Published online Jun 14, 2023. doi: 10.3748/wjg.v29.i22.3482
Peer-review started: April 16, 2023
First decision: April 28, 2023
Revised: May 4, 2023
Accepted: May 11, 2023
Article in press: May 11, 2023
Published online: June 14, 2023
Processing time: 51 Days and 11.8 Hours
Due to the poor prognosis of gastric cancer (GC), early detection methods are urgently needed. Plasma exosomal circular RNAs (circRNAs) have been suggested as novel biomarkers for GC.
To identify a novel biomarker for early detection of GC.
Healthy donors (HDs) and GC patients diagnosed by pathology were recruited. Nine GC patients and three HDs were selected for exosomal whole-transcriptome RNA sequencing. The expression profiles of circRNAs were analyzed by bioinformatics methods and validated by droplet digital polymerase chain reaction. The expression levels and area under receiver operating characteristic curve values of plasma exosomal circRNAs and standard serum biomarkers were used to compare their diagnostic efficiency.
There were 303 participants, including 240 GC patients and 63 HDs, involved in the study. The expression levels of exosomal hsa_circ_0079439 were significantly higher in GC patients than in HDs (P < 0.0001). However, the levels of standard serum biomarkers were similar between the two groups. The area under the curve value of exosomal hsa_circ_0079439 was higher than those of standard biomarkers, including carcinoembryonic antigen, carbohydrate antigen (CA)19-9, CA72-4, alpha-fetoprotein, and CA125 (0.8595 vs 0.5862, 0.5660, 0.5360, 0.5082, and 0.5018, respectively). The expression levels of exosomal hsa_circ_0079439 were significantly decreased after treatment (P < 0.05). Moreover, the expression levels of exosomal hsa_circ_0079439 were obviously higher in early GC (EGC) patients than in HDs (P < 0.0001).
Our results suggest that plasma exosomal hsa_circ_0079439 is upregulated in GC patients. Moreover, the levels of exosomal hsa_circ_0079439 could distinguish EGC and advanced GC patients from HDs. Therefore, plasma exosomal hsa_circ_0079439 might be a potential biomarker for the diagnosis of GC during both the early and late stages.
Core Tip: Gastric cancer (GC) remains a prevalent disease worldwide. Due to its atypical clinical manifestations, GC is usually diagnosed at late stages in most patients and has a poor prognosis. Therefore, early detection of GC is of great importance. Based on the results of exosomal whole-transcriptome RNA sequencing, we performed bioinformatic analysis and droplet digital polymerase chain reaction (dd-PCR) tests. As validated by dd-PCR, exosomal hsa_circ_0079439 had a higher area under the curve value than standard biomarkers. Therefore, plasma exosomal hsa_circ_0079439 could serve as a novel biomarker for the early detection of GC.
- Citation: Li X, Lin YL, Shao JK, Wu XJ, Li X, Yao H, Shi FL, Li LS, Zhang WG, Chang ZY, Chai NL, Wang YL, Linghu EQ. Plasma exosomal hsa_circ_0079439 as a novel biomarker for early detection of gastric cancer. World J Gastroenterol 2023; 29(22): 3482-3496
- URL: https://www.wjgnet.com/1007-9327/full/v29/i22/3482.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i22.3482
Gastric cancer (GC) is the fifth most commonly diagnosed cancer and the fourth leading cause of cancer-related death; it accounted for 1089103 new cases and 768793 deaths in 2020 globally[1]. The prognosis of advanced GC (AGC) patients is poor[2]. Early detection of GC and identification of individuals who might benefit from a given treatment are of great importance. Unfortunately, the most common symptoms associated with GC are nonspecific, such as weight loss, poor appetite, and abdominal pain[2]. Based on the abovementioned conditions, the diagnosis of GC is often at a late stage[2]. Therefore, accurate early detection of GC is one of the most urgent issues in its management.
A previous study showed that carcinoembryonic antigen (CEA), carbohydrate antigen (CA)19-9, and CA72-4 levels are significantly associated with tumor stage and patient survival[3]. In addition, some studies have shown that CA125 and alpha-fetoprotein (AFP) are linked to GC[4,5]. However, the widespread use of less invasive detection methods has resulted in a number of inaccurate diagnoses due to their low sensitivity and specificity[6-8]. Therefore, it is necessary to identify novel biomarkers with higher accuracy for the early detection of GC.
Exosomes, a subset of nanometer-sized (50-150 nm) extracellular vesicles, can be secreted by almost all kinds of cells[9,10]. In addition, exosomes comprise diverse components, including proteins, DNAs, circular RNAs (circRNAs), long noncoding RNAs (lncRNAs), microRNAs (miRNAs), messenger RNAs (mRNAs), and lipids[10-13]. An increasing number of studies have shown that exosomes are involved in mediating intercellular communication and many pathological processes[9,10,14]. Therefore, they have great potential to become novel biomarkers in liquid biopsy. CircRNAs, a class of lncRNAs, were first discovered by electron microscopy more than 40 years ago[15-17]. Because of the exosomal phos
Furthermore, a number of studies have demonstrated that circRNAs play an important role in GC progression, invasion, and metastasis. However, the utility of exosomal circRNAs for early detection of GC is largely unknown. Profiling circRNAs in exosomes could be useful for identifying molecular markers for early detection and prognosis prediction for various types of cancers[14,18-20].
In this study, we analyzed the expression levels of plasma exosomal circRNAs from GC patients in different clinical stages. The findings of this study support the notion that plasma exosomal circRNAs could serve as biomarkers for the diagnosis of early GC (EGC). Moreover, they might be considered as new therapeutic targets as well as effective prognostic markers for GC.
EGC is defined as GC confined to the mucosal and submucosal layers. Locally AGC (LAGC) was defined as stages T2-4aN0-3M0 GC, corresponding to stages Ib to IIIc and excluding T1 or T4b tumors[21-23]. Metastatic GC (MGC) refers to the invasion of GC cells into other tissues or organs. AGC includes LAGC and MGC[24].
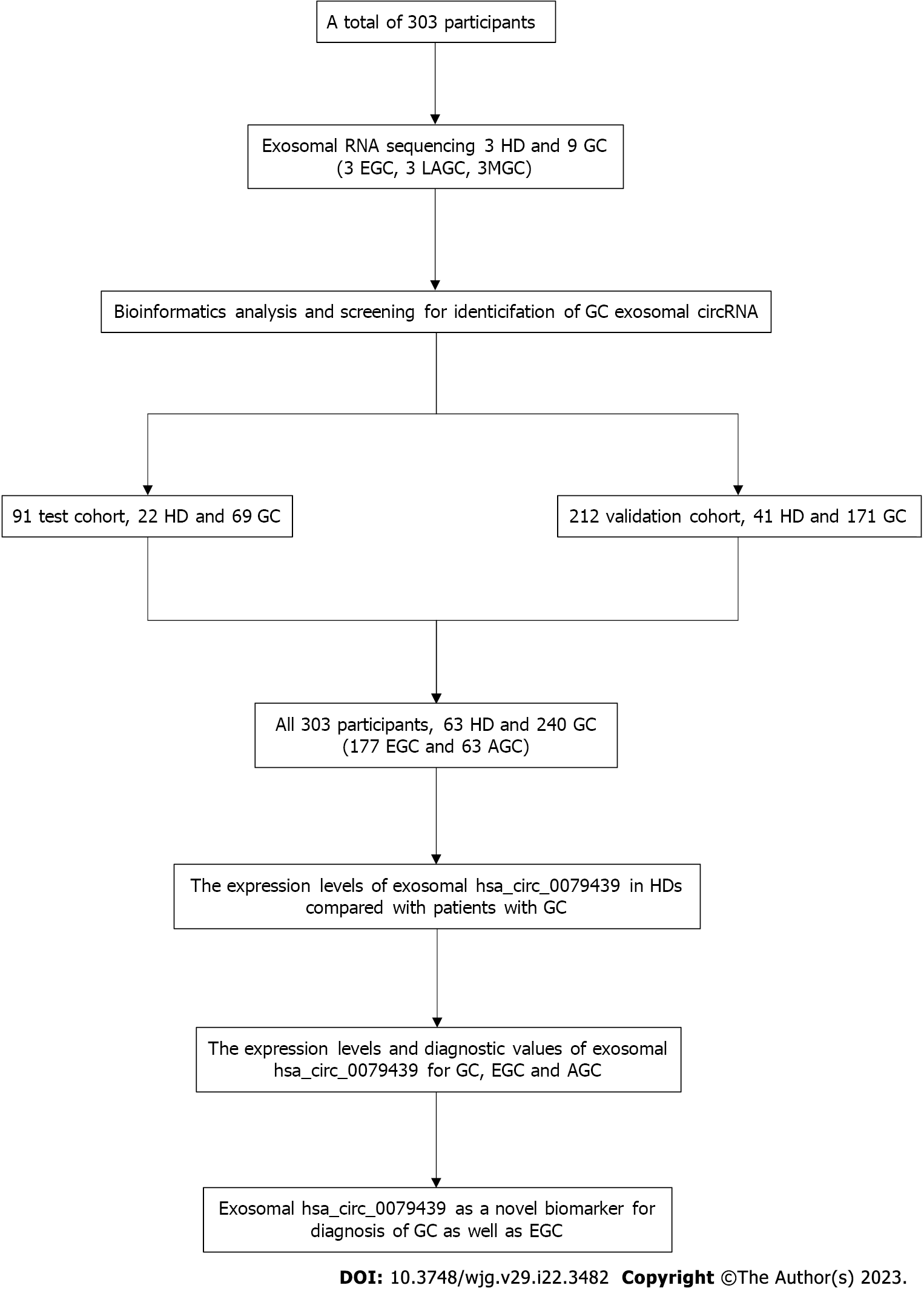
A total of 240 GC patients (including 177 EGC patients and 63 AGC patients) and 63 healthy donors (HDs) were enrolled in our study from April 2018 to October 2022 (Figure 1). All of the GC patients had been diagnosed with adenocarcinoma after surgery. Patients who had been treated by chemoradiotherapy, targeted therapy, or cytotoxic therapy were excluded. HDs had undergone gastric endoscopy, and they did not have GC or any other gastric disease. We selected plasma exosomal samples obtained from nine GC patients (3 EGC, 3 LAGC, and 3 MGC) and three HDs for whole transcriptome RNA sequencing (nine GC patients were included in the validation cohort, and the expression levels of circRNA were verified). None of these GC patients had received any previous treatment before their first blood samples were obtained.
All participants were divided into a test cohort and a validation cohort. There were 69 GC patients and 22 HDs in the test cohort from April 2018 to December 2018. Another 171 GC patients and 41 HDs were included in the validation cohort from January 2019 to October 2022. According to the American Joint Committee on Cancer 8th edition[22,23], the clinical stage and TNM classification of GC in all patients were assessed and analyzed. All patients underwent endoscopic submucosal dissection or minimally invasive surgery to remove the lesions, which meant that accurate pathology results could be confirmed. In addition, plasma samples were acquired from GC patients 7 d after treatment.
Blood samples were collected from GC patients or HDs in K2-EDTA Blood Collection Tubes (Becton, Dickinson and Company). Blood samples were centrifuged at 500 × g for 15 min, and the supernatant collected was plasma. Plasma was transferred to a new tube and a unique classification number was written on the tube, which was then stored at -80 °C.
GC cells (GCCs) (AGS and HGC-27) were obtained from Shanghai Gaining Biological Technology Company (http://www.shgnsw.com). KATO III was obtained from the American Type Culture Collection (ATCC, http://www.atcc.org). Other GCCs (SNU-1, SGC-7901, MGC-803, and BGC-823) were obtained from the BeNa Culture Collection (BNCC, http://www.bncc.org.cn). Normal gastric epithelial cells (NGECs) were obtained from the Shanghai Qincheng Biological Technology Company (http://www.shqcsw.com). The culture methods used for the cell lines are shown in Supplementary Table 1.
For whole-transcriptome RNA sequencing, exosomes were isolated by size-exclusion chromatography. We used a procedure described in a previous study with a few minor modifications[25]. PBS was used to perform a 1.5-fold dilution of 1 mL of each plasma sample (0.8 μm filtered). Then, the mixture was purified using Exosupur® columns (Echobiotech, China) and more PBS was used to perform elution, which resulted in a total of 2 mL of eluate. Finally, by using 100 kDa molecular weight cutoff Amicon® Ultra spin filters (Merck, Germany), the fractions obtained during the previous step were concentrated to 200 μL.
For droplet digital polymerase chain reaction (dd-PCR) test, plasma exosomes were isolated according to the manufacturer’s instructions for the ExoQuickTM Exosome Precipitation Solution (SBI). Briefly, 16 μL of thrombin was added to 1000 μL of plasma at a final concentration of 5 U/mL. The mixture was incubated at room temperature for 5 min and centrifuged at 10000 rpm for 5 min, and a visible pellet was observed at the bottom of the tube. The supernatant was transferred to a new tube and centrifuged at 3000 × g for 5 min to remove cells and other debris. The supernatant was transferred to a new tube; 63 μL of ExoQuick Exosome Precipitation Solution was added, and the mixture was incubated at 4 °C for 30 min. After centrifuging the mixture at 1500 × g for 30 min, the fluid was removed. The pellet at the bottom of the tube, which contained exosomes, was collected.
Exosomes isolated from plasma were diluted. Ten microliters of exosome solution was added to a copper mesh and incubated for 10 min. After cleaning the mesh with sterile distilled water, filter paper was used to absorb the liquid. Ten microliters of 2% phosphotungstic acid was added to the copper mesh and incubated for 1 min. Then, the copper mesh was dried with filter paper. The copper mesh was kept dry for several minutes at room temperature. And transmission electron microscopy (TEM) was performed.
After the instrument qualification test, the diameter and quantity of the exosomes isolated from plasma were measured by ZetaView PMX 110 (Particle Metrix, Meerbusch). The movement of particles was analyzed by nanoparticle tracking analysis (NTA) software (ZetaView 8.02.28) by taking 60 s of video at a rate of 30 frames/s.
After total protein extraction from exosomes, the protein concentration was measured with the Pierce BCA Protein Assay kit (Thermo). Western blot (WB) analysis was performed to measure protein expression in the extracts. The positive markers of exosomes that were used included CD9 (Abcam), TSG101 (Abcam), and CD81 (Abcam). The negative marker was calnexin (Abcam).
Total RNA was extracted from plasma exosomes and purified with the miRNeasy Serum/Plasma Advanced Kit (Qiagen). The concentration and purity were evaluated with the RNA Nano 6000 Assay Kit and an Agilent Bioanalyzer 2100 System (Agilent Technologies).
According to the instructions of the SMARTer Stranded Total RNA-Seq Kit V2 (Takara), a total amount of 250 pg-10 ng RNA per sample was used as input material for RNA-seq library preparation. For small RNA libraries, a total amount of 1 ng-500 ng RNA per sample was used as input material for the QIAseq miRNA Library Kit (Qiagen). The quality of RNA-seq libraries and small RNA libraries was assessed with an Agilent Bioanalyzer 2100 (Agilent Technologies). Based on the acBot Cluster Generation System, the index-coded samples were clustered using TruSeq PE Cluster Kitv3-cBot-HS (Illumina). Then, the prepared libraries were sequenced on an Illumina NovaSeq 6000 platform. Paired-end reads were generated at EchoBiotech Co. Ltd., Beijing, P. R. China.
The R package DESeq2 was used for the differential expression analysis of circRNAs, lncRNAs, miRNAs, and mRNAs between HDs and GC patients. The criteria were set as |log2FC| > 1 and P < 0.01. The R package pheatmap was used for clustering analysis of these differentially expressed circRNAs, lncRNAs, miRNAs, and mRNAs. The Cancer-Specific CircRNA Database (CSCD) and CircInteractome databases were used to predict the target miRNAs of circRNAs. The TargetScan, miRDB, and miRTarBase databases were used to predict the target genes of miRNAs. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis and Gene Ontology (GO) functional enrichment analysis were performed with R 4.1.1 software to identify the enriched genes.
Dd-PCR was performed on a QX200 Droplet System using dd-PCR EvaGreen Supermix (Bio-Rad). The divergent primers overlapping the splice junction of hsa_circ_0079439 were designed by Primer-BLAST (Supplementary Table 2).
Statistical analyses were performed with SPSS 26.0 and GraphPad Prism 8.0. The expression levels of plasma exosomal hsa_circ_0079439 and standard serum biomarkers, including CEA, CA19-9, CA72-4, AFP, and CA125, were compared between the test cohort and validation cohort, and the differences were analyzed by the t test. The data are shown as the mean (SD). The expression levels and area under the receiver operating characteristic (ROC) curve (AUC) values of plasma exosomal hsa_circ_0079439 and standard serum biomarkers were used to assess their diagnostic efficiency for GC, EGC, and AGC. The results with P < 0.05 were considered significant.
There were 303 participants, including 63 HDs and 240 GC patients, enrolled in our study (Table 1). There were 177 patients (73.75%) with GC at T1 stage, 7 (2.92%) with GC at T2 stage, 37 (15.42%) with GC at T3 stage, and 19 (7.91%) with GC at T4 stage. Among these GC patients, 47 (19.58%) had lymph node metastasis, and 9 (3.75%) had distant metastasis. Most GC cases were in stage I (180, 75.00%), and others were in stage II (16, 6.67%), stage III (35, 14.58%), and stage IV (9, 3.75%). The median age (range) of the HD group was 53 years (24-81 years) and that of the GC group was 64 years (34-94 years). The ratio of males to females was 43/20 in the HD group, and 187/53 in the GC group.
| Characteristic | Test cohort (n = 91) | Validation cohort (n = 212) | Total participants (n = 303) | |||
| HD (n = 22) | GC (n = 69) | HD (n = 41) | GC (n = 171) | HD (n = 63) | GC (n = 240) | |
| Gender | ||||||
| Male | 12 (54.55) | 51 (73.91) | 31 (75.61) | 136 (79.53) | 43 (68.25) | 187 (77.92) |
| Female | 10 (45.45) | 18 (26.09) | 10 (24.39) | 35 (20.47) | 20 (31.75) | 53 (22.08) |
| Age (yr), median (range) | 54 (32-77) | 66 (35-84) | 53 (24-81) | 64 (34-94) | 53 (24-81) | 64 (34-94) |
| T stage | / | / | / | |||
| T1 | 69 (100.00) | 108 (63.16) | 177 (73.75) | |||
| T2 | 0 | 7 (4.09) | 7 (2.92) | |||
| T3 | 0 | 37 (21.64) | 37 (15.42) | |||
| T4 | 0 | 19 (11.11) | 19 (7.91) | |||
| N stage | / | / | / | |||
| N0 | 69 (100.00) | 124 (72.51) | 193 (80.42) | |||
| N1 | 0 | 11 (6.43) | 11 (4.58) | |||
| N2 | 0 | 14 (8.19) | 14 (5.83) | |||
| N3 | 0 | 22 (12.87) | 22 (9.17) | |||
| M stage | / | / | / | |||
| M0 | 69 (100.00) | 162 (94.74) | 231 (96.25) | |||
| M1 | 0 | 9 (5.26) | 9 (3.75) | |||
| Clinical stage | / | / | / | |||
| I | 69 (100.00) | 111 (64.91) | 180 (75.00) | |||
| II | 0 | 16 (9.36) | 16 (6.67) | |||
| III | 0 | 35 (20.47) | 35 (14.58) | |||
| IV | 0 | 9 (5.26) | 9 (3.75) | |||
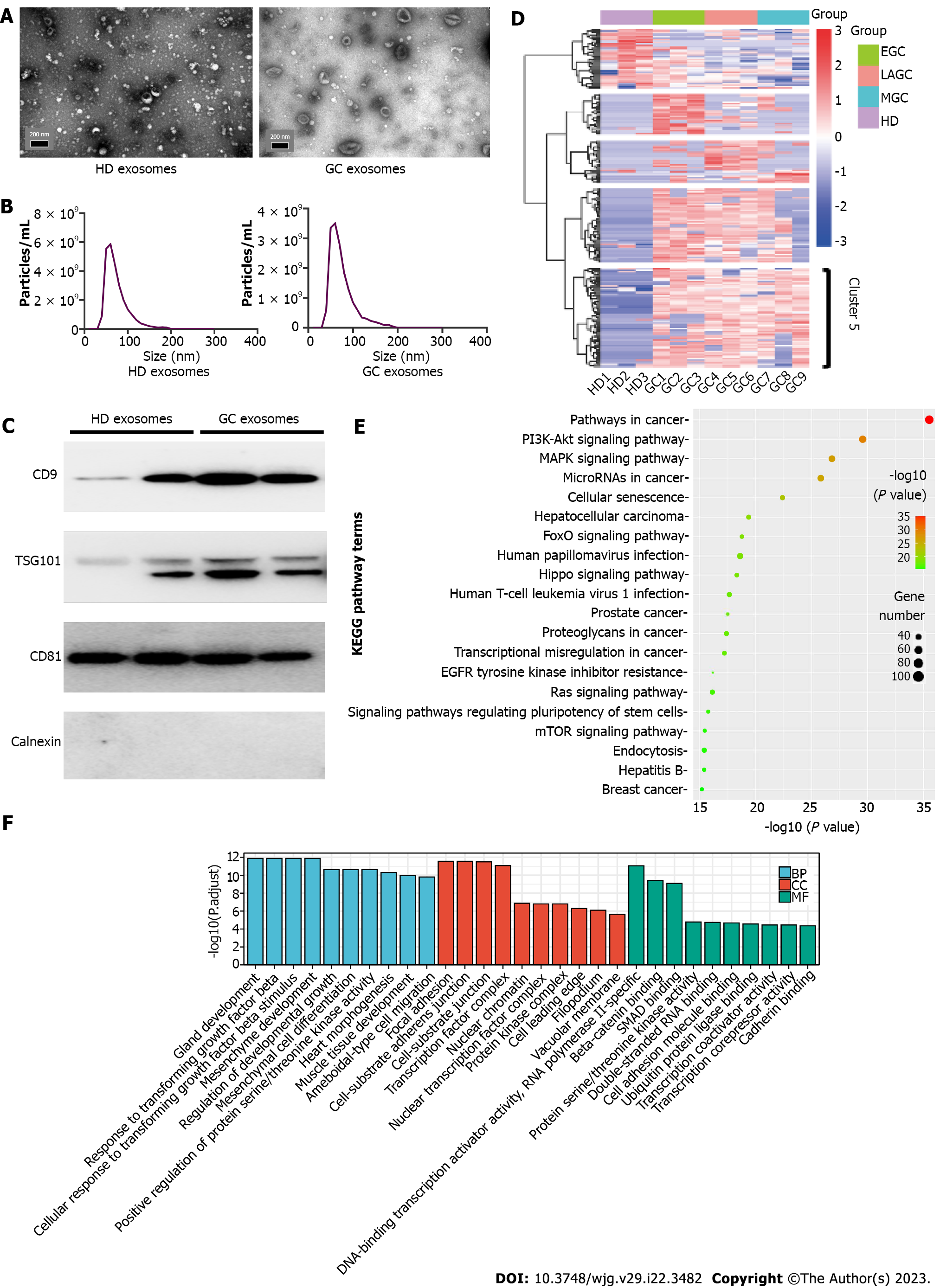
To characterize exosomes and confirm their isolation from GC patients and HDs, TEM, NTA, and WB analysis were performed. The exosomes appeared to be almost round in shape according to electron microscopy images (Figure 2A). NTA indicated that the diameter of exosomes ranged from 40 nm to 200 nm, which was consistent with previous research results[26] (Figure 2B). WB results showed that exosomes expressed typical markers, including CD9, TSG101, and CD81. The negative marker calnexin was not expressed (Figure 2C).
There were 14976 circRNAs, 3061 lncRNAs, 1629 miRNAs, and 12830 mRNAs identified by whole transcriptome RNA sequencing of exosomes. Clustering analysis was performed to reveal the differentially expressed circRNAs, lncRNAs, miRNAs, and mRNAs between HDs and GC patients. Unlike the relatively ambiguous clusters observed for the latter three RNA types (Supplementary Figure 1A-C), circRNAs were clearly clustered into five groups (Figure 2D). Based on data obtained from CSCD and CircInteractome, 2262 miRNA response elements were predicted. By intersecting the results obtained from TargetScan, miRDB, and miRTarBase, 8145 common potential target genes were identified. Then, we performed differential expression analyses of GC patient samples vs HD samples by using The Cancer Genome Atlas (TCGA) and identified 2512 genes (1072 upregulated, 1440 downregulated); 998 of these genes were also identified as potential targets in the abovementioned analysis. KEGG pathway analysis demonstrated that these genes were enriched in pathways in cancer, the cell cycle, and the PI3K-Akt signaling pathway (Supplementary Figure 2A). GO enrichment analysis indicated that these genes were associated with nuclear division, organelle fission, and cell cycle G1/S phase transition (Supplementary Figure 2B).
To further investigate the function of cluster 5 (68 circRNAs, marked in Figure 2D), similar analysis methods were used. These circRNAs were significantly upregulated in all GC samples, and 1731 target miRNAs were predicted. To identify potential miRNAs related to GC, we performed differential expression analyses based on TCGA and identified 64 miRNAs that were downregulated in GC samples vs HD samples. Thirty-one miRNAs were selected by taking the intersection of the two groups of results. These miRNAs were predicted to bind 1578 target genes. KEGG pathway analysis demonstrated that these genes were enriched in five pathways, including pathways in cancer, the PI3K-Akt signaling pathway, the MAPK signaling pathway, miRNAs in cancer, and cellular senescence (Figure 2E). GO enrichment analysis indicated that these genes were associated with nuclear division, organelle fission and cell cycle G1/S phase transition (Figure 2F).
We generated a circRNA-miRNA-mRNA regulatory network related to the genes enriched in the top 5 KEGG pathways. Ultimately, we found that six circRNAs (hsa_circ_0113953, hsa_circ_0076179, hsa_circ_0007353, hsa_circ_0012823, hsa_circ_0001470, and hsa_circ_0079439) were included in all the networks (Supplementary Figure 3A-E).
Similar to a previous study, the expression levels of ncRNAs in exosomes were consistent with those in GCCs or tissues[27]. Then, we assessed the expression levels of the six circRNAs in NGECs and GCCs (AGS, MGC-803, SGC-7901, HGC-27, SNU-1, KATO III, and BGC-823) (Supplementary Figure 4A-F). The results showed that the expression levels of hsa_circ_0079439 in the GCCs were obviously higher than those in NGECs. Thus, hsa_circ_0079439 was selected for further validation by dd-PCR. The primers used to measure the levels of the six circRNAs are shown in Supplementary Table 1.
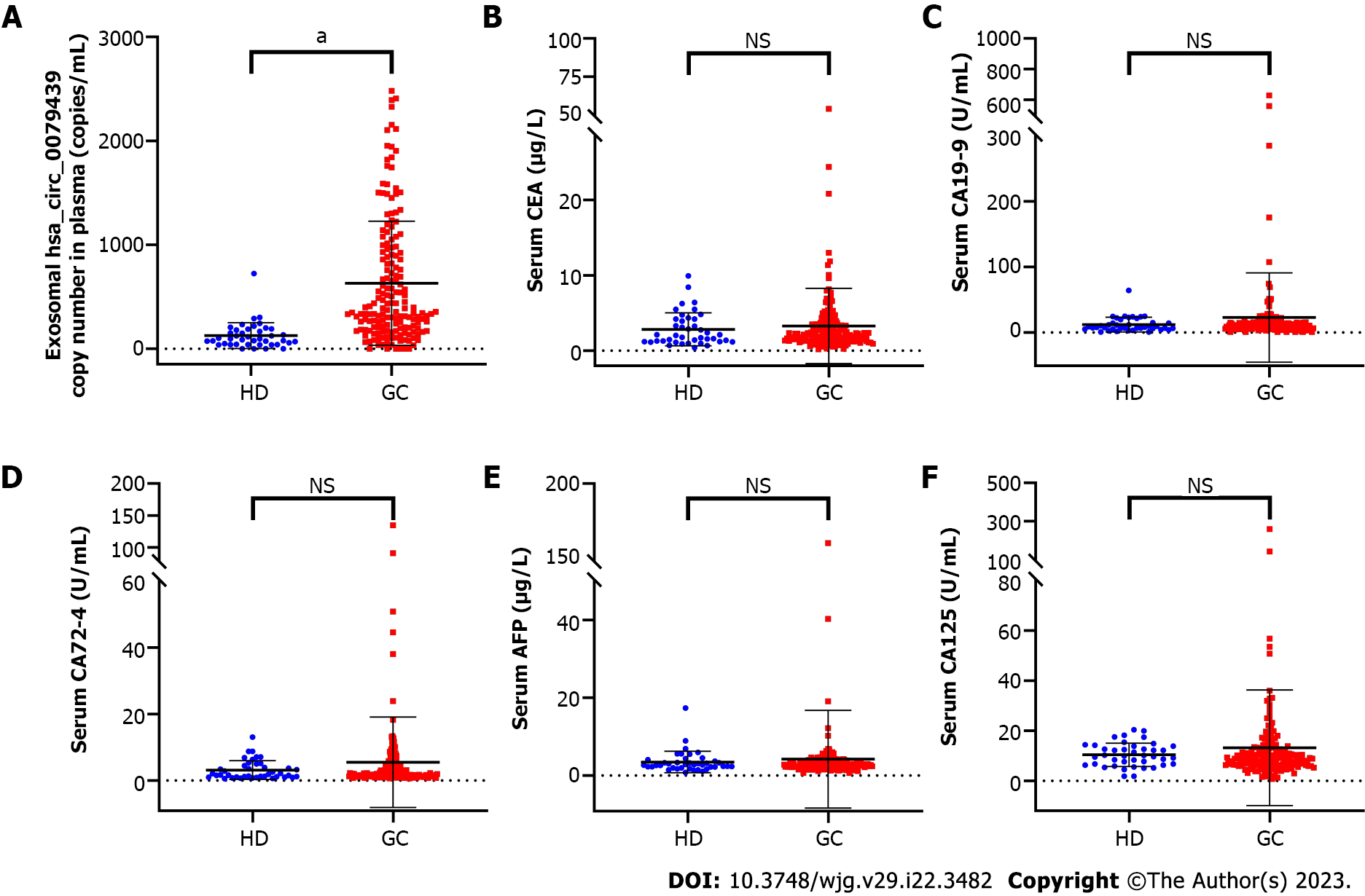
In the test cohort, the expression levels of exosomal hsa_circ_0079439 were significantly higher in GC patients than in HDs (P < 0.0001) (Figure 3A). In addition, the levels of serum CEA in GC patients were higher than those in HDs (P < 0.001) (Figure 3B). However, the expression levels of CA19-9, CA72-4, AFP, and CA125 were not significantly different between GC patients and HDs (Figure 3C-F).
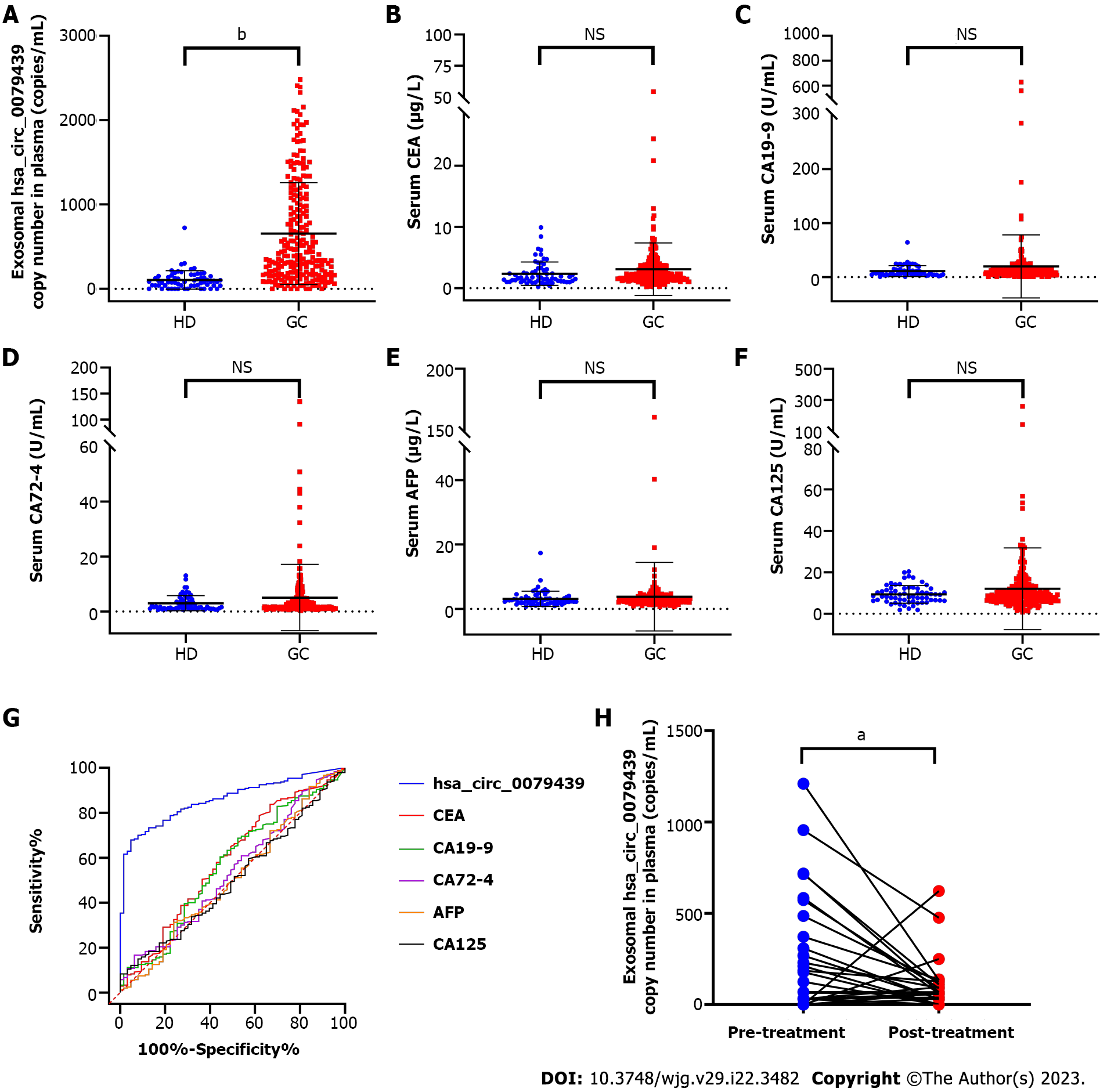
Furthermore, in the validation cohort, we again found that the expression levels of exosomal hsa_circ_0079439 were significantly higher in GC patients than in HDs (P < 0.0001) (Figure 4A). However, the expression levels of standard serum biomarkers between GC patients and HDs were not significantly different (Figure 4B-F).
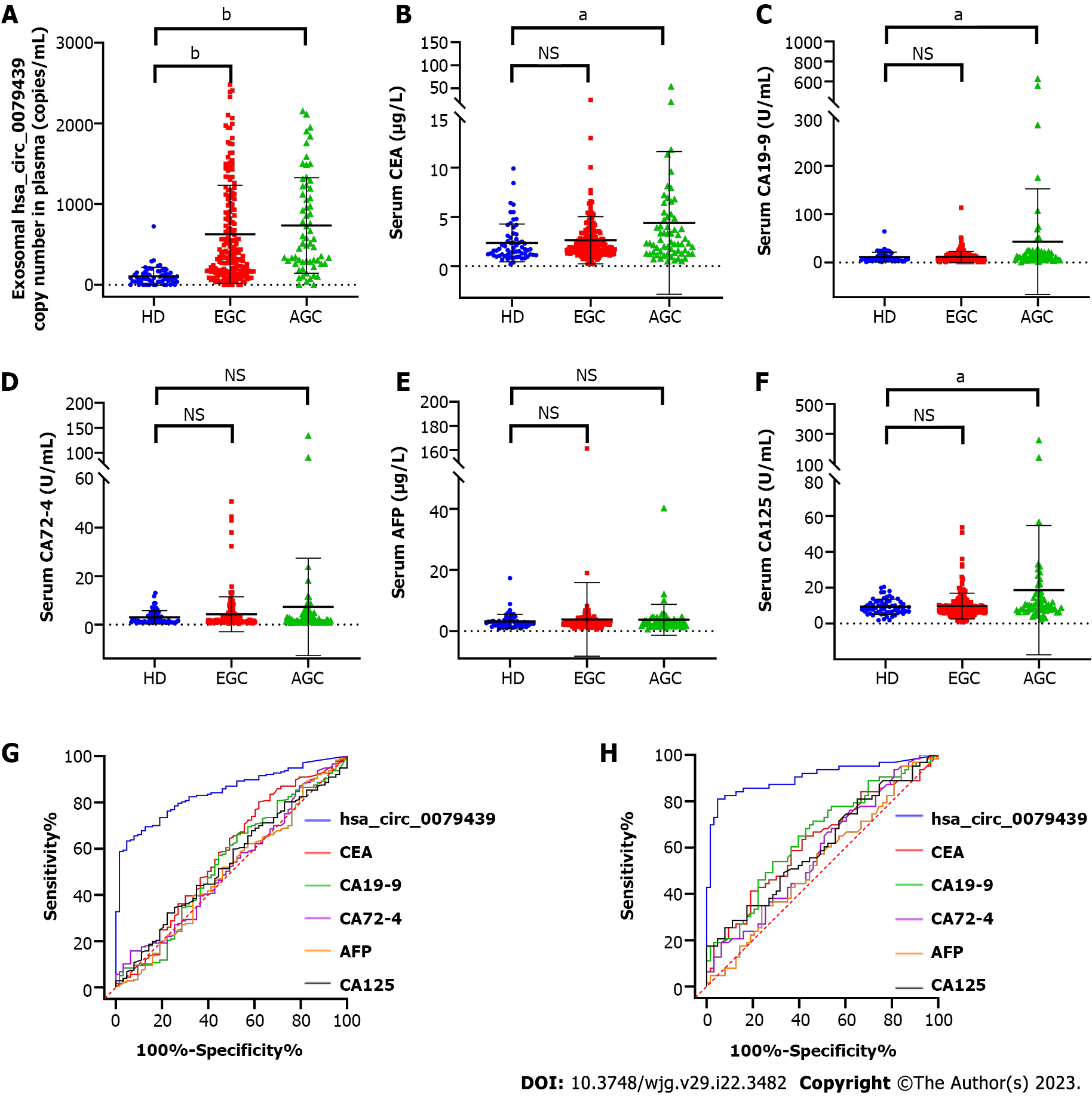
To evaluate the diagnostic value of exosomal hsa_circ_0079439 for GC, EGC, and AGC, we combined the data obtained from the test cohort and validation cohort. We compared the expression levels and analyzed the ROC curves of exosomal hsa_circ_0079439, CEA, CA19-9, CA72-4, AFP, and CA125. The expression levels of exosomal hsa_circ_0079439 were significantly higher in GC patients than in HDs (P < 0.0001) (Figure 5A). However, the levels of standard serum biomarkers showed no significant difference between GC patients and HDs (Figure 5B-F). The AUC value of exosomal hsa_circ_0079439 was higher than those of CEA, CA19-9, CA72-4, AFP, and CA125 (0.8595 vs 0.5862, 0.5660, 0.5360, 0.5082, and 0.5018, respectively) (Figure 5G). To investigate the changes in the expression levels of exosomal hsa_circ_0079439 before and after treatment, we analyzed 30 paired plasma samples obtained from GC patients. The expression levels of exosomal hsa_circ_0079439 were significantly decreased after treatment (Figure 5H) (P < 0.05).
To determine whether the results were similar in EGC patients, all GC patients were divided into two groups (EGC patients and AGC patients). The expression levels of exosomal hsa_circ_0079439 were obviously higher in EGC patients (P < 0.0001) and AGC patients (P < 0.0001) than in HDs (Figure 6A). However, the serum levels of CEA, CA19-9, CA72-4, AFP, and CA125 could not be used to distinguish EGC patients from HDs (Figure 6B-F). The AUC value of exosomal hsa_circ_0079439 was obviously higher than those of CEA, CA19-9, CA72-4, AFP, and CA125 (0.8433 vs 0.5732, 0.5362, 0.5203, 0.5024, and 0.5352, respectively) (Figure 6G).
In addition, the serum levels of CEA, CA19-9, and CA125 were apparently higher in AGC patients than in HDs (P < 0.05) (Figure 6B, C and F). However, when comparing the levels of CA72-4 and AFP, there was no significant difference between AGC patients and HDs (Figure 6D and E). The AUC value of exosomal hsa_circ_0079439 was obviously higher than those of CEA, CA19-9, CA72-4, AFP, and CA125 (0.9050 vs 0.6228, 0.6497, 0.5801, 0.5379, and 0.6058, respectively) (Figure 6H).
Although the incidence and mortality rates of GC have declined over the past century, GC is still a major health problem worldwide, especially in East Asian countries[2,28]. GC has a poor prognosis in most patients due to late detection. Currently, the overall survival rate of EGC patients treated by endoscopy can exceed 90%[28,29]. Nonetheless, the 5-year survival rate of AGC patients with distant metastases is lower than 5%[30]. However, a screening program carried out by the Shanghai Research Institute in ten medical centers showed that the rate of EGC diagnosis is only 9.61%[28].
The clinical symptoms of EGC are atypical in most patients, and these symptoms include anorexia, malnutrition, and stomachache[2]. The methods of early diagnosis of GC include detection of standard serum biomarkers, barium radiography, computerized tomography, and endoscopic biopsy[31]. Over time, the role of liquid biopsy in GC has received close attention. Furthermore, liquid biopsy has emerged as a revolutionary strategy for the early diagnosis of cancers, detection of therapeutic targets, prediction of prognosis, monitoring of therapeutic efficacy, and analysis of drug resistance; it also has the advantage of being minimally invasive[14,32-34]. Although numerous studies have investigated biomarkers for the early detection of GC, there are still many limitations regarding the use of these biomarkers for the early diagnosis of GC. For example, CEA and CA72-4 are standard clinical biomarkers, but both lack sensitivity and specificity.
In recent years, exosomes have been found in nearly all types of human body fluids, such as blood, urine, breast milk, saliva, ascites, and cerebrospinal fluid. Because of their phospholipid bilayer membrane structure, they can protect bioactive cargos from degradation. There are many metabolites and products in exosomes produced by all kinds of cells. These cargoes include nucleic acids, proteins, lipids, and metabolites[35].
CircRNAs are a class of single-stranded noncoding RNAs[36]. Initially, circRNAs were considered to be the result of splicing errors. However, with the development of bioinformatics and high-throughput sequencing technologies, more circRNAs have been identified, and they have been found to be involved in the occurrence and development of many human diseases. Many studies have demonstrated that circRNAs are abundant, stable and conserved rather than random products of RNA splicing[36]. Due to their high stability, circRNAs have many important biological functions, including noncoding and coding functions. The most widely studied function is their role in the regulation of miRNA target genes as miRNA sponges or decoys. They can protect target mRNAs from miRNA-dependent degradation. Moreover, it has been found that circRNAs may function as protein sponges or decoys, enhancers of protein function, protein scaffolds, protein recruiters, and templates for translation[17,37].
For instance, Zhang et al[38] reported that exosomal circNRIP1, as a tumor promoter, sponges miR-149-5p to regulate the expression level of AKT1 in GC. Similarly, exosomal circSHKBP1 may be a promising circulating biomarker for GC diagnosis and prognosis[39]. Exosomal circSHKBP1 regulates the miR-582-3p/HUR/vascular endothelial growth factor pathway and suppresses HSP90 degradation to promote GC progression[39]. In addition, Lu et al[40] found that exosomal circ-RanGAP1 facilitates GC invasion and metastasis by targeting miR-877-3p to regulate vascular endothelial growth factor-A expression, suggesting that circ-RanGAP1 may act as a tumor promoter in GC.
In this study, we performed cluster analysis of exosomal circRNAs, lncRNAs, miRNAs, and mRNAs based on the results of whole-transcriptome RNA sequencing. The results showed that circRNAs could be used to distinguish GC patients from HDs better than lncRNAs, miRNAs, and mRNAs. Then, we investigated the expression of exosomal hsa_circ_0079439 in HDs and GC patients, including EGC and AGC, by dd-PCR. The expression levels of exosomal hsa_circ_0079439 were significantly higher in GC patients.
In recent years, dd-PCR has become the latest technology used for DNA quantification. The reaction volume containing the nucleic acids is divided into thousands of nanosized droplets, each of which either contains the gene of interest or does not. After PCR amplification, each droplet is detected one by one. A droplet with a fluorescence signal is interpreted as a 1, and a droplet without a fluorescence signal is interpreted as a 0. According to the Poisson distribution principle and the number and proportion of positive droplets, the initial copy number or concentration of the gene of interest can be obtained. There is no suitable reference gene in exosomes for normalization of the expression levels of circRNAs in real-time PCR (RT-PCR) experiments. Previous studies have shown that dd-PCR can be used to accurately quantify the copy numbers of specific nucleotides in plasma without external calibrators and has higher sensitivity than RT-PCR[41-44]. Therefore, we compared the diagnostic efficiency of exosomal hsa_circ_0079439 in GC patients, especially EGC patients, with standard clinical biomarkers, by dd-PCR. We found that exosomal hsa_circ_0079439 had higher sensitivity and specificity than standard clinical biomarkers. The findings of this study support the notion that plasma exosomal circRNAs could serve as biomarkers for the diagnosis of EGC and might be new therapeutic targets as well as effective prognostic markers for GC.
However, there are still some limitations to our study. First, the main limitation of this study is that since it was conducted at a single center, selection bias may have affected the results. This may indicate that this method is suitable for the early diagnosis of GC in Asians but not necessarily in other populations. Second, some groups had relatively few samples, so the generalizability of our study results may be affected. Third, since there is no standard isolation method for exosomes, the use of different isolation methods may result in differences in detection efficiency for the early diagnosis of GC[45-47]. Fourth, although circulating exosomal circRNAs serve as good candidates for blood-based biomarkers, it will take some time before they can be applied in the clinic. Therefore, multicenter studies with larger sample sizes are encouraged to obtain more accurate results for the early diagnosis of GC.
Based on whole-transcriptome RNA resequencing data, we found that circRNAs have greater potential than other RNA types as novel biomarkers for the detection of GC, especially EGC. To our knowledge, this study is the first to discover and verify that the levels of exosomal hsa_circ_0079439 are higher in GC patients than in HDs. In addition, exosomal hsa_circ_0079439 performs better in distinguishing EGC patients from HDs than standard clinical biomarkers. Overall, exosomal hsa_circ_0079439 is a prospective biomarker for the diagnosis of GC, including EGC and AGC.
Gastric cancer (GC) is a major health problem worldwide, especially in East Asian countries, and GC has a poor prognosis in most patients due to late detection. Therefore, accurate early detection of GC is one of the most urgent issues in its management.
A number of studies have demonstrated that circular RNAs (circRNAs) play an important role in GC progression, invasion, and metastasis. However, the utility of exosomal circRNAs for early detection of GC is largely unknown.
To analyze the expression levels of plasma exosomal circRNAs from GC patients in different clinical stages to identify a novel biomarker for early detection of GC.
Healthy donors (HDs) and GC patients diagnosed by pathology were recruited. Based on the results of whole-transcriptome RNA sequencing, the expression profiles of circRNAs were analyzed by bioinformatics methods and validated by droplet digital polymerase chain reaction. Then we compared the diagnostic efficiency of plasma exosomal circRNAs and standard serum biomarkers.
The expression levels of exosomal hsa_circ_0079439 were obviously higher in GC patients than in HDs (P < 0.0001). And the area under the curve value of exosomal hsa_circ_0079439 was obviously higher than those of carcinoembryonic antigen, carbohydrate antigen (CA)19-9, CA72-4, alpha-fetoprotein, and CA125 (0.8433 vs 0.5732, 0.5362, 0.5203, 0.5024, and 0.5352, respectively).
Plasma exosomal hsa_circ_0079439 is a prospective biomarker for the diagnosis of GC.
Multicenter studies with larger sample sizes are encouraged to obtain more accurate results for the early diagnosis of GC.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author’s Membership in Professional Societies: Chinese Society of Digestive Endoscopy, President; Endoscopy Branch of Chinese Medical Association, Vice President; Digestive Endoscopy Branch, Beijing Medical Association, President; Digestive Endoscopy Branch, Beijing Physicians Association, President; Chinese Journal of Gastroenteroscopy Electronics, Editor-in-chief; Chinese Journal of Digestive Endoscopy, Vice President.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Liu L, China; Schmidt T, Germany S-Editor: Wang JJ L-Editor: Wang TQ P-Editor: Chen YL
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64362] [Article Influence: 16090.5] [Reference Citation Analysis (175)] |
| 2. | Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1150] [Cited by in RCA: 2827] [Article Influence: 565.4] [Reference Citation Analysis (5)] |
| 3. | Shimada H, Noie T, Ohashi M, Oba K, Takahashi Y. Clinical significance of serum tumor markers for gastric cancer: a systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer. 2014;17:26-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 367] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 4. | Feng F, Tian Y, Xu G, Liu Z, Liu S, Zheng G, Guo M, Lian X, Fan D, Zhang H. Diagnostic and prognostic value of CEA, CA19-9, AFP and CA125 for early gastric cancer. BMC Cancer. 2017;17:737. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 267] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 5. | Liu X, Cheng Y, Sheng W, Lu H, Xu Y, Long Z, Zhu H, Wang Y. Clinicopathologic features and prognostic factors in alpha-fetoprotein-producing gastric cancers: analysis of 104 cases. J Surg Oncol. 2010;102:249-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 107] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 6. | Guo X, Lv X, Ru Y, Zhou F, Wang N, Xi H, Zhang K, Li J, Chang R, Xie T, Wang X, Li B, Chen Y, Yang Y, Chen L. Circulating Exosomal Gastric Cancer-Associated Long Noncoding RNA1 as a Biomarker for Early Detection and Monitoring Progression of Gastric Cancer: A Multiphase Study. JAMA Surg. 2020;155:572-579. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 127] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 7. | Xu Y, Zhang P, Zhang K, Huang C. The application of CA72-4 in the diagnosis, prognosis, and treatment of gastric cancer. Biochim Biophys Acta Rev Cancer. 2021;1876:188634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 59] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 8. | Thanh Huong P, Gurshaney S, Thanh Binh N, Gia Pham A, Hoang Nguyen H, Thanh Nguyen X, Pham-The H, Tran PT, Truong Vu K, Xuan Duong N, Pelucchi C, La Vecchia C, Boffetta P, Nguyen HD, Luu HN. Emerging Role of Circulating Tumor Cells in Gastric Cancer. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 9. | Zhou R, Chen KK, Zhang J, Xiao B, Huang Z, Ju C, Sun J, Zhang F, Lv XB, Huang G. The decade of exosomal long RNA species: an emerging cancer antagonist. Mol Cancer. 2018;17:75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 127] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 10. | Kok VC, Yu CC. Cancer-Derived Exosomes: Their Role in Cancer Biology and Biomarker Development. Int J Nanomedicine. 2020;15:8019-8036. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 300] [Cited by in RCA: 293] [Article Influence: 58.6] [Reference Citation Analysis (0)] |
| 11. | Hannafon BN, Trigoso YD, Calloway CL, Zhao YD, Lum DH, Welm AL, Zhao ZJ, Blick KE, Dooley WC, Ding WQ. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18:90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 462] [Cited by in RCA: 437] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 12. | Xu H, Zhou J, Tang J, Min X, Yi T, Zhao J, Ren Y. Identification of serum exosomal lncRNA MIAT as a novel diagnostic and prognostic biomarker for gastric cancer. J Clin Lab Anal. 2020;34:e23323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 13. | Kim KU, Kim WH, Jeong CH, Yi DY, Min H. More than Nutrition: Therapeutic Potential of Breast Milk-Derived Exosomes in Cancer. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 14. | Yu D, Li Y, Wang M, Gu J, Xu W, Cai H, Fang X, Zhang X. Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer. 2022;21:56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 335] [Cited by in RCA: 478] [Article Influence: 159.3] [Reference Citation Analysis (0)] |
| 15. | Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6253] [Cited by in RCA: 6017] [Article Influence: 501.4] [Reference Citation Analysis (0)] |
| 16. | Shan C, Zhang Y, Hao X, Gao J, Chen X, Wang K. Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol Cancer. 2019;18:136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 172] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 17. | Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3273] [Cited by in RCA: 3135] [Article Influence: 522.5] [Reference Citation Analysis (0)] |
| 18. | Yu W, Hurley J, Roberts D, Chakrabortty SK, Enderle D, Noerholm M, Breakefield XO, Skog JK. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol. 2021;32:466-477. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 328] [Cited by in RCA: 546] [Article Influence: 136.5] [Reference Citation Analysis (0)] |
| 19. | Baassiri A, Nassar F, Mukherji D, Shamseddine A, Nasr R, Temraz S. Exosomal Non Coding RNA in LIQUID Biopsies as a Promising Biomarker for Colorectal Cancer. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 20. | Poulet C, Njock MS, Moermans C, Louis E, Louis R, Malaise M, Guiot J. Exosomal Long Non-Coding RNAs in Lung Diseases. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 21. | Yu J, Huang C, Sun Y, Su X, Cao H, Hu J, Wang K, Suo J, Tao K, He X, Wei H, Ying M, Hu W, Du X, Hu Y, Liu H, Zheng C, Li P, Xie J, Liu F, Li Z, Zhao G, Yang K, Liu C, Li H, Chen P, Ji J, Li G; Chinese Laparoscopic Gastrointestinal Surgery Study (CLASS) Group. Effect of Laparoscopic vs Open Distal Gastrectomy on 3-Year Disease-Free Survival in Patients With Locally Advanced Gastric Cancer: The CLASS-01 Randomized Clinical Trial. JAMA. 2019;321:1983-1992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 522] [Cited by in RCA: 524] [Article Influence: 87.3] [Reference Citation Analysis (1)] |
| 22. | Marano L, D'Ignazio A, Cammillini F, Angotti R, Messina M, Marrelli D, Roviello F. Comparison between 7th and 8th edition of AJCC TNM staging system for gastric cancer: old problems and new perspectives. Transl Gastroenterol Hepatol. 2019;4:22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 23. | He X, Wu W, Lin Z, Ding Y, Si J, Sun LM. Validation of the American Joint Committee on Cancer (AJCC) 8th edition stage system for gastric cancer patients: a population-based analysis. Gastric Cancer. 2018;21:391-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 24. | Digklia A, Wagner AD. Advanced gastric cancer: Current treatment landscape and future perspectives. World J Gastroenterol. 2016;22:2403-2414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 359] [Cited by in RCA: 402] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 25. | Böing AN, van der Pol E, Grootemaat AE, Coumans FA, Sturk A, Nieuwland R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles. 2014;3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 566] [Cited by in RCA: 853] [Article Influence: 77.5] [Reference Citation Analysis (0)] |
| 26. | Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 815] [Cited by in RCA: 1812] [Article Influence: 362.4] [Reference Citation Analysis (0)] |
| 27. | Li W, Gao YQ. MiR-217 is involved in the carcinogenesis of gastric cancer by down-regulating CDH1 expression. Kaohsiung J Med Sci. 2018;34:377-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 28. | Hu Y, Fang JY, Xiao SD. Can the incidence of gastric cancer be reduced in the new century? J Dig Dis. 2013;14:11-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 29. | Hatta W, Gotoda T, Oyama T, Kawata N, Takahashi A, Yoshifuku Y, Hoteya S, Nakagawa M, Hirano M, Esaki M, Matsuda M, Ohnita K, Yamanouchi K, Yoshida M, Dohi O, Takada J, Tanaka K, Yamada S, Tsuji T, Ito H, Hayashi Y, Nakaya N, Nakamura T, Shimosegawa T. A Scoring System to Stratify Curability after Endoscopic Submucosal Dissection for Early Gastric Cancer: "eCura system". Am J Gastroenterol. 2017;112:874-881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 221] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 30. | Mattiuzzi C, Lippi G. Current Cancer Epidemiology. J Epidemiol Glob Health. 2019;9:217-222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 321] [Cited by in RCA: 841] [Article Influence: 168.2] [Reference Citation Analysis (1)] |
| 31. | Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700-713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1159] [Cited by in RCA: 1324] [Article Influence: 120.4] [Reference Citation Analysis (0)] |
| 32. | Vaidyanathan R, Soon RH, Zhang P, Jiang K, Lim CT. Cancer diagnosis: from tumor to liquid biopsy and beyond. Lab Chip. 2018;19:11-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 33. | Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic - implementation issues and future challenges. Nat Rev Clin Oncol. 2021;18:297-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 737] [Article Influence: 184.3] [Reference Citation Analysis (0)] |
| 34. | Alix-Panabières C, Pantel K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021;11:858-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 576] [Article Influence: 144.0] [Reference Citation Analysis (0)] |
| 35. | Dai J, Su Y, Zhong S, Cong L, Liu B, Yang J, Tao Y, He Z, Chen C, Jiang Y. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct Target Ther. 2020;5:145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 256] [Cited by in RCA: 785] [Article Influence: 157.0] [Reference Citation Analysis (0)] |
| 36. | Chen L, Shan G. CircRNA in cancer: Fundamental mechanism and clinical potential. Cancer Lett. 2021;505:49-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 312] [Article Influence: 78.0] [Reference Citation Analysis (0)] |
| 37. | Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci. 2020;77:1661-1680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 277] [Article Influence: 55.4] [Reference Citation Analysis (0)] |
| 38. | Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, Xu P, Sun G, Xu J, Lv J, Xu Z. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 508] [Cited by in RCA: 610] [Article Influence: 101.7] [Reference Citation Analysis (0)] |
| 39. | Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F, Ma P, Jiang H, Wu X, Shu Y, Xu T. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer. 2020;19:112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 291] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 40. | Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie JW, Wang JB, Lin JX, Chen QY, Cao LL, Zheng CH, Li P, Huang CM. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 194] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 41. | Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, Emslie KR. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012;84:1003-1011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 679] [Cited by in RCA: 785] [Article Influence: 56.1] [Reference Citation Analysis (0)] |
| 42. | Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W, Yu R, Xiao B, Guo J. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl). 2018;96:85-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 207] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 43. | Sillence KA, Roberts LA, Hollands HJ, Thompson HP, Kiernan M, Madgett TE, Welch CR, Avent ND. Fetal Sex and RHD Genotyping with Digital PCR Demonstrates Greater Sensitivity than Real-time PCR. Clin Chem. 2015;61:1399-1407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 44. | Huggett JF, Cowen S, Foy CA. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem. 2015;61:79-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 325] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 45. | Nordin JZ, Lee Y, Vader P, Mäger I, Johansson HJ, Heusermann W, Wiklander OP, Hällbrink M, Seow Y, Bultema JJ, Gilthorpe J, Davies T, Fairchild PJ, Gabrielsson S, Meisner-Kober NC, Lehtiö J, Smith CI, Wood MJ, El Andaloussi S. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine. 2015;11:879-883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 483] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 46. | Van Deun J, Mestdagh P, Sormunen R, Cocquyt V, Vermaelen K, Vandesompele J, Bracke M, De Wever O, Hendrix A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J Extracell Vesicles. 2014;3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 535] [Cited by in RCA: 699] [Article Influence: 63.5] [Reference Citation Analysis (0)] |
| 47. | Tang XH, Guo T, Gao XY, Wu XL, Xing XF, Ji JF, Li ZY. Exosome-derived noncoding RNAs in gastric cancer: functions and clinical applications. Mol Cancer. 2021;20:99. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 119] [Article Influence: 29.8] [Reference Citation Analysis (0)] |