Published online Apr 28, 2023. doi: 10.3748/wjg.v29.i16.2359
Peer-review started: October 19, 2022
First decision: December 12, 2022
Revised: December 17, 2022
Accepted: April 7, 2023
Article in press: April 7, 2023
Published online: April 28, 2023
Processing time: 187 Days and 5.3 Hours
High incidence (10.2%) and mortality (9.2%) rates led to the ranking of colorectal cancer (CRC) as the second most malignant tumor spectrum worldwide in 2020. Treatment strategies are becoming highly dependent on the molecular characteristics of CRC. The classical theories accept two models depicting the origin of CRC: The progression of adenoma to cancer and transformation from serrated polyps to cancer. However, the molecular mechanism of CRC development is very complex. For instance, CRCs originating from laterally spreading tumors (LST) do not adhere to any of these models and exhibit extremely serious progression and poor outcomes. In this article, we present another possible pathway involved in CRC development, particularly from LST, with important molecular characteristics, which would facilitate the design of a novel strategy for targeted therapy.
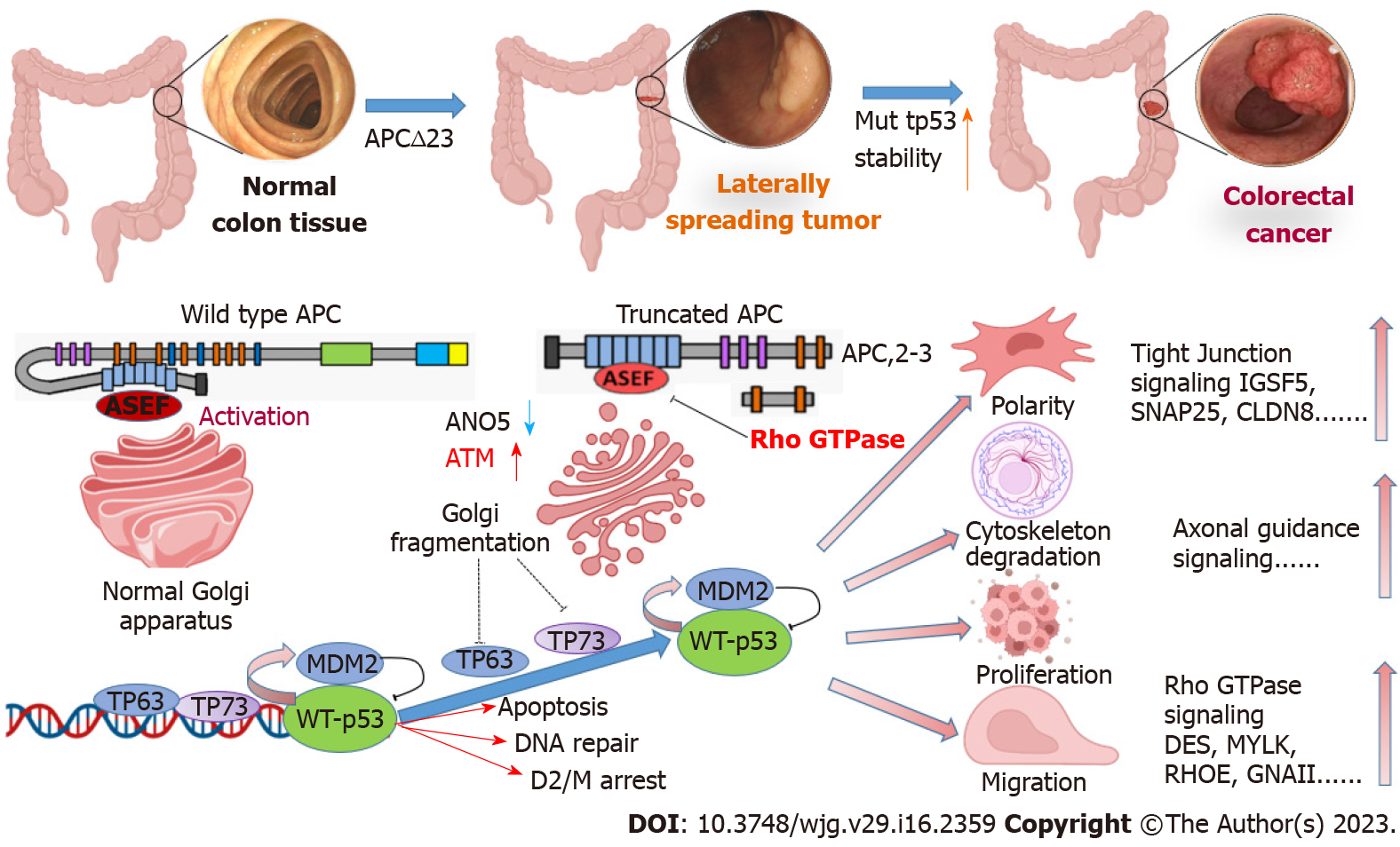
Core Tip: Although laterally spreading tumors (LST) are considered vital precancerous lesions of colorectal cancer (CRC), the mechanism mediating their transition to CRC development is unclear. Adenomatous polyposis coli (APC)-truncating mutations driven by Golgi fragmentation are very important cellular events that can abrogate the microtubule binding properties of APC. This effect reduces the stability of microtubules, impacts cell proliferation and survival, causes chromosomal instability, and increases migration. Downstream characteristics of Golgi fragmentation indicate alterations in Ataxia-telangiectasia mutated and anoctamin 5 expression, whereas their gene expression changes are significant in LST. This implies a novel pathway for CRC development from LST.
- Citation: Lu S, Jia CY, Yang JS. Future therapeutic implications of new molecular mechanism of colorectal cancer. World J Gastroenterol 2023; 29(16): 2359-2368
- URL: https://www.wjgnet.com/1007-9327/full/v29/i16/2359.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i16.2359
In 2020, the World Health Organization International Agency for Research on Cancer officially released the latest cancer data, showing a 10.2% and 12.2% incidence of colorectal cancer (CRC)[1,2]. Consequently, CRC was ranked the third and second highest in malignant tumor spectrum and in China, respectively, while its mortality of 9.2% ranked it the second highest among all cancers[1,2]. Most incidences of CRC develop from benign polyps (adenomas and serrated polyps) through a series of genetic and epigenetic changes that occur over 10 to 15 years[3,4].
A laterally spreading tumor (LST) is a special digestive tract tumor that is an important precancerous lesion of CRC. Its morphological features are hidden and, consequently, it is easily misdiagnosed. LST can develop into progressive CRC within 3 years with a very poor prognosis[5]. Large-scale controlled studies have shown that LST patients have an 8.4%-52.5% possibility of developing CRC, and benign LST lesions can develop into advanced CRC within 3 years[5].
Pathologically, LST has certain similarities to colorectal polyps and adenoma, and the molecular mechanisms underlying their progression to carcinoma have been clearly elucidated. However, studies on how LST develops into CRC are rare, and the molecular mechanism of the associated carcinogenesis is unclear. Therefore, systematically and comprehensively exploring the molecular mechanisms underlying the malignant transformation of LST to CRC is critical. Specifically, the exploration would have potential theoretical significance and clinical value for early diagnosis and precise treatment of CRC. Here, we present a potential alternative pathway mediating the development of CRC, particularly from LST, with critical molecular characteristics, which would facilitate the design of a novel strategy for targeted therapy.
Two classical models have been proposed for the development and progression of CRC from colon epithelial cells. The first model describes the process of transformation of adenoma to cancer, mainly initiated by mutations in the adenomatous polyposis coli (APC) gene. In this model, APC mutation is followed by mutations in Kirsten rat sarcoma viral oncogene homolog (KRAS)/neuroblastoma rat sarcoma viral oncogenes, mothers against decapentaplegic homolog 4, and finally, tumor protein p53 (TP53).
The other model describes the transformation from serrated polyps to cancer, with the driver mutation of catenin beta 1 secondary to mutations of KRAS/B-Raf proto-oncogene, serine/threonine kinase (BRAF), and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha[6]. These mutations eventually result in transforming growth factor-beta receptor type 2 (TGFRB2) overexpression[6]. The main signaling molecules involved are Wnt, mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K)/Akt, and TP53, which are hyperactivated[7]. Several other signaling pathways, involving hedgehog, erb-b2 receptor tyrosine kinase, ras homolog family member A, Notch, bone morphogenetic protein, Hippo, AMP-activated protein kinase, nuclear factor kB, and Jun N terminal kinase, also participate in the occurrence and development of CRC[8].
Several studies have indicated that APC mutations are extremely important in both the mutation frequency and different stages of CRC tumor development[9-11]. APC is the “gatekeeper” gene of the colorectal mucosal epithelium and the key molecule regulating colon epithelial cell homeostasis, polarity, and movement. APC acts as a tumor suppressor gene of CRC and 80% of sporadic CRCs harbor mutations, which are widely considered an early event in colorectal malignancy. Somatic point mutations of APC mainly occur in the mutation cluster region (MCR); however, APC contains several other mutations in its protein-coding region[9,10].
The most important cytological events after mutation in the MCR are the structural truncation of APC (amino acid sequence from 1362 to 1540, namely, the APC-2,3 repeats) and lack of axin, catenin, microtubules, and other binding sites[11]. Mutations in truncated APC result in the loss of the microtubule-binding properties of APC and further reduce microtubule stability. Truncating mutations cause APC to lose the properties of normal tumor suppressor genes and show “acquired” proto-oncogene properties, including abnormal cell proliferation and survival, chromosomal instability, and increased migration.
Truncated APC binds to and activates APC-stimulated guanine nucleotide exchange factor (ASEF), which is closely related to actin remodeling and movement and causes significant changes in cell structure and function[12]. Knockdown of ASEF or APC-truncating mutations markedly reduces cell migration; however, overexpression of full-length APC does not increase ASEF-mediated cell migration. The Golgi complex is a dynamic organelle that is essential for sorting, processing, and transporting proteins by which the stability of cellular structures is maintained.
Fragmentation of the Golgi apparatus is observed in age-related diseases including Alzheimer’s disease, Parkinson’s disease, and cancer[13]. The Golgi apparatus may be closely involved in the development of human diseases; however, the mechanisms and significance of its fragmentation are poorly understood. APC-truncating mutations induce fragmentation of the Golgi apparatus and lead to structural reorganization of cytoskeletal proteins and actin[13]. This causes cells to exhibit abnormal biological behavior, such as loss of polarity and increased migration[13]. Therefore, APC-truncating mutations driven by Golgi fragmentation are an important event in normal cells.
Mutation-rich regions of APC in the normal mucosa, LST, colorectal adenocarcinoma, and colorectal adenoma specimens were detected using polymerase chain reaction-single-strand conformation polymorphism[14]. The results of that study showed that while no APC mutations were observed in the normal mucosa of the large intestine, the mutation rates were 25%, 30%, and 27.8% in LST, colorectal adenocarcinoma, and colorectal adenoma, respectively[14]. The difference in rates between the specimens was not statistically significant and the results were consistent with other reported APC mutation rates of 15.5%-42.4% in LST specimens. Two recent studies showed APC mutation frequencies of 80% (10 samples) and 57% (14 samples) in LST[15]. Therefore, APC mutation may act as an important initiator in LST development[16].
TP53 and CRC are important intracellular tumor suppressor genes, and the main biological function of TP53 is the repair of cellular damage. Normal TP53 can be used to monitor the integrity of genomic DNA in real time. During DNA damage, TP53 stops cell division at the G1/S phase to allow cells to have enough time to repair the damage. For irreparable DNA damage, TP53 induces programmed apoptotic cell death, thereby inhibiting the generation of possible mutant cancerous cells[17,18].
TP53 mutations primarily occur during the middle and late stages of carcinogenesis. Numerous TP53 mutations reduce the proportion of wild-type TP53 and weaken its function in monitoring genomic DNA integrity, thereby allowing tumorigenesis[19]. Studies on TP53 and LST are scarce, and a recent comprehensive and unbiased screening of the genome, epigenome, and transcriptome was conducted based on the Cancer Genome Atlas (TCGA) database[14].
Bioinformatic data integrated from 11 LST samples and validated in an additional cohort of 84 benign colorectal injury samples, identified several high-frequency genetic, epigenetic, and transcriptional alterations[14]. Deletions occurred in chromosomes 1p, 5q, 14q, and 18, whereas doubling occurred in chromosomes 7, 8, 13, 19, and 20. Furthermore, these alterations were highly prevalent in the panel of the colorectal and rectal adenocarcinoma validation groups. The main signaling pathways associated with LST are axonal guidance, thyroid cancer, human embryonic stem cell pluripotency (Nanog homeobox), and Wnt/β-catenin.
Cohort validation studies compared 10 LSTs with 212 CRC samples with a focus on five major altered signaling pathways, Wnt, TGF β, PI3K, MAPK, and P53[20,21]. The results showed that the differences in the TGFβ and TP53 signaling pathways were significant[20,21]. The results suggested that ataxia-telangiectasia mutated (ATM), a very important molecule in the TP53 pathway, was significantly increased, which stabilized TP53 molecules. The expression of another very important gene, anoctamin 5 (ANO5), was significantly reduced, leading to mitochondrial fragmentation.
Our results based on TCGA data analysis also confirmed a significantly low expression level of ANO5 in LST. The expression of the Golgi fragmentation-related genes ATM and ANO5 was significantly different. This differential expression weakened the expression of the wild-type TP53 transcription factors MDM2 proto-oncogene (MDM2) and TP73, and subsequently modulated TP53 stability in the TP53 signaling pathway. These two important signaling molecules are closely related to the stability of TP53 mutants. These data suggest that TP53 mutation has a unique molecular mechanism that differs from that in polyps and adenomas in CRC development from LST.
In our opinion, the progression of LST to CRC is distinct from the development of common polyps or adenoma carcinogenesis. Therefore, we proposed a two-stage cascade mutation hypothesis from LST to CRC (Figure 1): The driver stage and cancerous stage. In the driver stage, APC-truncating mutations drive Golgi fragmentation, resulting in reorganization of cellular structural proteins, thereby leading to abnormal polarity and lateral growth. In the cancerous stage, Golgi fragmentation further affects the repair mechanism of TP53 base mismatch mediated by the heat shock proteins MDM2, TP63, and TP73, resulting in increased stability of mutant TP53 and promotion of LST progression to carcinoma.
Based on the mRNA data of 25711 samples from 980 healthy donors in the genotype-tissue expression (GTEx) v8 database, pan-cancer mRNA-sequencing (mRNA-seq, n = 11057) and survival (n = 10121) data were downloaded from the TCGA database. We then performed a differential expression analysis of ATM, ANO5, APC, and TP53 in pan-cancer and adjacent normal samples. Survival analysis of these genes was also performed in pan-cancers.
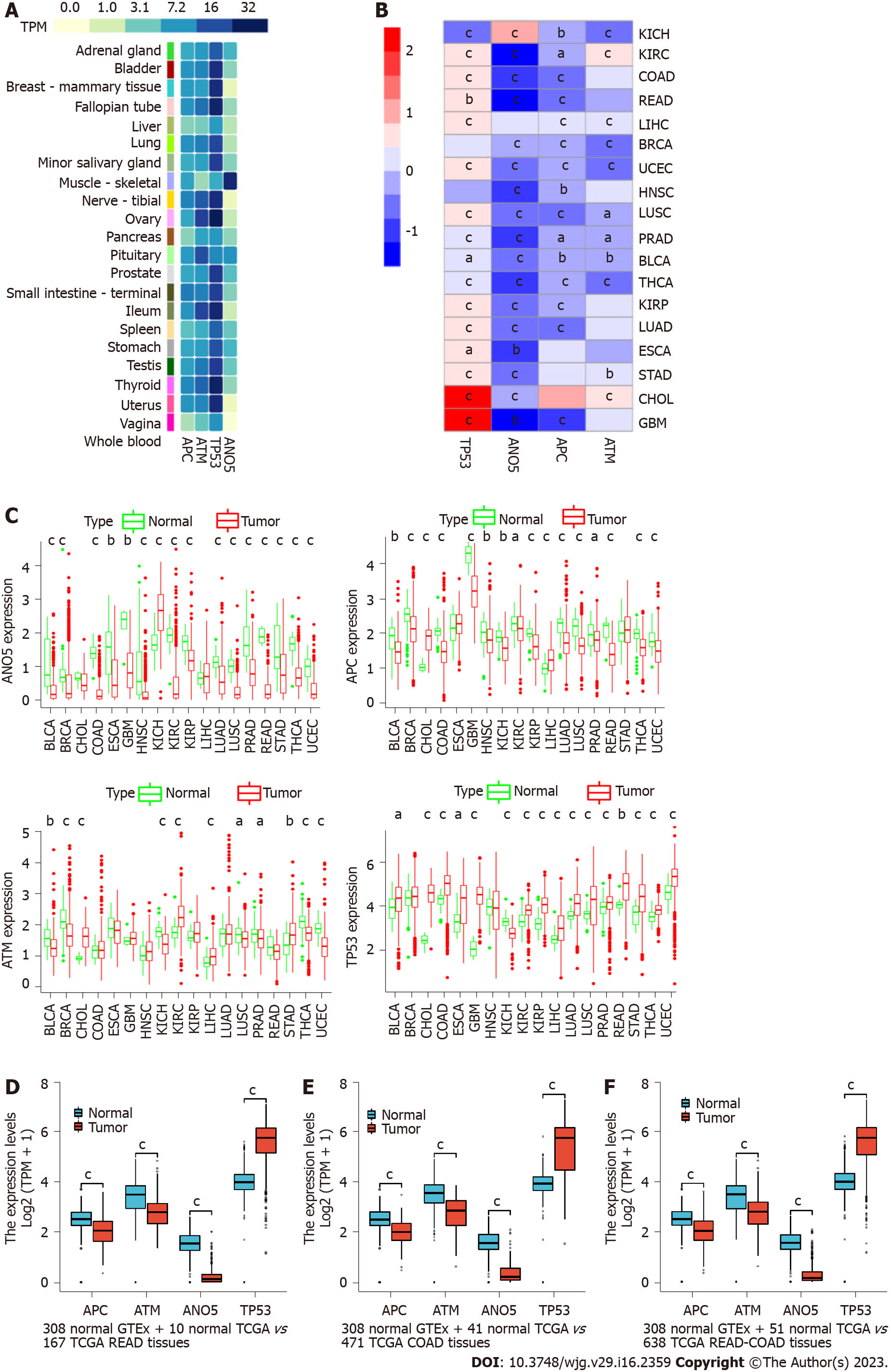
We analyzed the expression profiles of ATM, ANO5, APC, and TP53 in human normal and pan-cancer samples and found that ATM, ANO5, and APC were significantly downregulated in rectal adenocarcinoma (READ), colon adenocarcinoma (COAD), and READ-COAD samples. In contrast, TP53 showed an obviously higher level in READ, COAD, and READ-COAD samples than in normal samples.
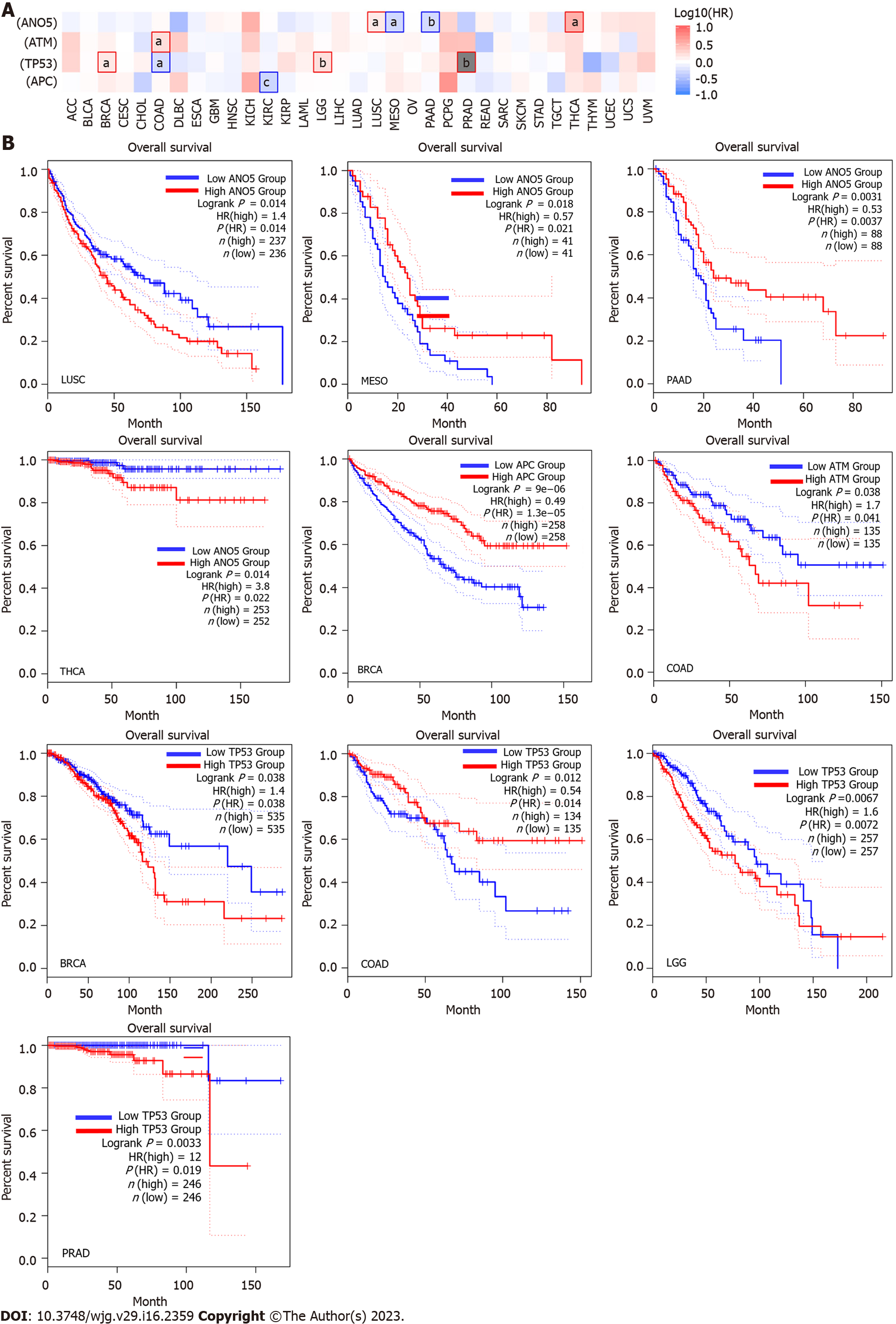
Furthermore, the survival analysis results indicated that ATM, ANO5, APC, and TP53 expression were correlated with the overall survival (OS) of lung squamous cell carcinoma (LUSC), thyroid carcinoma (THCA), mesothelioma (MESO), pancreatic adenocarcinoma (PAAD), COAD, brain lower grade glioma (LGG), and breast invasive carcinoma (BRCA) patients.
We analyzed the expression profiles of ATM, ANO5, APC, and TP53 in normal human tissues based on transcripts per million (TPM) values from the GTEx v8 database. The results indicated that ATM, ANO5, APC, and TP53 were highly or moderately expressed in most organs or tissues (1 < average TPM < 32), except that ANO5 expression was low in the whole blood, vagina, and breast (average TPM < 1, Figure 2A).
Then, we performed a differential expression analysis of mRNAs for these genes across 18 cancer types that had over five pairs of cancer-adjacent samples based on the log2[fragments per kilobase of exon per million mapped fragments (FPKM) + 1] data from TCGA. Figure 2B shows the heatmap of log2(FPKM + 1) expression status of ATM, ANO5, APC, and TP53 between cancer and cancer-adjacent samples where red and blue represent upregulation and downregulation, respectively]. Figure 2C shows the log2(FPKM + 1) expression status of ATM, ANO5, APC, and TP53 between cancer and cancer-adjacent samples.
Table 1 shows that the expression of TP53 was significantly upregulated in bladder cancer (BLCA), cholangiocarcinoma (CHOL), COAD, esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), LUSC, prostate adenocarcinoma (PRAD), READ, stomach adenocarcinoma (STAD), THCA, and uterine corpus endometrial carcinoma (UCEC) and downregulated in kidney chromophobe (KICH), compared with the expression levels in cancer-adjacent samples (|log2FC| > 0.19, 4.99E-12 < P < 0.03). The ANO5 level was significantly higher in KICH and obviously lower in BLCA, BRCA, COAD, ESCA, GBM, HNSC, KIRC, KIRP, LUAD, LUSC, PRAD, READ, STAD, THCA, and UCEC than in cancer-adjacent samples (|log2FC| > 0.46, 2.36E-26 < P < 0.003).
| Cancer type | APC | TP53 | ATM | ANO5 | ||||
| LogFC | P value | LogFC | P value | LogFC | P value | LogFC | P value | |
| BLCA | -0.28889 | 0.005445 | 0.314908 | 0.032102 | -0.27112 | 0.002207 | -0.75984 | 5.46E-07 |
| BRCA | -0.37268 | 1.46E-15 | 0.070558 | 0.261271 | -0.41783 | 2.30E-16 | -0.26734 | 2.03E-17 |
| CHOL | 0.798977 | 4.07E-06 | 2.043449 | 1.51E-07 | 0.718097 | 8.17E-06 | -0.08998 | 0.123426 |
| COAD | -0.57688 | 2.42E-15 | 0.505369 | 4.40E-09 | 0.086761 | 0.429393 | -1.16109 | 3.06E-24 |
| ESCA | 0.134442 | 0.480134 | 0.792589 | 0.011648 | -0.0696 | 0.629714 | -0.81744 | 0.003288 |
| GBM | -1.01588 | 0.000957 | 2.388451 | 0.000173 | 0.016551 | 0.761476 | -1.26367 | 0.001745 |
| HNSC | -0.18476 | 0.004628 | -0.08774 | 0.958273 | 0.116251 | 0.114312 | -0.82404 | 7.30E-09 |
| KICH | -0.32313 | 0.003139 | -0.63636 | 1.15E-07 | -0.42688 | 1.83E-05 | 0.889727 | 7.05E-07 |
| KIRC | -0.12313 | 0.033897 | 0.452895 | 2.19E-16 | 0.476935 | 2.14E-16 | -1.43405 | 4.66E-34 |
| KIRP | -0.34452 | 1.85E-05 | 0.784867 | 3.85E-12 | 0.110608 | 0.113455 | -0.50628 | 2.70E-09 |
| LIHC | 0.253395 | 8.34E-06 | 0.41186 | 5.80E-06 | 0.187905 | 0.000776 | 0.070468 | 0.537737 |
| LUAD | -0.4475 | 1.24E-13 | 0.430958 | 3.79E-09 | 0.14328 | 0.135749 | -0.46868 | 9.13E-13 |
| LUSC | -0.51847 | 1.39E-15 | 0.467462 | 0.000189 | -0.14078 | 0.016441 | -0.73565 | 5.71E-25 |
| PRAD | -0.17991 | 0.011276 | 0.193039 | 0.000387 | -0.12553 | 0.049659 | -0.89276 | 6.11E-19 |
| READ | -0.71729 | 3.89E-05 | 0.638392 | 0.007547 | -0.18753 | 0.144797 | -1.5861 | 1.99E-07 |
| STAD | 0.025202 | 0.628002 | 0.660337 | 1.23E-06 | 0.321073 | 0.001535 | -0.63734 | 2.77E-05 |
| THCA | -0.36112 | 1.91E-13 | 0.320993 | 4.99E-12 | -0.40106 | 1.15E-13 | -0.89401 | 2.36E-26 |
| UCEC | -0.34584 | 3.78E-06 | 0.552104 | 1.66E-08 | -0.56028 | 9.35E-14 | -0.63913 | 2.84E-15 |
APC expression was significantly upregulated in CHOL and LIHC relative to that in cancer-adjacent samples and an obvious downregulation was noticed in BLCA, BRCA, COAD, GBM, HNSC, KICH, KIRC, KIRP, LUAD, LUSC, PRAD, READ, THCA, and UCEC (|log2FC| > 0.25, 1.46E-15 < P < 0.03). In addition, the ATM expression was obviously upregulated in CHOL, KIRC, LIHC, and STAD compared to that in cancer-adjacent samples. In contrast, BLCA, BRCA, KICH, LUSC, PRAD, THCA, and UCEC (|log2FC| > 0.12, 2.14E-16 < P < 0.049) were obviously downregulated.
We also analyzed the difference in expression among COAD, READ, and their cancer-adjacent samples based on the merged and batch-normalized TPM expression data of the GTEx and TCGA. The results showed that ATM, ANO5, and APC were significantly downregulated, whereas TP53 showed an obviously higher expression in READ (Figure 2D), COAD (Figure 2E), and READ-COAD (Figure 2F) samples than in normal samples (P < 0.001).
To explore the role of ATM, ANO5, APC, and TP53 in pan-cancer prognosis, we conducted a survival analysis in pan-cancers based on the log2(FPKM + 1) data and clinical survival data of 33 cancer types. The survival map of ATM, ANO5, APC, and TP53 expression in pan-cancers indicated that the expression of these four genes was correlated with OS in LUSC, THCA, MESO, PAAD, COAD, LGG, and BRCA (Figure 3A). The relationship between ATM, ANO5, APC, and TP53 and the OS of cancer patients (aP < 0.05, bP < 0.01, and cP < 0.001) was identified.
Briefly, LUSC or THCA patients with ANO5 expression had a poor OS [1.40 < hazard ratio (HR) < 3.80, P = 0.014], whereas MESO or PAAD patients with ANO5 expression showed a good OS (0.53 < HR < 0.57, 0.0031 < P < 0.018). ATM expression was indicative of a poor OS in COAD (HR = 1.70, P = 0.038). TP53 expression was positively associated with a poor OS in BRCA (HR = 1.40, P = 0.038), LGG (HR = 1.60, P = 0.0067), and PAAD (HR = 12, P = 0.0033). In contrast, ATM expression exhibited a positive association with good OS in COAD (HR = 0.54, P = 0.012). Moreover, APC expression showed a good co-relation with OS in BRCA (HR = 0.49, P = 9E-06, Figure 3B).
APC-truncating mutations driven by Golgi fragmentation are a very important cellular event, which can cause loss of the microtubule binding properties of APC. This effect further reduces microtubule stability, resulting in abnormal cell proliferation and survival, chromosomal instability, and increased migration. Downstream characteristics of Golgi fragmentation include gene expression alterations with ATM upregulation and ANO5 downregulation, which are significant in LST. These observations suggest the existence of a unique and novel pathway for the development of CRC from LST, and future studies of potential CRC treatment should focus on this newly identified mechanism.
Provenance and peer review: Unsolicited article; Externally peer reviewed
Peer-review model: Single blind
Corresponding Author’s Membership in Professional Societies: The corresponding author is a member of the editorial board or peer reviewer of a BPG journal, 00068967.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kumar A, India; Rather AA, India S-Editor: Wang JJ L-Editor: Wang TQ P-Editor: Wang JJ
| 1. | Lauby-Secretan B, Vilahur N, Bianchini F, Guha N, Straif K; International Agency for Research on Cancer Handbook Working Group. The IARC Perspective on Colorectal Cancer Screening. N Engl J Med. 2018;378:1734-1740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 234] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 2. | Pinsky P, Rabeneck L, Lauby-Secretan B. The IARC Perspective on Colorectal Cancer Screening. N Engl J Med. 2018;379:301-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 3. | Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4616] [Cited by in RCA: 4464] [Article Influence: 120.6] [Reference Citation Analysis (0)] |
| 4. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8087] [Cited by in RCA: 7995] [Article Influence: 228.4] [Reference Citation Analysis (1)] |
| 5. | Yamada M, Saito Y, Sakamoto T, Nakajima T, Kushima R, Parra-Blanco A, Matsuda T. Endoscopic predictors of deep submucosal invasion in colorectal laterally spreading tumors. Endoscopy. 2016;48:456-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 63] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 6. | Ireland MJ, March S, Crawford-Williams F, Cassimatis M, Aitken JF, Hyde MK, Chambers SK, Sun J, Dunn J. A systematic review of geographical differences in management and outcomes for colorectal cancer in Australia. BMC Cancer. 2017;17:95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 7. | Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ, Watanabe T. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1014] [Cited by in RCA: 1114] [Article Influence: 111.4] [Reference Citation Analysis (0)] |
| 8. | Mahar AL, Compton C, Halabi S, Hess KR, Weiser MR, Groome PA. Personalizing prognosis in colorectal cancer: A systematic review of the quality and nature of clinical prognostic tools for survival outcomes. J Surg Oncol. 2017;116:969-982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 102] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 9. | Ni HB, Wang FY, Xu J, He XJ, Chen J, Wu Q, Wu JF, Sun YS. Screening and identification of a tumor specific methylation phenotype in the colorectal laterally spreading tumor. Eur Rev Med Pharmacol Sci. 2017;21:2611-2616. [PubMed] |
| 10. | Aitchison A, Hakkaart C, Day RC, Morrin HR, Frizelle FA, Keenan JI. APC Mutations Are Not Confined to Hotspot Regions in Early-Onset Colorectal Cancer. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 11. | Kohler EM, Derungs A, Daum G, Behrens J, Schneikert J. Functional definition of the mutation cluster region of adenomatous polyposis coli in colorectal tumours. Hum Mol Genet. 2008;17:1978-1987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 12. | Kim SB, Zhang L, Yoon J, Lee J, Min J, Li W, Grishin NV, Moon YA, Wright WE, Shay JW. Truncated Adenomatous Polyposis Coli Mutation Induces Asef-Activated Golgi Fragmentation. Mol Cell Biol. 2018;38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Petrosyan A. Onco-Golgi: Is Fragmentation a Gate to Cancer Progression? Biochem Mol Biol J. 2015;1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 90] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 14. | Hesson LB, Ng B, Zarzour P, Srivastava S, Kwok CT, Packham D, Nunez AC, Beck D, Ryan R, Dower A, Ford CE, Pimanda JE, Sloane MA, Hawkins NJ, Bourke MJ, Wong JW, Ward RL. Integrated Genetic, Epigenetic, and Transcriptional Profiling Identifies Molecular Pathways in the Development of Laterally Spreading Tumors. Mol Cancer Res. 2016;14:1217-1228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 15. | Nong Y, Zhang Y, Pan L, Chen J, Zhu C, Han L, Li A, Liu S. Analysis of genetic alterations identifies the frequent mutation of GNAS in colorectal laterally spreading tumors. Cancer Commun (Lond). 2020;40:636-640. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 16. | Cho KR, Oliner JD, Simons JW, Hedrick L, Fearon ER, Preisinger AC, Hedge P, Silverman GA, Vogelstein B. The DCC gene: structural analysis and mutations in colorectal carcinomas. Genomics. 1994;19:525-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 153] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 17. | Bykov VJN, Eriksson SE, Bianchi J, Wiman KG. Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer. 2018;18:89-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 659] [Article Influence: 94.1] [Reference Citation Analysis (0)] |
| 18. | McCarthy N. Metastasis: understanding the prowess of mutant p53. Nat Rev Cancer. 2014;14:385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 19. | Zhang X, Wang Q, Jia C, Li D, Lv Z, Yang J. Role of atrial natriuretic peptide receptor in inhibition of laterally spreading tumors via Wnt/β-catenin signaling. Arch Med Sci Atheroscler Dis. 2022;7:e104-e108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.7] [Reference Citation Analysis (1)] |
| 20. | Facciorusso A, Antonino M, Di Maso M, Barone M, Muscatiello N. Non-polypoid colorectal neoplasms: Classification, therapy and follow-up. World J Gastroenterol. 2015;21:5149-5157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 50] [Cited by in RCA: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 21. | Ikezawa N, Toyonaga T, Tanaka S, Yoshizaki T, Takao T, Abe H, Sakaguchi H, Tsuda K, Urakami S, Nakai T, Harada T, Miura K, Yamasaki T, Kostalas S, Morita Y, Kodama Y. Feasibility and safety of endoscopic submucosal dissection for lesions in proximity to a colonic diverticulum. Clin Endosc. 2022;55:417-425. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |