Published online Feb 14, 2022. doi: 10.3748/wjg.v28.i6.683
Peer-review started: July 18, 2021
First decision: August 15, 2021
Revised: August 21, 2021
Accepted: January 19, 2022
Article in press: January 19, 2022
Published online: February 14, 2022
Processing time: 205 Days and 11.5 Hours
The intra and extracellular pathways of hepatic injury by coronavirus disease 2019 (COVID-19) are still being studied. Understanding them is important to treat this viral disease and other liver and biliary tract disorders. Thus, this paper aims to present three hypotheses about liver injury caused by COVID-19: (1) The interactions between severe acute respiratory syndrome coronavirus 2 spike protein and membrane receptors in the hepatocyte; (2) The dysbiosis and “gut-liver axis” disruption in patients with serious clinical presentations of COVID-19; and (3) The inflammatory response exacerbated through the production of interleukins such as interleukin-6. However, despite these new perspectives, the pathophysiological process of liver injury caused by COVID-19 is still complex and multifactorial. Thus, understanding all these variables is a challenge to science but also the key to propose individualized and effective patient therapies.
Core Tip: This paper aimed to present new hypotheses on the pathophysiology of liver injury caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Interactions between SARS-CoV-2 spike protein and other membrane receptors in the liver; “gut-liver axis” disruption and dysbiosis; and increased inflammatory process mediated by interleukin-6 and AT1R-metalloprotease 17 seem to be factors that contribute to such injury.
- Citation: Gonçalves Júnior J. COVID-19, liver dysfunction and pathophysiology: A conceptual discussion. World J Gastroenterol 2022; 28(6): 683-688
- URL: https://www.wjgnet.com/1007-9327/full/v28/i6/683.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i6.683
I have read the work of Prof. Gracia-Ramos et al[1] about the clinical aspects of the relationship between liver dysfunction and coronavirus disease 2019 (COVID-19). The author aimed to summarize the pathophysiology, clinical importance, and management of COVID-19 in patients with or without preexisting liver disease.
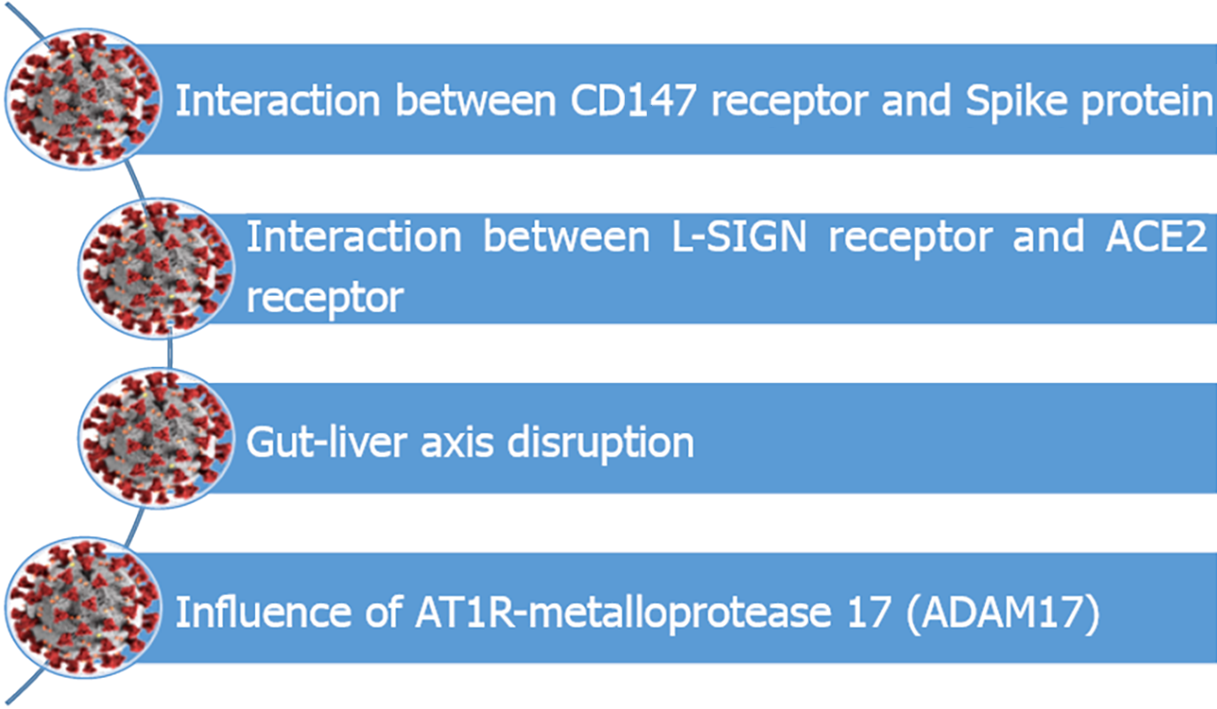
I would like to highlight some hypotheses for the pathophysiological impairment of the liver in COVID-19. To facilitate visualization, I have summarized the findings in Figure 1. I believe the information provided will enrich the current discussion and may enhance the results of the aforementioned paper[1].
The first theory states that liver cells have two receptors that have an affinity with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein. The first receptor is the Cluster of Differentiation 147 (CD147) or basigin (BSG) or Extracellular Matrix Metalloproteinase Inducer (EMMPRIN)[2], and the second receptor is the Liver/Lymph node-specific intercellular adhesion molecule-3-grabbing non-integrin (L-SIGN)[3].
CD147 is a transmembrane glycoprotein of the immunoglobulin superfamily overexpressed in an inflammatory process triggered by viral infections (e.g., Severe Acute Respiratory Syndrome in 2002), bacterial infections, and parasitic infections (e.g., Plasmodium falciparum)[2] (Figure 1). Evidence of CD147 protein expression in the liver tissue was found in 1999[4]. Recently, a United States publication in Nature journal highlighted the possibility that a chimeric anti-CD147 receptor would be a possible treatment for hepatocellular carcinoma[5]. Experimental research has shown affinity between CD147 and SARS-CoV-2 spike protein. A Chinese study published in Nature journal evaluating the in vitro association between the CD147 receptor and the SARS-CoV-2 spike protein by enzyme-linked immunosorbent assay and plasmon resonance demonstrated an affinity of 1.85 × 10–7 Michaelis between them. In parallel, the authors demonstrated that in cell cultures, when the CD147 protein is blocked by specific autoantibodies (e.g., meplazumab), SARS-CoV-2 amplification is inhibited. Additionally, the virus was able to enter into naturally non-susceptible cells (e.g., cells of baby hamster lineage) more easily when CD147 expression was induced in this population[6]. Moreover, increased expression of CD147 in tissues outside the lung has also been shown as an alternative pathway for SARS-CoV-2 infection in bioinformatics studies[7] and systematic reviews[8]. Therefore, an interesting hypothesis would be that the affinity between the CD147 receptor and the SARS-CoV-2 spike protein represents another way for the virus to infect liver cells.
L-SIGN is a liver-specific membrane receptor related to viral capture[3]. L-SIGN is already widely studied in diseases that affect the liver, such as diseases caused by the hepatitis C virus, the human immunodeficiency virus, the Rift Valley fever virus, the Uukuniemi virus, and the Toscana virus[9]. A recent study supports this hypothesis by suggesting that L-SIGN may provide a new way for SARS-CoV-2 to enter human cells[10]. In COVID-19, autopsy studies showed that SARS-CoV-2-infected hepatic sinusoid cells expressed more L-SIGN receptors compared to control groups[11]. Besides that, the literature has shown in vitro interactions between L-SIGN and the spike protein[12,13], in which this receptor binds to angiotensin II (ACE2), increasing the capacity of SARS-CoV-2 to infect liver cells[8,14] (Figure 1).
The second hypothesis (Figure 1) highlights the apparent “gut–liver axis disruption”[15] caused by COVID-19. More than half (60%) of the patients infected with SARS-CoV-2 developed liver injury[16], and some studies have already shown that the hepatic clearance of toxins is negatively impacted by COVID-19[17]. This shows there are varying degrees of dysfunction. Past infections by H1N1[15] and SARS-like viruses[18] led to severe cytopathic alterations in the gastrointestinal tract within 48 h of the beginning of the infectious process[19]. Thus, the disruption of this cross-talk would lead to two consequences: (1) Bacterial translocation stimulating septic shock; and (2) Perpetuation of the septic shock that would lead to a worsened ischemic state[15,20].
The literature has shown a decrease in commensal bacteria and pathogenic microorganisms in patients with COVID-19. This situation persists even after the absence of symptoms and an undetectable viral load by reverse transcription-polymerase chain reaction. Increased colony-forming units of opportunistic bacteria, such as Veillonella spp., Rothia spp., Actinomyces spp.[20], Faecalibacterium prausnitzii, Clostridium ramosum, and Clostridium hathewayi, in the fecal sample of patients with COVID-19, have been associated with more severe illness by SARS-CoV-2[19]. Furthermore, subspecies of Bacteroides sp. (which decrease ACE2 expression in murine intestine models), when present in human fecal samples, have been correlated with a lower viral load of SARS-CoV-2[21,22]. Besides that, epidemiological studies[23] and meta-analyses[24] have shown that COVID-19 can cause cellular dysfunction in enterocytes. More than half (54%) of the patients infected with COVID-19 had SARS-CoV-2 RNA in their fecal samples in a Chinese study[25]. A paper published by Mazza et al[26] demonstrated the presence of fecal calprotectin in a patient infected with COVID-19 showing direct damage to the gastric mucosa. Thus, the disruption of the gastric mucosa feeds back the “cytokine storm” caused by COVID-19 and can lead to hepatic tissue injury[27].
Parohan et al[28], when analyzing 3428 patients with COVID-19, demonstrated a significant increase in serum aspartate aminotransferase, alanine aminotransferase, and total bilirubin levels with lower levels of albumin in critically ill patients. An epidemiological survey showed that 62% of the patients admitted to intensive care units (ICUs) had increased liver enzymes. Furthermore, in these ICUs, patients had higher values of pro-inflammatory cytokines such as interleukins (IL) 10, 7, 2; monocyte chemoattractant protein-1 (MCP1); gamma induced protein 10 (IP-10); granulocyte colony-stimulating factor (GCSF); and tumor necrosis factor α (TNF-α) when compared to their controls not admitted to ICUs[29]. Indeed, autopsy studies of patients with severe acute respiratory syndrome caused by COVID-19 showed centrilobular sinusoidal dilation and lobular infiltration by small lymphocytes[30]. Percutaneous liver biopsy of patients infected with coronavirus showed histopathological findings suggestive of liver injury, such as acidophilic bodies, hepatocyte ballooning, and lobular activity without fibrin deposition or fibrosis[16].
The third theory is that SARS-CoV-2 endocytosis by immune system cells is caused by AT1R-metalloprotease 17 (ADAM17), which is also involved in the genesis of liver injury (Figure 1). The mechanism by which ADAM17 facilitates viral entry is not yet known. However, it is known that the increase in its activity can lead to the cleavage of pro-inflammatory molecules (e.g., IL-6; TNF-α), reinforcing the inflammatory process and injury to various organs, including the liver, during SARS-CoV-2 infection[31,32]. Additionally, ADAM17 breaks down several proteins that are responsible for liver regeneration/protection. Among ADAM17 substrates are the epidermal growth factor receptor (EGFR) ligand amphiregulin (AR), the heparin-binding-EGF-like growth factor (HB-EGF), and the hepatocyte growth factor (HGF). ADAM17 deletion in cell cultures of hepatocytes led to a decrease in EGFR and HB-EGF (responsible for preventing liver injury). These molecules increased the apoptosis of hepatocytes and decreased their proliferation[33].
Interestingly, studies have shown increased serum levels of ADAMS17 in comorbidities known to be risk factors for severe cases of COVID-19, such as heart failure[34], COPD[35], diabetes mellitus[36], kidney disease[37], and increasing age[34]. On the other hand, decreased ADAM17 activity is correlated with decreased ACE2 receptors, thus having a protective effect against SARS-CoV-2 infections[38].
Therefore, the pathophysiological process of liver injury caused by COVID-19 is complex, multifactorial, and extensive. There are many (intra and extracellular) inflammatory pathways we are not yet aware of, in addition to local and systemic environmental factors that interfere. Understanding all these variables is a challenge to science. Additionally, only with this understanding, we will be able to propose individualized and effective therapies.
The author is grateful to Hospital das Clínicas, Faculdade de Medicina, Universidade de São Paulo, SP, Brazil.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Ssekandi AM S-Editor: Fan JR L-Editor: Filipodia P-Editor: Fan JR
| 1. | Gracia-Ramos AE, Jaquez-Quintana JO, Contreras-Omaña R, Auron M. Liver dysfunction and SARS-CoV-2 infection. World J Gastroenterol. 2021;27:3951-3970. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 21] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 2. | Xiong L, Edwards CK 3rd, Zhou L. The biological function and clinical utilization of CD147 in human diseases: a review of the current scientific literature. Int J Mol Sci. 2014;15:17411-17441. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 167] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 3. | Jeffers SA, Tusell SM, Gillim-Ross L, Hemmila EM, Achenbach JE, Babcock GJ, Thomas WD Jr, Thackray LB, Young MD, Mason RJ, Ambrosino DM, Wentworth DE, Demartini JC, Holmes KV. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci U S A. 2004;101:15748-15753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 497] [Cited by in RCA: 467] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 4. | Frayne J, Ingram C, Love S, Hall L. Localisation of phosphatidylethanolamine-binding protein in the brain and other tissues of the rat. Cell Tissue Res. 1999;298:415-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 5. | Prince KC, Diviacco B. On "Coherent control in the extreme ultraviolet and attosecond regime by synchrotron radiation" by Hikosaka et al, Nat. Comm. 10, 4988 (2019). Nat Commun. 2021;12:3784. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 6. | Wang K, Chen W, Zhang Z, Deng Y, Lian JQ, Du P, Wei D, Zhang Y, Sun XX, Gong L, Yang X, He L, Zhang L, Yang Z, Geng JJ, Chen R, Zhang H, Wang B, Zhu YM, Nan G, Jiang JL, Li L, Wu J, Lin P, Huang W, Xie L, Zheng ZH, Zhang K, Miao JL, Cui HY, Huang M, Zhang J, Fu L, Yang XM, Zhao Z, Sun S, Gu H, Wang Z, Wang CF, Lu Y, Liu YY, Wang QY, Bian H, Zhu P, Chen ZN. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther. 2020;5:283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 408] [Cited by in RCA: 747] [Article Influence: 149.4] [Reference Citation Analysis (0)] |
| 7. | He C, Hua X, Sun S, Li S, Wang J, Huang X. Integrated Bioinformatic Analysis of SARS-CoV-2 Infection Related Genes ACE2, BSG and TMPRSS2 in Aerodigestive Cancers. J Inflamm Res. 2021;14:791-802. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 8. | Qiao J, Li W, Bao J, Peng Q, Wen D, Wang J, Sun B. The expression of SARS-CoV-2 receptor ACE2 and CD147, and protease TMPRSS2 in human and mouse brain cells and mouse brain tissues. Biochem Biophys Res Commun. 2020;533:867-871. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 9. | Léger P, Tetard M, Youness B, Cordes N, Rouxel RN, Flamand M, Lozach PY. Differential Use of the C-Type Lectins L-SIGN and DC-SIGN for Phlebovirus Endocytosis. Traffic. 2016;17:639-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 55] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 10. | Raghav PK, Kalyanaraman K, Kumar D. Human cell receptors: potential drug targets to combat COVID-19. Amino Acids. 2021;53:813-842. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 11. | Kondo Y, Larabee JL, Gao L, Shi H, Shao B, Hoover CM, McDaniel JM, Ho YC, Silasi-Mansat R, Archer-Hartmann SA, Azadi P, Srinivasan RS, Rezaie AR, Borczuk A, Laurence JC, Lupu F, Ahamed J, McEver RP, Papin JF, Yu Z, Xia L. L-SIGN is a receptor on liver sinusoidal endothelial cells for SARS-CoV-2 virus. JCI Insight. 2021;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 12. | Gao C, Zeng J, Jia N, Stavenhagen K, Matsumoto Y, Zhang H, Li J, Hume AJ, Mühlberger E, van Die I, Kwan J, Tantisira K, Emili A, Cummings RD. SARS-CoV-2 Spike protein interacts with multiple innate immune receptors. 2020 Preprint. Available from: bioRxiv:2020.07.29.227462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 13. | Zhang Q, Xiang R, Huo S, Zhou Y, Jiang S, Wang Q, Yu F. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct Target Ther. 2021;6:233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 206] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 14. | Gadanec LK, McSweeney KR, Qaradakhi T, Ali B, Zulli A, Apostolopoulos V. Can SARS-CoV-2 Virus Use Multiple Receptors to Enter Host Cells? Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 114] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 15. | Gu S, Chen Y, Wu Z, Gao H, Lv L, Guo F, Zhang X, Luo R, Huang C, Lu H, Zheng B, Zhang J, Yan R, Zhang H, Jiang H, Xu Q, Guo J, Gong Y, Tang L, Li L. Alterations of the Gut Microbiota in Patients With Coronavirus Disease 2019 or H1N1 Influenza. Clin Infect Dis. 2020;71:2669-2678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 324] [Cited by in RCA: 569] [Article Influence: 142.3] [Reference Citation Analysis (0)] |
| 16. | Chau TN, Lee KC, Yao H, Tsang TY, Chow TC, Yeung YC, Choi KW, Tso YK, Lau T, Lai ST, Lai CL. SARS-associated viral hepatitis caused by a novel coronavirus: report of three cases. Hepatology. 2004;39:302-310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 307] [Cited by in RCA: 305] [Article Influence: 14.5] [Reference Citation Analysis (1)] |
| 17. | Rodriguez-Morales AJ, Cardona-Ospina JA, Gutiérrez-Ocampo E, Villamizar-Peña R, Holguin-Rivera Y, Escalera-Antezana JP, Alvarado-Arnez LE, Bonilla-Aldana DK, Franco-Paredes C, Henao-Martinez AF, Paniz-Mondolfi A, Lagos-Grisales GJ, Ramírez-Vallejo E, Suárez JA, Zambrano LI, Villamil-Gómez WE, Balbin-Ramon GJ, Rabaan AA, Harapan H, Dhama K, Nishiura H, Kataoka H, Ahmad T, Sah R. Clinical, laboratory and imaging features of COVID-19: A systematic review and meta-analysis. Travel Med Infect Dis. 2020;34:101623. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1647] [Cited by in RCA: 1432] [Article Influence: 286.4] [Reference Citation Analysis (0)] |
| 18. | Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 691] [Cited by in RCA: 683] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 19. | Vodnar DC, Mitrea L, Teleky BE, Szabo K, Călinoiu LF, Nemeş SA, Martău GA. Coronavirus Disease (COVID-19) Caused by (SARS-CoV-2) Infections: A Real Challenge for Human Gut Microbiota. Front Cell Infect Microbiol. 2020;10:575559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 20. | Watson AJ, Hughes KR. TNF-α-induced intestinal epithelial cell shedding: implications for intestinal barrier function. Ann N Y Acad Sci. 2012;1258:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 103] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 21. | Amirian ES. Potential fecal transmission of SARS-CoV-2: Current evidence and implications for public health. Int J Infect Dis. 2020;95:363-370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 218] [Cited by in RCA: 197] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 22. | Zuo T, Zhang F, Lui GCY, Yeoh YK, Li AYL, Zhan H, Wan Y, Chung ACK, Cheung CP, Chen N, Lai CKC, Chen Z, Tso EYK, Fung KSC, Chan V, Ling L, Joynt G, Hui DSC, Chan FKL, Chan PKS, Ng SC. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology. 2020;159:944-955.e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 739] [Cited by in RCA: 1067] [Article Influence: 213.4] [Reference Citation Analysis (0)] |
| 23. | Jin X, Lian JS, Hu JH, Gao J, Zheng L, Zhang YM, Hao SR, Jia HY, Cai H, Zhang XL, Yu GD, Xu KJ, Wang XY, Gu JQ, Zhang SY, Ye CY, Jin CL, Lu YF, Yu X, Yu XP, Huang JR, Xu KL, Ni Q, Yu CB, Zhu B, Li YT, Liu J, Zhao H, Zhang X, Yu L, Guo YZ, Su JW, Tao JJ, Lang GJ, Wu XX, Wu WR, Qv TT, Xiang DR, Yi P, Shi D, Chen Y, Ren Y, Qiu YQ, Li LJ, Sheng J, Yang Y. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69:1002-1009. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 944] [Cited by in RCA: 871] [Article Influence: 174.2] [Reference Citation Analysis (0)] |
| 24. | Parasa S, Desai M, Thoguluva Chandrasekar V, Patel HK, Kennedy KF, Roesch T, Spadaccini M, Colombo M, Gabbiadini R, Artifon ELA, Repici A, Sharma P. Prevalence of Gastrointestinal Symptoms and Fecal Viral Shedding in Patients With Coronavirus Disease 2019: A Systematic Review and Meta-analysis. JAMA Netw Open. 2020;3:e2011335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 301] [Cited by in RCA: 302] [Article Influence: 60.4] [Reference Citation Analysis (0)] |
| 25. | Xie C, Jiang L, Huang G, Pu H, Gong B, Lin H, Ma S, Chen X, Long B, Si G, Yu H, Yang X, Shi Y, Yang Z. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int J Infect Dis. 2020;93:264-267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 225] [Cited by in RCA: 228] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 26. | Mazza S, Sorce A, Peyvandi F, Vecchi M, Caprioli F. A fatal case of COVID-19 pneumonia occurring in a patient with severe acute ulcerative colitis. Gut. 2020;69:1148-1149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 27. | Cardinale V, Capurso G, Ianiro G, Gasbarrini A, Arcidiacono PG, Alvaro D. Intestinal permeability changes with bacterial translocation as key events modulating systemic host immune response to SARS-CoV-2: A working hypothesis. Dig Liver Dis. 2020;52:1383-1389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 69] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 28. | Parohan M, Yaghoubi S, Seraji A. Liver injury is associated with severe coronavirus disease 2019 (COVID-19) infection: A systematic review and meta-analysis of retrospective studies. Hepatol Res. 2020;50:924-935. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 88] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 29. | Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35178] [Cited by in RCA: 30122] [Article Influence: 6024.4] [Reference Citation Analysis (3)] |
| 30. | Tian S, Xiong Y, Liu H, Niu L, Guo J, Liao M, Xiao SY. Pathological study of the 2019 novel coronavirus disease (COVID-19) through postmortem core biopsies. Mod Pathol. 2020;33:1007-1014. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 576] [Cited by in RCA: 656] [Article Influence: 131.2] [Reference Citation Analysis (0)] |
| 31. | Zipeto D, Palmeira JDF, Argañaraz GA, Argañaraz ER. ACE2/ADAM17/TMPRSS2 Interplay May Be the Main Risk Factor for COVID-19. Front Immunol. 2020;11:576745. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 201] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 32. | Li D, Ding X, Xie M, Tian D, Xia L. COVID-19-associated liver injury: from bedside to bench. J Gastroenterol. 2021;56:218-230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 33. | Al-Salihi M, Bornikoel A, Zhuang Y, Stachura P, Scheller J, Lang KS, Lang PA. The role of ADAM17 during liver damage. Biol Chem. 2021;402:1115-1128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 34. | Sama IE, Ravera A, Santema BT, van Goor H, Ter Maaten JM, Cleland JGF, Rienstra M, Friedrich AW, Samani NJ, Ng LL, Dickstein K, Lang CC, Filippatos G, Anker SD, Ponikowski P, Metra M, van Veldhuisen DJ, Voors AA. Circulating plasma concentrations of angiotensin-converting enzyme 2 in men and women with heart failure and effects of renin-angiotensin-aldosterone inhibitors. Eur Heart J. 2020;41:1810-1817. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 325] [Cited by in RCA: 336] [Article Influence: 67.2] [Reference Citation Analysis (0)] |
| 35. | Saheb Sharif-Askari N, Saheb Sharif-Askari F, Alabed M, Temsah MH, Al Heialy S, Hamid Q, Halwani R. Airways Expression of SARS-CoV-2 Receptor, ACE2, and TMPRSS2 Is Lower in Children Than Adults and Increases with Smoking and COPD. Mol Ther Methods Clin Dev. 2020;18:1-6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 209] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 36. | Fiorentino L, Vivanti A, Cavalera M, Marzano V, Ronci M, Fabrizi M, Menini S, Pugliese G, Menghini R, Khokha R, Lauro R, Urbani A, Federici M. Increased tumor necrosis factor alpha-converting enzyme activity induces insulin resistance and hepatosteatosis in mice. Hepatology. 2010;51:103-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 81] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 37. | Salem ES, Grobe N, Elased KM. Insulin treatment attenuates renal ADAM17 and ACE2 shedding in diabetic Akita mice. Am J Physiol Renal Physiol. 2014;306:F629-F639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 87] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 38. | Riera M, Anguiano L, Clotet S, Roca-Ho H, Rebull M, Pascual J, Soler MJ. Paricalcitol modulates ACE2 shedding and renal ADAM17 in NOD mice beyond proteinuria. Am J Physiol Renal Physiol. 2016;310:F534-F546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |