Published online Apr 14, 2023. doi: 10.3748/wjg.v29.i14.2134
Peer-review started: November 28, 2022
First decision: December 20, 2022
Revised: December 29, 2022
Accepted: March 9, 2023
Article in press: March 9, 2023
Published online: April 14, 2023
Processing time: 135 Days and 20.5 Hours
New drugs are urgently needed for the treatment of liver cancer, a feat that could be feasibly accomplished by finding new therapeutic purposes for marketed drugs to save time and costs. As a new class of national anti-infective drugs, carrimycin (CAM) has strong activity against gram-positive bacteria and no cross resistance with similar drugs. Studies have shown that the components of CAM have anticancer effects.
To obtain a deeper understanding of CAM, its distribution, metabolism and anti-inflammatory effects were assessed in the organs of mice, and its mechanism of action against liver cancer was predicted by a network pharmacology method.
In this paper, the content of isovaleryl spiramycin III was used as an index to assess the distribution and metabolism of CAM and its effect on inflammatory factors in various mouse tissues and organs. Reverse molecular docking technology was utilized to determine the target of CAM, identify each target protein based on disease type, and establish a target protein-disease type network to ascertain the effect of CAM in liver cancer. Then, the key action targets of CAM in liver cancer were screened by a network pharmacology method, and the core targets were verified by molecular docking and visual analyses.
The maximum CAM concentration was reached in the liver, kidney, lung and spleen 2.5 h after intragastric administration. In the intestine, the maximum drug concentration was reached 0.5 h after administration. In addition, CAM signi
We determined that CAM inhibited inflammation. We also predicted the complex multitargeted effects of CAM that involve multiple pathways and the diversity of these effects in the treatment of liver cancer, which provides a basis and direction for further clinical research.
Core Tip: Although some studies have shown that carrimycin (CAM) has therapeutic effects on inflammation and liver cancer, there are few experimental studies on its component analysis and mechanism of action. The mechanism of action was predicted by reverse molecular docking between the liver cancer target and CAM. By establishing an inflammatory mouse model to assess the state of inflammatory factors after administration, we further proved the therapeutic effect of CAM on inflammation and its possible mechanism in the treatment of liver cancer.
- Citation: Li XY, Luo YT, Wang YH, Yang ZX, Shang YZ, Guan QX. Anti-inflammatory effect and antihepatoma mechanism of carrimycin. World J Gastroenterol 2023; 29(14): 2134-2152
- URL: https://www.wjgnet.com/1007-9327/full/v29/i14/2134.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i14.2134
Carrimycin (CAM), also known as bitespiramycin and shengjimycin, is a new national class I drug that was jointly developed by the Institute of Pharmaceutical Biotechnology of the Chinese Academy of Medical Sciences and Shenyang Tonglian Group Co., Ltd and was approved for listing in June 2019. CAM is a biosynthetic drug produced through genetic engineering technology that was designed according to the characteristics of bacterial drug resistance. CAM catalyzes the formation of isovaleryl side chains by introducing foreign genes, that is, the isovaleryltransferase gene introduced into Streptomyces spiralis, which encodes a sixteen-membered ring multicomponent macrolide antibiotic that was obtained by directional acylation of spiramycin (SPM) in microorganisms[1]. Its main components include isovaleryl SPM III, isovaleryl SPM II, isovaleryl SPM I and a certain amount of 9 mixed chemicals, including butyryl-, propionyl-, and acetylspiramycin III and propionyl- and acetylspiramycin II[2]. Structurally, CAM is a macrolide antibiotic. CAM can block the activity of peptidyl transferase in 50S ribosomes to inhibit bacterial protein synthesis to produce antibacterial effects. Moreover, CAM can combine with peroxide scavenging enzymes to induce peroxide generation, destroying biological macromolecules such as DNA and achieving sterilization. In addition, CAM can promote phagocytosis by damaged neutrophils and macrophages in the body. A completed phase III clinical trial showed that CAM is safe for administration, requires a low oral dose, has a low rate of adverse reactions, produces a wide range of antibacterial activities and shows no secondary drug resistance[3]. Thus, CAM has strong medicinal value.
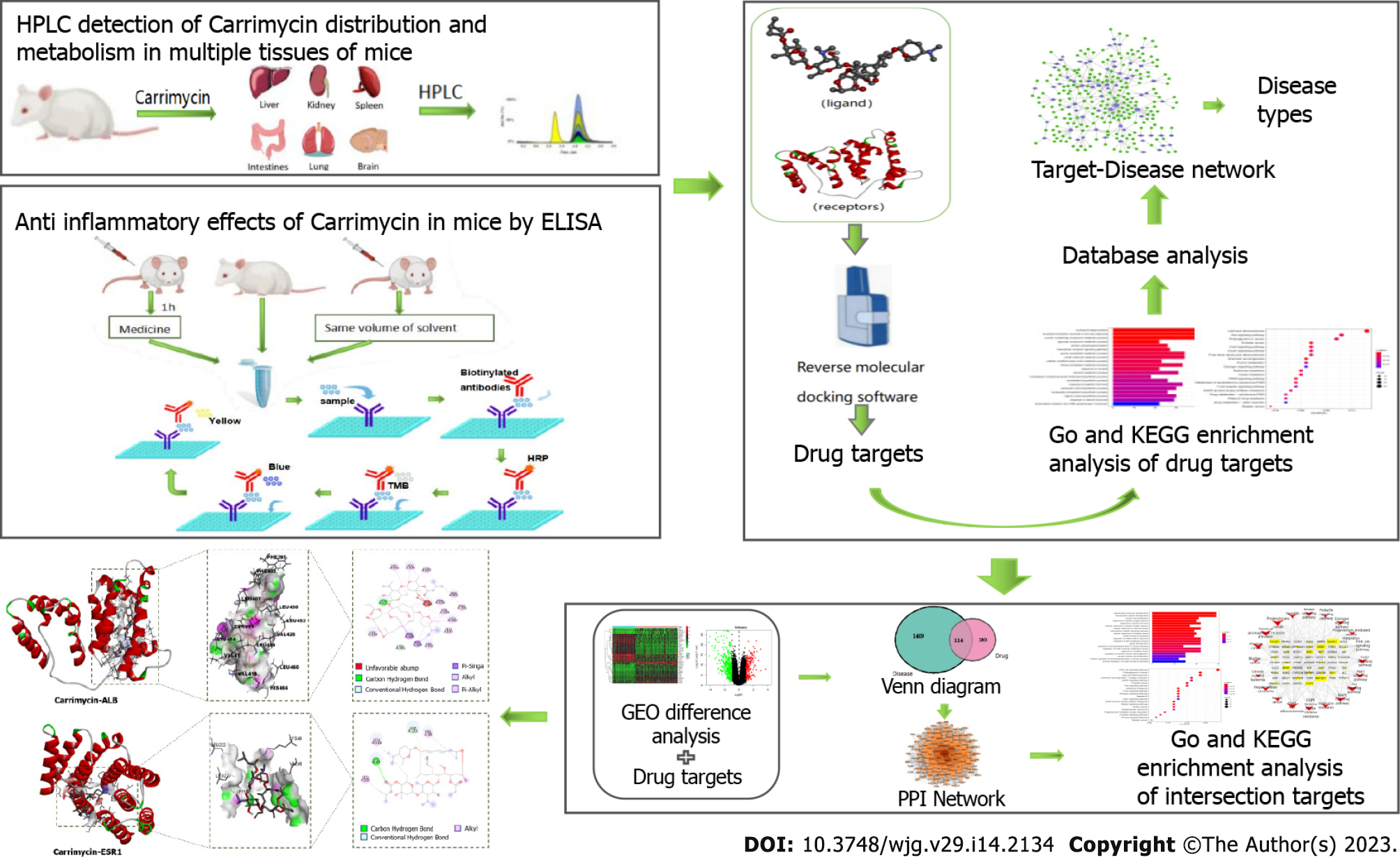
At present, there is a bottleneck in the research and development of new drugs, and using the traditional ‘multiple ligands-single target’ research and development method makes drug innovation difficult. The cost of discovering a new molecular entity and applying it to the market exceeds $2 billion USD[4]. However, the rate of return is far lower than the investment cost and the time needed. The research strategy of ‘multiple targets-single ligand’, namely, utilizing reverse molecular docking technology, has effectively solved these problems and become a favorable tool for drug repositioning. Starting from protein receptors and ligands with known chemical structures and using the principles of geometric complementarity and energy complementarity, researchers can calculate the force and affinity of the interaction between small molecule ligands and biological receptor macromolecules and score the results. These scoring results provide the optimal conformation in which a small molecule ligand binds to the biological target macromolecule, leading to prediction of the ideal binding mode between the two molecules. Finally, the corresponding biological targets can be enriched and analyzed to find the most likely disease spectrum. In this study, the distribution and metabolism of CAM in various tissues and organs of mice were assessed by hyphenated to liquid chromatography (HPLC), and the effect of CAM on inflammatory factors in these tissues and organs was detected by enzyme-linked immunosorbent assay (ELISA). Reverse molecular docking technology was used to screen the targets of CAM, and the predicted target was enriched and analyzed to obtain data on the related diseases. Then, the key targets and associated pathways for the treatment of liver cancer were analyzed by network pharmacology. The molecular mechanism of CAM and the related experimental research in disease treatment are presented in Figure 1. This study provides a reference for relevant research.
All chemicals used were of analytical grade. Chromatographic grade acetonitrile, methanol, and isopropanol, ethyl acetate, ammonium acetate, SPM, roxithromycin, and sodium carbonate were purchased from Aladdin (United States). NaCl, NaOH, and Na2HPO4 were purchased from Sinopharm Chemical Reagent Co., Ltd. CAM was provided by Shanghai Tonglian Pharmaceutical Co., Ltd.
Preparation of solution: A standard solution of CAM was prepared as follows. CAM was diluted to 1200 μL with acetonitrile to prepare a 500 μg/mL solution. This solution was then diluted to 400 μg/mL, 300 μg/mL, 200 μg/mL, 150 μg/mL, 100 μg/mL, 80 μg/mL, 60 μg/mL, 40 μg/mL, 20 μg/mL and 10 μg/mL with acetonitrile. Preparation of the control sample solution was performed as follows. The appropriate amount of roxithromycin was weighed, and acetonitrile was added to prepare a 10 μg/mL mother liquor solution. Then, a total of 980 μL of acetonitrile was added to 20 μL of the solution to prepare a 200 μg/mL sample solution.
HPLC conditions: HPLC (LC-20AT, Shimadzu, Japan) was used to analyze the chemical composition of the samples using the following parameters: Detector, spd-20a; chromatographic column, Zorbax Eclipse XdB C18 (Agilent, United States); mobile phases, phase A (10 mmol/L ammonium acetate) and phase B (acetonitrile), 50:50 (v/v); flow rate, 1 mL/min; column temperature, 30 °C; and detection wavelength, 232 nm.
Animals and drug administration: Kunming mice were purchased from the Experimental Animal Center of Jiangsu University using animal certificate No. 201927943. A total of 24 mice were used, and they were adapted to the environment in the laboratory before the experiment. The mice were randomly divided into two groups, and a standard curve was constructed. Mice in the administration group were given compound by gavage at one time point, and the second dose was double that of the first dose. The dose administered was 240 mg/kg. Three mice were randomly selected at 0.5, 2.5, 4.5, 12 and 24 h after administration, and their liver, spleen, lung, kidney, intestine and brain were dissected and stored at -80 °C. During the experiment, the mice ate and drank water normally.
Preparation of tissue homogenate: Tissue samples were weighed using LE104E/02 METTLER electronic scales (METTLER TOLEDO Instrument Co., Ltd, China) (lung, spleen, brain, and kidney, 0.3 g), 0.6 mL of acetonitrile was added, and a tissue homogenate instrument (Shanghai Jingxin Industrial Development Co., Ltd, China) was used to prepare the homogenate at a speed of 2800 rpm for 30 s. Pure water (0.3 mL) was added to the homogenate, which was then ultrasonicated (Ningbo Xinzhi Biotechnology Co., Ltd, China) and centrifuged (3500 r/min) for 10 min, and the supernatant was removed for analysis. Tissue samples (liver and intestine, 0.5 g) were used to prepare the homogenate. Pure water (0.5 mL) was added to the homogenate, which was then ultrasonicated and centrifuged (3500 r/min) for 10 min. The supernatant was removed for analysis.
Preparation of the standard curve: First, 100 μL of tissue homogenate was added to 200 μL of water, 50 μL of internal standard (200 μg/mL roxithromycin acetonitrile solution), and 50 μL of standard solution. Two hundred microliters of alkaline reagent (0.05 M sodium carbonate solution) and 2 mL of ethyl acetate-isopropanol (95:5, v/v) extraction solvent were added, and the solution was placed in a vortex shaker (Qilin Bell Instrument Manufacturing Co., Ltd, China). The solution was vortexed for 3 min and shaken at 50 rpm in the dark for 10 min (Changzhou Langyue Instrument Manufacturing Co., Ltd, China), followed by centrifugation (Thermo, United States) for 10 min at 2000 rpm. Two milliliters of the upper organic phase was placed in a 2 mL EP tube (Jiangsu Shitai Experimental Equipment Co., Ltd, China), which was then placed in a metal bath (Scilogex, United States) at 40 °C and dried with nitrogen. The organic solvent was dissolved diluted with 100 μL of mobile phase, vortexed, and injected into the instrument after passing through a 0.45 μm nylon membrane (Jianhu Yadong Glass Instrument Factory, China). The chromatogram was recorded. The standard curve was drawn with the concentration as the abscissa and the peak area/internal standard peak area (AT/AS) as the ordinate.
Sample pretreatment to determine the drug content in tissue: A total of 600 μL of tissue homogenate was placed in a 50 mL centrifuge tube (Corning, United States), 1.2 mL of ultrapure water, 300 μL of internal standard (200 μg/mL roxithromycin acetonitrile solution), and 300 μL of acetonitrile were added, the solution was mixed, and 1.2 mL of alkaline reagent (0.05 M sodium carbonate solution) was then incorporated. Next, 12 mL of ethyl acetate-isopropyl alcohol (95:5, v/v) was added for extraction, and the solution was vortexed for 3 min and shaken at 50 rpm for 10 min. After centrifugation at 2000 rpm for 10 min, 12 mL of the upper organic phase was taken from 6 centrifuge tubes and combined in a new centrifuge tube. This tube was placed in a metal bath at 40 °C and volatilized with nitrogen for accelerated solvent extraction. When approximately 300-500 μL of liquid remained, the EP tube was vortexed for 10 s, and the liquid was added to a 2 mL EP tube and dried under nitrogen flow. The final sample was dissolved in 100 μL of mobile phase, vortexed, and injected into the instrument after passing through a 0.45 μm nylon membrane, and the chromatogram was recorded.
Data processing: According to the standard curve, we calculated the changes in drug content in various tissues and organs at 0, 0.5, 2.5, 4.5, 12 and 24 h after administration and drew a histogram of these changes in the tissues at different times after administration. Excel 2016 statistical, drawing and calculation software was used.
Drug preparation: Drug preparation was carried out as follows. Several doses of CAM were added to polyethylene glycol 400 and Tween 80, and the combination was mixed well by oscillation. Then, appropriate amounts of distilled water were added to prepare CAM solutions with high, medium and low concentrations. The model drug was prepared as follows. A 1.82 mg/mL solution of the model drug azithromycin (Aladdin Reagent Co., Ltd) was diluted with a small amount of anhydrous ethanol, and then water was added to produce a solution with a final ethanol content of 10%.
Reagent preparation: PBS (pH 7.35) and Tween 20 were used to prepare a washing solution composed of PBS with 0.05% Tween 20 (if crystals formed in the buffer concentrate, the solution was gently heated until they had completely dissolved). PBS (10 ×) was diluted 1:10 in deionized water to prepare coating buffer (1 ×). The concentrated diluent (5) was diluted 1:5 in deionized water to prepare a 5 × ELISA/ELISPOT diluent.
Standard preparation was carried out as follows. The recombinant mouse interleukin-β (IL-1β) standard was dissolved in distilled water, the volume of which was indicated on the label of the vial. The standard solution was prepared 10-30 min in advance and mixed well to ensure complete and uniform dissolution (concentration of recombinant standard = 1000 pg/mL). Antibody detection was performed by diluting the detection antibody (250 ×) 1:250 in ELISA/ELISPOT diluent (1 ×). The enzyme was prepared by diluting HRP concentrate (100 ×) 1:100 in ELISA/ELISPOT diluent (1 ×).
Establishment of the animal model: Kunming mice were purchased from the Experimental Animal Center of Jiangsu University using animal certificate No. 201929470. A total of 144 mice were used, and they were adapted to the environment in the laboratory before the experiment. Male Kunming mice weighing approximately 24 g were randomly divided into 6 groups, including the normal group, model group, low-dose group (30 mg/kg), middle-dose group (60 mg/kg), high-dose group (120 mg/kg), and azithromycin group (37.9 mg/kg). The effect of each component was assessed at 0 h, 0.5 h, 2.5 h, 4.5 h, 12 h, 24 h, 48 h and 72 h, and three mice were used at each time point. Staphylococcus aureus (S. aureus) was inoculated into 50 mL of culture medium (washed with PBS and resuspended in 1000 μL of PBS with normal saline. S. aureus (3 × 108 CFU·mL-1) was injected into the mice via the tail vein at 100 μL S. aureus·24 g-1 mouse body weight. Drugs were administered one hour later. The mice in the normal group were not administered drugs or injected with bacteria. The mice in the model group were not treated with drugs. The mice in the CAM and azithromycin groups were intragastrically administered 500 μL of the appropriate compound, with a double dose given on the first day. The mice in the model group were given the same volume of solvent. The mice were sacrificed in batches at different time points after administration.
Detection of inflammatory factors: The mice were sacrificed after orbital blood was taken, and each tissue and organ was weighed by an electronic balance. Fifty milligrams of each tissue was placed into a 1.5 mL EP tube, and then 1 mL of precooled PBS and magnetic beads were added. Then, the tissue was homogenized (300 Hz, 30 s). After standing on ice for 30 min, the mixture was centrifuged at 4 °C (10000 g, 10 min), and the supernatant was taken as the test sample. The IL-4 and IL-1β levels in the supernatant were measured according to the instructions of the kit (Thermo Fisher Scientific, China).
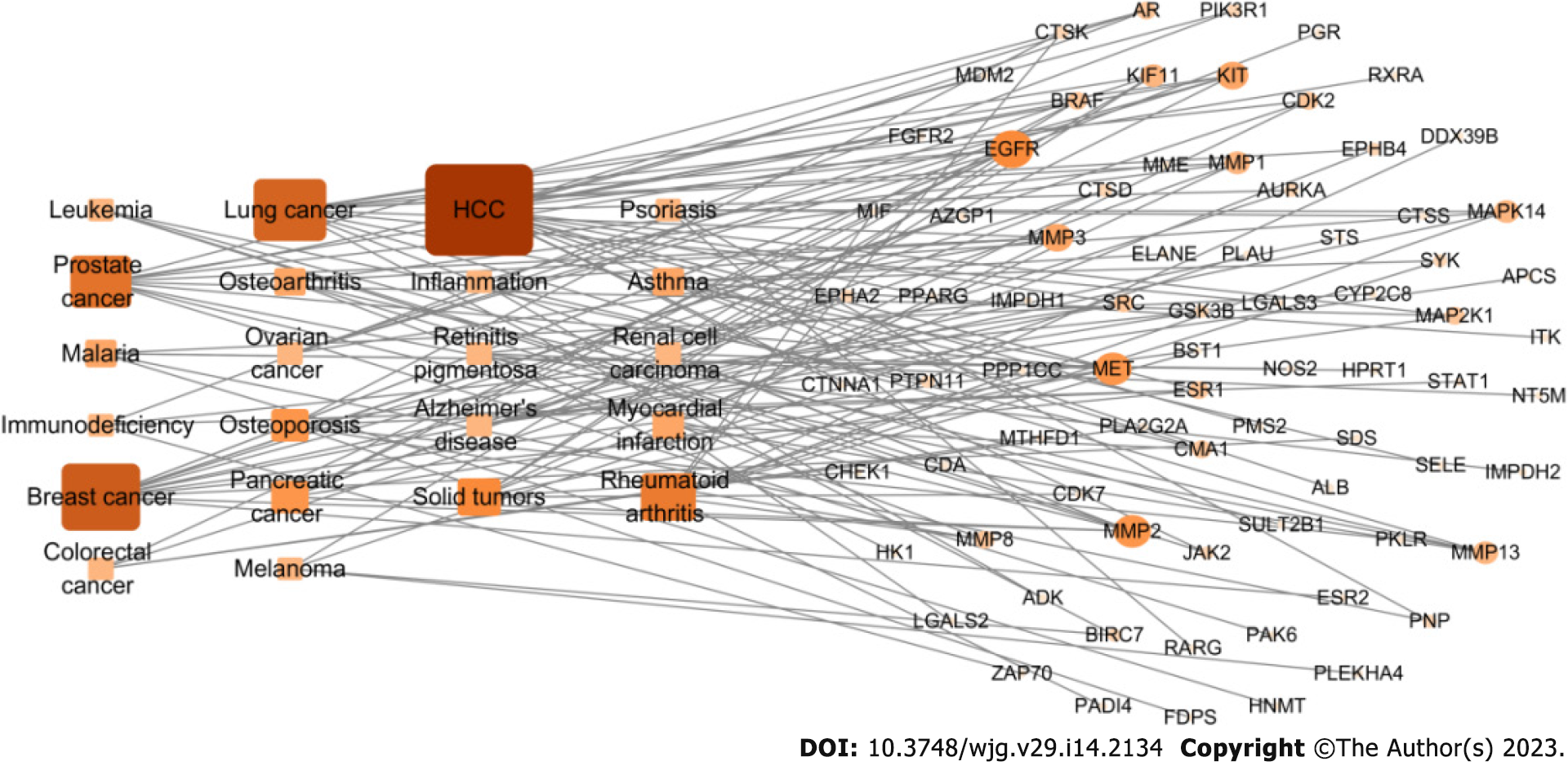
Screening for drug targets and related diseases: First, multiple data platforms, such as the PDB (https://www.rcsb.org) and TTD (http://db.idrblab.net/tt), were used to construct the target protein library of CAM. Docking software, such as INVDOCK and tarfisdock, was used to connect CAM with all targets in the protein library to predict its target. Additionally, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed. Combining these data with those from GeneCard (https://www.genecards.org), UniProt (https://www.UniProt.org), TCMSP (https://old.tcmsp-e.com/tcmsp.php) and other databases, the predicted target proteins were analyzed one by one, and a relationship diagram was drawn with Cytoscape. This established the target protein-disease type network (degree > 3) to determine the most relevant diseases, which were consistent with the results from enrichment analysis.
Screening for disease targets and drug-disease intersection targets: The most relevant disease from the above analyses was liver cancer, which was selected for further study. Using the GEO database (https://www.ncbi.nlm.nih.gov/geo/) with ‘liver cancer’ as the search term, the GSE121248 dataset and GPL570 platform were downloaded. logFC ≥ 1 and adj P value < 0.05 were the screening conditions to identify the differentially expressed genes. Moreover, the liver cancer-related targets were integrated with the search term ‘liver cancer’ in the GeneCards database (https://www.genecards.org/), and the genes differentially expressed in liver cancer were combined to obtain a comprehensive list of liver cancer targets. The target gene of CAM in liver cancer was determined by intersection of the target of CAM and the target of liver cancer.
Construction of the protein-protein interaction network and enrichment analysis: The intersecting target gene of CAM and liver cancer was introduced into the STRING database (https://string-db.org) to obtain protein interaction information, and the research species was set as Homo sapiens. The lowest interaction score used to obtain the protein-protein interaction (PPI) network was 0.009. Then, Cytoscape 3.7.2 topology analysis of the PPI network was performed, and the core target of CAM in liver cancer was obtained. Then, GO and KEGG enrichment analyses were performed by R software (P < 0.05).
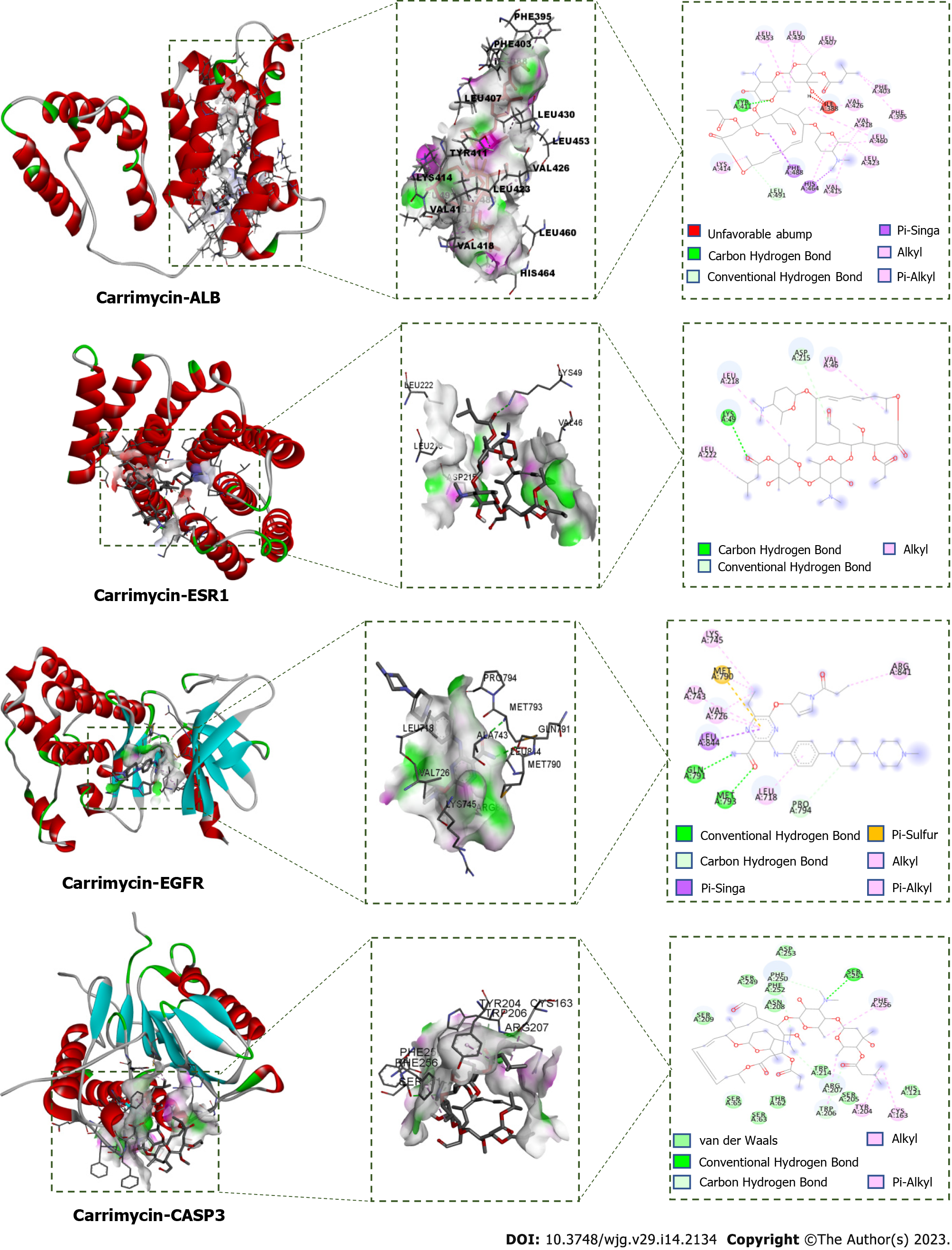
Molecular docking verification and visual analysis: ChemOffice software was used to generate the 3D structure of CAM, which was loaded into AutoDock Tools 1.5.6 as the docking ligand. The crystal structures of albumin (ALB) (PDB ID: 1YSX), estrogen receptor 1 (ESR1) (PDB ID: 3HA8), epidermal growth factor receptor (EGFR) (PDB ID: 5Y9T), caspase 3 (CASP3) (PDB ID: 4JJE), and androgen receptor (AR) (PDB ID: 2PIT) were downloaded from the PDB database (http://www.rcsb.org). AutoDock Tools 1.5.6 software was used to remove the water and hydrogen atoms and to perform ligand separation, and the resulting protein structures were stored as the acceptors. AutoDock Vina 1.1.2 was used for molecular docking. The conformation with the lowest binding energy from all docking runs was selected as the docking conformation, and Discovery Studio 2016 Client software was used to visualize the docking conformation.
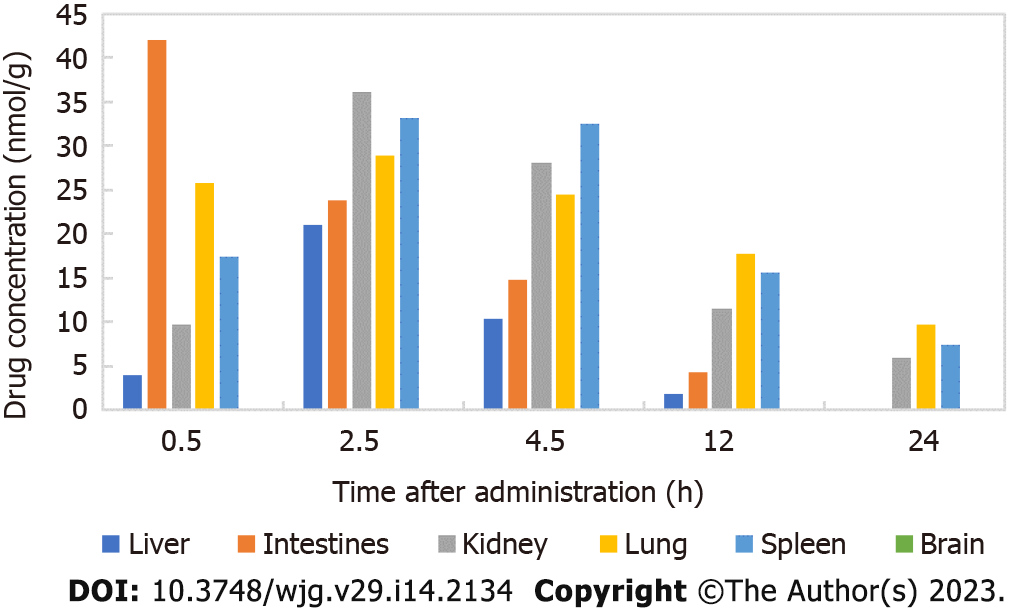
There was a good linear relationship between isovaleryl SPM III in the approximate concentration range of 0.1-200.0 μg/mL. The regression equations of the correlation curves in the liver, intestine, kidney, lung, spleen and brain were Y = 0.1977C-0.1072 (R2 = 0.9953), Y = 0.1929C-0.3317 (R2 = 0.9970), Y = 0.1447C-0.3909 (R2 = 0.9982), Y = 0.2512C-0.7149 (R2 = 0.9942), Y = 0.1748C-0.7313 (R2 = 0.9966) and Y = 0.2100C-0.9697 (R2 = 0.9957), respectively, each with an RSD value less than 2%. Thus, the results are reliable and reproducible. The contents of CAM in different tissues and organs 24 h after intragastric administration to mice is shown in Figure 2. The results showed that 2.5 h after CAM treatment, the maximum drug concentration was reached in the liver, kidney, lung and spleen. In the intestine, the maximum drug concentration was reached 0.5 h after administration, and CAM was not detected in the brain.
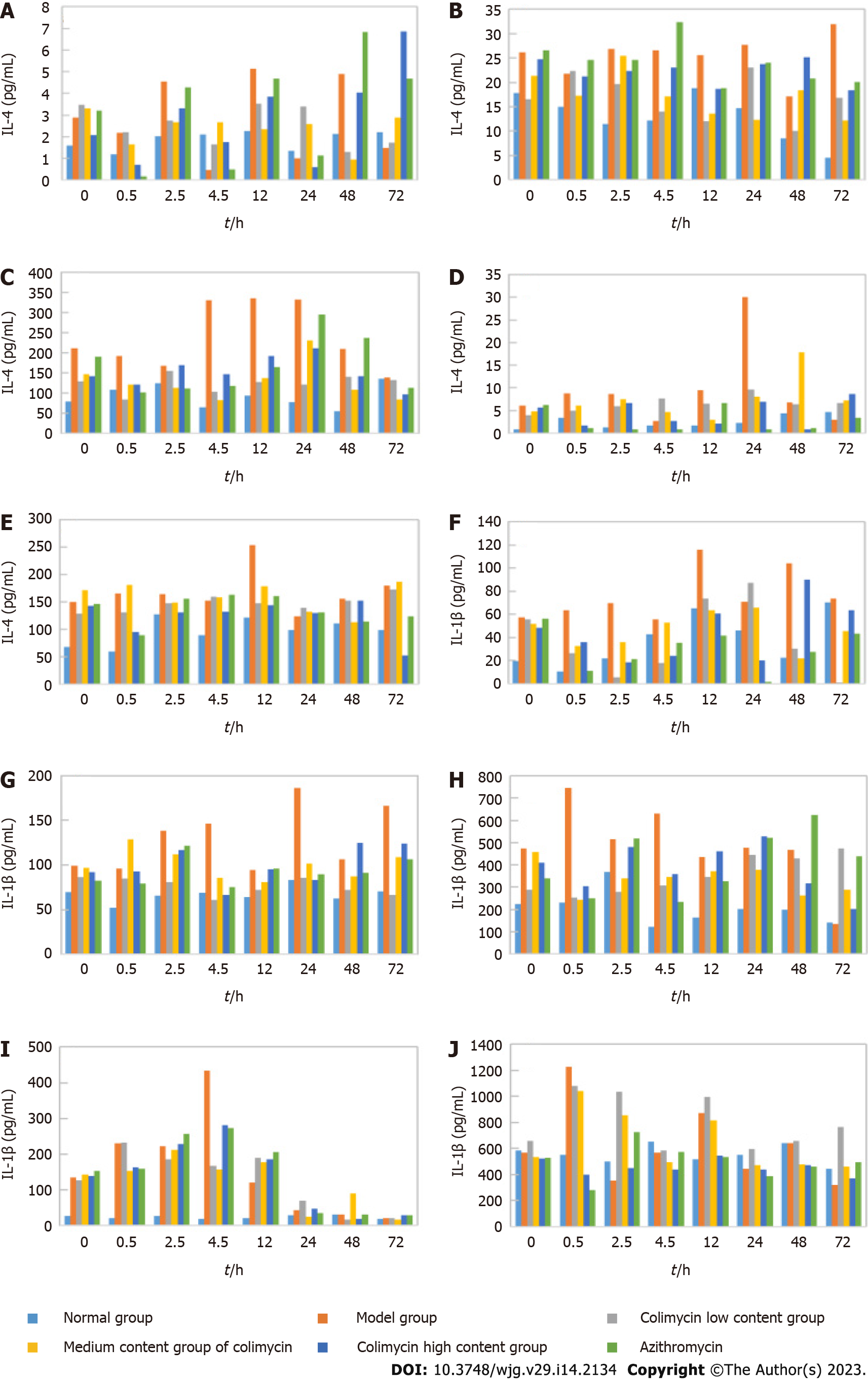
IL-4 and IL-1β are common inflammatory cytokines; thus, ELISA was used to analyze the IL-4 and IL-1β levels in different mouse organs and tissues to determine the anti-inflammatory effect of CAM (Figure 3). CAM significantly reduced IL-4 levels in the lung and kidney and especially the liver and spleen. CAM also significantly reduced IL-1β levels in the spleen, liver, and kidney and particularly the small intestine and lung.
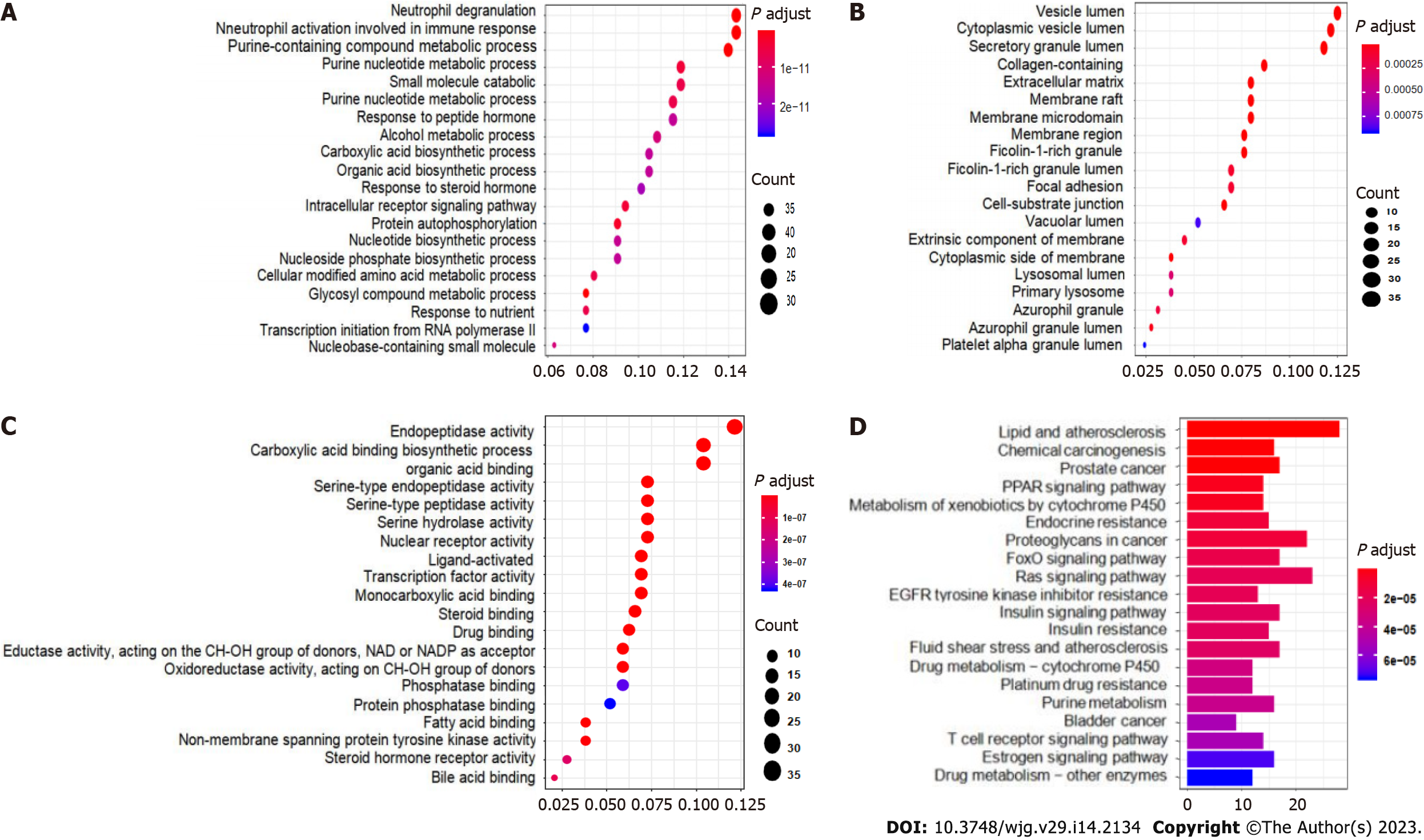
From the conformation analysis and scoring results, the targets of CAM were selected. After excluding the ineffective and repeated data, 293 targets were obtained. The 293 predicted target proteins were analyzed by GO and KEGG enrichment analyses. The output GO enrichment analysis results included three pathways: Biological processes (BP) (Figure 4A), cellular components (CC) (Figure 4B), and molecular functions (MF) (Figure 4C). The core targets of CAM are mainly involved in the response to neutrophil degranulation and the immune response and in neutrophil activation, purine-containing compound metabolism, glycosyl compound metabolism and protein autophosphorylation. The molecular functions mainly involve carboxylic acid binding, organic acid binding, protein tyrosine kinase activity and serine endopeptidase activity. In terms of cell composition, the main targets involved vesicle cavity, cytoplasmic vesicle cavity, and collagen-containing extracellular matrix (ECM) pathways. The KEGG enrichment pathway analysis results in Figure 4D demonstrate that the pathways in which CAM participates in disease treatment mainly include the Ras, FoxO, and insulin signaling pathways.
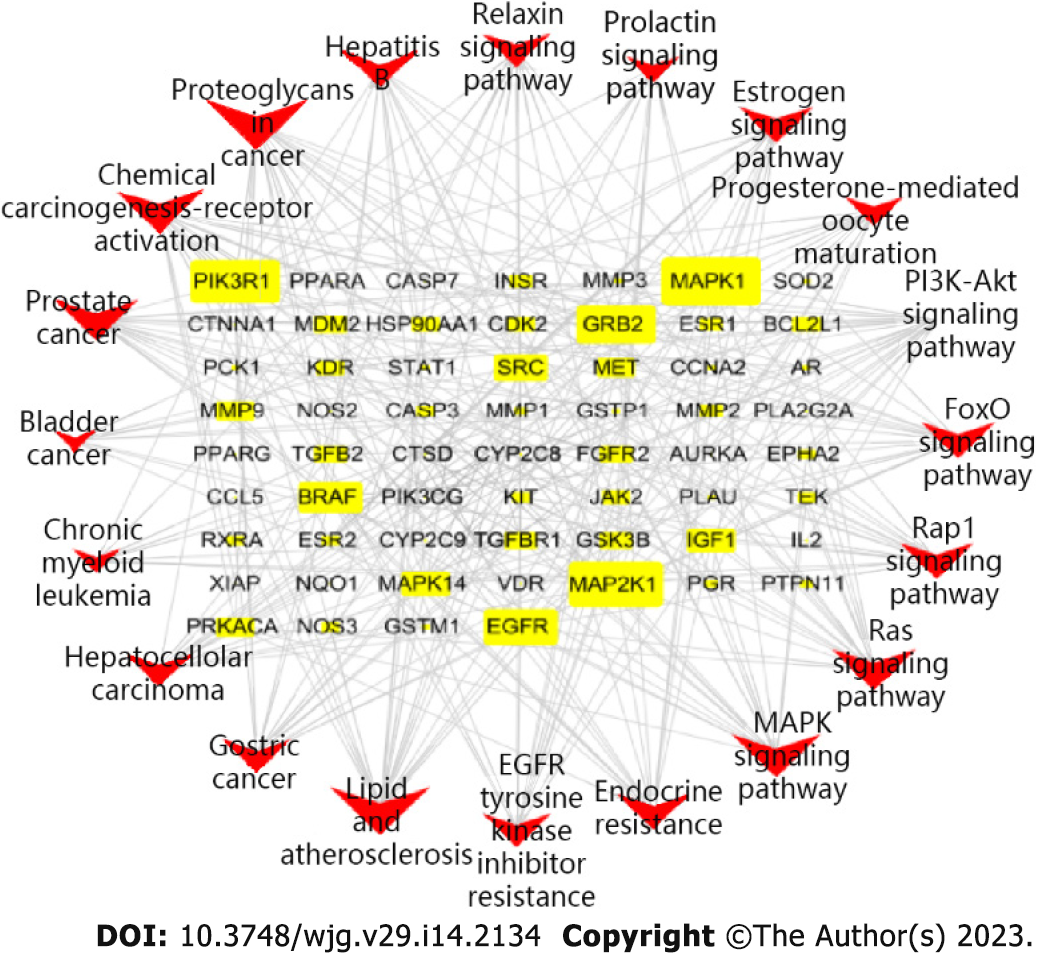
The 293 predicted target proteins were analyzed one by one, and the data from multiple databases were combined to screen 77 main targets (circles) and 22 main diseases (rectangles) (Figure 5). The targets with the highest degrees were EGFR (degree = 7), mesenchymal epithelial cell transforming (MET) (degree = 7), matrix metalloproteinas (MMP)-2 (degree = 7), and ESR1 (degree = 6). The main diseases involved were liver cancer (degree = 18), breast cancer (degree = 13), lung cancer (degree = 12), and prostate cancer (degree = 10).
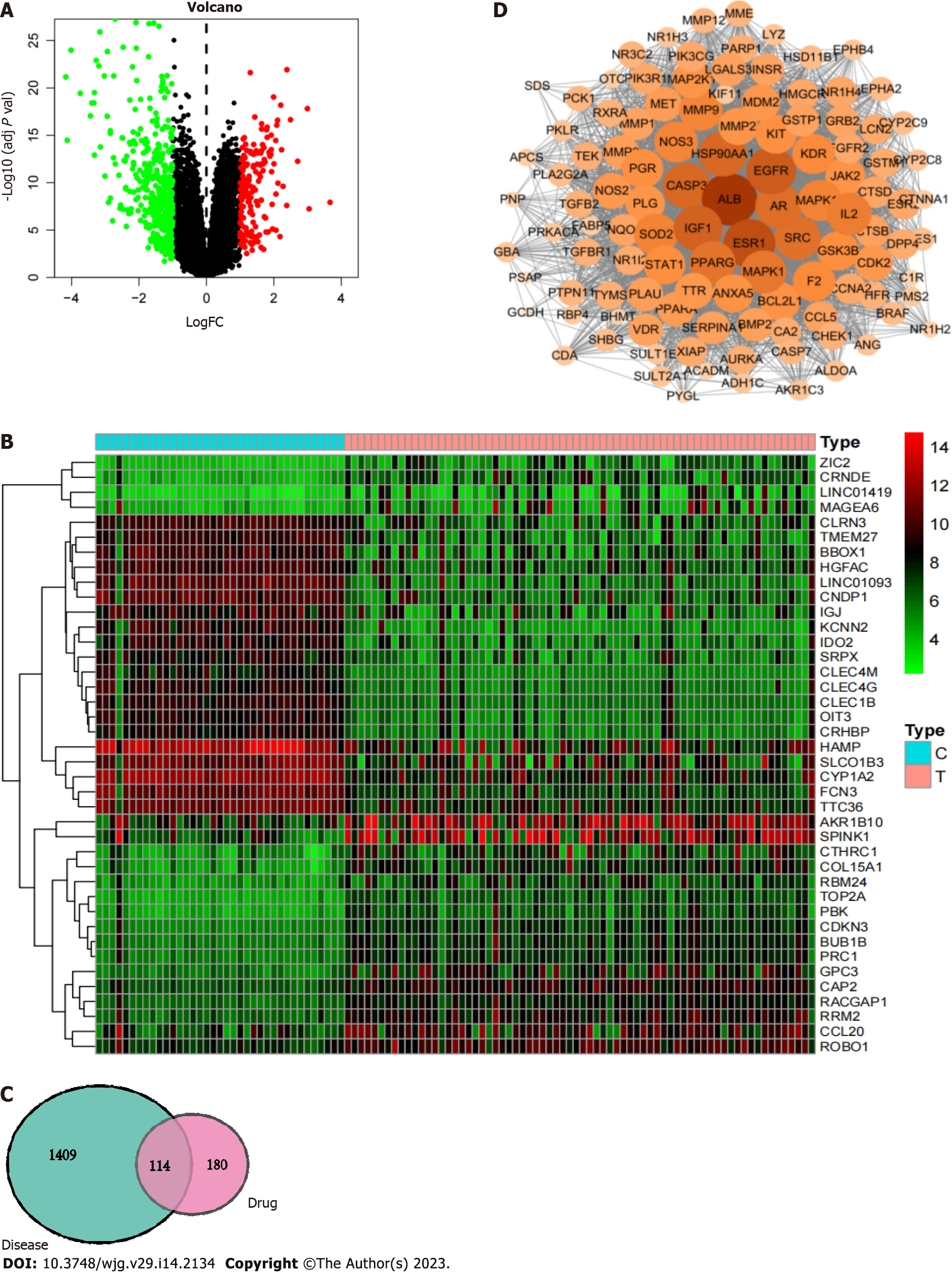
First, 580 differentially expressed genes in liver cancer were screened using the GEO database to construct a volcano plot (Figure 6A). Genes with significant upregulation and downregulation were selected to draw a heatmap (Figure 6B). Among them, 167 differentially expressed genes were upregulated in hepatocellular carcinoma, such as AKR1B10, SPINK1, ROBO1, CCL20, and RRM2, and 413 differentially expressed genes were downregulated in hepatocellular carcinoma, such as SRPX, CLEC4 M, CLEC4G, CLEC1B, OIT3, and CRHBP. After integration with the liver cancer-related targets from the GeneCards database, a total of 943 liver cancer-related targets were retrieved, and duplicate values were removed from the combined dataset to obtain 1523 liver cancer disease targets.
R software was used to match and map 294 CAM target genes with the 1523 liver cancer disease targets, resulting in 114 intersecting target genes, including transthyretin, bone morphogenetic protein 2, ALB, Glutathione S-transferase P1, PNP, and PLG. These results are shown in the Venn diagram in Figure 6C.
The obtained PPI network is shown in Figure 6D. The PPI network was used with Network Analyzer in Cytoscape v3.7.2 for degree of freedom, mediation and centrality algorithm topology analysis to select the greater than the average targets as the key targets. Only the top 15 targets were retained (Table 1). Finally, we screened the core target proteins of CAM in liver cancer: ALB, Insulin-like growth factor 1 (IGF1) (somatomedin C), ESR1, EGFR, AR, mitogen-activated protein kinase 1 (MAPK1), MAP2K1 (gene encoding mitogen-activated protein kinase 1), CASP3, etc.
| Gene name | Degree | Betweenness centrality | Closeness |
| ALB | 110 | 0.04074405 | 0.97413793 |
| ESR1 | 102 | 0.02219136 | 0.91129032 |
| EGFR | 97 | 0.01545146 | 0.87596899 |
| IGF1 | 95 | 0.01457077 | 0.86259542 |
| HSP90AA1 | 93 | 0.02011442 | 0.84962406 |
| CASP3 | 93 | 0.01307351 | 0.84962406 |
| AR | 91 | 0.01215915 | 0.83703704 |
| SRC | 90 | 0.01137002 | 0.83088235 |
| PPARG | 89 | 0.01582869 | 0.82481752 |
| MAPK1 | 88 | 0.00943128 | 0.81884058 |
| IL2 | 86 | 0.01154328 | 0.80714286 |
| SOD2 | 84 | 0.01534439 | 0.79577465 |
| MAPK14 | 84 | 0.00918383 | 0.79577465 |
| NOS3 | 83 | 0.0095422 | 0.79020979 |
| MMP9 | 83 | 0.00820256 | 0.79020979 |
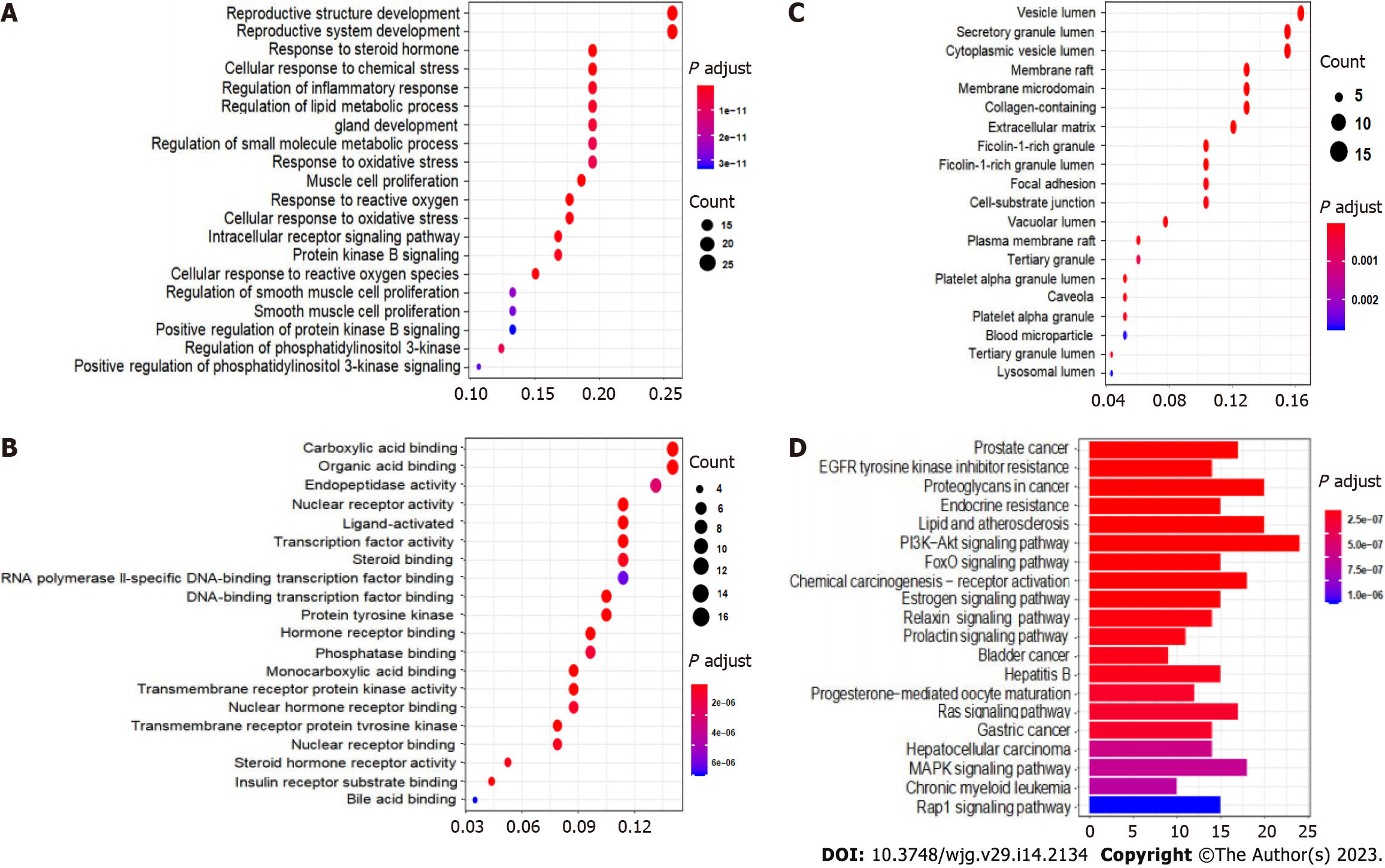
The GO enrichment analysis results included the terms BP (Figure 7A), MF (Figure 7B) and CC (Figure 7C). The core targets of the active components of CAM were mainly enriched in processes including reproductive structure development, reproductive system development, muscle cell proliferation, response to reactive oxygen species, cell response to reactive oxygen species, molecular functions, nuclear receptor activity, transcription factor activity, and ligand activation, while the cellular components included vesicle cavity, secretion granules, cytoplasm vesicle cavity, membrane microregion, membrane region and ECM.
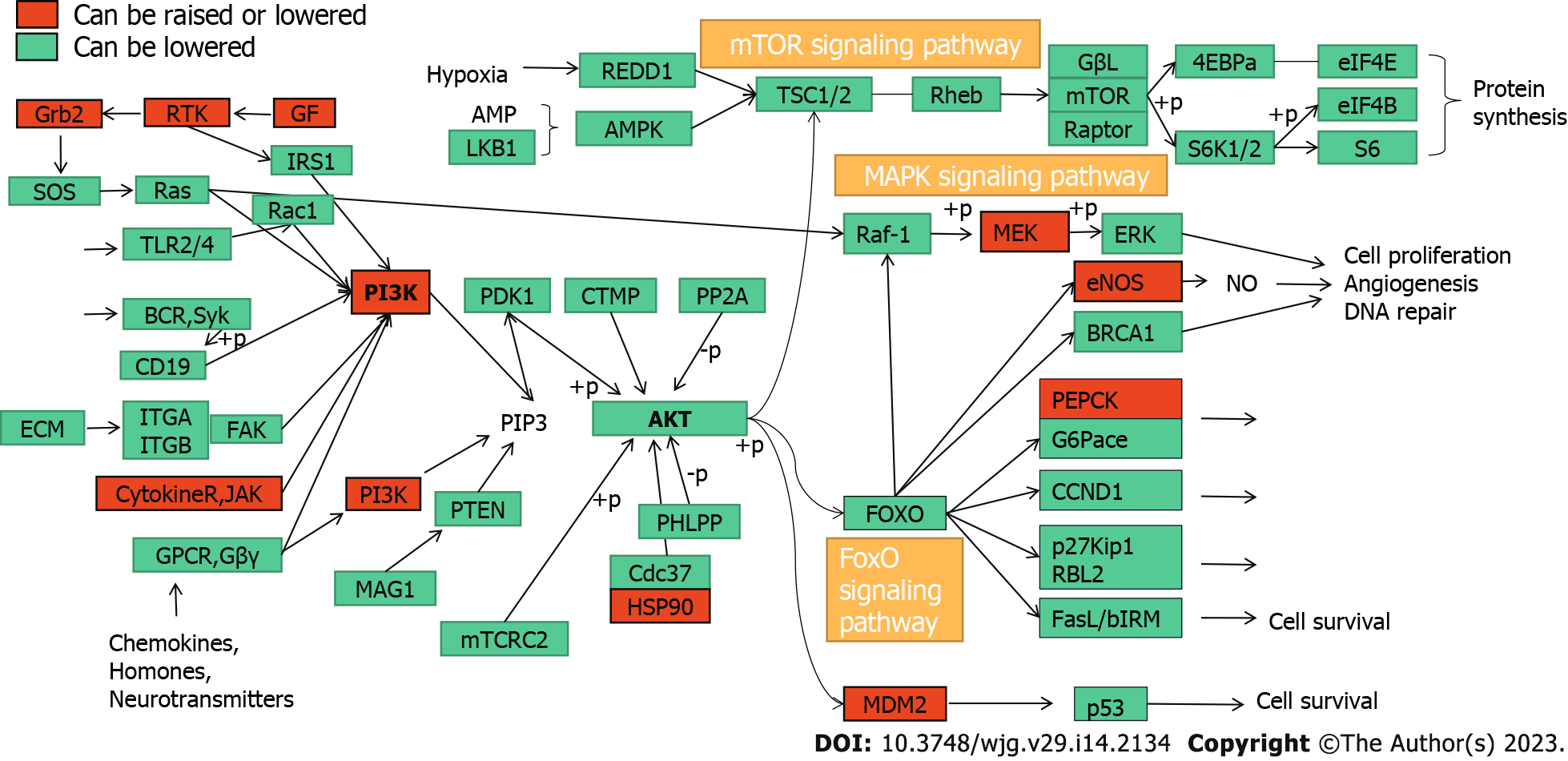
KEGG pathway enrichment analysis of the key targets of CAM in liver cancer performed in R resulted in 140 signaling pathways (P < 0.05). The top 20 pathways were selected to create a bubble map (Figure 7D). The main pathways involved in CAM treatment of liver cancer include the PI3K-Akt, MAPK, Ras, FoxO, estrogen, and hepatitis B pathways and others. CAM acts on multiple targets in the PI3K-Akt signaling pathway to produce effects against liver cancer by activating or inhibiting certain proteins in this pathway (Figure 8A).
According to the relationships between the key targets of action and the signaling pathways, a key target-pathway network map was constructed using R language and Cytoscape v3.7.2 (Figure 9). The graph has 80 nodes and 307 edges. The nodes include 60 related targets indicated by yellow rectangles and 20 signaling pathways indicated in red type V. The node size corresponds to the degree value; the larger the node is, the more important this factor is the network. Therefore, CAM can regulate multiple biological pathways and treat liver cancer by acting on multiple targets, such as MAPK1, MAP2K1, phosphoinositide-3-kinase, regulatory subunit 1, EGFR and MET.
In molecular docking, it is generally thought that a stronger affinity between a ligand and a receptor gives a lower binding energy, and hence, a greater probability of interaction. When the obtained binding energy is less than zero, the receptor and ligand can bind spontaneously. The binding energies for each receptor-CAM interaction are shown in Table 2. All of the binding energies are lower than -5.0 kcal/mol, indicating that CAM has a good ability to bind to these targets. To further study these interactions, the relationships between CAM and these targets were plotted and analyzed (Figure 10).
| Key target | Binding energy (kcal/mol) |
| ALB | -9.3 |
| ESR1 | -5.8 |
| EGFR | -6.5 |
| CASP3 | -6.1 |
| AR | -5.6 |
| HSP90AA1 | -5.0 |
| IGF1 | -5.1 |
| MAPK1 | -6.7 |
| MAP2K1 | -5.2 |
| IL2 | -5.8 |
In this study, reverse molecular docking and network pharmacology were used to screen the targets of action and related diseases of CAM, analyze the function and pathway enrichment of the targets of action, and establish a network of the key target proteins in disease to determine the most relevant disease types from the enrichment analysis results. In addition, the distribution and metabolism of CAM and its effect on inflammatory factors in various tissues and organs were examined, and the mechanism by which CAM can treat disease was systematically explored.
According to the HPLC analysis of the distribution and metabolism of CAM in mice, the maximum concentration of CAM in the intestine was reached 0.5 h after administration and that in the liver, kidney, lung and spleen occurred 2.5 h after administration. CAM was eliminated faster from the liver and intestine than from the kidney, lung and spleen. CAM was not detected at any time point in the brain, indicating that this compounds does not easily cross the blood-brain barrier.
IL-4 is a common inflammatory cytokine that is involved in the occurrence of inflammation, and the inflammatory microenvironment is closely related to the deterioration of tumor cells[5]. IL-4 inhibits the inflammatory response in advanced liver cancer, mainly because an increase in its levels can weaken the extent of the inflammatory response and viral replication in the liver[6]. IL-1β is a proinflammatory cytokine derived from monocytes and macrophages that can participate in the processes of infection, inflammation, immune injury, tissue destruction and so on[7]. Moreover, relevant studies have shown that IL-1β may be involved in the pathogenesis and evolution of tumors[8]. Therefore, the effects of CAM on inflammatory factors in various tissues and organs were examined. CAM was found in the lung, kidney, liver and spleen and had particularly affected the IL-1β levels in the spleen, liver, and kidney and especially the small intestine and lung.
The network pharmacology results showed that CAM is involved in disease treatment through processes including neutrophil degranulation, neutrophil activation, immune response participation, purine-containing compound metabolism, glycosyl compound metabolism and protein autophosphorylation. Its molecular functions mainly entail carboxylic acid binding, organic acid binding, protein tyrosine kinase activity, endopeptidase activity and serine endopeptidase activity. In terms of cell composition, CAM is mainly involved in vesicle cavity, cytoplasmic vesicle cavity, secretory granule cavity and collagen-containing ECM.
Neutrophils are a kind of natural immune cell, and with the deepening of research, the relationship between neutrophils and tumors has attracted attention. Many studies have shown that neutrophils have potential antitumor functions[9] and cytotoxic effects on tumor cells[10]. Neutrophils are also an important member of the human immune system, playing an important role in the defense mechanisms of the human body[11]. Neutrophils release proteolytic substances such as myeloperoxidase (MPO), neutrophil elastase (NE), and tumor necrosis factor (TNF)-related apoptosis inducing ligand (TRAIL) and active substances with biological effects related to their degranulation functions[12]. MPO and NE are present in the celestial green granules of neutrophils, and their extracellular release via degranulation produces biological effects[13]. MPO can catalyze the formation of HOCl in vivo, and HOCl and NE can inhibit tumor growth at certain concentrations[14]. TRAIL is a member of the TNF superfamily, whose receptors are expressed and distributed on the surfaces of many tumor cells. Therefore, for some tumor cells, TRAIL can not only induce tumor cell apoptosis but also inhibit tumor angiogenesis through selective killing effects[8].
Purines are heterocyclic aromatic compounds. Since Noell and Robins[15] discovered 2-amino-6-alkyl thiopurine derivatives as antitumor drugs in 1962, considerable efforts have been made to develop purine derivatives with antitumor activity over the last few decades[16]. Purine compounds, such as mercaptopurine, which was discovered earlier, and nelarabine and clofarabine, which were discovered later, have been used as antitumor drugs for many years[17]. Therefore, the metabolic processes of purine-containing compounds have inhibitory effects on tumors.
Protein phosphorylation and dephosphorylation regulate almost all life activities, such as cell signal transduction, differentiation, growth, and apoptosis. Approximately 30% of the proteins in the body can undergo different types of phosphorylation[18]. Abnormal protein phosphorylation often leads to abnormal cellular activities and even cell damage or carcinogenesis[19]. Abnormalities in cellular activities, such as those involved in tumor growth, differentiation, apoptosis and movement, are all accompanied by abnormalities in protein phosphorylation in their corresponding cellular signal transduction pathways.
KEGG analysis of the enriched pathways of the CAM targets showed that the pathways involved in the treatment of diseases mainly included the Ras, FoxO, insulin, estrogen, and peroxisome proliferator-activated receptor (PPAR) signaling pathways. The Ras signaling pathway was named because this protein is a common mediator of the signal transduction processes of many growth factors. It has been reported that the Ras family diabetes-related gene RRAD can inhibit the proliferation, aerobic glycolysis and apoptosis of hepatocellular carcinoma cells by downregulating ACTG1 levels[20]. FoxO transcription factors shuttle through and outside the nucleus to regulate pathways involved in cell differentiation, proliferation, and stress, cell cycle metabolism, and tumor inhibition. The pathogenesis of hepatitis B-related liver cancer is closely related to activation of the FOXM1/FoxO3a signaling pathway[21]. Insulin signaling affects energy metabolism and growth, and this signal is transmitted through the Akt branch. The Akt signaling pathway can inhibit tumor cell proliferation, induce apoptosis and downregulate the expression of Bcl-2[22]. Akt is a regulatory molecule downstream of PI3K. Akt is activated after being phosphorylated and can affect the expression of the downstream metastasis-related gene MMP-9, the apoptosis-related gene Bcl-2 and the proliferation-related gene cyclin D1[23]. Akt plays an important role in regulating cell proliferation, metastasis and apoptotic processes, and its hyperactivation is closely related to the development of various tumors, including liver cancer[24]. Dysregulation of the estrogen signaling pathway is an important factor in the development of many malignant tumors, such as those found in breast cancer, liver cancer and lung cancer. Estrogen exerts its physiological and pathological functions by binding to the estrogen receptor (ESR). ESR-α36 expression is upregulated in hepatocellular carcinoma, so ESR-α36 may be involved in the rapid estrogen signal transduction that mediates hepatocarcinogenesis[25]. Abnormal expression of ESR-α36 in the liver is related to hepatocyte proliferation. Estrogen passes through the ESR-α36 and EGFR/SRC axes to induce MAPK/ESR-K phosphorylation and promote the expression of cyclin D1. ESR-α36 interacts with the EGFR/SRC/SHC complex through the EGFR/ESR-α36/ERK signaling axis and mediates rapid estrogen signal transduction in hepatoma cells. Thus, the ESR-α36/EGFR signaling loop is a potential target for the development of a new approach to treat liver cancer, as the ESR-α36/EGFR positive regulatory loop plays an important role in the maintenance and positive regulation of hepatoma cells[26]. PPAR-γ is an extremely important regulator of fat and glucose metabolism and plays an important role in autoimmune responses and inflammatory processes[27]. PPAR-γ is expressed in bronchial airway epithelial cells in situ and can inhibit the activity of these cells, activate endogenous PPAR-γ, and antagonize the inflammatory response. Zhu et al[28] suggested that PPAR-γ can affect the transport of macrophages and increase their phagocytosis, promoting the occurrence of a practical immune response. PPAR-γ ligand stimulation in tumor cells and tissues can have a major antitumor effect[29].
According to the analysis of CAM targets, EGFR intersects with multiple signaling pathways, including those in healthy tissues and liver cancer, breast cancer, kidney cancer, colon cancer and other tumor cells[30]. When EGFR binds to its ligand, the receptor can dimerize specific tyrosine residues can be phosphorylated. These phosphorylated tyrosine residues can recruit other signaling molecules to initiate the downstream signal transduction pathway that mediates the proliferation, metastasis and drug resistance of tumor cells. In liver cancer tissues, the expression of EGFR is significantly upregulated. For example, when EGFR is activated by its ligand, it can mediate the growth, proliferation and metastasis of liver cancer cells through multiple signaling pathways, such as the PI3K-Akt, JAK/STAT3, and Ras/MEK/ERK pathways[31]. In organisms, a variety of proteases participate in the degradation of the basement membrane and ECM, among which MMPs play the most prominent role. MMPs are the main targets of drugs against tumor invasion and metastasis. MMP-2 can specifically degrade the main components of the basement membrane and intercellular matrix (type IV collagen) and improve tumor cell invasiveness and metastasis. Therefore, MMP-2 is considered one of the most important MMPs in these processes[32]. It has been reported that a low level of MMP-2 expression can promote both the activation of nuclear factor-kappa B protein signaling and the invasion and metastasis of lung cancer[33]. MMP-3 is one of the key enzymes involved in ECM synthesis and the regulation of catabolic activity. MMP-3 is also closely related to the invasion and metastasis of tumor cells[34].
After analysis of the PPI network constructed with the intersecting targets of CAM and liver cancer, it was found that ALB, IGF1, ESR1, EGFR, AR, MAPK1, SRC, MAPK14, KIT, MMP-2, and CASP3 are both involved in the pathological process of liver cancer and also key targets of CAM. Molecular docking showed that CAM could bind well to ALB, ESR1, EGFR, CASP3 and other targets through hydrogen, conjugated and other bonding interactions, which further confirmed the therapeutic effect of CAM on liver cancer. Studies have shown that ALB is a serum protein that is almost exclusively expressed in liver tissues and that ALB genes have a closed conformation in other tissues[35]. Dai et al[36] showed that ESR1 expression is a candidate tumor suppressor gene in hepatocellular carcinoma (HCC). Further, promoter hypermethylation may be a mechanism by which expression of ESR1 is repressed, and the extent of hypermethylation of ESR1 may be a marker for HCC status and progression. EGFR is at the intersection of multiple signaling pathways and also an important mediator of physiological activities such as cell growth, proliferation, differentiation and adhesion. EGFR expression levels were found to be low in healthy tissues but significantly high in liver, breast, kidney, colon and other tumor cells[30]. When EGFR binds to its ligand, the receptor can dimerize specific tyrosine residues can be phosphorylated. These phosphorylated tyrosine residues can recruit other signaling molecules to initiate the downstream signal transduction pathway that mediates the proliferation, metastasis and drug resistance of tumor cells. The expression of EGFR was found to be significantly upregulated in hepatocellular carcinoma, and high EGFR expression is also closely related to the malignant transformation of hepatocellular carcinoma[37]. A large number of studies have reported the role of EGFR in the occurrence and development of liver cancer. For example, when EGFR is activated by its ligand, it can mediate the growth, proliferation and metastasis of liver cancer cells through multiple signaling pathways, such as the PI3K-Akt, JAK/STAT3, and Ras/MEK/ERK pathways[31]. CASP3 is a protease that can specifically cleave poly ADP ribose polymerase and acetyl-devd-7-amino-4-methylcoumarin, resulting in DNA cleavage and the promotion of apoptosis, both of which play a core roles in the executive stage of apoptosis. Xie et al[38] have confirmed activation of macrophages plays a critical role in liver injury and fibrogenesis during hepatitis B virus (HBV) infection. And hepatitis Be antigen (HBeAg) can obviously induce the production of macrophage inflammatory cytokines. During HBV-induced macrophage activation, the ESR-K/CREB signaling pathway promotes miR-212-3p expression. MiR-212-3p can inhibit the production of inflammatory cytokines induced by HBeAg by regulating the target gene MAPK1.
According to GO analysis of the intersecting genes of CAM and liver cancer targets, CAM was found to be involved in the regulation of the inflammatory response and protein kinase B signaling associated with cancer. KEGG analysis of the effects of CAM on the key targets of liver cancer showed that many signaling pathways, such as the FoxO, PI3K-Akt, estrogen, Ras, and MAPK signaling pathways, coincide with the pathways of the predicted targets of CAM, which proves that CAM can indeed affect liver cancer cells through the activities of multiple targets and multiple pathways.
In this study, we examined the distribution and metabolism of CAM and its effects on inflammatory factors in various tissues and organs of mice. Using reverse molecular docking technology, we screened for diseases related to CAM activity in liver cancer. Moreover, we systematically analyzed the targets and biological pathways of CAM in the treatment of liver cancer by network pharmacology and molecular docking to further explore the mechanism by which CAM is involved in disease treatment. However, network pharmacology methods are based on data research, so there are some remaining limitations, such as incomplete database data, imperfect screening criteria for components and targets and diverse pathway enrichment methods, which will affect the accuracy of the research results. Therefore, follow-up experiments need to be carried out on the material basis of the pharmacodynamic pathways to verify the research results here, which can provide a theoretical and experimental basis for treating liver cancer or inflammation with CAM.
As a new national first-class drug, carrimycin (CAM) has much potential medicinal value. The clinical results show that its antibacterial activity and pharmacodynamic activity are significantly greater than those of similar antibiotics. Through the combination of animal experiments and network pharmacology, this study demonstrates that CAM plays a positive role in the treatment of inflammation and some cancers.
At present, the mechanism of CAM in the treatment of liver cancer is not clear, so the mechanism can be speculated by network pharmacology.
To gain a deeper understanding of CAM, the distribution, metabolism and anti-inflammatory effects of CAM in organs were assessed, and the anti-liver cancer mechanism of CAM was analyzed.
Taking the content of isovaleryl spiramycin III as the index, the distribution and metabolism of CAM in various tissues and organs of mice and its effect on inflammatory factors in various tissues and organs of mice were detected. The target of CAM was determined by reverse molecular docking technology, the disease type corresponding to each target protein was selected, the target protein disease type network was established, and the key targets of CAM in liver cancer were screened by network pharmacological methods. The core target was verified by molecular docking and visual analysis.
The maximum CAM concentration was reached in the liver, kidney, lung and spleen 2.5 h after intragastric administration. In the intestine, the maximum drug concentration was reached 0.5 h after administration. CAM is predicted to regulate related pathways by acting on many targets, such as albumin, estrogen receptor 1, epidermal growth factor receptor and caspase 3, to treat cancer, inflammation and other diseases.
CAM had an inhibitory effect on inflammation. It also predicts the multitarget, complexity of CAM involving multiple pathways and the diversity of CAM effects in the treatment of liver cancer, which provides a basis and direction for further clinical research.
We examined the distribution and metabolism of CAM in murine tissues and organs and the effect of CAM on inflammatory factors and performed a systematic analysis of the targets and biological pathways involved in the treatment of liver cancer with CAM by network pharmacology, which led to further insights into the mechanism of treatment of the disease with CAM.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ahn KS, South Korea; Shroff RT, United States S-Editor: Wang JJ L-Editor: A P-Editor: Yuan YY
| 1. | Liu J, Zhang Y, He W. [Construction of a novel carrimycin-producing strain by using CRISPR-Cas9 and ribosome engineering techniques]. Sheng Wu Gong Cheng Xue Bao. 2021;37:2116-2126. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 2. | Lu Z, Zhang X, Dai J, Wang Y, He W. Engineering of leucine-responsive regulatory protein improves spiramycin and bitespiramycin biosynthesis. Microb Cell Fact. 2019;18:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 3. | Liang SY, Zhao TC, Zhou ZH, Ju WT, Liu Y, Tan YR, Zhu DW, Zhang ZY, Zhong LP. Anti-tumor effect of carrimycin on oral squamous cell carcinoma cells in vitro and in vivo. Transl Oncol. 2021;14:101074. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 4. | Kharkar PS, Warrier S, Gaud RS. Reverse docking: a powerful tool for drug repositioning and drug rescue. Future Med Chem. 2014;6:333-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 86] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 5. | Tian SB, Yu JC, Liu YQ, Kang WM, Ma ZQ, Ye X, Yan C. MiR-30b suppresses tumor migration and invasion by targeting EIF5A2 in gastric cancer. World J Gastroenterol. 2015;21:9337-9347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 35] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 6. | Njoku DB. Suppressive and pro-inflammatory roles for IL-4 in the pathogenesis of experimental drug-induced liver injury: a review. Expert Opin Drug Metab Toxicol. 2010;6:519-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 7. | Peiró T, Patel DF, Akthar S, Gregory LG, Pyle CJ, Harker JA, Birrell MA, Lloyd CM, Snelgrove RJ. Neutrophils drive alveolar macrophage IL-1β release during respiratory viral infection. Thorax. 2018;73:546-556. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 8. | Razeghian E, Suksatan W, Sulaiman Rahman H, Bokov DO, Abdelbasset WK, Hassanzadeh A, Marofi F, Yazdanifar M, Jarahian M. Harnessing TRAIL-Induced Apoptosis Pathway for Cancer Immunotherapy and Associated Challenges. Front Immunol. 2021;12:699746. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 9. | Smith JA. Neutrophils, host defense, and inflammation: a double-edged sword. J Leukoc Biol. 1994;56:672-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 592] [Cited by in RCA: 595] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 10. | Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol. 2006;6:173-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1905] [Cited by in RCA: 2071] [Article Influence: 109.0] [Reference Citation Analysis (0)] |
| 11. | John B, Hunter CA. Immunology. Neutrophil soldiers or Trojan Horses? Science. 2008;321:917-918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 12. | Aratani Y. Myeloperoxidase: Its role for host defense, inflammation, and neutrophil function. Arch Biochem Biophys. 2018;640:47-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 647] [Article Influence: 92.4] [Reference Citation Analysis (0)] |
| 13. | Witko-Sarsat V, Rieu P, Descamps-Latscha B, Lesavre P, Halbwachs-Mecarelli L. Neutrophils: molecules, functions and pathophysiological aspects. Lab Invest. 2000;80:617-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 743] [Cited by in RCA: 746] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 14. | Shapiro SD. Neutrophil elastase: path clearer, pathogen killer, or just pathologic? Am J Respir Cell Mol Biol. 2002;26:266-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 78] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 15. | Noell CW, Robins RK. The antitumor activity of 2-amino-6-alkylthio-9-(beta-dribofuranosylpurines and related derivatives of 2-amino-6-purinethiol (thioguanine). J Med Pharm Chem. 1962;91:1074-1085. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 16. | Seley KL, Januszczyk P, Hagos A, Zhang L, Dransfield DT. Synthesis and antitumor activity of thieno-separated tricyclic purines. J Med Chem. 2000;43:4877-4883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 43] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 17. | Gandhi V, Plunkett W. Clofarabine and nelarabine: two new purine nucleoside analogs. Curr Opin Oncol. 2006;18:584-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 18. | Raggiaschi R, Gotta S, Terstappen GC. Phosphoproteome analysis. Biosci Rep. 2005;25:33-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 48] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 19. | Yang F, Stenoien DL, Strittmatter EF, Wang J, Ding L, Lipton MS, Monroe ME, Nicora CD, Gristenko MA, Tang K, Fang R, Adkins JN, Camp DG 2nd, Chen DJ, Smith RD. Phosphoproteome profiling of human skin fibroblast cells in response to low- and high-dose irradiation. J Proteome Res. 2006;5:1252-1260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 88] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 20. | Yan Y, Xu H, Zhang L, Zhou X, Qian X, Zhou J, Huang Y, Ge W, Wang W. RRAD suppresses the Warburg effect by downregulating ACTG1 in hepatocellular carcinoma. Onco Targets Ther. 2019;12:1691-1703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 21. | He L, Yang X, Cao X, Liu F, Quan M, Cao J. Casticin induces growth suppression and cell cycle arrest through activation of FOXO3a in hepatocellular carcinoma. Oncol Rep. 2013;29:103-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 22. | Tan M, Cheng Y, Zhong X, Yang D, Jiang S, Ye Y, Ding M, Guan G, Zhao X. LNK promotes granulosa cell apoptosis in PCOS via negatively regulating insulin-stimulated AKT-FOXO3 pathway. Aging (Albany NY). 2021;13:4617-4633. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 23. | Wang D, Chen Q, Tan Y, Liu B, Liu C. Ellagic acid inhibits human glioblastoma growth in vitro and in vivo. Oncol Rep. 2017;37:1084-1092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 24. | An T, Dong T, Zhou H, Chen Y, Zhang J, Zhang Y, Li Z, Yang X. The transcription factor Krüppel-like factor 5 promotes cell growth and metastasis via activating PI3K/AKT/Snail signaling in hepatocellular carcinoma. Biochem Biophys Res Commun. 2019;508:159-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 25. | Wang R, Chen J, Yu H, Wei Z, Ma M, Ye X, Wu W, Chen H, Fu Z. Downregulation of estrogen receptor-α36 expression attenuates metastasis of hepatocellular carcinoma cells. Environ Toxicol. 2022;37:1113-1123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 26. | You H, Meng K, Wang ZY. The ER-α36/EGFR signaling loop promotes growth of hepatocellular carcinoma cells. Steroids. 2018;134:78-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 27. | Zhou XM, Hou G, Gu DX, Wang QY, Zhao L. Peroxisome proliferator-activated receptor-γ in induced sputum is correlated with MMP-9/TIMP-1 imbalance and formation of emphysema in COPD patients. J Thorac Dis. 2017;9:3703-3710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 28. | Zhu J, Tang H, Zhang Z, Zhang Y, Qiu C, Zhang L, Huang P, Li F. Kaempferol slows intervertebral disc degeneration by modifying LPS-induced osteogenesis/adipogenesis imbalance and inflammation response in BMSCs. Int Immunopharmacol. 2017;43:236-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 29. | Grommes C, Landreth GE, Heneka MT. Antineoplastic effects of peroxisome proliferator-activated receptor gamma agonists. Lancet Oncol. 2004;5:419-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 350] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 30. | Guo FY, Yang J, Xiong SM, Zhu MQ, Gao S, Li JP. [Clinical significance of epidermal growth factor receptor and thymidylate synthase expression in primary liver cancer]. Zhonghua Gan Zang Bing Za Zhi. 2018;26:666-669. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 31. | Ellina MI, Bouris P, Aletras AJ, Theocharis AD, Kletsas D, Karamanos NK. EGFR and HER2 exert distinct roles on colon cancer cell functional properties and expression of matrix macromolecules. Biochim Biophys Acta. 2014;1840:2651-2661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 32. | Taniwaki K, Fukamachi H, Komori K, Ohtake Y, Nonaka T, Sakamoto T, Shiomi T, Okada Y, Itoh T, Itohara S, Seiki M, Yana I. Stroma-derived matrix metalloproteinase (MMP)-2 promotes membrane type 1-MMP-dependent tumor growth in mice. Cancer Res. 2007;67:4311-4319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 33. | Chandrashekar N, Selvamani A, Subramanian R, Pandi A, Thiruvengadam D. Baicalein inhibits pulmonary carcinogenesis-associated inflammation and interferes with COX-2, MMP-2 and MMP-9 expressions in-vivo. Toxicol Appl Pharmacol. 2012;261:10-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 34. | Liu HQ, Song S, Wang JH, Zhang SL. Expression of MMP-3 and TIMP-3 in gastric cancer tissue and its clinical significance. Oncol Lett. 2011;2:1319-1322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 35. | Derman E, Krauter K, Walling L, Weinberger C, Ray M, Darnell JE Jr. Transcriptional control in the production of liver-specific mRNAs. Cell. 1981;23:731-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 502] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 36. | Dai B, Geng L, Yu Y, Sui C, Xie F, Shen W, Zheng T, Yang J. Methylation patterns of estrogen receptor α promoter correlate with estrogen receptor α expression and clinicopathological factors in hepatocellular carcinoma. Exp Biol Med (Maywood). 2014;239:883-890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 37. | Hu W, Zheng S, Guo H, Dai B, Ni J, Shi Y, Bian H, Li L, Shen Y, Wu M, Tian Z, Liu G, Hossain MA, Yang H, Wang D, Zhang Q, Yu J, Birnbaumer L, Feng J, Yu D, Yang Y. PLAGL2-EGFR-HIF-1/2α Signaling Loop Promotes HCC Progression and Erlotinib Insensitivity. Hepatology. 2021;73:674-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 58] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 38. | Xie X, Lv H, Liu C, Su X, Yu Z, Song S, Bian H, Tian M, Qin C, Qi J, Zhu Q. HBeAg mediates inflammatory functions of macrophages by TLR2 contributing to hepatic fibrosis. BMC Med. 2021;19:247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |