Published online Jul 21, 2023. doi: 10.3748/wjg.v29.i27.4317
Peer-review started: March 28, 2023
First decision: April 27, 2023
Revised: May 10, 2023
Accepted: June 2, 2023
Article in press: June 2, 2023
Published online: July 21, 2023
Processing time: 106 Days and 22.9 Hours
Gastric cancer (GC) is one of the most common cancer types worldwide, and its prevention and treatment methods have garnered much attention. As the active ingredient of licorice, 18β-glycyrrhetinic acid (18β-GRA) has a variety of pharmacological effects. The aim of this study was to explore the effective target of 18β-GRA in the treatment of GC, in order to provide effective ideas for the clinical prevention and treatment of GC.
To investigate the mechanism of 18β-GRA in inhibiting cell proliferation and promoting autophagy flux in GC cells.
Whole transcriptomic analyses were used to analyze and screen differentially expressed microRNAs (miRNAs) in GC cells after 18β-GRA intervention. Lentivirus-transfected GC cells and the Cell Counting Kit-8 were used to detect cell proliferation ability, cell colony formation ability was detected by the clone formation assay, and flow cytometry was used to detect the cell cycle and apoptosis. A nude mouse transplantation tumor model of GC cells was constructed to verify the effect of miR-328-3p overexpression on the tumorigenicity of GC cells. Tumor tissue morphology was observed by hematoxylin and eosin staining, and microtubule-associated protein light chain 3 (LC3) expression was detected by immunohistochemistry. TransmiR, STRING, and miRWalk databases were used to predict the relationship between miR-328-3p and signal transducer and activator of transcription 3 (STAT3)-related information. Expression of STAT3 mRNA and miR-328-3p was detected by quantitative polymerase chain reaction (qPCR) and the expression levels of STAT3, phosphorylated STAT3 (p-STAT3), and LC3 were detected by western blot analysis. The targeted relationship between miR-328-3p and STAT3 was detected using the dual-luciferase reporter gene system. AGS cells were infected with monomeric red fluorescent protein-green fluorescent protein-LC3 adenovirus double label. LC3 was labeled and autophagy flow was observed under a confocal laser microscope.
The expression of miR-328-3p was significantly upregulated after 18β-GRA intervention in AGS cells (P = 4.51E-06). Overexpression of miR-328-3p inhibited GC cell proliferation and colony formation ability, arrested the cell cycle in the G0/G1 phase, promoted cell apoptosis, and inhibited the growth of subcutaneous tumors in BALB/c nude mice (P < 0.01). No obvious necrosis was observed in the tumor tissue in the negative control group (no drug intervention or lentivirus transfection) and vector group (the blank vector for lentivirus transfection), and more cells were loose and necrotic in the miR-328-3p group. Bioinformatics tools predicted that miR-328-3p has a targeting relationship with STAT3, and STAT3 was closely related to autophagy markers such as p62. After overexpressing miR-328-3p, the expression level of STAT3 mRNA was significantly decreased (P < 0.01) and p-STAT3 was downregulated (P < 0.05). The dual-luciferase reporter gene assay showed that the luciferase activity of miR-328-3p and STAT3 3’ untranslated regions of the wild-type reporter vector group was significantly decreased (P < 0.001). Overexpressed miR-328-3p combined with bafilomycin A1 (Baf A1) was used to detect the expression of LC3 II. Compared with the vector group, the expression level of LC3 II in the overexpressed miR-328-3p group was downregulated (P < 0.05), and compared with the Baf A1 group, the expression level of LC3 II in the overexpressed miR-328-3p + Baf A1 group was upregulated (P < 0.01). The expression of LC3 II was detected after intervention of 18β-GRA in GC cells, and the results were consistent with the results of miR-328-3p overexpression (P < 0.05). Additional studies showed that 18β-GRA promoted autophagy flow by promoting autophagosome synthesis (P < 0.001). qPCR showed that the expression of STAT3 mRNA was downregulated after drug intervention (P < 0.05). Western blot analysis showed that the expression levels of STAT3 and p-STAT3 were significantly downregulated after drug intervention (P < 0.05).
18β-GRA promotes the synthesis of autophagosomes and inhibits GC cell proliferation by regulating the miR-328-3p/STAT3 signaling pathway.
Core Tip: 18β-glycyrrhetinic acid (18β-GRA) is an important bioactive component of glycyrrhiza liquorice. Our results showed that 18β-GRA upregulated the expression of miR-328-3p in gastric cancer (GC) cells. Overexpression of miR-328-3p inhibited GC cell proliferation and colony formation, arrested the cell cycle, promoted apoptosis, inhibited subcutaneous tumor formation, and inhibited signal transducer and activator of transcription 3 (STAT3) expression. Dual-luciferase reporter assay showed that miR-328-3p targeted the regulation of STAT3. Confocal microscopy and western blotting indicated that 18β-GRA promotes autophagy flow by promoting autophagosome synthesis. Thus, 18β-GRA appears to promote cell autophagy and inhibit GC cell proliferation by regulating miR-328-3p/STAT3.
- Citation: Yang Y, Nan Y, Du YH, Huang SC, Lu DD, Zhang JF, Li X, Chen Y, Zhang L, Yuan L. 18β-glycyrrhetinic acid promotes gastric cancer cell autophagy and inhibits proliferation by regulating miR-328-3p/signal transducer and activator of transcription 3. World J Gastroenterol 2023; 29(27): 4317-4333
- URL: https://www.wjgnet.com/1007-9327/full/v29/i27/4317.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i27.4317
Gastric cancer (GC) is the fifth most common type of cancer and the fourth leading cause of cancer-related death worldwide[1]. GC is a local disease whose leading causes include diet, environmental and genetic factors, and Helicobacter pylori infection[2]. The current standard treatment for GC is surgical excision of operable tumor tissue combined with local radiotherapy, chemotherapy and conventional anticancer drugs. However, many GC cells appear to have significant invasion and metastasis after surgery. Although chemotherapy is an important management approach for preventing invasion and metastasis, it tends to substantially decrease the patients' quality of life due to severe toxic and side effects (e.g., hair loss, bone marrow transplantation, gastrointestinal reactions) and drug resistance[3,4]. Therefore, there is an urgent need to find natural, effective, and less toxic anti-GC drugs.
Glycyrrhiza uralensis Fisch. is a traditional Chinese medicine, and 18β-glycyrrhetinic acid (18β-GRA; Figure 1A), an important bioactive component of licorice, with many pharmacological effects including anti-inflammatory, liver protection, antiviral, anti-allergic, antitumor, antioxidant, and immunomodulatory effects[5-8]. In recent years, it has been found that 18β-GRA has excellent antitumor activity against human malignant tumors such as breast cancer, glioma, lung cancer, and prostate cancer[8-12]. In GC, 18β-GRA regulates the migration of SGC-7901 cells by regulating the reactive oxygen species (ROS)/protein kinase C alpha/extracellular signal-regulated kinase signaling pathway. It also inhibits the activities of matrix metalloproteinase 2 (MMP2) and MMP9 in a dose-dependent manner[13]. Expression of the antioncogene potassium-transporting ATPase alpha chain 1 (ATP4a) is downregulated in GC, and 18-GRA can activate ATP4a, thus inhibiting the occurrence of GC[14]. In transgenic mice, 18β-GRA was found to improve the severity of gastritis and inhibit the occurrence of GC. Also, studies have shown that 18β-GRA can improve the tumor inflammatory microenvironment by downregulating the expression of cyclooxygenase-2 and inhibiting the expression of the direct target Wnt-1 by upregulating the antioncogene miR-149-3p and thus inhibiting the occurrence and development of GC[15]. Cao et al[16] reported that 18β-GRA regulates toll-like receptor 2 (TLR2) expression through methylation and inhibits the proliferation, energy metabolism, and carcinogenesis of TLR2-activated GC cells. Our previous study found that 18β-GRA could inhibit GC cell proliferation by regulating the mitochondrial ribosome protein L35[17]. These results suggest that 18β-GRA may be an effective drug for preventing and treating GC.
MicroRNAs (miRNAs) is a novel endogenous, highly conserved non-coding RNA molecule with a length of 18-25 nucleotides[18]. miRNAs have an important role in cancer, and to date, many human miRNA sequences have been identified to participate in cancer pathogenesis and regulate protein expression levels in a complementary manner to their target mRNAs. miRNAs regulate and participate in the regulation of tumorigenesis, cellular processes, and pathways such as proliferation, differentiation, and apoptosis[19]. Expression profiles of miRNAs differ between cancerous and noncancerous tissues, different types and subtypes of cancer, and early and advanced cancers, depending on the aggressiveness of the disease[20-22]. Current studies have found that some miRNAs not only exist as oncogenes but also inhibit the occurrence of tumors. For example, miR-381 is upregulated in glioma, osteosarcoma and other cancers, while overexpression of miR-381 in GC and colon cancer can inhibit cell migration and invasion and promote cell apoptosis[23-26]. miRNAs also have a dual role in inducing or inhibiting tumorigenesis in GC. Previous studies have shown that the expression level of miR-148a in the plasma of GC patients is lower than that of healthy individuals, and miR-148a overexpression can inhibit proliferation, migration, cell invasion, and the epithelial-mesenchymal transition (EMT)[27]. Li et al[28] reported that low expression of miRNA-149 and overexpression of forkhead box C1 (FOXC1) were found in GC tissues and cells. Overexpression of miRNA-149 inhibits GC cell proliferation and migration, arrests the cell cycle, and promotes cell apoptosis. FOXC1 silencing can inhibit the biological functions of AGS and MKN28 cells in GC[28]. Overexpression of miR-140-3p can inhibit the migration and invasion ability of GC cells and directly downregulate the expression of B-cell lymphoma 2 (BCL2), thus inhibiting cell apoptosis. Downregulation of BCL2 further activates autophagy induced by Beclin 1, an autophagy marker, and inhibits the EMT[29]. An increasing number of GC-associated miRNAs have been found to participate in the formation and development of GC.
Autophagy is essential in many physiological and pathological processes, such as development, metabolism, inflammation, and cancer[30]. According to the mode of transport to lysosomes, autophagy can be divided into microautophagy, macroautophagy, and chaperone-mediated autophagy. Autophagy is closely related to the diagnosis, treatment, and prognosis of GC. In their study, Wang et al[31] found that there were 10 autophagy-related (ATG) genes with significant differences in expression between GC and paracancerous tissue. It was also found that genes such as ATG14 were significantly associated with the tumor-node-metastasis stage, and the expression of these genes was significantly lower in early T, indicating that autophagy may participate in early GC progression[31]. The expression of miR-183 is significantly decreased in GC tissues compared with paracancerous tissues, whereas the mammalian target of rapamycin (mTOR) is significantly increased. By targeting the expression of mTOR, miR-183 inhibits cell proliferation and promotes cell apoptosis and autophagy[32]. Sec62 homolog, preprotein translocation factor (Sec62), is an endoplasmic reticulum membrane protein highly expressed in GC. Overexpression of Sec62 can increase the expression of eukaryotic translation initiation factor 2 alpha kinase 3/cyclic AMP-dependent transcription factor ATF-4, bind to light chain 3 II (LC3 II), and activate the focal adhesion kinase family interacting protein of 200 kD/Beclin 1/ATG5 pathway to activate autophagy. Therefore, dual inhibition of Sec62 and autophagy may be an effective therapeutic strategy for GC metastasis[33]. It can also be concluded that autophagy is a “double-edged sword” that promotes and inhibits cell survival[34].
Based on previous studies, this study further analyzed the differential expression of miRNAs after the intervention of 18β-GRA. Cell function and animal experiments were used to detect the cell phenotype and subcutaneous tumor formation in nude mice. We detected the targeting relationship between miRNA and signal transducer and activator of transcription 3 (STAT3) and the changes in autophagy flow rate. An important finding was that 18β-GRA could affect autophagy flow and inhibit GC cell proliferation by regulating the miR-328-3p/STAT3 signaling pathway, which may contribute to the clinical development of effective therapies.
Human AGS cell lines were purchased from GeneChem (Cat. No. GCC-ST0003RT; Shanghai, China). They were cultured in Dulbecco’s Modified Eagle Medium (DMEM)/F12 medium (Cat. No. SH30023.01; Hyclone, Logan, UT, United States) containing 100 mL/L fetal bovine serum (Cat. No. 35-081-CV; Corning, Corning, NY, United States), and 10 mL/L penicillin and streptomycin (Cat. No. P1400; Solarbio, Beijing, China). The culture flasks were placed in an incubator at a constant temperature of 37 °C, with 50 mL/L carbon dioxide and saturated humidity.
Human gastric adenocarcinoma AGS cells were cultured and treated with 18β-GRA (purity > 97%, Cat. No. G10105-10G; Sigma, St. Louis, MO, United States) for 24 h. Cells in the negative control (NC) group and the 18β-GRA group were collected, and total RNA was extracted. Agarose gel electrophoresis was used to analyze the degradation degree of RNA and whether there was contamination. The NanoDrop 2000 spectrophotometer and Qubit 2.0 (Thermo Fisher Scientific, Waltham, MA, United States) were used to determine the purity and concentration of total RNA, and the Agilent 2100 Bioanalyzer (Agilent, Savage, MD, United States) was used to accurately detect the RNA integrity. After the samples were tested as qualified, a library was constructed with a Small RNA Sample Prep Kit. Using the 3’-end hydroxyl structure of the small RNA and the complete 5’-end phosphoric acid group, total RNA was taken as the starting sample, the two ends of the small RNA were added, and reverse transcription was performed to synthesize the cDNA. After polymerase chain reaction (PCR) amplification, polyacrylamide gel electrophoresis was used to isolate the target DNA fragment, after which the cDNA library was recovered.
After the library was successfully constructed, Qubit2.0 was used for preliminary quantification and Agilent 2100 was used to determine the library insert size. Once the insert size reached the expected value, quantitative PCR (qPCR) was used to accurately quantify the effective library concentration and ensure library quality (effective library concentration > 2 nmol/L). After the library was approved, different libraries were pooled into the Illumina SE50 sequencer (Illumina, San Diego, CA, United States) as needed.
TransmiR v2.0 database (http://www.cuilab.cn/transmir) was used to predict miR-328-3p-related proteins, and the STRING database (https://cn.string-db.org/) was used to predict the relationship between miR-328-3p- and autophagy-related proteins. Finally, the binding sites of miRNA-related proteins were searched on the miRWalk website (http://129.206.7.150/).
Trizol reagent was used for total RNA extraction (Cat. No. DP424; TIANGEN Biochemical Technology, Beijing, China). cDNA reverse transcription was conducted with the PrimeScript™ RT Reagent Kit with gDNA Eraser (Cat. No. RR04B; TaKaRa Bio Inc., Shiga, Japan) and the BlazeTaq SYBR Green qPCR Mix 2.0 (Cat. No. P031-S; GeneCopoeia, Rockville, MD, United States). The primer sequences were: miR-328-3p: forward: ATATCTGGCCCTCTCTGCCCTTC, reverse: AGTGCAGGGTCCGAGGTATT; U6: forward: CTCGCTTCGGCAGCACA, reverse: AACGCTTCACGAATTTGCGT; STAT3: forward: ACCAGCAGTATAGCCGCTTC, reverse: GCCACAATCCGGGCAATCT; GAPDH: forward: CACCCACTCCTCCACCTTTGA, reverse: TCTCTCTTCCTCTTGTGCTCTTGC. All experimental groups were studied in triplicate.
AGS cells were cultured in a T25 cell breathable culture flask. After 24 h, GC cells were transfected with a green fluorescent protein (GFP)-labeled overexpressed miR-328-3p lentivirus (GeneChem, Shanghai, China). The virus volume = (multiplicity of infection [MOI] × number of cells)/virus titer. At 8 h after transfection, a complete fresh medium was replaced and observed under a fluorescence microscope. The lentivirus transfection efficiency was determined according to the cells with GFP fluorescence in bright and GFP fields. Follow-up experiments were performed when the transfection efficiency reached about 90%.
Lentivirus-transfected AGS cells and normal AGS cells were inoculated into 96-well plates and cultured for 24 h, 48 h and 72 h. Next, the Cell Counting Kit-8 (CCK-8) reagent (Cat. No. CK04; DOJINDO Laboratories, Kumamoto, Japan) was added to each well, and absorbance (optical density [OD]) was measured at 450 nm with an enzyme marker. Cell viability = (ODexperiment - ODblank)/(ODcontrol - ODblank) × 100%.
The AGS cells were inoculated in a 6-well plate with 600 cells per well (three wells in each group were prepared for further culture), and the cells were observed under a microscope. When the number of clones of most single cells was > 50, and the diameter was 0.3-1.0 mm, the experiment was terminated. Cells were fixed in 4 mL/L paraformaldehyde (Cat. No. D16013; Saint-Bio, Shanghai, China) for 15 min, followed by incubation in crystal indigo water solution for 4-5 min. Next, they were washed with normal saline, left to dry, photographed and visually counted.
The AGS cells transfected with lentivirus were inoculated in 6-well plates for 24 h, digested, centrifuged at 3000 r/min for 5 min, and washed once with phosphate-buffered saline (PBS). The cell concentration was adjusted to 1 × 106/mL, and 0.5 mL cold ethanol was added to fix overnight. After 16 h, the cells were washed once with PBS, and then 500 μL was added to the staining solution prepared according to Rnase A: propidium iodide = 1:9 (Cat. No. KGA511; Keygen, Changchun, China) and incubated for 30 min. Flow cytometry was used to measure the cell cycle.
The AGS cells were collected, the concentration was adjusted to 4 × 105/mL, 1 mL cell suspension was centrifuged, 500 mL of Binding Buffer was added, and 5 μL Annexin V-APC was added. The mixing process was not too forceful. Next, 5 μL 7-aminoactinomycin D staining solution (Cat. No. KGA1023; Keygen) was added, mixed, and incubated for 20 min. Apoptosis was detected with a machine.
The BALB/c nude mice were kept in polypropylene cages with a humidity of 50% ± 5% and temperature of 22 ± 1 °C for 1 d at a 12 h/12 h light/dark cycle, with ad libitum access to food and water. After 2 wk of adaptive feeding, the mice were randomly grouped. The AGS cells and lentivirus-transfected GC cells were collected, and the cell concentration of each group was adjusted to 4 × 106/200 μL. The 200 μL cell suspension was absorbed by microinjection and slowly injected into the subcutaneous area of the right back of nude mice. The subcutaneous tumor-bearing conditions of nude mice were observed daily. The long diameter (L) and short diameter (W) of the tumor were measured with an electronic vernier caliper. The formula for calculating the tumor volume (V) was as follows: V = (W2 × L)/2. When the tumor volume of nude mice grew to a certain size, all animals were euthanized with 1000 mL/L carbon dioxide, and the tumor was removed and photographed. The animal protocols were approved by the Institutional Animal Care and Use Committee of Ningxia Medical University (IACUC-NYLAC-2022-108; Yinchuan, China).
Tissue samples fixed in 4 mL/L paraformaldehyde were dehydrated in 75 mL/L, 85 mL/L, 90 mL/L ethanol, and anhydrous ethanol and then embedded in paraffin. Paraffin sections 3-5 μm thick were taken from the microtome, baked at 65 °C for 1 h, and quickly sliced into xylene and ethanol for dewaxing treatment. The water was double steamed for 3 min, stained with hematoxylin dye (Cat. No. BA-4041; BaSO Diagnostics Inc., Zhuhai, China) for 5 min, double steamed with water for 3 min, incubated with acid alcohol fast differentiation solution (Cat. No. C0163M; Beyotime, Beijing, China) for 5 s, incubated with eosin dye solution (Cat. No. BA-4024; BaSO Diagnostics Inc.) for 5 min, and washed for 5 min. Next, the slices were dehydrated in 85 mL/L, 95 mL/L ethanol, and anhydrous ethanol. The dehydrated paraffin sections were placed in xylene transparent for 10 min and sealed with neutral gum (Cat. No. Zli-9555; ZSGB-Bio, Beijing, China). The cell structure and morphology were observed by optical microscope (BX43; Olympus, Tokyo, Japan).
Paraffin sections were routinely dewaxed, and the sections were repaired with citric acid repair solution (pH = 6.8) under high pressure for 10 min and washed with distilled water for 5 min. The sections were incubated with 3 mL/L hydrogen peroxide for 20 min, washed with water for 5 min, and soaked in PBS for 1 min. Sections were incubated with primary antibody (LC3, 1:50, Cat. No. 12741T from Cell Signaling Technology [CST], Danvers, MA, United States; STAT3, 1:90, Cat. No. AF6294 from Affinity Biosciences, Melbourne, Australia; p-STAT3, 1:100, Cat. No. AF3293 from Affinity Biosciences) at 4 °C overnight and washed with PBS three times, 5 min each time. Next, sections were incubated with secondary antibody at 37 °C for 20 min, followed by washing with PBS, DAB (Cat. No. ZLI-9018; ZSGB-Bio) color development, and washing with tap water. Hematoxylin was used for staining, gradient alcohol was used for dehydration, and neutral gum was used for sealing. The images were observed and acquired with the Olympus Imaging System (UC90).
The protein lysis buffer included 10 μL phosphatase inhibitor, 1 μL protease inhibitor, and 5 μL 100 mmol/L protease inhibitor (Cat. No. KGP2100; Keygen). The cells were collected, and the protein lysate was added to extract the whole protein. The protein content was determined with the bicinchoninic acid assay (Cat. No. KGPBCA; Keygen). Appropriate protein loading buffer (Cat. No. DL101; TransGen Biotech Co., Ltd., Beijing, China) was added and boiled at 100 °C for 10 min. According to the experimental design, the sample was added to prefabricated glue (Cat. No. M00659; GenScript, Nanjing, China), and constant pressure electrophoresis was carried out at 120 V until the marker strip with the lowest molecular weight appeared. After electrophoresis, the proteins were electrotransferred to a PVDF membrane (Cat. No. ISEQ00010; Millipore, Burlington, MA, United States) and incubated overnight at 4 °C with primary antibody, followed by washing and then incubation with secondary antibody for 1 h. The luminescent solution (Cat. No. KF005; Affinity Biosciences) was prepared according to the manufacturer’s instructions, and the PVDF membrane was soaked in it for 2-3 min and then placed in the intelligent chemiluminescence imaging system (iBright 1500; Invitrogen, Waltham, MA, United States) to obtain protein bands. Protein bands were analyzed using image J.
Retrieval of hsa-miR-328-3p sequence from NCBI Nucleotide website: UGGCCCUCUCUGCCCUUCCGU, the binding sites of miR-328-3p and STAT3 were retrieved from the miRWalk website, and the binding sites with high binding energy were screened based on the scores.
AGS cells were cultured in 24-well plates with 2 × 104 cells per well, and the degree of cell confluence reached about 40%. The experimental group included: Luc-STAT3-3’ untranslated region (3’UTR) Mutant (Mut)-NC + miR-328-3p-NC, Luc-STAT3-3’UTR Mut-NC + miR-328-3p, Luc-STAT3-3’UTR Wild Type (WT) + miR-328-3p-NC, Luc-STAT3-3’UTR WT + miR-328-3p, Luc-STAT3-3’UTR Mut + miR-328-3p-NC, and Luc-STAT3-3’UTR Mut + miR-328-3p. The plasmid and transfection reagent (Cat. No. IV1216075; Invigentech, Irvine, CA, United States) were mixed 1:1 to prepare the complex, after which the complex was added and mixed, and the culture was continued for 24 h. Next, the liquid was changed and the culture was continued.
After 48 h of co-transfection with plasmids, the 24-well plates were removed and balanced to room temperature. Extraction 250 μL medium was removed from each well, and Passive lysis buffer 100 μL was added to each well and shaken for 15 min for lysis. The lysate was added into 1.5 mL EP tubes and then into black 96-well plates with 20 μL per well and 3 multiple Wells per group. Next, Luciferase Assay Reagent II 100 μL was added. Fluorescence values were detected at 560 nm with multifunctional reagent II. Then 100 μL of Stop & Glo® reagent (Cat. No. E1910; Promega, Madison, WI, United States) was added to each well, and the fluorescence value was detected at 465 nm. Luciferase activity = ODfirefly luciferase/ODrenilla luciferase.
AGS cells with a concentration of 4 × 104 cells/mL were inoculated in a 15 mm confocal dish (Biosharp Biotechnology Inc., Hefei, Anhui, China) and cultured in the incubator. When the cells were fused to 40%, the fresh medium volume was changed to 1 mL. Monomeric Red Fluorescent Protein (mRFP)-GFP-LC3 tandem fluorescent protein adenovirus (Cat. No. P21071905; HanBio Therapeutics, Shanghai, China) was added, and the volume of virus = MOI × cell number/virus titer × 103, and then 1 mL culture medium was added and placed in the incubator for 4 h. After 16 h of infection, the culture medium was changed into a fresh medium for further culture. At 24 h and 48 h, the cell state and infection situation were observed under fluorescence microscope. The infection efficiency was determined by observing the fluorescence expression level of GFP.
The AGS cells infected with adenovirus were synchronously treated for 6 h, after which the corresponding drug treatment was given (the NC group was replaced with fresh medium). Next, they were placed in an incubator for 24 h, fixed with 4 mL/L paraformaldehyde for 20 min, and observed with a laser confocal microscope. Cells infected with adenovirus using mRFP-GFP-LC3 tandem fluorescent protein were identified by observing the fluorescence of mRFP and GFP after microscopic imaging.
Whole transcriptomics was used to analyze and screen the differentially expressed miRNAs in AGS cells after the 18β-GRA intervention. Through the selection criteria of log2 |fold change| > 1, P < 0.05, 283 differentially expressed miRNAs were obtained, of which 120 were upregulated and 163 were downregulated. Notably, the expression of miR-328-3p was upregulated after drug treatment, log2(fold change) = 3.04871, the P value was 4.51E-06 (Figure 1B and C).
qRT-PCR was used to verify whether the intervention process of 18β-GRA on AGS cells was related to miR-328-3p. Our results showed that the expression of miR-328-3p in AGS cells treated with 18β-GRA was significantly upregulated compared with the NC group (P < 0.01) (Figure 1D).
The expression level of miR-328-3p after transfection was detected by qRT-PCR. miR-328-3p in the overexpression group was significantly higher than in the vector group (Figure 1E). Cell status and transfection efficiency were observed under the Cytation 5 microscope (Figure 1F), and cell survival rate was detected. The results showed that, compared with the vector group, the survival rate of cells in the overexpressed miR-328-3p group decreased significantly (P < 0.01) (Figure 1G).
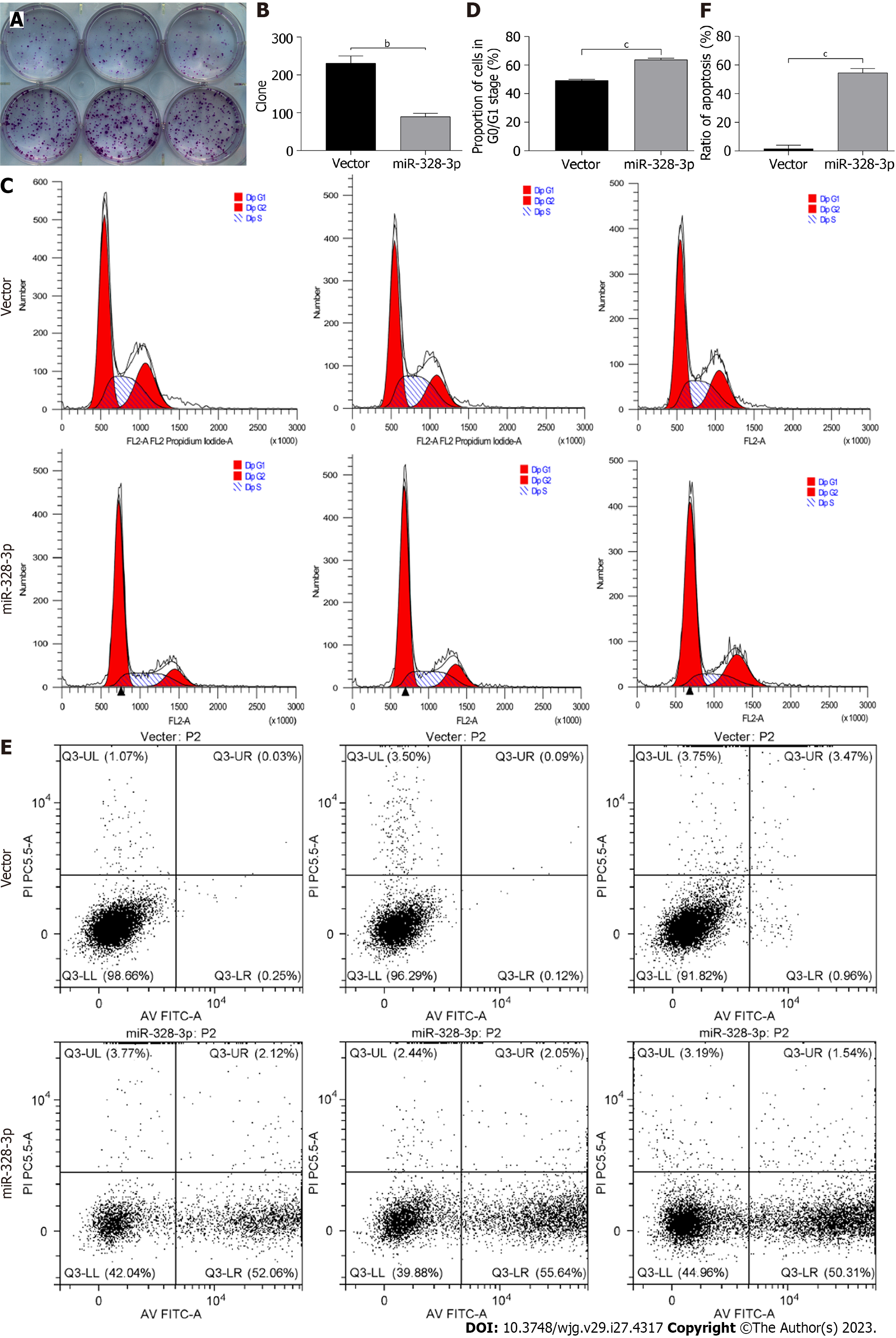
The results of the cloning formation experiment showed that compared with the vector group, the crystal violet staining positive of the overexpressing miR-328-3p group was significantly decreased (Figure 2A), and the cloning formation rate of AGS cells was significantly reduced (P < 0.01) (Figure 2B). These results indicated that overexpression of miR-328-3p could inhibit the colony formation of GC cells.
The cell cycle results showed that the average percentage of G0/G1 phase, S phase, and G2/M phase cells in the vector group was 49.23%, 29.41%, and 21.37%, respectively. The average percentage of G0/G1 phase, S phase, and G2/M phase cells in the overexpressed miR-328-3p group was 63.73%, 20.19%, and 16.08%, respectively. Compared with the vector group, the percentage of G0/G1 phase cells in the overexpressing miR-328-3p group was significantly higher (P < 0.001) (Figure 2C and D).
The results of the cell apoptosis experiment showed that the cell fragments in the vector group were increased, indicating that lentivirus transfection had a certain damage effect on AGS cells. Also, the early apoptosis rate of cells overexpressing miR-328-3p was significantly higher than that of the vector group (P < 0.001) (Figure 2E and F).
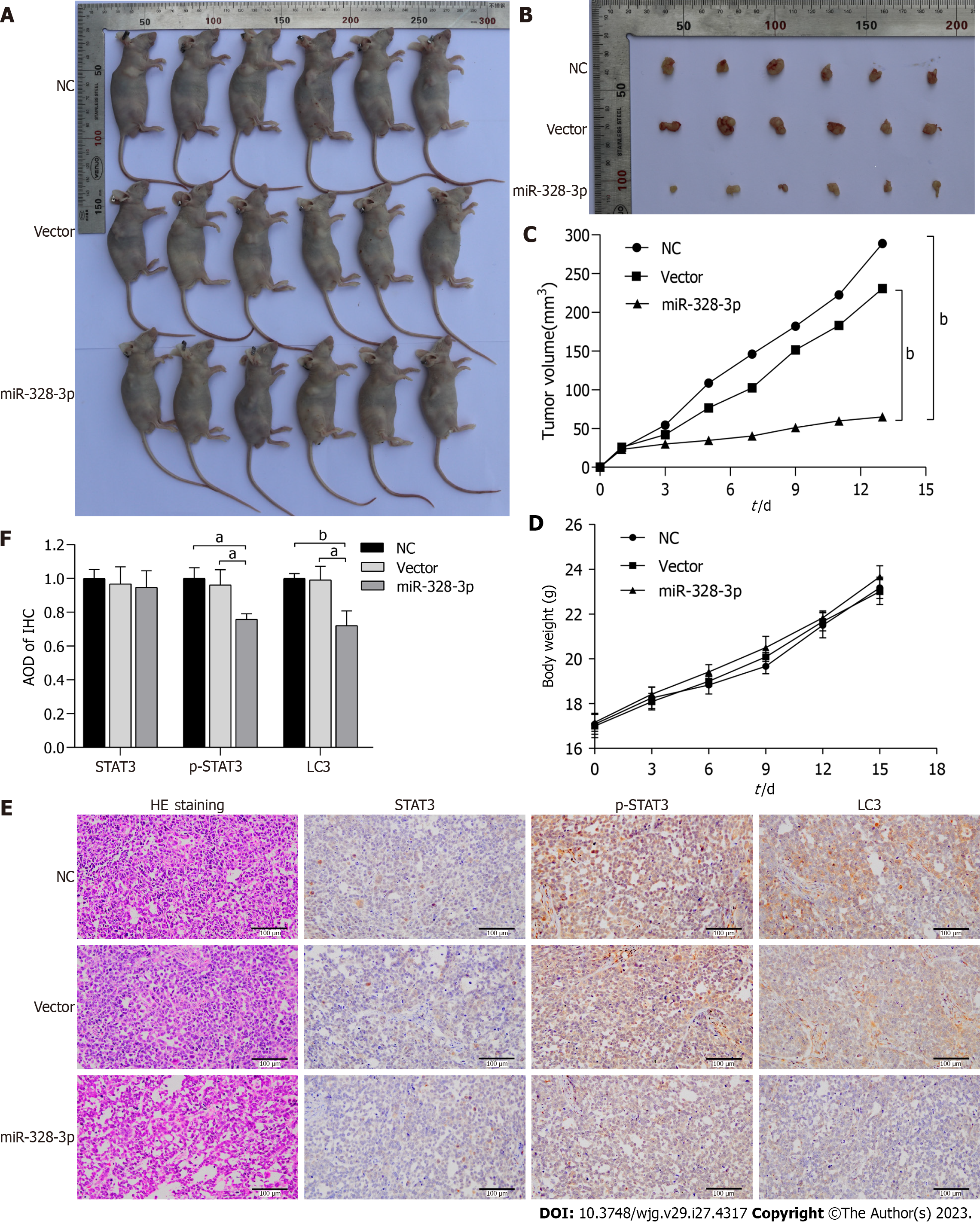
The results of the subcutaneous tumor experiment with nude mice showed that the subcutaneous tumor volume in the back of nude mice overexpressed with miR-328-3p was significantly smaller than that in the NC and vector groups (n = 6, Figure 3A and B). The tumor growth curve also showed that the tumor growth rate of the overexpressed miR-328-3p group was significantly lower compared to the vector group and the NC group (P < 0.01) (Figure 3C). There was no significant difference in body weight among the groups during the whole duration of the experiment(Figure 3D).
Hematoxylin and eosin staining showed that tumor cells in the NC and vector groups grew strongly, with different sizes, certain atypia, and larger nuclei. Also, there was no obvious necrosis in the tumor tissue. In the overexpress group, the cell arrangement was loose, the tissue structure was irregular, and there were more necrotic cells (Figure 3E).
Immunohistochemical staining results showed that compared with the vector and NC groups, the expression of p-STAT3 and LC3 in the miR-328-3p group was significantly decreased (P < 0.05), and there was no significant difference in the positive rate of STAT3 expression in transplanted tumor tissues among all groups (Figure 3E and F).
TransmiR v2.0 database predicted miR-328-3p related proteins and found that miR-328 was associated with STAT3 (Figure 4A). The relationship between STAT3 and autophagy-related proteins LC3, p62, BECN1, and ATG13 was subsequently predicted in the STRING database (Figure 4B). The above prediction results suggest that STAT3 is a target gene of miR-328-3p.
qRT-PCR was used to detect the expression level of STAT3 mRNA in transfected lentivirus in AGS cells. Our results showed that the expression level of STAT3 mRNA in the overexpressed miR-328-3p group was significantly lower than that in the vector group (P < 0.01) (Figure 4C).
We also detected STAT3 and p-STAT3 in AGS cells overexpressed with miR-328-3p by western blot (Figure 4D). The experimental results showed that the expression level of STAT3 in the miR-328-3p group did not significantly change, and the expression of p-STAT3 was downregulated (P < 0.05). After the ratio of p-STAT3 to STAT3, the p-STAT3/STAT3 ratio in the overexpressed miR-328-3p group was significantly down-regulated (P < 0.01) (Figure 4E).
The miRWalk online tool was used to find the binding site of miR-328-3p and STAT3, suggesting that there is a targeting relationship between miR-328-3p and STAT3 (Figure 4F). Dual-luciferase reporter assay was used to detect the relationship between miR-328-3p and STAT3 in AGS cells (Figure 4G). Our results showed that the luciferase activity of the STAT3 WT + miR-328-3p NC group was lower than that of the STAT3 WT + miR-328-3p group, and the difference was statistically significant (P < 0.001) (Figure 4H). We found that miR-328-3p could bind specifically to the 3’UTR of STAT3.
In order to further investigate the effect of miR-328-3p targeted regulation of STAT3 on the autophagy flow of AGS cells, western blot was used in combination with 0.5 μmoL/L Baf A1 to detect the expression of LC3 II in AGS cells after overexpression of miR-328-3p (Figure 5A). Our experimental results showed that the expression level of LC3 II in the overexpressing miR-328-3p group was significantly downregulated compared with the vector group (P < 0.05). Compared with the Baf A1 group, the expression level of LC3 II in miR-328-3p combined with the Baf A1 group was significantly upregulated (P < 0.01) (Figure 5B).
To further investigate the effect of 18β-GRA on autophagic flow in AGS cells, we treated AGS cells with different drugs. The results showed that compared with the NC group, LC3 II expression level in the 18β-GRA group was significantly downregulated (P < 0.05). Compared with the Baf A1 group, LC3 II in 18β-GRA + Baf A1 group was significantly upregulated (P < 0.05) (Figure 5C and D).
mRFP-GFP-LC3 tandem fluorescent protein adenovirus was also used to infect AGS cells and locally tag LC3 to detect autophagy flow. First, we explored the MOI of mRFP-GFP-LC3 tandem fluorescent protein adenovirus-infected AGS cells. The NC, MOI = 30, MOI = 50, and MOI = 80 groups were set up separately. Corresponding virus volumes were given for transfection, and photographs were taken under the fluorescence microscope at 24 h and 48 h. The results showed that, compared with MOI = 50, the virus transfection efficiency was lower when MOI = 30, while the cell state was poorer when MOI = 80. When MIO = 50 and the transfection time was 48 h, both transfection efficiency and cell state were good. Therefore, an MOI of 50 was determined as the MOI for subsequent experimental infection (Figure 5E and F). We used mRFP-GFP-LC3 tandem fluorescent protein adenovirus (MOI = 50) was used to infect AGS cells for 48 h and gave corresponding drug treatment. The experimental results showed that the number of yellow spots (GFP/mRFP) in the 18β-GRA group increased significantly compared with the NC group (P < 0.001). Separate red signals were also significantly increased (P < 0.01) (Figure 5G), suggesting that 18β-GRA could increase the number of early autophagosomes and late autolysosomes. Compared with the Baf A1 group, the number of yellow spots in the 18β-GRA + Baf A1 group increased significantly (P < 0.001), and the single red signal showed no significant change, indicating that the late autophagy inhibitor caused the blocking of late autophagic lysosome degradation. 18β-GRA promoted autophagosome synthesis, increasing the number of yellow spots (Figure 5H).
To investigate whether 18β-GRA promotes autophagy flow through the miR-328-3p/STAT3 signaling pathway, we first used qRT-PCR to collect 18β-GRA intervention AGS cells to detect the expression level of STAT3 mRNA. Our results showed that STAT3 mRNA expression was significantly downregulated in AGS cells after 18β-GRA intervention compared with the NC group (P < 0.05) (Figure 5I). Then, western blot results showed that the expression levels of STAT3 and p-STAT3 in AGS cells after 18β-GRA intervention were downregulated, and the p-STAT3/STAT3 ratio was also significantly downregulated (P < 0.05) (Figure 5J and K).
The present study found that 18β-GRA could regulate the expression of miR-328-3p, inhibit the proliferation of GC cells, arrest the cell cycle, and promote cell apoptosis. The dual-luciferase reporter gene experiment confirmed that miR-328-3p targeted the regulation of STAT3 and participated in promoting autophagy. Our results suggest that 18β-GRA promoted the synthesis of autophagosomes and inhibited cell proliferation by regulating the miR-328-3p/STAT3 signaling pathway.
miR-328-3p expression was found to be significantly inhibited by palmitate-induced endoplasmic reticulum stress in liver cell lines[35]. In tumors, overexpression of miR-328-3p can inhibit the proliferation and invasion of hepatocellular carcinoma cells, promote cell apoptosis, and have an excellent targeting relationship with endoplasmic reticulum metalloproteinases[36]. Overexpression of miR-328-3p inhibited proliferation, migration, and invasion of colorectal cancer cells and inactivated the phosphatidylinositol 3 kinase/protein kinase B (Akt) signaling pathway[37]. Xu et al[38] analyzed the miRNA expression profiles of 389 GC patients, using forward and reverse variable selection and multiple Cox regression analysis models, finding that miR-328-3p exists in GC patients as a protective miRNA. However, Xiao et al[39] defined miR-328-3p as an oncogene for the first time and found that miR-328-3p promotes the progression of GC by targeting the kelch-like ECH-associated protein 1/nuclear factor erythroid 2-related factor 2 axis. Although there is some controversy about the effect of the miR-328-3p expression on GC progression, it also suggests that miR-328-3p may be a target for GC drug therapy.
Autophagy (“self-eating”) usually refers to the process in which cells wrap their own pathological organelles into double-layer vesicles to form autophagosomes when they resist stress such as hunger and then deliver them to lysosomes and combine with them to form autolysosome to degrade the contents and produce energy for recycling and utilization to maintain cell survival[40]. During autophagy, the autophagy containing misfolded proteins and damaged organelles is degraded in the lysosome, which has a limiting membrane to prevent the leakage of its degrading enzymes. The autophagy process is fast and frequent and involves relatively complex membrane dynamics[41]. It is generally believed that the conversion of LC3 I to LC3 II detected in Western blot, or the increased expression level of LC3 II represents the activation of autophagy flow, and after the autophagy flow is blocked, the expression level of LC3 II is decreased (the conversion of LC3I to LC3 II was blocked) or LC3 II is excessively degraded by autolysosome[42]. In this study, autophagy marker protein LC3 was detected by combining with autophagy inhibitor Baf A1 to explore the effects of drugs and miR-328-3p on the autophagy of GC cells, revealing that both drugs and miR-328-3p could promote the occurrence of autophagy flow.
To further explore whether drugs promote the synthesis of autophagosomes or inhibit the degradation of autophagosomes, we infected GC cells with mRFP-GFP-LC3 autophagic double-labeled adenovirus, and the mRFP and GFP were used to label LC3. Because GFP fluorescent protein is sensitive to acidic environments, green fluorescence is quenched after the fusion of autophagosome and lysosome, and only mRFP fluorescence can be detected. The yellow signal composed of GFP and mRFP fluorescence signals co-located in the cell represents the early autophagosome, while the mRFP fluorescence signal alone represents the late autophagosome. The increased yellow and red signals indicate an increased autophagic flow level[43]. Using a dual verification approach, we found that drugs promoted autophagy flow by promoting autophagy synthesis at the causal level.
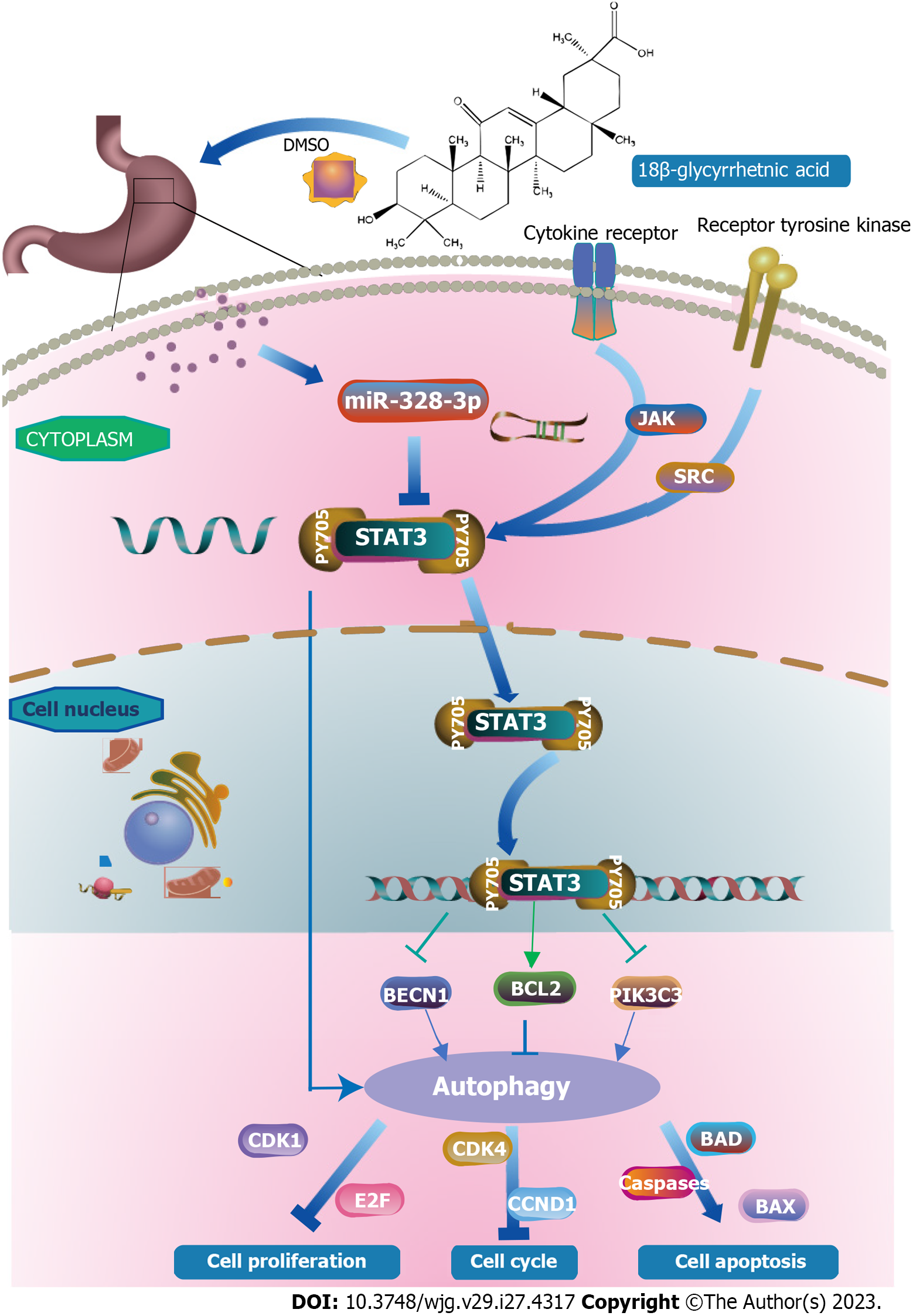
Several studies have found a close relationship between STAT3 and autophagy[44]. This study validated the targeting relationship between miR-328-3p and STAT3 by dual-luciferase reporter gene experiments. In conclusion, regulating the miR-328-3p/STAT3 signaling pathway promotes the synthesis of autophagosomes, and 18β-GRA inhibits the proliferation of GC cells. Typically, STAT3 monomers in the cytoplasm are phosphorylated to form STAT3 dimers, which are then shuttled into the nucleus and bind to specific DNA elements to transcriptionally activate or inhibit the expression of target genes, thereby inhibiting cell proliferation, arresting the cell cycle, and promoting autophagy[45]. At the same time, STAT3 in the unphosphorylated cytoplasm activates some autophagy-related genes, such as serine/threonine-protein kinase ULK2 and ATG12, via FOXO1 and FOXO3 transcription[46]. STAT3 monomer can also be transferred to mitochondria and interact with complex I/II of mitochondria to inhibit the production of reactive oxygen ROS[47] (Figure 6).
The present study showed that 18-GRA could inhibit GC cell proliferation, arrest cell cycle in G0/G1, promote apoptosis, and promote autophagosome synthesis of GC cells by regulating the miR-328-3p/STAT3 signaling pathway. Our future research program will be divided into three following phases: (1) Electrophoretic mobility shift assay was used to detect the binding ability of the STAT3 promoter regulatory region and miR-328-3p, to explore whether the transcriptional regulation of miR-328-3p on STAT3 is directly binding or indirectly binding; (2) the interaction mechanism between miR-328-3p and STAT3 proteins will be verified, and chromatin immuno-precipitation technology will be considered to further verify the binding regulation of the promoter region of the transcription factor STAT3 and miR-328-3p; (3) using Cre mice, adeno-associated virus was used for genome editing in vivo, and conditional gene targeting of miR-328-3p was performed in gastric mucosal epithelial cells to explore the overall phenotype and physiological status of Cre mice. This study mainly provided the scientific basis for the clinical treatment of GC and also sought effective drug targets for the prevention and treatment of GC.
18β-GRA can promote the synthesis of autophagosomes in GC cells, inhibit cell proliferation, arrest cell cycle and promote cell apoptosis by regulating miR-328-3p/STAT3 signaling pathway.
Gastric cancer(GC) is among the most severe gastrointestinal malignancies, with high morbidity and mortality. In recent years, increasing evidence has shown that natural products can prevent and inhibit the development of GC by regulating miRNA and other genes, showing great therapeutic potential.
Targeting miRNA and other genes in GC with natural drugs is a promising strategy, which provides a valid reference to further elucidate the molecular mechanism of natural products in the treatment of GC.
The purpose of this study was to investigate the molecular mechanism of 18β-glycyrrhetinic acid (18β-GRA) regulating the miR-328-3p/ signal transducer and activator of transcription 3 (STAT3) signaling pathway, promoting the autophagy flow of GC cells and inhibiting cell proliferation.
The differentially expressed miRNAs were screened by full transcriptomic analysis. The cells were transfected with lentivirus, and the functional indices of the cells were determined by Cell Counting Kit-8, clone forming method, and flow cytometry. The effect of overexpression of miR-328-3p on the tumorigenicity of GC cells was detected in the transplanted tumor model of nude mice. Hematoxylin-eosin staining and immunohistochemistry were used to observe tumor tissue morphology and detect protein expression, respectively. Bioinformatics analysis was performed using TransmiR, STRING, and miRWalk databases. Real-time quantitative polymerase chain reaction and western blot were used to detect mRNA and protein expression levels. Dual luciferase reporter system was used to verify the targeting relationship between genes. The autophagy flow of monomeric red fluorescent protein- green fluorescent protein - light chain 3 adenovirus double-labeled infected cells was observed under confocal laser microscopy.
18β-GRA could upregulate the expression of miR-328-3p in AGS cells. Overexpression of miR-328-3p inhibited the cell proliferation and colony formation ability, arrested the cell cycle, promoted apoptosis, inhibited the growth of subcutaneous tumors, and led to GC tissue cell necrosis increase. miR-328-3p can target and regulate STAT3. 18β-GRA intervention in GC cells and overexpression of miR-328-3p could downregulate the expression level of STAT3 mRNA. Compared with the vector group, the expression level of LC3 II was downregulated in the overexpressed miR-328-3p + bafilomycin A1 (Baf A1) group and upregulated in the overexpressed miR-328-3p + Baf A1 group. The number of yellow spots and separate red signals in the 18β-GRA group were significantly increased compared with the negative control group, and the number of yellow spots in the 18β-GRA + Baf A1 group was significantly increased compared with the Baf A1 group.
18β-GRA promotes GC cells' autophagosome synthesis and inhibits cell proliferation by regulating the miR-328-3p/STAT3 signaling pathway.
MiR-328-3p/STAT3 can be regulated by 18β-GRA and can be used as an effective drug target for the prevention and treatment of GC, thus providing a scientific basis for the clinical treatment of GC.
The authors would like to acknowledge Li-Qun Wang for statistical analysis assistance, as well as Joanna Japhet Tibenda for polishing the language.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Senchukova M, Russia; Shah OJ, India S-Editor: Wang JJ L-Editor: Filipodia P-Editor: Zhao S
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64295] [Article Influence: 16073.8] [Reference Citation Analysis (174)] |
| 2. | Lyons K, Le LC, Pham YT, Borron C, Park JY, Tran CTD, Tran TV, Tran HT, Vu KT, Do CD, Pelucchi C, La Vecchia C, Zgibor J, Boffetta P, Luu HN. Gastric cancer: epidemiology, biology, and prevention: a mini review. Eur J Cancer Prev. 2019;28:397-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 98] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 3. | Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1150] [Cited by in RCA: 2819] [Article Influence: 563.8] [Reference Citation Analysis (5)] |
| 4. | Jácome AA, Coutinho AK, Lima EM, Andrade AC, Dos Santos JS. Personalized medicine in gastric cancer: Where are we and where are we going? World J Gastroenterol. 2016;22:1160-1171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 34] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 5. | Wang CY, Kao TC, Lo WH, Yen GC. Glycyrrhizic acid and 18β-glycyrrhetinic acid modulate lipopolysaccharide-induced inflammatory response by suppression of NF-κB through PI3K p110δ and p110γ inhibitions. J Agric Food Chem. 2011;59:7726-7733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 153] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 6. | Feng Y, Mei L, Wang M, Huang Q, Huang R. Anti-inflammatory and Pro-apoptotic Effects of 18beta-Glycyrrhetinic Acid In Vitro and In Vivo Models of Rheumatoid Arthritis. Front Pharmacol. 2021;12:681525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 7. | Huo X, Meng X, Zhang J, Zhao Y. Hepatoprotective effect of different combinations of 18α-and 18β-Glycyrrhizic acid against CCl(4)-induced liver injury in rats. Biomed Pharmacother. 2020;122:109354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 8. | Luo YH, Wang C, Xu WT, Zhang Y, Zhang T, Xue H, Li YN, Fu ZR, Wang Y, Jin CH. 18β-Glycyrrhetinic Acid Has Anti-Cancer Effects via Inducing Apoptosis and G2/M Cell Cycle Arrest, and Inhibiting Migration of A549 Lung Cancer Cells. Onco Targets Ther. 2021;14:5131-5144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 9. | Ye L, Xu Y, Wang L, Zhang C, Hu P, Tong S, Liu Z, Tian D. Downregulation of CYP2E1 is associated with poor prognosis and tumor progression of gliomas. Cancer Med. 2021;10:8100-8113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 10. | Wang X, Tan Y, Zhang Y, Xu Z, Xu B, Lei H, Ding C, Cheng S, Wang X, Wei P, Wang Z, Mao Q, Ai C, Hua Q. The novel glycyrrhetinic acid-tetramethylpyrazine conjugate TOGA induces anti-hepatocarcinogenesis by inhibiting the effects of tumor-associated macrophages on tumor cells. Pharmacol Res. 2020;161:105233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 11. | Sun Y, Jiang M, Park PH, Song K. Transcriptional suppression of androgen receptor by 18β-glycyrrhetinic acid in LNCaP human prostate cancer cells. Arch Pharm Res. 2020;43:433-448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 12. | Shukla A, Tyagi R, Meena S, Datta D, Srivastava SK, Khan F. 2D- and 3D-QSAR modelling, molecular docking and in vitro evaluation studies on 18β-glycyrrhetinic acid derivatives against triple-negative breast cancer cell line. J Biomol Struct Dyn. 2020;38:168-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 13. | Cai H, Chen X, Zhang J, Wang J. 18β-glycyrrhetinic acid inhibits migration and invasion of human gastric cancer cells via the ROS/PKC-α/ERK pathway. J Nat Med. 2018;72:252-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 14. | Cao D, Zhao D, Jia Z, Su T, Zhang Y, Wu Y, Wu M, Tsukamoto T, Oshima M, Jiang J, Cao X. Reactivation of Atp4a concomitant with intragenic DNA demethylation for cancer inhibition in a gastric cancer model. Life Sci. 2020;242:117214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 15. | Cao D, Jia Z, You L, Wu Y, Hou Z, Suo Y, Zhang H, Wen S, Tsukamoto T, Oshima M, Jiang J, Cao X. 18β-glycyrrhetinic acid suppresses gastric cancer by activation of miR-149-3p-Wnt-1 signaling. Oncotarget. 2016;7:71960-71973. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 16. | Cao D, Wu Y, Jia Z, Zhao D, Zhang Y, Zhou T, Wu M, Zhang H, Tsukamoto T, Oshima M, Jiang J, Cao X. 18β-glycyrrhetinic acid inhibited mitochondrial energy metabolism and gastric carcinogenesis through methylation-regulated TLR2 signaling pathway. Carcinogenesis. 2019;40:234-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 17. | Yuan L, Yang Y, Li X, Zhou X, Du YH, Liu WJ, Zhang L, Yu L, Ma TT, Li JX, Chen Y, Nan Y. 18β-glycyrrhetinic acid regulates mitochondrial ribosomal protein L35-associated apoptosis signaling pathways to inhibit proliferation of gastric carcinoma cells. World J Gastroenterol. 2022;28:2437-2456. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 5] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (1)] |
| 18. | Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2112] [Cited by in RCA: 2436] [Article Influence: 162.4] [Reference Citation Analysis (0)] |
| 19. | Coradduzza D, Cruciani S, Arru C, Garroni G, Pashchenko A, Jedea M, Zappavigna S, Caraglia M, Amler E, Carru C, Maioli M. Role of miRNA-145, 148, and 185 and Stem Cells in Prostate Cancer. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 20. | Brase JC, Johannes M, Schlomm T, Fälth M, Haese A, Steuber T, Beissbarth T, Kuner R, Sültmann H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128:608-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 427] [Cited by in RCA: 458] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 21. | Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23:3-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 346] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 22. | Naeini MM, Ardekani AM. Noncoding RNAs and Cancer. Avicen J Med Biotechnol. 2009;1:55-70. |
| 23. | Yin Y, Li X, Guo Z, Zhou F. MicroRNA381 regulates the growth of gastric cancer cell by targeting TWIST1. Mol Med Rep. 2019;20:4376-4382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 24. | He X, Wei Y, Wang Y, Liu L, Wang W, Li N. MiR-381 functions as a tumor suppressor in colorectal cancer by targeting Twist1. Onco Targets Ther. 2016;9:1231-1239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 25. | Wang Z, Yang J, Xu G, Wang W, Liu C, Yang H, Yu Z, Lei Q, Xiao L, Xiong J, Zeng L, Xiang J, Ma J, Li G, Wu M. Targeting miR-381-NEFL axis sensitizes glioblastoma cells to temozolomide by regulating stemness factors and multidrug resistance factors. Oncotarget. 2015;6:3147-3164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 26. | Li Y, Zhao C, Yu Z, Chen J, She X, Li P, Liu C, Zhang Y, Feng J, Fu H, Wang B, Kuang L, Li L, Lv G, Wu M. Low expression of miR-381 is a favorite prognosis factor and enhances the chemosensitivity of osteosarcoma. Oncotarget. 2016;7:68585-68596. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 27. | Komatsu S, Imamura T, Kiuchi J, Takashima Y, Kamiya H, Ohashi T, Konishi H, Shiozaki A, Kubota T, Okamoto K, Otsuji E. Depletion of tumor suppressor miRNA-148a in plasma relates to tumor progression and poor outcomes in gastric cancer. Am J Cancer Res. 2021;11:6133-6146. [PubMed] |
| 28. | Li D, Zhang Y, Li Y, Wang X, Wang F, Du J, Zhang H, Shi H, Wang Y, Gao Y, Feng Y, Yan J, Xue Y, Yang Y, Zhang J. miR-149 Suppresses the Proliferation and Metastasis of Human Gastric Cancer Cells by Targeting FOXC1. Biomed Res Int. 2021;2021:1503403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 29. | Chen J, Cai S, Gu T, Song F, Xue Y, Sun D. MiR-140-3p Impedes Gastric Cancer Progression and Metastasis by Regulating BCL2/BECN1-Mediated Autophagy. Onco Targets Ther. 2021;14:2879-2892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 30. | Wen X, Klionsky DJ. At a glance: A history of autophagy and cancer. Semin Cancer Biol. 2020;66:3-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 73] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 31. | Wang M, Jing J, Li H, Liu J, Yuan Y, Sun L. The expression characteristics and prognostic roles of autophagy-related genes in gastric cancer. PeerJ. 2021;9:e10814. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 32. | Wei Y, Hong D, Zang A, Wang Z, Yang H, Zhang P, Wang Y. miR-183 Enhances Autophagy of GC Cells by Targeted Inhibition of mTOR. Ann Clin Lab Sci. 2021;51:837-843. [PubMed] |
| 33. | Su S, Shi YT, Chu Y, Jiang MZ, Wu N, Xu B, Zhou H, Lin JC, Jin YR, Li XF, Liang J. Sec62 promotes gastric cancer metastasis through mediating UPR-induced autophagy activation. Cell Mol Life Sci. 2022;79:133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 34. | Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102-1109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1556] [Cited by in RCA: 1657] [Article Influence: 92.1] [Reference Citation Analysis (0)] |
| 35. | Miyamoto Y, Mauer AS, Kumar S, Mott JL, Malhi H. Mmu-miR-615-3p regulates lipoapoptosis by inhibiting C/EBP homologous protein. PLoS One. 2014;9:e109637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 36. | Lu H, Hu J, Li J, Lu W, Deng X, Wang X. miR-328-3p overexpression attenuates the malignant proliferation and invasion of liver cancer via targeting Endoplasmic Reticulum Metallo Protease 1 to inhibit AKT phosphorylation. Ann Transl Med. 2020;8:754. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 37. | Pan S, Ren F, Li L, Liu D, Li Y, Wang A, Li W, Dong Y, Guo W. MiR-328-3p inhibits cell proliferation and metastasis in colorectal cancer by targeting Girdin and inhibiting the PI3K/Akt signaling pathway. Exp Cell Res. 2020;390:111939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 38. | Xu J, Wen J, Li S, Shen X, You T, Huang Y, Xu C, Zhao Y. Immune-Related Nine-MicroRNA Signature for Predicting the Prognosis of Gastric Cancer. Front Genet. 2021;12:690598. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 39. | Xiao Z, Zheng YB, Dao WX, Luo JF, Deng WH, Yan RC, Liu JS. MicroRNA-328-3p facilitates the progression of gastric cancer via KEAP1/NRF2 axis. Free Radic Res. 2021;55:720-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 40. | Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 591] [Cited by in RCA: 655] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 41. | Mizushima N, Levine B. Autophagy in Human Diseases. N Engl J Med. 2020;383:1564-1576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 749] [Article Influence: 149.8] [Reference Citation Analysis (0)] |
| 42. | LV X, Hu Z. [Methods for detecting autophagy flow]. Acta Pharmaceutica Sinica. 2016;51:45-51. |
| 43. | Funakoshi T, Aki T, Unuma K, Uemura K. Lysosome vacuolation disrupts the completion of autophagy during norephedrine exposure in SH-SY5Y human neuroblastoma cells. Brain Res. 2013;1490:9-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 44. | Pietrocola F, Izzo V, Niso-Santano M, Vacchelli E, Galluzzi L, Maiuri MC, Kroemer G. Regulation of autophagy by stress-responsive transcription factors. Semin Cancer Biol. 2013;23:310-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 206] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 45. | You L, Wang Z, Li H, Shou J, Jing Z, Xie J, Sui X, Pan H, Han W. The role of STAT3 in autophagy. Autophagy. 2015;11:729-739. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 255] [Cited by in RCA: 404] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 46. | Xu S, Huang P, Yang J, Du H, Wan H, He Y. Calycosin alleviates cerebral ischemia/reperfusion injury by repressing autophagy via STAT3/FOXO3a signaling pathway. Phytomedicine. 2023;115:154845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 49] [Reference Citation Analysis (0)] |
| 47. | Gilardini Montani MS, Santarelli R, Granato M, Gonnella R, Torrisi MR, Faggioni A, Cirone M. EBV reduces autophagy, intracellular ROS and mitochondria to impair monocyte survival and differentiation. Autophagy. 2019;15:652-667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |