Published online Jun 14, 2023. doi: 10.3748/wjg.v29.i22.3385
Peer-review started: January 25, 2023
First decision: March 15, 2023
Revised: March 23, 2023
Accepted: May 4, 2023
Article in press: May 4, 2023
Published online: June 14, 2023
Processing time: 132 Days and 14.8 Hours
Clostridioides difficile (formerly called Clostridium difficile, C. difficile) infection (CDI) is listed as an urgent threat on the 2019 antibiotic resistance threats report in the United States by the Centers for Disease Control and Prevention. Early detection and appropriate disease management appear to be essential. Meanwhile, although the majority of cases are hospital-acquired CDI, community-acquired CDI cases are also on the rise, and this vulnerability is not limited to immunocompromised patients. Gastrointestinal treatments and/or gastrointestinal tract surgeries may be required for patients diagnosed with digestive diseases. Such treatments could suppress or interfere with the patient’s immune system and disrupt gut flora homeostasis, creating a suitable microecosystem for C. difficile overgrowth. Currently, stool-based non-invasive screening is the first-line approach to CDI diagnosis, but the accuracy is varied due to different clinical microbiology detection methods; therefore, improving reliability is clearly required. In this review, we briefly summarised the life cycle and toxicity of C. difficile, and we examined existing diagnostic approaches with an emphasis on novel biomarkers such as microRNAs. These biomarkers can be easily detected through non-invasive liquid biopsy and can yield crucial information about ongoing pathological phenomena, particularly in CDI.
Core Tip:Clostridioides difficile infection (CDI) is listed as an urgent threat, and early detection and appropriate disease management from hospital-acquired or community-acquired CDI appear to be essential. Currently, stool-based non-invasive screening is the first-line approach to CDI diagnosis, but the accuracy is varied due to different clinical microbiology detection methods. Therefore, improving reliability is clearly required. This review summarised the life cycle and toxicity of Clostridioides difficile and examined existing diagnostic potentials on microRNA as novel biomarkers. MicroRNAs can be easily detected through non-invasive liquid biopsy and can yield crucial information about ongoing pathological phenomena, particularly in CDI.
- Citation: Bocchetti M, Ferraro MG, Melisi F, Grisolia P, Scrima M, Cossu AM, Yau TO. Overview of current detection methods and microRNA potential in Clostridioides difficile infection screening. World J Gastroenterol 2023; 29(22): 3385-3399
- URL: https://www.wjgnet.com/1007-9327/full/v29/i22/3385.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i22.3385
Clostridioides difficile (formerly called Clostridium difficile, C. difficile) infection (CDI) was classified as an urgent threat in the 2019 antibiotic resistance report in the United States by the Centers for Disease Control and Prevention. Early detection and disease management are urgently needed to reduce public health spending[1]. In addition, of those infected with a medically-related CDI, 1 in 11 people over the age of 65 dies within a month[2]. The acute inflammation caused by the bacteria triggers cytokine production, neutrophil recruitment, mucosal permeability and fluid secretion, leading to colonic tissue damage, nasal diarrhoea and colitis[3,4], which is similar to inflammatory bowel disease[5].
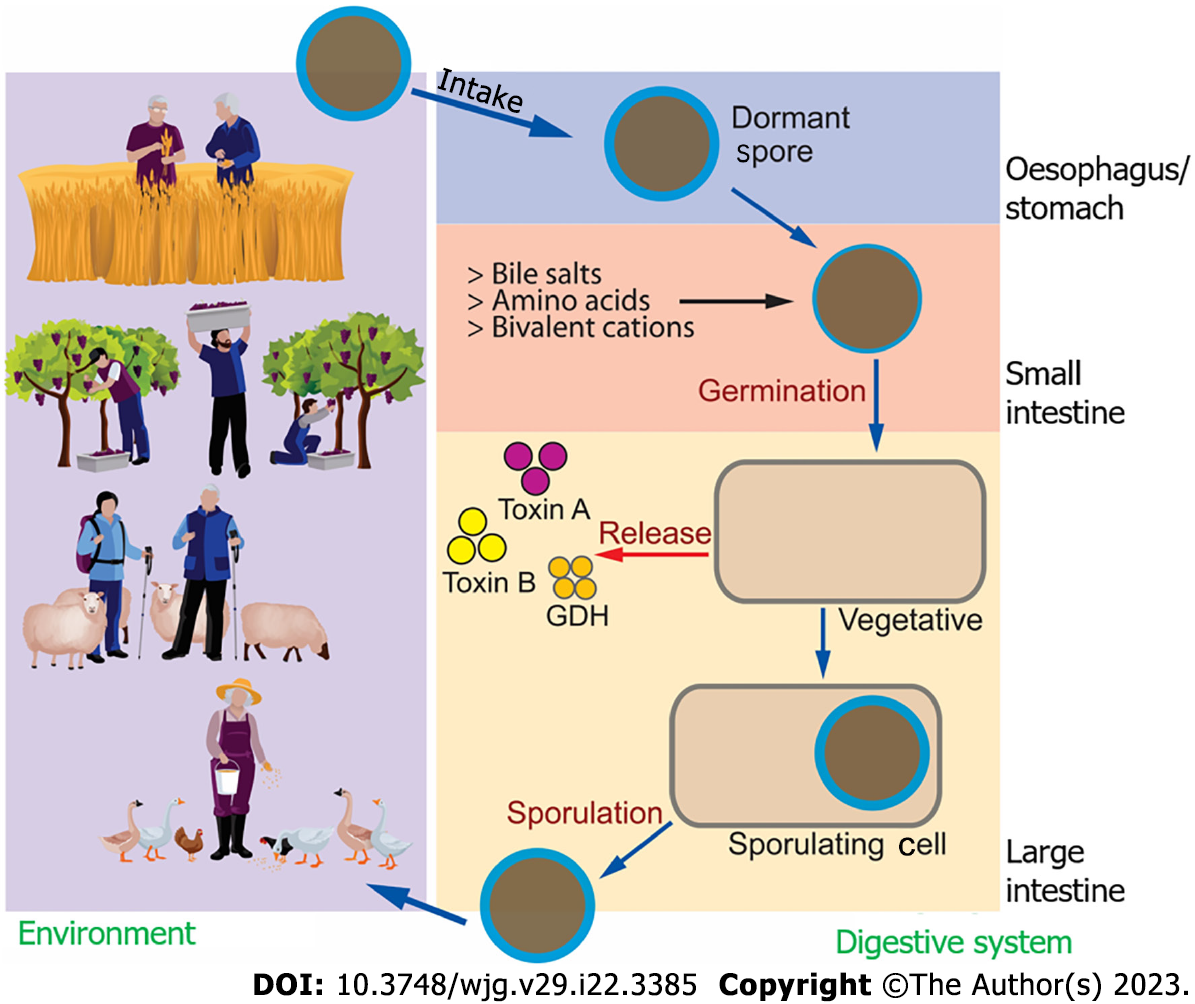
Presently, although most cases are hospital-acquired CDI, community-acquired CDI cases are rising in frequency[6]. Exposure to C. difficile in the community might come from various factors, including pets, water, soil, livestock, farms, food processing and production[7]. Previous studies focused on the hospitalisation length and indicated that environmental exposure to animals would be associated with a greater possibility of C. difficile colonisation[8]. A population-based study in India found that the composition of the gut microbiota was primarily associated with several geographical factors rather than body mass index and that these changes extended to circulating immunometabolic profiles such as serum N-glycans, immunoglobulins and short-chain fatty acid profiles[9]. These factors may also affect the infection rate.
Once patients have been diagnosed with digestive diseases, gastrointestinal treatments, including gastric-acid suppressing agents, broad-spectrum antibiotics, chemotherapy and/or gastrointestinal tract surgery may be required[10,11]. Such therapies could suppress or interfere with the patient’s immune system and disrupt gut flora homeostasis, creating a suitable microecosystem for C. difficile overgrowth[12]. Antibiotics such as metronidazole, vancomycin and fidaxomicin have been approved for treatment of patients with CDI, while C. difficile strains resistant to various antibiotics have been reported that do not respond to the treatments[13-15]. Faecal microbiota transplantation (FMT) is an alternative treatment strategy for CDI patients; however, it is still in clinical trials because of the treatment safety concern[16].
To date, there are several diagnostic methods for CDI in medical laboratories. However, due to technical limitations and the difficulty of distinguishing symptomatic infections from asymptomatic C. difficile colonisation, the accuracy and turnaround time of the test varies[17]. As a result, searching for better tools may increase the accuracy of the detection. In this light, microRNA (miRNA) expression profiling can be helpful for prognosis and diagnosis.
This article reviewed this proposition by briefly presenting the C. difficile life cycle and describing the toxins produced. We then described the current laboratory-based diagnostic tools for CDI patient screening. We discussed the potential use of miRNA to monitor and improve the management of CDI patients.
C. difficile is a Gram-positive, anaerobic bacterium belonging to the phylum Firmicutes. There are three main stages that define the C. difficile life cycle: spore formation; germination; and growth (vegetative). At the spore stage (dormant phase), it is typically harmless in a balanced gut flora microenvironment. C. difficile can resist oxygen, heat and many other environmental insults, including ethanol-based disinfectants (Figure 1)[18].
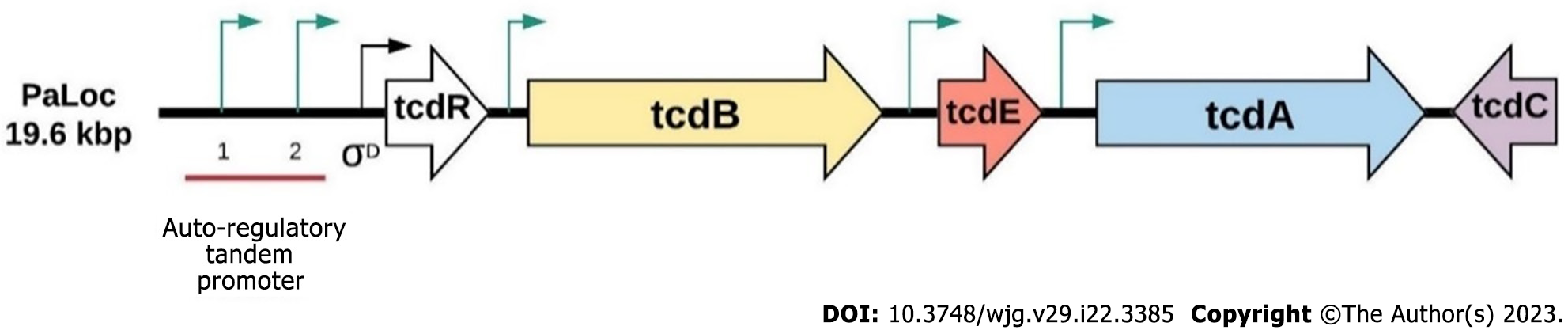
Once set in a stable environment, spores germinate rapidly and produce two major toxins, toxin A and toxin B encoded by TcdA and TcdB, which are located at the 19.6 kbp long pathogenicity locus region (Figure 2)[19]. Toxin A and toxin B trigger cytosol translocation of target host cells and inactivate small GTP-binding proteins (such as CDC42, Rho and Rac) through monoglucosylation, leading to actin condensation, cytoskeleton disintegration, cell rounding and apoptosis[20]. tcdR is an RNA polymerase sigma factor that initiates tcdA and tcdB translation via its two tandem promoters[21,22] and is involved in the final stages of flagellar assembly[23]. Some C. difficile strains, such as ribotype 027 and ribotype 078 are able to produce C. difficile transferase toxin, an actin-specific ADP-ribosyltransferase homologous to iota-toxin from Clostridium perfringens and Clostridium spiroforme toxin, and potentially enhances C. difficile virulence and disease severity[24,25].
Similar to colorectal cancer screening, CDI can be detected by examining the colon through flexible sigmoidoscopy or colonoscopy to look for pseudomembranes and inflamed areas. X-ray abdominal imaging or computerised tomography scan can also be applied on a case-by-case basis. In addition to the invasive approaches, the cost effective, non-invasive faecal-based screening is the first-line approach for CDI diagnosis. Still, it varies widely due to different clinical microbiology methods and their different accuracy and variance[26,27]. These methods include cell cytotoxicity neutralisation assay, toxigenic culture (TC), enzyme immunoassay (EIAs) [including toxins and glutamate dehydrogenase (GDH)] and nucleic acid amplification test (NAAT) and each of the methods has advantages and disadvantages in terms of turn-around time and the screening performance (Table 1).
| Test
| Abbreviation | Sensitivity | Specificity | Turn-around time | Target substance |
| Culture-based | |||||
| Cell cytotoxicity neutralisation assay | CCTN | High | High | < 24 h | Toxins |
| Toxigenic culture | TC | High | Low1 | > 3 d | C. difficile vegetative cells or spores |
| DNA-based | |||||
| Nucleic acid amplification test2 | NAAT | High | Low/moderate | < 4 h | Toxin genes |
| Protein-based | |||||
| Glutamate dehydrogenase | GDH-EIA | High | Low1 | < 2 h | C. difficile antigens |
| Toxin A and B enzyme immunoassays | EIA | Low | Moderate | < 2 h | Toxins |
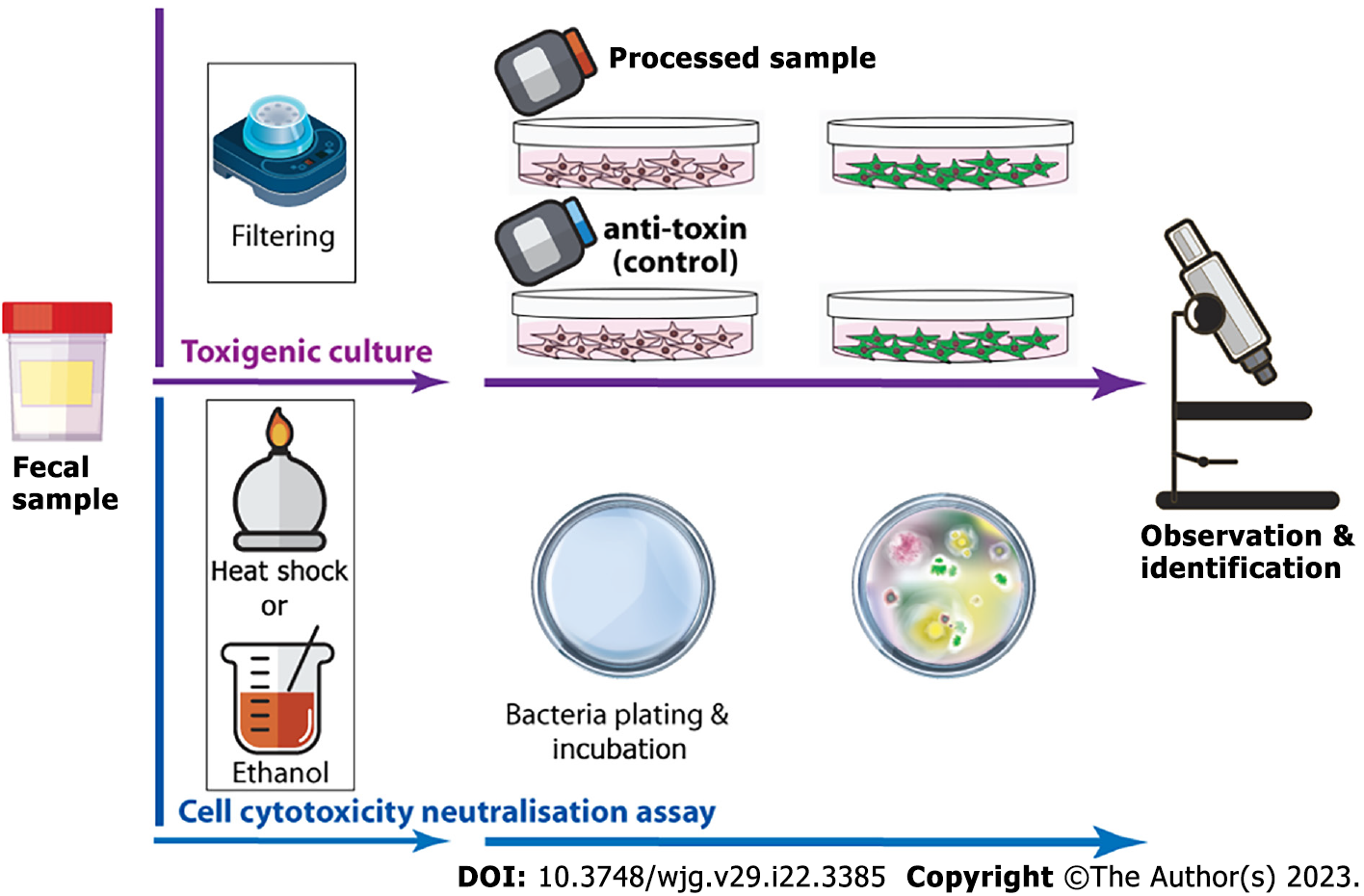
The cell cytotoxicity neutralisation assay is the first-line faecal-based diagnostic test for CDI[28,29]. It requires multiple steps in order to isolate C. difficile toxins from faeces and at least 24 h of cell culture (mainly human fibroblasts). The cytopathic effect is characterised by rounding and morphologic changes of the cultured cells[29,30], and the phenomena must also be reverted by C. difficile or Clostridium sordellii anti-toxin used as a control to prove that the cytopathic effect is not related to nonspecific substances in the faeces. In contrast, toxigenic culture requires faeces inoculation to selective cycloserine-cefoxitin-fructose, chromogenic or similar agar plates for a specific incubation period; suspicious colonies are selected for further bacteria culture and/or C. difficile toxin test to confirm the finding (Figure 3)[31-33]. It is important to remember that there is no standard to eliminate or reduce nonspecific bacteria colonisation from C. difficile culture. Heat shock and alcohol shock are the traditional preculture approaches, while a culture media containing bacteria “suppressors” such as antibiotics may also be applied[34-38]. Both culture-based assays are time-consuming, labour-intensive and require a certain level of laboratory skill. As such, it is unlikely to be used for first-line clinical screening and is commonly used as the reference method for research and outbreak investigations, even though it is considered the gold standard[31,32,38].
The nucleic acid amplification test is a quantitative real-time (qRT)-PCR-based diagnostic assay able to rapidly detect C. difficile toxin genes such as tcdA, tcdB, tcdBv, cdt and/or tcdCΔ117 at the DNA level. There is a wide range of United States Food and Drug Administration-approved detection assays, from C. difficile specific point-of-care testing to high-volume, high-throughput multiple gastrointestinal pathogens laboratory tests. The majority of PCR detection approaches for C. difficile screening are probe-based qRT-PCR (Table 2). The use of probes increases amplification specificity during the PCR cycle since the additional sequence of the probe is specific for and binds to the C. difficile DNA sequence. Additionally, it is possible to perform multiplex qRT-PCR by using different fluorescent dyes[39]. The nucleic acid-based C. difficile toxin screening has over ten times higher sensitivity than a cytotoxin assay[40,41], but the screening specificity is relatively low due to high false positive cases from asymptomatic infection. As a result, optimisation is required, especially on PCR threshold cycle settings[42,43]. In addition, patient preselection based on clinical symptoms and intestinal inflammation biomarkers (i.e. faecal calprotectin, lactoferrin and cytokines) appears to be necessary to reduce the risk of a false positive[27,43-47].
| Assay name | Developer | Target | Method | Sensitivity/specificity (%) |
| AmpliVue C. difficile assay[70] | Quidel | tcdA | Isothermal nucleic acid amplification | 93.6/94.1 |
| ARIES C. difficile assay[71,72] | Luminex | tcdA and tcdB | qRT-PCR | 90-98/92-98 |
| ARTUS C. difficile QS-RGQ MDX Kit[73] | QIAGEN, GMBH | tcdA and tcdB (+ tcdBv) | qRT-PCR | 100/90-100 |
| BD Diagnostics BD MAX C. diff assay[71,74-77] | GeneOhm Sciences | tcdB | TaqMan probe-based qRT-PCR | 86-98/89-100 |
| BD GeneOhm C. diff assay[78-82] | BD Diagnostics/GeneOhm Sciences | tcdB | Beacon probe-based PCR | 91-95/96-100 |
| Cobas C. diff Nucleic Acid Test For Use On The Cobas Liat System[83,84] | Roche | tcdB | TaqMan probe-based qRT-PCR | 93/99 |
| GenePOC C. diff[85] | GenePOC | tcdB of toxigenic C | TaqMan probe-based qRT-PCR | 81/97 |
| ICEPlex C. diff Kit1[86] | PrimeraDx | tcdB | qRT-PCR + capillary electrophoresis-based detection | 90/97 |
| Illumigene C. diff DNA Amplification assay[77] | Meridian Bioscience | tcdA | Loop-mediated isothermal DNA amplification | 82-100/94-100 |
| IMDx C. difficile for Abbott m2000[76,77] | Intelligent Medical Devices | tcdA and tcdB (+ tcdBv) | Probe-based qRT-PCR | 62-84/94-99 |
| Portrait Toxigenic C. difficile assay[87,88] | Great Basin Scientific | tcdB | Primer-mediated helicase-dependent amplification + chip-based detection | 98.2/92.8 |
| ProGastro Cd assay[81,89] | Prodesse | tcdB | Probe-based qRT-PCR | 77-100/94-99 |
| Quidel Molecular Direct C. difficile assay[70,90] | Quidel | tcdA and tcdB | TaqMan probe-based qRT-PCR | 82-96/97-100 |
| Simplexa C. difficile Universal Direct assay[70,91] | Focus Diagnostics | tcdB | qRT-PCR + bifunctional fluorescent primer-probes | 87-98/99-100 |
| Solana C. difficile assay1[92] | Quidel | tcdA | Helicase-dependent amplification | 93/99 |
| Verigene C. difficile Nucleic acid Test[91,93,94] | Nanosphere | tcdA, tcdB, cdt and tcdCΔ117 | PCR + nanoparticle-based array | 94-96/96-98 |
| X/Pert C. difficile/Epi[71,77,95,96] | Cepheid | tcdA, tcdB, cdt and tcdCΔ117 | TaqMan probe-based qRT-PCR | 90-100/93-99 |
EIAs utilise antibodies to detect the presence of antigens. It is still relatively common to detect toxin A, toxin B and/or GDH in faeces for CDI. There are several types of EIAs available for CDI, including microplates (enzyme-linked immunosorbent assay), membrane EIA, chemiluminescence immunoassay, enzyme-linked fluorescent assay and chromatographic immunoassay (Table 3). A meta-analysis published in 2016 evaluated the major commercially available C. difficile diagnosis assays compared to the gold standards. The pooled sensitivities were 83% [95% confidence interval (CI): 76%-88%] and 57% (95%CI: 51%-63%) compared to cell culture cytotoxicity assay and toxigenic bacterial culture, respectively, at a specificity of 99%[48]. This may be because these EIAs were developed in the early 21st century and are less sensitive to low toxin(s) levels, and immunocompromised CDI patients have a lower concentration trigger point[49]. This meta-analysis also indicated that GDH detection had a range of 94% (95%CI: 86%-97%) for sensitivity and 96% (95%CI: 92%-98%) for specificity compared to toxigenic bacterial culture and cell culture cytotoxicity assay together[48]. However, a few studies reported sensitivities below 90% for GDH assays[38,50,51]. Due to the known limitations and relatively poor accuracy, stand-alone toxin or GDH EIA tests are not recommended by professional medical societies, including the European Society of Clinical Microbiology and Infectious Diseases and the Infectious Diseases Society of America[27,48,52].
| Methods/assays | Target(s) | Developer | Cell cytotoxicity assay | Toxigenic culture | ||
| Sen, % | Spe, % | Sen, % | Spe, % | |||
| Microwell enzyme immunoassay (enzyme-linked immunosorbent assay) | ||||||
| GA C. diff Antigen[97] | Toxin A + B | The Binding Site | 76.8 | 90.0 | 68.8 | 91.4 |
| Premier toxin A+B[97-106] | Toxin A + B | Meridian Bioscience | 58-99 | 94-100 | 40-86 | 91-100 |
| Ridascreen toxin A/B[97,107,108] | Toxin A + B | Ridascreen | 57-67 | 95-97 | 52-60 | 96-98 |
| ProSpecT toxin A/B[97,109] | Toxin A + B | Remel/Oxoid (Thermo Scientific) | 90-91 | 93-97 | 82 | 93 |
| Techlab C. diff Chek-60 (GDH)[53,97,110] | GDH | Techlab | 92.0-93.5 | 94.0-98.0 | 88.0-93.0 | 94.0-97.0 |
| TechLab toxin A/B II[81,97,100,109-111] | Toxin A + B | TechLab | 72-91 | 87-100 | 58-85 | 96-99 |
| Membrane enzyme immunoassay | ||||||
| ImmunoCard toxins A/B[41,97,109,112,113] | Toxin A + B | Meridian Bioscience | 85-96 | 97-99 | 41-69 | 93-99 |
| Quick Chek Complete[40,105,112,114-117] | Toxin A + B | TechLab | 50-73 | 100 | 29-79 | 89-100 |
| Remel X/pect C. diff toxin A/B[97,113,118] | Toxin A + B | Remel | 44-83 | 99-100 | 48-69 | 95-99 |
| Tox A/B Quick Check[97,115,119-121] | Toxin A + B | TechLab | 61-84 | 99 | 40-74 | 94-100 |
| Chemiluminescence immunoassay | ||||||
| Liaison C. difficile toxins A and B[122,123] | Toxin A + B | DiaSorin | 88 | 95 | 69-88 | 95-99 |
| Enzyme-linked fluorescent assay | ||||||
| Vidas toxin A and B[97,116,123-128] | Toxin A + B | Vidas | 53-98 | 99-100 | 44-80 | 95-99 |
| Vidas GDH[40,129] | GDH | Vidas | 97 | 87 | 56 | 100 |
As a result, several multiple-step diagnostic algorithms have been proposed to improve assay performance and detection accuracy. For example, a two-step algorithm initially detects GDH, and positive GDH cases are followed up with a toxin A and/or toxin B immunoassay. If the toxin immunoassay result is negative, a bacterial culture will be performed[53]. In addition, new protein-based assays such as lateral flow assay are under development and may potentially yield a result in 15 min[54].
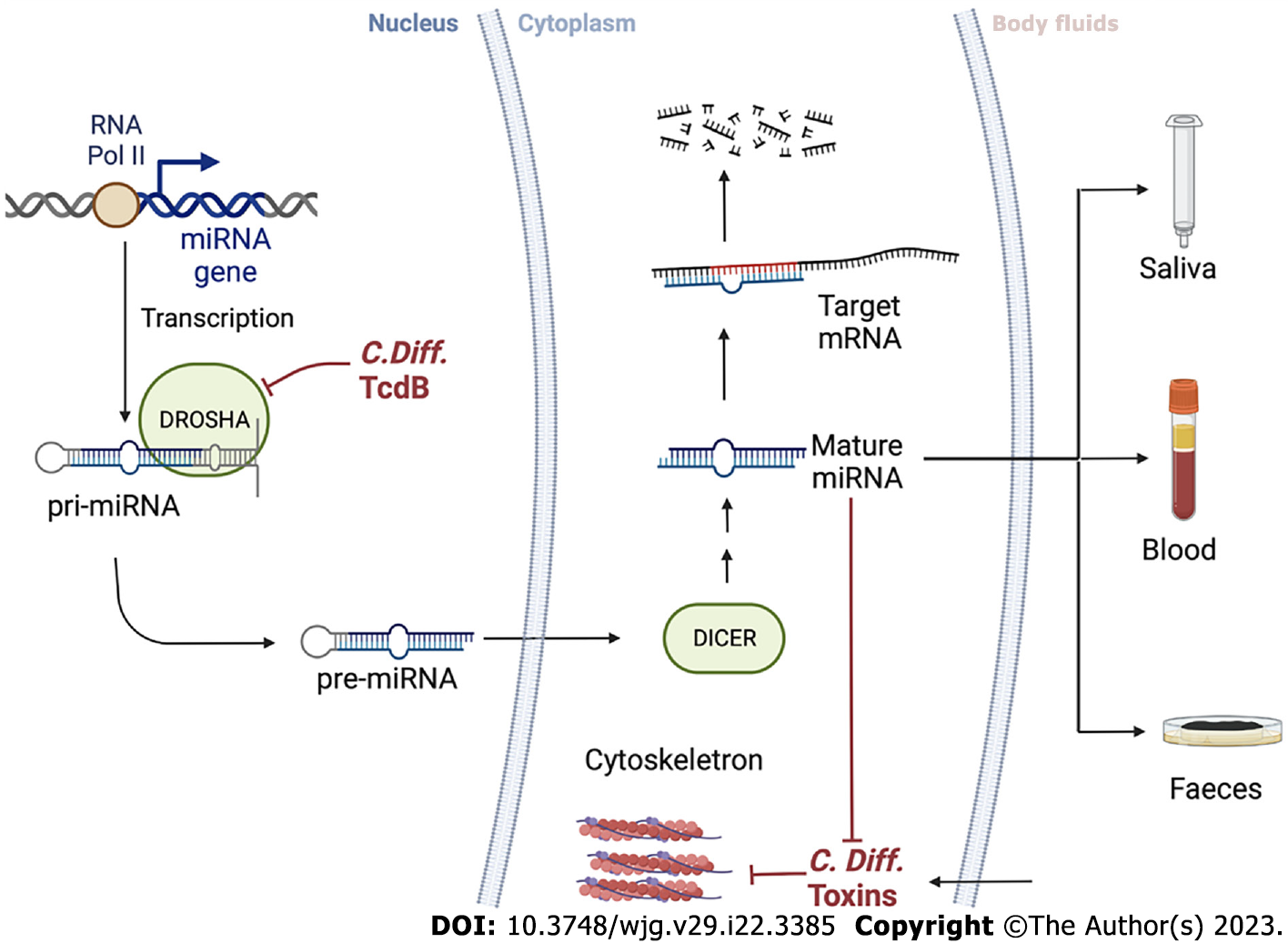
miRNAs are small single-stranded endogenous RNA molecules that are key regulators of gene expression and silencing at the post-transcriptional level. miRNA complex biosynthesis begins in the nucleus where primary miRNAs are produced by transcription of miRNAs from DNA sequences or miRNA genes. Precursor miRNAs are transcripts of approximately 60-110 nucleotides in length with a shorter stem-loop structure, produced from primary miRNAs by RNase type III enzymes (DROSHA) and undergo several maturation processes inside and outside the nucleus to form mature miRNAs, which are transported into the cytoplasm[55].
The stability of miRNA-specific target mRNAs is disturbed when RNA-induced silencing complexes possess loaded miRNAs. A portion of the miRNA, the seed sequence, which is two to eight nucleotides long, pairs with a specific sequence on the target mRNA and is referred to as an miRNA response element that results in translational repression and degradation of the target mRNA due to the binding of miRNAs in the 3’ untranslated region. miRNAs primarily repress genes by inhibiting protein synthesis, preventing elongation and ribosome decline and disrupting mRNAs through the processes of demethylation and decap, resulting in their degradation[56].
miRNAs have the potential to act via molecular mechanisms at every step of CDI, inhibiting specific transcripts or inflammatory molecule transcription, thereby influencing the pathology grade. The imbalance of these biomarkers can be measured and exploited for diagnosis, coupled with standard methods to strengthen the results. In particular, miRNAs are easily detectable (through sequencing, RT-qPCR, etc) in body fluids such as saliva, blood and even faecal material, and their levels correlate with target transcript alterations or non-physiological events[57]. These biomarkers are contributing increasingly to the establishment of less invasive “liquid biopsies,” which is important and appealing for patient compliance compared to normal, invasive biopsies that require a long time for results. Although, it cannot fully substitute canonical diagnosis methods in most cases currently, it will undoubtedly happen in the near future.
Numerous studies have shown that miRNAs can be used to detect diseases and for their mana
FMT has been proven in the treatment of recurrent CDI[61,62]. It is intended to restore colonic microbiota through introducing “healthy” bacteria via colonoscopy, enema or oral capsules that contain bacteria in a powder form. However, the safety concerns of FMT could be an obstacle to extending the application as a regular treatment strategy[63,64]. To monitor treatment conditions, detecting a panel of miRNA markers in circulation could help physicians make a more accurate decision[65,66]. Another study showed that 71 different circulating miRNAs were found to be expressed in 126 sera from 42 patients at 4 wk and 12 wk after FMT treatment[65]. The authors used qRT-PCR and 3’ untranslated region luciferase reporter assays to validate the top miRNA candidates and confirmed that hsa-miR-23a-3p, hsa-miR-150-5p, hsa-miR-26b-5p and hsa-miR-28-5p expression levels inversely correlated with the sera protein and cell-free circulating mRNA on several inflammation-related biomarkers, such as IL-12B, IL-18, FGF21 and TNFRSF9[65].
In a mouse model of relapsing CDI, qRT-PCR analyses of faecal and sera RNA extracts revealed inhibition of these miRNAs, while the FMT treatment enabled the recovery of their inhibitory effect. This study also showed that toxin B (TcdB) mediates the inhibitory effect of CDI on miRNA via DROSHA, based on the human colonoids and the mice colon models, where miR-23a and miR-150 were used to demonstrate the cytoprotective effects against TcdB[65]. A small, in-depth phenomics study of four adults treated with sequential FMT for severe or fulminant CDI found that miR-451a and miR-16 from the serum samples were upregulated in the responders vs the non-responders on average across all timepoints[66].
Using miRNAs as a diagnostic tool for CDI presents both opportunities and challenges. On one hand, as miRNAs play a role in controlling and influencing gene expression, changes in their activity and expression levels can be associated with different pathological events. This makes miRNA expression patterns a potentially powerful diagnostic tool, with the potential for use in therapeutic applications. Monitoring the response to FMT with miRNA can be an additional indicator for personalised treatment, and miRNA detection from non-invasive sources, such as blood, faeces, urine and saliva, can provide convenient and longitudinal measurements of miRNA levels for CDI. On the other hand, the fact that miRNA can be derived from blood cells released by different pathological events makes detecting CDI specifically a challenging task. Therefore, the use of miRNAs as a diagnostic tool for CDI requires further research and validation. Figure 4 provides a schematic summary of the intracellular mechanisms discussed and the potential use of miRNA profiling from a variety of body fluids, such as liquid biopsies, for diagnostic purposes as demonstrated in various research studies related to colorectal cancer screening[67-69].
To date, current laboratory-based CDI assays have varied in their detection accuracy. The expression of miRNAs has become increasingly important as novel biomarkers for assessment. The different signature profiles obtained through the differential expression of these small non-coding RNAs are essential for early diagnosis and prediction of therapeutic response and disease management, these are also including cancer and infection. Notably, this can be accomplished by using liquid biopsy, enabling a non-invasive and low-cost screening approach. This approach may also be used to detect C. difficile infections, while further research and validation are clearly needed. These reported miRNA studies can be used in conjunction with current diagnostic tools to improve diagnostic accuracy aiding in patient management for both symptomatic and asymptomatic CDI patients.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United Kingdom
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lopitz-Otsoa F, Spain; Zhou B, China S-Editor: Fan JR L-Editor: Filipodia P-Editor: Yu HG
| 1. | U. S. Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2019. Atlanta, Georgia. Available from: https://stacks.cdc.gov/view/cdc/82532. |
| 2. | Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372:825-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2023] [Cited by in RCA: 1968] [Article Influence: 196.8] [Reference Citation Analysis (1)] |
| 3. | Karlsson S, Dupuy B, Mukherjee K, Norin E, Burman LG, Akerlund T. Expression of Clostridium difficile toxins A and B and their sigma factor TcdD is controlled by temperature. Infect Immun. 2003;71:1784-1793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 66] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 4. | Adams SD, Mercer DW. Fulminant Clostridium difficile colitis. Curr Opin Crit Care. 2007;13:450-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 70] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 5. | Yau TO, Vadakekolathu J, Foulds GA, Du G, Dickins B, Polytarchou C, Rutella S. Hyperactive neutrophil chemotaxis contributes to anti-tumor necrosis factor-α treatment resistance in inflammatory bowel disease. J Gastroenterol Hepatol. 2022;37:531-541. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Reference Citation Analysis (2)] |
| 6. | Fu Y, Luo Y, Grinspan AM. Epidemiology of community-acquired and recurrent Clostridioides difficile infection. Therap Adv Gastroenterol. 2021;14:17562848211016248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 63] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 7. | Kachrimanidou M, Tzika E, Filioussis G. Clostridioides (Clostridium) Difficile in Food-Producing Animals, Horses and Household Pets: A Comprehensive Review. Microorganisms. 2019;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 8. | Muñoz-Price LS, Hanson R, Singh S, Nattinger AB, Penlesky A, Buchan BW, Ledeboer NA, Beyer K, Namin S, Zhou Y, Pezzin LE. Association Between Environmental Factors and Toxigenic Clostridioides difficile Carriage at Hospital Admission. JAMA Netw Open. 2020;3:e1919132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 9. | Monaghan TM, Biswas RN, Nashine RR, Joshi SS, Mullish BH, Seekatz AM, Blanco JM, McDonald JAK, Marchesi JR, Yau TO, Christodoulou N, Hatziapostolou M, Pucic-Bakovic M, Vuckovic F, Klicek F, Lauc G, Xue N, Dottorini T, Ambalkar S, Satav A, Polytarchou C, Acharjee A, Kashyap RS. Multiomics Profiling Reveals Signatures of Dysmetabolism in Urban Populations in Central India. Microorganisms. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 10. | Janarthanan S, Ditah I, Adler DG, Ehrinpreis MN. Clostridium difficile-associated diarrhea and proton pump inhibitor therapy: a meta-analysis. Am J Gastroenterol. 2012;107:1001-1010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 365] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 11. | Roughead EE, Chan EW, Choi NK, Griffiths J, Jin XM, Lee J, Kimura M, Kimura T, Kubota K, Lai EC, Man KK, Nguyen TA, Ooba N, Park BJ, Sato T, Shin JY, Wang T, Wong IC, Yang YK, Pratt NL. Proton pump inhibitors and risk of Clostridium difficile infection: a multi-country study using sequence symmetry analysis. Expert Opin Drug Saf. 2016;15:1589-1595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 12. | Rineh A, Kelso MJ, Vatansever F, Tegos GP, Hamblin MR. Clostridium difficile infection: molecular pathogenesis and novel therapeutics. Expert Rev Anti Infect Ther. 2014;12:131-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 13. | Lyras D, Adams V, Lucet I, Rood JI. The large resolvase TnpX is the only transposon-encoded protein required for transposition of the Tn4451/3 family of integrative mobilizable elements. Mol Microbiol. 2004;51:1787-1800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 33] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 14. | Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Tárraga AM, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38:779-786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 693] [Cited by in RCA: 708] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 15. | Maxwell-Scott HG, Goldenberg SD. Existing and investigational therapies for the treatment of Clostridium difficile infection: A focus on narrow spectrum, microbiota-sparing agents. Med Mal Infect. 2018;48:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 16. | Hensgens MP, Goorhuis A, Dekkers OM, Kuijper EJ. Time interval of increased risk for Clostridium difficile infection after exposure to antibiotics. J Antimicrob Chemother. 2012;67:742-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 249] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 17. | Bagdasarian N, Rao K, Malani PN. Diagnosis and treatment of Clostridium difficile in adults: a systematic review. JAMA. 2015;313:398-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 343] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 18. | Wilcox MH. Gastrointestinal disorders and the critically ill. Clostridium difficile infection and pseudomembranous colitis. Best Pract Res Clin Gastroenterol. 2003;17:475-493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 66] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 19. | Isidro J, Mendes AL, Serrano M, Henriques AO, Oleastro M. Overview of Clostridium difficile Infection: Life Cycle, Epidemiology, Antimicrobial Resistance and Treatment. In: Clostridium Difficile-A Comprehensive Overview. InTech. 2017;. [DOI] [Full Text] |
| 20. | Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev. 2005;18:247-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 815] [Cited by in RCA: 845] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 21. | Mani N, Dupuy B. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc Natl Acad Sci U S A. 2001;98:5844-5849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 194] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 22. | Mani N, Lyras D, Barroso L, Howarth P, Wilkins T, Rood JI, Sonenshein AL, Dupuy B. Environmental response and autoregulation of Clostridium difficile TxeR, a sigma factor for toxin gene expression. J Bacteriol. 2002;184:5971-5978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 98] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 23. | El Meouche I, Peltier J, Monot M, Soutourina O, Pestel-Caron M, Dupuy B, Pons JL. Characterization of the SigD regulon of C. difficile and its positive control of toxin production through the regulation of tcdR. PloS One. 2013;8:e83748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 87] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 24. | Perelle S, Gibert M, Bourlioux P, Corthier G, Popoff MR. Production of a complete binary toxin (actin-specific ADP-ribosyltransferase) by Clostridium difficile CD196. Infect Immun. 1997;65:1402-1407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 219] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 25. | Barth H, Aktories K, Popoff MR, Stiles BG. Binary bacterial toxins: biochemistry, biology, and applications of common Clostridium and Bacillus proteins. Microbiol Mol Biol Rev. 2004;68:373-402, table of contents. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 318] [Cited by in RCA: 290] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 26. | Lübbert C, John E, von Müller L. Clostridium difficile infection: guideline-based diagnosis and treatment. Dtsch Arztebl Int. 2014;111:723-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 27. | McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, Dubberke ER, Garey KW, Gould CV, Kelly C, Loo V, Shaklee Sammons J, Sandora TJ, Wilcox MH. Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis. 2018;66:e1-e48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1397] [Cited by in RCA: 1381] [Article Influence: 197.3] [Reference Citation Analysis (0)] |
| 28. | Bartlett JG, Chang TW, Gurwith M, Gorbach SL, Onderdonk AB. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N Engl J Med. 1978;298:531-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 957] [Cited by in RCA: 839] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 29. | Chang TW, Lauermann M, Bartlett JG. Cytotoxicity assay in antibiotic-associated colitis. J Infect Dis. 1979;140:765-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 162] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 30. | Tichota-Lee J, Jaqua-Stewart MJ, Benfield D, Simmons JL, Jaqua RA. Effect of age on the sensitivity of cell cultures to Clostridium difficile toxin. Diagn Microbiol Infect Dis. 1987;8:203-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 31. | Perry JD. A Decade of Development of Chromogenic Culture Media for Clinical Microbiology in an Era of Molecular Diagnostics. Clin Microbiol Rev. 2017;30:449-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 92] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 32. | Housman ST, Banevicius MA, Lamb LM, Nicolau DP. Isolation and quantitation of Clostridium difficile in aqueous and fecal matter using two types of selective media. J Microbiol Immunol Infect. 2016;49:445-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 33. | Binning M, John MA, Schieven BC, Austin TW, Lannigan R, Hussain Z. Comparison of culture, cytotoxin assay and two EIA tests with clinical diagnosis of Clostridium difficile-associated diarrhea. Can J Infect Dis. 1994;5:163-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 34. | Tyrrell KL, Citron DM, Leoncio ES, Merriam CV, Goldstein EJ. Evaluation of cycloserine-cefoxitin fructose agar (CCFA), CCFA with horse blood and taurocholate, and cycloserine-cefoxitin mannitol broth with taurocholate and lysozyme for recovery of Clostridium difficile isolates from fecal samples. J Clin Microbiol. 2013;51:3094-3096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 35. | Hink T, Burnham CA, Dubberke ER. A systematic evaluation of methods to optimize culture-based recovery of Clostridium difficile from stool specimens. Anaerobe. 2013;19:39-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 56] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 36. | Mundy LS, Shanholtzer CJ, Willard KE, Gerding DN, Peterson LR. Laboratory detection of Clostridium difficile. A comparison of media and incubation systems. Am J Clin Pathol. 1995;103:52-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 37. | Bliss DZ, Johnson S, Clabots CR, Savik K, Gerding DN. Comparison of cycloserine-cefoxitin-fructose agar (CCFA) and taurocholate-CCFA for recovery of Clostridium difficile during surveillance of hospitalized patients. Diagn Microbiol Infect Dis. 1997;29:1-4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 26] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 38. | Burnham CA, Carroll KC. Diagnosis of Clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin Microbiol Rev. 2013;26:604-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 298] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 39. | Angione SL, Sarma AA, Novikov A, Seward L, Fieber JH, Mermel LA, Tripathi A. A novel subtyping assay for detection of Clostridium difficile virulence genes. J Mol Diagn. 2014;16:244-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 40. | Xiao Y, Liu Y, Qin X. Comparative Study of Clostridium difficile Clinical Detection Methods in Patients with Diarrhoea. Can J Infect Dis Med Microbiol. 2020;2020:8753284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 41. | van den Berg RJ, Bruijnesteijn van Coppenraet LS, Gerritsen HJ, Endtz HP, van der Vorm ER, Kuijper EJ. Prospective multicenter evaluation of a new immunoassay and real-time PCR for rapid diagnosis of Clostridium difficile-associated diarrhea in hospitalized patients. J Clin Microbiol. 2005;43:5338-5340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 92] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 42. | Crobach MJT, Duszenko N, Terveer EM, Verduin CM, Kuijper EJ. Nucleic Acid Amplification Test Quantitation as Predictor of Toxin Presence in Clostridium difficile Infection. J Clin Microbiol. 2018;56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 43. | Anikst VE, Gaur RL, Schroeder LF, Banaei N. Organism burden, toxin concentration, and lactoferrin concentration do not distinguish between clinically significant and nonsignificant diarrhea in patients with Clostridium difficile. Diagn Microbiol Infect Dis. 2016;84:343-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 44. | Dubberke ER, Han Z, Bobo L, Hink T, Lawrence B, Copper S, Hoppe-Bauer J, Burnham CA, Dunne WM Jr. Impact of clinical symptoms on interpretation of diagnostic assays for Clostridium difficile infections. J Clin Microbiol. 2011;49:2887-2893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 152] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 45. | Buckel WR, Avdic E, Carroll KC, Gunaseelan V, Hadhazy E, Cosgrove SE. Gut check: Clostridium difficile testing and treatment in the molecular testing era. Infect Control Hosp Epidemiol. 2015;36:217-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 46. | Popiel KY, Gheorghe R, Eastmond J, Miller MA. Usefulness of Adjunctive Fecal Calprotectin and Serum Procalcitonin in Individuals Positive for Clostridium difficile Toxin Gene by PCR Assay. J Clin Microbiol. 2015;53:3667-3669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 47. | Crobach MJT, Baktash A, Duszenko N, Kuijper EJ. Diagnostic Guidance for C. difficile Infections. Adv Exp Med Biol. 2018;1050:27-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 48. | Crobach MJ, Planche T, Eckert C, Barbut F, Terveer EM, Dekkers OM, Wilcox MH, Kuijper EJ. European Society of Clinical Microbiology and Infectious Diseases: update of the diagnostic guidance document for Clostridium difficile infection. Clin Microbiol Infect. 2016;22 Suppl 4:S63-S81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 392] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 49. | Erb S, Frei R, Strandén AM, Dangel M, Tschudin-Sutter S, Widmer AF. Low sensitivity of fecal toxin A/B enzyme immunoassay for diagnosis of Clostridium difficile infection in immunocompromised patients. Clin Microbiol Infect. 2015;21:998.e9-998.e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 50. | Terveer EM, Crobach MJ, Sanders IM, Vos MC, Verduin CM, Kuijper EJ. Detection of Clostridium difficile in Feces of Asymptomatic Patients Admitted to the Hospital. J Clin Microbiol. 2017;55:403-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 51. | Walkty A, Lagacé-Wiens PR, Manickam K, Adam H, Pieroni P, Hoban D, Karlowsky JA, Alfa M. Evaluation of an algorithmic approach in comparison with the Illumigene assay for laboratory diagnosis of Clostridium difficile infection. J Clin Microbiol. 2013;51:1152-1157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 52. | Bouza E, Aguado JM, Alcalá L, Almirante B, Alonso-Fernández P, Borges M, Cobo J, Guardiola J, Horcajada JP, Maseda E, Mensa J, Merchante N, Muñoz P, Pérez Sáenz JL, Pujol M, Reigadas E, Salavert M, Barberán J. Recommendations for the diagnosis and treatment of Clostridioides difficile infection: An official clinical practice guideline of the Spanish Society of Chemotherapy (SEQ), Spanish Society of Internal Medicine (SEMI) and the working group of Postoperative Infection of the Spanish Society of Anesthesia and Reanimation (SEDAR). Rev Esp Quimioter. 2020;33:151-175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 53. | Fenner L, Widmer AF, Goy G, Rudin S, Frei R. Rapid and reliable diagnostic algorithm for detection of Clostridium difficile. J Clin Microbiol. 2008;46:328-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 110] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 54. | Qi H, Sun Q, Ma Y, Wu P, Wang J. Advantages of Lateral Flow Assays Based on Fluorescent Submicrospheres and Quantum Dots for Clostridium difficile Toxin B Detection. Toxins (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 55. | Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016-3027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1468] [Cited by in RCA: 1567] [Article Influence: 74.6] [Reference Citation Analysis (0)] |
| 56. | Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1145] [Cited by in RCA: 1416] [Article Influence: 141.6] [Reference Citation Analysis (0)] |
| 57. | Cossu AM, Scrima M, Lombardi A, Grimaldi A, Russo M, Ottaiano A, Caraglia M, Bocchetti M. Future directions and management of liquid biopsy in non-small cell lung cancer. Explor Target Antitumor Ther. 2020;1:239-252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 58. | Viladomiu M, Hontecillas R, Pedragosa M, Carbo A, Hoops S, Michalak P, Michalak K, Guerrant RL, Roche JK, Warren CA, Bassaganya-Riera J. Modeling the role of peroxisome proliferator-activated receptor γ and microRNA-146 in mucosal immune responses to Clostridium difficile. PloS One. 2012;7:e47525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 59. | Imad K, Hussein K, Cécile L, Jean-christophe M. Use of the mir-27a-5p microRNA for treating Clostridium difficile-induced bowel inflammation. France patent EP3876948A1. September 15, 2021. |
| 60. | Verdier J, Breunig IR, Ohse MC, Roubrocks S, Kleinfeld S, Roy S, Streetz K, Trautwein C, Roderburg C, Sellge G. Faecal Micro-RNAs in Inflammatory Bowel Diseases. J Crohns Colitis. 2020;14:110-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 61. | Drekonja D, Reich J, Gezahegn S, Greer N, Shaukat A, MacDonald R, Rutks I, Wilt TJ. Fecal Microbiota Transplantation for Clostridium difficile Infection: A Systematic Review. Ann Intern Med. 2015;162:630-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 269] [Article Influence: 26.9] [Reference Citation Analysis (1)] |
| 62. | Hensley-McBain T, Zevin AS, Manuzak J, Smith E, Gile J, Miller C, Agricola B, Katze M, Reeves RK, Kraft CS, Langevin S, Klatt NR. Effects of Fecal Microbial Transplantation on Microbiome and Immunity in Simian Immunodeficiency Virus-Infected Macaques. J Virol. 2016;90:4981-4989. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 63. | Xu Y, Li X, Zhu B, Liang H, Fang C, Gong Y, Guo Q, Sun X, Zhao D, Shen J, Zhang H, Liu H, Xia H, Tang J, Zhang K, Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26:502-505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 992] [Cited by in RCA: 1050] [Article Influence: 210.0] [Reference Citation Analysis (0)] |
| 64. | Alang N, Kelly CR. Weight gain after fecal microbiota transplantation. Open Forum Infect Dis. 2015;2:ofv004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 270] [Cited by in RCA: 299] [Article Influence: 29.9] [Reference Citation Analysis (1)] |
| 65. | Monaghan TM, Seekatz AM, Markham NO, Yau TO, Hatziapostolou M, Jilani T, Christodoulou N, Roach B, Birli E, Pomenya O, Louie T, Lacy DB, Kim P, Lee C, Kao D, Polytarchou C. Fecal Microbiota Transplantation for Recurrent Clostridioides difficile Infection Associates With Functional Alterations in Circulating microRNAs. Gastroenterology. 2021;161:255-270.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 66. | Monaghan TM, Duggal NA, Rosati E, Griffin R, Hughes J, Roach B, Yang DY, Wang C, Wong K, Saxinger L, Pučić-Baković M, Vučković F, Klicek F, Lauc G, Tighe P, Mullish BH, Blanco JM, McDonald JAK, Marchesi JR, Xue N, Dottorini T, Acharjee A, Franke A, Li Y, Wong GK, Polytarchou C, Yau TO, Christodoulou N, Hatziapostolou M, Wang M, Russell LA, Kao DH. A Multi-Factorial Observational Study on Sequential Fecal Microbiota Transplant in Patients with Medically Refractory Clostridioides difficile Infection. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 67. | Yau TO, Wu CW, Dong Y, Tang CM, Ng SS, Chan FK, Sung JJ, Yu J. microRNA-221 and microRNA-18a identification in stool as potential biomarkers for the non-invasive diagnosis of colorectal carcinoma. Br J Cancer. 2014;111:1765-1771. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 68. | Yau TO, Wu CW, Tang CM, Chen Y, Fang J, Dong Y, Liang Q, Ng SS, Chan FK, Sung JJ, Yu J. MicroRNA-20a in human faeces as a non-invasive biomarker for colorectal cancer. Oncotarget. 2016;7:1559-1568. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 69. | Yau TO, Tang CM, Harriss EK, Dickins B, Polytarchou C. Faecal microRNAs as a non-invasive tool in the diagnosis of colonic adenomas and colorectal cancer: A meta-analysis. Sci Rep. 2019;9:9491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 70. | Deak E, Miller SA, Humphries RM. Comparison of Illumigene, Simplexa, and AmpliVue Clostridium difficile molecular assays for diagnosis of C. difficile infection. J Clin Microbiol. 2014;52:960-963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 71. | Juretschko S, Manji R, Khare R, Das S, Dunbar S. Performance Evaluation of the Luminex Aries C. difficile Assay in Comparison to Two Other Molecular Assays within a Multihospital Health Care Center. J Clin Microbiol. 2019;57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 72. | Pancholi P, Young S, Widen R, Silbert S, Schmitt B, Dunn R, Drain A, Weissfeld SA. A multicenter evaluation of a sample to answer real-time PCR assay for toxigenic C. difficile in symptomatic subjects. Diagn Microbiol Infect Dis. 2020;96:114920. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 73. | Jazmati N, Wiegel P, Ličanin B, Plum G. Evaluation of the Qiagen artus C. difficile QS-RGQ Kit for Detection of Clostridium difficile Toxins A and B in Clinical Stool Specimens. J Clin Microbiol. 2015;53:1942-1944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 74. | Hirvonen JJ, Kaukoranta SS. Comparison of BD Max Cdiff and GenomEra C. difficile molecular assays for detection of toxigenic Clostridium difficile from stools in conventional sample containers and in FecalSwabs. Eur J Clin Microbiol Infect Dis. 2015;34:1005-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 75. | Dalpke AH, Hofko M, Zorn M, Zimmermann S. Evaluation of the fully automated BD MAX Cdiff and Xpert C. difficile assays for direct detection of Clostridium difficile in stool specimens. J Clin Microbiol. 2013;51:1906-1908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 76. | Stellrecht KA, Espino AA, Maceira VP, Nattanmai SM, Butt SA, Wroblewski D, Hannett GE, Musser KA. Premarket evaluations of the IMDx C. difficile for Abbott m2000 Assay and the BD Max Cdiff Assay. J Clin Microbiol. 2014;52:1423-1428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 77. | Shin BM, Yoo SM, Shin WC. Evaluation of Xpert C. difficile, BD MAX Cdiff, IMDx C. difficile for Abbott m2000, and Illumigene C. difficile Assays for Direct Detection of Toxigenic Clostridium difficile in Stool Specimens. Ann Lab Med. 2016;36:131-137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 78. | Terhes G, Urbán E, Sóki J, Nacsa E, Nagy E. Comparison of a rapid molecular method, the BD GeneOhm Cdiff assay, to the most frequently used laboratory tests for detection of toxin-producing Clostridium difficile in diarrheal feces. J Clin Microbiol. 2009;47:3478-3481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 49] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 79. | Stamper PD, Alcabasa R, Aird D, Babiker W, Wehrlin J, Ikpeama I, Carroll KC. Comparison of a commercial real-time PCR assay for tcdB detection to a cell culture cytotoxicity assay and toxigenic culture for direct detection of toxin-producing Clostridium difficile in clinical samples. J Clin Microbiol. 2009;47:373-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 150] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 80. | Kvach EJ, Ferguson D, Riska PF, Landry ML. Comparison of BD GeneOhm Cdiff real-time PCR assay with a two-step algorithm and a toxin A/B enzyme-linked immunosorbent assay for diagnosis of toxigenic Clostridium difficile infection. J Clin Microbiol. 2010;48:109-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 106] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 81. | Karre T, Sloan L, Patel R, Mandrekar J, Rosenblatt J. Comparison of two commercial molecular assays to a laboratory-developed molecular assay for diagnosis of Clostridium difficile infection. J Clin Microbiol. 2011;49:725-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 82. | Shin BM, Mun SJ, Yoo SJ, Kuak EY. Comparison of BD GeneOhm Cdiff and Seegene Seeplex ACE PCR assays using toxigenic Clostridium difficile culture for direct detection of tcdB from stool specimens. J Clin Microbiol. 2012;50:3765-3767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 83. | Garg SK, Lu K, Duncan J, Peterson LR, Liesenfeld O. Equivalent Performance of the Cobas(®) Cdiff Test for Use on the Cobas(®) Liat(®) System and the Cobas(®) 4800 System. Eur J Microbiol Immunol (Bp). 2017;7:310-318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 84. | Peterson LR, Young SA, Davis TE Jr, Wang ZX, Duncan J, Noutsios C, Liesenfeld O, Osiecki JC, Lewinski MA. Evaluation of the cobas Cdiff Test for Detection of Toxigenic Clostridium difficile in Stool Samples. J Clin Microbiol. 2017;55:3426-3436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 85. | Mashock MJ, Faron ML, Carroll KC, Dang C, Lewis S, Salimnia H, Lephart P, Loo VG, Schmitt BH, Young S, Buchan BW, Ledeboer NA. A Multicenter Study of the Revogene C. difficile System for Detection of the Toxin B Gene from Unformed Stool Specimens. J Clin Microbiol. 2020;58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 86. | U. S. Food and Drug Administration. 510(k) Summary ICEPlex C. difficile Kit on the ICEPlex System 2013. Available from: https://www.fda.gov/. |
| 87. | Buchan BW, Mackey TL, Daly JA, Alger G, Denys GA, Peterson LR, Kehl SC, Ledeboer NA. Multicenter clinical evaluation of the portrait toxigenic C. difficile assay for detection of toxigenic Clostridium difficile strains in clinical stool specimens. J Clin Microbiol. 2012;50:3932-3936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 88. | Denys GA. Portrait Toxigenic Clostridium difficile assay, an isothermal amplification assay detects toxigenic C. difficile in clinical stool specimens. Expert Rev Mol Diagn. 2014;14:17-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 89. | Stamper PD, Babiker W, Alcabasa R, Aird D, Wehrlin J, Ikpeama I, Gluck L, Carroll KC. Evaluation of a new commercial TaqMan PCR assay for direct detection of the clostridium difficile toxin B gene in clinical stool specimens. J Clin Microbiol. 2009;47:3846-3850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 90. | Beck ET, Buchan BW, Riebe KM, Alkins BR, Pancholi P, Granato PA, Ledeboer NA. Multicenter evaluation of the Quidel Lyra Direct C. difficile nucleic acid amplification assay. J Clin Microbiol. 2014;52:1998-2002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 91. | Gilbreath JJ, Verma P, Abbott AN, Butler-Wu SM. Comparison of the Verigene Clostridium difficile, Simplexa C. difficile Universal Direct, BD MAX Cdiff, and Xpert C. difficile assays for the detection of toxigenic C. difficile. Diagn Microbiol Infect Dis. 2014;80:13-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 92. | U. S. Food and Drug Administration. 510(K) Substantial Equivalence Determination Decision Summary Assay and Instrument Combination Template 2018. Available from: https://www.baidu.com/link?url=bxZ2qu2ujOxmV4JObs_gYXHE5P8RRA1m4gfponKdX2KcgYUD6hwTI2h16v_7mP6lA3faJYql3jOpwCrN7NcpQPR98wNFSfu76enVjhfex2a&wd=&eqid=ae7ede620006e9e8000000066447ba8e. |
| 93. | Carroll KC, Buchan BW, Tan S, Stamper PD, Riebe KM, Pancholi P, Kelly C, Rao A, Fader R, Cavagnolo R, Watson W, Goering RV, Trevino EA, Weissfeld AS, Ledeboer NA. Multicenter evaluation of the Verigene Clostridium difficile nucleic acid assay. J Clin Microbiol. 2013;51:4120-4125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 94. | Kosai K, Iwanaga Y, Akamatsu N, Okada Y, Kaku N, Uno N, Morinaga Y, Hasegawa H, Miyazaki T, Izumikawa K, Mukae H, Yanagihara K. Performance evaluation of the Verigene(®)Clostridium difficile nucleic acid test, an automated multiplex molecular testing system for detection of C. difficile toxin. J Infect Chemother. 2017;23:674-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 95. | Bai Y, Sun X, Jin Y, Wang Y, Li J. Accuracy of Xpert Clostridium difficile assay for the diagnosis of Clostridium difficile infection: A meta analysis. PloS One. 2017;12:e0185891. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 96. | Moon HW, Kim HN, Hur M, Shim HS, Kim H, Yun YM. Comparison of Diagnostic Algorithms for Detecting Toxigenic Clostridium difficile in Routine Practice at a Tertiary Referral Hospital in Korea. PloS One. 2016;11:e0161139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 97. | Eastwood K, Else P, Charlett A, Wilcox M. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol. 2009;47:3211-3217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 261] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 98. | O’Connor D, Hynes P, Cormican M, Collins E, Corbett-Feeney G, Cassidy M. Evaluation of methods for detection of toxins in specimens of feces submitted for diagnosis of Clostridium difficile-associated diarrhea. J Clin Microbiol. 2001;39:2846-2849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 74] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 99. | Drudy D, Harnedy N, Fanning S, O’Mahony R, Kyne L. Isolation and characterisation of toxin A-negative, toxin B-positive Clostridium difficile in Dublin, Ireland. Clin Microbiol Infect. 2007;13:298-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 60] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 100. | Planche TD, Davies KA, Coen PG, Finney JM, Monahan IM, Morris KA, O’Connor L, Oakley SJ, Pope CF, Wren MW, Shetty NP, Crook DW, Wilcox MH. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C difficile infection. Lancet Infect Dis. 2013;13:936-945. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 317] [Cited by in RCA: 342] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 101. | Novak-Weekley SM, Marlowe EM, Miller JM, Cumpio J, Nomura JH, Vance PH, Weissfeld A. Clostridium difficile testing in the clinical laboratory by use of multiple testing algorithms. J Clin Microbiol. 2010;48:889-893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 172] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 102. | Barkin JA, Nandi N, Miller N, Grace A, Barkin JS, Sussman DA. Superiority of the DNA amplification assay for the diagnosis of C. difficile infection: a clinical comparison of fecal tests. Dig Dis Sci. 2012;57:2592-2599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 103. | Babady NE, Stiles J, Ruggiero P, Khosa P, Huang D, Shuptar S, Kamboj M, Kiehn TE. Evaluation of the Cepheid Xpert Clostridium difficile Epi assay for diagnosis of Clostridium difficile infection and typing of the NAP1 strain at a cancer hospital. J Clin Microbiol. 2010;48:4519-4524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 79] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 104. | Leitner E, Einetter M, Grisold AJ, Marth E, Feierl G. Evaluation of the BD MAX Cdiff assay for the detection of the toxin B gene of Clostridium difficile out of faecal specimens. Diagn Microbiol Infect Dis. 2013;76:390-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 105. | Ota KV, McGowan KL. Clostridium difficile testing algorithms using glutamate dehydrogenase antigen and C. difficile toxin enzyme immunoassays with C. difficile nucleic acid amplification testing increase diagnostic yield in a tertiary pediatric population. J Clin Microbiol. 2012;50:1185-1188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 106. | van den Berg RJ, Vaessen N, Endtz HP, Schülin T, van der Vorm ER, Kuijper EJ. Evaluation of real-time PCR and conventional diagnostic methods for the detection of Clostridium difficile-associated diarrhoea in a prospective multicentre study. J Med Microbiol. 2007;56:36-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 81] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 107. | Vanpoucke H, De Baere T, Claeys G, Vaneechoutte M, Verschraegen G. Evaluation of six commercial assays for the rapid detection of Clostridium difficile toxin and/or antigen in stool specimens. Clin Microbiol Infect. 2001;7:55-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 108. | Mattner F, Winterfeld I, Mattner L. Diagnosing toxigenic Clostridium difficile: new confidence bounds show culturing increases sensitivity of the toxin A/B enzyme immunoassay and refute gold standards. Scand J Infect Dis. 2012;44:578-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 109. | Musher DM, Manhas A, Jain P, Nuila F, Waqar A, Logan N, Marino B, Graviss EA. Detection of Clostridium difficile toxin: comparison of enzyme immunoassay results with results obtained by cytotoxicity assay. J Clin Microbiol. 2007;45:2737-2739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 57] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 110. | Snell H, Ramos M, Longo S, John M, Hussain Z. Performance of the TechLab C. DIFF CHEK-60 enzyme immunoassay (EIA) in combination with the C. difficile Tox A/B II EIA kit, the Triage C. difficile panel immunoassay, and a cytotoxin assay for diagnosis of Clostridium difficile-associated diarrhea. J Clin Microbiol. 2004;42:4863-4865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 85] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 111. | Massey V, Gregson DB, Chagla AH, Storey M, John MA, Hussain Z. Clinical usefulness of components of the Triage immunoassay, enzyme immunoassay for toxins A and B, and cytotoxin B tissue culture assay for the diagnosis of Clostridium difficile diarrhea. Am J Clin Pathol. 2003;119:45-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 112. | Bruins MJ, Verbeek E, Wallinga JA, Bruijnesteijn van Coppenraet LE, Kuijper EJ, Bloembergen P. Evaluation of three enzyme immunoassays and a loop-mediated isothermal amplification test for the laboratory diagnosis of Clostridium difficile infection. Eur J Clin Microbiol Infect Dis. 2012;31:3035-3039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 113. | Sloan LM, Duresko BJ, Gustafson DR, Rosenblatt JE. Comparison of real-time PCR for detection of the tcdC gene with four toxin immunoassays and culture in diagnosis of Clostridium difficile infection. J Clin Microbiol. 2008;46:1996-2001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 170] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 114. | Hart J, Putsathit P, Knight DR, Sammels L, Riley TV, Keil A. Clostridium difficile infection diagnosis in a paediatric population: comparison of methodologies. Eur J Clin Microbiol Infect Dis. 2014;33:1555-1564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 115. | Kawada M, Annaka M, Kato H, Shibasaki S, Hikosaka K, Mizuno H, Masuda Y, Inamatsu T. Evaluation of a simultaneous detection kit for the glutamate dehydrogenase antigen and toxin A/B in feces for diagnosis of Clostridium difficile infection. J Infect Chemother. 2011;17:807-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 116. | Swindells J, Brenwald N, Reading N, Oppenheim B. Evaluation of diagnostic tests for Clostridium difficile infection. J Clin Microbiol. 2010;48:606-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 120] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 117. | Kim H, Kim WH, Kim M, Jeong SH, Lee K. Evaluation of a rapid membrane enzyme immunoassay for the simultaneous detection of glutamate dehydrogenase and toxin for the diagnosis of Clostridium difficile infection. Ann Lab Med. 2014;34:235-239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 118. | de Boer RF, Wijma JJ, Schuurman T, Moedt J, Dijk-Alberts BG, Ott A, Kooistra-Smid AM, van Duynhoven YT. Evaluation of a rapid molecular screening approach for the detection of toxigenic Clostridium difficile in general and subsequent identification of the tcdC Δ117 mutation in human stools. J Microbiol Methods. 2010;83:59-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 119. | Yoldaş Ö, Altındiş M, Cufalı D, Aşık G, Keşli R. A Diagnostic Algorithm for the Detection of Clostridium difficile-Associated Diarrhea. Balkan Med J. 2016;33:80-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 120. | Le Guern R, Herwegh S, Grandbastien B, Courcol R, Wallet F. Evaluation of a new molecular test, the BD Max Cdiff, for detection of toxigenic Clostridium difficile in fecal samples. J Clin Microbiol. 2012;50:3089-3090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 121. | Reller ME, Alcabasa RC, Lema CA, Carroll KC. Comparison of two rapid assays for Clostridium difficile Common antigen and a C difficile toxin A/B assay with the cell culture neutralization assay. Am J Clin Pathol. 2010;133:107-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 122. | Benedek O, Podbielski A, Warnke P. Laboratory Experience with the Liaison Analyzer in the Diagnosis of Clostridium Difficile-Associated Diarrhea. Eur J Microbiol Immunol (Bp). 2016;6:215-218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 123. | Makristathis A, Zeller I, Mitteregger D, Kundi M, Hirschl AM. Comprehensive evaluation of chemiluminescent immunoassays for the laboratory diagnosis of Clostridium difficile infection. Eur J Clin Microbiol Infect Dis. 2017;36:1253-1259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 124. | Shin BM, Kuak EY, Lee EJ, Songer JG. Algorithm combining toxin immunoassay and stool culture for diagnosis of Clostridium difficile infection. J Clin Microbiol. 2009;47:2952-2956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 125. | Shin BM, Lee EJ, Kuak EY, Yoo SJ. Comparison of VIDAS CDAB and CDA immunoassay for the detection of Clostridium difficile in a tcdA- tcdB+ C. difficile prevalent area. Anaerobe. 2009;15:266-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 126. | Kim H, Jeong SH, Kim M, Lee Y, Lee K. Detection of Clostridium difficile toxin A/B genes by multiplex real-time PCR for the diagnosis of C. difficile infection. J Med Microbiol. 2012;61:274-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 127. | Shin S, Kim M, Lim H, Kim H, Lee K, Chong Y. Evaluation of the Xpert Clostridium difficile assay for the diagnosis of Clostridium difficile infection. Ann Lab Med. 2012;32:355-358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 128. | de Jong E, de Jong AS, Bartels CJ, van der Rijt-van den Biggelaar C, Melchers WJ, Sturm PD. Clinical and laboratory evaluation of a real-time PCR for Clostridium difficile toxin A and B genes. Eur J Clin Microbiol Infect Dis. 2012;31:2219-2225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 129. | Shin BM, Lee EJ, Moon JW, Lee SY. Evaluation of the VIDAS glutamate dehydrogenase assay for the detection of Clostridium difficile. Anaerobe. 2016;40:68-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |