Published online Sep 21, 2022. doi: 10.3748/wjg.v28.i35.5188
Peer-review started: April 12, 2022
First decision: July 13, 2022
Revised: July 26, 2022
Accepted: August 22, 2022
Article in press: August 22, 2022
Published online: September 21, 2022
Processing time: 151 Days and 7.2 Hours
The microbes and metabolomics of microbiota dysbiosis in the gut in the different phases of hepatitis B virus (HBV) infection are not fully understood.
To investigate the specific gut microbiota and metabolites of the immune-tolerant (IT) and immune-active (IA) phases of chronic hepatitis B (CHB).
Clinical fecal samples from healthy individuals and patients in the IT and IA phases of HBV infection were collected. Next, non-target metabolomics, bioinfor
A total of 293 different metabolites in 14 phyla, 22 classes, 29 orders, 51 families, and 190 genera were identified. The four phyla of Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria were the most abundant, accounting for 99.72%, 99.79%, and 99.55% in the healthy controls, IT-phase patients, and IA-phase patients, respectively. We further identified 16 genera with different richness in the IT phase and IA phase of HBV infection. Of the 134 named metabolites, 57 were upregulated and 77 were downregulated. A total of 101 different metabolic functions were predicted in this study, with 6 metabolic pathways having the highest enrichments, namely carbohydrate metabolism (14.85%), amino acid metabolism (12.87%), lipid metabolism (11.88%), metabolism of cofactors and vitamins (11.88%), xenobiotic biodegradation (9.9%), and metabolism of terpenoids and polyketides (7.92%).
These findings provide observational evidence of compositional alterations of the gut microbiome and some related metabolites in patients with IT-phase or IA-phase HBV infection. Further studies should investigate whether microbiota modulation can facilitate the progression of CHB and the cause-effect relationship between the gut microbiota and CHB.
Core Tip: This article provided observational evidence of compositional alterations of gut microbiome and some related metabolite in patients with immune-tolerant phase hepatitis B virus (HBV) infection and immune-active phase HBV infection.
- Citation: Li YN, Kang NL, Jiang JJ, Zhu YY, Liu YR, Zeng DW, Wang F. Gut microbiota of hepatitis B virus-infected patients in the immune-tolerant and immune-active phases and their implications in metabolite changes. World J Gastroenterol 2022; 28(35): 5188-5202
- URL: https://www.wjgnet.com/1007-9327/full/v28/i35/5188.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i35.5188
As a disease of public health significance, hepatitis B often leads to liver cirrhosis or hepatocellular carcinoma (HCC). Currently, there are few effective treatments and strategies for eliminating hepatitis B virus (HBV) infection. The immune-tolerant (IT) phase is the first stage of HBV infection, and recent studies suggest that HBV-infected patients in this phase suffer from a high risk of HCC and death[1-3]. However, anti-HBV treatment is not generally recommended at this stage. The IT status of patients largely determines the patients’ outcomes. Thus, the identification of factors that affect the consequences of HBV infection is crucial for eliminating this disease.
Recent studies indicate that the gut microbiota is involved in chronic hepatitis B (CHB) as well as other liver diseases[4-6]. Some commensal bacteria regulate the host metabolic pathways by improving food-derived energy or modulating the host-derived compounds that alter metabolism[7-9]. In addition, there are various gut microbiota in patients with CHB, hepatitis B-related liver cirrhosis, or HCC[10-12]. To date, information about the gut microbiota in patients at the initial phase of HBV infection remains largely unknown. We believe that studies on the gut microbiota of HBV-infected patients in the IT phase would help to elucidate the underlying mechanism of HBV IT as it will provide valuable information about the immune environment for HBV and its long-term existence.
Most studies on the microbiome have been cross-sectional with samples collected at a single time point, and only some studies have been performed with samples from different stages of HBV infection[12-15]; moreover, few comparisons of HBV-infected patients and healthy individuals have been carried out in these studies. CHB patients may develop recurrent active hepatitis, while the IT phase may change into the immune-active (IA) phase at a later stage in patients[16], which can lead to a substantial change in the composition of the gut microbiota. Therefore, understanding how the gut microbiota change from the IT phase to the IA phase will be important for the development of potential microbiome-targeting therapeutic drugs to combat HBV infection. Furthermore, this dynamic variation in gut microbiota in HBV-infected patients may help us to identify unique bacterial taxa that would contribute to postponing disease progression.
The present study focused on the dynamic profiles of the gut microbiota in IT-phase patients and IA-phase HBV-infected patients without liver fibrosis by 16S rDNA sequencing and analysis. We aimed to identify changes of bacteria involved in the transition from the IT phase to the IA phase in HBV-infected patients and to illustrate the regulation of both microbiota and metabolites. The results will provide a new perspective for the noninvasive diagnosis and treatment of IT-phase HBV-infected patients.
HBV-infected patients in the IT phase or the IA phase were recruited from The First Affiliated Hospital of Fujian Medical University from January 2018 to December 2020. All HBV-infected patients were at least 18-years-old, hepatitis B surface antigen positive for ≥ 6 mo, and hepatitis B virus e-antigen positive; moreover, the IT patients had a normal alanine aminotransferase level (< 40 IU/L), HBV DNA > 1 million IU/mL, and no fibrosis found by FibroScan analysis[17]. The IA patients had an HBV DNA serum concentration of > 20000 IU/mL as well as elevated alanine aminotransferase and/or aspartate aminotransferase levels[17]. However, patients who suffered from other non-HBV diseases/infections were excluded from the study. Samples from healthy volunteers were blindly collected, and participants who had received antiviral therapy, immunotherapy, probiotics, or antibiotics within 8 wk before enrollment were excluded. Verbal informed consent was provided by all participants. The experimental protocol and participant enrollment procedure were approved by the Ethics Review Committee of the university and were conducted according to the Declaration of Helsinki guidelines.
Liver stiffness measurements were conducted using a FibroScan instrument. Measurements with more than ten successful acquisitions were obtained (with a rate > 60% and an interquartile range < 30%). In addition, the alanine aminotransferase, aspartate aminotransferase, glutamyl transpeptidase, total bilirubin, albumin, globulin, and alpha-fetoprotein levels were measured. HBV serological testing was performed with the Architect platform (Abbott Laboratories, Chicago, IL, United States). HBV DNA was tested by a quantitative PCR assay (PG Company, Shenzhen, China).
Fecal samples were obtained from all participants, filtered with a 2-mm sieve to remove interferents, and then stored at -80 °C for the following experiments. Total bacterial DNA extraction was performed with a PowerSoil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, United States), and the purity and quality of the genomic DNA were checked by electrophoresis on 0.8% agarose gels. The V3-4 hypervariable region of bacterial 16S rDNA was amplified by previously reported conserved primers: 338F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’)[18]. For each sample, the 10-digit barcode sequence was added to the 5’-end of the forward and reverse primers (provided by Allwegene Company, Beijing, China). The PCR was carried out on a Mastercycler Gradient (Eppendorf, Germany) using a reaction volume of 25 μL, containing 12.5 μL of KAPA 2G Robust Hot Start Ready Mix, 1 µL of forward primer (5 µmol/L), 1 µL of reverse primer (5 µmol/L), 5 µL of DNA (total template quantity of 30 ng), and 5.5 µL of H2O. The cycling parameters were 95 °C for 5 min, followed by 28 cycles of 95 °C for 45 s, 55 °C for 50 s, and 72 °C for 45 s, with a final extension at 72 °C for 10 min.
Three PCR products per sample were pooled to mitigate reaction-level PCR biases. The PCR products were purified using a QIAquick Gel Extraction Kit (QIAGEN, Germany), quantified using real-time PCR, and sequenced at Allwegene Company, Beijing, China. Deep sequencing was done with the Miseq platform at the Allwegene Company (Beijing, China). The raw data were first screened, and sequences were removed from consideration if they were shorter than 200 bp, had a low quality score (≤ 20), contained ambiguous bases, or did not exactly match the primer sequences and barcode tags. Qualified reads were separated using the sample-specific barcode sequences and trimmed with Illumina Analysis Pipeline Version 2.6. Thereafter, the dataset after cleaning was analyzed using the Quantitative Insights into Microbial Ecology (QIIME) platform.
All sequences were clustered into operational taxonomic units (OTUs) at a similarity level of 97%[19] to generate rarefaction curves. The Ribosomal Database Project classifier tool was used to analyze different taxonomic groups[20]. The Venn diagram was built by the R package program. Shared taxa presented in all groups were defined as the core microbiota. Clustering analysis and principal component analysis were performed by using the R package, as described previously[21]. The evolution distances were analyzed with the unweighted pair group method and an arithmetic mean clustering tree[22]. Heatmaps of the top 20 OTUs were generated using Mothur, as described in a previous study[23].
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) analysis based on the 16S rDNA was performed. The OTU table derived from QIIME was compared using the Kyoto Encyclopedia of Genes and Genomes and MetaboAnalyst (http://www.metaboanalyst.ca/) databases, and the metabolic function of the gut microbiota was predicted based on the findings. The abundances of functional genes were visualized as heatmaps by the R package.
For metabolite extraction, a 10-μL aliquot of each sample was mixed with 990 μL of the extraction solvent (acetonitrile/methanol/water, 2:2:1), and the mixture was vortexed for 30 s, incubated at -20 °C for 1 h, and then centrifuged at 12000 rpm and 4 °C for 15 min. Finally, the supernatant was diluted 10 times for ultra-high-performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS) analysis[24]. UHPLC separation was performed on an Agilent 1290 Infinity II series UHPLC system (Agilent Technologies). Mobile phase A included both 10 mmol/L ammonium formate and 10 mmol/L ammonia, while mobile phase B was acetonitrile. The temperatures for the column and the autosampler were set at 35 °C and 4 °C, respectively[25].
One-way analysis of variance was performed to compare continuous variables between two groups. The Wilcoxon signed rank test, Kruskal–Wallis test, χ2 test, or Student’s t test was used to compare categorical variables between groups. All statistical analyses were calculated either in the R package (version 3.6.1) or SPSS (version 26.0). P values less than 0.05 were considered as significant differences.
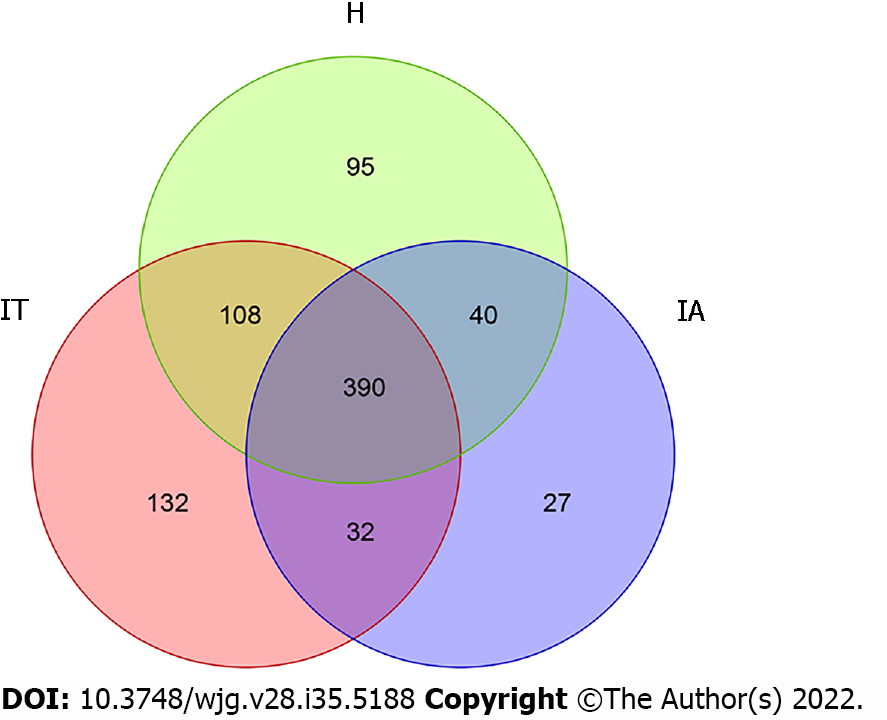
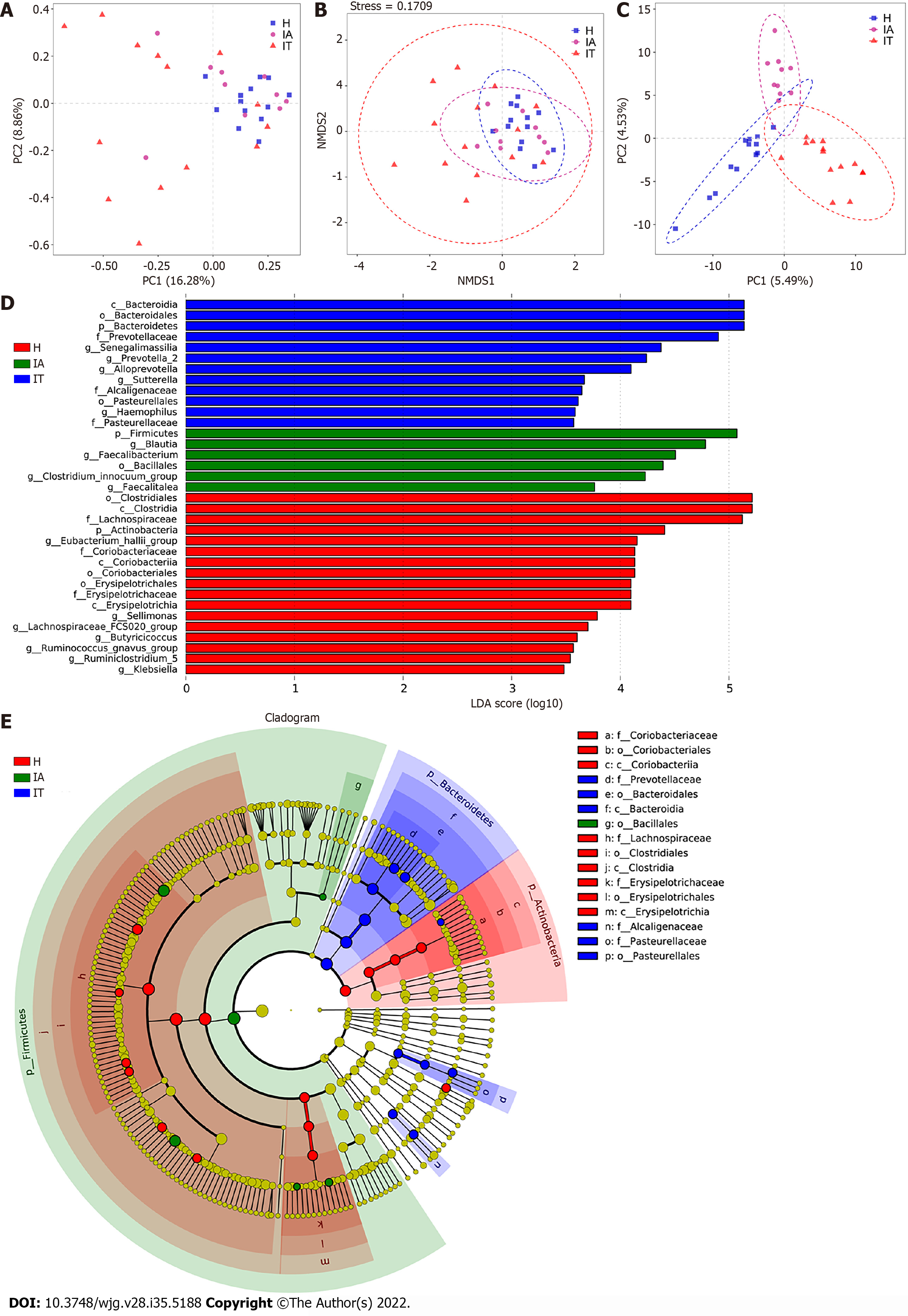
To differentiate the characteristics of the gut microbiota in IT-phase HBV-infected patients, IA-phase HBV-infected patients, and healthy individuals, 16S rDNA gene sequencing was performed on the stool samples from 14 IT-phase HBV-infected patients, 10 IA-phase HBV-infected patients, and 13 healthy donors (Table 1). A total of 1521813, 411189, and 1109236 sequences were acquired for the IT, IA, and healthy groups, respectively, after excluding low-quality reads. A total of 19556 clean tags were obtained, of which 824 OTUs were matched. After applying strict trimming criteria to exclude low-quality clean tags, the numbers of OTUs for healthy individuals, IT-phase patients, and IA-phase patients were 633, 662, and 489, respectively, as shown in Table 2. A total of 95, 132, and 27 OTUs existed independently in the healthy controls, IT-phase patients, and IA-phase patients, respectively (Figure 1). There were no significant differences in the Chao1, Shannon, and Simpson indices among the IT, IA, and healthy groups (all P values > 0.05, Table 2). The bacterial communities in the healthy individuals were relatively more heterogeneous than those found in the IT- or IA-phase patients. The IT and IA patients could be separated from the healthy individuals by non-metric multidimensional scaling and principal component analysis (Figure 2A and B). Partial least squares discriminant analysis showed structural differences in the gut bacterial community structure among the groups, indicating that the classification model was effective (Figure 2C).
| Characteristic | H, n = 13 | IT, n = 14 | IA, n = 10 | P value |
| Sex, M/F | 9/4 | 9/5 | 7/3 | 0.949 |
| Age in yr | 26.4 ± 0.8 | 26.9 ± 5.8 | 36.2 ± 11.3 | 0.004 |
| HBsAg as log10 IU/mL | NA | 4.62 ± 0.18 | 3.83 ± 0.81 | < 0.001 |
| HBeAg as log10 S/Co | NA | 3.18 ± 0.08 | 1.21 ± 1.70 | < 0.001 |
| HBV DNA as log10 IU/mL | NA | 7.81 ± 0.80 | 6.28 ± 1.70 | < 0.001 |
| AFP in ng/mL | 2.06 ± 0.82 | 2.49 ± 1.05 | 22.61 ± 40.82 | 0.045 |
| TBIL in μmol/L | 5.9 ± 3.1 | 12.7 ± 5.3 | 19.5 ± 9.2 | < 0.001 |
| ALB in g/L | 48.8 ± 3.5 | 45.8 ± 2.9 | 43.0 ± 4.9 | 0.002 |
| GLO in g/L | 24.4 ± 4.0 | 26.6 ± 4.4 | 29.3 ± 4.3 | 0.017 |
| ALT in U/L | 22.0 (18.0, 29.0) | 29.5 (23.7, 33.0) | 277.5 (101.5, 410.8) | < 0.001 |
| AST in U/L | 16.0 (12.0, 22.0) | 23.0 (19.5, 27.8) | 111.0 (67.0, 143.5) | 0.006 |
| GGT in U/L | 25.7 ± 11.5 | 18.5 ± 9.4 | 72.7 ± 64.9 | 0.001 |
| TBA in μmol/L | 2.3 (1.9, 3.4) | 5.6 (4.2, 8.4) | 13.9 (5.5, 31.1) | 0.012 |
| LSM in kPa | 4.99 ± 0.99 | 5.11 ± 1.01 | 6.38 ± 0.72 | 0.026 |
| Group | Healthy controls, n = 14 | IT phase HBV infection, n = 13 | IA phase HBV infection, n = 10 | P value |
| OTUs | 633 | 662 | 489 | |
| α-diversity indexes, median | ||||
| Chao1 | 264.310 | 233.825 | 227.000 | 0.924 |
| Shannon | 5.020 | 4.720 | 4.705 | 1.739 |
| Simpson | 0.940 | 0.885 | 0.915 | 1.666 |
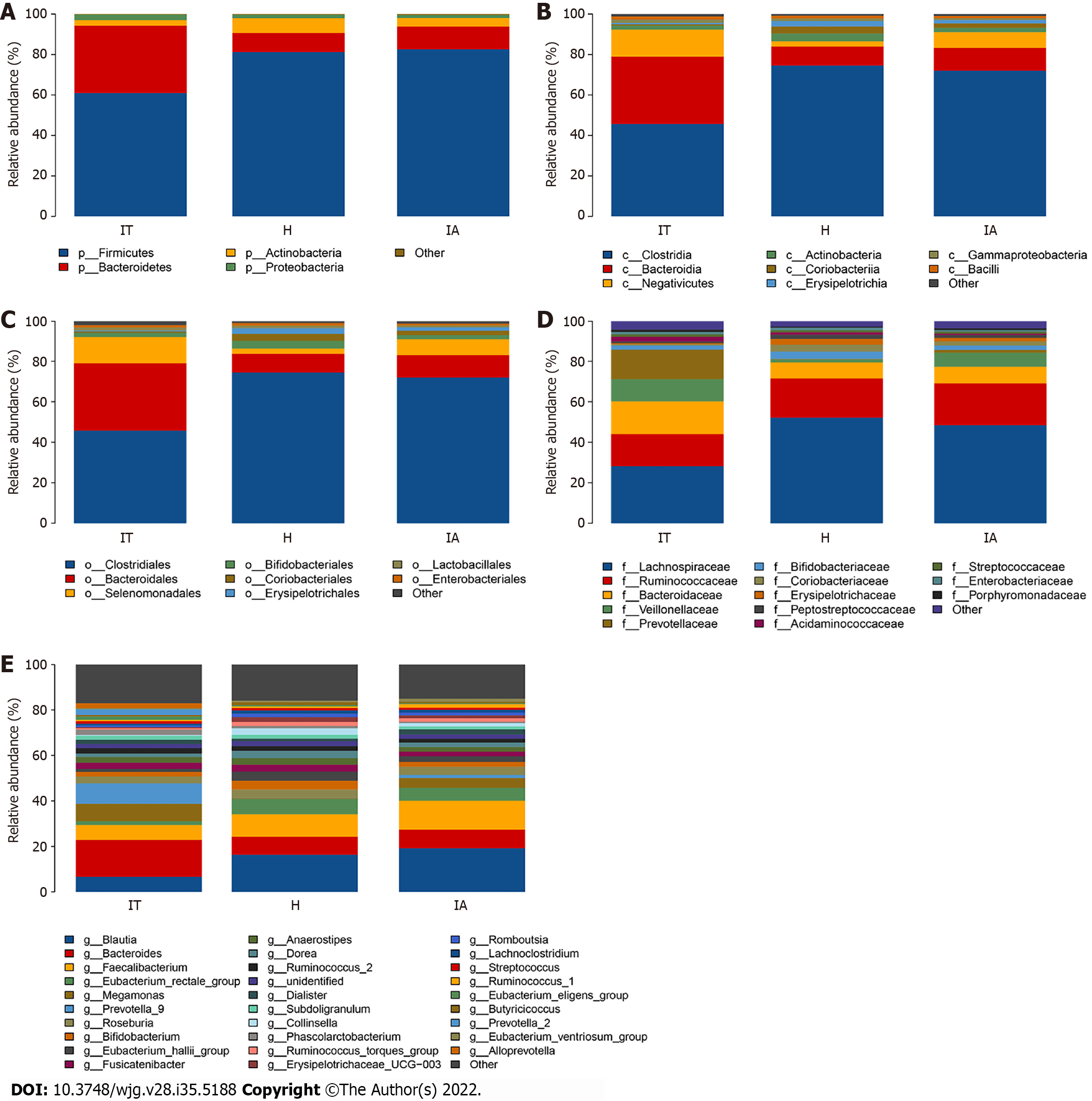
To show the abundance of bacteria of all fecal samples, 14 phyla, 22 classes, 29 orders, 51 families, and 190 genera were identified, and the dominant gut microbiota are shown in Supplementary Table 1 and Figure 3. The four phyla of Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria were the most abundant, accounting for 99.72%, 99.79%, and 99.55% in the healthy controls, IT-phase patients, and IA-phase patients, respectively. In detail, the highest relative abundance of Actinobacteria, which reached 7.32%, was found in the healthy group. Meanwhile, compared with the healthy control and the IA-phase patients, the IT patients had the highest abundance of Bacteroidetes (33.27%). The IT-phase patients had less Firmicutes (61.00%) and Actinobacteria (2.71%) than the IA-phase patients. Clostridiales, Bacteroidales, Selenomonadales, Bifidobacteriales, Coriobacteriales, Erysipelotrichales, Lactobacillales, and Enterobacteriales were the most abundant bacteria at the order level in all three groups.
For the genus level, an abundance of Bacteroides (16.19%), Prevotella 9 (9.04%), and Megamonas (7.66%) was found in the IT-phase patients. However, Blautia (19.20%) and Faecalibacterium (12.68%) were enriched in the IA-phase patients. Interestingly, the healthy groups had more Eubacterium rectale (6.69%), Eubacterium hallii (4.03%), Bifidobacterium (3.87%), and Dorea (3.23%). In addition, Faecalibacterium and Blautia accounted for a high proportion in the IA-phase patients; these bacteria are involved in butyrate short-chain fatty acid metabolism and inhibit inflammation.
Linear discriminant analysis (LDA) effect size modeling was applied to identify specific bacterial taxa associated with different stages of CHB (Figure 2D and E). There were markedly significant differences in the community compositions in CHB patients compared with the healthy individuals. There were 12 and 6 significantly different taxa in the IT- and IA-phase patients, respectively. The five most enriched genera in the IT-phase patients were Senegalimassilia (LDA score = 4.38, P < 0.05), Prevotella 2 (LDA score = 4.24, P < 0.01), Alloprevotella (LDA score = 4.09, P < 0.05), Sutterella (LDA score = 3.67, P < 0.001), and Haemophilus (LDA score = 3.58, P < 0.05). The four most enriched genera in the IA-phase patients included Blautia (LDA score = 4.78, P < 0.01), Faecalibacterium (LDA score = 4.51, P < 0.05), Clostridium innocuum group (LDA score = 4.23, P < 0.01), and Faecalitalea (LDA score = 3.76, P < 0.05) (Supple
Furthermore, the Wilcoxon and Kruskal-Wallis rank sum tests showed the abundance at different taxonomic levels with P < 0.05 and FDR q < 0.1 (Supplementary Table 3). The top 20 gut microbiota at the genus level consisted of Bacteroides, Prevotella 9, Megamonas, Blautia, Faecalibacterium, Roseburia, Fusicatenibacter, Anaerostipes, Prevotella 2, Ruminococcus 2, Phascolarctobacterium, Alloprevotella, Bifidobacterium, Subdoligranulum, Dialister, Eubacterium rectale, Eubacterium eligens, Dorea, Eubacterium hallii, and Streptococcus. Moreover, the shared species between the significantly different gut microbiota and the top 20 gut microbiota were Blautia, Faecalibacterium, Fusicatenibacter, Prevotella 2, Alloprevotella, and Eubacterium hallii group, which might be highly associated with the outcomes of CHB in different phases. The differences found in this study revealed the dysbiosis involved in the development of CHB and the aberrant ecological networks of microbial communities during infection.
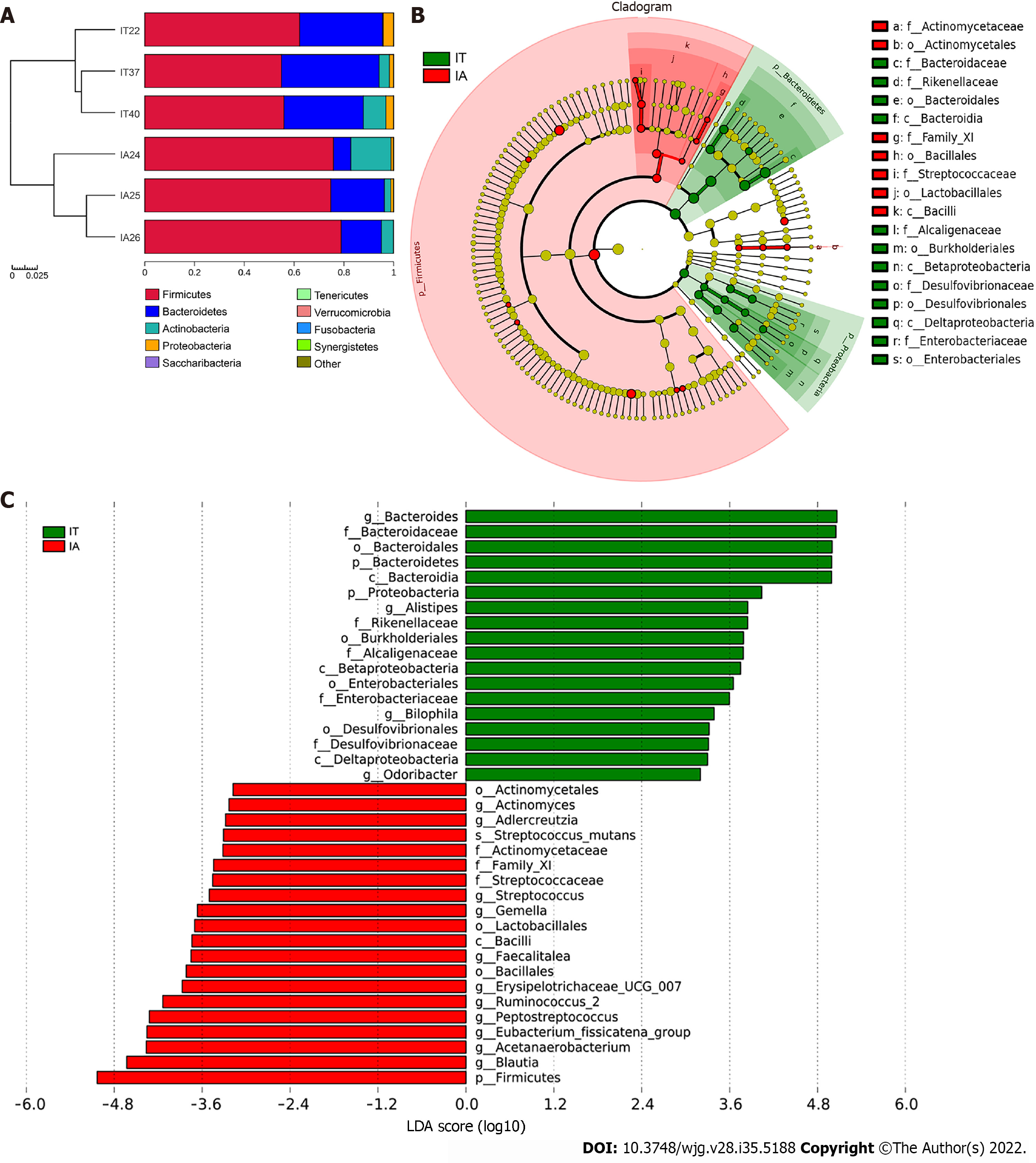
Longitudinal follow-up analysis showed that 3 patients progressed from the IT phase to the IA phase and required medical treatments. The change in the microbiota of these 3 patients during treatment was studied. The abundance of Firmicutes and Actinobacteria increased, while the abundance of Bacteroidetes and Proteobacteria decreased at the phylum level (Figure 4A). Moreover, Bacteroides, Alistipes, and Bilophila were mostly abundant in the IT phase, while Actinomyces, Adlercreutzia, and Streptococcus were more abundant in the IA phase (Figure 4B and C). A decreased ratio of Bacteroidetes to Firmicutes was observed, which might be involved in inflammatory disorders in the IA phase.
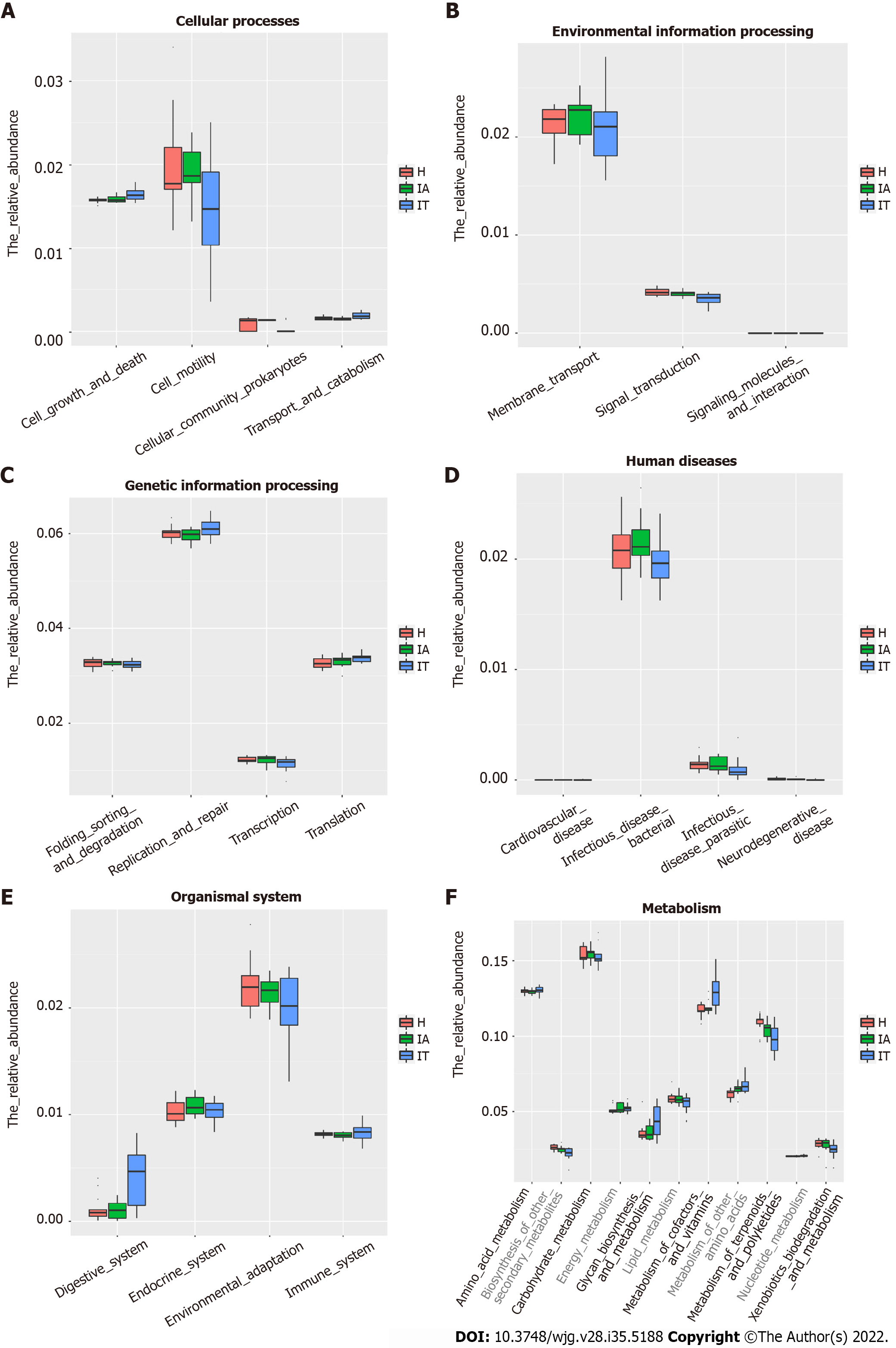
To identify the bacterial functional alteration in IT and IA patients, we analyzed the functional potential of gut microbiota with PICRUSt analysis. A total of 101 different metabolic functions were predicted in this study, with 6 metabolic pathways having the highest enrichments, namely carbohydrate metabolism (14.85%), amino acid metabolism (12.87%), lipid metabolism (11.88%), metabolism of cofactors and vitamins (11.88%), xenobiotic biodegradation (9.90%), and metabolism of terpenoids and polyketides (7.92%). In addition, the functional roles of the bacteria were highly related to infectious diseases, including bacterial infection (50.00%) and parasitic infection (33.33%) as well as cardiovascular disease (10.00%) and neurodegenerative disease (10.00%) (Supplementary Table 4). The parasitic infectious disease-related gene subgroups (P = 0.03) in the IA-phase patients were highly enriched (Figure 5). The cell motility-related genes were highly enriched in the IA-phase patients (Figure 5A), while the transport and catabolism-related genes (P = 0.03) were significantly enriched in the IT-phase patients. However, signal transduction-related genes were significantly enriched in the healthy controls (P < 0.01) (Figure 5B). The replication repair-related genes and digestive system-related genes in the IT-phase patients were more abundant than in the other groups, but the difference between the three groups was not statistically significant (Figure 5C and E). Moreover, the digestive system disease-related gene subset (P = 0.02) was highly enriched in the IT-phase patients (Figure 5D). Genes that are related to the amino acid metabolism glycosyl biosynthesis cofactors and vitamin metabolism were more enriched in the IT-phase patients than in the healthy controls (P < 0.01) (Figure 5F). These observed results suggested that changes in the bacterial composition can significantly alter gene function, which may contribute to the development of CHB.
As shown in the co-occurrence network analysis using Cytoscape software (Supplementary Figure 1), five genera (Dorea, Bifidobacterium, Bacteroides, Blautia, and Romboutsia) were highly positively correlated. Dorea and Bifidobacterium exhibited the highest degree of linkage, while Dorea and Bacteroides had the least degree of linkage.
A total of 293 substantially different metabolites were identified, among which 134 metabolites had an MS2.name provided by the mass spectrometry qualitative matching analysis. A total of 57 metabolites were upregulated, while 77 were downregulated. The different metabolites are shown in Supple
To date, there are limited studies showing the role of dysbiosis of the gut microbiota in HBV-infected patients during the different immune phases of infection. This study revealed the profiles of the gut microbiota during HBV infection from the IT phase to the IA phase. The results suggested that the diversity, composition, and functionality of the microbiota changed from the IT phase to the IA phase and were related to the progression of CHB.
In our study, analysis of α-diversity using the Chao1, Shannon, and Simpson diversity indices showed no significant differences among the three groups. Nevertheless, one study has reported that the α-diversity is increased in cirrhosis patients compared to healthy controls and positively correlated to the Child-Pugh score[26]. In our study, a decreased ratio of Bacteroidetes to Firmicutes was observed as patients progressed from the IT phase to the IA phase, while another study has demonstrated that the ratio of Bacteroidetes to Firmicutes increased in patients with HBV-related cirrhosis compared to that of healthy individuals[27]. These results in different studies indicate that the results may vary greatly depending on the grouping. In addition, our results suggest that the composition of the gut microbiota had changed in the early stages of HBV infection. Furthermore, metabolic changes at different stages of HBV infection were also observed by PICRUSt analysis. Consistent with another study[27], these results suggest that changes in the composition of the gut microbiota can significantly alter gene function, which may have a potential role in HBV-infected patients.
When compared to the healthy controls, there was a correlation between some gut microbiota in patients at the IT and IA phases. Notably, the IT group presented a high relative abundance of Senegalimassilia, Prevotella 2, Alloprevotella, Sutterella, and Haemophilus; while the IA group showed a high relative abundance of Blautia, Faecalibacterium, Clostridium innocuum group, and Faecalitalea. Some Prevotella strains have been reported as potentially clinically important pathobionts in human diseases by increasing the levels of interleukin (IL)-23, IL-1, IL-8, IL-6, and C-C motif chemokine ligand 20[28]. In addition, Alloprevotella is enriched in fecal samples from patients with chronic kidney disease[29]. Moreover, the gut microbiota in prediabetic individuals was found to be aberrant, with decreased Clostridium and increased Sutterella levels[30]. Furthermore, Sutterella has the capacity to degrade IgA, causing failure of therapeutics treating ulcerative colitis[31]. These findings reveal that Prevotella 2, Alloprevotella, and Sutterella are involved in chronic human diseases. Our results also indicate that Prevotella 2, Alloprevotella, and Sutterella are specifically related to HBV infection in the IT phase, suggesting their role of mediating viral escape from the host immune response.
Blautia, Faecalibacterium, Clostridium innocuum group, and Faecalitalea were identified as signature gut microbiota related to HBV infection in the IA phase. Notably, an elevated abundance of Blautia has been shown to alleviate the severity of lethal acute graft-versus-host disease[32]. In addition, Benítez-Páez et al[33] proposed that Blautia luti and Blautia wexlerae might help to reduce the inflammation that is linked to obesity-related complications. Moreover, Blautia is positively associated with the pathophysiology of type 2 diabetes[34]. Furthermore, Faecalibacterium prausnitzii, belonging to Faecalibacterium, is a major constituent of the gut microbiota in healthy individuals. This bacterium, which has anti-inflammatory activity, is decreased in patients with inflammatory bowel diseases such as Crohn’s disease. Inte
Additionally, a recent finding indicated that in patients with early HBV-related HCC, genera belonging to butyrate-producing bacterial families, including Ruminococcus, Oscillibacter, Faecalibacterium, Clostridium IV, and Coprococcus decreased, while the lipopolysaccharide-producing bacteria Klebsiella and Haemophilus increased compared to the levels in cirrhosis patients[37]. Herein, the proportions of Blautia, Faecalibacterium, and Clostridium innocuum group were higher in the IA-phase patients than in the controls, indicating that enrichment of these three signature microbial taxa might be a sign of severe inflammation or exacerbation of disease. Notably, these beneficial gut microbiota might help to clear viral infection in the IA phase, while some gut microbiota in the IT phase might help the virus to escape from host immune responses. Taken together, these findings highlight an important role for the composition of the gut microbiota in the progression of HBV infection, which has significant clinical implications.
Based on the composition and structure of the gut microbiota, the metabolic function of the microflora was further analyzed through PICRUSt. In this study, the genes involved in glycan biosynthesis and metabolism as well as cofactor and vitamin metabolism were more enriched in the IT-phase patients than in the healthy controls. One explanation for this finding is that Senegalimassilia, Prevotella 2, Alloprevotella, Sutterella, and Haemophilus are enriched in the IT phase and consume glycan as their energy source; the genes found in these bacteria are related to cofactor and vitamin metabolism. In contrast, terpenoid and polyketide metabolism-related genes were more abundant in the IA phase, suggesting that Senegalimassilia, Prevotella 2, Alloprevotella, Sutterella, and Haemophilus participate in the metabolism of terpenoids and polyketides. These results may indicate that the gut microbiota and metabolites contribute to the abnormal metabolism status in CHB.
Previous research has shown that the gut microbiota composition is closely related to the occurrence of various chronic liver diseases[38-40]. Our current study demonstrated that potential links exist between certain bacteria and different pathological mechanisms during CHB progression from the IT phase to the IA phase.
Studies using mouse models have uncovered that the gut microbiota have a vital role in overcoming the IT phase of viral infection at different ages[41] and that gut microbiota lead to Kupffer cell-mediated T cell suppression, which is associated with HBV persistence[42]. In addition, fecal microbiota transplantation therapy has been shown to improve the clearance rate of HBV antigens in CHB patients after long-term therapy[43]. Our study demonstrated that the gut microbiota composition changed at different phases of HBV infection. Changes in the gut microbiota composition could be a biological factor for the progression of CHB. We believe that the gut microbiome of CHB patients may provide a useful prognostic marker for disease progression, outcome prediction, and treatment and that further microbiome-based study may provide new insights into the pathogenesis of CHB, the etiology of its progression, and novel therapeutic strategies such as the use of probiotics or fecal microbiota trans
Although this study uncovered some insightful findings, some limitations remain. First, all patients enrolled in this study were from different families. Having different genetic backgrounds and dietary habits might affect the gut microbiota composition. In addition, the healthy donors should have had a similar age to the patients; however, the controls were not age-matched to the patients in this study. Second, only 3 patients were followed up from the IT phase to the IA phase. A larger longitudinal sample size will strengthen the results. Third, a real-world cross-sectional study would be beneficial to understand the impact of the microbiota on the progression of liver disease. Fourth, the methods we used have some limitations. Although transcriptomics also contributes to gene expression, the findings could be further confirmed if we used metagenomic sequencing rather than 16S rDNA gene sequencing. Metagenomic sequencing not only sequences the 16S rDNA genes, but it also sequences the whole genome of each bacterium. Thus, only by doing metagenomic sequencing, one can get the complete picture. Fifth, the functional studies of the gut microbiota are based on statistical predictions; therefore, further studies using germ-free animal models and related functional studies are required to investigate the specific roles of the gut microbiota in CHB. Furthermore, the IT phase is an important phase that needs more attention.
These findings provide observational evidence of compositional alterations of the gut microbiome and some related metabolites in patients with IT-phase or IA-phase HBV infection. Further studies should investigate whether microbiota modulation can facilitate the progression of CHB and the cause-effect relationship between the gut microbiota and CHB.
Chronic hepatitis B virus (HBV) infection (CHB) represents a public health problem that may progress to cirrhosis and hepatocellular carcinoma (HCC). HBV-infected individuals in immune tolerance (IT) are generally not recommended for anti-HBV treatment, owing to the absence of curative treatment and the evidence of a low risk of progressive liver injury in the IT patients. However, recent studies indicated hepatocarcinogenesis may occur in IT-phase patients; the body’s IT status is a key factor affecting the outcome of the disease. Therefore, finding out the factors that affect the prognosis of HBV infection is of paramount significance for the rapid elimination of virus and the reduction of CHB.
The bacterial diversity level and composition varied between CHB, hepatitis B-related liver cirrhosis, and HCC. Gut microbiota of healthy controls is more consistent, whereas those of CHB, hepatitis B-related liver cirrhosis and HCC varied substantially. As the first phase of HBV progression, IT phase provides a favorable immune environment for HBV invasion and long-term existence, then transitions to immune clearance in the third decade. Therefore, in-depth study of the disease status and changes in the body during the IT phase is conducive to the development of new antiviral treatment methods to break the IT of the body and improve the antiviral efficacy and thus improve the long-term prognosis.
This study aimed to find some potential bacteria, linking different pathological mechanisms of IT phase HBV infection and some related metabolites to the IT phase of CHB infection.
Clinical fecal samples from healthy individuals and patients in the IT and IA phases of HBV infection were collected. Next, non-target metabolomics, bioinformatics, and 16S rDNA sequencing analyses were performed.
A total of 293 different metabolites in 14 phyla, 22 classes, 29 orders, 51 families, and 190 genera were identified. The four phyla of Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria were the most abundant, accounting for 99.72%, 99.79%, and 99.55% in the healthy controls, IT-phase patients, and IA-phase patients, respectively. We further identified 16 genera with different richness in the IT phase and IA phase of HBV infection. Of the 134 named metabolites, 57 were upregulated and 77 were downregulated. A total of 101 different metabolic functions were predicted in this study, with 6 metabolic pathways having the highest enrichments, namely carbohydrate metabolism (14.85%), amino acid metabolism (12.87%), lipid metabolism (11.88%), metabolism of cofactors and vitamins (11.88%), xenobiotic biodegradation (9.9%), and metabolism of terpenoids and polyketides (7.92%).
The composition of the gut microbiota changed in the early stages of HBV infection, and changes in the composition of the gut microbiota can significantly alter gene function, which may have a potential role in HBV-infected patients.
It is relatively difficult to fully understand the causal relationship between gut microbiota and HBV-induced chronic liver disease at different stages in this real-world cross-sectional study. Nevertheless, it should be noted that germ-free animals are good models to study the effect of gut microbiota on human diseases. In a future study, it is imperative to use germ-free animal models and additional biofunctional assays to reveal the cause-effect relationship between gut microbiota and chronic HBV infection.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Infectious diseases
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Fraga RS, Brazil; Nath L, India S-Editor: Zhang H L-Editor: Filipodia P-Editor: Li X
| 1. | Mason WS, Gill US, Litwin S, Zhou Y, Peri S, Pop O, Hong ML, Naik S, Quaglia A, Bertoletti A, Kennedy PT. HBV DNA Integration and Clonal Hepatocyte Expansion in Chronic Hepatitis B Patients Considered Immune Tolerant. Gastroenterology. 2016;151:986-998.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 318] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 2. | Kennedy PTF, Litwin S, Dolman GE, Bertoletti A, Mason WS. Immune Tolerant Chronic Hepatitis B: The Unrecognized Risks. Viruses. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 57] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 3. | Kim GA, Lim YS, Han S, Choi J, Shim JH, Kim KM, Lee HC, Lee YS. High risk of hepatocellular carcinoma and death in patients with immune-tolerant-phase chronic hepatitis B. Gut. 2018;67:945-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 192] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 4. | Xu D, Huang Y, Wang J. Gut microbiota modulate the immune effect against hepatitis B virus infection. Eur J Clin Microbiol Infect Dis. 2015;34:2139-2147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 5. | Wang B, Li L. Who determines the outcomes of HBV exposure? Trends Microbiol. 2015;23:328-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 6. | Maslennikov R, Ivashkin V, Efremova I, Poluektova E, Kudryavtseva A, Krasnov G. Gut dysbiosis and small intestinal bacterial overgrowth as independent forms of gut microbiota disorders in cirrhosis. World J Gastroenterol. 2022;28:1067-1077. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 18] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (2)] |
| 7. | Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2779] [Cited by in RCA: 3081] [Article Influence: 237.0] [Reference Citation Analysis (0)] |
| 8. | Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262-1267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2838] [Cited by in RCA: 3234] [Article Influence: 248.8] [Reference Citation Analysis (0)] |
| 9. | Hernández-Ceballos W, Cordova-Gallardo J, Mendez-Sanchez N. Gut Microbiota in Metabolic-associated Fatty Liver Disease and in Other Chronic Metabolic Diseases. J Clin Transl Hepatol. 2021;9:227-238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 10. | Liu Q, Li F, Zhuang Y, Xu J, Wang J, Mao X, Zhang Y, Liu X. Alteration in gut microbiota associated with hepatitis B and non-hepatitis virus related hepatocellular carcinoma. Gut Pathog. 2019;11:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 146] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 11. | Wang J, Wang Y, Zhang X, Liu J, Zhang Q, Zhao Y, Peng J, Feng Q, Dai J, Sun S, Zhao L, Zhang Y, Hu Y, Zhang M. Gut Microbial Dysbiosis Is Associated with Altered Hepatic Functions and Serum Metabolites in Chronic Hepatitis B Patients. Front Microbiol. 2017;8:2222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 141] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 12. | Zeng Y, Chen S, Fu Y, Wu W, Chen T, Chen J, Yang B, Ou Q. Gut microbiota dysbiosis in patients with hepatitis B virus-induced chronic liver disease covering chronic hepatitis, liver cirrhosis and hepatocellular carcinoma. J Viral Hepat. 2020;27:143-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 13. | Li R, Yi X, Yang J, Zhu Z, Wang Y, Liu X, Huang X, Wan Y, Fu X, Shu W, Zhang W, Wang Z. Gut Microbiome Signatures in the Progression of Hepatitis B Virus-Induced Liver Disease. Front Microbiol. 2022;13:916061. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 14. | Chen Z, Xie Y, Zhou F, Zhang B, Wu J, Yang L, Xu S, Stedtfeld R, Chen Q, Liu J, Zhang X, Xu H, Ren J. Featured Gut Microbiomes Associated With the Progression of Chronic Hepatitis B Disease. Front Microbiol. 2020;11:383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 15. | Yang XA, Lv F, Wang R, Chang Y, Zhao Y, Cui X, Li H, Yang S, Li S, Zhao X, Mo Z, Yang F. Potential role of intestinal microflora in disease progression among patients with different stages of Hepatitis B. Gut Pathog. 2020;12:50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 16. | Sarin SK, Kumar M, Lau GK, Abbas Z, Chan HL, Chen CJ, Chen DS, Chen HL, Chen PJ, Chien RN, Dokmeci AK, Gane E, Hou JL, Jafri W, Jia J, Kim JH, Lai CL, Lee HC, Lim SG, Liu CJ, Locarnini S, Al Mahtab M, Mohamed R, Omata M, Park J, Piratvisuth T, Sharma BC, Sollano J, Wang FS, Wei L, Yuen MF, Zheng SS, Kao JH. Asian-Pacific clinical practice guidelines on the management of hepatitis B: a 2015 update. Hepatol Int. 2016;10:1-98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1985] [Cited by in RCA: 1959] [Article Influence: 217.7] [Reference Citation Analysis (0)] |
| 17. | European Association for the Study of the Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67:370-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3745] [Cited by in RCA: 3800] [Article Influence: 475.0] [Reference Citation Analysis (1)] |
| 18. | Munyaka PM, Eissa N, Bernstein CN, Khafipour E, Ghia JE. Antepartum Antibiotic Treatment Increases Offspring Susceptibility to Experimental Colitis: A Role of the Gut Microbiota. PLoS One. 2015;10:e0142536. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 111] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 19. | Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996-998. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8951] [Cited by in RCA: 9972] [Article Influence: 831.0] [Reference Citation Analysis (0)] |
| 20. | Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141-D145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3794] [Cited by in RCA: 3483] [Article Influence: 217.7] [Reference Citation Analysis (0)] |
| 21. | Wang Y, Sheng HF, He Y, Wu JY, Jiang YX, Tam NF, Zhou HW. Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl Environ Microbiol. 2012;78:8264-8271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 515] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 22. | Jiang XT, Peng X, Deng GH, Sheng HF, Wang Y, Zhou HW, Tam NF. Illumina sequencing of 16S rRNA tag revealed spatial variations of bacterial communities in a mangrove wetland. Microb Ecol. 2013;66:96-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 256] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 23. | Jami E, Israel A, Kotser A, Mizrahi I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013;7:1069-1079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 511] [Cited by in RCA: 632] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 24. | Zhou B, Xiao JF, Tuli L, Ressom HW. LC-MS-based metabolomics. Mol Biosyst. 2012;8:470-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 388] [Cited by in RCA: 388] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 25. | Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN, Nicholls AW, Wilson ID, Kell DB, Goodacre R; Human Serum Metabolome (HUSERMET) Consortium. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6:1060-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2495] [Cited by in RCA: 2105] [Article Influence: 150.4] [Reference Citation Analysis (0)] |
| 26. | Sanduzzi Zamparelli M, Rocco A, Compare D, Nardone G. The gut microbiota: A new potential driving force in liver cirrhosis and hepatocellular carcinoma. United European Gastroenterol J. 2017;5:944-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 27. | Shu W, Shanjian C, Jinpiao L, Qishui O. Gut microbiota dysbiosis in patients with hepatitis B virus-related cirrhosis. Ann Hepatol. 2022;27:100676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 28. | Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151:363-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 472] [Cited by in RCA: 888] [Article Influence: 111.0] [Reference Citation Analysis (0)] |
| 29. | Li F, Wang M, Wang J, Li R, Zhang Y. Alterations to the Gut Microbiota and Their Correlation With Inflammatory Factors in Chronic Kidney Disease. Front Cell Infect Microbiol. 2019;9:206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 195] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 30. | Allin KH, Tremaroli V, Caesar R, Jensen BAH, Damgaard MTF, Bahl MI, Licht TR, Hansen TH, Nielsen T, Dantoft TM, Linneberg A, Jørgensen T, Vestergaard H, Kristiansen K, Franks PW; IMI-DIRECT consortium, Hansen T, Bäckhed F, Pedersen O. Aberrant intestinal microbiota in individuals with prediabetes. Diabetologia. 2018;61:810-820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 316] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 31. | Kaakoush NO. Sutterella Species, IgA-degrading Bacteria in Ulcerative Colitis. Trends Microbiol. 2020;28:519-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 129] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 32. | Jenq RR, Taur Y, Devlin SM, Ponce DM, Goldberg JD, Ahr KF, Littmann ER, Ling L, Gobourne AC, Miller LC, Docampo MD, Peled JU, Arpaia N, Cross JR, Peets TK, Lumish MA, Shono Y, Dudakov JA, Poeck H, Hanash AM, Barker JN, Perales MA, Giralt SA, Pamer EG, van den Brink MR. Intestinal Blautia Is Associated with Reduced Death from Graft-versus-Host Disease. Biol Blood Marrow Transplant. 2015;21:1373-1383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 587] [Cited by in RCA: 572] [Article Influence: 57.2] [Reference Citation Analysis (0)] |
| 33. | Benítez-Páez A, Gómez Del Pugar EM, López-Almela I, Moya-Pérez Á, Codoñer-Franch P, Sanz Y. Depletion of Blautia Species in the Microbiota of Obese Children Relates to Intestinal Inflammation and Metabolic Phenotype Worsening. mSystems. 2020;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 221] [Article Influence: 44.2] [Reference Citation Analysis (0)] |
| 34. | Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, Shulzhenko N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020;51:102590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 501] [Cited by in RCA: 1075] [Article Influence: 215.0] [Reference Citation Analysis (0)] |
| 35. | Lopez-Siles M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 2017;11:841-852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 612] [Cited by in RCA: 549] [Article Influence: 68.6] [Reference Citation Analysis (0)] |
| 36. | Abaidullah M, Peng S, Kamran M, Song X, Yin Z. Current Findings on Gut Microbiota Mediated Immune Modulation against Viral Diseases in Chicken. Viruses. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 37. | Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H, Xie H, Chen X, Shao L, Zhang R, Xu S, Zhang H, Cui G, Sun R, Wen H, Lerut JP, Kan Q, Li L, Zheng S. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. 2019;68:1014-1023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 542] [Cited by in RCA: 508] [Article Influence: 84.7] [Reference Citation Analysis (0)] |
| 38. | Aron-Wisnewsky J, Vigliotti C, Witjes J, Le P, Holleboom AG, Verheij J, Nieuwdorp M, Clément K. Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat Rev Gastroenterol Hepatol. 2020;17:279-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 690] [Article Influence: 138.0] [Reference Citation Analysis (0)] |
| 39. | Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, Zhou J, Ni S, Liu L, Pons N, Batto JM, Kennedy SP, Leonard P, Yuan C, Ding W, Hu X, Zheng B, Qian G, Xu W, Ehrlich SD, Zheng S, Li L. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1230] [Cited by in RCA: 1536] [Article Influence: 139.6] [Reference Citation Analysis (40)] |
| 40. | Albhaisi SAM, Bajaj JS, Sanyal AJ. Role of gut microbiota in liver disease. Am J Physiol Gastrointest Liver Physiol. 2020;318:G84-G98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 95] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 41. | Chou HH, Chien WH, Wu LL, Cheng CH, Chung CH, Horng JH, Ni YH, Tseng HT, Wu D, Lu X, Wang HY, Chen PJ, Chen DS. Age-related immune clearance of hepatitis B virus infection requires the establishment of gut microbiota. Proc Natl Acad Sci U S A. 2015;112:2175-2180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 161] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 42. | Zhou W, Luo J, Xie X, Yang S, Zhu D, Huang H, Yang D, Liu J. Gut Microbiota Dysbiosis Strengthens Kupffer Cell-mediated Hepatitis B Virus Persistence through Inducing Endotoxemia in Mice. J Clin Transl Hepatol. 2022;10:17-25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 43. | Ren YD, Ye ZS, Yang LZ, Jin LX, Wei WJ, Deng YY, Chen XX, Xiao CX, Yu XF, Xu HZ, Xu LZ, Tang YN, Zhou F, Wang XL, Chen MY, Chen LG, Hong MZ, Ren JL, Pan JS. Fecal microbiota transplantation induces hepatitis B virus e-antigen (HBeAg) clearance in patients with positive HBeAg after long-term antiviral therapy. Hepatology. 2017;65:1765-1768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 117] [Article Influence: 14.6] [Reference Citation Analysis (0)] |