Published online Jun 14, 2023. doi: 10.3748/wjg.v29.i22.3469
Peer-review started: March 28, 2023
First decision: April 10, 2023
Revised: April 19, 2023
Accepted: May 16, 2023
Article in press: May 16, 2023
Published online: June 14, 2023
Processing time: 70 Days and 21.3 Hours
Wild rats have the potential to hold zoonotic infectious agents that can spread to humans and cause disease.
To better understand the composition of gut bacterial communities in rats is essential for preventing and treating such diseases. As a tropical island located in the south of China, Hainan province has abundant rat species. Here, we examined the gut bacterial composition in wild adult rats from Hainan province.
Fresh fecal samples were collected from 162 wild adult rats, including three species (Rattus norvegicus, Leopoldamys edwardsi, and Rattus losea), from nine regions of Hainan province between 2017-2018.
We analyzed the composition of gut microbiota using the 16S rRNA gene amplicon sequencing. We identified 4903 bacterial operational taxonomic units (30 phyla, 175 families, and 498 genera), which vary between samples of different rat species in various habitats at various times of the year. In general, Firmicutes were the most abundant phyla, followed by Bacteroidetes (15.55%), Proteobacteria (6.13%), and Actinobacteria (4.02%). The genus Lactobacillus (20.08%), unidentified_Clostridiales (5.16%), Romboutsia (4.33%), unidentified_Ruminococcaceae (3.83%), Bacteroides (3.66%), Helicobacter (2.40%) and Streptococcus (2.37%) were dominant.
The composition and abundance of the gut microbial communities varied between rat species and locations. This work provides fundamental information to identify microbial communities useful for disease control in Hainan province.
Core Tip: Several researches have investigated the microbial communities in arthropods but few researches have investigated microbial diversity in wild rats’ gut in Hainan province. Here, by 16S rRNA gene amplicon sequencing we compared the gut bacterial communities in fecal samples from 162 wild rats of three species and nine geographic locations in Hainan.
- Citation: Niu LN, Zhang GN, Xuan DD, Lin C, Lu Z, Cao PP, Chen SW, Zhang Y, Cui XJ, Hu SK. Comparative analysis of the gut microbiota of wild adult rats from nine district areas in Hainan, China. World J Gastroenterol 2023; 29(22): 3469-3481
- URL: https://www.wjgnet.com/1007-9327/full/v29/i22/3469.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i22.3469
The laboratory rat has been used as a useful experimental model for biomedical sciences since the last century. Rats were domesticated as early as the Edo period[1]. Gut microbiota is composed of myriad microbes (archaea, bacteria, fungi, and viruses) that interact with each other and with their host, playing important roles in the host’s metabolism, physiology, homeostasis, health, and disease development[2]. There is an increase in studies of the laboratory rat gut microbiota[3-9], but very few on wild rats[10].
Wild rats pose a serious threat to public health because they are reservoirs for a number of zoonotic pathogens[11-13] and frequently and closely interact with people, domestic animals, and other animals.
Only a small number of researchers have looked into the microbial diversity in the guts of wild rats in the province of Hainan, compared to the number of researchers who have looked into the microbial communities in arthropods. Here, by 16S rRNA gene amplicon sequencing we compared the gut bacterial communities in fecal samples from 162 wild rats of three species and nine geographic locations in Hainan.
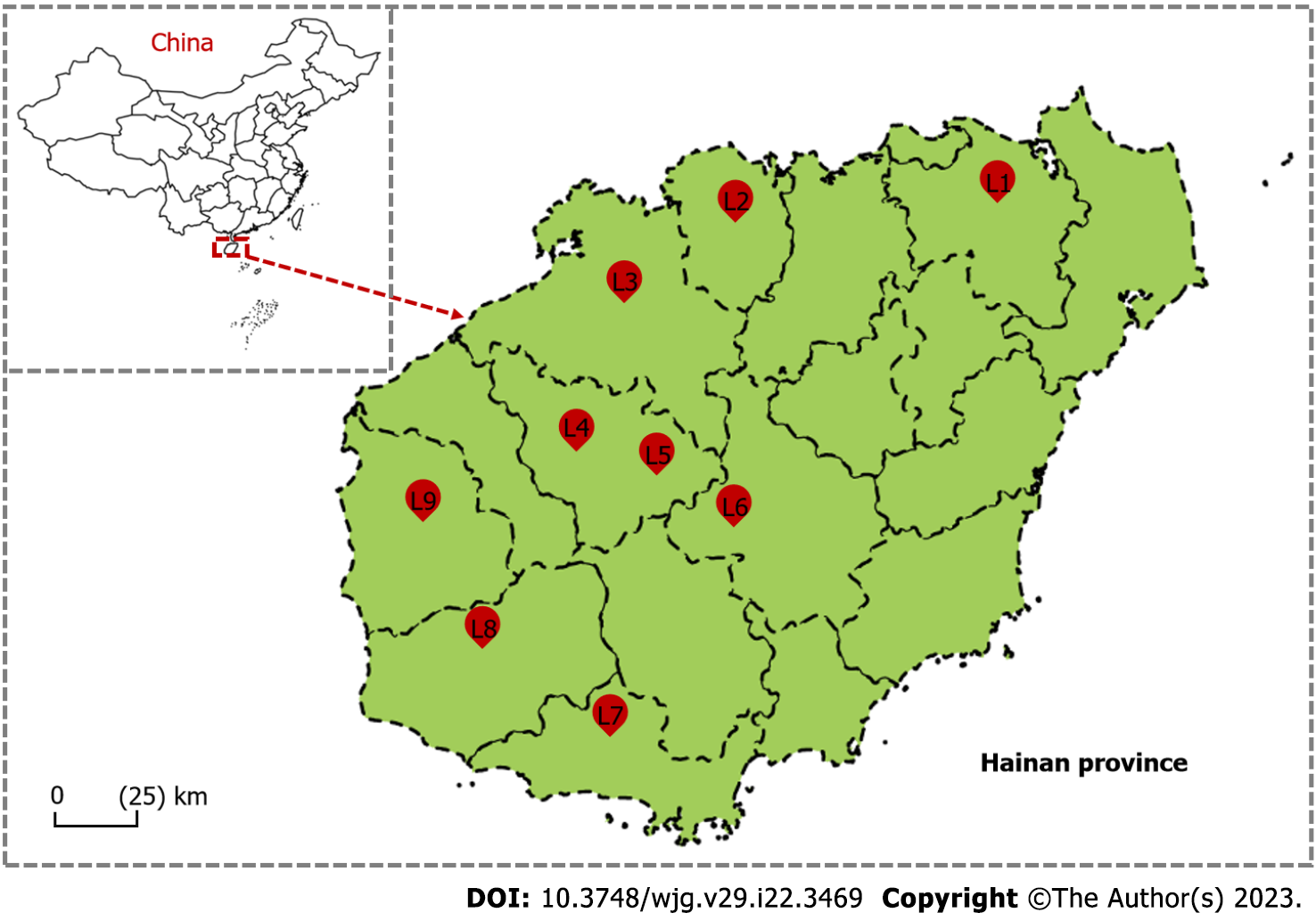
Between December 2017 and November 2018, wild adult rats were collected using mousetraps in rat cages from nine geographic locations in Hainan province (L1-L9: L1 = Yinggeling; L2 = Huan
The SDS method was used for DNA extraction. The PCR assay was performed to identify rat species using the primer 5′-TACCATGAGGACAAATATCATTCTG-3′ and 5′-CCTCCTAGTTTGTTAGGGATTGATCG-3′[14]. Amplification was performed in 25 μL total volume, including DNA template (2 μL), each primer (0.5 μL), 10 × Gene Amp PCR Buffer (2.5 μL), dNTP (1.25 mmol/L) (2 μL), rTaq DNA polymerase (5 U/μL) (0.5 μL) and double distilled water (17 μL). The PCR procedure was as follows: (1) Initial denaturation 95 ℃ (5 min); (2) 35 cycles each of denaturation 94 ℃ (30 s); (3) primer annealing 51 ℃ (30 s); and (4) extension 72 ℃ (30 s). It took 5 min to complete the extension step's 35th cycle. The mouse species was determined through a blast to the NCBI nucleotide database, and the amplification products were identified using Sanger sequencing.
CO2 was used for euthanizing rats. Intestinal tissue was dissected for each rat species, rinsing three times in sterile normal saline. In the meantime, samples of feces were taken from the intestinal tissues. Then, samples were immediately collected in a brain heart infusion medium with glycerol and subsequently stored at -80 ℃ until analysis.
Fecal samples weighing about 50 mg were used for 16S rRNA microbial profiling analyses. Following the manufacturer's instructions, microbial DNA was extracted from the fecal samples using a QIAamp Fast DNA Stool Mini Kit (Qiagen, Germany). The PCR products were examined using electrophoresis. Thermo GeneJET Gel Extraction Kit was used to recover the PCR products after running them on 2% agarose gels in TAE buffer. Until analyses, all of the extracted DNA was kept at -20 °C.
The hypervariable regions V3-V4 of the bacterial 16S rRNA were amplified with primers 5′-CCTAYGGGRBGCASCAG-3′ and 5′-GGACTACNNGGGTATCTAAT-3′. PCR reactions were performed in a 30 μL volume consisting of Phusion® High-Fidelity PCR Master Mix with GC buffer (New England Biolabs) (15 μL), forward and reverse primers (0.2 μM), and template DNA (10 ng). The PCR reaction was done with the following conditions: (1) Initial denaturation at 98 ℃ for 1 min; (2) 30 cycles of denaturation at 98 ℃ for 10 s; (3) annealing at 50 ℃ for 30 s; (4) elongation at 72 ℃ for 30 s; and (5) final extension 72 ℃ for 5 min. Sequencing libraries were made with an Ion Plus Fragment Library Kit (48 rxns) (Thermo Scientific) according to the manufacturer’s protocols. The Qubit 2.0 Fluorometer (Thermo Scientific) was used to evaluate the library's quality. Sequencing was done on the Ion S5TM XL platform last.
Based on the unique barcode, single-end reads were assigned to samples and truncated by cutting off the barcode and primer sequence. The Cutadapt quality control process involved treating the raw reads with quality filtering under specific filtering conditions in order to produce high-quality clean reads[15]. Then, we compared the reads with the Silva database, and reference database[16] using UCHIME Algorithm[17] to detect and remove chimera sequences[18] to obtain clean reads.
Uparse software performed analyses of microbial profiling[19]. Sequences with a 97% or higher similarity to one another were found to the same Operational taxonomic unit (OTU). We screened the representative sequence of each OTU for further annotation. Taxonomic data was annotated at various taxonomic levels using the Silva Database, which uses the Mothur algorithm[16]. We used the MUSCLE software to conduct multiple sequence alignment to study the difference between the dominant species in different groups[20]. We normalized OTUs abundance using the standard sequence number of the sample with the least sequences and summarized the OTU abundance in a table. Subsequently, we analyzed the alpha diversity (α-diversity) and beta diversity (β-diversity) based on this normalized data.
In the complexity of species diversity, we applied α-diversity through 6 indices (Observed-species, Chao1, ACE, Shannon, Simpson, Good’s coverage). Chao1 and ACE were selected as the Chao1 and ACE estimators to determine richness. The Shannon index and Simpson index were used to calculating Shannon diversity and Simpson diversity. Using Good's coverage, Coverage was chosen to describe the depth of the Sequencing.
We applied β-diversity for evaluating the similarity of different samples rather than describing within-sample diversity like α-diversity. We calculated β-diversity on both Weighted and Unweighted unifrac. The dimension of the original variables was reduced using principal component analysis. Principal coordinate analysis (PCoA) was performed to visualize complex, multidimensional data and was displayed to visualize microbial community structure relationships. Firstly, we obtained a distance matrix among samples in weighted or unweighted unifrac. Secondly, we transformed the distance matrix to a new set of orthogonal axes. Finally, the first principal coordinate demonstrated the maximum variation factor, and the second principal coordinate demonstrated the second maximum one, and so on. We performed Adonis analysis to know to what extent location explained variation in microbial composition. For significant difference analysis, either the USEARCH software (https://www.drive5.com/usearch) or R software (version 4.0.3) was used.
Among the 162 wild rats collected by traps from nine district areas which covered almost half of Hainan province (Figure 1), three rat species (Rattus norvegicus, Leopoldamys edwardsi, and Rattus losea) were predominant (Table 1 and Supplementary Table 1). These rat species showed distinct geographic distributions. Rattus norvegicus (brown rat) was mainly found in farmland (Danzhou, Dongfang, Baisha, and Sanya) and residential areas (Haikou); Leopoldamys edwardsi (Edward’s long-tailed rat) was exclusively found in mountain areas in Yinggeling, Huangjingjiaoling, and Jianfengling; Rattus losea (Lesser rice-field rat) was only found from mountains in Lingao area (Table 1).
| Site | Rat species | Number of midgut samples | Sampling season | Trap sites |
| YGL | LE | 32 | 2018.11 | Mountains |
| HJJL | LE | 25 | 2018.11 | Mountains |
| DZ | RN | 12 | 2017.12 | Farmland |
| DF | RN | 12 | 2018.01 | Farmland |
| HK | RN | 13 | 2017.12 | Residential areas |
| JFL | LE | 13 | 2018.06 | Mountains |
| BS | RN | 12 | 2018.01 | Farmland |
| SY | RN | 23 | 2018.05 | Farmland |
| LG | RL | 20 | 2018.11 | Mountains |
A total of 162 fecal samples (one from each rat) were collected from the field-collected wild adult rats and subjected to bacterial profiling analyses, and estimating the bacterial community composition of these samples involved 16S rRNA gene amplicon sequencing. A total of 13326753 raw reads (mean ± SE = 82263.91 ± 8277.45 per sample) were obtained from the 162 samples that were sequenced (Supplementary Table 2). Among them, 12537453 clean reads (mean ± SE = 77391.69 ± 7604.97 per sample) remained for subsequent analyses after quality control processing. These sequences were divided into 4903 OTUs that belonged to 30 phyla, 175 families, and 498 genera (SupplementaryTable
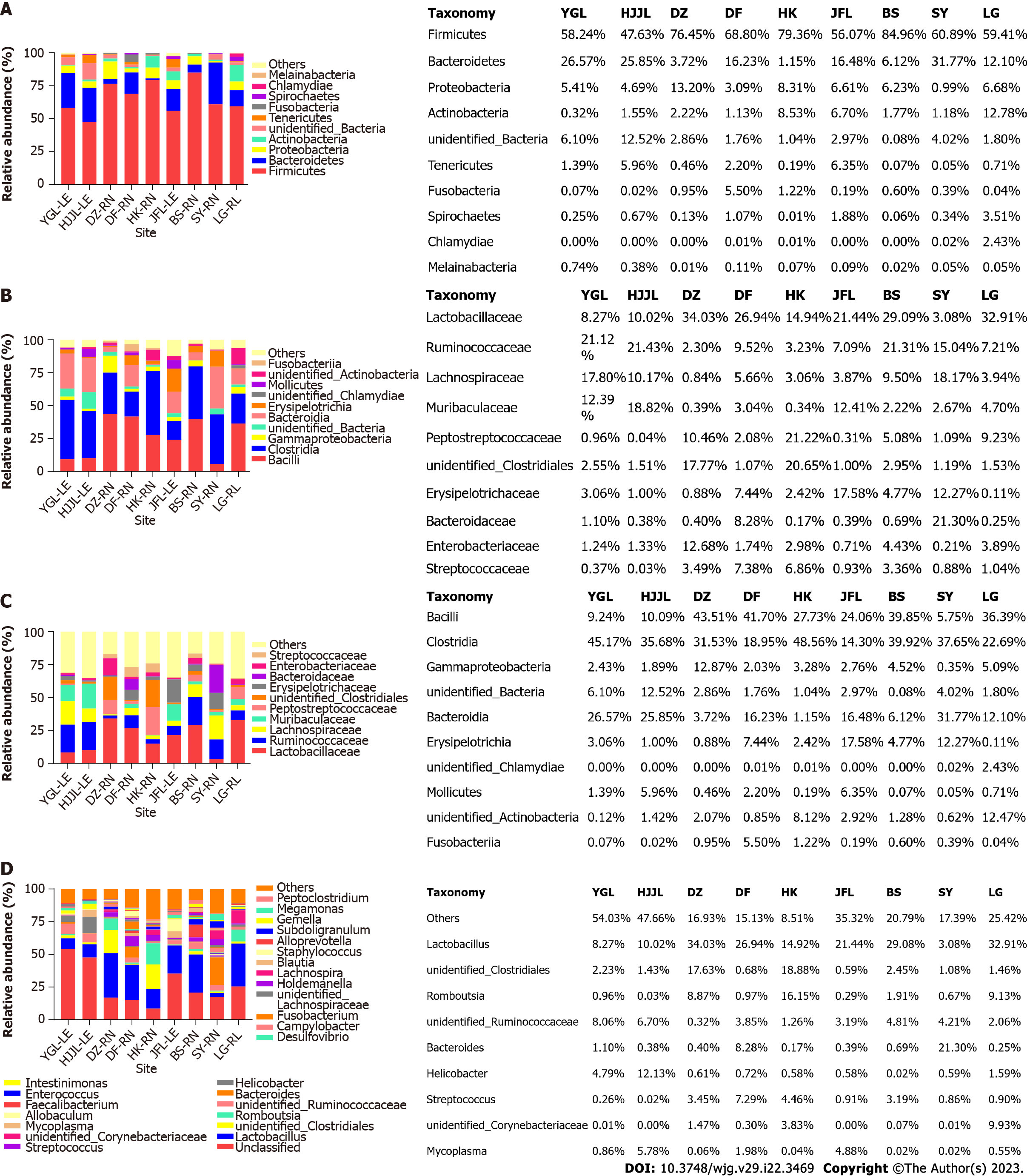
As shown in Figure 2A, sequences originating from Firmicutes (average 65.76%, SupplementaryTable 4) were dominant, followed by Bacteroidetes (15.55%), Proteobacteria (6.13%), Actinobacteria (4.02%), Unclassified bacteria (3.68%), Tenericutes (1.93%), Fusobacteria (1.00%), Spirochaetes (0.88%), Chlamydiae (0.27%), Melainabacteria (0.17%), and so on (detailed in Supplementary Table 4, those < 0.15% were pooled together as “Others” in Figure 2A). Although Bacteroidetes were the second most abundant on average, this was not the case for samples from Danzhou, Haikou, Baisha, and Lingao (Supplementary Table 4).
Following a similar trend, Clostridia (averaged 32.72%) and Bacilli (averaged 26.48%) were the top two classes of bacteria in most samples (Figure 2B), but Clostridia only ranked fourth in samples from the Jianfengling area, and Bacteroidia was the 2nd most abundant in samples from Yinggeling, Huangjingjiaoling and Sanya (Supplementary Table 5).
According to the average value, the most abundant family was Lactobacillaceae (20.08%), followed by Ruminococcaceae (12.03%), Lachnospiraceae (8.11%), Muribaculaceae (6.33%), Peptostreptococcaceae (5.61%), unidentified_Clostridiales (5.58%), Erysipelotrichaceae (5.50%), Bacteroidaceae (3.66%), Enterobacteriaceae (3.25%), and Streptococcaceae (2.71%) (detailed in Supplementary Table 6, those < 2.5% were pooled together as “Others” in Figure 2C). Compared to the profiles for the bacterial phylum and class, the profiles for the actual values were more complex. Lactobacillaceae was the most abundant in samples from five areas (Danzhou, Dongfang, Jianfengling, Baisha, and Lingao), Ruminococcaceae was most abundant in samples from Yinggeling and Huangjingjiaoling, and Peptostreptococcaceae and Bacteroidaceae were the richest in samples from Haikou and Sanya, respectively (SupplementaryTable 6).
As illustrated in Figure 2D and detailed in Supplementary Table 7, the genus with the highest OTU abundance in the feces of rats was those (with relative abundance less than 0.5%) designated as ‘others’ (average 26.80%), followed by Lactobacillus (20.08%), unidentified_Clostridiales (5.16%), Romboutsia (4.33%), and so on. In samples from eight different areas, "Others" or Lactobacillus ranked first and second, but not in samples from the Haikou area, which had the highest concentrations of unidentified Clostridiales and Romboutsia.
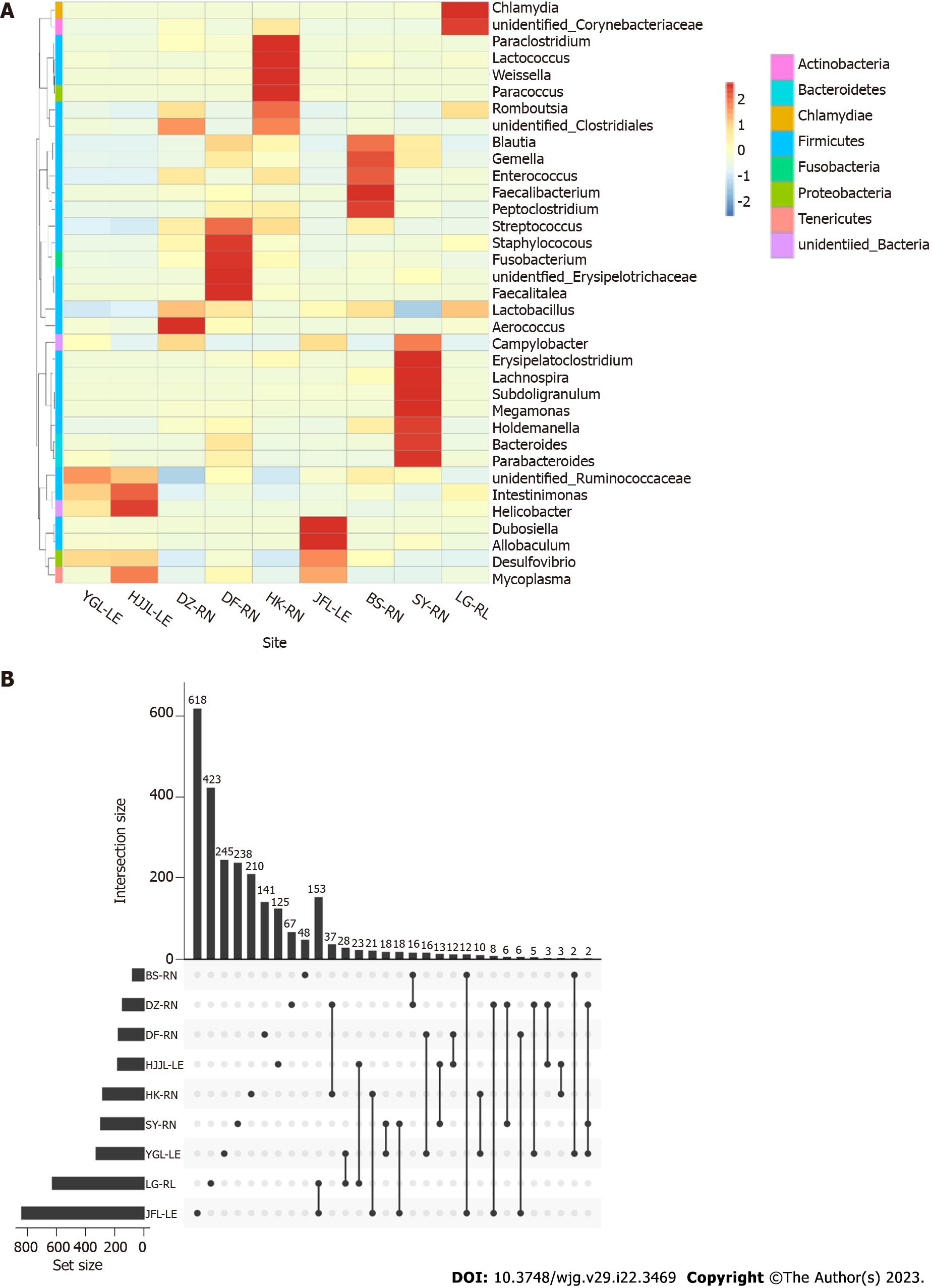
As shown in Figure 3, the heatmap color-coded the different OUT compositions in samples of the nine groups (Figure 3A); all of the samples contained unique bacterial OTUs, according to the upset plot (Figure 3B), but those from the Jianfengling region had the highest numbers of both (618 for unique and 153 for common with the Lingao area).
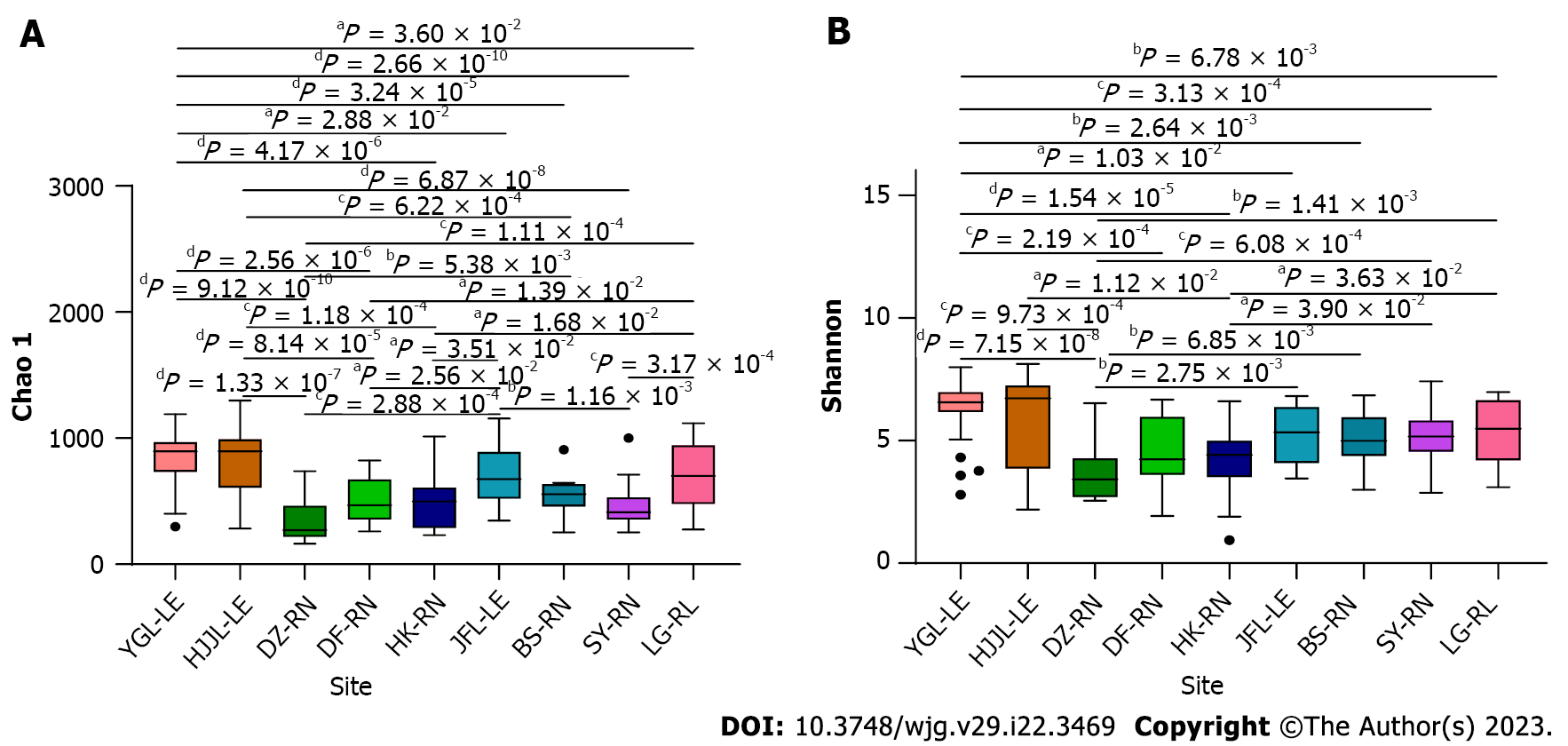
Alpha diversity indices were performed to detect whether bacterial diversity and richness significantly varied among samples of different rat species and geographic locations. The bacterial species richness, represented by Chao1 and ace denoting the number of species rather than the abundance of each species was highest for samples from the Yinggeling area (Table 2). Statistics were conducted for the Chao1 values because the values of both indices (Chao1 and ace) changed in a remarkably similar way across the nine groups. Except for Lingao and Jianfengling, samples from Yinggeling and Huangjingjiaoling, the two highest among all, had significantly more bacteria than the other five samples (Figure 4A). Taking Yinggeling as an example, it was most significantly higher (P < 0.0001) than those from Danzhou, Dongfang, Haikou, Sanya and Baisha, and significant (P < 0.05) compared to Lingao and Jianfengling (Figure 4A). In addition, samples from the Jianfengling area had higher bacterial richness compared to those from Danzhou and Sanya, and those from the Lingao area had higher richness than those from Danzhou and Sanya. There was no significant difference between other groups if not mentioned above.
| Group | Observed_species | Chao1 | Ace | Shannon | Simpson |
| YGL-LE | 662.16 ± 185.6 | 853.76 ± 201.05 | 862.86 ± 191.94 | 6.30 ± 1.19 | 0.95 ± 0.05 |
| HJJL-LE | 654.16 ± 207.22 | 827.96 ± 227.51 | 835.31 ± 217.91 | 5.83 ± 1.88 | 0.88 ± 0.15 |
| DZ-RN | 213.42 ± 128.99 | 340.60 ± 168.92 | 348.04 ± 161.47 | 3.70 ± 1.14 | 0.82 ± 0.10 |
| DF-RN | 369.50 ± 177.23 | 490.55 ± 186.68 | 511.53 ± 179.36 | 4.59 ± 1.39 | 0.87 ± 0.12 |
| HK-RN | 346.85 ± 188.71 | 492.23 ± 226.98 | 511.22 ± 234.13 | 4.19 ± 1.59 | 0.80 ± 0.21 |
| JFL-LE | 533.77 ± 209.11 | 696.02 ± 238.19 | 700.85 ± 227.48 | 5.25 ± 1.17 | 0.89 ± 0.07 |
| BS-RN | 424.17 ± 126.51 | 550.92 ± 164.77 | 574.32 ± 159.53 | 5.04 ± 1.07 | 0.89 ± 0.09 |
| SY-RN | 342.91 ± 129.91 | 458.86 ± 162.56 | 466.06 ± 153.41 | 5.12 ± 1.01 | 0.92 ± 0.06 |
| LG-RL | 532.85 ± 221.85 | 715.35 ± 259.97 | 747.67 ± 259.51 | 5.30 ± 1.31 | 0.90 ± 0.09 |
Species diversity, represented by Shannon diversity and Simpson diversity, indicates species richness and evenness in the community. As the two indices changed accordingly (Table 2), values of the Shannon diversity were used to do the statistics. In comparison to samples from Danzhou (P < 0.0001), Dongfang (P < 0.01), Haikou (P < 0.001), and Sanya (P < 0.05), samples from the Yinggeling area showed significant differences. Although similar in its bacterial composition pattern to those from the Yinggeling area, the species diversity of samples from the Huangjingjiaoling area was less significant with samples from Danzhou (P < 0.001) and Haikou (P < 0.05). In contrast to the insignificant results among sample groups not mentioned above, the species diversity of samples from the Danzhou area was significantly different (P < 0.05) from those from the Lingao area (Figure 4B).
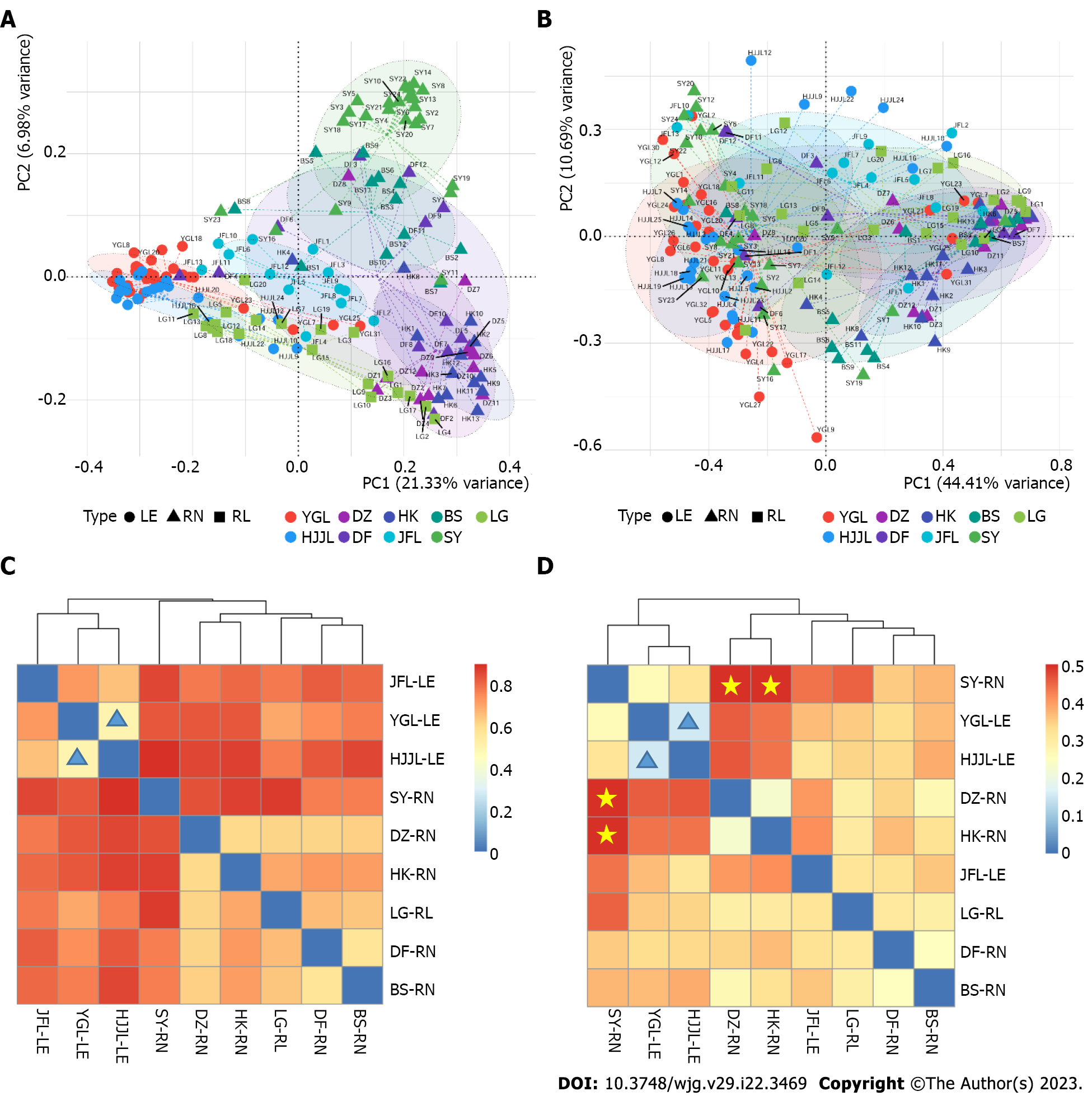
Distance metrics were used to estimate differences between rat species and geographical locations in bacterial community profiles. The results showed a significant difference in microbiota among the three rat species and nine locations based on unweighted unifrac and weighted unifrac distances. When analyzing the fecal microbiota from various study sites, we used PCoA plots to visualize its composition. Using Unweighted Unifrac distance, two PCoA coordinates percent variation explained PCo1 (21.33%) and PCo2 (6.98%) of the total variation, respectively. We observed significant differences in fecal microbial β-diversity among communities from different locations, including Leopoldamys edwardsi from Yinggeling, Huangjingjiaoling, and Jianfengling; Rattus norvegicus from Danzhou, Baisha, and Sanya. Rattus norvegicus from the Dongfang area does not cluster tightly. There were no observable differences between Rattus norvegicus from Danzhou and Haikou (Figure 5A). Two PCoA coordinates based on Weighted Unifrac distance percent variation revealed PCo1 (44.41%) and PCo2 (10.69%), respectively (Figure 5B). These differences were mainly due to the geographical location, which included a plethora of variables (rat species, habitat, season, diet, etc.).
Heatmaps based on Bray-Curtis distances and Weighted unifrac distances, respectively, showed the diversity and similarity of microbial communities (Figure 5C and D). As shown in Figure 5C, the most similar (0.508, the lowest in Supplementary Table 8) intestinal bacterial community structure was between samples from Jianfengling and Huangjingjiaoling (highlighted with blue triangles in Figure 5C), followed by that between Baisha and Dongfang (0.57), and those of Danzhou area (0.61-0.63, Supplementary Table 8) with Baisha, Dongfang, Haikou, and Lingao (Figure 5C). Conversely, samples from the Sanya area were most dissimilar to those from Huangjingjiaoling (0.91), Lingao (0.89), and Haikou (0.88). As shown in Figure 5D, the lowest difference (0.16, Supplementary Table 9) was observed between samples from Yinggeling and Huangjingjiaoling (highlighted by the blue triangles), suggesting the bacteria in these two groups of samples were evolutionary more similar. Conversely, the bacteria in the samples from the Sanya area were evolutionary most distant (highlighted by the stars in Figure 5D) to those from Danzhou and Haikou (0.51 and 0.57, respectively; Supplementary Table 9).
Through the same procedure from sample collection and processing to sequencing, the gut bacterial communities differed at all levels (phylum, class, family, and genus) among fecal samples of wild rats from nine geographical locations, three different habitats, and rat species, and two seasons (winter vs summer). According to scant research, diet uncontrollable for wild rats is a significant determinant of gut bacterial composition[21-23], which differs for rats living in diverse habitats in every location at all seasons. There were quite a few striking findings from our study. As the only representative samples of residential area (Haikou) rats, their gut bacterial composition was different at the family and genus level, richest with Peptostreptococcaceae and unidentified_Clostridiales at the family level, and with unidentified_Clostridiales and Romboutsia at the genus level. Although collected from the same rat species (Rattus norvegicus) during the same winter season, the bacterial profiles of the four groups of samples from different areas (Baisha, Dongfang, Danzhou, and Haikou) were distinctly different at the class, family, and genus level, revealing the complexity of factors (e.g., habitat, farmland vs residential area) behind the bacterial community. The general bacterial profile of samples taken from the same species of rat (Leopoldamys edwardsi) in the summer in one mountain area (Yinggeling) varied from samples taken in the winter from the other two mountain areas (Huangjingjiaoling and Jianfengling), indicating that season (and thus a variety of factors) played a significant role in determining their gut bacterial composition.
Several studies report that Firmicutes, Bacteroidetes, and Proteobacteria are the most abundant phyla, and Lactobacillus is the predominant genus in fecal samples from both wild (Rattus norvegicus) and laboratory (Sprague-Dawley) rats[7,9], a pattern similar to our results. Fusobacteria was interestingly the fourth most prevalent phylum in[10], which is most consistent with findings from one of our nine groups (Dongfang) (Figure 2B). Although of the same species (Rattus norvegicus), sample size (12), and similar climate, rats from the Dongfang area of this study were trapped in January from farmland on an isolated island contrasting to all year round (one mouse per month) from a residential area in an inland metropolitan city[10].
Although we identified some potential bacterial pathogens by their species names and reputation in the samples (Supplementary Table 3), a profile distinct from the results of other studies[11,12], they still need to be tested for virulence and pathogenesis. Because wild rats carry some bacteria (Bacillus anthracis, Vibrio cholerae, and Yersinia pestis)[11,12] and this study] can cause a serious outbreak or even a pandemic, it would be prudent to start a regular survey program at some strategic points to monitor the distribution of sentinel rodents and the target pathogens the animals may transmit.
The ideal comparison should only have one factor, but this seems like an impossible task for our current research. Admittedly our study had some limitations, to name a few: (1) As some variables, like sex, age, and so on, were difficult, if not impossible, to control as a baseline investigation; (2) despite the interesting findings (having the highest Corynebacteriaceae in all samples) of the gut bacterial composition in Rattus losea, it was from only one area (Lingao); (3) the summer samples were collected from two different rat species (Rattus norvegicus vs Leopoldamys edwardsi,) in two different habitats (farmland vs mountain) from only one area each, making the results less conclusive and comparable; and (4) samples from residential rats were only from Haikou area, making some of the results less comparable.
The study revealed the structure of gut bacterial communities by sampling wild rats’ feces. In general, the gut microbiota was different in both composition and abundance in samples from the nine areas of Hainan of three wild rat species, three habitats (mountain, farmland, and residential area), and two seasons (summer and winter). This study provides fundamental information for identifying microbial communities that will be useful for disease control in Hainan Province.
Wild rats are potential reservoirs for zoonotic infectious agents which can be transmitted to and cause diseases in humans.
As a tropical island locating in the south of China, Hainan province has abundant rat species.
To better understand the composition of gut bacterial communities in rats is essential for preventing and treating such diseases. Here, we examined the gut bacterial composition in wild adult rats from Hainan province.
Fresh faecal samples were collected from 162 wild adult rats, including three species (Rattus norvegicus, Leopoldamys edwardsi, and Rattus losea), from nine regions of Hainan province between 2017-2018.
We analyzed the composition of gut microbiota using the 16S rRNA gene amplicon sequencing. We identified 4,903 bacterial operational taxonomic units (30 phyla, 175 families, and 498 genera), which is different among samples of different rat species in different habitat during different season. In general, Firmicutes were the most abundant phyla, followed by Bacteroidetes (15.55%), Proteobacteria (6.13%) and Actinobacteria (4.02%). The genus Lactobacillus (20.08%), unidentified_Clostridiales (5.16%), Romboutsia (4.33%), unidentified_Ruminococcaceae (3.83%), Bacteroides (3.66%), Helicobacter (2.40%) and Streptococcus (2.37%) were dominant.
Among locations for rat species, the gut microbial communities were different in composition and abundance. This work provides fundamental information to identify microbial communities useful for disease control in Hainan province.
The study revealed the structure of gut bacterial communities by sampling wild rats’ feces. In general, the gut microbiota was different in both composition and abundance in samples from the nine areas of Hainan of three wild rat species, three habitats (mountain, farmland and residential area), and of two seasons (summer and winter). This work provides fundamental information to identify microbial communities which will be useful for disease control in Hainan province.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Calinescu AM, Switzerland; Landberg G, Sweden S-Editor: Fan JR L-Editor: A P-Editor: Fan JR
| 1. | Hulme-Beaman A, Orton D, Cucchi T. The origins of the domesticate brown rat (Rattus norvegicus) and its pathways to domestication. Anim Front. 2021;11:78-86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 2. | Chen Y, Zhou J, Wang L. Role and Mechanism of Gut Microbiota in Human Disease. Front Cell Infect Microbiol. 2021;11:625913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 293] [Article Influence: 73.3] [Reference Citation Analysis (0)] |
| 3. | Brooks SP, McAllister M, Sandoz M, Kalmokoff ML. Culture-independent phylogenetic analysis of the faecal flora of the rat. Can J Microbiol. 2003;49:589-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 58] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 4. | Manichanh C, Reeder J, Gibert P, Varela E, Llopis M, Antolin M, Guigo R, Knight R, Guarner F. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res. 2010;20:1411-1419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 242] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 5. | Ferrario C, Statello R, Carnevali L, Mancabelli L, Milani C, Mangifesta M, Duranti S, Lugli GA, Jimenez B, Lodge S, Viappiani A, Alessandri G, Dall'Asta M, Del Rio D, Sgoifo A, van Sinderen D, Ventura M, Turroni F. How to Feed the Mammalian Gut Microbiota: Bacterial and Metabolic Modulation by Dietary Fibers. Front Microbiol. 2017;8:1749. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 6. | Flemer B, Gaci N, Borrel G, Sanderson IR, Chaudhary PP, Tottey W, O'Toole PW, Brugère JF. Fecal microbiota variation across the lifespan of the healthy laboratory rat. Gut Microbes. 2017;8:428-439. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 81] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 7. | Li D, Chen H, Mao B, Yang Q, Zhao J, Gu Z, Zhang H, Chen YQ, Chen W. Microbial Biogeography and Core Microbiota of the Rat Digestive Tract. Sci Rep. 2017;8:45840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 115] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 8. | Nagpal R, Wang S, Solberg Woods LC, Seshie O, Chung ST, Shively CA, Register TC, Craft S, McClain DA, Yadav H. Comparative Microbiome Signatures and Short-Chain Fatty Acids in Mouse, Rat, Non-human Primate, and Human Feces. Front Microbiol. 2018;9:2897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 188] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 9. | Pan H, Guo R, Zhu J, Wang Q, Ju Y, Xie Y, Zheng Y, Wang Z, Li T, Liu Z, Lu L, Li F, Tong B, Xiao L, Xu X, Li R, Yuan Z, Yang H, Wang J, Kristiansen K, Jia H, Liu L. A gene catalogue of the Sprague-Dawley rat gut metagenome. Gigascience. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 10. | He WQ, Xiong YQ, Ge J, Chen YX, Chen XJ, Zhong XS, Ou ZJ, Gao YH, Cheng MJ, Mo Y, Wen YQ, Qiu M, Huo ST, Chen SW, Zheng XY, He H, Li YZ, You FF, Zhang MY, Chen Q. Composition of gut and oropharynx bacterial communities in Rattus norvegicus and Suncus murinus in China. BMC Vet Res. 2020;16:413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Himsworth CG, Parsons KL, Jardine C, Patrick DM. Rats, cities, people, and pathogens: a systematic review and narrative synthesis of literature regarding the ecology of rat-associated zoonoses in urban centers. Vector Borne Zoonotic Dis. 2013;13:349-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 244] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 12. | Easterbrook JD, Kaplan JB, Vanasco NB, Reeves WK, Purcell RH, Kosoy MY, Glass GE, Watson J, Klein SL. A survey of zoonotic pathogens carried by Norway rats in Baltimore, Maryland, USA. Epidemiol Infect. 2007;135:1192-1199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 136] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 13. | Zhao W, Zhou H, Yang L, Ma T, Zhou J, Liu H, Lu G, Huang H. Prevalence, genetic diversity and implications for public health of Enterocytozoon bieneusi in various rodents from Hainan Province, China. Parasit Vectors. 2020;13:438. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 14. |
Verma SK, Singh L, Novel universal primers establish identity of an enormous number of animal species for forensic application.
|
| 15. | Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17. [RCA] [DOI] [Full Text] [Cited by in Crossref: 13233] [Cited by in RCA: 18855] [Article Influence: 1346.8] [Reference Citation Analysis (0)] |
| 16. | Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590-D596. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15104] [Cited by in RCA: 17920] [Article Influence: 1493.3] [Reference Citation Analysis (0)] |
| 17. | Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194-2200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9953] [Cited by in RCA: 9715] [Article Influence: 693.9] [Reference Citation Analysis (0)] |
| 18. | Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ; Human Microbiome Consortium, Petrosino JF, Knight R, Birren BW. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494-504. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2862] [Cited by in RCA: 2383] [Article Influence: 170.2] [Reference Citation Analysis (0)] |
| 19. | Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996-998. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8951] [Cited by in RCA: 9922] [Article Influence: 826.8] [Reference Citation Analysis (0)] |
| 20. | Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792-1797. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28603] [Cited by in RCA: 29875] [Article Influence: 1422.6] [Reference Citation Analysis (0)] |
| 21. | Queipo-Ortuño MI, Seoane LM, Murri M, Pardo M, Gomez-Zumaquero JM, Cardona F, Casanueva F, Tinahones FJ. Gut microbiota composition in male rat models under different nutritional status and physical activity and its association with serum leptin and ghrelin levels. PLoS One. 2013;8:e65465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 283] [Cited by in RCA: 354] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 22. | Liu X, Blouin JM, Santacruz A, Lan A, Andriamihaja M, Wilkanowicz S, Benetti PH, Tomé D, Sanz Y, Blachier F, Davila AM. High-protein diet modifies colonic microbiota and luminal environment but not colonocyte metabolism in the rat model: the increased luminal bulk connection. Am J Physiol Gastrointest Liver Physiol. 2014;307:G459-G470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 23. | Qi X, Xu W, Guo M, Chen S, Liu Y, He X, Huang K. Rice- or pork-based diets with similar calorie and content result in different rat gut microbiota. Int J Food Sci Nutr. 2017;68:829-839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |