Published online Oct 7, 2022. doi: 10.3748/wjg.v28.i37.5420
Peer-review started: November 22, 2021
First decision: January 11, 2022
Revised: January 24, 2022
Accepted: September 12, 2022
Article in press: September 12, 2022
Published online: October 7, 2022
Processing time: 311 Days and 9 Hours
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide. The identification of novel diagnostic and prognostic biomarkers for CRC is a key research imperative. Immunohistochemical analysis has revealed high expression of centromere protein K (CENPK) in CRC. However, the role of CENPK in the progression of CRC is not well characterized.
To evaluate the effects of knockdown of CENPK and overexpression of Cullin 4A (CUL4A) in RKO and HCT116 cells.
Human colon cancer samples were collected and tested using a human gene expression chip. We identified CENPK as a potential oncogene for CRC based on bioinformatics analysis. In vitro experiments verified the function of this gene. We investigated the expression of CENPK in RKO and HCT116 cells using quantitative polymerase chain reaction (qPCR), western blot, and flow cytometry. The effect of short hairpin RNA (shRNA) virus-infected RKO cells on tumor growth was evaluated in vivo using quantitative analysis of fluorescence imaging. To evaluate the effects of knockdown of CENPK and overexpression of CUL4A in RKO and HCT116 cells, we performed a series of in vitro experiments, using qPCR, western blot, MTT assay, and flow cytometry.
We demonstrated overexpression of CENPK in human colon cancer samples. CENPK was an independent risk factor in patients with CRC. The downstream genes FBX32, CUL4A, and Yes-associated protein isoform 1 were examined to evaluate the regulatory action of CENPK in RKO cells. Significantly delayed xenograft tumor emergence, slower growth rate, and lower final tumor weight and volume were observed in the CENPK short hairpin RNA virus infected group compared with the CENPK negative control group. The CENPK gene interference inhibited the proliferation of RKO cells in vitro and in vivo. The lentivirus-mediated shRNA interference of CENPK inhibited the proliferation of RKO and HCT116 colon cancer cells, with overexpression of the CUL4A.
We indicated a potential role of CENPK in promoting tumor proliferation, and it may be a novel diagnostic and prognostic biomarker for CRC.
Core Tip: High expression of centromere protein K (CENPK) in colorectal cancer (CRC) was found by immunohistochemistry. We demonstrated overexpression of CENPK in human colon cancer samples. CENPK was an independent risk factor in patients with CRC. Our findings indicate a potential role of CENPK in promoting tumor proliferation. CENPK may serve as a novel diagnostic and prognostic biomarker in patients with CRC.
- Citation: Li X, Han YR, Xuefeng X, Ma YX, Xing GS, Yang ZW, Zhang Z, Shi L, Wu XL. Lentivirus-mediated short hairpin RNA interference of CENPK inhibits growth of colorectal cancer cells with overexpression of Cullin 4A. World J Gastroenterol 2022; 28(37): 5420-5443
- URL: https://www.wjgnet.com/1007-9327/full/v28/i37/5420.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i37.5420
Colorectal cancer (CRC) is the third most common cancer in the Western Hemisphere, and the incidence increases with age. RNA interference has been developed to silence genes of interest. Short hairpin RNA (shRNA) expression vectors are useful in gene silencing[1]. The shRNA-mediated silencing of MASTL expression in colon cancer cells induced cell cycle arrest and apoptosis in vitro and xenograft-tumor growth in vivo[2]. Emerging studies have suggested that shRNAs play a crucial role in CRC tumorigenesis and progression[3]. Inducible gene knockdown systems based on lentivirus-mediated gene transfer were developed to regulate colon cancer progression[1]. This property of the shRNA system offers unique applications to study gene function in animals that cannot be achieved using knockout technologies[4]. Although lentivirus vectors have been used for several years, the use of Tet-on lentiviral vectors expressing shRNA as a therapeutic tool for CRC has not been clearly explored[5].
The role of centromere protein K (CENPK) in cancer is an emerging research hotspot. CENPK is overexpressed in several tumor types, and it promotes tumor progression. Research on the role of CENPK in the progression of hepatocellular carcinoma (HCC) has shown that CENPK knockdown significantly inhibits proliferation, migration, invasion, and epithelial-mesenchymal transition in HCC cells[6]. The expression of CENPK has been silenced and promoted via lentivirus-mediated transfection with shRNA sequences in differentiated thyroid carcinomas, such as two pore channel 1 and FTC-133 cells[7]. CENPK is specifically upregulated in ovarian cancer tissues and cell lines, and its overexpression is associated with a poor prognosis in patients with ovarian cancer[8]. Overexpression of CENPK promotes expression of oncogenic cell cycle regulators[9]. However, there has been little research on the role of CENPK in the progression of CRC.
The ubiquitin ligase Cullin 4A (CUL4A) is highly expressed in CRC. CUL4A promotes proliferation and inhibits apoptosis of CRC cells by regulating the Hippo pathway[10]. CUL4A significantly promotes the migration of CRC cells in vitro, which suggests that CUL4A acts as an oncogene in CRC and may become a potential therapeutic target[11]. Therefore, CUL4A expression positively correlates with the prognosis of CRC[12]. Despite extensive fundamental studies, the role of CUL4A expression and lentivirus-mediated transfection with shRNA for CENPK in CRC is not clear. The present study identified CENPK as a novel proto-oncogene in CRC. Increased CENPK levels were found in CRC specimens and negatively correlated with survival rate. Knockdown of CENPK in RKO and HCT116 CRC cells induced apoptosis and suppressed cell proliferation and xenograft tumor formation. Lentivirus-mediated transfection with shRNA for CENPK and CUL4A overexpression played major regulatory roles in CRC.
Fourteen patients with CRC were enrolled from 2016 to 2019, and five tumor-adjacent normal tissues were taken from an area > 10 cm away from the primary neoplasm. The median age of the patients was 59 years (range: 49-68 years) at the time of surgery, and the median follow-up was 31 mo postoperatively (range: 25-37 mo). The Ethics Committee of The Affiliated Hospital of Inner Mongolia Medical University approved the study. Written informed consent was obtained from all patients.
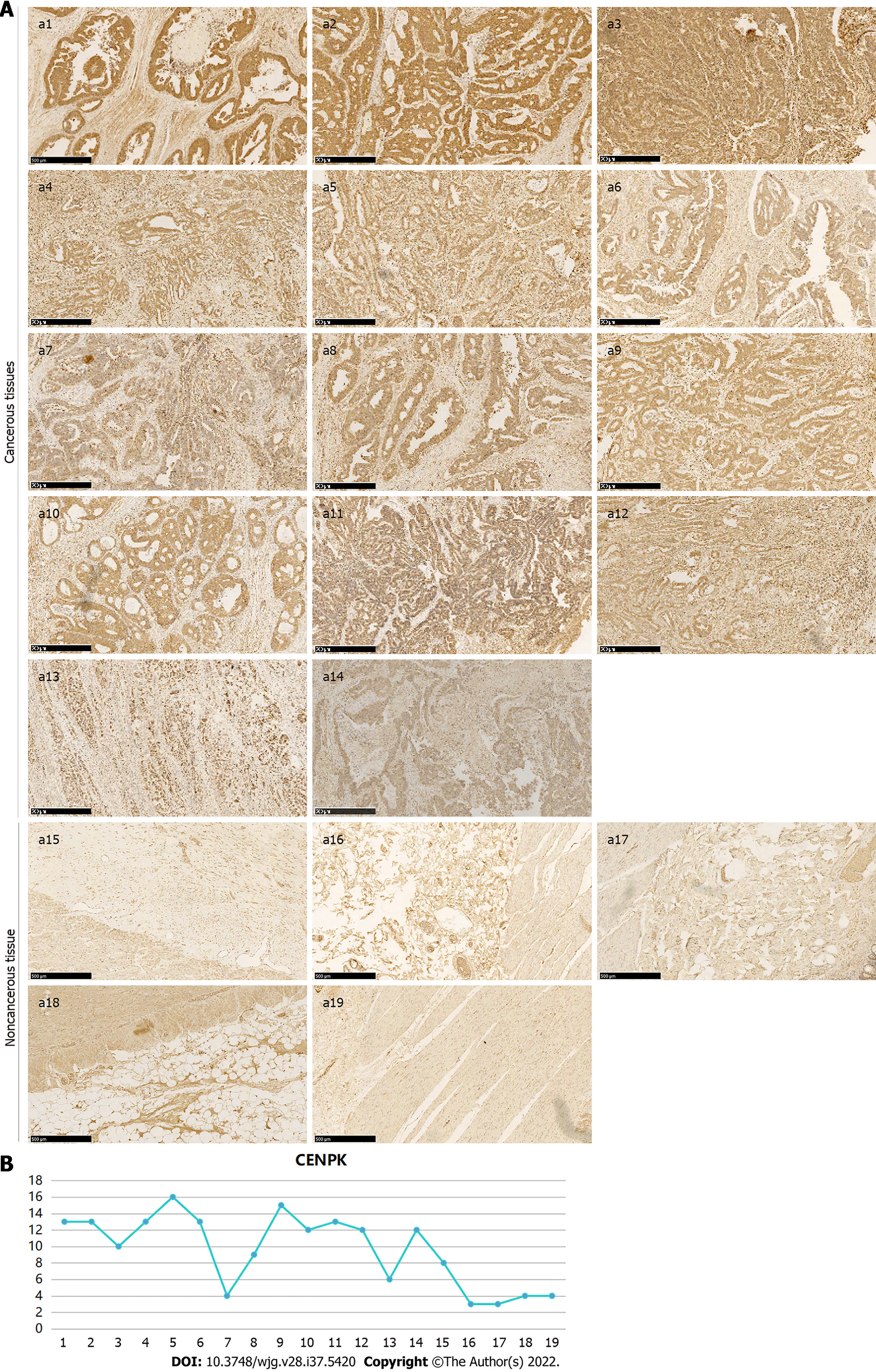
The expression and localization of CENPK protein in 14 cancerous and five noncancerous tissue samples were detected using immunohistochemistry. An antibody against CENPK (26208-1-AP, 1:100; SIGMA) was used for analysis. Vulcan Fast Red Chromogen kit 2 staining was performed for 15-20 min to stop the reaction until 3,3-diaminobenzidine staining was performed to obtain a yellow color to complete the reaction. The staining intensity and positive rate of cytosolic and membrane staining of the antibody (0/1+/2+/3+) in the cancer and adjacent tissue (epithelium) were interpreted. The intensity of the staining score was 0 (negative), 1 (weak), 2 (middle), and 3 (strong). The positive rate of staining was as follows: 0 (negative), 1 (1%-25%), 2 (26%-50%), 3 (51%-75%), and 4 (76%-100%).
Gene expression in a human colon cancer tissue chip (C06161) was detected. The sample was divided into male (age < 57 years) and female (age > 57 years) groups by T stage, N stage, pathological grade, and clinical stage. For the chip assay, total RNA was extracted from tissue using the TRIzol method and measured using a Nanodrop 2000. Qualified samples were used in the chip experiment. The quality checking standard was 1.7 < A260/A280 < 2.2 on Thermo NanoDrop 2000 and RNA integrity number (RIN) ≥ 7.0 and 28S/18S > 0.7 on the Agilent 2100 Bioanalyzer. The quality check results are shown in Table 1. The human gene expression chip type was GeneChip PrimeView Human (100 format, 902487; Affymetrix). Chip hybridization was performed in a GeneChip Hybridization Oven 645 ChIP, washing and dyeing were performed using a GeneChip Fluidics Station 450, and ChIP scanning was performed using a GeneChip Scanner 3000. The reagent used was a GeneChip Hybridization Wash and Stain Kit.
| Order number | Sample number | Sample name | Thermo NanoDrop 2000 | 2100 result | Results | ||
| Concentration (ng/μL) | A260/A280 | RIN | 28S/18S | ||||
| 1 | Y9148-1 | NC | 739.969 | 1.951 | 10 | 2.1 | Pass |
| 2 | Y9148-2 | NC | 688.748 | 1.979 | 10 | 2.1 | Pass |
| 3 | Y9148-3 | NC | 668.435 | 1.944 | 10 | 1.9 | Pass |
| 4 | Y9149-1 | KD | 676.797 | 1.93 | 10 | 1.7 | Pass |
| 5 | Y9149-2 | KD | 697.671 | 1.958 | 10 | 1.7 | Pass |
| 6 | Y9149-3 | KD | 678.338 | 1.971 | 10 | 1.7 | Pass |
For the chip detection, gradient dilution of poly-A RNA controls was prepared to synthesize the first-chain cDNA and second-chain cDNA, and then cRNA of synthetic markers was synthesized via transcription in vitro, which was followed by cRNA purification and quantification to obtain cRNA fragment markers. The expression analysis of the CENPK gene in human colon cancer tissue in the chip (C06161) and clinical data are shown in Table 2. A statistical analysis of CRC tissue chips was performed.
| Number | Variable | Group 1 | Group 2 |
| 1 | Age (yr) | < 57 | > 57 |
| 2 | Sex | Male | Female |
| 3 | T stage | T1/2/3 | T4 |
| 4 | N metastasis | N0 | N1/2 |
| 5 | Pathological grade | 1/2 | 3 |
| 6 | Clinical stage | I/II | III/IV |
The human CRC cell lines RKO and HCT 116 were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Corning) and 1% penicillin and streptomycin solution (Corning) at 37 °C with 5% CO2[13].
Total RNA was extracted from RKO cells with TRIzol reagent (Thermo Fisher Scientific). A 1-mg RNA sample was reverse transcribed into first-strand cDNA using the Revert Aid First Strand cDNA Synthesis kit (Thermo Fisher Scientific). Quantitative polymerase chain reaction (qPCR) was performed using the Taq-Man Gene Expression assay (Thermo Fisher Scientific) on an Mx3000P real-time PCR system (Agilent Technologies). The primer pairs were designed and synthesized by Sangon Biotech (GAPDH: Forward, TGACTTCAACAGCGACACCCA and reverse, CACCCTGTTGCTGTAGCCAAA; CENPK: Forward, ATGGTACTGTCCACTAAGGAGTC and reverse, TGTTCATCCAACCACCGTTGT). The mRNA levels were normalized to the internal reference gene GAPDH, with the mRNA relative expression calculated using the 2∆∆Ct method.
After protein extraction from RKO cells, the protein concentrations of different groups were quantified using a commercial BCA kit. Electrophoresis was performed for 2 h at 4 °C and 300 mA, and the protein was transferred to polyvinylidene fluoride (PVDF) membranes (IPVH00010; Millipore) for immunostaining. The PVDF membranes were incubated in TBST solution containing 5% skimmed milk at room temperature for 1 h. The antibodies were diluted in the blocking solution and then incubated with the PVDF membranes for 2 h at room temperature. The antibodies applied in the present study included those against Yes-associated protein isoform 1 (YAP1) (ab52771; Abcam), CENPK (ab13939; Abcam), CUL4A (ab92554; Abcam), F-box protein 32 (FBX32) (ab168372; Abcam), X chromosome-linked inhibitor of apoptosis (XIAP) (ab28151; Abcam), heat shock protein 90 family class A member 1 (HSP90AA1) (4877; Cell Signaling Technology), class III-tubulin (TUBB3) (#5568; Abcam), and mitogen-activated protein kinase kinase kinase 7 (MAP3K7) (ab109526; Abcam). After primary antibody incubation, the membranes were washed three times with TBST (10 min each) and incubated with the secondary antibodies mouse immunoglobulin G (IgG) (1:2000, sc-2005; Santa Cruz Biotechnology) and rabbit IgG (1:2000, sc-2004; Santa Cruz Biotechnology) at room temperature for 2 h. The Pierce ECL Western Blotting Substrate kit (Thermo) was used for X-ray photography.
The Ethics Committee of The Affiliated Hospital of Inner Mongolia Medical University approved the study. RKO cells with sufficient logarithmic growth were digested with trypsin and suspended in culture medium with the final cell density adjusted to 2.0 × 107/mL. To establish the tumor model, 4.0 × 106 cells were subcutaneously injected into mice. The tumor size and animal body weight were measured 5-20 d later. The tumor volume was calculated as π/6 × L × W × W, where L represents the long diameter, and W and W represent the short diameter. Cancer progression was imaged at 10 d after tumor formation (animals were fasted for 1 d in advance to reduce fluorescence interference) with intraperitoneal injection of 10 μL/g D-luciferin (Bench Mark 15 mg/mL) into mice. After anesthesia by intraperitoneal injection of 10 μL/g pentobarbital solution (BM 7 mg/mL), the mice underwent imaging examination, quantitative analysis, and total fluorescence examination to determine the CENPK function in vivo. After imaging, the experimental animals were killed with an overdose of 2% sodium pentobarbital (0.5 mL) and were in a complete coma before removal of the cervical spine to confirm death. Animals were dissected with medical scissors and tweezers to observe the lungs, liver, and other organs and tissues, which were removed for imaging to observe biological changes and measure the fluorescence.
293T cells for lentivirus packaging were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS. The scramble order for the RNAi negative control was TTCTCCGAACGTGTCACGT. Vector enzyme digestion products were obtained for agarose gel electrophoresis to recover bands of interest. The framework of the viral vector was constructed, and single-strand primers were syn
The lentivirus-infected HCT116 cells are shown in Table 3. RKO and HCT 116 cells (2 × 105 cell/mL) were cultured in RPMI 1640 medium containing 10% FBS with polybrene in six-well plates. The following experimental groups were used: (1) Cells (RKO or HCT 116 cells) infected with CENPK negative control virus [NC (KD)]; (2) Cells with lentivirus-mediated shRNA interference of CENPK (KD); (3) Cells with overexpression and negative control virus interference [NC (OE)]; (4) Cells virally infected with CUL4A (OE); (5) Cells with shRNA interference of CENPK and overexpression of negative control virus [KD + NC (OE)]; and (6) Cells with lentivirus-mediated shRNA interference of CENPK and CUL4A (KD + OE).
| Virus number | Experimental group | Virus titer | Virus infection dose |
| psc3741 | NC (KD) | 2.0E + 08 TU/mL | 10.00 μL |
| LVpGCSIL-004PSC45249-1 | KD | 3.0E + 08 TU/mL | 6.67 μL |
| KL9688-21 | NC (OE) | 4.0E + 8 TU/mL | 5.00 μL |
| LVKL6733-2 | OE | 2.0E + 08 TU/mL | 10.00 μL |
| LVpGCSIL-004PSC45249-1+ KL9688-21 | KD+NC (OE) | 3.0E + 08 + 2E + 8 TU/mL | 3.33 + 5.00 μL |
| LVpGCSIL-004PSC45249-1+ LVKL6733-2 | KD+OE | 3.0E + 08 + 2E + 8 TU/mL | 3.33 + 5.00 μL |
For the lentiviral infection, media for adherent cell inoculation and viral infection were used at a 1:1 ratio in 96-well plates. The culture medium was replaced, and the optimal amount of virus was added for the infection. The most appropriate time point after infection was selected for the replacement of conventional medium and continued culture, 8-12 h after infection. When fluorescently labeled lentiviral infection occurred, the green fluorescent protein reporter signal was observed by fluorescence microscopy 72 h after infection. After infection efficiency reached 80%, 0.5 mL of puromycin was used for cell function experiments. Western blot and qPCR analysis were performed to confirm the knockdown of CENPK.
The proliferation potential of RKO and HCT 116 cells was analyzed by cell growth curve and MTT assay. For the cell growth curve assay, RKO and HCT 116 cells of different groups [shCtrl and shCENPK or NC, NC (KD), NC (OE), OE, KD + NC (OE), and KD + OE] were collected, replated in 24-well plates at 2000 cells/well, and cultured at 37 °C in a 5% CO2 incubator for 5 d. Beginning on day 2, the optical absorption value of each group was checked once daily and the number of fluorescent cells calculated. A growth curve was drawn to show cell proliferation.
For the MTT assay, RKO and HCT 116 cells were replated in 24-well plates at 2000 cells/well and cultured at 37 °C in a 5% CO2 incubator for 24 h. Subsequently, 20 μL MTT (5 mg/mL) was added to the wells. After incubation for 4 h, reactions were stopped with 100 μL DMSO. After shaking for 2-5 min, the OD value at 490/570 nm was determined (M2009PR, Tecan Infinite).
Apoptosis was analyzed by flow cytometry (fluorescence-activated cell sorting; FACS) with a commercial Annexin V-APC kit (88-8007, eBioscience). RKO and HCT 116 cells of each group were collected after washing in 1 × binding buffer, and centrifuged at 1300 rpm for 3 min. After centrifugation, the cell suspension was prepared by adding 200 μL of 1 × binding buffer. Subsequently, 10 μL of Annexin V-APC staining solution was added to the suspension. After incubation in the dark at room temperature for 10-15 min, 400 μL of 1 × binding buffer was added to the cell suspension, and FACS was conducted for apoptosis detection.
Caspases 3/7 were detected in RKO and HCT 116 cells using a commercial Caspase-Glo3/7 detection kit (G8091, Promega). For the preparation of Caspase-Glo reaction solution, 10 mL of caspase-Glo3/7 buffer was added and vortexed until the dissolve of substrates. After collection and counting, the RKO and HCT 116 cells at a final density of 104 cells/well were replated in 96-well plates, and an empty control group containing culture medium without RKO or HCT 116 cells was established. Next, 100 μL of Caspase-Glo reaction solution was added to each well and shaken for 30 min at 500 rpm at room temperature for 2 h. The signal strength was measured using a microplate reader (M2009PR, Tecan Infinite).
RKO cells were incubated with 1 mL of 1 × cell lysis buffer (Cell Signaling Technology) containing 1 mmol/L PMSF on ice for 2 min. Then, 100 μL of blocking solution was added to RKO cells with the cell lysate samples shaken at room temperature for 15 min. After incubation at room temperature for 2 h, the cell lysate samples were washed with 1 × cleaning solution on a horizontal shaker for 5 min. After three washes, 75 μL of 1 × antibody was added to the cell lysate samples and incubated on a horizontal shaker for 1 h. Then, 75 μL of 1 × HRP-linked streptavidin solution was added, followed by incubation on a horizontal shaker for 1 h. 1 × array cleaning solution was added to the cell lysate samples, which were then washed on a horizontal shaking bed four times (5 min each). The slides were placed in 1 × cleaning solution and then in exposure liquid (9 mL deionized water + 0.5 mL lumiGlo + 0.5 mL hydrogen peroxide) for 1-2 s, followed by observation with a chemiluminescence imaging system (ChemiScope 5300; Clinx Science Instruments).
Statistical analyses were performed using SPSS version 19.0 (IBM Corp.). Data are presented as the mean ± SD. Data comparing two samples were analyzed using student’s t-test. Comparisons of datasets containing > 3 groups were evaluated by one-way repeated-measures ANOVA followed by the Bonferroni post hoc test. Gene expression of cancer and noncancer tissue groups was evaluated by the Mann-Whitney U test. P < 0.05 was considered to indicate a significant difference.
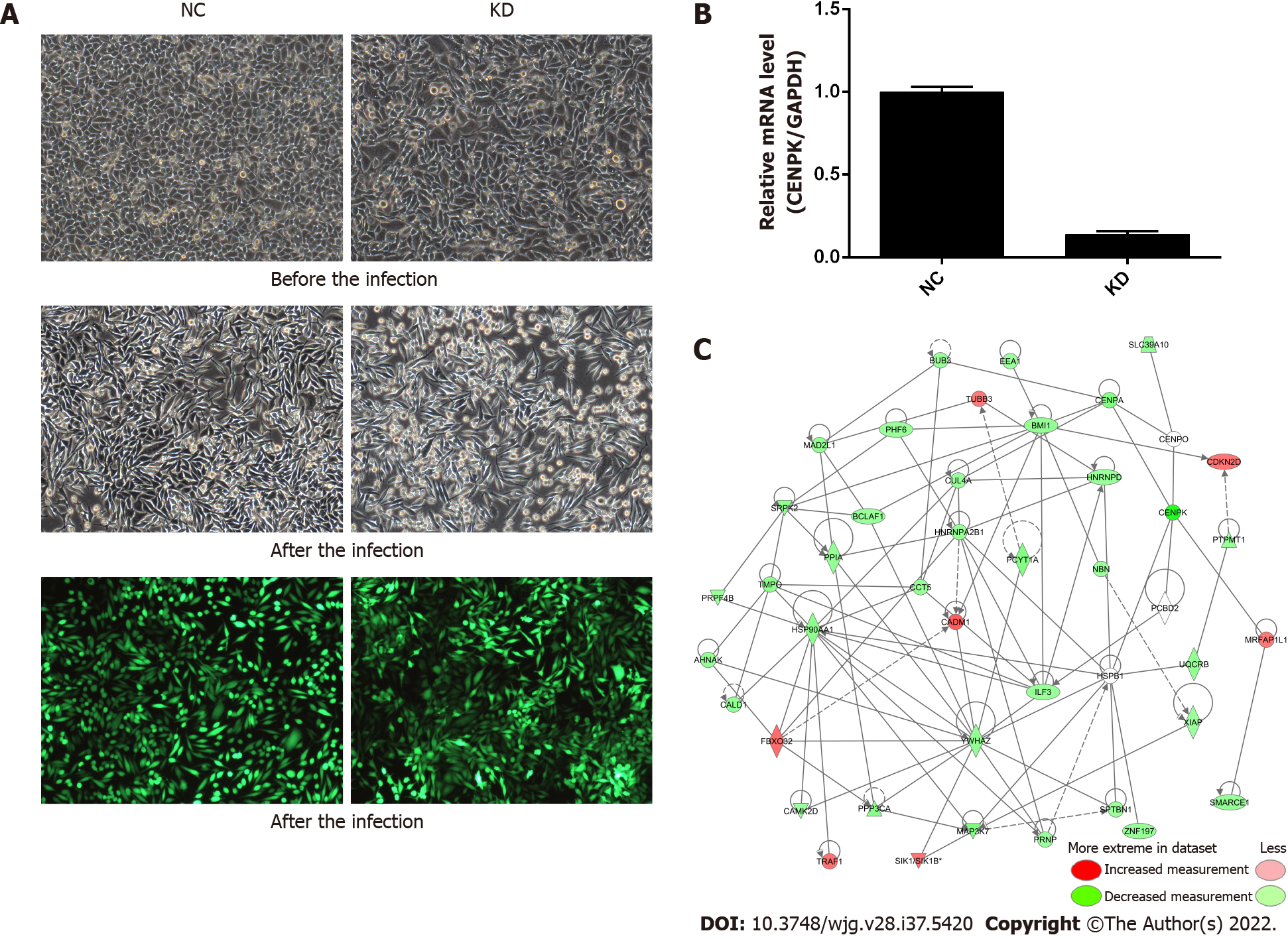
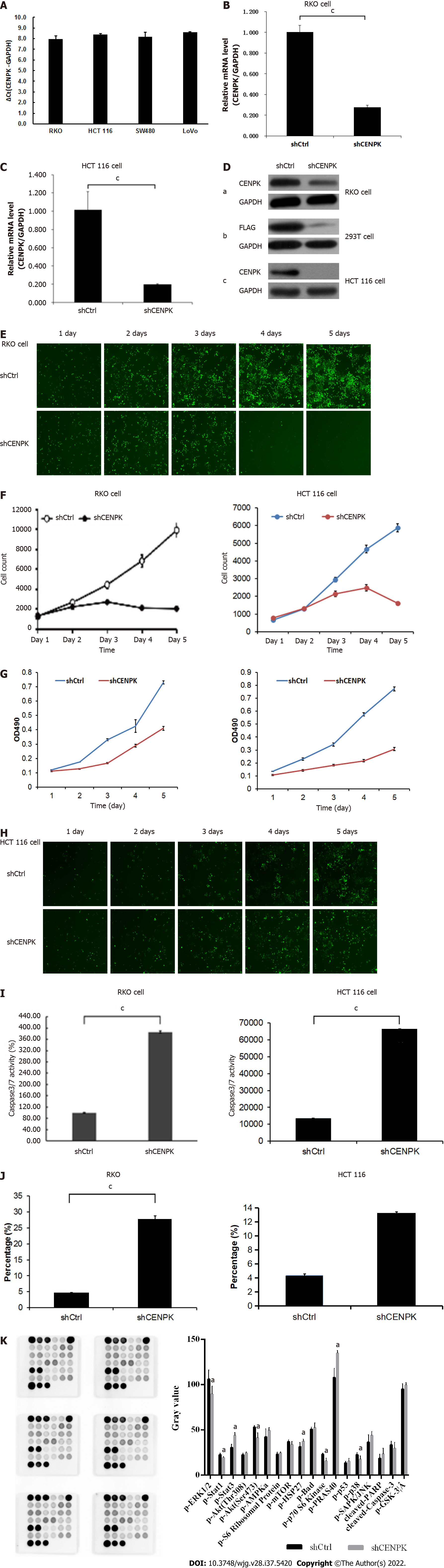
For lentiviral infection marked with antibiotic resistance genes, the antibiotics (such as puromycin) were used to screen cells after 72 h, and the infection efficiency was approximately 80% for fluorescence labeled lentivirus infection (Figure 1A). CENPK knockdown efficiency in RKO cells in the KD group reached 86.1% compared to the NC group (23.4%) (Figure 1B). This analysis revealed the genes related to tumor cell proliferation and invasion. The genes that inhibited tumor cell proliferation or tumor invasion were selected, together with the target genes of concern, to develop a gene relationship network diagram. This analysis suggested that CENPK affected the proliferation and invasion of cells by acting on YWHAZ and BMI1, which affected the performance of downstream genes. The gene interaction network diagram shows a network of interaction relationships between molecules within a determined functional region. Different shapes represent genes, proteins, and chemicals in the network maps (Figure 1C).
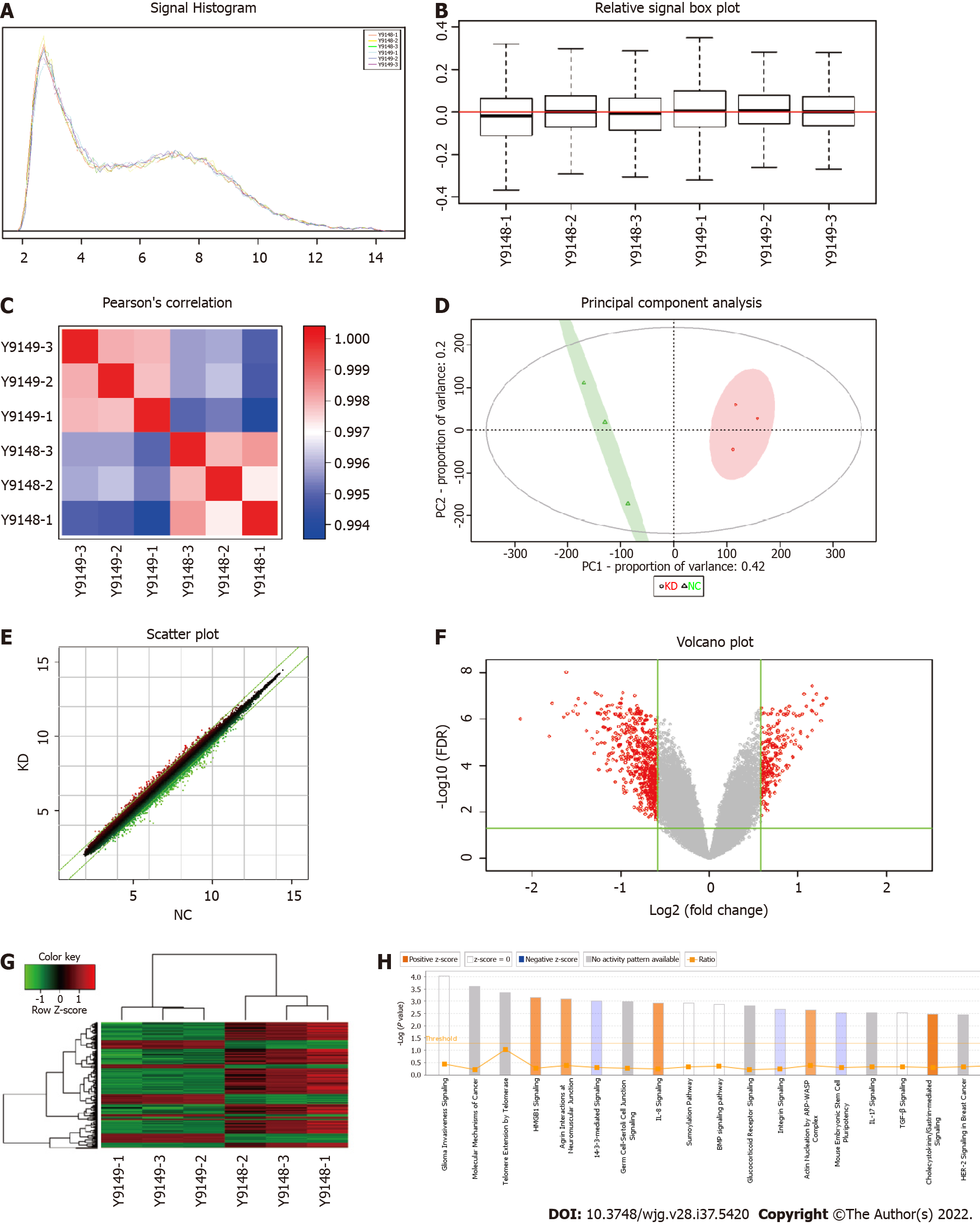
The original quality evaluation for the quality control data and the remaining data meeting the filter standards, including significant difference analysis and functional analysis of differential genes, was used to evaluate the tumor occurrence, development, metastasis, and other molecular mechanisms using chip information analysis for target screening and drug design. Figure 2A shows the signal strength distribution of the entire chip probes. Better signal intensity distribution curves of different samples indicated higher reliability of the chip experiment. The average Z score of all samples within one signal strength range was < 2, which supported the reliability of all chip results meeting the continuing analysis criteria. Figure 2B shows the distribution of the relative logarithmic signal strength for all chips. Closer distribution of the relative logarithmic signal strength box diagram indicated better repeatability of the data. The median Z score for all samples was < 2, which indicated good repeatability of the chip experiments in the project that met the continued analysis criteria. The Pearson correlation coefficient distribution diagram shows the level of correlation of the signal strength between all chips, where each lattice represents the degree of correlation between two samples corresponding to the ordinate and transverse (Figure 2C). The group sample gene expression patterns were typically similar and demonstrated a high correlation coefficient, as shown in red in the correlation coefficient distribution diagram. The gene expression pattern difference between groups demonstrated a low correlation coefficient, which is shown in blue in the correlation coefficient distribution diagram. The correlation coefficient in the KD and NC groups was > 0.99, which indicated a high similarity of gene expression in the same group. The intergroup correlation coefficient was significantly lower than the group correlation coefficient, which indicated a large difference between groups that met the continuing analysis standard.
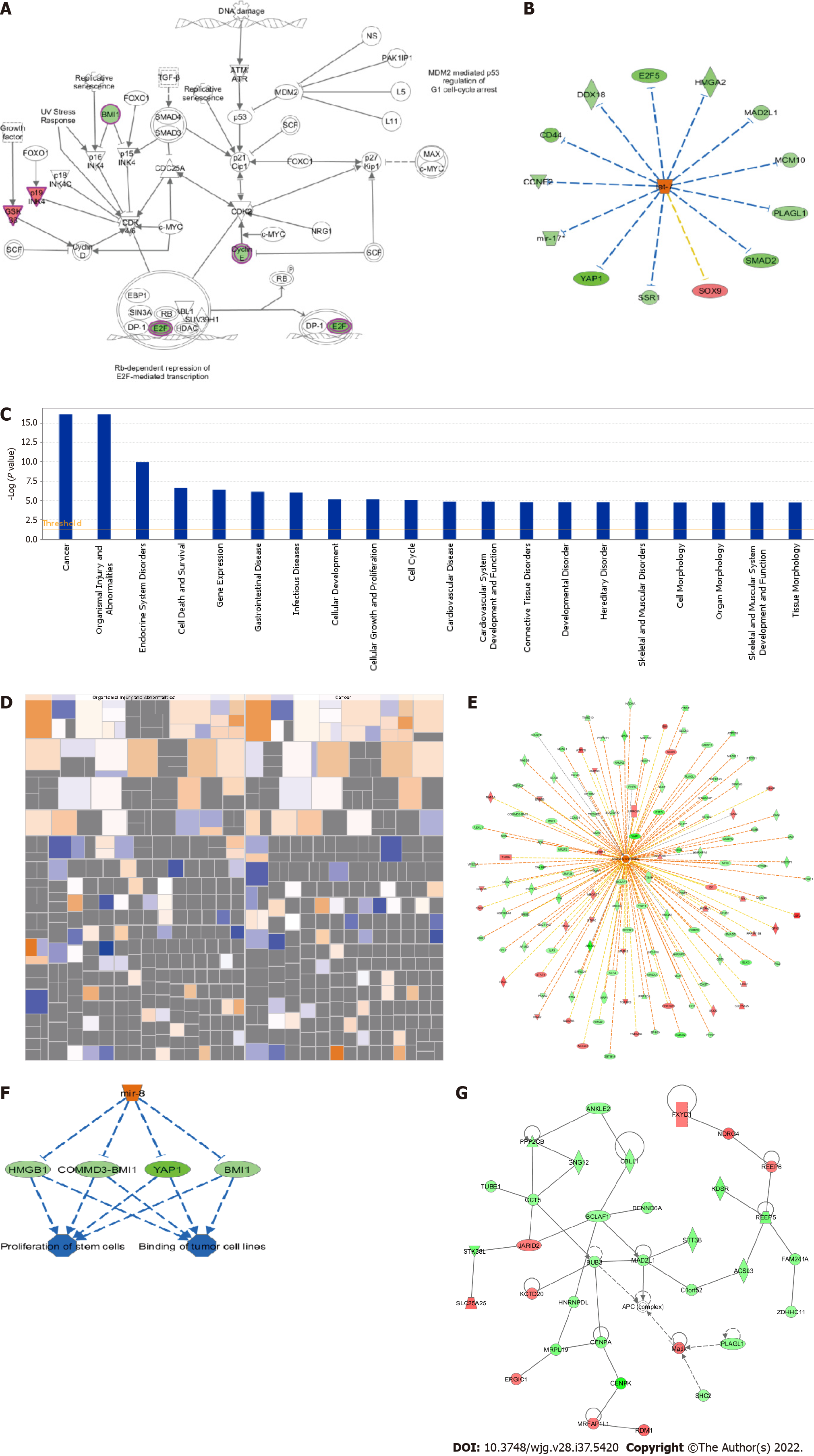
From the PCA score chart (Figure 2D), there was an obvious sample aggregation trend in the PC1 and PC2 dimensions in the KD and NC groups and an obvious intergroup separation trend, which indicated similar samples and significant differences between groups that met the standard of the continuing analysis. The cross ordinate of each point in Figure 2E represents the signal strength of a probe group in the NC and KD groups. The parallel green solid line is the reference line difference, and the points in the interval within the reference line represent the probe group without a significant change. The red point outside the interval represents the relatively elevated probe group in the KD group, and the green point represents the relatively elevated probe group in the NC group. A volcano graph was drawn using gene expression differences (fold change) and false discovery rate (FDR) tests between the two samples to show the significant differences between the two samples (Figure 2F). Thermal mapping of KD and NC samples was performed using the differential expression spectrum of genes screened by |fold change| ≥ 1.5 and FDR < 0.05 (Figure 2G). Figure 2H shows the significant enrichment of differentially expressed genes in the classical pathway. The orange notation indicates an activated pathway (Z > 0); the blue label indicates inhibition of the pathway (Z < 0); shades of orange and blue (or absolute Z values) represent the degree of activation or inhibition (Z ≥ 2 represents a significant activation and standard, and Z ≤ -2 denotes significant suppression); and the ratio represents the ratio of the difference of the gene number in this signaling pathway to all genes contained in the pathway. G1/S checkpoint regulation was activated in the cell cycle, and Z was 2.236.
G1/S checkpoint regulation was significantly activated. Red represents significant upregulation, and green represents the genes. Figure 3A shows the changing trend of gene expression in the pathway, which provided the basis for the study of the molecular mechanisms involved in the changes between sample groups. The upstream regulator network diagram (Figure 3B) shows the interaction between upstream regulators and downstream molecules that were directly associated in the dataset. Let-7 significantly suppressed SOX9 mRNA levels, and consistent trends were observed between let-7 and signal sequence receptor 1. Figure 3C shows the significant enrichment of differentially expressed genes in disease and function. According to the internal algorithms and standards of IPA, Z ≥ 2 denoted significant activation of the disease or function, and Z ≤ -2 indicated significant suppression of the disease or function. Significantly activated diseases or functions included morbidity or mortality (Z = 4.216) and organismal death (Z = 4.164). Significantly suppressed diseases or functions included infection by human immunodeficiency virus 1 (Z = -3.335) and viral infection (Z = -3.2) (Figure 3D). The network contains all the differentially expressed genes associated with the specified disease or function, and demonstrates possible interactions and changes in expression trends between genes based on the ingenuity knowledge base (Figure 3E). The top ranked regulatory network in this regulatory effect analysis suggested that the dataset was inhibitory due to the regulator miR-8 in the binding of tumor cell lines and the proliferation of stem cells via BMI1, COMMD3-BMI1, HMGB1, and YAP1 (Figure 3F). The network of interactions between molecules of a disease or function-related relationship is shown in Figure 3G.
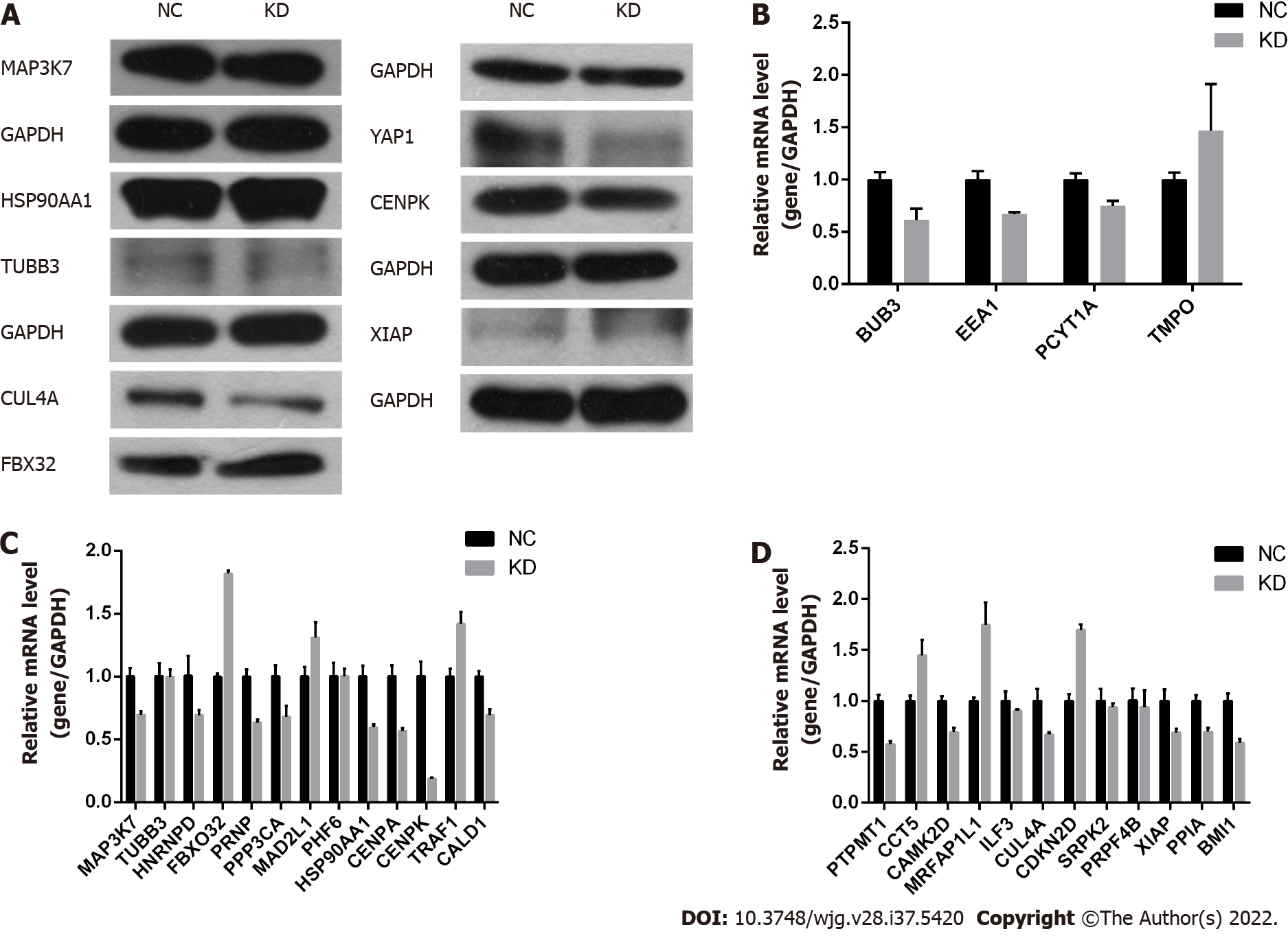
Based on the above biological analysis, we speculated that CENPK was more likely to function in CRC by regulating the expression of the above-mentioned genes. Therefore, these genes were selected for western blot validation. Western blot analyses and qPCR showed that expression of FBX32 was upregulated, and CUL4A and YAP1 were downregulated (Figures 4A-D). Taken together, these results suggested that CENPK played a role in CRC by regulating FBX32, CUL4A, YAP1, and other genes in RKO cells.
The expression of CENPK in CRC tissue was measured using immunohistochemistry to detect target protein expression and localization in tissue cells. Expression of CENPK was higher in cancerous than in paracancerous tissues (Figure 5A). Positive expression of CENPK in the cancerous and noncancerous tissues exhibited a differential multiple of 2.61, which was a significant difference (Figure 5B). Expression and proportional statistics of the CENPK gene in cancerous and noncancerous tissue are shown in Table 4.
| Case group | P value | ||
| Cancerous tissues | Noncancerous tissue | ||
| Higher expression | 4 | 5 | 0.008 |
| Lower expression | 10 | 0 | |
CENPK gene expression in cancerous and noncancerous tissues is shown in Table 5. The differential expression of CENPK gene in cancerous and noncancerous tissues was analyzed using the Mann-Whitney U test. The significantly higher expression of CENPK in cancerous than in noncancerous tissues (P < 0.05) supports CENPK as an effective diagnostic marker for CRC. The expression and proportion of CENPK genes in cancerous tissues are shown in Table 6. The CENPK gene expression in different types of clinical experiment, including sex, age, T stage, and N metastases, was analyzed to compare its low expression and high expression. Expression of CENPK gene in cancerous tissue differed significantly according to T stage, N stage, and clinical stage, which suggested that CENPK expression was associated with these pathological indicators. The correlation between CENPK expression and clinical data (T stage, N stage, and clinical stage) in cancerous tissues was examined using Spearman’s test, and the potential role of these clinical data in the development of CRC was examined (Table 7). Expression of the CENPK gene in cancerous tissues was positively correlated with T stage, N stage, and clinical stage, which suggested that low CENPK expression in the early stage of cancer gradually increased with the disease course.
| CENPK gene expression group | Total | P value | |||
| Low expression | High expression | ||||
| Tissue type group | Cancerous tissues | 146 | 160 | 306 | |
| Noncancerous tissue | 27 | 1 | 28 | 0.000 | |
| Total | 173 | 161 | 334 | ||
| CENPK gene expression group | Total | P value | |||
| Low expression | High expression | ||||
| Sex | Male | 88 | 103 | 191 | 0.460 |
| Female | 58 | 57 | 115 | ||
| Total | 146 | 160 | 306 | ||
| Age | < 57 | 75 | 82 | 157 | 0.983 |
| > 57 | 71 | 78 | 149 | ||
| Total | 146 | 160 | 306 | ||
| T stage | T1/2/3 | 108 | 98 | 206 | 0.018 |
| T4 | 38 | 62 | 100 | ||
| Total | 146 | 160 | 306 | ||
| N metastasis | N0 | 117 | 108 | 225 | 0.012 |
| N1/2 | 29 | 52 | 81 | ||
| Total | 146 | 160 | 306 | ||
| Pathological grade | 1/2 | 111 | 114 | 225 | 0.130 |
| 3 | 29 | 45 | 74 | ||
| Total | 140 | 159 | 299 | ||
| Clinical stage | I/II | 116 | 108 | 224 | 0.019 |
| III/IV | 30 | 52 | 82 | ||
| Total | 146 | 160 | 306 | ||
| Gene expression | T stage | N metastasis | Pathological grade | ||
| Gene expression | Correlation coefficient | 1.000 | 0.135 | 0.143 | 0.135 |
| Significance (two-tailed) | 0.000 | 0.018 | 0.012 | 0.018 | |
| T stage | Correlation coefficient | 0.135 | 1.000 | 0.000 | 0.000 |
| Significance (two-tailed) | 0.018 | 0.000 | 0.000 | 0.000 | |
| N metastasis | Correlation coefficient | 0.143 | 0.000 | 1.000 | 0.000 |
| Significance (two-tailed) | 0.012 | 0.000 | 0.000 | 0.000 | |
| Pathological grade | Correlation coefficient | 0.135 | 0.000 | 0.000 | 1.000 |
| Significance (two-tailed) | 0.018 | 0.000 | 0.000 | 0.000 | |
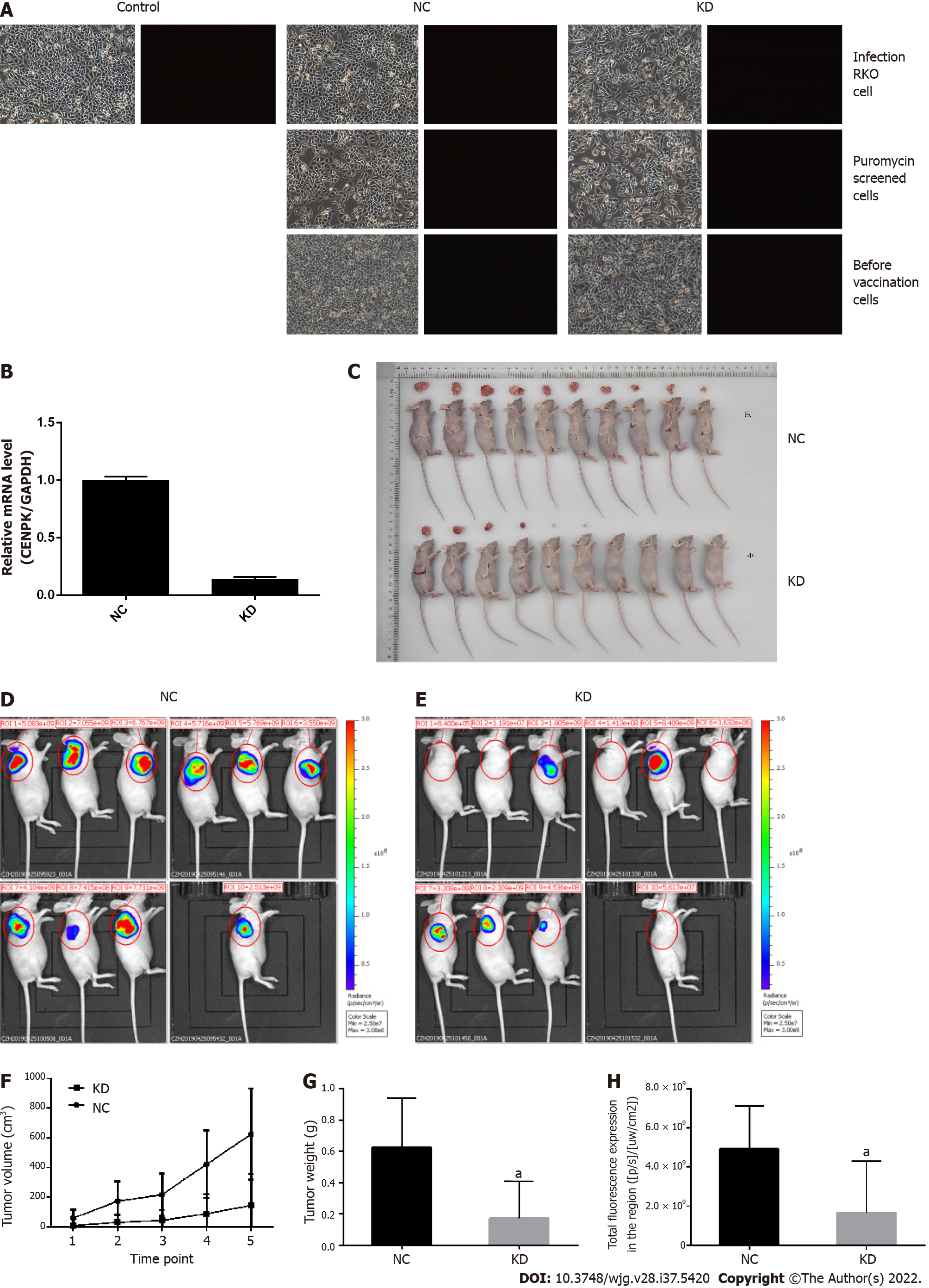
RKO cells infected by viruses were subcutaneously injected into nude mice for analysis of tumor growth conditions (Figure 6). RKO cells were infected with lentivirus, and there was no significant cell death, especially in the NC group, which was comparable to the control group. In contrast, the infection efficiency reached approximately 80% in the KD group (Figure 6A). The infection efficiency in the target screening experiment was > 90% in the KD group. Therefore, the good cell conditions and qualified infection efficiency up to 80% were used for downstream testing. Here, RKO cells were injected subcutaneously into the nude mice to analyze the tumor growth conditions and detect the infection efficiency of RKO cells. Based on the PCR results, CENPK gene knockdown efficiency in RKO cells reached 83.2% in the KD group compared to the NC group. The tumor status (the tumor formed and size) was good 1 d after subcutaneous injection. Over 20 d, the tumors gradually decreased and even disappeared in the KD group, but they persisted in the NC group (Figure 6C). Tumors in mice were analyzed using live imaging. The total fluorescence expression volume in ten nude mice in the KD and NC groups was measured via quantitative analysis of fluorescence (Figures 6D and 6E). Total fluorescence expression in the region was significantly decreased in the KD group compared to the NC group (Figure 6H). Tumor weight and volume were significantly decreased in the KD group compared to the NC group (P < 0.05) (Figures 6F and 6G). Therefore, lentiviral infection of RKO cells inhibited the growth of tumor cells in vivo.
Expression of CENPK gene in CRC cells is shown in Figure 7A, and the target gene expression abundance was relatively low. Based on the mRNA level, the CENPK gene reduction efficiency was examined using qPCR. Expression of the CENPK gene in RKO and HCT116 cells in the experimental group was inhibited after 3 d of shRNA lentivirus infection (Figures 7B and 7C). Target reduction of foreign proteins due to CENPK gene expression was detected using western blot, which showed that shCENPK had a significant knockdown effect on the exogenous expression of CENPK gene at the protein level in 293T cells (Figure 7D). Endogenous protein expression of CENPK showed that shCENPK had a significant knockdown effect on endogenous CENPK expression at the protein level in RKO cells (Figure 7D). Endogenous expression of CENPK was knocked down at the protein level in HCT116 cells (Figure 7D).
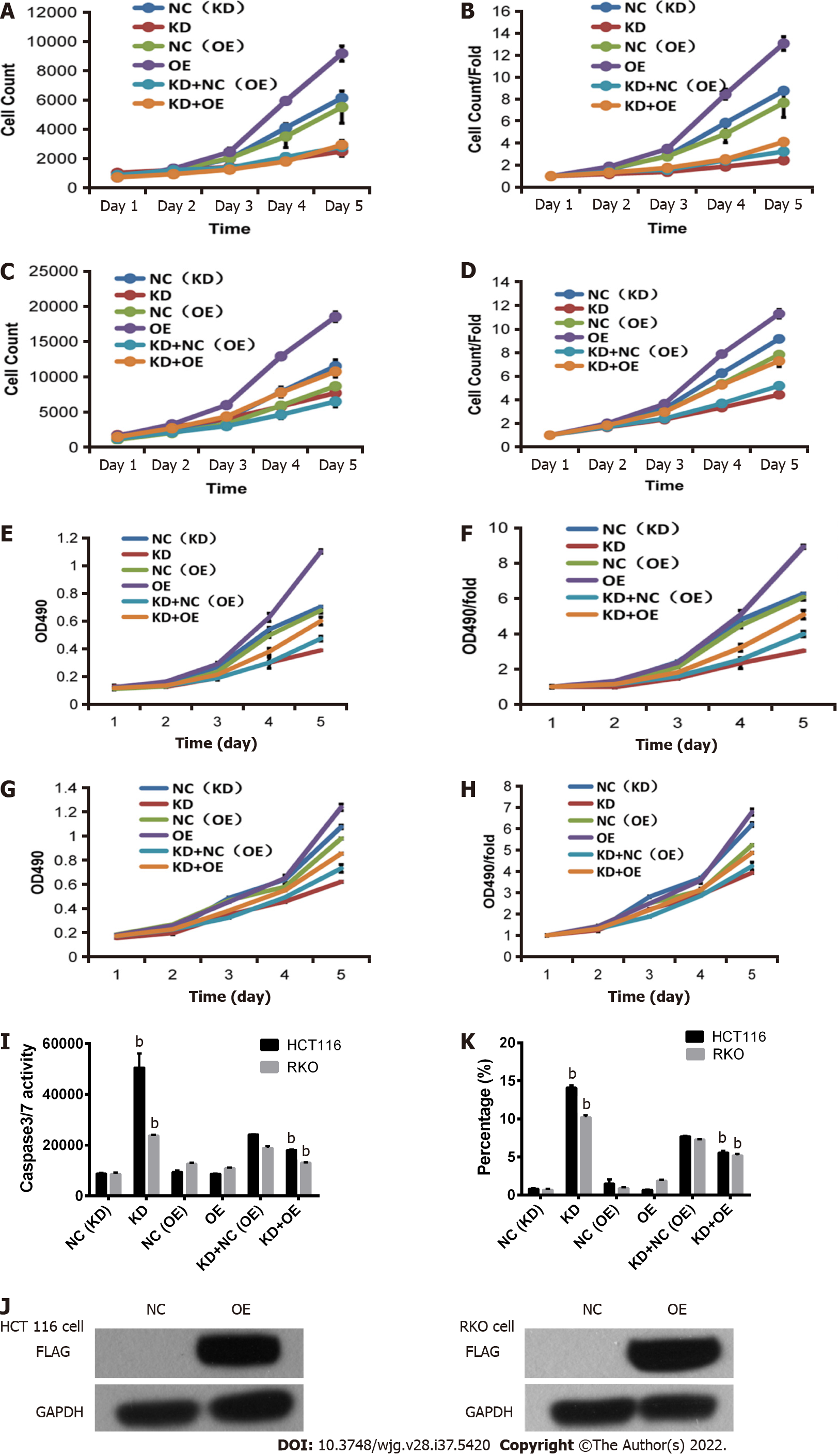
After 3 d of shRNA lentivirus infection, 1500 cells were seeded in 96-well plates. The effect of CENPK on cell proliferation via Celigo detection showed that proliferation of RKO cells was significantly inhibited in the experimental group. Celigo continuous detection for 5 d suggested that the CENPK gene was significantly associated with RKO and HCT116 cell proliferation (Figures 7E-H). To examine the effects of gene reduction on cell proliferation after 3 d of shRNA lentivirus infection, the cells were seeded in 96-well plates. After 5 d, the proliferation rate of RKO and HCT116 cells in the experimental group was significantly inhibited, which suggests a significant correlation between CENPK gene and RKO and HCT116 cell proliferation (Figure 7G). Detection of the effect of CENPK gene reduction on apoptosis using FACS showed that after 5 d of shRNA lentivirus infection, RKO and HCT116 cells in the experimental group increased significantly, which suggested that the CENPK gene was significantly related to apoptosis of RKO and HCT116 cells (Figure 7J, Supplementary Figures 1 and 2). Based on the detection of CENPK for apoptosis, caspase 3/7 activity was increased after 3 d of shRNA lentivirus infection in the experimental group, which suggested that CENPK gene was significantly related to apoptosis of RKO and HCT116 cells (Figure 7I). Therefore, CENPK gene inhibited proliferation of RKO cells. After RNAi of the CENPK gene in RKO cells, associated genes in the signaling pathway were detected using PathScan. The protein expression levels in the p-ERK1/2, p-Stat1, p-Stat3, p-Akt (Ser473), p-HSP27, p-p70 S6 kinase, p-PRAS40, and p-p38 signaling pathways were significantly different (P < 0.05) (Figure 7K).
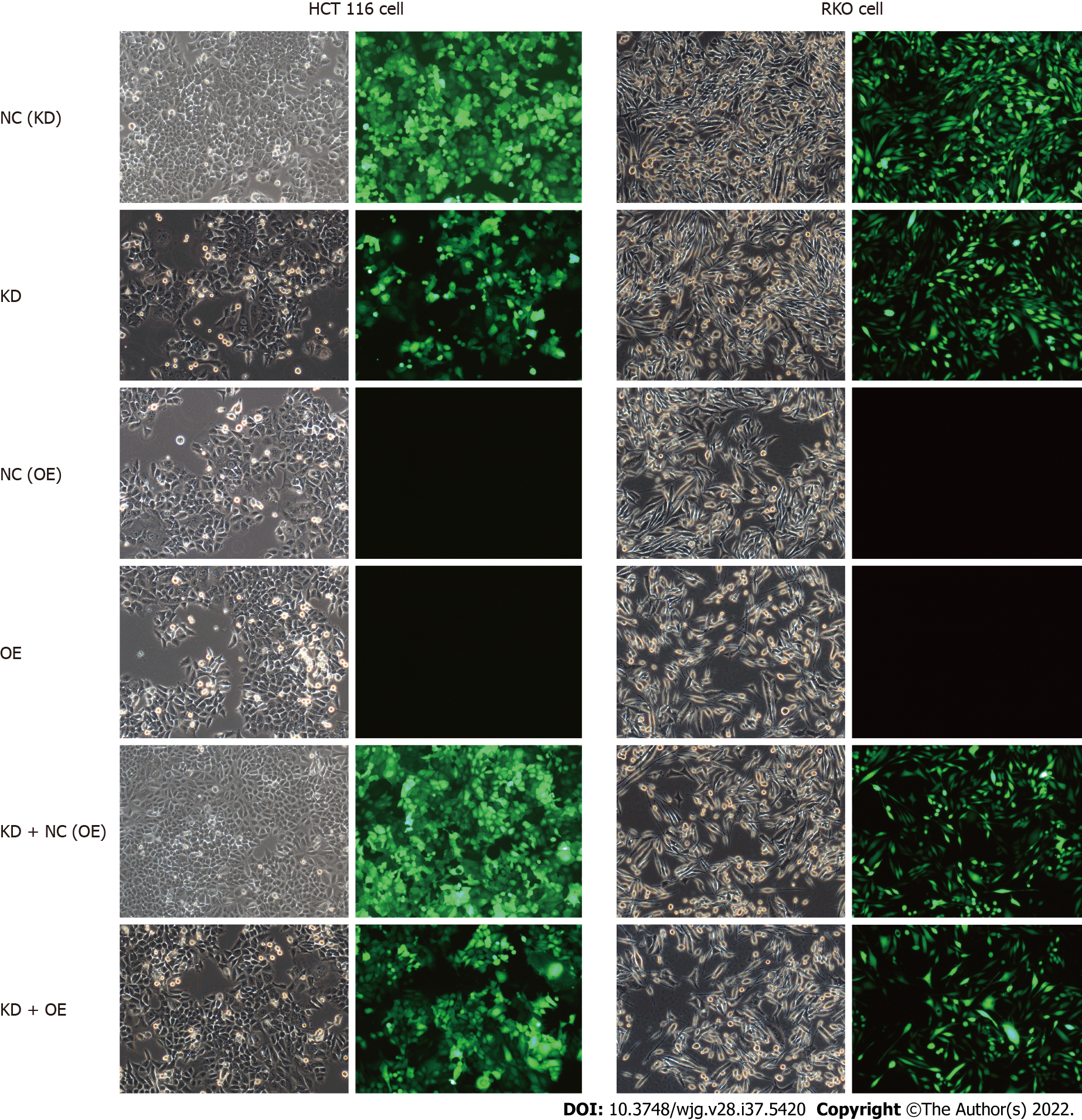
Lentivirus-mediated shRNA interference of CENPK in HCT116 cells and RKO cells was observed (Figure 8). The control and target lentivirus-infected cells (HCT116 cells and RKO cells) were observed after 72 h under a microscope, and cell infection efficiency reached > 80%, and the cell state was normal. The effects of CENPK gene reduction and CUL4A gene overexpression on HCT 116 and RKO cell proliferation using Celigo detection are shown in Figures 9A-D. At 3 d after lentiviral infection, the number of HCT 116 cells was 1000, and the number of RKO cells was 2000. After 5 d, the KD group exhibited slower cell proliferation compared to the NC (KD) group. Cell proliferation increased rapidly in the KD + OE and NC (OE) groups compared to the KD + NC (OE) and OE groups in HCT 116 cells (Figures 8A and 8B, Supplementary Figures 3 and 4) and RKO cells (Figures 6C and 6D). HCT 116 and RKO cell proliferation was measured using the MTT assay (Figures 9E-H). Cell proliferation increased slowly in the KD group compared to the NC (KD) group, but it increased rapidly in the OE group compared to the NC (OE) group. However, RKO cell proliferation was not consistently increased in the KD + OE group compared with the KD + NC (OE) group, and HCT 116 cell proliferation was obviously increased in the KD + OE group. HCT 116 and RKO cell apoptosis was induced by CENPK gene reduction and CUL4A gene overexpression; therefore, caspase 3/7 activity and apoptotic cells were measured after lentiviral infection for 5 d. Based on the findings for HCT 116 and RKO cells shown in Figure 9I, caspase 3/7 activity and apoptotic cells were increased in the KD group compared to the NC (KD) group. Caspase 3/7 activity did not change significantly in the OE group, and apoptotic cells did not change significantly compared with the NC (OE) group. Similar to the KD + NC (OE) group, the KD + OE groups exhibited decreased caspase 3/7 activity and apoptosis. HCT 116 and RKO cell apoptosis via CENPK gene reduction and CUL4A gene overexpression was detected using FACS (Figure 9K, Supplementary Figures 5 and 6). HCT 116 and RKO cell apoptosis was increased significantly in the KD group (P < 0.01) compared to the NC (KD) group. Apoptosis was decreased significantly in the KD + OE group (P < 0.01) compared to the KD + NC (OE) group. However, no significant change in apoptosis occurred in the OE group. The FLAG protein level after CUL4A overexpression was detected using western blot (Figure 9J). The proliferation rate in the OE group compared to the NC group after CUL4A overexpression in HCT116 and RKO cells was detected by FLAG. These results suggested that lentivirus-mediated shRNA interference of CENPK inhibited the proliferation of RKO and HCT116 cells, and overexpression of the CUL4A gene reversed this effect in RKO and HCT 116 cells with CENPK gene knockdown.
CRC is one of the most common malignant tumors. Increasing evidence suggests that CENPK plays a key role in promoting carcinogenesis[14]. To the best of our knowledge, this study is the first to clarify the clinical value of CENPK in human CRC tissues. However, the underlying mechanism is largely unknown. The expression of CENPK in CRC tissues was quantified using western blot and quantitative real-time PCR. CENPK was highly expressed in CRC tissue and positively correlated with large tumor size and advanced tumor stage. CENPK was found to be frequently downregulated in CRC primary tumor samples compared to adjacent normal tissues, using bioinformatics analysis and experiments in vivo. Expression of the CENPK gene was significantly different in cancerous and noncancerous tissues, which supports the use of CENPK for diagnosis of CRC and patient selection for therapy. Tissue chip detection showed that CENPK expression was associated with pathological indicators, such as T stage, N stage, and clinical stage. This confirmed the low level of CENPK expression in early cancer and increasing expression during the course of the disease. We used in vitro and in vivo models to demonstrate that lentivirus-mediated shRNA interference of CENPK regulated the proliferation of CRC cells. The CENPK gene may affect the expression of a series of downstream genes by acting on YWHAZ and BMI1, which affect cell proliferation and invasion. Therefore, CENPK gene functions in the progression of CRC by altering the expression of CUL4A, CENPK, XIAP, FBX32, YAP1, MAP3K7, TUBB3, and HSP90AA1. Western blot analysis showed that FBX32 was upregulated and CUL4A and YAP1 were downregulated.
YAP1 is an effector of the Hippo pathway, which is critical for regulating organ size, cell proliferation, and tumor growth in mammals[15]. Research on YAP1 expression in CRC tissues showed the effect of silencing expression of the YAP1 gene on the proliferation and invasion of SW620 CRC cells[16]. Another study showed that FBX32 and the mRNA levels of several proteasome-related genes were significantly upregulated by methionine limitation[17]. Moreover, CUL4A is highly expressed in colon cancer and promotes the proliferation and inhibits the apoptosis of colon cancer cells by regulating the Hippo pathway[10]. Therefore, the mechanism of the effect of CUL4A expression on cell proliferation and apoptosis in CRC by regulating the CENPK pathway was further investigated.
Lentivirus-mediated shRNA interference of CENPK was successfully and stably induced in RKO cells, which lays a foundation for further study of the role of the CENPK gene in the carcinogenesis and progression of CRC. With regard to therapeutic options for CRC, increased CENPK expression inhibited CRC cell proliferation and induced apoptosis of RKO cells with lentiviral infection, which suggests that CENPK inhibits CRC progression. A xenograft mouse model also confirmed the tumor-suppressive function of CENPK. We used RKO cells with lentiviral infection to control these processes and inhibit tumor growth. Specifically, we found that the CENPK gene regulated tumor growth, and the total fluorescence in the tumor was significantly decreased compared to the controls. We analyzed whether lentivirus-mediated shRNA interference of CENPK was involved in cell proliferation and apoptosis. The results indicated that interference with the CENPK gene inhibited RKO cell proliferation. Taken together, these findings support the hypothesis that lentivirus-mediated shRNA suppresses CENPK expression via lentivirus epigenetically. Overexpression of CENPK has been observed in various tumor types[8,18,19].
CUL4A plays a critical role in cellular proliferation and tumor invasion[20,21]. In vitro experiments were performed to verify the function of the CUL4A gene. To investigate whether CUL4A gene was involved in CENPK-induced CRC cell proliferation, we performed rescue experiments, which indicated that knockdown of CENPK and overexpression of CUL4A partially rescued proliferation in RKO and HCT116 cells. These findings support the hypothesis that interference with the CENPK gene inhibits RKO and HCT116 cell proliferation. Overexpression of CUL4A had a response effect on RKO and HCT 116 cells.
The functional effects of CUL4A proteins on their targets have been well characterized and are involved in the proliferation of cancer cells[22]. CUL4A may be a prognostic marker of precancerous lesions and a potential therapeutic target in cancer[23]. Consistent with these reports, overexpression of CUL4A was observed in 20 pairs of CRC tissues, and knockdown of CUL4A inhibited cell proliferation. A previous study showed that CUL4A was highly expressed in CRC and promoted proliferation and inhibited apoptosis of CRC cells by regulating the Hippo pathway[10]. Our study demonstrated that restoration of CUL4A expression suppressed CRC cell proliferation. These results indicate that CUL4A acts as a tumor suppressor in CRC. It has been found that high CUL4A protein expression is an independent prognostic marker in CRC[12].
Our study demonstrated that overexpression of CUL4A led to CRC tumorigenesis, and knockdown of the CENPK gene affected CRC progression and development in target cells. To the best of our knowledge, this study is the first to investigate CUL4A protein expression and its potential interference with CENPK gene interactions in CRC. We demonstrated the critical role of the CUL4A gene in the progression of CRC. However, due to the complexity of shRNA biological function, further analysis is required to achieve a more comprehensive understanding of CRC diagnosis and therapy.
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide. Immunohistochemistry has found high expression of centromere protein (CENPK) in CRC. However, the role of CENPK in the progression of CRC is not well characterized.
To explore the role of Cullin (CUL)4A expression and lentivirus-mediated transfection with short hairpin RNA (shRNA) for CENPK in CRC.
We performed a series of in vitro experiments, such as quantitative polymerase chain reaction (qPCR), western blot, MTT assay, and flow cytometry, to evaluate the knockdown behavior of CENPK and overexpression of CUL4A in RKO and HCT 116 CRC cells.
We identified CENPK as a potential new oncogene for CRC based on bioinformatics analysis. In vitro experiments verified the function of this gene. We investigated the expression of CENPK in RKO and HCT 116 cells using virus infection and analyzed datasets from qPCR, western blot, and flow cytometry. The effect of RKO cells infected with virus on tumor growth was evaluated in vivo using quantitative analysis of fluorescence imaging.
The downstream genes FBX32, CUL4A, and YAP1 were examined to evaluate the regulatory action of CENPK in RKO cells. Significantly delayed xenograft emergence, slower growth, and lower final tumor weight and volume were observed in the RKO and HCT 116 with lentivirus-mediated shRNA interference of CENPK. Interference of CENPK inhibited the proliferation rate of RKO cells in vitro and in vivo. The shRNA interference of CENPK inhibited the proliferation of RKO and HCT 116 cells, and overexpression of CUL4A gene responded to RKO and HCT 116 cells with CENPK silencing.
Our findings indicate a potential role of CENPK in promoting tumor proliferation, and it may serve as a novel diagnostic and prognostic biomarker in patients with CRC.
An investigation for lentivirus-mediated transfection with shRNA for CENPK and overexpression of CUL4A demonstrated major regulatory roles in CRC. Further analysis is required to achieve a more comprehensive understanding of CRC diagnosis and therapy.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Fouad MA, Egypt; Mohamed SY, Egypt S-Editor: Wang JJ L-Editor: Wang TQ P-Editor: Wang JJ
| 1. | Matsushita N, Matsushita S, Hirakawa S, Higashiyama S. Doxycycline-dependent inducible and reversible RNA interference mediated by a single lentivirus vector. Biosci Biotechnol Biochem. 2013;77:776-781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 2. | Uppada SB, Gowrikumar S, Ahmad R, Kumar B, Szeglin B, Chen X, Smith JJ, Batra SK, Singh AB, Dhawan P. MASTL induces Colon Cancer progression and Chemoresistance by promoting Wnt/β-catenin signaling. Mol Cancer. 2018;17:111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 66] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 3. | Li Y, Yang Z, Wu N, Xu L and Leng Z. Effects and Mechanism of shRNA Interfering with KLF4 Expression on the Migration and Invasion of Lgr5 + Colorectal Cancer Stem Cells. Med J Wuhan University. 2018;39:899-904. [DOI] [Full Text] |
| 4. | Sacca R, Engle SJ, Qin W, Stock JL, McNeish JD. Genetically engineered mouse models in drug discovery research. Methods Mol Biol. 2010;602:37-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 5. | Zhao B, Yang C, Yang S, Gao Y, Wang J. Construction of conditional lentivirus-mediated shRNA vector targeting the human Mirk gene and identification of RNAi efficiency in rhabdomyosarcoma RD cells. Int J Oncol. 2013;43:1253-1259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 6. | Wang J, Li H, Xia C, Yang X, Dai B, Tao K, Dou K. Downregulation of CENPK suppresses hepatocellular carcinoma malignant progression through regulating YAP1. Onco Targets Ther. 2019;12:869-882. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 7. | Li Q, Liang J, Zhang S, An N, Xu L, Ye C. Overexpression of centromere protein K (CENPK) gene in Differentiated Thyroid Carcinoma promote cell Proliferation and Migration. Bioengineered. 2021;12:1299-1310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 8. | Lee YC, Huang CC, Lin DY, Chang WC, Lee KH. Overexpression of centromere protein K (CENPK) in ovarian cancer is correlated with poor patient survival and associated with predictive and prognostic relevance. PeerJ. 2015;3:e1386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 9. | Jia B, Dao J, Han J, Huang Z, Sun X, Zheng X, Xiang S, Zhou H, Liu S. LINC00958 promotes the proliferation of TSCC via miR-211-5p/CENPK axis and activating the JAK/STAT3 signaling pathway. Cancer Cell Int. 2021;21:147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 10. | Yi LJ, Yi LJ, Ding N, Ren J. CUL4A promotes proliferation and inhibits apoptosis of colon cancer cells via regulating Hippo pathway. Eur Rev Med Pharmacol Sci. 2020;24:10518-10525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 11. | Sui X, Zhou H, Zhu L, Wang D, Fan S, Zhao W. CUL4A promotes proliferation and metastasis of colorectal cancer cells by regulating H3K4 trimethylation in epithelial-mesenchymal transition. Onco Targets Ther. 2017;10:735-743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 12. | Li C, Bu J, Liao Y, Zhang J, Han J, Zhang H, Xing H, Li Z, Wu H, Liang L, Wang M, Qin W, Yang T. High Expressions of CUL4A and TP53 in Colorectal Cancer Predict Poor Survival. Cell Physiol Biochem. 2018;51:2829-2842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | Wu XL, Yang ZW, He L, Dong PD, Hou MX, Meng XK, Zhao HP, Wang ZY, Wang F, Baoluri, Wurenqimuge, Agudamu, Jia YF, Shi L. RRS1 silencing suppresses colorectal cancer cell proliferation and tumorigenesis by inhibiting G2/M progression and angiogenesis. Oncotarget. 2017;8:82968-82980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 14. | Wang Y, Wang Y, Ren C, Wang H, Zhang Y, Xiu Y. Upregulation of centromere protein K is crucial for lung adenocarcinoma cell viability and invasion. Adv Clin Exp Med. 2021;30:691-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 15. | Ou C, Sun Z, Li X, Ren W, Qin Z, Zhang X, Yuan W, Wang J, Yu W, Zhang S, Peng Q, Yan Q, Xiong W, Li G, Ma J. Corrigendum to "MiR-590-5p, a density-sensitive microRNA, inhibits tumorigenesis by targeting YAP1 in colorectal cancer", [Canc. Lett. 399 (2017) 53-63]. Cancer Lett. 2018;420:260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 16. | Zhao YP, Han ZW, Xie Y. Expression and significance of YAP1 in colorectal cancer tissues. Chin J Curr Advan Gene Sur. 2019;. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 17. | Belghit I, Skiba-Cassy S, Geurden I, Dias K, Surget A, Kaushik S, Panserat S, Seiliez I. Dietary methionine availability affects the main factors involved in muscle protein turnover in rainbow trout (Oncorhynchus mykiss). Br J Nutr. 2014;112:493-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 18. | Huang H, Cheng S, Ding M, Wen Y, Ma M, Zhang L, Li P, Cheng B, Liang X, Liu L, Du Y, Zhao Y, Kafle OP, Han B, Zhang F. Integrative analysis of transcriptome-wide association study and mRNA expression profiles identifies candidate genes associated with autism spectrum disorders. Autism Res. 2019;12:33-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 19. | Parolin C, Corso AD, Alberghina L, Porro D, Branduardi P. Heterologous production of five Hepatitis C virus-derived antigens in three Saccharomyces cerevisiae host strains. J Biotechnol. 2005;120:46-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 20. | Wang Y, Wen M, Kwon Y, Xu Y, Liu Y, Zhang P, He X, Wang Q, Huang Y, Jen KY, LaBarge MA, You L, Kogan SC, Gray JW, Mao JH, Wei G. CUL4A induces epithelial-mesenchymal transition and promotes cancer metastasis by regulating ZEB1 expression. Cancer Res. 2014;74:520-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 175] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 21. | Pan Y, Wang B, Yang X, Bai F, Xu Q, Li X, Gao L, Ma C, Liang X. CUL4A facilitates hepatocarcinogenesis by promoting cell cycle progression and epithelial-mesenchymal transition. Sci Rep. 2015;5:17006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 22. | Ashok C, Owais S, Srijyothi L, Selvam M, Ponne S, Baluchamy S. A feedback regulation of CREB activation through the CUL4A and ERK signaling. Med Oncol. 2019;36:20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 23. | Feng M, Wang Y, Bi L, Zhang P, Wang H, Zhao Z, Mao JH, Wei G. CRL4ADTL degrades DNA-PKcs to modulate NHEJ repair and induce genomic instability and subsequent malignant transformation. Oncogene. 2021;40:2096-2111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |