Published online Nov 14, 2021. doi: 10.3748/wjg.v27.i42.7324
Peer-review started: April 27, 2021
First decision: June 13, 2021
Revised: June 14, 2021
Accepted: October 25, 2021
Article in press: October 25, 2021
Published online: November 14, 2021
Processing time: 196 Days and 11.6 Hours
Recent evidences have shown a relationship between prion protein (PrPc) expression and pancreatic ductal adenocarcinoma (PDAC). Indeed, PrPc could be one of the markers explaining the aggressiveness of this tumor. However, studies investigating the specific compartmentalization of increased PrPc expression within PDAC cells are lacking, as well as a correlation between ultrastructural evidence, ultrastructural morphometry of PrPc protein and clinical data. These data, as well as the quantitative stoichiometry of this protein detected by immuno-gold, provide a significant advancement in understanding the biology of disease and the outcome of surgical resection.
To analyze quantitative stoichiometry and compartmentalization of PrPc in PDAC cells and to correlate its presence with prognostic data
Between June 2018 and December 2020, samples from pancreatic tissues of 45 patients treated with pancreatic resection for a preoperative suspicion of PDAC at our Institution were collected. When the frozen section excluded a PDAC diagnosis, or the nodules were too small for adequate sampling, patients were ruled out from the present study. Western blotting was used to detect, quantify and compare the expression of PrPc in PDAC and control tissues, such as those of non-affected neighboring pancreatic tissue of the same patient. To quantify the increase of PrPc and to detect the subcellular compartmentalization of PrPc within PDAC cells, immuno-gold stoichiometry within specific cell compartments was analyzed with electron microscopy. Finally, an analysis of quantitative PrPc expression according to prognostic data, such as cancer stage, recurrence of the disease at 12 mo after surgery and recurrence during adjuvant chemotherapy was made.
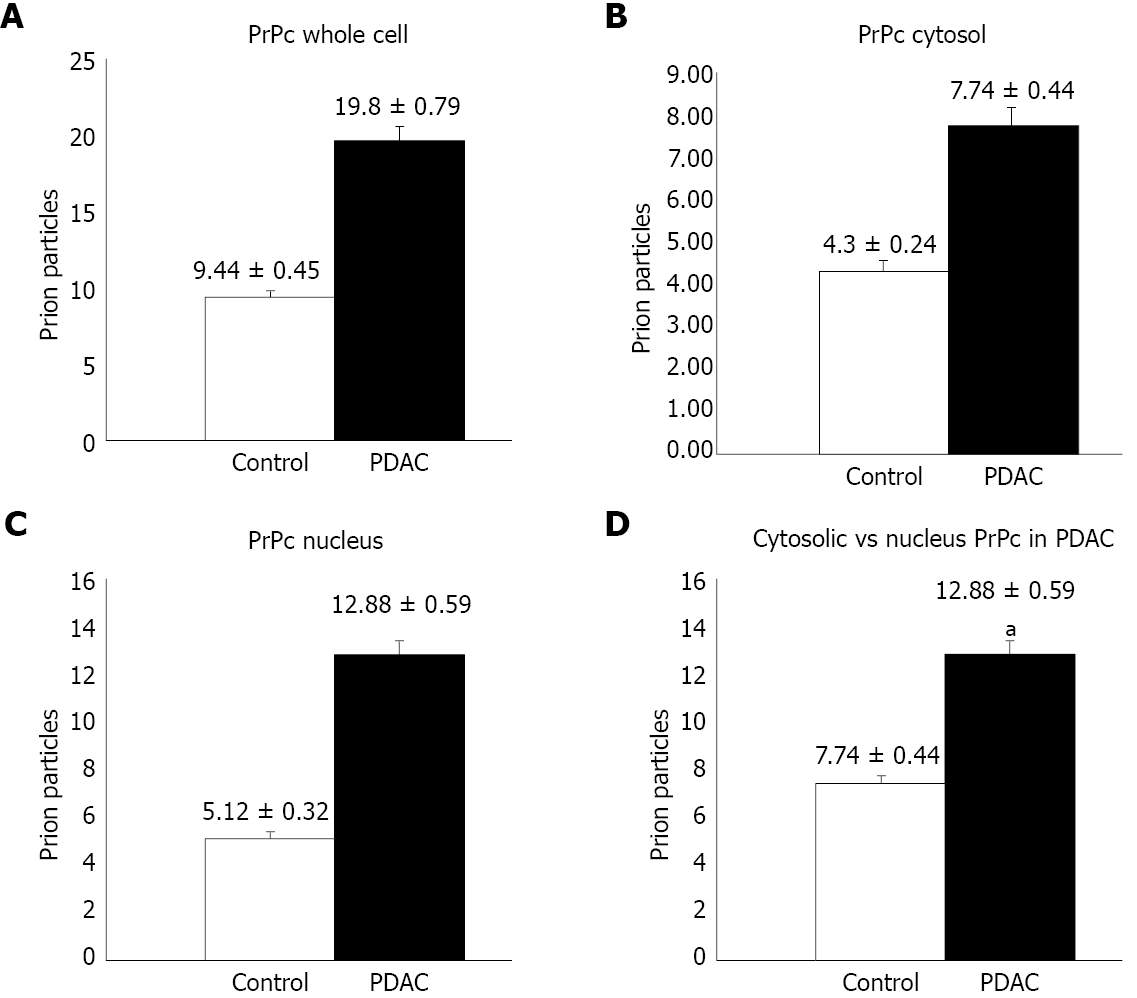
The amount of PrPc within specimen from 38 out of 45 patients was determined by semi-quantitative analysis by using Western blotting, which indicates that PrPc increases almost three-fold in tumor pancreatic tissue compared with healthy pancreatic regions [242.41 ± 28.36 optical density (OD) vs 95 ± 17.40 OD, P < 0.0001]. Quantitative morphometry carried out by using immuno-gold detection at transmission electron microscopy confirms an increased PrPc expression in PDAC ductal cells of all patients and allows to detect a specific compartmentalization of PrPc within tumor cells. In particular, the number of immuno-gold particles of PrPc was significantly higher in PDAC cells respect to controls, when considering the whole cell (19.8 ± 0.79 particles vs 9.44 ± 0.45, P < 0.0001). Remarkably, considering PDAC cells, the increase of PrPc was higher in the nucleus than cytosol of tumor cells, which indicates a shift in PrPc compartmentalization within tumor cells. In fact, the increase of immuno-gold within nuclear compartment exceeds at large the augment of PrPc which was detected in the cytosol (nucleus: 12.88 ± 0.59 particles vs 5.12 ± 0.32, P < 0.0001; cytosol: 7.74. ± 0.44 particles vs 4.3 ± 0.24, P < 0.0001).
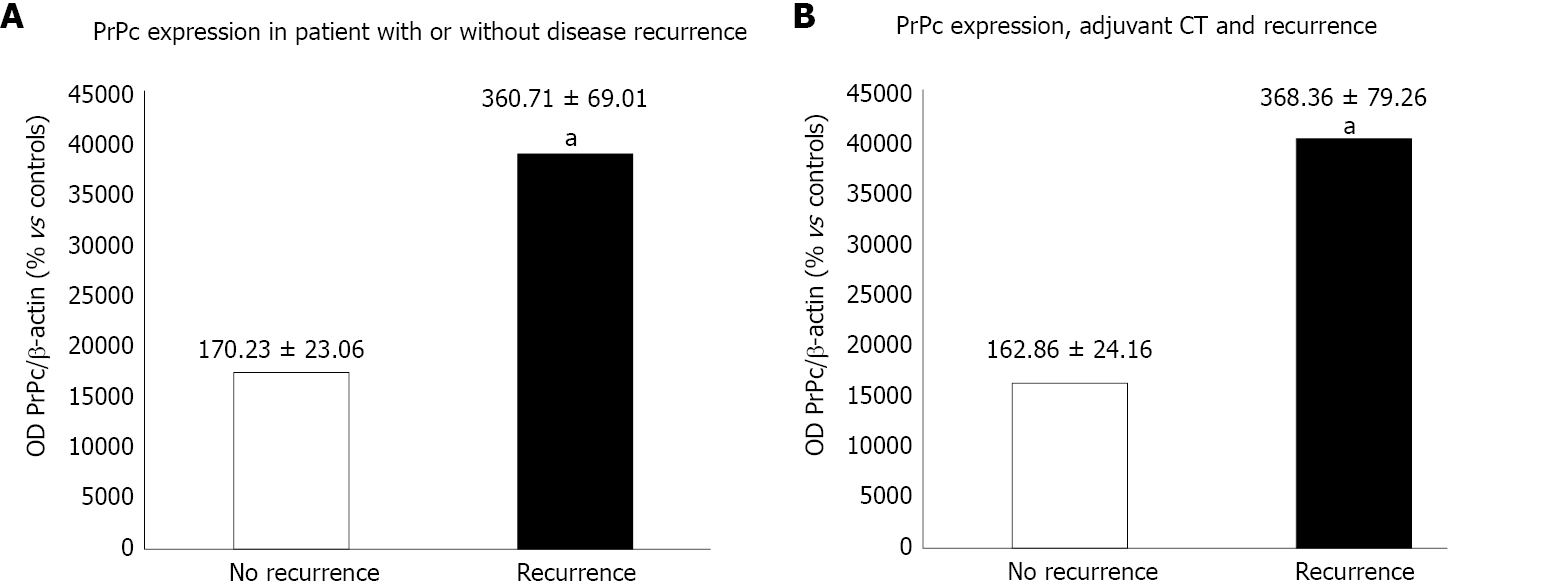
In order to analyze the prognostic impact of PrPc, we found a correlation between PrPc expression and cancer stage according to pathology results, with a significantly higher expression of PrPc for advanced stages. Moreover, 24 patients with a mean follow-up of 16.8 mo were considered. Immuno-blot analysis revealed a significantly higher expression of PrPc in patients with disease recurrence at 12 mo after radical surgery (360.71 ± 69.01 OD vs 170.23 ± 23.06 OD, P = 0.023), also in the subgroup of patients treated with adjuvant CT (368.36 ± 79.26 OD in the recurrence group vs 162.86 ± 24.16 OD, P = 0.028), which indicates a correlation with a higher chemo-resistance.
Expression of PrPc is significantly higher in PDAC cells compared with control, with the protein mainly placed in the nucleus. Preliminary clinical data confirm the correlation with a poorer prognosis.
Core Tip: Pancreatic ductal adenocarcinoma (PDAC) is an extremely lethal cancer and we are far away from understanding its biology. Recent in vitro evidence hypothesizes some role of cellular prion protein (PrPc) in PDAC carcinogenesis. We found that this protein is over-expressed in vivo within PDAC tissue of surgically resected patients, here we quantify the stoichiometric increase and provide evidence for a shift towards a nuclear compartmentalization. Such a protein amount is associated with poorer post-surgical prognosis and neuro-invasion. Thus, PrPc might be a key in the puzzled evidence of PDAC biology leading to better comprehend and cure such an aggressive disorder.
- Citation: Bianchini M, Giambelluca MA, Scavuzzo MC, Di Franco G, Guadagni S, Palmeri M, Furbetta N, Gianardi D, Funel N, Ricci C, Gaeta R, Pollina LE, Falcone A, Vivaldi C, Di Candio G, Biagioni F, Busceti CL, Morelli L, Fornai F. Detailing the ultrastructure’s increase of prion protein in pancreatic adenocarcinoma. World J Gastroenterol 2021; 27(42): 7324-7339
- URL: https://www.wjgnet.com/1007-9327/full/v27/i42/7324.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i42.7324
Pancreatic cancer is currently the fourth most frequent cause of death among cancers with a prevalence, which is increasing, mostly in western countries, where it is estimated to become the second prevalent cause of death from cancer by 2030[1]. The overall median survival of patients with pancreatic ductal adenocarcinoma (PDAC) is 6 mo and the 5-year survival rate is less than 10%[2,3].
Surgical resection is still the only approach with a curative intent, but it is feasible for less than 20% of patients at diagnosis, while 80% of cases are considered too advanced for surgery, and this is based on regional infiltration or distant metastasis[4]. Even in surgically eligible cases, the 5-years survival rate is extremely low, since patients experience early local or distant relapse. The aggressiveness of the disease is mainly related to the extensive local infiltration and to the early lymphatic and hematogenous spread, which rely on the high propensity of these cells to produce neuro-invasion.
In this scenario, an in-depth knowledge of the biology of the disease is fundamental. The comprehension of the mechanisms involved in tumorigenesis is needed, in order to discover early diagnostic tools, and novel therapeutic strategies to improve patients’ prognosis. When comparing PDAC with other tumors, the biology of the disease is scarcely investigated, and it remains largely unknown.
Recent evidence demonstrates that cellular Prion Protein (PrPc) is overexpressed either in vitro and in vivo in PDAC cells and that it interacts with several pathways, enhancing cellular growth, tumoral proliferation and invasion[5-7].
Still, the occurrence of increased PrPc was described so far by using quite general approaches, in the absence of subcellular localization of the protein and without a quantitative, stoichiometric count of protein particles within cancer cells. Thus, we felt it mandatory to provide a quantitative measurement of the increase in PrPc, by using ultrastructural stoichiometry, which is more reliable in protein analytical detection compared with immunohistochemistry. Again, understanding cell compartmentalization of PrPc, and whether this is shifted within PDAC cells is key to establish its potential role in the biology of disease. This is expected to improve the knowledge about the intrinsic role of PrPc in PDAC pathogenesis. So far, studies focusing on these aspects based on an in vivo approach are lacking. Again, it is fundamental to strengthen whether a higher expression of PrPc in PDAC cells does associate with specific clinical phenotypes of PDAC. Therefore, the aim of this study is to quantify the increase in PrPc and analyze the subcellular localization of PrPc in PDAC cells from surgically resected patients. These findings are correlated with clinical data, in order to dissect a potential role of PrPc as a biological marker to predict disease severity.
Samples from tumors of patients surgically treated with pancreatic resections at our Institution were collected between January 2018 and December 2020. Written informed consent was obtained from patients to use their surgical specimens and clinical pathological data for research purposes.
All patients had a preoperative suspicion of PDAC. Preoperative evaluation included medical history, physical, laboratory and radiological examinations, computed tomography (CT) and magnetic resonance imaging, often with magnetic resonance cholangiopancreatography. In addition, abdominal ultrasound with and without contrast, endoscopic ultrasonography (EUS), and fine-needle aspiration (FNA) during EUS were also performed in selected patients. Preoperative data included age and gender.
All the specimens were frozen intraoperatively and further sliced and scored for histology. Tissue from pancreatic nodules other than adenocarcinomas were ruled out from the present study.
Similarly, we could not proceed when tumor specimens were too small. When PDAC diagnosis was confirmed, the pathologist took specimens from the pancreatic tumor and from normal pancreatic tissue. From each specimen (either from controls or tumor tissues) a part was fixed and kept in glutaraldehyde and paraformaldehyde for electron microscopy analysis, while the other one was rapidly frozen and kept at -80°C for storage until for western blotting analyses (SDS-PAGE immunoblotting) to be carried out.
Histological data included: (1) Histological phenotype of the tumor; (2) Grade of differentiation; (3) Tumor size; (4) Number of harvested lymph nodes; (5) Number of metastatic lymph nodes; and (6) Occurrence of angio-invasion and peri-neural infiltration.
Patients were staged according to the T and N definitions proposed for the AJCC 8th edition[8]. Proposed T-stage definitions are the following: T1 ≤ 2 cm maximal diameter, T2 > 2 ≤ 4 cm maximal diameter, T3 > 4 cm maximal diameter, T4 = locally unresectable. Extra-pancreatic extension was not included in T-stage definitions. The N-staging included the following: N0 = node negative, N1 = 1-3 nodes positive for metastatic disease, N2 ≥ 4 nodes positive for metastatic disease.
Pathology, post-operative disease outcome and oncologic follow-up were prospectively retrieved and organized in a specific database. For our clinical analysis patients with at least 12 mo from surgical resection were considered. The degree of PrPc expression in PDAC was reported and compared also on the basis of cancer stage according to AJCC 8th edition.
Pancreatic tissue to be immunoblotted was homogenized in ice-cold lysis buffer containing 50 mmol/L Tris-HCl (pH = 7.5), 150 mmol/L NaCl, 5 mmol/L EDTA, 1% SDS, 0.1% IGEPAL (NP40) and Complete Protease Inhibitor Cocktail Tablet (leupeptin, pepstatin, aprotinin) (Santa Cruz Biotechnology, Dallas, TX, United States). Then tissue was sonicated, and homogenates were centrifuged at 5000 x g for 5 min. An aliquot of supernatant was used to determine the protein concentration by a protein assay kit (Sigma-Aldrich, St. Louis, MO).
Samples (25 μg) were separated on 4%-20% sodium dodecyl sulfate-polyacrylamide gel. Following electrophoresis, proteins were transferred to a nitrocellulose membrane (Biorad; Milano, Italy). The membrane was then immersed in blocking solution containing PBS with 0.05% Tween-20 (PBS-T) and 5% not fat dried milk (Sigma) for 1 h at room temperature. Then the membrane was incubated overnight at 4°C with primary antibody anti-protein PrPc (1:2000, Abcam, Cambridge, United Kingdom) diluted in PBS-T containing 2% BSA (Sigma). The blots were washed three times with PBS-T and incubated for 1 h with goat anti-rabbit horseradish peroxidase-labelled secondary antibody (1:2000; KPL, Maryland, United States) diluted in PBS-T containing 2% not fat dried milk (Sigma).
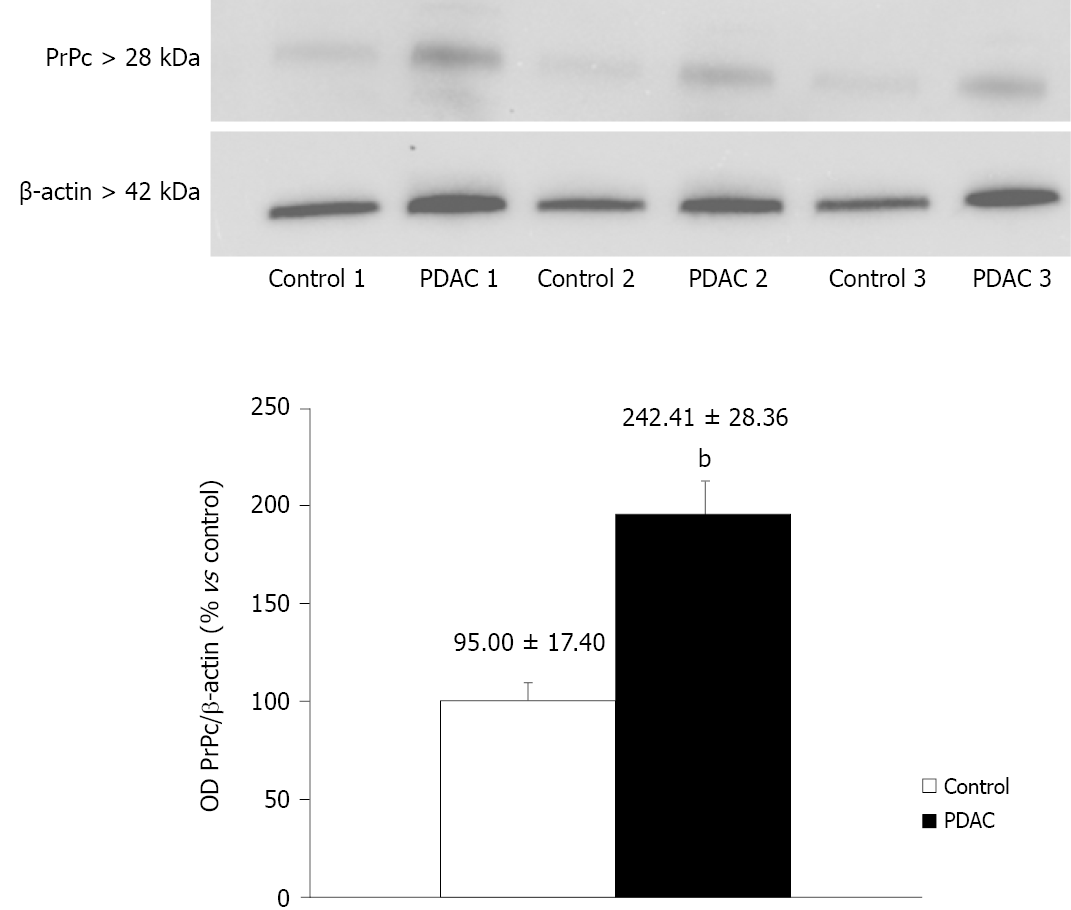
The bands were visualized with enhanced chemiluminescence reagents (Immuno-Star HRP Substrate; Bio-RadLaboratories) and image analysis was carried out by ChemiDoc System (Bio-RadLaboratories). β-Actin was used as an internal standard for semi-quantitative protein measurement, so-called “house-keeping protein”. Densitometric analysis was performed with ImageJ software and the unit of measure was the optical density (OD). Western blotting of PDAC tissues were compared with control tissues.
When preparing embedded pancreas tissue blocks for electron microscopy analysis, firstly we carried out semi-thin sections in order to better focus on those areas of the tissue where ductal and parenchymal area could be evidenced. Each semi-thin section owning a thickness range of 3-6 μm was cut at ultra-microtome. After cutting, slices were picked up with an iron loop 1 cm long and 2 mm thick. By using the loop, the slice was moved into a drop of distilled water lying on a glass slide and it was then placed on a hot plate at approximately 60°C to be dried. Then, 1 or 2 drops of a toluidine blue staining solution were added on the semi-thin slice. When the edge of the staining drop switched the color from blue to metallic gold, the slide was removed quickly from hot plate to be rinsed with distilled water. Finally, these slides were mounted by using the mounting medium DPX to be analyzed by a Nikon Eclipse 80i light microscope, which was connected to the NIS Elements software for image analysis (Nikon, Tokyo, Japan).
For electron microscopy small fragments of normal and tumoral pancreatic tissue were fixed in 0.1% glutaraldehyde and 2 % paraformaldehyde in phosphate buffer pH = 7.4 for 90 min, creating a fixing solution minimally covering antigen epitope, while fairly preserving tissue architecture.
After washing for 10 min in buffer, samples were post-fixed in 1% OsO4 buffered solution for 1 h at 4°C. Then samples were dehydrated in a series of increasing ethanol concentration (50%, 70%, 90% 95%, 100%) followed by propylene oxide for 20 min. Afterward, samples were embedded in a mixture of Epon-Araldite and propylene oxide (ratio of 1:1 overnight at room temperature) and finally they were embedded in pure Epon-Araldite resin for 72 h at 60°C. Ultra-thin sections were stained with uranyl acetate and lead citrate and examined with Jeol Jem 100SX transmission electron microscope (TEM) (Jeol, Tokyo, Japan) at an acceleration voltage of 80 kV.
Post-embedding procedure was carried out on ultrathin sections collected on nickel grids. Grids were washed in PBS and incubated in a blocking solution containing 10% goat serum and 0.2% saponin for 20 min, at room temperature, then they were incubated with a primary antibody solution containing anti-rabbit Prion protein PrPc (Abcam, diluted 1:50), with 0.2% saponin and 1% goat serum in a humidified chamber, overnight, at 4°C. After washing in PBS, grids were incubated with secondary anti-rabbit antibodies conjugated with gold particles (20 nm mean diameter, BB International Crumlin, United Kingdom) which were diluted 1:40 in PBS containing 0.2 % saponin and 1% goat antiserum for 1 h, at room temperature. Slices used as methodological controls were incubated with secondary antibody only.
After washing in PBS, grids were incubated on droplet of 1% glutaraldehyde for 3 min; an additional extensive washing of grids with distilled water was carried out to remove an excess of salt traces. Sections were stained with uranyl acetate and lead citrate and examined at TEM.
From each experimental group (Controls and PDAC) 20 grids were observed each one containing a mean of 5 cells: In particular we selected the region in which the cellular ducts were present. In order to measure the distribution of the immuno-gold particles, first we counted the total number of gold particles within each cell, then their numbers in nucleus and in cytosol. TEM analysis was performed at a magnification of 6000-8000x which allowed the concomitant visualization of immuno-gold particles and all cell organelles, using higher magnification when it was necessary to better visualize the immuno-gold particles and lower magnification when an ensemble view of the whole ultrastructure was requested for our analysis. The expression of PrPc was revealed by counting the immuno-gold particles in whole cells, nucleus and cytosol both in control and PDAC groups.
Continuous variables with normal distribution are expressed as the mean ± SD of the mean and they were compared by using Student’s t test. A P value equal or lower than 0.05 was arbitrarily considered as not due to biological variability (H0 hypothesis rejected). The software used for such a statistical analysis was SPSS (Statistical Production and Service Solution for Windows, SPSS Inc., Chicago, IL, United States), version 23.
Surgical specimens of 45 patients were collected. 7 of them were ruled out because of the small dimension of the tumor that prevented an adequate sampling or because the diagnosis of PDAC was excluded at frozen section.
The samples from 38 PDAC patients which were included in the analysis, 19 (50%) were from male, while 19 (50%) from female. The mean age was 72.7 ± 7.9 years (range 52-87). Histological exam confirmed the presence of PDAC in all these 38 cases. The grading of the pancreatic tumor was “moderately differentiated” in 35/38 cases (92.1%), “poorly differentiated” in 3/38 (7.9%). The mean tumor size was 3.2 ± 1.1 cm (range 1.5-6.5 cm). The mean harvested lymph nodes were 34.2 ± 15 (range 14-79). Metastatic lymph-nodes occurred in 30/38 cases (78.9%) with a number of metastatic lymph-nodes of 4.5 ± 4.9 (range 1-23). The presence of angio-invasion was reported in 5/38 cases (13%), while the presence of peri-neural infiltration was reported in 33/38 cases (86.8%). Three cancer stage groups were identified according to pTNM, stage I (n = 8, 21.1%), stage II (n = 14, 36.8%) and stage III (n = 16, 42.1%). These data are shown in Table 1.
| Number of patients, n | 38 |
| Pancreatic ductal adenocarcinoma, n (%) | 38 (100) |
| Mean tumor dimension, cm | 3.2 1.1 (1.5-6.5) |
| Mean harvest lymph nodes, n | 34.2 15 (14-79) |
| Mean metastatic lymph nodes, n | 4.5 4.9 (1-23) |
| Angioinvasion, n (%) | 5 (13) |
| Perineural infiltration, n (%) | 33 (86.8) |
| T status, n (%) | |
| T1 | 2 (8.7) |
| T2 | 14 (60.9) |
| T3 | 7 (30.4) |
| T status, n (%) | |
| T1 | 3 (7.9) |
| T2 | 24 (63.2) |
| T3 | 11 (28.9) |
| N status, n (%) | |
| N0 | 7 (18.4) |
| N1 | 15 (39.5) |
| N2 | 16 (42.1) |
| Stage, n (%) | |
| I | 8 (21.1) |
| II | 14 (36.8) |
| III | 16 (42.1) |
| Patients with available follow-up, n | 24 |
| Patients with disease recurrence at 12 months, n (%) | 13 (54.1) |
| Patients treated with adjuvant CT, n (%) | 21 (87.5) |
| Recurrence during CT, n (%) | 11 (52.4) |
PrPc was markedly expressed in tumor pancreatic tissues, while the expression in non-cancer tissues was scarce, with a significant difference (242.41 ± 28.36 OD vs 95 ± 17.40 OD, P < 0.0001) (Figure 1).
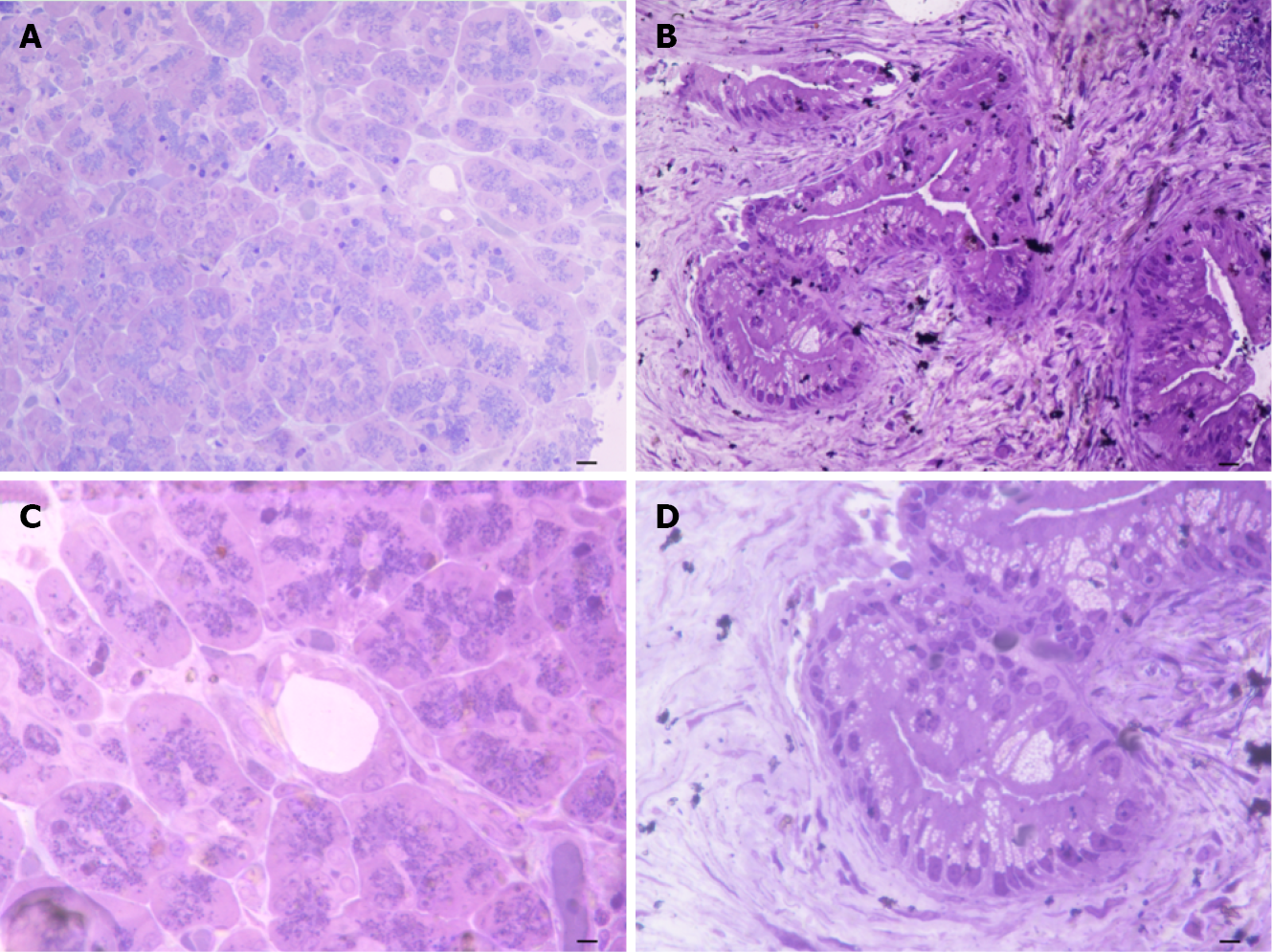
When observing representative semi-thin sections at different magnification (Figure 2) the difference between control and PDAC pancreas was strikingly evident, mostly at the level of ductal tissue. In detail, the pancreas from control at low magnification possesses a quite homogeneous staining at toluidine blue, where ductal areas are simply evident as empty roundish areas surrounded by cell staining as much as those in the neighboring parenchyma (Figure 2A). This was further evidenced at higher magnification showing a pale toluidine staining of ductal cells (Figure 2B). Conversely, in the pancreas affected by PDAC, low magnification showed a highly non-homogeneous tissue where ductal regions were markedly stained. Also, neighboring areas possess a scattered toluidine staining (Figure 2C). At high magnification, the ductal cells from PDAC are overwhelmed and they tend to occlude the ductal lumen with multiple cell layers with an abnormal shape and abundant cell protrusions (Figure 2D). This ductal tissue was the topographical reference to proceed with electron microscopy analysis.
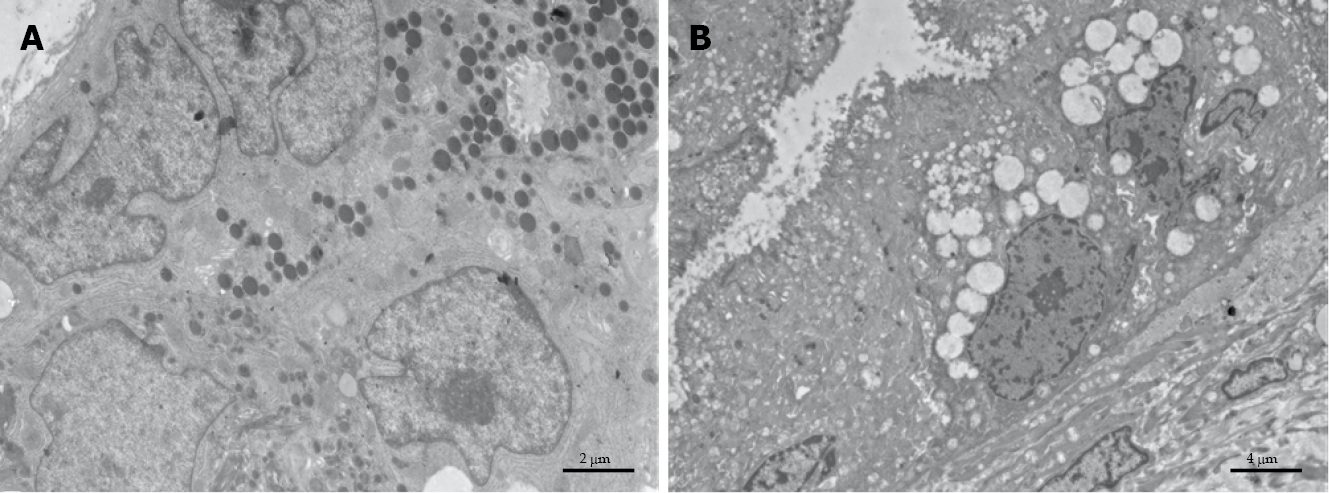
The ultrastructure of pancreatic healthy cells owns normal architecture with well-preserved cell compartments and well-defined membranes. The zymogen granules maintain their integrity, which suggests fair biochemical activity (Figure 3A). Conversely, PDAC cells (Figure 3B) possess severe cell pathology, which is concomitant with a marked derangement of vacuolar compartment and damaged organelles.
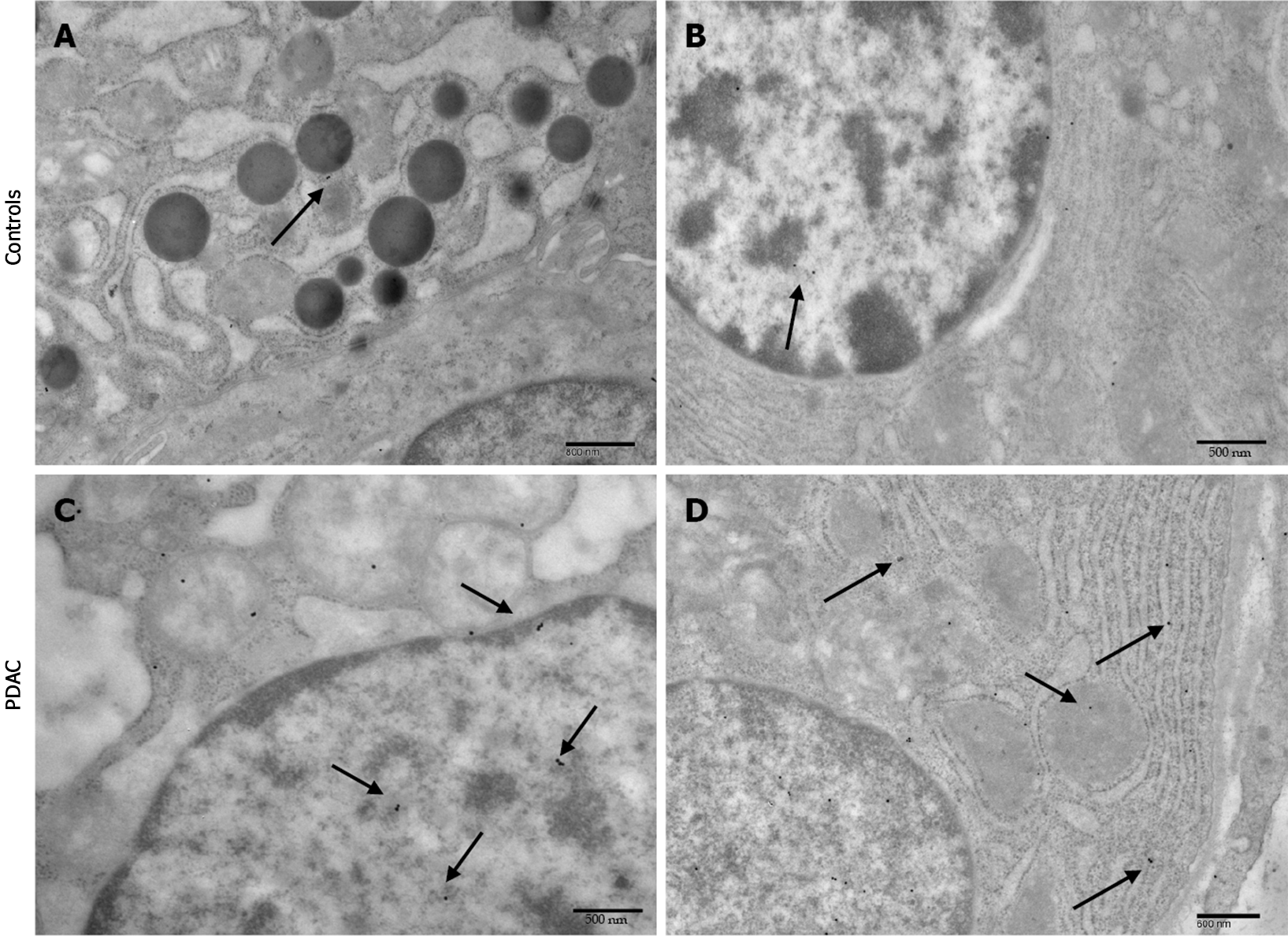
Immuno-cytochemistry shows increased PrPc expression in PDAC cells of all patients. In particular the number of immuno-gold particles was significantly higher in PDAC cells compared with controls (Figure 4) either considering the whole cell (19.8 ± 0.79 particles vs 9.44 ± 0.45, P < 0.0001) (Figure 5A), or the nuclear compartment (12.88 ± 0.59 particles vs 5.12 ± 0.32, P < 0.0001) (Figure 5C) or the cytosol (7.74. ± 0.44 particles and 4.3 ± 0.24, P < 0.0001, respectively) (Figure 5B).
Within PDAC cells, PrPc was more abundant in the nucleus compared with the cytosol (12.88 ± 0.59 particles vs 7.74. ± 0.44, P < 0.0001) (Figure 5D). Conversely, in normal cells no difference was noticeable for the placement of PrPc within nucleus and cytosol (5.12 ± 0.32 particles and 4.3 ± 0.24, P > 0.05, respectively).
When correlating within PDAC group the amount of PrPc expression with specific cancer stages, a significantly higher expression of PrPc for advanced stages was detected. In particular, PrPc expression at Western Blotting was 161.69 ± 63.92 OD in stage I, 173.25 ± 76.5 OD in stage II and 346.86 ± 55.26 OD in stage III (P = 0.0042).
When considering clinical data retrieved from our follow-up, patients surgically treated in the last 12 mo were ruled out from correlation with disease prognosis. Overall, 27 patients were considered, of which 3 were ruled out for missing information.
For the remaining 24 patients, the mean follow-up was 16.8 mo (range 5.6-34.4 mo). Median overall survival and disease-free survival were 15.9 mo and 11.2 mo, respectively. The 12-mo recurrence rate was 54.1% (n = 13). Comparing patients with relapse at 1 year with those without evidence of the disease, the difference in PrPc expression at immuno-blotting was statistically significant between the two groups (360.71 ± 69.01 OD vs 170.23 ± 23.06 OD, P = 0.023) (Figure 6A).
Moreover, 21 patients out of 24 received adjuvant chemotherapy (CT). Of these, 10/21 (47.6%) were without evidence of disease relapse at 12 mo, while 11/21 (52.4%) experienced a relapse of the disease. Detailing our analysis of PrPc expression correlated with disease recurrence a significant higher PrPc expression was detected for patients who experienced a relapse, despite the administration of adjuvant CT compared with those receiving CT without evidence of disease at follow-up (368.36 ± 79.26 OD vs 162.86 ± 24.16 OD, respectively, P = 0.028) (Figure 6B).
Cellular prion protein is a cell surface glycoprotein, firstly discovered as the normal isoform of the scrapie prion protein (PrPSc), the infectious agent of Transmissible Spongiform Encephalopathies. Despite the normal isoform is a highly conserved glycoprotein present in all vertebrates, which indicates some intrinsic and fundamental roles in cell homeostasis, studies about the physiology of PrPc have long been overlooked.
Only recently some authors focused on the role of PrPc and many paths have been opened[11]. PrPc was proposed to protect neurons against cell death and oxidative stress[12], to control copper metabolism[13], to regulate cell cycle[14], synaptic transmission[15], cell adhesion[16], and activation of the immuno-system[17]. It is remarkable that PrPc plays a role in pluripotency and differentiation of embryonic stem cells[18], cell proliferation and differentiation[19-24] and muscle cell regeneration[25].
In detail, the discovery of a role of this protein in regulating the cell cycle led some authors to focus on studying a relationship between PrPc and carcinogenesis. It is now well established that PrPc may sustain cancer cell proliferation in various types of cancers[26]: Gastric[27], colon cancer[28-30], as well as glioblastoma[31-33]. Overall, the contribution of PrPc to cancer cell proliferation fully fits with a gain of its physiological function in normal cells, where it controls the activation of several effectors associated with cell growth[24,34]. A second field of investigation focuses on the correlation between PrPc and chemo-resistance. High PrPc expression levels are indeed associated with increased resistance to various types of agents in glioblastoma[35], gastric[27,36-38], breast[39-41], and colon cancer[29,42,43]. According to several studies this effect could be related to the activation of several pathways that lead to over expression of MDR1 (multidrug-resistance protein 1)[44]. For instance it was recently found that PrPc confers doxorubicin resistance to breast cancer cells[45].
PrPc also promotes cancer cells invasion/migration. In fact, elevated PrPc levels confer enhanced invasive properties to glioblastoma[46], gastric[47], breast[40,48], colon[49], lung[50] and melanoma[51] cell lines.
Finally, PrPc directly induces proliferation and confers resistance to apoptosis in cancer stem cells[18], by dysregulating their interactions with surrounding environment and thus causing cancer stem cells proliferation[52]. As with proliferation, the contribution of PrPc to cancer stem cells self-renewal may be envisioned as a diversion of its physiological role into normal stem cell maintenance[18].
Collectively, the involvement of PrPc in various aspects of cancer progression may be viewed as directly related to its physiological role in normal cells. Prion-mediated changes may represent initiating events that promote the emergence of the hallmarks of cancer, including self-sufficiency in growth signals, insensitivity to antigrowth signals, tissue invasion and metastasis, limitless replicative potential and inhibition of apoptosis[53]. Moreover, over-expression of PrPc has been found to be related to a higher chemoresistance[54]. This is likely to have clinical implications: from a therapeutic perspective, reducing PrPc expression may be beneficial, as it was documented for glioblastoma[55] or colon cancer[56]. Besides, alternative opportunities may ensue from a better knowledge of the signals upregulating PrPc expression in cancer cells.
In particular, looking to PDAC, a deeper comprehension of its role may add a significant piece of evidence in the puzzle of this highly aggressive pathology, with a potentially relevant clinical implication. The present study indicates a higher expression of PrPc in PDAC tissues in vivo, and provides the first evidence of the cellular localization of this protein. A higher amount of PrPc specifically within PDAC cells is now confirmed in a wider pool of patients compared with our recent in vivo study[7].
Moreover, the present study provides a stoichiometric measurement of the protein is situ within PDAC cells and indicates a shift in its sub-cellular placement in PDAC. Such a latter novelty was provided by ultrastructural morphometry, and immuno-gold staining under transmission electron microscopy. For instance, such an approach allowed to detect a misplacement of PrPc towards the nuclear compartment, which is in line with its strong effects on cancer-related gene expression. In particular, in PDAC cells the number of PrPc immuno-gold particles was two-fold higher compared with controls, although such an increase is more relevant in the cell nucleus, where it rises up to three-fold of the amount measured in controls. When comparing PrPc compartmentalization in PDAC cells and normal ductal pancreatic cells, the nuclear concentration of PrPc is 1.65-fold higher compared with cytosol, while in the normal cells there is no significant difference between nucleus and cytosol.
From one hand, this may be due to a marked ongoing over-expression of PrPc gene (PRNP) in PDAC cells. In fact, the first hint of a link between PrPc and pancreatic cancer dates back to the early 2000s, when PRNP was identified as one of the 30 genes mostly expressed in pancreatic cancer cell lines when compared with normal cells[57]. On the other hand, such a preferential nuclear compartmentalization may disclose a specific role of PrPc in the biology of PDAC. So far, the evidences from PDAC cell cultures show that PrPc act as a cell surface glycoprotein to activate specific intracellular pathways and signaling that bring to a proliferative effect[5]. However, the detection of a peculiar and prominent concentration in the nucleus suggests an involvement of PrPc in regulating directly gene expression, acting as a nucleo-junctional interplay in order to modulate the transcriptional activity of different pathways involved in carcinogenesis. In fact, PrPc could have a role in signaling complexes that contribute to a regulation of proliferation and cell-to-cell adhesion. Recently an association has been found in enterocytes between nuclear PrPc and Wnt and Hippo pathways, which are modulated by cell contacts and are de-regulated at high frequency in many human cancers. In this way, PrPc should be considered as an actor in oncogenic processes through its role in the dynamics of cell-to-cell junctions because its nuclear localization could lead to modulate transcriptional activity of Wnt and Hippo effectors, some of the pathways clearly involved in carcinogenesis[58].
The relevance of these findings in clinical setting is supported by the evidence of a more aggressive behavior of PDAC depending on the amount of PrPc expression. As already demonstrated in our previous study[7], the expression of PrPc correlates with predicted patients’ prognosis based on cancer stage according to pathology results. These data are encouraging, since they are confirmed also in a wider group of patients. Moreover, by further collecting early clinical data, we were able to validate such a hypothesis. In fact, in the subgroup of patients with a relapse of the disease after a surgical R0 resection, the expression of PrPc was significantly higher compared with those patients without evidence of relapse.
Moreover, these data indicate a relationship between PrPc and chemo-resistance, since the relapse during adjuvant CT was significantly higher in patients according to their expression of PrPc at western blot.
This is the first project that investigates the sub-cellular nuclear compartmentalization of increased PrPc expression with electron microscopy in vivo in patients with PDAC treated with surgery. Our findings could open new research avenues: in fact, being PrPc markedly expressed in the nuclear compartment, studies should investigate whether this protein is involved in specific activation of some unknown nuclear pathways that can lead to a higher cancer aggressiveness and de-differentiation. Similarly, the correlation of PrPc expression with a chemo-resistant phenotype could be used not only for prognostic purposes. For instance, patients may be stratified for prognosis according to PrPc expression in the resected specimen. This may involve also the development of novel therapeutic outcomes. In fact, specific therapeutic agents designed to target PrPc metabolism should be able to reduce a pathological cell growth and to revert chemo-resistance.
The study confirms our previous data, highlighting in vivo in patients PDAC tissue the role of PrPc in PDAC aggressiveness. The evidence of a peculiar nuclear compartmentalization of this protein in the cellular nuclei of PDAC cells is in line with in vitro data from literature showing over-expression of PRNP gene in PDAC cells, and suggests the presence of some, still unknown, molecular pathways triggered by PrPc in the nuclear compartment. This extends the influence of PrPc beyond its role as cell-membrane glycoprotein. Finally, PrPc expression seems to be associated with a greater risk of relapse after radical surgery and in shifting the cancer towards a phenotype with a higher chemo-resistance. These data provide a step forward in the comprehension of PDAC biology, confirming that PrPc is likely to represent a marker of disease severity and a determinant of PDAC biology. Further studies are needed to validate these results and to investigate the molecular mechanism of PrPc in PDAC pathogenesis and its potential clinical application.
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal human cancers, but its molecular basis is still poorly understood. Among the several new targets for the comprehension of its biology, cellular Prion protein (PrPc) deserves particular mention, since it seems to be involved also in tumorigenesis.
Recent evidences have shown a relationship between PrPc expression and PDAC, with a possible role of this protein in the molecular basis of PDAC aggressiveness itself.
The present study aimed to further analyze the occurrence of PrPc within PDAC tissues, by investigating the specific compartmentalization of PrPc within PDAC cells, which is a fundamental aspect in order to provide a significant advancement in understanding the biology of disease. Moreover, we aimed to correlate the presence of PrPc with clinical data in order to find an association with patients’ prognosis.
Samples from pancreatic tissues of 45 patients treated with pancreatic resection PDAC at a single institution were collected. Immune-gold stoichiometry within specific cell compartments was analyzed with electron microscopy in order to elucidate the subcellular compartmentalization of PrPc within PDAC cells. Western blotting was used to detect, quantify and compare the expression of PrPc in PDAC and control tissues, such as those of non-affected neighboring pancreatic tissue of the same patient. Data from western blot analysis where also used to perform a correlation with prognostic data from patients’ follow-up.
Immune-electron microscopy highlighted an increased PrPc expression in PDAC ductal cells of all patients and allowed to detect a peculiar compartmentalization of PrPc within tumor cells, with a specific increase of PrPc in the nucleus. Furthermore, semi-quantitative analysis by using Western blotting, showed that PrPc increased almost three-fold in tumor pancreatic tissue compared with healthy pancreatic regions, with a significantly higher expression of PrPc in patients with disease recurrence at 12 mo after radical surgery, also in the subgroup of patients treated with adjuvant CT, thus revealing a possible higher chemoresistance.
Our study provides evidence for a correlation between PrPc expression in PDAC and a worse biological behavior, with a higher recurrence rate and chemo-resistance. Moreover, it provides for the first-time the evidence of a peculiar subcellular compartmentalization of PrPc itself within PDAC cells.
PrPc could be a molecular marker associated to PDAC aggressiveness and ominous prognosis. Its nuclear compartmentalization suggests the activation of specific, but still unknown, molecular pathways involved in the biology of the disease and further studies in this sense are necessary, since specific therapeutic agents designed to target PrPc metabolism should be able to reduce a pathological cell growth and to revert chemo-resistance.
The authors thank King SB for the Language Editing.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Șurlin VM, Yamanaka K S-Editor: Wu YXJ L-Editor: A P-Editor: Wu YXJ
| 1. | Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the united states. Cancer Res. 2014;. [RCA] [DOI] [Full Text] [Cited by in Crossref: 5379] [Cited by in RCA: 5136] [Article Influence: 466.9] [Reference Citation Analysis (0)] |
| 2. | Warshaw AL, Fernández-del Castillo C. Pancreatic carcinoma. N Engl J Med. 1992;326:455-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1056] [Cited by in RCA: 1015] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 3. | Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605-1617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2078] [Cited by in RCA: 2205] [Article Influence: 147.0] [Reference Citation Analysis (2)] |
| 4. | Weledji EP, Enoworock G, Mokake M, Sinju M. How Grim is Pancreatic Cancer? Oncol Rev. 2016;10:294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 5. | Wang Y, Yu S, Huang D, Cui M, Hu H, Zhang L, Wang W, Parameswaran N, Jackson M, Osborne B, Bedogni B, Li C, Sy MS, Xin W, Zhou L. Cellular Prion Protein Mediates Pancreatic Cancer Cell Survival and Invasion through Association with and Enhanced Signaling of Notch1. Am J Pathol. 2016;186:2945-2956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 6. | Sy MS, Li C, Yu S, Xin W. The fatal attraction between pro-prion and filamin A: prion as a marker in human cancers. Biomark Med. 2010;4:453-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 7. | Bianchini M, Giambelluca MA, Scavuzzo MC, Di Franco G, Guadagni S, Palmeri M, Furbetta N, Gianardi D, Funel N, Pollina LE, Di Candio G, Fornai F, Morelli L. The occurrence of prion protein in surgically resected pancreatic adenocarcinoma. Pancreatology. 2020;20:1218-1225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | Amin MB, Edge SB; American Joint Committee on Cancer. AJCC cancer staging manual [Internet]. [cited 17 September 2019]. Available from: https://www.springer.com/gp/book/9783319406176. |
| 9. | Funel N, Giovannetti E, Del Chiaro M, Mey V, Pollina LE, Nannizzi S, Boggi U, Ricciardi S, Del Tacca M, Bevilacqua G, Mosca F, Danesi R, Campani D. Laser microdissection and primary cell cultures improve pharmacogenetic analysis in pancreatic adenocarcinoma. Lab Invest. 2008;88:773-784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 10. | Ricci C, Mota C, Moscato S, D'Alessandro D, Ugel S, Sartoris S, Bronte V, Boggi U, Campani D, Funel N, Moroni L, Danti S. Interfacing polymeric scaffolds with primary pancreatic ductal adenocarcinoma cells to develop 3D cancer models. Biomatter. 2014;4:e955386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 11. | Manni G, Lewis V, Senesi M, Spagnolli G, Fallarino F, Collins SJ, Mouillet-Richard S, Biasini E. The cellular prion protein beyond prion diseases. Swiss Med Wkly. 2020;150:w20222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 12. | McLennan NF, Brennan PM, McNeill A, Davies I, Fotheringham A, Rennison KA, Ritchie D, Brannan F, Head MW, Ironside JW, Williams A, Bell JE. Prion protein accumulation and neuroprotection in hypoxic brain damage. Am J Pathol. 2004;165:227-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 182] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 13. | Brown DR, Schulz-Schaeffer WJ, Schmidt B, Kretzschmar HA. Prion protein-deficient cells show altered response to oxidative stress due to decreased SOD-1 activity. Exp Neurol. 1997;146:104-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 327] [Cited by in RCA: 323] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 14. | Liang J, Pan Y, Zhang D, Guo C, Shi Y, Wang J, Chen Y, Wang X, Liu J, Guo X, Chen Z, Qiao T, Fan D. Cellular prion protein promotes proliferation and G1/S transition of human gastric cancer cells SGC7901 and AGS. FASEB J. 2007;21:2247-2256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 80] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 15. | Bremer J, Baumann F, Tiberi C, Wessig C, Fischer H, Schwarz P, Steele AD, Toyka K V, Nave K-A, Weis J, Aguzzi A. Axonal prion protein is required for peripheral myelin maintenance. Nat Neurosci. 13:310-318. [RCA] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 305] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 16. | Málaga-Trillo E, Solis GP, Schrock Y, Geiss C, Luncz L, Thomanetz V, Stuermer CA. Regulation of embryonic cell adhesion by the prion protein. PLoS Biol. 2009;7:e55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 166] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 17. | Mabbott N, Turner M. Prions and the blood and immune systems. Haematologica. 2005;90:542-548. [PubMed] |
| 18. | Martin-Lannerée S, Hirsch TZ, Hernandez-Rapp J, Halliez S, Vilotte JL, Launay JM, Mouillet-Richard S. PrP(C) from stem cells to cancer. Front Cell Dev Biol. 2014;2:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 19. | Steele AD, Emsley JG, Ozdinler PH, Lindquist S, Macklis JD. Prion protein (PrPc) positively regulates neural precursor proliferation during developmental and adult mammalian neurogenesis. Proc Natl Acad Sci U S A. 2006;103:3416-3421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 215] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 20. | Zhang CC, Steele AD, Lindquist S, Lodish HF. Prion protein is expressed on long-term repopulating hematopoietic stem cells and is important for their self-renewal. Proc Natl Acad Sci U S A. 2006;103:2184-2189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 181] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 21. | Lima FR, Arantes CP, Muras AG, Nomizo R, Brentani RR, Martins VR. Cellular prion protein expression in astrocytes modulates neuronal survival and differentiation. J Neurochem. 2007;103:2164-2176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 95] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 22. | Lee YJ, Baskakov I V. Treatment with normal prion protein delays differentiation and helps to maintain high proliferation activity in human embryonic stem cells. J Neurochem. 114:362-373. [RCA] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 23. | Panigaj M, Glier H, Wildova M, Holada K. Expression of prion protein in mouse erythroid progenitors and differentiating murine erythroleukemia cells. PLoS One. 2011;6:e24599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 24. | Santos TG, Silva IR, Costa-Silva B, Lepique AP, Martins VR, Lopes MH. Enhanced neural progenitor/stem cells self-renewal via the interaction of stress-inducible protein 1 with the prion protein. Stem Cells. 2011;29:1126-1136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 25. | Stella R, Massimino ML, Sandri M, Sorgato MC, Bertoli A. Cellular prion protein promotes regeneration of adult muscle tissue. Mol Cell Biol. 2010;30:4864-4876. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 26. | Ryskalin L, Biagioni F, Busceti CL, Giambelluca MA, Morelli L, Frati A, Fornai F. The role of cellular prion protein in promoting stemness and differentiation in cancer. Cancers (Basel). 2021;. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 27. | Liang J, Luo G, Ning X, Shi Y, Zhai H, Sun S, Jin H, Liu Z, Zhang F, Lu Y, Zhao Y, Chen X, Zhang H, Guo X, Wu K, Fan D. Differential expression of calcium-related genes in gastric cancer cells transfected with cellular prion protein. Biochem Cell Biol. 2007;85:375-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 28. | Le Corre D, Ghazi A, Balogoun R, Pilati C, Aparicio T, Martin-Lannerée S, Marisa L, Djouadi F, Poindessous V, Crozet C, Emile JF, Mulot C, Le Malicot K, Boige V, Blons H, de Reynies A, Taieb J, Ghiringhelli F, Bennouna J, Launay JM, Laurent-Puig P, Mouillet-Richard S. The cellular prion protein controls the mesenchymal-like molecular subtype and predicts disease outcome in colorectal cancer. EBioMedicine. 2019;46:94-104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 29. | Chieng CK, Say YH. Cellular prion protein contributes to LS 174T colon cancer cell carcinogenesis by increasing invasiveness and resistance against doxorubicin-induced apoptosis. Tumour Biol. 2015;36:8107-8120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 30. | Li QQ, Sun YP, Ruan CP, Xu XY, Ge JH, He J, Xu ZD, Wang Q, Gao WC. Cellular prion protein promotes glucose uptake through the Fyn-HIF-2α-Glut1 pathway to support colorectal cancer cell survival. Cancer Sci. 2011;102:400-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 31. | Ryskalin L, Busceti CL, Biagioni F, Limanaqi F, Familiari P, Frati A, Fornai F. Prion protein in glioblastoma multiforme. Int J Mol Sci. 2019;. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 32. | Lopes MH, Santos TG, Rodrigues BR, Queiroz-Hazarbassanov N, Cunha IW, Wasilewska-Sampaio AP, Costa-Silva B, Marchi FA, Bleggi-Torres LF, Sanematsu PI, Suzuki SH, Oba-Shinjo SM, Marie SK, Toulmin E, Hill AF, Martins VR. Disruption of prion protein-HOP engagement impairs glioblastoma growth and cognitive decline and improves overall survival. Oncogene. 2015;34:3305-3314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 33. | Corsaro A, Bajetto A, Thellung S, Begani G, Villa V, Nizzari M, Pattarozzi A, Solari A, Gatti M, Pagano A, Würth R, Daga A, Barbieri F, Florio T. Cellular prion protein controls stem cell-like properties of human glioblastoma tumor-initiating cells. Oncotarget. 2016;7:38638-38657. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 34. | Halliez S, Martin-Lannerée S, Passet B, Hernandez-Rapp J, Castille J, Urien C, Chat S, Laude H, Vilotte JL, Mouillet-Richard S, Béringue V. Prion protein localizes at the ciliary base during neural and cardiovascular development, and its depletion affects α-tubulin post-translational modifications. Sci Rep. 2015;5:17146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 35. | Zhuang D, Liu Y, Mao Y, Gao L, Zhang H, Luan S, Huang F, Li Q. TMZ-induced PrPc/par-4 interaction promotes the survival of human glioma cells. Int J Cancer. 2012;130:309-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 36. | Zhao Y, You H, Liu F, An H, Shi Y, Yu Q, Fan D. Differentially expressed gene profiles between multidrug resistant gastric adenocarcinoma cells and their parental cells. Cancer Lett. 2002;185:211-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 98] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 37. | Du J, Pan Y, Shi Y, Guo C, Jin X, Sun L, Liu N, Qiao T, Fan D. Overexpression and significance of prion protein in gastric cancer and multidrug-resistant gastric carcinoma cell line SGC7901/ADR. Int J Cancer. 2005;113:213-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 81] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 38. | Luo G, Wang W, Wu Q, Lu Y, Su T, Gu N, Li K, Wang J, Du R, Zhao X, Li X, Fan R, Zhang H, Nie Y, Zhou X, Shi Y, Liang J, Wang X, Fan D. MGr1-Antigen/37 kDa laminin receptor precursor promotes cellular prion protein induced multi-drug-resistance of gastric cancer. Oncotarget. 2017;. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 39. | Diarra-Mehrpour M, Arrabal S, Jalil A, Pinson X, Gaudin C, Piétu G, Pitaval A, Ripoche H, Eloit M, Dormont D, Chouaib S. Prion protein prevents human breast carcinoma cell line from tumor necrosis factor alpha-induced cell death. Cancer Res. 2004;64:719-727. [PubMed] |
| 40. | Cheng Y, Tao L, Xu J, Li Q, Yu J, Jin Y, Chen Q, Xu Z, Zou Q, Liu X. CD44/cellular prion protein interact in multidrug resistant breast cancer cells and correlate with responses to neoadjuvant chemotherapy in breast cancer patients. Mol Carcinog. 2014;53:686-697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 41. | Meslin F, Conforti R, Mazouni C, Morel N, Tomasic G, Drusch F, Yacoub M, Sabourin JC, Grassi J, Delaloge S, Mathieu MC, Chouaib S, Andre F, Mehrpour M. Efficacy of adjuvant chemotherapy according to Prion protein expression in patients with estrogen receptor-negative breast cancer. Ann Oncol. 2007;18:1793-1798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 42. | Atkinson CJ, Kawamata F, Liu C, Ham S, Győrffy B, Munn AL, Wei MQ, Möller A, Whitehall V, Wiegmans AP. EGFR and Prion protein promote signaling via FOXO3a-KLF5 resulting in clinical resistance to platinum agents in colorectal cancer. Mol Oncol. 2019;13:725-737. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 43. | Park JY, Jeong JK, Lee JH, Moon JH, Kim SW, Lee YJ, Park SY. Induction of cellular prion protein (PrPc) under hypoxia inhibits apoptosis caused by TRAIL treatment. Oncotarget. 2015;6:5342-5353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 44. | Liang J, Ge F, Guo C, Luo G, Wang X, Han G, Zhang D, Wang J, Li K, Pan Y, Yao L, Yin Z, Guo X, Wu K, Ding J, Fan D. Inhibition of PI3K/Akt partially leads to the inhibition of PrP(C)-induced drug resistance in gastric cancer cells. FEBS J. 2009;276:685-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 45. | Wiegmans AP, Saunus JM, Ham S, Lobb R, Kutasovic JR, Dalley AJ, Miranda M, Atkinson C, Foliaki ST, Ferguson K, Niland C, Johnstone CN, Lewis V, Collins SJ, Lakhani SR, Al-Ejeh F, Möller A. Secreted cellular prion protein binds doxorubicin and correlates with anthracycline resistance in breast cancer. JCI Insight. 2019;5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 46. | Iglesia RP, Prado MB, Cruz L, Martins VR, Santos TG, Lopes MH. Engagement of cellular prion protein with the co-chaperone Hsp70/90 organizing protein regulates the proliferation of glioblastoma stem-like cells. Stem Cell Res Ther. 2017;8:76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 47. | Pan Y, Zhao L, Liang J, Liu J, Shi Y, Liu N, Zhang G, Jin H, Gao J, Xie H, Wang J, Liu Z, Fan D. Cellular prion protein promotes invasion and metastasis of gastric cancer. FASEB J. 2006;20:1886-1888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 100] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 48. | Gil M, Kim YK, Kim KE, Kim W, Park CS, Lee KJ. Cellular prion protein regulates invasion and migration of breast cancer cells through MMP-9 activity. Biochem Biophys Res Commun. 2016;470:213-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 49. | de Lacerda TC, Costa-Silva B, Giudice FS, Dias MV, de Oliveira GP, Teixeira BL, Dos Santos TG, Martins VR. Prion protein binding to HOP modulates the migration and invasion of colorectal cancer cells. Clin Exp Metastasis. 2016;33:441-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 50. | Lin SC, Lin CH, Shih NC, Liu HL, Wang WC, Lin KY, Liu ZY, Tseng YJ, Chang HK, Lin YC, Yeh YC, Minato H, Fujii T, Wu YC, Chen MY, Chou TY. Cellular prion protein transcriptionally regulated by NFIL3 enhances lung cancer cell lamellipodium formation and migration through JNK signaling. Oncogene. 2020;39:385-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 51. | Li C, Yu S, Nakamura F, Pentikäinen OT, Singh N, Yin S, Xin W, Sy MS. Pro-prion binds filamin A, facilitating its interaction with integrin beta1, and contributes to melanomagenesis. J Biol Chem. 2010;285:30328-30339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 52. | Rezza A, Sennett R, Rendl M. Adult Stem Cell Niches [Internet]. In: Current topics in developmental biology. 2014 [cited 2019 Sep 17]. page 333–372 Available from: http://www.ncbi.nlm.nih.gov/pubmed/24439812. [PubMed] |
| 53. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 47117] [Article Influence: 3365.5] [Reference Citation Analysis (5)] |
| 54. | Mehrpour M, Codogno P. Prion protein: From physiology to cancer biology. Cancer Lett. 2010;290:1-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 71] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 55. | Barbieri G, Palumbo S, Gabrusiewicz K, Azzalin A, Marchesi N, Spedito A, Biggiogera M, Sbalchiero E, Mazzini G, Miracco C, Pirtoli L, Kaminska B, Comincini S. Silencing of cellular prion protein (PrPC) expression by DNA-antisense oligonucleotides induces autophagy-dependent cell death in glioma cells. Autophagy. 2011;7:840-853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 56. | Du L, Rao G, Wang H, Li B, Tian W, Cui J, He L, Laffin B, Tian X, Hao C, Liu H, Sun X, Zhu Y, Tang DG, Mehrpour M, Lu Y, Chen Q. CD44-positive cancer stem cells expressing cellular prion protein contribute to metastatic capacity in colorectal cancer. Cancer Res. 2013;73:2682-2694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 57. | Han H, Bearss DJ, Browne LW, Calaluce R, Nagle RB, Von Hoff DD. Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res. 2002;62:2890-2896. [PubMed] |
| 58. | Rousset M, Leturque A, Thenet S. The nucleo-junctional interplay of the cellular prion protein: A new partner in cancer-related signaling pathways? Prion. 2016;10:143-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |