Published online Jan 14, 2023. doi: 10.3748/wjg.v29.i2.310
Peer-review started: September 22, 2022
First decision: October 18, 2022
Revised: November 2, 2022
Accepted: December 21, 2022
Article in press: December 21, 2022
Published online: January 14, 2023
Processing time: 105 Days and 12.2 Hours
Inflammatory bowel diseases (IBDs) comprising ulcerative colitis, Crohn’s disease and microscopic colitis are characterized by chronic inflammation of the gastrointestinal tract. IBD has spread around the world and is becoming more prevalent at an alarming rate in developing countries whose societies have become more westernized. Cell therapy, intestinal microecology, apheresis therapy, exosome therapy and small molecules are emerging therapeutic options for IBD. Currently, it is thought that low-molecular-mass substances with good oral bio-availability and the ability to permeate the cell membrane to regulate the action of elements of the inflammatory signaling pathway are effective therapeutic options for the treatment of IBD. Several small molecule inhibitors are being developed as a promising alternative for IBD therapy. The use of highly efficient and time-saving techniques, such as computational methods, is still a viable option for the development of these small molecule drugs. The computer-aided (in silico) discovery approach is one drug development technique that has mostly proven efficacy. Computational approaches when combined with traditional drug development methodology dramatically boost the likelihood of drug discovery in a sustainable and cost-effective manner. This review focuses on the modern drug discovery approaches for the design of novel IBD drugs with an emphasis on the role of computational methods. Some computational approaches to IBD genomic studies, target identification, and virtual screening for the discovery of new drugs and in the repurposing of existing drugs are discussed.
Core Tip: For the design of small molecule drugs to treat inflammatory bowel disease (IBD), highly effective and time-saving approaches, such as computational methods, are still a viable choice. By complementing experimental studies with computational approaches, the probability of successful drug discovery is increased while simultaneously reducing associated costs. This article provides a summary of the current drug discovery pipeline for IBD, with special emphasis on the part played by computational methods. The use of in silico genomic studies, target identification, and virtual screening to find new drugs and repurpose existing drugs for the treatment of IBD are discussed.
- Citation: Johnson TO, Akinsanmi AO, Ejembi SA, Adeyemi OE, Oche JR, Johnson GI, Adegboyega AE. Modern drug discovery for inflammatory bowel disease: The role of computational methods. World J Gastroenterol 2023; 29(2): 310-331
- URL: https://www.wjgnet.com/1007-9327/full/v29/i2/310.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i2.310
Inflammatory bowel disease (IBD) refers to a group of disorders characterized by chronic inflammation of the gastrointestinal tract that is oftentimes triggered by unknown causes. Crohn’s disease (CD) and ulcerative colitis (UC) are the two most prevalent IBDs[1]. The clinical courses are characterized by consistent relapses and recoveries, linked to destructive idiopathic inflammation of the digestive tract. Despite the fact that the precise causes of IBD are still being investigated, it is believed to be the result of an intricate relationship between various parameters, including immune system dysfunction, microbial dysbiosis, a genetic susceptibility, and some environmental factors[2,3].
As the etiology of IBD continues to be a “crossword” to solve, its prevalence and significant global impact continue to rise[4-6]. Globally, the prevalence of IBD increased by 85.1% between 1990 and 2017, and the number of people diagnosed with the disease increased by 95%, as reported by the Global Burden of Disease 2017 Inflammatory Bowel Disease Collaborators. Since 1990, the incidence of IBD has risen in newly industrialized countries in Africa, Asia and South America, including Brazil, to the extent that at the turn of the 21st century this idiopathic disease[7] has become a global disease with accelerating incidence in countries whose societies have become more westernized[8]. Also of interest is the increasing prevalence of microscopic colitis, a subtype of IBD characterized by chronic watery diarrhea caused by colon inflammation. The disease has recently gained more attention due to the availability of more information about its pathogenesis, diagnosis and therapy. The incidence of microscopic colitis is progressively on the rise, approaching that of CD and UC in some populations[9].
Treatments options for IBDs, especially CD and UC, have made significant progress in the last few years[5]. A wide range of anti-inflammatory and symptom-relieving drugs is available for patients with UC. Recent treatments aim to improve the patient’s quality of life by alleviating symptoms like abdominal pain and diarrhea and bringing the disease under control as a whole[10]. Conventional treatments include aminosalicylates, corticosteroids, immunomodulators and biologics as part of the pharmacotherapy to control symptoms. When necessary, they also include some general procedures, including surgery. The advent of biologics and other therapeutic advancements in recent years has altered not only the treatment modalities but also the perception of IBD therapy. While symptom remission, complication avoidance and complication treatment are all important, mucosal healing is also a major target. Healing of the mucosa occurs when inflammation in that area subsides and the mucosal lining is returned to its normal structure[11]. Emerging evidence suggests that mucosal healing is linked to improved long-term outcomes, including lower rates of clinical recurrence, hospitalization, surgery, and disability[12,13].
Many patients with IBD have seen a dramatic improvement in their long-term outcomes, both regarding disability and quality of life since the introduction of monoclonal antibodies targeting tumor necrosis factor (TNF) about 20 years ago. However, despite these developments, there are still many unfulfilled needs. For instance, less than a third of treated patients who begin a biologic therapy achieve and maintain drug remission at 1 year. Even in cases when clinical and endoscopic criteria for remission have been met, symptoms such as diarrhea, stomach discomfort, joint problems, and exhaustion may still be experienced[14]. Also, the lack of responsiveness to biologic therapies, which can be caused in part by the protein's immunogenicity upon administration, and the need to discontinue drugs because of intolerance or side effects show that a better generation of therapeutic alternatives is still needed. To achieve the desired results of immune homeostasis restoration and better symptom control, additional progress is still required. Emerging treatment options for IBD include cell therapy, intestinal microecology, apheresis therapy, exosome therapy, and small molecules[15]. Low-molecular-mass compounds with excellent oral bioavailability and the capacity to cross the cell membrane to modulate the functions of parts of the inflammatory signaling pathway are now regarded to be promising therapeutic alternatives for the treatment of IBD. Several small molecule inhibitors are being explored as a possible treatment option for IBD, such as inhibitors of Janus kinase (JAK) enzymes[1]. For the development of these small molecule drugs, the use of highly effective and time-saving techniques, such as computational methods, remain a viable option.
Owing to the intricate nature of the molecular mechanisms involved in disease pathogenesis, the process of developing small molecule drugs is complicated[16,17]. The traditional methods of drug design and development require time-consuming, costly and laborious scientific procedures. On the other hand, computational tools hold great promise as a viable option for the design and development of new small molecules of biomedical relevance[18]. Computer-aided drug design (CADD) employs computer processing power, statistics, mathematics and three-dimensional (3D) graphics to elucidate the affinity and binding mode of small molecules against therapeutic targets.
As a whole, computational methods aid in the identification of candidate target proteins for drug screening and design by searching through large amounts of genomic data for the most important genes[19,20]. In combination with experimental data, protein structures can be predicted with some accuracy[21]. In order to find drug binding sites and understand how drugs work, it is necessary to conduct research into the structural and thermodynamic features of target proteins, both of which can be accomplished with biomolecular simulations using multiscale models[22]. Next, chemical libraries are explored using virtual screening to identify potential drug candidates based on the ligand binding sites on target proteins[23]. With a significantly reduced number of potential drug candidates, in vitro and/or in vivo experiments can be used to further assess the effectiveness of these molecules. Another CADD approach, besides virtual screening, is provided by de novo drug design techniques, which produce highly specific small molecules with good synthetic accessibility[24].
Medicinal chemists use CADD methods to help in rationalizing the selection of hit compounds and in hit-to-lead optimization. CADD has been used as an efficient method for identifying potential lead compounds toward the development of drugs for a wide range of diseases, including IBDs. A variety of computational approaches, such as molecular docking, molecular dynamic (MD) simulation, quantitative structure-activity relationship (QSAR) and pharmacophore modeling, are used to rationally design potent therapeutics with higher efficacies and fewer side effects[25]. The availability of more information about the molecular basis of IBD pathogenesis has further enhanced the use of computational methods in the design of small molecule drugs for IBD treatment. In the pathogenesis of IBD, there is an interplay between various factors such that any stimulus can lead to a cascade of events or even a vicious cycle. This gives a variety of therapeutic targets for which IBD drugs can be designed.
An interplay of genetic, epithelial and immune factors in the presence of intestinal microbiota are believed to be responsible for the development of the IBD[26].
The genetic factors causing IBD were initially established through family and twin studies. The disease was observed to cluster in families, with a relative risk of 13-36 for siblings of CD patients and 7-17 for UC patients[27]. Hence, the prevalence of IBD is much higher among first-degree relatives of those with IBD than it is among the general population, suggesting a role for genetic factors in the pathogenesis of the condition. Monozygotic twins exhibit a much higher rate of disease concordance than dizygotic twins, and there are significant differences in the incidence and prevalence of IBD among various populations, according to the twin studies[27]. In CD, the concordance rate for monozygotic twins is approximately 50% compared to 15% for dizygotic twins. Meanwhile, the concordance of monozygotic twins for UC is only 16%, suggesting that genetic factors are less dominant in this form of IBD. In general, there is a growing number of genetic markers linked with the pathogenesis of IBD at various levels including innate immunity, epithelial integrity, antigen presentation, cell adhesion, and drug transporter. Along with environmental and immune system factors, the genetic markers at the different levels play a major role in genetic susceptibility to IBD[28].
Nucleotide-binding oligomerization domain-containing protein 2 (NOD2) is a cytoplasmic protein that is induced in epithelial cells[29] and is constitutively expressed in neutrophils, macrophages, and dendritic cells[30]. Specifically, the leucine-rich repeat domain of NOD2 is necessary for recognition of muramyl dipeptide, a fragment of peptidoglycan present in bacterial cell walls. Ultimately, muramyl dipeptide causes nuclear factor kappa B activation and the induction of proinflammatory cytokines[31,32]. Activation of the innate and adaptive mucosal immune responses as a result of NOD2 poly
Evidence from genome-wide association studies (GWAS)[33,34] also shows the involvement of hepatocyte nuclear factor 4α (HNF4α) in the pathogenesis of UC. HNF4α is a member of the nuclear receptor superfamily of ligand-dependent transcription factors that is highly conserved and well expressed in liver and gastrointestinal organs. Several studies have linked HNF4α to intestinal epithelial differentiation, lipid metabolism and epithelial junctions[27,35], which are important components in colon development[36]. A zebrafish model suggests that the gut microbiota negatively regulates expression levels of HNF4α[37]. There is evidence linking alterations in HNF4 expression and activity, as well as germline variants, to IBD and colorectal cancer[38,39]. In UC, polymorphisms of HNF4α result in defects of epithelial tight junctions and intestinal permeability. Due to these epithelial barrier defects, there is transepithelial influx of bacteria, activation of the immune responses, release of cytokines, and further increase in epithelial junction permeability[40-42].
The development of UC has also been linked to a decrease in fatty acid oxidation in the colonic mucosal epithelium. Carnitine is a necessary cofactor in lipid metabolism and may be transported by the organic cation transporter (OCTN), a family of transporter proteins for organic cations. Carnitine helps shuttle long-chain fats into the mitochondria. Both OCTN1 and OCTN2 have been linked to CD-causing mutations: SLC22A4 1672C/T for OCTN1 and SLC22A5 +207G/C for OCTN2. The TC haplotype, formed by the presence of one or more of these mutations, is linked to the development of ileal, colonic and perineal symptoms and the requirement for surgical treatment of CD[28,43].
A number of tissues including the colon, small intestine, placenta, liver, heart and skeletal muscle contain high levels of Drosophila long disc homologue 5. The Drosophila long disc homologue 5 gene belongs to the membrane-associated guanylate kinase gene family, which encodes cell scaffolding proteins. It is thought to be involved in the maintenance of epithelial cell integrity and in signal transduction. Mutations in this gene have been linked to an increase in intestinal permeability[28]. The D haplotypes were found to be associated with UC and CD in a European cohort[44]. CD patients in Japan also showed another form of this gene (rs37462)[45]. Patients with IBD were found to have an excess transmission of haplotype D, which is defined by the haplotype-tagging single nucleotide polymorphisms (SNPs) G113A[28].
In normal intestinal mucosal immune response, the presence of gut microbiota has conditioned the dendritic cells present in the epithelium so that there is activation of T regulator cells that produce anti-inflammatory cytokines interleukin (IL)-10 and transforming growth factor-beta (TGF-β) as well as suppression of T effector cells. However, in IBD, the dendritic cells respond to gut microbiota by activating CD4+ T effector cells that differentiate into T helper (Th1, Th2 and Th17) cells depending on the IBD type[40,42]. Recently, the proinflammatory cytokine IL-12 family, which includes IL-22, IL-23, IL-25, and IL-27, has been linked to the pathophysiology of CD and other immune-mediated disorders[46]. The IL-12 family is responsible for the differentiation of Th cells into Th1 cells. IL-12 and IL-23 are heterodimeric proteins with a unique subunit linked to a shared p40 subunit. Patients with CD have elevated levels of these subunits[47,48]. Ustekinumab, a Food and Drug Administration (FDA)-approved drug for moderate-severe CD, inhibits IL-12 and IL-23 receptors on T cells, natural killer cells, and antigen-presenting cells[49]. In addition, several monoclonal antibodies have been discovered that target the Th1/Th17 pathway via IL-23. However, none of them have progressed to phase 3 trials[50,51].
The upregulation of IL-13, another crucial cytokine in the Th2 immune response, is considered to be a major trigger of mucosal inflammation in UC patients[52,53]. Research in mice revealed an increase in IL-13 in colitis, and this increase could be mitigated by inhibiting the ability of IL-13 to bind to its signaling receptor[54]. Tralokinumab is a human immunoglobulin G4 monoclonal antibody that binds to IL-13 and inhibits its activity.
Molecular studies have also identified NOD2 as a susceptibility gene in CD. The gene encodes a protein that transduces signals to activate nuclear factor kappa B in monocytes and functions as an intracellular receptor for bacterial products. Muramyl dipeptide activates NOD2, leading to autophagy in dendritic cells. Defective autophagy induction and decreased bacterial localisation in autophagolysosomes are observed in CD patients with susceptibility polymorphisms in the NOD2 gene. Genetic analyses have identified two other genes that are involved in autophagy and intracellular bacteria clearance, namely IRGM and ATG16L1, demonstrating the importance of autophagy in IBD immune responses. Other genes related with autoimmune disease, such as IL23R and PTPN2, reveal an additional feature of CD pathophysiology[38,40].
Another major player in IBD pathogenesis is TGF-β1, an essential factor in the healing of intestinal cells and the reduction of inflammation[55]. It is an immunosuppressive cytokine with the ability to inhibit pathogenic T cell activity and antigen-presenting cell responses. Increased levels of Mothers against decapentaplegic homolog 7 (SMAD7), an intracellular protein that binds TGF-β1 receptor and prevents TGF-β1- and SMAD-associated signaling, are believed to account for the deficiency in TGF-β1 activity observed in IBD patients. Mongersen, an oral medication that inhibits SMAD7 activity after administration to the terminal ileum and right colon is being investigated for efficacy in CD[56]. In its current form, the site of action is limited to the terminal ileum and right colon, hence it may not be beneficial for patients with more complicated CD or post-operative recurrence[57].
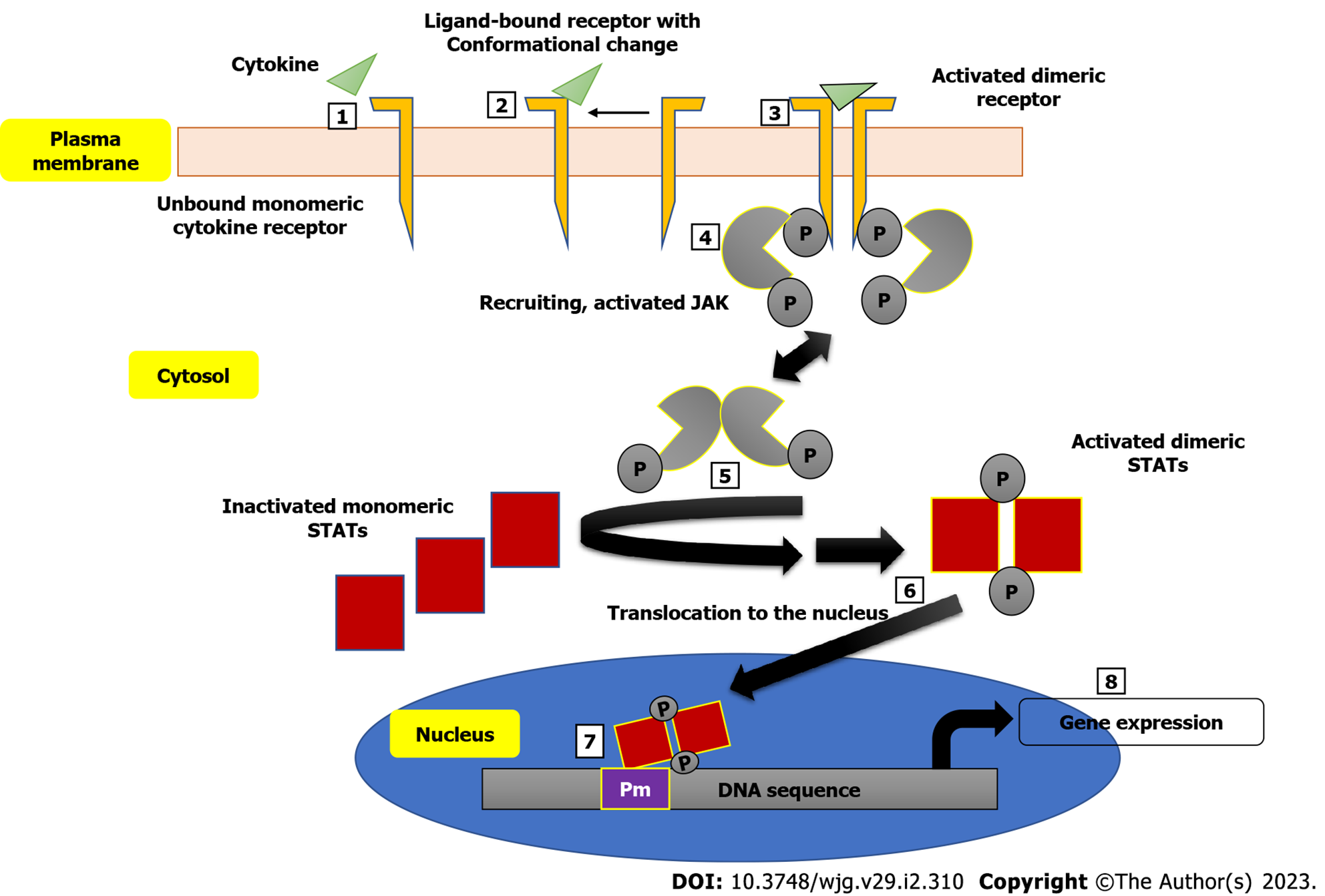
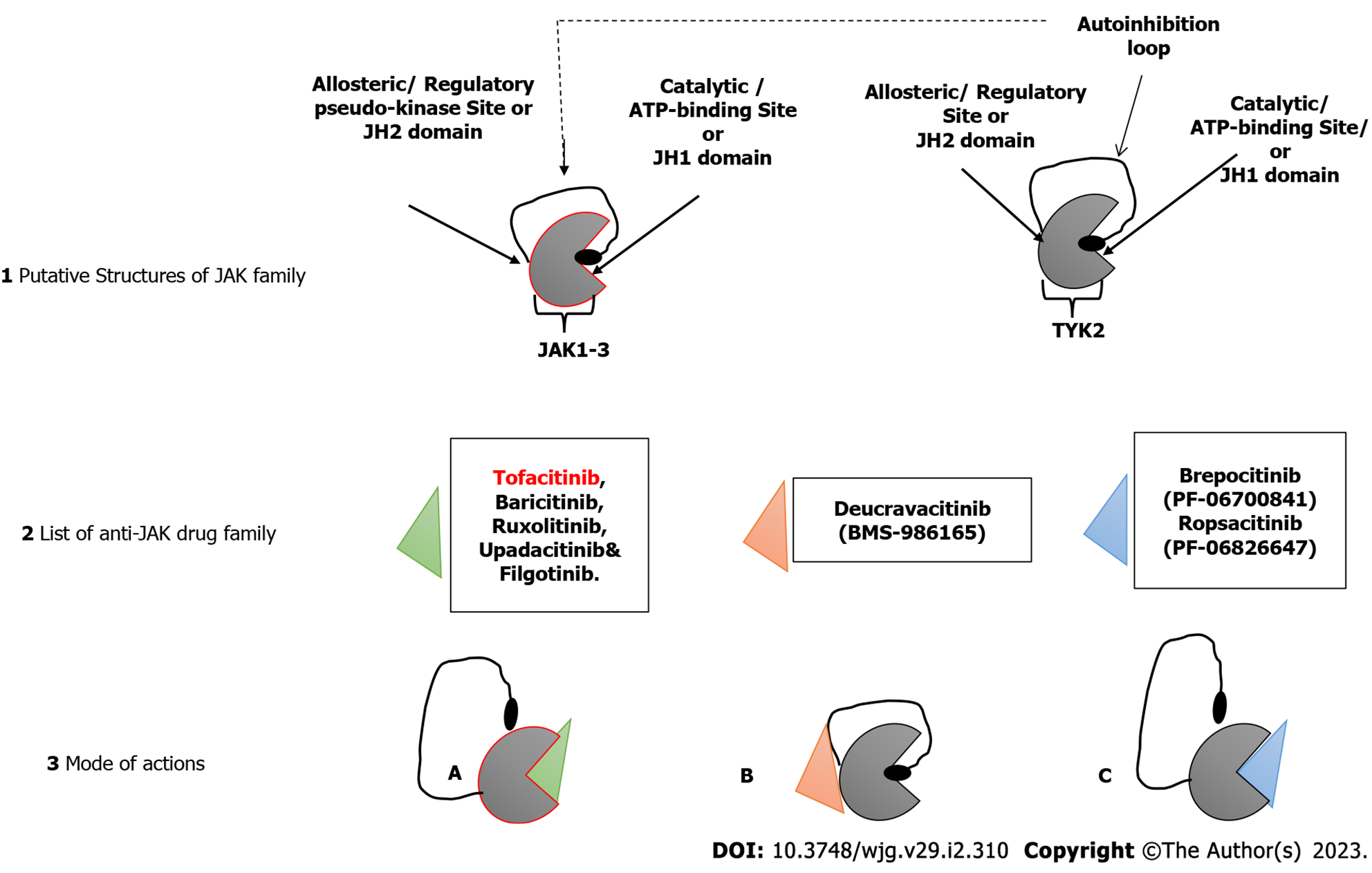
The involvement of the JAK family of intracellular non-receptor tyrosine protein kinases in the pathogenesis of a number of autoimmune diseases has also been demonstrated[58]. Many autoimmune diseases, including IBD, have been linked to specific SNPs in the genome, and the, JAK/signal transducer and activator of transcription proteins (JAK/STAT) signaling pathway has yielded several cytokines and receptors as potential treatment targets[59]. The JAK/STAT pathway interferes with a couple of inflammatory pathways and development of disease as seen in CD and UC patients is characterized by imbalanced effector T cells, leading to increased effector Th cells (Th1, Th2 and Th17 cells), thus mediating the inflammation[60]. Figure 1 shows a schematic description of the JAK/STAT pathway leading to the expression of genes encoding some inflammatory markers. Inhibition of JAK leads to the inhibition of signaling of a specific subset of cytokines, which are implicated in inflammation[61]. Some small molecule inhibitors of the JAK/STAT pathway with good oral bioavailability have been developed. A very good example is tofacitinib. When compared to monoclonal antibodies, JAK inhibitors have the therapeutic advantage of being able to target multiple downstream signaling pathways induced by inflammatory cytokines, whereas monoclonal antibodies can only block a single molecule[62].
The successful development of tofacitinib, as well as the promising results of other JAK inhibitors in both UC and CD (as shown in Figure 2), demonstrate that JAK inhibition has a role in the treatment of IBD. However, long-term safety studies in people with rheumatoid arthritis and UC who took tofacitinib showed a higher risk of reactivation of herpes zoster, especially at higher doses[63]. This increased risk is likely a class effect of JAK inhibitors and related to IFN and IL-15 inhibition. Also, there might be a possible thrombogenic risk, as shown by patients with rheumatoid arthritis[64]. More selective JAK-1, JAK-3 or tyrosine kinase 2 inhibitors are expected to improve safety while maintaining efficacy. However, these drugs are still systemic; a gut-selective JAK inhibitor with high intestinal exposure and target engagement but no systemic effects are still needed for treating people with IBD. Continued progress is being made in all of these areas[65].
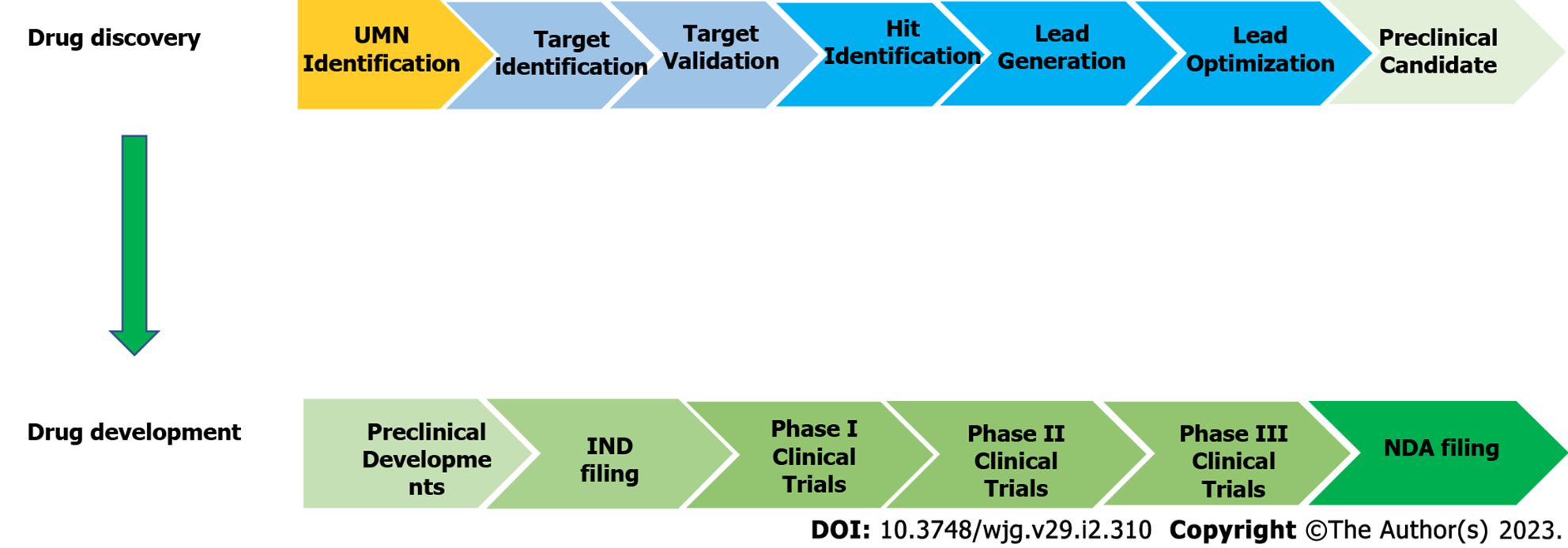
The process of identifying a therapeutic agent for extensive study as a potential drug candidate is known as drug discovery[66]. In general, the modern drug discovery process involves identifying a condition to be treated and its unfulfilled medical necessity, selecting a druggable molecular target and validating it, developing in vitro assays followed by high-throughput screening of compound libraries against the target to identify hits and optimizing hits to create lead compounds with acceptable potency and selectivity towards the biological target and demonstrated efficacy in an animal model. Following that, the lead compounds are further refined to improve their effectiveness and pharmacokinetics before moving further with drug development (Figure 3). The discovery and development of innovative drugs is a complex process that takes around 12 years and an average of $1.8 billion to bring a new medication to market[66,67].
The process of IBD drug discovery begins with target discovery and selection followed by biological confirmation in cellular and animal models[68]; this is usually the pre-clinical phase. Several therapeutic targets of IBD have been identified, and some are listed in Table 1. Target identification in IBDs is based on qualitative relevance to the pathogenesis of the disease as well as increased magnitude of expression[68]. Biochemical pathways and specific proteins are targeted, and the drugs are developed based on the understanding of the mechanism of action of these targets. This is followed by pharmaceutical developments such as screening for safety, toxicity, pharmacokinetics and efficacy. At the preclinical stage, drugs are tested with targets to investigate levels of interactions and outcomes. Models and methods used differ between laboratories based on specific targets of interest.
| Target name | Abbreviation | Description | Disease implication | Modulatory effect of drug |
| Integrin alpha-4 | ITGA4 | A member of the family of integrins. Integrins alpha-4/beta-1 (VLA-4) and alpha-4/beta-7 are fibronectin and VCAM1 receptors. Integrin alpha-4/beta-7 is also a MADCAM1 receptor. On activated endothelial cells, VLA-4 integrin induces homotypic aggregation in the majority of VLA-4-positive leukocyte cell lines. ITGA4: ITGB1 binds fractalkine (CX3CL1) and may function as its coreceptor in fractalkine signaling dependent on CX3CR1[123] | ITGA4 upregulated in irritable bowel disease (IBD) | Inhibition |
| Interleukin 12B | IL12B | IL12B is also known as natural killer cell stimulatory factor 2 or p40, and it associates with IL23A to form IL23, a known stimulator of the JAK/signal transducer and activator of transcription (STAT) signaling pathway and a pathway with proven importance in IBD[124] | IL12B upregulated in IBD | Inhibition |
| Tumor necrosis factor | TNF | A type of cytokine, which binds to TNFRSF1A/TNFR1 and TNFRSF1B/TNFBR. It is secreted by macrophages and is capable of triggering cell death of most tumor cell lines, although capable of promoting cell proliferation and induce cell differentiation under certain conditions[123] | TNF upregulated in IBD | Inhibition |
| Janus kinase 2 | JAK2 | A class of kinase, a non-receptor kinase that phosphorylates specific tyrosine residues on the cytoplasmic tails of the receptor. In the cytoplasm, JAK2 plays a pivotal role in signal transduction via its association with type I receptors such as growth hormone (GHR), prolactin (PRLR), leptin (LEPR), erythropoietin (EPOR), thrombopoietin (THPO) or type II receptors including IFN-alpha, IFN-beta, IFN-gamma, and multiple interleukins. It stimulates cell growth, development, differentiation or histone modification[123] | JAK2 upregulated in IBD | Inhibition |
| Prostaglandin-endoperoxide synthase 1 and 2 | PTGS1/2 | Also referred to as cyclooxygenase; are the primary enzymes involved in the synthesis of prostaglandin. They act both as a dioxygenase and as peroxidase, having two isozymes PTGS1 and PTGS2. This gene encodes the PTGS2 inducible isozyme. Its involvement in prostanoid-dependent inflammation and mitogenesis can be related to their regulation by specific stimulation[123] | PTGS1/2 upregulated in IBD | Inhibition |
| Peroxisome proliferator activated receptor gamma | PPARγ | A nuclear receptor. It consists of a group of approximately 50 transcription factors involved in many biological processes. It controls some regulatory genes involved in lipid metabolism and insulin sensitization as well as in inflammation and cell proliferation. It is highly expressed in the colon and majorly involved in bacterial-induced inflammation, also mediating the common aminosalicylate activities in IBD[125]. It acts as a critical regulator of gut homeostasis by suppressing nuclear factor-kappa B-mediated proinflammatory responses | PPARγ downregulated in IBD, mostly ulcerative colitis | Activation |
| Integrin subunit beta 7 | ITGB7 | Integrin alpha-4/beta-7 is an adhesion molecule that mediates lymphocyte migration and homing to gut-associated lymphoid tissue (GALT). The vascular endothelium of the gastrointestinal tract expresses MADCAM1, an adhesion molecule, which is the medium integrin alpha-4/beta-7 interacts with the gastrointestinal tract. VCAM1 and fibronectin found on the extracellular matrix of the cell also interacts with the integrin. Interaction with fibronectin is due to the CS-1 region[123] | ITGB7 upregulated in IBD | Inhibition |
| Nuclear receptor subfamily 3 group C member 1 | NR3C1 | This is a receptor recognized by glucocorticoids. It modulates the activities of cortisol and acts as a transcription factor that modulates the expression of its target genes[126]. It modulates inflammatory responses, cellular proliferation and differentiation in target tissues | NR3C1 downregulated in IBD | Activation |
| Janus kinase 3 | JAK3 | Non-receptor tyrosine kinase involved in signal transduction in the cytoplasm via its association with type I receptors sharing the common subunit gamma such as IL2R, IL4R, IL7R, IL9R, IL15R, and IL21R. It also plays a vital role in STAT5 activation. IBD pathology is associated with receptor-mediated signaling through the JAK and STAT DNA-binding families of proteins[127] | JAK3 upregulated in IBD | Inhibition |
| Arachidonate 5-lipoxygenase | ALOX5 | ALOX5, an important member of the lipoxygenase gene family, exclusively involved in IBD development[128]. Catalyzes the oxygenation of arachidonate (5Z,8Z,11Z,14Z)- eicosatetraenoate) to 5-hydroperoxyeicosatetraenoate (5-HPETE) followed by the dehydration to 5,6- epoxyeicosatetraenoate (leukotriene A4/LTA4), the steps in the biosynthesis of leukotrienes, that mediates inflammation[123] | ALOX5 upregulated in IBD, especially ulcerative colitis | Inhibition |
| Tyrosine kinase 2 | TYK2 | A non-receptor kinase that carries out both structural and catalytic roles in numerous cytokines and interferons signaling. TYK2 plays a key role in mediating signaling and functional responses downstream of the IL-12, IL-23, and type I interferon (IFN) receptors and TYK2-mediated IL-12, IL-23 and type I IFN signaling activates STAT-dependent transcription, which promotes chronic inflammation[129] | TYK2 upregulated in IBD | Inhibition |
| Phosphoribosyl pyrophosphate aminotransferase | PPAT | PPAT is a key rate-limiting reaction in purine biosynthesis, transferring gamma-nitrogen from glutamine to 5-phosphoribosyl pyrophosphate (PRPP)[130] | PPAT upregulated in IBD | Inhibition |
| Vitamin D receptor | VDR | A nuclear, ligand-dependent transcription factor that regulates the expression of T cells and genes involved in different physiological functions when in complex with hormonally active vitamin D, 1,25(OH)2D3[131]. VDR plays a multifaceted role in the pathogenesis of IBD and is crucial in regulating autophagy, immune response, tight junctions, gut microbiome, and metabolites[132] | VDR downregulated in Crohn’s disease | Activation |
| Matrix metallopeptidase 1 | MMP1 | An interstitial collagenase, that digests the spiral structure of collagen types I, II, III and X, subjecting them to hydrolysis by gelatinase and are major players in extracellular matrix degradation[123] | MMP1 upregulated in IBD | Inhibition |
| Matrix metallopeptidase 7 | MMP7 | A metallopeptidase member necessary for neutrophil migration into the airspaces by cleaving syndecan-1, a heparin sulfate proteoglycan necessary for the establishment of a neutrophil chemokine gradient[133]. Degrades casein, gelatins I, III, IV and V, and fibronectin and is responsible for the activation of procollagenase[123] | MMP7 upregulated in IBD | Inhibition |
| Dihydrofolate reductase | DHFR | An enzyme that converts dihydrofolate to tetrahydrofolate in folate metabolism and involved in purine and mitochondrial thymidylate biosynthesis[123] | DHFR upregulated in Crohn’s disease | Inhibition |
| Matrix metallopeptidase 13 | MMP13 | A member of the family of collagenases. Matrix substrates of MMP13 include native collagen, gelatin and aggrecan. Lipopolysaccharide (LPS)-induced shock and dioctyl sodium sulfosuccinate (DSS)-induced colitis induce MMP13 upregulation in the gut, which results in MMP13-mediated shedding of transmembrane-bound TNF and release of bioactive, soluble TNF, thus triggering a cascade that leads to leakage of intestinal components, which increases systemic inflammation and subsequent organ damage[134] | MMP13 upregulated in IBD | Inhibition |
| Sphingosine-1-phosphate receptor 1 | S1PR1 | A type of G-protein-coupled receptor. S1P binds to the S1PR1, which triggers angiogenesis, endothelial barrier enhancement, blood vessel constriction, heart rate modulation and lymphocyte trafficking[135] | S1PR1 downregulated in IBD | Activation |
| ATPase H+/K+ transporting subunit alpha | ATP4A | A P-type cation-transporting ATPase. The gastric H+, K+-ATPase is a heterodimer made of high molecular, weight catalytic alpha subunit with a glycosylated beta subunit. It is a proton pump that catalyzes the hydrolysis of ATP coupled with the exchange of H (+) for K (+) ions across the plasma membrane and also responsible for gastric acid secretion due to its ability to generate proton gradient across the membrane[123] | ATP4A upregulated in IBD | Inhibition |
For instance, in IBD, targets such as JAK/STAT and inflammatory mediator interleukins such as IL-12, IL-23, IL-6, IL-22Fc[69], tyrosine kinase and toll-like receptors[70] are explored using biologic agents at different stages of development. Following the in vivo studies using murine and human T cells, JAK/STAT inhibition by tofacitinib[71] has led to further application of the drug in several clinical trials. Integrins, a class of receptors that facilitate T lymphocyte trafficking into the gut[72] are targets as well. Integrins facilitate adhesive interactions between lymphocytes and endothelial cells, which leads to the trafficking[73] and can lead to T cell-dependent chronic intestinal inflammation in IBD[70]. Etrolizumab, an anti-adhesion agent approved by the FDA for treatment of IBD was subjected to in vitro testing, where it internalized β7 integrin using cell culture models[74]. Etrolizumab selectively inhibits α4β7 and αEβ7, which are involved in T lymphocyte homing in the gut[75]. Although there are challenges involved in the use of in vivo and in vitro methods in establishing IBD[68], conditions for the studies of potential drugs are optimized. The molecules with desired preclinical effect on the target go into clinical trial.
In clinical trials, drugs that scale through preclinical studies are expected to maintain remission in long-term mucosal healing. Clinical trial phases are regarded as long and extensive periods that take years from the point of research to the stage of approval of a candidate drug. Prior to these trials, detailed protocols that show the characteristics of the studies are provided[76]. The clinical phase comes after certainty has been established that the drugs/molecules are effective and non-toxic. Clinical trials involve three main phases before approval, known as phase I, phase II, and phase III. The first two phases are early phases.
The phase I is the stage in drug development when testing and pharmacokinetics studies are carried out using healthy or ill subjects[76] to ascertain safety level of the drugs. The pharmacokinetics and pharmacodynamics of the drugs are determined using volunteer patients or patients who are not responsive to previous treatments in order to determine drug safety alongside desired clinical activities and an Absorption, Digestion, Metabolism and Excretion (ADME) profile[68]. For instance, a phase I study of Japanese patients with UC treated with vedolizumab, an anti-integrin antibody, showed an appreciable level of tolerance in the desired clinical activity of the drug as well as a satisfactory level of pharmacodynamics and pharmacokinetics[77]. The outcome of this phase determines whether the drug scales through to the next phase. The phase I studies can last for up to many years and part of the process involves identification of adverse effects or events other than the occurrence(s) observed in the preclinical stage. Foreknowledge of a possible mechanism of action of a drug can also guide expectations in clinical trials.
The phase II is the stage that involves dose ranging. The efficacy of the drugs are determined alongside the optimum dose that would be considered safe[76]. At the point of dose determination, the outcome of the effect of different doses would guide the decision to proceed to the next phase. A phase II study of varied doses of ozanimod, an agent that targets sphingosine-1-phosphate receptors 1 and 5, was used to treat adult patients with moderate to severe CD based on an uncontrolled multicenter trial within 12 wk and the effects of different doses at different intervals were observed[78]. A drug that scales through is launched into the third phase.
Phase III is the stage whereby the efficacy of the drug is compared with that of an already established standard. Patients recruited for this phase are usually from the general population compared to phase I and II. The side effects of the drugs are studied as well as their effectiveness, comparing them with common treatments to help guide safety and proper use. Currently, etrolizumab, is undergoing a phase III clinical trial for treatment of IBD using six multicenter prospective randomized controlled trials and two open-label extension studies[75]. Upon conclusion of the studies, the drugs are considered for approval based on FDA standards. Drugs are reviewed by the FDA based on the evidence of preclinical and clinical outcomes and approved for marketing based on the terms of the FDA.
The use of computational methods has become a crucial part of the drug discovery process[18]. A number of commercially available medications as well as several clinical candidates have been discovered or improved with the help of molecular modeling techniques. CADD combines multiple chemical-molecular and quantum approaches to the discovery, design and development of drugs. Structure-activity relationships form the basis of many CADD methods. The objectives of CADD are multidisciplinary in nature, with the end goal being the modification of bioactive molecules, the formation of therapeutic alternatives and the knowledge of biological events at the molecular level[79].
From drug identification to pharmacological role discovery and from preclinical testing to drug marketing, computers have played a fundamental role in the entire drug discovery and development process[15]. In addition, CADD opens the door to the possibility of drug repurposing, which is the systematic identification of new potential uses from drugs that have already been approved for other indications[79]. According to the results of a computational drug repurposing study conducted by Bai et al[80], atorvastatin shows promise as a novel treatment approach for improving symptoms in patients with UC. They developed a framework for systematically integrating publicly accessible heterogeneous molecular data with clinical data on a large scale in order to repurpose FDA-approved drugs for a wide variety of human diseases.
Computational methods are useful in various aspects of modern drug discovery for IBD. These include but are not limited to genetic studies for the identification of pathways linked to IBD pathogenesis, target identification, and virtual screening.
Computational methods have made tremendous contributions to the field of IBD genetic studies. As mentioned earlier, genetic factors contribute to the pathogenesis of IBD and the understanding of the molecular events are important for target identification. The advent of GWAS is a turning point in the history of genetic research into complex human disease. GWAS is a technique for identifying genetic markers that are linked to an increased probability of developing a disease or exhibiting a certain trait. The strategy entails analyzing the genomes of a large number of people to identify genetic variations that are more common in people who have a disease or a particular trait than in those who do not. Following the discovery of such genomic variants, researchers often use GWAS to search for other potentially causal variants located in close proximity[81]. IBD is a prime example of the usefulness of GWAS and related analyses. Using GWAS based on SNPs, researchers were able to identify 163 genetic loci and numerous signaling pathways that are linked to IBD[82]. Many of these loci have pleiotropic effects, and risk prediction models were developed using a wide variety of genetic variations. Key contributions of the gut microbiome and important interactions between genes and the environment are emerging as a result of these studies[83].
The development of computational methods has contributed significantly to the identification of the vast quantity of genetic determinants. There are a number of different software packages available for use in GWAS research[84], and a common example is PLINK. In addition to standard GWAS features like quality control filtering and SNP association testing, PLINK also includes more advanced tools like gene-based analysis, annotation and epistasis testing in a compact, user-friendly package. PLINK initially discovered the actin-related protein 2/3 complex subunit 2[85], IL10, HNF4a, cadherin-3, cadherin-1 and laminin subunit beta-1[33] loci and recently discovered 163 IBD loci[82]. In the majority of GWAS-based genetic studies of IBD, PLINK has served as the primary analytical tool. A linear mixed model, which accounts for both fixed and random effects, has been used in GWAS in recent years. Factored Spectrally Transformed Linear Mixed Models[86], Efficient Mixed-Model Association expedited[87] and Genome-wide efficient mixed-model association (GEMMA)[88] are all examples of linear mixed model-based software. They are able to adjust for both overt and covert connections within a population simultaneously. Multiple correlated phenotypes can be analyzed with GEMMA as well.
There is a plethora of pathway analysis software packages available, many of which are based on GWAS[89]. The development and implementation of GWAS-based pathway software has frequently been illustrated with CD as an example. To examine the CD dataset from the Wellcome Trust Case Control Consortium, Wang et al[90] used the GenGen software to find significant associations of the IL12/IL23 pathway with CD status in multiple cohorts genotyped with different SNP chips and from different ethnic backgrounds[91]. This was among the first analyses of IBD pathways based on GWAS data. IL3 activation and signaling pathway were also linked to CD, based on the research by Torkamani et al[92]. They used the MetaCore program, which is a for-profit product created by GeneGo Inc. (St. Joseph, MI, United States).
Using the statistical technique of Simes/false discovery rate, Peng et al[93] discovered significant enrichment of the JAK/STAT signaling pathway, as well as the cytokine-cytokine receptor interaction. With a model-based strategy, Carbonetto et al[94] found a similar result for a number of cytokine signaling pathways. Holmans et al[95] used the software ALIGATOR to discover the involvement of MHC genes in addition to IL/cytokine signaling. Similarly, Jostins et al[82] examined the largest IBD GWAS to date for enrichment of canonical pathways or Gene Ontology terms and discovered significant enrichment of Gene Ontology terms pertaining to the regulation of cytokine production, lymphocyte activation, and the JAK/STAT signaling pathway.
It is possible that the differences between the studies can be attributed to the different statistical methods and/or the different pathway databases used in their studies. However, all of these pathways were related to interleukins and the immune system. Torkamani et al[92] found a significant association between calcium signaling and the carbohydrate response element binding protein regulation pathway; Peng et al[93] found a significant association between ABC transporters and the extracellular matrix receptor interaction. Numerous user-friendly web-based software programs, such as improved Gene Set Enrichment Analysis for (i-GSEA4) GWAS[96] and GSEA-SNP[97], have been developed for pathway analysis using GWAS data.
Following the discovery of the biological basis of a disease, the next step in drug discovery is the identification of potential drug targets. The most promising targets for drug discovery are hypothesized to be highly prevalent in the disease-affected population, to have a well-established function in the underlying pathology and to be directly linked to the disease of interest. Potential drug targets are defined as disease-modifying rather than disease-causing. In the present day, a wide range of methods, both experimental and computational, are used to identify drug targets. The experimental methods rely heavily on comparative genomics, with phenotype and gene association analysis serving as complementary tools. All experimental methods yield credible findings, but they have significant drawbacks, including the high cost and extensive scientific labor needed to experimentally probe the entire space of chemical compounds to identify viable drug targets[98]. In light of these drawbacks, researchers and pharmaceutical companies increasingly rely on computational methods for initial investigations before turning to experimental approaches for validation and other purposes.
Various bioinformatics resources are available for the identification of drug targets as illustrated in Table 2. These programs efficiently process a large volume of data from genomic, transcriptomic and proteomic databases and ultimately provide potential drug targets in a short period of time and at a low cost. Several computational methods are currently accessible, each of which makes use of a unique type of molecular information, such as a gene or genomic sequence, molecular interaction data or the 3D structure of a protein[98]. There are strong connections between most of these methods.
| Tool/Database | Description |
| Open Targets Platform | To facilitate systematic target identification and prioritization for drug discovery based on underlying evidence, the Open Targets Platform offers users a searchable knowledgebase and user interface[123] |
| SELF-BLM | A self-training support vector machine-based bipartite local model that predicts drug-target interactions[136] |
| iDTIESBoost | A model for detecting drug-target interactions based on evolutionary and structural features[137] |
| GEO | Database that stores array- and sequence-based transcriptomics data that can be applied to functional genomics[138] |
| DASPfind | Predicts drug-target protein interactions that stem from shared structural features[139] |
| NetCBP | Network methods for predicting drug-target interactions. Furthermore, it suggests new drugs even when no data on their interactions with their targets are available[140] |
| DbMDR | Offers a database of multidrug resistance (MDR) genes and their orthologs, which could be used to develop new treatments[141] |
| TDR targets | Drug development molecular target identification and prioritization[142] |
| DrugBank | An extensive drug database with annotations covering drug targets and mechanisms of action[143] |
| PDTD | Database of potential proteins for in silico drug target identification[144] |
| DEG | Contains all known essential genes from different organisms[145] |
| TTD | Publicly accessible cross-links database that provides inclusive information about known therapeutic targets with related information, i.e. pathway information and the corresponding drugs/ligands[146] |
| KEGG | Offers information about the pathway, gene and ligands in three different databases, i.e. Pathway, Gene and Ligand[147] |
| Genecards | Officially known as Genecards: The Human Gene Database, it is an all-inclusive, authoritative compilation of annotative information about human genes[148] |
| DisGeNET | A public resource that houses a massive database of genetic variants and their links to human disease[149] |
| CTD | The Comparative Toxicogenomics Database is a vast, freely accessible database with the objective of increasing understanding of the effects of environmental exposures on human health. It includes information on chemical-gene/protein interactions, chemical-disease relationships and gene-disease links that has been curated by humans[150] |
| UniProt | The Universal Protein Resource is the world’s most comprehensive, high-quality and freely accessible database of protein sequence and functional information[151] |
Mohan et al[99] reported using genetic databases to find new molecular targets for IBD. They used four different genetic databases to categorize the protein-coding genes associated with UC (3783 genes), CD (3980 genes), uveitis (1043 genes), arthritis (5583 genes), primary sclerosing cholangitis (1313 genes), and pyoderma gangrenosum (119 genes). The databases used were Genecards: The Human Gene Database, DisGeNET, the Comparative Toxicogenomics Database, and the Universal Protein Resource. Then, they used Network Data Exchange to map a distinct signal pathway based on the identified common genes underlying the aforementioned diseases. Across UC, CD, uveitis, arthritis, pyoderma gangrenosum and primary sclerosing cholangitis, they identified a distinct set of 20 genes with the highest probability of overlap. Different disease processes were linked to some unique immune modulators. IL-25 and monensin-resistant homolog 2 were observed in UC, CD, pyoderma gan
Drug discovery relies heavily on the physical screening of large chemical libraries for biological targets in order to find new lead compounds. High-throughput screening is a method for finding active molecules in experiments by analyzing more than a million compounds biochemically. However, developing and deploying this technology takes a long time and a sizable investment. Therefore, virtual high-throughput screening was developed as a more affordable and effective calculation method. This technique has seen extensive use in the earliest stages of drug discovery. The goal is to search through huge compound libraries to find the structure of a novel, active small molecule. To some extent, this supports the goals of high-throughput screening. Virtual screening saves money by reducing the number of compounds used to measure pharmacological activity, while high-throughput screening uses all compounds in the database[18,24,100]. In this sub-section, we will go over the various virtual screening techniques that are commonly used in IBD drug discovery.
Molecular docking: Molecular docking is commonly used in the drug discovery and development process because of its ability to predict interaction patterns between proteins and small molecules as well as proteins[18,101,102]. The principle behind this phenomenon proposes that ligand and receptor recognition is predicated on a similarity in spatial shape and energy. Understanding the action mechanism of a drug requires first establishing its binding conformation to a specific protein receptor[24]. The goal of docking is to accurately assess the strength of binding by fitting the structure of a ligand within the requirements of a receptor binding site[103]. The most frequently cited molecular docking software packages for use in drug discovery are summarized in Table 3.
| Program | Description | Website |
| AutoDock[152] | A docking toolkit. It is meant to foretell the binding mode of small molecules to a receptor with a known 3D structure, such as a substrate or a drug candidate. There have been multiple engines developed, and it has undergone constant evolution and refinement over the years to incorporate new features | https://autodock.scripps.edu/ |
| AutoDock Vina[153] | One of the AutoDock Suite’s docking engines. It is a free and open-source molecular docking software. Dr. Oleg Trott of The Scripps Research Institute’s Molecular Graphics Lab (now CCSB) created and initially implemented the system. The most recent version of AutoDock Vina is v.1.2.0 | https://vina.scripps.edu/ |
| Hex[154] | Invented by Dave Ritchie and is a program for molecular superposition and protein docking. Hex can read protein and DNA structures in the Protein Data Bank format as well as small-molecule SDF files. It has been downloaded over 40000 times as of December 2015 | http://hex.loria.fr/ |
| MOE[155] | Integrated computer-aided molecular design platform for small molecule and biological therapeutics. Common platform for chemists, biologists and crystallographers. Small Molecules - Peptides – Biologics | https://www.chemcomp.com/ |
| Glide Schrodinger[156] | Provides a full range of speed vs accuracy options, ranging from the high-throughput virtual screening mode that efficiently enriches million compound libraries to the standard precision) mode that reliably docks tens to hundreds of thousands of ligands with high accuracy to the extra precision mode that eliminates false positives by sampling more extensively and using more advanced scoring, resulting in even higher enrichment | https://www.schrodinger.com/ |
Keretsu et al[104] used computational methods to design new JAK1 inhibitors from a series of pyrrolopyridine derivatives. Autodock 4.2 was utilized to predict the protein-ligand binding. Tofacitinib, an established ligand, which was co-crystallized with the structure of JAK1 (protein data bank ID: 3EYG) obtained from the protein data bank (www.rcsb.org), was used as a reference compound to validate the docking procedure. The docked conformation of tofacitinib closely matched that of one obtained in the crystal structure. The binding affinity of compound 42 for JAK1 was found to be -10.2 kcal/mol. The F958 and L959 residues of the protein formed H-bond interactions with the pyrrolopyridine moiety. As a result, the methyl group of the methyl piperidine moiety protruded beyond the binding pocket and into the bulk solvent. The chlorobenzyl group slid into the hydrophobic pocket made by the residues of the activation loop, the α-helix and the P-loop. The selected binding conformation was prepared for further MD simulation study. The finding of the study was presented as a possible guide in the design of more effective JAK1 inhibitors[104].
In order to identify the bioactive compounds of the Phyllanthus nivosus leaf responsible for its activity against UC for further drug design, Johnson et al[101] combined molecular docking with an in vivo study. Levels of TNF-α, IL-6, nitric oxide, malondialdehyde, reduced glutathione, superoxide dismutase and catalase in the serum of rats experimentally induced with UC and treated with Phyllanthus nivosus were monitored. The bioactive ingredients of the most active fraction were identified using gas chromatography-mass spectrometry followed by molecular docking against IL-1b converting enzyme (caspase-1), beta-2 adrenergic receptor, cyclooxygenase-2 and TNF-α. The study led to the identification of ethyl iso-allocholate cholest-22-ene-21-ol, 3,5-dehydro-6-methoxy-, pivalate and alpha-cadinol as promising compounds for further development into drugs for the treatment of UC[101].
In another study, Halder et al[105] used molecular docking in combination with other in silico techniques for the repurposing of FDA-approved medications and provided a framework for drug exploration and computational methods in the discovery of drugs for the treatment of IBD. After being imported and processed using Protein Preparation Wizard and LigPrep, respectively, the molecular target TNF (protein data bank ID: 2AZ5) with a small molecule inhibitor and the FDA-approved drugs (from the Zinc database) were subjected to molecular docking, ADMET analysis and binding free-energy calculation [Molecular mechanics with generalised Born and surface area solvation (MMGBSA)]. Following that, the medications were ranked based on docking score, ADMET parameters and MMGBSA dG binding score. The selected drugs were then subjected to an induced-fit docking study. The MD simulation study was conducted on the two most promising compounds, iopromide (ZINC000003830957) and deferoxamine (ZINC000003830635). Finally, the bioisosteric substitution was applied to enhance the ADMET properties of these compounds[105].
Pharmacophore modelling: Virtual screening of databases with the pharmacophore model has become one of the most important ways to find new lead compounds as compound databases and computing power have advanced. A pharmacophore is a conceptual description of the molecular features required for molecular recognition of a ligand by a biological macromolecule, which describes how structurally distinct ligands can bind to the same receptor site. To achieve therapeutic efficacy, drug molecules adopt an active conformation that is both geometrically and energetically complementary to that of the target macromolecule.
Medicinal chemists have discovered that modifications to specific chemical groups in drug molecules greatly affect the interaction between drugs and targets, while modifications to other groups have little to no effect[106]. Additionally, it was discovered that molecules exhibiting the same activity share common properties. Accordingly, Ehrlich proposed the idea of pharmacophores in 1909[107], which referred to the molecular framework of atoms with active essential characteristics. In 1977, Gund[108] provided further elaboration on the concept of pharmacophores as a class of molecules that recognize receptors and form structural features of molecular biological activity.
Pharmacophores can be discovered using one of two common approaches. If the structure of the target molecule is known, then the structure of the pharmacophore can be inferred using techniques like conformational analysis and molecular folding[109]. The pharmacophore recognition procedure will then choose an active compound that can be used to create the model. Conversely, pharmacophore studies are conducted on a number of compounds when either the structure of the target or its action mechanism is still unknown; this allows for a summary of data on certain groups that are crucial to the activity of the compound[24].
Babu et al[110] combined ligand-based pharmacophore modeling with virtual screening and molecular docking to find JAK1 inhibitors with high potency and selectivity. In the first step, they developed ligand-based pharmacophore models and checked them for accuracy with potency and selectivity validation techniques. A pharmacophore-based virtual screening was carried out on eight selected pharmacophore models using six different databases. ADME prediction and molecular docking were used to narrow down the hits found during screening. Docking results were verified using the binding free-energy calculation and induced fit docking techniques. A cross docking analysis was then performed to determine which lead compounds are selective for JAK1. In the end, five promising compounds were chosen and subjected to further investigation using MD and density functional theory. T5923555 and T5923531 were identified as the most promising leads among the five compounds and will be pursued for additional validation via in vitro and in vivo techniques.
QSAR: QSAR is frequently used in the drug discovery process to find compounds with desirable inhibitory effects on target proteins, with minimal side effects (nonspecific activity). The QSAR model is a quantitative investigation into the relationships that exist between small organic molecules and large biological macromolecules. The calculated properties of molecules (such as the absorption, distribution and metabolism of small organic molecules in living organisms) are correlated with their biological activity, as determined experimentally[111]. QSAR is the most precise and efficient approach to drug design when the structure of the receptor being targeted is unknown.
In the 1980s, 3D structural information was incorporated into the quantitative structure-activity relationship to form the 3D-QSAR method. Since the 1990s, structure-based drug design has increasingly replaced QSAR in the area of drug design due to the increase in computational power and the availability of the 3D structure of many biomolecules. However, QSAR with its advantages of small amount of calculation and good predictive ability[112], continues to play an important role in pharmaceutical studies.
3D-QSAR enables the investigation of the three-dimensional structure of bioactive compounds and the correct representation of energy changes and interaction patterns between bioactive molecules and receptors. Different drugs are evaluated by fitting their physicochemical and 3D structural parameters to the quantitative relationship. The structures of the newly created compounds are subsequently predicted and improved upon. 3D-QSAR analysis is a research method that integrates QSAR with computational chemistry and molecular graphics. It is an effective method for determining the nature of drug-target interactions, creating hypothetical images of simulated targets, determining the correlation between drug structure and activity and developing new medications. In addition, predictive 2D-QSAR models, 3D-QSAR models and 3D target and ligand-based approaches have all been developed for the purpose of finding IBD drugs[24].
Yang et al[113] incorporated molecular docking and QSAR study in the design of a series of TNF-α converting enzyme (TACE) inhibitors with the ability to bind in the S1’ pocket of the enzyme. A total of 12 analogues were synthesized by altering the chain length and alkylation pattern on the aromatic ring of the side chain. Most compounds inhibited cellular TNF-α production and TACE in vitro. The most promising compound from in vitro and in vivo pharmacokinetic studies had a moderate systemic clearance and a good oral bioavailability of 42%. It was also tested in a rat model of carrageenan-induced paw edema and found to be effective at reducing edema in the animal’s paws. The series of α-alkoxyaryl alkyl substituted chromen-based analogues was then validated by means of a QSAR study and docking. Coumarin core TACE inhibitors with long, bulky α-substituent groups are able to enter the S1’ and S3’ pockets, where they form van der Waals interactions, with increased inhibitory activity. The docking study of the compound demonstrated its dual-function inhibitory activity toward TACE and matrix metallopeptidase-3. Based on the resulting QSAR descriptors, new α-substituted chromen-based TACE inhibitors with enhanced TACE inhibitory activity can be developed.
MD simulation in IBD drug discovery: MD simulation[102,114-116] is another popular approach to studying biomolecules; it is based on Newtonian mechanics and applies empirical molecular mechanics force fields. The drug discovery process can be aided by using explicit/implicit solvent models, which allow for simulations of time and space, and all-atom, united-atom and coarse-grained MD simulations[117,118]. MD simulations have typically been used to identify potential drug binding sites on target proteins, calculate the binding free energy between proteins and ligands, determine the mechanism of action of drug molecules, and more[119,120].
Taldaev et al[121] used MD to create structural representations of the arrangement of binding sites for the JAK family enzymes in order to elucidate the selectivity of upadacitinib for JAK1 among other isoforms. They found that the high affinity of upadacitinib was due to its ability to form four hydrogen bonds with amino acid residues in the hinge region of JAK1 as opposed to just two with other JAK isoforms. Structural features of the JAK1 binding site, including the unique residues S963 and E966, are responsible for stabilizing the molecule at the hinge region, as proposed by the authors. Hydrogen bonding with the JAK1 (E883) and JAK2 (N859) amino acid residues in the glycine loops was reported to increase the affinity. The research findings were presented as having the potential to direct the creation of more selective and effective next-generation JAK inhibitors, thereby enhancing the treatment of a wide range of cytokine-mediated diseases.
In another MD simulation research conducted by Du et al[122], the interaction mechanism between oncostatin M (OSM) and its receptor (OSMR) at the atomic level was predicted. Binding of OSM to OSMR is said to be implicated in the pathogenesis of IBD. The OSMR interaction domain was built using the homology modeling approach. Docking was used to establish the near-native structure of the OSM-OSMR complex, and long-time scale MD simulation in an explicit solvent was used to sample the conformations when OSM binds to OSMR. Following the equilibration of the simulated system, the per-residue energy contribution was determined to describe the key residues for the formation of the OSM-OSMR complex. Premised on these key residues, eight residues (OSM: Arg100, Leu103, Phe160, and Gln161; OSMR: Tyr214, Ser223, Asp262 and Trp267) were identified as “hot spots” by computational alanine mutagenesis analysis and confirmed by further MD simulation of the R100A (one of the discovered “hotspots”) mutant. Furthermore, the FTMap analysis revealed six cavities at the OSM-OSMR interface, which were proposed as key binding sites. The predicted 3D structure of the OSM-OSMR complex and the discovered “hotspots” provide useful information in understanding the OSM-OSMR interactions, and the identified locations serve as potential targets in designing small molecules to inhibit the interactions[122].
The global prevalence of IBD is increasing, with developing countries experiencing an increase due to modernization. There is still a need for a new generation of alternative therapies due to the loss of response to biologic drugs, which can be caused in part by the immunogenicity of the administered protein as well as the need to discontinue drugs due to intolerance or side effects. The development of small molecule drugs is difficult because of the complexity of the molecular pathways involved in the progression of disease. The conventional drug design and development processes are lengthy, expensive and filled with arduous scientific procedures. However, computational tools show considerable promise as a practical means of creating new small molecules with biomedical application. There have been a number of groundbreaking successes with drugs for several diseases developed using CADD. Many of these drugs are either FDA-approved or under clinical trials. However, computational methods appear to be underutilized for IBD drug research, as observed during this literature search. To hasten efforts in finding treatments for IBD, more scientists need to turn to computational methods. The modern drug discovery process for IBD makes use of a wide range of computational tools, some of which are geared toward specific tasks such as genomic studies, target identification, and virtual screening.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Nigeria
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Wu SC, China; Zhang YW, China S-Editor: Liu GL L-Editor: A P-Editor: Liu GL
| 1. | Furfaro F, Ragaini E, Peyrin-Biroulet L, Danese S. Novel Therapies and Approaches to Inflammatory Bowel Disease (IBD). J Clin Med. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 2. | Lee M, Chang EB. Inflammatory Bowel Diseases (IBD) and the Microbiome-Searching the Crime Scene for Clues. Gastroenterology. 2021;160:524-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 392] [Article Influence: 98.0] [Reference Citation Analysis (0)] |
| 3. | Zhang YZ, Li YY. Inflammatory bowel disease: pathogenesis. World J Gastroenterol. 2014;20:91-99. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 751] [Cited by in RCA: 1038] [Article Influence: 94.4] [Reference Citation Analysis (18)] |
| 4. | Ho SM, Lewis JD, Mayer EA, Plevy SE, Chuang E, Rappaport SM, Croitoru K, Korzenik JR, Krischer J, Hyams JS, Judson R, Kellis M, Jerrett M, Miller GW, Grant ML, Shtraizent N, Honig G, Hurtado-Lorenzo A, Wu GD. Challenges in IBD Research: Environmental Triggers. Inflamm Bowel Dis. 2019;25:S13-S23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 5. | Falloon KA, Fiocchi C. Current Therapy in Inflammatory Bowel Disease: Why and How We Need to Change? EMJ Innov. 2022;6:40-49. [DOI] [Full Text] |
| 6. | Kuenzig ME, Fung SG, Marderfeld L, Mak JWY, Kaplan GG, Ng SC, Wilson DC, Cameron F, Henderson P, Kotze PG, Bhatti J, Fang V, Gerber S, Guay E, Kotteduwa Jayawarden S, Kadota L, Maldonado D F, Osei JA, Sandarage R, Stanton A, Wan M; InsightScope Pediatric IBD Epidemiology Group, Benchimol EI. Twenty-first Century Trends in the Global Epidemiology of Pediatric-Onset Inflammatory Bowel Disease: Systematic Review. Gastroenterology. 2022;162:1147-1159.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 334] [Article Influence: 111.3] [Reference Citation Analysis (0)] |
| 7. | Baumgart DC, Carding SR. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369:1627-1640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1299] [Cited by in RCA: 1508] [Article Influence: 83.8] [Reference Citation Analysis (2)] |
| 8. | Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, Sung JJY, Kaplan GG. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390:2769-2778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2677] [Cited by in RCA: 4102] [Article Influence: 512.8] [Reference Citation Analysis (110)] |
| 9. | Mihaly E, Patai Á, Tulassay Z. Controversials of Microscopic Colitis. Front Med (Lausanne). 2021;8:717438. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 10. | Currò D, Pugliese D, Armuzzi A. Frontiers in Drug Research and Development for Inflammatory Bowel Disease. Front Pharmacol. 2017;8:400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 11. | Neurath MF. New targets for mucosal healing and therapy in inflammatory bowel diseases. Mucosal Immunol. 2014;7:6-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 255] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 12. | Schnitzler F, Fidder H, Ferrante M, Noman M, Arijs I, Van Assche G, Hoffman I, Van Steen K, Vermeire S, Rutgeerts P. Mucosal healing predicts long-term outcome of maintenance therapy with infliximab in Crohn's disease. Inflamm Bowel Dis. 2009;15:1295-1301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 533] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 13. | Huang S, Li L, Ben-Horin S, Mao R, Lin S, Qiu Y, Feng R, He Y, Chen B, Zeng Z, Chen M, Zhang S. Mucosal Healing Is Associated With the Reduced Disabling Disease in Crohn's Disease. Clin Transl Gastroenterol. 2019;10:e00015. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 14. | Ben-Horin S, Kopylov U, Chowers Y. Optimizing anti-TNF treatments in inflammatory bowel disease. Autoimmun Rev. 2014;13:24-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 307] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 15. | García-Domenech R, Gálvez-Llompart M, Zanni R, Recio MC, Gálvez J. QSAR methods for the discovery of new inflammatory bowel disease drugs. Expert Opin Drug Discov. 2013;8:933-949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 16. | Mouchlis VD, Melagraki G, Zacharia LC, Afantitis A. Computer-Aided Drug Design of β-Secretase, γ-Secretase and Anti-Tau Inhibitors for the Discovery of Novel Alzheimer's Therapeutics. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 17. | Cummings J, Lee G, Ritter A, Sabbagh M, Zhong K. Alzheimer's disease drug development pipeline: 2019. Alzheimers Dement (N Y). 2019;5:272-293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 514] [Cited by in RCA: 516] [Article Influence: 86.0] [Reference Citation Analysis (0)] |
| 18. | Johnson TO, Adegboyega AE, Iwaloye O, Eseola OA, Plass W, Afolabi B, Rotimi D, Ahmed EI, Albrakati A, Batiha GE, Adeyemi OS. Computational study of the therapeutic potentials of a new series of imidazole derivatives against SARS-CoV-2. J Pharmacol Sci. 2021;147:62-71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 19. | Yamanishi Y, Araki M, Gutteridge A, Honda W, Kanehisa M. Prediction of drug-target interaction networks from the integration of chemical and genomic spaces. Bioinformatics. 2008;24:i232-i240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 681] [Cited by in RCA: 668] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 20. | Bakheet TM, Doig AJ. Properties and identification of human protein drug targets. Bioinformatics. 2009;25:451-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 202] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 21. | Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A. Critical assessment of methods of protein structure prediction (CASP)-Round XII. Proteins. 2018;86 Suppl 1:7-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 220] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 22. | Ayton GS, Noid WG, Voth GA. Multiscale modeling of biomolecular systems: in serial and in parallel. Curr Opin Struct Biol. 2007;17:192-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 308] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 23. | Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc. 2016;11:905-919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 785] [Cited by in RCA: 1461] [Article Influence: 162.3] [Reference Citation Analysis (0)] |
| 24. | Lin X, Li X, Lin X. A Review on Applications of Computational Methods in Drug Screening and Design. Molecules. 2020;25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 298] [Cited by in RCA: 290] [Article Influence: 58.0] [Reference Citation Analysis (0)] |
| 25. | Baig MH, Ahmad K, Rabbani G, Danishuddin M, Choi I. Computer Aided Drug Design and its Application to the Development of Potential Drugs for Neurodegenerative Disorders. Curr Neuropharmacol. 2018;16:740-748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 85] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 26. | Goyette P, Labbé C, Trinh TT, Xavier RJ, Rioux JD. Molecular pathogenesis of inflammatory bowel disease: genotypes, phenotypes and personalized medicine. Ann Med. 2007;39:177-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 93] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 27. | Bonen DK, Cho JH. The genetics of inflammatory bowel disease. Gastroenterology. 2003;124:521-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 289] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 28. | Yamamoto-Furusho JK. Genetic factors associated with the development of inflammatory bowel disease. World J Gastroenterol. 2007;13:5594-5597. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 23] [Cited by in RCA: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 29. | Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem. 2001;276:4812-4818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1034] [Cited by in RCA: 1040] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 30. | Ogura Y, Lala S, Xin W, Smith E, Dowds TA, Chen FF, Zimmermann E, Tretiakova M, Cho JH, Hart J, Greenson JK, Keshav S, Nuñez G. Expression of NOD2 in Paneth cells: a possible link to Crohn's ileitis. Gut. 2003;52:1591-1597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 326] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 31. | Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, Foster SJ, Moran AP, Fernandez-Luna JL, Nuñez G. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J Biol Chem. 2003;278:5509-5512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1255] [Cited by in RCA: 1251] [Article Influence: 56.9] [Reference Citation Analysis (0)] |
| 32. | Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869-8872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1755] [Cited by in RCA: 1771] [Article Influence: 80.5] [Reference Citation Analysis (0)] |
| 33. | UK IBD Genetics Consortium; Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, Phillips A, Wesley E, Parnell K, Zhang H, Drummond H, Nimmo ER, Massey D, Blaszczyk K, Elliott T, Cotterill L, Dallal H, Lobo AJ, Mowat C, Sanderson JD, Jewell DP, Newman WG, Edwards C, Ahmad T, Mansfield JC, Satsangi J, Parkes M, Mathew CG; Wellcome Trust Case Control Consortium 2, Donnelly P, Peltonen L, Blackwell JM, Bramon E, Brown MA, Casas JP, Corvin A, Craddock N, Deloukas P, Duncanson A, Jankowski J, Markus HS, Mathew CG, McCarthy MI, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Samani N, Trembath RC, Viswanathan AC, Wood N, Spencer CC, Barrett JC, Bellenguez C, Davison D, Freeman C, Strange A, Donnelly P, Langford C, Hunt SE, Edkins S, Gwilliam R, Blackburn H, Bumpstead SJ, Dronov S, Gillman M, Gray E, Hammond N, Jayakumar A, McCann OT, Liddle J, Perez ML, Potter SC, Ravindrarajah R, Ricketts M, Waller M, Weston P, Widaa S, Whittaker P, Deloukas P, Peltonen L, Mathew CG, Blackwell JM, Brown MA, Corvin A, McCarthy MI, Spencer CC, Attwood AP, Stephens J, Sambrook J, Ouwehand WH, McArdle WL, Ring SM, Strachan DP. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330-1334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 359] [Cited by in RCA: 417] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 34. | van Sommeren S, Visschedijk MC, Festen EA, de Jong DJ, Ponsioen CY, Wijmenga C, Weersma RK. HNF4α and CDH1 are associated with ulcerative colitis in a Dutch cohort. Inflamm Bowel Dis. 2011;17:1714-1718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 35. | Stegmann A, Hansen M, Wang Y, Larsen JB, Lund LR, Ritié L, Nicholson JK, Quistorff B, Simon-Assmann P, Troelsen JT, Olsen J. Metabolome, transcriptome, and bioinformatic cis-element analyses point to HNF-4 as a central regulator of gene expression during enterocyte differentiation. Physiol Genomics. 2006;27:141-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 86] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 36. | Garrison WD, Battle MA, Yang C, Kaestner KH, Sladek FM, Duncan SA. Hepatocyte nuclear factor 4alpha is essential for embryonic development of the mouse colon. Gastroenterology. 2006;130:1207-1220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 132] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 37. | Davison JM, Lickwar CR, Song L, Breton G, Crawford GE, Rawls JF. Microbiota regulate intestinal epithelial gene expression by suppressing the transcription factor Hepatocyte nuclear factor 4 alpha. Genome Res. 2017;27:1195-1206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 38. | Loddo I, Romano C. Inflammatory Bowel Disease: Genetics, Epigenetics, and Pathogenesis. Front Immunol. 2015;6:551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 303] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 39. | Yeh MM, Bosch DE, Daoud SS. Role of hepatocyte nuclear factor 4-alpha in gastrointestinal and liver diseases. World J Gastroenterol. 2019;25:4074-4091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 33] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 40. | Kumar V, Abbas AK, Aster JC. Robbins basic pathology. 10th ed. Elsevier. 2017; 910p. Available from: https://www.elsevier.com/books/robbins-basic-pathology/kumar/978-0-323-35317-5. |
| 41. | Babeu JP, Boudreau F. Hepatocyte nuclear factor 4-alpha involvement in liver and intestinal inflammatory networks. World J Gastroenterol. 2014;20:22-30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 130] [Cited by in RCA: 139] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 42. | Ince MN, Elliott DE. Immunologic and molecular mechanisms in inflammatory bowel disease. Surg Clin North Am. 2007;87:681-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 42] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 43. | Noble CL, Nimmo ER, Drummond H, Ho GT, Tenesa A, Smith L, Anderson N, Arnott ID, Satsangi J. The contribution of OCTN1/2 variants within the IBD5 Locus to disease susceptibility and severity in Crohn's disease. Gastroenterology. 2005;129:1854-1864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 90] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 44. | Stoll M, Corneliussen B, Costello CM, Waetzig GH, Mellgard B, Koch WA, Rosenstiel P, Albrecht M, Croucher PJ, Seegert D, Nikolaus S, Hampe J, Lengauer T, Pierrou S, Foelsch UR, Mathew CG, Lagerstrom-Fermer M, Schreiber S. Genetic variation in DLG5 is associated with inflammatory bowel disease. Nat Genet. 2004;36:476-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 315] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 45. | Yamazaki K, Takazoe M, Tanaka T, Ichimori T, Saito S, Iida A, Onouchi Y, Hata A, Nakamura Y. Association analysis of SLC22A4, SLC22A5 and DLG5 in Japanese patients with Crohn disease. J Hum Genet. 2004;49:664-668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 85] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 46. | Engel T, Kopylov U. Ustekinumab in Crohn's disease: evidence to date and place in therapy. Ther Adv Chronic Dis. 2016;7:208-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 47. | Strober W, Fuss IJ. Proinflammatory cytokines in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1756-1767. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 887] [Cited by in RCA: 852] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 48. | Tuskey A, Behm BW. Profile of ustekinumab and its potential in patients with moderate-to-severe Crohn's disease. Clin Exp Gastroenterol. 2014;7:173-179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 49. | Feagan BG, Sandborn WJ, Gasink C, Jacobstein D, Lang Y, Friedman JR, Blank MA, Johanns J, Gao LL, Miao Y, Adedokun OJ, Sands BE, Hanauer SB, Vermeire S, Targan S, Ghosh S, de Villiers WJ, Colombel JF, Tulassay Z, Seidler U, Salzberg BA, Desreumaux P, Lee SD, Loftus EV Jr, Dieleman LA, Katz S, Rutgeerts P; UNITI–IM-UNITI Study Group. Ustekinumab as Induction and Maintenance Therapy for Crohn's Disease. N Engl J Med. 2016;375:1946-1960. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1467] [Cited by in RCA: 1342] [Article Influence: 149.1] [Reference Citation Analysis (0)] |
| 50. | Panaccione R, Sandborn WJ, Gordon GL, Lee SD, Safdi A, Sedghi S, Feagan BG, Hanauer S, Reinisch W, Valentine JF, Huang B, Carcereri R. Briakinumab for treatment of Crohn's disease: results of a randomized trial. Inflamm Bowel Dis. 2015;21:1329-1340. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 51. | Holleran G, Lopetuso L, Petito V, Graziani C, Ianiro G, McNamara D, Gasbarrini A, Scaldaferri F. The Innate and Adaptive Immune System as Targets for Biologic Therapies in Inflammatory Bowel Disease. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 52. | Fuss IJ, Heller F, Boirivant M, Leon F, Yoshida M, Fichtner-Feigl S, Yang Z, Exley M, Kitani A, Blumberg RS, Mannon P, Strober W. Nonclassical CD1d-restricted NK T cells that produce IL-13 characterize an atypical Th2 response in ulcerative colitis. J Clin Invest. 2004;113:1490-1497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 516] [Cited by in RCA: 576] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 53. | Heller F, Florian P, Bojarski C, Richter J, Christ M, Hillenbrand B, Mankertz J, Gitter AH, Bürgel N, Fromm M, Zeitz M, Fuss I, Strober W, Schulzke JD. Interleukin-13 is the key effector Th2 cytokine in ulcerative colitis that affects epithelial tight junctions, apoptosis, and cell restitution. Gastroenterology. 2005;129:550-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 54. | Jovani M, Fiorino G, Danese S. Anti-IL-13 in inflammatory bowel disease: from the bench to the bedside. Curr Drug Targets. 2013;14:1444-1452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 55. | Monteleone G, Kumberova A, Croft NM, McKenzie C, Steer HW, MacDonald TT. Blocking Smad7 restores TGF-beta1 signaling in chronic inflammatory bowel disease. J Clin Invest. 2001;108:601-609. [PubMed] [DOI] [Full Text] |
| 56. | Monteleone G, Neurath MF, Ardizzone S, Di Sabatino A, Fantini MC, Castiglione F, Scribano ML, Armuzzi A, Caprioli F, Sturniolo GC, Rogai F, Vecchi M, Atreya R, Bossa F, Onali S, Fichera M, Corazza GR, Biancone L, Savarino V, Pica R, Orlando A, Pallone F. Mongersen, an oral SMAD7 antisense oligonucleotide, and Crohn's disease. N Engl J Med. 2015;372:1104-1113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 314] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 57. | Dulai PS, Sandborn WJ. Next-Generation Therapeutics for Inflammatory Bowel Disease. Curr Gastroenterol Rep. 2016;18:51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 58. | Bravatà I, Fiorino G, Allocca M, Repici A, Danese S. New targeted therapies such as anti-adhesion molecules, anti-IL-12/23 and anti-Janus kinases are looking toward a more effective treatment of inflammatory bowel disease. Scand J Gastroenterol. 2015;50:113-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 59. | Danese S, Fiocchi C. Ulcerative colitis. N Engl J Med. 2011;365:1713-1725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 812] [Cited by in RCA: 915] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 60. | Coskun M, Salem M, Pedersen J, Nielsen OH. Involvement of JAK/STAT signaling in the pathogenesis of inflammatory bowel disease. Pharmacol Res. 2013;76:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 257] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 61. | Olivera P, Danese S, Peyrin-Biroulet L. JAK inhibition in inflammatory bowel disease. Expert Rev Clin Immunol. 2017;13:693-703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 69] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 62. | Nielsen OH, Seidelin JB, Ainsworth M, Coskun M. Will novel oral formulations change the management of inflammatory bowel disease? Expert Opin Investig Drugs. 2016;25:709-718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 63. | Olivera PA, Lasa JS, Bonovas S, Danese S, Peyrin-Biroulet L. Safety of Janus Kinase Inhibitors in Patients With Inflammatory Bowel Diseases or Other Immune-mediated Diseases: A Systematic Review and Meta-Analysis. Gastroenterology. 2020;158:1554-1573.e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 219] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 64. | Verden A, Dimbil M, Kyle R, Overstreet B, Hoffman KB. Analysis of Spontaneous Postmarket Case Reports Submitted to the FDA Regarding Thromboembolic Adverse Events and JAK Inhibitors. Drug Saf. 2018;41:357-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 112] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 65. | Panés J, Vermeire S. JAK Inhibitors: Back to Small Molecules for the Treatment of IBD. J Crohns Colitis. 2020;14:S711-S712. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 66. | Sinha S, Vohora D. Drug Discovery and Development. In: Pharmaceutical Medicine and Translational Clinical Research. Elsevier, 2018: 19–32. Available from: https://www.sciencedirect.com/science/article/pii/B978012802103300002X. |
| 67. | Zhou SF, Zhong WZ. Drug Design and Discovery: Principles and Applications. Molecules. 2017;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 68. | Danese S, Fiocchi C, Panés J. Drug development in IBD: from novel target identification to early clinical trials. Gut. 2016;65:1233-1239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 69. | Al-Bawardy B, Shivashankar R, Proctor DD. Novel and Emerging Therapies for Inflammatory Bowel Disease. Front Pharmacol. 2021;12:651415. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 111] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 70. | Cohen NA, Rubin DT. New targets in inflammatory bowel disease therapy: 2021. Curr Opin Gastroenterol. 2021;37:357-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 71. | De Vries LCS, Wildenberg ME, De Jonge WJ, D'Haens GR. The Future of Janus Kinase Inhibitors in Inflammatory Bowel Disease. J Crohns Colitis. 2017;11:885-893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 86] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 72. | Gorfu G, Rivera-Nieves J, Ley K. Role of beta7 integrins in intestinal lymphocyte homing and retention. Curr Mol Med. 2009;9:836-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 244] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 73. | Ghosh S, Panaccione R. Anti-adhesion molecule therapy for inflammatory bowel disease. Therap Adv Gastroenterol. 2010;3:239-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 91] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 74. | Lichnog C, Klabunde S, Becker E, Fuh F, Tripal P, Atreya R, Klenske E, Erickson R, Chiu H, Reed C, Chung S, Neufert C, Atreya I, McBride J, Neurath MF, Zundler S. Cellular Mechanisms of Etrolizumab Treatment in Inflammatory Bowel Disease. Front Pharmacol. 2019;10:39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 75. | Sandborn WJ, Vermeire S, Tyrrell H, Hassanali A, Lacey S, Tole S, Tatro AR; Etrolizumab Global Steering Committee. Etrolizumab for the Treatment of Ulcerative Colitis and Crohn's Disease: An Overview of the Phase 3 Clinical Program. Adv Ther. 2020;37:3417-3431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 76. | D'Amico F, Danese S, Peyrin-Biroulet L. Adaptive Designs: Lessons for Inflammatory Bowel Disease Trials. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 77. | Kobayashi K, Suzuki Y, Watanabe K, Oda K, Mukae M, Yamada A, Yamagami H, Nishimura A, Okamoto H. A Phase 1, Multiple-Dose Study of Vedolizumab in Japanese Patients With Ulcerative Colitis. J Clin Pharmacol. 2019;59:271-279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 78. | Feagan BG, Sandborn WJ, Danese S, Wolf DC, Liu WJ, Hua SY, Minton N, Olson A, D'Haens G. Ozanimod induction therapy for patients with moderate to severe Crohn's disease: a single-arm, phase 2, prospective observer-blinded endpoint study. Lancet Gastroenterol Hepatol. 2020;5:819-828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 79. | Prieto-Martínez FD, López-López E, Eurídice Juárez-Mercado K, Medina-Franco JL. Computational Drug Design Methods—Current and Future Perspectives. Silico Drug Des, 2019: 19–44 Available from: https://www.sciencedirect.com/science/article/pii/B978012816125800002X. |
| 80. | Bai L, Scott MKD, Steinberg E, Kalesinskas L, Habtezion A, Shah NH, Khatri P. Computational drug repositioning of atorvastatin for ulcerative colitis. J Am Med Inform Assoc. 2021;28:2325-2335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 81. | Hutter GM. Genome-Wide Association Studies (GWAS). 2022 [cited 2022 May 10]. Database: genome [Internet]. Available from: https://www.genome.gov/genetics-glossary/Genome-Wide-Association-Studies. |
| 82. | Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Büning C, Cohain A, Cichon S, D'Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H; International IBD Genetics Consortium (IIBDGC), Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119-124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3979] [Cited by in RCA: 3601] [Article Influence: 277.0] [Reference Citation Analysis (0)] |
| 83. | Li J, Wei Z, Hakonarson H. Application of computational methods in genetic study of inflammatory bowel disease. World J Gastroenterol. 2016;22:949-960. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 4] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 84. | Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24221] [Cited by in RCA: 24223] [Article Influence: 1345.7] [Reference Citation Analysis (0)] |
| 85. | Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, Domingues FS, Albrecht M, Nothnagel M, Ellinghaus D, Sina C, Onnie CM, Weersma RK, Stokkers PC, Wijmenga C, Gazouli M, Strachan D, McArdle WL, Vermeire S, Rutgeerts P, Rosenstiel P, Krawczak M, Vatn MH; IBSEN study group, Mathew CG, Schreiber S. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319-1323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 486] [Cited by in RCA: 471] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 86. | Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. FaST linear mixed models for genome-wide association studies. Nat Methods. 2011;8:833-835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 807] [Cited by in RCA: 781] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 87. | Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348-354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2365] [Cited by in RCA: 1943] [Article Influence: 129.5] [Reference Citation Analysis (0)] |
| 88. | Zhou X, Stephens M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat Methods. 2014;11:407-409. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 637] [Cited by in RCA: 542] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 89. | Mooney MA, Nigg JT, McWeeney SK, Wilmot B. Functional and genomic context in pathway analysis of GWAS data. Trends Genet. 2014;30:390-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 90. | Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet. 2007;81:1278-1283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 657] [Cited by in RCA: 683] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 91. | Wang K, Zhang H, Kugathasan S, Annese V, Bradfield JP, Russell RK, Sleiman PM, Imielinski M, Glessner J, Hou C, Wilson DC, Walters T, Kim C, Frackelton EC, Lionetti P, Barabino A, Van Limbergen J, Guthery S, Denson L, Piccoli D, Li M, Dubinsky M, Silverberg M, Griffiths A, Grant SF, Satsangi J, Baldassano R, Hakonarson H. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am J Hum Genet. 2009;84:399-405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 227] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 92. | Torkamani A, Topol EJ, Schork NJ. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics. 2008;92:265-272. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 291] [Cited by in RCA: 270] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 93. | Peng G, Luo L, Siu H, Zhu Y, Hu P, Hong S, Zhao J, Zhou X, Reveille JD, Jin L, Amos CI, Xiong M. Gene and pathway-based second-wave analysis of genome-wide association studies. Eur J Hum Genet. 2010;18:111-117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 191] [Cited by in RCA: 187] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 94. | Carbonetto P, Stephens M. Integrated enrichment analysis of variants and pathways in genome-wide association studies indicates central role for IL-2 signaling genes in type 1 diabetes, and cytokine signaling genes in Crohn's disease. PLoS Genet. 2013;9:e1003770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 95. | Holmans P, Green EK, Pahwa JS, Ferreira MA, Purcell SM, Sklar P; Wellcome Trust Case-Control Consortium, Owen MJ, O'Donovan MC, Craddock N. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am J Hum Genet. 2009;85:13-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 330] [Cited by in RCA: 308] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 96. | Zhang K, Cui S, Chang S, Zhang L, Wang J. i-GSEA4GWAS: a web server for identification of pathways/gene sets associated with traits by applying an improved gene set enrichment analysis to genome-wide association study. Nucleic Acids Res. 2010;38:W90-W95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 150] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 97. | Holden M, Deng S, Wojnowski L, Kulle B. GSEA-SNP: applying gene set enrichment analysis to SNP data from genome-wide association studies. Bioinformatics. 2008;24:2784-2785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 135] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 98. | Katara P. Computational Approaches for Drug Target Identification. In: Singh DB, editor. Computer-Aided Drug Design. Singapore: Springer Nature Singapore Pte Ltd, 2020: 163–185. Available from: https://link.springer.com/chapter/10.1007/978-981-15-6815-2_8. |
| 99. | Mohan S, Mok S, Judge T. Identification of Novel Therapeutic Molecular Targets in Inflammatory Bowel Disease by Using Genetic Databases. Clin Exp Gastroenterol. 2020;13:467-473. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 100. | Gimeno A, Ojeda-Montes MJ, Tomás-Hernández S, Cereto-Massagué A, Beltrán-Debón R, Mulero M, Pujadas G, Garcia-Vallvé S. The Light and Dark Sides of Virtual Screening: What Is There to Know? Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 158] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 101. | Johnson TO, Odoh KD, Nwonuma CO, Akinsanmi AO, Adegboyega AE. Biochemical evaluation and molecular docking assessment of the anti-inflammatory potential of Phyllanthus nivosus leaf against ulcerative colitis. Heliyon. 2020;6:e03893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 102. | Johnson TO, Adegboyega AE, Ojo OA, Yusuf AJ, Iwaloye O, Ugwah-Oguejiofor CJ, Asomadu RO, Chukwuma IF, Ejembi SA, Ugwuja EI, Alotaibi SS, Albogami SM, Batiha GE, Rajab BS, Conte-Junior CA. A Computational Approach to Elucidate the Interactions of Chemicals From Artemisia annua Targeted Toward SARS-CoV-2 Main Protease Inhibition for COVID-19 Treatment. Front Med (Lausanne). 2022;9:907583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 103. | Singh DB. Computer-Aided Drug Design. Springer Singapore. 2020; 978-981-15-6815-2. Available from: https://link.springer.com/book/10.1007/978-981-15-6815-2. |
| 104. | Keretsu S, Ghosh S, Cho SJ. Computer aided designing of novel pyrrolopyridine derivatives as JAK1 inhibitors. Sci Rep. 2021;11:23051. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 105. | Halder D, Das S, Joseph A, Jeyaprakash RS. Molecular docking and dynamics approach to in silico drug repurposing for inflammatory bowels disease by targeting TNF alpha. J Biomol Struct Dyn. 2022;1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 106. | Seidel T, Bryant SD, Ibis G, Poli G, Langer T. 3D Pharmacophore Modeling Techniques in Computer-Aided Molecular Design Using LigandScout. In: Tutorials in Chemoinformatics. Chichester, UK: John Wiley & Sons, Ltd, 2017: 279–309. Available from: https://onlinelibrary.wiley.com/doi/10.1002/9781119161110.ch20. |
| 107. | Ehrlich P. Über den jetzigen Stand der Chemotherapie. Berichte der Dtsch Chem Gesellschaft. 1909;42:17-47. [RCA] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 174] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 108. | Gund P. Three-Dimensional Pharmacophoric Pattern Searching. In: Hahn, F.E., Kersten, H., Kersten, W., Szybalski W, editor. Progress in Molecular and Subcellular Biology. Vol 5. Springer, Berlin, Heidelberg, 1977: 117–143. Available from: https://link.springer.com/chapter/10.1007/978-3-642-66626-1_4. |
| 109. | Kaserer T, Beck KR, Akram M, Odermatt A, Schuster D. Pharmacophore Models and Pharmacophore-Based Virtual Screening: Concepts and Applications Exemplified on Hydroxysteroid Dehydrogenases. Molecules. 2015;20:22799-22832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 115] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 110. | Babu S, Nagarajan SK, Sathish S, Negi VS, Sohn H, Madhavan T. Identification of Potent and Selective JAK1 Lead Compounds Through Ligand-Based Drug Design Approaches. Front Pharmacol. 2022;13:837369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 111. | Vucicevic J, Nikolic K, Mitchell JBO. Rational Drug Design of Antineoplastic Agents Using 3D-QSAR, Cheminformatic, and Virtual Screening Approaches. Curr Med Chem. 2019;26:3874-3889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 112. | Kumar A, Rathi E, Kini SG. Identification of potential tumour-associated carbonic anhydrase isozyme IX inhibitors: atom-based 3D-QSAR modelling, pharmacophore-based virtual screening and molecular docking studies. J Biomol Struct Dyn. 2020;38:2156-2170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 113. | Yang JS, Chun K, Park JE, Cho M, Seo J, Song D, Yoon H, Park CH, Joe BY, Choi JH, Kim MH, Han G. Structure based optimization of chromen-based TNF-α converting enzyme (TACE) inhibitors on S1' pocket and their quantitative structure-activity relationship (QSAR) study. Bioorg Med Chem. 2010;18:8618-8629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 114. | Buchete NV, Hummer G. Peptide folding kinetics from replica exchange molecular dynamics. Phys Rev E Stat Nonlin Soft Matter Phys. 2008;77:030902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 90] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 115. | Hernández-Rodríguez M, Rosales-Hernández MC, Mendieta-Wejebe JE, Martínez-Archundia M, Basurto JC. Current Tools and Methods in Molecular Dynamics (MD) Simulations for Drug Design. Curr Med Chem. 2016;23:3909-3924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 71] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 116. | Takada S, Kanada R, Tan C, Terakawa T, Li W, Kenzaki H. Modeling Structural Dynamics of Biomolecular Complexes by Coarse-Grained Molecular Simulations. Acc Chem Res. 2015;48:3026-3035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 102] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 117. | Durrant JD, McCammon JA. Molecular dynamics simulations and drug discovery. BMC Biol. 2011;9:71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 629] [Cited by in RCA: 803] [Article Influence: 57.4] [Reference Citation Analysis (0)] |
| 118. | Borhani DW, Shaw DE. The future of molecular dynamics simulations in drug discovery. J Comput Aided Mol Des. 2012;26:15-26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 191] [Cited by in RCA: 192] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 119. | Hou T, Wang J, Li Y, Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model. 2011;51:69-82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2159] [Cited by in RCA: 1985] [Article Influence: 141.8] [Reference Citation Analysis (0)] |
| 120. | Wang Y, Lupala CS, Liu H, Lin X. Identification of Drug Binding Sites and Action Mechanisms with Molecular Dynamics Simulations. Curr Top Med Chem. 2018;18:2268-2277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 121. | Taldaev A, Rudnev VR, Nikolsky KS, Kulikova LI, Kaysheva AL. Molecular Modeling Insights into Upadacitinib Selectivity upon Binding to JAK Protein Family. Pharmaceuticals (Basel). 2021;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 122. | Du Q, Qian Y, Xue W. Molecular Simulation of Oncostatin M and Receptor (OSM-OSMR) Interaction as a Potential Therapeutic Target for Inflammatory Bowel Disease. Front Mol Biosci. 2020;7:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 123. | Ochoa D, Hercules A, Carmona M, Suveges D, Gonzalez-Uriarte A, Malangone C, Miranda A, Fumis L, Carvalho-Silva D, Spitzer M, Baker J, Ferrer J, Raies A, Razuvayevskaya O, Faulconbridge A, Petsalaki E, Mutowo P, Machlitt-Northen S, Peat G, McAuley E, Ong CK, Mountjoy E, Ghoussaini M, Pierleoni A, Papa E, Pignatelli M, Koscielny G, Karim M, Schwartzentruber J, Hulcoop DG, Dunham I, McDonagh EM. Open Targets Platform: supporting systematic drug-target identification and prioritisation. Nucleic Acids Res. 2021;49:D1302-D1310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 261] [Article Influence: 65.3] [Reference Citation Analysis (0)] |
| 124. | Dubinsky MC, Kugathasan S, Kwon S, Haritunians T, Wrobel I, Wahbeh G, Quiros A, Bahar R, Silber G, Farrior S, Stephens M, Teleten N, Panikkath D, Ippoliti A, Vasiliauskas E, Fleshner P, Williams C, Landers C, Rotter JI, Targan SR, Taylor KD, McGovern DP. Multidimensional prognostic risk assessment identifies association between IL12B variation and surgery in Crohn's disease. Inflamm Bowel Dis. 2013;19:1662-1670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 125. | Dubuquoy L, Rousseaux C, Thuru X, Peyrin-Biroulet L, Romano O, Chavatte P, Chamaillard M, Desreumaux P. PPARgamma as a new therapeutic target in inflammatory bowel diseases. Gut. 2006;55:1341-1349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 340] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 126. | Schote AB, Jäger K, Kroll SL, Vonmoos M, Hulka LM, Preller KH, Meyer J, Grünblatt E, Quednow BB. Glucocorticoid receptor gene variants and lower expression of NR3C1 are associated with cocaine use. Addict Biol. 2019;24:730-742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 127. | Salas A, Hernandez-Rocha C, Duijvestein M, Faubion W, McGovern D, Vermeire S, Vetrano S, Vande Casteele N. JAK-STAT pathway targeting for the treatment of inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17:323-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 425] [Article Influence: 85.0] [Reference Citation Analysis (0)] |
| 128. | Fain JA. Insulin resistance and the use of U-500 insulin: a case report. Diabetes Educ. 1987;13:386-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 129. | Danese S, Peyrin-Biroulet L. IBD in 2013: enriching the therapeutic armamentarium for IBD. Nat Rev Gastroenterol Hepatol. 2014;11:84-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 130. | Chu Q, Gu X, Zheng Q, Wang J, Zhu H. Phosphoribosyl Pyrophosphate Amido Transferase: A New Prognostic Biomarker for Hepatocellular Carcinoma. Int J Gen Med. 2022;15:353-358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 131. | Kongsbak M, Levring TB, Geisler C, von Essen MR. The vitamin d receptor and T cell function. Front Immunol. 2013;4:148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 218] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 132. | Chatterjee I, Zhang Y, Zhang J, Lu R, Xia Y, Sun J. Overexpression of Vitamin D Receptor in Intestinal Epithelia Protects Against Colitis via Upregulating Tight Junction Protein Claudin 15. J Crohns Colitis. 2021;15:1720-1736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 133. | Meyer NJ, Gattinoni L, Calfee CS. Acute respiratory distress syndrome. Lancet. 2021;398: 622-637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 529] [Cited by in RCA: 670] [Article Influence: 167.5] [Reference Citation Analysis (0)] |
| 134. | Vandenbroucke RE, Dejonckheere E, Van Hauwermeiren F, Lodens S, De Rycke R, Van Wonterghem E, Staes A, Gevaert K, López-Otin C, Libert C. Matrix metalloproteinase 13 modulates intestinal epithelial barrier integrity in inflammatory diseases by activating TNF. EMBO Mol Med. 2013;5:1000-1016. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 109] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 135. | Jin J, Ji M, Fu R, Wang M, Xue N, Xiao Q, Hu J, Wang X, Lai F, Yin D, Chen X. Sphingosine-1-Phosphate Receptor Subtype 1 (S1P1) Modulator IMMH001 Regulates Adjuvant- and Collagen-Induced Arthritis. Front Pharmacol. 2019;10:1085. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 136. | Keum J, Nam H. SELF-BLM: Prediction of drug-target interactions via self-training SVM. PLoS One. 2017;12:e0171839. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 137. | Rayhan F, Ahmed S, Shatabda S, Farid DM, Mousavian Z, Dehzangi A, Rahman MS. iDTI-ESBoost: Identification of Drug Target Interaction Using Evolutionary and Structural Features with Boosting. Sci Rep. 2017;7:17731. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 68] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 138. | Clough E, Barrett T. The Gene Expression Omnibus Database. Methods Mol Biol. 2016;1418:93-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 869] [Cited by in RCA: 1441] [Article Influence: 160.1] [Reference Citation Analysis (0)] |
| 139. | Ba-Alawi W, Soufan O, Essack M, Kalnis P, Bajic VB. DASPfind: new efficient method to predict drug-target interactions. J Cheminform. 2016;8:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 140. | Chen H, Zhang Z. A semi-supervised method for drug-target interaction prediction with consistency in networks. PLoS One. 2013;8:e62975. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 141. | Gupta S, Mishra M, Sen N, Parihar R, Dwivedi GR, Khan F, Sharma A. DbMDR: a relational database for multidrug resistance genes as potential drug targets. Chem Biol Drug Des. 2011;78:734-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 142. | Magariños MP, Carmona SJ, Crowther GJ, Ralph SA, Roos DS, Shanmugam D, Van Voorhis WC, Agüero F. TDR Targets: a chemogenomics resource for neglected diseases. Nucleic Acids Res. 2012;40:D1118-D1127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 82] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 143. | Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901-D906. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1752] [Cited by in RCA: 1889] [Article Influence: 104.9] [Reference Citation Analysis (0)] |
| 144. | Gao Z, Li H, Zhang H, Liu X, Kang L, Luo X, Zhu W, Chen K, Wang X, Jiang H. PDTD: a web-accessible protein database for drug target identification. BMC Bioinformatics. 2008;9:104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 194] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 145. | Zhang R, Ou HY, Zhang CT. DEG: a database of essential genes. Nucleic Acids Res. 2004;32:D271-D272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 373] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 146. | Chen X, Ji ZL, Chen YZ. TTD: Therapeutic Target Database. Nucleic Acids Res. 2002;30:412-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 486] [Cited by in RCA: 463] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 147. | Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18868] [Cited by in RCA: 24713] [Article Influence: 988.5] [Reference Citation Analysis (0)] |
| 148. | Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, Sirota-Madi A, Olender T, Golan Y, Stelzer G, Harel A, Lancet D. GeneCards Version 3: the human gene integrator. Database (Oxford). 2010;2010:baq020. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 718] [Cited by in RCA: 1276] [Article Influence: 85.1] [Reference Citation Analysis (0)] |
| 149. | Piñero J, Saüch J, Sanz F, Furlong LI. The DisGeNET cytoscape app: Exploring and visualizing disease genomics data. Comput Struct Biotechnol J. 2021;19:2960-2967. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 225] [Cited by in RCA: 318] [Article Influence: 79.5] [Reference Citation Analysis (0)] |
| 150. | Davis AP, Grondin CJ, Johnson RJ, Sciaky D, Wiegers J, Wiegers TC, Mattingly CJ. Comparative Toxicogenomics Database (CTD): update 2021. Nucleic Acids Res. 2021;49:D1138-D1143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 619] [Cited by in RCA: 692] [Article Influence: 173.0] [Reference Citation Analysis (0)] |
| 151. | UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480-D489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4025] [Cited by in RCA: 4265] [Article Influence: 1066.3] [Reference Citation Analysis (0)] |
| 152. | Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: applications of AutoDock. J Mol Recognit. 1996;9:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 153. | Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455-461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26714] [Cited by in RCA: 14540] [Article Influence: 969.3] [Reference Citation Analysis (0)] |
| 154. | Ritchie DW. Evaluation of protein docking predictions using Hex 3.1 in CAPRI rounds 1 and 2. Proteins. 2003;52:98-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 141] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 155. | Vilar S, Cozza G, Moro S. Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Curr Top Med Chem. 2008;8:1555-1572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 501] [Cited by in RCA: 718] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 156. | Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J Med Chem. 2004;47:1739-1749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5637] [Cited by in RCA: 7099] [Article Influence: 338.0] [Reference Citation Analysis (0)] |