Published online Apr 7, 2023. doi: 10.3748/wjg.v29.i13.1942
Peer-review started: November 29, 2022
First decision: December 19, 2022
Revised: December 25, 2022
Accepted: March 20, 2023
Article in press: March 20, 2023
Published online: April 7, 2023
Processing time: 128 Days and 20 Hours
The tremendous public health and economic impact of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a huge challenge globally. There is increasing evidence that SARS-CoV-2 induces intestinal infections. Type III interferon (IFN-λ) has an antiviral role in intestinal infection, with focused, long-lasting, and non-inflammatory characteristics. This review presents a summary of the structure of SARS-CoV-2, including its invasion and immune escape mechanisms. Emphasis was placed on the gastrointestinal impact of SARS-CoV-2, including changes to the intestinal microbiome, activation of immune cells, and inflammatory responses. We also describe the comprehensive functions of IFN-λ in anti-enteric SARS-CoV-2 infection, and discuss the potential application of IFN-λ as a therapeutic agent for COVID-19 with intestinal symptoms.
Core Tip: The tremendous public health and economic impact of the coronavirus disease 2019 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a huge challenge for countries across the whole world. Increased evidences have revealed that SARS-CoV-2 also induced intestinal infection. Type III interferon (IFN), also called IFN-λ, plays an antiviral role in intestinal infection, and has focused, long-lasting, as well as non-inflammatory characteristics. In this review, we summarized the invasion and immune escape mechanisms of the SARS-CoV-2. Furthermore, we concerned about the gastrointestinal impact of the SARS-CoV-2 and the comprehensive role of IFN-λ, also called type III IFN, in anti-enteric SARS-CoV-2 infection.
- Citation: Pan YY, Wang LC, Yang F, Yu M. Interferon-lambda: New role in intestinal symptoms of COVID-19. World J Gastroenterol 2023; 29(13): 1942-1954
- URL: https://www.wjgnet.com/1007-9327/full/v29/i13/1942.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i13.1942
Coronavirus disease 2019 (COVID-19) was first documented in December 2019, as an outbreak of acute community-acquired atypical pneumonia occurred in Wuhan, China, with unknown etiology. At that time, COVID-19 represented a newly emerged respiratory disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which subsequently became a pandemic[1].
Similar to the flu virus, SARS-CoV-2 spreads in small liquid particles from the mouth or nose of an infected person when they cough, sneeze, speak, sing, or breath. These particles range from larger respiratory droplets to smaller aerosols[2]. It is widely reported that coronavirus has the largest non-segmented genome (close to 30 kb) of all RNA virus. This unique genome size enhances the plasticity of CoVs, which alter through mutations and recombination[3]. The first SARS-CoV-2 variant, Alpha B.1.1.7, was reported in the United Kingdom in September 2020[4]. Increasing numbers of variants subsequently appeared, including Beta B.1.351, Gamma P.1, Delta B.1.617.2, and Omicron B.1.1.529[5]. By November 2021, Omicron B.1.1529 had become the current major SARS-CoV-2 variant globally[6]. Because of this strong plasticity in CoV genes, generating high genetic diversity and a high risk of cross transmission, specific treatments remain elusive.
As a systemic disease, SARS-CoV-2 can infect multiple organs, resulting in multiorgan dysfunction in COVID-19 patients. Organs that are infected by SARS-CoV-2 include the lungs, small intestine, gallbladder, kidneys, testes, thyroid, adipose tissue, heart muscle, vagina, breasts, ovaries, and pancreas[7]. The symptoms of COVID-19 include pneumonia, fever, fatigue, intestinal ischemia, and diarrhea[8,9]. Many studies have reported gastrointestinal (GI) symptoms in COVID-19 patients, including diarrhea, nausea, vomiting, anorexia, abdominal pain, acid reflux, upper GI bleeding, haematochezia, constipation, and melena[10]. Besides, angiotensin-converting enzyme 2 (ACE2), the receptor of SARS-CoV-2, is highly expressed in intestinal tissue. Moreover, fecal samples from COVID-19 patients have tested positive for SARS-CoV-2 RNA testing. Thus, there is potential for fecal-oral transmission, with SARS-CoV-2 Likely impacting the GI tract[2].
As a part of the innate immune system, type III interferon (IFN-λ) mediates antiviral responses in the epithelial barrier. Moreover, IFN-λ receptor (IFN-λR)1 is preferentially expressed by the epithelial cells of the respiratory, intestinal, and reproductive tracts. The antiviral role of IFN-λ in intestinal infection has focused, long-lasting, and non-inflammatory characteristics. Consequently, many studies have explored the potential of applying IFN-λ in COVID-19[11]. Continuing research on SARS-CoV-2 showed that it is also expressed in the intestinal tract. In particular, IFN-λ might have crucial roles in SARS-CoV-2 intestinal infection.
In this review, we summarize the invasion and escape mechanisms of SARS-CoV-2, and both the direct and indirect influence of SARS-CoV-2 on intestinal homeostasis. We also consider how IFN-λ impacts intestinal infections of SARS-CoV-2, and focus on the new functions of IFN-λ in COVID-19 with intestinal symptoms.
Coronaviruses belong to the subfamily coronavirinae of the coronaviridae family, and have caused three pandemic outbreaks globally[12]. The subfamily coronavirinae consists of four genera; α, β, γ, and δ-coronaviruses. α and β-coronaviruses usually cause respiratory illness in humans and gastroenteritis in animals. In comparison, γ and δ-coronaviruses mostly infect birds, and, sometimes, mammals. According to genome sequence analysis, SARS-CoV-2, which was the third virus to create a pandemic, is classified in the β-coronavirus genome[12].
The four structural proteins of SARS-CoV-2 are essential for infections to occur. They consist of the spike (S) surface glycoprotein, membrane (M) protein, envelope (E) protein, and nucleocapsid (N) protein. The S protein is cleaved by host proteases, and allows SARS-CoV-2 to attach to host cells[13]. Both E and M proteins induce S protein to reside in the Golgi/ER Golgi intermediate compartment (ERGIC) compartment, and subsequently, together, regulate N-glycosylation maturation of the S protein[14]. N structural proteins are responsible for viral gene binding, compression, and packaging. In addition, they contribute to RNA transcription, and achieve the innate immune escape of the host through the N protein directly targeting and sequestering G3BP1 and host mRNA[15]. Once infected, SARS-CoV-2 mainly induces T cell responses; however, the generation of high-titer neutralizing antibodies by B-cell expansion and maturation might be limited[16,17]. To explore potential treatment directions of SARS-CoV-2, it is important to learn its route of infection and mechanisms of immune evasion.
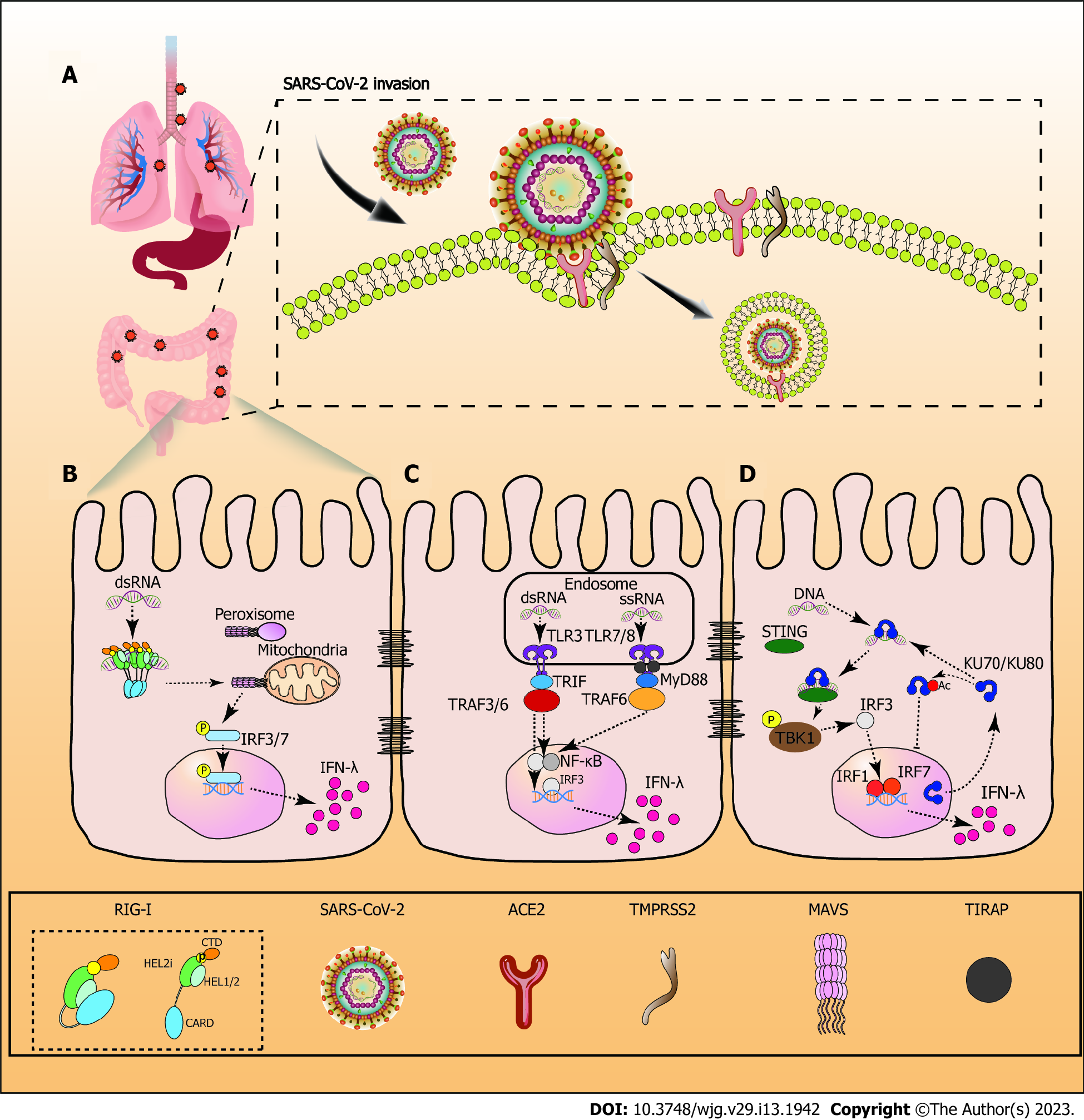
Zhou et al[18] first confirmed that SARS-CoV-2 only infects HeLa cells that express ACE2, which is used as a SARS-CoV-2 receptor in patients[18]. Although there are two independent ways to mediate the infection of SARS-CoV[19,20], the entry of SARS-CoV-2 involves ACE2-receptor-mediated transmembrane serine protease 2 (TMPRSS2)-dependent membrane fusion[21] (Figure 1).
SARS-CoV-2 is an enveloped virus containing a large N encapsidated positive sense RNA genome. S trimers with closed and open prefusion structures usually protrude from the lipid bilayer of SARS-CoV-2. They are distributed randomly on the viral surface to bind to the receptor, ACE2, and, subsequently, mediate viral uptake and fusion. The receptor binding domain (RBD) site is occluded by three copies of the N-terminal domain when in the closed prefusion. In the open prefusion, one or multiple RBDs lift up to expose the receptor binding site[22]. There are two main subunits, S1 and S2. S1 binds ACE2, while S2 triggers membrane fusion of the virus to the cell. In a typical SARS-CoV-2 infection process, S trimers must be activated by cellular protease-mediated cleavage at two distinct sites, S1/S2 and S2’. S2’ is responsible for triggering virus-cell membrane fusion. TMPRSS2 is expressed on the cell surface, and activates S trimers, which promote the virus to enter the plasma membrane in a pH-independent way[23]. Finally, the encapsidated genome buds into the ERGIC to form virions, which are trafficked to the plasma membrane and released[22] (Figure 1).
The first immune evasion of SARS-CoV-2 is related to the delayed occurrence of IFN-I[24]. As the first line of host defense against virus infections, IFN-I response is initiated by the recognition of pathogen-associated molecular patterns (PAMPs) via host pattern recognition receptors (PRRs). SARS-CoV-2 RNA is detected by various cytosolic sensors, including retinotic acid-inducible gene 1 (RIG-I/DExD/H-box helicase 58) and melanoma differentiation-associated gene 5 (IFN induced with helicase C domain 1). These sensors induce an antiviral signaling cascade to occur[25]. A subset of IFN-stimulated genes (ISG) is produced by the IFN-I response. These ISGs, along with other downstream molecules controlled by IFN-I, directly inhibit virus replication and recruitment, and activate diverse immune cells[26]. However, viral proteins (NSP3/papain-like protease) encoded by the SARS-CoV-2 genome selectively cleave interferon regulatory factors (IRF) 3 protein directly during SARS-CoV-2 infection, resulting in a blunted Type-I IFN response[27]. M proteins on SARS-CoV-2 also target mitochondrial antiviral signaling proteins (MAVS), and impair the aggregation and recruitment of downstream signaling components[28]. In parallel, stress granule analogues, formed by the N protein, competitively bind G3BP1 to inhibit the innate immune response[15]. Furthermore, SARS-CoV-2 nsp13 (helicase), nsp14 (exonuclease), nsp15 (endoribonuclease), and accessory protein orf6 block IFN-I production by antagonizing upstream signaling pathways[29,30].
The second method of evasion is the upregulation of human inhibitory receptors, which modulate natural killer (NK) cell-mediated cytotoxicity. This process causes the exhaustion of NK cells, reducing their ability to clear viral infection[31].
A recent meta-analysis of COVID-19 patients showed that physical activity could reduce the risk of COVID-19 infection. Physical activity strengthens the immune system, and allows cardiopulmonary and musculoskeletal adaptations[32]. Moreover, adherence to a high-quality healthy diet has been linked to a reduced risk of COVID-19 infection and lower hospitalization rates. Mediterranean-style dietary patterns are associated with a lower risk of death from respiratory infections, including COVID-19, while plant-based diets have been associated with lower COVID-19 infection rates[33]. Age is also an important factor in severe COVID-19 illness and its adverse consequences. Statistically, the case fatality ratio for COVID-19 increases with age[34]. There has also been widespread concern about whether pregnant women are more susceptible to SARS-CoV-2. However, to date, there are no data supporting an increased susceptibility to SARS-CoV-2 due to pregnancy. Nevertheless, the risk of death in pregnant women with COVID-19 is significantly increased[35]. There is also a nonlinear association between ambient temperature and the risk of infection with SARS-CoV-2. Specifically, the risk of infection with SARS-CoV-2 increases significantly at temperatures between 0 and 10 ℃[36].
As a receptor of SARS-CoV-2, ACE2 is expressed in most organs. Consequently, COVID-19 is capable of infecting multiple organs, in addition to the respiratory tract[37]. It has been confirmed that ACE2 is abundantly expressed in GI glandular epithelial cells[38]. There is also clinical evidence that the intestine is another target organ. When SARS-CoV-2 infects the GI tract, it disrupts gut microbiota and induces other GI symptoms. Current research on SARS-CoV-2 invasion of the gut is focused on (1) changes to the gut microbiome; and (2) whether GI symptoms are caused by direct infection of SARS-CoV-2 or the consequence of a systemic immune activation.
The gut microbiome consists of bacteria, fungi, viruses, and other microorganisms that dynamically interact with environmental factors to shape the intestinal mucosal immune system[39]. The intestinal bacteria of healthy individuals are mainly composed of Actinobacteria, Firmicutes, Proteobacteria, and Bacteroidetes. Intestinal microbiota is highly resilient to external disturbances, enabling the host to retain key species for a long time and maintain intestinal homeostasis. However, severe external influences cause microbial ecosystems to move from a stable state to an unhealthy stable state associated with disease[40]. A comparison of gut microbiota in COVID-19 patients and healthy people showed that the relative abundance of gut bacterial groups was higher in COVID-19 patients (including Ruminococcus gnavus, Clostridium ramosum, Coprobacillus, Akkermansia muciniphila, and Eggerthella lenta). Furthermore, the abundance of Alistipes shahii was comparatively lower in COVID-19 patients, along with several butyrate producers (including Roseburia intestinalis, Eubacterium hallii, Ruminococcus bromii, and Faecalibacterium prausnitzii)[41]. Gut microorganisms regulate mucosal sites in the distal part of the intestine via metabolites. These metabolites access other organs through the bloodstream, to induce immunomodulatory, immunoglobulin, and anti-inflammatory effects[42]. One cohort study showed that the lower abundance of B. adolescentis, F. prausnitzii, E. rectale, R. (Blautia) obeum and D. formicigenerans in the COVID-19 cohort might be associated with the decreased secretion of anti-inflammatory cytokines or increased secretion of inflammatory cytokines[43]. Another clinical study found that GI infection with SARS-CoV-2 differed to that without infection. Significant dysbiosis of the microbiome was documented in COVID-19 patients, with high levels of pathogenic bacteria[44]. In general, the intestinal microbiota of active GI infections with SARS-CoV-2 were mainly characterized by the enrichment of opportunistic pathogens and reduction of beneficial bacteria[45]. One study of COVID-19 patients showed that changes to the gut microbiome are correlated with the severity of COVID-19. In particular, the abundance of Coprobacillus, Clostridium ramosum, and Clostridium hathewayi is correlated with the severity of COVID-19. Furthermore, Bacteroides dorei, Bacteroides thetaiotaomicron, Bacterodies massiliensis, and Bacteroides ovatus downregulate the expression of ACE2 in the murine gut, correlating with SARS-CoV-2 load in fecal samples[46].
In addition to SARS-CoV-2 directly infecting the GI tract, it affects the functioning of the GI tract through other organs. For instance, Dhar et al[47] showed that gut microbiota affected pulmonary health via vital cross-talk between the gut microbiome and lungs which is called the “gut-lung axis.” However, the gut-lung axis is bidirectional; consequently, when SARS-CoV-2 infects the lungs, the gut microbiome might also be affected, including disruption[47].
When infected by SARS-CoV-2, activated CD8+ IEL migrate to the intestinal mucosa[48]. The infected cells block IFN signaling, and exhibit strong pro-inflammatory responses by strongly activating the nuclear factor kB (NF-κB)/tumour necrosis factor pathway. In parallel, bystander cells not infected by SARS-CoV-2 mount an IFN-mediated response[49]. Although human plasmacytoid predendritic cells (pDCs) are not infected by SARS-CoV-2, pDCs may be activated by SARS-CoV-2 through IRAK4 and UNC93B1-dependent pathways, inducing high levels of type I and type III IFNS[50].
In healthy cells, CCL5, CXCL1, CXCL10, and CXCL11 recruit immune cells and pro-inflammatory chemokines. However, these four chemokines were significantly upregulated in SARS-CoV-2-infected epithelial cells in a trial of biomimetic human gut-on-chip system[51]. Interestingly, in a clinical cohort study, a subset of COVID-19 patients with GI symptoms had small intestinal epithelial cells infected with SARS-CoV-2. Compared to patients without GI symptoms, key inflammatory genes [including IFN-γ, CXCL8, CXCL2, and interleukin (IL)-1B] were downregulated in these cells, and the frequency of pro-inflammatory dendritic cells was reduced in immune subsets[52]. The authors also recorded lower mortality among COVID-19 patients with GI symptoms. Thus, a stronger immune response was induced in SARS-CoV-2 infected human intestinal tissue, reducing the efficiency of viral replication[53]. A recent study of rhesus monkeys infected with SARS-CoV-2 proposed that antiviral immunity was activated and the inflammatory response was enhanced at the early stage of intestinal infection. This response damages the intestinal barrier; however, the intestine was repaired and inflammatory response decreased at the later stage, leading to mild intestinal pathological damage[54]. Intestinal IFN-related antiviral response and neutrophil related pathways are activated at the early stage. This process damages Paneth cells and the intestinal barrier, enhancing the inflammatory response. Nevertheless, the increasing number of Paneth cells and B cells that inhibit neutrophil-chemotactic cytokines CXCL8 and IL-1B at the later stage, repair the homeostasis of the intestinal barrier, inhibiting inflammation and tissue damage[54].
IFNs interfere with viral replication and induce inflammation, and are essential for mobilizing immune responses to SARS-CoV-2 infection[55]. There are three types of IFNs, and each has its own distinctive receptors. Type I IFN consists of at least 32 functional subtypes, including IFN-α subtypes (13 in humans and 14 in mice), IFN-β, IFN-ε, IFN-κ, IFN-ω, and IFN-ξ[56]. The type I IFN receptor (IFNR) is mainly expressed in immune cells. IFN-α/β responses to viral infections result in immunopathology. However, type I IFN also induces the infiltration of inflammation[57]. The mechanism of SARS-CoV-2 innate immune evasion depends on dysregulated IFN-I production, which contributes to robust early SARS-CoV-2 replication[31]. Type II IFN (IFN-γ) is a proinflammatory cytokine[58-60]. IFN-λ consists of four subtypes in humans (λ1, λ2, λ3, λ4), and its receptor is localized to epithelial cells and a subset of immune cells, including neutrophils. IFN-λ is widely believed to control pathologic microbes in the intestinal epithelium, and promotes the healing of the colonic epithelium without inducing an excessively strong inflammatory response[61].
Immune responses triggered by PAMPs are key factors for pathogen defense. Differences in the location of PAMP recognized by some PRRs affect the type of IFN produced. PRRs in cytosol include RIG-I like receptors (RLRs), cyclic guanosine monophosphate-adenosine monophosphate synthase (cGAS), and Ku70, whereas PPRs in the endosome are toll-like receptors (TLRs)[62]. cGAS mainly induces the expression of pro-inflammatory cytokines and the IFN-I gene through the stimulator of interferon genes (STING)-TANK-binding kinase 1 (TBK1)-IRF3/NF-κB signaling pathway. In comparison, RLRs, TLRs, and Ku70 induce the IFN-λ gene[63]. The downstream signaling pathways of IFN-λ is similar to IFN-I, which generates an antiviral response through the JAK-signal transducer and activator of transcription (STAT) pathway. In mitochondria or peroxisome, RLRs interacts with the MAVS, resulting IRF 3 and 7 being recruited into the nucleus. IRF3/7 binds to the promotor of IFN-λ to initiate the gene expression of the IFN-λ family. Activated TLR7, 8 and 3 signal is transduced by Myeloid differentiation primary response 88 and TIR-domain-containing adapter-inducing interferon-β. This process results in the recruitment of IRFs and nuclear factor ‘kappa-light-chain-enhancer’ of activated B-cells (NF-κB) in the nucleus. Subsequently, the expression of IFN-λ gene is enhanced[64]. The cytoplasmic translocation of Ku70 is the first step in initiating IFN-λ1 secretion, which facilitates the interaction between STING and Ku70. Activated STING promotes TBK1 phosphorylation, which activates IRF3. Since IRF3 is activated first, it produces the strong expression of IRF1 and IRF7, which are not endogenously expressed in cells. Subsequently IFN-λ1 is induced in the nucleus[65] (Figure 1).
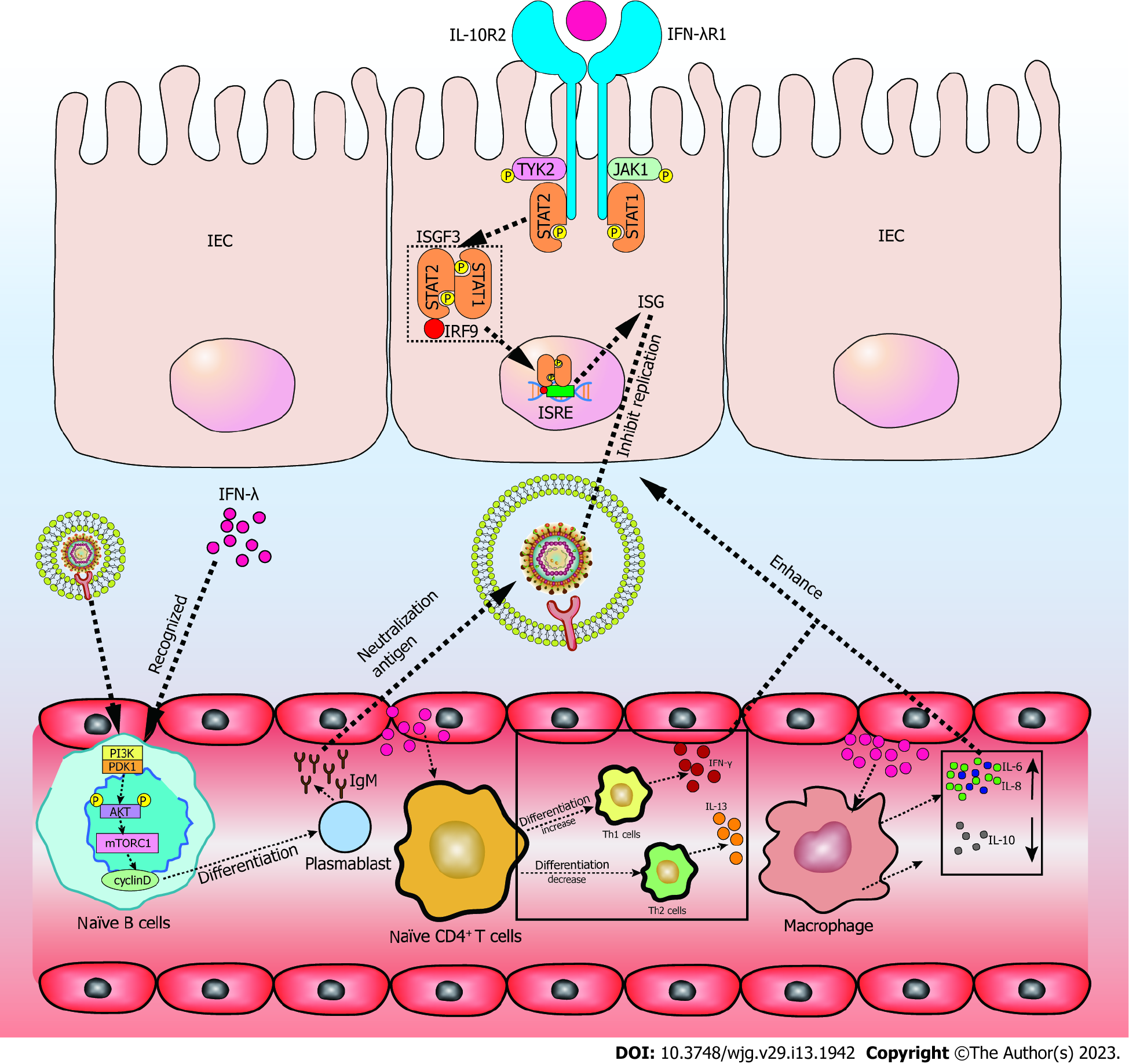
In contrast to the type I IFNR, the IFN-λR is preferentially expressed by epithelial cells and neutrophils. The IFN-λR consists of IFN-λR1 (also called IL-28Rα) and IL-10R2. IFN-λR1 specifically binds IFN-λ, whereas IL-10R2 binds with cytokines in the IL-10 family. When IFN-λ signaling is activated by IFN-λ, the receptor-associated kinases JAK1 and TYK2 phosphorylate STAT1 and STAT2. IFN-stimulated gene factor 3 (ISGF3) is activated by IFN-λ, and is a trimeric complex that is formed by phosphorylated STAT1, STAT2, and IRF9. ISGF3 binds IFN-stimulated response elements in the nucleus to promote the transcription of hundreds of antiviral effector ISGs[66]. In addition to the STAT1-dependent pathway, IFN-λ promotes tissue healing by the upregulation of collagens via the MAPK-dependent pathway[67] (Figure 2).
Diarrhea is the most common GI symptom in COVID-19 patients. It is associated with changes to gut microbiota, malabsorption, and inflammation. It is also associated with the release of virulent proteins and toxins, and viral-induced intestinal fluid and electrolyte secretion. Of note, calprotectin, an inflammatory hallmark, was secreted by infiltrated neutrophils in COVID-19 patients with diarrhea[10].
The rapid initiation of the innate immune response after pathogen encounter is the result of infection and survival of the host. Highly expressed IFNs induce the secretion of ISGs, which is a hallmark of the innate immune response. ISGs induce various cell-intrinsic antiviral responses, such as blocking the translation and induction of apoptosis, while recruiting of immune cells and stimulating effector functions[68]. Compared to IFN-I, the effects of IFN-λ are delayed but last longer. IFN-λ is transcribed and translated at higher rates in intestinal organoids. Furthermore, IFN-λ is preferentially induced on the mucosal surfaces of both the intestine and lung[69]. Furthermore, cells cultivated under polarizing conditions exhibit high IFN-λ responsiveness[70]. Pretreatment with IFN-λ in Vero E6 cells showed dose-dependent inhibition of SARS-CoV-2[71]. IFN-λ promotes epithelial healing in patients infected with SARS-CoV-2 with acute colonic inflammation and tissue damage[72]. Moreover, IFN-λ treatment directly improves the proliferation and regeneration ability of differentiated GI epithelial cells through Lgr5+ intestinal stem cells (ISC), leading to the recovery of intestinal barrier integrity[73]. Furthermore, IFN-λ protects epithelial cells from enteric viral infections, indicating that IFN-λ could be used to treat SARS-CoV-2 infection of the GI tract.
IFN has been confirmed to be the first line of innate immunity, producing an antiviral response when the virus infects the intestinal tract. SARS-CoV-2 infects, replicates, and implements de novo infectious virus production in human intestinal epithelial cells (IECs). Furthermore, IFN-λ has huge potential for treating SARS-CoV-2. One study showed that the JAK-STAT1 dependent pathway of IFN-λ is more critical for anti-SARS-CoV-2 activity than IFN-I. Pretreatment IECs with IFN-λ resulted in a substantial decrease in the number of SARS-CoV-2 infected cells and viral replication[74]. Type I and II IFN upregulate antiviral ISGs when the virus infects cells, with ACE2 mRNA expression simultaneously increasing. Nevertheless, IFN-λ upregulates antiviral ISGs, whereas ACE2 mRNA is only marginally elevated[75]. Furthermore, one of the mechanisms by which IFNs restrict viral replication is to inhibit the translation process, which inhibits the expression of structural proteins, with SARS-CoV-2 being more sensitive to IFN-λ. Compared to type I IFN, IFN-λ maintained a more consistent anti-SARS-CoV-2 infection state, with viral replication being inhibited for more than 72 h after drug withdrawal[76]. As for exogenous IFN-λ, pretreated IFN-λ significantly reduces the SARS-CoV-2 burden in the air-liquid interface of the lung model. The therapeutic administration of IFN-λ is also effective at restricting SARS-CoV-2 production in cultured human lung cells[77,78]. Another vitro experiment showed that recombinant bovine IFN-λ produced in HEK-293 cells effectively prevents SARS-CoV-2 infection toward VERO cells[79].
It remains unclear whether IFN-λ directly or indirectly regulates the immune response after SARS-CoV-2 infection. To date, IFN-λ1-4 is considered to regulate the immune response, including the upregulation of IL-6, IL-8, and the downregulation of IL-10 in peripheral blood mononuclear cells (PBMCs)[79]. It also alters Th1/Th2 T-cell balance, reduces IL-13 production by T cells[80], and induces ISGs in B cells and plasmacytoid dendritic cells, as well as in CD8 T cells, especially effector memory cells (TEMRA)[81] (Figure 2). As an anti-inflammatory factor, IL-10 induction in macrophages provides a powerful pathogen immune escape mechanism, which causes chronic infection[82]. IFN-λ stimulates a stronger innate immune response by upregulating IL-6 and IL-8, and by reducing IL-10 secretion in PBMCs, which promote an antiviral response[79]. In vivo, IFN-λ promotes the differentiation of initial T cells to Th1 cells, rather than Th2 cells, thus activating a stronger immune response, which combats the invasion of the gut by pathogens[80]. Furthermore, in the intestinal tract (unlike IFN-I), IFN-λ exhibits a compartmentalized response to virus infections because of the restricted expression of IFN-λR, which is preferentially expressed on IECs. IFN-λR expression has also been reported in NK cells, T cells, B cells, and pDCs[83]. Interestingly, IFN-λ induces the mechanistic target of rapamycin complex 1 pathway, leading to the differentiation of naïve B cells in plasmablasts by activating phosphoinositide 3-kinase via JAKs in an IRS-dependent, but STAT-independent, manner[84]. Thus, IFN-λ likely promotes the release of antibodies and increases the capability of antiviruses. These immune cells activate strong immune responses in the body to resist pathogen invasion and maintain intestinal homeostasis under IFN-λ regulation (Figure 2).
As a hallmark of intestinal inflammation in COVID-19 patients who have diarrhea, neutrophils are rapidly recruited from the blood after IEC infection. Neutrophils also restrict pathogen invasion through phagocytosis, reactive oxygen species (ROS), and the degranulation and release of cytotoxic antimicrobial molecules[85]. In contrast, excessive recruitment of neutrophils promotes inflammation. Thus, it is important to control the recruitment of neutrophils. Published studies show that IFN-λ provides a better balance of antiviral protection, with minimal inflammation and tissue damage. Specifically, IFN-λ in neutrophils inhibits the activation and phosphorylation of kinase AKT, which prevents the assembly of nicotinamide adenine dinucleotide phosphate oxidase complex NOX2 and decrease ROS in a JAK2-dependent way[86]. The effects of IFN-λ in neutrophils are independent of protein synthesis and transcription, only depending on the activation of AKT being inhibited[87]. Another study showed that human neutrophils optimally respond to IFN-λ in vivo, potentially inducing ISG levels to rise during inflammatory conditions by up-regulating IFN-λR1[88]. Mutual regulation between IFN-λ and neutrophils promotes intestinal resistance to SARS-CoV-2 invasion, and reduces the damage of intestinal tissue caused by inflammation.
IFN-λ potentially has a protective role in COVID-19 patients with intestinal infections. However, there is increasing evidence that IFN-λ has a complex dual role in the intestinal tract. Exposure to IFN-λ might increase intestinal permeability, which is associated with the disruption of junctional proteins[89]. Elevated IFN-λ is also related to lower numbers of Paneth cells and increased apoptosis of epithelial cells in the small intestinal[90]. In contrast, IFN-λ has been reported to exhibit an antiviral effect in the intestinal tract, and promotes the healing of IECs[71-73]. Although IFN-λ has an anti-SARS-CoV-2 effect in the intestinal tract, the mechanism remains unclear. Furthermore, how IFN-λ affects other intestinal cells (Paneth cells, goblet cells, and ISCs) and tight junctions in the intestine of SARS-CoV-2 patients requires investigation. Although IFN-λ has strong potential for use in COVID-19 intervention strategies, the mechanisms need to be elucidated first.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Al-Ani RM, Iraq; Rahmati M, Iran S-Editor: Fan JR L-Editor: A P-Editor: Fan JR
| 1. | To KK, Sridhar S, Chiu KH, Hung DL, Li X, Hung IF, Tam AR, Chung TW, Chan JF, Zhang AJ, Cheng VC, Yuen KY. Lessons learned 1 year after SARS-CoV-2 emergence leading to COVID-19 pandemic. Emerg Microbes Infect. 2021;10:507-535. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 175] [Cited by in RCA: 209] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 2. | Sharma A, Ahmad Farouk I, Lal SK. COVID-19: A Review on the Novel Coronavirus Disease Evolution, Transmission, Detection, Control and Prevention. Viruses. 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 245] [Cited by in RCA: 324] [Article Influence: 81.0] [Reference Citation Analysis (0)] |
| 3. | Amoutzias GD, Nikolaidis M, Tryfonopoulou E, Chlichlia K, Markoulatos P, Oliver SG. The Remarkable Evolutionary Plasticity of Coronaviruses by Mutation and Recombination: Insights for the COVID-19 Pandemic and the Future Evolutionary Paths of SARS-CoV-2. Viruses. 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 61] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 4. | Lopez Bernal J, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S, Stowe J, Tessier E, Groves N, Dabrera G, Myers R, Campbell CNJ, Amirthalingam G, Edmunds M, Zambon M, Brown KE, Hopkins S, Chand M, Ramsay M. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N Engl J Med. 2021;385:585-594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2223] [Cited by in RCA: 1887] [Article Influence: 471.8] [Reference Citation Analysis (0)] |
| 5. | Sanches PRS, Charlie-Silva I, Braz HLB, Bittar C, Freitas Calmon M, Rahal P, Cilli EM. Recent advances in SARS-CoV-2 Spike protein and RBD mutations comparison between new variants Alpha (B.1.1.7, United Kingdom), Beta (B.1.351, South Africa), Gamma (P.1, Brazil) and Delta (B.1.617.2, India). J Virus Erad. 2021;7:100054. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 61] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 6. | Kannan SR, Spratt AN, Sharma K, Chand HS, Byrareddy SN, Singh K. Omicron SARS-CoV-2 variant: Unique features and their impact on pre-existing antibodies. J Autoimmun. 2022;126:102779. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 188] [Cited by in RCA: 154] [Article Influence: 51.3] [Reference Citation Analysis (0)] |
| 7. | Synowiec A, Szczepański A, Barreto-Duran E, Lie LK, Pyrc K. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): a Systemic Infection. Clin Microbiol Rev. 2021;34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 149] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 8. | Renu K, Prasanna PL, Valsala Gopalakrishnan A. Coronaviruses pathogenesis, comorbidities and multi-organ damage - A review. Life Sci. 2020;255:117839. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 134] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 9. | Norsa L, Bonaffini PA, Indriolo A, Valle C, Sonzogni A, Sironi S. Poor Outcome of Intestinal Ischemic Manifestations of COVID-19. Gastroenterology. 2020;159:1595-1597.e1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 10. | Guo M, Tao W, Flavell RA, Zhu S. Potential intestinal infection and faecal-oral transmission of SARS-CoV-2. Nat Rev Gastroenterol Hepatol. 2021;18:269-283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 198] [Cited by in RCA: 212] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 11. | Park A, Iwasaki A. Type I and Type III Interferons - Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe. 2020;27:870-878. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 730] [Cited by in RCA: 676] [Article Influence: 135.2] [Reference Citation Analysis (0)] |
| 12. | Li H, Liu SM, Yu XH, Tang SL, Tang CK. Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int J Antimicrob Agents. 2020;55:105951. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 692] [Cited by in RCA: 582] [Article Influence: 116.4] [Reference Citation Analysis (0)] |
| 13. | Wang MY, Zhao R, Gao LJ, Gao XF, Wang DP, Cao JM. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front Cell Infect Microbiol. 2020;10:587269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 474] [Cited by in RCA: 522] [Article Influence: 104.4] [Reference Citation Analysis (0)] |
| 14. | Boson B, Legros V, Zhou B, Siret E, Mathieu C, Cosset FL, Lavillette D, Denolly S. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J Biol Chem. 2021;296:100111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 198] [Cited by in RCA: 217] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 15. | Lu S, Ye Q, Singh D, Cao Y, Diedrich JK, Yates JR 3rd, Villa E, Cleveland DW, Corbett KD. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat Commun. 2021;12:502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 266] [Cited by in RCA: 301] [Article Influence: 75.3] [Reference Citation Analysis (0)] |
| 16. | Woldemeskel BA, Kwaa AK, Garliss CC, Laeyendecker O, Ray SC, Blankson JN. Healthy donor T cell responses to common cold coronaviruses and SARS-CoV-2. J Clin Invest. 2020;130:6631-6638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 17. | Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, Muecksch F, Rutkowska M, Hoffmann HH, Michailidis E, Gaebler C, Agudelo M, Cho A, Wang Z, Gazumyan A, Cipolla M, Luchsinger L, Hillyer CD, Caskey M, Robbiani DF, Rice CM, Nussenzweig MC, Hatziioannou T, Bieniasz PD. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1088] [Cited by in RCA: 1031] [Article Influence: 206.2] [Reference Citation Analysis (0)] |
| 18. | Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15248] [Cited by in RCA: 14104] [Article Influence: 2820.8] [Reference Citation Analysis (1)] |
| 19. | Wang H, Yang P, Liu K, Guo F, Zhang Y, Zhang G, Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290-301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 478] [Cited by in RCA: 531] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 20. | Heurich A, Hofmann-Winkler H, Gierer S, Liepold T, Jahn O, Pöhlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2014;88:1293-1307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 602] [Cited by in RCA: 668] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 21. | Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271-280.e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11946] [Cited by in RCA: 14242] [Article Influence: 2848.4] [Reference Citation Analysis (0)] |
| 22. | Ke Z, Oton J, Qu K, Cortese M, Zila V, McKeane L, Nakane T, Zivanov J, Neufeldt CJ, Cerikan B, Lu JM, Peukes J, Xiong X, Kräusslich HG, Scheres SHW, Bartenschlager R, Briggs JAG. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 917] [Cited by in RCA: 857] [Article Influence: 171.4] [Reference Citation Analysis (0)] |
| 23. | Khan H, Winstone H, Jimenez-Guardeño JM, Graham C, Doores KJ, Goujon C, Matthews DA, Davidson AD, Rihn SJ, Palmarini M, Neil SJD, Malim MH. TMPRSS2 promotes SARS-CoV-2 evasion from NCOA7-mediated restriction. PLoS Pathog. 2021;17:e1009820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 24. | Xia H, Cao Z, Xie X, Zhang X, Chen JY, Wang H, Menachery VD, Rajsbaum R, Shi PY. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 777] [Cited by in RCA: 716] [Article Influence: 143.2] [Reference Citation Analysis (0)] |
| 25. | Zheng Y, Zhuang MW, Han L, Zhang J, Nan ML, Zhan P, Kang D, Liu X, Gao C, Wang PH. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct Target Ther. 2020;5:299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 235] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 26. | Sa Ribero M, Jouvenet N, Dreux M, Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16:e1008737. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 388] [Cited by in RCA: 369] [Article Influence: 73.8] [Reference Citation Analysis (0)] |
| 27. | Moustaqil M, Ollivier E, Chiu HP, Van Tol S, Rudolffi-Soto P, Stevens C, Bhumkar A, Hunter DJB, Freiberg AN, Jacques D, Lee B, Sierecki E, Gambin Y. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerg Microbes Infect. 2021;10:178-195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 184] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 28. | Fu YZ, Wang SY, Zheng ZQ, Yi Huang, Li WW, Xu ZS, Wang YY. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cell Mol Immunol. 2021;18:613-620. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 149] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 29. | Yuen CK, Lam JY, Wong WM, Mak LF, Wang X, Chu H, Cai JP, Jin DY, To KK, Chan JF, Yuen KY, Kok KH. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect. 2020;9:1418-1428. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 299] [Cited by in RCA: 399] [Article Influence: 79.8] [Reference Citation Analysis (0)] |
| 30. | Miorin L, Kehrer T, Sanchez-Aparicio MT, Zhang K, Cohen P, Patel RS, Cupic A, Makio T, Mei M, Moreno E, Danziger O, White KM, Rathnasinghe R, Uccellini M, Gao S, Aydillo T, Mena I, Yin X, Martin-Sancho L, Krogan NJ, Chanda SK, Schotsaert M, Wozniak RW, Ren Y, Rosenberg BR, Fontoura BMA, García-Sastre A. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc Natl Acad Sci U S A. 2020;117:28344-28354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 268] [Cited by in RCA: 401] [Article Influence: 80.2] [Reference Citation Analysis (0)] |
| 31. | Bouayad A. Innate immune evasion by SARS-CoV-2: Comparison with SARS-CoV. Rev Med Virol. 2020;30:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 32. | Rahmati M, Shamsi MM, Khoramipour K, Malakoutinia F, Woo W, Park S, Yon DK, Lee SW, Shin JI, Smith L. Baseline physical activity is associated with reduced mortality and disease outcomes in COVID-19: A systematic review and meta-analysis. Rev Med Virol. 2022;32:e2349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 37] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 33. | Rahmati M, Fatemi R, Yon DK, Lee SW, Koyanagi A, Il Shin J, Smith L. The effect of adherence to high-quality dietary pattern on COVID-19 outcomes: A systematic review and meta-analysis. J Med Virol. 2023;95:e28298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 25] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 34. | Chen Y, Klein SL, Garibaldi BT, Li H, Wu C, Osevala NM, Li T, Margolick JB, Pawelec G, Leng SX. Aging in COVID-19: Vulnerability, immunity and intervention. Ageing Res Rev. 2021;65:101205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 664] [Cited by in RCA: 598] [Article Influence: 149.5] [Reference Citation Analysis (0)] |
| 35. | Rasmussen SA, Jamieson DJ. COVID-19 and Pregnancy. Infect Dis Clin North Am. 2022;36:423-433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 36. | Yang HY, Lee JKW. The Impact of Temperature on the Risk of COVID-19: A Multinational Study. Int J Environ Res Public Health. 2021;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 37. | Qi F, Qian S, Zhang S, Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun. 2020;526:135-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 523] [Cited by in RCA: 740] [Article Influence: 148.0] [Reference Citation Analysis (0)] |
| 38. | Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology. 2020;158:1831-1833.e3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1985] [Cited by in RCA: 1993] [Article Influence: 398.6] [Reference Citation Analysis (1)] |
| 39. | Shi N, Li N, Duan X, Niu H. Interaction between the gut microbiome and mucosal immune system. Mil Med Res. 2017;4:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 223] [Cited by in RCA: 402] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 40. | Fassarella M, Blaak EE, Penders J, Nauta A, Smidt H, Zoetendal EG. Gut microbiome stability and resilience: elucidating the response to perturbations in order to modulate gut health. Gut. 2021;70:595-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 363] [Article Influence: 90.8] [Reference Citation Analysis (70)] |
| 41. | Sarkar A, Harty S, Moeller AH, Klein SL, Erdman SE, Friston KJ, Carmody RN. The gut microbiome as a biomarker of differential susceptibility to SARS-CoV-2. Trends Mol Med. 2021;27:1115-1134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 42. | Donati Zeppa S, Agostini D, Piccoli G, Stocchi V, Sestili P. Gut Microbiota Status in COVID-19: An Unrecognized Player? Front Cell Infect Microbiol. 2020;10:576551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 76] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 43. | Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY, Chung AC, Cheung CP, Tso EY, Fung KS, Chan V, Ling L, Joynt G, Hui DS, Chow KM, Ng SSS, Li TC, Ng RW, Yip TC, Wong GL, Chan FK, Wong CK, Chan PK, Ng SC. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698-706. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 995] [Cited by in RCA: 861] [Article Influence: 215.3] [Reference Citation Analysis (0)] |
| 44. | Vodnar DC, Mitrea L, Teleky BE, Szabo K, Călinoiu LF, Nemeş SA, Martău GA. Coronavirus Disease (COVID-19) Caused by (SARS-CoV-2) Infections: A Real Challenge for Human Gut Microbiota. Front Cell Infect Microbiol. 2020;10:575559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 45. | Zuo T, Liu Q, Zhang F, Lui GC, Tso EY, Yeoh YK, Chen Z, Boon SS, Chan FK, Chan PK, Ng SC. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut. 2021;70:276-284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 262] [Article Influence: 65.5] [Reference Citation Analysis (0)] |
| 46. | Villapol S. Gastrointestinal symptoms associated with COVID-19: impact on the gut microbiome. Transl Res. 2020;226:57-69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 194] [Cited by in RCA: 217] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 47. | Dhar D, Mohanty A. Gut microbiota and Covid-19- possible link and implications. Virus Res. 2020;285:198018. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 330] [Cited by in RCA: 395] [Article Influence: 79.0] [Reference Citation Analysis (0)] |
| 48. | Lehmann M, Allers K, Heldt C, Meinhardt J, Schmidt F, Rodriguez-Sillke Y, Kunkel D, Schumann M, Böttcher C, Stahl-Hennig C, Elezkurtaj S, Bojarski C, Radbruch H, Corman VM, Schneider T, Loddenkemper C, Moos V, Weidinger C, Kühl AA, Siegmund B. Human small intestinal infection by SARS-CoV-2 is characterized by a mucosal infiltration with activated CD8(+) T cells. Mucosal Immunol. 2021;14:1381-1392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 60] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 49. | Triana S, Metz-Zumaran C, Ramirez C, Kee C, Doldan P, Shahraz M, Schraivogel D, Gschwind AR, Sharma AK, Steinmetz LM, Herrmann C, Alexandrov T, Boulant S, Stanifer ML. Single-cell analyses reveal SARS-CoV-2 interference with intrinsic immune response in the human gut. Mol Syst Biol. 2021;17:e10232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 71] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 50. | Onodi F, Bonnet-Madin L, Meertens L, Karpf L, Poirot J, Zhang SY, Picard C, Puel A, Jouanguy E, Zhang Q, Le Goff J, Molina JM, Delaugerre C, Casanova JL, Amara A, Soumelis V. SARS-CoV-2 induces human plasmacytoid predendritic cell diversification via UNC93B and IRAK4. J Exp Med. 2021;218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 105] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 51. | Guo Y, Luo R, Wang Y, Deng P, Song T, Zhang M, Wang P, Zhang X, Cui K, Tao T, Li Z, Chen W, Zheng Y, Qin J. SARS-CoV-2 induced intestinal responses with a biomimetic human gut-on-chip. Sci Bull (Beijing). 2021;66:783-793. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 102] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 52. | Livanos AE, Jha D, Cossarini F, Gonzalez-Reiche AS, Tokuyama M, Aydillo T, Parigi TL, Ladinsky MS, Ramos I, Dunleavy K, Lee B, Dixon RE, Chen ST, Martinez-Delgado G, Nagula S, Bruce EA, Ko HM, Glicksberg BS, Nadkarni G, Pujadas E, Reidy J, Naymagon S, Grinspan A, Ahmad J, Tankelevich M, Bram Y, Gordon R, Sharma K, Houldsworth J, Britton GJ, Chen-Liaw A, Spindler MP, Plitt T, Wang P, Cerutti A, Faith JJ, Colombel JF, Kenigsberg E, Argmann C, Merad M, Gnjatic S, Harpaz N, Danese S, Cordon-Cardo C, Rahman A, Schwartz RE, Kumta NA, Aghemo A, Bjorkman PJ, Petralia F, van Bakel H, Garcia-Sastre A, Mehandru S. Intestinal Host Response to SARS-CoV-2 Infection and COVID-19 Outcomes in Patients With Gastrointestinal Symptoms. Gastroenterology. 2021;160:2435-2450.e34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 124] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 53. | Chu H, Chan JF, Wang Y, Yuen TT, Chai Y, Shuai H, Yang D, Hu B, Huang X, Zhang X, Hou Y, Cai JP, Zhang AJ, Zhou J, Yuan S, To KK, Hung IF, Cheung TT, Ng AT, Hau-Yee Chan I, Wong IY, Law SY, Foo DC, Leung WK, Yuen KY. SARS-CoV-2 Induces a More Robust Innate Immune Response and Replicates Less Efficiently Than SARS-CoV in the Human Intestines: An Ex Vivo Study With Implications on Pathogenesis of COVID-19. Cell Mol Gastroenterol Hepatol. 2021;11:771-781. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 54. | Zheng H, Chen Y, Li J, Li H, Zhao X, Yang F, Li Y, Liu C, Qin L, Zuo Y, Zhang Q, He Z, Shi H, Li Q, Liu L. Longitudinal analyses reveal distinct immune response landscapes in lung and intestinal tissues from SARS-CoV-2-infected rhesus macaques. Cell Rep. 2022;39:110864. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 55. | Negishi H, Taniguchi T, Yanai H. The Interferon (IFN) Class of Cytokines and the IFN Regulatory Factor (IRF) Transcription Factor Family. Cold Spring Harb Perspect Biol. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 285] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 56. | Rojas JM, Alejo A, Martín V, Sevilla N. Viral pathogen-induced mechanisms to antagonize mammalian interferon (IFN) signaling pathway. Cell Mol Life Sci. 2021;78:1423-1444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 57. | Snell LM, McGaha TL, Brooks DG. Type I Interferon in Chronic Virus Infection and Cancer. Trends Immunol. 2017;38:542-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 344] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 58. | Langer V, Vivi E, Regensburger D, Winkler TH, Waldner MJ, Rath T, Schmid B, Skottke L, Lee S, Jeon NL, Wohlfahrt T, Kramer V, Tripal P, Schumann M, Kersting S, Handtrack C, Geppert CI, Suchowski K, Adams RH, Becker C, Ramming A, Naschberger E, Britzen-Laurent N, Stürzl M. IFN-γ drives inflammatory bowel disease pathogenesis through VE-cadherin-directed vascular barrier disruption. J Clin Invest. 2019;129:4691-4707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 177] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 59. | Sayoc-Becerra A, Krishnan M, Fan S, Jimenez J, Hernandez R, Gibson K, Preciado R, Butt G, McCole DF. The JAK-Inhibitor Tofacitinib Rescues Human Intestinal Epithelial Cells and Colonoids from Cytokine-Induced Barrier Dysfunction. Inflamm Bowel Dis. 2020;26:407-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 74] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 60. | Lo Presti E, Mocciaro F, Mitri RD, Corsale AM, Di Simone M, Vieni S, Scibetta N, Unti E, Dieli F, Meraviglia S. Analysis of colon-infiltrating γδ T cells in chronic inflammatory bowel disease and in colitis-associated cancer. J Leukoc Biol. 2020;108:749-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 61. | Wallace JW, Constant DA, Nice TJ. Interferon Lambda in the Pathogenesis of Inflammatory Bowel Diseases. Front Immunol. 2021;12:767505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 62. | Hemann EA, Gale M Jr, Savan R. Interferon Lambda Genetics and Biology in Regulation of Viral Control. Front Immunol. 2017;8:1707. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 63. | Hopfner KP, Hornung V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat Rev Mol Cell Biol. 2020;21:501-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 1218] [Article Influence: 243.6] [Reference Citation Analysis (0)] |
| 64. | Krammer S, Sicorschi Gutu C, Grund JC, Chiriac MT, Zirlik S, Finotto S. Regulation and Function of Interferon-Lambda (IFNλ) and Its Receptor in Asthma. Front Immunol. 2021;12:731807. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 65. | Sui H, Hao M, Chang W, Imamichi T. The Role of Ku70 as a Cytosolic DNA Sensor in Innate Immunity and Beyond. Front Cell Infect Microbiol. 2021;11:761983. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 66. | Kim YM, Shin EC. Type I and III interferon responses in SARS-CoV-2 infection. Exp Mol Med. 2021;53:750-760. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 190] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 67. | Alase AA, El-Sherbiny YM, Vital EM, Tobin DJ, Turner NA, Wittmann M. IFNλ Stimulates MxA Production in Human Dermal Fibroblasts via a MAPK-Dependent STAT1-Independent Mechanism. J Invest Dermatol. 2015;135:2935-2943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 68. | Nice TJ, Robinson BA, Van Winkle JA. The Role of Interferon in Persistent Viral Infection: Insights from Murine Norovirus. Trends Microbiol. 2018;26:510-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 69. | Ingle H, Peterson ST, Baldridge MT. Distinct Effects of Type I and III Interferons on Enteric Viruses. Viruses. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 70. | Bhushal S, Wolfsmüller M, Selvakumar TA, Kemper L, Wirth D, Hornef MW, Hauser H, Köster M. Cell Polarization and Epigenetic Status Shape the Heterogeneous Response to Type III Interferons in Intestinal Epithelial Cells. Front Immunol. 2017;8:671. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 71. | Felgenhauer U, Schoen A, Gad HH, Hartmann R, Schaubmar AR, Failing K, Drosten C, Weber F. Inhibition of SARS-CoV-2 by type I and type III interferons. J Biol Chem. 2020;295:13958-13964. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 194] [Cited by in RCA: 188] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 72. | McElrath C, Espinosa V, Lin JD, Peng J, Sridhar R, Dutta O, Tseng HC, Smirnov SV, Risman H, Sandoval MJ, Davra V, Chang YJ, Pollack BP, Birge RB, Galan M, Rivera A, Durbin JE, Kotenko SV. Critical role of interferons in gastrointestinal injury repair. Nat Commun. 2021;12:2624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 73. | Henden AS, Koyama M, Robb RJ, Forero A, Kuns RD, Chang K, Ensbey KS, Varelias A, Kazakoff SH, Waddell N, Clouston AD, Giri R, Begun J, Blazar BR, Degli-Esposti MA, Kotenko SV, Lane SW, Bowerman KL, Savan R, Hugenholtz P, Gartlan KH, Hill GR. IFN-λ therapy prevents severe gastrointestinal graft-versus-host disease. Blood. 2021;138:722-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 74. | Stanifer ML, Kee C, Cortese M, Zumaran CM, Triana S, Mukenhirn M, Kraeusslich HG, Alexandrov T, Bartenschlager R, Boulant S. Critical Role of Type III Interferon in Controlling SARS-CoV-2 Infection in Human Intestinal Epithelial Cells. Cell Rep. 2020;32:107863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 258] [Article Influence: 51.6] [Reference Citation Analysis (0)] |
| 75. | Busnadiego I, Fernbach S, Pohl MO, Karakus U, Huber M, Trkola A, Stertz S, Hale BG. Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2. mBio. 2020;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 123] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 76. | Metz-Zumaran C, Kee C, Doldan P, Guo C, Stanifer ML, Boulant S. Increased Sensitivity of SARS-CoV-2 to Type III Interferon in Human Intestinal Epithelial Cells. J Virol. 2022;96:e0170521. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 77. | Vanderheiden A, Ralfs P, Chirkova T, Upadhyay AA, Zimmerman MG, Bedoya S, Aoued H, Tharp GM, Pellegrini KL, Manfredi C, Sorscher E, Mainou B, Lobby JL, Kohlmeier JE, Lowen AC, Shi PY, Menachery VD, Anderson LJ, Grakoui A, Bosinger SE, Suthar MS. Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures. J Virol. 2020;94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 242] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 78. | Sohn SY, Hearing J, Mugavero J, Kirillov V, Gorbunova E, Helminiak L, Mishra S, Mackow E, Hearing P, Reich NC, Kim HK. Interferon-Lambda Intranasal Protection and Differential Sex Pathology in a Murine Model of SARS-CoV-2 Infection. mBio. 2021;12:e0275621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 79. | Cardoso NP, Mansilla FC, Benedetti E, Turco CS, Barone LJ, Iserte JA, Soria I, Baumeister E, Capozzo AV. Bovine Interferon Lambda Is a Potent Antiviral Against SARS-CoV-2 Infection in vitro. Front Vet Sci. 2020;7:603622. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 80. | Egli A, Santer DM, O'Shea D, Tyrrell DL, Houghton M. The impact of the interferon-lambda family on the innate and adaptive immune response to viral infections. Emerg Microbes Infect. 2014;3:e51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 122] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 81. | Coto-Llerena M, Lepore M, Spagnuolo J, Di Blasi D, Calabrese D, Suslov A, Bantug G, Duong FH, Terracciano LM, De Libero G, Heim MH. Interferon lambda 4 can directly activate human CD19(+) B cells and CD8(+) T cells. Life Sci Alliance. 2021;4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 82. | Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol. 2011;29:71-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1100] [Cited by in RCA: 1414] [Article Influence: 101.0] [Reference Citation Analysis (0)] |
| 83. | Lee S, Baldridge MT. Interferon-Lambda: A Potent Regulator of Intestinal Viral Infections. Front Immunol. 2017;8:749. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 84. | Syedbasha M, Bonfiglio F, Linnik J, Stuehler C, Wüthrich D, Egli A. Interferon-λ Enhances the Differentiation of Naive B Cells into Plasmablasts via the mTORC1 Pathway. Cell Rep. 2020;33:108211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 85. | Chen K, Shao LH, Wang F, Shen XF, Xia XF, Kang X, Song P, Wang M, Lu XF, Wang C, Hu QY, Liu S, Guan WX. Netting Gut Disease: Neutrophil Extracellular Trap in Intestinal Pathology. Oxid Med Cell Longev. 2021;2021:5541222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 86. | Hemann EA, Schwerk J, Savan R. IFN-λ 'guts' neutrophil-mediated inflammation. Nat Immunol. 2017;18:1061-1062. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 87. | Broggi A, Tan Y, Granucci F, Zanoni I. IFN-λ suppresses intestinal inflammation by non-translational regulation of neutrophil function. Nat Immunol. 2017;18:1084-1093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 88. | Santer DM, Minty GES, Golec DP, Lu J, May J, Namdar A, Shah J, Elahi S, Proud D, Joyce M, Tyrrell DL, Houghton M. Differential expression of interferon-lambda receptor 1 splice variants determines the magnitude of the antiviral response induced by interferon-lambda 3 in human immune cells. PLoS Pathog. 2020;16:e1008515. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 89. | Xu P, Becker H, Elizalde M, Pierik M, Masclee A, Jonkers D. Interleukin-28A induces epithelial barrier dysfunction in CD patient-derived intestinal organoids. Am J Physiol Gastrointest Liver Physiol. 2021;320:G689-G699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 90. | Günther C, Ruder B, Stolzer I, Dorner H, He GW, Chiriac MT, Aden K, Strigli A, Bittel M, Zeissig S, Rosenstiel P, Atreya R, Neurath MF, Wirtz S, Becker C. Interferon Lambda Promotes Paneth Cell Death Via STAT1 Signaling in Mice and Is Increased in Inflamed Ileal Tissues of Patients With Crohn's Disease. Gastroenterology. 2019;157:1310-1322.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 91. | Thoresen D, Wang W, Galls D, Guo R, Xu L, Pyle AM. The molecular mechanism of RIG-I activation and signaling. Immunol Rev. 2021;304:154-168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 147] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 92. | Bayer AL, Alcaide P. MyD88: At the heart of inflammatory signaling and cardiovascular disease. J Mol Cell Cardiol. 2021;161:75-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 55] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 93. | Fitzgerald KA, Kagan JC. Toll-like Receptors and the Control of Immunity. Cell. 2020;180:1044-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 529] [Cited by in RCA: 1395] [Article Influence: 279.0] [Reference Citation Analysis (0)] |
| 94. | Sui H, Chen Q, Imamichi T. Cytoplasmic-translocated Ku70 senses intracellular DNA and mediates interferon-lambda1 induction. Immunology. 2021;163:323-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |