Published online Jun 21, 2023. doi: 10.3748/wjg.v29.i23.3622
Peer-review started: March 20, 2023
First decision: April 10, 2023
Revised: April 24, 2023
Accepted: May 17, 2023
Article in press: May 17, 2023
Published online: June 21, 2023
Processing time: 87 Days and 16.4 Hours
Gastric cancer (GC) is a common gastrointestinal malignancy worldwide. Based on cancer-related mortality, the current prevention and treatment strategies for GC still show poor clinical results. Therefore, it is important to find effective drug treatment targets.
To explore the molecular mechanism of 18β-glycyrrhetinic acid (18β-GRA) regulating the miR-345-5p/TGM2 signaling pathway to inhibit the proliferation of GC cells.
CCK-8 assay was used to determine the effect of 18β-GRA on the survival rate of GES-1 cells and AGS and HGC-27 cells. Cell cycle and apoptosis were detected by flow cytometry, cell migration was detected by a wound healing assay, the effect of 18β-GRA on subcutaneous tumor growth in BALB/c nude mice was inves
18β-GRA could inhibit GC cells viability, promote cell apoptosis, block cell cycle, reduce cell wound healing ability, and inhibit the GC cells growth in vivo. MDC staining results showed that 18β-GRA could promote autophagy in GC cells. By TMT proteomic analysis and miRNAs transcriptome analysis, it was concluded that 18β-GRA could down-regulate TGM2 expression and up-regulate miR-345-5p expression in GC cells. Subsequently, we verified that TGM2 is the target of miR-345-5p, and that overexpression of miR-345-5p significantly inhibited the protein expression level of TGM2. Western blot showed that the expression of autophagy-related proteins of TGM2 and p62 was significantly reduced, and LC3II, ULK1 and AMPK expression was significantly increased in GC cells treated with 18β-GRA. Overexpression of miR-345-5p not only inhibited the expression of TGM2, but also inhibited the proliferation of GC cells by promoting cell apoptosis and arresting cell cycle.
18β-GRA inhibits the proliferation of GC cells and promotes autophagy by regulating the miR-345-5p/TGM2 signaling pathway.
Core Tip: Gastric cancer (GC) is a global health problem that seriously endangers human life, so it is urgent to find drugs to treat it. 18β-glycyrrhetinic acid (18β-GRA), as one of the main components of glycyrrhiza, has a strong antitumor effect. In this paper, the inhibitory effect of 18β-GRA on GC was verified by in vitro and in vivo experiments. In addition, it was found that 18β-GRA promoted autophagy and inhibited the proliferation of GC cells through the miR-345-5p/TGM2 signaling pathway. These findings provide the theoretical basis for the GC clinical treatment of 18β-GRA.
- Citation: Li X, Ma XL, Nan Y, Du YH, Yang Y, Lu DD, Zhang JF, Chen Y, Zhang L, Niu Y, Yuan L. 18β-glycyrrhetinic acid inhibits proliferation of gastric cancer cells through regulating the miR-345-5p/TGM2 signaling pathway. World J Gastroenterol 2023; 29(23): 3622-3644
- URL: https://www.wjgnet.com/1007-9327/full/v29/i23/3622.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i23.3622
The fourth most common cause of cancer-related death worldwide is gastric cancer (GC)[1]. Envi
18β-glycyrrhetinic acid (18β-GRA) is a compound extracted from licorice[4]. Studies have revealed that 18β-GRA has multiple pharmacological effects, like anti-inflammatory, anti-viral, anti-tumor, liver protection and so on[5-9]. In recent years, the therapeutic effect of 18β-GRA on lung cancer[7], breast cancer[10], ovarian cancer[11], prostate cancer[12] and other cancers has been confirmed. Cao et al[13] revealed that 18β-GRA inhibited GC cells proliferation, energy metabolism and carcinogenesis by down-regulating toll-like receptor 2. Through reactive oxygen species/protein kinase C-α/extracellular signal-regulated kinase pathway, as well as matrix metalloproteinase MMP2 and MMP9 activity, 18β-GRA inhibited GC cells migration and invasion[14]. And our previous study found that 18β-GRA inhibited GC cells proliferation by regulating MRPL35[15]. These results indicated that 18β-GRA may be a useful drug for the prevention and treatment of GC.
MicroRNAs (miRNAs) are highly conserved non-coding RNA molecules, that take part in the occurrence and development of cancer[16]. Many sequences of human miRNAs associated with cancer mechanisms have been identified. They regulate protein expression levels by their target mRNA, and participate in vital cellular processes and pathways[17]. There is growing evidence that the difference in miRNAs expression exists not only between normal and cancer tissues and between different cancer types and subtypes, but also between early and advanced cancers[18,19]. Komatsu et al[20] found that up-regulation of miR-148a inhibited the GC cells proliferation, invasion and EMT. The researchers revealed that Helicobacter pylori infection reduced miR-1298-5p expression in GC cells, while low-expressed miR-1298-5p could promote GC cells proliferation, migration and invasion[21].
Autophagy is a biological process that occurs in cells[22]. A lot of research has found that it plays a key role in physiological processes and disease occurrences, including development, metabolism, inflammation and cancer[23]. Dysfunctional autophagy results in incorrect organelles and protein breakdown, which kill autophagic cells and affect tumor cell survival. Wei et al[24] revealed that miR-183 induced GC cells autophagy and inhibited proliferation via targeted inhibition of mechanistic target of rapamycin (mTOR) expression.
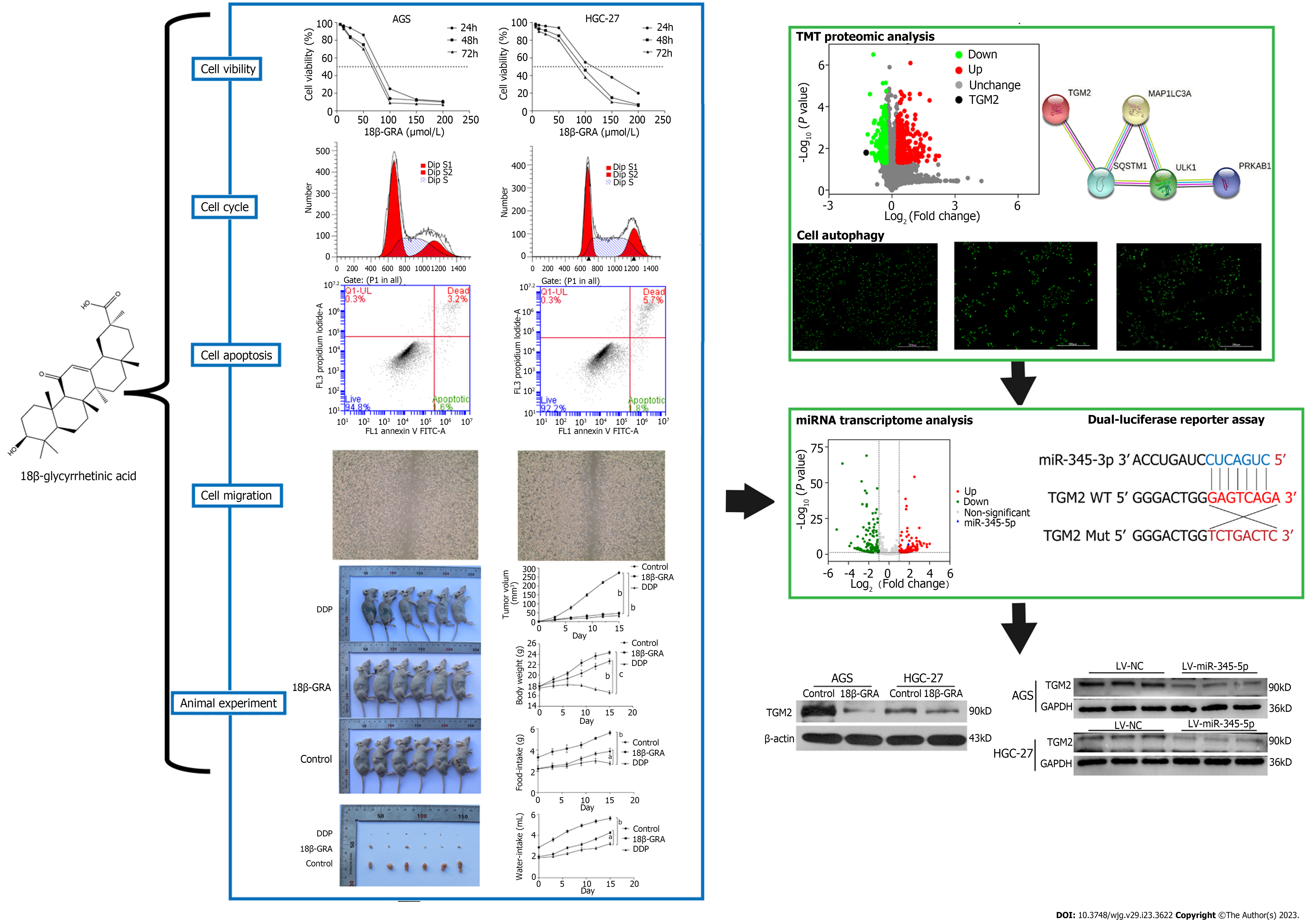
In our study, we revealed 18β-GRA’s effect on GC cells phenotype and tumor formation in nude mice, and 18β-GRA’s effect on GC cells autophagy. We used TMT proteomic analysis and the STRING database to predict the differentially expressed autophagy-related proteins and their interactions. The differentially expressed miRNAs were analyzed using miRNAs transcriptome analysis, and the corresponding miRNAs and complementary binding sites of autophagy-related proteins were predicted using the miRBase and TargetScan databases. Later, we verified the link between miR-345-5p and TGM2 using a dual-luciferase reporter assay. Quantitative real-time polymerase chain reaction (qRT-PCR) was used to detect expression of miR-345-5p, western blot was used to detect autophagy-related proteins, and lentivirus transfection technique verified the effect of miR-345-5p on TGM2. We concluded that 18β-GRA can inhibit GC cells proliferation and promote autophagy via regulating the miR-345-5p/TGM2 signaling pathway, which may afford a theoretical basis for GC treatment with 18β-GRA. The research idea is shown in Figure 1.
Human gastric epithelial cell GES-1 was purchased from BNCC (Cat. No. BNCC353464, Beijing, China). Human GC cell lines AGS, HGC-27 and MKN-45 were purchased from Procell (Cat. No. CL-0022/CL-0107/CL-0292, Wuhan, China). Fetal bovine serum (FBS) was purchased from Corning (Cat. No. 35-076-CV, United States). DMEM, DMEM/F-12 and RPIM-1640 mediums were purchased from Gibco. The dual-luciferase reporter assay system was purchased from Promega (United States). Shanghai Gene Biotechnology Co., Ltd. (China) provided the plasmid and lentiviral expression vector. The MDC kit, cell apoptosis kit and cell cycle kit (Cat. No. KGATG001/KGA1026/KGA512, Jiangsu, China) were purchased from Jiangsu KeyGEN Bio TECH Corp., Ltd. TaKaRa provided the PrimeScriptTM RT reagent kit and TB Green Premix Ex Taq II (Cat. No. RR047A/RR82LR, Japan). Thermofisher provided Trizol. MedChemExpress provided cisplatin (DDP) and 18β-GRA (Cat. No. HY17394/HY-N0180, United States). Immunoway provided TGM2, AMPK, p62, ULK1, LC3I/II, GAPDH and β-actin antibodies, while CST provided anti-mouse/rabbit immunoglobulin G antibodies.
BABL/c nude mice (male, 18-22g, SPF) were provided by the Animal Laboratory Center of Ningxia Medical University. All animals were fed standard laboratory feed and water in 12 h light/dark cycle environment. The animal protocols (IACUC-NYLAC-2022-108) were approved by the Institutional Animal Care and Use Committee of Ningxia Medical University. All animals were euthanized by CO2.
GES-1 cells were cultured in DMEM medium, HGC-27 cells were cultured in RPIM-1640 medium, and AGS and MKN-45 cells were cultured in DMEM/F-12 medium, and 10% FBS and 1% penicillin-streptomycin liquid were added into all mediums to. The cell culture flask was placed in an incubator with a constant temperature of 37 °C and 5% CO2. The GFP-labeled miR-345-5p overexpression lentiviral vector (LV-miR-345-5p) and empty lentiviral vector (LV-NC) were then transfected into HGC-27 and AGS cells using the tool virus user manual as a guide.
The cells were inoculated into 96-well plates at 4000 cells per well and treated with 18β-GRA (12.5 μmol/L-200 μmol/L) for 24 h, 48 h and 72 h. Next, 10 μL CCK-8 was added to each well, and the absorbance was detected at 450 nm.
The cells were treated with 18β-GRA at concentrations of 30, 60 and 120 μmol/L or 45, 90 and 135 μmol/L for 48 h, respectively. Next, the cells were collected and fixed with 70% pre-cooled ethanol. The fixative solution was washed off with PBS. 500 μL pre-configured working solution (RNase A:PI = 1:9) was added. The percentage of cell cycle was determined by flow cytometry after 30 min. The cells in each group were collected, 500 μL binding buffer, 5 μL Annexin V-FITC and 5 μL PI or 500 μL binding buffer, 5 μL Annexin V-APC and 5 μL 7-AAD were added and mixed. Cell apoptosis was determined after 15 min using flow cytometry.
The cells were inoculated into the 6-well plates at the appropriate concentration and cultured for 24 h. Next, we made a scratch in the central part of each hole, and 18β-GRA at concentrations of 30, 60, 120 μmol/L or 45, 90, 135 μmol/L was added for 48 h. Finally, the cells were washed and photographed with a microscope. Cell migration area was calculated by Image J.
The cells were treated with 18β-GRA at concentrations of 30, 60 and 120 μmol/L or 45, 90 and 135 μmol/L for 48 h, respectively. Next, the cells were gently washed twice with a wash buffer, stained with MDC and incubated away from light for 15-45 min. Photographs were taken under a fluorescence microscope after washing twice with 1 × wash buffer.
The BALB/c nude mice were fed for 1 wk, then 200 μL MKN-45 cells suspension containing 4 × 106 cells were inoculated subcutaneously into nude mice using a microsyringe. The experiment was divided into three groups: Control, cisplatin group (DDP) and 18β-GRA groups. After 5 d of subcutaneous inoculation, the 18β-GRA group was intraperitoneally injected with 50 mg/kg[13,25], the DDP group with 2 mg/kg[26], and the control group was given normal saline. V = (L × W2)/2 was used to calculate tumor volumes (V, volume; W, width; L, length). After 14 d of continuous administration, all nude mice were killed by CO2 inhalation and photographed.
GC cells were treated with 18β-GRA, and differentially expressed proteins were detected and analyzed by China Gene Biotechnology Co., Ltd. All the obtained original data files were processed by Proteome Discoverer 2.2 (Thermo Fisher, United States) software. Proteins with expression ratio > 1.2 and P < 0.05 were considered as differentially expressed proteins.
Cellular processes, biological processes, and molecular functions of differentially expressed proteins were analyzed using Metascape (https://metascape.org/gp/index.html#/main/step1). Gene Ontology annotation, Kyoto Encyclopedia of Genes and Genomes pathway, and InterPro domain enrichment analysis were performed using Fisher accuracy test. WoLFPSOR was used to locate and predict the differentially expressed proteins. The protein expression levels are classified by Matplotlib to form hierarchical clustering heatmaps.
GC cells were treated with 18β-GRA at concentrations of 60 μmol/L and total RNA was extracted from it. Agarose gel electrophoresis, Nanodrop, Qubit 2.0, and Agilent 2100 were used to ensure that the quality of the collected RNA met the requirements of subsequent experiments. A small RNA Sample Pre kit was used to construct the library, and specific enzymes were used to connect the PCR primer connectors at the 3’ and 5’ ends. Then the cDNA synthesis can be completed efficiently and rapidly by reverse transcription. The single strand of DNA is combined with the primer and the PCR amplification stage begins. After PAGE electrophoresis, a cDNA library (effective concentration > 2 nM) was obtained. Qubit 2.0 and Agilent 2100 were combined with qPCR to ensure the quality of the library. The optical signal was transformed into a sequence by computer analysis. The original sequencing data was further assessed for quality with P < 0.05 and |Log2 (fold change)| > 1 as a filter condition, with a certain number of differential expressions of miRNAs eventually selected.
We predicted protein-protein interaction (PPI) networks from the STRING database (https://string-db.org/). The miRBase website (http://www.mirbase.org/) was used to predict the corresponding miRNAs of autophagy-related proteins and the TargetScan database (https://www.targetscan.org/) was used to predict the binding sites of miRNA and mRNA.
TargetScan predicted the binding sites for miR-345-5p and TGM2. The fragments, including the wild 3’UTR regions or mutant 3’UTR regions of TGM2, were inserted into GV716 with a firefly and renilla luciferase reporter gene. The overexpressed miR-345-5p plasmid was then individually transfected into AGS cells. After 48 h of transfection, the relative luciferase activity was detected using a dual-luciferase reporter assay system.
The cells were cultured and treated 18β-GRA at concentrations of 30, 60 and 120 μmol/L or 45, 90 and 135 μmol/L for 48 h, respectively. Total RNA was extracted using Trizol, and cDNA was synthesized using PrimeScript™ RT reagent kit. The expression level was detected with TB Green Premix Ex Taq II. The primer sequences are as follows: miR-345-5p, forward, 5’-GCTGACTCCTAGTCCAGGGCTC-3’ and reverse, 5’-GGCCAACCGCGAGAAGATG-3’; U6, forward, 5’-CTGCGCAAGGATGACACGCAAATT-3’ and reverse, 5’-GGCCAACCGCGAGAAGATG-3’. U6 as a housekeeping gene. The relative expre
The cells were cultured and treated 18β-GRA at concentrations of 60 μmol/L or 90 μmol/L for 48 h, respectively. The cells were collected and the total protein was extracted, the protein content was detected with BCA method. The protein was isolated and transferred onto a PVDF membrane, which is then sealed with 5% skim milk powder, soaked in primary antibodies and incubated overnight. The next day, the PVDF membrane was cleaned with TBST and soaked in secondary antibodies for 2 h. The protein was detected using an ECL solution, and ultimately the grey values were measured using ImageJ.
The statistical methods of this study were reviewed by Li-Qun Wang, Department of Epidemiology and Medical Statistics, Institute of Public Health and Management, Ningxia Medical University. All data were statistically analyzed using GraphPad Prism 7. All data were shown as mean ± SD, and the differences between different groups were analyzed using a one-way ANOVA or t-test. The significance level was set at P < 0.05.
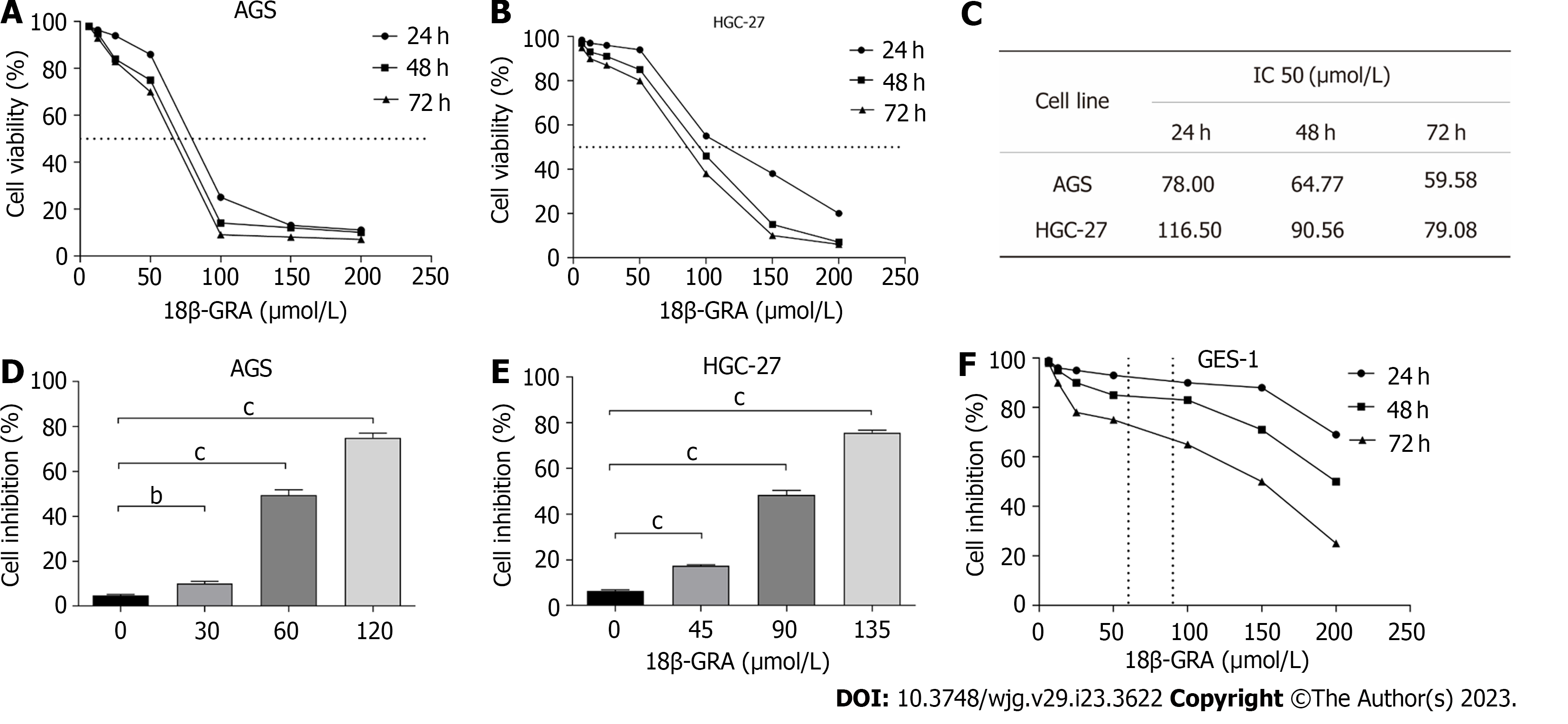
The results demonstrated that when the 18β-GRA concentration was 12.5 μmol/L-200 μmol/L, the inhibitory effect of 18β-GRA on AGS and HGC-27 cells viability was enhanced as the 18β-GRA dose was increased (Figures 2A and B). Next, 18β-GRA treated AGS and HGC-27 cells for 24 h, 48 h and 72 h, IC50 value was shown in Figure 2C. Therefore, we determined that the low, medium and high concentrations of 18β-GRA treated AGS cells for 48 h were 30, 60 and 120 μmol /L, and the low, medium and high concentrations of 18β-GRA treated HGC-27 cells for 48 h were 45, 90 and 135 μmol/L. AGS and HGC-27 cells viability was inhibited at low, medium and high doses of 18β-GRA when compared with the control group (0 μmol/L) (P < 0.01) (Figures 2D and E).
Subsequently, we investigated 18β-GRA’s effect on GES-1 cells viability. The results demonstrated that GES-1 cells viability was slightly affected when 18β-GRA concentration was 12.5 μmol/L-100 μmol/L (Figure 2F). When the 18β-GRA concentration was less than 150 μmol/L, the GES-1 cells viability remained above 71% at 48 h.
18β-GRA’s effect on GC cells cycle results demonstrated that the percentage of G0/G1 phase (49.15%, 56.23% and 73.07%) in AGS cells treated with low, medium and high doses of 18β-GRA was higher than the control group (33.71%) (Figures 3A and B). The percentage of G0/G1 phase (46.58%, 51.69% and 77.11%) in HGC-27 cells treated with low, medium and high doses of 18β-GRA was higher than the control group (41.67%) (Figures 3C and D).
18β-GRA’s effect on GC cells apoptosis results demonstrated that the apoptosis rates of AGS cells treated with low, medium and high doses of 18β-GRA were 7.50%, 19.80% and 72. 00%, which were obviously higher than the control group (4. 80%) (P < 0.01) (Figures 3E and F). The apoptosis rates of HGC-27 cells treated with low, medium and high doses of 18β-GRA were 6.50%, 8.10% and 15.40%, which were obviously higher than the control group (6.40%) (P < 0.05) (Figures 3G-H).
The wound healing assay revealed that both high and medium doses of 18β-GRA can inhibit AGS and HGC-27 cells’ wound healing abilities (P < 0.001) (Figures 4A-D). The inhibitory effect of 18β-GRA on wound healing ability was gradually enhanced as the 18β-GRA dose was increased in AGS and HGC-27 cells.
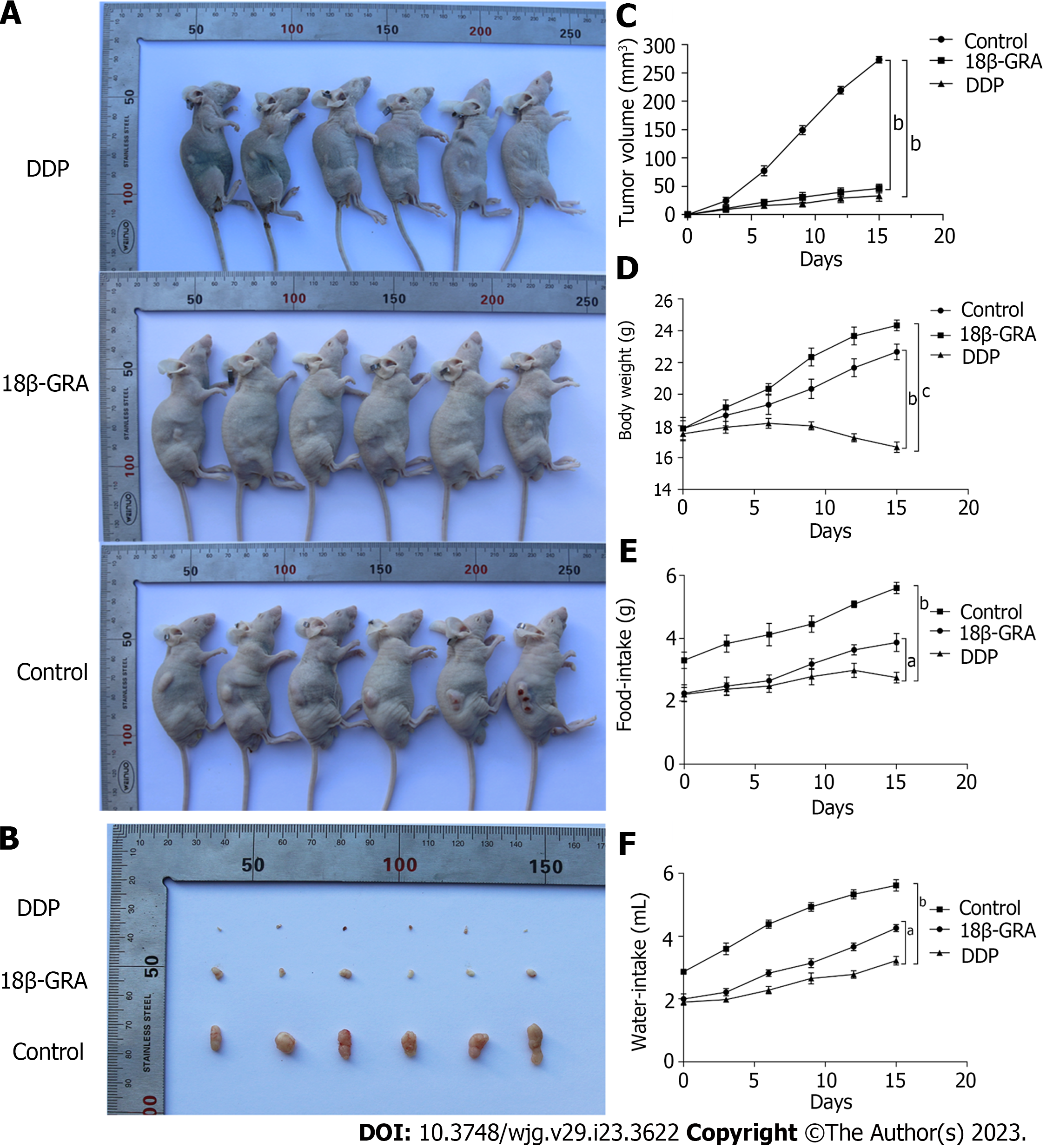
Through the subcutaneous tumor formation experiment in BALB/c nude mice, we observed that the back tumors in the control group were larger than those in the DDP and 18β-GRA groups (P < 0.01) (Figures 5A and B). The tumor volume was measured every 3 d and the tumor growth curve was plotted. In comparison to the control group, tumor size and growth rate were slower in the 18β-GRA and DDP groups (P < 0.001) (Figure 5C). Additionally, the body weight of DDP group was lower than the control and 18β-GRA groups. Diet and water intake were lower in the 18β-GRA and DDP groups when compared with the control group (P < 0.05) (Figures 5D-F).
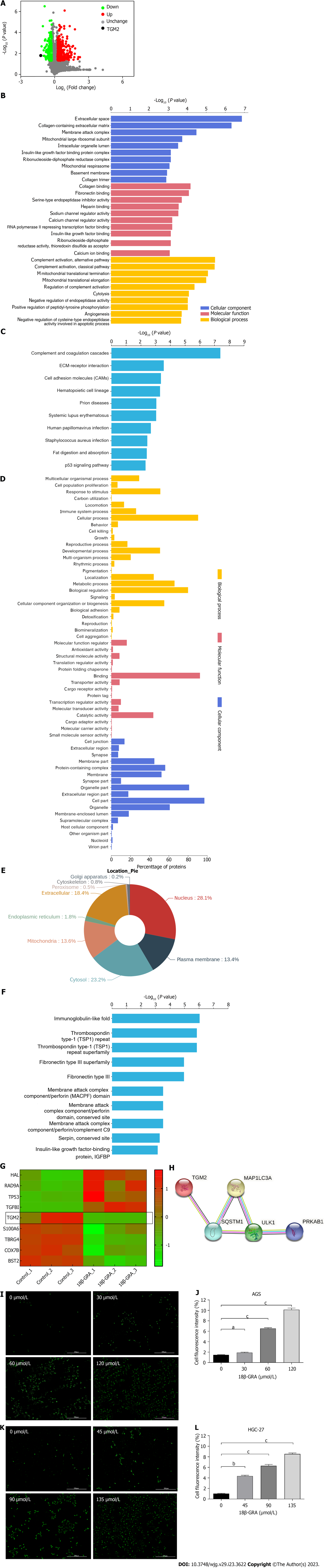
Part of the data of TMT proteomics analysis comes from the previous research of our team (Figures 6A-F)[15]. Cluster analysis was performed for differential proteins and heat map was drawn (Figure 6G), and the expression of TGM2 was down-regulated (P = 0.01593178). We used the STRING database to construct the PPI network, and the interaction diagram between the differentially expressed autophagy-related protein TGM2 and the autophagy marker proteins ULK1, p62 (SQSTM1), LC3I/II (MAPILC3) and AMPK (PRKAB1) was shown in Figure 6H, which indicated TGM2 was closely related to autophagy marker proteins. The results of MDC staining showed that AGS and HGC-27 cells displayed various degrees of autophagy after 18β-GRA intervention, and the number of autophagy cells increased as the dose of 18β-GRA was increased (P < 0.05) (Figures 6I-L).
Western blot analysis was used to determine 18β-GRA’s influence on autophagy-related proteins in the AGS and HGC-27 cells. The findings demonstrated that, in contrast to the control group, the 18β-GRA group’s protein expressions of TGM2 and p62 were lowered, whereas LC3II, ULK1 and AMPK were elevated (P < 0.01) (Figures 7A-E).
283 miRNAs were identified as differentially expressed by miRNAs transcriptome analysis, of which 163 miRNAs were down-regulated and 120 miRNAs were up-regulated, and the selection criteria for |Log2 (fold change)| > 1, P < 0.05. Among them, the expression of miR-345-5p was up-regulated (P = 5.68E-08) (Figure 8A). The miRBase website was used to predict the corresponding miRNAs of autophagy-related proteins, and then we intersected the miRNAs corresponding to autophagy-related proteins with the differentially expressed miRNAs screened by miRNA transcriptome. Among the obtained miRNAs, miR-345-5p was expressed most significantly.
We used qRT-PCR to determine the expression changes of miR-345-5p in AGS and HGC-27 cells treated with 18β-GRA. The outcomes demonstrated that miR-345-5p was highly up-regulated in AGS and HGC-27 cells after 18β-GRA treatment (P < 0.01) (Figures B and 8C). This verified the results of miRNAs transcriptome.
We utilized TargetScan database to identify complementary binding sites between miR-345-5p and TGM2 (Figure 8D). According to dual-luciferase reporter analyses, overexpression of miR-345-5p reduced luciferase expression in the TGM2-wild-type reporter but not in the mutant reporter (P < 0.05) (Figure 8E). These results suggest that miR-345-5p can regulate the expression of TGM2.
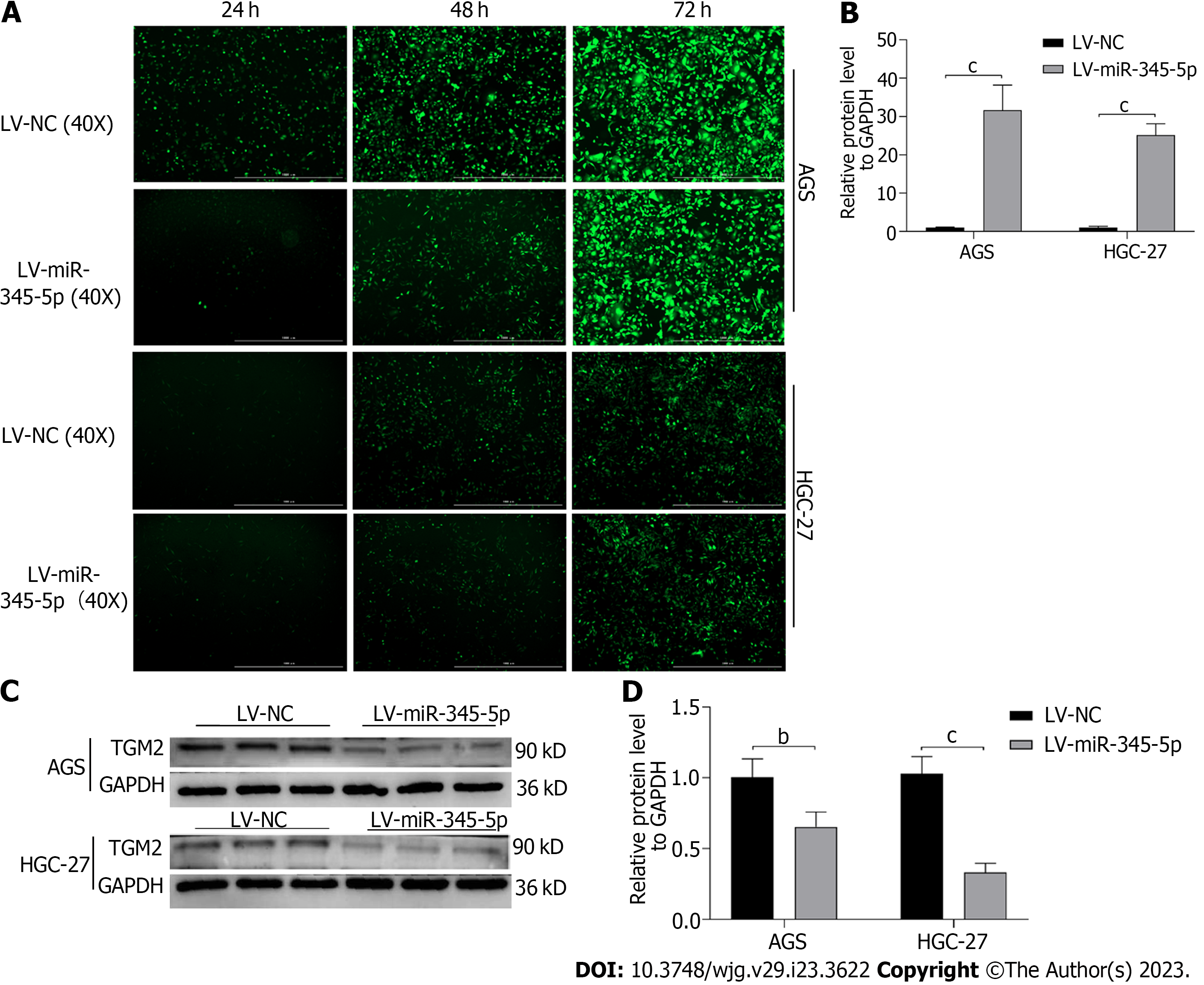
Lentivirus transfection technique caused miR-345-5p overexpression on AGS and HGC-27 cells, and the transfection rate reached about 90% after 72 h (Figure 9A). Then, qRT-PCR discovered that miR-345-5p expression in the LV-miR-345-5p group was higher compared to the LV-NC group on AGS and HGC-27 cells (P < 0.001) (Figure 9B). Meanwhile, overexpression of miR-345-5p inhibited the protein expression of TGM2 (P < 0.01) (Figures 9C and D).
Some researchers found that the expression of miR-345-5p in GC tissues was significantly lower than that in para-carcinoma tissue. which was correlated with aggressive stage and grade[27]. We evaluated the impact of miR-345-5p overexpression on cell viability in GC cells through CCK-8 assay. Our findings indicated that miR-345-5p overexpression resulted in a significant inhibition of the cell viability on AGS and HGC-27 cells (P < 0.01) (Figures 10A and B).
In order to further investigate the impart of miR-345-5p overexpression on GC cells, we conducted flow cytometry analysis to examine its effect on cell apoptosis and cell cycle. Our results showed the cell apoptosis rates in the miR-345-5p overexpression group were significantly higher than that in the LV-NC group on AGS and HGC-27 cells (P < 0.001) (Figures 10C and D). Moreover, the cell cycle experiment revealed that miR-345-5p overexpression caused cell cycle arrest in the G0/G1 phase on AGS and HGC-27 cells (P < 0.001) (Figures 10E and F). These results suggested that miR-345-5p overexpression could inhibit GC cells proliferation by promoting cell apoptosis and arresting cell cycle.
In this study, 18β-GRA treated GC cells phenotypic alterations were identified in vitro. The findings demonstrated that 18β-GRA had a minimal effect on the survival rate of normal gastric epithelial GES-1 cells, but dramatically reduced the AGS and HGC-27 cells viability. Next, we found that 18β-GRA could block the AGS and HGC-27 cells in G0/G1 phase and promote cell apoptosis. Both DNA replication and mitosis crucially depend on the G0/G1 phase. According to studies, after drug intervention, cells can either irreversibly end the cell cycle through senescence or apoptosis[28], or they can reversibly exit the cell cycle by beginning immobilization. We looked at how 18β-GRA affected the GC cells apoptosis and verified the results of cell cycle, which led us to conclude that 18β-GRA made GC cells exit the cell cycle selectively. It has been demonstrated that cells can exit the cell cycle reversibly by initiating immobilization, or irreversibly by senescence or apoptosis. Our findings confirmed the conclusion that 18β-GRA can induce GC cells to exit the cell cycle selectively.
18β-GRA decreased the migration ability of AGS and HGC-27 cells, according to a wound healing assay. At the same time, we also performed cell invasion experiments, but the results were not statistically significant. Studies have shown that TGM2 could promote GC cells’ proliferation, migration and invasion by activating extracellular signal-regulated kinase 1/2[29]. In addition, TGM2 was thought to be involved in EMT processes in breast cancer[30], colorectal cancer[31], and hepatocellular carcinoma[32], which are highly correlated with invasion phenotypes. These findings imply that TGM2 is involved in cancer cell invasion. In contrast, 18β-GRA had no impact on AGS and HGC-27 cells invasion during our studies.
In vivo tumorigenesis experiment was performed on immunodeficient nude mice. The results showed that 18β-GRA and DDP significantly inhibited the subcutaneous tumor volume in nude mice. At the same time, we monitored the body weight, diet and water intake of nude mice during the experiment. It was found that the body weight of nude mice in the DDP group was lighter than that in 18β-GRA and control groups, showing obvious adverse reactions. On the contrary, the body weight of nude mice in 18β-GRA group was slightly higher than that in the control group and the nude mice were in good health during the experiment. In terms of diet and water intake, nude mice administered with 18β-GRA and DDP had reduced diet and water intake. Overall, the experimental results demonstrated 18β-GRA had fewer side effects.
We further investigated the mechanism of GC treatment with 18β-GRA. MDC staining results showed that 18β-GRA can promote autophagy of AGS and HGC-27 cells. TMT proteomic analysis and miRNAs transcriptome analysis revealed that TGM2 was the target of miR-345-5p, which was verified by a dual-luciferase reporter assay. The STRING database was used to build the PPI network, and it was found that TGM2 was closely related to autophagy marker proteins ULK1, p62, LC3I/II and AMPK. TGM2 induces autophagy, differentiation and inhibition of angiogenesis, and its role in cancer is very complex. In fact, it has been proven that TGM2 participates in all aspects of cancer progression by activating mTOR[33]. At the beginning phase of autophagy, AMPK promotes autophagy occurrence by directly activating ULK1 or by negatively regulating mTOR to block its inhibition of ULK1[34,35]. After the formation of pre-autophage, it enters the second stage, which is the stage of autophagy extending into nucleus. ULK1 activates p62 and promotes the binding of p62 mediated substrate proteins to autophagic bodies[36]. At the same time, the substrate protein passes through the LC3 domain of p62 into the degradation phase of the autophagic vesicle, where it interacts with LC3, which is the third stage of autophagy. Thus, when the p62 content decreases, it indicates that autophagic lysosomal degradation is inhibited[37,38]. Therefore, when the content of p62 decreases, it indicates that autophagic lysosome degradation is inhibited. LC3I/II, one of the homologues of ATG8, is located in the cytoplasm. LC3I is formed after exposure to glycine at the carboxyl terminal, which is catalyzed to combine with phosphatidyl ethanolamine to form LC3II. The increased amount of LC3II can reflect the increased number of autophagosomes caused by enhanced autophagy activity[39] (Figure 11). Western blot results proved that TGM2 and p62 were reduced, while AMPK, ULK1, and LC3II were increased in AGS and HGC-27 cells treated with 18β-GRA. Additionally, we employed a lentiviral vector to miR-345-5p overexpress in GC cells and observed the expression of TGM2. Our findings indicate that miR-345-5p overexpression significantly reduced the protein expression of TGM2. At the same time, we also found that miR-345-5p overexpression could inhibit GC cells proliferation by promoting cell apoptosis and arresting cell cycle. Based on the aforementioned findings, we draw the conclusion that 18β-GRA may play a role in the occurrence and progression of GC via the miR-345-5p/TGM2 signaling pathway.
To sum up, this study proved that 18β-GRA can inhibit GC cells viability, induce cells apoptosis, block cell cycle, inhibit cell wound healing ability, and induce cell autophagy by regulating the miR-345-5p/TGM2 signaling pathway, thereby inhibiting the GC cells proliferation, which could provide theoretical basis for the research of 18β-GRA in GC treatment. However, our study has some limitations and more experiments are needed to support our future research. Hence, we will carry on exploring the relationship between 18β-GRA and GC in the future. First, GC cells were infected with RFP-GFP-LC3 double-labeled adenoviruses to research 18β-GRA’s effect on the autophagic. Secondly, 18β-GRA’s effect when combined with chemotherapeutic drugs on chemotherapeutic drug sensitivity will be investigated. Thirdly, the molecular mechanism of 18β-GRA treatment for GC was further studied by gene overexpression, CO-IP and other methods.
Gastric cancer (GC) is a common gastrointestinal malignancy worldwide. Based on the cancer-related mortality, the current prevention and treatment strategies for GC still show poor clinical results. Therefore, it is important to find effective drug treatment targets.
At present, the treatment of GC is mainly surgery, chemotherapy and radiotherapy, and the first-line treatment drugs are harmful to side effects.
The purpose of this study was to explore the molecular mechanism of 18β-glycyrrhetinic acid (18β-GRA) regulating the miR-345-5p/TGM2 signaling pathway to inhibit the proliferation of GC cells.
The effects of 18β-GRA on GC cell phenotype and tumor growth in vivo were studied. TMT proteomic analysis and microRNAs (miRNAs) transcriptome analysis were used to screen for targets, and targeted connections were validated using a dual-luciferase report assay. Finally, the prediction was confirmed by experiment in vitro.
Our experiment confirmed that 18β-GRA inhibited GC cells growth both in vitro and in vivo, and MDC staining showed that 18β-GRA promoted GC cell autophagy. By TMT proteomic analysis and miRNAs transcriptomic analysis, we found that 18β-GRA down-regulates TGM2 expression and up-regulates miR-345-5p expression in GC cells. Subsequently, TGM2 was verified as the target of miR-345-5p by a dual-luciferase report assay. In 18β-GRA treated GC cells, the expressions of autophagy-related proteins TGM2 and p62 were significantly decreased, while the expressions of LC3II, ULK1 and AMPK were significantly increased. In addition, overexpression of miR-345-5p not only inhibited TGM2 expression, but also inhibited GC cell proliferation by promoting apoptosis and blocking cell cycle.
These observations indicate that 18β-GRA can promote autophagy and inhibit GC cells proliferation via regulating the miR-345-5p/TGM2 signaling pathway.
MiR-345-5p can be used for targeted therapy of GC, and can also be used as a new biomarker for GC.
The authors would like to acknowledge Li-Qun Wang for statistical analysis assistance. Thanks to Joanna Tibenda for revising the article.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kotelevets SM, Russia; Liu YQ, United States S-Editor: Wang JJ L-Editor: A P-Editor: Cai YX
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64647] [Article Influence: 16161.8] [Reference Citation Analysis (176)] |
| 2. | Abbas M, Habib M, Naveed M, Karthik K, Dhama K, Shi M, Dingding C. The relevance of gastric cancer biomarkers in prognosis and pre- and post- chemotherapy in clinical practice. Biomed Pharmacother. 2017;95:1082-1090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 3. | Duan R, Li X, Zeng D, Chen X, Shen B, Zhu D, Zhu L, Yu Y, Wang D. Tumor Microenvironment Status Predicts the Efficacy of Postoperative Chemotherapy or Radiochemotherapy in Resected Gastric Cancer. Front Immunol. 2020;11:609337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 4. | Yang T, Zhou J, Fang L, Wang M, Dilinuer M, Ainiwaer A. Protection function of 18β-glycyrrhetinic acid on rats with high-altitude pulmonary hypertension based on (1)H NMR metabonomics technology. Anal Biochem. 2021;631:114342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 5. | Yang Y, Zhu Q, Zhong Y, Cui X, Jiang Z, Wu P, Zheng X, Zhang K, Zhao S. Synthesis, anti-microbial and anti-inflammatory activities of 18β-glycyrrhetinic acid derivatives. Bioorg Chem. 2020;101:103985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 6. | Wang LJ, Geng CA, Ma YB, Huang XY, Luo J, Chen H, Zhang XM, Chen JJ. Synthesis, biological evaluation and structure-activity relationships of glycyrrhetinic acid derivatives as novel anti-hepatitis B virus agents. Bioorg Med Chem Lett. 2012;22:3473-3479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 7. | Stecanella LA, Bitencourt APR, Vaz GR, Quarta E, Silva Júnior JOC, Rossi A. Glycyrrhizic Acid and Its Hydrolyzed Metabolite 18β-Glycyrrhetinic Acid as Specific Ligands for Targeting Nanosystems in the Treatment of Liver Cancer. Pharmaceutics. 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 8. | Wang Z, Ma J, He Y, Miu KK, Yao S, Tang C, Ye Y, Lin G. Nrf2-mediated liver protection by 18β-glycyrrhetinic acid against pyrrolizidine alkaloid-induced toxicity through PI3K/Akt/GSK3β pathway. Phytomedicine. 2022;102:154162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 9. | Liu J, Xu Y, Yan M, Yu Y, Guo Y. 18β-Glycyrrhetinic acid suppresses allergic airway inflammation through NF-κB and Nrf2/HO-1 signaling pathways in asthma mice. Sci Rep. 2022;12:3121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 28] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 10. | Yadav DK, Kalani K, Singh AK, Khan F, Srivastava SK, Pant AB. Design, synthesis and in vitro evaluation of 18β-glycyrrhetinic acid derivatives for anticancer activity against human breast cancer cell line MCF-7. Curr Med Chem. 2014;21:1160-1170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 11. | Li X, Liu Y, Wang N, Wang S, Wang H, Li A, Ren S. Synthesis and discovery of 18β-glycyrrhetinic acid derivatives inhibiting cancer stem cell properties in ovarian cancer cells. RSC Adv. 2019;9:27294-27304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 12. | Sun Y, Jiang M, Park PH, Song K. Transcriptional suppression of androgen receptor by 18β-glycyrrhetinic acid in LNCaP human prostate cancer cells. Arch Pharm Res. 2020;43:433-448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 13. | Cao D, Wu Y, Jia Z, Zhao D, Zhang Y, Zhou T, Wu M, Zhang H, Tsukamoto T, Oshima M, Jiang J, Cao X. 18β-glycyrrhetinic acid inhibited mitochondrial energy metabolism and gastric carcinogenesis through methylation-regulated TLR2 signaling pathway. Carcinogenesis. 2019;40:234-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 14. | Cai H, Chen X, Zhang J, Wang J. 18β-glycyrrhetinic acid inhibits migration and invasion of human gastric cancer cells via the ROS/PKC-α/ERK pathway. J Nat Med. 2018;72:252-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 15. | Yuan L, Yang Y, Li X, Zhou X, Du YH, Liu WJ, Zhang L, Yu L, Ma TT, Li JX, Chen Y, Nan Y. 18β-glycyrrhetinic acid regulates mitochondrial ribosomal protein L35-associated apoptosis signaling pathways to inhibit proliferation of gastric carcinoma cells. World J Gastroenterol. 2022;28:2437-2456. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 5] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (1)] |
| 16. | Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1765] [Cited by in RCA: 1701] [Article Influence: 189.0] [Reference Citation Analysis (0)] |
| 17. | Coradduzza D, Cruciani S, Arru C, Garroni G, Pashchenko A, Jedea M, Zappavigna S, Caraglia M, Amler E, Carru C, Maioli M. Role of miRNA-145, 148, and 185 and Stem Cells in Prostate Cancer. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 18. | Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, Robinson DR, Nesvizhskii AI, Chinnaiyan AM. The Landscape of Circular RNA in Cancer. Cell. 2019;176:869-881.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1086] [Cited by in RCA: 1186] [Article Influence: 197.7] [Reference Citation Analysis (0)] |
| 19. | Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23:3-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 346] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 20. | Komatsu S, Imamura T, Kiuchi J, Takashima Y, Kamiya H, Ohashi T, Konishi H, Shiozaki A, Kubota T, Okamoto K, Otsuji E. Depletion of tumor suppressor miRNA-148a in plasma relates to tumor progression and poor outcomes in gastric cancer. Am J Cancer Res. 2021;11:6133-6146. [PubMed] |
| 21. | Li X, Zhu M, Zhao G, Zhou A, Min L, Liu S, Zhang N, Zhu S, Guo Q, Zhang S, Li P. MiR-1298-5p level downregulation induced by Helicobacter pylori infection inhibits autophagy and promotes gastric cancer development by targeting MAP2K6. Cell Signal. 2022;93:110286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 22. | Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 591] [Cited by in RCA: 655] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 23. | Wen X, Klionsky DJ. At a glance: A history of autophagy and cancer. Semin Cancer Biol. 2020;66:3-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 24. | Wei Y, Hong D, Zang A, Wang Z, Yang H, Zhang P, Wang Y. miR-183 Enhances Autophagy of GC Cells by Targeted Inhibition of mTOR. Ann Clin Lab Sci. 2021;51:837-843. [PubMed] |
| 25. | Lin D, Zhong W, Li J, Zhang B, Song G, Hu T. Involvement of BID translocation in glycyrrhetinic acid and 11-deoxy glycyrrhetinic acid-induced attenuation of gastric cancer growth. Nutr Cancer. 2014;66:463-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 26. | Zhu Y, Zhou B, Hu X, Ying S, Zhou Q, Xu W, Feng L, Hou T, Wang X, Zhu L, Jin H. LncRNA LINC00942 promotes chemoresistance in gastric cancer by suppressing MSI2 degradation to enhance c-Myc mRNA stability. Clin Transl Med. 2022;12:e703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 86] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 27. | Zhang J, Wang C, Yan S, Yang Y, Zhang X, Guo W. miR-345 inhibits migration and stem-like cell phenotype in gastric cancer via inactivation of Rac1 by targeting EPS8. Acta Biochim Biophys Sin (Shanghai). 2020;52:259-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 28. | Xiang SS, Wang XA, Li HF, Shu YJ, Bao RF, Zhang F, Cao Y, Ye YY, Weng H, Wu WG, Mu JS, Wu XS, Li ML, Hu YP, Jiang L, Tan ZJ, Lu W, Liu F, Liu YB. Schisandrin B induces apoptosis and cell cycle arrest of gallbladder cancer cells. Molecules. 2014;19:13235-13250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 29. | Wang X, Yu Z, Zhou Q, Wu X, Chen X, Li J, Zhu Z, Liu B, Su L. Tissue transglutaminase-2 promotes gastric cancer progression via the ERK1/2 pathway. Oncotarget. 2016;7:7066-7079. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 30. | He W, Sun Z, Liu Z. Silencing of TGM2 reverses epithelial to mesenchymal transition and modulates the chemosensitivity of breast cancer to docetaxel. Exp Ther Med. 2015;10:1413-1418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 31. | Shan H, Zhou X, Chen C. MicroRNA214 suppresses the viability, migration and invasion of human colorectal carcinoma cells via targeting transglutaminase 2. Mol Med Rep. 2019;20:1459-1467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 32. | Ma H, Xie L, Zhang L, Yin X, Jiang H, Xie X, Chen R, Lu H, Ren Z. Activated hepatic stellate cells promote epithelial-to-mesenchymal transition in hepatocellular carcinoma through transglutaminase 2-induced pseudohypoxia. Commun Biol. 2018;1:168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 33. | Wang F, Wang L, Qu C, Chen L, Geng Y, Cheng C, Yu S, Wang D, Yang L, Meng Z, Chen Z. Kaempferol induces ROS-dependent apoptosis in pancreatic cancer cells via TGM2-mediated Akt/mTOR signaling. BMC Cancer. 2021;21:396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 97] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 34. | Mack HI, Zheng B, Asara JM, Thomas SM. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy. 2012;8:1197-1214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 184] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 35. | Zibrova D, Vandermoere F, Göransson O, Peggie M, Mariño KV, Knierim A, Spengler K, Weigert C, Viollet B, Morrice NA, Sakamoto K, Heller R. GFAT1 phosphorylation by AMPK promotes VEGF-induced angiogenesis. Biochem J. 2017;474:983-1001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 79] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 36. | Lim J, Lachenmayer ML, Wu S, Liu W, Kundu M, Wang R, Komatsu M, Oh YJ, Zhao Y, Yue Z. Proteotoxic stress induces phosphorylation of p62/SQSTM1 by ULK1 to regulate selective autophagic clearance of protein aggregates. PLoS Genet. 2015;11:e1004987. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 200] [Cited by in RCA: 245] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 37. | Bjørkøy G, Lamark T, Pankiv S, Øvervatn A, Brech A, Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 754] [Cited by in RCA: 937] [Article Influence: 58.6] [Reference Citation Analysis (0)] |
| 38. | Zhang H, Zhang Y, Zhu X, Chen C, Zhang C, Xia Y, Zhao Y, Andrisani O, Kong L. DEAD Box Protein 5 Inhibits Liver Tumorigenesis by Stimulating Autophagy via Interaction with p62/SQSTM1. Hepatology. 2019;69:1046-1063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 90] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 39. | Yoshii SR, Mizushima N. Monitoring and Measuring Autophagy. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 552] [Cited by in RCA: 852] [Article Influence: 106.5] [Reference Citation Analysis (0)] |