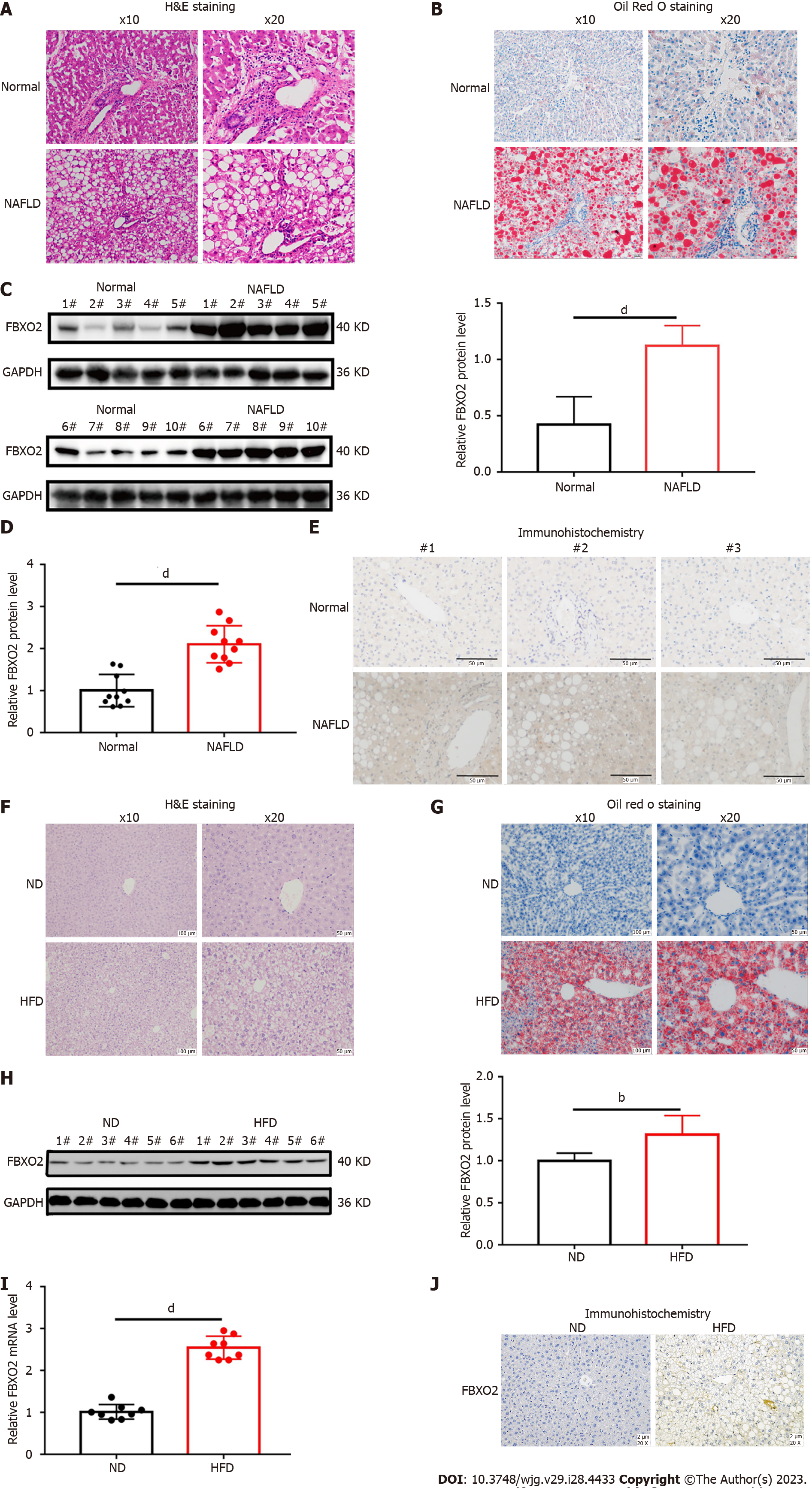
To investigate whether FBXO2 is involved in the occurrence and development of NAFLD, we analyzed its expression in liver tissues isolated from patients with NAFLD (Table 1, Figures 2A and B). We found that the mRNA and protein levels of FBXO2 were significantly higher in liver tissues of patients with NAFLD compared to that in control liver tissues via qRT-PCR analysis (Figure 2D, P < 0.0001), western blot analysis (Figure 2C, P < 0.0001), and immunohistochemical staining (Figure 2E). Moreover, we established a NAFLD mouse model by feeding C57BL6/J mice an HFD for 16 wk (Figures 2F and G, Supplementary Table 1). Similarly, we also found significantly higher FBXO2 mRNA and protein levels in liver samples obtained from HFD mice compared to ND mice via qRT-PCR (Figure 2I, P < 0.0001), western blot (Figure 2H, P < 0.01), and immunohistochemical staining (Figure 2J) analyses. Based on these results, we confirmed that FBXO2 levels in the liver of patients with NAFLD and the HFD-fed mouse model were significantly increased compared to those in non-steatosis samples.
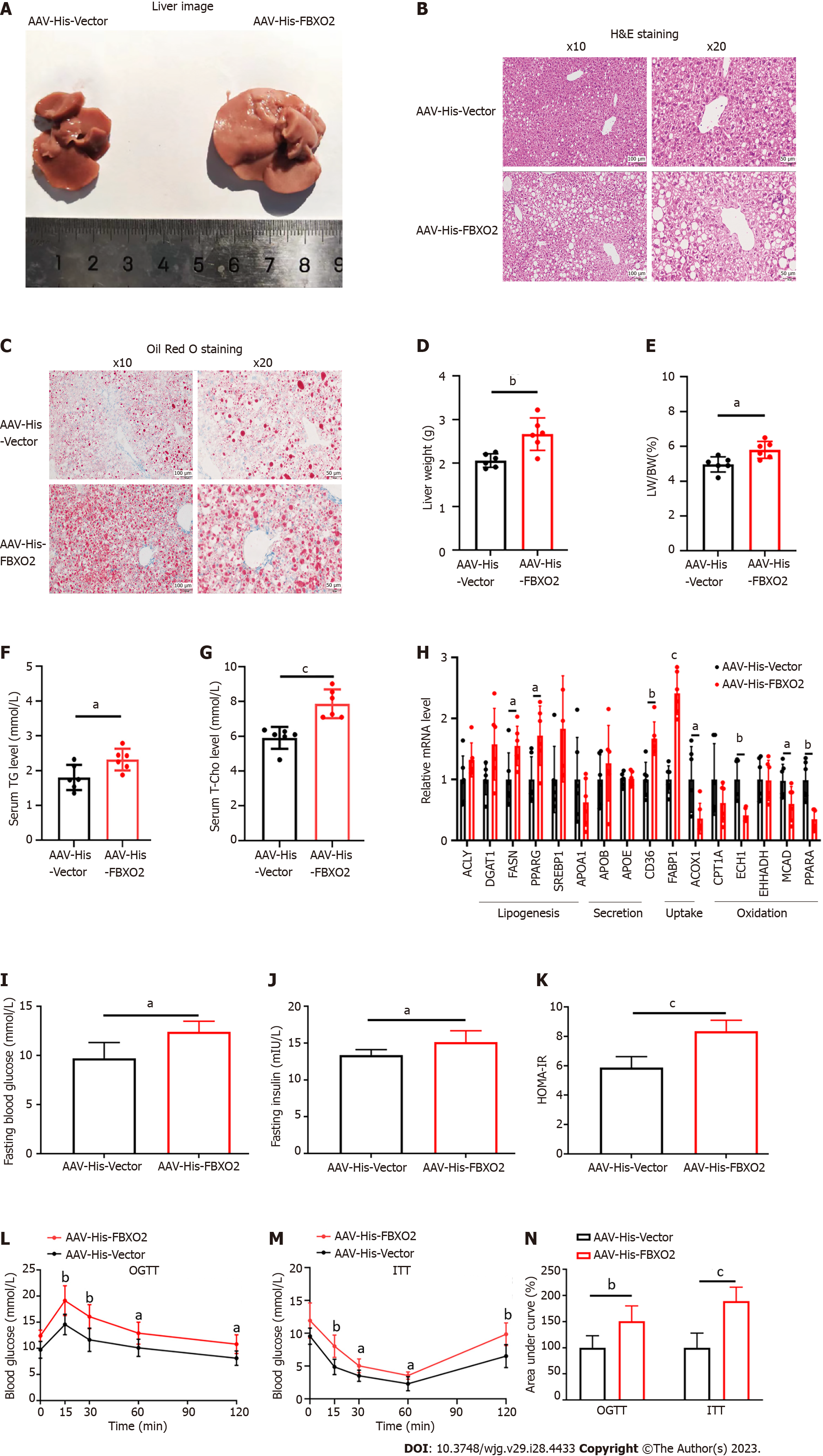
Given the close correlation between FBXO2 expression and a fatty liver, we constructed a hepatocyte-specific FBXO2-overexpressing mouse model (Supplementary Figures 2A and B) to investigate the role of FBXO2 in exacerbating the HFD-induced liver steatosis, insulin resistance, and abnormal glucose metabolism. As expected, AAV-His-FBXO2-treated mice exhibited a significant increase in body weight (Supplementary Figure 2C), liver weight (Figure 3D), and the ratio of liver weight to body weight (Figure 3E) compared to AAV-His-vector-treated mice. The deposition of lipids in the liver was noticeably enhanced in AAV-His-FBXO2 mice, as determined by liver observations (Figure 3A), H&E staining (Figure 3B), Oil Red O staining (Figure 3C), and TG and T-Cho levels (Figures 3F and G). The overexpression of FBXO2 in the livers of AAV-His-FBXO2-treated mice significantly promoted the expression of lipogenesis genes [FASN and peroxisome proliferator-activated receptor gamma (PPARγ)] and fatty acid uptake genes [cluster of differentiation 36 (CD36) and fatty acid binding protein-1 (FABP1)] and suppressed the expression of fatty acid β-oxidation-related genes [acyl-CoA oxidase 1 (ACOX1), enoyl coenzyme A hydratase 1 (ECH1), medium-chain acyl-coenzyme A dehydrogenase (MCAD), and PPARα] (Figure 3H).
NAFLD is often accompanied by glucose metabolic disorders and insulin resistance. Unsurprisingly, FBXO2 overexpression significantly increased fasting blood glucose and insulin levels (Figures 3I and J). The HOMA-IR index was also increased accordingly (Figure 3K). Notably, there was no significant difference in food intake between AAV-His-FBXO2 mice and AAV-His-vector mice (Supplementary Figure 2D). In addition, the AAV-His-FBXO2 mice exhibited significantly higher blood glucose levels than AAV-His-vector mice, as determined from the OGTT and ITT data (Figures 3L-N).
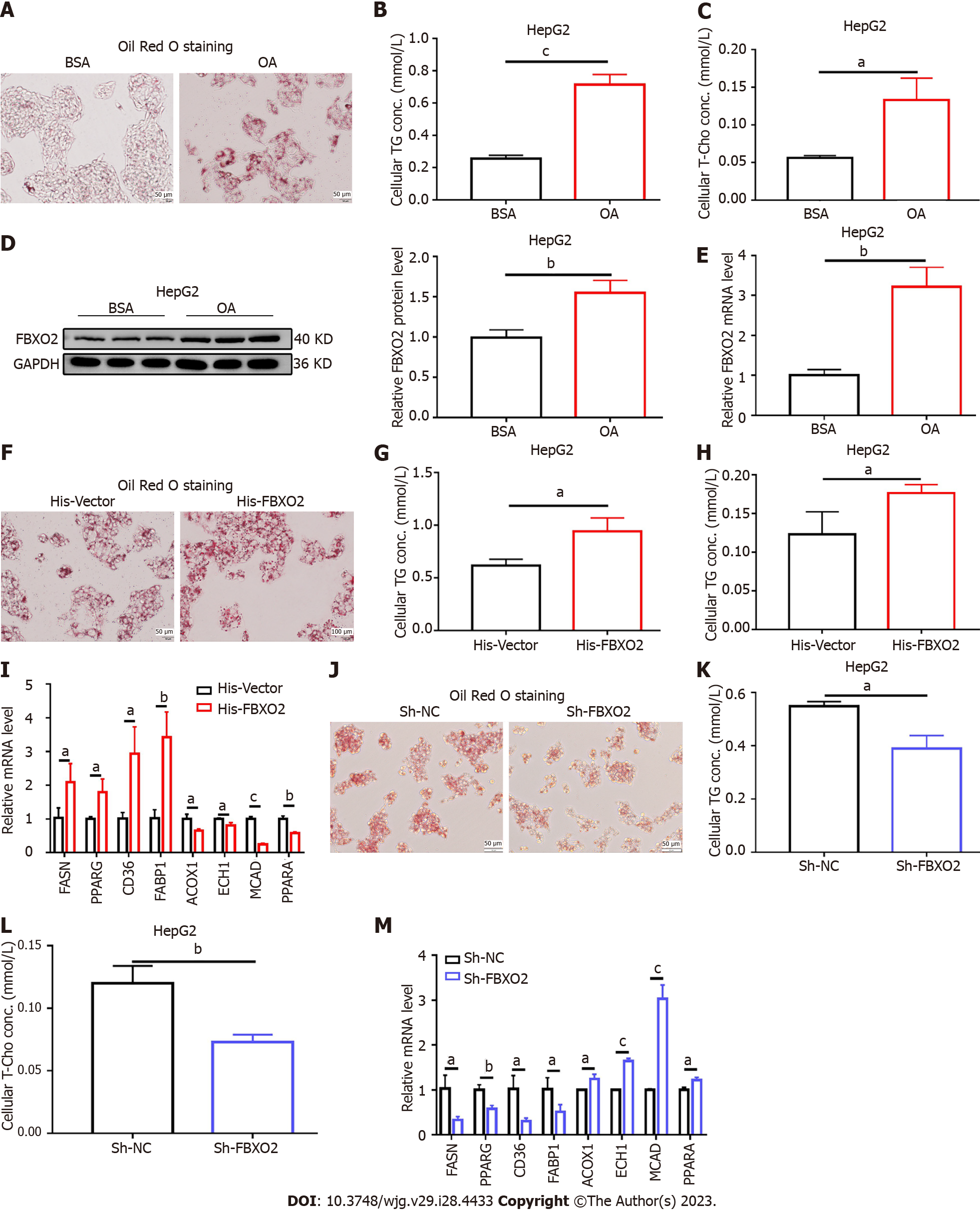
We also investigated the effects of FBXO2 overexpression on lipid metabolic disorders in vitro. HepG2 cells were treated with OA to simulate the NAFLD phenotype. An increase in the mRNA and protein levels of FBXO2 was observed after 24 h of OA administration (Figures 4D and E). Oil Red O staining revealed that the intracellular lipid deposition (Figure 4A) and TG and T-Cho levels in HepG2 cells (Figures 4B and C) increased after OA treatment. Next, the His-FBXO2 plasmid was transfected into HepG2 cells. qRT-PCR and western blotting revealed increased FBXO2 expression (Supplementary Figures 3A and B). The effect of FBXO2 overexpression on lipid deposition was also observed in OA-treated HepG2 cells. Specifically, FBXO2 overexpression after transfecting His-FBXO2 plasmids into HepG2 cells, greatly increased OA-induced lipid deposition (Figure 4F). FBXO2 overexpression also increased OA-induced cellular TG and T-Cho levels (Figures 4G and H). The overexpression of FBXO2 in OA-treated HepG2 cells also promoted the expression of FASN, PPARG, CD36, and FABP1 and suppressed the expression of ACOX1, ECH1, MCAD, and PPARα (Figure 4I). These data suggest that the up-regulation of FBXO2 expression in the liver exacerbated liver steatosis, HFD-triggered insulin resistance, and abnormal glucose metabolism.
After confirming that the overexpression of FBXO2 exacerbated liver steatosis, we examined the effect of FBXO2 knockdown on lipid metabolic disorder in hepatocytes. To achieve this, the Sh-FBXO2 plasmid was transfected into HepG2 cells. Both qRT-PCR and western blot results indicated that FBXO2 expression levels decreased after transfection, confirming successful FBXO2 knockdown (Supplementary Figures 3C and D). FBXO2 knockdown decreased the OA-induced lipid deposition (Figure 4J) and regulated OA-induced TG and T-Cho secretion (Figures 4K and L). The knockdown of FBXO2 in OA-treated HepG2 cells inhibited the expression of lipogenesis genes (FASN and PPARγ) and fatty acid uptake genes (CD36 and FABP1) and promoted the expression of fatty acid β-oxidation related genes (ACOX1, ECH1, MCAD, and PPARα) (Figure 4M). Consistent with these data, in vitro gene therapy using Sh-FBXO2 plasmids further demonstrated that the knockdown of FBXO2 could significantly alleviate OA-induced lipid accumulation in hepatocytes (hepatic steatosis).
We used a co-IP-based MS method to identify which proteins interact with endogenous FBXO2 in HepG2 cells. Based on peptide spectra matching protein score, HADHA was identified as a potential binding protein for FBXO2 in HepG2 cells (Figure 5A). In vitro interaction studies suggested that both exogenous and endogenous FBXO2 could bind directly to HADHA and vice versa. Additionally, the interaction between FBXO2 and HADHA was determined via a co-IP assay in HEK293T (Figures 5C and D) and HepG2 cells (Figures 5E and F). In vitro interaction tests suggested that both exogenous and endogenous FBXO2 could directly bind to HADHA and vice versa. Immunofluorescence staining also revealed the co-localization of FBXO2 and HADHA after the HEK293T cells were transfected with His-FBXO2 and Myc-HADHA plasmids (Figure 5B).
Next, we tested whether FBXO2 could activate the ubiquitination of HADHA. The expression of FBXO2 and HA-Ub together significantly enhanced the ubiquitination of HADHA in HEK293T cells, compared to control cells transfected with vectors expressing FBXO2 and HA-Ub alone (Figure 6A). Consistent with the results of the ubiquitination assays, gene expression analysis supported the reduction of HADHA protein levels in HepG2 cells and HFD-fed mouse models overexpressing FBXO2 (Figures 6B and D). Further, HADHA degradation was reduced after co-treatment with the proteasome inhibitor MG132 (SML1135; Sigma, United States) in HepG2 cells (Figure 6E). Moreover, FBXO2 knockdown increased the level protein of HADHA in HepG2 cells (Figure 6C). Notably, the mRNA levels of HADHA were not affected by FBXO2 overexpression or knockdown (Supplementary Figures 4A-C). Additionally, we analysed whether FBXO2 decreased the half-life of the HADHA protein. We transfected HepG2 cells with His-Vector or His-FBXO2 plasmids and treated them with the protein synthesis inhibitor cycloheximide (CHX) at different time points. The half-life of the HADHA protein was drastically reduced in FBXO2-overexpressing cells compared to the control cells (Figure 6F). These findings indicated that FBXO2 facilitates the degradation of HADHA via the ubiquitination machinery.
The above finding led us to determine whether HADHA alterations mediate the effect of FBXO2 on lipid accumulation in hepatocytes. First, FBXO2 overexpression increased OA-induced lipid deposition and TG and T-Cho secretion. Moreover, the overexpression of FBXO2 in OA- treated HepG2 cells enhanced the expression of FASN, PPARγ, CD36, and FABP1 and inhibited the expression of ACOX1, ECH1, MCAD, and PPARα. However, these effects were reversed after HepG2 cells were transfected with the Myc-HADHA plasmid to overexpress HADHA as indicated by the changes in lipid deposition (Figure 6G), TG and T-Cho secretion (Figures 6H and I), and mRNA expression of lipid metabolism-related genes (Figure 6J). FBXO2 and HADHA overexpression efficiency was also confirmed via western blot (Supplementary Figure 3E). Collectively, these indicated that the up-regulation of FBXO2 exacerbated OA-induced lipid accumulation by promoting the ubiquitination and degradation of HADHA in HepG2 cells.
FBXO2 is an E3 ligase that can regulate protein stability through ubiquitin-mediated protein-protein interaction and plays an indispensable role in the cell cycle regulation of the UPS[21]. It regulates cell proliferation, transcription, and apoptosis through the ubiquitination and degradation of target proteins, thereby affecting the occurrence and development of various diseases, such as malignant tumours, neuronal diseases, and inflammation[22,24,32,33]. However, the status of FBXO2 expression in NAFLD and its relationship with clinicopathological parameters has not been clarified. In the present study, we found that FBXO2 levels were significantly up-regulated in NAFLD liver tissues. Subsequently, we simulated NAFLD in HFD mice and OA-treated HepG2 cells, and the results were consistent with those obtained in human liver tissue. Our sequencing results of NAFLD liver tissue samples revealed changes in multiple metabolic pathways, for example the PI3K-AKT pathway. Recent studies conducted by our team found that the expression level of FBXO2 in NAFLD mouse models was downregulated after sleeve gastrectomy. Upregulation of FBXO2 could activate the PI3K-AKT pathway[31]. Based on the above results, we speculate that FBXO2 upregulation is related to NAFLD occurrence and development.
Next, we constructed a hepatocyte-specific FBXO2-overexpressing mouse model to further explore the relationship between FBXO2 and NAFLD. Lipid accumulation in the liver is a clinical feature of NAFLD and is caused by an imbalance between lipid input and output[34]. This imbalance is regulated by four pathways: Lipid lipogenesis, lipid uptake, lipid secretion, and fatty acid oxidation[27,35]. In the present study, we found that the deposition of liver lipids was visibly enhanced in hepatocyte-specific FBXO2-overexpressing mice models. FBXO2 overexpression in the liver significantly promoted the expression of lipogenesis genes (FASN and PPARγ) and fatty acid uptake genes (CD36 and FABP1) and inhibited the expression of fatty acid β-oxidation-related genes (ACOX1, ECH1, MCAD and PPARα) in AAV-His-FBXO2 mice. In contrast, FBXO2 knockdown in OA-treated HepG2 cells had the opposite effect on the expression of lipid accumulation and the expression of lipid metabolism-related genes. The alleviative effect of FBXO2 knockdown on lipid metabolism suggests its effect in treating NAFLD. In addition, combined with the effect of the overexpression and knockdown of FBXO2 on lipid metabolism, it is worth considering that FBXO2 may promote the ubiquitination of one or more proteins that can mediate lipid metabolism. In other words, the overexpression of FBXO2 can accelerate protein degradation, thus aggravating NAFLD. On the contrary, the knockdown of FBXO2 protects lipid metabolising enzymes from degradation, thus maintaining normal lipid metabolism.
To further explore the mechanism through which FBXO2 affects NAFLD, we screened the proteins that could interact with endogenous FBXO2 in HepG2 cells using a co-IP-based MS method. The mitochondrial trifunctional enzyme subunit-alpha (HADHA) was identified as a potential binding protein for FBXO2. HADHA, a mitochondrial β-oxidation-associated enzyme, encodes the α-subunit of the mitochondrial trifunctional protein (hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase). This enzyme complex catalyses the three steps of the β-oxidation of fatty acids in the mitochondria[14]. HADHA deficiency in mice has been reported to aggravate NAFLD[36-38]. Further, HADHA knockout can lead to hyperglycemia and consequently induce liver lipid deposition[18,39]. In vitro interaction tests in the present study suggested that both exogenous and endogenous FBXO2 could directly bind to HADHA and vice versa. In addition, the results of our ubiquitination assays showed that FBXO2 facilitates the degradation of HADHA via the ubiquitination machinery. We further confirmed the interaction between FBXO2 and HADHA in hepatocytes using MG132 and CHX. We also found that FBXO2 overexpression during OA-induced liver steatosis was reversed after Myc-HADHA plasmids were transfected into HepG2 cells to overexpress HADHA.
Another interesting observation is that in addition to the exacerbation of steatosis, the targeted upregulation of FBXO2 expression in HFD-fed mouse livers led to a significant exacerbation of hyperglycemia and insulin resistance, which could be caused by various reasons. Previous studies have shown that FBXO2 targets insulin receptors to achieve ubiquitin-dependent degradation, thus regulating insulin signaling integrity[24]. The overexpression of FBXO2 can stimulate severe insulin resistance in the liver and inhibit its normal function, promoting adipogenesis. Therefore, increased insulin resistance leads to excessive lipid accumulation in the liver[34]. Recent studies have shown that HADHA knockdown augments the glucagon response[14]. This study speculates that a decrease in HADHA levels might induce an increase in blood glucose levels after the overexpression of FBXO2. In addition, the increased expression of lipogenesis- and fatty acid uptake-associated genes and decreased expression of fatty acid β-oxidation-related genes might also contribute to the observed effects on glycemic control[40-44].
Liver FBXO2 expression is upregulated in NAFLD and FBXO2 is correlated with metabolic disorders. Moreover, upregulation of FBXO2 exacerbates liver steatosis by promoting the degradation of HADHA. Finally, our findings suggest that FBXO2 can be used as a feasible therapeutic target for NAFLD.
Non-alcoholic fatty liver disease (NAFLD) is a major health burden with an increasing global incidence. Unfortunately, the unavailability of knowledge underlying NAFLD pathogenesis inhibits effective preventive and therapeutic measures. The key issues affecting the treatment of patients with NAFLD are atherosclerotic cardio-cerebrovascular diseases, cirrhosis, and malignant tumors associated with the metabolic syndrome. The abnormal expression of F-box only protein 2 (FBXO2), an E3 ligase, in obese mice disrupts glucose homeostasis through ubiquitin-mediated insulin receptor degradation, however, the role of FBXO2 in NAFLD pathology remains unclear.
Recent studies conducted by our team found that the expression level of FBXO2 in NAFLD mouse models was downregulated after sleeve gastrectomy.
This study aimed to explore the molecular mechanism of NAFLD.
Whole genome sequencing (WGS) analysis was performed on liver tissues from patients with NAFLD (n = 6) and patients with normal metabolic conditions (n = 6) to identify the target genes. A NAFLD C57BL6/J mouse model induced by 16 wk of high-fat diet (HFD) feeding and a hepatocyte-specific FBXO2 overexpression mouse model were used for in vivo studies. Plasmid transfection, co-immunoprecipitation-based mass spectrometry assays, and ubiquitination in HepG2 cells and HEK293T cells were used for in vitro studies.
A total of 30982 genes were detected in WGS analysis, with 649 up-regulated and 178 down-regulated. Expression of FBXO2 was upregulated in the liver tissues of patients with NAFLD. Hepatocyte-specific FBXO2 overexpression facilitated NAFLD-associated phenotypes in mice. Overexpression of FBXO2 aggravated odium oleate (OA)-induced lipid accumulation in HepG2 cells, resulting in an abnormal expression of genes related to lipid metabolism, such as fatty acid synthase, peroxisome proliferator-activated receptor alpha, and so on. In contrast, knocking down FBXO2 in HepG2 cells significantly alleviated the oleate-induced lipid accumulation and aberrant expression of lipid metabolism genes. The hydroxyl CoA dehydrogenase alpha subunit (HADHA), a protein involved in oxidative stress, was a target of FBXO2-mediated ubiquitination. FBXO2 directly bound to HADHA and facilitated its proteasomal degradation in HepG2 and HEK293T cells. Supplementation with HADHA alleviated lipid accumulation caused by FBXO2 overexpression in HepG2 cells.
FBXO2 exacerbates lipid accumulation by targeting HADHA.
Our results shed light on the role of FBXO2 not only in NAFLD but also in other metabolic diseases involving lipid accumulation, supporting further investigation of the therapeutic potential of this molecule in further studies.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gasperetti A, Italy; Strong VE, United States S-Editor: Wang JJ L-Editor: A P-Editor: Yu HG
| 1. | Drescher HK, Weiskirchen S, Weiskirchen R. Current Status in Testing for Nonalcoholic Fatty Liver Disease (NAFLD) and Nonalcoholic Steatohepatitis (NASH). Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 97] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 2. | Hirode G, Vittinghoff E, Wong RJ. Increasing Clinical and Economic Burden of Nonalcoholic Fatty Liver Disease Among Hospitalized Adults in the United States. J Clin Gastroenterol. 2019;53:765-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 3. | Conway BN, Han X, Munro HM, Gross AL, Shu XO, Hargreaves MK, Zheng W, Powers AC, Blot WJ. The obesity epidemic and rising diabetes incidence in a low-income racially diverse southern US cohort. PLoS One. 2018;13:e0190993. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 4. | Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, George J, Bugianesi E. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15:11-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4054] [Cited by in RCA: 3795] [Article Influence: 542.1] [Reference Citation Analysis (2)] |
| 5. | Huby T, Gautier EL. Immune cell-mediated features of non-alcoholic steatohepatitis. Nat Rev Immunol. 2022;22:429-443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 358] [Article Influence: 119.3] [Reference Citation Analysis (0)] |
| 6. | Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24:908-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1376] [Cited by in RCA: 2918] [Article Influence: 416.9] [Reference Citation Analysis (1)] |
| 7. | Byrne CD, Targher G. NAFLD: a multisystem disease. J Hepatol. 2015;62:S47-S64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1516] [Cited by in RCA: 2140] [Article Influence: 214.0] [Reference Citation Analysis (0)] |
| 8. | Rinella ME, Sanyal AJ. Management of NAFLD: a stage-based approach. Nat Rev Gastroenterol Hepatol. 2016;13:196-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 284] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 9. | Li K, Zhang K, Wang H, Wu Y, Chen N, Chen J, Qiu C, Cai P, Li M, Liang X, Su D. Hrd1-mediated ACLY ubiquitination alleviate NAFLD in db/db mice. Metabolism. 2021;114:154349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 10. | Donne R, Saroul-Ainama M, Cordier P, Hammoutene A, Kabore C, Stadler M, Nemazanyy I, Galy-Fauroux I, Herrag M, Riedl T, Chansel-Da Cruz M, Caruso S, Bonnafous S, Öllinger R, Rad R, Unger K, Tran A, Couty JP, Gual P, Paradis V, Celton-Morizur S, Heikenwalder M, Revy P, Desdouets C. Replication stress triggered by nucleotide pool imbalance drives DNA damage and cGAS-STING pathway activation in NAFLD. Dev Cell. 2022;57:1728-1741.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 36] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 11. | Fang J, Ji YX, Zhang P, Cheng L, Chen Y, Chen J, Su Y, Cheng X, Zhang Y, Li T, Zhu X, Zhang XJ, Wei X. Hepatic IRF2BP2 Mitigates Nonalcoholic Fatty Liver Disease by Directly Repressing the Transcription of ATF3. Hepatology. 2020;71:1592-1608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | Chen Z, Yu Y, Cai J, Li H. Emerging Molecular Targets for Treatment of Nonalcoholic Fatty Liver Disease. Trends Endocrinol Metab. 2019;30:903-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 97] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 13. | Lonardo A, Mantovani A, Petta S, Carraro A, Byrne CD, Targher G. Metabolic mechanisms for and treatment of NAFLD or NASH occurring after liver transplantation. Nat Rev Endocrinol. 2022;18:638-650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 32] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 14. | Pan A, Sun XM, Huang FQ, Liu JF, Cai YY, Wu X, Alolga RN, Li P, Liu BL, Liu Q, Qi LW. The mitochondrial β-oxidation enzyme HADHA restrains hepatic glucagon response by promoting β-hydroxybutyrate production. Nat Commun. 2022;13:386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 39] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 15. | Le-Tian Z, Cheng-Zhang H, Xuan Z, Zhang Q, Zhen-Gui Y, Qing-Qing W, Sheng-Xuan W, Zhong-Jin X, Ran-Ran L, Ting-Jun L, Zhong-Qu S, Zhong-Hua W, Ke-Rong S. Protein acetylation in mitochondria plays critical functions in the pathogenesis of fatty liver disease. BMC Genomics. 2020;21:435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 16. | Nakamura MT, Yudell BE, Loor JJ. Regulation of energy metabolism by long-chain fatty acids. Prog Lipid Res. 2014;53:124-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 395] [Cited by in RCA: 568] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 17. | Ding J, Wu L, Zhu G, Zhu J, Luo P, Li Y. HADHA alleviates hepatic steatosis and oxidative stress in NAFLD via inactivation of the MKK3/MAPK pathway. Mol Biol Rep. 2023;50:961-970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 18. | Ma M, Zhang C, Cao R, Tang D, Sang X, Zou S, Wang X, Xu H, Liu G, Dai L, Tian Y, Gao X, Fu X. UBE2O promotes lipid metabolic reprogramming and liver cancer progression by mediating HADHA ubiquitination. Oncogene. 2022;41:5199-5213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 19. | Kodroń A, Mussulini BH, Pilecka I, Chacińska A. The ubiquitin-proteasome system and its crosstalk with mitochondria as therapeutic targets in medicine. Pharmacol Res. 2021;163:105248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 20. | Kwon YT, Ciechanover A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem Sci. 2017;42:873-886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 581] [Article Influence: 72.6] [Reference Citation Analysis (0)] |
| 21. | Ji J, Shen J, Xu Y, Xie M, Qian Q, Qiu T, Shi W, Ren D, Ma J, Liu W, Liu B. FBXO2 targets glycosylated SUN2 for ubiquitination and degradation to promote ovarian cancer development. Cell Death Dis. 2022;13:442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 22. | Zhao X, Guo W, Zou L, Hu B. FBXO2 modulates STAT3 signaling to regulate proliferation and tumorigenicity of osteosarcoma cells. Cancer Cell Int. 2020;20:245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 23. | Liu EA, Schultz ML, Mochida C, Chung C, Paulson HL, Lieberman AP. Fbxo2 mediates clearance of damaged lysosomes and modifies neurodegeneration in the Niemann-Pick C brain. JCI Insight. 2020;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 24. | Liu B, Lu H, Li D, Xiong X, Gao L, Wu Z, Lu Y. Aberrant Expression of FBXO2 Disrupts Glucose Homeostasis Through Ubiquitin-Mediated Degradation of Insulin Receptor in Obese Mice. Diabetes. 2017;66:689-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 25. | Weber M, Mera P, Casas J, Salvador J, Rodríguez A, Alonso S, Sebastián D, Soler-Vázquez MC, Montironi C, Recalde S, Fucho R, Calderón-Domínguez M, Mir JF, Bartrons R, Escola-Gil JC, Sánchez-Infantes D, Zorzano A, Llorente-Cortes V, Casals N, Valentí V, Frühbeck G, Herrero L, Serra D. Liver CPT1A gene therapy reduces diet-induced hepatic steatosis in mice and highlights potential lipid biomarkers for human NAFLD. FASEB J. 2020;34:11816-11837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 26. | Yang H, Deng Q, Ni T, Liu Y, Lu L, Dai H, Wang H, Yang W. Targeted Inhibition of LPL/FABP4/CPT1 fatty acid metabolic axis can effectively prevent the progression of nonalcoholic steatohepatitis to liver cancer. Int J Biol Sci. 2021;17:4207-4222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 54] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 27. | Ma N, Wang YK, Xu S, Ni QZ, Zheng QW, Zhu B, Cao HJ, Jiang H, Zhang FK, Yuan YM, Zhang EB, Chen TW, Xia J, Ding XF, Chen ZH, Zhang XP, Wang K, Cheng SQ, Qiu L, Li ZG, Yu YC, Wang XF, Zhou B, Li JJ, Xie D. PPDPF alleviates hepatic steatosis through inhibition of mTOR signaling. Nat Commun. 2021;12:3059. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 28. | Wang D, Day EA, Townsend LK, Djordjevic D, Jørgensen SB, Steinberg GR. GDF15: emerging biology and therapeutic applications for obesity and cardiometabolic disease. Nat Rev Endocrinol. 2021;17:592-607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 277] [Article Influence: 69.3] [Reference Citation Analysis (0)] |
| 29. | Hu Y, He W, Huang Y, Xiang H, Guo J, Che Y, Cheng X, Hu F, Hu M, Ma T, Yu J, Tian H, Tian S, Ji YX, Zhang P, She ZG, Zhang XJ, Huang Z, Yang J, Li H. Fatty Acid Synthase-Suppressor Screening Identifies Sorting Nexin 8 as a Therapeutic Target for NAFLD. Hepatology. 2021;74:2508-2525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 65] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 30. | Park HS, Song JW, Park JH, Lim BK, Moon OS, Son HY, Lee JH, Gao B, Won YS, Kwon HJ. TXNIP/VDUP1 attenuates steatohepatitis via autophagy and fatty acid oxidation. Autophagy. 2021;17:2549-2564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 139] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 31. | Chen N, Cao R, Zhang Z, Zhou S, Hu S. Sleeve Gastrectomy Improves Hepatic Glucose Metabolism by Downregulating FBXO2 and Activating the PI3K-AKT Pathway. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 32. | Yamada A, Hikichi M, Nozawa T, Nakagawa I. FBXO2/SCF ubiquitin ligase complex directs xenophagy through recognizing bacterial surface glycan. EMBO Rep. 2021;22:e52584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 33. | Che X, Jian F, Wang Y, Zhang J, Shen J, Cheng Q, Wang X, Jia N, Feng W. FBXO2 Promotes Proliferation of Endometrial Cancer by Ubiquitin-Mediated Degradation of FBN1 in the Regulation of the Cell Cycle and the Autophagy Pathway. Front Cell Dev Biol. 2020;8:843. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 34. | Xu M, Tan J, Dong W, Zou B, Teng X, Zhu L, Ge C, Dai X, Kuang Q, Zhong S, Lai L, Yi C, Tang T, Zhao J, Wang L, Liu J, Wei H, Sun Y, Yang Q, Li Q, Lou D, Hu L, Liu X, Kuang G, Luo J, Xiong M, Feng J, Zhang C, Wang B. The E3 ubiquitin-protein ligase Trim31 alleviates non-alcoholic fatty liver disease by targeting Rhbdf2 in mouse hepatocytes. Nat Commun. 2022;13:1052. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 30] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 35. | Yu M, Alimujiang M, Hu L, Liu F, Bao Y, Yin J. Berberine alleviates lipid metabolism disorders via inhibition of mitochondrial complex I in gut and liver. Int J Biol Sci. 2021;17:1693-1707. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 58] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 36. | Nassir F, Arndt JJ, Johnson SA, Ibdah JA. Regulation of mitochondrial trifunctional protein modulates nonalcoholic fatty liver disease in mice. J Lipid Res. 2018;59:967-973. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 37. | Malloy VL, Perrone CE, Mattocks DA, Ables GP, Caliendo NS, Orentreich DS, Orentreich N. Methionine restriction prevents the progression of hepatic steatosis in leptin-deficient obese mice. Metabolism. 2013;62:1651-1661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 91] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 38. | Dang Y, Xu J, Zhu M, Zhou W, Zhang L, Ji G. Gan-Jiang-Ling-Zhu decoction alleviates hepatic steatosis in rats by the miR-138-5p/CPT1B axis. Biomed Pharmacother. 2020;127:110127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 39. | Bhatia H, Pattnaik BR, Datta M. Inhibition of mitochondrial β-oxidation by miR-107 promotes hepatic lipid accumulation and impairs glucose tolerance in vivo. Int J Obes (Lond). 2016;40:861-869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 40. | Karagiannis F, Masouleh SK, Wunderling K, Surendar J, Schmitt V, Kazakov A, Michla M, Hölzel M, Thiele C, Wilhelm C. Lipid-Droplet Formation Drives Pathogenic Group 2 Innate Lymphoid Cells in Airway Inflammation. Immunity. 2020;52:620-634.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 41. | Cifarelli V, Appak-Baskoy S, Peche VS, Kluzak A, Shew T, Narendran R, Pietka KM, Cella M, Walls CW, Czepielewski R, Ivanov S, Randolph GJ, Augustin HG, Abumrad NA. Visceral obesity and insulin resistance associate with CD36 deletion in lymphatic endothelial cells. Nat Commun. 2021;12:3350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 81] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 42. | Manickam R, Tur J, Badole SL, Chapalamadugu KC, Sinha P, Wang Z, Russ DW, Brotto M, Tipparaju SM. Nampt activator P7C3 ameliorates diabetes and improves skeletal muscle function modulating cell metabolism and lipid mediators. J Cachexia Sarcopenia Muscle. 2022;13:1177-1196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 35] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 43. | Castaño C, Kalko S, Novials A, Párrizas M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc Natl Acad Sci U S A. 2018;115:12158-12163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 293] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 44. | Tu J, Zhu S, Li B, Xu G, Luo X, Jiang L, Yan X, Zhang R, Chen C. Gegen Qinlian Decoction Coordinately Regulates PPARγ and PPARα to Improve Glucose and Lipid Homeostasis in Diabetic Rats and Insulin Resistance 3T3-L1 Adipocytes. Front Pharmacol. 2020;11:811. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |