Published online Jan 28, 2022. doi: 10.3748/wjg.v28.i4.464
Peer-review started: August 8, 2021
First decision: November 7, 2021
Revised: November 15, 2021
Accepted: January 6, 2022
Article in press: January 6, 2022
Published online: January 28, 2022
Processing time: 166 Days and 22.7 Hours
Esophageal squamous cell carcinoma (ESCC) is one of the most prevalent malignancies that seriously threaten people’s health worldwide. DEAD-box helicase 51 (DDX51) is a member of the DEAD-box (DDX) RNA helicase family, and drives or inhibits tumor progression in multiple cancer types.
To determine whether DDX51 affects the biological behavior of ESCC.
The expression of DDX51 in ESCC tumor tissues and adjacent normal tissues was detected by Immunohistochemistry (IHC) analyses and quantitative PCR (qPCR). We knocked down DDX51 in ESCC cell lines by using a small interfering RNA (siRNA) transfection. The proliferation, apoptosis, and mobility of DDX51 siRNA-transfected cells were detected. The effect of DDX51 on the phosphoinositide 3-kinase (PI3K)/AKT pathway was investigated by western blot analysis. A mouse xenograft model was established to investigate the effects of DDX51 knockdown on ESCC tumor growth.
DDX51 exhibited high expression in ESCC tissues compared with normal tissues and represented a poor prognosis in patients with ESCC. Knockdown of DDX51 induced inhibition of ESCC cell proliferation and promoted apoptosis. Moreover, DDX51 siRNA-expressing cells also exhibited lower migration and invasion rates. Investigations into the underlying mechanisms suggested that DDX51 knock
Our study suggests for the first time that DDX51 promotes cancer cell proliferation by regulating the PI3K/AKT pathway; thus, DDX51 might be a therapeutic target for ESCC.
Core Tip: Our study revealed that DEAD-box helicase 51 was upregulated in esophageal squamous cell carcinoma (ESCC) tumor tissues and promoted tumor proliferation and development by upregulating the phosphorylation of phosphoinositide 3-kinase/AKT pathway members. These data extend our knowledge of the function and molecular mechanism of the DEAD-box family in tumor biology and provide a potential therapeutic target for ESCC treatment.
- Citation: Hu DX, Sun QF, Xu L, Lu HD, Zhang F, Li ZM, Zhang MY. Knockdown of DEAD-box 51 inhibits tumor growth of esophageal squamous cell carcinoma via the PI3K/AKT pathway. World J Gastroenterol 2022; 28(4): 464-478
- URL: https://www.wjgnet.com/1007-9327/full/v28/i4/464.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i4.464
Esophageal squamous cell carcinoma (ESCC) is the sixth leading cause of cancer-related deaths, causing approximately 300,000 deaths each year[1-3]. China has the highest incidence of ESCC globally, caused by multiple factors such as diet and the environment. At present, surgical treatment is the most effective means to cure ESCC. However, surgical treatment is highly challenging due to the close distance between the esophagus and life-supporting structures such as the respiratory system[4]. Chemotherapy and radiation therapy are commonly used as adjuvant treatments. The main problem with these two treatments is inherent resistance, and even local ESCC is often resistant[5,6]. Due to the extremely high tumor recurrence rate (about 40%) and frequent local lymph node metastases, the prognosis of ESCC patients is very poor, with a 5-year survival rate of only 10%-30%[1]. Molecular targeted therapy has shown good application prospects in ESCC patients in recent years. To improve the survival of ESCC patients, it is essential to explore effective molecular targets for ESCC treatment.
DEAD-box (DDX) RNA helicases, belonging to helicase superfamily 2, comprise the largest group of RNA helicases[7]. There are at least 31 members of DDX family in humans, including DEAD-box helicase 51 (DDX51). DDX proteins are highly conserved helicases that catalyze duplex unwinding and structural remodeling of RNA or ribonucleoprotein complexes[8]. They play important roles in nearly all aspects of RNA metabolism including RNA transcription, post-transcriptional splicing, ribosome assembly, and RNA degradation[9]. In recent years, DDX family members have been identified to be dysregulated and function as tumor oncogenes or suppressors in various tumor types[10]. For example, knockdown of DDX5 exerts inhibitory effects on the proliferation and epithelial-to-mesenchymal transition of ESCC cells by downregulating β-catenin, c-Myc, and cyclin D1[11]. DDX51 is a recently identified member of the DDX family. DDX51 is mainly responsible for maturation of the 3’ end of 28S and its proliferation-promoting activities have been demonstrated in non-small cell lung cancer (NSCLC) and breast cancer[12,13]. However, whether DDX51 plays a role in the progression of ESCC is still unclear.
In this study, we investigated the function of DDX51 in ESCC cell proliferation and tumor growth by using a small interfering RNA (siRNA) silencing approach.
ESCC tumor tissues and adjacent normal tissues (n = 118) were obtained from patients with ESCC between 2016 to 2020 in Shandong Provincial Hospital Affiliated to Shandong First Medical University (Shandong, China). The Research Ethics Committee approved all experimental procedures of Shandong Provincial Hospital Affiliated to Shandong First Medical University. Patients or their guardians wrote the informed consents. The clinicopathological parameters of ESCC patients were collected. Immunohistochemistry (IHC) staining was performed to investigate the expression of DDX51 in ESCC tissues and adjacent normal tissues, and the experimental procedure is described in a previous report[14].
The ESCC cell lines Eca109 and TE-1 were purchased from the Cell Bank of the Chinese Academy. The cells were cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, United States) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin in a 37 °C cell incubator. When cells were grown to 70% confluence, si-DDX51 or negative control siRNA (si-NC) was transfected into Eca109 and TE-1 cells using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, United States) according to the corresponding experimental groups. The stable DDX51 knockdown ESCC cell line was established by using lentivirus transfection. Lentivirus expressing si-DDX51 or si-NC was purchased from Genechem (Shanghai, China). After 48 h of transfection, cells were collected and analyzed for DDX51 expression using quantitative PCR (qPCR) and western blot analysis.
Total RNA was extracted from ESCC tumor tissues and cell lines using Trizol reagent (Thermo Fisher Scientific, Waltham, MA, United States). Then 1 μg total RNA was reverse-transcribed to cDNA using the PrimeScript RT Master Mix (Takara Biotechnology Co., Ltd., Dalian, China). The expression of DDX51 was determined via qPCR with the FastStart Universal SYBR Green Master (Roche, Germany). The experiment was performed on the C1000 Thermal Cycler (Bio-Rad, Hercules, CA, United States). The PCR reaction conditions were as follows: Pre-degeneration at 95 °C for 5 min, followed by 45 cycles of 95 °C for 5 s, 60 °C for 10s, and 72 °C for 10 s. GAPDH was used as an internal control.
After transfection, Eca109 and TE-1 cells were seeded into 96-well plates at a density of 3000 cells per well. Each group had three wells. The Cell Counting Kit-8 (CCK-8; Dojindo, Kyushu, Japan) was used to detect the cell viability of transfected cells. At specific time points (24, 48, 72, and 96 h), 10 μL CCK-8 reagent was added to each well and incubated at 37 °C for 2 h. Cell viability was represented by the absorbance at 450 nm, which was detected on a spectrophotometer (Molecular Devices, San Jose, CA, United States).
Cell apoptosis was detected by using the Annexin V-FITC/Propidium Iodide (PI) Apoptosis Detection Kit I (BD Biosciences, San Jose, CA, United States) following the manufacturer’s instructions. The transfected cells were digested with 0.25% EDTA-free trypsin and resuspended in binding buffer. Then 100 μL cell suspension was incubated with 5 μL AnnexinV-FITC and 5 μL PI for 30 min. The incubation was performed at 37 °C in the dark. The apoptosis percentage was analyzed by flow cytometry (Becton Dickinson, Franklin Lakes, NJ, United States).
A scratch assay evaluated the effect of cell transfection on its migration ability. Eca109 and TE-1 cells were seeded in a 6-well plate at a density of 1 × 105 cells/mL. Then cells were transfected as described as before. After 16 h, cell transfection was stopped and cells were continuously cultured to 100% confluence. Then a single-line scratch was created on the cell monolayer using a sterile micropipette tip. Phosphate-buffered saline was used to wash the cell debris, and serum-free medium was added to the plates. Cells were photographed at 0 and 24 h under an inverted fluorescence microscope (IX-71). The width of the scratch was measured. All experiments were performed in triplicate and repeated at least three times.
Cell invasion was detected using a transwell assay. Transwell inserts (6.5 mm, 8 μm pore size; CoStar Group, Washington, DC, United States) were added to the 24-well transwell plates. The inserts were coated with 50 μL of 1 mg/mL Matrigel matrix (BD Biosciences). Eca109 and TE-1 cells were transfected as described above and the cell density was adjusted to 106 cells/mL. Then 200 μL cell suspension was added to the upper chamber of the transwell inserts, while 600 μL medium with 10% FBS was added to the lower well. Cells were cultured at 37 °C for 24 h, followed by fixation and staining with 1% crystal violet. The invasive cells were observed under a light microscope, and five random fields were selected to take pictures.
Total proteins from transfected ESCC cells were extracted using radioimmunoprecipitation assay buffer with a 1% protease inhibitor. The protein concentration in the extraction solution was determined using the bicinchoninic acid assay. Then 20 μg total protein was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and proteins were electrotransferred to a PVDF membrane. The membrane was blocked in 5% bovine serum albumin containing Tris-buffered saline with Tween 20 (TBST) (10 mmol/L Tris–HCl, pH 7.4, 150 mmol/L NaCl, 0.05% Tween-20) at room temperature for 2 h, and then incubated with primary antibodies at 4 °C overnight. The primary antibodies included phosphatase and tensin homolog (PTEN) (1:1000, ab267787; Abcam, Cambridge, MA, United States), phosphoinositide 3-kinase (PI3K) (1:1000, CY5355; Abways, Shanghai, P.R. China), phosphorylated PI3K (p-PI3K) (1:1000, CY6427; Abways), AKT (1:1000, AF6261; Affinity Biosciences, Cincinnati, OH, United States), p-AKT (1:1000, AF016; Affinity Biosciences), mammalian target of rapamycin (mTOR) (1:1000, ab134903), p-mTOR (1:1000, ab137133; Abcam), and anti-GAPDH (1:5,000; ab8245; Abcam). After washing with TBST buffer four times every 5 min, the membrane was incubated with secondary antibodies (Santa Cruz, CA, United States) for 1 h at room temperature and washed with TBST another four times. Finally, proteins were visualized by chemiluminescence (Santa Cruz Biotechnology, Dallas, TX, United States).
Male nude mice (6-wk-old) were obtained from Shanghai SIPPR-BK Laboratory Animal Co. Ltd. (Shanghai, P.R. China). All animal experiments were approved by the Shandong Provincial Hospital Affiliated to Shandong First Medical University. The animals were maintained in specific pathogen-free conditions in accordance with the institutional animal care and use committee regulations. ESCC cells were transfected with lentivirus-NC or lentivirus-si-DDX51 for 48 h and then resuspended in a serum-free medium. The cell density was adjusted to 2 × 107/mL. Each mouse was subcutaneously injected with 5 × 106 of ESCC cells. The tumor length and width were monitored every other day and length × width2/2 represented tumor volume. On day 28, nude mice were sacrificed by carbon dioxide asphyxiation.
All experimental data are expressed as the mean ± standard deviation from three independent experiments. The difference between the two groups was analyzed using the Student’s t-test. The difference among multiple groups was evaluated using one-way analysis of variance. P < 0.05 was considered statistically significant. GraphPad Prism 5 Software (GraphPad, San Diego, CA, United States) was used for all statistical analyses. All data in this study satisfied parametric test assumptions.
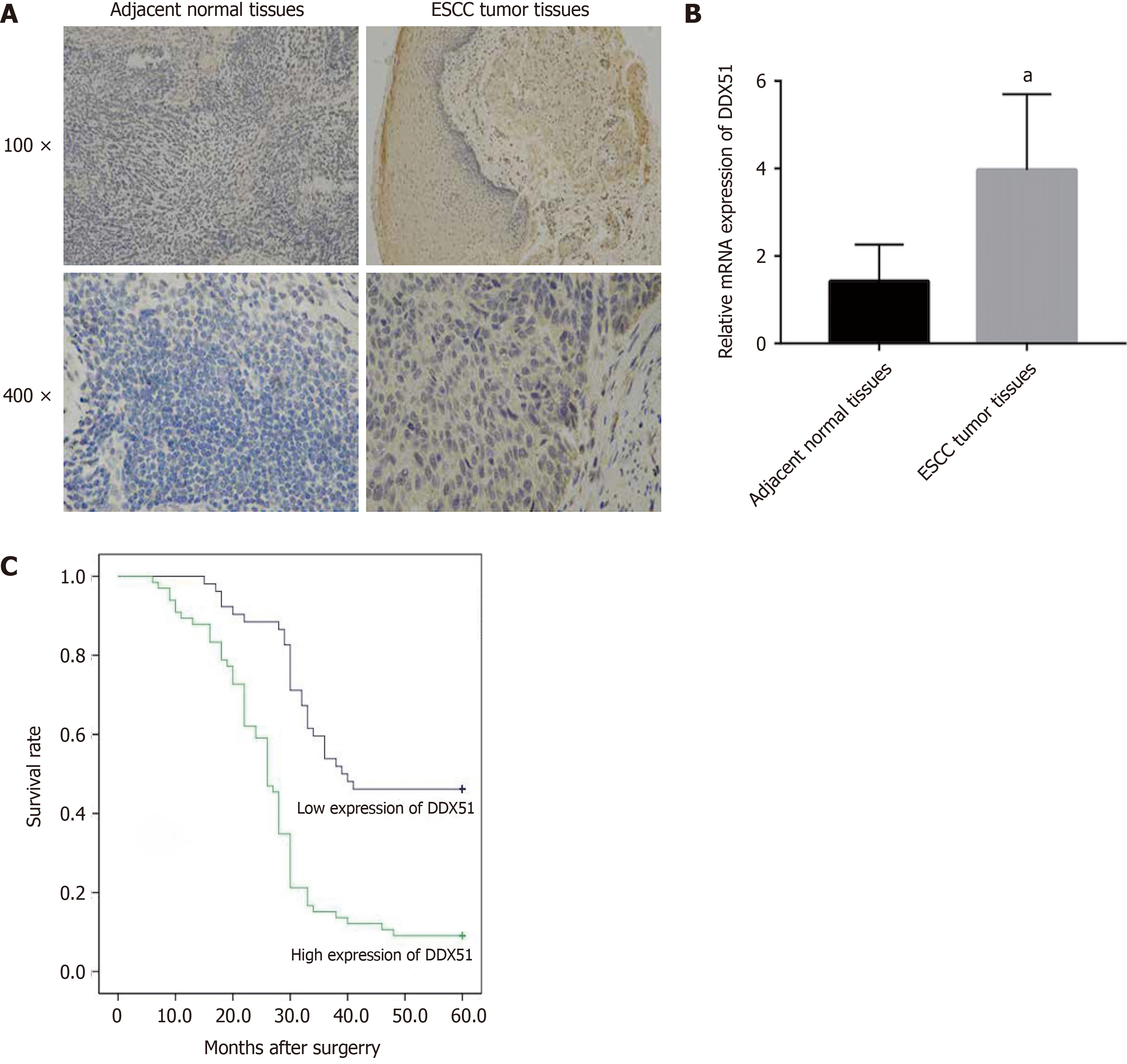
DDX51 is upregulated in breast cancer. Here, we detected the expression of DDX51 in ESCC tissues, which might provide insights into its role in ESCC. As shown in Figure 1A, we performed IHC analysis of DDX51 expression in the tumor tissues and adjacent normal tissues of patients with ESCC. The results showed strong IHC staining of ESCC tissues, which was very weak in the normal controls. Meanwhile, DDX51 expression was detected by qPCR, which also showed significantly higher expression in ESCC tumor tissues than in normal adjacent tissues (Figure 1B). Furthermore, the correlation between DDX51 expression with patients’ clinicopathological features was analyzed. As shown in Table 1, DDX51 expression did not correlate with patients’ age, sex, tumor size, or T stage, but was associated with tumor differentiation degree, N stage and tumor-node-metastasis (TNM) stage. Survival analysis results indicated that ESCC patients with high expression of DDX51 had lower survival rates compared to those with low expression of DDX51 (Figure 1C). These results suggest that DDX51 exhibits high expression in ESCC tumor tissues and may be associated with a poor prognosis in patients with ESCC (Table 2).
| Characteristics | Cases (n = 118) | DDX51 expression level | P value | |
| Low (n = 52) | High (n = 66) | |||
| Age (yr) | 0.740 | |||
| < 50 | 30 | 14 | 16 | |
| ≥ 50 | 88 | 38 | 50 | |
| Gender | 0.201 | |||
| Male | 91 | 43 | 48 | |
| Female | 27 | 9 | 18 | |
| Tumor size | 0.194 | |||
| < 30 mm | 20 | 12 | 8 | |
| 30-50 mm | 66 | 29 | 37 | |
| ≥ 50 mm | 32 | 11 | 21 | |
| Differentiation degree | 0.025a | |||
| High | 26 | 11 | 15 | |
| Moderate | 53 | 30 | 23 | |
| Low | 39 | 11 | 28 | |
| pT | 0.154 | |||
| T1 | 8 | 6 | 2 | |
| T2 | 51 | 23 | 28 | |
| T3 + T4 | 59 | 23 | 36 | |
| pN | 0.012a | |||
| N0 | 42 | 25 | 17 | |
| N1 + N2 | 76 | 27 | 49 | |
| pTNM | 0.002a | |||
| I + II | 37 | 24 | 13 | |
| III + IV | 81 | 28 | 53 | |
| B | SE | Wald | P value | HR | 95%CI | |
| Differentiation | 0.052 | 0.157 | 0.110 | 0.740 | 1.053 | 0.774-1.433 |
| pN | 0.029 | 0.451 | 0.004 | 0.949 | 1.029 | 0.425-2.491 |
| pTNM | 1.078 | 0.518 | 4.323 | 0.038 | 2.939 | 1.064-8.119 |
| DDX51 | 0.893 | 0.241 | 13.686 | < 0.01 | 2.443 | 1.522-3.921 |
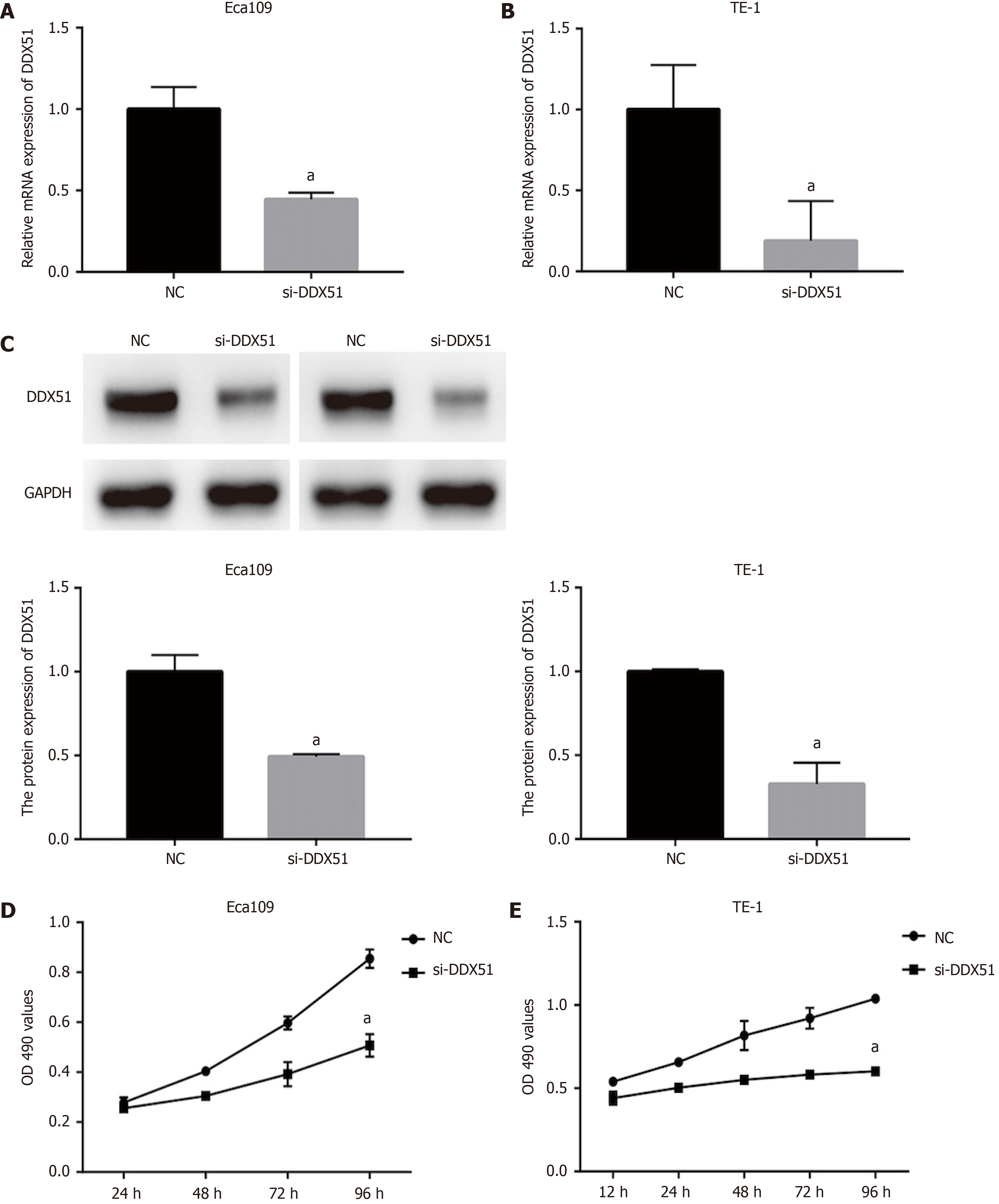
To investigate the function of DDX51 in ESCC progression, we performed a knockdown experiment in Eca109 and TE-1 cells using siRNA transfection. The transfection efficiencies are shown in Figure 2A-C. The qPCR results indicated that, compared with the NC group, the mRNA expression of DDX51 in the si-DDX51 group was decreased to 44.5% in Eca109 cells and 18.8% in TE-1 cells (Figure 2A and B, both
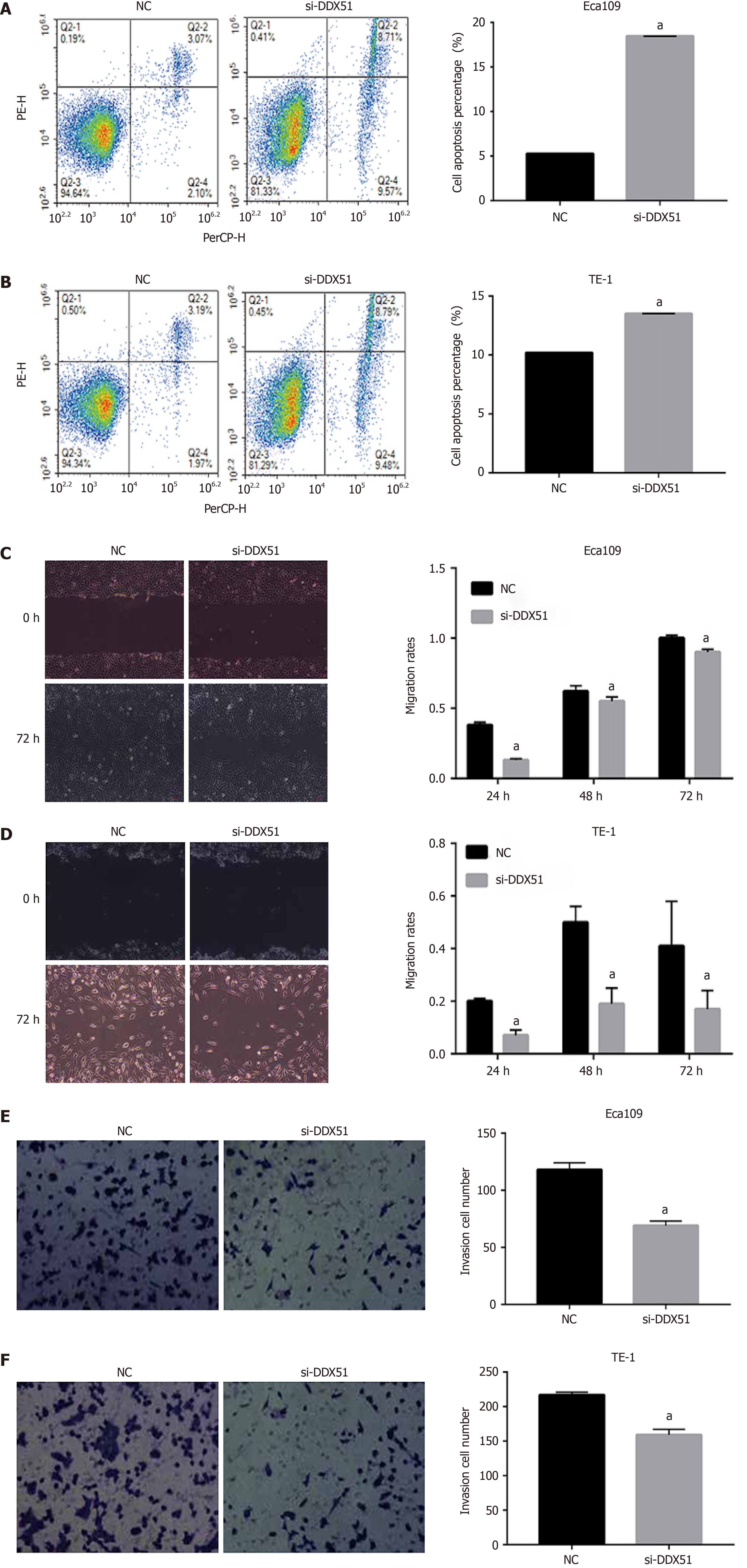
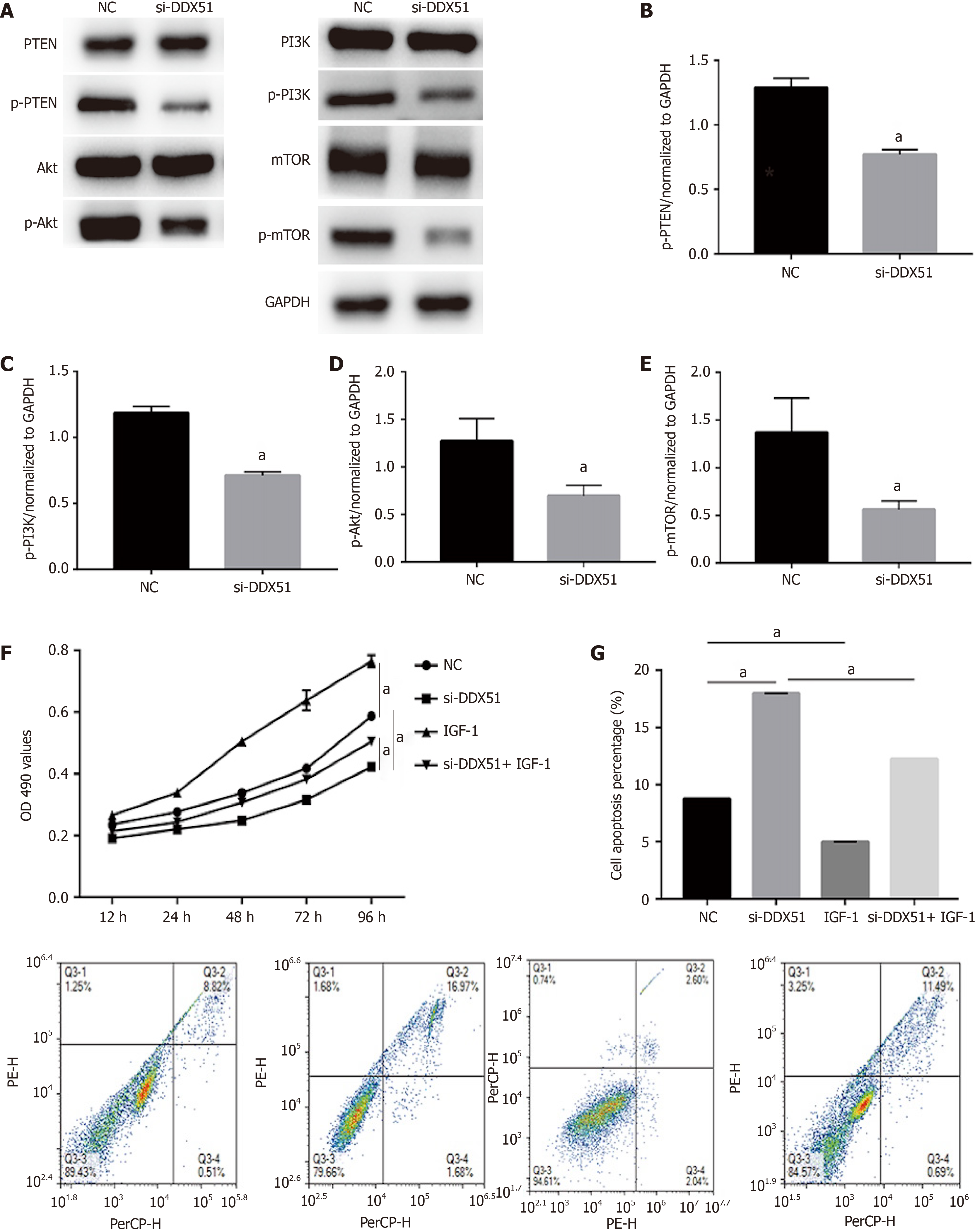
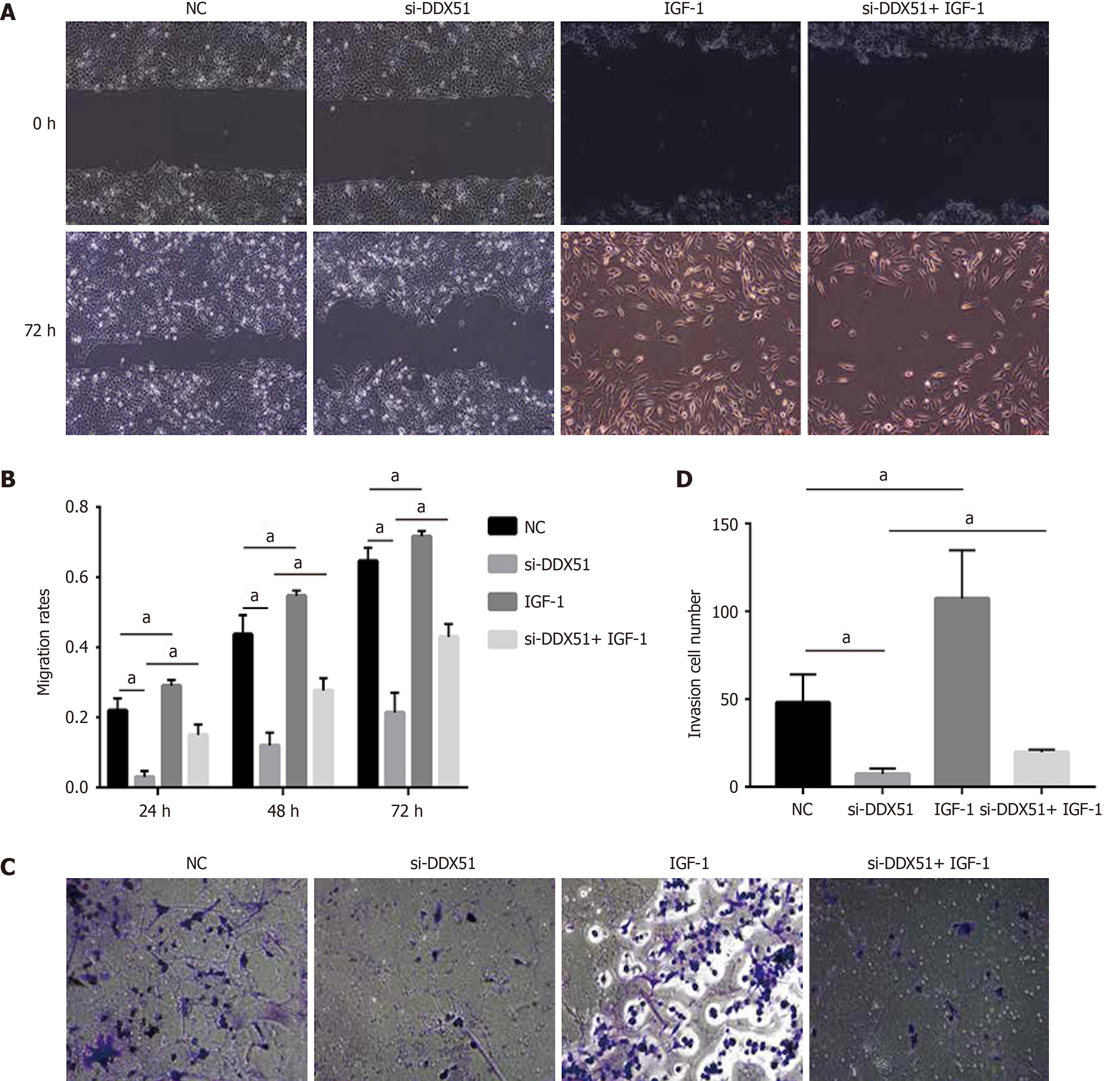
The PI3K/AKT signaling pathway is constitutively activated in multiple cancer types and extensively participates in tumor cell proliferation, apoptosis, metastasis, and other malignant phenotypes. Here, we determined whether the oncogenic function of DDX51 in ESCC involves the PI3K/AKT signaling pathway. As shown in Figure 4A-E, the expression of members in the PI3K/AKT signaling pathway including PTEN, p-PTEN, PI3K, p-PI3K, AKT, p-AKT, mTOR, and p-mTOR was evaluated by western blotting. The results showed that DDX51 knockdown induced a significant decrease in the phosphorylation levels of PTEN, PI3K, AKT, and mTOR, while the total protein was not affected, suggesting that the PI3K/AKT signaling pathway was inactivated in DDX51-knockdown ESCC cells. To further elucidate the role of the PI3K/AKT signaling pathway in the function of DDX51, we applied an AKT agonist insulin-like growth factor 1 (IGF-1), which induced persistent activation of the AKT pathway. IGF-1 treatment reversed the anti-proliferation and pro-apoptotic effects of DDX51 knockdown in ESCC cells (Figure 4F and G). Meanwhile, the migration and invasion abilities were also increased to the NC level when ESCC cells were treated with both si-DDX51 and IGF-1 (Figure 5). These results suggest that the anti-tumor effects of DDX51 knockdown in ESCC are, at least in part, mediated by the PI3K/AKT signaling pathway.
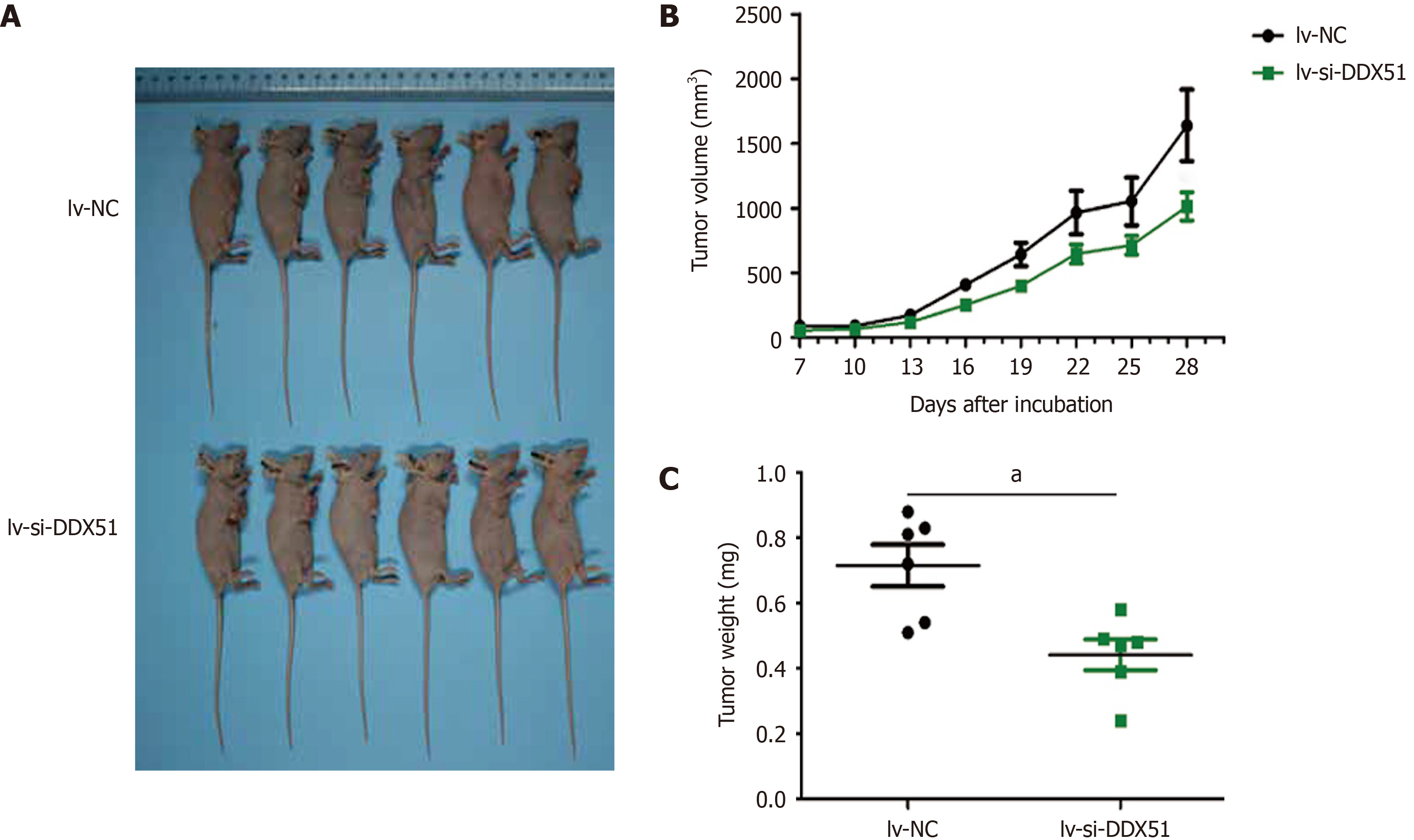
We further investigated the effects of DDX51 knockdown on ESCC tumor growth in vivo. TE-1 cells were transfected with lv-si-DDX51 or lv-NC for 48 h and subcutaneously injected into the right flanks of nude mice. Six repeats were designed for each group. The tumor volume was measured and analyzed every other day. At the end of the experiment, tumor weights were measured and DDX51 expression was analyzed by IHC analysis. The results are shown in Figure 6, which indicated that the tumor volume of the lv-si-DDX51 group (1639.35 ± 674.36 mm3) was significantly decreased compared with that of the NC group (1013.03 ± 264.51 mm3) on day 28. The weight of the NC group (0.442 ± 0.116 g) was also significantly higher than that of the lv-si-DDX51 group (0.715 ± 0.156 g). These results showed that DDX51 knockdown inhibited ESCC tumor growth in the mouse xenograft model.
The DDX family, which has activities of RNA unwinding and structure remodeling, regulates nearly all aspects of RNA-related biology, from synthesis to degradation[10]. Since mRNA serves as an intermediate between DNA genetic information and proteins, the DDX family plays crucial roles in multiple cellular processes such as cell proliferation, apoptosis, and malignant transformation[15-19]. DDX proteins ordinarily exert their function by forming a large multi-protein complex. Therefore, their exact function is probably impacted by their interacting partners and is profoundly context dependent[20,21]. The most important characteristic of a tumor is heterogeneity, which might explain the diverse functions of DDX proteins in tumors, as an oncogene or tumor suppressor[22,23]. DDX51 is a recently identified member of DDX family, which shows pro-proliferation activity in the progression of NSCLC and breast cancer[12,13].
Here we demonstrated that DDX51 was upregulated in ESCC tumor tissues and promoted tumor proliferation and metastasis. The results also suggested that the oncogenic function of DDX51 in ESCC was a result of activation of the PI3K/AKT signaling pathway, which is consistent with previous reports that revealed the regulatory action of other DDX proteins on the AKT pathway.
DDX family members are dysregulated in various tumor types, including ESCC[10]. For example, gene microarray data of breast cancer showed that DDX1 upregulation is associated with tumor occurrence and early recurrence and serves as an independent prognostic biomarker for patients’ survival[24]. Wang et al[12] investigated the expression of DDX51 in NSCLC patients. The results showed that DDX51 expression was associated with patient age but no other risk factors. In breast cancer, DDX51 also exhibits high expression in tumor tissues and predicts a high TNM stage and poor prognosis in patients[13]. Here, our results showed that DDX51 was upregulated in ESCC tumor tissues and was negatively correlated with patient survival, which was consistent with previous studies on DDX51 in tumors. Our study identified that the upregulated expression of DDX51 in ESCC is associated with the degree of differentiation, pathological N stage, and pathological TNM stage. The above findings suggest that DDX51 overexpression may serve as an independent prognostic factor in ESCC. However, it is currently unclear if this is an indication of the severity of the cancer itself. Further study should be performed to investigate the prognostic value of DDX51 in a larger number of specimens.
As RNA synthesis and decay involve the regulation of broad-spectrum protein expression, it is not surprising that the dysregulation of DDX proteins in tumors affects different signaling pathways. In a review reported in 2021, the signaling regulation network of DDX family in tumorigenesis was summarized including Wnt/β-catenin pathway, Snail/E-cadherin pathway, hypoxia inducible factor 1 alpha/DDX3/E-cadherin and so on[10]. For DDX51, microarray analyses revealed that DDX51 siRNA-expressing cells expressed higher levels of transforming growth factor beta receptor, interleukin 1 receptor, and c-FOS in NSCLC[12]. The Wnt/β-catenin pathway is also downregulated by DDX51 siRNA in breast cancer[13]. Here, we found that DDX51 siRNA resulted in the inactivation of the PI3K/AKT pathway, including decreased phosphorylation levels of PTEN, PI3K, AKT, and mTOR. The PI3K/AKT pathway is constitutively activated in many tumor types including ESCC, and participates in tumor cell proliferation, apoptosis, and progression[25-27]. In a previous study, DDX5 was demonstrated to occupy the AKT promoter with β-catenin as well as nuclear factor kappa B, and induces the mRNA and protein expression of AKT[28]. However, in gastric cancer miR-5590-3p targets DDX5, which further decreases the phosphorylation levels of AKT and does not affect AKT protein expression[29]. These results suggest that DDX proteins can regulate the AKT pathway through either protein expression or phosphorylation level. In the future, we intend to investigate the effects of DDX51 on transcriptome and proteome expression in tumor progression, which will provide more information on the mechanism of action of this protein.
This study had some limitations. First, in vivo experiments were not performed to further confirm the effect of DDX51 knockdown on lung and lymph node metastases. Second, the exact regulatory mechanism of DDX51 in ESCC was not explored. Future studies are required to further confirm the potential of DDX51 as a therapeutic target for the targeted treatment of ESCC.
Esophageal squamous cell carcinoma (ESCC) is one of the most prevalent malignancies that seriously threatens people’s health worldwide. DDX51 is a member of the DEAD-box (DDX) RNA helicase family, which drives or inhibits tumor progression in multiple cancer types.
To identify the role of DDX51 in ESCC and the molecular mechanisms involved.
To explore the effect of DDX51 on ESCC progression.
The expression of DDX51 in ESCC tumor tissues and adjacent normal tissues was detected by immunohistochemistry analysis and quantitative PCR (qPCR). We knocked down DDX51 in ESCC cell lines using small interfering RNA (siRNA) transfection. The proliferation, apoptosis, and mobility of DDX51 siRNA-transfected cells were detected. The effects of DDX51 on the phosphoinositide 3-kinase (PI3K)/AKT pathway were investigated using western blot analysis. A mouse xenograft model was established to investigate the effects of DDX51 knockdown on ESCC tumor growth.
DDX51 exhibited high expression in ESCC tissues compared with normal tissues and was associated with a poor prognosis in patients with ESCC. Knockdown of DDX51 induced inhibition of ESCC cell proliferation and promoted apoptosis. Moreover, DDX51 siRNA-expressing cells also exhibited lower migration and invasion rates. Investigation into the mechanism of action suggested that DDX51 knockdown induced inactivation of the PI3K/AKT pathway including decreased phosphorylation levels of PTEN, PI3K, AKT and mTOR. Rescue experiments demonstrated that the AKT activator insulin-like growth factor 1 could reverse the inhibitory effects of DDX51 on ESCC malignant development. Finally, we injected DDX51 siRNA transfected TE-1 cell into an animal model, which resulted in slower tumor growth.
Our study suggests for the first time that DDX51 contributes to ESCC cell proliferation by regulating the PI3K/AKT signaling pathway.
DDX51 may serve as a potential therapeutic target for the treatment of ESCC.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Anandan H, Aurello P, Swai J S-Editor: Wang LL L-Editor: Filipodia P-Editor: Li JH
| 1. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13206] [Article Influence: 1467.3] [Reference Citation Analysis (3)] |
| 2. | Rustgi A, El-Serag HB. Esophageal carcinoma. N Engl J Med. 2015;372:1472-1473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 3. | Liu L, Lin C, Liang W, Wu S, Liu A, Wu J, Zhang X, Ren P, Li M, Song L. TBL1XR1 promotes lymphangiogenesis and lymphatic metastasis in esophageal squamous cell carcinoma. Gut. 2015;64:26-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 87] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 4. | de Vos-Geelen J, Geurts SM, van Putten M, Valkenburg-van Iersel LB, Grabsch HI, Haj Mohammad N, Hoebers FJ, Hoge CV, Jeene PM, de Jong EJ, van Laarhoven HW, Rozema T, Slingerland M, Tjan-Heijnen VC, Nieuwenhuijzen GA, Lemmens VE. Trends in treatment and overall survival among patients with proximal esophageal cancer. World J Gastroenterol. 2019;25:6835-6846. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 12] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 5. | Wu J, Wang L, Du X, Sun Q, Wang Y, Li M, Zang W, Liu K, Zhao G. α-solanine enhances the chemosensitivity of esophageal cancer cells by inducing microRNA138 expression. Oncol Rep. 2018;39:1163-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 6. | Samson P, Robinson C, Bradley J, Lockhart AC, Puri V, Broderick S, Kreisel D, Krupnick AS, Patterson GA, Meyers B, Crabtree T. Neoadjuvant Chemotherapy vs Chemoradiation Prior to Esophagectomy: Impact on Rate of Complete Pathologic Response and Survival in Esophageal Cancer Patients. J Thorac Oncol. 2016;11:2227-2237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 7. | Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313-324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 810] [Cited by in RCA: 724] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 8. | Linder P, Lasko PF, Ashburner M, Leroy P, Nielsen PJ, Nishi K, Schnier J, Slonimski PP. Birth of the D-E-A-D box. Nature. 1989;337:121-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 539] [Cited by in RCA: 597] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 9. | Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol. 2004;5:232-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 567] [Cited by in RCA: 599] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 10. | Ali MAM. The DEAD-box protein family of RNA helicases: sentinels for a myriad of cellular functions with emerging roles in tumorigenesis. Int J Clin Oncol. 2021;26:795-825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 11. | Ma Z, Feng J, Guo Y, Kong R, Ma Y, Sun L, Yang X, Zhou B, Li S, Zhang W, Jiang J, Zhang J, Qiao Z, Cheng Y, Zha D, Liu S. Knockdown of DDX5 Inhibits the Proliferation and Tumorigenesis in Esophageal Cancer. Oncol Res. 2017;25:887-895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 12. | Wang X, Liu H, Zhao C, Li W, Xu H, Chen Y. The DEAD-box RNA helicase 51 controls non-small cell lung cancer proliferation by regulating cell cycle progression via multiple pathways. Sci Rep. 2016;6:26108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 13. | Sun W, Cang S, Lv X, Wang P, Lin Q, Zhang Q, Yan Z, Liu Z, Song Y. DDX51 gene promotes proliferation by activating Wnt/β-catenin signaling in breast cancer. Int J Clin Exp Pathol. 2017;10:10892-10900. [PubMed] |
| 14. | Chen Z, Gao Y, Huang X, Yao Y, Chen K, Zeng S, Mao W. Tissue-based metabolomics reveals metabolic biomarkers and potential therapeutic targets for esophageal squamous cell carcinoma. J Pharm Biomed Anal. 2021;197:113937-113937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 15. | Zhang Y, Baysac KC, Yee LF, Saporita AJ, Weber JD. Elevated DDX21 regulates c-Jun activity and rRNA processing in human breast cancers. Breast Cancer Res. 2014;16:449. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 16. | Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, McCombie WR, Wigler M, Hicks J, Hannon GJ, Powers S, Lowe SW. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852-864. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 363] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 17. | Yassin ER, Abdul-Nabi AM, Takeda A, Yaseen NR. Effects of the NUP98-DDX10 oncogene on primary human CD34 + cells: role of a conserved helicase motif. Leukemia. 2010;24:1001-1011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 42] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 18. | Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543-2555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 190] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 19. | Yang L, Lin C, Liu ZR. Phosphorylations of DEAD box p68 RNA helicase are associated with cancer development and cell proliferation. Mol Cancer Res. 2005;3:355-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 70] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 20. | Wilson BJ, Bates GJ, Nicol SM, Gregory DJ, Perkins ND, Fuller-Pace FV. The p68 and p72 DEAD box RNA helicases interact with HDAC1 and repress transcription in a promoter-specific manner. BMC Mol Biol. 2004;5:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 117] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 21. | Clark EL, Coulson A, Dalgliesh C, Rajan P, Nicol SM, Fleming S, Heer R, Gaughan L, Leung HY, Elliott DJ, Fuller-Pace FV, Robson CN. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res. 2008;68:7938-7946. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 161] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 22. | Hassan C, Afshinnekoo E, Li S, Wu S, Mason CE. Genetic and epigenetic heterogeneity and the impact on cancer relapse. Exp Hematol. 2017;54:26-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 23. | Morii E. Heterogeneity of tumor cells in terms of cancer-initiating cells. J Toxicol Pathol. 2017;30:1-6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 24. | Germain DR, Graham K, Glubrecht DD, Hugh JC, Mackey JR, Godbout R. DEAD box 1: a novel and independent prognostic marker for early recurrence in breast cancer. Breast Cancer Res Treat. 2011;127:53-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 57] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 25. | Tasioudi KE, Sakellariou S, Levidou G, Theodorou D, Michalopoulos NV, Patsouris E, Korkolopoulou P, Saetta AA. Immunohistochemical and molecular analysis of PI3K/AKT/mTOR pathway in esophageal carcinoma. APMIS. 2015;123:639-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 26. | Song Y, Liu H, Cui C, Peng X, Wang C, Tian X, Li W. Silencing of Peroxiredoxin 1 Inhibits the Proliferation of Esophageal Cancer Cells and Promotes Apoptosis by Inhibiting the Activity of the PI3K/AKT Pathway. Cancer Manag Res. 2019;11:10883-10890. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 27. | Li F, Zhang Z, Wang P, Wen P, Xu Q, Wang Y, Pan P, Ma L. ALC1 knockdown enhances cisplatin cytotoxicity of esophageal squamous cell carcinoma cells by inhibition of glycolysis through PI3K/Akt pathway. Life Sci. 2019;232:116679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 28. | Nyamao RM, Wu J, Yu L, Xiao X, Zhang FM. Roles of DDX5 in the tumorigenesis, proliferation, differentiation, metastasis and pathway regulation of human malignancies. Biochim Biophys Acta Rev Cancer. 2019;1871:85-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 29. | Wu N, Han Y, Liu H, Jiang M, Chu Y, Cao J, Lin J, Liu Y, Xu B, Xie X. miR-5590-3p inhibited tumor growth in gastric cancer by targeting DDX5/AKT/m-TOR pathway. Biochem Biophys Res Commun. 2018;503:1491-1497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |