Published online Dec 21, 2022. doi: 10.3748/wjg.v28.i47.6689
Peer-review started: September 18, 2022
First decision: October 30, 2022
Revised: November 7, 2022
Accepted: November 22, 2022
Article in press: November 22, 2022
Published online: December 21, 2022
Processing time: 91 Days and 23.1 Hours
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) poses a serious threat to global health. SARS-CoV-2 infects host cells primarily by binding to angiotensin-converting enzyme 2, which is coexpressed in alveolar type 2 cells and gut epithelial cells. It is known that COVID-19 often presents with gastrointestinal symptoms and gut dysbiosis, mainly characterized by an increase in opportunistic pathogens and a decrease in beneficial commensal bacteria. In recent years, multiple studies have comprehensively explored gut microbiota alterations in COVID-19 and highlighted the clinical correlation between dysbiosis and COVID-19. SARS-CoV-2 causes gastrointestinal infections and dysbiosis mainly through fecal-oral transmission and the circulatory and immune pathways. Studies have shown that the gut microbiota and its metabolites can regulate the immune response and modulate antiviral effects. In addition, the gut microbiota is closely related to gastrointestinal symptoms, such as diarrhea, a common gastrointestinal symptom among COVID-19. Therefore, the contribution of the gut microbiota in COVID-19 should not be overlooked. Strategies targeting the gut microbiota via probiotics, prebiotics and fecal microbiota transplantation should be considered to treat this patient population in the future. However, the specific alterations and mechanisms as well as the contributions of gut microbiota in COVID-19 should be urgently further explored.
Core Tip: Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a global health threat. SARS-CoV-2 infects host cells through binding to angiotensin-converting enzyme 2. COVID-19 patients exhibit gut dysbiosis. Here, the gut microbiota alterations in COVID-19 are summarized. The pathways and possible mechanisms of dysbiosis caused by SARS-CoV-2, as well as the impact of the gut microbiota and its metabolites on the inflammatory response and antiviral effects during the course of the disease are also described. Therefore, targeting the gut microbiota should be considered a promising strategy for COVID-19 prevention, treatment, and prognostic assessment.
- Citation: Xiang H, Liu QP. Alterations of the gut microbiota in coronavirus disease 2019 and its therapeutic potential. World J Gastroenterol 2022; 28(47): 6689-6701
- URL: https://www.wjgnet.com/1007-9327/full/v28/i47/6689.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i47.6689
Coronavirus disease 2019 (COVID-19) is a new acute infectious disease that is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which belongs to the Coronaviridae family[1]. SARS-CoV-2 infects host cells through the binding of the S protein to angiotensin-converting enzyme 2 (ACE2) and interacts with transmembrane serine protease 2 (TMPRSS2), which cleaves the viral S-protein, allowing efficient viral fusion[2,3]. Interestingly, ACE2 is coexpressed in alveolar type 2 cells and intestinal and colonic epithelial cells, especially in small intestinal enterocytes[4,5]. Correspondingly, in addition to infecting the respiratory system and causing respiratory symptoms (e.g., fever, dry cough, dyspnea, myalgia, headache, etc.), SARS-CoV-2 also infects the gastrointestinal tract, where it replicates abundantly[6]. Overwhelming evidence substantiates the detection of viral RNA in fecal samples or rectal swabs from COVID-19 patients[6-8]. It is estimated that up to 48.1% of COVID-19 patients had fecal samples positive for viral RNA[7], even when the virus was not detected in respiratory and/or sputum samples[9]. Moreover, researchers have found that the gut microbiota composition of COVID-19 patients exhibits significant alterations (dysbiosis), mainly characterized by an increase in the abundance of opportunistic pathogens and a decrease in the abundance of beneficial commensal bacteria[10]. Gut dysbiosis is closely associated with gastrointestinal symptoms and disease severity[11,12]. Therefore, the crosstalk between the gut microbiota and COVID-19 is gaining attention.
The gut microbiota has become a hot research topic in recent years. The resident microbial composition of the human gut mainly includes bacteria, archaea, viruses and fungi[13]. The human gut microbiota consists of more than 1014 bacteria and comprises approximately 500 to 1000 species. Gut bacteria in healthy individuals are mainly Actinobacteria, Firmicutes, Proteobacteria, and Bacteroidetes[13]. The complex gut microbiota communities have important genomic and enzymatic properties and perform a critical role in the immune system, which protects against pathogens and helps maintain gut microbiota homeostasis. Gut microbiota homeostasis is essential for maintaining human health. Conversely, dysbiosis can lead to metabolic disturbance, immune dysfunction and systemic inflammation and has been linked to various diseases[14].
Therefore, this review mainly summarizes the gut microbiota alterations and the possible mechanisms of dysbiosis in COVID-19. Furthermore, we highlight the theoretical basis that the gut microbiota can be considered a promising therapeutic target in COVID-19, potentially interfering with immune and inflammatory responses and antiviral effects. Finally, we also reviewed multiple interventions targeting the gut microbiota, such as prebiotics, probiotics and fecal microbiota transplantation (FMT), which could optimize COVID-19 treatment.
Gut dysbiosis in COVID-19 patients has received widespread attention in recent years (Table 1)[10,12,15-22]. The gut microbiota is significantly altered in COVID-19 patients receiving and not receiving medication compared to that in non-COVID-19 individuals[12]. Gut dysbiosis persists throughout the course of the disease and even after viral clearance[10,12]. Yeoh et al[12] suggested that members of the Bacteroidetes phylum and Actinobacteria were predominant in COVID-19 and non-COVID-19 individuals, respectively. In an American cohort study, Peptoniphilus, Corynebacterium, and Campylobacter were identified as the most enriched genera in COVID-19 patients[23]. Immunomodulatory gut bacteria, such as Faecalibacterium prausnitzii (F. prausnitzii), Bifidobacterium adolescentis and Bifidobacterium longum, were depleted in COVID-19, and their depletion was correlated with an elevation in the levels of inflammatory cytokines and markers (CXCL10, IL-10, TNF-α, CCL2, and CRP)[12]. However, Tao et al[16] found that COVID-19 patients had higher abundances of Clostridium, Veillonella, Streptococcus, Fusobacterium, Lactobacillus, Escherichia and Bifidobacterium and lower abundances of Bacteroidetes, Sutterella, Faecalibacterium, Coprococcus and Parabacteroides than controls. The increased richness of Streptococcus exacerbates the risk of opportunistic pathogenic infections[24].
| Study | Method | Increased abundance of gut microbiota | Decreased abundance of gut microbiota | Main conclusion |
| Gu et al[15], 2020 | 16S rrna | Streptococcus, Rothia, Veillonella, Actinomyces, Erysipelatoclostridium | Ruminococcaceae family, Lachnospiraceae family, Agathobacter, Fusicatenibacter, Roseburia | Gut microbiota has potential value as a diagnostic biomarker and therapeutic target for COVID-19 |
| Zuo et al[10], 2020 | Shotgun | Clostridium hathewayi, Actinomyces viscosus, Bacteroides nordii | Eubacterium, Faecalibacterium prausnitzii, Roseburia, Lachnospiraceae | Fecal microbiota alterations are associated with fecal virus levels and COVID-19 severity; symbionts were depletion and opportunistic pathogens were enrichment in COVID-19 patients; gut dysbiosis persists in COVID-19 patients after virus clearance |
| Tao et al[16], 2020 | 16S rrna | Streptococcus, Clostridium, Lactobacillus, Bifidobacterium | Bacteroidetes, Roseburia, Faecalibacterium, Coprococcus, Parabacteroides | IL-18 level was higher in the fecal samples from COVID-19 patients; dysbiosis may contribute to SARS-CoV-2-induced production of inflammatory cytokines and cytokine storm in the gut |
| Tang et al[17], 2020 | Q-PCR | Enterococcus, Enterobacteriaceae | Faecalibacterium prausnitzii, Clostridium butyricum, Clostridium leptum, Eubacterium rectale | Specific gut microbiota can be considered diagnostic biomarkers for COVID-19; the Ec/E ratio can be used to predict death in critically ill patients |
| Zuo et al[18], 2020 | Shotgun | Candida albicans, Candida auris, Aspergillus flavus | - | The guts of COVID-19 patients are accompanied by massive fungal blooms |
| Yeoh et al[12], 2021 | Shotgun | Actinobacteria, Ruminococcus gnavus, Ruminococcus torques, Bacteroides dorei | Bifidobacterium adolescentis, Faecalibacterium prausnitzii, Eubacterium rectale | Immunomodulatory gut bacteria were depleted in COVID-19 patients; gut dysbiosis persists in COVID-19 patients after virus clearance; gut microbiota composition was associated with disease severity |
| Wu et al[19], 2021 | 16S rrna | Streptococcus, Weissella, Enterococcus, Rothia, Lactobacillus, Actinomyces, Granulicatella | Blautia, Coprococcus, Collinsella | Personalized microbiome affects disease outcomes in COVID-19 patients; targeting the gut microbiota has potential to prevent and treat COVID-19 |
| Zuo et al[20], 2021 | Shotgun | Collinsella aerofaciens, Collinsella tanakaei, Streptococcus infantis, Morganella morganii | Parabacteroides merdae, Bacteroides stercoris,Alistipes onderdonkii, Lachnospiraceae bacterium 1_1_57FAA | Elimination of gut SARS-CoV-2 activity and modulation of gut microbiome composition should be considered new treatments for COVID-19; the 3’ end of SARS-CoV-2 genome was highly covered than the 5’ end |
| Lv et al[21], 2021 | ITS sequencing | - | Ascomycota (Aspergillaceae, Candida parapsilosis, Talaromyces wortmannii); Basidiomycota (Malassezia yamatoensis, Rhodotorula mucilaginosa, Moesziomyces aphidis, Trechispora sp. and Wallemia sebi) | Total gut fungal burden was significantly elevated in patients infected with SARS- CoV-2; altered gut fungi and microbiota are closely related to patient clinical characteristics |
| Suskun et al[22], 2022 | 16S rrna | Bifidobacterium adolescentis, Dorea formicigenerasus, Eubacterium dolichum, Eggerthella lenta | Faecalibacterium prausnitzii | First evaluate the microbiota composition in multisystem inflammatory syndrome in children cases |
Moreover, it is widely thought that gut dysbiosis is closely associated with COVID-19 severity. The basal gut microbiota of healthy individuals and its alterations during SARS-CoV-2 infection influence host susceptibility to SARS-CoV-2 and disease recovery. The abundances of Coprobacillus, Clostridium ramosum, and Clostridium hathewayi were significantly and positively associated with COVID-19 severity, while the abundances of Bifidobacterium bifiduz, Alistipes onderdonkii and F. prausnitzii were negatively associated[10,12]. However, even different members of the same phylum, such as Firmicutes, have opposing correlations in influencing disease severity and regulating ACE2 expression[10]. ACE2 presents a critical role in gut microbial ecology, gut inflammation and innate immunity[25]. Moreover, the abundances of other bacteria, including Bacteroides dorei (B. dorei), Bacteroides thetaiotaomicron (B. thetaiotaomicron), Bacteroides ovatus (B. ovatus), Ruminococcus, Clostridium citroniae, Bifidobacterium, and Haemophilus parainfluenzae, were negatively associated with viral load in fecal samples from SARS-CoV-2-infected patients[10]. Interestingly, B. dorei, B. thetaiotaomicron, and B. ovatus can downregulate ACE2 expression in the murine gut[10,26], suggesting that Bacteroides species may play a protective role against SARS-CoV-2 infection by interfering with ACE2 production. The abundances of Erysipelotrichaceae bacterium, Prevotella copri, and Eubacterium dolichum have been reported to be positively correlated with fecal viral load[10,19]. Furthermore, Prevotella, Enterococcus, Enterobacteriaceae, and Campylobacter can contribute to higher infectivity and worse prognosis in COVID-19[27]. For instance, Prevotella is associated with enhanced T helper 17 (Th17)-mediated mucosal inflammation, stimulating the production of cytokines and subsequently promoting neutrophil recruitment and inflammation[28]. Although it has been established that an altered gut microbiota composition is prevalent in COVID-19, conflicting results have been reported due to the heterogeneity of the gut microbiota itself, the sample size, and so on. Geographic and demographic differences also appear to affect the conclusions and dysbiosis recovery after SARS-CoV-2 infection[10,29]. Therefore, targeting specific gut microbiota alterations represents a potential strategy to alleviate disease severity in COVID-19.
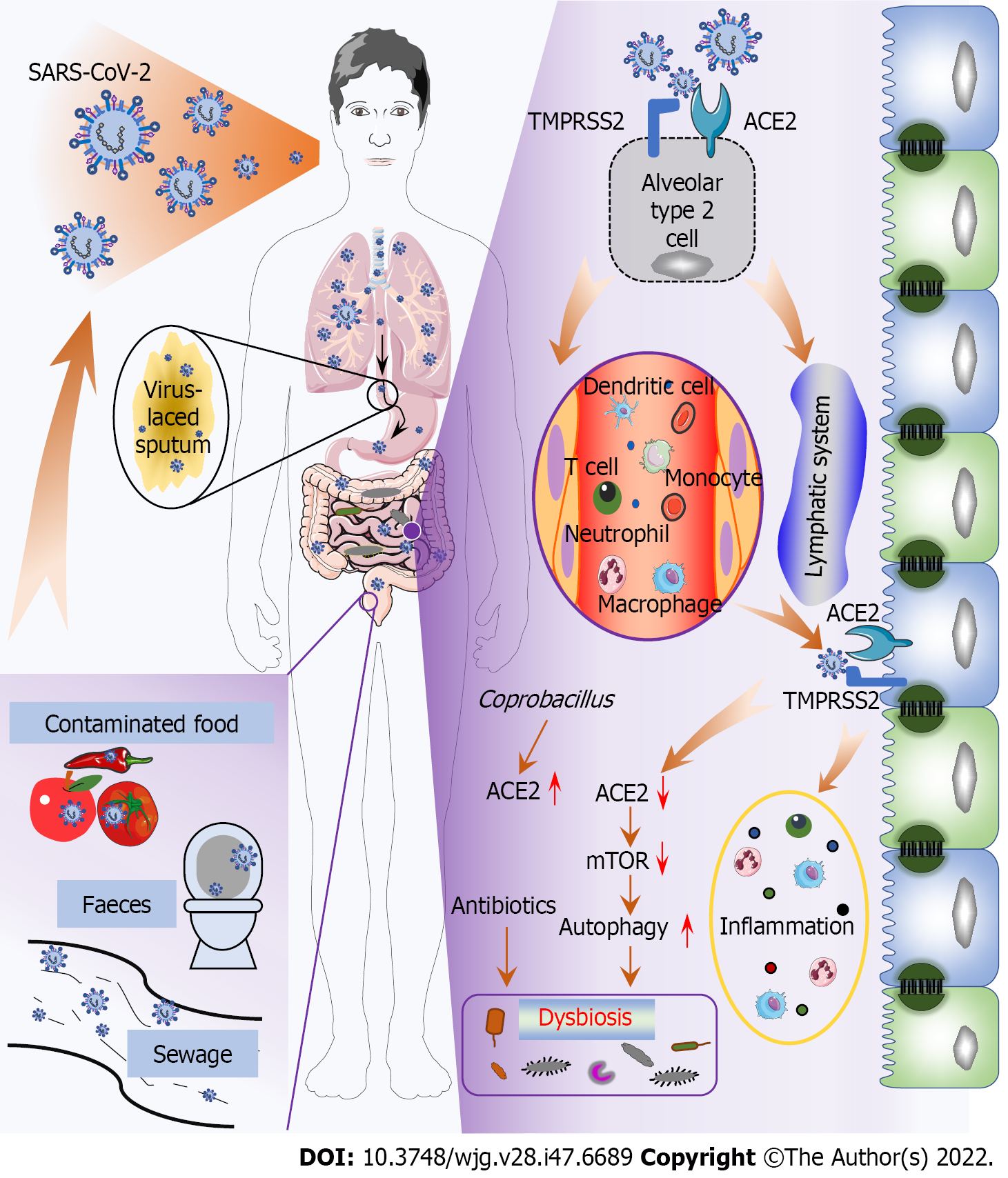
Generally, SARS-CoV-2 is principally transmitted via respiratory droplets and close contact, subsequently inducing diverse symptoms[30]; however, this paradigm does not explain how SARS-CoV-2 causes gut infection and dysbiosis. An in vitro study reported that the virus could be transmitted by the fecal-oral route through contaminated water, food, etc[30,31]. Since then, a new SARS-CoV-2 transmission route has been revealed (Figure 1). Notably, many viral activities are greatly diminished or even lost after passing through the gastrointestinal tract since gastric and intestinal fluids (low pH, rich in bile and digestive enzymes) can destroy the viral lipid envelope, inhibiting infectivity. For example, SARS-CoV has long been thought to be inactivated under acidic conditions (pH < 3). However, SARS-CoV-2 seems to overcome this obstacle since virus have been detected in the feces of infected individuals[30,31], and viruses isolated from feces can survive for an additional 1-2 d[32]. Therefore, SARS-CoV-2 may remain infectious in feces, particularly when patients have diarrhea[1]. Furthermore, virus-containing sputum swallowed by COVID-19 patients may be another pathway of gut infection because viscous sputum can protect the virions, preserving virus infectivity[33]. However, in the absence of evidence of fecal viral titers and viral viability in sewage and contaminated food, the capability of SARS-CoV-2 to be transmitted by the fecal-oral route requires further confirmation.
Moreover, circulatory and immune pathways are reportedly critical for SARS-CoV-2 to cause gut infection and dysbiosis (Figure 1). Studies conducted on SARS showed that coronaviruses damage lung tissue and then migrate to the systemic circulation, where they migrate to gut cells through the circulatory and lymphatic systems[34]. The virus can be found in the blood samples and gut of COVID-19 patients; in addition to those in epithelial cells, viral components are mainly present in intestinal lymphocytes and macrophages[31,35]. Therefore, it is speculated that SARS-CoV-2 may be transported from the lungs to other tissues, including the gastrointestinal tract, through transport via immune cells, similar to influenza virus[35,36]. Subsequently, SARS-CoV-2 invades gut epithelial cells by binding to AEC2[37], causing the release of cytokines and chemokines and triggering a gut inflammatory response characterized by neutrophil, macrophage, and T-cell infiltration, which further promotes dysbiosis[38-40]. In addition, the amino acid transport function of ACE2 is related to the microecology in the gut. B0AT1, a molecular ACE2 chaperone, mediates the absorption of neutral amino acids in the intestinal epithelium[41]. Studies have confirmed that SARS-CoV-2-induced downregulation of B0AT1 on gut epithelial cells contributes to gut barrier disruption and dysbiosis, promoting pathogen invasion and COVID-19 exacerbation[42-44]. de Oliveira et al[45] hypothesized that the internalization of SARS-CoV-2 causes ACE2 downregulation, leading to mechanistic target of rapamycin (mTOR) inhibition and intestinal autophagy activation. Interestingly, autophagy can regulate the gut microbiota, and increased autophagy has been associated with diarrhea[45,46]. There is currently no evidence, however, that the ACE2/mTOR/autophagy pathway is involved in the pathogenesis of COVID-19. Paradoxically, Coprobacillus, which is mostly correlated with the COVID-19 severity, has been shown to increase colonic expression of ACE2[26,47]. Therefore, the net expression of ACE2 in the gut remains largely unclear, warranting further investigation.
Misuse and overuse of antibiotics, common in initially treating COVID-19 patients[48], significantly impact the gut microbiota (Figure 1). Yeoh et al[12] found that COVID-19 patients treated with and without antibiotics had different gut microbiota composition. In addition, antibiotics attenuated the antiviral activity of commensal-enhanced type I interferons (IFN-I)[49]. Therefore, antibiotics are unlikely to improve patient outcomes without comorbid bacterial infections but instead exacerbate and prolong gut dysbiosis in this patient population.
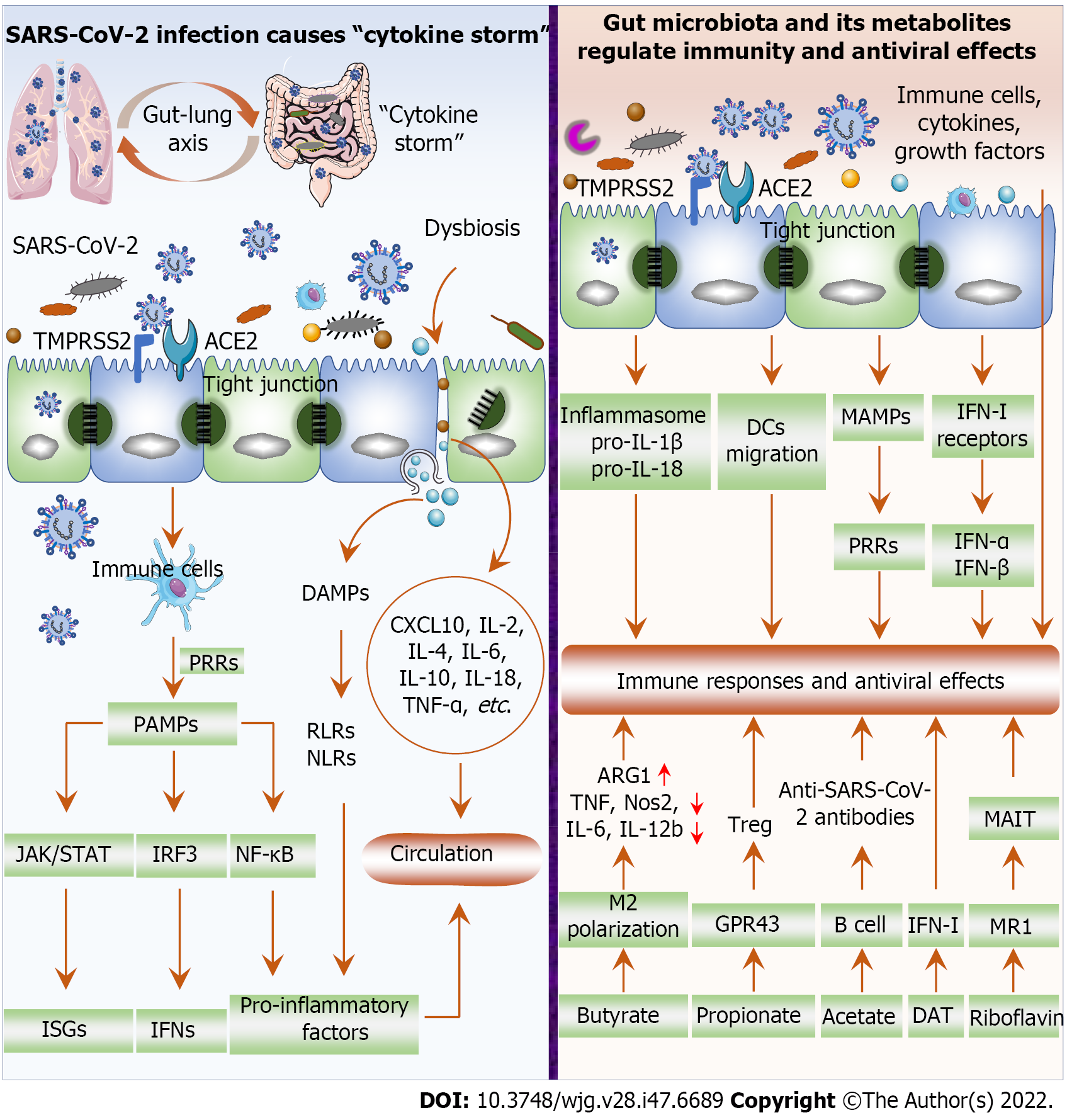
Immune and inflammatory responses are important pathophysiological mechanisms in the pathogenesis of COVID-19. SARS-CoV-2 invades cells through ACE2 and TMPRSS2, where it replicates rapidly, producing and releasing large numbers of viruses and inducing excessive inflammatory cytokine release “cytokine storm”, such as IL-6, IL-1β, TNF-α, and IFN[41,50] (Figure 2). Excessive production of proinflammatory cytokines is pathologically associated with acute respiratory distress syndrome, extensive tissue damage, and even death. Viral replication after SARS-CoV-2 invades cells induces immune cells to recognize and bind viral pathogen-associated molecular patterns through pattern recognition receptors (PRRs), followed by the activation of the NF-κB, IRF3 and JAK/STAT signaling pathways to induce the expression of proinflammatory factors, IFNs and IFN-stimulated genes (ISGs)[41,51,52]. In addition, killing or damage of cells by SARS-CoV-2 results in the release of danger-associated molecular patterns, activating RIG-I-like receptors and NOD-like receptors and subsequently facilitating proinflammatory factor expression[41,51]. Dysbiosis after SARS-CoV-2 infection further damages the gut barrier and promotes the production of inflammatory factors such as CXCL10, IL-2, IL-4, IL-6, IL-10, IL-18, and TNF-α[16,53,54]. For example, B. dorei and Akkermansia muciniphila were positively associated with IL-1β, IL-6 and CXCL8[12]. Subsequently, opportunistic pathogens and inflammatory factors infiltrate the circulation and cause systemic inflammation and infection[55]. Therefore, SARS-CoV-2 infection promotes gut inflammation to aggravate dysbiosis, which in turn exacerbates inflammation and disease progression, forming a vicious cycle.
The gut microbiota and its metabolites influence the host immune response, inflammation and the development and regression of pulmonary infectious diseases, such as influenza A virus and Streptococcus pneumonia[56]. A healthy microbiome protects against respiratory viral infections[57,58]. During respiratory viral infection, the gut microbiota regulates the host immune response via the gut-lung axis, which refers to the crosstalk between the gut microbiota and lung. Therefore, endotoxins and microbial metabolites can affect the lung through the blood circulation, and conversely, lung inflammation will affect the gut microbiota. It has been reported that gut dysbiosis significantly increases mortality from respiratory viral infections, which may be associated with dysregulated immune responses; increased secretion of IFN-γ, IL-6, and CCL2; and decreased Treg cell counts in the lung and gastrointestinal tract[59]. The gut microbiota facilitates inflammasome activation, pro-IL-1β and pro-IL-18 expression, and dendritic cell (DC) migration, which are critical for protective immunity post-influenza virus infection[60] (Figure 2). In addition, microbial-associated molecular patterns are transmitted to parenteral tissues to activate PRRs in immune cells and influence innate immune responses[61]. The gut microbiota reportedly regulates the expression of IFN-I receptors in respiratory epithelial cells that limit viral replication and increase resistance to viral infections through IFN-α and IFN-β[58,62]. The gut commensal microbiota promotes the IFN-I response and ISG expression by inducing IFN-β expression mediated by TLR4-TRIF signaling through colonic lamina propria DCs, thereby enhancing antiviral capacity[62]. The gut microbiota can also affect respiratory mucosal immunity through multiple mechanisms. On the one hand, activated immune cells within the mucosa can reach and affect distant mucosal sites, highlighting the beneficial effects of the gut microbiota during respiratory viral infections[63,64]. On the other hand, the gut microbiota affects the secretion of cytokines and growth factors by the gastrointestinal mucosa, which reach the systemic circulation and act on other mucosal tissues[63,65]. Furthermore, severe COVID-19 clinical symptoms or complications and higher mortality are more likely to arise in elderly individuals or in patients with concomitant underlying disease, such as cardiovascular disease, diabetes, and cancer[66]. Critically ill patients with COVID-19 often require invasive ventilator-assisted ventilation. However, gut microbiota intervention can reduce the demand for invasive mechanical ventilation in critically ill patients and reduce the incidence of ventilator-associated pneumonia[67], although this finding needs further validation in COVID-19 patients. Therefore, restoring gut microbiota homeostasis is essential for inhibiting the inflammatory response and enhancing antiviral effects.
Gut microbiota metabolites are signaling molecules and substrates for metabolic reactions[68]. Microbial metabolites are absorbed by the gut mucosa and participate in mucosal immune regulation, a process known as “metabolic reprogramming”[69,70]. For example, short-chain fatty acids (SCFAs, mainly including acetate, propionate, and butyrate) can reach distant organs through the bloodstream to exert immunomodulatory and immunoglobulin expression-inducing effects, as well as anti-inflammatory and antiviral effects[70,71] (Figure 2). Butyrate facilitates the polarization of M2 macrophage and exerts anti-inflammatory effects by increasing arginase 1 (ARG1) and decreasing TNF, Nos2, IL-6, and IL-12b expression[41,72]. Butyrate also inhibits histone deacetylase activity or increases the Foxp3 promoter transcription in naive T cells, thereby promoting the naive T cells differentiation into Treg cells[73,74]. Treg cells can suppress autoinflammatory responses and protect tissues from damage caused by pathological immune responses[41]. Furthermore, butyrate restored CD8+ T-cell function in mice and inhibited proinflammatory cytokine production as well as eosinophilic lung infiltration[49]. Unfortunately, a reduced richness of butyrate-producing bacteria such as F. prausnitzii and Clostridium species has been found in COVID-19 patients[15,17,20,75]. Propionate promotes Treg cell proliferation by activating G-protein-coupled receptor 43 (GPR43)[76]. Moreover, microbe-produced SCFAs (acetate) enhance B-cell metabolism and gene expression, promoting the production of anti-SARS-CoV-2 antibodies and thereby inhibiting disease progression[77]. Deaminotyrosine (DAT) produced by Clostridium orbiscindens augments IFN-I signaling to protect the host from viral infection[57]. In addition, riboflavin is a product of gut microbiota constituents, which can activate mucosal-associated T cells (MAIT) via restrictive major histocompatibility complex (MHC)-related protein-1 (MR1)[78,79]. MAIT cells, as innate sensors and mediators of the antiviral response, exert antiviral response to SARS-CoV-2 through the gut-lung axis[80]. Antimicrobial peptides secreted by bacteria can promote virolysis, block cell virus fusion, and induce adaptive immune responses, and thus they have been proposed as viable alternative therapeutic treatments for infections by viruses, such as MERS-CoV[81]. Therefore, COVID-19 severity is tightly correlated with the levels of gut microbiota metabolites. However, studies on the direct action of gut microbiota metabolites in COVID-19 remain scarce.
The gut microbiota is closely related to various human gastrointestinal diseases and symptoms, such as gastrointestinal cancer, stomachache, diarrhea, and flatulence[82,83]. For instance, Escherichia coli, Shigella, Salmonella, Campylobacter, Clostridium difficile, and Aeromonas are the main pathogens that cause diarrhea[84,85]. Reprogramming the gut microbiota via probiotics, prebiotics and FMT is well documented to effectively treat gastrointestinal diseases. Moreover, probiotic/prebiotic administration has been shown to protect against viral infections in multiple studies (e.g., those caused by influenza virus, rhinovirus, respiratory syncytial virus and coronaviruses). For example, Johnson et al[86] found that Bacillus subtilis peptidoglycans reduced the infectivity of coronavirus. Peptidoglycan-associated surfactin can disrupt virion integrity, such as in influenza, Ebola, Zika, and Mayaro[86]. Therefore, probiotics, prebiotics and FMT can restore ecological homeostasis by regulating the gut microbiota, representing an effective alternative approach to ameliorate or suppress COVID-19 severity.
Probiotics, such as Lactobacillus, yeast, Bifidobacterium, Enterococcus, and Bacillus, are live microorganisms that benefit the host by colonizing the human body. Probiotics maintain healthy gut homeostasis and exert antiviral effects through the gut-lung axis[87,88] and have been widely used to treat gastrointestinal diseases, including diarrhea (acute, antibiotic-associated and C. difficile-associated) and adult inflammatory bowel disease (IBD)[89-91]. On the one hand, probiotics can strengthen Treg cell and natural killer cell activity and suppress proinflammatory cytokines, such as TNF-a, CRP, IL-1b, IL2, IL-6, IL7, MCP1, and LDH[92,93]. Lactobacillus exerts antiviral activity through direct probiotic-virus interactions, production of antiviral metabolites, and stimulation of the immune system[94]. On the other hand, probiotics are beneficial for enhancing epithelial barrier function and improving gut microbial diversity. Furthermore, probiotics combat and block harmful bacterial strains in the gut or enhance beneficial signaling pathways[95,96]. Existing evidence shows that probiotic miRNA modulation and regulation of signaling pathways such as NF-kB and STAT1 ameliorate COVID-19 complications[97,98]. Probiotics have the potential to interact with ACE2; for example, some lactobacilli release peptides with a high affinity for ACE2[93,99]. Lactobacillus spp. and Bifidobacterium spp. exhibit the strongest anti-respiratory virus activity via an immunomodulatory mechanism[56]. In January 2020, relevant Chinese government departments recommended the addition of probiotics in COVID-19 treatment to improve gut microbiota homeostasis and protect against subsequent bacterial infections. However, evidence detailing the efficacy of probiotics in treating COVID-19 is limited, although relevant clinical trials are being conducted. Moreover, the safety of probiotics in COVID-19 patients should be emphasized to ensure that their use does not induce new gastrointestinal symptoms or secondary infections.
Prebiotics are substrates that are selectively utilized by host microorganisms to provide health benefits[100]. Unfortunately, little information is available on using prebiotics to treat respiratory infections. In a clinical trial of prebiotics, KB109 was found to regulate gut microbiome composition and increase the production of SCFAs in the gut (NCT04414124). For instance, butyrate and propionate obtained from the fermentation of prebiotics affect the differentiation or function of T cells, macrophages, and DCs[93]. In addition, prebiotics selectively stimulate beneficial bacterial growth or enhance the activity of indigenous probiotics[93]. Accordingly, the role of prebiotics in COVID-19 patients should be further emphasized.
FMT refers to the transplantation of a functional microbiota from healthy individuals into the gastrointestinal tract of patients to treat intestinal and extraintestinal diseases[84]. An expanding body of evidence suggests that FMT can effectively treat many diseases, such as cancer, liver diseases, C. difficile infection, irritable bowel syndrome (IBS), and IBD[101-103]. Since COVID-19 patients exhibit a depletion of beneficial commensal bacteria, FMT represents a theoretically promising strategy to mitigate disease severity. Regrettably, convincing clinical evidence for the effect of FMT in COVID-19 is lacking, and its safety and efficacy warrant further investigation.
COVID-19 was previously identified as a respiratory infectious disease. However, accumulating clinical studies have subsequently found that a large proportion of patients have gastrointestinal symptoms, such as abdominal pain, diarrhea, vomiting, and acid reflux, as well as significant gut microbiota alterations[7,30]. The gut microbiota not only significantly affects the COVID-19 development and disease severity but also reflects the susceptibility of COVID-19 patients to long-term complications[104]. Studies have confirmed that dysbiosis increases the poor prognosis of COVID-19[104]. However, controversial results exist regarding gut microbiota alterations in COVID-19 patients. A variety of factors, such as sex, age, basic health status, medication use, genetics, ethnicity, and geographic location, can affect the composition of the gut microbiota and lead to individual differences and varying responses to SARS-CoV-2 infection. Therefore, further exploration of the specific gut microbiota alterations in COVID-19 patients and the clinical correlation between the gut microbiota and COVID-19 will be a very challenging and valuable research direction in the future.
Unfortunately, direct evidence for the contribution of the gut microbiota in COVID-19 remains lacking. For example, the exact mechanism of dysbiosis remains unclear. Moreover, the regulation of host immune and inflammatory responses and antiviral effects by the gut microbiota during SARS-CoV-2 infection relies largely on inferences or conjectures from previous studies. Given that specific drugs for COVID-19 remain enigmatic, vaccines represent the most effective prevention and control strategy; however, the continuous mutation of the virus has exacerbated this conundrum. Therefore, gut microbiota intervention through probiotics, prebiotics and FMT is undoubtedly one of the promising cosupplementation strategies for the future treatment of COVID-19, but large-scale studies are lacking and there are no corresponding treatment guidelines. The therapeutic prospects of the gut microbiota in COVID-19 are promising, but there is still a long way to go to realize its potential.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chauhan S, United States; Mahmoud MZ, Saudi Arabia S-Editor: Chen YL L-Editor: A P-Editor: Chen YL
| 1. | Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, Liu Z, Brulois KF, Wang X, Greenberg HB, Diamond MS, Ciorba MA, Whelan SPJ, Ding S. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 809] [Cited by in RCA: 765] [Article Influence: 153.0] [Reference Citation Analysis (0)] |
| 2. | Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM, Feldman J, Muus C, Wadsworth MH 2nd, Kazer SW, Hughes TK, Doran B, Gatter GJ, Vukovic M, Taliaferro F, Mead BE, Guo Z, Wang JP, Gras D, Plaisant M, Ansari M, Angelidis I, Adler H, Sucre JMS, Taylor CJ, Lin B, Waghray A, Mitsialis V, Dwyer DF, Buchheit KM, Boyce JA, Barrett NA, Laidlaw TM, Carroll SL, Colonna L, Tkachev V, Peterson CW, Yu A, Zheng HB, Gideon HP, Winchell CG, Lin PL, Bingle CD, Snapper SB, Kropski JA, Theis FJ, Schiller HB, Zaragosi LE, Barbry P, Leslie A, Kiem HP, Flynn JL, Fortune SM, Berger B, Finberg RW, Kean LS, Garber M, Schmidt AG, Lingwood D, Shalek AK, Ordovas-Montanes J; HCA Lung Biological Network. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181:1016-1035.e19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1890] [Cited by in RCA: 1793] [Article Influence: 358.6] [Reference Citation Analysis (0)] |
| 3. | Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271-280.e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11946] [Cited by in RCA: 14257] [Article Influence: 2851.4] [Reference Citation Analysis (0)] |
| 4. | Dhar D, Mohanty A. Gut microbiota and Covid-19- possible link and implications. Virus Res. 2020;285:198018. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 330] [Cited by in RCA: 395] [Article Influence: 79.0] [Reference Citation Analysis (0)] |
| 5. | Qi F, Qian S, Zhang S, Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun. 2020;526:135-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 523] [Cited by in RCA: 741] [Article Influence: 148.2] [Reference Citation Analysis (0)] |
| 6. | Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology. 2020;158:1831-1833.e3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1985] [Cited by in RCA: 1993] [Article Influence: 398.6] [Reference Citation Analysis (1)] |
| 7. | Cheung KS, Hung IFN, Chan PPY, Lung KC, Tso E, Liu R, Ng YY, Chu MY, Chung TWH, Tam AR, Yip CCY, Leung KH, Fung AY, Zhang RR, Lin Y, Cheng HM, Zhang AJX, To KKW, Chan KH, Yuen KY, Leung WK. Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples From a Hong Kong Cohort: Systematic Review and Meta-analysis. Gastroenterology. 2020;159:81-95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1113] [Cited by in RCA: 1131] [Article Influence: 226.2] [Reference Citation Analysis (1)] |
| 8. | Lin W, Xie Z, Li Y, Li L, Wen C, Cao Y, Chen X, Ou X, Hu F, Li F, Tang X, Cai W. Association between detectable SARS-COV-2 RNA in anal swabs and disease severity in patients with coronavirus disease 2019. J Med Virol. 2021;93:794-802. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 9. | Xu Y, Li X, Zhu B, Liang H, Fang C, Gong Y, Guo Q, Sun X, Zhao D, Shen J, Zhang H, Liu H, Xia H, Tang J, Zhang K, Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26:502-505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 992] [Cited by in RCA: 1052] [Article Influence: 210.4] [Reference Citation Analysis (0)] |
| 10. | Zuo T, Zhang F, Lui GCY, Yeoh YK, Li AYL, Zhan H, Wan Y, Chung ACK, Cheung CP, Chen N, Lai CKC, Chen Z, Tso EYK, Fung KSC, Chan V, Ling L, Joynt G, Hui DSC, Chan FKL, Chan PKS, Ng SC. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology. 2020;159:944-955.e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 739] [Cited by in RCA: 1062] [Article Influence: 212.4] [Reference Citation Analysis (0)] |
| 11. | Villapol S. Gastrointestinal symptoms associated with COVID-19: impact on the gut microbiome. Transl Res. 2020;226:57-69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 194] [Cited by in RCA: 217] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 12. | Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY, Chung AC, Cheung CP, Tso EY, Fung KS, Chan V, Ling L, Joynt G, Hui DS, Chow KM, Ng SSS, Li TC, Ng RW, Yip TC, Wong GL, Chan FK, Wong CK, Chan PK, Ng SC. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698-706. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 995] [Cited by in RCA: 862] [Article Influence: 215.5] [Reference Citation Analysis (0)] |
| 13. | Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69:137-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 693] [Cited by in RCA: 815] [Article Influence: 62.7] [Reference Citation Analysis (0)] |
| 14. | Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19:55-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 729] [Cited by in RCA: 2651] [Article Influence: 530.2] [Reference Citation Analysis (1)] |
| 15. | Gu S, Chen Y, Wu Z, Gao H, Lv L, Guo F, Zhang X, Luo R, Huang C, Lu H, Zheng B, Zhang J, Yan R, Zhang H, Jiang H, Xu Q, Guo J, Gong Y, Tang L, Li L. Alterations of the Gut Microbiota in Patients With Coronavirus Disease 2019 or H1N1 Influenza. Clin Infect Dis. 2020;71:2669-2678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 324] [Cited by in RCA: 568] [Article Influence: 142.0] [Reference Citation Analysis (0)] |
| 16. | Tao W, Zhang G, Wang X, Guo M, Zeng W, Xu Z, Cao D, Pan A, Wang Y, Zhang K, Ma X, Chen Z, Jin T, Liu L, Weng J, Zhu S. Analysis of the intestinal microbiota in COVID-19 patients and its correlation with the inflammatory factor IL-18. Med Microecol. 2020;5:100023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 110] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 17. | Tang L, Gu S, Gong Y, Li B, Lu H, Li Q, Zhang R, Gao X, Wu Z, Zhang J, Zhang Y, Li L. Clinical Significance of the Correlation between Changes in the Major Intestinal Bacteria Species and COVID-19 Severity. Engineering (Beijing). 2020;6:1178-1184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 115] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 18. | Zuo T, Zhan H, Zhang F, Liu Q, Tso EYK, Lui GCY, Chen N, Li A, Lu W, Chan FKL, Chan PKS, Ng SC. Alterations in Fecal Fungal Microbiome of Patients With COVID-19 During Time of Hospitalization until Discharge. Gastroenterology. 2020;159:1302-1310.e5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 250] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 19. | Wu Y, Cheng X, Jiang G, Tang H, Ming S, Tang L, Lu J, Guo C, Shan H, Huang X. Altered oral and gut microbiota and its association with SARS-CoV-2 viral load in COVID-19 patients during hospitalization. NPJ Biofilms Microbiomes. 2021;7:61. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 134] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 20. | Zuo T, Liu Q, Zhang F, Lui GC, Tso EY, Yeoh YK, Chen Z, Boon SS, Chan FK, Chan PK, Ng SC. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut. 2021;70:276-284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 262] [Article Influence: 65.5] [Reference Citation Analysis (0)] |
| 21. | Lv L, Gu S, Jiang H, Yan R, Chen Y, Luo R, Huang C, Lu H, Zheng B, Zhang H, Xia J, Tang L, Sheng G, Li L. Gut mycobiota alterations in patients with COVID-19 and H1N1 infections and their associations with clinical features. Commun Biol. 2021;4:480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 62] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 22. | Suskun C, Kilic O, Yilmaz Ciftdogan D, Guven S, Karbuz A, Ozkaya Parlakay A, Kara Y, Kacmaz E, Sahin A, Boga A, Kizmaz Isancli D, Gulhan B, Kanik-Yuksek S, Kiral E, Bozan G, Arslanoglu MO, Kizil MC, Dinleyici M, Us T, Varis A, Kaya M, Vandenplas Y, Dinleyici EC. Intestinal microbiota composition of children with infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and multisystem inflammatory syndrome (MIS-C). Eur J Pediatr. 2022;181:3175-3191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 36] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 23. | Newsome RC, Gauthier J, Hernandez MC, Abraham GE, Robinson TO, Williams HB, Sloan M, Owings A, Laird H, Christian T, Pride Y, Wilson KJ, Hasan M, Parker A, Senitko M, Glover SC, Gharaibeh RZ, Jobin C. The gut microbiome of COVID-19 recovered patients returns to uninfected status in a minority-dominated United States cohort. Gut Microbes. 2021;13:1-15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 24. | Weiser JN, Ferreira DM, Paton JC. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol. 2018;16:355-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 535] [Cited by in RCA: 681] [Article Influence: 97.3] [Reference Citation Analysis (0)] |
| 25. | Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, Sigl V, Hanada T, Hanada R, Lipinski S, Wild B, Camargo SM, Singer D, Richter A, Kuba K, Fukamizu A, Schreiber S, Clevers H, Verrey F, Rosenstiel P, Penninger JM. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477-481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 787] [Cited by in RCA: 985] [Article Influence: 75.8] [Reference Citation Analysis (0)] |
| 26. | Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, Yanortsang TB, Yang L, Jupp R, Mathis D, Benoist C, Kasper DL. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell. 2017;168:928-943.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 397] [Cited by in RCA: 543] [Article Influence: 67.9] [Reference Citation Analysis (0)] |
| 27. | Hung YP, Lee CC, Lee JC, Tsai PJ, Ko WC. Gut Dysbiosis during COVID-19 and Potential Effect of Probiotics. Microorganisms. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 28. | Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151:363-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 472] [Cited by in RCA: 884] [Article Influence: 110.5] [Reference Citation Analysis (0)] |
| 29. | Chen Y, Gu S, Chen Y, Lu H, Shi D, Guo J, Wu WR, Yang Y, Li Y, Xu KJ, Ding C, Luo R, Huang C, Yu L, Xu M, Yi P, Liu J, Tao JJ, Zhang H, Lv L, Wang B, Sheng J, Li L. Six-month follow-up of gut microbiota richness in patients with COVID-19. Gut. 2022;71:222-225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 139] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 30. | Guo M, Tao W, Flavell RA, Zhu S. Potential intestinal infection and faecal-oral transmission of SARS-CoV-2. Nat Rev Gastroenterol Hepatol. 2021;18:269-283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 198] [Cited by in RCA: 213] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 31. | Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020;323:1843-1844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1894] [Cited by in RCA: 2658] [Article Influence: 531.6] [Reference Citation Analysis (0)] |
| 32. | Chan KH, Sridhar S, Zhang RR, Chu H, Fung AY, Chan G, Chan JF, To KK, Hung IF, Cheng VC, Yuen KY. Factors affecting stability and infectivity of SARS-CoV-2. J Hosp Infect. 2020;106:226-231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 172] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 33. | Hirose R, Nakaya T, Naito Y, Daidoji T, Watanabe Y, Yasuda H, Konishi H, Itoh Y. Mechanism of Human Influenza Virus RNA Persistence and Virion Survival in Feces: Mucus Protects Virions From Acid and Digestive Juices. J Infect Dis. 2017;216:105-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 34. | Aktas B, Aslim B. Gut-lung axis and dysbiosis in COVID-19. Turk J Biol. 2020;44:265-272. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 92] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 35. | Qian Q, Fan L, Liu W, Li J, Yue J, Wang M, Ke X, Yin Y, Chen Q, Jiang C. Direct Evidence of Active SARS-CoV-2 Replication in the Intestine. Clin Infect Dis. 2021;73:361-366. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 119] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 36. | Choi SM, Xie H, Campbell AP, Kuypers J, Leisenring W, Boudreault AA, Englund JA, Corey L, Boeckh M. Influenza viral RNA detection in blood as a marker to predict disease severity in hematopoietic cell transplant recipients. J Infect Dis. 2012;206:1872-1877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 37. | Hikmet F, Méar L, Edvinsson Å, Micke P, Uhlén M, Lindskog C. The protein expression profile of ACE2 in human tissues. Mol Syst Biol. 2020;16:e9610. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 631] [Cited by in RCA: 709] [Article Influence: 141.8] [Reference Citation Analysis (0)] |
| 38. | Harmer D, Gilbert M, Borman R, Clark KL. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532:107-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 585] [Cited by in RCA: 655] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 39. | Dang AT, Marsland BJ. Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. 2019;12:843-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 633] [Article Influence: 105.5] [Reference Citation Analysis (0)] |
| 40. | Katz-Agranov N, Zandman-Goddard G. Autoimmunity and COVID-19 - The microbiotal connection. Autoimmun Rev. 2021;20:102865. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 41. | Wang B, Zhang L, Wang Y, Dai T, Qin Z, Zhou F. Alterations in microbiota of patients with COVID-19: potential mechanisms and therapeutic interventions. Signal Transduct Target Ther. 2022;7:143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 120] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 42. | Viana SD, Nunes S, Reis F. ACE2 imbalance as a key player for the poor outcomes in COVID-19 patients with age-related comorbidities - Role of gut microbiota dysbiosis. Ageing Res Rev. 2020;62:101123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 121] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 43. | Penninger JM, Grant MB, Sung JJY. The Role of Angiotensin Converting Enzyme 2 in Modulating Gut Microbiota, Intestinal Inflammation, and Coronavirus Infection. Gastroenterology. 2021;160:39-46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 103] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 44. | Kowalczuk S, Bröer A, Tietze N, Vanslambrouck JM, Rasko JE, Bröer S. A protein complex in the brush-border membrane explains a Hartnup disorder allele. FASEB J. 2008;22:2880-2887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 163] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 45. | de Oliveira AP, Lopes ALF, Pacheco G, de Sá Guimarães Nolêto IR, Nicolau LAD, Medeiros JVR. Premises among SARS-CoV-2, dysbiosis and diarrhea: Walking through the ACE2/mTOR/autophagy route. Med Hypotheses. 2020;144:110243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 46. | Megyeri K, Dernovics Á, Al-Luhaibi ZII, Rosztóczy A. COVID-19-associated diarrhea. World J Gastroenterol. 2021;27:3208-3222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 33] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (2)] |
| 47. | Enaud R, Prevel R, Ciarlo E, Beaufils F, Wieërs G, Guery B, Delhaes L. The Gut-Lung Axis in Health and Respiratory Diseases: A Place for Inter-Organ and Inter-Kingdom Crosstalks. Front Cell Infect Microbiol. 2020;10:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 372] [Cited by in RCA: 471] [Article Influence: 94.2] [Reference Citation Analysis (0)] |
| 48. | Langford BJ, So M, Raybardhan S, Leung V, Westwood D, MacFadden DR, Soucy JR, Daneman N. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect. 2020;26:1622-1629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1053] [Cited by in RCA: 978] [Article Influence: 195.6] [Reference Citation Analysis (0)] |
| 49. | Gasmi A, Tippairote T, Mujawdiya PK, Peana M, Menzel A, Dadar M, Benahmed AG, Bjørklund G. The microbiota-mediated dietary and nutritional interventions for COVID-19. Clin Immunol. 2021;226:108725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 50. | Sims JT, Krishnan V, Chang CY, Engle SM, Casalini G, Rodgers GH, Bivi N, Nickoloff BJ, Konrad RJ, de Bono S, Higgs RE, Benschop RJ, Ottaviani S, Cardoso A, Nirula A, Corbellino M, Stebbing J. Characterization of the cytokine storm reflects hyperinflammatory endothelial dysfunction in COVID-19. J Allergy Clin Immunol. 2021;147:107-111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 126] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 51. | Khanmohammadi S, Rezaei N. Role of Toll-like receptors in the pathogenesis of COVID-19. J Med Virol. 2021;93:2735-2739. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 257] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 52. | Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20:355-362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1356] [Cited by in RCA: 1805] [Article Influence: 361.0] [Reference Citation Analysis (0)] |
| 53. | Effenberger M, Grabherr F, Mayr L, Schwaerzler J, Nairz M, Seifert M, Hilbe R, Seiwald S, Scholl-Buergi S, Fritsche G, Bellmann-Weiler R, Weiss G, Müller T, Adolph TE, Tilg H. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut. 2020;69:1543-1544. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 223] [Cited by in RCA: 226] [Article Influence: 45.2] [Reference Citation Analysis (0)] |
| 54. | Tian W, Zhang N, Jin R, Feng Y, Wang S, Gao S, Gao R, Wu G, Tian D, Tan W, Chen Y, Gao GF, Wong CCL. Immune suppression in the early stage of COVID-19 disease. Nat Commun. 2020;11:5859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 170] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 55. | Velikova T, Snegarova V, Kukov A, Batselova H, Mihova A, Nakov R. Gastrointestinal mucosal immunity and COVID-19. World J Gastroenterol. 2021;27:5047-5059. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 56. | Donati Zeppa S, Agostini D, Piccoli G, Stocchi V, Sestili P. Gut Microbiota Status in COVID-19: An Unrecognized Player? Front Cell Infect Microbiol. 2020;10:576551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 76] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 57. | Steed AL, Christophi GP, Kaiko GE, Sun L, Goodwin VM, Jain U, Esaulova E, Artyomov MN, Morales DJ, Holtzman MJ, Boon ACM, Lenschow DJ, Stappenbeck TS. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science. 2017;357:498-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 409] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 58. | Bradley KC, Finsterbusch K, Schnepf D, Crotta S, Llorian M, Davidson S, Fuchs SY, Staeheli P, Wack A. Microbiota-Driven Tonic Interferon Signals in Lung Stromal Cells Protect from Influenza Virus Infection. Cell Rep. 2019;28:245-256.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 196] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 59. | Grayson MH, Camarda LE, Hussain SA, Zemple SJ, Hayward M, Lam V, Hunter DA, Santoro JL, Rohlfing M, Cheung DS, Salzman NH. Intestinal Microbiota Disruption Reduces Regulatory T Cells and Increases Respiratory Viral Infection Mortality Through Increased IFNγ Production. Front Immunol. 2018;9:1587. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 60. | Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108:5354-5359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 967] [Cited by in RCA: 1137] [Article Influence: 81.2] [Reference Citation Analysis (0)] |
| 61. | Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228-231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 793] [Cited by in RCA: 880] [Article Influence: 58.7] [Reference Citation Analysis (0)] |
| 62. | Stefan KL, Kim MV, Iwasaki A, Kasper DL. Commensal Microbiota Modulation of Natural Resistance to Virus Infection. Cell. 2020;183:1312-1324.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 187] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 63. | Kitazawa H, Villena J. Modulation of Respiratory TLR3-Anti-Viral Response by Probiotic Microorganisms: Lessons Learned from Lactobacillus rhamnosus CRL1505. Front Immunol. 2014;5:201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 64. | Zelaya H, Alvarez S, Kitazawa H, Villena J. Respiratory Antiviral Immunity and Immunobiotics: Beneficial Effects on Inflammation-Coagulation Interaction during Influenza Virus Infection. Front Immunol. 2016;7:633. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 88] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 65. | Zhang D, Li S, Wang N, Tan HY, Zhang Z, Feng Y. The Cross-Talk Between Gut Microbiota and Lungs in Common Lung Diseases. Front Microbiol. 2020;11:301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 294] [Cited by in RCA: 270] [Article Influence: 54.0] [Reference Citation Analysis (0)] |
| 66. | Lithander FE, Neumann S, Tenison E, Lloyd K, Welsh TJ, Rodrigues JCL, Higgins JPT, Scourfield L, Christensen H, Haunton VJ, Henderson EJ. COVID-19 in older people: a rapid clinical review. Age Ageing. 2020;49:501-515. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 161] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 67. | Zeng J, Wang CT, Zhang FS, Qi F, Wang SF, Ma S, Wu TJ, Tian H, Tian ZT, Zhang SL, Qu Y, Liu LY, Li YZ, Cui S, Zhao HL, Du QS, Ma Z, Li CH, Li Y, Si M, Chu YF, Meng M, Ren HS, Zhang JC, Jiang JJ, Ding M, Wang YP. Effect of probiotics on the incidence of ventilator-associated pneumonia in critically ill patients: a randomized controlled multicenter trial. Intensive Care Med. 2016;42:1018-1028. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 119] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 68. | Krautkramer KA, Fan J, Bäckhed F. Gut microbial metabolites as multi-kingdom intermediates. Nat Rev Microbiol. 2021;19:77-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 789] [Article Influence: 157.8] [Reference Citation Analysis (0)] |
| 69. | Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, Marsland BJ. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1604] [Cited by in RCA: 2035] [Article Influence: 185.0] [Reference Citation Analysis (0)] |
| 70. | Villena J, Kitazawa H. The Modulation of Mucosal Antiviral Immunity by Immunobiotics: Could They Offer Any Benefit in the SARS-CoV-2 Pandemic? Front Physiol. 2020;11:699. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 71. | Sencio V, Barthelemy A, Tavares LP, Machado MG, Soulard D, Cuinat C, Queiroz-Junior CM, Noordine ML, Salomé-Desnoulez S, Deryuter L, Foligné B, Wahl C, Frisch B, Vieira AT, Paget C, Milligan G, Ulven T, Wolowczuk I, Faveeuw C, Le Goffic R, Thomas M, Ferreira S, Teixeira MM, Trottein F. Gut Dysbiosis during Influenza Contributes to Pulmonary Pneumococcal Superinfection through Altered Short-Chain Fatty Acid Production. Cell Rep. 2020;30:2934-2947.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 237] [Article Influence: 59.3] [Reference Citation Analysis (0)] |
| 72. | Scott NA, Andrusaite A, Andersen P, Lawson M, Alcon-Giner C, Leclaire C, Caim S, Le Gall G, Shaw T, Connolly JPR, Roe AJ, Wessel H, Bravo-Blas A, Thomson CA, Kästele V, Wang P, Peterson DA, Bancroft A, Li X, Grencis R, Mowat AM, Hall LJ, Travis MA, Milling SWF, Mann ER. Antibiotics induce sustained dysregulation of intestinal T cell immunity by perturbing macrophage homeostasis. Sci Transl Med. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 232] [Article Influence: 38.7] [Reference Citation Analysis (0)] |
| 73. | Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451-455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2516] [Cited by in RCA: 3424] [Article Influence: 285.3] [Reference Citation Analysis (0)] |
| 74. | Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2951] [Cited by in RCA: 3780] [Article Influence: 315.0] [Reference Citation Analysis (0)] |
| 75. | McNabney SM, Henagan TM. Short Chain Fatty Acids in the Colon and Peripheral Tissues: A Focus on Butyrate, Colon Cancer, Obesity and Insulin Resistance. Nutrients. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 361] [Cited by in RCA: 346] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 76. | Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282-1286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2007] [Cited by in RCA: 2396] [Article Influence: 149.8] [Reference Citation Analysis (0)] |
| 77. | Kim M, Qie Y, Park J, Kim CH. Gut Microbial Metabolites Fuel Host Antibody Responses. Cell Host Microbe. 2016;20:202-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 609] [Article Influence: 67.7] [Reference Citation Analysis (0)] |
| 78. | Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, Chen Z, Reantragoon R, Meehan B, Cao H, Williamson NA, Strugnell RA, Van Sinderen D, Mak JY, Fairlie DP, Kjer-Nielsen L, Rossjohn J, McCluskey J. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509:361-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 533] [Cited by in RCA: 680] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 79. | Legoux F, Salou M, Lantz O. MAIT Cell Development and Functions: the Microbial Connection. Immunity. 2020;53:710-723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 120] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 80. | Parrot T, Gorin JB, Ponzetta A, Maleki KT, Kammann T, Emgård J, Perez-Potti A, Sekine T, Rivera-Ballesteros O; Karolinska COVID-19 Study Group, Gredmark-Russ S, Rooyackers O, Folkesson E, Eriksson LI, Norrby-Teglund A, Ljunggren HG, Björkström NK, Aleman S, Buggert M, Klingström J, Strålin K, Sandberg JK. MAIT cell activation and dynamics associated with COVID-19 disease severity. Sci Immunol. 2020;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 151] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 81. | Mustafa S, Balkhy H, Gabere MN. Current treatment options and the role of peptides as potential therapeutic components for Middle East Respiratory Syndrome (MERS): A review. J Infect Public Health. 2018;11:9-17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 82. | Weingarden AR, Vaughn BP. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes. 2017;8:238-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 331] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 83. | Ford AC, Sperber AD, Corsetti M, Camilleri M. Irritable bowel syndrome. Lancet. 2020;396:1675-1688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 448] [Article Influence: 89.6] [Reference Citation Analysis (2)] |
| 84. | Li Y, Xia S, Jiang X, Feng C, Gong S, Ma J, Fang Z, Yin J, Yin Y. Gut Microbiota and Diarrhea: An Updated Review. Front Cell Infect Microbiol. 2021;11:625210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 117] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 85. | Levine MM, Nasrin D, Acácio S, Bassat Q, Powell H, Tennant SM, Sow SO, Sur D, Zaidi AKM, Faruque ASG, Hossain MJ, Alonso PL, Breiman RF, O'Reilly CE, Mintz ED, Omore R, Ochieng JB, Oundo JO, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ahmed S, Qureshi S, Quadri F, Hossain A, Das SK, Antonio M, Saha D, Mandomando I, Blackwelder WC, Farag T, Wu Y, Houpt ER, Verweiij JJ, Sommerfelt H, Nataro JP, Robins-Browne RM, Kotloff KL. Diarrhoeal disease and subsequent risk of death in infants and children residing in low-income and middle-income countries: analysis of the GEMS case-control study and 12-month GEMS-1A follow-on study. Lancet Glob Health. 2020;8:e204-e214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 124] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 86. | Johnson BA, Hage A, Kalveram B, Mears M, Plante JA, Rodriguez SE, Ding Z, Luo X, Bente D, Bradrick SS, Freiberg AN, Popov V, Rajsbaum R, Rossi S, Russell WK, Menachery VD. Peptidoglycan-Associated Cyclic Lipopeptide Disrupts Viral Infectivity. J Virol. 2019;93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 87. | Anand S, Mande SS. Diet, Microbiota and Gut-Lung Connection. Front Microbiol. 2018;9:2147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 167] [Cited by in RCA: 276] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 88. | Tiwari SK, Dicks LMT, Popov IV, Karaseva A, Ermakov AM, Suvorov A, Tagg JR, Weeks R, Chikindas ML. Probiotics at War Against Viruses: What Is Missing From the Picture? Front Microbiol. 2020;11:1877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 89. | Wen B, Taibi A, Villa CR, Lee SH, Sagaidak S, Comelli EM. Effects of Bifidobacterium bifidum in Mice Infected with Citrobacter rodentium. Microorganisms. 2019;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 90. | Azagra-Boronat I, Massot-Cladera M, Knipping K, Garssen J, Ben Amor K, Knol J, Franch À, Castell M, Rodríguez-Lagunas MJ, Pérez-Cano FJ. Strain-Specific Probiotic Properties of Bifidobacteria and Lactobacilli for the Prevention of Diarrhea Caused by Rotavirus in a Preclinical Model. Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 91. | Gareau MG, Sherman PM, Walker WA. Probiotics and the gut microbiota in intestinal health and disease. Nat Rev Gastroenterol Hepatol. 2010;7:503-514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 707] [Cited by in RCA: 623] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 92. | Rocha-Ramírez LM, Hernández-Ochoa B, Gómez-Manzo S, Marcial-Quino J, Cárdenas-Rodríguez N, Centeno-Leija S, García-Garibay M. Evaluation of Immunomodulatory Activities of the Heat-Killed Probiotic Strain Lactobacillus casei IMAU60214 on Macrophages In Vitro. Microorganisms. 2020;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 93. | Hu J, Zhang L, Lin W, Tang W, Chan FKL, Ng SC. Review article: Probiotics, prebiotics and dietary approaches during COVID-19 pandemic. Trends Food Sci Technol. 2021;108:187-196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 63] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 94. | Al Kassaa I, Hober D, Hamze M, Chihib NE, Drider D. Antiviral potential of lactic acid bacteria and their bacteriocins. Probiotics Antimicrob Proteins. 2014;6:177-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 133] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 95. | Murch SH. Toll of allergy reduced by probiotics. Lancet. 2001;357:1057-1059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 63] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 96. | Jobin C. Probiotics and ileitis: could augmentation of TNF/NFκB activity be the answer? Gut Microbes. 2010;1:196-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 97. | Din AU, Hassan A, Zhu Y, Zhang K, Wang Y, Li T, Wang G. Inhibitory effect of Bifidobacterium bifidum ATCC 29521 on colitis and its mechanism. J Nutr Biochem. 2020;79:108353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 98. | Resta-Lenert S, Barrett KE. Probiotics and commensals reverse TNF-alpha- and IFN-gamma-induced dysfunction in human intestinal epithelial cells. Gastroenterology. 2006;130:731-746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 230] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 99. | Li J, Zhao J, Wang X, Qayum A, Hussain MA, Liang G, Hou J, Jiang Z, Li A. Novel Angiotensin-Converting Enzyme-Inhibitory Peptides From Fermented Bovine Milk Started by Lactobacillus helveticus KLDS.31 and Lactobacillus casei KLDS.105: Purification, Identification, and Interaction Mechanisms. Front Microbiol. 2019;10:2643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 100. | Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, Verbeke K, Reid G. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14:491-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2256] [Cited by in RCA: 3179] [Article Influence: 397.4] [Reference Citation Analysis (0)] |
| 101. | El-Salhy M, Hatlebakk JG, Gilja OH, Bråthen Kristoffersen A, Hausken T. Efficacy of faecal microbiota transplantation for patients with irritable bowel syndrome in a randomised, double-blind, placebo-controlled study. Gut. 2020;69:859-867. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 318] [Article Influence: 63.6] [Reference Citation Analysis (0)] |
| 102. | Lima SF, Gogokhia L, Viladomiu M, Chou L, Putzel G, Jin WB, Pires S, Guo CJ, Gerardin Y, Crawford CV, Jacob V, Scherl E, Brown SE, Hambor J, Longman RS. Transferable Immunoglobulin A-Coated Odoribacter splanchnicus in Responders to Fecal Microbiota Transplantation for Ulcerative Colitis Limits Colonic Inflammation. Gastroenterology. 2022;162:166-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 106] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 103. | Khoruts A, Staley C, Sadowsky MJ. Faecal microbiota transplantation for Clostridioides difficile: mechanisms and pharmacology. Nat Rev Gastroenterol Hepatol. 2021;18:67-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 103] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 104. | Liu Q, Mak JWY, Su Q, Yeoh YK, Lui GC, Ng SSS, Zhang F, Li AYL, Lu W, Hui DS, Chan PK, Chan FKL, Ng SC. Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome. Gut. 2022;71:544-552. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 319] [Cited by in RCA: 359] [Article Influence: 119.7] [Reference Citation Analysis (1)] |