Published online Apr 7, 2023. doi: 10.3748/wjg.v29.i13.2001
Peer-review started: December 12, 2022
First decision: January 22, 2023
Revised: February 1, 2023
Accepted: March 20, 2023
Article in press: March 20, 2023
Published online: April 7, 2023
Processing time: 116 Days and 2.9 Hours
Macrotrabecular-massive hepatocellular carcinoma (MTM-HCC) is closely related to aggressive phenotype, gene mutation, carcinogenic pathway, and immunohistochemical markers and is a strong independent predictor of early recurrence and poor prognosis. With the development of imaging technology, successful applications of contrast-enhanced magnetic resonance imaging (MRI) have been re
To establish and verify a nomogram for preoperatively identifying MTM-HCC by comparing different machine learning algorithms.
This retrospective study enrolled 232 (training set, 162; test set, 70) hepatocellular carcinoma patients from April 2018 to September 2021. A total of 3111 radiomics features were extracted from dynamic contrast-enhanced MRI, followed by dimension reduction of these features. Logistic regression (LR), K-nearest neighbour (KNN), Bayes, Tree, and support vector machine (SVM) algorithms were used to select the best radiomics signature. We used the relative standard deviation (RSD) and bootstrap methods to quantify the stability of these five algorithms. The algorithm with the lowest RSD represented the best stability, and it was used to construct the best radiomics model. Multivariable logistic analysis was used to select the useful clinical and radiological features, and different predictive models were established. Finally, the predictive performances of the different models were assessed by evaluating the area under the curve (AUC).
The RSD values based on LR, KNN, Bayes, Tree, and SVM were 3.8%, 8.6%, 4.3%, 17.7%, and 17.4%, respectively. Therefore, the LR machine learning algorithm was selected to construct the best radiomics signature, which performed well with AUCs of 0.766 and 0.739 in the training and test sets, respectively. In the multivariable analysis, age [odds ratio (OR) = 0.956, P = 0.034], alpha-fetoprotein (OR = 10.066, P < 0.001), tumour size (OR = 3.316, P = 0.002), tumour-to-liver apparent diffusion coefficient (ADC) ratio (OR = 0.156, P = 0.037), and radiomics score (OR = 2.923, P < 0.001) were independent predictors of MTM-HCC. Among the different models, the predictive performances of the clinical-radiomics model and radiological-radiomics model were significantly improved compared to those of the clinical model (AUCs: 0.888 vs 0.836, P = 0.046) and radiological model (AUCs: 0.796 vs 0.688, P = 0.012), respectively, in the training set, highlighting the improved predictive performance of radiomics. The nomogram performed best, with AUCs of 0.896 and 0.805 in the training and test sets, respectively.
The nomogram containing radiomics, age, alpha-fetoprotein, tumour size, and tumour-to-liver ADC ratio revealed excellent predictive ability in preoperatively identifying the MTM-HCC subtype.
Core Tip: Radiomics features can be used to predict the macrotrabecular-massive hepatocellular carcinoma (MTM-HCC) subtype. The logistic regression algorithm can improve the accuracy and stability of predicting MTM-HCC. Age, alpha-fetoprotein, tumour size, tumour-to-liver apparent diffusion coefficient ratio, and radiomics score were significant independent predictors of MTM-HCC. The nomogram based on radiomics, clinical and radiological features can serve as a noninvasive biomarker to preoperatively identify MTM-HCC.
- Citation: Zhang Y, He D, Liu J, Wei YG, Shi LL. Preoperative prediction of macrotrabecular-massive hepatocellular carcinoma through dynamic contrast-enhanced magnetic resonance imaging-based radiomics. World J Gastroenterol 2023; 29(13): 2001-2014
- URL: https://www.wjgnet.com/1007-9327/full/v29/i13/2001.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i13.2001
Hepatocellular carcinoma (HCC) is one of the leading causes of cancer-related death worldwide, with a 5-year recurrence rate of 70% after surgical resection[1]. The poor prognosis of patients with HCC is closely related to histopathological subtypes[2]. Recently, a newly identified histopathological subtype was named "macrotrabecular-massive HCC (MTM-HCC)" and was officially included in the new classification of HCC by the World Health Organization in 2019[3]. As the most common subtype with metastatic potential[4], MTM-HCC is closely related to gene mutation, carcinogenic pathway, and immunohistochemical markers[5] and is a strong independent predictor of early recurrence and poor prognosis[6,7].
Early diagnosis and appropriate treatment of MTM-HCC are beneficial to prevent early recurrence and improve prognosis. Current studies have shown that radiofrequency ablation is not recommended for patients with aggressive HCC, while performing resection with wide margins or anatomical hepatectomy and shorter follow-up intervals may be recommended for monitoring[8,9]. MTM-HCC shows an aggressive phenotype[10]. Therefore, an accurate preoperative diagnosis of MTM-HCC can provide the best individualized treatment plan.
With the development of imaging technology, successful applications of magnetic resonance imaging (MRI) have been reported in identifying the MTM-HCC subtype[11]. Several studies[11-14] have reported that MTM-HCC has characteristic MRI features, such as intratumor substantial necrosis and intratumor fat. However, these studies mainly focused on the qualitative analysis of imaging features. Radiomics, as an objective and beneficial method for tumour evaluation, is used to convert medical images into high-throughput quantification features that greatly push the development of precision medicine[15]. A recent study has also shown that radiomics is a superior tool for predicting MTM-HCC. However, this study only enrolled 88 patients and lacked a validation set, and there was limited reproducibility of the results. In addition, a highly accurate and reliable radiomics model can be constructed by comparing different machine learning algorithms. Therefore, we hypothesize that valuable MRI-based radiomics features could be extracted and a noninvasive and comprehensive model could be constructed through machine learning to better predict MTM-HCC.
In this study, we aimed to apply MRI-based radiomics to preoperatively predict MTM-HCC using different machine learning algorithms to build the best radiomics signature and to establish and validate the nomogram by combining preoperative clinical and radiological features to improve the decision-making process in clinical practice.
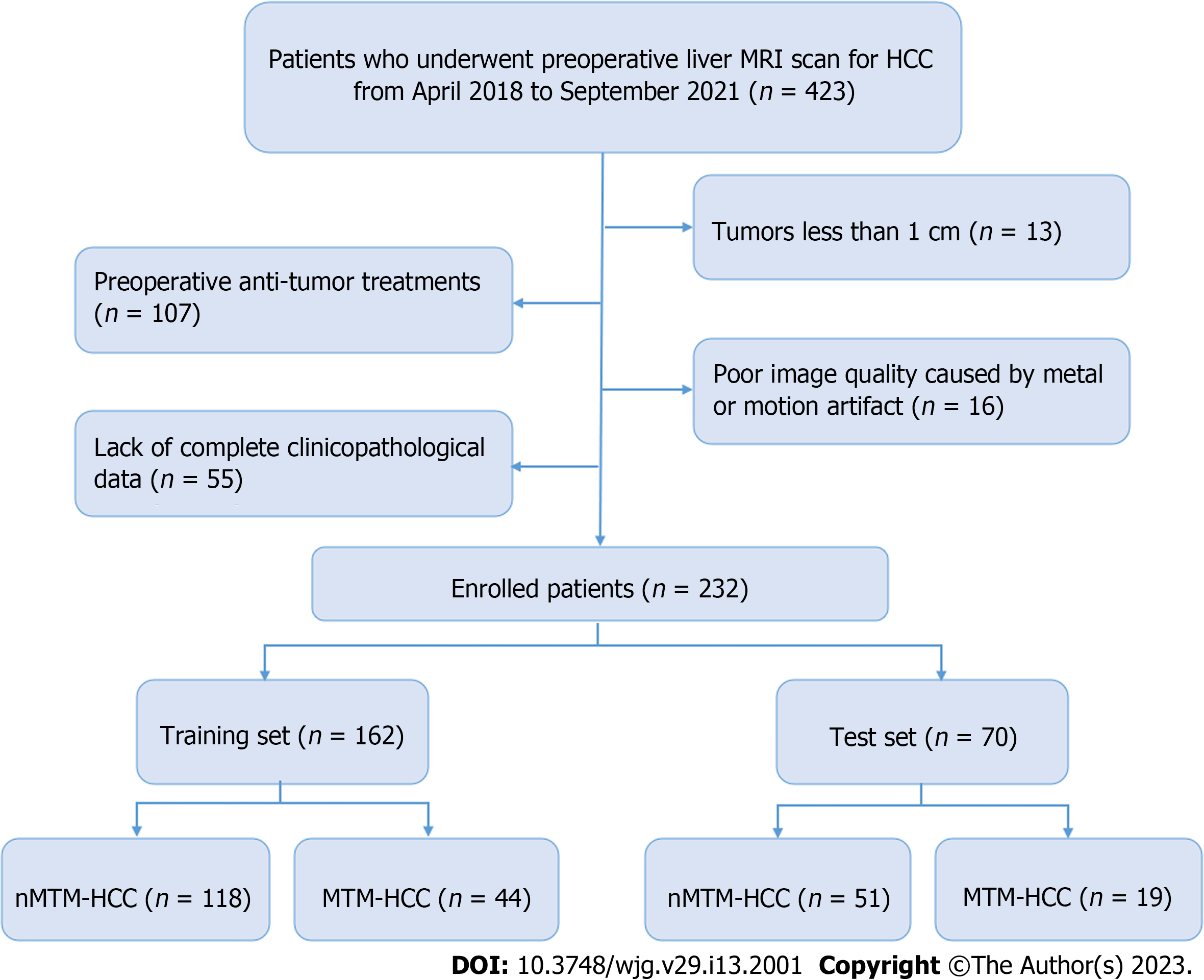
This retrospective study was approved by our institutional review board, and the requirement for written informed consent was waived. Between April 2018 and September 2021, patients with suspected HCCs who underwent preoperative liver MRI examinations were included. The inclusion criteria included the following: (1) Pathologically proven primary HCC; (2) received liver MRI examinations one month before surgery; and (3) underwent curative hepatectomy. The exclusion criteria included: (1) Tumours less than 1 cm; (2) preoperative antitumour treatments; (3) poor image quality caused by metal or motion artifacts; and (4) lack of complete clinicopathological data. Finally, 232 patients were enrolled and divided into training (n = 162) and test (n = 70) sets at a ratio of 7:3. The patient recruitment process is shown in Figure 1.
All MRI examinations were performed on a 3.0 T MRI scanner (Discovery MR 750, GE Healthcare, Waukesha, WI, United States). Our liver MRI protocol included the following sequence: (1) Axial T2-weighted imaging with fat suppression: Repetition time (TR)/echo time (TE), 13000/75 msec; field of view (FOV), 360 mm × 360 mm; matrix, 320 × 320; slice thickness, 5 mm; (2) dual-echo (in-phase and opposed-phase) T1-weighted imaging: TR/TE, 3.7/1.7 msec; FOV, 360 mm × 288 mm; matrix, 260 × 224; slice thickness, 5 mm; (3) diffusion-weighted imaging (DWI) and apparent diffusion coefficient (ADC): TR/TE, 8000/50 msec; FOV, 360 mm × 288 mm; matrix, 128×96; and slice thickness, 5 mm. DWI was obtained using respiratory triggering, a single-shot echo-planar imaging pulse sequence with b values of 0 and 800 s/mm2; and (4) The dynamic contrast-enhanced (DCE) three-dimensional fast-spoiled gradient-recalled echo sequences were as follows: TR/TE, 3.7/1.7 msec; FOV, 360 mm × 288 mm; matrix, 260 × 224; and slice thickness, 5 mm. The DCE sequences were acquired at 15-20 s (arterial phase, AP), 50-55 s (portal phase, PP) and 85-90 s (delayed phase, DP) after contrast-agent injection.
Clinical data included age, sex, hepatitis B surface antigen (HBsAg) status (positive or negative), serum alpha-fetoprotein (AFP) level (recorded as > 400 µg/L or ≤ 400 µg/L), carcinoembryonic antigen (CEA), carbohydrate antigen 19-9 (CA19-9), platelet count (PLT), total bilirubin (TBIL), direct bilirubin (DBIL), indirect bilirubin (IBIL), albumin, aspartate transaminase (AST), alanine transferase (ALT) and AST/ALT.
Radiological data of all patients were retrospectively analysed by two radiologists who were unaware of the clinicopathological findings and included liver cirrhosis, tumour size, tumour shape, intratumor fat, intratumor necrosis, intratumor haemorrhage, enhancing capsule and tumour-to-liver ADC ratio. For qualitative data, an agreement was reached by negotiation when there was disagreement between the two radiologists. For quantitative data, the measurements from these two radiologists were averaged. Detailed descriptions of the radiological features are shown in Supplementary Table 1.
All enrolled patients underwent hepatectomy, and pathological evaluations were performed in consensus by two experienced pathologists. MTM-HCC was defined as a predominant macrotrabecular architectural pattern (> 6 cells thick) involving more than 50% of the entire tumour[7]. nMTM-HCC was defined as non-MTM-HCC. The following pathological features were recorded: tumour differentiation according to the Edmondson-Steiner grade (I-II or III-IV), presence of microvascular invasion, satellite nodules, and biliary invasion.
Image segmentation: Image segmentation was performed with an open-source software named ITK-SNAP. The volume of interest (VOI) was defined by manually outlining the whole tumour border in AP, PP, and DP sequences slice-by-slice for each patient by a radiologist with five years of experience in liver MRI. Then, the segmentation results were validated by another radiologist with more than 10 years of experience in liver MRI using the intraclass correlation coefficient (ICC) on a cohort of 30 randomly selected patients. The image segmentation process is shown in Supplementary Figure 1.
Radiomics feature extraction: All segmented VOIs were loaded into the Pyradiomics-based PHIgo software (GE Healthcare, V1.2.0, China) for feature extraction, which complies with the image biomarker standardization initiative (IBSI)[16]. Before that, all images were subjected to standardized preprocessing, including resampling the images at the same resolution (1 mm × 1 mm × 1 mm) and classifying the grayscale into 1-10 Levels. Then, 1037 radiomics features were extracted from each VOI of each sequence, including the first-order features (first order: 18 features), shape-based features (shape: 14 features), gray-level run length matrix features (GLRLM: 16 features), gray-level size zone matrix (GLSZM: 16 features), neighbourhood gray tone difference matrix (NGTDM: 5 features), gray-level co-occurrence matrix features (GLCM: 24 features), gray-level dependence matrix (GLDM: 14 features), LoG transform features (LoG: 186 features), and wavelet transform features (wavelet: 744 features). The DCE sequences, including AP, PP, and DP, were scanned, affording 3111 radiomics features per patient.
Radiomics feature selection: The ICCs of the measurements from the two radiologists were applied to evaluate the interobserver reliability and reproducibility. The features with ICCs > 0.80 were considered robust features. Then, dimension reduction was performed using analysis of variance, correlation analysis, and gradient boosting decision tree (GBDT) to reduce data redundancy and to further select the most significant radiomics features based on DCE sequences.
Radiomics signature development: Five machine learning algorithms, including logistic regression (LR), K-nearest neighbour (KNN), Bayes, Tree, and support vector machine (SVM), were used to construct radiomics signatures based on the retained significant features. Then, we used the relative standard deviation (RSD) and bootstrap methods to quantify the stability of these five algorithms. The algorithm with the lowest RSD represented the best stability, which was used to construct the best radiomics model. Finally, the radiomics score (rad-score) was calculated via a linear combination of the remaining features that were weighted by their respective coefficients to quantify the discriminability of the radiomics model. Details of the RSD can be found in the Supplementary materials.
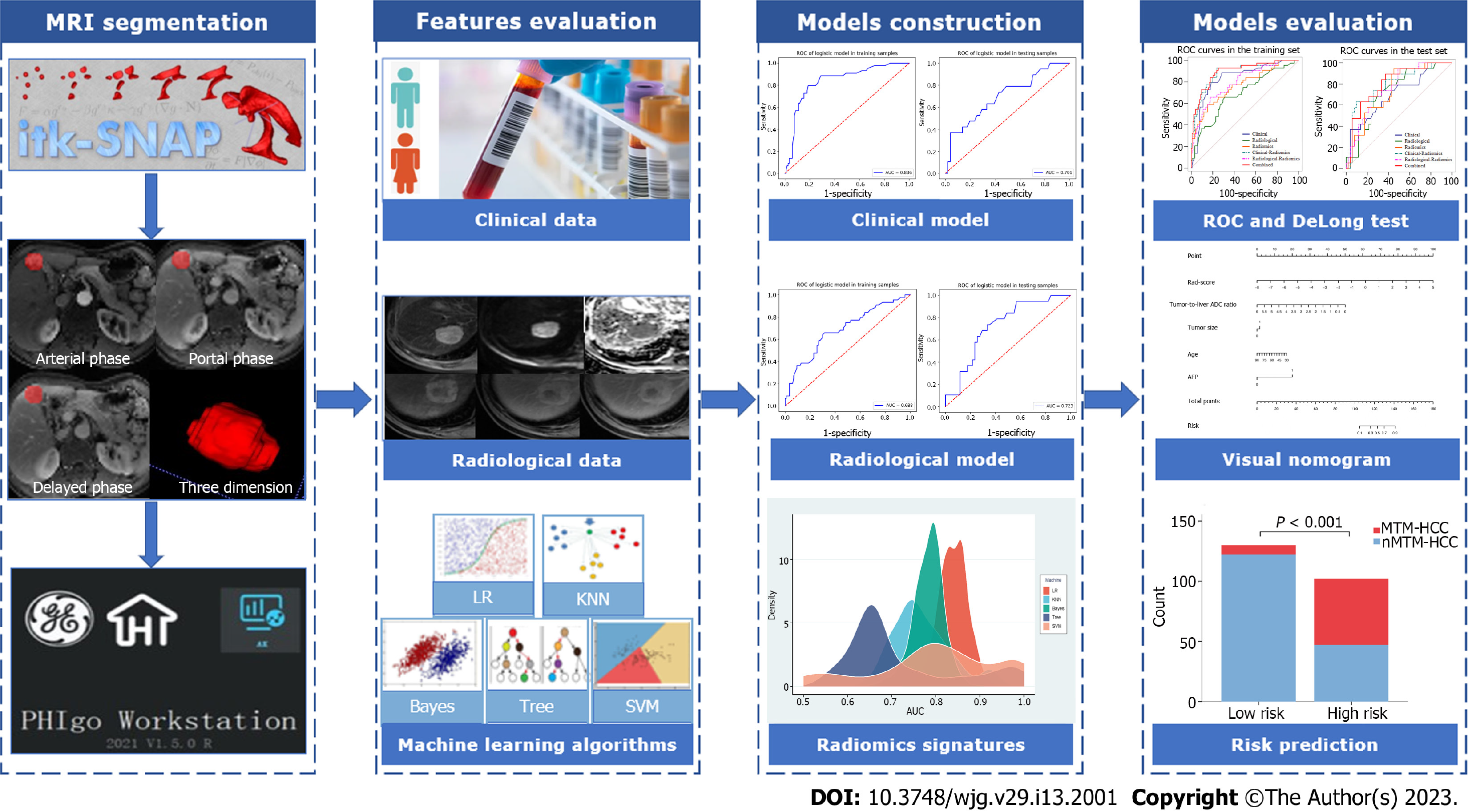
Univariate logistic analysis was performed on variables, including the abovementioned clinical features, radiological features, and rad-score. Variables with P < 0.05 were included in the multivariate logistic analysis to determine the potential independent predictors of MTM-HCC, based on which combined model was built. In addition, the clinical model was constructed based on the final selected clinical features, and the radiological model was constructed based on the final selected radiological features. To verify the improvement in the performance of the model after including radiomics, we integrated the selected independent predictors to construct different fusion models. The area under the receiver operating characteristic (ROC) curve (AUC) and the DeLong test were used to evaluate the performance of the different models for predicting MTM-HCC. The Hosmer–Lemeshow test was used to assess the goodness-of-fit of the combined model. A nomogram based on the combined model was established for easy use to generate a probability of MTM-HCC. Then, the patients were classified into high-risk and low-risk groups according to the nomogram. The flowchart of the model construction and evaluation is shown in Figure 2.
Statistical analyses were performed with SPSS software (version 24.0, Chicago, IL, United States) and R software (version 3.4.1, Vienna, Austria). Continuous variables were expressed as the mean ± SD or median (interquartile range). Categorical variables are presented as numbers (percentages). Continuous variables were analysed using a two-sample t test or Mann-Whitney U test if not normally distributed. The chi-squared test or Fisher’s exact test was used for categorical variables. Statistical significance was set at a two-sided P < 0.05.
Clinical, pathological, and radiological features of the HCC patients are shown in Tables 1 and 2. In terms of the MTM-HCC and nMTM-HCC groups, age, PLT, tumour size, intratumor haemorrhage, and tumour-to-liver ADC ratio were significantly different in both the training and test sets (P < 0.05). In addition, AFP > 400 µg/L (P < 0.001), CA19-9 (P = 0.021), AST (P = 0.004), ALT (P = 0.023), tumour shape (P = 0.024), and intratumor necrosis (P = 0.005) also differed significantly between the two groups in the training set. The Edmondson-Steiner grade (P = 0.048) differed significantly between the two groups in the test set. There were no significant differences in any of the features between the training and test sets (P > 0.05).
| Characteristics | Training set (n = 162) | Test set (n = 70) | P value | ||||
| nMTM-HCC (n = 118) | MTM-HCC (n = 44) | P value | nMTM-HCC (n = 51) | MTM-HCC (n = 19) | P value | ||
| Age, mean ± SD | 60.6 ± 10.9 | 52.4 ± 9.9 | < 0.001 | 60.7 ± 10.4 | 54.7 ± 9.7 | 0.034 | 0.647 |
| Sex (men, %) | 102 (86.4) | 42 (95.5) | 0.179 | 39 (76.5) | 18 (94.7) | 0.161 | 0.125 |
| HBsAg (positive, %) | 91 (77.1) | 33 (75.0) | 0.777 | 38 (74.5) | 14 (73.7) | 0.944 | 0.712 |
| AFP > 400 µg/L, n (%) | 18 (15.3) | 31 (70.5) | < 0.001 | 12 (23.5) | 9 (47.4) | 0.053 | 0.970 |
| CEA (ug/L) | 2.6 (2.1) | 2.7 (1.9) | 0.719 | 2.2 (2.0) | 2.2 (1.2) | 0.543 | 0.332 |
| CA19-9 (U/mL) | 15.0 (19.3) | 28.0 (25.4) | 0.021 | 16.7 (17.1) | 16.0 (34.4) | 0.687 | 0.770 |
| PLT (× 109/L) | 135.0 (83.0) | 174.0 (88.5) | 0.027 | 135.0 (57.0) | 175.0 (127.0) | 0.001 | 0.970 |
| TBIL (µmol/L) | 16.6 (10.8) | 20.7 (9.8) | 0.067 | 15.7 (10.3) | 14.4 (7.6) | 0.916 | 0.509 |
| DBIL (µmol/L) | 3.8 (2.6) | 4.8 (2.9) | 0.060 | 3.2 (2.1) | 3.3 (2.7) | 0.620 | 0.419 |
| IBIL (µmol/L) | 12.5 (7.0) | 15.0 (7.4) | 0.136 | 12.2 (7.8) | 10.9 (4.5) | 0.712 | 0.061 |
| Albumin, mean ± SD | 37.8 ± 5.0 | 37.0 ± 5.8 | 0.387 | 37.8 ± 5.0 | 38.3 ± 5.1 | 0.715 | 0.647 |
| AST (U/L) | 32.0 (23.5) | 43.0 (68.8) | 0.004 | 33.0 (29.0) | 43.0 (61.0) | 0.135 | 0.980 |
| ALT (U/L) | 31.5 (34.0) | 46.0 (64.0) | 0.023 | 28.0 (35.0) | 57.0 (96.0) | 0.074 | 0.748 |
| AST/ALT | 1.1 (0.6) | 1.1 (0.5) | 0.702 | 1.3 (0.5) | 1.1 (0.7) | 0.207 | 0.842 |
| Edmondson-Steiner grade (III-IV, %) | 38 (32.2) | 18 (40.9) | 0.300 | 14 (27.5) | 10 (52.6) | 0.048 | 0.967 |
| Microvascular invasion, n (%) | 56 (47.5) | 20 (45.5) | 0.820 | 18 (35.3) | 10 (52.6) | 0.188 | 0.331 |
| Satellite nodules, n (%) | 9 (7.6) | 5 (11.4) | 0.452 | 1 (2.0) | 1 (5.3) | 0.461 | 0.110 |
| Biliary invasion, n (%) | 4 (3.4) | 1 (2.3) | 0.715 | 2 (3.9) | 1 (5.3) | 0.805 | 0.646 |
| Radiological features | Training set (n = 162) | Test set (n = 70) | P value | ||||
| nMTM-HCC (n = 118) | MTM-HCC (n = 44) | P value | nMTM-HCC (n = 51) | MTM-HCC (n = 19) | P value | ||
| Liver cirrhosis (positive, %) | 69 (58.5) | 29 (65.9) | 0.389 | 34 (66.7) | 15 (78.9) | 0.319 | 0.168 |
| Tumour size > 5 cm, n (%) | 29 (24.6) | 22 (50.0) | 0.002 | 12 (23.5) | 10 (52.6) | 0.020 | 0.994 |
| Tumour shape (irregular, %) | 25 (21.2) | 17 (38.6) | 0.024 | 17 (33.3) | 7 (36.8) | 0.783 | 0.195 |
| Intratumor fat, n (%) | 24 (20.3) | 10 (22.7) | 0.740 | 11 (21.6) | 1 (5.3) | 0.210 | 0.500 |
| Intratumor necrosis, n (%) | 34 (28.8) | 23 (52.3) | 0.005 | 13 (25.5) | 8 (42.1) | 0.177 | 0.443 |
| Intratumor hemorrhage, n (%) | 24 (20.3) | 17 (38.6) | 0.017 | 7 (13.7) | 8 (42.1) | 0.025 | 0.526 |
| Enhancing capsule, n (%) | 85 (72.0) | 35 (79.5) | 0.332 | 39 (76.5) | 15 (78.9) | 0.826 | 0.620 |
| Tumour-to-liver ADC ratio | 0.9 (0.2) | 0.8 (0.3) | 0.018 | 0.9 (0.1) | 0.8 (0.2) | 0.045 | 0.312 |
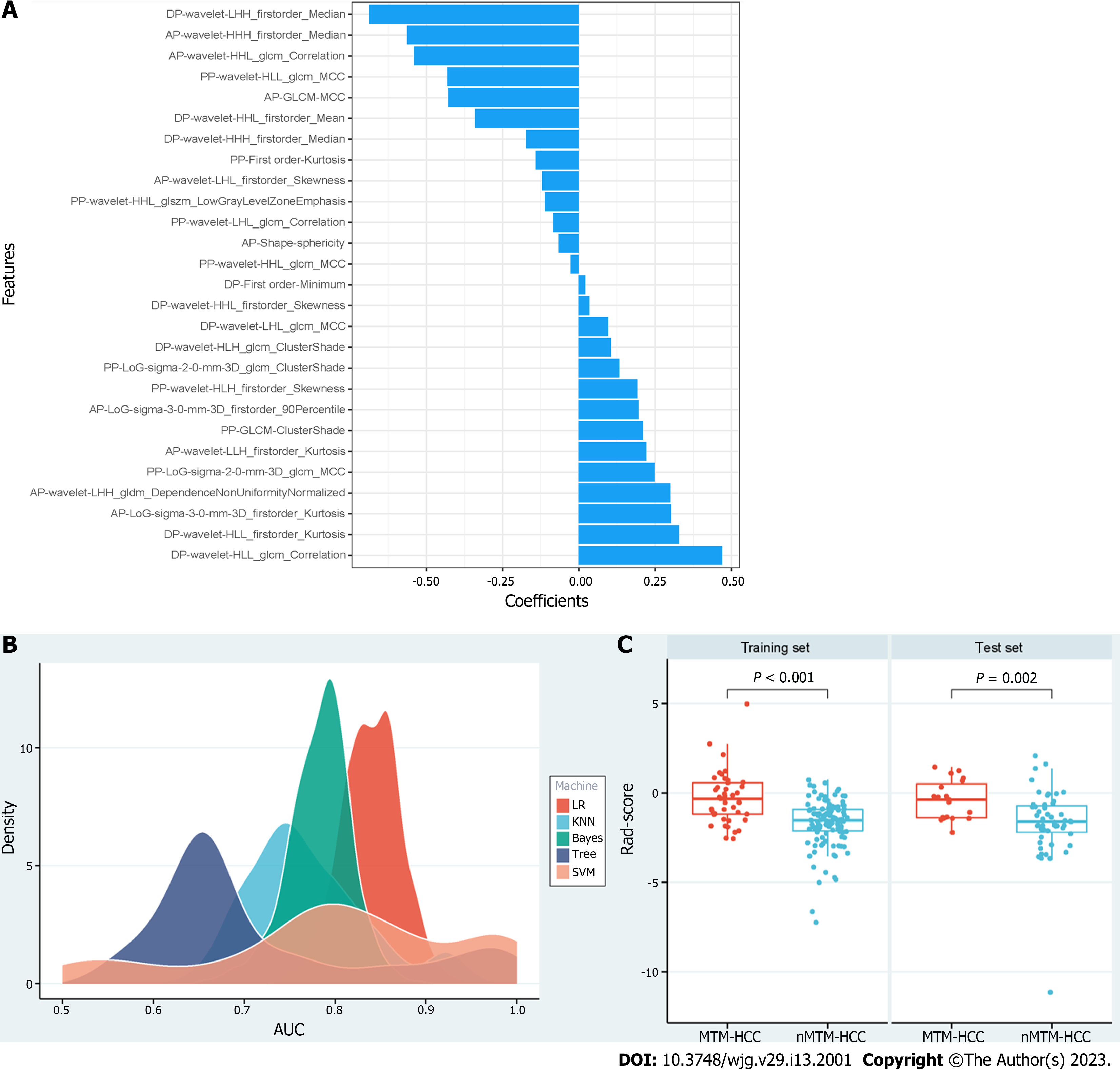
A total of 3111 radiomics features were extracted for each patient based on the DCE sequences. Then, 2417 features with ICCs > 0.80 were obtained as robust features. After feature selection by analysis of variance and correlation analysis, 75 features were selected. Following GBDT, 27 features were ultimately retained, as shown in Figure 3A. The RSD values based on LR, KNN, Bayes, Tree, and SVM were 3.8%, 8.6%, 4.3%, 17.7%, and 17.4%, respectively. Therefore, the LR machine learning algorithm was chosen to construct the best radiomics signature (Figure 3B). The corresponding rad-score was calculated and was significantly different between the MTM-HCC and nMTM-HCC groups in the training set (P < 0.001) and test set (P = 0.002), as shown in Figure 3C. Details of the retained radiomics features and rad-score are shown in Supplementary Table 2 and Supplementary Figure 2.
The multivariate logistic analysis showed that age (OR = 0.956, P = 0.034), AFP (OR = 10.066, P < 0.001), tumour size (OR = 3.316, P = 0.002), tumour-to-liver ADC ratio (OR=0.156, P = 0.037), and rad-score (OR = 2.923, P < 0.001) were independent predictors of MTM-HCC (Table 3). A clinical model was constructed based on age and AFP. A radiological model was constructed based on tumour size and the tumour-to-liver ADC ratio. To verify the improvement in the performance of the model after including radiomics, we constructed different fusion models, including a clinical-radiomics model and a radiological-radiomics model. Finally, we integrated all five of the selected independent predictors to build the combined model. The corresponding model score of the combined model was calculated, as shown in Supplementary Figure 3.
| Variables | Univariate logistic regression | Multivariate logistic regression | ||
| OR (95%CI) | P value | OR (95%CI) | P value | |
| Clinical features | ||||
| Age | 0.930 (0.896, 0.964) | < 0.001 | 0.956 (0.918, 0.997) | 0.034 |
| AFP > 400 µg/L | 13.248 (5.839, 30.058) | < 0.001 | 10.066 (4.304, 23.541) | < 0.001 |
| PLT | 1.006 (1.002, 1.011) | 0.009 | NA | NA |
| Radiological features | ||||
| Tumour size > 5 cm | 3.069 (1.487, 6.333) | 0.002 | 3.316 (1.579, 6.962) | 0.002 |
| Tumour shape | 2.342 (1.106, 4.961) | 0.026 | NA | NA |
| Intratumor necrosis | 2.706 (1.326, 5.521) | 0.006 | NA | NA |
| Intratumor hemorrhage | 2.466 (1.160, 5.244) | 0.019 | NA | NA |
| Tumour-to-liver ADC ratio | 0.183 (0.035, 0.972) | 0.046 | 0.156 (0.027, 0.894) | 0.037 |
| Radiomics | ||||
| Rad-score | 2.718 (1.809, 4.084) | < 0.001 | 2.923 (1.740, 4.911) | < 0.001 |
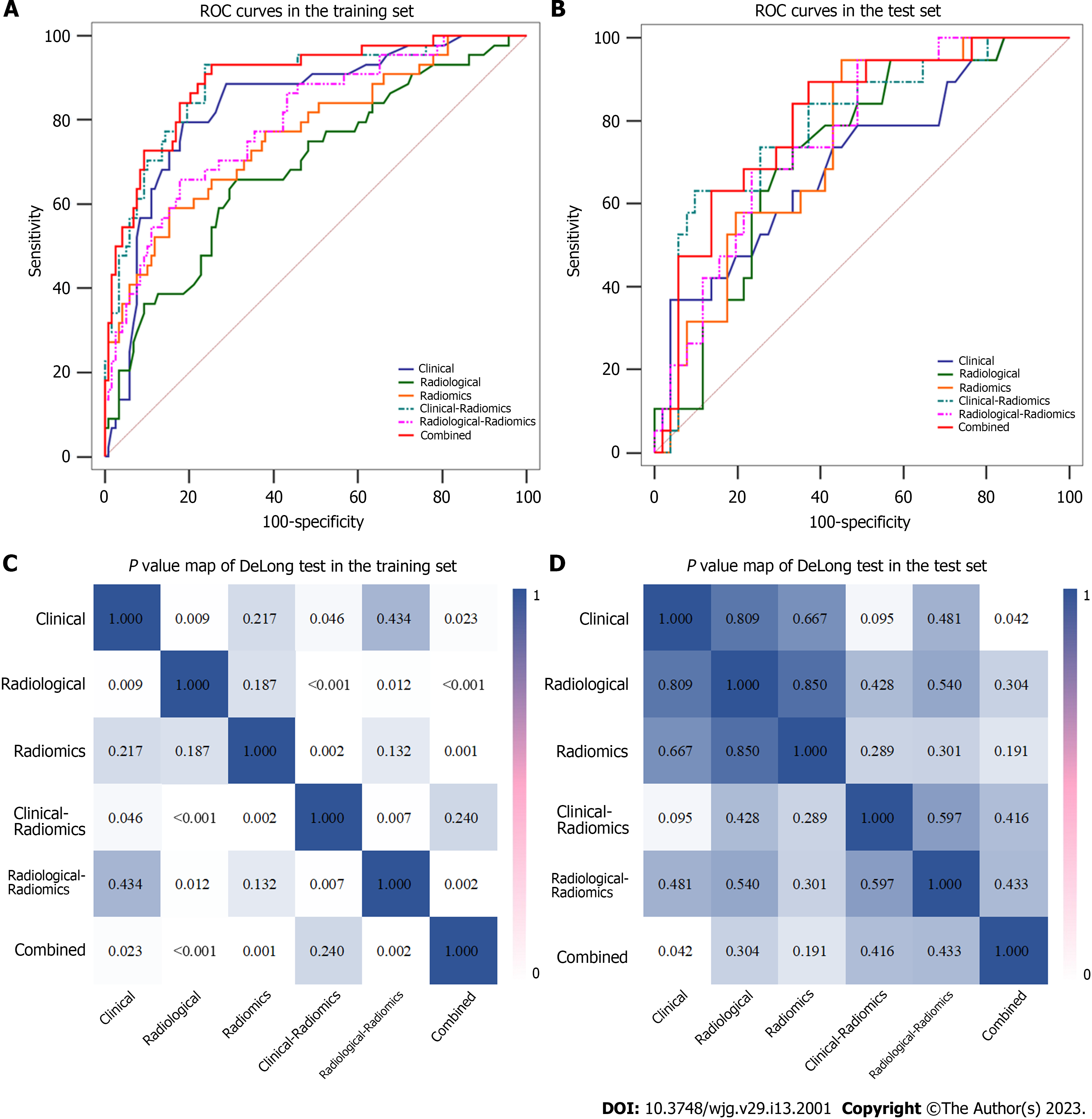
Among the six different models, the combined model performed best, with AUCs of 0.896 and 0.805 in the training and test sets, respectively (Table 4). The DeLong test showed that the predictive performances of the clinical-radiomics model and radiological-radiomics model were significantly improved compared to those of the clinical model (AUCs: 0.888 vs 0.836, P = 0.046) and radiological model (AUCs: 0.796 vs 0.688, P = 0.012), respectively, in the training set, highlighting the improved predictive performance of radiomics. In addition, the combined model performed better than the radiological model (P < 0.001), radiomics model (P = 0.001), and radiological-radiomics model (P = 0.002) in the training set, and the combined model was significantly different from the clinical model in both the training set (P = 0.023) and the test set (P = 0.042) (Table 4 and Figure 4).
| Models | Training set | Test set | ||||||
| AUC (95%CI) | Accuracy | Sensitivity | Specificity | AUC (95%CI) | Accuracy | Sensitivity | Specificity | |
| Clinical | 0.836 (0.773-0.888) | 0.802 | 0.750 | 0.822 | 0.701 (0.575-0.814) | 0.671 | 0.579 | 0.706 |
| Radiological | 0.688 (0.604-0.769) | 0.685 | 0.636 | 0.703 | 0.723 (0.610-0.829) | 0.700 | 0.632 | 0.725 |
| Radiomics | 0.766 (0.692-0.836) | 0.772 | 0.568 | 0.847 | 0.739 (0.634-0.837) | 0.743 | 0.579 | 0.804 |
| Clinical-radiomics | 0.888 (0.835-0.934) | 0.802 | 0.909 | 0.763 | 0.793 (0.682-0.893) | 0.686 | 0.737 | 0.667 |
| Radiological-radiomics | 0.796 (0.725-0.858) | 0.772 | 0.636 | 0.822 | 0.764 (0.661-0.859) | 0.729 | 0.632 | 0.765 |
| Combined | 0.896 (0.847-0.939) | 0.796 | 0.932 | 0.746 | 0.805 (0.704, 0.895) | 0.700 | 0.895 | 0.628 |
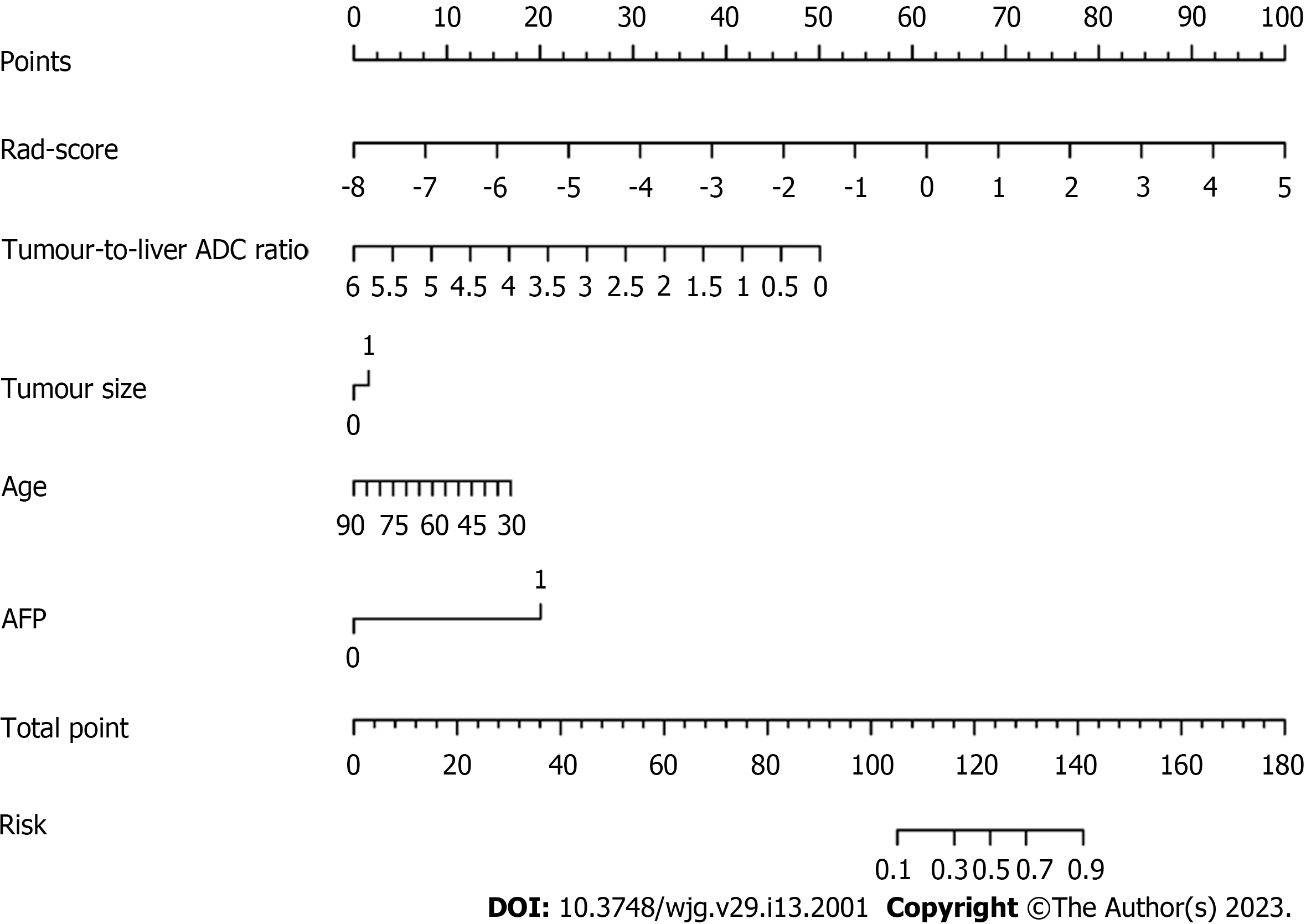
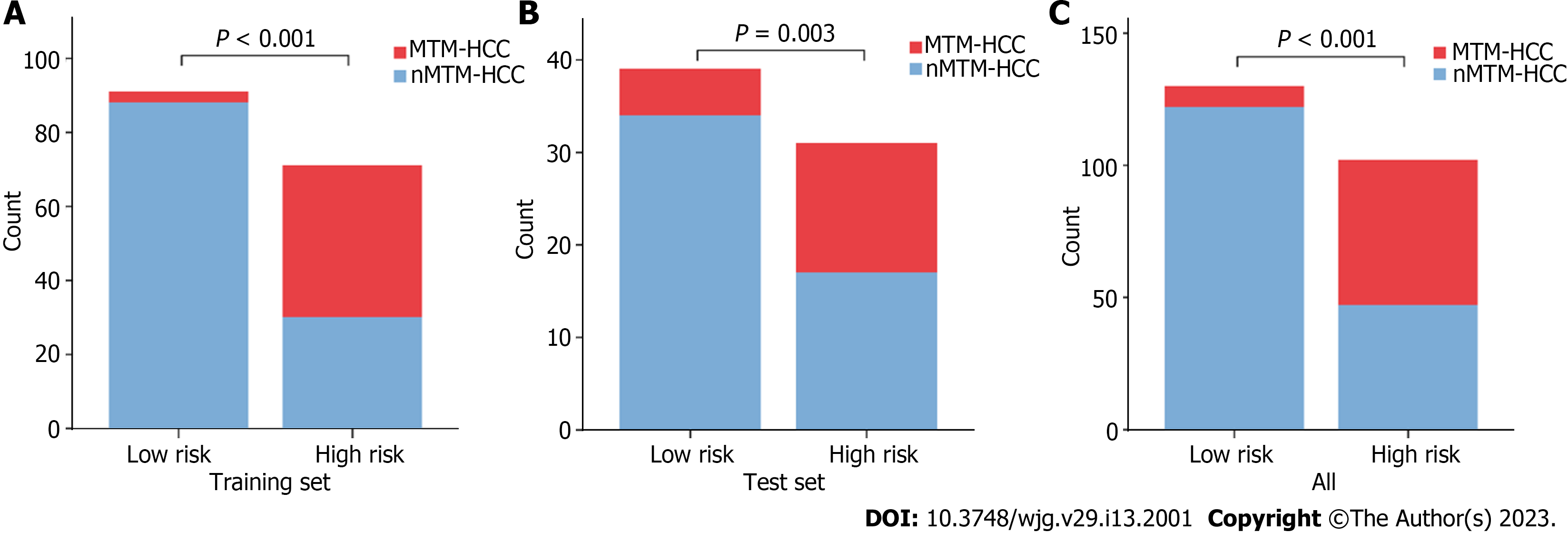
Based on the combined model, we developed an intuitive, simple-to-use nomogram for individual risk prediction of MTM-HCC (Figure 5). The Hosmer–Lemeshow test exhibited good calibration of the nomogram in the training set (P = 0.995) and test set (P = 0.466). According to the optimal cut-off value of -1.663, the patients were stratified into low-risk and high-risk groups. As shown in Figure 6, there were significant differences in the number of patients who were predicted to have MTM-HCC between the low-risk and high-risk groups in the training set (P < 0.001), test set (P = 0.003), and all study populations (P < 0.001), indicating the clinical applicability of the nomogram.
In this study, we comprehensively evaluated clinical and radiological features and found that age, AFP, tumour size, and tumour-to-liver ADC ratio were significant independent predictors of MTM-HCC. By comparing five different machine learning algorithms (LR, KNN, Bayes, Tree, and SVM), we finally selected the LR algorithm with the best stability to construct the radiomics signature. Finally, by further comparing the predictive performance of the different models, the optimal combined model was selected, and a visual nomogram was constructed, which provided a reliable theoretical basis for the development of a simple, easy-to-use, and accurate assessment tool and indicated that it had great potential in the field of preoperative noninvasive prediction of MTM-HCC.
Although some previous studies have focused on the clinical and radiological features of MTM-HCC, there is no consensus on the best biomarker for predicting MTM-HCC. Our study showed that MTM-HCC patients presented with large tumour sizes and high AFP loads, which were consistent with previous findings[17-20]. Interestingly, this study found that the age of patients with MTM-HCC was lower than that of the patients with nMTM-HCC. This may be because the activation of angiogenesis is the reason for the unique invasive biological characteristics of such tumours. Overexpression of angiopoietin 2 and vascular endothelial growth Factor A can cooperatively promote new angiogenesis in MTM-HCC, and these factors are indicators of poor prognosis in solid tumours[6,21]. This also confirmed that the aggressiveness of MTM-HCC may be related to molecular factors. Various studies have explored the relationship between PLT and MTM-HCC, but no consensus has been reached[22]. Our study showed that PLT was not an independent predictor. The multivariate logistic analysis showed that a low tumour-to-liver ADC ratio was an independent predictor of MTM-HCC, which was consistent with the results of Chen et al[12]. This is because the cellular structure is increased, the arterial supply is reduced, and diffusion is more restricted in more aggressive HCC[5]. In addition, previous studies[11,20] have suggested that intratumor necrosis can be used as an independent predictor of MTM-HCC. However, in our study, although the univariate logistic analysis showed that intratumor necrosis was more common and statistically significant in MTM-HCC, it was not an independent predictor, which was also consistent with the results of Zhu et al[14]. This may be because intratumor necrosis is not a commonly reported finding of aggressiveness or poor prognosis in HCC and lacks high specificity. This finding was more common in non-HCC malignancies, such as intrahepatic cholangiocarcinoma or metastasis, and therefore was considered a feature of non-HCC malignancies[23].
At present, there is no research on the value of different machine learning algorithms in predicting MTM-HCC. In this study, we compared the performance of radiomics signatures constructed by five machine learning algorithms, and the results showed that the radiomics signature constructed by the LR algorithm had the best stability, with an RSD of 3.8%. The radiomics model based on the LR algorithm performed well in predicting MTM-HCC, with AUCs of 0.766 and 0.739 and specificities of 0.847 and 0.804 in the training and test sets, respectively. Therefore, radiomics is very useful for timely capturing and reflecting the underlying histopathological features. In addition, 27 features were finally retained in this study, of which 22 (81.5%) features (4 LoG and 18 wavelet features) were extracted from the original and derived images[24]. For example, this study confirmed that "cluster shade", "correlation" and "MCC" were the most meaningful features among GLCM, showing differences in the regional signal intensity distribution and linear dependency. Among the first-order features, “Mean” and “Median” describe the mean and median grey intensity of the voxel intensity within the tumour region, respectively, and the differences in the grey intensity distribution are represented by “Kurtosis” and “Skewness”. “GISZM_LowGrayLevelZoneEmphasis” and “GLDM_DependenceNonUniformityNormalized” represent the heterogeneity of the tumour. These results are consistent with some radiomics features extracted from previous studies on the pathological and survival prediction of HCC[25-27], indicating that LoG and wavelet features can represent signal intensity distribution or grey distribution in tissues, which can better reflect the biological characteristics and heterogeneity of tumours.
Our study found that the clinical model based on age and AFP (AUC, 0.836; sensitivity, 0.750; specificity, 0.822) had a significantly higher diagnostic efficiency than the clinical model based on AST, PT, and AFP proposed by Shan et al[28] (AUC, 0.723; sensitivity, 0.711; specificity, 0.607) in predicting MTM-HCC. The limitation of our clinical model was that it had poor performance in the test set, with an AUC of 0.701, a sensitivity of 0.579, and a specificity of 0.706. However, when the radiomics signature was introduced, the predictive performances of the clinical-radiomics model and radiological-radiomics model were improved compared to the clinical model (AUCs: 0.888 vs 0.836 in the training set, 0.793 vs 0.701 in the test set) and radiological model (AUCs: 0.796 vs 0.688, 0.764 vs 0.723 in the test set), respectively, highlighting the improved predictive performance of radiomics. This is due to the unprecedented opportunities for extracting potential quantitative information from images provided by advanced radiomics analysis. Traditional imaging diagnosis usually relies on morphological changes observed by the naked eye. However, it often takes a long time for the appearance of observable morphological image changes of tumours caused by pathological changes. This also indicates that it is valuable for clinical applications to extract the quantitative radiomics features behind the images.
Since the diagnosis of HCC does not require biopsy, preoperative noninvasive identification of the MTM-HCC subtype is critical for treatment and prognosis. There was only one previous study on radiomics in the prediction of MTM-HCC, and the nomogram established based on radiomics and intratumor fat showed satisfactory prediction performance, with an AUC of 0.785[14]. However, that small sample study (n = 88) lacked a test set, and the reproducibility of the results was limited. It is worth mentioning that our nomogram performed best in the preoperative noninvasive prediction of MTM-HCC, with AUCs of 0.896 and 0.805 in the training and test sets, respectively. This study achieves the first step towards the noninvasive evaluation of MTM-HCC using radiomics and clinical and radiological features in clinical practice and may guide the selection of patients for whom targeted therapies would be effective. This again suggested the application value of radiomics in predicting MTM-HCC. Integrating multidimensional features is of great significance for building a powerful prediction model.
Some limitations should be noted. First, this retrospective study only included surgically resected lesions, which may cause selection bias. Therefore, the results of this study may not be representative of the entire clinical spectrum. Second, all patients were recruited from a single center, which may limit the external validation. Therefore, further studies using larger, multicenter samples are needed to verify our findings.
In this study, we found that age, AFP, tumour size, and tumour-to-liver ADC ratio were significant independent predictors of MTM-HCC. We compared and selected the optimal LR machine learning algorithm to construct the radiomics signature. The nomogram showed excellent predictive ability in preoperatively identifying MTM-HCC and showed great potential in clinical application, which was helpful to guide individualized treatment and improve the long-term survival outcomes of HCC patients.
Macrotrabecular-massive hepatocellular carcinoma (MTM-HCC) shows an aggressive phenotype. Early diagnosis of MTM-HCC is beneficial to prevent early recurrence and improve prognosis. Radiomics can convert medical images into high-throughput quantification features, which greatly push the development of precision medicine.
Currently, magnetic resonance imaging (MRI) features have been successfully applied to identify MTM-HCC but have mainly focused on the qualitative analysis of imaging features. In this study, we systematically analysed radiomics, clinical and radiological features to build a more comprehensive prediction model. We aimed to develop a noninvasive model for the preoperative prediction of MTM-HCC.
In this study, we aimed to establish and verify a nomogram based on contrast-enhanced MRI for preoperatively identifying MTM-HCC by comparing different machine learning algorithms.
A total of 232 (training set, 162; test set, 70) hepatocellular carcinoma patients were enrolled. Radiomics features were extracted from contrast-enhanced MRI, followed by dimension reduction. Logistic regression (LR), K-nearest neighbour, Bayes, Tree, and support vector machine algorithms were used to construct radiomics signatures. The relative standard deviation (RSD) was used to quantify the stability of these five algorithms. Multivariable logistic analysis was used to select the useful clinical and radiological features, and different predictive models were established. The performances of the different models were assessed using the area under the curve (AUC).
The LR algorithm with the smaller RSD (3.8%) was used to construct the best radiomics signature, which performed well with AUCs of 0.766 and 0.739 in the training and test sets, respectively. Age, alpha-fetoprotein, tumour size, tumour-to-liver apparent diffusion coefficient ratio, and radiomics score were identified as independent predictors of MTM-HCC to build the nomogram, which performed best with AUCs of 0.896 and 0.805 in the training and test sets, respectively.
The nomogram is a reliable tool for preoperatively identifying the MTM-HCC subtype.
More precise and reliable tools are urgently needed to predict the MTM-HCC subtype. Radiomics is a new method to convert medical images into high-throughput quantification features. In this study, we aimed to develop a dynamic contrast-enhanced MRI-based nomogram for preoperatively identifying the MTM-HCC subtype.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Rompianesi G, Italy; Sahin TT, Turkey S-Editor: Yan JP L-Editor: A P-Editor: Cai YX
| 1. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4235] [Cited by in RCA: 11434] [Article Influence: 3811.3] [Reference Citation Analysis (4)] |
| 2. | Nault JC, Martin Y, Caruso S, Hirsch TZ, Bayard Q, Calderaro J, Charpy C, Copie-Bergman C, Ziol M, Bioulac-Sage P, Couchy G, Blanc JF, Nahon P, Amaddeo G, Ganne-Carrie N, Morcrette G, Chiche L, Duvoux C, Faivre S, Laurent A, Imbeaud S, Rebouissou S, Llovet JM, Seror O, Letouzé E, Zucman-Rossi J. Clinical Impact of Genomic Diversity From Early to Advanced Hepatocellular Carcinoma. Hepatology. 2020;71:164-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 143] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 3. | Nagtegaal ID, Odze RD, Klimstra D, Paradis V, Rugge M, Schirmacher P, Washington KM, Carneiro F, Cree IA; WHO Classification of Tumours Editorial Board. The 2019 WHO classification of tumours of the digestive system. Histopathology. 2020;76:182-188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2554] [Cited by in RCA: 2441] [Article Influence: 488.2] [Reference Citation Analysis (3)] |
| 4. | Kumar D, Hafez O, Jain D, Zhang X. Can primary hepatocellular carcinoma histomorphology predict extrahepatic metastasis? Hum Pathol. 2021;113:39-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 5. | Yoneda N, Matsui O, Kobayashi S, Kitao A, Kozaka K, Inoue D, Yoshida K, Minami T, Koda W, Gabata T. Current status of imaging biomarkers predicting the biological nature of hepatocellular carcinoma. Jpn J Radiol. 2019;37:191-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 6. | Calderaro J, Couchy G, Imbeaud S, Amaddeo G, Letouzé E, Blanc JF, Laurent C, Hajji Y, Azoulay D, Bioulac-Sage P, Nault JC, Zucman-Rossi J. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol. 2017;67:727-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 537] [Article Influence: 67.1] [Reference Citation Analysis (0)] |
| 7. | Ziol M, Poté N, Amaddeo G, Laurent A, Nault JC, Oberti F, Costentin C, Michalak S, Bouattour M, Francoz C, Pageaux GP, Ramos J, Decaens T, Luciani A, Guiu B, Vilgrain V, Aubé C, Derman J, Charpy C, Zucman-Rossi J, Barget N, Seror O, Ganne-Carrié N, Paradis V, Calderaro J. Macrotrabecular-massive hepatocellular carcinoma: A distinctive histological subtype with clinical relevance. Hepatology. 2018;68:103-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 173] [Article Influence: 24.7] [Reference Citation Analysis (1)] |
| 8. | Lee S, Kang TW, Song KD, Lee MW, Rhim H, Lim HK, Kim SY, Sinn DH, Kim JM, Kim K, Ha SY. Effect of Microvascular Invasion Risk on Early Recurrence of Hepatocellular Carcinoma After Surgery and Radiofrequency Ablation. Ann Surg. 2021;273:564-571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 217] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 9. | Zhang EL, Cheng Q, Huang ZY, Dong W. Revisiting Surgical Strategies for Hepatocellular Carcinoma With Microvascular Invasion. Front Oncol. 2021;11:691354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 10. | Calderaro J, Ziol M, Paradis V, Zucman-Rossi J. Molecular and histological correlations in liver cancer. J Hepatol. 2019;71:616-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 376] [Article Influence: 62.7] [Reference Citation Analysis (0)] |
| 11. | Mulé S, Galletto Pregliasco A, Tenenhaus A, Kharrat R, Amaddeo G, Baranes L, Laurent A, Regnault H, Sommacale D, Djabbari M, Pigneur F, Tacher V, Kobeiter H, Calderaro J, Luciani A. Multiphase Liver MRI for Identifying the Macrotrabecular-Massive Subtype of Hepatocellular Carcinoma. Radiology. 2020;295:562-571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 103] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 12. | Chen J, Xia C, Duan T, Cao L, Jiang H, Liu X, Zhang Z, Ye Z, Wu Z, Gao R, Shi Y, Song B. Macrotrabecular-massive hepatocellular carcinoma: imaging identification and prediction based on gadoxetic acid-enhanced magnetic resonance imaging. Eur Radiol. 2021;31:7696-7704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 13. | Rhee H, Cho ES, Nahm JH, Jang M, Chung YE, Baek SE, Lee S, Kim MJ, Park MS, Han DH, Choi JY, Park YN. Gadoxetic acid-enhanced MRI of macrotrabecular-massive hepatocellular carcinoma and its prognostic implications. J Hepatol. 2021;74:109-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 85] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 14. | Zhu Y, Weng S, Li Y, Yan C, Ye R, Wen L, Zhou L, Gao L. A radiomics nomogram based on contrast-enhanced MRI for preoperative prediction of macrotrabecular-massive hepatocellular carcinoma. Abdom Radiol (NY). 2021;46:3139-3148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 15. | Chen S, Feng S, Wei J, Liu F, Li B, Li X, Hou Y, Gu D, Tang M, Xiao H, Jia Y, Peng S, Tian J, Kuang M. Pretreatment prediction of immunoscore in hepatocellular cancer: a radiomics-based clinical model based on Gd-EOB-DTPA-enhanced MRI imaging. Eur Radiol. 2019;29:4177-4187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 124] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 16. | Zwanenburg A, Vallières M, Abdalah MA, Aerts HJWL, Andrearczyk V, Apte A, Ashrafinia S, Bakas S, Beukinga RJ, Boellaard R, Bogowicz M, Boldrini L, Buvat I, Cook GJR, Davatzikos C, Depeursinge A, Desseroit MC, Dinapoli N, Dinh CV, Echegaray S, El Naqa I, Fedorov AY, Gatta R, Gillies RJ, Goh V, Götz M, Guckenberger M, Ha SM, Hatt M, Isensee F, Lambin P, Leger S, Leijenaar RTH, Lenkowicz J, Lippert F, Losnegård A, Maier-Hein KH, Morin O, Müller H, Napel S, Nioche C, Orlhac F, Pati S, Pfaehler EAG, Rahmim A, Rao AUK, Scherer J, Siddique MM, Sijtsema NM, Socarras Fernandez J, Spezi E, Steenbakkers RJHM, Tanadini-Lang S, Thorwarth D, Troost EGC, Upadhaya T, Valentini V, van Dijk LV, van Griethuysen J, van Velden FHP, Whybra P, Richter C, Löck S. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology. 2020;295:328-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2090] [Cited by in RCA: 2319] [Article Influence: 463.8] [Reference Citation Analysis (0)] |
| 17. | Sessa A, Mulé S, Brustia R, Regnault H, Galletto Pregliasco A, Rhaiem R, Leroy V, Sommacale D, Luciani A, Calderaro J, Amaddeo G. Macrotrabecular-Massive Hepatocellular Carcinoma: Light and Shadow in Current Knowledge. J Hepatocell Carcinoma. 2022;9:661-670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 18. | Cannella R, Dioguardi Burgio M, Beaufrère A, Trapani L, Paradis V, Hobeika C, Cauchy F, Bouattour M, Vilgrain V, Sartoris R, Ronot M. Imaging features of histological subtypes of hepatocellular carcinoma: Implication for LI-RADS. JHEP Rep. 2021;3:100380. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 19. | Mulé S, Serhal A, Pregliasco AG, Nguyen J, Vendrami CL, Reizine E, Yang GY, Calderaro J, Amaddeo G, Luciani A, Miller FH. MRI features associated with HCC histologic subtypes: a western American and European bicenter study. Eur Radiol. 2023;33:1342-1352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 20. | Feng Z, Li H, Zhao H, Jiang Y, Liu Q, Chen Q, Wang W, Rong P. Preoperative CT for Characterization of Aggressive Macrotrabecular-Massive Subtype and Vessels That Encapsulate Tumor Clusters Pattern in Hepatocellular Carcinoma. Radiology. 2021;300:219-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 84] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 21. | Gerald D, Chintharlapalli S, Augustin HG, Benjamin LE. Angiopoietin-2: an attractive target for improved antiangiogenic tumor therapy. Cancer Res. 2013;73:1649-1657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 166] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 22. | Aryal B, Yamakuchi M, Shimizu T, Kadono J, Furoi A, Gejima K, Komokata T, Hashiguchi T, Imoto Y. Deciphering Platelet Kinetics in Diagnostic and Prognostic Evaluation of Hepatocellular Carcinoma. Can J Gastroenterol Hepatol. 2018;2018:9142672. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 23. | Fraum TJ, Tsai R, Rohe E, Ludwig DR, Salter A, Nalbantoglu I, Heiken JP, Fowler KJ. Differentiation of Hepatocellular Carcinoma from Other Hepatic Malignancies in Patients at Risk: Diagnostic Performance of the Liver Imaging Reporting and Data System Version 2014. Radiology. 2018;286:158-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 103] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 24. | van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin JC, Pieper S, Aerts HJWL. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017;77:e104-e107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1922] [Cited by in RCA: 3907] [Article Influence: 488.4] [Reference Citation Analysis (0)] |
| 25. | Fang S, Lai L, Zhu J, Zheng L, Xu Y, Chen W, Wu F, Wu X, Chen M, Weng Q, Ji J, Zhao Z, Tu J. A Radiomics Signature-Based Nomogram to Predict the Progression-Free Survival of Patients With Hepatocellular Carcinoma After Transcatheter Arterial Chemoembolization Plus Radiofrequency Ablation. Front Mol Biosci. 2021;8:662366. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 26. | Che F, Xu Q, Li Q, Huang ZX, Yang CW, Wang LY, Wei Y, Shi YJ, Song B. Radiomics signature: A potential biomarker for β-arrestin1 phosphorylation prediction in hepatocellular carcinoma. World J Gastroenterol. 2022;28:1479-1493. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 8] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 27. | Fan Y, Yu Y, Wang X, Hu M, Du M, Guo L, Sun S, Hu C. Texture Analysis Based on Gd-EOB-DTPA-Enhanced MRI for Identifying Vessels Encapsulating Tumor Clusters (VETC)-Positive Hepatocellular Carcinoma. J Hepatocell Carcinoma. 2021;8:349-359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 28. | Shan Y, Yu X, Yang Y, Sun J, Wu S, Mao S, Lu C. Nomogram for the Preoperative Prediction of the Macrotrabecular-Massive Subtype of Hepatocellular Carcinoma. J Hepatocell Carcinoma. 2022;9:717-728. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |