Published online Jun 14, 2022. doi: 10.3748/wjg.v28.i22.2417
Peer-review started: February 16, 2022
First decision: March 9, 2022
Revised: March 22, 2022
Accepted: April 15, 2022
Article in press: April 15, 2022
Published online: June 14, 2022
Processing time: 113 Days and 12.1 Hours
Chronic hepatitis C virus (HCV) infection is the principal etiology of cirrhosis and, ultimately, hepatocellular carcinoma (HCC). At present, approximately 71 million people are chronically infected with HCV, and 10%–20% of these are expected to develop severe liver complications throughout their lifetime. Scientific evidence has clearly shown the causal association between miRNAs, HCV infection and HCC. Although it is not completely clear whether miRNA dysregulation in HCC is the cause or the consequence of its development, variations in miRNA patterns have been described in different liver diseases, including HCC. Many studies have analyzed the importance of circulating miRNAs and their effect on cell proliferation and apoptosis. In this Review, we aim to summarize current knowledge on the association between miRNA, HCV and HCC from a diagnostic point of view, and also the potential implications for therapeutic approaches.
Core tip: Hepatocellular carcinoma (HCC) is one of the most life-threatening cancers worldwide. Among other factors, HCC is frequently caused by chronic hepatitis C virus infection. Currently, the mechanisms responsible for its neoplastic transformation are not completely understood. Regardless of the recent discovery and therapeutic use of latest-generation direct-acting antivirals, HCC remains an unresolved problem, even after viral eradication. In this scenario, miRNAs can become both a diagnostic tool and, hopefully, a novel therapeutic approach.
- Citation: Badami E, Busà R, Douradinha B, Russelli G, Miceli V, Gallo A, Zito G, Conaldi PG, Iannolo G. Hepatocellular carcinoma, hepatitis C virus infection and miRNA involvement: Perspectives for new therapeutic approaches. World J Gastroenterol 2022; 28(22): 2417-2428
- URL: https://www.wjgnet.com/1007-9327/full/v28/i22/2417.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i22.2417
Chronic hepatitis C virus (HCV) infection is a well-known risk factor for hepatocellular carcinoma (HCC)[1,2]. The virus belongs to the Flaviviridae family, and is the only member of the hepacivirus genus[3-6], and is presently classified in seven main genotypes, although 11 genotypes and at least 67 confirmed subtypes are known[7-10].
HCV is a single-stranded positive-sense RNA virus consisting of an icosahedral symmetrical nucleocapsid, surrounded by a double-layer lipid envelope in which the envelope glycoproteins E1 and E2 are inserted[6]. Its genome of 9.6 kb harbors an open reading frame encoding a polyprotein precursor of about 3000 amino acids, which is processed through proteolytic cleavage by viral and host proteases into smaller molecules, including structural (core, E1 and E2), p7, and nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A and NS5B)[11].
The total global prevalence of chronic HCV infection is estimated at 71 million people by the World Health Organization (WHO)[12], and varies according to health conditions, sociodemographic characteristics, and the presence of risk factors for transmission that can alter the efficiency of the transmission routes[9,13]. HCV is transmitted primarily parenterally, through large or repeated direct percutaneous exposures to infected blood[9,13-15]. Major high-risk populations include people who inject drugs (PWIDs), men who have sex with men, and prisoners[16].
The outcome of HCV infection depends strictly on the strength and breadth of the host response during the acute phase. Around 30% of HCV-infected people spontaneously clear the virus within 6 mo of infection, although the remaining 60%–80% of people, despite developing an efficient antiviral immune response, are unable to clear the virus, resulting in persistent infection[17,18]. Activation of the immune response against HCV contributes importantly to the establishment of long-lasting inflammation and consequent liver damage. The deregulation of cytotoxic cells and the continued activation of an apoptosis pathway results in scarring and progression to cirrhosis that, in 25% of HCV infected patients, can result in HCC over a period of 20–30 years post-infection[2,19,20].
Until the last decade, interferon (IFN) and ribavirin (RBV) were the only available therapies against chronic HCV infection, although they were accompanied by significant side effects. Moreover, IFN-based therapies had only limited efficacy, as the response was genotype dependent[21-24]. The recent introduction of direct-acting antivirals (DAAs) has resulted in remarkable therapeutic improvements. These are currently the standard therapeutic choice, and a sustained virology response (SVR) > 90% is attained after 12 wk of treatment. However, some patients show relapse of HCV infection, even after DAA treatment, and achieving SVR does not completely rule out the risk of developing HCC[25]. Also, following DAA treatment and subsequent viral clearance, HCV reinfections remain a problem among individuals with high-risk behaviors, e.g., PWIDs[26,27].
Moreover, access to DAA therapy is not simple for all patients, and only the 62% of HCV-diagnosed patients have been treated with DAAs (WHO) (https://www.who.int/news-room/fact-sheets/detail/hepatitis-c).
Although the pathogenesis of HCV infection has not been fully elucidated, interactions between structural and nonstructural viral proteins and host cell components, such as miRNAs, have been reported by many groups. It has been shown that host miRNAs are involved in many steps of the biological cycle of HCV, such as infection and replication. Likewise, HCV infection regulates the expression of many cellular miRNAs involved, for example in liver fibrosis, hepatocarcinogenesis and HCC progression[28-31]. Due to such mutual interactions, miRNAs can be used for risk assessment and prognosis of HCV-related HCC, and could be considered for diagnostic approaches and new therapeutic strategies.
HCC is the main type of liver cancer, and the most life-threatening cancer worldwide. HCC onset consists of several processes involving multiple risk factors, but most often it presents in people with chronic liver diseases and cirrhosis[32].
The most common etiological factors that lead to liver cirrhosis, therefore predisposing to HCC transformation, are chronic infection with hepatitis B virus (HBV), HCV, or hepatitis D virus, alcoholic liver disease, and nonalcoholic steatohepatitis/nonalcoholic fatty liver disease (NAFLD). The less common causes are hereditary hemochromatosis, 1 antitrypsin deficiency, autoimmune hepatitis, porphyria, disorders of steroid hormones, Wilson’s disease, and dietary aflatoxins[33-36].
Approximately 50% of all liver cancers are related to chronic viral hepatitis, which can develop into cirrhosis and HCC[37]. In particular, HCV causes chronic infections in 70%–80% of cases, while HBV leads to chronicity in only 10% of infected people[38]. Among HCV chronically infected patients, about 20% develop liver cirrhosis within 20–30 years and, once cirrhosis is established, the rate of HCC development increases by 1%–4% per year. Moreover, chronic HCV infection is associated with a 20–30-fold increased risk of developing into HCC compared to healthy individuals[39]. HCV-related HCC is mediated both by virus-related factors and host-induced immunological responses. In addition, HCV damage is a gradual and continual process, characterized by recurrent infection that induces the immune system to attack liver cells, provoking repeated damage to the genomic material that can lead to mistakes during proofreading repair[38]. Recent studies suggest that HCV core protein can promote initial development of HCC, acting on cell signaling pathways. Indeed, HCV core protein may directly inhibit the tumor suppressor genes and the cell cycle checkpoints, inducing the activation of signaling pathways that upregulate growth and cell division[40]. The specific tumor suppressor genes inhibited by HCV core protein include retinoblastoma protein and p53 tumor suppressor, which, if synergistically lost, lead to a higher degree of carcinogenesis[41]. Repeated cell cycles are associated with the accumulation of mutations that may transform hepatocytes into cancer cells. Among the genes most mutated are telomerase reverse transcriptase, tumor protein 53, and β-catenin. These mutations not only threaten telomere maintenance, but also lead to increased oxidative stress on hepatocytes, inducing chronic inflammation secondary to HCV, thus promoting HCC progression[42].
Over the last few years, in an attempt to identify new therapies and more accurate biomarkers for early diagnosis and treatment of HCC, the pivotal role played by miRNAs in the development and progression of cancer has emerged[43].
Recently, progress has been made in the study of miRNAs in HCC, with the discovery that some of them are upregulated or downregulated in HCC. Aberrant expression of miRNAs has been linked to HCC proliferation, apoptosis and invasion, but also metastasis formation, epithelial–mesenchymal transition, angiogenesis, drug resistance, and autophagy. In addition, some miRNAs can also be potential diagnostic and prediction markers for HCC[44].
The principal goal of anti-HCV therapy is to eradicate HCV infection, thus resolving the liver disease. In the past, the standard of care for eradication of chronic HCV treatment was IFN-based therapy; most commonly in combination with RBV. However, this combination therapy had an outcome that depended on the viral genotype, was poorly tolerated, and also associated with severe side effects such as depression, flu-like symptoms, fatigue, diarrhea, and hematological toxicity[45]. These unwanted adverse effects were the main reason for patients abandoning therapy. With the introduction of the first NS5B polymerase inhibitor, sofosbuvir, in 2014, IFN-free treatment became available[46]. Hence, DAAs were introduced in clinical practice to treat HCV+ patients, obtaining SVR rates above 90%[47,48], where SVR corresponds to a cure of the HCV infection, as late relapse occurs in less than 0.2% of cases beyond 6 mo of follow-up[49]. While patients with cirrhosis are often not treatable with IFN-based treatment, DAAs, including NS5B and NS5A inhibitors, are effective for patients with any stage of liver fibrosis, including those with advanced liver disease and decompensated cirrhosis, which predispose to developing HCC at a higher degree. This has allowed a significant increase in the number of treatable patients, with excellent safety profiles[50].
Clinically, SVR leads to stabilization and improvement of the liver function, and is thus associated with a lower risk of hepatic dysfunction, a reduced need for liver transplantation, and lower overall mortality. It has been demonstrated that DAA therapy attains much higher SVR rates than IFN-based treatment by reducing the inflammatory cargo in the liver, fibrosis, and neoplastic formation[51].
With more successful SVR obtained using DAAs in the clinic, there were higher expectations in terms of a significant reduction in the incidence of HCC. However, such expectations were not always met. In fact, despite the documented positive impact on HCV infection clearance, the effect of DAA treatment on the development and/or recurrence of HCC in patients treated for liver cirrhosis remains hard to define[52].
In a meta-analysis, Frazzoni et al[53] compared different trials based either on the use of DAAs or IFN[53]. The aim of this study was to define the long-term incidence and occurrence of HCC after the achievement of SVR using either IFN-free or IFN-based therapies. This investigation showed the higher safety profile of DAA therapy, accompanied by a lower risk of HCC recurrence[53]. A controversial subject is the effect of DAA-based SVR on the recurrence of HCC following curative treatment of early HCC. It has also been found that about 30% of patients with a history of curative treatment for HCC undergoing DAA therapy for HCV infection develop HCC recurrence, raising many surgical and clinical questions[54].
Some studies initially suggested that DAA treatment might increase the recurrence rate for HCC, with consequent doubts on the use of this therapy for patients with previous HCC[55]. However, these studies were not confirmed by further trials, and new guidelines were provided recommending the use of DAAs for patients with a history of HCC[56]. At present, patients with HCV-related cirrhosis who have undergone resection or ablation for HCC should not be dissuaded from receiving DAA therapy to prevent the progression of the liver disease.
The putative correlation between DAA treatment and de novo HCC occurrence is still a matter of debate. Some studies have shown how HCV+ patients diagnosed as negative for HCC develop aggressive and fatal forms of liver cancer after DAA treatment[57-61]. These studies highlighted the importance of the surveillance of patients before and during DAA treatment, pointing to a possible correlation between the stage of fibrosis and the pre-existence of decompensated cirrhosis, with increased susceptibility to the development of de novo HCC[58,61,62].To conclude, patients failing to respond to antiviral treatment are at high risk and often need to be monitored during therapy[63]. In light of these observations, it appears clear how the use of biomarkers such as HCV/HCC/liver-related miRNA are important for early HCC diagnosis before and during DAA treatment.
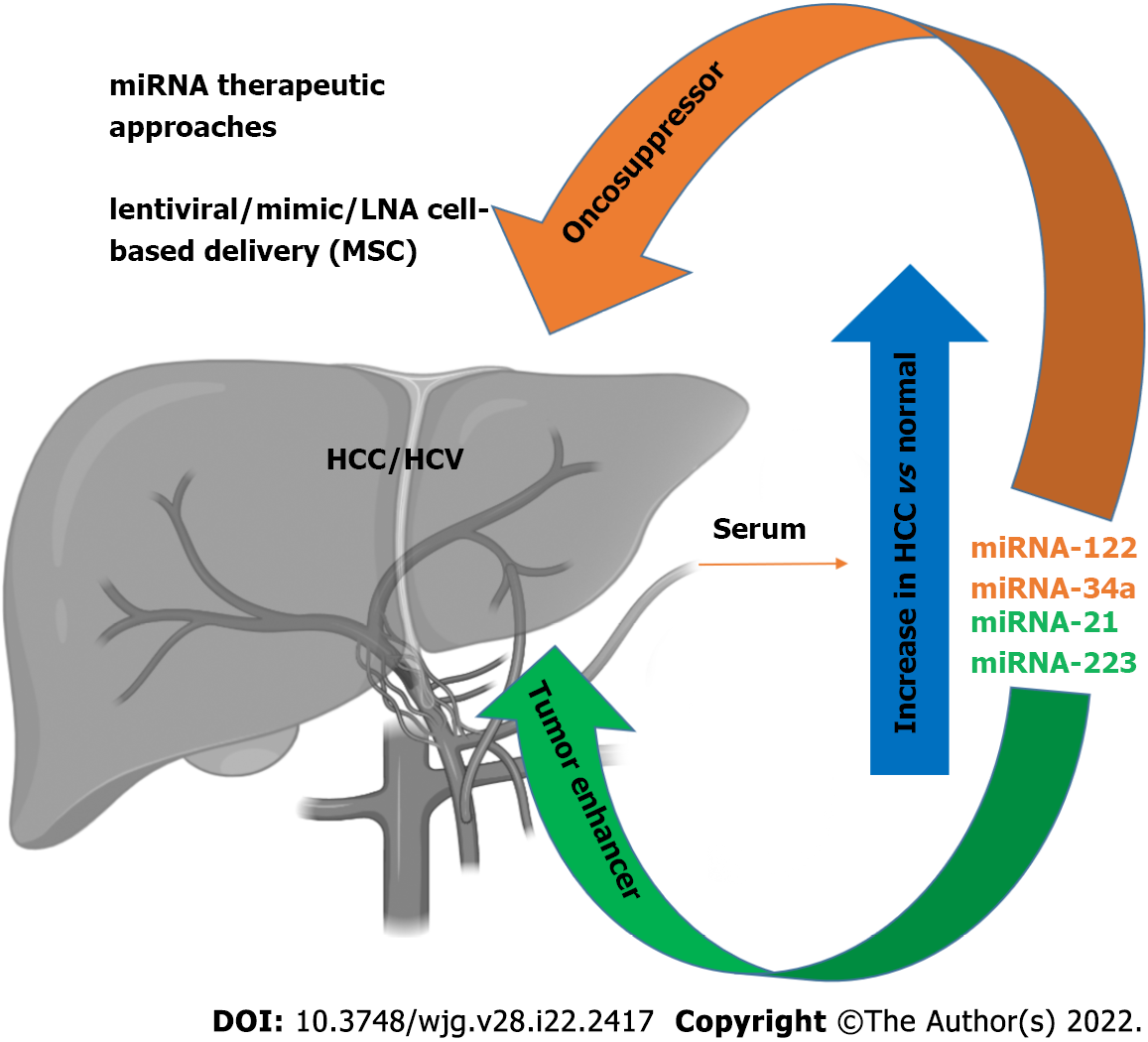
As discussed above, miRNAs are active players in tumor initiation and progression for their involvement in the regulation of expression for proteins implicated in the pathophysiological mechanisms of cancer development, including cell growth, apoptosis, and metastasis[64-66]. In the liver, it has been shown that liver-enriched transcription factors such as HNF1A, HNF3A and HNF3B regulate miR-122 expression, which can function as a tumor suppressor factor by inhibiting angiogenesis, HCC growth/invasion, and high levels of HCC apoptosis and cell cycle arrest[67,68]. This miRNA represents 70% of the total miRNAs in hepatocytes[69,70], and has been therefore nicknamed hepatic miRNA. It has been found that HCV RNA possesses two tandem miR-122 complementary sites at the 5’ end of the viral genome[71], responsible for viral RNA accumulation[72]. The miR-122 binding with the HCV RNA has been described at multiple levels: miR-122 affects folding of the viral internal ribosomal entry site (IRES)[73], required for viral translation, and modulates viral replication and polyprotein translation[74]. In addition, it has been found that the stability of HCV RNA is mediated by RNA-induced silencing-like complex (RISC-like), suggesting the coordination of Ago2 and miR-122 in stabilizing and protecting the viral genome from 5′ exonuclease activity[75]. Furthermore, HCV RNA acts as a sponge for miR-122, leading to de-repression of host mRNAs normally targeted by miR-122, thus providing a fertile environment for the long-term oncogenic potential of HCV[76]. The serum levels of miR-122 are clearly higher in HCV-infected patients with HCC[77-79]. During the course of HCV infection, miR-122 is coupled with miR-34a[80] (Figure 1). While several studies have reported a downregulation of miR-34a in neoplastic transformation in various tissues[81], miR-34a was found increased in HCC[82]. Moreover, we recently reported an increase of miR-34a in HCC cells after HCV infection, the effect of which can be modulated by p53 induction[31]. It has recently been proposed that flaviviruses can upregulate expression of miRNAs that inhibit viral replication in target cells[31], demonstrating that miR34 overexpression induces the IFN-mediated response in dengue virus, West Nile virus and Japanese encephalitis virus infection[83]. In HCC cells, miR-34a induces cell cycle arrest and apoptosis. Concomitantly, other groups have also demonstrated that miR-34 inhibits fibrosis in stellate cells by regulating the TGF-β1/Smad3 pathway[84].
Expression of miR-21 increases in patients with HCV-related HCC[85], while miR-223 expression is increased in patients with advanced fibrosis compared to moderate/minimal fibrosis[86]. In contrast, during HCV-related chronic hepatitis, miR-21 and miR-223 display a pro-fibrotic/tumorigenic effect[85] (Figure 1).
Depending on the role played in viral infection, some miRNAs have been classified as pro- or antiviral. For example, miR-135a has been described as proviral miRNA because of its ability to enhance HCV RNA replication in hepatocytes. In addition, miR-135a inhibits the expression of host restriction factors, such as CXCL2, MyD88 and IRPK2, which are involved in antiviral immunity[87]. Likewise, miR-146a-5p enhances HCV infection by playing an immunoregulatory role through the downregulation of the inflammatory signaling, and by turning off the immune response in hepatocytes. Moreover, miR-146a-5p promotes the late steps of the HCV replication cycle, likely by modulating HCV assembly[88]. Another example is represented by miR-21-5p, which triggers HCC growth and metastasis by modulating a PTEN-dependent pathway[89].
Conversely, let-7 family miRNAs have demonstrated strong anti-HCV activity, thus being classified as antiviral miRNA[90]. It has been found that let-7b potentially reduces HCV replication through the targeting of IGF2BP1 required for HCV replication[90], and increases the cell apoptosis rate[91]. Similar to other antiviral miRNAs, mir-199a* is a HCV RNA binder in IRES, targeting the HCV 5′-UTR[64,92].
The mechanisms that drive liver injury during HCV infection have not been fully elucidated. Several studies have attempted to understand the interactions between viral proteins and the cellular host machinery[93]. An important effort has been made to address and understand the complex interconnections between HCV infection and miRNAs, aimed at finding new therapeutic and diagnostic tools.
It has recently been shown that the expression of specific patterns of miRNAs in HCC patients may be used for diagnostic purposes, with a sufficient level of reliability, thus highlighting a potential role of miRNAs as biomarkers[94]. Specific circulating miRNA profiles are associated with several diseases, including HCV infection. These miRNAs have been proposed as biomarkers of HCV-related HCC pathophysiology and prognosis. Many studies have focused on setting miRNA panels to be tested in HCV-related HCC patients, and used to discriminate healthy individuals from ill patients; above all, during the early phases of disease. For example, Ali et al[95] described and validated a panel of nine liver-associated miRNAs (miR-21, miR-30c, miR-93, miR-122, miR-125b, miR-126, miR-130a, miR-193b and miR-222)[95], while Zekri et al[96] identified miR-122, miR-885-5p and miR-29b as associated with fluctuations in the levels of the diagnostic liver-specific biomarker -fetoprotein (AFP)[96]. Wahb et al[97] suggested the use of circulating miR-9-3p and endocan as novel biomarkers for risk assessment in the early diagnosis of HCV-related HCC[97]. It has been shown that circulating extracellular vesicles (EVs) containing specific patterns of noncoding RNAs are highly related to disease progression of HCV-associated HCC[98]. Recent reports have highlighted the importance of serum miR-34a and miR-122 as biomarkers of HCV-related HCC[80]. We recently showed that HCV infection induces an increase in miR-34a expression in the HCC cell line HuH7.5, and acts as a tumor suppressor[31]. We also found that HCV infection induces the release of EVs containing miR-34a, and these EVs can exert a growth inhibition effect and induce apoptosis[31]. We were able to demonstrate that miR-34 action is not limited only to the infected cells, as EV release has a paracrine effect on neighboring hepatocytes, thus underscoring the potential use of this miRNA for therapeutic purposes[31] (Figure 1). A newly generated polymer-based nanosystem redox-sensitive, oligopeptide-guided, self-assembling, and efficiency-enhanced (ROSE) loaded with miR-34a inhibits HCC proliferation in vitro and in vivo, reducing the epithelial mesenchymal transition[99].
Conversely, it has been demonstrated that miR-21 can act as an oncogene by stimulating HCC growth, invasion, and migration[89,100]. Indeed, it has been shown that the inhibition of miR-21 suppresses HCC growth both in vitro and in vivo[100].
It is worth mentioning that miR-122 is known to potentiate HCV replication by inhibiting the degradation of the viral genome[70]. Several clinical trials are testing the ability of reducing HCV replication using locked nucleic acid inhibition of miR-122[101-103], perhaps evidence for the efficacy of this treatment, which is devoid of adverse effects[64]. For example, miR-122 antagonists can be used as a therapeutic approach in synergistic association with DAA therapy to obtain an enhancement of the clinical outcome. These results point out the potential therapeutic interest of miR-122, in particular, for patients who do not respond to antiviral agents[104]. Recently, the use of EVs to treat HCC has become an interesting topic, but one that requires further studies. Lou et al[105] revealed that the injection of EVs derived from miR-122-modified mesenchymal stem cells (MSCs) can significantly improve chemotherapeutic sensitivity of HCC, and increase the efficacy of sorafenib treatment[105] (Figure 1). Wei et al[106] found that there are different miRNA expression patterns between HCC cells and their EVs, suggesting a self-modulating mechanism whereby HCC-cell-derived EVs were able to shuttle miRNAs to recipient cells, promote cell growth, and migration and invasion of HCC cells[106]. EVs contain both oncogenic and tumor suppressor miRNAs, and their deregulated expression in HCC tissues can promote HCC development. Therefore, EV-mediated miRNA transfer might represent a crucial mechanism, exploitable on the one hand as a diagnostic tool by identifying circulating EV-derived miRNA, and, on the other, EV-derived miRNAs could be a useful target to inhibit HCC growth. In agreement, Zhang et al[107] generated virus-like vectors containing both miRNA-21-sponge and pre-miRNA-122, and demonstrated that virus-like particles were able to correct HCC miRNA dysregulation, and to decrease proliferation, migration, and invasion of HCC[107].
Currently, 20 clinical trials (Table 1) are investigating both tissue-specific and circulating miRNAs as a diagnostic/prognostic tool for HCC or for monitoring HCC during a specific treatment (clinicaltrial.gov). Future clinical trials will be needed to provide appropriate analysis of the performance of miRNAs as biomarkers, and to discover new miRNA-related HCC targets for implementation of new treatments.
| Study title | Phase | Status | NCT number |
| MicroRNAs as diagnostic biomarkers in hepatocellular carcinoma among Somali patients | NA | Unknown | NCT03227510 |
| Prospective cohort study of changes in circulatory microRNA of resected hepatocellular carcinoma | NA | Recruiting | NCT05148572 |
| Clinical significance of hepatic and circulating microRNAs miR-221 and miR-222 in hepatocellular carcinoma | NA | Unknown | NCT02928627 |
| Study of microRNAs as a diagnostic tool for HCV-related hepatocellular carcinoma | NA | Unknown | NCT03429530 |
| Impact of IL-28B rs12979860 and rs4803217 gene polymorphisms associated with miRNAs deregulation on HCV-related hepatocellular carcinoma | NA | Unknown | NCT02507882 |
| MiRNA as a diagnostic and prognostic biomarker of hepatocellular carcinoma | NA | Not yet recruiting | NCT02448056 |
| Early Detection of Hepatocellular Carcinoma in a High-risk Prospective Cohort (ELEGANCE) | NA | Recruiting | NCT04965259 |
| Blood sample collection for experimental blood test to track liver cancer | NA | Recruiting | NCT04720430 |
| Different genetic features associated with hepatic carcinogenesis | NA | Unknown | NCT01247506 |
| Genetic profiling of liver cancer in patients undergoing liver transplantation | NA | Active, not recruiting | NCT02412579 |
| A study of individualized radiotherapy based on a prediction model of lymph node metastasis in hepatocellular carcinoma | NA | Unknown | NCT03416803 |
| Fingerprint characterization of sorafenib treated HCC | NA | Unknown | NCT03958669 |
| Risk of HCC in cirrhotic patients post DAAs treatment | NA | Unknown | NCT03414554 |
| Risk of hepatocellular carcinoma in patient with liver cirrhosis | NA | Unknown | NCT03083002 |
| Axitinib for the treatment of advanced hepatocellular carcinoma | 2 | Completed | NCT01210495 |
| The phase III study of icaritin versus sorafenib in PD-L1 positive advanced hepatocellular carcinoma subjects | 3 | Recruiting | NCT03236649 |
| The phase III study of icaritin versus HUACHANSU PIAN in hepatocellular carcinoma subjects | 3 | Recruiting | NCT03236636 |
| A study of rSIFN-co in subjects with advanced solid tumors | 1 | Completed | NCT02387307 |
| Stereotactic body radiation therapy and transarterial chemoembolization in treating patients with liver cancer that cannot be removed by surgery | 1 | Completed | NCT02507765 |
| Nal-IRI with 5-fluorouracil (5-FU) and leucovorin or gemcitabine plus cisplatin in advanced biliary-tract cancer | 2 | Active, not recruiting | NCT03044587 |
As described above, there is a strong correlation between miRNA expression and HCC. However, some of the regulated miRNAs are not only related to HCV infection, but also with other pathological liver conditions. In particular, alterations in some miRNAs have been also found in NAFLD[108-110]. Among the identified miRNAs, a significant upregulation of miR-122 in the serum of NAFLD patients was noted. This variation is comparable to the upregulation of the same miRNA observed in the serum of HCC/HCV patients. Another group reported a significant increase of miR-34a expression in the serum of NAFLD patients[110], in agreement with our observations in HCV HCC cells infections[31]. These results indicate that these miRNAs could be considered potential markers of pathological liver conditions induced by inflammation or transformation (Figure 1). The most direct implication of this finding is the possibility of having new, noninvasive, and reliable biomarkers for early HCC detection that could be applied in the near future. In particular, a specific circulating miRNA expression, in association with conventional tumor markers such as AFP, protein induced by vitamin K absence/antagonist II (PIVKA-II), and classical clinical parameters has been observed[111,112]. The definition of an miRNA expression signature (88 miRNAs) provides a substantial increase of accuracy in the detection of HCC (up to 99.5%), associated with a strong sensitivity (100%), while AFP evaluation as a tumor biomarker has an accuracy and a sensitivity of 76.5% and 63.8%, respectively[111]. The strength of these results refers mostly to the detection of small HCC tumors (< 3 cm). This analysis also revealed that an miRNA expression signature shows specific variations for HCC compared to other pathological liver conditions, where cirrhotic patients or chronic hepatitis B infection can be distinguished by a characteristic alteration of expression pattern[110]. These results strengthen the possibility of using miRNAs as a new approach for early tumor detection and subsequent early intervention, particularly considering that, after HCV DAA treatment, there is still the need to monitor HCC occurrence after viral eradication, as also suggested elsewhere[31,113,114].
The introduction of DAA therapy signals a milestone in the history of HCV treatment, with achievement of SVR at high rates. However, whether the development of HCC is related to pre-existing HCV infections after DAA treatment is disputed. Not all HCV patients are eligible for DAA treatment, and HCV reinfection in high-risk populations frequently occurs. In light of this, understanding the molecular mechanisms of HCV viral infection that lead to development of liver cancer in the host is fundamental.
It has been reported that cellular miRNAs may contribute to HCV pathogenesis, either by direct or indirect interactions with the viral genome or proteins. Several miRNAs and their targets have been shown to be associated with HCV and HCC progression, and might represent a diagnostic biomarker for the early prognosis of HCC. Early HCC prognosis acquires relevance during DAA treatment to minimize the risks of HCC development in HCV+ patients. Likewise, many miRNAs could be used also for therapeutic purposes. However, further studies are needed to elucidate the molecular interplay between miRNA and HCC, and the development of novel therapeutic strategies for the treatment of HCC is a major critical goal to be achieved.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kumar R, India; Zhang C, China S-Editor: Fan JR L-Editor: Kerr C P-Editor: Fan JR
| 1. | IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Biological agents. Volume 100 B. A review of human carcinogens. IARC Monogr Eval Carcinog Risks Hum. 2012;100:1-441. [PubMed] |
| 2. | McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of Hepatocellular Carcinoma. Hepatology. 2021;73 Suppl 1:4-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 820] [Cited by in RCA: 1346] [Article Influence: 336.5] [Reference Citation Analysis (1)] |
| 3. | Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J Gen Virol. 2000;81:1631-1648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 450] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 4. | Bartenschlager R, Lohmann V. Novel cell culture systems for the hepatitis C virus. Antiviral Res. 2001;52:1-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 146] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 5. | Goutagny N, Fatmi A, De Ledinghen V, Penin F, Couzigou P, Inchauspé G, Bain C. Evidence of viral replication in circulating dendritic cells during hepatitis C virus infection. J Infect Dis. 2003;187:1951-1958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 119] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 6. | Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 973] [Cited by in RCA: 1006] [Article Influence: 55.9] [Reference Citation Analysis (0)] |
| 7. | Horner SM. Activation and evasion of antiviral innate immunity by hepatitis C virus. J Mol Biol. 2014;426:1198-1209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 8. | Jackowiak P, Kuls K, Budzko L, Mania A, Figlerowicz M. Phylogeny and molecular evolution of the hepatitis C virus. Infect Genet Evol. 2014;21:67-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | Preciado MV, Valva P, Escobar-Gutierrez A, Rahal P, Ruiz-Tovar K, Yamasaki L, Vazquez-Chacon C, Martinez-Guarneros A, Carpio-Pedroza JC, Fonseca-Coronado S, Cruz-Rivera M. Hepatitis C virus molecular evolution: transmission, disease progression and antiviral therapy. World J Gastroenterol. 2014;20:15992-16013. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 33] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (5)] |
| 10. | Sharma SD. Hepatitis C virus: molecular biology & current therapeutic options. Indian J Med Res. 2010;131:17-34. [PubMed] |
| 11. | Paul D, Madan V, Bartenschlager R. Hepatitis C virus RNA replication and assembly: living on the fat of the land. Cell Host Microbe. 2014;16:569-579. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 195] [Cited by in RCA: 204] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 12. | Rabaan AA, Al-Ahmed SH, Bazzi AM, Alfouzan WA, Alsuliman SA, Aldrazi FA, Haque S. Overview of hepatitis C infection, molecular biology, and new treatment. J Infect Public Health. 2020;13:773-783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 13. | Papatheodoridis G, Hatzakis A. Public health issues of hepatitis C virus infection. Best Pract Res Clin Gastroenterol. 2012;26:371-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 14. | Lemoine M, Thursz M. Hepatitis C, a global issue: access to care and new therapeutic and preventive approaches in resource-constrained areas. Semin Liver Dis. 2014;34:89-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 15. | Liang TJ. Current progress in development of hepatitis C virus vaccines. Nat Med. 2013;19:869-878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 127] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 16. | Averhoff FM, Glass N, Holtzman D. Global burden of hepatitis C: considerations for healthcare providers in the United States. Clin Infect Dis. 2012;55 Suppl 1:S10-S15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 215] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 17. | Seeff LB. The history of the "natural history" of hepatitis C (1968-2009). Liver Int. 2009;29 Suppl 1:89-99. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 172] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 18. | Zeisel MB, Fafi-Kremer S, Fofana I, Barth H, Stoll-Keller F, Doffoel M, Baumert TF. Neutralizing antibodies in hepatitis C virus infection. World J Gastroenterol. 2007;13:4824-4830. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 35] [Cited by in RCA: 33] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Spengler U, Nattermann J. Immunopathogenesis in hepatitis C virus cirrhosis. Clin Sci (Lond). 2007;112:141-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 68] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 20. | Zeisel MB, Fafi-Kremer S, Robinet E, Habersetzer F, Baumert TF, Stoll-Keller F. Adaptive immunity to hepatitis C virus. Viruses. 2009;1:276-297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 21. | Boyer N, Marcellin P. Pathogenesis, diagnosis and management of hepatitis C. J Hepatol. 2000;32:98-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 111] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 22. | Farci P, Strazzera R, Alter HJ, Farci S, Degioannis D, Coiana A, Peddis G, Usai F, Serra G, Chessa L, Diaz G, Balestrieri A, Purcell RH. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc Natl Acad Sci U S A. 2002;99:3081-3086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 165] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 23. | Feeney ER, Chung RT. Antiviral treatment of hepatitis C. BMJ. 2014;348:g3308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 157] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 24. | McHutchison JG, Gordon SC, Schiff ER, Shiffman ML, Lee WM, Rustgi VK, Goodman ZD, Ling MH, Cort S, Albrecht JK. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1485-1492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2509] [Cited by in RCA: 2435] [Article Influence: 90.2] [Reference Citation Analysis (0)] |
| 25. | Aleman S, Rahbin N, Weiland O, Davidsdottir L, Hedenstierna M, Rose N, Verbaan H, Stål P, Carlsson T, Norrgren H, Ekbom A, Granath F, Hultcrantz R. A risk for hepatocellular carcinoma persists long-term after sustained virologic response in patients with hepatitis C-associated liver cirrhosis. Clin Infect Dis. 2013;57:230-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 195] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 26. | Shoukry NH. Hepatitis C Vaccines, Antibodies, and T Cells. Front Immunol. 2018;9:1480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 27. | Heo YA, Deeks ED. Sofosbuvir/Velpatasvir/Voxilaprevir: A Review in Chronic Hepatitis C. Drugs. 2018;78:577-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 28. | Lee CH, Kim JH, Lee SW. The role of microRNAs in hepatitis C virus replication and related liver diseases. J Microbiol. 2014;52:445-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 29. | Tamori A, Murakami Y, Kubo S, Itami S, Uchida-Kobayashi S, Morikawa H, Enomoto M, Takemura S, Tanahashi T, Taguchi YH, Kawada N. MicroRNA expression in hepatocellular carcinoma after the eradication of chronic hepatitis virus C infection using interferon therapy. Hepatol Res. 2016;46:E26-E35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 30. | Xu G, Zhang Y, Wei J, Jia W, Ge Z, Zhang Z, Liu X. MicroRNA-21 promotes hepatocellular carcinoma HepG2 cell proliferation through repression of mitogen-activated protein kinase-kinase 3. BMC Cancer. 2013;13:469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 80] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 31. | Badami E, Carcione C, Chinnici CM, Tinnirello R, Conaldi PG, Iannolo G. HCV Interplay With Mir34a: Implications in Hepatocellular Carcinoma. Front Oncol. 2021;11:803278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 32. | Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J Carcinog. 2017;16:1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 413] [Cited by in RCA: 533] [Article Influence: 66.6] [Reference Citation Analysis (0)] |
| 33. | El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2881] [Cited by in RCA: 3088] [Article Influence: 220.6] [Reference Citation Analysis (0)] |
| 34. | Sukocheva OA. Estrogen, estrogen receptors, and hepatocellular carcinoma: Are we there yet? World J Gastroenterol. 2018;24:1-4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 54] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 35. | Miceli V, Cervello M, Azzolina A, Montalto G, Calabrò M, Carruba G. Aromatase and amphiregulin are correspondingly expressed in human liver cancer cells. Ann N Y Acad Sci. 2009;1155:252-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 36. | Granata OM, Cocciadiferro L, Miceli V, Polito LM, Campisi I, Carruba G. Metabolic profiles of androgens in malignant human liver cell lines. Ann N Y Acad Sci. 2006;1089:262-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 0.8] [Reference Citation Analysis (1)] |
| 37. | Bruix J, Sherman M; American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020-1022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5972] [Cited by in RCA: 6573] [Article Influence: 469.5] [Reference Citation Analysis (1)] |
| 38. | Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5:215-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1202] [Cited by in RCA: 1212] [Article Influence: 60.6] [Reference Citation Analysis (0)] |
| 39. | El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264-1273.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2183] [Cited by in RCA: 2508] [Article Influence: 192.9] [Reference Citation Analysis (2)] |
| 40. | Okuda M, Li K, Beard MR, Showalter LA, Scholle F, Lemon SM, Weinman SA. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122:366-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 690] [Cited by in RCA: 680] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 41. | Lemon SM, McGivern DR. Is hepatitis C virus carcinogenic? Gastroenterology. 2012;142:1274-1278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 42. | Axley P, Ahmed Z, Ravi S, Singal AK. Hepatitis C Virus and Hepatocellular Carcinoma: A Narrative Review. J Clin Transl Hepatol. 2018;6:79-84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 133] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 43. | O'Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne). 2018;9:402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2111] [Cited by in RCA: 3380] [Article Influence: 482.9] [Reference Citation Analysis (0)] |
| 44. | Xu X, Tao Y, Shan L, Chen R, Jiang H, Qian Z, Cai F, Ma L, Yu Y. The Role of MicroRNAs in Hepatocellular Carcinoma. J Cancer. 2018;9:3557-3569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 131] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 45. | Younossi ZM, Stepanova M, Nader F, Lam B, Hunt S. The patient's journey with chronic hepatitis C from interferon plus ribavirin to interferon- and ribavirin-free regimens: a study of health-related quality of life. Aliment Pharmacol Ther. 2015;42:286-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 80] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 46. | Lawitz E, Poordad FF, Pang PS, Hyland RH, Ding X, Mo H, Symonds WT, McHutchison JG, Membreno FE. Sofosbuvir and ledipasvir fixed-dose combination with and without ribavirin in treatment-naive and previously treated patients with genotype 1 hepatitis C virus infection (LONESTAR): an open-label, randomised, phase 2 trial. Lancet. 2014;383:515-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 429] [Cited by in RCA: 443] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 47. | Bourlière M, Gordon SC, Flamm SL, Cooper CL, Ramji A, Tong M, Ravendhran N, Vierling JM, Tran TT, Pianko S, Bansal MB, de Lédinghen V, Hyland RH, Stamm LM, Dvory-Sobol H, Svarovskaia E, Zhang J, Huang KC, Subramanian GM, Brainard DM, McHutchison JG, Verna EC, Buggisch P, Landis CS, Younes ZH, Curry MP, Strasser SI, Schiff ER, Reddy KR, Manns MP, Kowdley KV, Zeuzem S; POLARIS-1 and POLARIS-4 Investigators. Sofosbuvir, Velpatasvir, and Voxilaprevir for Previously Treated HCV Infection. N Engl J Med. 2017;376:2134-2146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 399] [Cited by in RCA: 418] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 48. | Poordad F, Hezode C, Trinh R, Kowdley KV, Zeuzem S, Agarwal K, Shiffman ML, Wedemeyer H, Berg T, Yoshida EM, Forns X, Lovell SS, Da Silva-Tillmann B, Collins CA, Campbell AL, Podsadecki T, Bernstein B. ABT-450/r-ombitasvir and dasabuvir with ribavirin for hepatitis C with cirrhosis. N Engl J Med. 2014;370:1973-1982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 698] [Cited by in RCA: 683] [Article Influence: 62.1] [Reference Citation Analysis (0)] |
| 49. | Sarrazin C, Isakov V, Svarovskaia ES, Hedskog C, Martin R, Chodavarapu K, Brainard DM, Miller MD, Mo H, Molina JM, Sulkowski MS. Late Relapse Versus Hepatitis C Virus Reinfection in Patients With Sustained Virologic Response After Sofosbuvir-Based Therapies. Clin Infect Dis. 2017;64:44-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 84] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 50. | Guarino M, Morisco F, Valvano MR, Ippolito AM, Librandi M, Andriulli N, Greco M, Amoruso A, Iacobellis A, Niro G, Caporaso N, Andriulli A. Systematic review: interferon-free regimens for patients with HCV-related Child C cirrhosis. Aliment Pharmacol Ther. 2017;45:1193-1200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 51. | Probst A, Dang T, Bochud M, Egger M, Negro F, Bochud PY. Role of hepatitis C virus genotype 3 in liver fibrosis progression--a systematic review and meta-analysis. J Viral Hepat. 2011;18:745-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 124] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 52. | Roche B, Coilly A, Duclos-Vallee JC, Samuel D. The impact of treatment of hepatitis C with DAAs on the occurrence of HCC. Liver Int. 2018;38 Suppl 1:139-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 108] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 53. | Frazzoni L, Sikandar U, Metelli F, Sadalla S, Mazzella G, Bazzoli F, Fuccio L, Azzaroli F. Hepatocellular Carcinoma Recurrence after Hepatitis C Virus Therapy with Direct-Acting Antivirals. A Systematic Review and Meta-Analysis. J Clin Med. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 54. | Miyasaka A, Yoshida Y, Suzuki A, Sawara K, Takikawa Y. A Novel Standard for Hepatocellular Carcinoma Screening Intensity After Hepatitis C Elimination. Int J Gen Med. 2021;14:8935-8943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 55. | Conti F, Buonfiglioli F, Scuteri A, Crespi C, Bolondi L, Caraceni P, Foschi FG, Lenzi M, Mazzella G, Verucchi G, Andreone P, Brillanti S. Early occurrence and recurrence of hepatocellular carcinoma in HCV-related cirrhosis treated with direct-acting antivirals. J Hepatol. 2016;65:727-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 708] [Cited by in RCA: 699] [Article Influence: 77.7] [Reference Citation Analysis (0)] |
| 56. | European Association for the Study of the Liver; Clinical Practice Guidelines Panel: Chair; EASL Governing Board representative; Panel members. EASL recommendations on treatment of hepatitis C: Final update of the series☆. J Hepatol. 2020;73:1170-1218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 567] [Cited by in RCA: 779] [Article Influence: 155.8] [Reference Citation Analysis (0)] |
| 57. | Abdelaziz AO, Nabil MM, Abdelmaksoud AH, Shousha HI, Cordie AA, Hassan EM, Omran DA, Leithy R, Elbaz TM. De-novo versus recurrent hepatocellular carcinoma following direct-acting antiviral therapy for hepatitis C virus. Eur J Gastroenterol Hepatol. 2018;30:39-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 58. | Kuftinec G, Loehfelm T, Corwin M, Durbin-Johnson B, Candido M, Hluhanich R, Sarkar S. De novo hepatocellular carcinoma occurrence in hepatitis C cirrhotics treated with direct-acting antiviral agents. Hepat Oncol. 2018;5:HEP06. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 59. | Nishijima N, Nasu A, Kimura T, Osaki Y. Two Cases of Hepatocellular Carcinoma Occurring Immediately after Direct-acting Antiviral Agents against Hepatitis C Virus. Intern Med. 2019;58:225-231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 60. | Tapia-Sosa R, Hernández-Cabral F, Gabutti A, Páez-Zayas VM, García-Juárez I. Hepatocellular carcinoma associated with direct-acting antiviral therapy for hepatitis C virus: A report of two cases. Rev Gastroenterol Mex (Engl Ed). 2021;86:197-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 61. | Tayyab GUN, Rasool S, Nasir B, Rubi G, Abou-Samra AB, Butt AA. Hepatocellular carcinoma occurs frequently and early after treatment in HCV genotype 3 infected persons treated with DAA regimens. BMC Gastroenterol. 2020;20:93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 62. | Flisiak R, Zarębska-Michaluk D, Janczewska E, Łapiński T, Rogalska M, Karpińska E, Mikuła T, Bolewska B, Białkowska J, Flejscher-Stępniewska K, Tomasiewicz K, Karwowska K, Pazgan-Simon M, Piekarska A, Berak H, Tronina O, Garlicki A, Jaroszewicz J. Five-Year Follow-Up of Cured HCV Patients under Real-World Interferon-Free Therapy. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 63. | Finkelmeier F, Dultz G, Peiffer KH, Kronenberger B, Krauss F, Zeuzem S, Sarrazin C, Vermehren J, Waidmann O. Risk of de novo Hepatocellular Carcinoma after HCV Treatment with Direct-Acting Antivirals. Liver Cancer. 2018;7:190-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 64. | Gallo A, Miceli V, Bulati M, Iannolo G, Contino F, Conaldi PG. Viral miRNAs as Active Players and Participants in Tumorigenesis. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 65. | Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1765] [Cited by in RCA: 1702] [Article Influence: 189.1] [Reference Citation Analysis (0)] |
| 66. | Sartorius K, Makarova J, Sartorius B, An P, Winkler C, Chuturgoon A, Kramvis A. The Regulatory Role of MicroRNA in Hepatitis-B Virus-Associated Hepatocellular Carcinoma (HBV-HCC) Pathogenesis. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 67. | Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122--a key factor and therapeutic target in liver disease. J Hepatol. 2015;62:448-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 399] [Cited by in RCA: 490] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 68. | Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526-3536. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 585] [Cited by in RCA: 590] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 69. | Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, Zaret KS, Taylor JM. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 607] [Cited by in RCA: 657] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 70. | Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577-1581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1993] [Cited by in RCA: 1987] [Article Influence: 99.4] [Reference Citation Analysis (0)] |
| 71. | Jopling CL, Schütz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 338] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 72. | Jopling CL. Regulation of hepatitis C virus by microRNA-122. Biochem Soc Trans. 2008;36:1220-1223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 103] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 73. | Schult P, Roth H, Adams RL, Mas C, Imbert L, Orlik C, Ruggieri A, Pyle AM, Lohmann V. microRNA-122 amplifies hepatitis C virus translation by shaping the structure of the internal ribosomal entry site. Nat Commun. 2018;9:2613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 100] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 74. | Masaki T, Arend KC, Li Y, Yamane D, McGivern DR, Kato T, Wakita T, Moorman NJ, Lemon SM. miR-122 stimulates hepatitis C virus RNA synthesis by altering the balance of viral RNAs engaged in replication versus translation. Cell Host Microbe. 2015;17:217-228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 111] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 75. | Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci U S A. 2012;109:941-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 304] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 76. | Luna JM, Scheel TK, Danino T, Shaw KS, Mele A, Fak JJ, Nishiuchi E, Takacs CN, Catanese MT, de Jong YP, Jacobson IM, Rice CM, Darnell RB. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 2015;160:1099-1110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 276] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 77. | Amr KS, Elmawgoud Atia HA, Elazeem Elbnhawy RA, Ezzat WM. Early diagnostic evaluation of miR-122 and miR-224 as biomarkers for hepatocellular carcinoma. Genes Dis. 2017;4:215-221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 78. | Li G, Shen Q, Li C, Li D, Chen J, He M. Identification of circulating MicroRNAs as novel potential biomarkers for hepatocellular carcinoma detection: a systematic review and meta-analysis. Clin Transl Oncol. 2015;17:684-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 79. | Li J, Jin B, Wang T, Li W, Wang Z, Zhang H, Song Y, Li N. Serum microRNA expression profiling identifies serum biomarkers for HCV-related hepatocellular carcinoma. Cancer Biomark. 2019;26:501-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 80. | Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 423] [Cited by in RCA: 467] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 81. | Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12:414-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 352] [Cited by in RCA: 409] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 82. | Mizuguchi Y, Mishima T, Yokomuro S, Arima Y, Kawahigashi Y, Shigehara K, Kanda T, Yoshida H, Uchida E, Tajiri T, Takizawa T. Sequencing and bioinformatics-based analyses of the microRNA transcriptome in hepatitis B-related hepatocellular carcinoma. PLoS One. 2011;6:e15304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 83. | Smith JL, Jeng S, McWeeney SK, Hirsch AJ. A MicroRNA Screen Identifies the Wnt Signaling Pathway as a Regulator of the Interferon Response during Flavivirus Infection. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 103] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 84. | Feili X, Wu S, Ye W, Tu J, Lou L. MicroRNA-34a-5p inhibits liver fibrosis by regulating TGF-β1/Smad3 pathway in hepatic stellate cells. Cell Biol Int. 2018;42:1370-1376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 85. | Bihrer V, Waidmann O, Friedrich-Rust M, Forestier N, Susser S, Haupenthal J, Welker M, Shi Y, Peveling-Oberhag J, Polta A, von Wagner M, Radeke HH, Sarrazin C, Trojan J, Zeuzem S, Kronenberger B, Piiper A. Serum microRNA-21 as marker for necroinflammation in hepatitis C patients with and without hepatocellular carcinoma. PLoS One. 2011;6:e26971. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 116] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 86. | Shaker OG, Senousy MA. Serum microRNAs as predictors for liver fibrosis staging in hepatitis C virus-associated chronic liver disease patients. J Viral Hepat. 2017;24:636-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 87. | Sodroski C, Lowey B, Hertz L, Jake Liang T, Li Q. MicroRNA-135a Modulates Hepatitis C Virus Genome Replication through Downregulation of Host Antiviral Factors. Virol Sin. 2019;34:197-210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 88. | Bandiera S, Pernot S, El Saghire H, Durand SC, Thumann C, Crouchet E, Ye T, Fofana I, Oudot MA, Barths J, Schuster C, Pessaux P, Heim MH, Baumert TF, Zeisel MB. Hepatitis C Virus-Induced Upregulation of MicroRNA miR-146a-5p in Hepatocytes Promotes Viral Infection and Deregulates Metabolic Pathways Associated with Liver Disease Pathogenesis. J Virol. 2016;90:6387-6400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 89. | Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647-658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2191] [Cited by in RCA: 2184] [Article Influence: 121.3] [Reference Citation Analysis (0)] |
| 90. | Cheng M, Si Y, Niu Y, Liu X, Li X, Zhao J, Jin Q, Yang W. High-throughput profiling of alpha interferon- and interleukin-28B-regulated microRNAs and identification of let-7s with anti-hepatitis C virus activity by targeting IGF2BP1. J Virol. 2013;87:9707-9718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 91. | Jamalidoust M, Shafaati M, Kalani M, Zare M, Ziyeayan M. MicroRNA let-7b inhibits hepatitis C virus and induces apoptosis in human hepatoma cells. Mol Biol Rep. 2022;49:1273-1280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 92. | Murakami Y, Aly HH, Tajima A, Inoue I, Shimotohno K. Regulation of the hepatitis C virus genome replication by miR-199a. J Hepatol. 2009;50:453-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 169] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 93. | Pascut D, Hoang M, Nguyen NNQ, Pratama MY, Tiribelli C. HCV Proteins Modulate the Host Cell miRNA Expression Contributing to Hepatitis C Pathogenesis and Hepatocellular Carcinoma Development. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 94. | Xu J, An P, Winkler CA, Yu Y. Dysregulated microRNAs in Hepatitis B Virus-Related Hepatocellular Carcinoma: Potential as Biomarkers and Therapeutic Targets. Front Oncol. 2020;10:1271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 95. | Ali HEA, Abdel Hameed R, Effat H, Ahmed EK, Atef AA, Sharawi SK, Ali M, Abd Elmageed ZY, Abdel Wahab AH. Circulating microRNAs panel as a diagnostic tool for discrimination of HCV-associated hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2017;41:e51-e62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 96. | Zekri AN, Youssef AS, El-Desouky ED, Ahmed OS, Lotfy MM, Nassar AA, Bahnassey AA. Serum microRNA panels as potential biomarkers for early detection of hepatocellular carcinoma on top of HCV infection. Tumour Biol. 2016;37:12273-12286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 75] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 97. | Wahb AMSE, El Kassas M, Khamis AK, Elhelbawy M, Elhelbawy N, Habieb MSE. Circulating microRNA 9-3p and serum endocan as potential biomarkers for hepatitis C virus-related hepatocellular carcinoma. World J Hepatol. 2021;13:1753-1765. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 98. | Wang SC, Li CY, Chang WT, Cheng WC, Yen CH, Tu WY, Lin ZY, Lin CC, Yeh ML, Huang CF, Huang JF, Dai CY, Chuang WL, Chen YL, Yu ML. Exosome-derived differentiation antagonizing non-protein coding RNA with risk of hepatitis C virus-related hepatocellular carcinoma recurrence. Liver Int. 2021;41:956-968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 99. | Hu Q, Wang K, Sun X, Li Y, Fu Q, Liang T, Tang G. A redox-sensitive, oligopeptide-guided, self-assembling, and efficiency-enhanced (ROSE) system for functional delivery of microRNA therapeutics for treatment of hepatocellular carcinoma. Biomaterials. 2016;104:192-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 100. | Wagenaar TR, Zabludoff S, Ahn SM, Allerson C, Arlt H, Baffa R, Cao H, Davis S, Garcia-Echeverria C, Gaur R, Huang SM, Jiang L, Kim D, Metz-Weidmann C, Pavlicek A, Pollard J, Reeves J, Rocnik JL, Scheidler S, Shi C, Sun F, Tolstykh T, Weber W, Winter C, Yu E, Yu Q, Zheng G, Wiederschain D. Anti-miR-21 Suppresses Hepatocellular Carcinoma Growth via Broad Transcriptional Network Deregulation. Mol Cancer Res. 2015;13:1009-1021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 63] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 101. | Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685-1694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1665] [Cited by in RCA: 1702] [Article Influence: 141.8] [Reference Citation Analysis (0)] |
| 102. | Ottosen S, Parsley TB, Yang L, Zeh K, van Doorn LJ, van der Veer E, Raney AK, Hodges MR, Patick AK. In vitro antiviral activity and preclinical and clinical resistance profile of miravirsen, a novel anti-hepatitis C virus therapeutic targeting the human factor miR-122. Antimicrob Agents Chemother. 2015;59:599-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 184] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 103. | van der Ree MH, de Vree JM, Stelma F, Willemse S, van der Valk M, Rietdijk S, Molenkamp R, Schinkel J, van Nuenen AC, Beuers U, Hadi S, Harbers M, van der Veer E, Liu K, Grundy J, Patick AK, Pavlicek A, Blem J, Huang M, Grint P, Neben S, Gibson NW, Kootstra NA, Reesink HW. Safety, tolerability, and antiviral effect of RG-101 in patients with chronic hepatitis C: a phase 1B, double-blind, randomised controlled trial. Lancet. 2017;389:709-717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 207] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 104. | Liu F, Shimakami T, Murai K, Shirasaki T, Funaki M, Honda M, Murakami S, Yi M, Tang H, Kaneko S. Efficient Suppression of Hepatitis C Virus Replication by Combination Treatment with miR-122 Antagonism and Direct-acting Antivirals in Cell Culture Systems. Sci Rep. 2016;6:30939. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 105. | Lou G, Song X, Yang F, Wu S, Wang J, Chen Z, Liu Y. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J Hematol Oncol. 2015;8:122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 387] [Cited by in RCA: 556] [Article Influence: 55.6] [Reference Citation Analysis (0)] |
| 106. | Wei JX, Lv LH, Wan YL, Cao Y, Li GL, Lin HM, Zhou R, Shang CZ, Cao J, He H, Han QF, Liu PQ, Zhou G, Min J. Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology. 2015;61:1284-1294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 146] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 107. | Zhang J, Li D, Zhang R, Peng R, Li J. Delivery of microRNA-21-sponge and pre-microRNA-122 by MS2 virus-like particles to therapeutically target hepatocellular carcinoma cells. Exp Biol Med (Maywood). 2021;246:2463-2472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 108. | Gim JA, Bang SM, Lee YS, Lee Y, Yim SY, Jung YK, Kim H, Kim BH, Kim JH, Seo YS, Yim HJ, Yeon JE, Um SH, Byun KS. Evaluation of the severity of nonalcoholic fatty liver disease through analysis of serum exosomal miRNA expression. PLoS One. 2021;16:e0255822. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 109. | Zhang JW, Pan HT. microRNA profiles of serum exosomes derived from children with nonalcoholic fatty liver. Genes Genomics. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 110. | Liu CH, Ampuero J, Gil-Gómez A, Montero-Vallejo R, Rojas Á, Muñoz-Hernández R, Gallego-Durán R, Romero-Gómez M. miRNAs in patients with non-alcoholic fatty liver disease: A systematic review and meta-analysis. J Hepatol. 2018;69:1335-1348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 123] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 111. | Long XR, Zhang YJ, Zhang MY, Chen K, Zheng XFS, Wang HY. Identification of an 88-microRNA signature in whole blood for diagnosis of hepatocellular carcinoma and other chronic liver diseases. Aging (Albany NY). 2017;9:1565-1584. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 112. | Caviglia GP, Abate ML, Gaia S, Petrini E, Bosco C, Olivero A, Rosso C, Ciancio A, Pellicano R, Saracco GM, Rizzetto M, Smedile A. Risk of hepatocellular carcinoma in HBV cirrhotic patients assessed by the combination of miR-122, AFP and PIVKA-II. Panminerva Med. 2017;59:283-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 113. | Sapena V, Enea M, Torres F, Celsa C, Rios J, Rizzo GEM, Nahon P, Mariño Z, Tateishi R, Minami T, Sangiovanni A, Forns X, Toyoda H, Brillanti S, Conti F, Degasperi E, Yu ML, Tsai PC, Jean K, El Kassas M, Shousha HI, Omar A, Zavaglia C, Nagata H, Nakagawa M, Asahina Y, Singal AG, Murphy C, Kohla M, Masetti C, Dufour JF, Merchante N, Cavalletto L, Chemello LL, Pol S, Crespo J, Calleja JL, Villani R, Serviddio G, Zanetto A, Shalaby S, Russo FP, Bielen R, Trevisani F, Cammà C, Bruix J, Cabibbo G, Reig M. Hepatocellular carcinoma recurrence after direct-acting antiviral therapy: an individual patient data meta-analysis. Gut. 2022;71:593-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 72] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 114. | Ahumada A, Rayón L, Usón C, Bañares R, Alonso Lopez S. Hepatocellular carcinoma risk after viral response in hepatitis C virus-advanced fibrosis: Who to screen and for how long? World J Gastroenterol. 2021;27:6737-6749. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 7] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |