Published online Dec 28, 2022. doi: 10.3748/wjg.v28.i48.6875
Peer-review started: September 5, 2022
First decision: November 5, 2022
Revised: November 7, 2022
Accepted: November 27, 2022
Article in press: November 27, 2022
Published online: December 28, 2022
Processing time: 112 Days and 16.1 Hours
The coronavirus disease 2019 (COVID-19) pandemic is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Hepatic involvement is common in SARS-CoV-2-infected individuals. It is currently accepted that the direct and indirect hepatic effects of SARS-CoV-2 infection play a significant role in COVID-19. In individuals with pre-existing infectious and non-infectious liver disease, who are at a remarkably higher risk of developing severe COVID-19 and death, this pathology is most medically relevant. This review emphasizes the current pathways regarded as contributing to the gastrointestinal and hepatic ailments linked to COVID-19-infected patients due to an imbalanced interaction among the liver, systemic inflammation, disrupted coagulation, and the lung.
Core Tip: Clinical manifestations of coronavirus disease 2019 (COVID-19) may be triggered by the presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in the liver. SARS-CoV-2 genomic RNA and its replicative intermediates were found in liver tissues. SARS-CoV-2 causes direct cholangiocyte damage. Systemic inflammation due to COVID-19 correlated with the degree of acute liver injury as revealed by the rise in aspartate aminotransferase levels. SARS-CoV-2 infection increased the risk of morbidity and mortality in patients with a history of advanced liver disease.
- Citation: Quarleri J, Delpino MV. Molecular mechanisms implicated in SARS-CoV-2 liver tropism. World J Gastroenterol 2022; 28(48): 6875-6887
- URL: https://www.wjgnet.com/1007-9327/full/v28/i48/6875.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i48.6875
The coronavirus disease 2019 (COVID-19) pandemic is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Globally, as of 5 August 2022, there have been 579119505 confirmed cases of COVID-19, including 6457101 deaths, as reported by the World Health Organization[1].
Coronaviruses are enveloped with crown-shaped spike glycoprotein, positive-sense single-stranded RNA viruses that include three genera: alphacoronavirus, betacoronavirus, and gammacoronavirus, mainly related to respiratory infections. SARS-CoV-2 employs receptor recognition mechanisms comparable to those used by preceding virulent coronaviruses such as SARS-CoV, responsible for the SARS epidemic of 2003[2-5].
In addition to common respiratory symptoms, COVID-19 patients also present with gastrointestinal symptoms comprising diarrhea, nausea, and vomiting[6]. Anal swabs from COVID-19 patients test positive for genomic SARS-CoV-2 RNA, and the virus can be isolated from stool samples[7,8]. However, the possibility of fecal-oral transmission cannot be completely ruled out[9,10].
The entry of the virus into target cells is mediated by the coronavirus spike protein (S) that engages angiotensin-converting enzyme 2 (ACE2) expressed in multiple cell types allowing SARS-CoV-2 to infect different organs such as the nasopharynx, lungs, lymph nodes, kidney, stomach, small intestine, spleen, brain, and liver leading to multiple organ damage[11]. Cell entry also requires the trans-membrane serine protease 2 or other proteases[12]. The binding efficiency of the virus to ACE2 is a key determinant of transmissibility[13]. Different reports have revealed that SARS-CoV-2 has higher binding affinity to ACE2 than the previous SARS-CoV. This finding may in part explain the increased transmissibility of SARS-CoV-2, organ tropism, and ultimately multi-organ damage and mortality[14-16]. The mechanisms that could be involved in the multi-organ injury due to infection with SARS-CoV-2 comprise the damage of endothelial cells, dysregulation of the immune response, and an imbalance in the renin-angiotensin-aldosterone system (RAAS). The relative significance of these mechanisms in the pathophysiology of COVID-19 is at present not completely known. Although some of these mechanisms comprising ACE2-mediated viral entry and tissue injury with misbalance of the RAAS may be specific to COVID-19, immune pathogenesis produced by systemic delivery of cytokines and impaired microcirculatory function can also take place due to viral sepsis[17].
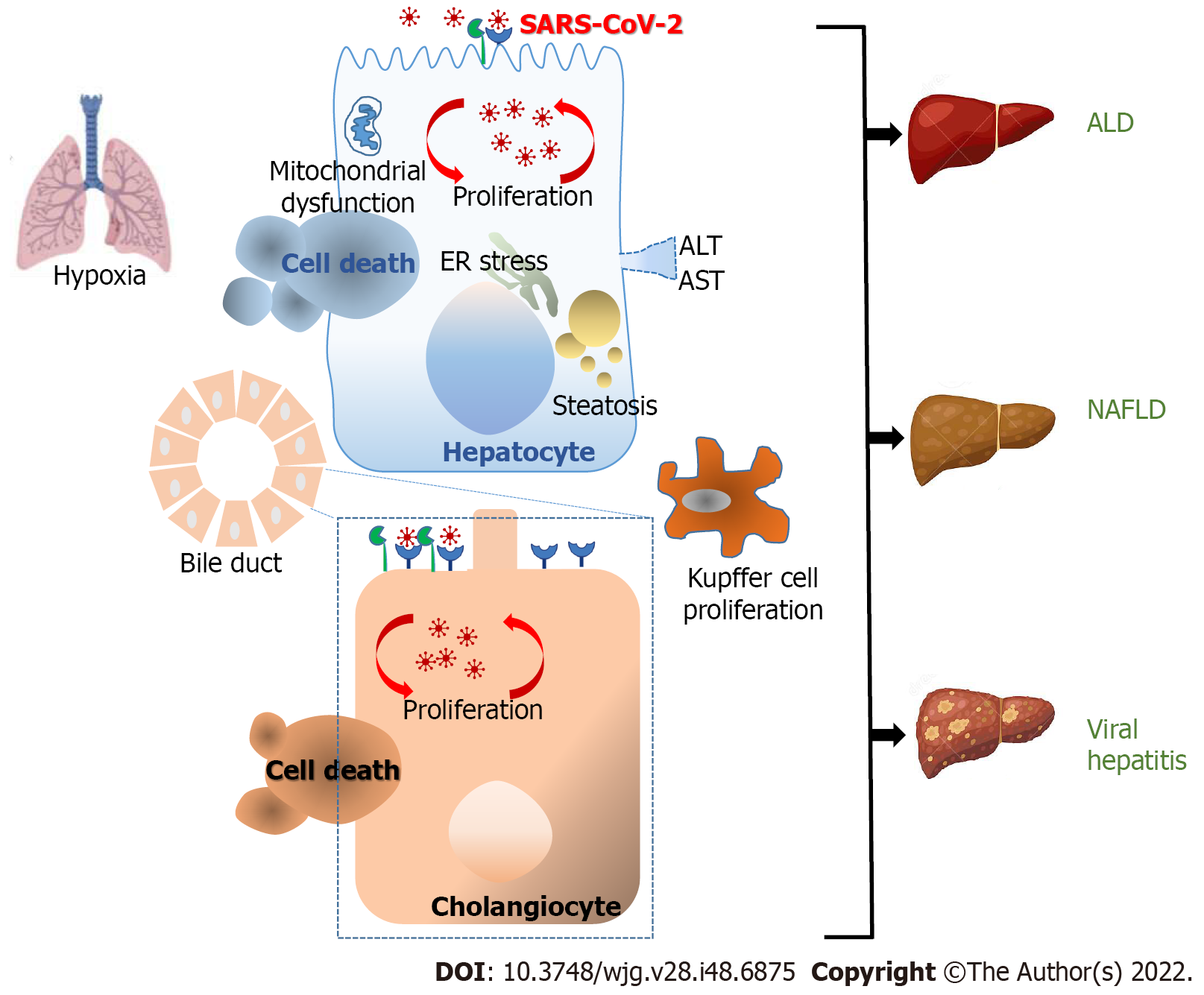
Hepatic involvement is common in SARS-CoV-2-infected patients. In cases with severe COVID-19 and to a lesser extent in mild/moderate COVID-19, some authors have reported a significant increase in alanine aminotransferase and aspartate aminotransferase (AST) indicating abnormal liver function[18]. Likewise, albumin decreased while alkaline phosphatase, gamma-glutamyl transferase (GGT), C-reactive protein (CRP), procalcitonin, ferritin, lactate dehydrogenase (LDH) as well as bilirubin levels were also significantly higher in severe than in other cases[19-21]. Patients without a history of liver illness were found to have abnormal liver test results[20]. These data suggests a direct connection between SARS-CoV-2 infection and digestive tract impairment. This review emphasizes the pathways currently regarded as contributing to the gastrointestinal and hepatic ailments linked to COVID-19.
To recognize the relevant literature, we employed a search and screening strategy. This process consisted of an extensive search of the online scientific database on the PubMed webpage using the most frequent synonyms to detect all possibly pertinent studies. In the following steps, references were analyzed to eliminate papers that were not relevant, and the remaining references were organized into categories for additional evaluation.
The liver coordinates an essential role in the host-microbe defense by assembling the portal and systemic circulation. Changes in the liver such as ductular fibrosis, hepatic steatosis, cholestasis, acute liver necrosis, and central vein thrombosis with concomitant lymphocytic infiltrate were detected in autopsies from COVID-19 deceased patients[22,23].
In line with previous reports, ACE2 is mostly expressed in cholangiocytes and to a lesser extent in hepatocytes. Accordingly, formalin-embedded liver tissues contain the SARS-CoV-2 RNA genome[24]. Other studies have stated that 2 of 3 autopsy livers carry the infectious virus, and 31 of 45 postmortem liver tissues contain the SARS-CoV-2 RNA genome[25]. Such studies that demonstrate the presence of SARS-CoV-2 RNA and proteins in liver cells are significant, as these may be found in the vascular lumen of blood vessels feeding the liver but also in portal vein endothelial cells, suggesting that the virus can also invade the liver through the circulation[26]. Postmortem liver biopsies have shown the presence of typical coronavirus particles in the cytoplasm of hepatocytes by electron microscopy[27], while hepatic parenchymal cells have also shown the presence of SARS-CoV-2 RNA[25]. Furthermore, the viral nucleocapsid protein has been detected in hepatic stem cells, hepatocytes, and cholangiocytes[28]. The presence of the SARS-CoV-2 RNA genome and its replicative intermediates in liver tissues has also been reported[28]. Most importantly, the nucleocapsid and the spike protein have been found in the liver 6 mo after recovery from COVID-19[29].
There is some proof that COVID-19 may be triggered by the presence of SARS-CoV-2 infection in the liver. However, other histological examinations revealed non-detectable viral particles in the liver and signs of significant hepatic damage[23,30]. This supports the notion that both direct and indirect pathways contribute to liver damage in the context of SARS-CoV-2 infection; more investigation is required to assess the importance of each pathway.
The concept that SARS-CoV-2 can reach the liver cells through the ACE2 receptor is supported by the fact that ACE2 is present in liver and bile duct cells[11]. Furthermore, recent research found that 59.7% of cholangiocytes and 2.6% of hepatocytes express ACE2. Likely, SARS-CoV-2 might infect cholangiocytes and cause liver damage since the amount of ACE2 found in cholangiocytes is similar to that reported in type 2 alveolar cells of the lung[31]. SARS-CoV-2-associated liver damage may be related to the presence of ACE2 receptors in cholangiocytes rather than hepatocytes[32]. Moreover, given the rich supply of blood to the liver from the small bowel, the SARS-CoV-2 virus can use the gut-liver way across the liver reticular system to reach the liver[33,34]. Taking into account that ACE2 was regarded as an interferon-inducible gene in human epithelial cells from respiratory tissues, the hepatocyte permissiveness for SARS-CoV-2 might be also modified when the viral receptor expression is increased after liver injury[35], but it would be because of the shortened isoform of ACE2, identified as deltaACE2, instead of the functional viral receptor[36].
In homeostasis, the bile acid released by hepatocytes into bile ducts is transported by cholangiocytes. The tight junction between cholangiocytes conserves the barrier function of the bile ductal epithelial cells, allowing bile acid collection and excretion. Besides, cholangiocytes play an important function in liver renewal and immune response[37]. Thus, the disruption of the cholangiocyte function can induce hepatobiliary injury. Previous reports have demonstrated that SARS-CoV-2 infection impairs the barrier through the modulation of tight junctions[38]. This effect could be attributed to the direct viral cytopathic effect on cholangiocytes. Likewise, SARS-CoV-2 infection induces the expression of apoptotic factors, including cluster of differentiation 40 (CD40), caspase recruitment domain family member 8, and serine/threonine kinase 4 in cholangiocytes[38]. Thus, SARS-CoV-2 causes direct cholangiocyte damage with subsequent liver homeostasis disruption in COVID-19 patients.
Liver biopsies from SARS-CoV-2 infected patients have revealed fatty degeneration, cellular infiltration, hepatocellular necrosis, increased ballooned hepatocytes, and mitotic cells. These findings are in line with the idea that liver damage in COVID-19 patients is caused by an indirect effect as a result of direct viral cholangiocyte damage and subsequent bile acid accumulation. However, it remains uncertain whether hepatic involvement during SARS-CoV-2 infection reveals direct cytopathic effects of the virus, ischemia and hypoxic reperfusion-related injury, exacerbated immune response responsible for the systemic inflammatory response syndrome, or a combination of mechanisms that have not been completely elucidated until now[39].
SARS-CoV-2 infection is related with an acute phase response characterized by the secretion of very high levels of proinflammatory cytokines in conjunction with CRP and ferritin[40]. The mechanisms involved in this “cytokine storm” are not completely elucidated. The proinflammatory response appears to be associated with the ability of the SARS-CoV-2 spike protein to activate C-type lectins and 20 family member 2 on innate immune cells[41]. A transcriptome analysis of 284 samples from 196 SARS-CoV-2-infected patients revealed that diverse peripheral immune alterations are associated with clinical characteristics comprising severity and disease phase of COVID-19. The increased expression and release of S100A8/A9 during inflammation exerts a critical role in controlling the inflammatory reaction by inducing leukocyte recruitment and stimulating cytokine release. These are calcium-binding proteins constitutively expressed as a heterodimer in neutrophils and monocytes, and they are the most important platelet-derived activators of endothelial cells. Additionally, SARS-CoV-2 RNA was detected in different epithelial and immune cells, followed by transcription alterations within virus-positive cells[42].
One of the mechanisms that encourage platelet adhesion to inflamed vascular endothelium is the early adhesive events that stimulate platelets to “roll” along endothelium. Among them is the interaction between the main platelet membrane receptor, GP1b (CD42)-1X-V, and von Willebrand factor secreted by endothelial cells. This phenomenon is strengthened by interactions between upregulated CD62P expression, which is present in both cell types and its counter receptor, P selectin glycoprotein ligand-1, which is also present in both cell types, although weakly in platelets. In a similar manner, the SARS-CoV-2 spike protein bound to the CD42b receptor to activate platelets via two different signaling pathways and stimulated platelet-monocyte interaction by engaging P selectin glycoprotein ligand-1 (PGSL-1) and CD40 ligand (CD40L)/CD40, which causes monocytes to produce proinflammatory cytokines. These findings demonstrate the correlation between hypercoagulation, monocyte activation, and a cytokine storm in patients severely affected.
The contact between the spike protein of SARS-CoV-2 and CD42 activates platelets and stimulates platelet-monocyte interactions through CD40L/CD40 and P-selectin/PGSL-1, contributing to hypercytokine secretion by monocytes[43]. Additionally, systemic inflammation is evidenced by a complement activation induced by the interferon (IFN)-Janus Kinase 1/2 (JAK1/2)-signal transducer and activator of transcription 1 (STAT1) signaling pathway[44].
The significance of the IFN pathways has been revealed in postmortem analyses of deceased patients due to severe COVID-19, where the livers displayed a significant induction of type I and II IFN responses and their related-JAK-STAT signaling pathways[25].
Moreover, the inflammation and cytokine stimulation observed in SARS-CoV-2 disease can contribute to endothelial injury and vascular damage, revealed by hypercoagulation, and arterial and venous embolism with the activation of immune cells and platelets, leading to clot formation[45,46]. High levels of CRP, which are indicative of acute liver injury, are correlated with the degree of systemic inflammation[47,48]. Increased levels of high-sensitive CRP, interleukin 6 (IL-6), and ferritin in COVID-19 patients are indicative of systemic inflammation, which correlates with the degree of acute liver injury as determined by the rise in AST levels[49]. Through the downregulation of hepatobiliary uptake and deficiencies in the excretory systems, the massive systemic inflammation contributes to hepatocellular cholestasis in the late stages of the disease[50]. Thus, it can be conclusively stated that SARS-CoV-2 infection is accompanied by a “cytokine storm” with a high proinflammatory cytokine profile that causes hepatic injury. It is obvious that systemic inflammation may contribute to acute liver damage, but SARS-CoV-2 infection cannot be ruled out directly. This theory is supported by the observation that liver damage appears early in the course of infection, as indicated by an increase in AST levels.
Since the onset of the COVID-19 pandemic, an increased risk of morbidity and mortality was observed among SARS-CoV-2-infected patients with a history of advanced liver disease[51]. Data obtained from multicenter studies have revealed a higher possibility of hospitalization and risk of death compared to patients without chronic liver disease[52]. To date, the mechanisms linking both pathologies are unknown. However, it has been proposed that the prothrombotic alteration caused by COVID-19 upsets the delicate homeostatic balance of cirrhotic patients, leading to venous microthrombosis and parenchymal dysfunction along with subsequent respiratory failures[53] (Figure 1).
Patients with viral hepatitis are more likely to experience liver damage, according to data from previous SARS-CoV infections. The severity of liver disease and a worse prognosis are associated with SARS-CoV-2 superinfection in patients with chronic hepatitis B virus (HBV) infection. According to a study of 105 patients with both chronic HBV infection and SARS-CoV-2, the risk of complications such as acute chronic liver failure, acute cardiac injury, and shock was higher in co-infected patients than in patients who only had the SARS-CoV-2 virus[54]. Consequently, patients with liver damage had a higher mortality rate (28.6%) than patients without liver damage (3.3%), and the treatment of COVID-19 in co-infected individuals had a significant negative impact on liver function[54]. A higher risk of abnormal liver function was found in inactive HBV carriers in a retrospective study that included 133 hospitalized patients with confirmed COVID-19, 116 of whom tested negative for serum hepatitis B antigen, and 17 were HBV inactive carriers. Hepatocytes and the immune response, as shown by the production of IL-6, D-dimer, and LDH are involved in the liver damage observed in SARS-CoV-2/HBV[55]. Moreover, chronic HBV infection-induced immune dysfunction may be a key factor in the development of COVID-19. Due to persistent viral antigens, studies have shown that chronic HBV infection is linked to the depletion of virus-specific CD4+ and CD8+ T cells[56]. Interleukin (IL-2) and tumor necrosis factor alpha (TNF-α) release are particularly hampered by HBV-specific exhausted T lymphocytes, resulting in progressively diminished antiviral function[57]. IL-2, IL-6, and TNF-α are among the proinflammatory cytokines overproduced as a result of the excessive immunological response to SARS-CoV-2 infection (cytokine storm), which is to our knowledge a significant factor associated with disease severity and mortality[58]. In this situation, it is conceivable that immunosuppression and depletion of HBV-specific T lymphocytes might prevent an excessive immunological response to SARS-CoV-2 and lessen the cytokine storm, resulting in milder illness. Although HBV reactivation is a potential side result of SARS-CoV-2 infection, the overall risk is minimal. Reactivation is primarily described as an abrupt and quick rise in HBV DNA levels in individuals with a history of detectable HBV DNA or recurrence of HBV DNA viremia in those with previous undetectable viral DNA[59]. Immunosuppressive therapy such as IL-6 receptor antagonists, IL-1 receptor antagonist, and high-dose corticosteroids are frequently used to treat HBV reactivation[60]. These treatments may be utilized in severe COVID-19 patients to manage the cytokine storm and to lessen the immune-mediated multiorgan damage. The impaired balance between the host’s immune system and viral replication is the main cause of the progression to HBV reactivation after infection with SARS-CoV-2. The dosage of glucocorticoids or immunosuppressive medications is a major risk factor for the reactivation of HBV during the treatment of COVID-19, together with the host baseline virological markers[61].
The COVID-19 pandemic may delay the global commitment to eradicate HBV infection for several years because of the decrease in chronic hepatitis B prevention, diagnosis, and treatment. According to estimates, during the COVID-19 pandemic, half of low- and middle-income families were unable to access healthcare facilities for the diagnosis, clinical evaluation, and treatment of HBV. This was primarily due to travel restrictions, job and income losses, and patients’ fear of contracting SARS-CoV-2[62]. Only 18% of the patients with hepatocellular carcinoma (HCC) and 32% of those with decom-pensated cirrhosis had continuity of care, while 23% of the health centers postponed HBV infection therapy initiation[63].
Regarding hepatitis C virus (HCV), some studies have revealed that corticosteroid-treated individuals might experience significant viral reactivation, which is mostly caused by immunosuppression[64-66]. Steroid therapy can lead to HCV reactivation through two different mechanisms: first, they increase the capacity of the virus to replicate by upregulating two essential HCV entry factors: occludin and scavenger receptor class B type I; and second, they do so by inhibiting the immune response against the virus, including T lymphocytes and plasmacytoid dendritic cells[67-69]. In individuals with persistent HCV infection, corticosteroid therapy can aggravate the course. Evidence suggests that HCV viremia rises in response to corticosteroids and falls back to normal levels in response to their cessation[70]. Therefore, it is best to avoid using corticosteroids in individuals with HCV infection[71].
However, the synergism between SARS-CoV-2 and chronic viral HBV and HCV has not been clearly elucidated. Hence, more widespread studies are required to evaluate the use of this therapy.
Currently, decompensated liver disease and HCC cause 2 million deaths annually as a result of liver cirrhosis, which affects 112 million people globally[72]. High COVID-19 death rates in cirrhotic patients have been found in numerous recent reports[73,74]. The baseline Child-Pugh score also showed to be substantially correlated with death. The most frequent cause of mortality in COVID-19 patients is lung damage. Liver dysfunction is a possible cause of persistent lung damage. Indeed, liver failure plays a significant role in individuals with bacterial chest sepsis[75]. Cirrhosis and SARS-CoV-2 may have a fatal synergy because of alterations in the immune system caused by viral infection and coagulation. Dysregulation of pulmonary dynamics has been attributed to a number of factors, including ascites or deteriorating encephalopathy, immunological dysfunction in viral infection, a rise in the burden of venous thromboembolic illness, and concurrent lung disease. According to Marjot et al[52], mortality rates in patients with cirrhosis was 32% compared to 8% in those without it, while the mortality rate rose in connection with the Child-Pugh class [A (19%), B (35%), and C (51%)] in patients with cirrhosis. The principal cause of decease was respiratory failure (71%).
There is very little effect of COVID-19 on patients with alcoholic hepatitis or alcoholic liver disease[74,76]. However, research on cirrhotic patients has revealed that, like other cirrhotic patients, those with alcohol-related cirrhosis have higher mortality rates[74,76]. Chronic kidney disease, obesity, and diabetes are common comorbidities among patients with alcohol-related cirrhosis, leading to higher risks of COVID-19 complications[77]. In a study involving 867 patients with COVID-19 and chronic liver disease, Kim et al[78] found that alcohol-associated liver disease (ALD) was independently associated with a higher risk of poor survival and a higher COVID-19 mortality rate. ALD is connected to the inflammatory state prompted by danger-associated molecular patterns which trigger the secretion of pro-inflammatory cytokines by particular immune cells[74,79]. It was hypothesized that the superimposed cytokine storm produced by SARS-CoV-2 in patients with ALD could exacerbate the heightened inflammatory process, leading to worse outcomes[80].
Diabetes, hypertension, cardiovascular disease, and obesity are well-known risk factors for severe COVID-19[81]. These metabolic comorbidities are closely related to non-alcoholic fatty liver disease (NAFLD).
Wide-ranging effects of the course of COVID-19 make distinguishing the independent effect on NAFLD a challenge. The difficulty is raised by a number of confounding cofactors, reverse causality from steatosis caused by SARS-CoV-2, as well as population heterogeneity and the diagnostic conditions studied. Consequently, results from clinical studies have been ambiguous. In a retrospective study of 202 SARS-CoV-2 infected patients, 35% were individuals with NAFLD. When compared to SARS-CoV-2 infected patients without NAFLD, patients with NAFLD displayed a higher risk of severe COVID-19, as evidenced by a higher probability of liver abnormalities in the hospitalized patient and long-term viral shedding[33].
Seventy-one consecutive COVID-19 patients from a different case-control study were divided into groups based on whether or not they had NAFLD. After reviewing all medical records, including demographic, clinical, and laboratory data, this study concluded that NAFLD poses a significant risk for developing severe COVID-19[82]. Patients with NAFLD were more probable to be admitted to the intensive care unit, according to a retrospective multicenter study with a cohort of hospitalized adults with COVID-19[83]. These results were supported by two thorough systematic reviews, as well as a meta-analysis[84,85].
On the other hand, NAFLD was not linked to severe COVID-19, as shown by a study conducted by Mushtaq et al[86] in a Middle Eastern cohort. The authors revealed that gene variants associated with NAFLD (transmembrane 6 superfamily 2 [TM6SF2], patatin-like phospholipase domain-containing protein 3 [PNPLA3], glucokinase regulator, and membrane-bound O-acyltransferase domain-containing protein 7 [MBOAT7]) and the severity of COVID-19 disease (TM6SF2, PNPLA3, and MBOAT7) is associated with genetic variation as a mechanism to establish a genetic risk score[87]. Additionally, other studies have concluded that some PNPLA3 allelic variants may act protectively by lowering the risk of COVID-19 mortality[88]. Finally, a study that used a Mendelian randomization approach to investigate the correlations between COVID-19 severity and NAFLD found scant evidence supporting such a relationship[89].
At the beginning of the SARS-CoV-2 pandemic, serious measures were taken to preserve cancer patients from morbidity and mortality by restraining hospital presence and submitting anti-cancer therapy. The European Association for the Study of the liver recommended postponing treatments and evaluating the possibility of gradually removing anti-cancer immunological therapy[90].
In the context of rapidly escalating viral transmission, a series of precautionary principles were dictated. These measures rested on the hypothesis that cancer and active anti-cancer therapy have an unfavorable effect on the consequences of SARS-CoV-2 infection. It is widely known that cancer patients are commonly immunosuppressed as a consequence of chemotherapy. Therefore, different studies indicated that patients with subjacent cancer were at major risk of acquiring SARS-CoV-2 infection and severe outcomes could eventually evolve[91,92].
Recently, a multicenter retrospective revision including 250 non-vaccinated patients with liver cancer and COVID-19 infection showed that the mortality rate was 12.96% in patients with a diagnosis of HCC simultaneously with SARS-CoV-2 infection, and 20.25% in individuals with HCC history[74].
However, the prevalence of SARS-CoV-2 infection did not display a real rise in patients with chronic liver disease or in HCC patients. Nevertheless, HCC cirrhotic patients with COVID-19 may have a worse prognosis than the general population regarding severe disease, complications of SARS-CoV-2 infection, and mortality. Hereafter, the significance of applying actions to decrease the possibility of infection in these patients[93].
Managing liver transplantation in the curse of SARS-CoV-2 pandemic was difficult as numerous hospitals had to essentially stop or drastically scale back their transplantation operations owing to a sudden drop in donor numbers and were forced to convert several care facilities into COVID-19 units. Due to the limited information available and the necessity of continuing immunosuppressive medication in these patients, the medical staff faced difficult challenges to manage post-liver transplant receivers in the course of COVID-19 pandemic while patients were at risk for a more severe COVID-19 infection and possible continued viral shedding. Qin et al[94] described the first instance of SARS-CoV-2 infection in a patient with hepatocellular cancer who had undergone liver transplantation, and discovered a higher viral load with an increasing immunosuppressive dosage. Immunosuppressive medications had no effect on the frequency of COVID-19 severity, according to Bhoori et al[95]. Early studies from Italy claimed that transplant patients experienced low death rates of less than 5%[96]. However, later assessments revealed that liver recipients and other solid organ transplants experienced mortality rates of over 25%[97,98]. Recently, findings from a prospective European trial comprising 57 liver transplant patients with proven SARS-CoV-2 infection and 19 transplant facilities were released. These results are consistent with is consistent with the projected mortality rate because patients with severe COVID-19 infection had overall and in-hospital case fatality rates of 12% and 17%, respectively. Five of the seven patients who passed away had a cancer history at the time of their deaths[99]. The evidence currently available does not support the idea that transplantation or certain immunosuppressive therapies have a significant impact on the likelihood of disease severity. Nevertheless, patients with underlying malignancies may need special care[97]. A number of COVID-19 vaccines have lately approved and have demonstrated effectiveness in healthy individuals. However, careful assessment and immunization of immunocompetent individuals are still required due to the potential immunological imbalance brought on by their illness or immunosuppressive medication. According to Boyarsky et al[100], solid organ transplant recipients who were fully vaccinated with the mRNA vaccine showed an appropriate humoral response, while the subpar response was linked to the use of antimetabolite immunosuppression.
It is widely acknowledged that the effects of SARS-CoV-2 infection on the liver have played a significant role during the COVID-19 pandemic. In patients with pre-existing infectious and non-infectious liver disease, who are at an extra high risk of developing severe COVID-19 and death, this feature is most medically relevant. This review aims to provide an overview of the current research on the molecular mediators responsible for inflammatory liver injury during SARS-CoV-2 infection. To fully comprehend the pathogenic pathways that cause clinical deterioration of patients with COVID-19 due to an imbalanced interaction between the liver, systemic inflammation, disrupted coagulation, and the lung, further research should be conducted.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Infectious diseases
Country/Territory of origin: Argentina
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Shekouhi R, Iran; Wang CY, Taiwan S-Editor: Chen YL L-Editor: Filipodia P-Editor: Chen YL
| 1. | World Health Organization. WHO Coronavirus (COVID-19) Dashboard. 2022. [cited 25 August 2022]. Available from: https://covid19.who.int/. |
| 2. | Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3499] [Cited by in RCA: 4379] [Article Influence: 875.8] [Reference Citation Analysis (0)] |
| 3. | Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221-224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3079] [Cited by in RCA: 2759] [Article Influence: 551.8] [Reference Citation Analysis (0)] |
| 4. | Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;183:1735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 383] [Cited by in RCA: 519] [Article Influence: 103.8] [Reference Citation Analysis (0)] |
| 5. | Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450-454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4113] [Cited by in RCA: 4598] [Article Influence: 209.0] [Reference Citation Analysis (0)] |
| 6. | Parasa S, Desai M, Thoguluva Chandrasekar V, Patel HK, Kennedy KF, Roesch T, Spadaccini M, Colombo M, Gabbiadini R, Artifon ELA, Repici A, Sharma P. Prevalence of Gastrointestinal Symptoms and Fecal Viral Shedding in Patients With Coronavirus Disease 2019: A Systematic Review and Meta-analysis. JAMA Netw Open. 2020;3:e2011335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 301] [Cited by in RCA: 301] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 7. | Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, Diaz G, Cohn A, Fox L, Patel A, Gerber SI, Kim L, Tong S, Lu X, Lindstrom S, Pallansch MA, Weldon WC, Biggs HM, Uyeki TM, Pillai SK; Washington State 2019-nCoV Case Investigation Team. First Case of 2019 Novel Coronavirus in the United States. N Engl J Med. 2020;382:929-936. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4155] [Cited by in RCA: 3820] [Article Influence: 764.0] [Reference Citation Analysis (1)] |
| 8. | Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS; China Medical Treatment Expert Group for Covid-19. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382:1708-1720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19202] [Cited by in RCA: 18856] [Article Influence: 3771.2] [Reference Citation Analysis (7)] |
| 9. | Zhong P, Xu J, Yang D, Shen Y, Wang L, Feng Y, Du C, Song Y, Wu C, Hu X, Sun Y. COVID-19-associated gastrointestinal and liver injury: clinical features and potential mechanisms. Signal Transduct Target Ther. 2020;5:256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 141] [Article Influence: 28.2] [Reference Citation Analysis (1)] |
| 10. | Ning T, Liu S, Xu J, Yang Y, Zhang N, Xie S, Min L, Zhang S, Zhu S, Wang Y. Potential intestinal infection and faecal-oral transmission of human coronaviruses. Rev Med Virol. 2022;32:e2363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 11. | Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631-637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3643] [Cited by in RCA: 4149] [Article Influence: 197.6] [Reference Citation Analysis (0)] |
| 12. | Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271-280.e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11946] [Cited by in RCA: 14244] [Article Influence: 2848.8] [Reference Citation Analysis (0)] |
| 13. | Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864-1868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1488] [Cited by in RCA: 1551] [Article Influence: 77.6] [Reference Citation Analysis (0)] |
| 14. | Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260-1263. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5792] [Cited by in RCA: 6469] [Article Influence: 1293.8] [Reference Citation Analysis (0)] |
| 15. | Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, Wang Q, Zhou H, Yan J, Qi J. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181:894-904.e9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2090] [Cited by in RCA: 2209] [Article Influence: 441.8] [Reference Citation Analysis (0)] |
| 16. | Lei C, Qian K, Li T, Zhang S, Fu W, Ding M, Hu S. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat Commun. 2020;11:2070. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 268] [Cited by in RCA: 312] [Article Influence: 62.4] [Reference Citation Analysis (0)] |
| 17. | Li H, Liu L, Zhang D, Xu J, Dai H, Tang N, Su X, Cao B. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet. 2020;395:1517-1520. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 772] [Cited by in RCA: 886] [Article Influence: 177.2] [Reference Citation Analysis (0)] |
| 18. | Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, Huang H, Zhang L, Du C, Zhang Y, Song J, Wang S, Chao Y, Yang Z, Xu J, Chen D, Xiong W, Xu L, Zhou F, Jiang J, Bai C, Zheng J, Song Y. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4960] [Cited by in RCA: 5515] [Article Influence: 1103.0] [Reference Citation Analysis (1)] |
| 19. | Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35178] [Cited by in RCA: 30086] [Article Influence: 6017.2] [Reference Citation Analysis (3)] |
| 20. | Fan Z, Chen L, Li J, Cheng X, Yang J, Tian C, Zhang Y, Huang S, Liu Z, Cheng J. Clinical Features of COVID-19-Related Liver Functional Abnormality. Clin Gastroenterol Hepatol. 2020;18:1561-1566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 559] [Cited by in RCA: 554] [Article Influence: 110.8] [Reference Citation Analysis (0)] |
| 21. | Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J, Fan Y, Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20:425-434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2493] [Cited by in RCA: 2309] [Article Influence: 461.8] [Reference Citation Analysis (0)] |
| 22. | Lax SF, Skok K, Zechner P, Kessler HH, Kaufmann N, Koelblinger C, Vander K, Bargfrieder U, Trauner M. Pulmonary Arterial Thrombosis in COVID-19 With Fatal Outcome : Results From a Prospective, Single-Center, Clinicopathologic Case Series. Ann Intern Med. 2020;173:350-361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 600] [Cited by in RCA: 617] [Article Influence: 123.4] [Reference Citation Analysis (0)] |
| 23. | Peiris S, Mesa H, Aysola A, Manivel J, Toledo J, Borges-Sa M, Aldighieri S, Reveiz L. Pathological findings in organs and tissues of patients with COVID-19: A systematic review. PLoS One. 2021;16:e0250708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 71] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 24. | Chornenkyy Y, Mejia-Bautista M, Brucal M, Blanke T, Dittmann D, Yeldandi A, Boike JR, Lomasney JW, Nayar R, Jennings LJ, Pezhouh MK. Liver Pathology and SARS-CoV-2 Detection in Formalin-Fixed Tissue of Patients With COVID-19. Am J Clin Pathol. 2021;155:802-814. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 25. | Wanner N, Andrieux G, Badia-I-Mompel P, Edler C, Pfefferle S, Lindenmeyer MT, Schmidt-Lauber C, Czogalla J, Wong MN, Okabayashi Y, Braun F, Lütgehetmann M, Meister E, Lu S, Noriega MLM, Günther T, Grundhoff A, Fischer N, Bräuninger H, Lindner D, Westermann D, Haas F, Roedl K, Kluge S, Addo MM, Huber S, Lohse AW, Reiser J, Ondruschka B, Sperhake JP, Saez-Rodriguez J, Boerries M, Hayek SS, Aepfelbacher M, Scaturro P, Puelles VG, Huber TB. Molecular consequences of SARS-CoV-2 liver tropism. Nat Metab. 2022;4:310-319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 122] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 26. | Sonzogni A, Previtali G, Seghezzi M, Grazia Alessio M, Gianatti A, Licini L, Morotti D, Zerbi P, Carsana L, Rossi R, Lauri E, Pellegrinelli A, Nebuloni M. Liver histopathology in severe COVID 19 respiratory failure is suggestive of vascular alterations. Liver Int. 2020;40:2110-2116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 218] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 27. | Wang Y, Liu S, Liu H, Li W, Lin F, Jiang L, Li X, Xu P, Zhang L, Zhao L, Cao Y, Kang J, Yang J, Li L, Liu X, Li Y, Nie R, Mu J, Lu F, Zhao S, Lu J, Zhao J. SARS-CoV-2 infection of the liver directly contributes to hepatic impairment in patients with COVID-19. J Hepatol. 2020;73:807-816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 353] [Cited by in RCA: 457] [Article Influence: 91.4] [Reference Citation Analysis (0)] |
| 28. | Kaltschmidt B, Fitzek ADE, Schaedler J, Förster C, Kaltschmidt C, Hansen T, Steinfurth F, Windmöller BA, Pilger C, Kong C, Singh K, Nierhaus A, Wichmann D, Sperhake J, Püschel K, Huser T, Krüger M, Robson SC, Wilkens L, Schulte Am Esch J. Hepatic Vasculopathy and Regenerative Responses of the Liver in Fatal Cases of COVID-19. Clin Gastroenterol Hepatol. 2021;19:1726-1729.e3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 29. | Cheung CCL, Goh D, Lim X, Tien TZ, Lim JCT, Lee JN, Tan B, Tay ZEA, Wan WY, Chen EX, Nerurkar SN, Loong S, Cheow PC, Chan CY, Koh YX, Tan TT, Kalimuddin S, Tai WMD, Ng JL, Low JG, Yeong J, Lim KH. Residual SARS-CoV-2 viral antigens detected in GI and hepatic tissues from five recovered patients with COVID-19. Gut. 2022;71:226-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 128] [Article Influence: 42.7] [Reference Citation Analysis (0)] |
| 30. | Delorey TM, Ziegler CGK, Heimberg G, Normand R, Yang Y, Segerstolpe Å, Abbondanza D, Fleming SJ, Subramanian A, Montoro DT, Jagadeesh KA, Dey KK, Sen P, Slyper M, Pita-Juárez YH, Phillips D, Biermann J, Bloom-Ackermann Z, Barkas N, Ganna A, Gomez J, Melms JC, Katsyv I, Normandin E, Naderi P, Popov YV, Raju SS, Niezen S, Tsai LT, Siddle KJ, Sud M, Tran VM, Vellarikkal SK, Wang Y, Amir-Zilberstein L, Atri DS, Beechem J, Brook OR, Chen J, Divakar P, Dorceus P, Engreitz JM, Essene A, Fitzgerald DM, Fropf R, Gazal S, Gould J, Grzyb J, Harvey T, Hecht J, Hether T, Jané-Valbuena J, Leney-Greene M, Ma H, McCabe C, McLoughlin DE, Miller EM, Muus C, Niemi M, Padera R, Pan L, Pant D, Pe'er C, Pfiffner-Borges J, Pinto CJ, Plaisted J, Reeves J, Ross M, Rudy M, Rueckert EH, Siciliano M, Sturm A, Todres E, Waghray A, Warren S, Zhang S, Zollinger DR, Cosimi L, Gupta RM, Hacohen N, Hibshoosh H, Hide W, Price AL, Rajagopal J, Tata PR, Riedel S, Szabo G, Tickle TL, Ellinor PT, Hung D, Sabeti PC, Novak R, Rogers R, Ingber DE, Jiang ZG, Juric D, Babadi M, Farhi SL, Izar B, Stone JR, Vlachos IS, Solomon IH, Ashenberg O, Porter CBM, Li B, Shalek AK, Villani AC, Rozenblatt-Rosen O, Regev A. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. 2021;595:107-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 590] [Cited by in RCA: 552] [Article Influence: 138.0] [Reference Citation Analysis (0)] |
| 31. | Kumar P, Sharma M, Kulkarni A, Rao PN. Pathogenesis of Liver Injury in Coronavirus Disease 2019. J Clin Exp Hepatol. 2020;10:641-642. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 32. | Qi F, Qian S, Zhang S, Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun. 2020;526:135-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 523] [Cited by in RCA: 740] [Article Influence: 148.0] [Reference Citation Analysis (0)] |
| 33. | Ji D, Qin E, Xu J, Zhang D, Cheng G, Wang Y, Lau G. Non-alcoholic fatty liver diseases in patients with COVID-19: A retrospective study. J Hepatol. 2020;73:451-453. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 336] [Cited by in RCA: 408] [Article Influence: 81.6] [Reference Citation Analysis (2)] |
| 34. | Pirola CJ, Sookoian S. SARS-CoV-2 virus and liver expression of host receptors: Putative mechanisms of liver involvement in COVID-19. Liver Int. 2020;40:2038-2040. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 35. | Chua RL, Lukassen S, Trump S, Hennig BP, Wendisch D, Pott F, Debnath O, Thürmann L, Kurth F, Völker MT, Kazmierski J, Timmermann B, Twardziok S, Schneider S, Machleidt F, Müller-Redetzky H, Maier M, Krannich A, Schmidt S, Balzer F, Liebig J, Loske J, Suttorp N, Eils J, Ishaque N, Liebert UG, von Kalle C, Hocke A, Witzenrath M, Goffinet C, Drosten C, Laudi S, Lehmann I, Conrad C, Sander LE, Eils R. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol. 2020;38:970-979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 596] [Cited by in RCA: 791] [Article Influence: 158.2] [Reference Citation Analysis (0)] |
| 36. | Onabajo OO, Banday AR, Stanifer ML, Yan W, Obajemu A, Santer DM, Florez-Vargas O, Piontkivska H, Vargas JM, Ring TJ, Kee C, Doldan P, Tyrrell DL, Mendoza JL, Boulant S, Prokunina-Olsson L. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat Genet. 2020;52:1283-1293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 206] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 37. | Banales JM, Huebert RC, Karlsen T, Strazzabosco M, LaRusso NF, Gores GJ. Cholangiocyte pathobiology. Nat Rev Gastroenterol Hepatol. 2019;16:269-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 340] [Article Influence: 56.7] [Reference Citation Analysis (1)] |
| 38. | Zhao B, Ni C, Gao R, Wang Y, Yang L, Wei J, Lv T, Liang J, Zhang Q, Xu W, Xie Y, Wang X, Yuan Z, Zhang R, Lin X. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell. 2020;11:771-775. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 268] [Cited by in RCA: 311] [Article Influence: 62.2] [Reference Citation Analysis (0)] |
| 39. | Sahu T, Pande B, Pl M, Verma HK. Liver dysfunction during COVID-19 pandemic: Contributing role of associated factors in disease progression and severity. World J Hepatol. 2022;14:1099-1110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.7] [Reference Citation Analysis (1)] |
| 40. | Pedersen SF, Ho YC. SARS-CoV-2: a storm is raging. J Clin Invest. 2020;130:2202-2205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 869] [Cited by in RCA: 852] [Article Influence: 170.4] [Reference Citation Analysis (0)] |
| 41. | Lu Q, Liu J, Zhao S, Gomez Castro MF, Laurent-Rolle M, Dong J, Ran X, Damani-Yokota P, Tang H, Karakousi T, Son J, Kaczmarek ME, Zhang Z, Yeung ST, McCune BT, Chen RE, Tang F, Ren X, Chen X, Hsu JCC, Teplova M, Huang B, Deng H, Long Z, Mudianto T, Jin S, Lin P, Du J, Zang R, Su TT, Herrera A, Zhou M, Yan R, Cui J, Zhu J, Zhou Q, Wang T, Ma J, Koralov SB, Aifantis I, Segal LN, Diamond MS, Khanna KM, Stapleford KA, Cresswell P, Liu Y, Ding S, Xie Q, Wang J. SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2. Immunity. 2021;54:1304-1319.e9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 120] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 42. | Ren X, Wen W, Fan X, Hou W, Su B, Cai P, Li J, Liu Y, Tang F, Zhang F, Yang Y, He J, Ma W, Wang P, Cao Q, Chen F, Chen Y, Cheng X, Deng G, Deng X, Ding W, Feng Y, Gan R, Guo C, Guo W, He S, Jiang C, Liang J, Li YM, Lin J, Ling Y, Liu H, Liu J, Liu N, Liu SQ, Luo M, Ma Q, Song Q, Sun W, Wang G, Wang F, Wang Y, Wen X, Wu Q, Xu G, Xie X, Xiong X, Xing X, Xu H, Yin C, Yu D, Yu K, Yuan J, Zhang B, Zhang P, Zhang T, Zhao J, Zhao P, Zhou J, Zhou W, Zhong S, Zhong X, Zhang S, Zhu L, Zhu P, Zou B, Zou J, Zuo Z, Bai F, Huang X, Zhou P, Jiang Q, Huang Z, Bei JX, Wei L, Bian XW, Liu X, Cheng T, Li X, Wang FS, Wang H, Zhang Z, Qu K, Wang X, Chen J, Jin R. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184:1895-1913.e19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 335] [Cited by in RCA: 498] [Article Influence: 124.5] [Reference Citation Analysis (0)] |
| 43. | Li T, Yang Y, Li Y, Wang Z, Ma F, Luo R, Xu X, Zhou G, Wang J, Niu J, Lv G, Crispe IN, Tu Z. Platelets mediate inflammatory monocyte activation by SARS-CoV-2 spike protein. J Clin Invest. 2022;132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 56] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 44. | Yan B, Freiwald T, Chauss D, Wang L, West E, Mirabelli C, Zhang CJ, Nichols EM, Malik N, Gregory R, Bantscheff M, Ghidelli-Disse S, Kolev M, Frum T, Spence JR, Sexton JZ, Alysandratos KD, Kotton DN, Pittaluga S, Bibby J, Niyonzima N, Olson MR, Kordasti S, Portilla D, Wobus CE, Laurence A, Lionakis MS, Kemper C, Afzali B, Kazemian M. SARS-CoV-2 drives JAK1/2-dependent local complement hyperactivation. Sci Immunol. 2021;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 145] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 45. | Merad M, Blish CA, Sallusto F, Iwasaki A. The immunology and immunopathology of COVID-19. Science. 2022;375:1122-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 498] [Article Influence: 166.0] [Reference Citation Analysis (0)] |
| 46. | Buso G, Becchetti C, Berzigotti A. Acute splanchnic vein thrombosis in patients with COVID-19: A systematic review. Dig Liver Dis. 2021;53:937-949. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 47. | Wang M, Yan W, Qi W, Wu D, Zhu L, Li W, Wang X, Ma K, Ni M, Xu D, Wang H, Chen G, Yu H, Ding H, Xing M, Han M, Luo X, Chen T, Guo W, Xi D, Ning Q. Clinical characteristics and risk factors of liver injury in COVID-19: a retrospective cohort study from Wuhan, China. Hepatol Int. 2020;14:723-732. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 48. | Da BL, Kushner T, El Halabi M, Paka P, Khalid M, Uberoi A, Lee BT, Perumalswami PV, Rutledge SM, Schiano TD, Friedman SL, Saberi B. Liver Injury in Patients Hospitalized with Coronavirus Disease 2019 Correlates with Hyperinflammatory Response and Elevated Interleukin-6. Hepatol Commun. 2021;5:177-188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 49. | Effenberger M, Grander C, Grabherr F, Griesmacher A, Ploner T, Hartig F, Bellmann-Weiler R, Joannidis M, Zoller H, Weiss G, Adolph TE, Tilg H. Systemic inflammation as fuel for acute liver injury in COVID-19. Dig Liver Dis. 2021;53:158-165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 65] [Article Influence: 16.3] [Reference Citation Analysis (2)] |
| 50. | Shafran N, Issachar A, Shochat T, Shafran IH, Bursztyn M, Shlomai A. Abnormal liver tests in patients with SARS-CoV-2 or influenza - prognostic similarities and temporal disparities. JHEP Rep. 2021;3:100258. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 51. | McConnell MJ, Kondo R, Kawaguchi N, Iwakiri Y. Covid-19 and Liver Injury: Role of Inflammatory Endotheliopathy, Platelet Dysfunction, and Thrombosis. Hepatol Commun. 2022;6:255-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 42] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 52. | Marjot T, Moon AM, Cook JA, Abd-Elsalam S, Aloman C, Armstrong MJ, Pose E, Brenner EJ, Cargill T, Catana MA, Dhanasekaran R, Eshraghian A, García-Juárez I, Gill US, Jones PD, Kennedy J, Marshall A, Matthews C, Mells G, Mercer C, Perumalswami PV, Avitabile E, Qi X, Su F, Ufere NN, Wong YJ, Zheng MH, Barnes E, Barritt AS 4th, Webb GJ. Outcomes following SARS-CoV-2 infection in patients with chronic liver disease: An international registry study. J Hepatol. 2021;74:567-577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 399] [Cited by in RCA: 383] [Article Influence: 95.8] [Reference Citation Analysis (0)] |
| 53. | Russo FP, Burra P, Zanetto A. COVID-19 and liver disease: where are we now? Nat Rev Gastroenterol Hepatol. 2022;19:277-278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 54. | Zou X, Fang M, Li S, Wu L, Gao B, Gao H, Ran X, Bian Y, Li R, ShanshanYu, Ling J, Li D, Tian D, Huang J. Characteristics of Liver Function in Patients With SARS-CoV-2 and Chronic HBV Coinfection. Clin Gastroenterol Hepatol. 2021;19:597-603. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 67] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 55. | Lin Y, Yuan J, Long Q, Hu J, Deng H, Zhao Z, Chen J, Lu M, Huang A. Patients with SARS-CoV-2 and HBV co-infection are at risk of greater liver injury. Genes Dis. 2021;8:484-492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 56. | Anugwom CM, Aby ES, Debes JD. Inverse Association Between Chronic Hepatitis B Infection and Coronavirus Disease 2019 (COVID-19): Immune Exhaustion or Coincidence? Clin Infect Dis. 2021;72:180-182. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 57. | Brooks DG, Teyton L, Oldstone MB, McGavern DB. Intrinsic functional dysregulation of CD4 T cells occurs rapidly following persistent viral infection. J Virol. 2005;79:10514-10527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 192] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 58. | Sapir T, Averch Z, Lerman B, Bodzin A, Fishman Y, Maitra R. COVID-19 and the Immune Response: A Multi-Phasic Approach to the Treatment of COVID-19. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 59. | Papatheodoridis GV, Lekakis V, Voulgaris T, Lampertico P, Berg T, Chan HLY, Kao JH, Terrault N, Lok AS, Reddy KR. Hepatitis B virus reactivation associated with new classes of immunosuppressants and immunomodulators: A systematic review, meta-analysis, and expert opinion. J Hepatol. 2022;77:1670-1689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 63] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 60. | Chang Y, Jeong SW, Jang JY. Hepatitis B Virus Reactivation Associated With Therapeutic Interventions. Front Med (Lausanne). 2021;8:770124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 25] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 61. | Loomba R, Liang TJ. Hepatitis B Reactivation Associated With Immune Suppressive and Biological Modifier Therapies: Current Concepts, Management Strategies, and Future Directions. Gastroenterology. 2017;152:1297-1309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 440] [Article Influence: 55.0] [Reference Citation Analysis (0)] |
| 62. | Gerussi A, Rigamonti C, Elia C, Cazzagon N, Floreani A, Pozzi R, Pozzoni P, Claar E, Pasulo L, Fagiuoli S, Cristoferi L, Carbone M, Invernizzi P. Coronavirus Disease 2019 in Autoimmune Hepatitis: A Lesson From Immunosuppressed Patients. Hepatol Commun. 2020;4:1257-1262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 63. | Sun J, Aghemo A, Forner A, Valenti L. COVID-19 and liver disease. Liver Int. 2020;40:1278-1281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 219] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 64. | Pan Q, Tilanus HW, Metselaar HJ, Janssen HL, van der Laan LJ. Virus-drug interactions--molecular insight into immunosuppression and HCV. Nat Rev Gastroenterol Hepatol. 2012;9:355-362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 65. | Torres HA, Hosry J, Mahale P, Economides MP, Jiang Y, Lok AS. Hepatitis C virus reactivation in patients receiving cancer treatment: A prospective observational study. Hepatology. 2018;67:36-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 89] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 66. | Lee HL, Bae SH, Jang B, Hwang S, Yang H, Nam HC, Sung PS, Lee SW, Jang JW, Choi JY, Han NI, Song BJ, Lee JW, Yoon SK. Reactivation of Hepatitis C Virus and Its Clinical Outcomes in Patients Treated with Systemic Chemotherapy or Immunosuppressive Therapy. Gut Liver. 2017;11:870-877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 67. | Ciesek S, Steinmann E, Iken M, Ott M, Helfritz FA, Wappler I, Manns MP, Wedemeyer H, Pietschmann T. Glucocorticosteroids increase cell entry by hepatitis C virus. Gastroenterology. 2010;138:1875-1884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 62] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 68. | Dumortier J, Boillot O, Scoazec JY. Natural history, treatment and prevention of hepatitis C recurrence after liver transplantation: past, present and future. World J Gastroenterol. 2014;20:11069-11079. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 20] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 69. | Rosen HR, Hinrichs DJ, Gretch DR, Koziel MJ, Chou S, Houghton M, Rabkin J, Corless CL, Bouwer HG. Association of multispecific CD4(+) response to hepatitis C and severity of recurrence after liver transplantation. Gastroenterology. 1999;117:926-932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 123] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 70. | Magrin S, Craxi A, Fabiano C, Simonetti RG, Fiorentino G, Marino L, Diquattro O, Di Marco V, Loiacono O, Volpes R. Hepatitis C viremia in chronic liver disease: relationship to interferon-alpha or corticosteroid treatment. Hepatology. 1994;19:273-279. [PubMed] |
| 71. | Shokri S, Mahmoudvand S. The possibility of hepatitis C reactivation in COVID-19 patients treated with corticosteroids. Ann Hepatol. 2022;27:100704. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 72. | Asrani SK, Devarbhavi H, Eaton J, Kamath PS. Burden of liver diseases in the world. J Hepatol. 2019;70:151-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1382] [Cited by in RCA: 2282] [Article Influence: 380.3] [Reference Citation Analysis (0)] |
| 73. | Qi X, Liu Y, Wang J, Fallowfield JA, Li X, Shi J, Pan H, Zou S, Zhang H, Chen Z, Li F, Luo Y, Mei M, Liu H, Wang Z, Li J, Yang H, Xiang H, Liu T, Zheng MH, Liu C, Huang Y, Xu D, Kang N, He Q, Gu Y, Zhang G, Shao C, Liu D, Zhang L, Kawada N, Jiang Z, Wang F, Xiong B, Takehara T, Rockey DC; COVID-Cirrhosis-CHESS Group. Clinical course and risk factors for mortality of COVID-19 patients with pre-existing cirrhosis: a multicentre cohort study. Gut. 2021;70:433-436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 71] [Article Influence: 17.8] [Reference Citation Analysis (1)] |
| 74. | Iavarone M, D'Ambrosio R, Soria A, Triolo M, Pugliese N, Del Poggio P, Perricone G, Massironi S, Spinetti A, Buscarini E, Viganò M, Carriero C, Fagiuoli S, Aghemo A, Belli LS, Lucà M, Pedaci M, Rimondi A, Rumi MG, Invernizzi P, Bonfanti P, Lampertico P. High rates of 30-day mortality in patients with cirrhosis and COVID-19. J Hepatol. 2020;73:1063-1071. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 276] [Article Influence: 55.2] [Reference Citation Analysis (2)] |
| 75. | Shah BA, Ahmed W, Dhobi GN, Shah NN, Khursheed SQ, Haq I. Validity of pneumonia severity index and CURB-65 severity scoring systems in community acquired pneumonia in an Indian setting. Indian J Chest Dis Allied Sci. 2010;52:9-17. [PubMed] |
| 76. | Moon AM, Webb GJ, Aloman C, Armstrong MJ, Cargill T, Dhanasekaran R, Genescà J, Gill US, James TW, Jones PD, Marshall A, Mells G, Perumalswami PV, Qi X, Su F, Ufere NN, Barnes E, Barritt AS, Marjot T. High mortality rates for SARS-CoV-2 infection in patients with pre-existing chronic liver disease and cirrhosis: Preliminary results from an international registry. J Hepatol. 2020;73:705-708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 203] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 77. | Kushner T, Cafardi J. Chronic Liver Disease and COVID-19: Alcohol Use Disorder/Alcohol-Associated Liver Disease, Nonalcoholic Fatty Liver Disease/Nonalcoholic Steatohepatitis, Autoimmune Liver Disease, and Compensated Cirrhosis. Clin Liver Dis (Hoboken). 2020;15:195-199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 78. | Kim D, Adeniji N, Latt N, Kumar S, Bloom PP, Aby ES, Perumalswami P, Roytman M, Li M, Vogel AS, Catana AM, Wegermann K, Carr RM, Aloman C, Chen VL, Rabiee A, Sadowski B, Nguyen V, Dunn W, Chavin KD, Zhou K, Lizaola-Mayo B, Moghe A, Debes J, Lee TH, Branch AD, Viveiros K, Chan W, Chascsa DM, Kwo P, Dhanasekaran R. Predictors of Outcomes of COVID-19 in Patients With Chronic Liver Disease: US Multi-center Study. Clin Gastroenterol Hepatol. 2021;19:1469-1479.e19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 174] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 79. | Sarin SK, Choudhury A, Lau GK, Zheng MH, Ji D, Abd-Elsalam S, Hwang J, Qi X, Cua IH, Suh JI, Park JG, Putcharoen O, Kaewdech A, Piratvisuth T, Treeprasertsuk S, Park S, Wejnaruemarn S, Payawal DA, Baatarkhuu O, Ahn SH, Yeo CD, Alonzo UR, Chinbayar T, Loho IM, Yokosuka O, Jafri W, Tan S, Soo LI, Tanwandee T, Gani R, Anand L, Esmail ES, Khalaf M, Alam S, Lin CY, Chuang WL, Soin AS, Garg HK, Kalista K, Batsukh B, Purnomo HD, Dara VP, Rathi P, Al Mahtab M, Shukla A, Sharma MK, Omata M; APASL COVID Task Force, APASL COVID Liver Injury Spectrum Study (APCOLIS Study-NCT 04345640). Pre-existing liver disease is associated with poor outcome in patients with SARS CoV2 infection; The APCOLIS Study (APASL COVID-19 Liver Injury Spectrum Study). Hepatol Int. 2020;14:690-700. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 217] [Cited by in RCA: 216] [Article Influence: 43.2] [Reference Citation Analysis (1)] |
| 80. | Jose RJ, Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir Med. 2020;8:e46-e47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 789] [Cited by in RCA: 877] [Article Influence: 175.4] [Reference Citation Analysis (0)] |
| 81. | Cai Q, Chen F, Wang T, Luo F, Liu X, Wu Q, He Q, Wang Z, Liu Y, Liu L, Chen J, Xu L. Obesity and COVID-19 Severity in a Designated Hospital in Shenzhen, China. Diabetes Care. 2020;43:1392-1398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 387] [Cited by in RCA: 419] [Article Influence: 83.8] [Reference Citation Analysis (0)] |
| 82. | Mahamid M, Nseir W, Khoury T, Mahamid B, Nubania A, Sub-Laban K, Schifter J, Mari A, Sbeit W, Goldin E. Nonalcoholic fatty liver disease is associated with COVID-19 severity independently of metabolic syndrome: a retrospective case-control study. Eur J Gastroenterol Hepatol. 2021;33:1578-1581. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 66] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 83. | Hashemi N, Viveiros K, Redd WD, Zhou JC, McCarty TR, Bazarbashi AN, Hathorn KE, Wong D, Njie C, Shen L, Chan WW. Impact of chronic liver disease on outcomes of hospitalized patients with COVID-19: A multicentre United States experience. Liver Int. 2020;40:2515-2521. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 117] [Article Influence: 23.4] [Reference Citation Analysis (2)] |
| 84. | Singh A, Hussain S, Antony B. Non-alcoholic fatty liver disease and clinical outcomes in patients with COVID-19: A comprehensive systematic review and meta-analysis. Diabetes Metab Syndr. 2021;15:813-822. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 63] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 85. | Hegyi PJ, Váncsa S, Ocskay K, Dembrovszky F, Kiss S, Farkas N, Erőss B, Szakács Z, Hegyi P, Pár G. Metabolic Associated Fatty Liver Disease Is Associated With an Increased Risk of Severe COVID-19: A Systematic Review With Meta-Analysis. Front Med (Lausanne). 2021;8:626425. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 86. | Mushtaq K, Khan MU, Iqbal F, Alsoub DH, Chaudhry HS, Ata F, Iqbal P, Elfert K, Balaraju G, Almaslamani M, Al-Ejji K, AlKaabi S, Kamel YM. NAFLD is a predictor of liver injury in COVID-19 hospitalized patients but not of mortality, disease severity on the presentation or progression - The debate continues. J Hepatol. 2021;74:482-484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 63] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 87. | Valenti L, Jamialahmadi O, Romeo S. Lack of genetic evidence that fatty liver disease predisposes to COVID-19. J Hepatol. 2020;73:709-711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 88. | Innes H, Buch S, Barnes E, Hampe J, Marjot T, Stickel F. The rs738409 G Allele in PNPLA3 Is Associated With a Reduced Risk of COVID-19 Mortality and Hospitalization. Gastroenterology. 2021;160:2599-2601.e2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 89. | Liu D, Zhang Q, Bai P, Zhao J. Assessing causal relationships between COVID-19 and non-alcoholic fatty liver disease. J Hepatol. 2022;76:740-742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 90. | Boettler T, Marjot T, Newsome PN, Mondelli MU, Maticic M, Cordero E, Jalan R, Moreau R, Cornberg M, Berg T. Impact of COVID-19 on the care of patients with liver disease: EASL-ESCMID position paper after 6 months of the pandemic. JHEP Rep. 2020;2:100169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 121] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 91. | Sharma R, Pinato DJ. Management of Hepatocellular Cancer in the time of SARS-CoV-2. Liver Int. 2020;40:1823-1825. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 92. | Dai M, Liu D, Liu M, Zhou F, Li G, Chen Z, Zhang Z, You H, Wu M, Zheng Q, Xiong Y, Xiong H, Wang C, Chen C, Xiong F, Zhang Y, Peng Y, Ge S, Zhen B, Yu T, Wang L, Wang H, Liu Y, Chen Y, Mei J, Gao X, Li Z, Gan L, He C, Shi Y, Qi Y, Yang J, Tenen DG, Chai L, Mucci LA, Santillana M, Cai H. Patients with Cancer Appear More Vulnerable to SARS-CoV-2: A Multicenter Study during the COVID-19 Outbreak. Cancer Discov. 2020;10:783-791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1142] [Cited by in RCA: 969] [Article Influence: 193.8] [Reference Citation Analysis (0)] |
| 93. | Guarino M, Cossiga V, Capasso M, Mazzarelli C, Pelizzaro F, Sacco R, Russo FP, Vitale A, Trevisani F, Cabibbo G; The Associazione Italiana Per Lo Studio Del Fegato AISF HCC Special Interest Group. Impact of SARS-CoV-2 Pandemic on the Management of Patients with Hepatocellular Carcinoma. J Clin Med. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 94. | Qin J, Wang H, Qin X, Zhang P, Zhu L, Cai J, Yuan Y, Li H. Perioperative Presentation of COVID-19 Disease in a Liver Transplant Recipient. Hepatology. 2020;72:1491-1493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 88] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 95. | Bhoori S, Rossi RE, Citterio D, Mazzaferro V. COVID-19 in long-term liver transplant patients: preliminary experience from an Italian transplant centre in Lombardy. Lancet Gastroenterol Hepatol. 2020;5:532-533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 198] [Cited by in RCA: 205] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 96. | D'Antiga L. Coronaviruses and Immunosuppressed Patients: The Facts During the Third Epidemic. Liver Transpl. 2020;26:832-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 479] [Cited by in RCA: 492] [Article Influence: 98.4] [Reference Citation Analysis (0)] |
| 97. | Belli LS, Duvoux C, Karam V, Adam R, Cuervas-Mons V, Pasulo L, Loinaz C, Invernizzi F, Patrono D, Bhoori S, Ciccarelli O, Morelli MC, Castells L, Lopez-Lopez V, Conti S, Fondevila C, Polak W. COVID-19 in liver transplant recipients: preliminary data from the ELITA/ELTR registry. Lancet Gastroenterol Hepatol. 2020;5:724-725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 98. | Lee BT, Perumalswami PV, Im GY, Florman S, Schiano TD; COBE Study Group. COVID-19 in Liver Transplant Recipients: An Initial Experience From the US Epicenter. Gastroenterology. 2020;159:1176-1178.e2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 99. | Becchetti C, Zambelli MF, Pasulo L, Donato MF, Invernizzi F, Detry O, Dahlqvist G, Ciccarelli O, Morelli MC, Fraga M, Svegliati-Baroni G, van Vlierberghe H, Coenraad MJ, Romero MC, de Gottardi A, Toniutto P, Del Prete L, Abbati C, Samuel D, Pirenne J, Nevens F, Dufour JF; COVID-LT group. COVID-19 in an international European liver transplant recipient cohort. Gut. 2020;69:1832-1840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 122] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 100. | Boyarsky BJ, Werbel WA, Avery RK, Tobian AAR, Massie AB, Segev DL, Garonzik-Wang JM. Antibody Response to 2-Dose SARS-CoV-2 mRNA Vaccine Series in Solid Organ Transplant Recipients. JAMA. 2021;325:2204-2206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 780] [Cited by in RCA: 801] [Article Influence: 200.3] [Reference Citation Analysis (0)] |