Published online Nov 7, 2021. doi: 10.3748/wjg.v27.i41.7134
Peer-review started: May 14, 2021
First decision: July 14, 2021
Revised: July 21, 2021
Accepted: August 30, 2021
Article in press: August 30, 2021
Published online: November 7, 2021
Processing time: 175 Days and 20.5 Hours
It remains unclear which factors, such as tumor volume and tumor invasion, influence circulating tumor DNA (ctDNA), and the origin of ctDNA in liquid biopsy is always problematic. To use liquid biopsies clinically, it will be very important to address these questions.
To assess the origin of ctDNA, clarify the dynamics of ctDNA levels, assess ctDNA levels by using a xenograft mouse after treatment, and to determine whether tumor volume and invasion are related to ctDNA levels.
Tumor xenotransplants were established by inoculating BALB/c-nu/nu mice with the TE11 cell line. Groups of mice were injected with xenografts at two or four sites and sacrificed at the appropriate time point after xenotransplantation for ctDNA analysis. Analysis of ctDNA was performed by droplet digital PCR, using the human telomerase reverse transcriptase (hTERT) gene.
Mice given two-site xenografts were sacrificed for ctDNA at week 4 and week 8. No hTERT was detected at week 4, but it was detected at week 8. However, in four-site xenograft mice, hTERT was detected both at week 4 and week 6. These experiments revealed that both tumor invasion and tumor volume were asso
We clarified the origin and dynamics of ctDNA, showing that tumor volume is an important factor. We also found that when the tumor was completely resected, ctDNA was absent after one or more days.
Core Tip: We clarified the origin and dynamics of circulating tumor DNA (ctDNA), showing that not only tumor invasion but also tumor volume was an important factor. The possibility of detecting ctDNA in early-stage cancers with shallow depth was demonstrated. Also, ctDNA could be measured at 1 d after tumor resection to evaluate the residuals, and the half-life of ctDNA was estimated to be 1.8-3.2 h.
- Citation: Terasawa H, Kinugasa H, Nouso K, Yamamoto S, Hirai M, Tanaka T, Takaki A, Okada H. Circulating tumor DNA dynamics analysis in a xenograft mouse model with esophageal squamous cell carcinoma. World J Gastroenterol 2021; 27(41): 7134-7143
- URL: https://www.wjgnet.com/1007-9327/full/v27/i41/7134.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i41.7134
Liquid biopsy, a molecular biological diagnostic method for blood and body fluids, has progressed dramatically in recent years. Circulating tumor DNA (ctDNA), one of the targets of liquid biopsy, is expected to be a useful method for screening and detection of cancer, monitoring therapy, prediction of prognosis, and personalized medicine[1-3]. Therefore, in addition to direct biopsy, which is the basis of conventional cancer diagnosis, a hybrid method, which includes non-invasive liquid biopsy, is becoming the mainstream.
Cell-free DNA (cfDNA), which includes ctDNA, is derived from apoptotic or necrotic cells[4,5]. Theoretically, it could be applied regardless of the stage. However, reports of its usefulness for early stages of cancer are controversial. Bettegowda et al[6] revealed that the rate of ctDNA detection is generally high in advanced stages of cancer, but ctDNA levels are generally lower in early stages of cancer. On the other hand, some reports indicated that ctDNA was useful for detecting early-stage cancers[6-9]. It remains unclear which factors, such as tumor volume and tumor invasion, influence ctDNA, and the origin of ctDNA in liquid biopsy is always problematic. To use liquid biopsies clinically, it will be very important to address these questions.
In this study, we used a xenograft mouse model to assess the origin of ctDNA, clarify the dynamics of ctDNA levels, assess ctDNA levels after treatment, and to determine whether tumor volume and invasion are related to ctDNA levels.
The human esophageal squamous cell carcinoma cell line TE11 was used because we established an experimental system for TE11 previously[10] and used it to show that liquid biopsy is useful in esophageal cancer cells as well as other gastrointestinal cancers. Cells were grown in RPMI 1640 (Thermo Fisher Scientific, Tokyo, Japan) containing 10% fetal bovine serum and 1% penicillin/streptomycin (Sigma-Aldrich, Tokyo, Japan) at 37.0 °C in a 5% CO2 atmosphere. Appropriate passages were made such that confluency did not exceed 70% prior to xenotransplantation. A Countess Automated Cell Counter (Thermo Fisher Scientific, Tokyo, Japan) was used to count cells, and 0.2% Trypan blue dye was used to exclude dead cells.
Xenograft mouse experimental protocols were approved by the Ethical Committee of Okayama University (OKU-2019276). Six-week-old female nude mice (BALB/c-nu/nu) (Charles River Laboratories, Japan) were used. Mice were raised in the animal facility of Okayama University and given food and water. The physical conditions of the mice, including the presence or absence of body movement or the availability of food and drink, were monitored daily. Mice were euthanized with isoflurane if mice stopped moving or eating.
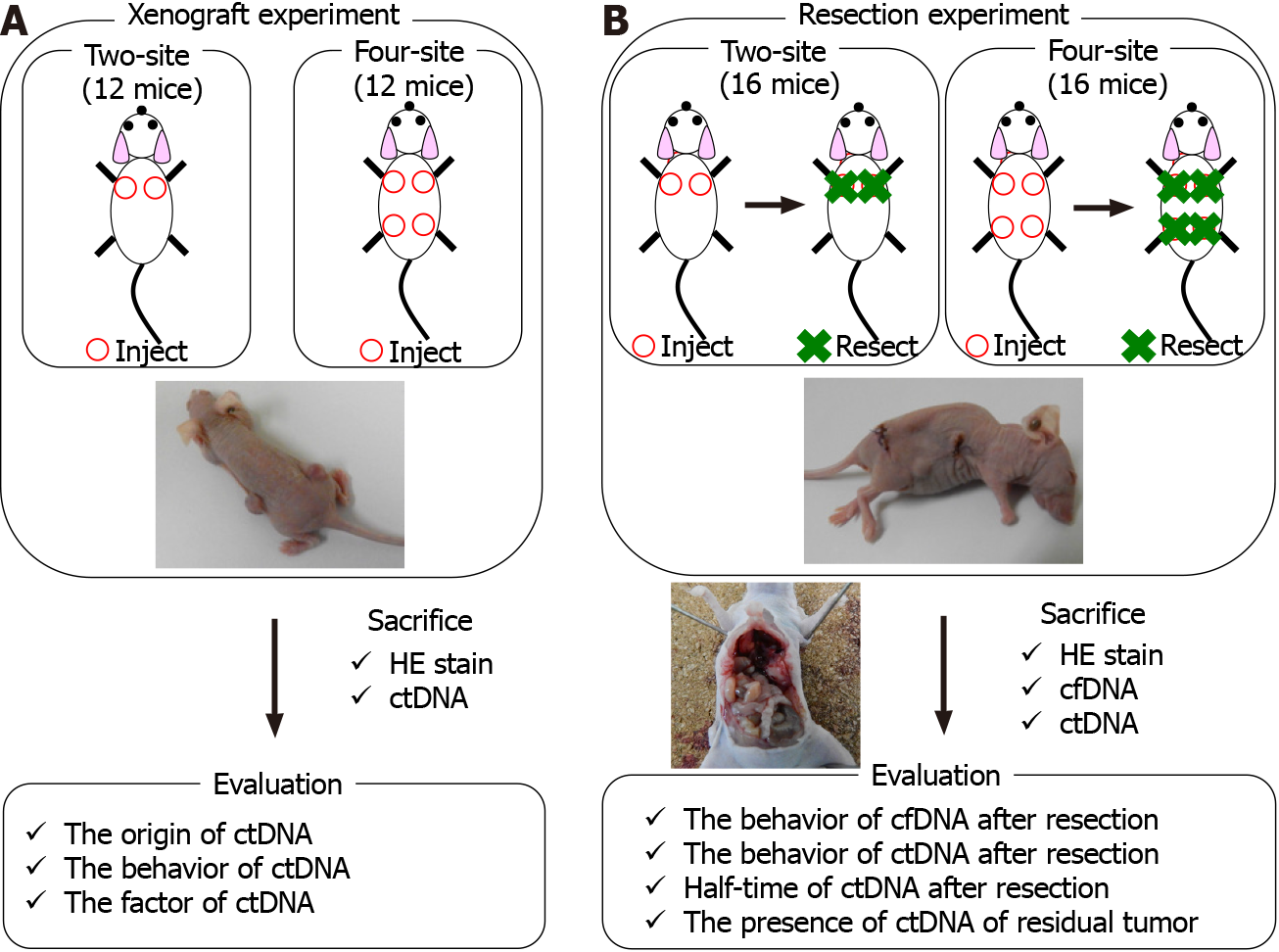
Tumor xenotransplants were established in mice by inoculation in the shoulders or flanks with 1 × 106 TE11 cells suspended in 50 μL medium plus 50 μL Matrigel (Corning Product No. 356234). Inoculation was performed at two sites (i.e., both shoulders, two-site xenograft mouse group, 28 mice) or at four sites (i.e., both shoul
Tumor formation was confirmed in all xenograft mice; although, the changes in size varied. Differences in tumor volume were evaluated over time. Two-site and four-site xenograft mouse groups were sacrificed for ctDNA analysis at the appropriate time point after xenotransplantation. To minimize the effects of differences in tumor size, four mice were used for each ctDNA time point analysis.
A sample size calculation using power analysis determined 24 mice were needed in xenograft experiments and 32 mice were needed in resection experiments.
Twelve mice received two-site xenografts, and 12 received four-site xenografts. Tumor size was measured every week after xenotransplantation, and ctDNA was evaluated at two time points: 4 wk and 8 wk after xenotransplantation (Figure 1).
Sixteen mice received two-site xenografts, and 16 mice received four-site xenografts. All tumors were resected at week 7 after xenotransplantation in the two-site xenograft group or at week 5 in the four-site xenograft group. cfDNA and ctDNA were evalu
For ctDNA analysis, whole blood was collected in BD Vacutainer tubes (Becton, Dickinson and Company, Franklin Lakes, NJ), and processed within 1 h after co
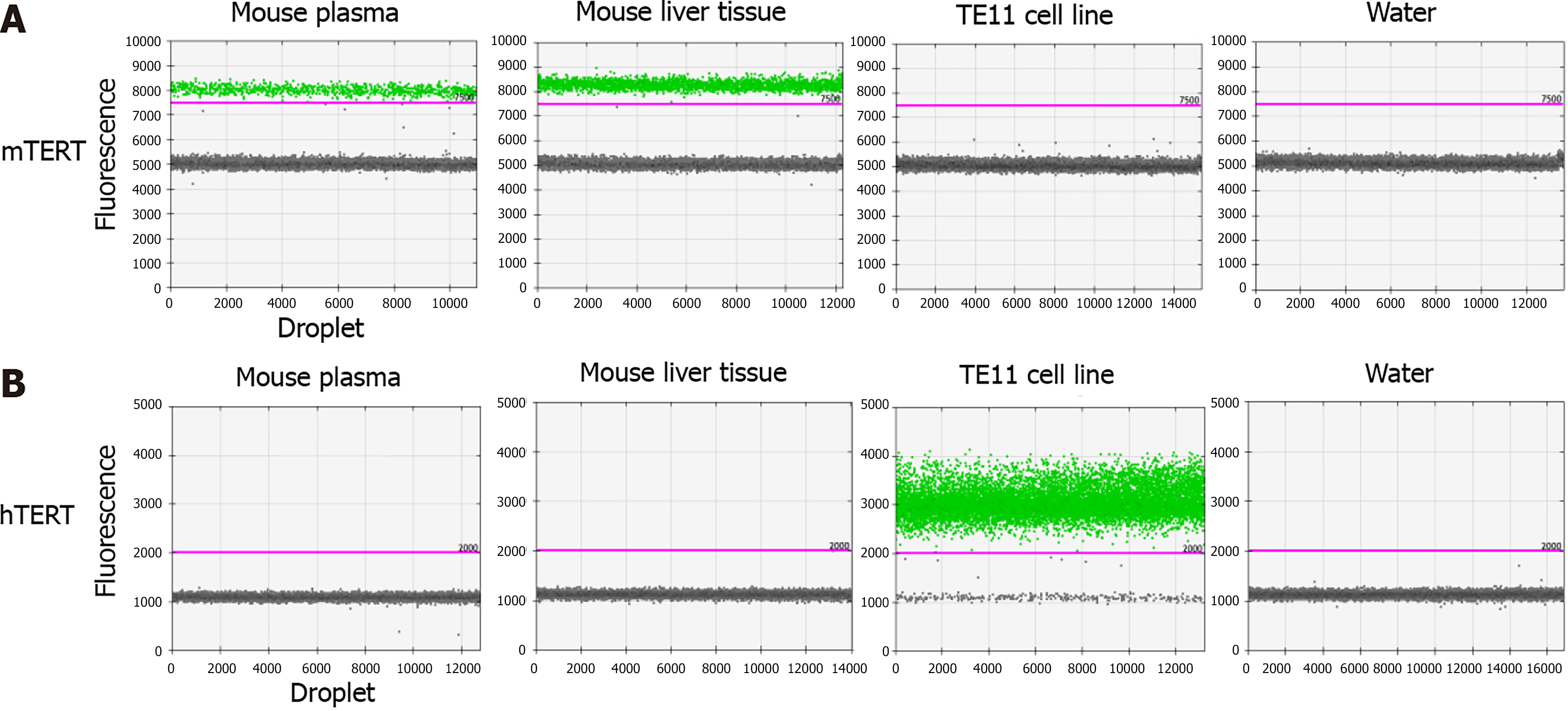
The wild-type telomerase reverse transcriptase (TERT) gene was analyzed by a mouse TERT (mTERT) assay (Thermo Fisher Scientific, Tokyo, Japan) or human TERT (hTERT) assay (Bio-Rad Laboratories, Hercules, CA, United States of America) to take advantage of the differences between mTERT and hTERT genes. The verification experiments using a droplet digital PCR (QX200 system; Bio-Rad Laboratories, Hercules, CA, United States of America) was performed.
To evaluate ctDNA, hTERT was detected via droplet digital polymerase chain reaction (PCR) according to the following protocol. DNA eluent (5 μL) from plasma was combined with Droplet PCR Supermix (10 μL; Bio-Rad Laboratories, Hercules, CA, United States of America), primer/probe mixture (1 μL), 5M Betaine (2 μL), 80 mmol/L EDTA (0.25 μL), CviQl enzyme (0.25 μL), and sterile DNase- and RNase-free water (3.5 μL). The mixture (22 μL) was added to Droplet Generation Oil (70 μL; Bio-Rad Laboratories, Hercules, CA, United States of America) to produce droplets. Thermal cycling of the emulsion was as follows: an initial denaturation at 95 °C for 10 min, followed by 50 cycles of 96 °C for 30 s and 62 °C for 1 min. After a final enzyme deactivation step of 98 °C for 10 min, the reaction mixtures were analyzed using a droplet reader (Bio-Rad Laboratories, Hercules, CA, United States of America). For quantification, the fluorescence signal was acquired with QuantaSoft software (Bio-Rad Laboratories, Hercules, CA, United States of America). We set the threshold fluorescence intensity at 7500 (mTERT) or 2000 (hTERT), according to positive and negative controls in this study, i.e., plasma and tissue of healthy human, control mouse, or TE11 cell line.
We used JMP version 14.0 (SAS Institute, Cary, NC, United States of America) for statistical analysis and set the threshold of significance at P < 0.05. Continuous data were analyzed using the non-parametric Wilcoxon test, and categorical data were analyzed using a Chi-squared test.
In verification experiments using a droplet digital PCR (QX200 system; Bio-Rad Laboratories, Hercules, CA, United States of America), we confirmed that the mTERT gene was detected in tissue and plasma of control mice, but not in TE11 genomic DNA, whereas the hTERT gene was detected in TE11 genomic DNA, but not in the tissue or plasma of control mice (Figure 2).
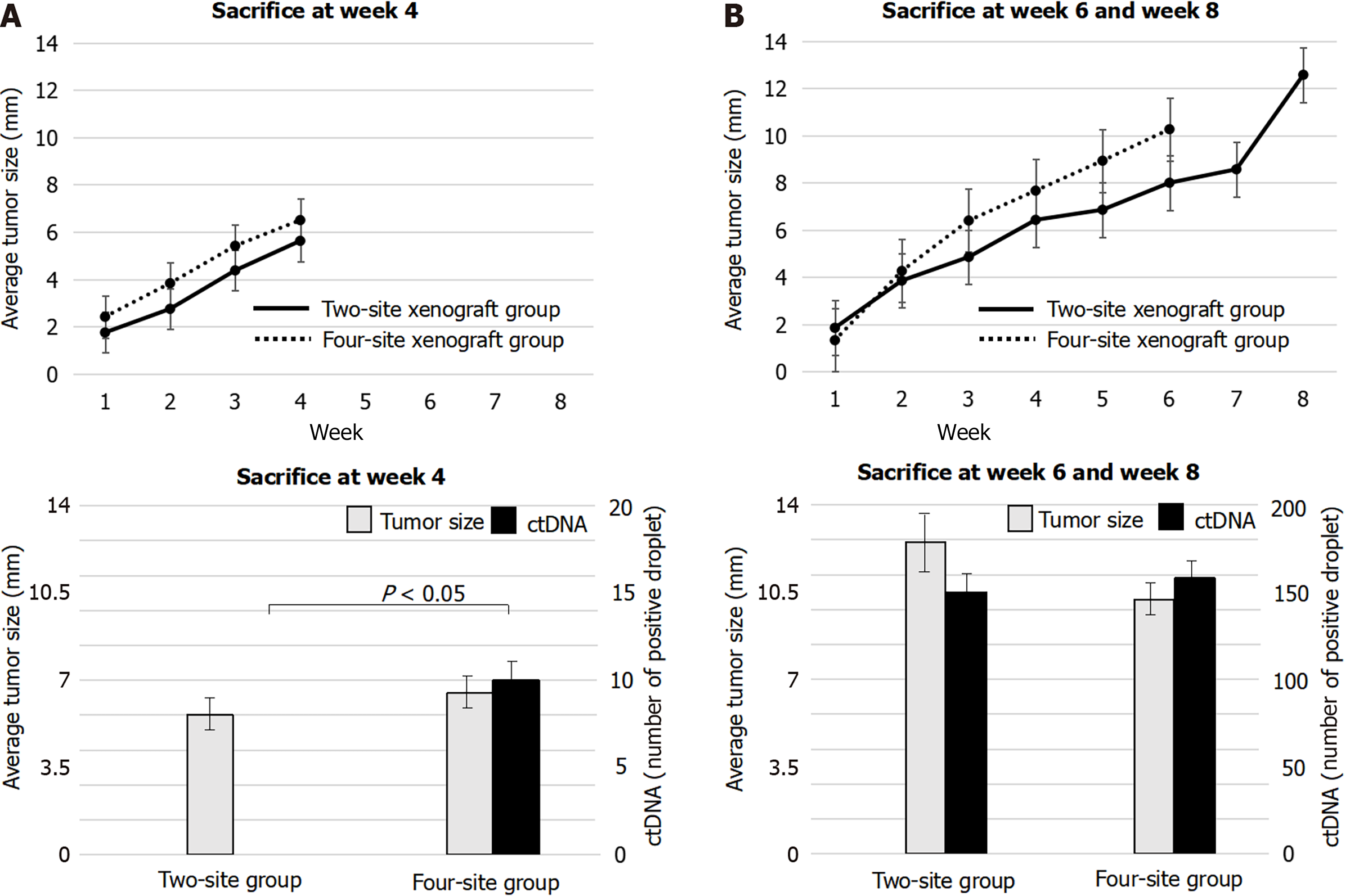
Xenograft experiments were designed to reveal the origin of ctDNA and factors contributing to ctDNA increase. Average tumor sizes measured in the two-site xenograft group 1, 2, 3, 4, 5, 6, 7, and 8 wk after xenotransplantation were 1.8, 3.2, 4.6, 6.0, 6.8, 8.0, 8.5, and 12.5 mm, respectively. Two-site xenograft mice were sacrificed 4 or 8 wk after xenotransplantation to evaluate ctDNA. No hTERT was detected at week 4, but hTERT was detected at week 8 (Figure 3). These results indicated that ctDNA was associated with tumor growth.
In four-site xenograft mice, the average tumor sizes at week 1, 2, 3, 4, 5, and 6 after xenotransplantation were 1.8, 4.0, 5.9, 7.1, 8.9, and 10.2 mm. The 8 wk evaluation planned for this group was revised to occur at week 6, because the tumor in one mouse had grown rapidly to cause thoracic invasion, and it was unlikely to survive to week 8. Four-site xenograft mice were sacrificed for ctDNA at week 4 and week 6. hTERT was detected both at week 4 and at week 6 in this group (Figure 3). These results indicated that ctDNA was associated with tumor growth as well as those of two-site xenograft mice. There were no other unexpected adverse events.
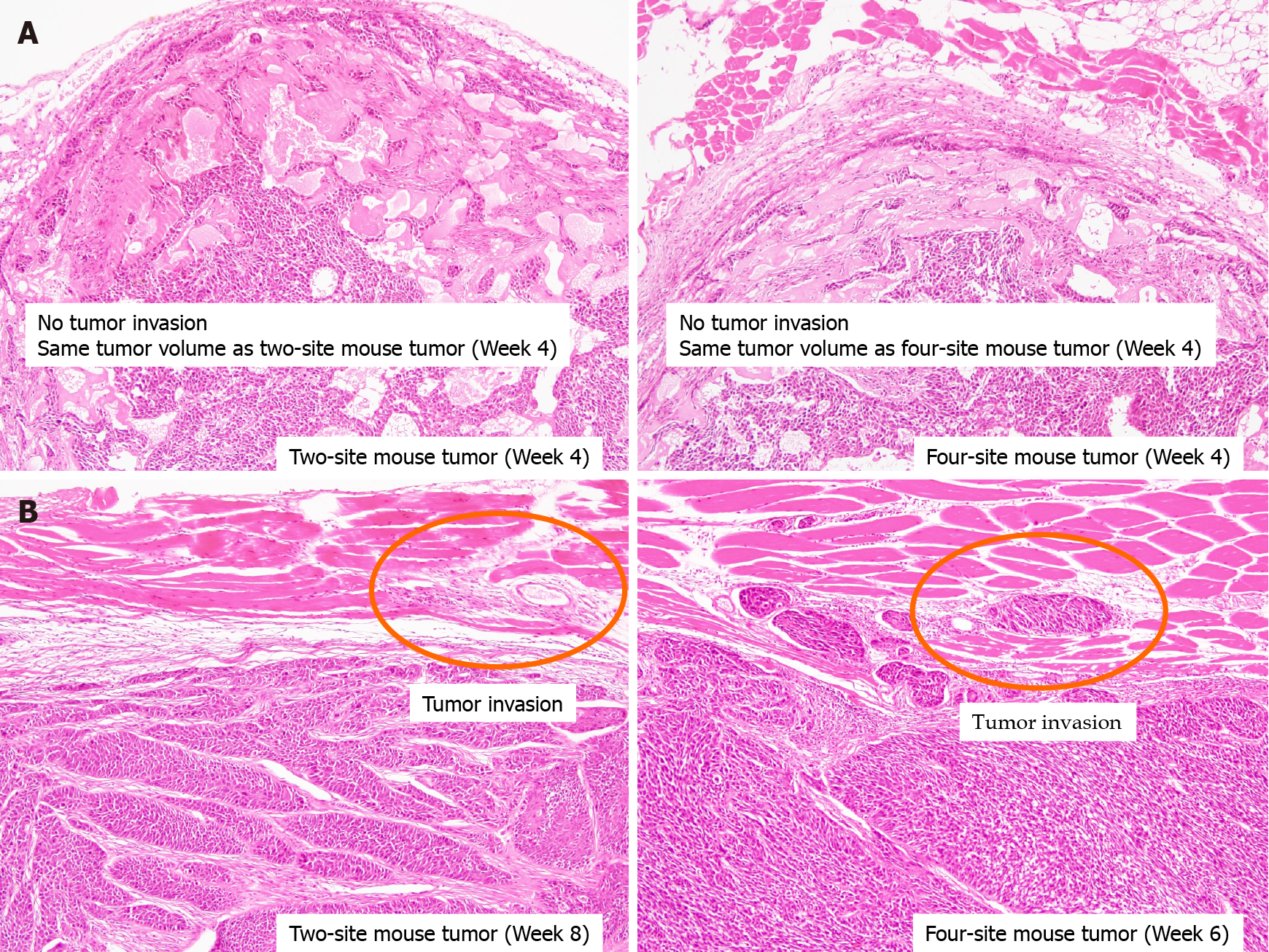
Histopathology of tumors at week 4 showed no invasion in either the two-site or four-site xenograft group, while tumors showed invasion into muscle both at week 8 in the two-site xenograft mice (P = 0.02) and at week 6 in the four-site xenograft mice (Figure 4; P = 0.03). These results indicated that ctDNA was associated with tumor invasion.
The rates of tumor size increase were similar between the two-site xenograft group and the four-site group. Interestingly, the two groups showed similar tumor diameters (P = 0.25) and invasion at week 4 (Figures 3 and 4), but a clear difference in the ctDNA detection rate (Figure 3; P = 0.02). These findings showed that not only invasion but also tumor volume might be related to the rate of ctDNA detection.
Resection experiments were designed to clarify responses of ctDNA to tumor re
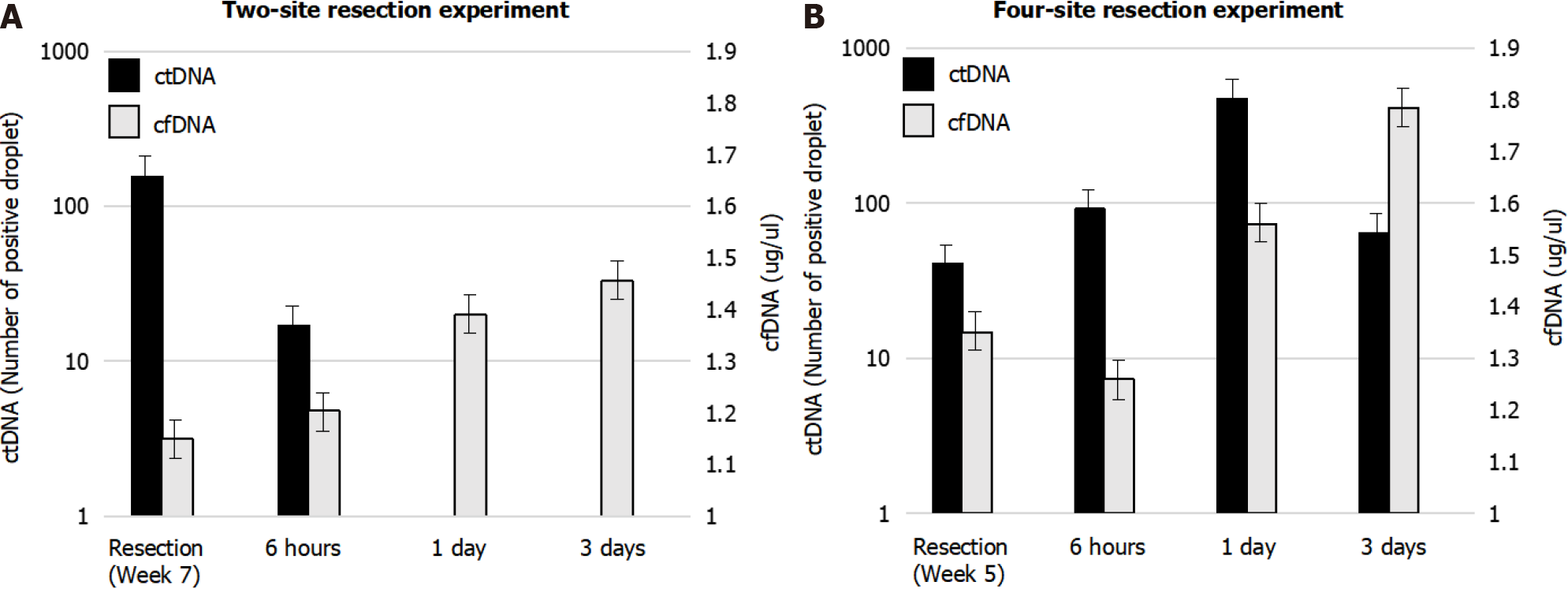
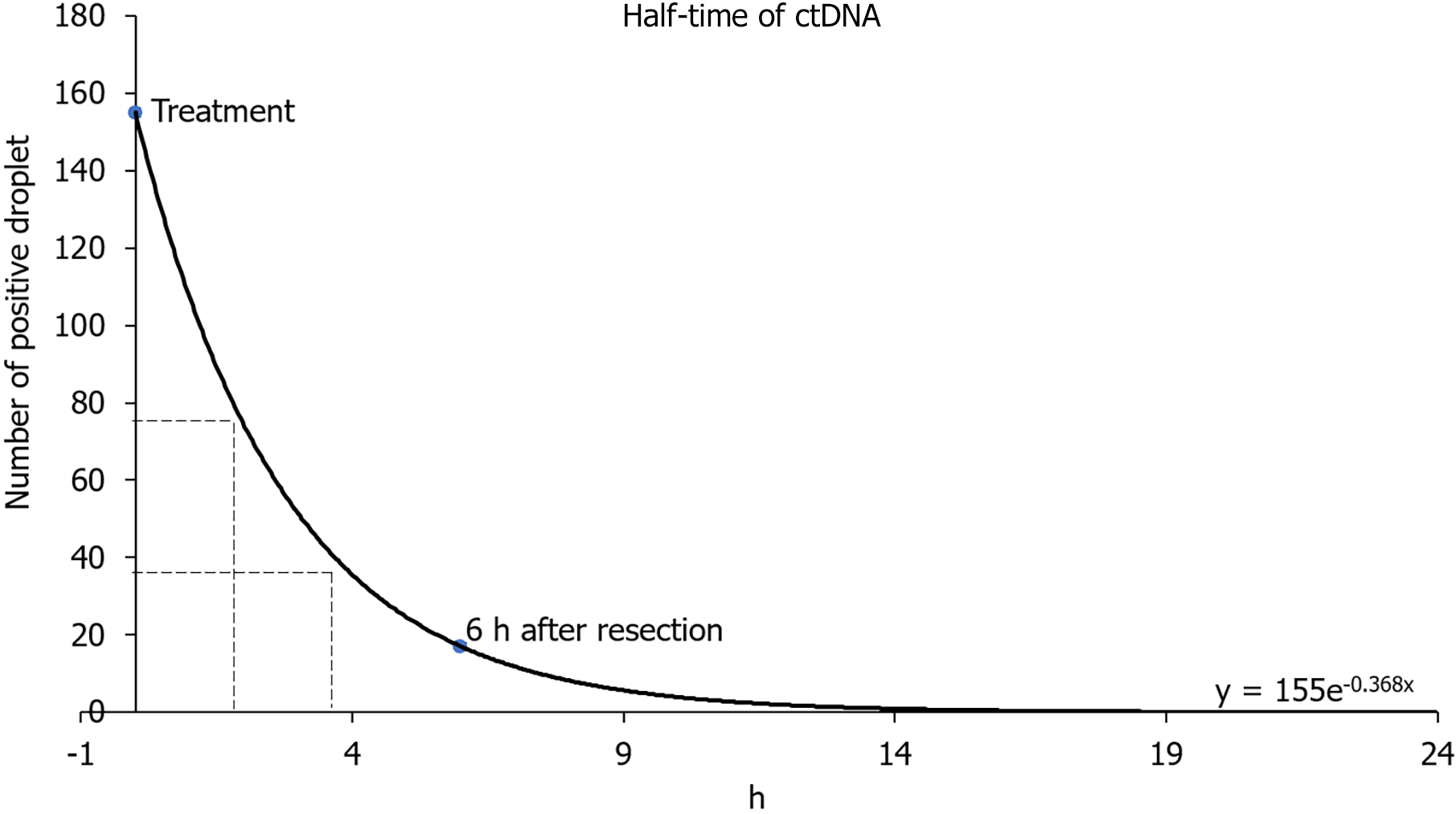
In two-site xenograft mice, tumor resection was performed at week 7. The average tumor size in the control group was 10.3 mm at the time of resection, and the average tumor sizes measured 6 h, 1 d, or 3 d at the time of resection were 10.1, 10.3, and 10.2 mm, respectively (P = 0.98). We detected hTERT at resection (control), but hTERT had decreased by 6 h, and was undetectable 1 d or 3 d after resection (Figure 5). The control cfDNA concentration was 1.1 μg/mL at the time of resection, and was 1.2, 1.3, and 1.4 μg/mL measured 6 h, 1 d, and 3 d after resection. Pathological autopsy confirmed the absence of macroscopic residual tumor at each evaluation in this experiment. Using data for the number of positive droplets measured 0 and 6 h after tumor resection in the two-site xenograft resection experiment, the half-life of ctDNA may be calculated from y = 155e - 0.368x. In our study, the half-life of ctDNA was estimated to be 1.8–3.2 h (Figure 6).
In four-site xenograft mice, tumor resection was performed at week 5. The average tumor size in the control group was 9.7 mm at the time of resection, while average tumor sizes measured 6 h, 1 d, or 3 d at the time of resection were 11.4, 10.6, and 10.2 mm, respectively (P = 0.34). In this experiment, hTERT was detected in all groups (Figure 5). The control cfDNA concentration was 1.3 μg/mL at resection and 1.2, 1.5, and 1.7 μg/mL measured 6 h, 1 d, and 3 d, respectively, after resection. Here, pathological autopsy revealed the presence of macroscopic residual tumor at each resection evaluation, with tumor invasion and intrathoracic metastasis in all mice. This experiment revealed that residual ctDNA was associated with incomplete resection and metastasis.
Because the TERT gene sequence differs between human and mouse, we were able to determine the origin and dynamics of ctDNA in a xenograft mouse model in which human-derived esophageal cancer cells were injected into the epidermis of mice. This model allowed assessment of ctDNA, which has traditionally been difficult to assess in the human body, due to tumor heterogeneity and the influence of other cells. In our experiment, tumor volume was involved in increases or decreases in ctDNA. In addition, if ctDNA was present over 1 d after resection, the presence of residual tumor is suspected.
Although studies of liquid biopsy using xenograft mouse model have been reported mainly in circulating tumor cells [11], we focused on ctDNA in this study. This model seems to be an ideal method because clinical samples contain a variety of cellular information as well as limitations such as ethical issues. Our report is also extremely valuable in providing direct evidence of the origin of plasma ctDNA, which we assessed in the xenograft mouse model by assaying mTERT and hTERT. Based on this ctDNA confirmation, other factors affecting ctDNA dynamics were examined. In our xenograft experiments, the average tumor sizes 4 wk after two-site and four-site xenografts were very similar (5.6 mm and 6.5 mm), and histology showed similar degrees of tumor invasion (Figure 4). However, ctDNA was detected in four-site xenograft mice but not in two-site xenograft mice. These findings revealed that tumor volume may influence ctDNA detection. In both groups, increasing ctDNA with tumor progression was confirmed at week 8 and week 6. The amount and detection rate of ctDNA correlated with tumor progression in a previous clinical study[6], and our results may support that finding. Although detailed studies on the association between tumor volume or invasion and ctDNA have not been conducted, ctDNA is assumed to be detectable in early cancer once the tumor reaches a certain volume.
The presence of ctDNA after surgical resection is observed in clinical samples from cancer patients, and evaluation during the perioperative period is useful for prediction of prognosis[12-14]. Detection of ctDNA after surgery suggests some residual disease[15]. However, these clinical studies may inevitably detect circulating DNA from sources other than tumor cells, and there have been no reports to indicate when liquid biopsy should be used. Regarding this point, our resection experiments demonstrated reduced hTERT at 6 h and its absence 1 to 3 d after resection, indicating that ctDNA evaluation 1 d after resection might be useful to detect residual tumor in clinical cases. These experiments also revealed tumor volume was involved in the increase or decrease of ctDNA and that post-tumor resection evaluation requires an interval of one day or more after resection.
The half-life of ctDNA was reported as approximately 2 h in one study[16], but another study found the half-life to be 16 min[17]. The metabolism and excretion of cfDNA is affected by liver and kidney function[18], and ctDNA levels might be re
cfDNA is derived from apoptotic or necrotic cells[19,20], and its increase is con
Carcinoembryonic antigen (CEA) and squamous cell carcinoma antigen (SCC-Ag) are biomarkers for esophageal cancer. However, the usefulness of these biomarkers in the early diagnosis of esophageal cancer has not been established. Currently, upper endoscopy is the most useful examination to pick up early-stage esophageal cancer. However, since this examination is invasive, the development of non-invasive me
Our study had the following limitations. First, the artificial implantation of tumor under the skin in the xenograft model differs from the physiology of actual tumor development. Second, individual mice exhibit differences in tumor growth rates, and therefore, our comparative analyses in the present study used the average values for four animals per group. Third, regarding residual tumor, although pathological autopsies were performed on all mice, complete certainty with respect to residual disease is impossible. Forth, TE11 cell line alone is not necessarily sufficient, other cell lines should be examined as well. Fifth, comparison with conventional biomarkers such as CEA and SCC-Ag needs to be shown.
We clarified the origin and dynamics of ctDNA in the xenograft mouse model. We showed that tumor volume was an important factor in ctDNA, and that if the tumor volume was sufficiently large, ctDNA can be detected even in early-stage or superficial cancers. We also found that, upon complete tumor resection, ctDNA disappeared after at least 1 d, unless residual tumor remained. These findings may indicate future clinical uses of liquid biopsy.
The clinical application of liquid biopsy is becoming more widespread. However, it remains unclear which factors, such as tumor volume and tumor invasion, influence circulating tumor DNA (ctDNA), and the origin of ctDNA in liquid biopsy is always problematic.
It will be very important to address the origin and dynamics of ctDNA for further clinical application of liquid biopsy.
A xenograft mouse model was used to assess the origin of ctDNA, clarify the dy
Tumor xenotransplants were established by inoculating BALB/c-nu/nu mice with the TE11 cell line (esophageal squamous cell carcinoma). Analysis of ctDNA was per
Mice given two-site xenografts were sacrificed for ctDNA at week 4 and week 8. No hTERT was detected at week 4, but it was detected at week 8. However, in four-site xenograft mice, hTERT was detected both at week 4 and week 6. These experiments revealed that both tumor invasion and tumor volume were associated with the detection of ctDNA. In resection experiments, hTERT was detected at resection, but had decreased by 6 h, and was no longer detected 1 and 3 d after resection. The half-life of ctDNA was estimated to be 1.8-3.2 h.
We clarified the origin and dynamics of ctDNA, showing that not only tumor invasion but also tumor volume was an important factor. Also, ctDNA could be measured at 1 d after tumor resection to evaluate the residuals.
In the clinical application of liquid biopsy, early-stage cancers could be targeted, and post-treatment monitoring should be performed 1 d after treatment.
We thank all staff in the animal facility of Okayama University, Shinya Ohashi (MD, PhD; Department of Therapeutic Oncology, Kyoto University) and Hiroshi Nakagawa (MD, PhD; Department of Medicine, Columbia University).
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and Hepatology
Country/Territory of origin: Japan
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Norčič G S-Editor: Wang LL L-Editor: A P-Editor: Yuan YY
| 1. | Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1634] [Cited by in RCA: 1752] [Article Influence: 219.0] [Reference Citation Analysis (0)] |
| 2. | Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579-586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1413] [Cited by in RCA: 1678] [Article Influence: 152.5] [Reference Citation Analysis (0)] |
| 3. | Kinugasa H, Nouso K, Miyahara K, Morimoto Y, Dohi C, Tsutsumi K, Kato H, Matsubara T, Okada H, Yamamoto K. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer. 2015;121:2271-2280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 181] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 4. | Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659-1665. [PubMed] |
| 5. | Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1954] [Cited by in RCA: 2135] [Article Influence: 152.5] [Reference Citation Analysis (0)] |
| 6. | Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong SM, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SK, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Sartore-Bianchi A, Schmidt K, Shih lM, Oba-Shinjo SM, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang TL, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N, Diaz LA Jr. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2770] [Cited by in RCA: 3518] [Article Influence: 319.8] [Reference Citation Analysis (0)] |
| 7. | Rolfo C, Russo A. Liquid biopsy for early stage lung cancer moves ever closer. Nat Rev Clin Oncol. 2020;17:523-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 8. | Alix-Panabières C, Pantel K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021;11:858-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 566] [Article Influence: 141.5] [Reference Citation Analysis (0)] |
| 9. | Kinugasa H, Hiraoka S, Nouso K, Yamamoto S, Hirai M, Terasawa H, Yasutomi E, Oka S, Ohmori M, Yamasaki Y, Inokuchi T, Takahara M, Harada K, Tanaka T, Okada H. Liquid biopsy for patients with IBD-associated neoplasia. BMC Cancer. 2020;20:1188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 10. | Natsuizaka M, Kinugasa H, Kagawa S, Whelan KA, Naganuma S, Subramanian H, Chang S, Nakagawa KJ, Rustgi NL, Kita Y, Natsugoe S, Basu D, Gimotty PA, Klein-Szanto AJ, Diehl JA, Nakagawa H. IGFBP3 promotes esophageal cancer growth by suppressing oxidative stress in hypoxic tumor microenvironment. Am J Cancer Res. 2014;4:29-41. [PubMed] |
| 11. | Vishnoi M, Liu NH, Yin W, Boral D, Scamardo A, Hong D, Marchetti D. The identification of a TNBC liver metastasis gene signature by sequential CTC-xenograft modeling. Mol Oncol. 2019;13:1913-1926. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 12. | Lee B, Lipton L, Cohen J, Tie J, Javed AA, Li L, Goldstein D, Burge M, Cooray P, Nagrial A, Tebbutt NC, Thomson B, Nikfarjam M, Harris M, Haydon A, Lawrence B, Tai DWM, Simons K, Lennon AM, Wolfgang CL, Tomasetti C, Papadopoulos N, Kinzler KW, Vogelstein B, Gibbs P. Circulating tumor DNA as a potential marker of adjuvant chemotherapy benefit following surgery for localized pancreatic cancer. Ann Oncol. 2019;30:1472-1478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 155] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 13. | Nakano Y, Kitago M, Matsuda S, Nakamura Y, Fujita Y, Imai S, Shinoda M, Yagi H, Abe Y, Hibi T, Fujii-Nishimura Y, Takeuchi A, Endo Y, Itano O, Kitagawa Y. KRAS mutations in cell-free DNA from preoperative and postoperative sera as a pancreatic cancer marker: a retrospective study. Br J Cancer. 2018;118:662-669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 76] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 14. | Chen K, Zhao H, Shi Y, Yang F, Wang LT, Kang G, Nie Y, Wang J. Perioperative Dynamic Changes in Circulating Tumor DNA in Patients with Lung Cancer (DYNAMIC). Clin Cancer Res. 2019;25:7058-7067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 155] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 15. | Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, Silliman N, Tacey M, Wong HL, Christie M, Kosmider S, Skinner I, Wong R, Steel M, Tran B, Desai J, Jones I, Haydon A, Hayes T, Price TJ, Strausberg RL, Diaz LA, Jr. , Papadopoulos N, Kinzler KW, Vogelstein B, Gibbs P. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 729] [Cited by in RCA: 1027] [Article Influence: 128.4] [Reference Citation Analysis (0)] |
| 16. | Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, Kinzler KW, Vogelstein B, Diaz LA Jr. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985-990. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2240] [Cited by in RCA: 2096] [Article Influence: 123.3] [Reference Citation Analysis (0)] |
| 17. | Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999;64:218-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 812] [Cited by in RCA: 810] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 18. | Tsumita T, Iwanaga M. Fate of injected deoxyribonucleic acid in mice. Nature. 1963;198:1088-1089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 78] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 19. | Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001;313:139-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 516] [Cited by in RCA: 570] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 20. | van der Vaart M, Pretorius PJ. Circulating DNA. Its origin and fluctuation. Ann N Y Acad Sci. 2008;1137:18-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 145] [Article Influence: 8.5] [Reference Citation Analysis (0)] |