Published online Jul 14, 2022. doi: 10.3748/wjg.v28.i26.3063
Peer-review started: January 10, 2022
First decision: April 16, 2022
Revised: April 21, 2022
Accepted: June 16, 2022
Article in press: June 16, 2022
Published online: July 14, 2022
Processing time: 184 Days and 2.2 Hours
Crohn’s disease (CD) is driven by the loss of tolerance to intestinal microbiota and excessive production of pro-inflammatory cytokines. These pro-inflammatory cytokines are produced by macrophages and dendritic cells (DCs) upon sensing the intestinal microbiota by the pattern recognition receptors (PRRs). Impaired activation of PRR-mediated signaling pathways is associated with chronic gastrointestinal inflammation, as shown by the fact that loss-of-function mutations in the nucleotide-binding oligomerization domain 2 gene increase the risk of CD development. Autophagy is an intracellular degradation process, during which cytoplasmic nutrients and intracellular pathogens are digested. Given that impaired reaction to intestinal microbiota alters signaling pathways mediated by PRRs, it is likely that dysfunction of the autophagic machinery is involved in the development of CD. Indeed, the loss-of-function mutation T300A in the autophagy related 16 like 1 (ATG16L1) protein, a critical regulator of autophagy, increases susceptibility to CD. Recent studies have provided evidence that ATG16L1 is involved not only in autophagy, but also in PRR-mediated signaling pathways. ATG16L1 negatively regulates pro-inflammatory cytokine responses of macrophages and DCs after these cells sense the intestinal microbiota by PRRs. Here, we discuss the molecular mechanisms underlying the develo
Core Tip: The loss-of-function mutation T300A in autophagy related 16 like 1 (ATG16L1) increases the risk of Crohn’s disease (CD). ATG16L1 is a multifunctional protein involved in autophagy and innate immunity. The CD-associated ATG16L1 mutation T300A leads to overgrowth of intestinal microbiota and enhanced pro-inflammatory cytokine responses, which disrupt intestinal immune homeostasis. In this minireview article, we have summarized the immunopathogenesis of CD in the presence of ATG16L1 mutation.
- Citation: Okai N, Watanabe T, Minaga K, Kamata K, Honjo H, Kudo M. Alterations of autophagic and innate immune responses by the Crohn’s disease-associated ATG16L1 mutation. World J Gastroenterol 2022; 28(26): 3063-3070
- URL: https://www.wjgnet.com/1007-9327/full/v28/i26/3063.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i26.3063
Pro-inflammatory cytokines, such as tumor necrosis factor (TNF)-α, interleukin (IL)-6, IL-12, and IL-23, underlie the immunopathogenesis of Crohn’s disease (CD), as evidenced by the clinical efficacy of targeting these cytokines for the treatment of patients[1,2]. These colitogenic cytokines are produced by macrophages and dendritic cells (DCs) upon sensing the intestinal microbiota by the pattern recognition receptors (PRRs), which are classified into Toll-like receptors (TLRs), nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), and retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs)[3-6]. Thus, excessive pro-inflammatory cytokine responses caused by PRR activation play critical roles in the development of CD. This notion is fully supported by the identification of loss-of-function mutations in NOD2 as one of the strongest risk factors for CD. NOD2 is an intracellular PRR that senses muramyl dipeptide (MDP) derived from bacterial cell wall components and negatively regulates TLR-mediated pro-inflammatory cytokine responses[5,6].
Autophagy refers to the process during which cytoplasmic components and intracellular pathogens are delivered to the lysosome for degradation in the form of double-membrane-bound autophagosomes[7]. The autophagy related 16 like 1 (ATG16L1) protein plays an indispensable role in the initiation and completion of the autophagic process. In addition to its role in autophagy, ATG16L1 has been shown to be involved in PRR-mediated innate immunity. ATG16L1 negatively regulates pro-inflammatory and type I interferon (IFN-I) responses mediated by TLRs, NLRs, and RLRs[8]. More importantly, the loss-of-function mutation T300A in ATG16L1 has been identified as a risk factor for CD in parallel with mutations in NOD2[6]. In this minireview article, we summarize the molecular mechanisms by which the T300A mutation in ATG16L1 predisposes the host to CD development by focusing on the regulatory role of ATG16L1 in PRR-mediated signaling pathways.
ATG16L1 is an indispensable molecule for autophagic responses (Table 1). The autophagy process includes vesicle nucleation, vesicle elongation, vesicle completion, fusion with lysosome, degradation, and recycling[9]. Autophagy dysfunction is associated with neurodegenerative diseases, microbial infections, and aging[7]. Although autophagy has been identified as the primary cell response to the lack of nutrients, recent studies have highlighted the importance of autophagy in microbial infection and immune responses[9]. The autophagy process is negatively regulated by growth factors, which activate the mechanistic target of rapamycin (mTOR) and the phosphoinositide 3-kinase (PI3K)-AKT pathways[7,9]. On the contrary, nutrient starvation or rapamycin treatment promotes the autophagic process through the inhibition of mTOR. Thus, the PI3K–AKT–mTOR pathway negatively regulates autophagic process. On the molecular level, mTOR activation controls the initiation of autophagy by suppressing the activation of the primary initiation complex of autophagy, called Unc-51 Like autophagy activating kinase 1 (ULK1) complex, composed of ULK1/2, ATG101, ATG13, and RB1CC1/FIP200[9]. The formed ULK1 complex translocates to the site of the second complex, called the PI3K complex[9]. The latter PI3K complex recruits a number of ATG proteins to promote elongation and expansion of the autophagosome.
| Function | Cell type | Ref. |
| Positive regulation | ||
| Autophagy | ECs, DCs, macrophages | [9,10] |
| Regulatory T cell responses | DCs | [22] |
| Negative regulation | ||
| IFN-I production by RLRs | MEFs | [11,12] |
| IFN-I production by TLR3 and TLR4 | Macrophages | [13] |
| IL-1β production by TLR4 | Macrophages | [16,17,19] |
| IL-6 and IL-12 production by TLR2 | DCs | [27] |
Two ubiquitin-like conjugation systems, the ATG5–ATG12–ATG16L1 conjugation system and the microtubule-associated protein 1 Light chain 3 (LC3) conjugation system, play important roles in the elongation and expansion of the autophagosome[7,9]. The conjugation of the membrane lipid phosphatidylethanolamine with the soluble form of LC3 and formation of the ATG5–ATG12–ATG16L1 complex is necessary for the maturation of autophagosomes[7,9,10]. Matured autophagosomes are fused with lysosomes for the degradation of cellular materials. Vesicles containing ATG16L1 are necessary for membrane trafficking and autophagosome formation[7,9,10]. Thus, ATG16L1 is an essential protein for the induction and completion of autophagic responses.
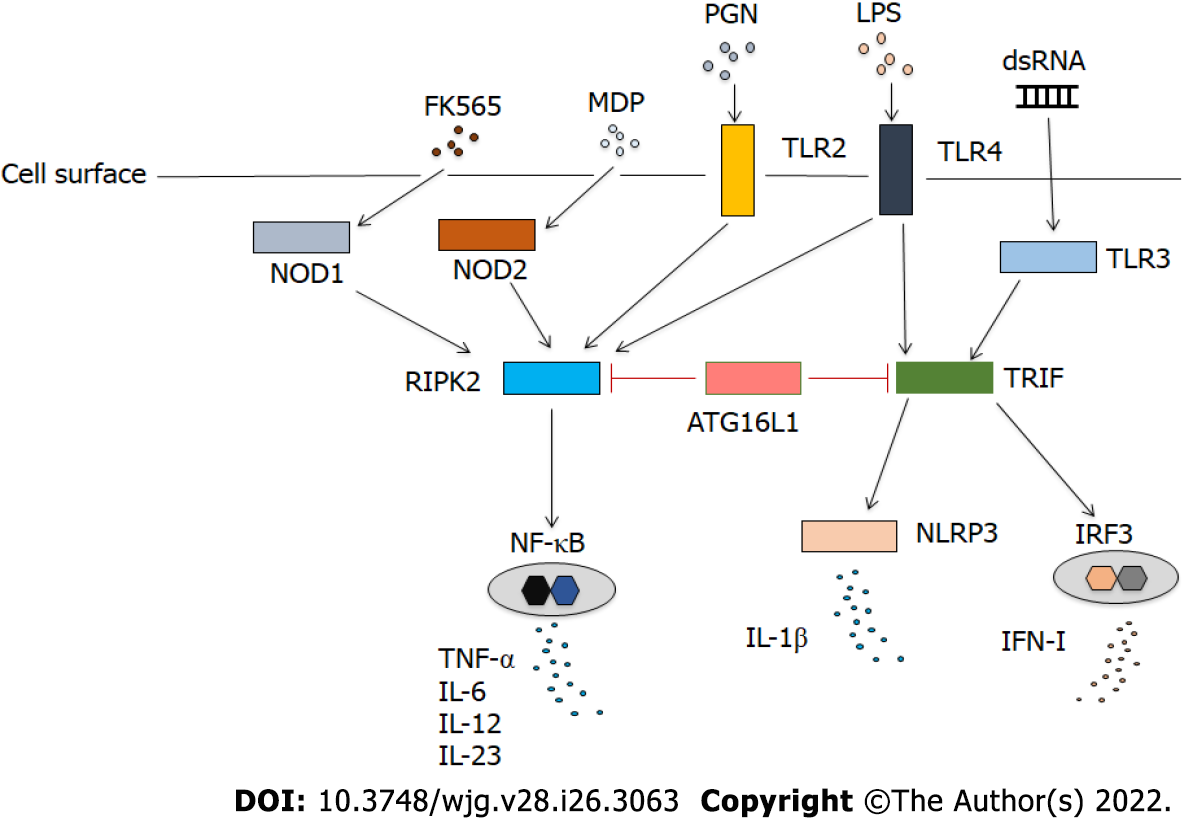
ATG16L1 has been shown to attenuate proinflammatory cytokine responses in innate immunity (Table 1)[8]. RLRs, including RIG-I and melanoma differentiation-associated gene 5 (MDA5), are sensors for RNA viruses[3]. IFN-I, which is produced after viral RNA is sensed by RLRs, plays a protective role in host defense[3]. Mouse embryonic fibroblasts deficient in ATG5 displayed enhanced production of IFN-I after exposure to vesicular stomatitis virus due to enhanced activation of IFN regulatory factor 3[11]. Enhanced production of IFN-I is associated with reduced viral load[11]. ATG16L1 is involved in the regulation of IFN-I mediated by RLRs. Two mitochondrial proteins, NLRX1 and its binding partner, Tu translation elongation factor, interact with ATG5, ATG12, and ATG16L1, and suppress RLR-induced IFN-I production and thereby promoting autophagy[12]. In addition, ATG16L1 has been shown to regulate IFN-I production by interacting with TLR3 and TLR4[13]. Samie et al[13] have provided evidence that macrophages deficient in ATG16L1 produced large amounts of IFN-I after stimulation with TLR3 and TLR4 Ligands (Figure 1). Mechanistically, the loss of ATG16L1 resulted in the accumulation of the toll-IL-1 receptor domain-containing adaptor inducing IFN-β protein (TRIF), leading to the excessive activation of TLR3- and TLR4-mediated signaling pathways. Interestingly, macrophages isolated from individuals bearing the CD-associated ATG16L1 T300A variant also exhibited enhanced IFN-I production upon stimulation with TLR3 and TLR4 Ligands[13]. Thus, ATG16L1 functions as a negative regulator of IFN-I production induced by TLR activation. Excessive activation of IFN-I signaling caused by ATG16L1 deficiency may protect against microbial infection. In fact, ATG16L1 hypomorphic mice displayed enhanced IFN-I signaling upon challenge with Citrobacter rodentium, which conferred protection from enteric pathogen infection[14]. This protection was mediated by mitochondrial antiviral signaling (MAVS) and stimulator of interferon genes (STING) proteins, because mice with hypomorphic ATG16L1 expression and deficient in MAVS or STING were not protected from the C. rodentium infection. Similarly, the clearance of Salmonella typhimurium from the intestine was augmented in mice with myeloid cell-specific ATG16L1 deficiency in an IFN-I-dependent manner[13]. IL-22 is a barrier protective cytokine that stimulates antimicrobial responses in the intestine[15]. IL-22 induces STING-dependent IFN-I signaling, which is augmented in the absence of ATG16L1[15]. Such enhanced IFN-I signaling promotes TNF-α production, leading to ileal inflammation, suggesting that ATG16L1 deficiency mediates pro-inflammatory TNF-α responses through cooperative interaction between IL-22 and IFN-I[15]. Taken together, these studies suggest that ATG16L1 dampens IFN-I production mediated by RLRs and TLRs. In turn, the lack of negative regulation of IFN-I signaling owing to the absence of ATG16L1 or the presence of ATG16L1 T300A variant mediates protection from microbial infection in the gastrointestinal tract in an IFN-I-dependent manner.
In addition to attenuating IFN-I production, ATG16L1 also suppresses IL-1β production by macrophages[16,17]. Macrophages expressing ATG16L1 that lacks the coiled-coil domain produced large amounts of IL-1β upon stimulation with lipopolysaccharide (LPS) (Figure 1)[17]. Pro-IL-1β is processed into the mature form of IL-1β by caspase-1[18]. Accumulation of TRIF is involved in enhanced IFN-I production in the absence of ATG16L1 or presence of the ATG16L1 T300A mutation[13]. Similarly, TRIF-dependent activation of caspase-1 leads to increased production of IL-1β in macrophages lacking ATG16L1[17]. In a murine model of urinary tract infection, ATG16L1 deficiency promoted clearance of uropathogenic Escherichia coli through excessive production of IL-1β[19]. Thus, ATG16L1 negatively regulates pro-inflammatory pathways mediated not only by IFN-I, but also by IL-1β.
Regulatory T cells (Tregs) expressing forkhead box P3 (FOXP3) are a specialized T cell population that is indispensable for the establishment and maintenance of immunological self-tolerance[20]. Impaired activation of Tregs leads to the development of autoimmune disorders. Bacteroides fragilis (B. fragilis) has been considered to stimulate beneficial immunoregulatory functions through induction of Tregs[21]. Chu et al[22] provided evidence that ATG16L1 expressed in DCs was required for the induction of Tregs expressing FOXP3 upon exposure to outer membrane vesicles (OMVs) of B. fragilis. Oral administration of OMVs from B. fragilis protected wild-type mice from experimental colitis[22], and this effect was accompanied by increased proportions of Tregs expressing FOXP3 and IL-10. Such protective effect of oral administration of OMVs was not seen in mice with DC-specific ATG16L1 deficiency. Thus, ATG16L1 is involved in the maintenance of immune homeostasis through induction of Tregs expressing FOXP3.
Mutations in NOD2 are the strongest risk factor for the development of CD[5,6]. MDP, a bacterial component derived from intestinal bacteria, is a prototypical NOD2 ligand[23,24]. Activation of NOD2 by MDP induces autophagy in macrophages, DCs, and fibroblasts, but not in cells harboring CD-associated NOD2 mutations[25]. Physical interaction between NOD2 and ATG16L1 is induced by the stimulation with MDP[25,26]. Thus, MDP activation of NOD2 mediates bactericidal effects in an ATG16L1-dependent manner, and the presence of CD-associated NOD2 mutations promotes overgrowth of intestinal bacteria, leading to excessive production of pro-inflammatory cytokines.
Receptor-interacting serine/threonine-protein kinase 2 (RIPK2) is a signaling molecule downstream of NOD2 and TLRs[23,24]. It remains unclear whether ATG16L1 binds to RIPK2 after activation of NOD2. In this regard, we confirmed that ATG16L1 binds to the kinase domain of RIPK2 in overexpression studies[26,27]. In human DCs, ATG16L1 interacted with RIPK2 after the stimulation with MDP and this interaction suppressed NF-κB-dependent proinflammatory responses mediated by TLRs[26,27]. Transfection of intact ATG16L1, but not of ATG16L1 with the T300A mutation, reduced TLR2-mediated NF-κB activation in human embryonic kidney cells. In the physiological setting, NF-κB activation, as assessed by the degradation of IκBα and expression of phospho-IκBα, was markedly suppressed in human DCs stimulated with TLR2 and NOD2 ligands as compared to the effect of stimulation with a TLR2 ligand alone[26,27]. Furthermore, knockdown of ATG16L1 by its specific siRNA increased IL-6 and IL-12p40 production by human DCs upon exposure to TLR2 and NOD2 ligands as compared to the levels of those cytokines in cells transfected with control siRNA[26,27]. These studies strongly suggest that ATG16L1 functions as a negative regulator of TLR2-mediated pro-inflammatory cytokine responses (Figure 1).
NF-κB activation mediated by TLRs and NOD2 is tightly regulated by Lys (K63)- linked polyubiquitination of RIPK2[23,24,27,28]. As for the molecular mechanisms accounting for the suppression of TLR2-mediated NF-κB activation and pro-inflammatory cytokine production, ATG16L1 has been shown to inhibit polyubiquitination of RIPK2[26,28]. NOD2 activation by MDP also inhibited polyubiquitination of RIPK2 through the induction of interferon regulatory factor 4 (IRF4)[23,24]. Overexpression studies revealed that ATG16L1 and IRF4 act cooperatively to suppress K63-linked polyubiquitination of RIPK2[27]. Given that physical interaction between RIPK2 and IRF4 or ATG16L1 is induced after NOD2 activation by MDP, it is likely that NOD2 downregulates TLR-mediated proinflammatory cytokine responses through binding of ATG16L1 and IRF4 to RIPK2. This idea is fully supported by the fact that RIPK2 expression level is markedly elevated in the colonic mucosa of patients with CD and ulcerative colitis (UC), and it corelates with the levels of pro-inflammatory cytokines, such as TNF-α and IL-6[29]. Furthermore, the percentages of lamina propria DCs expressing ATG16L1 and IRF4 in the colon inversely correlate with the expression levels of TNF-α and IL-6[27]. Collectively, these studies support the idea that ATG16L1 acts in concert with NOD2 to suppress excessive pro-inflammatory cytokine responses mediated by TLRs and thereby maintains intestinal homeostasis.
The polymorphism Thr300Ala (or T300A) in the coding region of the ATG16L1 gene confers increased risk for the development of CD[6,10,16]. This polymorphism is a loss-of-function mutation, which affects the induction of autophagy against invading bacteria and is associated with gut bacterial overgrowth and pro-inflammatory cytokine responses[6,10,16]. Recent studies have successfully elucidated some of the molecular mechanisms accounting for the development of CD in the presence of the ATG16L1 T300A variant. Given that ATG16L1 is constitutively expressed in epithelial cells, especially Paneth cells and myeloid cells, these studies have highlighted the importance of ATG16L1-mediated signaling pathways in innate immune cells for the immunopathogenesis of CD[17,25,28,30,31].
Paneth cells are localized at the base of the crypts in the ileum, and they contribute to the maintenance of intestinal homeostasis through the secretion of antimicrobial peptides (AMPs) and inhibition of intestinal bacterial overgrowth[32]. Mice with hypomorphic expression of ATG16L1 and ATG16L1 T300A-knockin (KI) mice exhibit increased proportions of Paneth cells with abnormal phenotypes, as assessed by lysozyme localization and granule morphology[30-32]. Moreover, Paneth cells from patients with CD carrying ATG16L1 T300A have unusual granule morphology and accumulation of AMPs, with both having been observed also in mice deficient in ATG16L1 or expressing ATG16L1 T300A[32]. Furthermore, defective function of Paneth cells in the absence of ATG16L1 or the presence of the ATG16L1 T300A mutation led to higher susceptibility to TNF-α-mediated necroptosis and accumulation of the endoplasmic reticulum stress sensor IRE1a, indicating that necroptosis and endoplasmic reticulum stress are involved in the pathogenesis of CD[33]. Thus, the ileal mucosa of patients and mice bearing ATG16L1 T300A is characterized by the defective function of Paneth cells, which results in the overgrowth of intestinal bacteria. This notion is supported by the fact that CD patients bearing the ATG16L1 T300A mutation display impaired clearance of pathogenic bacteria in the ileal mucosa[34]. It is well established that CD occurs as a result of the interplay between genetic susceptibility and environmental factors. Cigarette smoking is a risk factor for developing CD[35]. Interestingly, cigarette smoking has been suggested to amplify effects of the ATG16L1 T300A mutation, triggering Paneth cell defects, thereby causing chronic intestinal inflammation[31].
Pro-inflammatory cytokine responses play an important role in the development of CD[1]. The ATG16L1 T300A mutation has been shown to enhance pro-inflammatory cytokine responses in the intestine. Mice lacking ATG16L1 in hematopoietic cells were susceptible to dextran sodium sulfate (DSS)-induced colitis[17]. Aggravated DSS-induced colitis in mice lacking ATG16L1 was alleviated by blocking IL-1β-mediated signaling pathways[17]. Furthermore, macrophages lacking ATG16L1 produced more IL-1β upon stimulation with LPS[17]. As for the molecular mechanisms accounting for enhanced production of IL-1β in the absence of ATG16L1, Saitoh et al[17] showed that ATG16L1 deficiency resulted in increased production of this cytokine through the TRIF-dependent activation of caspase-1. Thus, ATG16L1 deficiency predisposed mice to DSS-induced colitis by activating IL-1β-mediated signaling pathways. In line with these data obtained in mice lacking ATG16L1 in hematopoietic cells, ATG16L1 T300A-KI mice displayed enhanced production of IL-1β upon exposure to LPS[16]. These studies, which utilized ATG16L1-deficient and ATG16L1 T300A-KI mice, support the idea that intact ATG16L1-medaited signaling pathways limit pro-inflammatory cytokine responses triggered by activation of TLRs. In this regard, we and others have reported that ATG16L1 negatively regulates pro-inflammatory cytokine responses mediated by RIPK2, a downstream signaling molecule of TLRs and NLRs[27,28]. Binding of ATG16L1 to the kinase domain of RIPK2 inhibits polyubiquitination of RIPK2, followed by suppression of NF-κB activation[27,28]. These studies strongly suggest that ATG16L1 activation maintains intestinal homeostasis and attenuates reactions against microbiota by inhibiting TLR-mediated pro-inflammatory cytokine responses in macrophages and DCs. Strong support for this idea also comes from the observations that colonic pro-inflammatory cytokine expression inversely correlates with the percentage of CD11c+ DCs expressing ATG16L1 in patients with CD and that induction of remission is accompanied by accumulation of CD11c+ DCs expressing ATG16L1 in the gastrointestinal tract of patients with CD[27].
ATG16L1 negatively regulates IFN-I responses mediated by RLRs and TLRs[11-14]. Isolated macrophages from patients with CD bearing the ATG16L1 T300A mutation produced more IFN-I upon stimulation with TLR3 and TLR4 ligands than macrophages from patients with intact ATG16L1[13,36]. Excessive production of IFN-I is involved in the immunopathogenesis of CD and UC. Expression levels of the IFN-stimulated genes was shown to be higher in the inflamed colonic mucosa of patients with CD or UC than in healthy controls[13]. Moreover, expression levels of IFN-stimulated genes rapidly declined in response to infliximab treatment. Although the presence of the ATG16L1 T300A variant is associated with colitogenic IFN-I responses, the enhanced production of IFN-I may improve survival of patients with colorectal cancer[36].
Similar to the molecular mechanisms of chronic inflammation in the presence of CD-associated mutations in NOD2, the ATG16L1 T300A mutation promotes the development of CD by causing impaired production of AMPs in Paneth cells and excessive secretion of TLR-mediated pro-inflammatory cytokines by macrophages and DCs. MDP activation of NOD2 induces robust production of AMPs from Paneth cells, thereby preventing bacterial overgrowth in the intestine[5]. Paneth cells deficient in NOD2 or bearing CD-associated NOD2 mutations fail to produce AMPs[5]. With regard to the pro-inflammatory cytokine responses, activation of intact NOD2 by MDP negatively regulates the production of TLR-mediated pro-inflammatory cytokines through the induction of IRF4[23,24]. In the absence of intact NOD2 or the presence of CD-associated NOD2 mutations, pro-inflammatory cytokine responses by DCs are markedly enhanced upon exposure to TLR ligands derived from the intestinal microbiota[5]. Thus, impaired function of Paneth cells and excessive pro-inflammatory cytokine responses by TLRs underlie the immunopathogenesis of CD in the presence of ATG16L1 and NOD2 mutations.
The autophagic protein ATG16L1 plays an indispensable role in the maintenance of intestinal homeostasis. The ATG16L1 T300A mutation confers an increased risk for the development of CD as it is associated with increased bacterial burden and excessive pro-inflammatory cytokine responses in the gastrointestinal tract. Elucidation of the molecular mechanisms by which the ATG16L1 T300A variant leads to the development of CD has provided new insights into the immunopathogenesis of CD induced by impaired induction of autophagy.
We appreciate Ms. Yukiko Ueno for her secretarial support.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: The Japanese Society of Gastroenterology, No. 34410.
Specialty type: Immunology
Country/Territory of origin: Japan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Huang X, China; Kaczmarek-Ryś M S-Editor: Ma YJ L-Editor: A P-Editor: Ma YJ
| 1. | Strober W, Fuss IJ. Proinflammatory cytokines in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1756-1767. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 887] [Cited by in RCA: 852] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 2. | Neurath MF. Current and emerging therapeutic targets for IBD. Nat Rev Gastroenterol Hepatol. 2017;14:269-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 315] [Cited by in RCA: 454] [Article Influence: 56.8] [Reference Citation Analysis (115)] |
| 3. | Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 681] [Cited by in RCA: 755] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 4. | Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2255] [Cited by in RCA: 2378] [Article Influence: 118.9] [Reference Citation Analysis (0)] |
| 5. | Strober W, Asano N, Fuss I, Kitani A, Watanabe T. Cellular and molecular mechanisms underlying NOD2 risk-associated polymorphisms in Crohn's disease. Immunol Rev. 2014;260:249-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 88] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 6. | Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 666] [Cited by in RCA: 688] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 7. | Klionsky DJ, Petroni G, Amaravadi RK, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cadwell K, Cecconi F, Choi AMK, Choi ME, Chu CT, Codogno P, Colombo MI, Cuervo AM, Deretic V, Dikic I, Elazar Z, Eskelinen EL, Fimia GM, Gewirtz DA, Green DR, Hansen M, Jäättelä M, Johansen T, Juhász G, Karantza V, Kraft C, Kroemer G, Ktistakis NT, Kumar S, Lopez-Otin C, Macleod KF, Madeo F, Martinez J, Meléndez A, Mizushima N, Münz C, Penninger JM, Perera RM, Piacentini M, Reggiori F, Rubinsztein DC, Ryan KM, Sadoshima J, Santambrogio L, Scorrano L, Simon HU, Simon AK, Simonsen A, Stolz A, Tavernarakis N, Tooze SA, Yoshimori T, Yuan J, Yue Z, Zhong Q, Galluzzi L, Pietrocola F. Autophagy in major human diseases. EMBO J. 2021;40:e108863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 555] [Cited by in RCA: 983] [Article Influence: 245.8] [Reference Citation Analysis (0)] |
| 8. | Hamaoui D, Subtil A. ATG16L1 functions in cell homeostasis beyond autophagy. FEBS J. 2022;289:1779-1800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 9. | Kabat AM, Pott J, Maloy KJ. The Mucosal Immune System and Its Regulation by Autophagy. Front Immunol. 2016;7:240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 71] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 10. | Salem M, Ammitzboell M, Nys K, Seidelin JB, Nielsen OH. ATG16L1: A multifunctional susceptibility factor in Crohn disease. Autophagy. 2015;11:585-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 104] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 11. | Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, Okuda K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A. 2007;104:14050-14055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 476] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 12. | Lei Y, Wen H, Yu Y, Taxman DJ, Zhang L, Widman DG, Swanson KV, Wen KW, Damania B, Moore CB, Giguère PM, Siderovski DP, Hiscott J, Razani B, Semenkovich CF, Chen X, Ting JP. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity. 2012;36:933-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 245] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 13. | Samie M, Lim J, Verschueren E, Baughman JM, Peng I, Wong A, Kwon Y, Senbabaoglu Y, Hackney JA, Keir M, Mckenzie B, Kirkpatrick DS, van Lookeren Campagne M, Murthy A. Selective autophagy of the adaptor TRIF regulates innate inflammatory signaling. Nat Immunol. 2018;19:246-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 108] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 14. | Martin PK, Marchiando A, Xu R, Rudensky E, Yeung F, Schuster SL, Kernbauer E, Cadwell K. Autophagy proteins suppress protective type I interferon signalling in response to the murine gut microbiota. Nat Microbiol. 2018;3:1131-1141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 15. | Aden K, Tran F, Ito G, Sheibani-Tezerji R, Lipinski S, Kuiper JW, Tschurtschenthaler M, Saveljeva S, Bhattacharyya J, Häsler R, Bartsch K, Luzius A, Jentzsch M, Falk-Paulsen M, Stengel ST, Welz L, Schwarzer R, Rabe B, Barchet W, Krautwald S, Hartmann G, Pasparakis M, Blumberg RS, Schreiber S, Kaser A, Rosenstiel P. ATG16L1 orchestrates interleukin-22 signaling in the intestinal epithelium via cGAS-STING. J Exp Med. 2018;215:2868-2886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 130] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 16. | Lassen KG, Kuballa P, Conway KL, Patel KK, Becker CE, Peloquin JM, Villablanca EJ, Norman JM, Liu TC, Heath RJ, Becker ML, Fagbami L, Horn H, Mercer J, Yilmaz OH, Jaffe JD, Shamji AF, Bhan AK, Carr SA, Daly MJ, Virgin HW, Schreiber SL, Stappenbeck TS, Xavier RJ. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci U S A. 2014;111:7741-7746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 292] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 17. | Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1523] [Cited by in RCA: 1664] [Article Influence: 97.9] [Reference Citation Analysis (0)] |
| 18. | Place DE, Kanneganti TD. Recent advances in inflammasome biology. Curr Opin Immunol. 2018;50:32-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 263] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 19. | Symington JW, Wang C, Twentyman J, Owusu-Boaitey N, Schwendener R, Núñez G, Schilling JD, Mysorekar IU. ATG16L1 deficiency in macrophages drives clearance of uropathogenic E. coli in an IL-1β-dependent manner. Mucosal Immunol. 2015;8:1388-1399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 20. | Kitagawa Y, Sakaguchi S. Molecular control of regulatory T cell development and function. Curr Opin Immunol. 2017;49:64-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 84] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 21. | Caruso R, Lo BC, Núñez G. Host-microbiota interactions in inflammatory bowel disease. Nat Rev Immunol. 2020;20:411-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 479] [Article Influence: 95.8] [Reference Citation Analysis (0)] |
| 22. | Chu H, Khosravi A, Kusumawardhani IP, Kwon AH, Vasconcelos AC, Cunha LD, Mayer AE, Shen Y, Wu WL, Kambal A, Targan SR, Xavier RJ, Ernst PB, Green DR, McGovern DP, Virgin HW, Mazmanian SK. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116-1120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 475] [Article Influence: 52.8] [Reference Citation Analysis (0)] |
| 23. | Watanabe T, Asano N, Murray PJ, Ozato K, Tailor P, Fuss IJ, Kitani A, Strober W. Muramyl dipeptide activation of nucleotide-binding oligomerization domain 2 protects mice from experimental colitis. J Clin Invest. 2008;118:545-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 176] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 24. | Watanabe T, Asano N, Meng G, Yamashita K, Arai Y, Sakurai T, Kudo M, Fuss IJ, Kitani A, Shimosegawa T, Chiba T, Strober W. NOD2 downregulates colonic inflammation by IRF4-mediated inhibition of K63-linked polyubiquitination of RICK and TRAF6. Mucosal Immunol. 2014;7:1312-1325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 25. | Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 760] [Cited by in RCA: 821] [Article Influence: 51.3] [Reference Citation Analysis (0)] |
| 26. | Honjo H, Watanabe T, Kamata K, Minaga K, Kudo M. RIPK2 as a New Therapeutic Target in Inflammatory Bowel Diseases. Front Pharmacol. 2021;12:650403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 56] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 27. | Honjo H, Watanabe T, Arai Y, Kamata K, Minaga K, Komeda Y, Yamashita K, Kudo M. ATG16L1 negatively regulates RICK/RIP2-mediated innate immune responses. Int Immunol. 2021;33:91-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 28. | Sorbara MT, Ellison LK, Ramjeet M, Travassos LH, Jones NL, Girardin SE, Philpott DJ. The protein ATG16L1 suppresses inflammatory cytokines induced by the intracellular sensors Nod1 and Nod2 in an autophagy-independent manner. Immunity. 2013;39:858-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 158] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 29. | Watanabe T, Minaga K, Kamata K, Sakurai T, Komeda Y, Nagai T, Kitani A, Tajima M, Fuss IJ, Kudo M, Strober W. RICK/RIP2 is a NOD2-independent nodal point of gut inflammation. Int Immunol. 2019;31:669-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 30. | Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HW 4th. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259-263. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1256] [Cited by in RCA: 1244] [Article Influence: 73.2] [Reference Citation Analysis (0)] |
| 31. | Liu TC, Kern JT, VanDussen KL, Xiong S, Kaiko GE, Wilen CB, Rajala MW, Caruso R, Holtzman MJ, Gao F, McGovern DP, Nunez G, Head RD, Stappenbeck TS. Interaction between smoking and ATG16L1T300A triggers Paneth cell defects in Crohn's disease. J Clin Invest. 2018;128:5110-5122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 32. | Stappenbeck TS, McGovern DPB. Paneth Cell Alterations in the Development and Phenotype of Crohn's Disease. Gastroenterology. 2017;152:322-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 33. | Tschurtschenthaler M, Adolph TE, Ashcroft JW, Niederreiter L, Bharti R, Saveljeva S, Bhattacharyya J, Flak MB, Shih DQ, Fuhler GM, Parkes M, Kohno K, Iwawaki T, Janneke van der Woude C, Harding HP, Smith AM, Peppelenbosch MP, Targan SR, Ron D, Rosenstiel P, Blumberg RS, Kaser A. Defective ATG16L1-mediated removal of IRE1α drives Crohn's disease-like ileitis. J Exp Med. 2017;214:401-422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 147] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 34. | Sadaghian Sadabad M, Regeling A, de Goffau MC, Blokzijl T, Weersma RK, Penders J, Faber KN, Harmsen HJ, Dijkstra G. The ATG16L1-T300A allele impairs clearance of pathosymbionts in the inflamed ileal mucosa of Crohn's disease patients. Gut. 2015;64:1546-1552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 72] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 35. | Georgiou AN, Ntritsos G, Papadimitriou N, Dimou N, Evangelou E. Cigarette Smoking, Coffee Consumption, Alcohol Intake, and Risk of Crohn's Disease and Ulcerative Colitis: A Mendelian Randomization Study. Inflamm Bowel Dis. 2021;27:162-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 36. | Grimm WA, Messer JS, Murphy SF, Nero T, Lodolce JP, Weber CR, Logsdon MF, Bartulis S, Sylvester BE, Springer A, Dougherty U, Niewold TB, Kupfer SS, Ellis N, Huo D, Bissonnette M, Boone DL. The Thr300Ala variant in ATG16L1 is associated with improved survival in human colorectal cancer and enhanced production of type I interferon. Gut. 2016;65:456-464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |