Published online Feb 14, 2021. doi: 10.3748/wjg.v27.i6.470
Peer-review started: October 20, 2020
First decision: December 8, 2020
Revised: December 21, 2020
Accepted: December 29, 2020
Article in press: December 29, 2020
Published online: February 14, 2021
Processing time: 108 Days and 8 Hours
Liver cancer is a common cancer and the main cause of cancer-related deaths worldwide. Liver cancer is the sixth most common cancer in the world. Although miR-34a and palmitoyl membrane palmitoylated protein (MPP2) are reportedly involved in various cell processes, their precise roles in liver cancer are still unclear.
To investigate the expression of micro RNA 34a (miR-34a), methylation of the miR-34a promoter and the expression of MPP2 in liver cancer cells and their related mechanisms.
Together, 78 cases of liver cancer tissues and 78 cases of adjacent tissues were collected. The methylation degree of miR-34a promoter in liver cancer/ paracancerous tissue and liver cancer cells/normal liver cells, and the expression levels of miR-34a and MPP2 in the above samples were detected. Demethylation of liver cancer cells or transfection of liver cancer cells with miR-34a mimetic was performed. The MPP2 overexpression vector was used to transfect liver cancer cells, and the changes in proliferation, invasion, apoptosis, migration, and other biological functions of liver cancer cells after the above interventions were observed. Double luciferase reporter genes were used to detect the targeting relationship between miR-34a and MPP2.
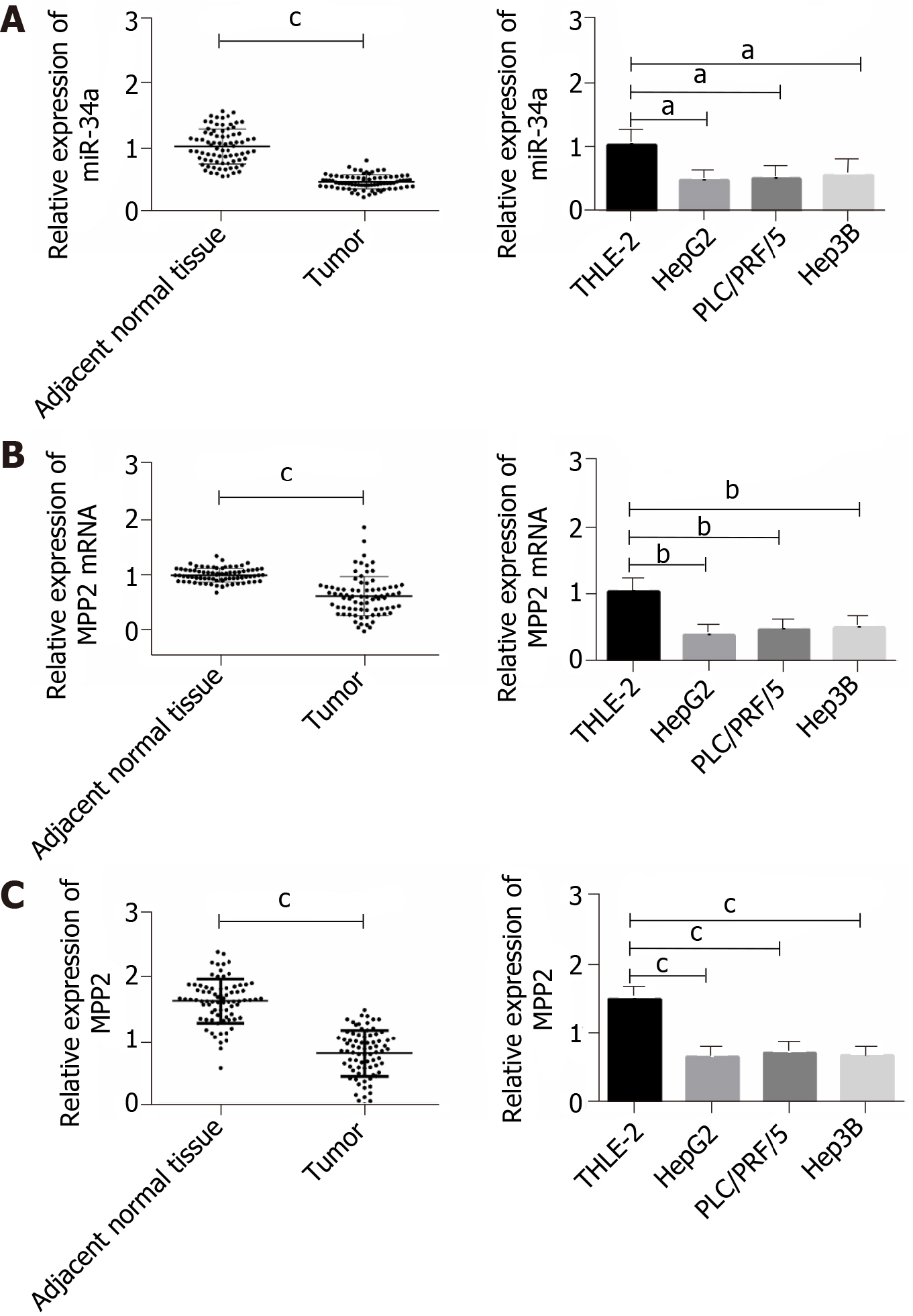
Clinical samples showed that the expression levels of miR-34a and MPP2 in liver cancer tissues were lower than those in the normal tissues. The methylation degree of miR-34a promoter region in liver cancer cells was higher than that in normal liver cells. After miR-34a demethylation/mimetic transfection/MPP2 overexpression, the apoptosis of liver cancer cells was increased; the proliferation, invasion and migration capabilities were decreased; the expression levels of caspase 3, caspase 9, E-cadherin, and B-cell lymphoma 2 (Bcl-2)-associated X protein were increased; and the expression levels of Bcl-2, N-cadherin, and β-catenin were decreased. Double luciferase reporter genes confirmed that MPP2 is targeted by miR-34a. Rescue experiments showed that small interfering MPP2 could counteract the promoting effect of miR-34a demethylation on apoptosis and the inhibitory effect on cell proliferation, invasion, and migration.
miR-34a demethylation upregulates the expression level of MPP2 in liver cancer cells and promotes the apoptosis of liver cancer cells. miR-34a demethylation is a potential method for liver cancer treatment.
Core Tip: This study confirmed the relationship of microRNA 34a (miR-34a) hypermethylation with membrane palmitoylated protein (MPP2) expression by investigating the changes of biological function and MPP2 expression level of liver cancer cells after miR-34a demethylation. miR-34a can inhibit the occurrence of liver cancer by upregulating MPP2, and its demethylation in liver cancer cells is a potential method of liver cancer treatment. In essence, miR-34a methylation is located upstream of its binding site with p53. Therefore, it is valuable to study the role of miR-34a methylation/demethylation on the expression level of upstream regulatory factors such as p53, and to explore its function in the future.
- Citation: Li FY, Fan TY, Zhang H, Sun YM. Demethylation of miR-34a upregulates expression of membrane palmitoylated proteins and promotes the apoptosis of liver cancer cells. World J Gastroenterol 2021; 27(6): 470-486
- URL: https://www.wjgnet.com/1007-9327/full/v27/i6/470.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i6.470
Liver cancer is the sixth most common cancer and second cause of cancer-related deaths worldwide[1]. In China, the population with a higher incidence of liver cancer is aged 50 or above[2]. Hepatitis B virus and hepatitis C virus are the main factors causing liver cancer[3]. The occurrence of liver cancer is a cumulative process. For example, liver cirrhosis mass develops from low to high, which can be regarded as the possible occurrence of liver cancer[4]. In the early stage of liver cancer, tumor resection, liver transplantation, local ablation and other treatment methods have relatively optimistic curative effects, but the recurrence rate of the above treatment in 5 years is relatively high[5]. MicroRNA (miRNA) influences the occurrence of tumors by mediating post-transcriptional regulation of gene expression[4], so targeted administration of miRNA has considerable prospects in tumor treatment.
miRNA is a key member of the non-coding RNA family[6], and can bind to the 3'-untranslated region (UTR) of mRNA to regulate the expression of mRNA[7]. miR-34a plays an important role in cell apoptosis. Inhibition of miR-34a can promote cell proliferation[8]. Yamakuchi et al[9] found that miR-34a binds to the 3'-UTR segment of sirtuin 1 (SIRT1) in human colon cancer cells to inhibit SIRT1 expression and ultimately regulate cell apoptosis. Chang et al[10] believed that p53 trans-activates miR-34a, thus causing gene re-expression and promoting cell apoptosis. miR-34a is involved in the development of liver cancer. Cui et al[11] proved that the low expression of miR-34a is related to the large tumor of liver cancer patients and the high serum alpha-fetoprotein level. miR-34a was considered a predictor of early recurrence of liver cancer. In liver cancer, miR-34 was upregulated by enhancing emodin protein[12].
Membrane palmitoylated protein (MPP) contains many domains for cell-cell interaction[13]. MPP promotes the polarization of epithelial cells and the formation of tight junctions by forming complexes with human Discs Large 1[14]. At the same time, MPP also interconnects the cell membrane-cytoskeleton[15]. MPP may be a specific exosome accessory factor required for the maturation of ribosomal RNA[16], indicating that MPP may affect protein synthesis. Moreover, MPP2 can control cell growth and interfere with intracellular protein by enhancing E7 protein[17].
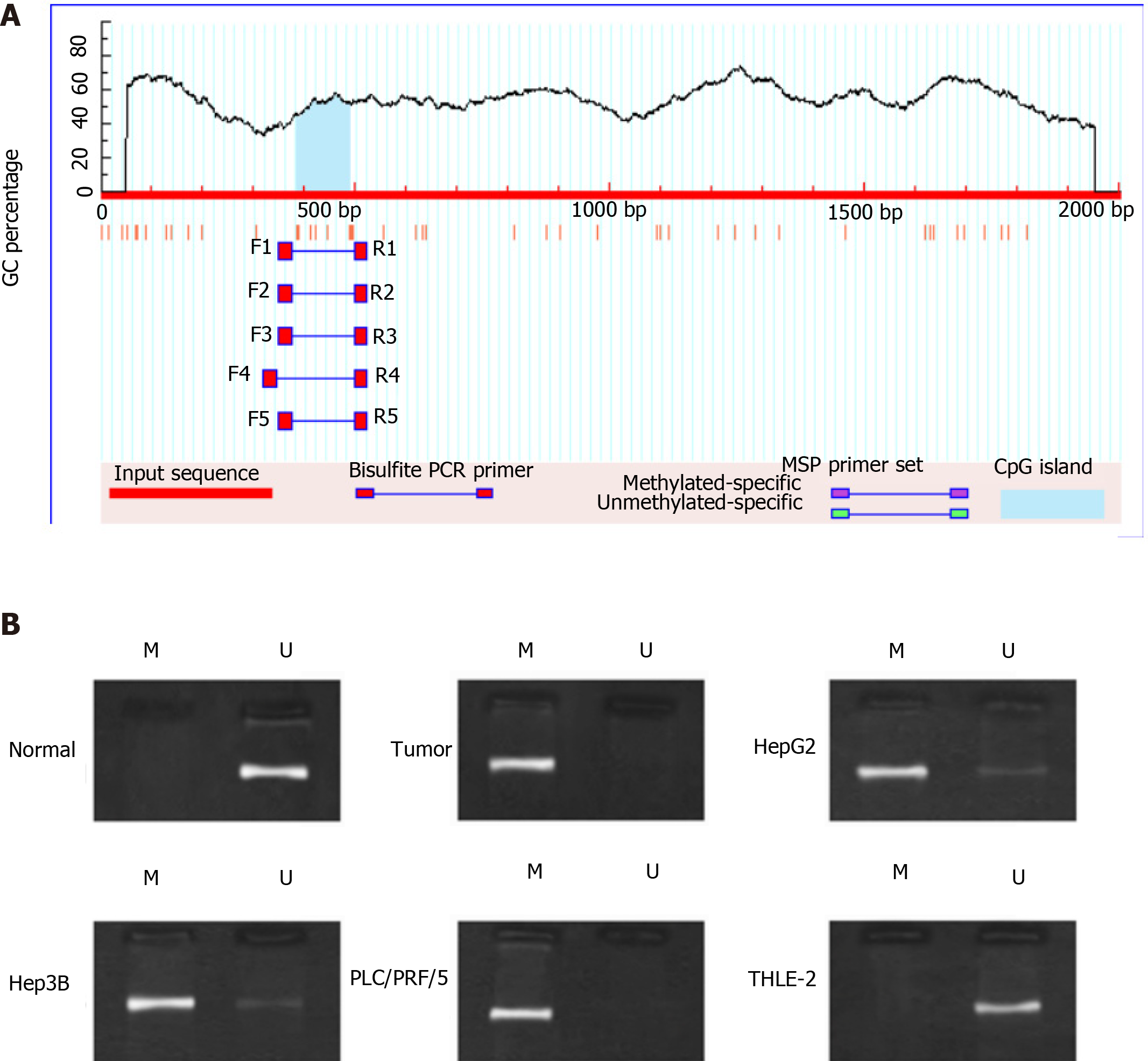
Comparing the expression levels of miR-34a in cancer tissues and paracancerous tissues, we found that miR-34a expression is low in liver cancer. Since abnormal DNA methylation at promoters is frequently involved in tumorigenesis, we explored the methylation of miR-34a promotor. Methprimer predicted that the promoter region of miR-34a has CpG binding sites. Methylation-specific polymerase chain reaction (PCR) found that miR-34a is hypermethylated in liver cancer. Based on this, we speculated that methylation may be the cause of low miR-34a expression in liver cancer, and we designed experiments for confirmation. miRDB and TargetScan databases revealed that MPP2 is downstream of miR-34a through exploration of the molecular mechanism of miR-34a methylation and MPP2 in liver cancer cells.
Tissue samples were collected from 78 patients with liver cancer in Shandong Cancer Hospital affiliated to Shandong University (Shandong, China), including 42 males and 36 females. Inclusion criterion was as follows[4]: Liver cancer was confirmed using blood tests, imaging tests, and liver biopsy. Exclusion criteria were as follows[4]: mentally ill patients; patients with other tumors; patients with a treatment history of surgery, chemotherapy, radiotherapy or antibiotic treatment; and patients who did not cooperate with the treatment. The patients were fully informed of the study, and the study was approved by the Shandong Cancer Hospital affiliated to Shandong University Ethics Committee. Tissue samples and sections were stored in liquid nitrogen for later use.
Hepatocellular carcinoma cell lines (HepG2, Hep3B, PLC/PRG/5) and human normal hepatocytes (THLE-2) were purchased from ATCC (Manassas, VA, United States). The cells were cultured in a cell incubator in 37 ℃ with 5% CO2. The culture medium system was as follows: Dulbecco’s Modified Eagle Medium (DMEM; Hyclone, Logan, UT, United States) + 10% fetal bovine serum (FBS) solution (Gibco, Gaithersburg, MD, United States) + 1% penicillin/streptomycin solution (100X, Solarbio, Beijing, China). Subsequent experiments were carried out when the cell fusion reached 80%-90%.
Before transfection, the medium was replaced without FBS to eliminate interference. Transfected cells were inoculated into 6-well plates with 1 × 105 cells per well. MPP2 small interfering RNA (siRNA) (siMPP2), MPP2-short hairpin RNA (shRNA) (sh-MPP2), negative control (NC) shRNA, miR-34a mimics (pre-miR 34a), and NC mimics (NC pre) were all purchased from Shanghai Sangon Biotech (Shanghai, China). Lipofectamine 2000 Transfection Kit (Invitrogen, Carlsbad, CA, United States) was used for cell transfection. After transfection for 8 h, fresh culture medium was applied and cultured in a humidity incubator including 5% CO2 with constant temperature for 48 h at 37 ℃.
Another group of cells with good growth status were treated with 5-Aza methylation inhibitor (10 mol/L). After 48 h, fresh culture medium was applied. The culture condition was 37 ℃ and 5% CO2.
The tissue sample was cut, milled, and prepared into a cell suspension. Then 1 mL Trizol was added to the cell suspension, and a pipette was used for repeated inhalation, and blow-out of cell suspension was performed to facilitate full cell lysis and extract total RNA from the tissue samples. The concentration and purity of total RNA at 260-280 nm were detected using an ultraviolet spectrophotometer, and those with OD260/OD280 more than 1.8 were used in subsequent experiments. The FastQuant RT Super Mix (KR108) kit (Tiangen Biotech, Beijing, China) was selected for reverse transcription. Then 5 μL of 4 × FQ-RT Super Mix, 1 μg total RNA, and RNase-Free ddH2O were used to replenish the system to 20 μL. Reverse transcription reaction was carried out for 15 min at 42 ℃ and enzyme inactivation was carried out for 3 min at 95 ℃.
Methprimer was used to predict the CpG site of miR-34a promoter. DNA was treated with sulfite using EZ DNA Methylation-GoldTM Kit (Zymo Research, Irvine, CA, United States). Methylation-specific PCR (MSP) detection was carried out using the MSP Kit (Tiangen). The miR-34a MSP primer was designed and synthesized by Shanghai Sangon Biotech. Upstream primer of miR-34a-MSP (5' to 3'): TGTTAGTTTTTTCGGGGAGTTTTCGG; downstream primer of miR-34a-MSP (5' to 3'): ACGCCAACTCCTCCCCCGTCCCGAAC; upstream primer of miR-34a-UMSP (5' to 3'): TAGTTTTTTTGGGGAGTTTTTGGTTT; downstream primer of miR-34a-UMSP (5' to 3'): TAACACCAACTCCTCCCCCATCCCAA.
miR-34a and MPP2 mRNA were quantified using FastFire qPCR PreMix SYBR Green Kit (Tiangen) and ABI PRISM 7000 (Applied Biosystems, Foster City, CA, United States) instruments. The primers were designed and synthesized by Shanghai Sangon Biotech. The upstream primer of miR-34a (5' to 3'): TCCTTCCTACTCGTACCACCAAA, and the downstream primer of miR-34a (5' to 3'): AGGTGAGGAGATGTCGTTGT; upstream primer of MPP2 mRNA (5' to 3'): AGATCT TCTTCTTCAAGGACCGGTT; downstream primer of MPP2 mRNA (5' to 3'): GGCTGGTCA GTGGCTTGGGGTA. The reaction system referred to the kit specification (50 μL). The reaction system was as follows: 1.5 μL upstream primer, 1.5 μL downstream primer, 25 μL of 2 × FastFire qPCR PreMix, 5 μL cDNA template, 5 μL of 50 × ROX Reference Dye, and RNase-Free ddH2O was added to make the total reaction system to 50 μL. The reaction process was as follows: pre-denaturation at 95 ℃ for 3 min, for 1 cycle; denaturation at 95 ℃ for 15 s, annealing at 60 ℃ for 15 s, for a total of 40 cycles. The results were analyzed by ABI PRISM 7000 instrument. The internal reference genes were U6 and GAPDH, which were standardized by 2-△ CT method. The upstream primer of GAPDH (5' to 3'): GTCTCCTCTGACTTCAACAGCG, downstream (5' to 3'): GTCTCCTCTGACTTCAACAGCG. The upstream primer of U6 (5' to 3'): CGCTTCGGCAGCACATATAC, downstream (5' to 3'): AAAATATGGAACGCTTCACGA.
The protein was extracted and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, electrotransferred to nitrocellulose filter membrane, and placed at room temperature for 1 h (closed with 5% of skim milk -phosphate-buffered solution [PBS] solution). MPP2 (at 2 g/mL), caspase 3 (1:5000), caspase 9 (1:10000), E-cadherin (1:10000), B-cell lymphoma 2 (Bcl-2)-associated X protein (Bax) (1:10000), Bcl-2 (1:2000), N-cadherin (1:10000), β-catenin (1:10000) and β-actin (1:10000) primary antibodies were added and placed overnight at 4 ℃. The nitrocellulose membrane was washed with PBS solution for three times, then goat anti-rabbit secondary antibody (horseradish peroxidase [HRP] cross-linked) was added, and the mixture was placed for 1h at room temperature. Finally, the nitrocellulose membrane was washed with PBS solution and visualized using enhanced chemiluminescence method. The internal reference protein was β-actin, and the relative expression level of the protein to be detected was normalized to the intensity of β-actin band.
Caspase 9, caspase 3, E-cadherin, Bax, Bcl-2, N-cadherin, β-catenin, β-actin primary antibody and secondary antibody goat anti-rabbit (HRP cross-linked) were purchased from Shanghai Abcam Company (Shanghai, China).
Trypsin enzymolysis was added to cells, centrifugation was performed to remove trypsin solution, fresh culture medium was added, and repeated inhalation and blow-out of cell suspension were carried out to prepare the cell suspension. Four 96-well plates were taken and cells were inoculated into the well (5 × 103 cells/100 μL per well), with three wells in each group. One well plate was taken out every 24 h, 5 mg/mL MTT solution was added with 10 μL/well, the cells were continued to culture for 1 h, then the culture medium was removed, and the OD value was measured at 570 nm with a microplate reader. Since the number of living cells is proportional to the absorbance, the absorbance value is applied as the cell activity. The experiment was repeated three times to visualize the cell activity-time curve.
Trypsin enzymolysis was carried out to prepare the cell suspension. Cells were inoculated into the migration upper chamber (containing 200 μL 10% FBS +1% DMEM) with 2 × 104 cells/well, and DMEM (containing 10% FBS with a total volume of 500 μL) was added into the lower chamber. After 24 h of cell culture, the upper chamber fluid was removed and the cells on the chamber wall were wiped off. A 4% of polymethanol was used to fix the Transwell opposite cells for 20 min. Crystal violet was used for staining for 15 min, and PBS was used to wash the Transwell chamber. Images of cell migration were collected under a 200-fold microscope. The cell number was calculated by randomly selecting three fields of view, and the average value was taken as the number of transmembrane cells. The experiment was repeated three times. Invasion was paved with 8% matrix glue on the above steps, and the number of cells per well was increased to 5 × 104.
After amplification, MPP2 was cloned into pmirGLO vector to construct pmirGLO-MPP2-wt, and GeneArt TM Site-Directed Mutagenesis PLUS System (Thermo Fisher Scientific, Waltham, MA, United States) to construct pmirGLO-MPP2-mut vector, which was co-transfected with miR-34a, Ncmics, NC pre and pre-miR-34a into cells respectively. After 48 h of transfection, the luciferase intensity was detected using double luciferase reporter genes (Promega, Madison, WI, United States). The operation steps were carried out strictly in accordance with the instructions.
SPSS 20.0 (AsiaAnalytics, Shanghai, China) was applied for index data analysis, and GraphPad Prism 6.0 was used for statistical analysis. Each experiment was repeated three times. The measurement data were expressed by mean ± SD. The comparison between groups was performed using one-way analysis of variance and t-test. Pearson correlation analysis was applied to discuss the correlation between miR-34a and MPP2. All data were tested using two-tailed test. The value of 95% was taken as confidence interval, and the difference was considered statistically significant at aP < 0.05.
In this study, 78 cases of liver cancer tissues and adjacent normal tissues were randomly selected as research objects. qPCR and Western blotting were used to determine the expression of miR-34a and MPP2 (Figure 1). Compared with adjacent normal tissues, miR-34a and MPP2 were lower in liver cancer tissues. Subsequently, the expression levels of miR-34a and MPP2 in three liver cancer cells (HepG2, Hep3B, PLC/PRG/5) and human normal liver cells (THLE-2) were detected. Compared with THLE-2, miR-34a and MPP2 were poorly expressed in liver cancer cells. The above results indicate that miR-34a and MPP2 are expressed at lower levels in liver cancer.
To determine the cause of low expression of miR-34a in liver cancer, methylation of the miR-34a promoter was studied. First, Methprimer was used to predict the CpG site in the miR-34a promoter region. The results showed that there was a CpG site near the promoter 500 bp (Figure 2A). Then the MSP method was used to detect the methylation of miR-34a in tissue samples and cells. The results showed that methylated PCR bands appeared in liver cancer tissues and cells (Figure 2B). Based on this, we speculate that miR-34a expression is low in liver cancer due to methylation.
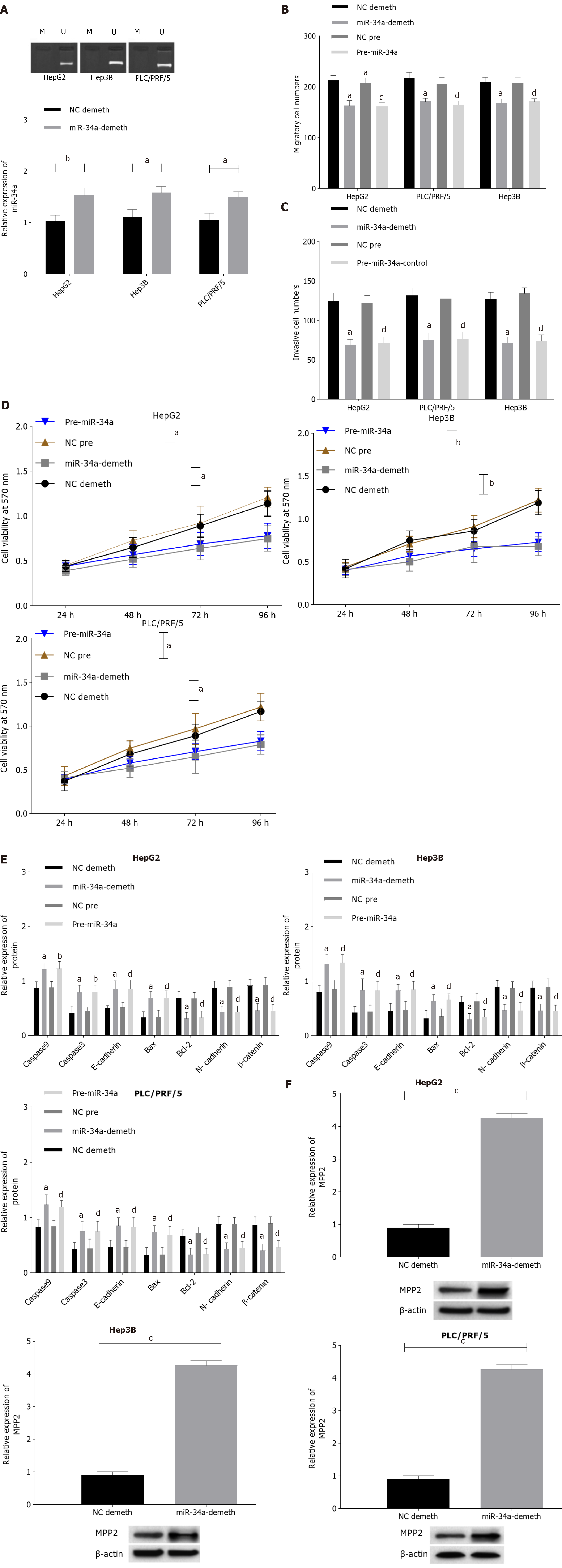
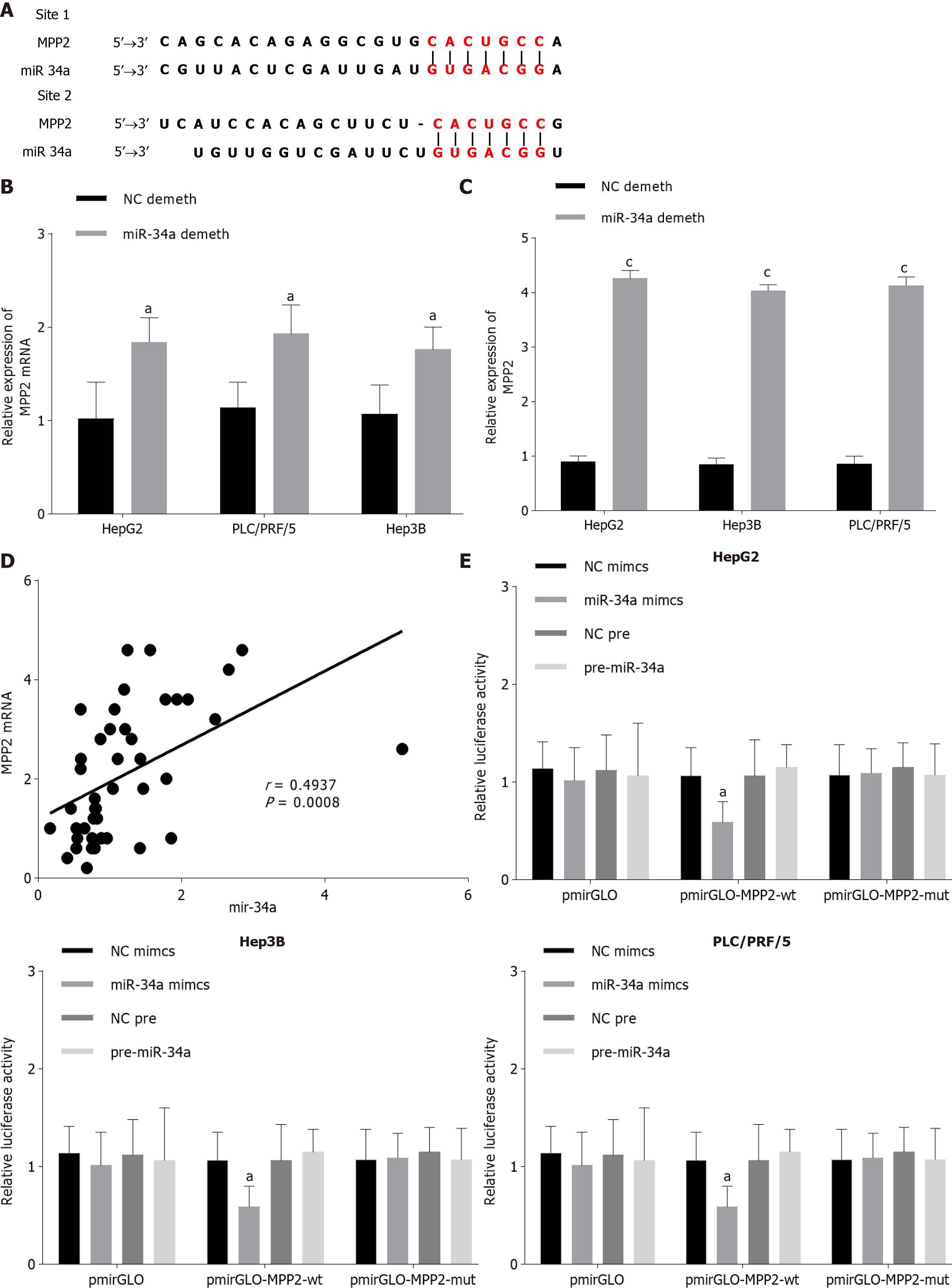
DNA methylation inhibitor 5-Aza was used to demethylate liver cancer cells, the MSP method was applied to detect the methylation of miR-34a, qPCR was applied to detect the expression level of miR-34a, and Western blotting was applied to detect the expression levels of MPP2, caspase 3, caspase 9, E-cadherin, Bax, Bcl-2, N- cadherin, and β-catenin. The MTT method was used to detect the changes in cell activity at different time points and concentrations. The Transwell method was used to detect the invasion and migration ability of cells. The specific results are shown in Figure 3 and Supplementary Figures 1 and 2. After 5-Aza intervention, the methylation level of miR-34a in cells was decreased and the expression level was increased. Compared with the NC demethylation group, the cell activity, cell migration, and proliferation were decreased; the expression of Bcl-2, N-cadherin and β-catenin was downregulated; and the expression levels of caspase 9, caspase 3, E-cadherin, and Bax were upregulated. Compared with the NC pre-group, the pre-miR-34a group also showed a similar change trend in miR-34a demethylation. After miR-34a demethylation or promotion of miR-34a expression level, MPP2 expression level was upregulated in liver cancer cells. The above results indicated that miR-34a expression level is upregulated after miR-34a demethylation, which promotes the expression and apoptosis of MPP2, caspase 9, caspase 3, E-cadherin, Bax, and Bcl-2 and inhibits cell proliferation, migration, and invasion.
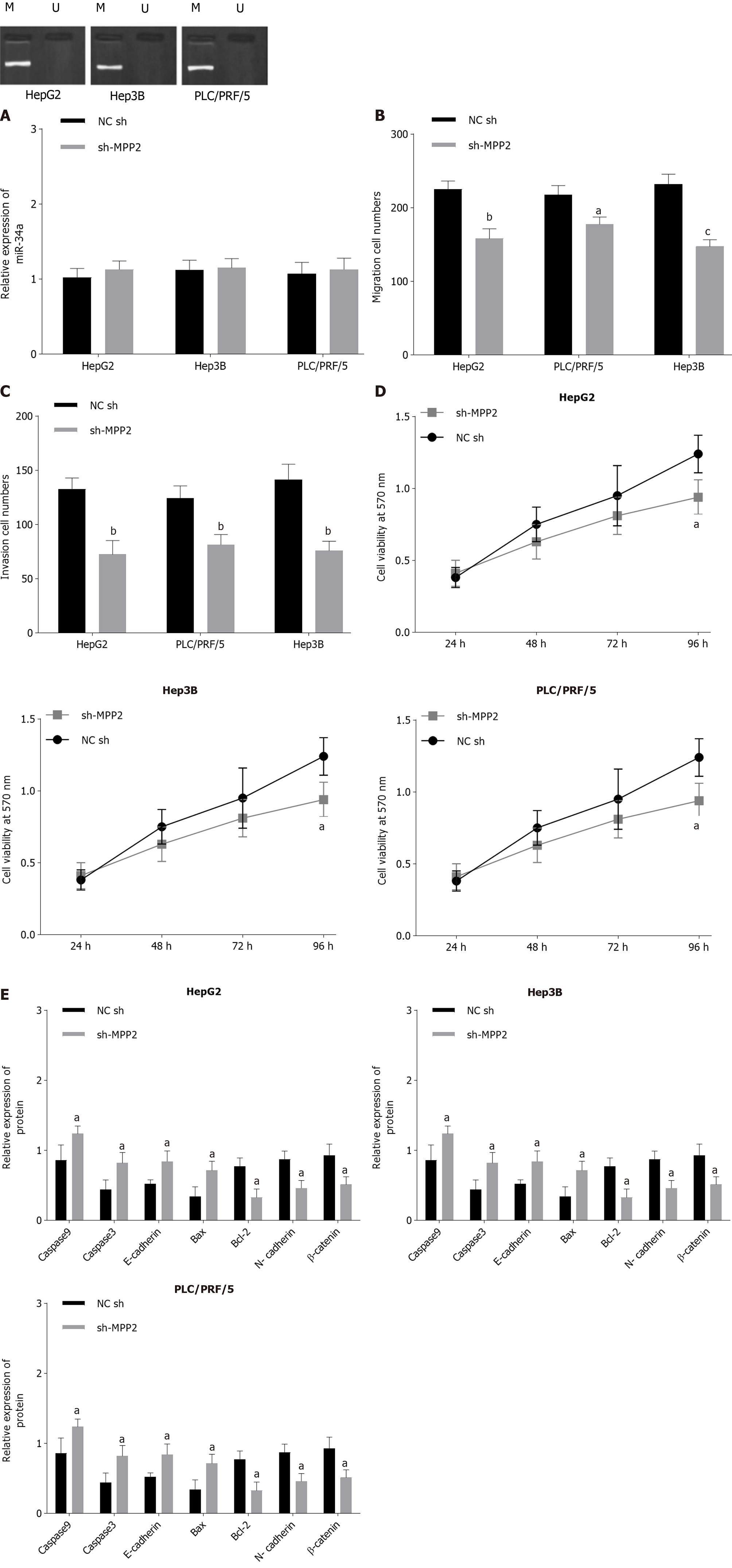
Since upregulation of miR-34a can change the expression level of MPP2, the effect of MMP2 overexpression on the biological function of liver cancer cells was studied. The MPP2 results showed that overexpression of MPP2 did not change the methylation and expression level of miR-34a (Figure 4A). However, the Transwell experiment showed that compared with the NC-sh group, the cell migration number (Figure 4B) and invasion number (Figure 4C) of the sh-MPP2 group decreased significantly. The MTT assay showed that compared with the NC-sh group, the sh-MPP2 group showed a decrease in cell activity (Figure 4D). Western blot analysis showed that compared with the N- sh group, the expression levels of Bcl-2, N-cadherin, and β-catenin in the sh-MPP2 group were downregulated, whereas the expression levels of caspase 9, caspase 3, E-cadherin, Bax, and Bcl-2 were upregulated (Figure 4E). The above results indicate that upregulation of MPP2 promotes cell apoptosis and inhibits cell proliferation, migration, and invasion.
Both the Targetscan and miRDB databases predicted that MPP2 was the target gene of miR-34a (Figure 5A). The upregulation or downregulation of MPP2 expression level was closely related to the upregulation or downregulation of miR-34a expression level (Figure 5B and C). Pearson analysis showed that there was a positive correlation between miR-34a and MPP2 (Figure 5D). To verif the targeted relationship between inflammatory miR-34a and MPP2, a double luciferase reporting experiment was carried out. The results showed that when miR-34a mimics and pmirGLO-MPP2-wt were co-transfected, fluorescence was decreased (Figure 5E). Therefore, miR-34a it appears can target and promote MPP2 expression.
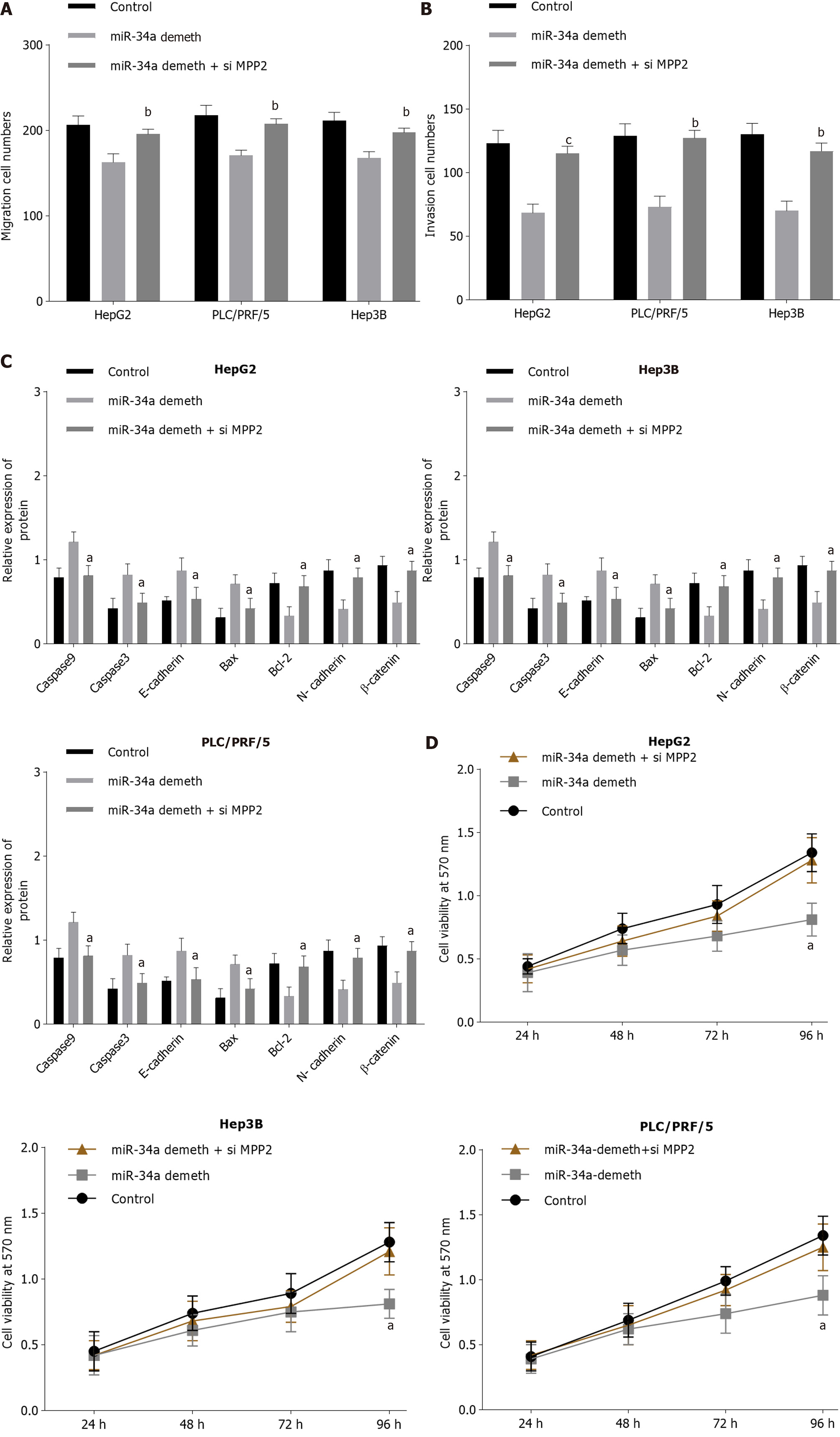
The liver cancer cells were transfected with miR-34a demethylation and miR-34a demethylation + si MMP2 respectively, and corresponding MTT, Western blot analysis, and Transwell experiments were conducted (Figure 6, Supplementary Figures 3-5). Compared with the miR-34a demethylation group, cell migration and proliferation in the miR-34a demethylation + si MPP2 group were increased; cell activity was increased; the expression levels of Bcl-2, N-cadherin, and β-catenin were upregulated; and the expression levels of caspase 9, caspase 3, E-cadherin, and Bax were downregulated. The miR-34a demethylation + si MPP2 group had no significant difference with the control group. The above results show that inhibition of MPP2 can counteract the effect of miR-34a demethylation on cell biological function.
miRNA plays an important role in the process of cell biology, and also affects cell processes such as cell proliferation, differentiation, apoptosis, and growth by regulating the expression level of many genes[18]. miRNA shows dynamic changes in different stages of cells. The high expression level of miRNA has specificity in different stages of cell development, thus playing a specific function in a certain period[19]. As the expression level of miRNA in normal tissues and tumor tissues is quite different[18] and can reflect the development and differentiation of tumor[20], how to regulate miRNA expression level has become a new research direction in cancer therapy. Some studies have claimed that miRNA is strictly regulated by DNA methylation[21]. Oshima et al[22] found that DNA methylation could downregulate miRNA encoded by 14q32 gene, thus inhibiting tumor development. Ning et al[23] showed that DNA (cytosine-5)-methyltransferase 1 and enhancer of zeste homolog 2 mediate DNA methylation and silence miR-200b, miR-200a and miR429, eventually promoting the development of gastric cancer and glioblastoma. Due to the reversibility of methylation and the amplification effect of miRNA, methylated miRNA can be used as a diagnostic index for cancer[24].
The miR-34a promoter has a CpG site near 400 bp to 500 bp (Figure 1B), so its expression level may be regulated by DNA methylation. The results showed that the expression level of miR-34a in liver cancer cells was lower than that in normal liver cells, whereas the methylation degree was higher than that in normal liver cells. After DNA demethylation, the expression level of miR-34a in liver cancer cells was increased; the apoptosis of cells was increased; and the migration, invasion, and proliferation ability were decreased. Many studies[25,26] have suggested that miR-34a can inhibit the proliferation and invasion of liver cancer cells and promote cell apoptosis by acting on different target proteins. The above results indicate that miR-34a is regulated by downregulation or silencing of CpG methylation[27], thus leading to the occurrence of liver cancer. The expression level of miR-34a in liver cancer cells was increased after demethylation, thus playing the role of tumor suppressor to prevent the subsequent development of liver cancer.
The essential action mechanism of miR-34a was to bind with downstream target protein mRNA, thus regulating transcription and translation of target gene and ultimately affecting cell biological function. To discuss the molecular mechanism of miR-34a on regulating cell biological functions, downstream studies of miR-34a were carried out. It was found that miR-34a had an MPP2 mRNA-binding site (Figure 5A), so miR-34a was considered a target on MPP2. The results showed that miR-34a could promote MPP2 expression in a targeted manner, thus causing increased apoptosis and decreased proliferation, migration and invasion of liver cancer cells. MPP2 belongs to the membrane-associated guanylate kinase p55 subgroup and plays an important role in cell-cell interaction[28]. The deletion and downregulation of MPP2 can lead to disorder of the SK2 channel and destroy Ca2+/K+ diffusion, thus affecting cell signal transmission[29]. Baumgartner et al[30] showed that MPP2 plays a role as a tumor suppressor in tumorigenesis, which can inhibit the expression level of c-Src and affect cell proliferation, migration and invasion. Many studies[31-33] have confirmed that the miRNA regulation of genes is not limited to inhibitory effects, as miRNA can also promote gene expression by binding with mRNA. In this study, the effect of miR-34a on liver cancer may be that miR-34a binds to MPP2 mRNA, enhances mRNA transcription, and upregulates MPP2 expression. Upregulation of MPP2 will inhibit the expression level of downstream proteins such as c-Src, promote the recovery of pathways such as SK2, lead to changes in cytoskeleton of liver cancer cells, and eventually lead to weakening of malignant functions such as proliferation, migration and invasion. Cell adhesion plays an important role in cell migration and invasion, that is, cell adhesion inhibits epithelial-mesenchymal transition, which is the basis of cancer cell migration and invasion. MPP2 is a protein closely related to cell adhesion. In this study, the expression levels of intercellular adhesion molecules E-cadherin and N-cadherin were detected, which can be used to evaluate cell adhesion. Cancer cells are often accompanied by downregulation of E-cadherin and upregulation of N-cadherin. In this paper, the demethylation of miR-34a or the upregulation of MPP2 could lead to the upregulation of E-cadherin and downregulation of N-cadherin. However, when miR-34a is demethylated and MPP2 is down-regulated, E-cadherin is downregulated and N-cadherin is upregulated. The above results indicate that demethylated miR-34a may promote cell adhesion through MPP2, and finally inhibit cell migration and invasion. Therefore, the relationship between miR-34a methylation and MPP2 expression can be used as a research point for liver cancer treatment. For the development of miR-34a, demethylation drugs are expected to become a potential treatment for hepatocellular carcinoma. Due to time and cost reasons, this paper failed to launch a suitable in vivo tumor experiment to further study the regulation of demethylated miR-34a/MPP2 axis in solid hepatocellular carcinoma. In addition, miRDB and TargetScan databases predicted that MPP2 might be the downstream target protein of miR-34a, and the double luciferase reporter gene verified that there was a targeting relationship between the two. However, other downstream target genes of miR-34a were not mentioned in this study. This is also the limitation of this paper. Other downstream targets of miR-34a will be discussed in future studies. According to Figure 1, miR-34a and MPP2 are statistically different between normal adjacent tissues and tumor tissues, indicating that miR-34a and MPP2 expression seem to be related to the distance between them and tumor. The lack of exploration on this point is the deficiency of this paper. Therefore, this "expression-distance" relationship will be discussed in depth in future research.
This study identified the relationship of miR-34a hypermethylation with MPP2 expression by studying the changes of biological function and MPP2 expression of liver cancer cells after miR-34a demethylation. It was shown that miR-34a can inhibit the occurrence of liver cancer by upregulating MPP2, and its demethylation in liver cancer cells is a potential method for liver cancer treatment. In essence, the miR-34a methylation site is located on the upstream of its binding site with p53[27]. Therefore, it is valuable to study the role of miR-34a methylation/demethylation on the expression of upstream regulatory factors such as p53, and to explore its function in the future.
Liver cancer is the sixth most common cancer in the world. Although miR-34a and palmitoyl membrane protein (MPP2) are reportedly involved in various cell processes, their precise roles in liver cancer are unknown.
miR-34a expression is low in liver cancer. The promoter region of miR-34a has CpG-binding sites and miR-34a is hypermethylated in liver cancer. MPP2 was downstream of miR-34a via miRDB and TargetScan databases prediction.
This study investigated the expression of miR-34a, the methylation of miR-34a promoter, and the expression of MPP2 in liver cancer cells and their related mechanisms.
The methylation degree of miR-34a promoter and the expression levels of miR-34a and MPP2 in the above samples were detected. miR-34a was demethylated by 5’-Za. miR-34a or MPP2 was upregulated by miR-34a mimics or MPP2 shRNA. The changes in proliferation, invasion, apoptosis, migration and other biological functions of liver cancer cells were observed. Double luciferase reporter genes were used to detect the targeting relationship between miR-34a and MPP2.
miR-34a and MPP2 were downregulated in liver cancer and miR-34a was highly methylated in liver cancer. After miR-34a demethylation/mimetic transfection/MPP2 overexpression, liver cancer cells apoptosis was increased, but the proliferation, invasion and migration capabilities were decreased. MPP2 was targeted by miR-34a.
miR-34a demethylation upregulates the expression of MPP2 in liver cancer cells and promotes the apoptosis of liver cancer cells.
The demethylation of miR-34a is a potential method for liver cancer treatment.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Gassler N, Gupta S, Zhang X S-Editor: Zhang L L-Editor: Filipodia P-Editor: Wang LL
| 1. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20108] [Cited by in RCA: 20504] [Article Influence: 2050.4] [Reference Citation Analysis (20)] |
| 2. | Bosch FX, Ribes J, Díaz M, Cléries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5-S16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1799] [Cited by in RCA: 1816] [Article Influence: 86.5] [Reference Citation Analysis (0)] |
| 3. | Tunissiolli NM, Castanhole-Nunes MMU, Biselli-Chicote PM, Pavarino EC, da Silva RF, da Silva RC, Goloni-Bertollo EM. Hepatocellular Carcinoma: a Comprehensive Review of Biomarkers, Clinical Aspects, and Therapy. Asian Pac J Cancer Prev. 2017;18:863-872. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 60] [Reference Citation Analysis (0)] |
| 4. | Sulas P, Di Tommaso L, Novello C, Rizzo F, Rinaldi A, Weisz A, Columbano A, Roncalli M. A Large Set of miRNAs Is Dysregulated from the Earliest Steps of Human Hepatocellular Carcinoma Development. Am J Pathol. 2018;188:785-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 5. | Xu X, Tao Y, Shan L, Chen R, Jiang H, Qian Z, Cai F, Ma L, Yu Y. The Role of MicroRNAs in Hepatocellular Carcinoma. J Cancer. 2018;9:3557-3569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 131] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 6. | Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov. 2016;6:235-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 401] [Cited by in RCA: 519] [Article Influence: 57.7] [Reference Citation Analysis (0)] |
| 7. | Shukla GC, Singh J, Barik S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol Cell Pharmacol. 2011;3:83-92. [PubMed] |
| 8. | Zhu M, Wu J, Ma X, Huang C, Wu R, Zhu W, Li X, Liang Z, Deng F, Zhu J, Xie W, Yang X, Jiang Y, Wang S, Geng S, Xie C, Zhong C. Butyl benzyl phthalate promotes prostate cancer cell proliferation through miR-34a downregulation. Toxicol In Vitro. 2019;54:82-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 9. | Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci. 2008;105:13421-13426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 950] [Cited by in RCA: 1079] [Article Influence: 63.5] [Reference Citation Analysis (0)] |
| 10. | Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745-752. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1580] [Cited by in RCA: 1575] [Article Influence: 87.5] [Reference Citation Analysis (0)] |
| 11. | Cui X, Wu Y, Wang Z, Liu X, Wang S, Qin C. MicroRNA-34a expression is predictive of recurrence after radiofrequency ablation in early hepatocellular carcinoma. Tumour Biol. 2015;36:3887-3893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 12. | Bai J, Wu J, Tang R, Sun C, Ji J, Yin Z, Ma G, Yang W. Emodin, a natural anthraquinone, suppresses liver cancer in vitro and in vivo by regulating VEGFR2 and miR-34a. Invest New Drugs. 2020;38:229-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 13. | Conte I, Lestingi M, den Hollander A, Miano MG, Alfano G, Circolo D, Pugliese M, Testa F, Simonelli F, Rinaldi E, Baiget M, Banfi S, Ciccodicola A. Characterization of MPP4, a gene highly expressed in photoreceptor cells, and mutation analysis in retinitis pigmentosa. Gene. 2002;297:33-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 14. | Stucke VM, Timmerman E, Vandekerckhove J, Gevaert K, Hall A. The MAGUK protein MPP7 binds to the polarity protein hDlg1 and facilitates epithelial tight junction formation. Mol Biol Cell. 2007;18:1744-1755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 72] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 15. | Stöhr H, Stojic J, Weber BH. Cellular localization of the MPP4 protein in the mammalian retina. Invest Ophthalmol Vis Sci. 2003;44:5067-5074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 16. | Schilders G, Raijmakers R, Raats JM, Pruijn GJ. MPP6 is an exosome-associated RNA-binding protein involved in 5.8S rRNA maturation. Nucleic Acids Res. 2005;33:6795-6804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 89] [Article Influence: 4.5] [Reference Citation Analysis (1)] |
| 17. | Lüscher-Firzlaff JM, Westendorf JM, Zwicker J, Burkhardt H, Henriksson M, Müller R, Pirollet F, Lüscher B. Interaction of the fork head domain transcription factor MPP2 with the human papilloma virus 16 E7 protein: enhancement of transformation and transactivation. Oncogene. 1999;18:5620-5630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 88] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 18. | Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, Gallagher WM, Eccles SA, Croce CM, Esteller M. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci. 2008;105:13556-13561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 795] [Cited by in RCA: 822] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 19. | Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 355] [Cited by in RCA: 373] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 20. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7369] [Article Influence: 368.5] [Reference Citation Analysis (0)] |
| 21. | Han L, Witmer PD, Casey E, Valle D, Sukumar S. DNA methylation regulates MicroRNA expression. Cancer Biol Ther. 2007;6:1284-1288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 226] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 22. | Oshima G, Poli EC, Bolt MJ, Chlenski A, Forde M, Jutzy JMS, Biyani N, Posner MC, Pitroda SP, Weichselbaum RR, Khodarev NN. DNA Methylation Controls Metastasis-Suppressive 14q32-Encoded miRNAs. Cancer Res. 2019;79:650-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 23. | Ning X, Shi Z, Liu X, Zhang A, Han L, Jiang K, Kang C, Zhang Q. DNMT1 and EZH2 mediated methylation silences the microRNA-200b/a/429 gene and promotes tumor progression. Cancer Lett. 2015;359:198-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 145] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 24. | Ezzat WM, Amr KS, Elhosary YA, Hegazy AE, Fahim HH, Eltaweel NH, Kamel RR. Detection of DNA methylated microRNAs in hepatocellular carcinoma. Gene. 2019;702:153-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (1)] |
| 25. | Zhou Y, Liu K, Liu Y, Tan L. MicroRNA-34a inhibit hepatocellular carcinoma progression by repressing hexokinase-1. J Cell Biochem. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 26. | Han R, Chen X, Li Y, Zhang S, Li R, Lu L. MicroRNA-34a suppresses aggressiveness of hepatocellular carcinoma by modulating E2F1, E2F3, and Caspase-3. Cancer Manag Res. 2019;11:2963-2976. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 27. | Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Körner H, Knyazev P, Diebold J, Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591-2600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 590] [Cited by in RCA: 615] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 28. | Rademacher N, Schmerl B, Lardong JA, Wahl MC, Shoichet SA. MPP2 is a postsynaptic MAGUK scaffold protein that links SynCAM1 cell adhesion molecules to core components of the postsynaptic density. Sci Rep. 2016;6:35283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 29. | Kim G, Luján R, Schwenk J, Kelley MH, Aguado C, Watanabe M, Fakler B, Maylie J, Adelman JP. Membrane palmitoylated protein 2 is a synaptic scaffold protein required for synaptic SK2-containing channel function. Elife. 2016;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 30. | Baumgartner M, Weiss A, Fritzius T, Heinrich J, Moelling K. The PDZ protein MPP2 interacts with c-Src in epithelial cells. Exp Cell Res. 2009;315:2888-2898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 31. | Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716-7729. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 177] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 32. | Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 918] [Cited by in RCA: 1019] [Article Influence: 59.9] [Reference Citation Analysis (0)] |
| 33. | Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, Yu A, Shuman M, Yu J, Li LC. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. 2012;40:1695-1707. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 235] [Article Influence: 16.8] [Reference Citation Analysis (0)] |