Published online Jul 14, 2022. doi: 10.3748/wjg.v28.i26.3164
Peer-review started: December 6, 2021
First decision: January 8, 2022
Revised: January 12, 2022
Accepted: June 16, 2022
Article in press: June 16, 2022
Published online: July 14, 2022
Processing time: 218 Days and 19 Hours
Inflammatory bowel disease (IBD) is caused by an abnormal immune response. Programmed cell death 1 (PD-1) is an immunostimulatory molecule, which interacts with PD ligand (PD-L1) playing a prime important role among autoimmune diseases. Bifidobacterium infantis (B. infantis) can promote the differentiation of CD (cluster of differentiation) 4+ T cells into regulatory T cells (Tregs). Tregs participate in the development of IBD and may be related to disease activity. B. infantis amplify the expression level of PD-1, PD-L1 and Tregs’ nuclear transcription factor forkhead box protein 3 (Foxp3). But the mechanism of B. infantis on PD-1/PD-L1 signaling remains unclear.
To explore the mechanism of B. infantis regulating the immune response in IBD.
Forty-eight-week-old BALB/c mice were randomly divided into five groups: The control group, dextran sulphate sodium (DSS) model group, DSS + B. infantis group, DSS + B. infantis + anti-PD-L1 group, and DSS + anti-PD-L1 group. The control group mice were given drinking water freely, the other four groups were given drinking water containing 5% DSS freely. The control group, DSS model group, and DSS + anti-PD-L1 group were given normal saline (NS) 400 μL daily by gastric lavage, and the DSS + B. infantis group and DSS + B. infantis + anti-PD-L1 group were given NS and 1 × 109 colony-forming unit of B. infantis daily by gastric lavage. The DSS + B. infantis + anti-PD-L1 group and DSS + anti-PD-L1 group were given 200 μg of PD-L1 blocker intraperitoneally at days 0, 3, 5, and 7; the control group, DSS + anti-PD-L1 group, and DSS + B. infantis group were given an intraperitoneal injection of an equal volume of phosphate buffered saline (PBS). Changes in PD-L1, PD-1, Foxp3, interleukin (IL)-10, and transforming growth factor β (TGF-β) 1 protein and gene expression were observed. Flow cytometry was used to observe changes in CD4+, CD25+, Foxp3+ cell numbers in the blood and spleen.
Compared to the control group, the expression of PD-1, Foxp3, IL-10, and TGF-β1 was significantly decreased in the intestinal tract of the DSS mice (P < 0.05). Compared to the control group, the proportion of CD4+, CD25+, Foxp3+ cells in spleen and blood of DSS group was visibly katabatic (P < 0.05). B. infantis upgraded the express of PD-L1, PD-1, Foxp3, IL-10, and TGF-β1 (P < 0.05) and increased the proportion of CD4+, CD25+, Foxp3+ cells both in spleen and blood (P < 0.05). After blocking PD-L1, the increase in Foxp3, IL-10, and TGF-β1 protein and gene by B. infantis was inhibited (P < 0.05), and the proliferation of CD4+, CD25+, Foxp3+ cells in the spleen and blood was also inhibited (P < 0.05). After blocking PD-L1, the messenger ribonucleic acid and protein expression of PD-1 were invariant.
It is potential that B. infantis boost the proliferation of CD4+, CD25+, Foxp3+ T cells in both spleen and blood, as well as the expression of Foxp3 in the intestinal tract by activating the PD-1/PD-L1 pathway.
Core Tip: Bifidobacterium infantis (B. infantis) can be used as a therapeutic agent to treat inflammatory bowel disease. It regulates the intestinal microbiota, alleviates inflammation, and regulates the immune response. We found that B. infantis increases the expression of forkhead box protein 3 (Foxp3) and the proliferation of Foxp3+ T cells, and activates the programmed cell death 1 (PD-1)/ PD ligand 1 pathway.
- Citation: Zhou LY, Xie Y, Li Y. Bifidobacterium infantis regulates the programmed cell death 1 pathway and immune response in mice with inflammatory bowel disease. World J Gastroenterol 2022; 28(26): 3164-3176
- URL: https://www.wjgnet.com/1007-9327/full/v28/i26/3164.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i26.3164
Inflammatory bowel disease (IBD) results from the aberrant immune responses or the disruption of immune tolerance to intestinal antigens. Several factors, including the immune system, infections, and genetic and environmental factors, may remarkably contribute to the development of IBD[1-4]. To maintain immune tolerance to the intestinal environment, the intestinal immune system monitors changes in the bacterial microbiota and the expression of antigens on the surface of the intestinal mucosa[5,6]. Antigen-presenting cells, including dendritic cells and intestinal epithelial cells, present intestinal antigens to CD4+ T cells and induce their differentiation into regulatory T cells (Tregs), which maintain tolerance to the intestinal microbiota. Hyperactive T cell responses to the intestinal microbiota contribute to the inflammatory response observed in IBD[7].
Programmed cell death protein 1 (PD-1) and PD-ligand 1 (PD-L1), belong to the CD28/B7 super
Tregs are a subpopulation of T lymphocytes with immunoregulatory functions[14]. They can inhibit the activation and proliferation of autoreactive T cells by secreting cytokines such as interleukin (IL)-10 and transforming growth factor β (TGF-β), downregulating the function of auxiliary T cells, and maintaining intestinal homeostasis and immune tolerance[15,16]. Tregs are divided into two categories: Natural Tregs (nTregs) and induced Tregs (iTregs). The nTregs mature in the thymus and are positive for CD4, CD25, and Foxp3. The iTregs are induced by specific antigen stimulation of CD4+ T cells in the presence of IL-2 and TGF-β1, in the intestine, spleen, and other peripheral sites[17]. Animal studies have shown that injecting T cells with no CD4 and Tregs expressing CD25 into T cell-deficient mice can induce the development of autoimmune colitis, whereas the injection of T cells expressing CD4 with that of CD25+ Tregs can inhibit colitis development. These results suggest that CD4+ and CD25+ Tregs are vital for the inhibition of the intestinal immune response[18]. Maul et al[19] found that the percentage of Tregs in the peripheral blood of patients with IBD decreased in the active phase and increased in the remission phase of the disease; however, the number of Tregs in the intestinal epithelium increased in the active phase of the disease but was still significantly lower than that observed in patients with diverticulitis. This suggests a role of Tregs in IBD development.
The adoptive transfer of immature CD4+ T cells into wild-type rag-/- mice and PD-L1-/-rag-/- mice significantly decreases the number of Tregs in PD-L1-/-rag-/- mice, suggesting a dominant role of PD-L1 in Treg differentiation[20]. PD-L1 can enhance Treg function and promote the production of IL-10 by Tregs[21]. Treg differentiation depends on the PD-L1 signaling pathway. Higher levels of PD-L1 expression in hepatodendritic cells result in greater induction of Tregs which maintain the tolerance toward transplanted organs.
We have previously found that Bifidobacterium infantis (B. infantis) can alleviate intestinal epithelial injury and maintain intestinal immune tolerance in a mouse model of IBD and may have therapeutic implications for the immunological injuries observed in IBD. Bifidobacterium infantis notably increased the expression levels of PD-L1 and PD-1 in the intestine and promoted the expression of nuclear transcription factors and of anti-inflammatory factors (IL-10 and TGF-β1) in Tregs[22,23]. Therefore, this study aimed to explore the mechanism of action of B. infantis in the PD-1/PD-L1 signaling pathway and the differentiation and function of Tregs.
Dextran sulfate sodium (DSS; molecular weight 36000-50000) was purchased from MP Biomedicals (Irvine, CA, United States). B. infantis freeze-dried powder, containing 1.6 × 1011 colony-forming units (CFU)/g, was provided by Shandong Kexing Biological Products Co., Ltd. (Batch No. 2017012, Shandong Province, China). Invivomab anti-mouse PD-L1 was purchased from BIOX Cell (Lebanon, NH, United States). BALB/c mice were purchased from Huafukang Biotechnology (Beijing, China). Allophycocyanin (APC) rat anti-mouse Cd4, Bb515 rat anti-mouse, P-phycoerythrin (PE) rat anti-mouse Foxp3, and a transcription factor buffer set were purchased from BD Biosciences (Franklin Lakes, NJ, United States). Antibodies against PD-L1, PD-1, and Foxp3 were purchased from Proteintech Group (Rosemont, IL, United States). Antibodies against IL-10 and TGF-β1 were purchased from Abcam (Cambridge, United Kingdom). Real-time quantitative PCR was performed using the following reagents: TRIzol (Invitrogen, Thermo Fisher, Waltham, MA, United States), PrimescriptTM RT Regent kit with gDNA eraser, quick response training (qRT) PCR kit SYBR® premier ex taqTM II (Tli RNaseH Plus, Takara, Japan), and the specific primers (Biotechnology Co., Ltd., China).
Forty-eight-week-old BALB/c mice, male and female, weighing 20 g ± 2 g, were raised under pathogen-free conditions in the standalone animal experimental center affiliated with the Shengjing Hospital of China Medical University. The mice were kept at 20 °C-26 °C and in an atmosphere with a relative humidity of 40%-70%, with a 12 h light/dark cycle. Sterilized water and standard feed were provided for free consumption by the animals. The experimental protocol was approved by the ethics committee of the hospital (No. 2017PS353K). The operators ensured that suitable measures were taken to reduce malaise and injury to the animals during experiments.
Forty mice were randomly divided into five groups: Control, DSS, DSS + B. infantis, DSS + B. infantis + anti-PD-L1, and DSS + anti-PD-L1. Mice in the control group were given free access drinking water for 7 d. The other four groups were administered sterilized water containing 5% DSS for 7 d. The drinking. Drinking water was changed daily. The control, DSS, and DSS + anti-PD-L1 groups were administered 400 μL normal saline (NS) via gavage daily, and the DSS + B. infantis and DSS + B. infantis + anti-PD-L1 groups were administered 400 μL NS via gavage and B. infantis (1 × 109 CFU) daily. The DSS + B. infantis + anti-PD-L1 and DSS + anti-PD-L1 groups were administered an intraperitoneal injection of PD-L1 blocker (200 μg), and the control, DSS model, and DSS + B. infantis groups were intraperitoneally injected with phosphate-buffered phosphate buffered saline (PBS) on days 0, 3, 5, and 7.
General characteristics of the mice: During the experimental period, temperament, reactivity, activity, hair color, weight, eating, and defecation of each mouse were observed seriatim and recorded in detail daily.
Peripheral blood collection: On day 8, all animals were anesthetized via isoflurane inhalation, and the beards were removed. Blood was collected through retro-orbital bleeding and placed in blood collection vessels containing EDTA. The blood and EDTA were mixed and stored on ice.
Extraction of single cells from mouse spleen: After blood collection, the sacrificed mice were dissected along the midline, and the spleen was fully exposed. After blunt dissection, the spleen was removed, placed in PBS, and transported on ice. The spleen was then transferred to a glass dish containing RPMI 1640 medium and mashed with ground glass. The cells were then transferred to a centrifuge tube and centrifuged at 1200 rpm for 5 min, and the supernatant was discarded. Next, 2 mL of RBC lysate was added to each sample. PBS (3 mL) was added to dilute and stop the lysis, and the samples were centrifuged again at 400 × g for 5 min at 4 °C. The supernatant was discarded and 3 mL of PBS was added. The cells were filtered and centrifuged for 10 min. The supernatant was discarded, and PBS was added to obtain a single-cell suspension. All the procedures were performed at 4 °C to ensure cell viability.
Acquisition of mouse colon: After splenectomy, the colon was exposed and the colon from the blind part to the anus was removed, washed with pre-cooled NS, and divided into four parts. The samples were then transferred to a -80 °C ultra-low-temperature refrigerator in liquid nitrogen for long-term preservation.
Spleen: Splenic CD4+, CD25+, and Foxp3+ T cells were detected using flow cytometry. The prepared single-cell spleen suspension (100 μL) was aliquoted into labeled flow tubes. Anti-CD4+ and anti-CD25+ antibodies were then added to the tubes and the tubes were incubated at 4 °C in the dark for 30 min. Next, 1 mL of 1X fix/perm working solution was added to each sample and the samples were incubated at 4 °C in the dark for 40 min to permeabilize the nucleus. Anti-Foxp3 antibody was then added and the resulting solution was incubated at 4 °C in the dark for 40 min. Excess antibodies were removed, and the samples were run on a flow cytometer (FACSCalibur, BD Bioscience).
Peripheral blood: After RBC lysis, flow cytometry was performed on the peripheral blood samples using the protocol described above.
Total protein was extracted from the colon, and the protein concentration was determined. The samples were subjected to electrophoresis at 60 V. After marker separation, the voltage was adjusted to 80 V. After 30 min, the voltage was adjusted to 100 V. Electrophoresis was terminated when the target protein with the lowest molecular weight reached the end of the gel. A voltage of 100 V was used to transfer proteins to the membrane. Proteins with a molecular weight < 25 kDa were transferred for 25 min, and proteins weighing 26-70 kDa were transferred for 70 min. The membrane was blocked with 2.5% bovine serum albumin (BSA) at room temperature for 1.5 h. Primary antibodies against PD-L1 (1:750), PD-1 (1:500), Foxp3 (1:1000), IL-10 (1:800), TGF-β1 (1:500), and GAPDH (1:10000) were added and the membrane was incubated at 4 °C overnight. Then, goat anti-rabbit IgG secondary antibody labeled with horseradish peroxidase was added to the membrane, followed by incubation at room temperature for 1.5 h. In a dark room, a chemiluminescence imaging analysis system was used to visualize the membranes. GelPro software was used to analyze the images and to perform quantitative analysis using the following formula: Protein content = grey value of the target protein of the sample/grey value of the same sample.
The experiment consists of 5 steps: (1) Ribonucleic acid (RNA) purification: DSS can reduce the purity of RNA, so extra purification of the colon RNA was necessary. RNA purification was performed as follows: 30 µL lithium chloride (8 mol/L) + 270 µL ddH2O was added to 10 µL RNA and placed on ice for 2 h. The samples were then centrifuged at 14000 × g for 30 min. The supernatant was then discarded, and the RNA was dissolved in 90 µL ddH2O. Next, 10 µL sodium acetate (3 mol/L) + 200 µL anhydrous ethanol was added to the RNA and incubated at -20 °C for 30 min to precipitate the RNA. The samples were then centrifuged at 14000 × g for 30 min at 4 °C. The supernatant was then discarded, 500 µL 75% ethanol was added, and the samples were gently blown with a pipette to clean the RNA. Next, the samples were centrifuged at 800 × g for 10 min at 4 °C. The supernatant was discarded, and the RNA was dissolved with 10 µL ddH2O. Finally, the samples were transferred to -80 °C on ice for preservation; (2) detection of RNA concentration: The ratio of A260/A280 was eligible for all of the samples, which indicates that the purity of the RNA was high and suitable for further experiments; (3) preparation of cDNA by RNA reverse transcription: The gDNA was removed, and the specimens were heated to 42 °C for 2 min. For reverse transcription, the reaction solution was prepared according to Table 1. The samples were heated at 37 °C for 15 min and 85 °C for 5 s. The reaction was then stopped and cooled down to 4 °C; (4) concentration and purity of cDNA: After zero adjustment, 1 μL of the cDNA sample to be tested was dropped onto the detection probe to determine the concentration and purity. The probe was washed with ddH2O between the evaluation of the two samples; and (5) qRT-PCR: qRT-PCR was carried out as follows: PCR amplification reaction, denaturation at 95 °C for 5 min, PCR reaction at 95 °C for 10 s, and 60 °C for 30 s for 45 cycles.
| Reagent | Dose (µL) |
| 5 × prime script buffer | 4 |
| Prime sript RT enzyme mix | 1 |
| RT primer xix | 1 |
| ddH2O | 4 |
Data are presented as mean ± SD. Differences among the groups were analyzed using the analysis of variance. SPSS (version 23.0; IBM, Armonk, NY, United States) and GraphPad 7.0 (Software, CA, United States) statistical software were used to perform statistical analyses. Two-tailed P values were calculated, and statistical significance was set as P < 0.05.
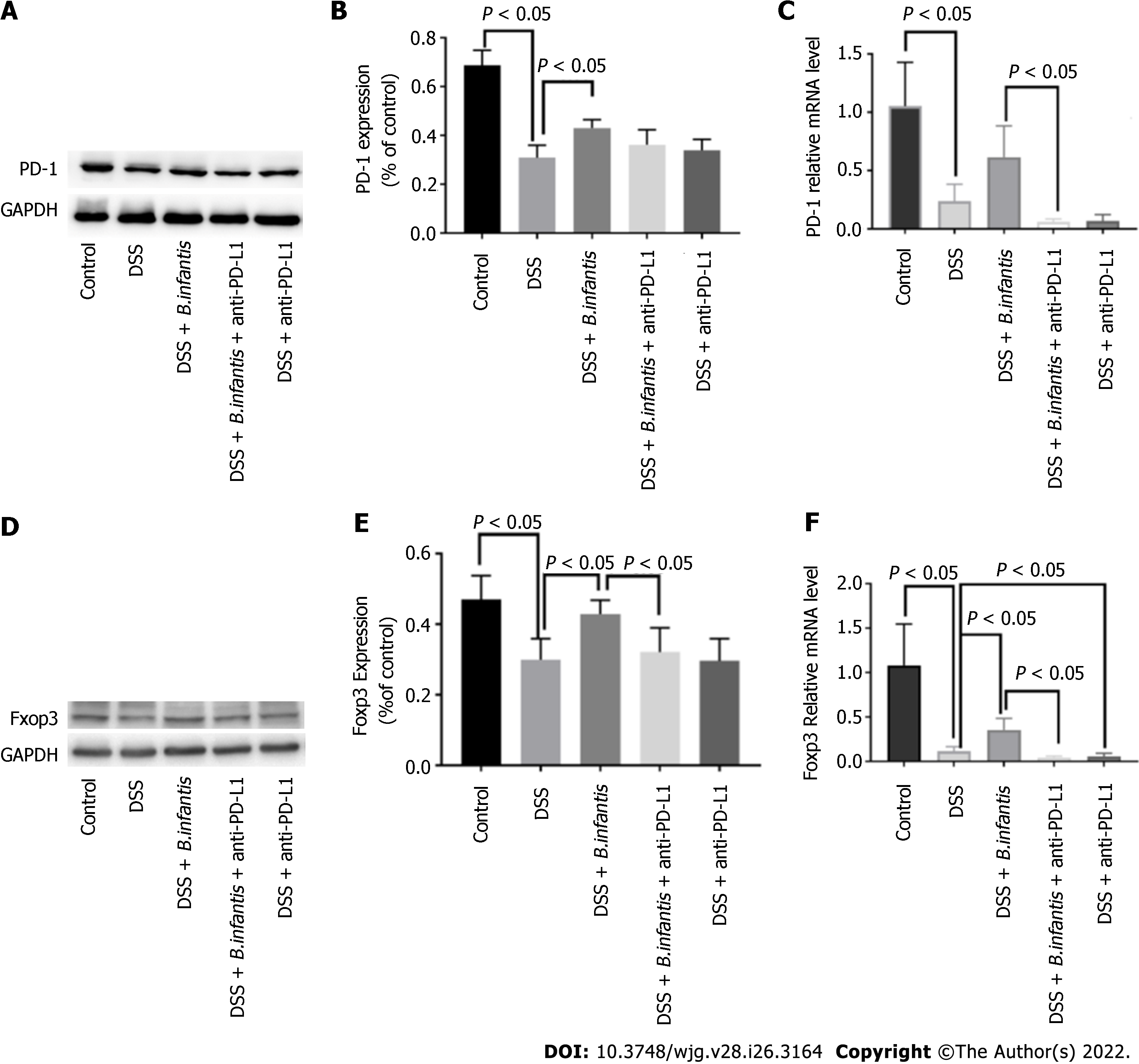
Western blot results: Compared to the DSS + B. infantis group, PD-1 protein in the DSS + B. infantis + anti-PD-L1 group decreased, but the difference was not statistically significant (P = 0.07). Compared to DSS model group, the expression of PD-1 was no significant distinction in DSS + anti-PD-L1 group (P = 0.62) (Figure 1A-C).
qRT-PCR results: In contrast to control group, PD-1 messenger ribonucleic acid (mRNA) in DSS group decreased significantly (P < 0.05). In constrast to DSS group, the expression of DSS + B. infantis group increased, but the difference was not statistically significant (P = 0.36). Compared to the B. infantis group, PD-1 RNA decreased significantly in the DSS + B. infantis + anti-PD-L1 group (P < 0.05) (Figure 1A-C).
Western blot results: Compared to the DSS + B. infantis group, Foxp3 protein decreased in DSS + B. infantis + anti-PD-L1 group, and the difference was statistically significant (P < 0.05). There was no significant difference in Foxp3 protein expression between the DSS model group and DSS + anti-PD-L1 group (P = 0.99) (Figure 1D-F).
RT-qPCR results: In contrast to control group, Foxp3 mRNA in DSS model group decreased significantly (P < 0.05); Foxp3 mRNA in DSS + B. infantis group was significantly higher than that in DSS model group (P < 0.05). Compared to the B. infantis group, the expression of Foxp3 mRNA decreased significantly in the DSS + B. infantis + anti-PD-L1 group (P < 0.05). In comparison with DSS model group, Foxp3 mRNA in DSS + anti-PD-L1 group was also visible distinction (P < 0.05) (Figure 1D-F).
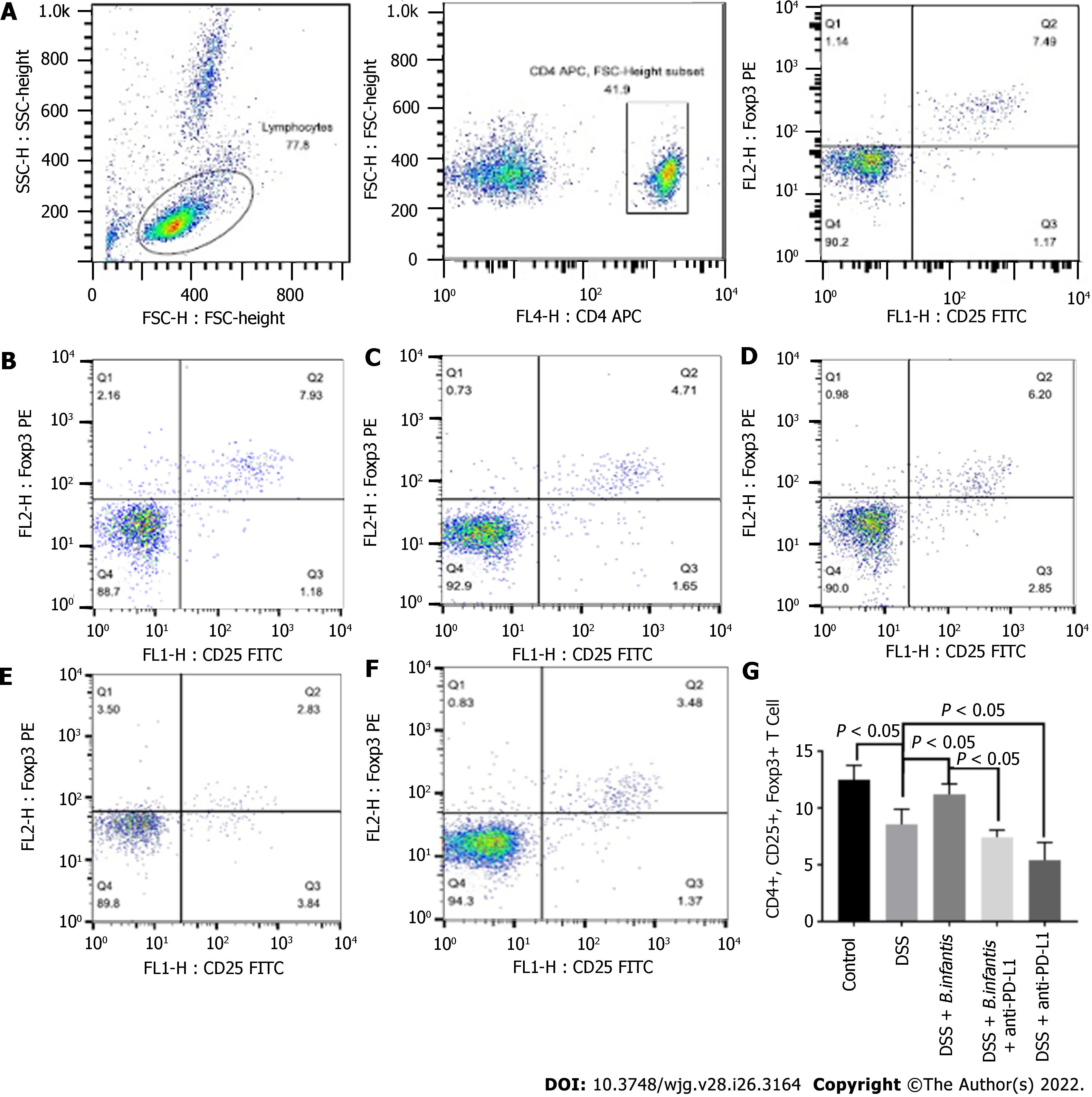
Flow cytometry of peripheral blood: Compared to control group, the proportion of peripheral CD4+, CD25+, Foxp3+ cells decreased visibly (P < 0.05) of DSS group and increased visibly in the blood of the DSS + B. infantis group (P < 0.05). Compared to the DSS + B. infantis group, the proportion of CD4+, CD25+, Foxp3+ cells in the peripheral blood of DSS + B. infantis + anti-PD-L1 group was significantly lower (P < 0.05). The proportion of CD4+, CD25+, Foxp3+ cells in the blood of the DSS + anti-PD-L1 group was also distinctly lower compared to the DSS group (P < 0.05) (Figure 2).
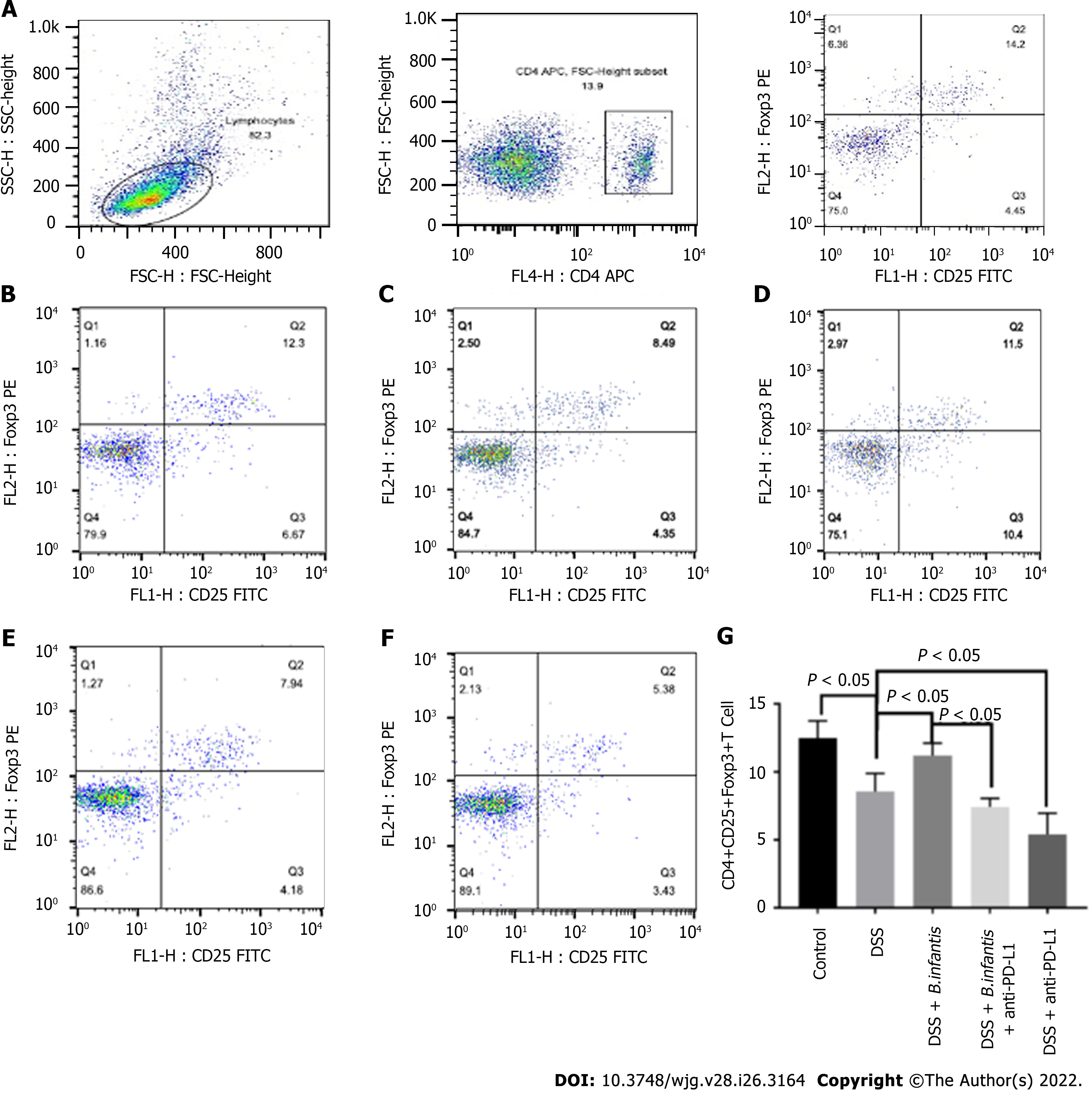
Flow cytometry of splenocytes: The ratio of splenic CD4+, CD25+, Foxp3+ cells in DSS model group was significantly lower (P < 0.05), comparing to control group. The ratio of CD4+, CD25+, Foxp3+ cells in the DSS + B. infantis group was significantly higher than that in the DSS model group (P < 0.05). Compared to the DSS + B. infantis group, the proportion of CD4+, CD25+, Foxp3+ cells in the DSS + B. infantis + anti-PD-L1 group decreased significantly (P < 0.05). The proportion of CD4+, CD25+, Foxp3+ cells in the DSS + anti-PD-L1 group also decreased significantly compared to the DSS group (P < 0.05) (Figure 3).
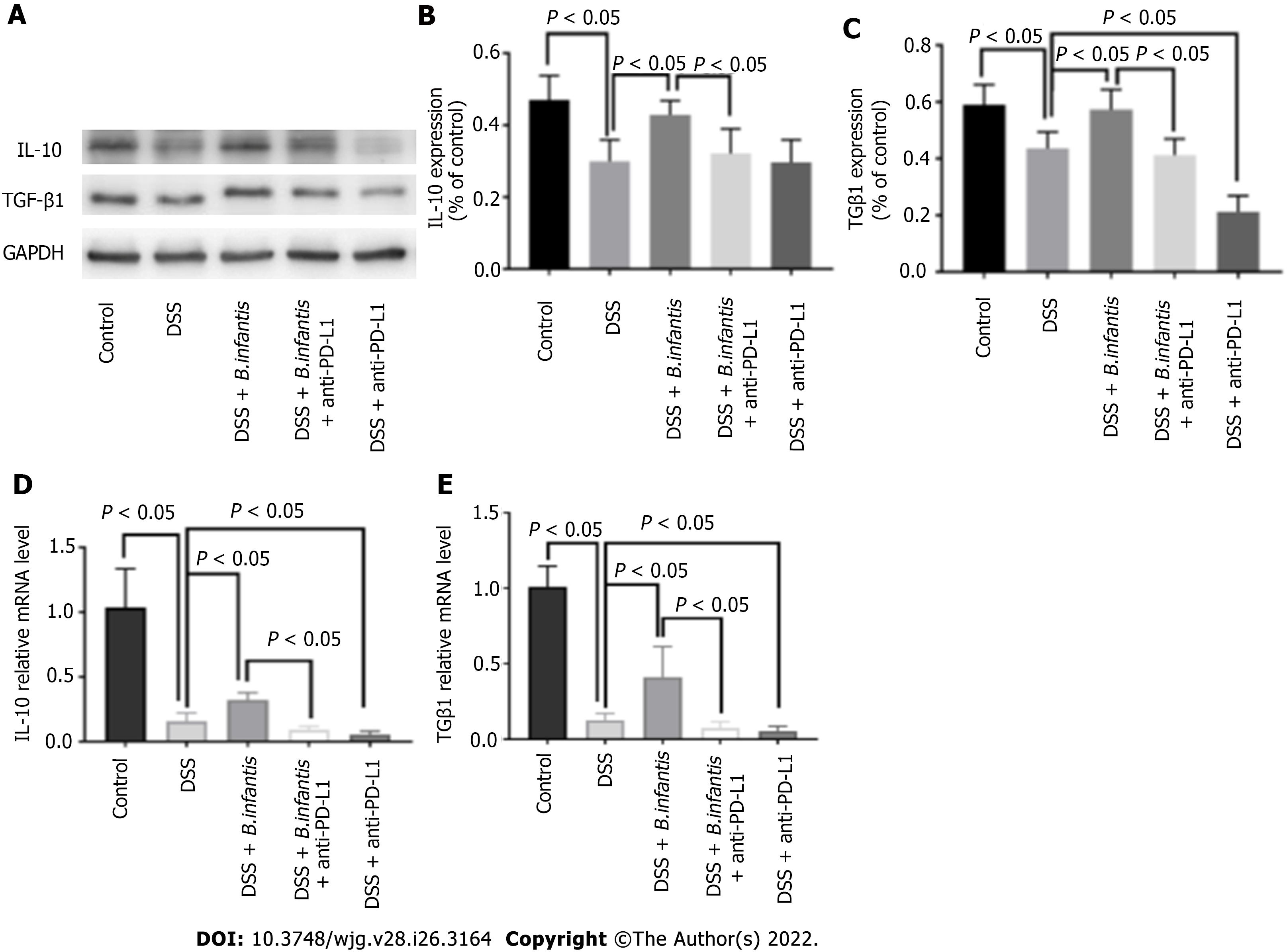
Western blot results: The expression of IL-10 and TGF-β1 protein in the DSS + B. infantis group was lower than that in DSS + B. infantis + anti-PD-L1 group (P < 0.05). Compared to the DSS model group, there was no apparent distinction in the express of IL-10 (P = 0.99) or TGF-β1 in the DSS + anti-PD-L1 group (P < 0.05) (Figure 4).
Real time PCR results: In comparison with control group, mRNA of IL-10 and TGF-β1 in the DSS model group decreased (P < 0.05), and mRNA of IL-10 and TGF-β1 in the DSS + B. infantis group increased clearly (P < 0.05). IL-10 and TGF-β1 mRNA expression in DSS + B. infantis + anti-PD-L1 group decreased clearly (P < 0.05) contrsating to DSS + B. infantis group. Compared to the DSS model group, IL-10 and TGF-β1 mRNA in DSS + anti-PD-L1 group were also statistically distinction (P < 0.05) (Figure 4).
The specific pathogenesis of IBD remains unclear; however, abnormal inflammatory responses and continuous inflammatory damage to the intestine are recognized as the basic mechanisms of IBD pathogenesis[24,25]. Intestinal immunity is a complex and interactive process, involving several immune factors such as intestinal mucosal immunity, T cells, cytokines, and intestinal microecology[26]. The intestinal mucosal immune system is responsible for monitoring the intestinal microbiota and surface antigens[27], presenting antigens to CD4+ T cells, promoting the interaction between PD-1 and PD-L1, establishing immune tolerance, and preventing the occurrence of autoimmunity. Several inflammatory mediators including interferon γ, tumor necrosis factor α, IL-10, and other cytokines, are involved in the pathogenesis of DSS colitis, suggesting that inflammatory immune responses play a critical role in the pathogenesis of IBD[28-30].
A recently discovered immunostimulatory molecule, PD-1 interacts with its ligand PD-L1 to regulate T cell-mediated immunity and induce immune tolerance, thereby playing a critical role in autoimmune diseases (such as IBD), tumor immunity, and the acceptance of transplanted organs. Studies have shown that PD-1 knockout results in autoimmune diseases in animal models[31-33]. Activation of the PD-1/PD-L1 signaling pathway can induce the differentiation of Tregs[34] and the release of cytokines, such as IL-10 and TGF-β1, to inhibit the activation and proliferation of reactive T cells, thus maintaining intestinal immune tolerance. Additionally, the inhibition of the PD-1/PD-L1 pathway may reduce the proportion of Tregs[35]. These findings indicate that PD-1/PD-L1 signaling plays a critical role in immune tolerance[36]. In this study, we found that PD-L1 inhibition did not alter the PD-1 protein and mRNA levels in the intestine of DSS-induced mice, suggesting that PD-L1 did not affect the transcription or translation of PD-1 in the intestine of the IBD mouse model. However, although the expression of PD-1 mRNA in the intestinal tracts of DSS mice significantly decreased, the expression of PD-1 protein did not change after B. infantis administration. We, therefore, speculate that PD-L1 inhibition may indirectly inhibit the B. infantis-induced PD-1 gene transcription, but not the post-transcriptional modification and translation of PD-1 protein. This transcriptional inhibition of PD-1 may be due to a negative feedback mechanism caused by the high PD-1 protein levels. Further studies are needed to determine how PD-L1 inhibition suppresses the transcription and translation of PD-1 and the underlying mechanism of action of the effect of B. infantis on PD-1 after PD-L1 inhibition.
B. infantis can be used to treat IBD by regulating the intestinal microbiota, alleviating inflammation, and regulating the immune response. B. infantis can reduce the intestinal wall permeability, edema, and neutrophil infiltration in IBD mice, and alleviate intestinal inflammatory responses[37]. We have previously shown that a combined administration of B. infantis with Clostridium butyricum increases the abundance of probiotic bacteria (such as members of the genera Bifidobacterium and Lactobacillus) in the intestinal microbiota of patients with IBD, reduces the prevalence of enterococci, improves clinical symptoms, and promotes the healing of colonic mucosa[38]. B. infantis also plays an important role in immune regulation by promoting the proliferation of Tregs[39] and increasing the expression of IL-10 and TGF-β1. In vivo and in vitro experiments have revealed that B. infantis can significantly accelerate the differentiation of CD4+ T cells into Tregs by inducing the maturation of resistant dendritic cells and further inhibiting the inflammatory response induced by reactive T cell activation. Furthermore, we have previously revealed that, compared to the observations in normal mice, the number of CD4+, CD25+, and Foxp3+ T cells in the blood and spleen of DSS mice and the expression of Foxp3 mRNA in their intestines showed a decrease, suggesting that the differentiation and proliferation of Tregs may be correlated to the pathogenesis of IBD[40]. The number of Tregs in the colon was reported to be related to the intestinal microbiota. Treg populations in the colons of sterile mice are significantly low; however, feeding sterile mice with feces collected from specific pathogen-free mice significantly increases the number of Tregs in the colon[41], indicating that Treg population is dependent on the intestinal microbiota. In patients with IBD, the proportion of normal intestinal bacteria decreases, resulting in intestinal microbiota-associated disorders[42]. Therefore, improving the composition of the intestinal microbiota can help increase the number of Tregs; however, further studies are needed for the elucidation of the underlying mechanism. In the present study, we found that B. infantis promotes the proliferation of CD4+, CD25+, and Foxp3+ T cells in the blood and spleen, as well as the expression of Foxp3 mRNA in the intestine. PD-L1 inhibition significantly reduced the numbers of CD4+, CD25+, and Foxp3+ T cells in the blood and spleen, and decreased the expression of Foxp3 protein and mRNA in the intestine. Therefore, B. infantis promotes Treg proliferation by activating the PD-L1/PD-1 pathway.
In addition, our results indicate that B. infantis promoted the mRNA expression of IL-10 and TGF-β1 in the mouse intestine. PD-L1 inhibition significantly reduced the protein and mRNA expression levels of IL-10 and TGF-β1 in the intestine. These results further indicate that B. infantis affected IL-10 and TGF-β1 expression through the PD-1/PD-L1 pathway. Notably, Tregs mainly secrete TGF-β1 and IL-10 to inhibit inflammatory responses. Further studies are needed to confirm the immunosuppressive effects of B. infantis in patients with IBD. Additionally, Francisco et al[20] found that PD-L1 can downregulate Protein kinase B (Akt), mammalian target of rapamycin (mTOR), and extracellular regulated protein kinases (ERK2), while upregulating phosphatase and tensin homolog deleted on chromosome ten (PTEN) expression in Tregs; however, whether B. infantis can accelerate the differentiation and proliferation of Tregs by activating the PD-1/PD-L1 pathway and regulating Akt, mTOR, or PTEN expression requires further investigation.
In conclusion, B. infantis may accelerate the proliferation of CD4+, CD25+, and Foxp3+ T cells in the spleen and peripheral blood, and the expression of Foxp3 in the intestine by activating the PD-1/PD-L1 signaling pathway. It can also promote the expression of IL-10 and TGF-β1 to reduce the intestinal inflammatory response, which has a therapeutic effect on IBD mice. We aim to further investigate the role of the PD-1/PD-L1 pathway in IBD and the possible therapeutic effect of B. infantis on patients with IBD in future studies.
The immune-inflammatory response plays an important role in the pathogenesis and development of inflammatory bowel disease (IBD). Bifidobacterium infantis (B. infantis) can repair the acute intestinal mucosal injury and maintain autoimmune tolerance in mice with IBD.
The specific mechanism of action of B. infantis in the treatment of IBD is unclear. Understanding this underlying mechanism will help in the treatment of IBD.
To explore if B. infantis can promote regulatory T cell Treg differentiation through the programmed cell death 1 (PD-1)/PD ligand (PD-L1) pathway to promote the expression of forkhead box protein 3 (Foxp3), interleukin (IL)-10 and transforming growth factor β (TGF-β) 1, and reduce the inflammatory response.
We blocked the expression of PD-L1 in the intestine and performed western blotting and real-time qPCR to observe the effects of B. infantis on PD-1, Foxp3, IL-10, and TGF-β1. Flow cytometry was used to examine the changes in the differentiation of CD4+, CD25+, and Foxp3+ cells in the blood and spleen after blocking PD-L1.
PD-L1 inhibition reduced the promoting effects of B. infantis on intestinal PD-1, Foxp3, IL-10, and TGF-β1. The promoting effect of B. infantis on the differentiation of CD4+, CD25+, and Foxp3+ cells was also reduced.
B. infantis mediates Foxp3 expression through the PD-1/PD-L1 pathway, thereby promoting Treg differentiation and improving IL-10 and TGF-β1 expression to reduce the immune and inflammatory response in IBD. B. infantis may act as a therapeutic agent for IBD.
To explore the mechanism of action of B. infantis in the treatment of IBD at the cellular level. Further experiments are essential to determine whether B. infantis inhibits the immune response through the PD-1/PD-L1 pathway in the patients with IBD.
The authors thank Professor Liu DY for her guidance during the experiments.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bozkurt HS, Turkey; Rodrigues AT, Brazil; Serban ED, Romania S-Editor: Chen YL L-Editor: A P-Editor: Cai YX
| 1. | Burisch J, Zammit SC, Ellul P, Turcan S, Duricova D, Bortlik M, Andersen KW, Andersen V, Kaimakliotis IP, Fumery M, Gower-Rousseau C, Girardin G, Valpiani D, Goldis A, Brinar M, Čuković-Čavka S, Oksanen P, Collin P, Barros L, Magro F, Misra R, Arebi N, Eriksson C, Halfvarson J, Kievit HAL, Pedersen N, Kjeldsen J, Myers S, Sebastian S, Katsanos KH, Christodoulou DK, Midjord J, Nielsen KR, Kiudelis G, Kupcinskas L, Nikulina I, Belousova E, Schwartz D, Odes S, Salupere R, Carmona A, Pineda JR, Vegh Z, Lakatos PL, Langholz E, Munkholm P; Epi-IBD group. Disease course of inflammatory bowel disease unclassified in a European population-based inception cohort: An Epi-IBD study. J Gastroenterol Hepatol. 2019;34:996-1003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 2. | Olendzki B, Bucci V, Cawley C, Maserati R, McManus M, Olednzki E, Madziar C, Chiang D, Ward DV, Pellish R, Foley C, Bhattarai S, McCormick BA, Maldonado-Contreras A. Dietary manipulation of the gut microbiome in inflammatory bowel disease patients: Pilot study. Gut Microbes. 2022;14:2046244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 36] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 3. | Lo Sasso G, Phillips BW, Sewer A, Battey JND, Kondylis A, Talikka M, Titz B, Guedj E, Peric D, Bornand D, Dulize R, Merg C, Corciulo M, Ouadi S, Yanuar R, Tung CK, Ivanov NV, Peitsch MC, Hoeng J. The reduction of DSS-induced colitis severity in mice exposed to cigarette smoke is linked to immune modulation and microbial shifts. Sci Rep. 2020;10:3829. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 4. | Algethmi W, Baumann C, Alnajjar W, Sroji A, Alsahafi M, Jawa H, Qari Y, Peyrin-Biroulet L, Saadah OI, Mosli M. Environmental exposures and the risk of inflammatory bowel disease: a case-control study from Saudi Arabia. Eur J Gastroenterol Hepatol. 2020;32:358-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 5. | Illiano P, Brambilla R, Parolini C. The mutual interplay of gut microbiota, diet and human disease. FEBS J. 2020;287:833-855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 219] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 6. | Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17:223-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 1199] [Article Influence: 239.8] [Reference Citation Analysis (0)] |
| 7. | Sartor RB. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1158] [Cited by in RCA: 1277] [Article Influence: 67.2] [Reference Citation Analysis (2)] |
| 8. | Li J, Jie HB, Lei Y, Gildener-Leapman N, Trivedi S, Green T, Kane LP, Ferris RL. PD-1/SHP-2 inhibits Tc1/Th1 phenotypic responses and the activation of T cells in the tumor microenvironment. Cancer Res. 2015;75:508-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 189] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 9. | Pauken KE, Sammons MA, Odorizzi PM, Manne S, Godec J, Khan O, Drake AM, Chen Z, Sen DR, Kurachi M, Barnitz RA, Bartman C, Bengsch B, Huang AC, Schenkel JM, Vahedi G, Haining WN, Berger SL, Wherry EJ. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 2016;354:1160-1165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 691] [Cited by in RCA: 985] [Article Influence: 109.4] [Reference Citation Analysis (0)] |
| 10. | Garcia-Bates TM, Palma ML, Shen C, Gambotto A, Macatangay BJC, Ferris RL, Rinaldo CR, Mailliard RB. Contrasting Roles of the PD-1 Signaling Pathway in Dendritic Cell-Mediated Induction and Regulation of HIV-1-Specific Effector T Cell Functions. J Virol. 2019;93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 11. | Zamani MR, Aslani S, Salmaninejad A, Javan MR, Rezaei N. PD-1/PD-L and autoimmunity: A growing relationship. Cell Immunol. 2016;310:27-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 221] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 12. | Kuol N, Stojanovska L, Nurgali K, Apostolopoulos V. PD-1/PD-L1 in disease. Immunotherapy. 2018;10:149-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 93] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 13. | Long Y, Xia C, Sun Y, Ma Y, Xu L, Song Y, Liu C. Increased circulating PD-1hiCXCR5- peripheral helper T cells are associated with disease severity of active ulcerative colitis patients. Immunol Lett. 2021;233:2-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 14. | Li JY, Yu M, Pal S, Tyagi AM, Dar H, Adams J, Weitzmann MN, Jones RM, Pacifici R. Parathyroid hormone-dependent bone formation requires butyrate production by intestinal microbiota. J Clin Invest. 2020;130:1767-1781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 121] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 15. | Hartwig T, Zwicky P, Schreiner B, Yawalkar N, Cheng P, Navarini A, Dummer R, Flatz L, Conrad C, Schlapbach C, Becher B. Regulatory T Cells Restrain Pathogenic T Helper Cells during Skin Inflammation. Cell Rep. 2018;25:3564-3572.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 16. | Barbi J, Pardoll D, Pan F. Treg functional stability and its responsiveness to the microenvironment. Immunol Rev. 2014;259:115-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 196] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 17. | Vitale A, Strisciuglio C, Vitale S, Santopaolo M, Bruzzese D, Micillo T, Scarpato E, Miele E, Staiano A, Troncone R, Matarese G, Gianfrani C. Increased frequency of regulatory T cells in pediatric inflammatory bowel disease at diagnosis: a compensative role? Pediatr Res. 2020;87:853-861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 18. | O'Garra A, Vieira P. Regulatory T cells and mechanisms of immune system control. Nat Med. 2004;10:801-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 604] [Cited by in RCA: 588] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 19. | Maul J, Loddenkemper C, Mundt P, Berg E, Giese T, Stallmach A, Zeitz M, Duchmann R. Peripheral and intestinal regulatory CD4+ CD25(high) T cells in inflammatory bowel disease. Gastroenterology. 2005;128:1868-1878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 470] [Cited by in RCA: 497] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 20. | Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK, Sharpe AH. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med. 2009;206:3015-3029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1346] [Cited by in RCA: 1622] [Article Influence: 101.4] [Reference Citation Analysis (0)] |
| 21. | Ding Q, Lu L, Zhou X, Zhou Y, Chou KY. Human PD-L1-overexpressing porcine vascular endothelial cells induce functionally suppressive human CD4+CD25hiFoxp3+ Treg cells. J Leukoc Biol. 2011;90:77-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 22. | Zhou L, Liu D, Xie Y, Yao X, Li Y. Bifidobacterium infantis Induces Protective Colonic PD-L1 and Foxp3 Regulatory T Cells in an Acute Murine Experimental Model of Inflammatory Bowel Disease. Gut Liver. 2019;13:430-439. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 23. | Chaudhary CL, Gurung P, Jang S, Banskota S, Nam TG, Kim JA, Jeong BS. Synthesis, activity and mechanism of alkoxy-, carbamato-, sulfonamido-, thioureido-, and ureido-derivatives of 2,4,5-trimethylpyridin-3-ol against inflammatory bowel disease. J Enzyme Inhib Med Chem. 2020;35:1-20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 24. | Lee KH, Ahn BS, Cha D, Jang WW, Choi E, Park S, Park JH, Oh J, Jung DE, Park H, Suh Y, Jin D, Lee S, Jang YH, Yoon T, Park MK, Seong Y, Pyo J, Yang S, Kwon Y, Jung H, Lim CK, Hong JB, Park Y, Shin JI, Kronbichler A. Understanding the immunopathogenesis of autoimmune diseases by animal studies using gene modulation: A comprehensive review. Autoimmun Rev. 2020;19:102469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 25. | Eichele DD, Kharbanda KK. Dextran sodium sulfate colitis murine model: An indispensable tool for advancing our understanding of inflammatory bowel diseases pathogenesis. World J Gastroenterol. 2017;23:6016-6029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 621] [Cited by in RCA: 581] [Article Influence: 72.6] [Reference Citation Analysis (15)] |
| 26. | Yan PG, Li JN. Advances in the understanding of the intestinal micro-environment and inflammatory bowel disease. Chin Med J (Engl). 2020;133:834-841. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 27. | Aden K, Rehman A, Waschina S, Pan WH, Walker A, Lucio M, Nunez AM, Bharti R, Zimmerman J, Bethge J, Schulte B, Schulte D, Franke A, Nikolaus S, Schroeder JO, Vandeputte D, Raes J, Szymczak S, Waetzig GH, Zeuner R, Schmitt-Kopplin P, Kaleta C, Schreiber S, Rosenstiel P. Metabolic Functions of Gut Microbes Associate With Efficacy of Tumor Necrosis Factor Antagonists in Patients With Inflammatory Bowel Diseases. Gastroenterology. 2019;157:1279-1292.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 191] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 28. | Yan Y, Kolachala V, Dalmasso G, Nguyen H, Laroui H, Sitaraman SV, Merlin D. Temporal and spatial analysis of clinical and molecular parameters in dextran sodium sulfate induced colitis. PLoS One. 2009;4:e6073. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 264] [Cited by in RCA: 311] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 29. | Hall LJ, Faivre E, Quinlan A, Shanahan F, Nally K, Melgar S. Induction and activation of adaptive immune populations during acute and chronic phases of a murine model of experimental colitis. Dig Dis Sci. 2011;56:79-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 87] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 30. | Fukata M, Michelsen KS, Eri R, Thomas LS, Hu B, Lukasek K, Nast CC, Lechago J, Xu R, Naiki Y, Soliman A, Arditi M, Abreu MT. Toll-like receptor-4 is required for intestinal response to epithelial injury and limiting bacterial translocation in a murine model of acute colitis. Am J Physiol Gastrointest Liver Physiol. 2005;288:G1055-G1065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 377] [Cited by in RCA: 393] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 31. | Chen L, Pai V, Levinson R, Sharpe AH, Freeman GJ, Braun J, Gordon LK. Constitutive neuronal expression of the immune regulator, programmed death 1 (PD-1), identified during experimental autoimmune uveitis. Ocul Immunol Inflamm. 2009;17:47-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 32. | Roosenboom B, Horjus Talabur Horje CS, Smids C, Leeuwis JW, van Koolwijk E, Groenen MJM, Wahab PJ, van Lochem EG. Distribution of mucosal PD-1 expressing T cells in patients with colitis of different etiologies. Scand J Gastroenterol. 2021;56:671-679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 33. | Bodhankar S, Galipeau D, Vandenbark AA, Offner H. PD-1 Interaction with PD-L1 but not PD-L2 on B-cells Mediates Protective Effects of Estrogen against EAE. J Clin Cell Immunol. 2013;4:143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 34. | Gianchecchi E, Fierabracci A. Inhibitory Receptors and Pathways of Lymphocytes: The Role of PD-1 in Treg Development and Their Involvement in Autoimmunity Onset and Cancer Progression. Front Immunol. 2018;9:2374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 158] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 35. | O'Shaughnessy MJ, Murray KS, La Rosa SP, Budhu S, Merghoub T, Somma A, Monette S, Kim K, Corradi RB, Scherz A, Coleman JA. Systemic Antitumor Immunity by PD-1/PD-L1 Inhibition Is Potentiated by Vascular-Targeted Photodynamic Therapy of Primary Tumors. Clin Cancer Res. 2018;24:592-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 36. | Lo YC, Price C, Blenman K, Patil P, Zhang X, Robert ME. Checkpoint Inhibitor Colitis Shows Drug-Specific Differences in Immune Cell Reaction That Overlap With Inflammatory Bowel Disease and Predict Response to Colitis Therapy. Am J Clin Pathol. 2021;156:214-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (1)] |
| 37. | Elian SD, Souza EL, Vieira AT, Teixeira MM, Arantes RM, Nicoli JR, Martins FS. Bifidobacterium longum subsp. infantis BB-02 attenuates acute murine experimental model of inflammatory bowel disease. Benef Microbes. 2015;6:277-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 38. | Li Hui, Gong Yi, Li Yan. Clinical efficacy of mesalazine combined with Clostridium butyricum in the treatment of active mild to moderate ulcerative colitis. Shiyong Yaowu Yu Linchuang. 2016;19:1389-1393. [DOI] [Full Text] |
| 39. | Mi H, Dong Y, Zhang B, Wang H, Peter CCK, Gao P, Fu H, Gao Y. Bifidobacterium Infantis Ameliorates Chemotherapy-Induced Intestinal Mucositis Via Regulating T Cell Immunity in Colorectal Cancer Rats. Cell Physiol Biochem. 2017;42:2330-2341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 69] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 40. | Fu L, Song J, Wang C, Fu S, Wang Y. Bifidobacterium infantis Potentially Alleviates Shrimp Tropomyosin-Induced Allergy by Tolerogenic Dendritic Cell-Dependent Induction of Regulatory T Cells and Alterations in Gut Microbiota. Front Immunol. 2017;8:1536. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 79] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 41. | Bozkurt HS, Kara B. A new treatment for ulcerative colitis: Intracolonic Bifidobacterium and xyloglucan application. Eur J Inflamm. 2020;18:1-7. [RCA] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 42. | Zuo T, Ng SC. The Gut Microbiota in the Pathogenesis and Therapeutics of Inflammatory Bowel Disease. Front Microbiol. 2018;9:2247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 380] [Cited by in RCA: 403] [Article Influence: 57.6] [Reference Citation Analysis (0)] |