Published online Jun 14, 2022. doi: 10.3748/wjg.v28.i22.2457
Peer-review started: April 28, 2021
First decision: June 13, 2021
Revised: June 27, 2021
Accepted: April 29, 2022
Article in press: April 29, 2022
Published online: June 14, 2022
Processing time: 408 Days and 16.5 Hours
A convolutional neural network (CNN) is a deep learning algorithm based on the principle of human brain visual cortex processing and image recognition.
To automatically identify the invasion depth and origin of esophageal lesions based on a CNN.
A total of 1670 white-light images were used to train and validate the CNN system. The method proposed in this paper included the following two parts: (1) Location module, an object detection network, locating the classified main image feature regions of the image for subsequent classification tasks; and (2) Classification module, a traditional classification CNN, classifying the images cut out by the object detection network.
The CNN system proposed in this study achieved an overall accuracy of 82.49%, sensitivity of 80.23%, and specificity of 90.56%. In this study, after follow-up pathology, 726 patients were compared for endoscopic pathology. The misdiagnosis rate of endoscopic diagnosis in the lesion invasion range was approximately 9.5%; 41 patients showed no lesion invasion to the muscularis propria, but 36 of them pathologically showed invasion to the superficial muscularis propria. The patients with invasion of the tunica adventitia were all treated by surgery with an accuracy rate of 100%. For the examination of submucosal lesions, the accuracy of endoscopic ultrasonography (EUS) was approximately 99.3%. Results of this study showed that EUS had a high accuracy rate for the origin of submucosal lesions, whereas the misdiagnosis rate was slightly high in the evaluation of the invasion scope of lesions. Misdiagnosis could be due to different operating and diagnostic levels of endoscopists, unclear ultrasound probes, and unclear lesions.
This study is the first to recognize esophageal EUS images through deep learning, which can automatically identify the invasion depth and lesion origin of submucosal tumors and classify such tumors, thereby achieving good accuracy. In future studies, this method can provide guidance and help to clinical endoscopists.
Core Tip: Convolutional neural networks with self-learning abilities are an effective method in medical image classification, segmentation, and detection. Endoscopic ultrasonography plays an important role in the diagnosis and treatment of esophageal lesions. However, its operation and lesion identification are more difficult than ordinary endoscopy. Automatic identification technology is of great significance to its development.
- Citation: Liu GS, Huang PY, Wen ML, Zhuang SS, Hua J, He XP. Application of endoscopic ultrasonography for detecting esophageal lesions based on convolutional neural network. World J Gastroenterol 2022; 28(22): 2457-2467
- URL: https://www.wjgnet.com/1007-9327/full/v28/i22/2457.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i22.2457
Endoscopic ultrasonography (EUS) is a mature and widely used invasive examination technology. It is a combination of ultrasound and endoscopy, and the micro high-frequency ultrasound probe is placed on the top of the endoscope to directly observe gastrointestinal mucosal lesions when the endoscope enters the body. It can be used for preoperative staging of digestive tract tumors[1], estimating the invasion depth and range[2], assessing the nature of submucosal lesions of the digestive tract wall[3-4], and diagnosing bilio-pancreatic system tumors[5-6]. The esophagus is the narrowest part of the digestive tract, and its diameter is not uniform. EUS can be used for high-precision evaluation of the esophagus. At present, common lesions of the esophagus include submucosal tumors (including leiomyoma, stromal tumor, and lipoma), precancerous lesions, and esophageal cancer.
China has a high incidence of esophageal cancer, and the current 5-year survival rate of esophageal cancer is 15%-25%[7-10]. A 13.5-year follow-up study has shown that 74% of patients with high-grade intraepithelial neoplasia (HGIN) have developed esophageal cancer[11]. Surgical and endoscopic resection (ER) has been approved as the standard treatment for esophageal HGIN, and endoscopic therapy has become a widely used method because of its safety, efficiency, and low risk of complications[12-13]. Compared with surgical treatment, endoscopic therapy can reduce complications and preserve esophageal function[9]. EUS plays an important role in the evaluation of esophageal lesions and in the determination of the need for surgery.
With the popularization of endoscopic ultrasonography in clinic, several endoscopic physicians are needed. However, endoscopic ultrasonography is different from other ultrasound techniques in that ultrasonic examination is performed on the basis of digestive endoscopy; therefore, the examiners should be professional endoscopic physicians with ultrasonic training. In addition, with the increase of chronic diseases, ageing populations, and dwindling resources, we need to shift to models that can intelligently extract, analyze, interpret, and understand increasingly complex data. The rapidly evolving field of machine learning is a key to this paradigm shift. Artificial intelligence (AI) is a mathematical predictive technology that automatically learns and recognizes data patterns. Deep learning is an AI algorithm and an advanced machine learning method using neural networks[14]. This method aims to build a neural network that mimics brain’s mechanisms for interpreting data, such as images, sounds, and text. Deep learning combines low-level features to form abstract high-level representation attribute categories or features and discover distributed feature representation of data. It is often used in AI algorithms, and it has been applied to medical diagnosis[15-18]. At present, relevant studies on deep learning in ultrasound have been found, including the classification of thyroid nodules[19-20], the diagnosis of liver diseases[21], the identification of benign and malignant breast masses[22], the imaging interpretation of carotid artery intima media thickness (FEATURED by its length)[23], and the prediction of the pancreatic ductal papillary mucinous tumor[5]. However, the research on the identification and classification of esophageal lesions by endoscopic ultrasonography has not yet been found.
This study aimed to construct a framework of deep learning network and study the application of deep learning in esophageal EUS in identifying the origin of submucosal lesions and defining the scope of esophageal lesions.
The method proposed in this paper included the following two parts: (1) Location module, an object detection network, locating the main image feature regions of the image for subsequent classification tasks; and (2) Classification module, a traditional classification convolution neural network, classifying the images cut out by the object detection network. In the evaluation phase, all the metrics were calculated based on the ten-fold cross-validation results. The dataset was divided into the training (80%), testing (10%) and validating (10%) datasets, respectively. The detailed data statistics distribution from the esophageal lesion database is shown in Tables 1 and 2.
| Lesions invading the submucosa and muscularis mucosae | Lesions invading the muscularis propria | Lesions invading the serosal layer | |
| Train | 473 | 376 | 74 |
| Validation | 59 | 47 | 9 |
| Test | 59 | 47 | 9 |
| Total | 591 | 470 | 92 |
| Lesions from the muscularis propria | Lesions from the muscularis mucosae | |
| Train | 119 | 294 |
| Validation | 15 | 37 |
| Test | 15 | 37 |
| Total | 149 | 368 |
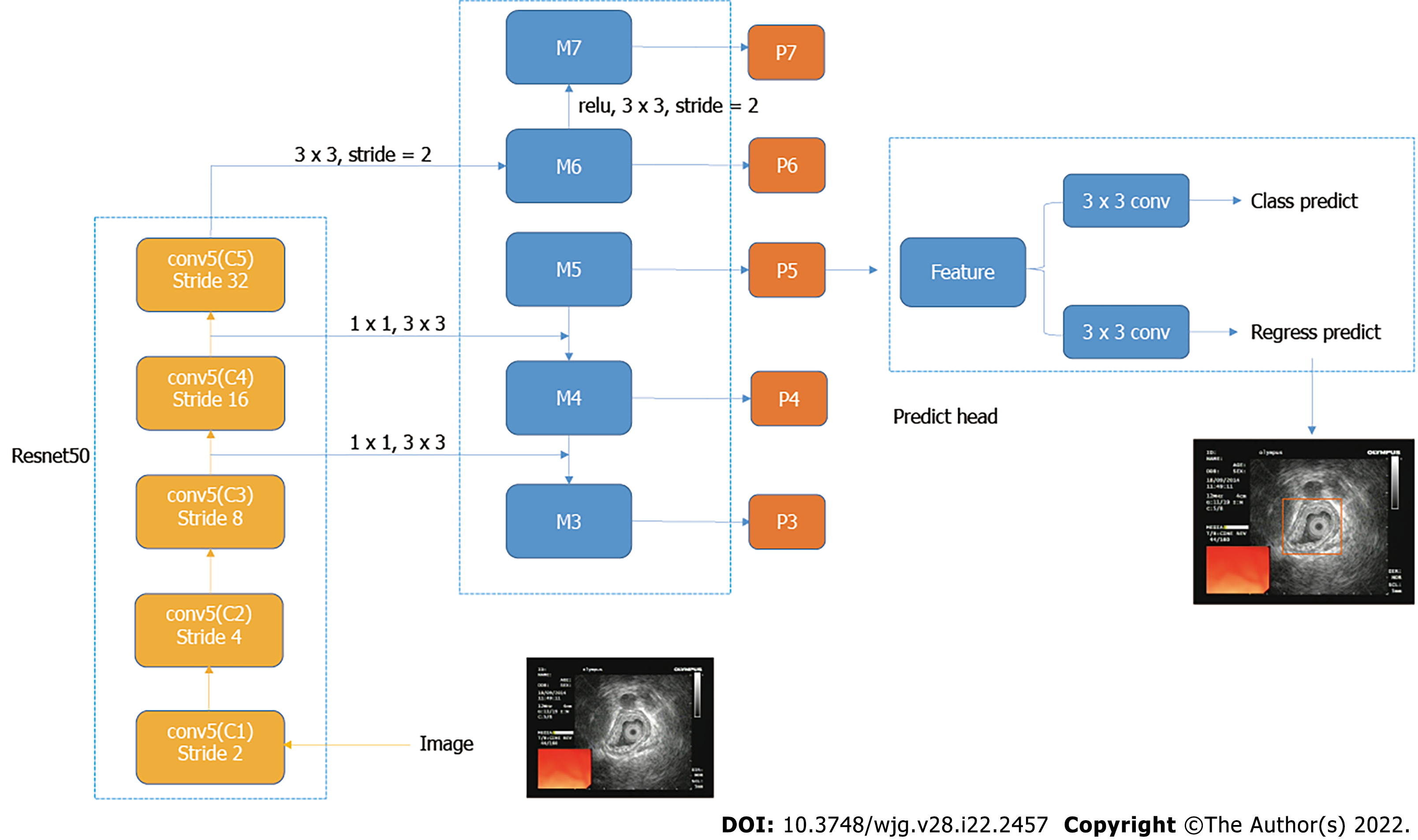
For gastric ultrasound images, identifying the classification of the whole image was not necessary because the introduction of background information can cause irrelevant features and affect the determination of the model. Therefore, the core area of the image was located, which can classify the image[24]. In this study, RetinaNet object detection network was used. RetinaNet network is a one-stage object detection method. It uses the focal loss cost function in the classification branch, which addresses the imbalance between positive and negative samples in one-stage object detection method.
Resnet50 network was used in the feature extraction part of RetinaNet network, and the FPN structure was added, that is, a top-down structure and lateral connection were adopted to detect the object of different scales. Finally, the features obtained were used to predict the classification branch and regression branch. The classification branch was retained; thus, the multitask training can improve the accuracy of the model, and it can be used to compare the experiment with the classification network (Figure 1).
For object detection anchor setting, this study adopted the default anchor setting methods, with anchor sizes of 32, 64, 128, 256, and 512 and aspect ratios of 1:2, 2:1, and 1:1.
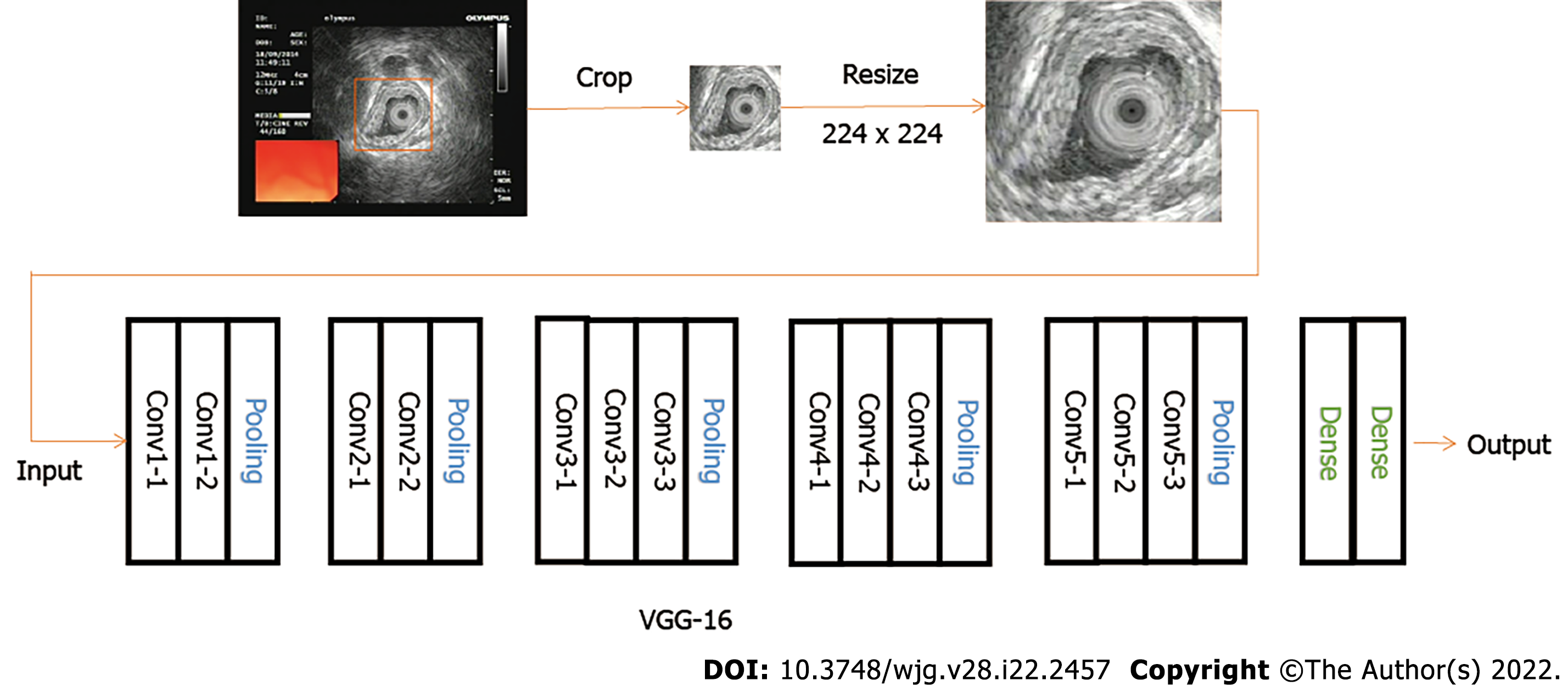
The object detection module cut out the core image block of the image and resize it to 224 × 224. Image augmentation artificially created training images through different ways of processing or combination of multiple processing, such as random rotation, shifts, shear, and flips, to address over-fitting and improve the generalization of the network (Table 3).
| Transformation type | Description |
| Rotation | Degree range for random rotations: -90°- 90° |
| Shift | Shift fraction of total height and total width: 0.1 |
| Zoom | Range for random zoom: 0.9-1.1 |
| Flip | Randomly flip image horizontally and vertically. |
| Shear | Shear intensity: 0.2 (shear angle in counter-clockwise direction in degrees) |
The classification convolution neural network used the VGGNet network. Compared with the previous network, the VGGNet network used small convolution and deeper network layers, which achieved better classification accuracy (ACC). The classification network was pre-trained on the ImageNet dataset. The last three fully connected layers of network were removed, and then two fully connected layers were added. The number of neurons was 256 and 5 (5 is the classification number of the dataset used in this paper; Figure 2).
During the experiment, GPU configuration was a NVIDIA Geforce GTX1070ti. All the codes were written in Python 3.6, and keras was used as the deep learning library.
In the object detection experiment, smooth L1 cost function was used for regression, and focal loss cost function was used for classification. Adam was used as the optimizer, and the learning rate, momentum, and weight decay were set to 1e-5, 0.9, and 0.0005, respectively. The short side of the input image was rescaled to 800, and the long side was rescaled in equal proportion, with the maximum of 1333.
In the classification experiment, the cross-entropy loss function was used. The optimizer used SGD and set the initial learning rate to 1e-3. When the loss did not decrease, the learning rate was multiplied by a factor of 0.5 until it reached 1e-6. The batch size was set to 16.
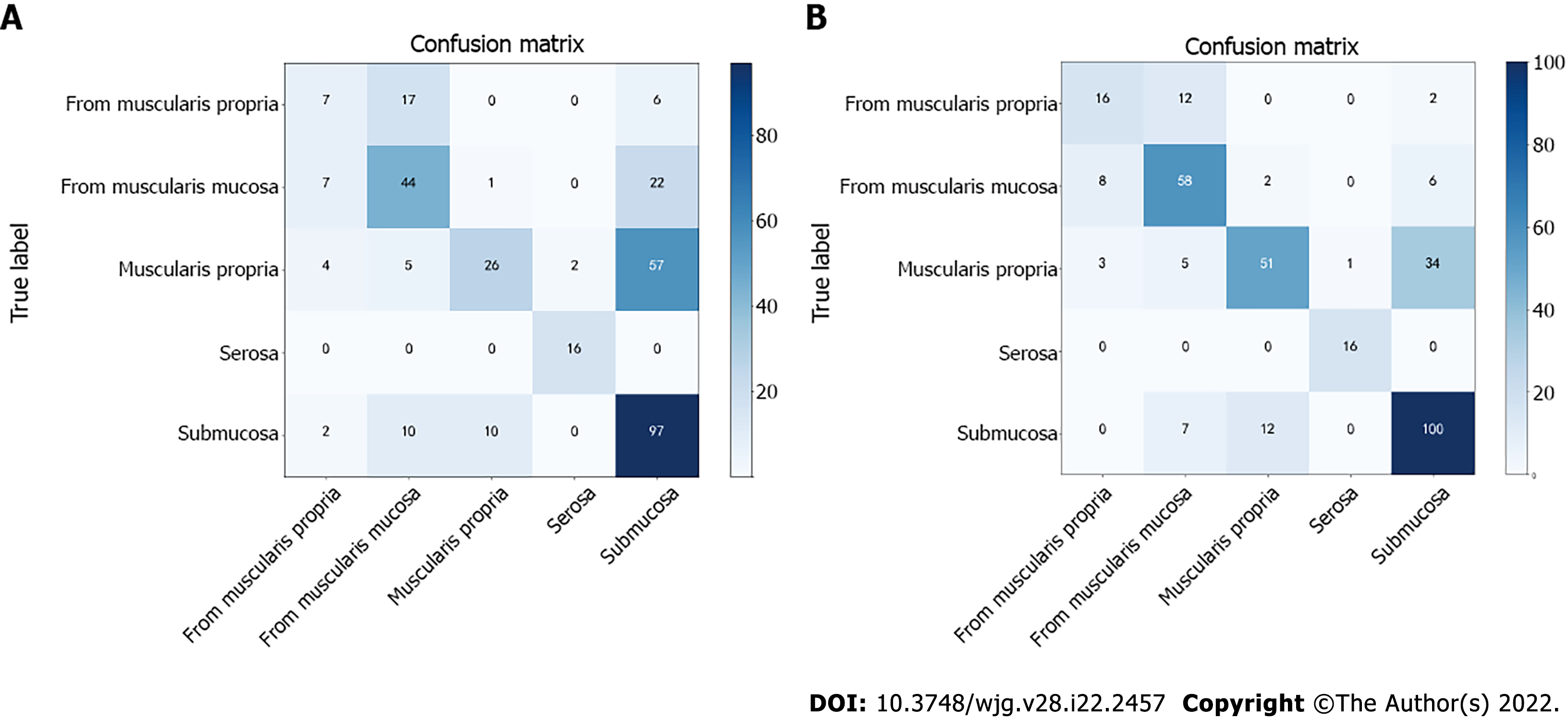
The quantitative and qualitative results of the proposed method were provided. A confusion matrix is a table that is often used to describe the performance of a classification model. Each row of the confusion matrix represents the instances of an actual class, and each column represents the instances of a predicted class. The diagonal values represent the number of correct classifications for each class, and the others represent the confusion number between every two classes. As shown in Figure 3, the ACC of using the classification network is improved compared with the classification branch of the object detection network, which is directly used for classification prediction. The classification ACC in the muscularis propria and muscularis mucosae increased significantly.
The classification performance was measured using sensitivity (SENS), specificity (SPEC), and ACC to clarify the evaluation metrics. Sensitivity measures how much a classifier can recognize positive examples. Specificity measures how much a classifier can recognize negative examples. ACC is the ratio of correct decisions made by a classifier. The sensitivity, specificity, and ACC were defined by equations (1)-(3):
SENS = TP / (TP + FN) (1)
SPEC = TN / (FP + TN) (2)
ACC = (TP + TN) / (TP + TN + FP + FN) (3)
Where TP is the amount of positive elements predicted as positive; TN is the amount of negative elements predicted as negative; FP is the amount of negative elements predicted as positive; FN is the amount of positive elements predicted as negative.
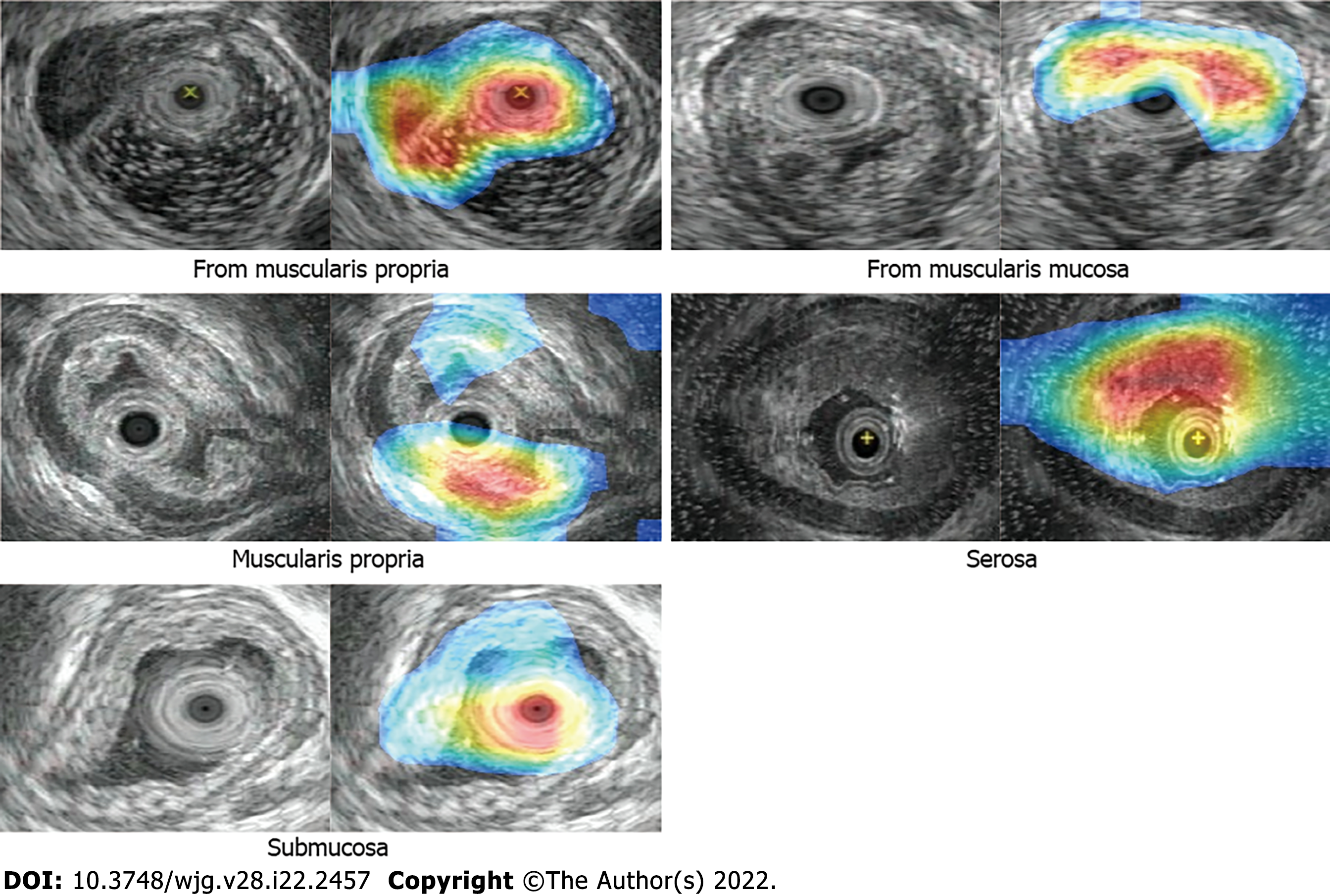
As shown in Table 4, the classification network outperforms the classification branch of the object detection network in SENS, SPEC, and ACC. Class activation maps (CAM) can be used to obtain the discriminative image regions using a convolutional neural network and identify a specific class in the image. Therefore, a CAM can identify the regions in the image that were relevant to this class. The results of CAM are shown in Figure 4.
| SENS | SPEC | ACC | |
| Object detection classify | 0.5839 | 0.8404 | 0.7861 |
| Classification network alone | 0.7400 | 0.9070 | 0.8733 |
A total of 1115 patients were included in this analysis, including 694 males and 421 females. The age ranged from 24 to 88 years, with the majority of patients aged over 45 years. In this study, patients who underwent surgery or endoscopic treatment were followed for pathology, and 726 of them were compared for ultrasound diagnosis and endoscopic pathology. In the diagnosis of endoscopic ultrasonography, lesions invading the muscularis propria were not detected in 41 patients, and the misdiagnosis rate was approximately 9.5%. In addition, 36 patients showed pathologic invasion of the superficial muscularis propria. All patients with serosal invasion were treated by surgery, and the ACC rate was 100%. For the detection of submucosal lesions by EUS, the ACC was approximately 99.3% (Table 5).
| Characteristic (n = 726) | ||
| Gender, male/female | 411/315 | |
| Age (yr), medina (range) | 54.5 (26-85) | |
| Invasion depth of esophageal lesion (n) | EUS diagnosis | Pathological diagnosis |
| Muscularis mucosa/submucosa | 188 | 147 |
| Muscularis propria | 197 | 238 |
| Serosa | 45 | 45 |
| Origin of esophageal lesions (n) | ||
| Muscularis mucosa | 148 | 150 |
| Muscularis propria | 148 | 146 |
Table 6 shows the ACC, SENS, and SPEC of each category in this study. The ACC of invasion of the serosa was 98.9%, and that of invasion of the muscularis propria and muscularis mucosa and submucosa was 73.4% and 77.11%, respectively. In submucosal lesions, the correct identification rate was 78.64% and 84.4%, respectively. The images of lesions originating from the muscularis mucosa were easily confused with the images of lesions invading the muscularis mucosa and submucosa.
| ACC | SENS | SPEC | |
| Lesions invading the submucosa and muscularis mucosae | 77.11 | 84.5 | 93.6 |
| Lesions invading the muscularis propria | 73.4 | 86.69 | 78.52 |
| Lesions invading the serosa | 98.9 | 72 | 99.14 |
| Lesions from the muscularis mucosa | 78.64 | 76.32 | 90.62 |
| Lesions from the muscularis propria | 84.4 | 81.64 | 90.92 |
| Average | 82.49 | 80.23 | 90.56 |
Traditionally, the treatment for benign and malignant esophageal lesions is mostly surgery or chemoradiotherapy, but the surgical trauma and sequelae are relatively large. With the development of gastrointestinal endoscopy in recent years, some lesions can be treated by endoscopy, such as ER, particularly submucosal dissection, which has become the standard treatment for esophageal mucosal cancer[25-27]. In Japan, ER of esophageal cancer refers to the tumor located in the mucosal epithelium (T1A-EP is equal to Tis in situ in the TNM stage[28]) or the proper mucosa (T1A-LPM) because the incidence of lymph node metastasis is low (≤ 5%)[29]. The recommended diagnostic methods are endoscopic ultrasonography[30], extended endoscopy, and narrow-band imaging (ME-NBI)[31].
Endoscopic ultrasonography plays an important role in the diagnosis and treatment of benign and malignant esophageal lesions. Xu et al[3] compared postoperative histological diagnosis with preoperative EUS diagnosis in patients with esophageal leiomyoma, and the diagnostic ACC of EUS was 88.6%. No treatment-related complications occurred in patients undergoing ER, and no recurrence was found in follow-up examination. In addition, Rana et al[32] found that small-probe EUS could predict the response to endoscopic dilation by describing the involvement degree of esophageal benign stenosis and measuring the thickness of the wall. Therefore, EUS plays an important role in the diagnosis of digestive tract diseases. In this study, after follow-up pathology, 726 of the patients were compared with endoscopic pathology. The misdiagnosis rate of endoscopic diagnosis in the lesion invasion range was approximately 9.5%; 41 patients did not show lesion invasion to the muscularis propria, but 36 of them pathologically showed invasion to the superficial muscularis propria. The patients with invasion of the tunica adventitia were all treated by surgery with an ACC rate of 100%. For the examination of submucosal lesions, the ACC of EUS was approximately 99.3%. The results of this study showed that endoscopic ultrasonography had a high ACC rate for the origin of submucosal lesions, and the misdiagnosis rate was slightly higher in the evaluation of the invasion scope of lesions. Misdiagnosis might be due to the different operating and diagnostic levels of endoscopists, unclear ultrasound probes, and unclear lesions. However, when we underestimated the depth of infiltration, the disadvantage became smaller because ER combined with additional chemoradiotherapy and surgical treatment could achieve good therapeutic effects[33-34]. However, the use of EUS is difficult for non-specialist endoscopist. Therefore, a more advanced measurement method is necessary to help non-specialists achieve diagnostic quality comparable to that of specialists without special techniques.
Deep learning is a new technique to evaluate image information objectively. In recent years, with the application of deep learning in various medical fields, remarkable progress has been made. AI is also becoming popular in ultrasonic examination. Kuwahara et al[5] predicted benign and malignant IPMNs by studying endoscopic ultrasonography of intraductal papillary mucinous tumors of the pancreas (IPMNs). The results showed that the SENS, SPEC, and ACC of AI for diagnosing malignant probability were 95.7%, 92.6%, and 94.0%, respectively, which were higher than those of human diagnosis. Although few studies have been conducted on AI in endoscopic ultrasonography, the diagnosis based on deep learning has shown its advantages. In this study, we found that deep learning algorithm could be used to clear the invasion scope and origin of lesions in endoscopic ultrasound images. In addition, VGGNet was used to perform classification tasks, and multiple superimposed filters were used to increase the nonlinearity of the whole function and reduce the number of parameters. The overall ACC, SENS, and SPEC were 82.49%, 80.23%, and 90.56%, respectively. However, in this study, the images of lesions invading the mucosal musculus and submucosa were easily confused with the images of lesions from the mucosal musculus, which may be caused by the lack of evident stratification due to tube wall compression or by the excessive noise in the ultrasound images caused by small lesions. No study has been conducted on the application of deep learning-based endoscopic ultrasonography in the esophagus or gastrointestinal tract, and the network framework applicable to endoscopic ultrasonography needs further screening and testing.
This study has some limitations. First, different probes or operators in endoscopic ultrasound images lead to uneven image quality levels, which may explain the low ACC in this study. Second, in this study, although the endoscopic ultrasonography probe could see seven layers of tube wall, 1-3 layers were classified into one category because the resolution of 1-3 layers was not clear in most images. Finally, the number of endoscopic ultrasound images was small, and all images were obtained from one center. Therefore, in the following study, we will obtain multi-center samples to increase the sample size and improve the image quality.
This study is the first to recognize esophageal endoscopic ultrasonography images through deep learning, which can automatically identify the lesion invasion depth and lesion source of submucosal tumors and classify such tumors, thereby achieving good ACC. In future studies, this method can provide guidance and help to clinical endoscopists.
The esophagus is the narrowest part of the digestive tract and its diameter is not uniform. Endoscopic ultrasonography (EUS) can be used for high-precision evaluation of the esophagus. With the popularization of endoscopic ultrasonography in clinic, more and more endoscopic physicians are needed. The rapidly evolving field of machine learning is key to this paradigm shift.
Endoscopic ultrasonography is different from other ultrasound modalities in that ultrasonic examination is performed on the basis of digestive endoscopy, so the examiners should be professional endoscopic physicians with ultrasonic training. And as we face the interplay of increasing chronic diseases, ageing populations, and dwindling resources, we need to shift to models that can intelligently extract, analyze, interpret, and understand increasingly complex data. However, the research on the identification and classification of esophageal lesions by endoscopic ultrasonography has not yet been found.
The purpose of this study was to construct a framework of deep learning network to study the application of deep learning in esophageal EUS in identifying the origin of submucosal lesions and defining the scope of esophageal lesions.
A total of 1670 white-light images were used to train and validate the convolutional neural network (CNN) system. In the study, VGGNet was used to perform classification tasks, and multiple superimposed filters were used to increase the nonlinearity of the whole function and reduce the number of parameters.
A total of 1115 patients were included in this analysis, including 694 males and 421 females. The overall accuracy, sensitivity, and specificity were 82.49%, 80.23%, and 90.56% respectively. The images of lesions originating from the muscularis mucosa were easily confused with the images of lesions invading the muscularis mucosa and submucosa.
This study constructed a CNN system which can automatically identify the lesion invasion depth and the lesion source of submucosal tumors, and classify them, achieving good accuracy.
In the future of medicine, artificial intelligence will reduce the workload of medical staff and make targeted tests more accurate, and in future studies, it can provide guidance and help to clinical endoscopists.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Popovic DD, Serbia; Zaghloul MS, Egypt S-Editor: Wu YXJ L-Editor: Wang TQ P-Editor: Wu YXJ
| 1. | Mortensen MB. Combined pretherapeutic endoscopic and laparoscopic ultrasonography may predict survival of patients with upper gastrointestinal tract cancer. Surg Endosc. 2012;26:891-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 2. | Rana SS, Sharma R, Gupta R. High-frequency miniprobe endoscopic ultrasonography for evaluation of indeterminate esophageal strictures. Ann Gastroenterol. 2018;31:680-684. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 3. | Xu GQ, Qian JJ, Chen MH, Ren GP, Chen HT. Endoscopic ultrasonography for the diagnosis and selecting treatment of esophageal leiomyoma. J Gastroenterol Hepatol. 2012;27:521-525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 4. | Sun LJ, Chen X, Dai YN, Xu CF, Ji F, Chen LH, Chen HT, Chen CX. Endoscopic Ultrasonography in the Diagnosis and Treatment Strategy Choice of Esophageal Leiomyoma. Clinics (Sao Paulo). 2017;72:197-201. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 5. | Kuwahara T, Hara K, Mizuno N, Okuno N, Matsumoto S, Obata M, Kurita Y, Koda H, Toriyama K, Onishi S, Ishihara M, Tanaka T, Tajika M, Niwa Y. Usefulness of Deep Learning Analysis for the Diagnosis of Malignancy in Intraductal Papillary Mucinous Neoplasms of the Pancreas. Clin Transl Gastroenterol. 2019;10:1-8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 127] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 6. | Sahakian AB, Aslanian HR, Mehra M, Rossi F, Laine L, Sanchez M, Ciarleglio MM, Adimoolam V, Siddiqui UD. The utility of esophagogastroduodenoscopy before endoscopic ultrasonography in patients undergoing endoscopic ultrasonography for pancreatico-biliary and mediastinal indications. J Clin Gastroenterol. 2013;47:857-860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 7. | Domper Arnal MJ, Ferrández Arenas Á, Lanas Arbeloa Á. Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol. 2015;21:7933-7943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 554] [Cited by in RCA: 748] [Article Influence: 74.8] [Reference Citation Analysis (15)] |
| 8. | Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598-5606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 736] [Cited by in RCA: 747] [Article Influence: 62.3] [Reference Citation Analysis (8)] |
| 9. | Lin Y, Totsuka Y, He Y, Kikuchi S, Qiao Y, Ueda J, Wei W, Inoue M, Tanaka H. Epidemiology of esophageal cancer in Japan and China. J Epidemiol. 2013;23:233-242. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 304] [Cited by in RCA: 429] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 10. | Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400-412. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1956] [Cited by in RCA: 1961] [Article Influence: 163.4] [Reference Citation Analysis (5)] |
| 11. | Wang GQ, Abnet CC, Shen Q, Lewin KJ, Sun XD, Roth MJ, Qiao YL, Mark SD, Dong ZW, Taylor PR, Dawsey SM. Histological precursors of oesophageal squamous cell carcinoma: results from a 13 year prospective follow up study in a high risk population. Gut. 2005;54:187-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 272] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 12. | Pech O, Bollschweiler E, Manner H, Leers J, Ell C, Hölscher AH. Comparison between endoscopic and surgical resection of mucosal esophageal adenocarcinoma in Barrett's esophagus at two high-volume centers. Ann Surg. 2011;254:67-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 213] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 13. | Das A, Singh V, Fleischer DE, Sharma VK. A comparison of endoscopic treatment and surgery in early esophageal cancer: an analysis of surveillance epidemiology and end results data. Am J Gastroenterol. 2008;103:1340-1345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 124] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 14. | LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36149] [Cited by in RCA: 20040] [Article Influence: 2004.0] [Reference Citation Analysis (0)] |
| 15. | Ting DSW, Cheung CY, Lim G, Tan GSW, Quang ND, Gan A, Hamzah H, Garcia-Franco R, San Yeo IY, Lee SY, Wong EYM, Sabanayagam C, Baskaran M, Ibrahim F, Tan NC, Finkelstein EA, Lamoureux EL, Wong IY, Bressler NM, Sivaprasad S, Varma R, Jonas JB, He MG, Cheng CY, Cheung GCM, Aung T, Hsu W, Lee ML, Wong TY. Development and Validation of a Deep Learning System for Diabetic Retinopathy and Related Eye Diseases Using Retinal Images From Multiethnic Populations With Diabetes. JAMA. 2017;318:2211-2223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1098] [Cited by in RCA: 1269] [Article Influence: 158.6] [Reference Citation Analysis (0)] |
| 16. | Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, Kim R, Raman R, Nelson PC, Mega JL, Webster DR. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA. 2016;316:2402-2410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3669] [Cited by in RCA: 3355] [Article Influence: 372.8] [Reference Citation Analysis (0)] |
| 17. | Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, van der Laak JAWM; the CAMELYON16 Consortium, Hermsen M, Manson QF, Balkenhol M, Geessink O, Stathonikos N, van Dijk MC, Bult P, Beca F, Beck AH, Wang D, Khosla A, Gargeya R, Irshad H, Zhong A, Dou Q, Li Q, Chen H, Lin HJ, Heng PA, Haß C, Bruni E, Wong Q, Halici U, Öner MÜ, Cetin-Atalay R, Berseth M, Khvatkov V, Vylegzhanin A, Kraus O, Shaban M, Rajpoot N, Awan R, Sirinukunwattana K, Qaiser T, Tsang YW, Tellez D, Annuscheit J, Hufnagl P, Valkonen M, Kartasalo K, Latonen L, Ruusuvuori P, Liimatainen K, Albarqouni S, Mungal B, George A, Demirci S, Navab N, Watanabe S, Seno S, Takenaka Y, Matsuda H, Ahmady Phoulady H, Kovalev V, Kalinovsky A, Liauchuk V, Bueno G, Fernandez-Carrobles MM, Serrano I, Deniz O, Racoceanu D, Venâncio R. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA. 2017;318:2199-2210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1871] [Cited by in RCA: 1557] [Article Influence: 194.6] [Reference Citation Analysis (0)] |
| 18. | Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5683] [Cited by in RCA: 5356] [Article Influence: 669.5] [Reference Citation Analysis (0)] |
| 19. | Chi J, Walia E, Babyn P, Wang J, Groot G, Eramian M. Thyroid Nodule Classification in Ultrasound Images by Fine-Tuning Deep Convolutional Neural Network. J Digit Imaging. 2017;30:477-486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 195] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 20. | Seo JK, Kim YJ, Kim KG, Shin I, Shin JH, Kwak JY. Differentiation of the Follicular Neoplasm on the Gray-Scale US by Image Selection Subsampling along with the Marginal Outline Using Convolutional Neural Network. Biomed Res Int. 2017;2017:3098293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 21. | Nishida N, Yamakawa M, Shiina T, Kudo M. Current status and perspectives for computer-aided ultrasonic diagnosis of liver lesions using deep learning technology. Hepatol Int. 2019;13:416-421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 22. | Choi JS, Han BK, Ko ES, Bae JM, Ko EY, Song SH, Kwon MR, Shin JH, Hahn SY. Effect of a Deep Learning Framework-Based Computer-Aided Diagnosis System on the Diagnostic Performance of Radiologists in Differentiating between Malignant and Benign Masses on Breast Ultrasonography. Korean J Radiol. 2019;20:749-758. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 23. | Zhou Z, Shin J, Feng R, Hurst RT, Kendall CB, Liang J. Integrating Active Learning and Transfer Learning for Carotid Intima-Media Thickness Video Interpretation. J Digit Imaging. 2019;32:290-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 24. | Al Jassim RA, Andrews FM. The bacterial community of the horse gastrointestinal tract and its relation to fermentative acidosis, laminitis, colic, and stomach ulcers. Vet Clin North Am Equine Pract. 2009;25:199-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 83] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 25. | Takahashi H, Arimura Y, Masao H, Okahara S, Tanuma T, Kodaira J, Kagaya H, Shimizu Y, Hokari K, Tsukagoshi H, Shinomura Y, Fujita M. Endoscopic submucosal dissection is superior to conventional endoscopic resection as a curative treatment for early squamous cell carcinoma of the esophagus (with video). Gastrointest Endosc. 2010;72:255-264, 264.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 243] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 26. | Repici A, Hassan C, Carlino A, Pagano N, Zullo A, Rando G, Strangio G, Romeo F, Nicita R, Rosati R, Malesci A. Endoscopic submucosal dissection in patients with early esophageal squamous cell carcinoma: results from a prospective Western series. Gastrointest Endosc. 2010;71:715-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 140] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 27. | Ishihara R, Iishi H, Uedo N, Takeuchi Y, Yamamoto S, Yamada T, Masuda E, Higashino K, Kato M, Narahara H, Tatsuta M. Comparison of EMR and endoscopic submucosal dissection for en bloc resection of early esophageal cancers in Japan. Gastrointest Endosc. 2008;68:1066-1072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 231] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 28. | Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J Clin. 2017;67:93-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2341] [Cited by in RCA: 4401] [Article Influence: 550.1] [Reference Citation Analysis (4)] |
| 29. | Kuwano H, Nishimura Y, Oyama T, Kato H, Kitagawa Y, Kusano M, Shimada H, Takiuchi H, Toh Y, Doki Y, Naomoto Y, Matsubara H, Miyazaki T, Muto M, Yanagisawa A. Guidelines for Diagnosis and Treatment of Carcinoma of the Esophagus April 2012 edited by the Japan Esophageal Society. Esophagus. 2015;12:1-30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 312] [Cited by in RCA: 345] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 30. | Oyama T, Inoue H, Arima M, Momma K, Omori T, Ishihara R, Hirasawa D, Takeuchi M, Tomori A, Goda K. Prediction of the invasion depth of superficial squamous cell carcinoma based on microvessel morphology: magnifying endoscopic classification of the Japan Esophageal Society. Esophagus. 2017;14:105-112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 223] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 31. | Esaki M, Matsumoto T, Moriyama T, Hizawa K, Ohji Y, Nakamura S, Hirakawa K, Hirahashi M, Yao T, Iida M. Probe EUS for the diagnosis of invasion depth in superficial esophageal cancer: a comparison between a jelly-filled method and a water-filled balloon method. Gastrointest Endosc. 2006;63:389-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 32. | Rana SS, Sharma R, Kishore K, Gupta R. High-frequency miniprobe endoscopic ultrasonography in the management of benign esophageal strictures. Ann Gastroenterol. 2020;33:25-29. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 33. | Shimizu Y, Kato M, Yamamoto J, Nakagawa S, Tsukagoshi H, Fujita M, Hosokawa M, Asaka M. EMR combined with chemoradiotherapy: a novel treatment for superficial esophageal squamous-cell carcinoma. Gastrointest Endosc. 2004;59:199-204. [RCA] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 53] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 34. | Saeki H, Watanabe M, Mine S, Shigaki H, Oya S, Ishiyama A, Tsuchida T, Fujisaki J, Baba H, Maehara Y, Sano T. Esophagectomy for superficial esophageal cancer after non-curative endoscopic resection. J Gastroenterol. 2015;50:406-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |