Published online Jul 7, 2023. doi: 10.3748/wjg.v29.i25.4053
Peer-review started: January 21, 2023
First decision: February 1, 2023
Revised: February 16, 2023
Accepted: May 23, 2023
Article in press: May 23, 2023
Published online: July 7, 2023
Processing time: 157 Days and 10.4 Hours
Cholangiocarcinoma (CCA) is a devastating malignancy and has a very poor prognosis if tumors spread outside the liver. Understanding the molecular mechanisms underlying the CCA progression will likely yield therapeutic approaches toward treating this deadly disease.
To determine the molecular pathogenesis in CCA progression.
In silico analysis, in vitro cell culture, CCA transgenic animals, histological, and molecular assays were adopted to determine the molecular pathogenesis.
The transcriptomic data of human CCA samples were retrieved from The Cancer Genome Atlas (TGCA, CHOL), European Bioinformatics Institute (EBI, GAD
In summary, our study comprehensively analyzed the gene expression pattern of CCA samples using publicly available datasets and identified the cell cycle and Notch pathways are potential therapeutic targets in this deadly disease.
Core Tip: Molecular profiling of cholangiocarcinoma (CCA) has been conducted using various cohorts. However, the identified targets vary among different cohorts. In the current study, we combined different cohorts of CCA RNA sequencing datasets and refined the potential therapeutic targets in human CCA malignancy. We validated the findings using human CCA cell lines, the KrasG12D and P53 mutation transgenic mouse model, and human CCA clinical data, thus supporting the potential of targeting the identified pathways in clinical trials.
- Citation: Liu D, Shi Y, Chen H, Nisar MA, Jabara N, Langwinski N, Mattson S, Nagaoka K, Bai X, Lu S, Huang CK. Molecular profiling reveals potential targets in cholangiocarcinoma. World J Gastroenterol 2023; 29(25): 4053-4071
- URL: https://www.wjgnet.com/1007-9327/full/v29/i25/4053.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i25.4053
Cholangiocarcinoma (CCA) is a devastating malignancy with “silent” clinical manifestations, genetically heterogeneous, anatomically distinct, but limited treatment options[1]. CCA can be classified as three subtypes depending on the origin of anatomical site, including intrahepatic (iCCA), perihilar (pCCA) and distal (dCCA) CCA[2]. If CCAs are diagnosed at the early stage, surgical resection may be a treatment option. However, CCA tumors are generally diagnosed at advanced stages due to lack of symptoms. Therefore, it is urgent to develop early detection markers and effective therapies for CCA patients. Understanding the molecular pathogenesis of CCAs may help identify early detection markers and potential therapies for this deadly disease.
Advances in whole-exome and transcriptome sequencing have paved the way for precision medicine in patients with advanced-stage or unresectable lesions[3-5]. Several genetic mutations have been identified in CCA patients[6]. Among them, IDH1 mutations have been well studied and the IDH1 mutation specific inhibitor has been developed for targeting malignant tumors with this mutation. In the ClarIDHy clinical trial, it was found that Ivosidenib-IDH1 mutation inhibitor could improve CCA patient overall survival (OS) and progression free survival[7,8], which eventually led to the FDA approval of Ivosidenib use in CCA patients with IDH1 mutation. Nevertheless, the 5-year survival of these patients treated with Ivosidenib is not significantly improved[7,8]. Besides, there is no effective therapy for those CCA patients with wild-type IDH1, despite that cisplatin and gemcitabine could extend the median OS for 3.6 mo but is not curative. Therefore, there is still an unmet need for identifying effective therapies in CCA patients.
In the current study, we utilized the transcriptomic profiling of CCA to explore the oncogenic driver events, analyzed the relevant genes expression, and identified the feasibility of target therapies with respect to those tumorigenic lesions.
A total of 2 archival tissue blocks from patients with eCCA were retrieved from the First Affiliated Hospital of Zhengzhou University (Zhengzhou, Henan Province, China) and the specimens were independently diagnosed by two hepatobiliary pathologists. This study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of the same institution (Approval No. 2022-KY-1514-002). Informed consents were obtained from all patients.
Human CCA cell lines, including H1 (OCUCh-LM1-H1)[9], HuCCT1[10], SSP-25[11], and ETK1[12] cell lines were kindly provided by Dr. Jack Wands. TFK-1 and RBE were purchased from RIKEN Cell Bank (Tsukuba, Japan). The non-malignant human bile duct cells hBD was purchased from Celprogen Inc (Torrance, CA, United States, Catalog Number: 36755-12). CCA cell lines were cultured in RPMI-1640 supplemented with 10% fetal bovine serum (FBS; GibcoTM, Thermo Scientific, Waltham, MA, United States; Cat# 10099141C), 100 μg/mL streptomycin, and 100 U/mL penicillin (HyClone™, Logan, UT, United States). The hBD cell line was grown in DMEM containing 10% FBS, 100 μg/mL strep. All cell lines were authenticated by STR and were negative for contamination of mycoplasma recently. Arcyriaflavin A, Flavopiridol, Roscovitine, and γ-secretase inhibitor, DAPT (N-[N-(3, 5-difluorophenacetyl)-l-alanyl]-s-phenylglycinet-butyl ester) were all purchased from Cayman Chemical (Ann Arbor, MI, United States).
RNA-Seq, DNA methylation, and clinicopathological data of 36 patients with cholangiocarcinoma (liver/normal bile duct = 9, iCCA = 30, pCCA = 4, dCCA = 2) were retrieved from The Cancer Genome Atlas (TCGA) database. The Genomic Data Commons mRNA quantifications were calculated by STAR[13], and then annotated by the reference gene model GENCODE v36. iCCA and the related para-cancerous normal livers from 30 patients, and their survival information were employed (GSE107943)[14]. Fastq files of RNA-Seq from 162 samples of CCA (iCCA = 122, pCCA = 14, dCCA = 26) and the associated survival status of patients were fetched from the European Genome-phenome Archive[15].
The Kaplan-Meier analysis was performed to evaluate the OS and disease-free survival (DFS) of patients from GEO and EBI cohorts. The optimal cutoff point for two groups was estimated based on maximally selected rank statistics[16], and then significance of survival analysis was calculated using log-rank test between the two groups.
Gene set enrichment analysis (GSEA) was conducted using R package “Pi”[17]. Hallmark (H) and KEGG subset of Canonical Pathways (CP) gene sets were selected for GSEA[18].
Single-sample GSEA was performed to analyze the tumor-infiltrating lymphocytes (TILs) based on the 28 immune cell types[19]. Correlations between the indicated genes expression and each infiltrating immune cells levels were calculated based on Spearman correlation (ρ).
All protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at Rhode Island Hospital, and all experiments were conducted in accordance with the guidelines of this IACUC. The specifical KrasG12D (LSL-KrasG12D), floxed-p53 (p53L/L), and albumin-Cre mutants strains were purchased from the Jackson Laboratory (Bar Harbor, ME, United States). They were intercrossed to achieve the mutant mice as indicated in the results section. The specific primers and genotyping protocol were performed following the instruction suggested by the Jackson Laboratory. Mice were bred in a pathogen-free environment and provided with free access to food and sterilized water Ad Libitum. Animals within experimental cohorts were monitored until the appearance of illness, including diminished activity, abdominal bloating, and cachexia, followed by full necropsy and the subsequent histopathological analysis.
Three CCA cell lines, including HuCCT1, H1, RBE, were seeded at 2 × 103 cells/well into 96-well plate in six replicates. And then the above-mentioned cells were treated with small molecular inhibitors targeting cell cycle (Arcyriaflavin a, Flavopiridol, and Roscovitine) and Notch (DAPT) pathways 24 h post-seeding, respectively. The treatments were changed as indicated at days 1, 3, 5, and 7. Cell proliferation was measured using MTT at the following day (day 0 and day 1) and every 48 h after (up to day 7). Absorbance at OD570 nm and 650 nm were read and then normalized to vehicle control (0.1% DMSO or the corresponding medium). The relative cell proliferation rates were calculated as OD570-OD650.
Total RNA was extracted from CCA cell lines using TRIzol™ (Invitrogen, Thermo Scientific; Cat# 15596026) according to the manufacturer’s protocol. 1 μg of total RNA was used to reverse transcription using iScript™ Reverse Transcription Supermix (Bio-Rad, Hercules, CA, United States, Cat#1708840). The qRT-PCR was conducted using SYBR® Green Realtime PCR Master Mix (Bio-Rad, Cat#1725270) by QuantStudio™ 5 Real-Time PCR System (Applied Biosystems). The relative mRNA expression was calculated based on 2-ΔΔCt protocol. All primers were listed in Supplementary Table 1.
Whole-cell protein extractions was acquired using RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific, Cat# 89901), supplemented with protease inhibitor (Thermo Fisher Scientific, Cat#78430). The concentration of protein was quantified using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Cat# J63283.QA) before mixed with protein loading buffer (3×). Then the protein samples were separated by 10% SDS-PAGE, and the detailed procedures conducted as previously described[20].
Antibodies including α-Tubulin, GAPDH, Cyclin D1, P21, PCNA, E2F1, CDK2, CDK4, CDK6, RB1, JAG1, JAG2, Caspase 8, BAX, XIAP, BCL-2L12, ATR, ATM, Rad50, Mre11, PIK3C3, LC3A/B, and ULK1 were purchased from Cell Signaling Technology (Danvers, MA, United States) and Santa Cruz Biotechnology (Dallas, TX, United States).
Statistical analyses were performed using STATA software 16.0 (StataCorp, College Station, TX, United States) and R software (version 4.2.0). Statistical analysis was performed using unpaired Student’s t and Mann-Whitney U test with respect to Gaussian distribution or not, respectively, unless otherwise indicated. Kruskal-wallis test was performed to compare the expression of genes examined by PCR, and the post-hoc tests within two groups were adjusted by Holm’s correction. Spearman correlation coefficient (ρ) is used to evaluate the correlations between DNA methylation levels of CpG islands and the associated genes expression. Wilcoxon signed-rank test was utilized to compare the expression level of cell cycle and Notch related genes between iCCA and its corresponding normal tissues. Two-way repeated measures Anova was utilized to compare optical densities of CCA cell lines treated with the serial gradient concentration of drugs, which measured more than once. Then simple effect at day 7 was calculated when there were interaction effects between time and dosages. P values were shown as: aP < 0.05, bP < 0.01, and cP < 0.001.
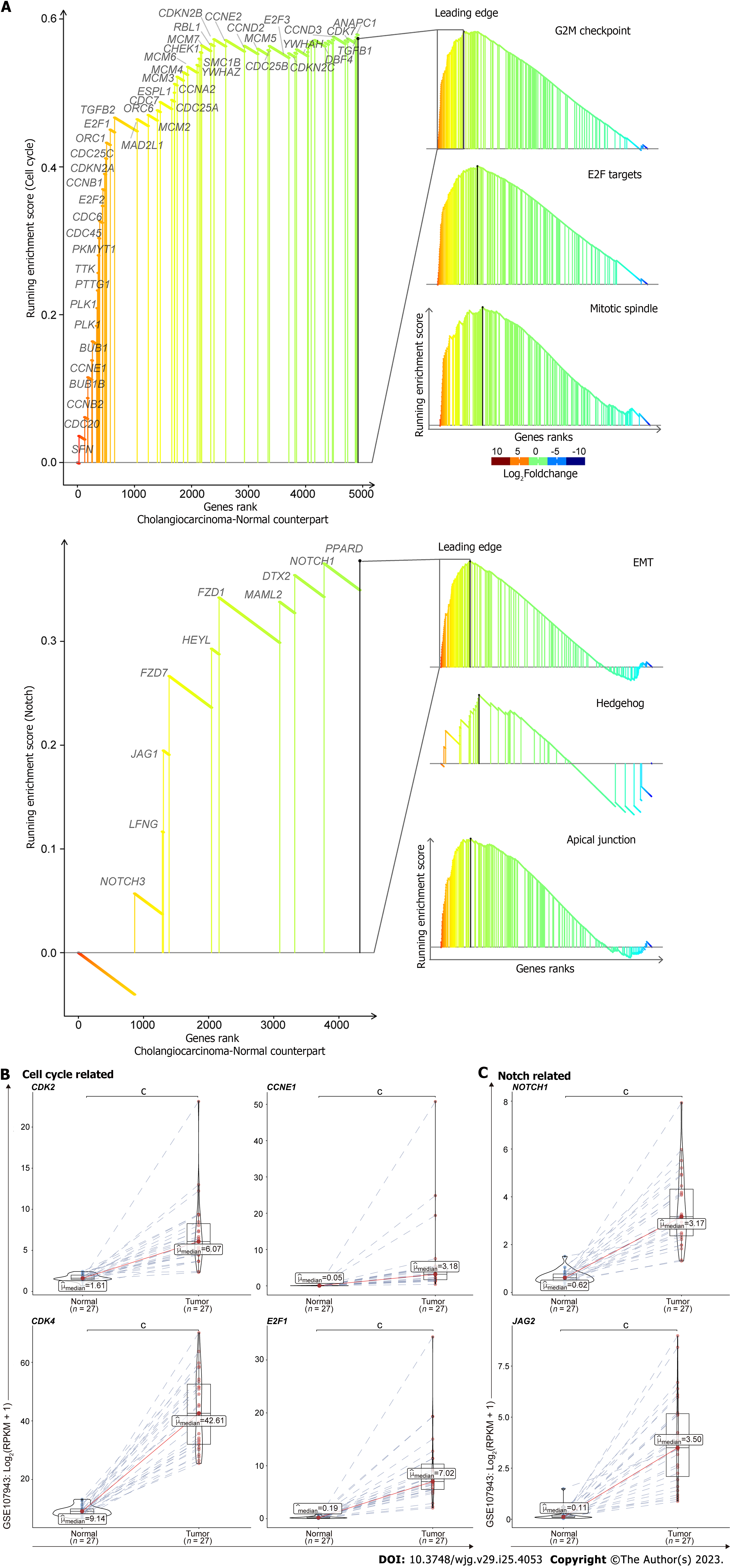
To determine what molecular events are up-regulated in cholangiocarcinogenesis, we downloaded the transcriptomic data of CHOL from the TCGA database. We performed the GSEA and found that cell cycle associated pathways were preferentially activated in tumors compared with the normal samples, such as genes involved in G2M checkpoint, E2F Targets, and Mitotic spindle (Figure 1A, upper). Besides, the pathways related with Notch were also induced in tumors, e.g., genes implicated in epithelial mesenchymal transition (EMT), Hedgehog, and Apical Junction (Figure 1A, lower). We retrieved the leading edge gene sets from those pathways, which drive the enrichment score and dominate the tumorigenicity of those pathways. Consistently, the genes from those leading edges are distinctly up-regulated in tumors compared with the normal samples, in line with the oncogenic properties (Figure 1B and C). Together, through the analysis of GSEA and differentially expressed genes, we found cell cycle and Notch pathways are up-regulated in CCA patients compared to non-malignant patient controls.
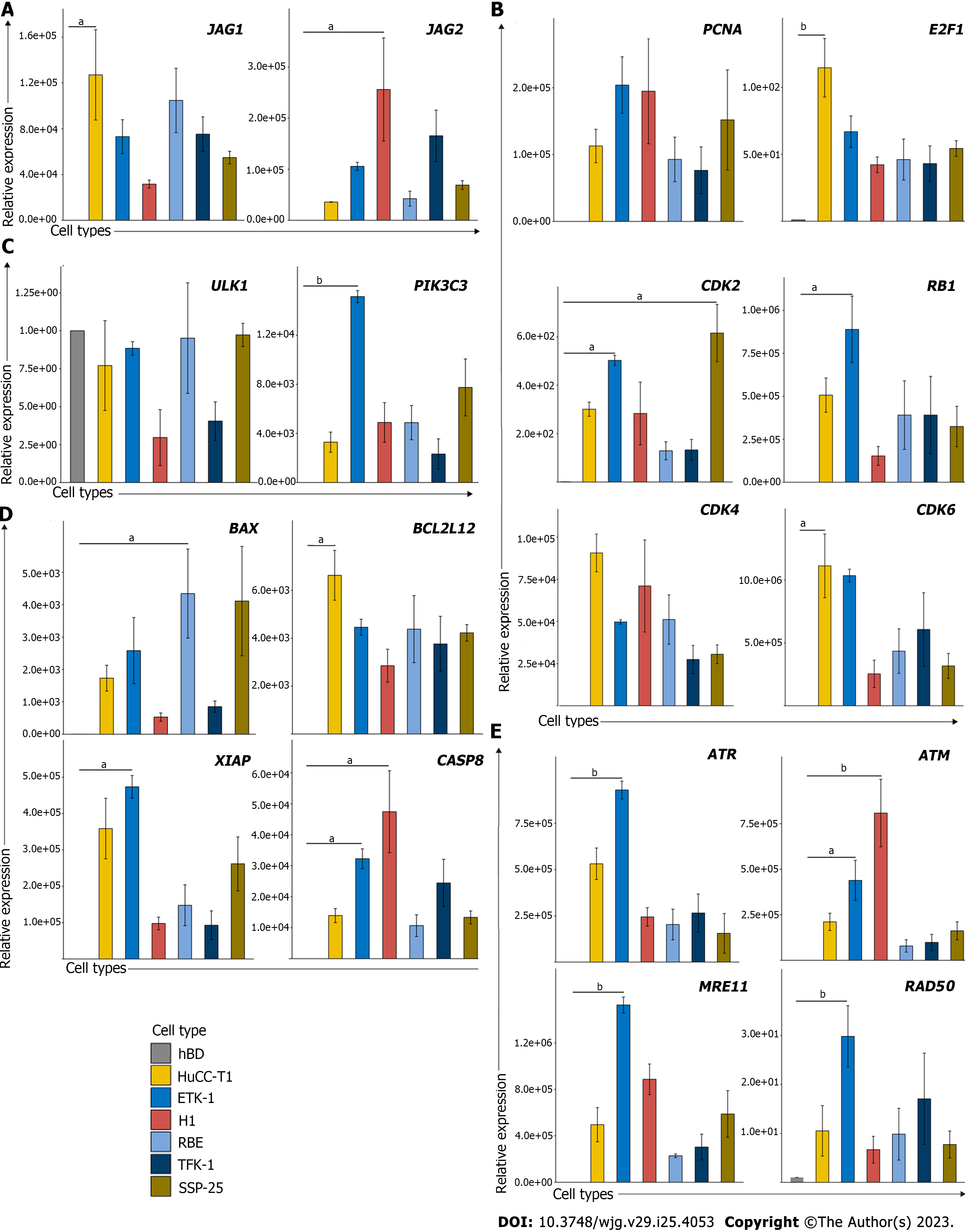
To facilitate the application of identified genes in preclinical studies, we must validate if these findings are reproducible in established normal biliary epithelial cells and CCA cells. In this case, we would be able to evaluate whether these pathways can be targeted using the in vitro cell culture system. Thus, we examined several leading-edge genes associated with these pathways in normal biliary epithelial cells and several CCA cell lines. The results suggest that several cell cycles associated genes were up-regulated in CCA cancer lines, including PCNA, E2F1, CDK2, CDK4, and CDK6 (Figure 2A). Besides, JAG1 and JAG2 which serve as the Notch ligand also substantially increased in CCA cell lines when compared with normal biliary epithelial cells (Figure 2B). We also examined several genes involved in autophagy, cell death, and DNA damage pathways since we wanted to illustrate if cell cycle associated pathways play a dominant role or cell death pathways dominate the CCA tumorigenesis. Interestingly, we found several up-regulated genes that are associated with DNA damage and cell death pathways, but not autophagy (Figure 2C-E). Moreover, the negative correlations between the methylation status (β value) and its expression of those genes suggested that the aberrant expression might be attributed to the hyper- or hypomethylation (Supplementary Figure 1).
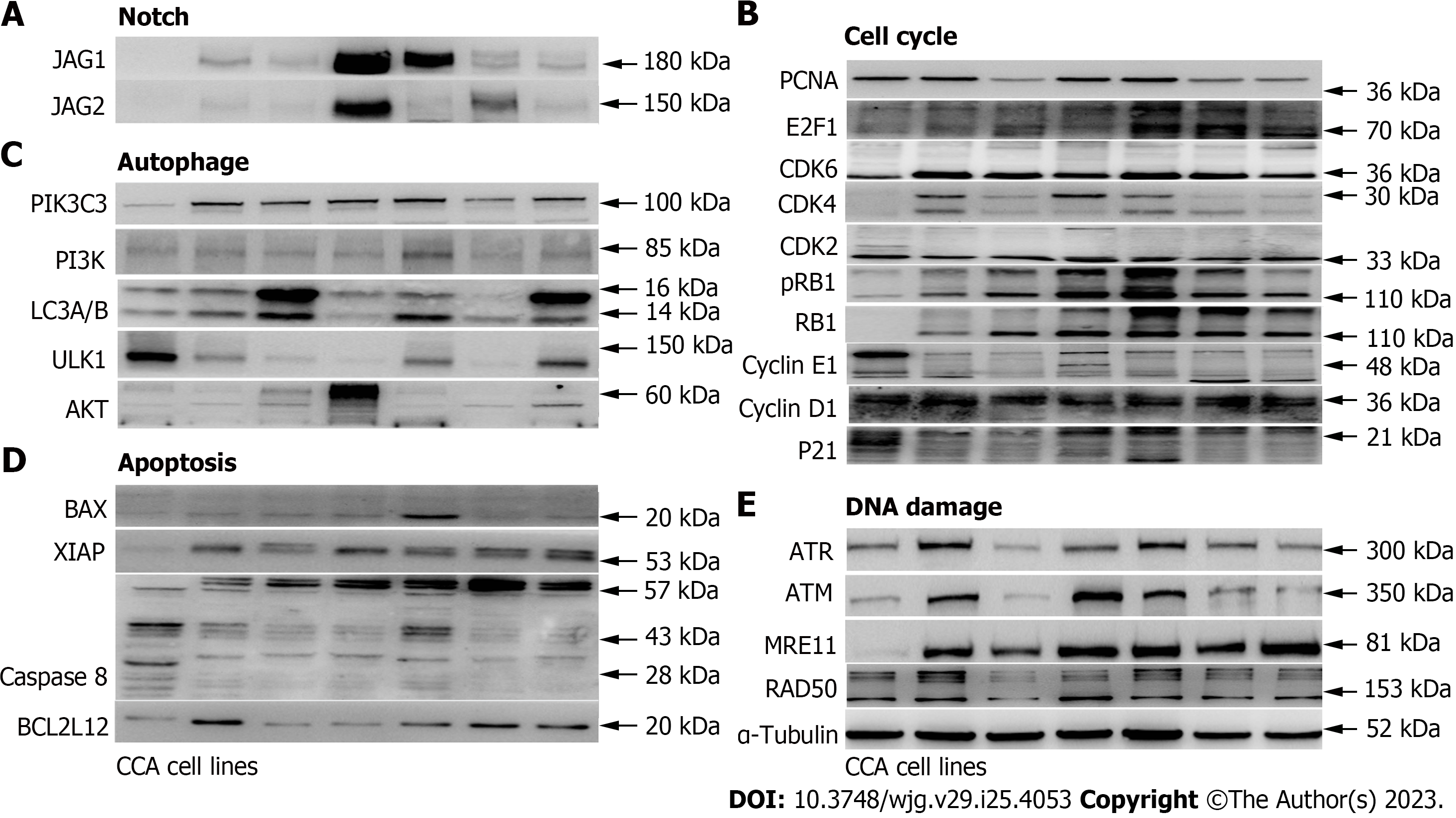
To determine if the identified genes have increased protein expression levels or promote other protein expression levels of the same pathway in CCA cell lines compared with the control biliary epithelial cells, we decided to examine protein expression of several major representative genes in each pathway (Figure 3, Supplementary Figure 2). As shown, several cell cycles associated genes were up-regulated in CCA cell lines when compared with the control biliary epithelial cells, including CDK2, CDK4, CDK6, and E2F1. Unexpectedly, several cyclins which include cyclin D1 and cyclin E1 were found decreased (Figure 3A). When we examined Notch ligands which include JAG1 and JAG2, we found JAG1 and JAG2 were all significantly increased in CCA cell lines compared to the biliary epithelial cells (Figure 3B). We further examined autophagy associated proteins and found no significant difference in autophagy between normal and malignant cells based on the autophagy definition (Figure 3C)[21]. In line with the mRNA change data, several apoptosis and DNA damage associated genes that were identified elevated in CCA cell lines were found increased as well (Figure 3D and E). Together, the mRNA and protein profile data demonstrated that it is feasible to identify potential target genes through mining public datasets and validating through in vitro human CCA cell lines.
It has been well characterized that KRAS and P53 are two of the most mutant genes in human CCA tumor samples[22]. Thus, a previous study has established the CCA transgenic mouse model by conditionally activating KrasG12D and depleting P53 (either P53 homozygous or P53 heterozygous knockout) in mouse liver using the Albumin promoter driven cre recombinase[23]. We also established the CCA transgenic mouse (Figure 4A) as evidenced by the genotyping data showing positive cre, KrasG12D, and heterozygous floxed P53 genes (Figure 4B). The gross images of representative mouse tumor were showed (Figure 4C). When comparing with human non-malignant bile duct and CCA tumor samples, the mouse CCA tumors present almost identical CCA histology to human CCA tumor samples (Figure 4C and D). To further determine whether mouse CCA and human CCA tumors have similar molecular profiles, we examined the genes that we identified using public human CCA dataset and validated using normal biliary epithelial cells and human CCA cell lines. Intriguingly, mouse and human CCA both have highly elevated genes associated with cell cycles and Notch, including CDK2, CDK4, CDK6, JAG1, and JAG2 (Figure 4E and Supplementary Figure 3). In line with CCA cell line data, several DNA damage genes, including RAD50 and MRE11, were found increased in mouse CCA tumor samples as well (Figure 4E). Given all these facts, our data demonstrated that the CCA tumors derived from the CCA transgenic mouse model have very high similarity to human CCA tissues and cell lines. Besides, the results of CCA transgenic mouse tumor data further demonstrated that refining the public human RNA sequencing data would help identify dysregulated pathways which may be druggable.
As we have found that cell cycle and Notch associated pathways are highly elevated in human CCA tumor samples and validated these findings in CCA cell lines, we proposed to determine whether targeting these pathways using small molecular inhibitors could achieve therapeutic potential in CCA cell lines, including H1, HuCCT1 (Kras and P53 mutations), and RBE (IDH1 mutation). It was found that the D1-dependent CDK4 inhibitor Arcyriaflavin a significantly suppressed cell growth in HuCCT1 and RBE CCA cell lines, but to a less extent in H1 (Figure 5A). The pan-CDK inhibitor Flavopiridol and the selected CDK inhibitor Roscovitine, which targets multiple CDKs, both displayed substantial growth inhibition in H1, HuCCT1, and RBE CCA cell lines (Figure 5B and C). It was noted that RBE CCA cells with IDH1 mutation are very vulnerable upon the treatments of CDK inhibitors (Figure 5A-C). Regarding the treatment using a Notch inhibitor, a γ-secretase inhibitor-DAPT was used to challenge these CCA cell lines. Although the Notch inhibitor could exert anti-tumor effects in CCA cell lines, the suppressive effects are not as strong as CDK inhibitors. Furthermore, the close relationships between the levels of TILs and those genes expressed indicated that cell cycle related pathways are associated with immune check point therapies (Supplementary Figure 4). Taken together, our results suggested that targeting cell cycle and/or Notch pathways could be considered as alternative modalities for patients with CCA.
It is essential to verify whether our findings are applicable to other datasets. Thus, we utilized another dataset (GSE107943) to validate the oncogenic traits of cell cycle and Notch related pathways in CCA. Consistently, the leading edge of GSEA showed pathways pertaining to cell cycle (Figure 6A, upper) and Notch (Figure 6A, lower) were significantly activated in tumors compared with the normal samples. Besides, the analysis of differentially expressed genes also revealed that upregulation of cell cycle related genes, including CDK2, CDK4, CCNE1, and E2F1, were observed in human CCA tumor samples compared with the adjacent normal liver tissues. Similarly, several Notch associated genes, including NOTCH1 and JAG2, were found up-regulated in CCA tumor samples as well (Figure 6C). Together, with the validation data derived from another dataset, it is therefore suggested that our findings are applicable to other clinical human CCA tumor samples.
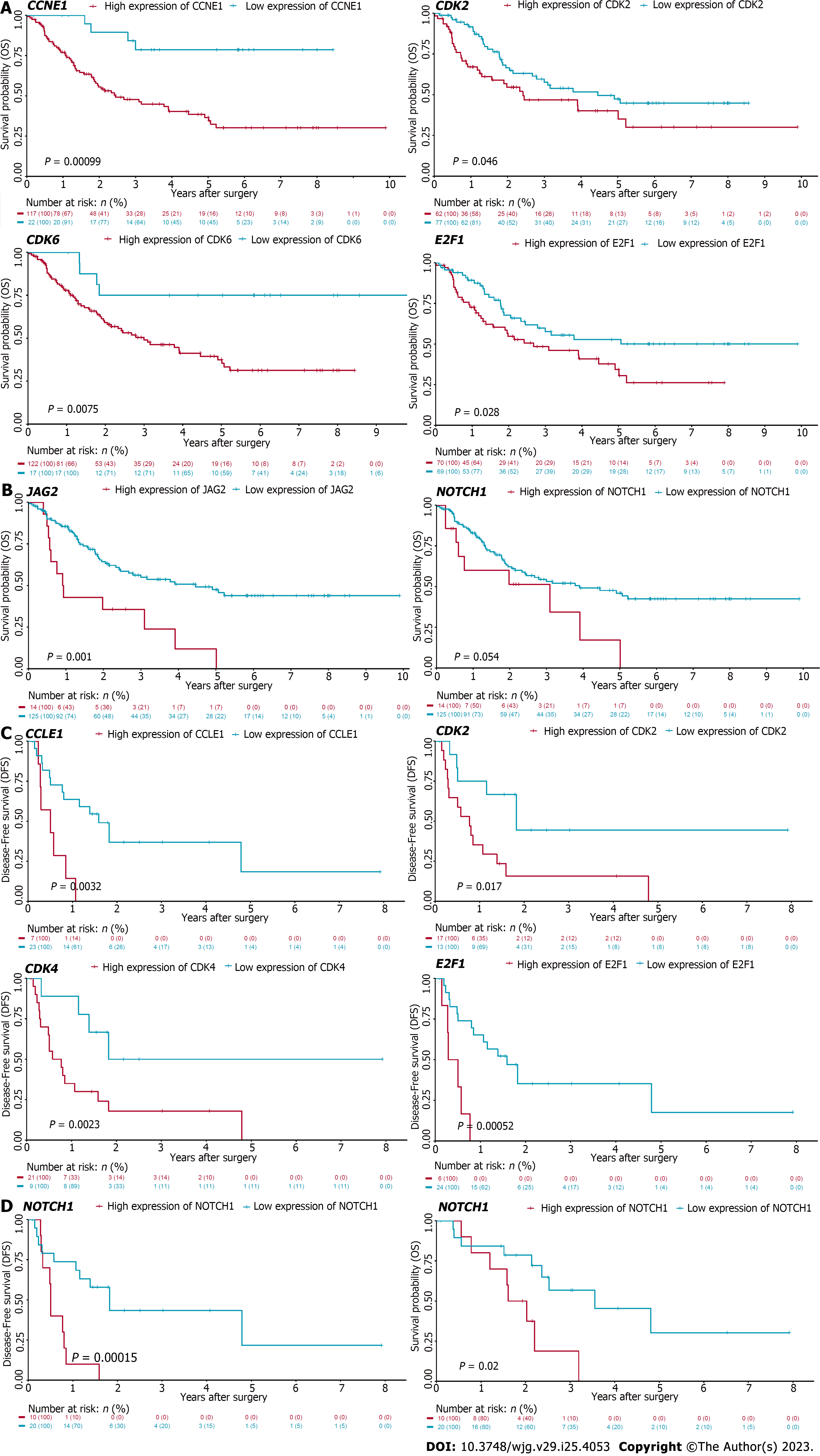
The cell cycle and Notch pathways were refined from the analysis of GSEA. These genes were found up-regulated in CCA tumor samples as well when compared with adjacent normal liver tissues. As we have found that targeting these pathways significantly suppressed CCA cell growth in vitro, we speculated that high expression of these genes may correlate with poor prognosis. To determine the correlation of these genes and CCA patient prognosis, we conducted the Kaplan-Meier survival analysis using the cell cycle and Notch pathways associated genes in CCA patients derived from two datasets, including GSE107943 and EBI (EGAD00001001076). It was found that high expression of cell cycle genes, including CDK2, CDK6, CCNE1, and E2F1, were negatively correlated with OS of CCA patients derived from the EBI database (EGAD00001001076) (Figure 7A). Similarly, high expression of Notch related genes, including NOTCH1 and JAG2, were associated with poor prognosis in these patients (Figure 7B). These findings were further validated using the GSE107943 database. Consistently, high CDK2, CDK4, CCNE1, and E2F1 were correlated with poor OS of CCA patients (Figure 7C). High NOTCH1 expression was associated with poor OS and DFS in these patients as well (Figure 7D). Collectively, these data demonstrated that refining the signaling pathways associated with tumorigenesis of human CCA using bioinformatics approach could identify potential therapeutic targets for patients with this deadly disease (Figure 8).
The current study demonstrated that cell cycle and notch associated pathways are elevated in human CCA patients compared with adjacent normal tissues. These findings were verified using human biliary epithelial cells and CCA cell lines. Consistently, cell cycle and Notch associated genes were found increased in CCA cell lines compared with non-malignant human biliary epithelial cells. As it has not been investigated if the cell cycle and Notch associated pathways are up-regulated in the CCA transgenic mice, our studies therefore disclose the similar but novel findings in the transgenic CCA mouse model. Cell cycle and Notch associated genes were up-regulated in mouse CCA tumors when compared with age-matched mouse normal bile ducts. Besides, we demonstrated that targeting cell cycle and Notch pathways could inhibit cell growth in human CCA cell lines. Furthermore, we validated the GSEA findings using another dataset. Most importantly, we demonstrated that high expression of several cell cycle and Notch associated genes are highly associated with poor prognosis in human CCA patients in two databases.
Refining RNA sequencing results have been well studied in liver malignancy, including hepatocellular carcinoma and CCA[3,4,24,25]. By combining whole genomic sequencing and RNA sequencing data, it was previously suggested that intrahepatic CCA could be classified as inflammation and proliferation groups which have different clinical outcomes, specifically worse prognosis in the proliferation group[26]. Recently, it was also suggested that extrahepatic CCA could be classified as immune, mesenchymal, metabolic, and proliferation groups. Further analysis revealed specific potential therapeutic targets correspondent to different groups, i.e., PD1/PD-L1 inhibitors were suggested to be potential therapies for the immune group of extrahepatic CCA patients[4]. Nevertheless, these potential therapeutic approaches have not been evaluated in preclinical models due to the possible reasons that no preclinical models could represent the individual group specifically. However, the recent study investigated the microenvironment-based classification of intrahepatic CCA and identified the inflammatory stroma class of intrahepatic CCA with T cell exhaustion and KRas mutation. Thus, they evaluated if the combination of KRas inhibitor and PD1 antibody could inhibit the malignant progression of CCA tumors established by using tail vein hydrodynamic injection of SB13 transposase, CRISPR/Cas9-sg-p19, pCaggs-KrasG12D. The model has high similarity to the inflammatory stroma intrahepatic CCA group. Their findings demonstrated that Kras inhibitor and PD1 antibody could substantially inhibit tumor malignant progression[3]. Taken together, it is very likely to identify potential therapeutic targets by refining publicly available RNA sequencing datasets.
By using this approach, we found that Notch and cell cycle associated pathways are potential therapeutic targets in CCA tumors. It has been previously demonstrated that specific overexpression of intracellular domain of Notch 1 could lead to CCA development possibly through cyclin E1[27]. The Notch1 Ligand, JAG1 could activate Notch1 signaling cascade to initiate cyclin D1 transcriptional expression[28]. Both JAG1 and Notch1 contain the epidermal growth factor like domain[29] which could be hydroxylated by aspartate beta-hydroxylase[30-32]. Indeed, previous studies have demonstrated that aspartate beta-hydroxylase could promote CCA progression through the Notch1-mediated cyclin D1 pathway[33]. Cell cycle is tightly controlled by the tumor suppressor retinoblastoma protein (RB1)[34]. Although RB1 mutation is not a common event in CCA patients[35], dysregulated cell cycle regulating genes are often found in these patients. Dysregulated cell cycle control would lead to cancer cell proliferation. As discussed, intrahepatic CCA patients could be classified as proliferative and inflammatory groups[26]. Previous studies also demonstrated the therapeutic potential of targeting CCA tumors by using the specific inhibitor targeting cyclin dependent kinases 4 and 6[36,37] which are important mediator controlling RB1 phosphorylation. Phosphorylation of RB1 prevents its binding from the E2F1 transcriptional factor, which finely modulates cell cycle regulating genes[38]. Thus, phosphorylation of RB1 would lead to inactivation of RB1 tumor suppressor function, in turn promoting cell cycle progression. It was previously reported that aspartate beta-hydroxylase could promote RB1 phosphorylation to disrupt cell cycle regulation in CCA tumors[39]. Interestingly, aspartate beta-hydroxylase was reported to be highly expressed in CCA but barely detectable in normal bile duct tissues[39], suggesting the linkage of identified Notch1 and cell cycle associated pathways to aspartate beta-hydroxylase.
The current study has several limitations. (1) We did not evaluate if targeting Notch1 and cell cycle pathways could inhibit CCA progression using preclinical models. Actually, the previous studies have demonstrated that Notch1 inhibitor could be a potential therapeutic approach[27] and that targeting cell cycle with CDK4/6 inhibitors could suppress CCA progression[36,37]. Although the current study was initiated from a different angle by using transcriptomic analysis, it ended up with similar findings that Notch1 and cell cycle pathways could be potential targets in CCA patients; (2) We did not separate intrahepatic and extrahepatic CCA patients. We pooled the available data by combining intrahepatic and extrahepatic transcriptomic datasets due to the limited datasets publicly available; and (3) our studies did not investigate if elevation of cell cycle and Notch associated pathways may be used as early detection markers. Future studies will be needed to further determine potential therapeutic approaches specifically for intrahepatic and extrahepatic CCA patients, respectively.
In conclusion, our data, through mRNA and protein profiling, identified very high similarity between CCA tumors derived from the CCA transgenic mice and human CCA tumors, thus providing a potential preclinical CCA model for investigating the CCA tumorigenesis. Besides, our data suggested that cell cycle and Notch associated pathways could be potential therapeutic targets in CCA patients.
The molecular pathogenesis of cholangiocarcinoma (CCA) remains largely unknown. Investigating the molecular mechanisms underlying CCA progression will potentially yield results toward identifying therapeutic targets.
Several mRNA sequencing (Seq) datasets in CCA samples are publicly available, but it remains unclear if the data can be validated in CCA cell lines, the transgenic CCA mouse model, and human CCA samples.
To summarize the mRNA Seq results and validate the findings in CCA cell lines, the transgenic CCA mouse model, and human CCA samples.
Bioinformatic analysis, cell culture studies, transgenic mouse model, human CCA samples, and molecular strategies were used to determine molecular pathogenesis of CCA.
Through bioinformatic analysis, we found that cell cycle and Notch associated pathways are up-regulated in human CCA samples, compared to the non-tumor controls. We validated the findings in human CCA cell lines and the transgenic CCA mouse model.
Our data, through mRNA and protein profiling, identified very high similarity between CCA tumors derived from the CCA transgenic mice and human CCA tumors, thus providing a potential preclinical CCA model for investigating CCA tumorigenesis. Besides, our data suggested that cell cycle and Notch associated pathways could be potential therapeutic targets in CCA patients.
Systemic chemotherapy, gemcitabine and cisplatin, is recommended for CCA patients. However, the 5-year survival of CCA patients has not been significantly improved, suggesting an urgent need to develop novel therapeutic approaches.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and Hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Amin A, United Arab Emirates; Han J, China; Wang CY, Taiwan S-Editor: Chang KL L-Editor: A P-Editor: Cai YX
| 1. | Banales JM, Marin JJG, Lamarca A, Rodrigues PM, Khan SA, Roberts LR, Cardinale V, Carpino G, Andersen JB, Braconi C, Calvisi DF, Perugorria MJ, Fabris L, Boulter L, Macias RIR, Gaudio E, Alvaro D, Gradilone SA, Strazzabosco M, Marzioni M, Coulouarn C, Fouassier L, Raggi C, Invernizzi P, Mertens JC, Moncsek A, Rizvi S, Heimbach J, Koerkamp BG, Bruix J, Forner A, Bridgewater J, Valle JW, Gores GJ. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol. 2020;17:557-588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1555] [Cited by in RCA: 1505] [Article Influence: 301.0] [Reference Citation Analysis (0)] |
| 2. | Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, Lind GE, Folseraas T, Forbes SJ, Fouassier L, Geier A, Calvisi DF, Mertens JC, Trauner M, Benedetti A, Maroni L, Vaquero J, Macias RI, Raggi C, Perugorria MJ, Gaudio E, Boberg KM, Marin JJ, Alvaro D. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol. 2016;13:261-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 731] [Cited by in RCA: 961] [Article Influence: 106.8] [Reference Citation Analysis (0)] |
| 3. | Martin-Serrano MA, Kepecs B, Torres-Martin M, Bramel ER, Haber PK, Merritt E, Rialdi A, Param NJ, Maeda M, Lindblad KE, Carter JK, Barcena-Varela M, Mazzaferro V, Schwartz M, Affo S, Schwabe RF, Villanueva A, Guccione E, Friedman SL, Lujambio A, Tocheva A, Llovet JM, Thung SN, Tsankov AM, Sia D. Novel microenvironment-based classification of intrahepatic cholangiocarcinoma with therapeutic implications. Gut. 2023;72: 736-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 85] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 4. | Montal R, Sia D, Montironi C, Leow WQ, Esteban-Fabró R, Pinyol R, Torres-Martin M, Bassaganyas L, Moeini A, Peix J, Cabellos L, Maeda M, Villacorta-Martin C, Tabrizian P, Rodriguez-Carunchio L, Castellano G, Sempoux C, Minguez B, Pawlik TM, Labgaa I, Roberts LR, Sole M, Fiel MI, Thung S, Fuster J, Roayaie S, Villanueva A, Schwartz M, Llovet JM. Molecular classification and therapeutic targets in extrahepatic cholangiocarcinoma. J Hepatol. 2020;73:315-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 192] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 5. | Nelson DR, Hrout AA, Alzahmi AS, Chaiboonchoe A, Amin A, Salehi-Ashtiani K. Molecular Mechanisms behind Safranal's Toxicity to HepG2 Cells from Dual Omics. Antioxidants (Basel). 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 42] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 6. | Chan-On W, Nairismägi ML, Ong CK, Lim WK, Dima S, Pairojkul C, Lim KH, McPherson JR, Cutcutache I, Heng HL, Ooi L, Chung A, Chow P, Cheow PC, Lee SY, Choo SP, Tan IB, Duda D, Nastase A, Myint SS, Wong BH, Gan A, Rajasegaran V, Ng CC, Nagarajan S, Jusakul A, Zhang S, Vohra P, Yu W, Huang D, Sithithaworn P, Yongvanit P, Wongkham S, Khuntikeo N, Bhudhisawasdi V, Popescu I, Rozen SG, Tan P, Teh BT. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat Genet. 2013;45:1474-1478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 385] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 7. | Abou-Alfa GK, Macarulla T, Javle MM, Kelley RK, Lubner SJ, Adeva J, Cleary JM, Catenacci DV, Borad MJ, Bridgewater J, Harris WP, Murphy AG, Oh DY, Whisenant J, Lowery MA, Goyal L, Shroff RT, El-Khoueiry AB, Fan B, Wu B, Chamberlain CX, Jiang L, Gliser C, Pandya SS, Valle JW, Zhu AX. Ivosidenib in IDH1-mutant, chemotherapy-refractory cholangiocarcinoma (ClarIDHy): a multicentre, randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 2020;21:796-807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 557] [Cited by in RCA: 704] [Article Influence: 140.8] [Reference Citation Analysis (0)] |
| 8. | Zhu AX, Macarulla T, Javle MM, Kelley RK, Lubner SJ, Adeva J, Cleary JM, Catenacci DVT, Borad MJ, Bridgewater JA, Harris WP, Murphy AG, Oh DY, Whisenant JR, Lowery MA, Goyal L, Shroff RT, El-Khoueiry AB, Chamberlain CX, Aguado-Fraile E, Choe S, Wu B, Liu H, Gliser C, Pandya SS, Valle JW, Abou-Alfa GK. Final Overall Survival Efficacy Results of Ivosidenib for Patients With Advanced Cholangiocarcinoma With IDH1 Mutation: The Phase 3 Randomized Clinical ClarIDHy Trial. JAMA Oncol. 2021;7:1669-1677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 299] [Article Influence: 74.8] [Reference Citation Analysis (0)] |
| 9. | Yamada N, Chung YS, Arimoto Y, Sawada T, Seki S, Sowa M. Establishment of a new human extrahepatic bile duct carcinoma cell line (OCUCh-LM1) and experimental liver metastatic model. Br J Cancer. 1995;71:543-548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 10. | Miyagiwa M, Ichida T, Tokiwa T, Sato J, Sasaki H. A new human cholangiocellular carcinoma cell line (HuCC-T1) producing carbohydrate antigen 19/9 in serum-free medium. In Vitro Cell Dev Biol. 1989;25:503-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 117] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 11. | Enjoji M, Sakai H, Nawata H, Kajiyama K, Tsuneyoshi M. Sarcomatous and adenocarcinoma cell lines from the same nodule of cholangiocarcinoma. In Vitro Cell Dev Biol Anim. 1997;33:681-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 12. | Enjoji M, Nakashima M, Honda M, Sakai H, Nawata H. Hepatocytic phenotypes induced in sarcomatous cholangiocarcinoma cells treated with 5-azacytidine. Hepatology. 1997;26:288-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 13. | Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22949] [Cited by in RCA: 32690] [Article Influence: 2514.6] [Reference Citation Analysis (0)] |
| 14. | Ahn KS, O'Brien D, Kang YN, Mounajjed T, Kim YH, Kim TS, Kocher JA, Allotey LK, Borad MJ, Roberts LR, Kang KJ. Prognostic subclass of intrahepatic cholangiocarcinoma by integrative molecular-clinical analysis and potential targeted approach. Hepatol Int. 2019;13:490-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 15. | Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, Hama N, Hosoda F, Urushidate T, Ohashi S, Hiraoka N, Ojima H, Shimada K, Okusaka T, Kosuge T, Miyagawa S, Shibata T. Genomic spectra of biliary tract cancer. Nat Genet. 2015;47:1003-1010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 651] [Cited by in RCA: 934] [Article Influence: 93.4] [Reference Citation Analysis (0)] |
| 16. | Wright MN, Dankowski T, Ziegler A. Unbiased split variable selection for random survival forests using maximally selected rank statistics. Stat Med. 2017;36:1272-1284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 17. | Fang H; ULTRA-DD Consortium, De Wolf H, Knezevic B, Burnham KL, Osgood J, Sanniti A, Lledó Lara A, Kasela S, De Cesco S, Wegner JK, Handunnetthi L, McCann FE, Chen L, Sekine T, Brennan PE, Marsden BD, Damerell D, O'Callaghan CA, Bountra C, Bowness P, Sundström Y, Milani L, Berg L, Göhlmann HW, Peeters PJ, Fairfax BP, Sundström M, Knight JC. A genetics-led approach defines the drug target landscape of 30 immune-related traits. Nat Genet. 2019;51:1082-1091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 142] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 18. | Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545-15550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27252] [Cited by in RCA: 37070] [Article Influence: 1853.5] [Reference Citation Analysis (0)] |
| 19. | Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017;18:248-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1446] [Cited by in RCA: 3144] [Article Influence: 393.0] [Reference Citation Analysis (0)] |
| 20. | Huang CK, Lee SO, Lai KP, Ma WL, Lin TH, Tsai MY, Luo J, Chang C. Targeting androgen receptor in bone marrow mesenchymal stem cells leads to better transplantation therapy efficacy in liver cirrhosis. Hepatology. 2013;57:1550-1563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 21. | Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1783] [Cited by in RCA: 2141] [Article Influence: 118.9] [Reference Citation Analysis (0)] |
| 22. | Farshidfar F, Zheng S, Gingras MC, Newton Y, Shih J, Robertson AG, Hinoue T, Hoadley KA, Gibb EA, Roszik J, Covington KR, Wu CC, Shinbrot E, Stransky N, Hegde A, Yang JD, Reznik E, Sadeghi S, Pedamallu CS, Ojesina AI, Hess JM, Auman JT, Rhie SK, Bowlby R, Borad MJ; Cancer Genome Atlas Network, Zhu AX, Stuart JM, Sander C, Akbani R, Cherniack AD, Deshpande V, Mounajjed T, Foo WC, Torbenson MS, Kleiner DE, Laird PW, Wheeler DA, McRee AJ, Bathe OF, Andersen JB, Bardeesy N, Roberts LR, Kwong LN. Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct IDH-Mutant Molecular Profiles. Cell Rep. 2017;18:2780-2794. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 333] [Cited by in RCA: 378] [Article Influence: 47.3] [Reference Citation Analysis (1)] |
| 23. | O'Dell MR, Huang JL, Whitney-Miller CL, Deshpande V, Rothberg P, Grose V, Rossi RM, Zhu AX, Land H, Bardeesy N, Hezel AF. Kras(G12D) and p53 mutation cause primary intrahepatic cholangiocarcinoma. Cancer Res. 2012;72:1557-1567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 169] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 24. | Craig AJ, Garcia-Lezana T, Ruiz de Galarreta M, Villacorta-Martin C, Kozlova EG, Martins-Filho SN, von Felden J, Ahsen ME, Bresnahan E, Hernandez-Meza G, Labgaa I, D'Avola D, Schwartz M, Llovet JM, Sia D, Thung S, Losic B, Lujambio A, Villanueva A. Transcriptomic characterization of cancer-testis antigens identifies MAGEA3 as a driver of tumor progression in hepatocellular carcinoma. PLoS Genet. 2021;17:e1009589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 25. | Montironi C, Castet F, Haber PK, Pinyol R, Torres-Martin M, Torrens L, Mesropian A, Wang H, Puigvehi M, Maeda M, Leow WQ, Harrod E, Taik P, Chinburen J, Taivanbaatar E, Chinbold E, Solé Arqués M, Donovan M, Thung S, Neely J, Mazzaferro V, Anderson J, Roayaie S, Schwartz M, Villanueva A, Friedman SL, Uzilov A, Sia D, Llovet JM. Inflamed and non-inflamed classes of HCC: a revised immunogenomic classification. Gut. 2023;72:129-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 144] [Article Influence: 72.0] [Reference Citation Analysis (0)] |
| 26. | Sia D, Hoshida Y, Villanueva A, Roayaie S, Ferrer J, Tabak B, Peix J, Sole M, Tovar V, Alsinet C, Cornella H, Klotzle B, Fan JB, Cotsoglou C, Thung SN, Fuster J, Waxman S, Garcia-Valdecasas JC, Bruix J, Schwartz ME, Beroukhim R, Mazzaferro V, Llovet JM. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013;144:829-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 431] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 27. | Zender S, Nickeleit I, Wuestefeld T, Sörensen I, Dauch D, Bozko P, El-Khatib M, Geffers R, Bektas H, Manns MP, Gossler A, Wilkens L, Plentz R, Zender L, Malek NP. A critical role for notch signaling in the formation of cholangiocellular carcinomas. Cancer Cell. 2013;23:784-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 151] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 28. | Cohen B, Shimizu M, Izrailit J, Ng NF, Buchman Y, Pan JG, Dering J, Reedijk M. Cyclin D1 is a direct target of JAG1-mediated Notch signaling in breast cancer. Breast Cancer Res Treat. 2010;123:113-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 129] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 29. | Bicker F, Vasic V, Horta G, Ortega F, Nolte H, Kavyanifar A, Keller S, Stankovic ND, Harter PN, Benedito R, Lutz B, Bäuerle T, Hartwig J, Baumgart J, Krüger M, Radyushkin K, Alberi L, Berninger B, Schmidt MHH. Neurovascular EGFL7 regulates adult neurogenesis in the subventricular zone and thereby affects olfactory perception. Nat Commun. 2017;8:15922. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 30. | Jia S, McGinnis K, VanDusen WJ, Burke CJ, Kuo A, Griffin PR, Sardana MK, Elliston KO, Stern AM, Friedman PA. A fully active catalytic domain of bovine aspartyl (asparaginyl) beta-hydroxylase expressed in Escherichia coli: characterization and evidence for the identification of an active-site region in vertebrate alpha-ketoglutarate-dependent dioxygenases. Proc Natl Acad Sci U S A. 1994;91:7227-7231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 31. | Jia S, VanDusen WJ, Diehl RE, Kohl NE, Dixon RA, Elliston KO, Stern AM, Friedman PA. cDNA cloning and expression of bovine aspartyl (asparaginyl) beta-hydroxylase. J Biol Chem. 1992;267:14322-14327. [PubMed] |
| 32. | Stenflo J, Holme E, Lindstedt S, Chandramouli N, Huang LH, Tam JP, Merrifield RB. Hydroxylation of aspartic acid in domains homologous to the epidermal growth factor precursor is catalyzed by a 2-oxoglutarate-dependent dioxygenase. Proc Natl Acad Sci U S A. 1989;86:444-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 66] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 33. | Huang CK, Iwagami Y, Aihara A, Chung W, de la Monte S, Thomas JM, Olsen M, Carlson R, Yu T, Dong X, Wands J. Anti-Tumor Effects of Second Generation β-Hydroxylase Inhibitors on Cholangiocarcinoma Development and Progression. PLoS One. 2016;11:e0150336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 34. | Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220-5227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 766] [Cited by in RCA: 868] [Article Influence: 45.7] [Reference Citation Analysis (0)] |
| 35. | Zou S, Li J, Zhou H, Frech C, Jiang X, Chu JS, Zhao X, Li Y, Li Q, Wang H, Hu J, Kong G, Wu M, Ding C, Chen N, Hu H. Mutational landscape of intrahepatic cholangiocarcinoma. Nat Commun. 2014;5:5696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 306] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 36. | Sittithumcharee G, Suppramote O, Vaeteewoottacharn K, Sirisuksakun C, Jamnongsong S, Laphanuwat P, Suntiparpluacha M, Matha A, Chusorn P, Buraphat P, Kakanaporn C, Charngkaew K, Silsirivanit A, Korphaisarn K, Limsrichamrern S, Tripatara P, Pairojkul C, Wongkham S, Sampattavanich S, Okada S, Jirawatnotai S. Dependency of Cholangiocarcinoma on Cyclin D-Dependent Kinase Activity. Hepatology. 2019;70:1614-1630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 37. | Song X, Liu X, Wang H, Wang J, Qiao Y, Cigliano A, Utpatel K, Ribback S, Pilo MG, Serra M, Gordan JD, Che L, Zhang S, Cossu A, Porcu A, Pascale RM, Dombrowski F, Hu H, Calvisi DF, Evert M, Chen X. Combined CDK4/6 and Pan-mTOR Inhibition Is Synergistic Against Intrahepatic Cholangiocarcinoma. Clin Cancer Res. 2019;25:403-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 38. | Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10:699-703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 680] [Cited by in RCA: 679] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 39. | Huang CK, Iwagami Y, Zou J, Casulli S, Lu S, Nagaoka K, Ji C, Ogawa K, Cao KY, Gao JS, Carlson RI, Wands JR. Aspartate beta-hydroxylase promotes cholangiocarcinoma progression by modulating RB1 phosphorylation. Cancer Lett. 2018;429:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |