Published online Jun 21, 2023. doi: 10.3748/wjg.v29.i23.3606
Peer-review started: February 1, 2023
First decision: February 12, 2023
Revised: February 25, 2023
Accepted: May 4, 2023
Article in press: May 4, 2023
Published online: June 21, 2023
Processing time: 135 Days and 3.3 Hours
Activated hepatic stellate cells (aHSCs) are the major source of cancer-associated fibroblasts in the liver. Although the crosstalk between aHSCs and colorectal cancer (CRC) cells supports liver metastasis (LM), the mechanisms are largely unknown.
To explore the role of BMI-1, a polycomb group protein family member, which is highly expressed in LM, and the interaction between aHSCs and CRC cells in promoting CRC liver metastasis (CRLM).
Immunohistochemistry was carried out to examine BMI-1 expression in LM and matched liver specimens of CRC. The expression levels of BMI-1 in mouse liver during CRLM (0, 7, 14, 21, and 28 d) were detected by Western blotting (WB) and the quantitative polymerase chain reaction (qPCR) assay. We overexpressed BMI-1 in HSCs (LX2) by lentivirus infection and tested the molecular markers of aHSCs by WB, qPCR, and the immunofluorescence assay. CRC cells (HCT116 and DLD1) were cultured in HSC-conditioned medium (LX2 NC CM or LX2 BMI-1 CM). CM-induced CRC cell proliferation, migration, epithelial-mesenchymal transition (EMT) phenotype, and transforming growth factor beta (TGF-β)/SMAD pathway changes were investigated in vitro. A mouse subcutaneous xenotransplantation tumor model was established by co-implantation of HSCs (LX2 NC or LX2 BMI-1) and CRC cells to investigate the effects of HSCs on tumor growth and the EMT phenotype in vivo.
Positive of BMI-1 expression in the liver of CRLM patients was 77.8%. The expression level of BMI-1 continued to increase during CRLM in mouse liver cells. LX2 overexpressed BMI-1 was activated, accompanied by increased expression level of alpha smooth muscle actin, fibronectin, TGF-β1, matrix metalloproteinases, and interleukin 6. CRC cells cultured in BMI-1 CM exhibited enhanced proliferation and migration ability, EMT phenotype and activation of the TGF-β/SMAD pathway. In addition, the TGF-βR inhibitor SB-505124 diminished the effect of BMI-1 CM on SMAD2/3 phosphorylation in CRC cells. Furthermore, BMI-1 overexpressed LX2 HSCs promoted tumor growth and the EMT phenotype in vivo.
High expression of BMI-1 in liver cells is associated with CRLM progression. BMI-1 activates HSCs to secrete factors to form a prometastatic environment in the liver, and aHSCs promote proliferation, migration, and the EMT in CRC cells partially through the TGF-β/SMAD pathway.
Core Tip: This study revealed that BMI-1 was upregulated in liver cells during colorectal cancer (CRC) liver metastasis. BMI-1-activated LX2 hepatic stellate cells (HSCs) promoted CRC cell proliferation, migration, and the epithelial-mesenchymal transition both in vitro and in vivo. Mechanistically, transforming growth factor beta 1 was increased in BMI-1 overexpressed LX2 HSCs, and triggered the phosphorylation of downstream SMAD2/3 in CRC cells and the interaction of activated HSCs and CRC cells, thereby further promoting CRC progression.
- Citation: Jiang ZY, Ma XM, Luan XH, Liuyang ZY, Hong YY, Dai Y, Dong QH, Wang GY. BMI-1 activates hepatic stellate cells to promote the epithelial-mesenchymal transition of colorectal cancer cells. World J Gastroenterol 2023; 29(23): 3606-3621
- URL: https://www.wjgnet.com/1007-9327/full/v29/i23/3606.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i23.3606
Colorectal cancer (CRC) is the third most common cancer and ranks second in terms of cancer mortality worldwide[1,2]. Liver metastasis (LM) is the primary cause of death in CRC patients, and the liver plays a major role in survival as it is often the only site of metastasis. In CRC patients, approximately 14%-35% have LM at diagnosis, and about 70% have LM in the late stage of the disease[3]. In colorectal cancer (CRC) liver metastasis (CRLM) patients, liver resection is the primary curative treatment option, with a 5-year survival rate of 20%-50%[4]. Although treatments have advanced in recent years, the rate of intrahepatic recurrence is still high.
During CRLM, CRC cells can orchestrate a premetastatic niche by changing the tumor microenvironment[5,6]. For example, tumor cells secrete cytokines and extracellular vesicles containing micro
BMI-1, one of the polycomb group protein family members, plays a critical role in negatively regulating the Ink4a/Arf locus which encodes p16INK4a and p19ARF[22]. BMI-1 regulates self-renewal of stem cells and participates in the carcinogenesis of human cancers including CRC[23-25]. We previously reported that BMI-1 was overexpressed in CRLM and contributes to LM by regulating the epithelial-mesenchymal transition (EMT) of CRC cells in vitro and in vivo[26]. Interestingly, we observed BMI-1-positive staining in liver cells both in human and mouse specimens of CRLM. Studies have shown that quiescent HSCs can be activated by cancer cells and transformed into CAFs, and in turn activated HSCs (aHSCs) promote cancer cell invasion and the EMT[8,19]. Thus, we speculated that BMI-1 plays a role in the crosstalk between HSCs and CRC cells during LM.
In this study, both CRLM patients and mice showed increased BMI-1 expression in liver cells. BMI-1 overexpressed HSCs were activated and transformed into CAFs. CRC cells cultured in conditioned medium (CM) from BMI-1 overexpressed HSCs showed enhanced proliferation, migration, and EMT ability. The experimental in vivo subcutaneous xenotransplantation tumor model also showed increased tumor proliferation and EMT of CRC cells cocultured with BMI-1 overexpressed HSCs. These findings may provide a potential new target for the treatment of CRLM.
LM and adjacent normal liver tissues from 18 clinically diagnosed patients with CRLM were collected from the Pathology Department (from 2013 to 2020), Sir Run Run Shaw Hospital, Hangzhou, China. The expression of BMI-1 was investigated by immunohistochemistry (IHC) staining. Anti-BMI-1 (1:100; Cell Signaling Technology, Danvers, MA, United States) was used as the primary antibody before secondary antibody incubation.
The high metastatic human CRC cell line (HCT116), low metastatic human CRC cell line (DLD1), mouse CRC cell line (CT26), and human HSC line (LX2) were purchased from the American Type Culture Collection (Manassas, VA, United States). Cells were cultured in Dulbecco’s Modified Eagle’s Medium (Gibco, Waltham, MA, United States) with 10% fetal bovine serum and 1% penicillin/streptomycin in a 5% carbon dioxide humidified incubator at 37 °C. The BMI-1 overexpressed lentiviral construct (pGC-FU-GFP-BMI-1) and negative control (pGC-FU-GFP) were obtained from Genechem (Shanghai, China). The control (LX2 NC) and stable BMI-1 overexpressed LX2 (LX2 BMI-1) cells were established by lentivirus transfection following the manufacturer’s instructions. In addition, to prepare the CM, we first cultured transfected LX2 cells in a 10 cm plate at a density of 1 × 106 cells. We then used 6 mL serum-free DMEM to culture the cells for 24 h. The supernatants were collected, centrifuged at 1000 × g for 10 min, and filtered through a 0.22 mm filter unit (Millex, Duluth, GA, United States). CM from NC LX2 (NC CM) and BMI-1 LX2 (BMI-1 CM) was prepared by mixing the different supernatant with complete medium (1:1). Then the CRC cells were cultured in CM for 24 h. All CM was used within 2 d. CRC cells were pretreated with SB-505124 (0.05 μM) for 1 h to inhibit SMAD2 phosphorylation, and then the cells were incubated with BMI-1 CM for 24 h.
Whole proteins from the tissue and cell samples were extracted using RIPA buffer with a 1% protease/phosphatase inhibitor. After centrifugation of the extraction solutions, the BCA Protein Concentration Assay Kit (Solarbio, Beijing, China) was used to quantify the proteins. Following sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the proteins were electrotransferred to polyvinylidene fluoride membranes. Antibodies against BMI-1, glyceraldehyde-3-phosphate dehydrogenase, β-actin, vimentin, SMAD2/3, phosphorylated SMAD3 (1:1000; Cell Signaling Technology), alpha smooth muscle actin (α-SMA), transforming growth factor beta 1 (TGF-β1), phosphorylated SMAD2 (1:1000; Abcam, Cambridge, MA, United States), fibronectin (1:1000; BD Biosciences, San Jose, CA, United States), E-cadherin, zinc finger E-box-binding homeobox 1 (ZEB-1), Twist-1 (1:1000; Novus Biologicals, Centennial, CO, United States), and Snail (1:1000; Proteintech Group, Rosemont, IL, United States) were used as primary antibodies. After washing with Tris-buffered saline with 0.1% Tween 20 three times every 10 min, goat anti-rabbit/mouse immunoglobulin G (Abcam) was used as the secondary antibody. The Bio-Rad CD Touch Detection System (Hercules, CA, United States) with enhanced chemiluminescence detection reagents was used to detect protein bands.
Whole RNAs from the tissue and cell samples were extracted using Trizol reagent (Cwbio, Dalian, China). HiScript II Q RT SuperMix (Vazyme Biotech, Nanjing, China) was subsequently used for reverse transcription. ChamQ Universal SYBR quantitative polymerase chain reaction (qPCR) Master Mix (Vazyme Biotech) was used for qPCR. The Bio-Rad CFX-96 Real-Time PCR System was utilized to analyze the results. The sequences of all primers are listed in Supplementary Table 1.
Immunofluorescence staining was performed in 96-well plates. Cells were fixed in paraformaldehyde and permeabilized with Triton X-100. Before incubation with secondary antibody, antibodies against vimentin (1:100; Cell Signaling Technology) and Snail (1:100; Proteintech Group) were used as primary antibodies. The Zeiss AXIO Observer A1 inverted fluorescence microscope system (Jena, Germany) was used to obtain images.
Cells were cultured in CM separately for 24, 48, 72, and 96 h. After treatment, Cell Counting Kit-8 (CCK-8) reagent (Yeasen Biotechnology, Shanghai, China) was added and incubated for 2 h. A spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) was used to detect the optical density value (450 nm).
CRC cells were cultured in CM for 2 wk. Cells were fixed in paraformaldehyde and stained with crystal violet. Colonies were viewed with the Olympus CKK53 microscope (Tokyo, Japan), and photos were taken. ImageJ software was utilized to measure the number of colonies.
CRC cells were cultured until they reached 90% confluence. Following the creation of a linear wound, cells were cultured in CM for 48 h. Wounds were viewed with the Olympus CKK53 microscope, and photos were taken at 0, 24, and 48 h. ImageJ software was used to measure the migration rate.
Transwell chambers were purchased from Corning Inc. (Corning, NY, United States). CRC cells were plated in the upper chambers with serum-free DMEM, while CM was placed in the lower chambers. After 48 h, cells in the upper chambers were removed, and cells on the lower membrane surface were fixed and stained. The migrated cells were viewed with the Olympus CKK53 microscope, and photos were taken. ImageJ software was used to measure the number of cells.
Tissue samples were fixed, dehydrated, paraffin-embedded, and sectioned. After deparaffinization, the sections were blocked in goat serum and incubated with primary antibodies for 24 h. The sections were incubated with secondary antibody for 30 min, followed by counterstaining with Mayer’s hematoxylin. The Olympus CKK53 microscope was used to view and photograph the stained sections.
All animal experimental procedures were approved by the Committee on the Ethics of Animal Experiments of Zhejiang University (No. ZJU20220447; Hangzhou, China).
Five-week-old male BALB/c mice (19-20 g) were randomly divided into five groups (n = 3). Isoflurane (inhalation) was utilized to anesthetize the mice. The spleen was then exteriorized through a left flank incision. CT26 cells (2 × 106) were injected intrasplenically to establish the tumor, and then the injection site was pressed for 5 min. Surgical thread was used to close the peritoneum and skin. The mice were sacrificed at 0, 7, 14, 21, and 28 d after inoculation of CT26 cells. Resected liver tissues were collected for qPCR, Western blotting (WB), and the IHC assay.
Five-week-old male BALB/c nude mice (17-18 g) were randomly divided into four groups (n = 5): (1) HCT116 cells (5 × 106) were mixed with LX2 NC cells (1 × 106); (2) HCT116 cells (5 × 106) were mixed with LX2 BMI-1 cells (1 × 106); (3) DLD1 cells (5 × 106) were mixed with LX2 NC cells (1 × 106); and (4) DLD1 cells (5 × 106) were mixed with LX2 BMI-1 cells (1 × 106). The mixed cells were subcutaneously injected into mouse flanks to establish the tumors. When measurable, tumor sizes were estimated every 2 d. The mice were killed 28 d after cell inoculation. Resected tumor tissues were collected for qPCR, WB, and the IHC assay. Total tumor volume (mm3) = L × W2/2 (L = length and W = width).
In all experiments, the data are shown as the mean ± standard deviation based on three independent experiments. The difference between the groups was analyzed by the χ2 test, Fisher’s exact probability, Student’s t-test, or one-way analysis of variance using GraphPad Prism 8 software or SPSS 25.0. P < 0.05 was considered statistically significant.
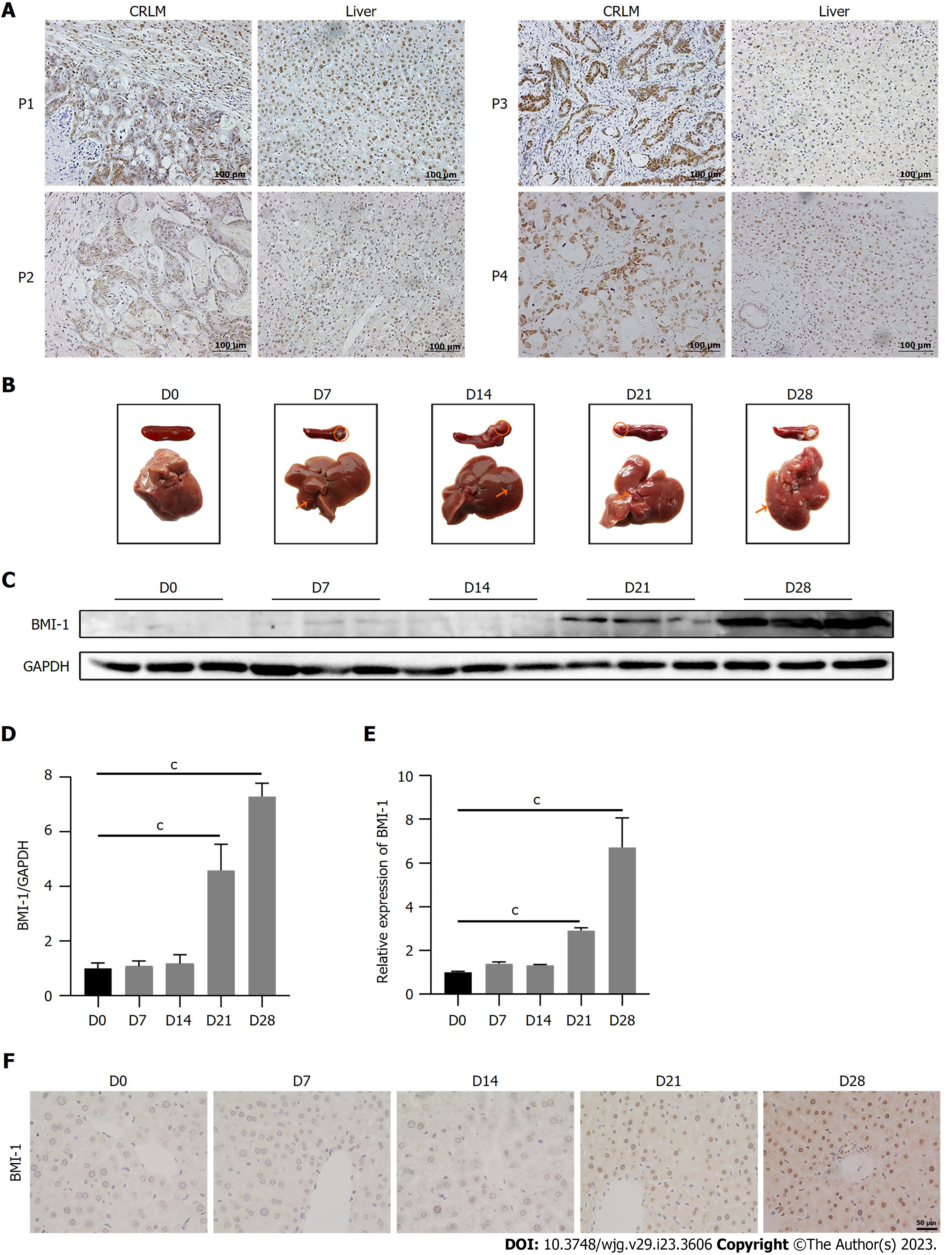
In our previous research, it was found that BMI-1 was overexpressed in LM of human CRC[26]. Interestingly, although normal liver cells were BMI-1-negative, 77.8% of liver cells from CRLM patients (14 of 18 samples; Figure 1A) were BMI-1-positive. The positive expression of BMI-1 was not strongly related to the clinical pathological characteristics (including tumor size and tumor number in the liver, differentiation degree, T stage, N stage) except the tumor site (Table 1). Positive BMI-1 expression in the liver cells of patients with rectal cancer LM was significantly higher (100%) than in patients with CRLM (60%). To investigate BMI-1 expression level in liver cells during CRLM, we measured BMI-1 expression at 0, 7, 14, 21, and 28 d following intrasplenic injection of CT26 cells in mice (Figure 1B). As shown in Figure 1C, small LM had formed by day 7. Compared with healthy control mice, the protein expression level of BMI-1 did not significantly change at 7 and 14 d, but markedly increased 4.58-fold at 21 d and continued increasing up to 7.29-fold at 28 d (Figure 1D). The mRNA expression level of BMI-1 accordingly increased 2.92-fold at 21 d and 6.71-fold at 28 d (Figure 1E). The IHC results also showed that few nuclei in liver cells were BMI-1-positive in CRLM mice from 14 d, but the BMI-1-positive rate and intensity increased markedly at 21 and 28 d (Figure 1F). These results suggest that BMI-1 was upregulated in liver cells after CRC cells metastasized to the liver, and may play a role in CRLM.
| Characteristics | Cases, n = 18 | BMI-1 expression | P value | |
| Negative, n = 4 | Positive, n = 14 | |||
| Age in yr | 0.051 | |||
| < 60 | 12 | 4 | 8 | |
| ≥ 60 | 6 | 0 | 6 | |
| Sex | 0.509 | |||
| Male | 11 | 3 | 8 | |
| Female | 7 | 1 | 6 | |
| Primary tumor site | 0.018a | |||
| Colon | 10 | 4 | 6 | |
| Rectum | 8 | 0 | 8 | |
| Tumor size in the liver in mm | 0.278 | |||
| < 30 | 8 | 3 | 5 | |
| 30-50 | 3 | 0 | 3 | |
| ≥ 50 | 5 | 1 | 5 | |
| Tumor numbers in the liver | 0.432 | |||
| 1-2 | 12 | 2 | 10 | |
| ≥ 3 | 6 | 2 | 4 | |
| Differentiation degree | 0.881 | |||
| Low | 4 | 1 | 3 | |
| Middle-high | 14 | 3 | 11 | |
| T stage | 1 | |||
| III | 18 | 4 | 14 | |
| N stage | 0.248 | |||
| N0 | 9 | 3 | 6 | |
| N1 + N2 | 7 | 1 | 8 | |
To investigate the effects of BMI-1 on liver cells, we overexpressed BMI-1 in LX2 HSCs and confirmed the overexpression by WB (Figure 2A). High expression of α-SMA, a myofibroblast marker, is a key marker of aHSCs. aHSCs highly secrete the key cytokine TGF-β1 and promote hepatic fibrosis with fibronectin expression[27]. Compared with control quiescent HSCs, BMI-1 overexpressed LX2 HSCs showed a significant increase in α-SMA expression. Fibronectin and TGF-β1 expression were also markedly increased in BMI-1 overexpressed LX2 HSCs. Immunofluorescence staining also confirmed that the expression of α-SMA and fibronectin was significantly increased in LX2 cells overexpressing BMI-1 (Figure 2B). It was also observed that the transcript levels of matrix metalloproteinases (MMP1, MMP2, MMP3, MMP7, MMP9) and inflammatory cytokines (TGF-β1 and interleukin 6 [IL-6]) were markedly increased (Figure 2C). Thus, LX2 HSCs were activated by the overexpression of BMI-1.
To examine the effect of BMI-1 on the interaction between HSCs and CRC cells, CRC cells (HCT116 and DLD1) were cultured in control LX2 CM (NC CM) or BMI-1 overexpressed LX2 CM (BMI-1 CM). We first measured cell proliferation using the CCK-8 assay. Compared with cells cultured in NC CM, the proliferation of CRC cells cultured in BMI-1 CM was markedly enhanced on day 4 (Figure 3A), and the colony formation ability of CRC cells cultured in BMI-1 CM also significantly increased (Figure 3B). In addition, CRC cells cultured in BMI-1 CM showed higher wound healing rates (Figure 3C). Transwell migration assays showed the same results (Figure 3D). Then we investigated EMT-related molecular changes. Consistent with the above results, the mRNA and protein expression levels of ZEB-1, Twist-1, vimentin, and Snail in CRC cells cultured in BMI-1 CM were significantly increased while E-cadherin was decreased (Figure 3E and F). Immunofluorescence staining further demonstrated that Snail and vimentin expression was obviously increased in CRC cells cultured in BMI-1 CM (Figure 3G). Therefore, BMI-1 may be an important regulator in aHSCs, which promoted the malignant phenotype of CRC cells.
As previously mentioned, TGF-β1 was upregulated in BMI-1 overexpressed LX2 HSCs (Figure 2A). The TGF-β/SMAD signal pathway plays an important role in metastasis by modulating the EMT process[28,29]; therefore, we hypothesized that BMI-1 CM induces EMT in CRC cells by activating SMADs. We first determined the expression levels of SMAD2/3. As shown in Figure 4A and B, although the mRNA and protein expression levels of SMAD2 and SMAD3 showed no changes in CRC cells cultured in BMI-1 CM, SMAD2 and SMAD3 phosphorylation was significantly increased compared with CRC cells cultured in NC CM. We then used SB-505124 (a TGF-β receptor [TGF-βR] inhibitor, which inhibits SMAD2 phosphorylation) to further confirm the involvement of SMADs in the pro-EMT effects of BMI-1 CM. SB-505124 markedly decreased BMI-1 CM-induced SMAD2 and SMAD3 phosphorylation in CRC cells (Figure 4C and D). In addition, the downstream target proteins (Snail and vimentin) of the TGF-β/SMAD pathway accordingly decreased in SB-505124-treated CRC cells cultured in BMI-1 CM (Figure 4C and D). These results confirmed that BMI-1 CM induces EMT in CRC cells partially by activating SMAD2/3.
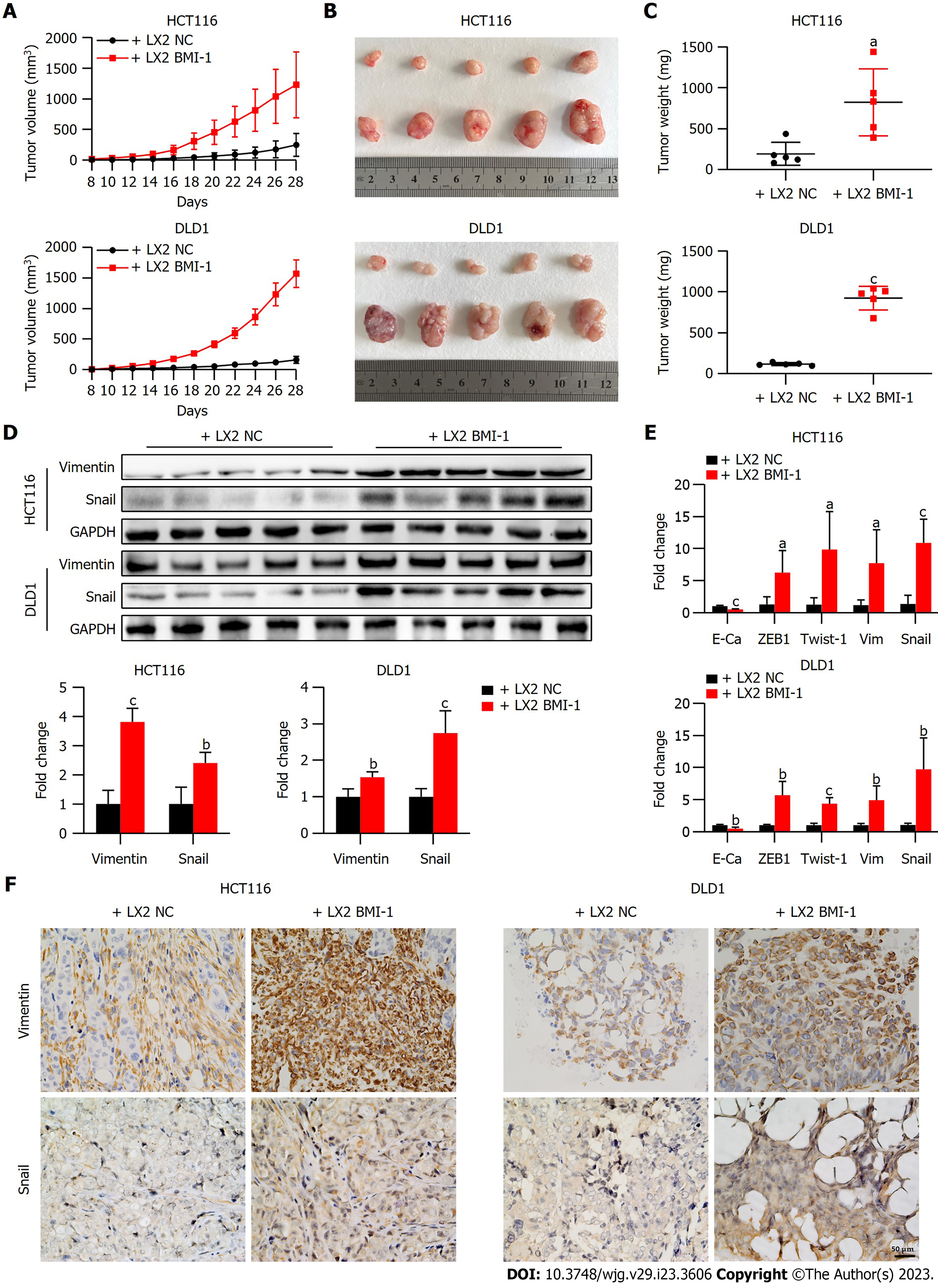
We further evaluated the effects of BMI-1 overexpressed LX2 HSCs on CRC cell growth in vivo. A subcutaneous xenotransplantation tumor model was established by co-implantation of HSCs (LX2 NC or LX2 BMI-1) and CRC cells (Figure 5A). Compared with mice co-implanted with LX2 NC, tumors grew faster and larger in mice co-implanted with LX2 BMI-1 HSCs and CRC cells as shown by increased tumor volume and weight (Figure 5B and C). Moreover, consistent with the in vitro results, we found that Snail and vimentin were upregulated in mice co-implanted with LX2 BMI-1 HSCs and CRC cells as shown by the qPCR and WB results (Figure 5D and E). Furthermore, the increased expression of Snail and vimentin in mouse liver co-implanted with LX2 BMI-1 HSCs and CRC cells was confirmed by IHC (Figure 5F). These results showed that LX2 BMI-1 HSCs promoted CRC tumor growth and EMT in the mouse model.
The liver is the most common target organ in terms of CRC metastases. Accumulating evidence suggests that the interactions between cancer cells and the liver microenvironment promote the progression of LM. In the liver microenvironment, HSC-derived CAFs are the main tumor-interacting population and play a vital role in cancer cell metastasis[8,30]. During the development of CRLM, HSC activation was found to be the most common biological process. Tumor-derived factors such as TGF-β and platelet-derived growth factor can activate HSCs[31]. BMI-1 has been proved to promote tumorigenesis and tumor progression in different types of cancers including CRC and HCC[24,26,32-34]. We previously reported that BMI-1 was abnormally highly expressed in CRLM and inhibition of BMI-1 in CRC cells dramatically reduced LM in vivo[26]. Normal liver cells were BMI-1-negative, and we observed that BMI-1-positive liver cells were common in CRLM samples from humans and mice. In particular, the mRNA and protein expression levels of BMI-1 in liver cells gradually increased during the development of CRLM in mice, suggesting that BMI-1 also participates in regulating liver cells during the course of CRLM. In this study, we found that BMI-1 had a novel function as a regulator in activating HSCs and plays an important role in the crosstalk between HSCs and CRC cells.
Quiescent HSCs normally resident in the space of Disse become activated with a myofibroblast-like phenotype (α-SMA+) in response to inflammatory stimuli and liver damage[27]. Studies have shown that cancer cells can also activate HSCs to release cytokines and chemokines to promote LM[8,35]. aHSCs reportedly orchestrate a premetastatic and prometastatic niche, which accelerate CRLM[5,6,36]. In this study, we found that BMI-1-positive liver cells appeared on day 14 and increased on days 21 and 28 after injection of CT26 cells, while small LM had formed on day 7. We further confirmed that BMI-1 overexpressed HSCs were activated to α-SMA+ myofibroblasts with increased expression of fibronectin, TGF-β1, MMPs, and IL-6. These secreted components can maintain the activated status of HSCs by enhancing the autocrine signaling loop[37]. aHSCs are the major source of ECM, and aHSC-secreted factors play a pivotal role in remodeling the ECM during tumor invasion and metastasis[38]. TGF-β1 is the most potent fibrogenic cytokine and is a fibrotic marker[39,40]. Fibronectin and MMPs are important ECM regulators in the liver fibrosis process[41]. ECM deposition by aHSCs provides a prometastatic environment. Thus, BMI-1 activates HSCs to secrete factors to form a prometastatic environment in the liver.
The interactions between cancer cells and aHSCs promote tumor growth and metastasis. CRC cells can stimulate aHSCs to release cytokines and chemokines, which in turn promote CRC growth and invasion[8,35]. In addition, aHSCs can secrete ECM components, induce angiogenesis, and modulate immunity to facilitate tumor growth and metastasis[31]. We found that high-metastatic HCT116 and low-metastatic DLD1 CRC cells cultured in CM from BMI-1 overexpressed HSCs showed the same enhanced proliferation and migration ability, and both HCT116 and DLD1 cells co-implanted with BMI-1 overexpressed HSCs showed increased tumor growth in vivo. These results demonstrated that BMI-1 aHSCs promote CRC cell proliferation and migration regardless of their ability to metastasize.
TGF-β signaling is important in tumor-stroma crosstalk. The TGF-β/SMAD pathway can act as both a tumor suppressor and promoter in CRLM[42,43]. The effect of the TGF-β/SMAD pathway in most malignant tumors is toward migration, the EMT or stemness, thereby promoting LM[44-46]. CRC cells are able to recruit and activate HSCs into CAFs by secreting TGF-β, and aHSCs lead to secretion of TGF-β which influences CRC cells[7]. LM has been found to be dependent on TGF-β signaling in liver stroma[47-50]. TGF-β1 is a key inducer of EMT transcription factors, which can promote EMT through the TGF-β/SMAD pathway[28,29]. The markers of EMT, including vimentin, Snail, ZEB-1, and Twist-1, were upregulated in CRC cells cultured in CM from BMI-1 overexpressed HSCs. The TGF-β/SMAD signaling pathway was activated and the phosphorylation of SMAD2/3 was induced in CRC cells cultured in CM from BMI-1 overexpressed HSCs, and treatment with the TGF-βR inhibitor SB-505124 attenuated the effect of BMI-1. Moreover, CRC cells co-implanted with BMI-1 overexpressed HSCs showed increased expression of EMT markers in vivo. Taken together, these findings indicate that BMI-1 overexpressed HSCs promote CRC cell growth and migration partly by activating the TGF-β/SMAD pathway and enhancing the EMT.
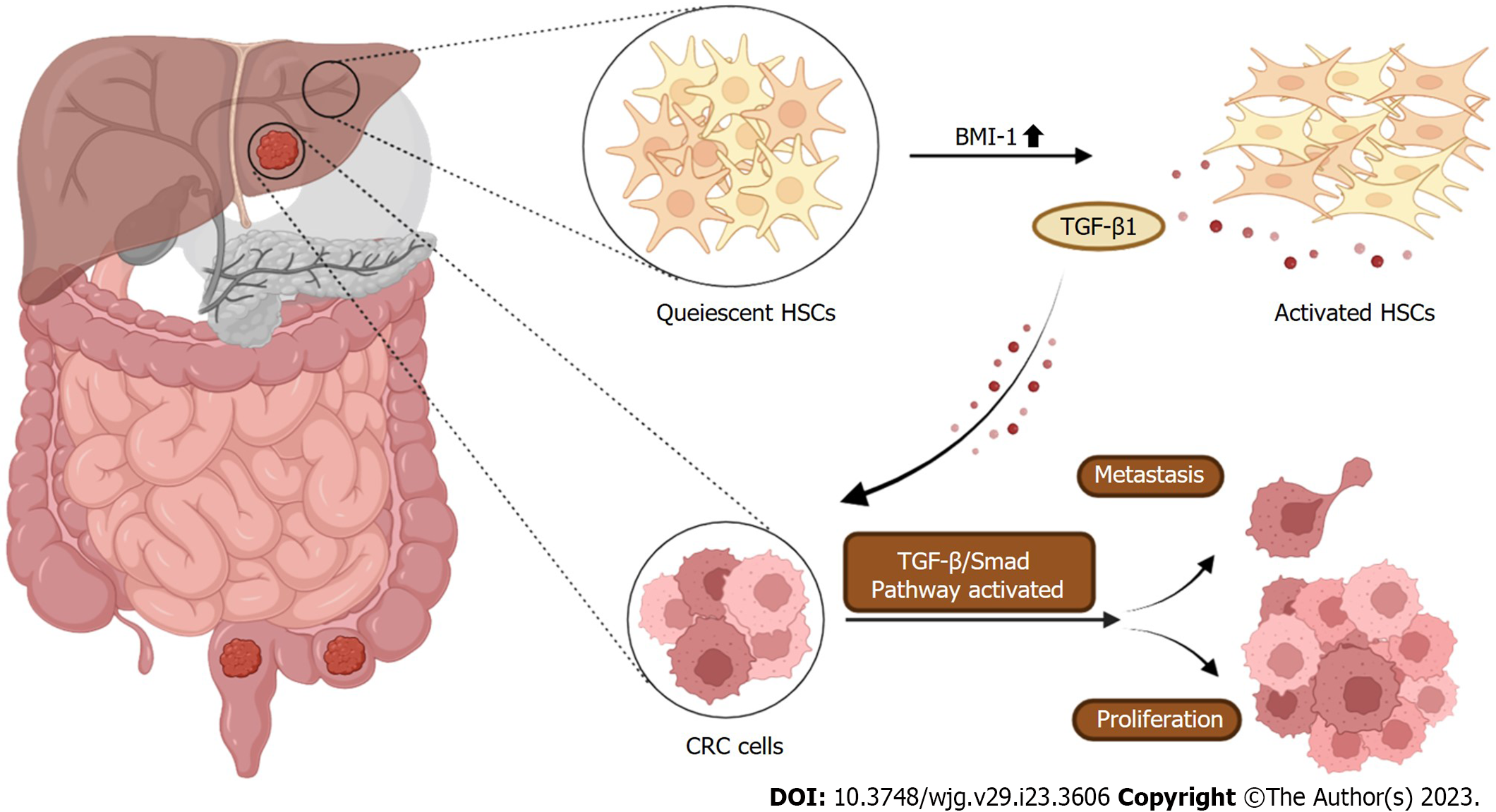
We propose a novel mechanism for BMI-1 involvement in promoting CRLM. BMI-1 was upregulated in liver cells during CRLM. BMI-1 aHSCs released cytokines and chemokines (fibronectin, TGF-β, MMPs, etc) to form a prometastatic environment in the liver, and promoted CRC cell proliferation, migration, and the EMT partially by activating the TGF-β/SMAD pathway (Figure 6). These findings provide a potential target for the treatment of CRLM.
Hepatic stellate cells (HSCs) are an important component of liver tissue and are a major source of cancer-associated fibroblasts. Cancer cells can activate HSCs, and in turn activated HSCs (aHSCs) promote tumor growth and metastasis. However, the mechanisms by which HSCs interact with colorectal cancer (CRC) cells to promote liver metastases (LM) are largely unknown.
We previously discovered that the expression of BMI-1 was abnormally high in LM of CRC. We speculated that BMI-1 plays an important role in the interaction between HSCs and CRC cells.
To examine the role of BMI-1 in HSC activation, and investigate the potential mechanisms of BMI-1 in the interaction between HSCs and CRC cells.
The expression of BMI-1 in liver tissue of patients with CRC liver metastasis (CRLM) was determined by immunohistochemistry analysis. We established a mouse LM model to assess the expression of BMI-1 during CRLM. We used lentiviral vectors to overexpress BMI-1 in LX2 HSCs (LX2 BMI-1) and evaluated the molecular markers of aHSCs. The proliferation and mobility of CRC cells in LX2 BMI-1 conditioned medium (CM) was detected. The epithelial-mesenchymal transition (EMT) and transforming growth factor beta (TGF-β)/SMAD pathway in CRC cells treated with LX2 BMI-1 CM were investigated by Western blot analysis and quantitative polymerase chain reaction. A mouse xenograft model was established to investigate the effects of co-implantation of LX2 BMI-1 and CRC cells on tumor growth, and the EMT phenotype of CRC.
BMI-1 expression was upregulated in liver cells during CRLM. Overexpression of BMI-1 induced the activation of LX2 HSCs. CRC cells cultured in LX2 BMI-1 CM showed higher proliferation, migration, and EMT ability compared with cells cultured in NC CM. LX2 BMI-1 CM induced activation of the TGF-β/SMAD pathway in CRC cells, and the TGF-βR inhibitor SB-505124 diminished the effect of BMI-1 CM. In addition, LX2 BMI-1 HSCs promoted CRC tumor growth and EMT in the mouse model.
BMI-1 aHSCs promoted the proliferation and migration of CRC partly via the TGF-β/SMAD pathway.
Targeting BMI-1 in liver tissues may have potential therapeutic implications for the treatment of CRLM.
We thank Xiao-Li Hong and Chao Bi from the Core Facilities of Zhejiang University School of Medicine for their technical support.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Emran TB, Bangladesh; Wu C, China S-Editor: Fan JR L-Editor: Filipodia P-Editor: Chen YX
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55839] [Article Influence: 7977.0] [Reference Citation Analysis (132)] |
| 2. | Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2268] [Cited by in RCA: 3272] [Article Influence: 654.4] [Reference Citation Analysis (2)] |
| 3. | Valderrama-Treviño AI, Barrera-Mera B, Ceballos-Villalva JC, Montalvo-Javé EE. Hepatic Metastasis from Colorectal Cancer. Euroasian J Hepatogastroenterol. 2017;7:166-175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 160] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 4. | Tsilimigras DI, Brodt P, Clavien PA, Muschel RJ, D'Angelica MI, Endo I, Parks RW, Doyle M, de Santibañes E, Pawlik TM. Liver metastases. Nat Rev Dis Primers. 2021;7:27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 302] [Article Influence: 75.5] [Reference Citation Analysis (0)] |
| 5. | Illemann M, Eefsen RH, Bird NC, Majeed A, Osterlind K, Laerum OD, Alpízar-Alpízar W, Lund IK, Høyer-Hansen G. Tissue inhibitor of matrix metalloproteinase-1 expression in colorectal cancer liver metastases is associated with vascular structures. Mol Carcinog. 2016;55:193-208. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 6. | Schütte M, Risch T, Abdavi-Azar N, Boehnke K, Schumacher D, Keil M, Yildiriman R, Jandrasits C, Borodina T, Amstislavskiy V, Worth CL, Schweiger C, Liebs S, Lange M, Warnatz HJ, Butcher LM, Barrett JE, Sultan M, Wierling C, Golob-Schwarzl N, Lax S, Uranitsch S, Becker M, Welte Y, Regan JL, Silvestrov M, Kehler I, Fusi A, Kessler T, Herwig R, Landegren U, Wienke D, Nilsson M, Velasco JA, Garin-Chesa P, Reinhard C, Beck S, Schäfer R, Regenbrecht CR, Henderson D, Lange B, Haybaeck J, Keilholz U, Hoffmann J, Lehrach H, Yaspo ML. Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nat Commun. 2017;8:14262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 198] [Cited by in RCA: 252] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 7. | Marvin DL, Heijboer R, Ten Dijke P, Ritsma L. TGF-β signaling in liver metastasis. Clin Transl Med. 2020;10:e160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 8. | Zhao S, Mi Y, Zheng B, Wei P, Gu Y, Zhang Z, Xu Y, Cai S, Li X, Li D. Highly-metastatic colorectal cancer cell released miR-181a-5p-rich extracellular vesicles promote liver metastasis by activating hepatic stellate cells and remodelling the tumour microenvironment. J Extracell Vesicles. 2022;11:e12186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 123] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 9. | Milette S, Sicklick JK, Lowy AM, Brodt P. Molecular Pathways: Targeting the Microenvironment of Liver Metastases. Clin Cancer Res. 2017;23:6390-6399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 92] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 10. | Brodt P. Role of the Microenvironment in Liver Metastasis: From Pre- to Prometastatic Niches. Clin Cancer Res. 2016;22:5971-5982. [PubMed] |
| 11. | Mao X, Xu J, Wang W, Liang C, Hua J, Liu J, Zhang B, Meng Q, Yu X, Shi S. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer. 2021;20:131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1314] [Cited by in RCA: 1330] [Article Influence: 332.5] [Reference Citation Analysis (0)] |
| 12. | Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16:582-598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2209] [Cited by in RCA: 2946] [Article Influence: 327.3] [Reference Citation Analysis (0)] |
| 13. | Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, Fearon D, Greten FR, Hingorani SR, Hunter T, Hynes RO, Jain RK, Janowitz T, Jorgensen C, Kimmelman AC, Kolonin MG, Maki RG, Powers RS, Puré E, Ramirez DC, Scherz-Shouval R, Sherman MH, Stewart S, Tlsty TD, Tuveson DA, Watt FM, Weaver V, Weeraratna AT, Werb Z. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020;20:174-186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1940] [Cited by in RCA: 2450] [Article Influence: 490.0] [Reference Citation Analysis (0)] |
| 14. | Higashi T, Friedman SL, Hoshida Y. Hepatic stellate cells as key target in liver fibrosis. Adv Drug Deliv Rev. 2017;121:27-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 530] [Cited by in RCA: 1059] [Article Influence: 132.4] [Reference Citation Analysis (0)] |
| 15. | Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125-172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2244] [Cited by in RCA: 2201] [Article Influence: 129.5] [Reference Citation Analysis (0)] |
| 16. | Barry AE, Baldeosingh R, Lamm R, Patel K, Zhang K, Dominguez DA, Kirton KJ, Shah AP, Dang H. Hepatic Stellate Cells and Hepatocarcinogenesis. Front Cell Dev Biol. 2020;8:709. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 105] [Article Influence: 21.0] [Reference Citation Analysis (1)] |
| 17. | Shiraha H, Iwamuro M, Okada H. Hepatic Stellate Cells in Liver Tumor. Adv Exp Med Biol. 2020;1234:43-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 18. | Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, Pradere JP, Schwabe RF. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 750] [Cited by in RCA: 1054] [Article Influence: 95.8] [Reference Citation Analysis (0)] |
| 19. | Makino Y, Hikita H, Kodama T, Shigekawa M, Yamada R, Sakamori R, Eguchi H, Morii E, Yokoi H, Mukoyama M, Hiroshi S, Tatsumi T, Takehara T. CTGF Mediates Tumor-Stroma Interactions between Hepatoma Cells and Hepatic Stellate Cells to Accelerate HCC Progression. Cancer Res. 2018;78:4902-4914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 82] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 20. | Biffi G, Tuveson DA. Diversity and Biology of Cancer-Associated Fibroblasts. Physiol Rev. 2021;101:147-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 641] [Cited by in RCA: 769] [Article Influence: 192.3] [Reference Citation Analysis (0)] |
| 21. | Affo S, Nair A, Brundu F, Ravichandra A, Bhattacharjee S, Matsuda M, Chin L, Filliol A, Wen W, Song X, Decker A, Worley J, Caviglia JM, Yu L, Yin D, Saito Y, Savage T, Wells RG, Mack M, Zender L, Arpaia N, Remotti HE, Rabadan R, Sims P, Leblond AL, Weber A, Riener MO, Stockwell BR, Gaublomme J, Llovet JM, Kalluri R, Michalopoulos GK, Seki E, Sia D, Chen X, Califano A, Schwabe RF. Promotion of cholangiocarcinoma growth by diverse cancer-associated fibroblast subpopulations. Cancer Cell. 2021;39:866-882.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 230] [Article Influence: 57.5] [Reference Citation Analysis (0)] |
| 22. | Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164-168. [PubMed] |
| 23. | Jiang L, Li J, Song L. Bmi-1, stem cells and cancer. Acta Biochim Biophys Sin (Shanghai). 2009;41:527-534. [PubMed] |
| 24. | Tateishi K, Ohta M, Kanai F, Guleng B, Tanaka Y, Asaoka Y, Tada M, Seto M, Jazag A, Lianjie L, Okamoto M, Isayama H, Yoshida H, Kawabe T, Omata M. Dysregulated expression of stem cell factor Bmi1 in precancerous lesions of the gastrointestinal tract. Clin Cancer Res. 2006;12:6960-6966. [PubMed] |
| 25. | Gil J, Bernard D, Peters G. Role of polycomb group proteins in stem cell self-renewal and cancer. DNA Cell Biol. 2005;24:117-125. [PubMed] |
| 26. | Xu Z, Zhou Z, Zhang J, Xuan F, Fan M, Zhou D, Liuyang Z, Ma X, Hong Y, Wang Y, Sharma S, Dong Q, Wang G. Targeting BMI-1-mediated epithelial-mesenchymal transition to inhibit colorectal cancer liver metastasis. Acta Pharm Sin B. 2021;11:1274-1285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 27. | Tsuchida T, Friedman SL. Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol. 2017;14:397-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1221] [Cited by in RCA: 2000] [Article Influence: 250.0] [Reference Citation Analysis (0)] |
| 28. | Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1766] [Cited by in RCA: 2180] [Article Influence: 136.3] [Reference Citation Analysis (0)] |
| 29. | Hao Y, Baker D, Ten Dijke P. TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 807] [Article Influence: 134.5] [Reference Citation Analysis (0)] |
| 30. | Kubo N, Araki K, Kuwano H, Shirabe K. Cancer-associated fibroblasts in hepatocellular carcinoma. World J Gastroenterol. 2016;22:6841-6850. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 116] [Cited by in RCA: 139] [Article Influence: 15.4] [Reference Citation Analysis (1)] |
| 31. | Kang N, Gores GJ, Shah VH. Hepatic stellate cells: partners in crime for liver metastases? Hepatology. 2011;54:707-713. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 144] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 32. | Effendi K, Mori T, Komuta M, Masugi Y, Du W, Sakamoto M. Bmi-1 gene is upregulated in early-stage hepatocellular carcinoma and correlates with ATP-binding cassette transporter B1 expression. Cancer Sci. 2010;101:666-672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 33. | Li X, Yang Z, Song W, Zhou L, Li Q, Tao K, Zhou J, Wang X, Zheng Z, You N, Dou K, Li H. Overexpression of Bmi-1 contributes to the invasion and metastasis of hepatocellular carcinoma by increasing the expression of matrix metalloproteinase (MMP)2, MMP-9 and vascular endothelial growth factor via the PTEN/PI3K/Akt pathway. Int J Oncol. 2013;43:793-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 85] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 34. | Xu Z, Tao J, Chen P, Chen L, Sharma S, Wang G, Dong Q. Sodium Butyrate Inhibits Colorectal Cancer Cell Migration by Downregulating Bmi-1 Through Enhanced miR-200c Expression. Mol Nutr Food Res. 2018;62:e1700844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 35. | Huang WH, Zhou MW, Zhu YF, Xiang JB, Li ZY, Wang ZH, Zhou YM, Yang Y, Chen ZY, Gu XD. The Role Of Hepatic Stellate Cells In Promoting Liver Metastasis Of Colorectal Carcinoma. Onco Targets Ther. 2019;12:7573-7580. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 36. | Eveno C, Hainaud P, Rampanou A, Bonnin P, Bakhouche S, Dupuy E, Contreres JO, Pocard M. Proof of prometastatic niche induction by hepatic stellate cells. J Surg Res. 2015;194:496-504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 37. | Watanabe T, Tajima H, Hironori H, Nakagawara H, Ohnishi I, Takamura H, Ninomiya I, Kitagawa H, Fushida S, Tani T, Fujimura T, Ota T, Wakayama T, Iseki S, Harada S. Sodium valproate blocks the transforming growth factor (TGF)-β1 autocrine loop and attenuates the TGF-β1-induced collagen synthesis in a human hepatic stellate cell line. Int J Mol Med. 2011;28:919-925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 38. | Ahmad SA, Berman RS, Ellis LM. Biology of colorectal liver metastases. Surg Oncol Clin N Am. 2003;12:135-150. [PubMed] |
| 39. | Xu F, Liu C, Zhou D, Zhang L. TGF-β/SMAD Pathway and Its Regulation in Hepatic Fibrosis. J Histochem Cytochem. 2016;64:157-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 569] [Article Influence: 63.2] [Reference Citation Analysis (0)] |
| 40. | Hellerbrand C, Stefanovic B, Giordano F, Burchardt ER, Brenner DA. The role of TGFbeta1 in initiating hepatic stellate cell activation in vivo. J Hepatol. 1999;30:77-87. [PubMed] |
| 41. | Klingberg F, Hinz B, White ES. The myofibroblast matrix: implications for tissue repair and fibrosis. J Pathol. 2013;229:298-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 418] [Cited by in RCA: 568] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 42. | Are C, Simms N, Rajput A, Brattain M. The role of transforming growth factor-beta in suppression of hepatic metastasis from colon cancer. HPB (Oxford). 2010;12:498-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 43. | Korkut A, Zaidi S, Kanchi RS, Rao S, Gough NR, Schultz A, Li X, Lorenzi PL, Berger AC, Robertson G, Kwong LN, Datto M, Roszik J, Ling S, Ravikumar V, Manyam G, Rao A, Shelley S, Liu Y, Ju Z, Hansel D, de Velasco G, Pennathur A, Andersen JB, O'Rourke CJ, Ohshiro K, Jogunoori W, Nguyen BN, Li S, Osmanbeyoglu HU, Ajani JA, Mani SA, Houseman A, Wiznerowicz M, Chen J, Gu S, Ma W, Zhang J, Tong P, Cherniack AD, Deng C, Resar L; Cancer Genome Atlas Research Network, Weinstein JN, Mishra L, Akbani R. A Pan-Cancer Analysis Reveals High-Frequency Genetic Alterations in Mediators of Signaling by the TGF-β Superfamily. Cell Syst. 2018;7:422-437.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 142] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 44. | Zhang B, Halder SK, Kashikar ND, Cho YJ, Datta A, Gorden DL, Datta PK. Antimetastatic role of Smad4 signaling in colorectal cancer. Gastroenterology. 2010;138:969-80.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 174] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 45. | Tsushima H, Ito N, Tamura S, Matsuda Y, Inada M, Yabuuchi I, Imai Y, Nagashima R, Misawa H, Takeda H, Matsuzawa Y, Kawata S. Circulating transforming growth factor beta 1 as a predictor of liver metastasis after resection in colorectal cancer. Clin Cancer Res. 2001;7:1258-1262. [PubMed] |
| 46. | Zhang C, Gao H, Li C, Tu J, Chen Z, Su W, Geng X, Chen X, Wang J, Pan W. TGFβ1 Promotes Breast Cancer Local Invasion and Liver Metastasis by Increasing the CD44(high)/CD24(-) Subpopulation. Technol Cancer Res Treat. 2018;17:1533033818764497. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 47. | Calon A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Céspedes MV, Sevillano M, Nadal C, Jung P, Zhang XH, Byrom D, Riera A, Rossell D, Mangues R, Massagué J, Sancho E, Batlle E. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22:571-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 745] [Cited by in RCA: 900] [Article Influence: 69.2] [Reference Citation Analysis (0)] |
| 48. | Tauriello DVF, Palomo-Ponce S, Stork D, Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, Sevillano M, Ibiza S, Cañellas A, Hernando-Momblona X, Byrom D, Matarin JA, Calon A, Rivas EI, Nebreda AR, Riera A, Attolini CS, Batlle E. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554:538-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 917] [Cited by in RCA: 1334] [Article Influence: 190.6] [Reference Citation Analysis (0)] |
| 49. | Zubeldia IG, Bleau AM, Redrado M, Serrano D, Agliano A, Gil-Puig C, Vidal-Vanaclocha F, Lecanda J, Calvo A. Epithelial to mesenchymal transition and cancer stem cell phenotypes leading to liver metastasis are abrogated by the novel TGFβ1-targeting peptides P17 and P144. Exp Cell Res. 2013;319:12-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 50. | Zhang B, Halder SK, Zhang S, Datta PK. Targeting transforming growth factor-beta signaling in liver metastasis of colon cancer. Cancer Lett. 2009;277:114-120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 114] [Article Influence: 7.1] [Reference Citation Analysis (0)] |