Published online Mar 14, 2022. doi: 10.3748/wjg.v28.i10.1067
Peer-review started: May 15, 2021
First decision: June 27, 2021
Revised: June 29, 2021
Accepted: February 9, 2022
Article in press: February 9, 2022
Published online: March 14, 2022
Processing time: 300 Days and 0.4 Hours
Gut dysbiosis and small intestinal bacterial overgrowth (SIBO) are commonly observed in patients with cirrhosis. Despite the substantial number of articles describing the relations between disorders of gut microbiota and various manifestations of cirrhosis, dysbiosis and SIBO were always studied separately.
To study the relationship of gut dysbiosis and SIBO in cirrhosis.
This observational study included 47 in-patients with cirrhosis. Stool microbiome was assessed using 16S rRNA gene sequencing. SIBO was assessed using the lactulose hydrogen breath test.
SIBO was found in 24/47 (51.1%) patients. Patients with SIBO had a higher abundance of Firmicutes (P = 0.017) and Fusobacteria (P = 0.011), and a lower abundance of Bacteroidetes (P = 0.013) than patients without SIBO. This increase in the abundance of Firmicutes occurred mainly due to an increase in the abundance of bacteria from the genus Blautia (P = 0.020) of the Lachnospiraceae family (P = 0.047), while the abundance of other major families of this phylum [Ruminococcaceae (P = 0.856), Peptostreptococcaceae (P = 0.066), Clostridiaceae (P = 0.463), Eubacteriaceae (P = 0.463), Lactobacillaceae (P = 0.413), and Veillonellaceae (P = 0.632)] did not differ significantly between the patients with and without SIBO. Reduced level of Bacteroidetes in samples from patients with SIBO was a result of the decrease in bacterial numbers from all the major families of this phylum [Bacteroidaceae (P = 0.014), Porphyromonadaceae (P = 0.002), and Rikenellaceae (P = 0.047)], with the exception of Prevotellaceae (P = 0.941). There were no significant differences in the abundance of taxa that were the main biomarkers of cirrhosis-associated gut dysbiosis [Proteobacteria (P = 0.790), Bacilli (P = 0.573), Enterobacteriaceae (P = 0.632), Streptococcaceae (P = 0.170), Staphylococcaceae (P = 0.450), and Enterococcaceae (P = 0.873)] between patients with and without SIBO.
Despite the differences observed in the gut microbiome between patients with and without SIBO, gut dysbiosis and SIBO are most likely independent disorders of gut microbiota in cirrhosis.
Core Tip: Patients with small intestinal bacterial overgrowth (SIBO) had a higher abundance of Firmicutes and Fusobacteria, and a lower abundance of Bacteroidetes than patients without SIBO. This increase in the abundance of Firmicutes occurred mainly due to an increase in the abundance of bacteria from the genus Blautia of the Lachnospiraceae family. There were no significant differences in the abundance of taxa that were the main biomarkers of cirrhosis-associated gut dysbiosis.
- Citation: Maslennikov R, Ivashkin V, Efremova I, Poluektova E, Kudryavtseva A, Krasnov G. Gut dysbiosis and small intestinal bacterial overgrowth as independent forms of gut microbiota disorders in cirrhosis. World J Gastroenterol 2022; 28(10): 1067-1077
- URL: https://www.wjgnet.com/1007-9327/full/v28/i10/1067.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i10.1067
Cirrhosis is one of the most serious problems in hepatology and is not only a disease of the liver, but it also affects other organs. The damage that occurs in these other organs can contribute to further progression of cirrhosis, forming a vicious circle. The examples of such organs are the gut and the community of microorganisms that reside within it (the gut microbiota). Two forms of disorders of gut microbiota have been described in cirrhosis: changes in its composition[1-5] (gut dysbiosis[6-10]) and an increase of bacterial number in the small intestine (small intestinal bacterial overgrowth, SIBO)[11-14]. It is believed that these disorders, in combination with increased intestinal permeability, lead to increased penetration of bacteria and their components into the intestinal wall, mesenteric lymph nodes, ascitic fluid, and portal and systemic bloodstreams[15]. This, in turn, leads to systemic inflammation, vasodilation, compensatory fluid retention, and worsening portal hypertension[16,17].
Despite the substantial number of articles describing the relations between disorders of gut microbiota and various manifestations of cirrhosis, dysbiosis and SIBO were always studied separately. Our research aims to study the relationship of gut dysbiosis and SIBO in cirrhosis.
In this observational study, 95 consecutive patients with cirrhosis were admitted to the Department of Hepatology of Clinic for Internal Diseases, Gastroenterology and Hepatology at Sechenov University (Moscow, Russia) and screened for inclusion. The study procedures were explained to potential participants, and written informed consent was obtained before enrollment. The present study was approved by the Ethics Committee of Sechenov University.
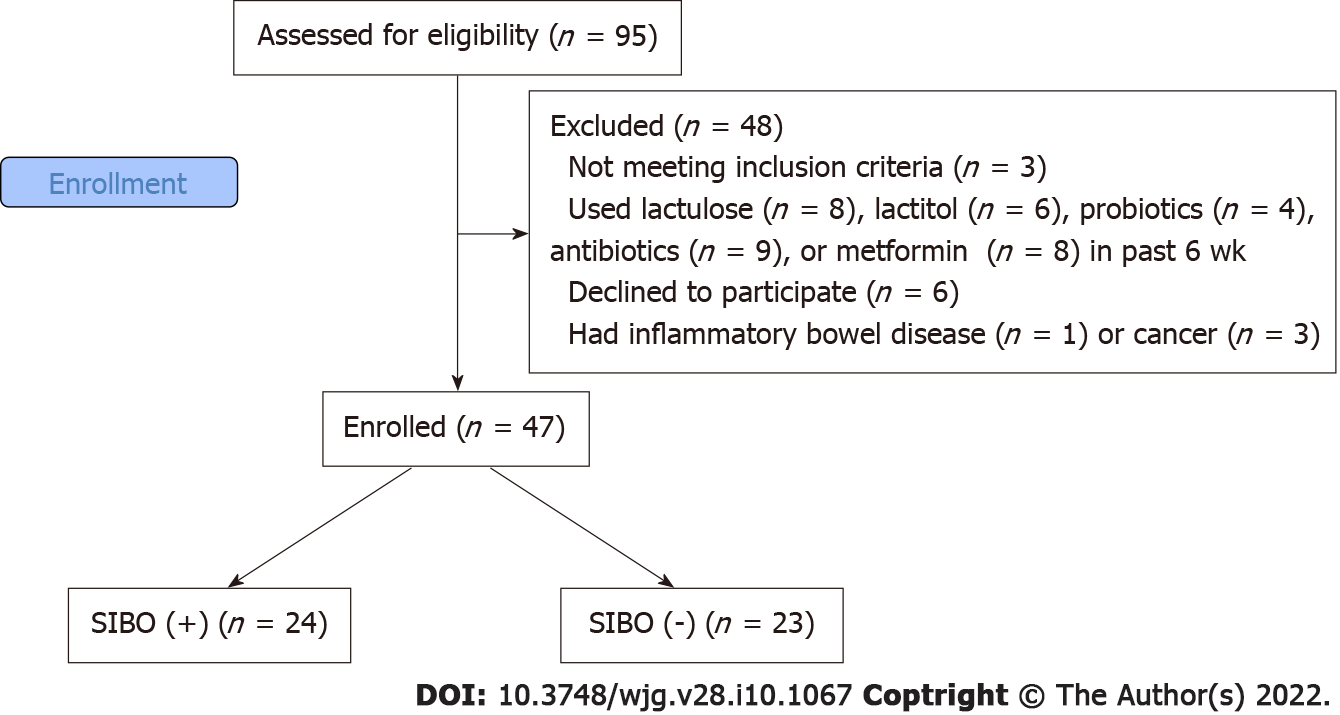
The inclusion criteria were as follows: diagnosis of cirrhosis verified by histology or clinical, biochemical, and ultrasound findings; and age between 18 and 70 years. The exclusion criteria were as follows: use of lactulose, lactitol, or other prebiotics, probiotics, antibiotics, or metformin in the past 6 wk; alcohol consumption in the past 6 wk; or inflammatory bowel disease, cancer, or any other serious disease. We used these criteria to exclude the influence of these factors on the composition of the gut microbiota. Of the original 95 patients screened for inclusion, 47 were enrolled in the study while 48 were excluded (Figure 1).
The participants of this study were 47 in-patients who were diagnosed with cirrhosis at admission. The morning after admission, SIBO was assessed using a lactulose hydrogen breath test (Gastrolyzer, Bedfont, The United Kingdom), following the manufacturer’s instructions and in accordance with the North American Consensus[18]. Participants were considered to have SIBO if breath hydrogen concentration increased by at least 20 ppm above the baseline value within 90 min[18].
The morning after admission, a stool sample was taken into a sterile disposable container and immediately frozen at -80 °C[19].
DNA from the stool was isolated using the MagNa Pure Compact Nucleic Acid Isolation Kit I (Roche, Basel, Switzerland) according to the manufacturer’s instructions. Libraries for sequencing were prepared by two rounds of polymerase chain reaction (PCR) amplification. In the first round, specific primers for the v3-v4 region of the 16S ribosomal RNA gene were used: 16S-F TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG and 16S-R GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC.
After amplification, the PCR product was purified using AMPure XP magnetic beads (Beckman Coulter, Brea, CA, United States). Then, a second round of PCR was performed to attach specific adapters and enable multiplexing of the samples. To begin, 5 μL of the first PCR product was added to the reaction after ball cleaning with primers containing Illumina indices (Nextera XT Index v2 Primers; San Diego, CA, United States) and adapter sequences as well as 2× KAPA HiFi HotStart ReadyMix. The amplification products were also purified using AMPure XP beads (Beckman Coulter). The concentrations of the prepared libraries were then measured using a Qubit 2.0 fluorimeter (London, United Kingdom) and quantitative PCR. The quality of the libraries was assessed using the Agilent 2100 Bioanalyzer (Santa Clara, CA, United States). The libraries were mixed in equal proportions and diluted to the required concentration to be run on a MiSeq (Illumina) device. Pair-end readings of 300 + 300 nucleotides were obtained. Reads were trimmed from the 3’-tail with Trimmomatic (Illumina) and then merged into a single amplicon with the MeFiT tool[20,21]. We did not perform operational taxonomic unit picking; instead, we classified amplicon sequences with the Ribosomal Database Project (RDP) classifier and RDP database[22].
Statistical analysis was performed with STATISTICA 10 (StatSoft Inc., Tulsa, OK, United States). The data are presented as medians (interquartile ranges). The abundance of taxa of the gut microbiome is presented as a percentage. Differences between continuous variables were assessed with the Mann-Whitney test. Fisher’s exact test was used to assess the differences between categorical variables. P values ≤ 0.05 were considered as statistically significant.
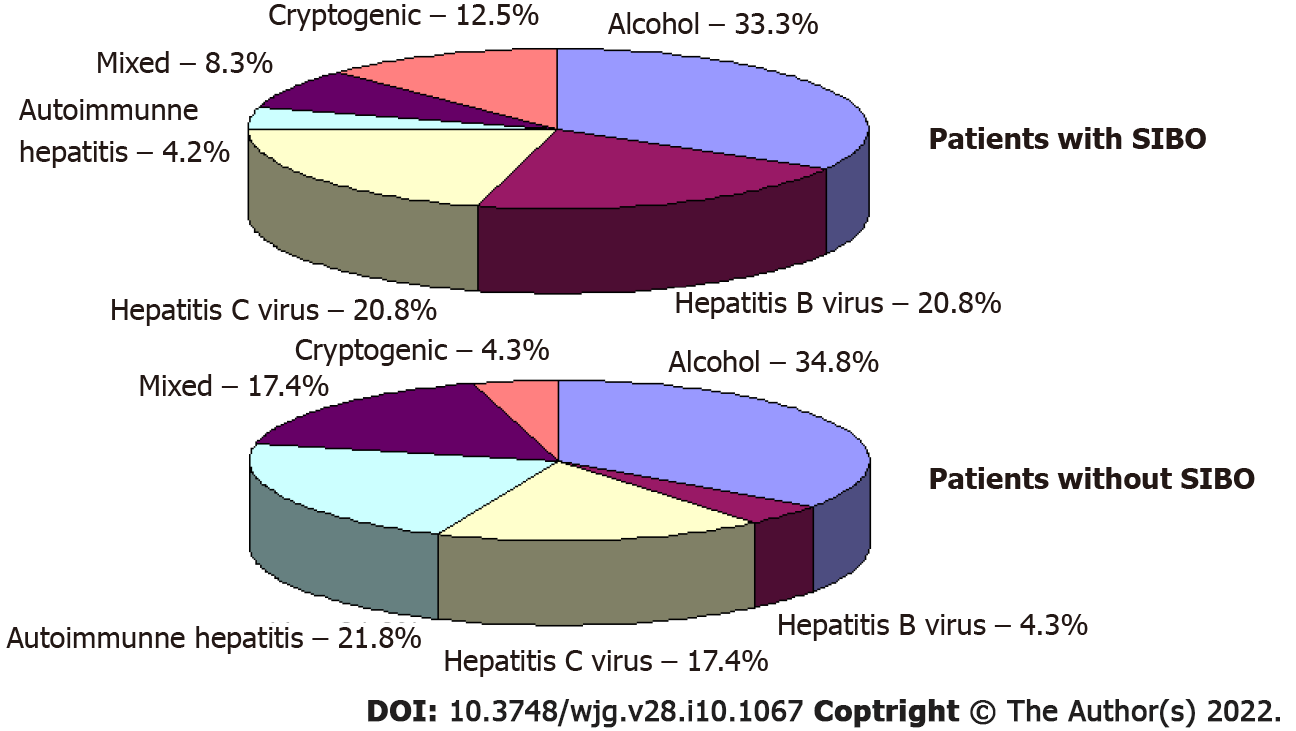
Out of the 47 participants in our study, the results from the lactulose hydrogen breath test revealed that 24 (51.1%) of patients met the criteria for SIBO. No significant differences in etiology (Figure 2), demographics, drugs used before inclusion, and the main manifestations of cirrhosis were observed between patients with and without SIBO, other than the value of C-reactive protein, which was higher in patients with SIBO. Patients with SIBO also displayed a trend toward having a larger spleen and higher incidence of ascites (Table 1).
| Patients with SIBO (n = 24) | Patients without SIBO (n = 23) | P value | |
| Age, yr | 50 (39-59) | 51 (37-58) | 0.949 |
| Male/female | 12/12 | 11/12 | 0.557 |
| Body mass index, kg/m2 | 25.0 (22.9-27.8) | 24.1 (22.3-26.1) | 0.302 |
| Red blood cells, 1012/L | 3.7 (3.3-4.2) | 4.0 (3.6-4.3) | 0.322 |
| White blood cells, 109/L | 3.6 (2.6-4.9) | 4.0 (3.2-5.6) | 0.437 |
| Platelets, 109/L | 78 (49-108) | 90 (59-115) | 0.229 |
| C-reactive protein, mg/L | 11.4 (1.6-17.1) | 2.1 (0.1-9.2) | 0.025 |
| Serum albumen, g/L | 35 (31-40) | 36 (31-41) | 0.617 |
| Serum total bilirubin, μmol/L | 36 (27-54) | 38 (23-64) | 0.848 |
| Prothrombin (Quick test), % | 57 (50-68) | 64 (53-70) | 0.233 |
| Ascites, n (%) | 17 (70.8) | 10 (43.4) | 0.054 |
| Esophageal varices, n (%) | 19 (79.2) | 19 (82.6) | 0.528 |
| Spleen length, cm | 16.2 (14.0-20.0) | 14.6 (13.3-16.2) | 0.080 |
| Portal vein diameter, mm | 13.0 (12.0-13.7) | 12.0 (11.0-13.5) | 0.263 |
| Overt hepatic encephalopathy, n (%) | 9 (37.5) | 8 (34.8) | 0.544 |
| Child-Pugh score | 8 (6-10) | 8 (6-9) | 0.551 |
| Drugs used by patients within 6 mo prior to study enrollment | |||
| Proton pump inhibitors, n (%) | 9 (37.5) | 8 (34.8) | 0.544 |
| Beta blockers, n (%) | 5 (20.8) | 3 (13.0) | 0.375 |
| L-ornithine-L-aspartate, n (%) | 4 (16.7) | 4 (17.4) | 0.625 |
| Diuretics, n (%) | 13 (54.2) | 8 (34.8) | 0.149 |
| Ursodeoxycholic acid, n (%) | 4 (16.7) | 5 (21.7) | 0.471 |
| Glucocorticoids, n (%) | 2 (8.3) | 6 (26.1) | 0.109 |
| Antiviral drugs, n (%) | 7 (29.2) | 4 (17.4) | 0.273 |
In terms of phyla, samples from patients with SIBO revealed a higher abundance of Firmicutes and Fusobacteria, and lower abundance of Bacteroidetes compared to those from patients without SIBO. There were no significant differences in the numbers of Proteobacteria and Actinobacteria as well as the main Firmicutes classes (Clostridia, Bacilli and Negativicutes, Table 2).
| Taxon | Patients with SIBO (n = 24) | Patients without SIBO (n = 23) | P value |
| Phyla | |||
| Firmicutes | 90.2 (77.7-94.3) | 80.0 (67.5-87.2) | 0.017 |
| Bacteroidetes | 5.3 (1.5-7.7) | 6.8 (5.6-1.3) | 0.013 |
| Proteobacteria | 1.3 (0.2-3.0) | 1.7 (0.2-4.1) | 0.790 |
| Actinobacteria | 0.5 (0.3-1.6) | 0.8 (0.4-3.8) | 0.343 |
| Fusobacteria | 0.01 (0.00-0.03) | 0.00 (0.00-0.00) | 0.011 |
| Main classes of Firmicutes | |||
| Clostridia | 77.9 (67.1-84.6) | 70.9 (61.6-83.1) | 0.151 |
| Bacilli | 2.4 (0.4-7.1) | 1.1 (0.4-7.3) | 0.573 |
| Negativicutes | 0.5 (0.2-0.8) | 0.4 (0.2-1.4) | 0.956 |
Our results reveal that the increase in the abundance of Firmicutes observed in patients with SIBO occurs mainly due to a larger population of bacterial species from the Lachnospiraceae family. The number of bacteria from the other major families of this phylum does not differ significantly between the samples taken from the two groups of patients (Table 3). After the exclusion of Lachnospiraceae from Firmicutes, the difference in the abundance of bacteria under this phylum was no longer significant [39.8 (32.3-53.3) vs 42.6 (25.1-54.3); P = 0.773].
| Family | Patients with SIBO (n = 24) | Patients without SIBO (n = 23) | P value |
| Families of Clostridia | |||
| Lachnospiraceae | 41.00 (29.71-55.82) | 31.30 (17.55-43.85) | 0.047 |
| Ruminococcaceae | 26.15 (13.79-35.50) | 21.41 (14.89-34.43) | 0.856 |
| Peptostreptococcaceae | 0.50 (0.18-2.72) | 0.17 (0.04-1.13) | 0.066 |
| Clostridiaceae | 0.30 (0.01-1.12) | 0.10 (0.01-0.47) | 0.463 |
| Eubacteriaceae | 0.00 (0.00-0.00) | 0.00 (0.00-0.01) | 0.463 |
| Families of Bacilli | |||
| Streptococcaceae | 1.46 (0.29-6.03) | 0.41 (0.10-3.42) | 0.170 |
| Lactobacillaceae | 0.23 (0.03-0.56) | 0.10 (0.01-0.38) | 0.413 |
| Staphylococcaceae | 0.02 (0.01-0.03) | 0.01 (0.01-0.02) | 0.450 |
| Enterococcaceae | 0.01 (0.00-0.04) | 0.01 (0.00-0.23) | 0.873 |
| Family of Negativicutes | |||
| Veillonellaceae | 0.43 (0.07-0.73) | 0.16 (0.05-1.15) | 0.632 |
| Families of Bacteroidetes | |||
| Bacteroidaceae | 1.13 (0.35-3.19) | 3.77 (1.05-4.96) | 0.014 |
| Prevotellaceae | 0.56 (0.01-3.12) | 0.27 (0.04-3.07) | 0.941 |
| Porphyromonadaceae | 0.11 (0.02-0.29) | 0.40 (0.23-0.66) | 0.002 |
| Rikenellaceae | 0.07 (0.01-0.74) | 0.45 (0.06-1.35) | 0.047 |
| Families of Proteobacteria | |||
| Enterobacteriaceae | 0.72 (0.02-2.13) | 1.51 (0.05-2.81) | 0.632 |
| Moraxellaceae | 0.04 (0.02-0.05) | 0.02 (0.01-0.04) | 0.014 |
| Pasteurellaceae | 0.01 (0.00-0.15) | 0.00 (0.00-0.01) | 0.025 |
| Sutterellaceae | 0.01 (0.00-0.06) | 0.02 (0.00-0.05) | 0.400 |
| Desulfovibrionaceae | 0.00 (0.00-0.01) | 0.01 (0.00-0.03) | 0.042 |
| Families of Actinobacteria | |||
| Bifidobacteriaceae | 0.46 (0.14-1.50) | 0.65 (0.12-3.22) | 0.366 |
| Coriobacteriaceae | 0.09 (0.07-0.14) | 0.08 (0.04-0.20) | 0.790 |
| Actinomycetaceae | 0.02 (0.01-0.04) | 0.01 (0.00-0.03) | 0.039 |
| Micrococcaceae | 0.02 (0.01-0.06) | 0.01 (0.00-0.01) | 0.045 |
The reduced population of Bacteroidetes in patients with SIBO results from a reduction in all major families of this phylum, with the exception of Prevotellaceae. Although there was no significant difference in the overall levels of Proteobacteria between the samples from the two patient groups, differences were detected in the minor families of the phylum. Specifically, samples from patients with SIBO revealed that the proportion of Pasteurellaceae and Moraxellaceae were higher, and Desulfovibrionaceae was lower, compared to those from patients without SIBO. Similarly, there was also no significant difference in the overall levels of bacteria in the Actinobacteria phylum, but the proportion of bacteria from the Actinomycetaceae and Micrococcaceae minor families were higher in the samples taken from patients with SIBO than in samples from patients without (Table 3).
At the genus level, analysis of samples from patients with SIBO revealed increased the abundance of Blautia, Fusicatenibacter, Acinetobacter, Oribacterium, and Haemophilus bacteria compared to the samples of patients without SIBO. In contrast, Bacteroides, Oscillibacter, Alistipes, Parabacteroides, Barnesiella, and Intestinimonas levels were lower (Table 4).
| Genus | Family | Patients with SIBO (n = 24) | Patients without SIBO (n = 23) | P value |
| Blautia | Lachnospiraceae | 13.38 (6.78-20.52) | 6.99 (3.22-13.47) | 0.020 |
| Faecalibacterium | Ruminococcaceae | 8.64 (3.65-19.55) | 7.82 (3.02-12.74) | 0.516 |
| Gemmiger | Ruminococcaceae | 2.33 (0.95-4.69) | 1.65 (0.03-2.86) | 0.237 |
| Ruminococcus | Ruminococcaceae | 1.80 (0.01-3.51) | 0.58 (0.12-1.83) | 0.617 |
| Roseburia | Lachnospiraceae | 1.66 (0.51-4.20) | 0.92 (0.13-3.42) | 0.377 |
| Streptococcus | Streptococcaceae | 1.36 (0.22-5.78) | 0.37 (0.08-3.42) | 0.151 |
| Dorea | Lachnospiraceae | 1.18 (0.63-2.51) | 0.69 (0.42-1.35) | 0.099 |
| Bacteroides | Bacteroidaceae | 1.15 (0.34-3.04) | 3.59 (1.08-5.05) | 0.012 |
| Fusicatenibacter | Lachnospiraceae | 0.79 (0.27-1.06) | 0.13 (0.04-0.31) | 0.003 |
| Prevotella | Prevotellaceae | 0.43 (0.01-2.27) | 0.12 (0.01-2.19) | 0.856 |
| Escherichia | Enterobacteriaceae | 0.38 (0.01-1.64) | 0.28 (0.01-1.31) | 0.890 |
| Bifidobacterium | Bifidobacteriaceae | 0.32 (0.11-1.15) | 0.47 (0.07-2.39) | 0.413 |
| Oscillibacter | Oscillospiraceae | 0.07 (0.01-0.15) | 0.21 (0.09-0.38) | 0.025 |
| Alistipes | Rikenellaceae | 0.06 (0.01-0.52) | 0.41 (0.06-1.25) | 0.025 |
| Anaerostipes | Lachnospiraceae | 0.62 (0.06-0.50) | 0.49 (0.08-0.67) | 0.170 |
| Parabacteroides | Porphyromonadaceae | 0.03 (0.00-0.11) | 0.16 (0.05-0.29) | 0.009 |
| Acinetobacter | Moraxellaceae | 0.03 (0.02-0.05) | 0.02 (0.01-0.03) | 0.004 |
| Akkermansia | Akkermansiaceae | 0.01 (0.00-0.71) | 0.01 (0.00-1.33) | 0.983 |
| Oribacterium | Lachnospiraceae | 0.01 (0.00-0.02) | 0.00 (0.00-0.00) | 0.040 |
| Barnesiella | Porphyromonadaceae | 0.00 (0.00-0.03) | 0.03 (0.00-0.06) | 0.025 |
| Intestinimonas | Oscillospiraceae | 0.00 (0.00-0.02) | 0.03 (0.01-0.06) | 0.015 |
| Haemophilus | Pasteurellaceae | 0.00 (0.00-0.10) | 0.00 (0.00-0.01) | 0.031 |
Further analysis demonstrated that Brautia was the main contributor to the increase in the abundances of the Lachnospiraceae family and the Firmicutes phylum in patients with SIBO. After subtracting the contribution of these bacteria, the statistical differences between the patient groups were no longer significant [Lachnospiraceae: 28.6 (19.6-35.0) vs 23.2 (10.7-33.2), P = 0.177; Firmicutes: 74.0 (62.2-79.5) vs 65.7 (55.9-78.6); P = 0.425].
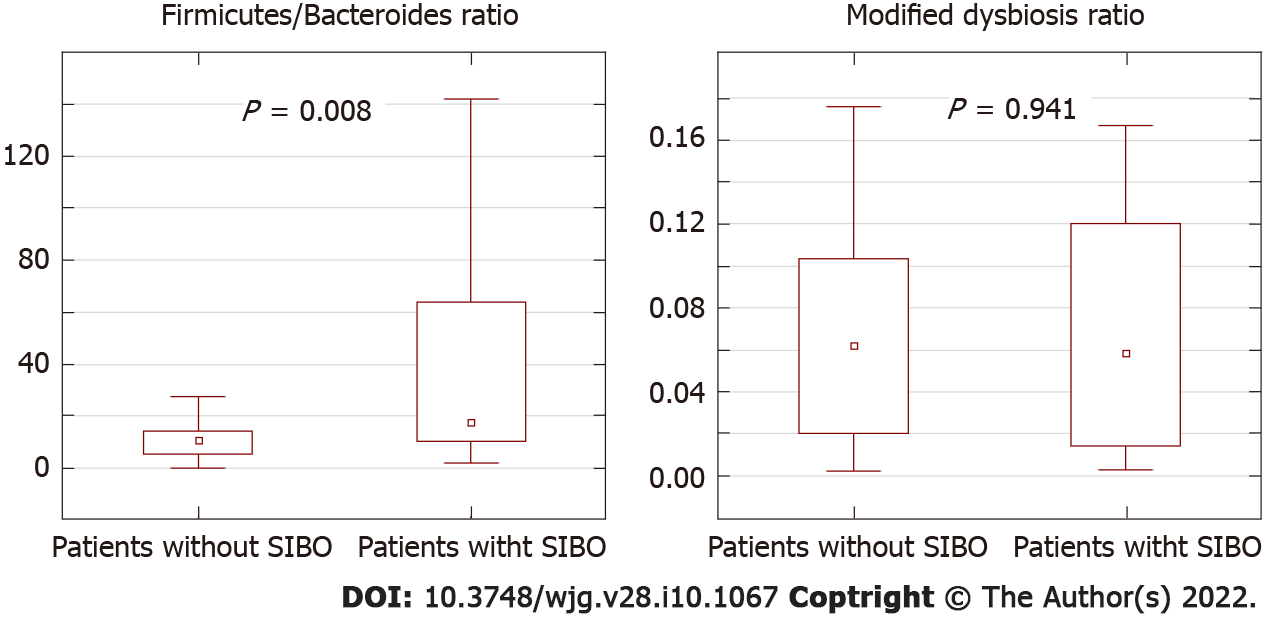
The Firmicutes/Bacteroides ratio was significantly higher in patients with SIBO than in patients without SIBO, while the modified dysbiosis ratio that determines the prognosis in cirrhosis[10] did not differ significantly between the two groups of patients (Figure 3).
We detect SIBO in half of the patients with cirrhosis, which is consistent with the data of previous studies[13]. SIBO is thought be caused by a slowdown in the motility of the small intestine, use of proton pump inhibitors, exocrine pancreatic insufficiency, immune and other disorders[23,24]. Furthermore, an association between delayed orocecal transit and SIBO has been reported in patients with cirrhosis[25]. Despite the presence of immune disorders[26], there are no studies on the association of them with the development of SIBO in cirrhosis. There was no difference in the frequency of using proton pump inhibitors between the patients with and without SIBO in our study.
Cirrhosis-associated gut dysbiosis is represented by an increase in the abundance of Proteobactria that produce active endoctoxin[1-5], and the facultative anaerobic bacteria Bacilli[4,6-8] that is capable of bacterial translocation[27]. This is accompanied by the decrease in the levels of beneficial bacteria from the major families under the Clostridia class[4,7,8,10]. Since the number of Clostridia and Bacilli bacteria can change in opposite directions, the abundance of Firmicutes, which include Clostridia and Bacilli, has been demonstrated to be either increased[9] or decreased[5] in cirrhosis. The change in levels of Bacteriodetes in cirrhosis has been reported to decrease[1,9], increase[2], not change[10], and be increased in compensated cirrhosis, while reduced to normal in decompensated cirrhosis[7].
The mechanisms that lead to gut dysbiosis in cirrhosis are not clear; it is suggested that they may be similar to those for SIBO. If SIBO and gut dysbiosis result from the same factors, it would be logical to assume that dysbiosis is more pronounced in patients with SIBO. However, the abundance of the main biomarkers of cirrhosis-associated dysbiosis (Proteobacteria, Bacilli and Clostridia) was not significantly different between patients with and without SIBO in our study. Moreover, the abundance of Lachnospiraceae, a decrease in which is also one of the biomarkers of cirrhosis-associated dysbiosis[6-8,10], on the contrary, was significantly higher in SIBO.
The gut microbiome has been evaluated depending on the presence of SIBO in patients who do not have cirrhosis in a small number of studies. It was shown that the composition of the small intestine[28,29] and fecal[30] microbiome does not depend significantly on the presence of SIBO. However, among patients with diarrheal variant of irritable bowel syndrome, the abundance of Prevotella were higher, while the abundance of Bacteroides were reduced in persons with SIBO compared with persons without SIBO[31]. Another study revealed that Firmicutes was increased in fecal microbiota of patients with SIBO[32]. Our research also showed an increase in the abundance of Firmicutes and a decrease in the abundance of Bacteroides but without a significant change in the abundance of Prevotella in fecal microbiota of patients with SIBO. Thus, changes in the gut microbiome of SIBO patients with cirrhosis are partly consistent with changes in it in SIBO patients with other diseases and do not coincide with the changes that are characteristic of cirrhosis-associated dysbiosis.
An increase in Firmicutes/Bacteroides ratio which was observed in our study in patients with SIBO is reported in patients with a significant slowdown in orosecal transit in cirrhosis[9]. Nevertheless, there were no differences in the abundance of Proteobacteria and Bacilli between cirrhotic patients with normal and delayed orocecal in that study. It can therefore be assumed that the slowing down of orocecal transit is a main factor in the development of SIBO, but it has a minimal effect on the development of cirrhosis-associated gut dysbiosis.
In our study, bacterial species that mainly increase in patients with SIBO belong to the Blautia genus. These bacteria have the ability to convert primary bile acids into secondary bile acids[33], for example, chenodeoxycholic acid to lithocholic acid. The ratio of lithocholic to chenodeoxycholic acid in feces strongly correlates with the content of Blautia in the gut microbiome of cirrhosis patients[8]. Furthermore, an increase in the content of bile acids in the feces is accompanied by an increase in the abundance of Blautia in persons on a cholerectic diet[34].
The changes in bile metabolism may explain the differences in the gut microbiome of cirrhosis patients with SIBO. The increase in the number of bacteria leads to the increased deconjugation of primary bile acids in the small intestine in patients with SIBO[35]. As deconjugated primary bile acids have a lower affinity for the proteins that carry them through the epithelium of the terminal ileum[36], more of these acids enter the large intestine. These bile acids may be a growth factor for Blautia and similar bacteria. The reduction in the abundance of Bacteroidetes may be the result of suppression their growth by bile acids or due to antagonism with Blautia. A larger study examining the amount and composition of bile acids in feces, the time of orocecal transit, the gut microbiome, and SIBO in cirrhosis should be performed to test this hypothesis.
The strength of our study is represented by the facts that it is the first that assesses the relationship between SIBO and gut dysbiosis in cirrhosis, as well as that we suggested the mechanism of development of changes in the gut microbiota in cirrhosis patients with SIBO and the idea for the next studies that can confirm or refute this hypothesis.
The limitation of our study is its small size, but this did not prevent significant results from being obtained.
In conclusion, we showed that despite the difference in the gut microbiome between patients with and without SIBO, gut dysbiosis and SIBO are most likely relatively independent forms of disorders of the gut microbiota in cirrhosis.
Gut dysbiosis and small intestinal bacterial overgrowth (SIBO) are commonly observed in patients with cirrhosis.
Despite the substantial number of articles describing the relations between disorders of gut microbiota and various manifestations of cirrhosis, dysbiosis and SIBO were always studied separately.
To study the relationship of gut dysbiosis and SIBO in cirrhosis.
This observational study included 47 in-patients with cirrhosis. Stool microbiome was assessed using 16S rRNA gene sequencing. SIBO was assessed using the lactulose hydrogen breath test.
Patients with SIBO had a higher abundance of Firmicutes and Fusobacteria, and a lower abundance of Bacteroidetes than patients without SIBO. This increase in the abundance of Firmicutes occurred mainly due to an increase in the abundance of bacteria from the genus Blautia of the Lachnospiraceae family, while the abundance of other major families of this phylum did not differ significantly between the patients with and without SIBO. There were no significant differences in the abundance of taxa that were the main biomarkers of cirrhosis-associated gut dysbiosis between patients with and without SIBO.
Despite the differences observed in the gut microbiome between patients with and without SIBO, gut dysbiosis and SIBO are most likely independent disorders of gut microbiota in cirrhosis.
Research perspectives are to study the mechanisms of development of SIBO and gut dysbiosis in patients with cirrhosis.
The authors are grateful to the staff of the Department of Hepatology: Zharkova M, Lapshin A, Ondos S, Fedosina E, Tkachenko P, Tikhonov I, and others.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Russia
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Almeida C, Li YL, Salvadori M S-Editor: Gao CC L-Editor: A P-Editor: Gao CC
| 1. | Jin M, Kalainy S, Baskota N, Chiang D, Deehan EC, McDougall C, Tandon P, Martínez I, Cervera C, Walter J, Abraldes JG. Faecal microbiota from patients with cirrhosis has a low capacity to ferment non-digestible carbohydrates into short-chain fatty acids. Liver Int. 2019;39:1437-1447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 102] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 2. | Zeng Y, Chen S, Fu Y, Wu W, Chen T, Chen J, Yang B, Ou Q. Gut microbiota dysbiosis in patients with hepatitis B virus-induced chronic liver disease covering chronic hepatitis, liver cirrhosis and hepatocellular carcinoma. J Viral Hepat. 2020;27:143-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 3. | Kajihara M, Koido S, Kanai T, Ito Z, Matsumoto Y, Takakura K, Saruta M, Kato K, Odamaki T, Xiao JZ, Sato N, Ohkusa T. Characterisation of blood microbiota in patients with liver cirrhosis. Eur J Gastroenterol Hepatol. 2019;31:1577-1583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 4. | Chen Z, Xie Y, Zhou F, Zhang B, Wu J, Yang L, Xu S, Stedtfeld R, Chen Q, Liu J, Zhang X, Xu H, Ren J. Featured Gut Microbiomes Associated With the Progression of Chronic Hepatitis B Disease. Front Microbiol. 2020;11:383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 5. | Zheng R, Wang G, Pang Z, Ran N, Gu Y, Guan X, Yuan Y, Zuo X, Pan H, Zheng J, Wang F. Liver cirrhosis contributes to the disorder of gut microbiota in patients with hepatocellular carcinoma. Cancer Med. 2020;9:4232-4250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 77] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 6. | Lapidot Y, Amir A, Nosenko R, Uzan-Yulzari A, Veitsman E, Cohen-Ezra O, Davidov Y, Weiss P, Bradichevski T, Segev S, Koren O, Safran M, Ben-Ari Z. Alterations in the Gut Microbiome in the Progression of Cirrhosis to Hepatocellular Carcinoma. mSystems. 2020;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 7. | Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, Noble NA, Unser AB, Daita K, Fisher AR, Sikaroodi M, Gillevet PM. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60:940-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 659] [Cited by in RCA: 837] [Article Influence: 76.1] [Reference Citation Analysis (0)] |
| 8. | Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM, White MB, Noble NA, Monteith P, Fuchs M, Thacker LR, Sikaroodi M, Bajaj JS. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949-955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 620] [Article Influence: 51.7] [Reference Citation Analysis (0)] |
| 9. | Liu Y, Jin Y, Li J, Zhao L, Li Z, Xu J, Zhao F, Feng J, Chen H, Fang C, Shilpakar R, Wei Y. Small Bowel Transit and Altered Gut Microbiota in Patients With Liver Cirrhosis. Front Physiol. 2018;9:470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 10. | Maslennikov R, Ivashkin V, Efremova I, Alieva A, Kashuh E, Tsvetaeva E, Poluektova E, Shirokova E, Ivashkin K. Gut dysbiosis is associated with poorer long-term prognosis in cirrhosis. World J Hepatol. 2021;13:557-570. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 11. | Ghosh G, Jesudian AB. Small Intestinal Bacterial Overgrowth in Patients With Cirrhosis. J Clin Exp Hepatol. 2019;9:257-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 12. | Shah A, Shanahan E, Macdonald GA, Fletcher L, Ghasemi P, Morrison M, Jones M, Holtmann G. Systematic Review and Meta-Analysis: Prevalence of Small Intestinal Bacterial Overgrowth in Chronic Liver Disease. Semin Liver Dis. 2017;37:388-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (1)] |
| 13. | Maslennikov R, Pavlov C, Ivashkin V. Small intestinal bacterial overgrowth in cirrhosis: systematic review and meta-analysis. Hepatol Int. 2018;12:567-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 14. | Maslennikov R, Pavlov C, Ivashkin V. Is small intestinal bacterial overgrowth a cause of hyperdynamic circulation in cirrhosis? Turk J Gastroenterol. 2019;30:964-975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 15. | Giannelli V, Di Gregorio V, Iebba V, Giusto M, Schippa S, Merli M, Thalheimer U. Microbiota and the gut-liver axis: bacterial translocation, inflammation and infection in cirrhosis. World J Gastroenterol. 2014;20:16795-16810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 156] [Cited by in RCA: 169] [Article Influence: 15.4] [Reference Citation Analysis (1)] |
| 16. | Simbrunner B, Mandorfer M, Trauner M, Reiberger T. Gut-liver axis signaling in portal hypertension. World J Gastroenterol. 2019;25:5897-5917. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 55] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 17. | Ponziani FR, Zocco MA, Cerrito L, Gasbarrini A, Pompili M. Bacterial translocation in patients with liver cirrhosis: physiology, clinical consequences, and practical implications. Expert Rev Gastroenterol Hepatol. 2018;12:641-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 94] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 18. | Rezaie A, Buresi M, Lembo A, Lin H, McCallum R, Rao S, Schmulson M, Valdovinos M, Zakko S, Pimentel M. Hydrogen and Methane-Based Breath Testing in Gastrointestinal Disorders: The North American Consensus. Am J Gastroenterol. 2017;112:775-784. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 395] [Cited by in RCA: 517] [Article Influence: 64.6] [Reference Citation Analysis (1)] |
| 19. | Fouhy F, Deane J, Rea MC, O'Sullivan Ó, Ross RP, O'Callaghan G, Plant BJ, Stanton C. The effects of freezing on faecal microbiota as determined using MiSeq sequencing and culture-based investigations. PLoS One. 2015;10:e0119355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 194] [Cited by in RCA: 226] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 20. | Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114-2120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30322] [Cited by in RCA: 41200] [Article Influence: 3745.5] [Reference Citation Analysis (1)] |
| 21. | Parikh HI, Koparde VN, Bradley SP, Buck GA, Sheth NU. MeFiT: merging and filtering tool for illumina paired-end reads for 16S rRNA amplicon sequencing. BMC Bioinformatics. 2016;17:491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 22. | Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261-5267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12842] [Cited by in RCA: 13190] [Article Influence: 732.8] [Reference Citation Analysis (0)] |
| 23. | Achufusi TGO, Sharma A, Zamora EA, Manocha D. Small Intestinal Bacterial Overgrowth: Comprehensive Review of Diagnosis, Prevention, and Treatment Methods. Cureus. 2020;12:e8860. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 24. | Bures J, Cyrany J, Kohoutova D, Förstl M, Rejchrt S, Kvetina J, Vorisek V, Kopacova M. Small intestinal bacterial overgrowth syndrome. World J Gastroenterol. 2010;16:2978-2990. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 383] [Cited by in RCA: 364] [Article Influence: 24.3] [Reference Citation Analysis (4)] |
| 25. | Lunia MK, Sharma BC, Sachdeva S. Small intestinal bacterial overgrowth and delayed orocecal transit time in patients with cirrhosis and low-grade hepatic encephalopathy. Hepatol Int. 2013;7:268-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 26. | Irvine KM, Ratnasekera I, Powell EE, Hume DA. Causes and Consequences of Innate Immune Dysfunction in Cirrhosis. Front Immunol. 2019;10:293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 125] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 27. | Berg RD, Garlington AW. Translocation of certain indigenous bacteria from the gastrointestinal tract to the mesenteric lymph nodes and other organs in a gnotobiotic mouse model. Infect Immun. 1979;23:403-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 572] [Cited by in RCA: 543] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 28. | Saffouri GB, Shields-Cutler RR, Chen J, Yang Y, Lekatz HR, Hale VL, Cho JM, Battaglioli EJ, Bhattarai Y, Thompson KJ, Kalari KK, Behera G, Berry JC, Peters SA, Patel R, Schuetz AN, Faith JJ, Camilleri M, Sonnenburg JL, Farrugia G, Swann JR, Grover M, Knights D, Kashyap PC. Small intestinal microbial dysbiosis underlies symptoms associated with functional gastrointestinal disorders. Nat Commun. 2019;10:2012. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 185] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 29. | Shin AS, Gao X, Bohm M, Lin H, Gupta A, Nelson DE, Toh E, Teagarden S, Siwiec R, Dong Q, Wo JM. Characterization of Proximal Small Intestinal Microbiota in Patients With Suspected Small Intestinal Bacterial Overgrowth: A Cross-Sectional Study. Clin Transl Gastroenterol. 2019;10:e00073. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 30. | Noh CK, Lee KJ. Fecal Microbiota Alterations and Small Intestinal Bacterial Overgrowth in Functional Abdominal Bloating/Distention. J Neurogastroenterol Motil. 2020;26:539-549. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 31. | Wu KQ, Sun WJ, Li N, Chen YQ, Wei YL, Chen DF. Small intestinal bacterial overgrowth is associated with Diarrhea-predominant irritable bowel syndrome by increasing mainly Prevotella abundance. Scand J Gastroenterol. 2019;54:1419-1425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 32. | Yang M, Zhang L, Hong G, Li Y, Li G, Qian W, Xiong H, Bai T, Song J, Hou X. Duodenal and rectal mucosal microbiota related to small intestinal bacterial overgrowth in diarrhea-predominant irritable bowel syndrome. J Gastroenterol Hepatol. 2020;35:795-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 33. | Ridlon JM, Devendran S, Alves JM, Doden H, Wolf PG, Pereira GV, Ly L, Volland A, Takei H, Nittono H, Murai T, Kurosawa T, Chlipala GE, Green SJ, Hernandez AG, Fields CJ, Wright CL, Kakiyama G, Cann I, Kashyap P, McCracken V, Gaskins HR. The 'in vivo lifestyle' of bile acid 7α-dehydroxylating bacteria: comparative genomics, metatranscriptomic, and bile acid metabolomics analysis of a defined microbial community in gnotobiotic mice. Gut Microbes. 2020;11:381-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 95] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 34. | Ocvirk S, Wilson AS, Posma JM, Li JV, Koller KR, Day GM, Flanagan CA, Otto JE, Sacco PE, Sacco FD, Sapp FR, Newton K, Brouard F, DeLany JP, Behnning M, Appolonia CN, Soni D, Bhatti F, Methé B, Fitch A, Morris A, Gaskins HR, Kinross J, Nicholson JK, Thomas TK, O'Keefe SJD. A prospective cohort analysis of gut microbial co-metabolism in Alaska Native and rural African people at high and low risk of colorectal cancer. Am J Clin Nutr. 2020;111:406-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 35. | Bala L, Ghoshal UC, Ghoshal U, Tripathi P, Misra A, Gowda GA, Khetrapal CL. Malabsorption syndrome with and without small intestinal bacterial overgrowth: a study on upper-gut aspirate using 1H NMR spectroscopy. Magn Reson Med. 2006;56:738-744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 52] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 36. | Trauner M, Boyer JL. Bile salt transporters: molecular characterization, function, and regulation. Physiol Rev. 2003;83:633-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 737] [Cited by in RCA: 701] [Article Influence: 31.9] [Reference Citation Analysis (0)] |