Published online Oct 28, 2021. doi: 10.3748/wjg.v27.i40.6927
Peer-review started: March 11, 2021
First decision: April 5, 2021
Revised: May 6, 2021
Accepted: September 1, 2021
Article in press: September 1, 2021
Published online: October 28, 2021
Processing time: 229 Days and 7.6 Hours
Quantitative hepatitis B core-related antigen (qHBcrAg) has a better correlation with intrahepatic hepatitis B virus (HBV) covalently closed circular DNA (cccDNA) than HBV DNA or hepatitis B e antigen (HBeAg), but data are still lacking for its clinical application.
The aim was to investigate serum qHBcrAg levels in patients with chronic hepatitis B and assess the correlation of serum qHBcrAg with pregenomic RNA (pgRNA), cccDNA, and HBeAg seroconversion.
This study was a secondary analysis of patients who underwent percutaneous liver biopsy between July 2014 and June 2019 in two multicenter randomized controlled clinical trials of peginterferon vs nucleos(t)ide analog (NUC)-based therapy (NCT03509688 and NCT03546530). Serum qHBcrAg, pgRNA, HBV DNA, hepatitis B core antigen, HBeAg, liver cccDNA, and HBV DNA were measured. The correlations of serum qHBcrAg with other biomarkers were analyzed.
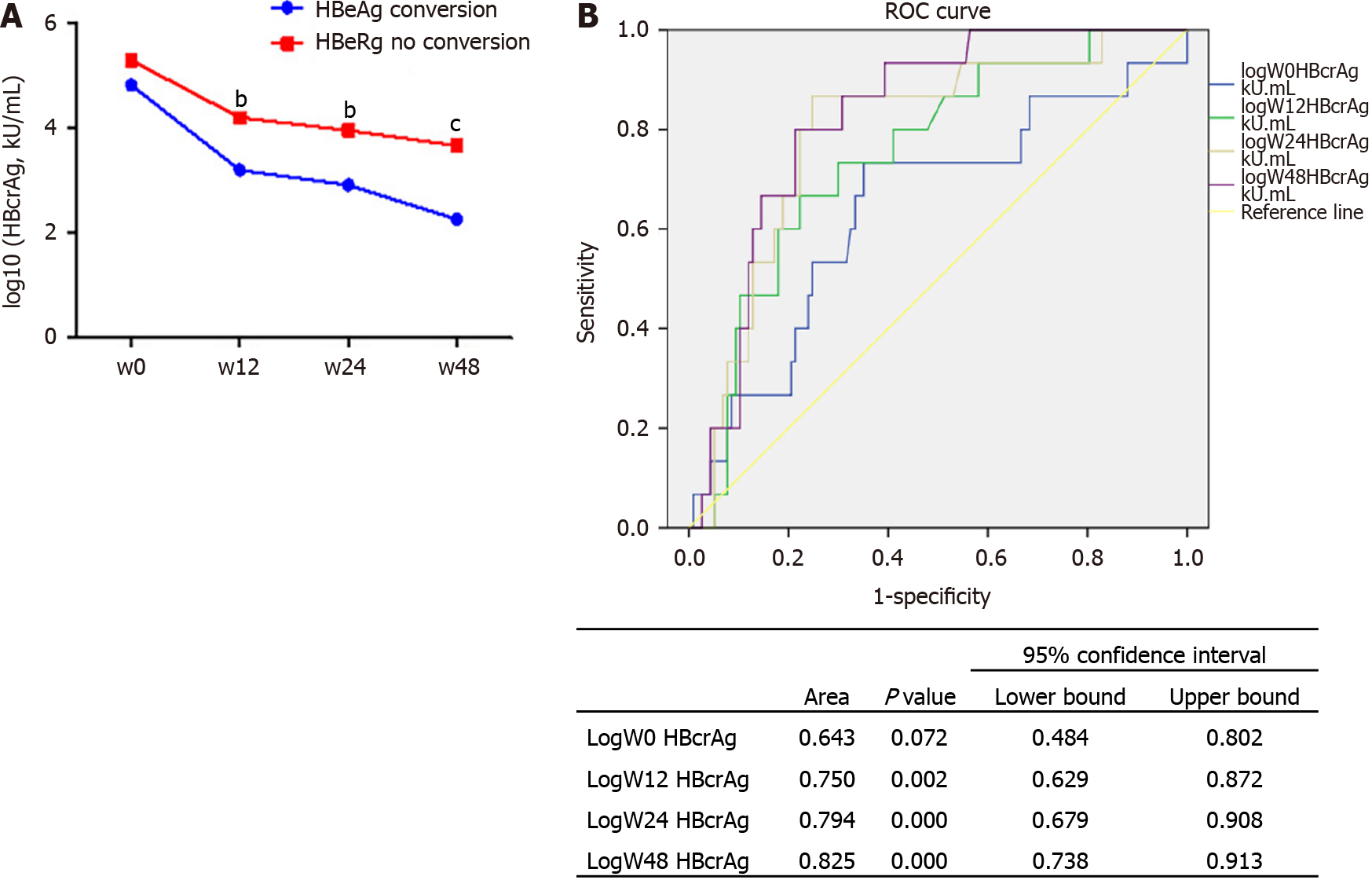
A total of 139 patients were included. The mean qHBcrAg levels were 5.32 ± 1.18 log10 U/mL at baseline and decreased during treatment (all P < 0.0001). Serum qHBcrAg levels were positively correlated with pgRNA (r = 0.597, P < 0.0001) and cccDNA (r = 0.527, P < 0.0001) levels. The correlation of serum qHBcrAg level and intrahepatic HBV DNA levels at baseline was weak but significant (r = 0.399, P < 0.0001). HBcrAg predicted HBeAg seroconversion, with areas under the receiver operating characteristics curve of 0.788 at 24 wk and 0.825 at 48 wk. Log HBcrAg at wk 24 and 48 was independently associated with HBeAg seroconversion [odds ratio (OR) = 2.402, 95% confidence interval (CI): 1.314-4.391, P = 0.004; OR = 3.587, 95%CI: 1.315-9.784, P = 0.013].
Serum HBcrAg levels were correlated with HBV virological markers and could be used to predict HBeAg seroconversion.
Core Tip: The mean quantitative hepatitis B core-related antigen (qHBcrAg) levels were decreased post treatment. Serum qHBcrAg levels were positively associated with pregenomic RNA and covalently closed circular DNA levels. qHBcrAg predicted hepatitis B e antigen (HBeAg) seroconversion with an area under the receiver ope
- Citation: Chi XM, Wang XM, Wang ZF, Wu RH, Gao XZ, Xu HQ, Ding YH, Niu JQ. Serum hepatitis B core-related antigen as a surrogate marker of hepatitis B e antigen seroconversion in chronic hepatitis B. World J Gastroenterol 2021; 27(40): 6927-6938
- URL: https://www.wjgnet.com/1007-9327/full/v27/i40/6927.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i40.6927
Chronic hepatitis B (CHB) is a liver disease caused by a chronic infection with the hepatitis B virus (HBV) and is potentially life-threatening. CHB is a global health problem that affects about 250 million people worldwide, with a prevalence of < 2% in the United States and other Western countries and > 5% in East Asia, Southeast Asia, and sub-Saharan Africa[1-3]. The disease is particularly endemic in China, where there are about 84 million individuals infected with HBV[4], with a prevalence as high as 6.3% in some rural areas[5]. All-cause mortality associated with HBV infection is about 6%-8%, and 15%-40% of untreated patients with CHB develop serious conditions such as cirrhosis and hepatocellular carcinoma[3,6].
The HBV genome exists in the nuclei of infected hepatocytes as a 3.2-kb double-stranded episomal DNA called covalently closed circular DNA (cccDNA). cccDNA is a key component in the HBV life cycle since it is the template for all viral genomic and subgenomic transcripts, including pregenomic RNA (pgRNA), and its level is correlated with the proliferative potential of HBV[7]. cccDNA serves as the template for pgRNA production, which is the main step in HBV replication[7]. The control of intrahepatic levels of HBV cccDNA and/or controlling the transcriptional activity of cccDNA are critical to prevent the occurrence of decompensated cirrhosis and hepatocellular carcinoma, which is the ultimate goal of anti-HBV therapies[8,9]. Hence, changes in the intrahepatic level of cccDNA can be used to monitor the efficacy of antiviral therapies and evaluate the possibility of viral rebound after stopping treatment[10-12]. The direct method to measure liver cccDNA levels is liver biopsy, but it is an invasive procedure not easily accepted by the patients[13]. Therefore, searching for surrogate indicators of intrahepatic HBV cccDNA is important to optimize patient management and quality of life.
Although sustained serum HBV DNA suppression and hepatitis B e antigen (HBeAg) seroconversion are associated with disease remission[8], their performance in reflecting the changes in intrahepatic cccDNA is poor[14,15]. Hepatitis B core-related antigen (HBcrAg) is an emerging marker of HBV DNA suppression[16,17]. HBcrAg consists of hepatitis B core antigen (HBcAg), HBeAg, and p22cr. HBcAg and HBeAg share the first 149 amino acids (aa) encoded by the core gene, overlapping the -10 to 183 aa region[18]. A study showed that quantitative HBcrAg (qHBcrAg) better reflects intrahepatic cccDNA levels than HBV DNA or HBeAg[15]. In addition, intrahepatic cccDNA can be determined with an enzyme immunoassay for HBcrAg[19,20]. Ne
Therefore, this study aimed to investigate the serum qHBcrAg levels of patients with CHB and assess the correlation of serum qHBcrAg with pgRNA, cccDNA, and HBeAg seroconversion. The results will add to the sparse literature about qHBcrAg and could eventually lead to its wider use in the clinical setting.
This study was a secondary analysis of a cohort of patients who underwent per
All patients were diagnosed with CHB according to the criteria of detectable HBV DNA ≥ 105 IU/mL, alanine aminotransferase (ALT) 1.5-10 times the upper limit of normal, and HBeAg positivity[3,6]. The inclusion criteria were: (1) Diagnosis of CHB; (2) Treatment with Peg-IFN or NUC-based therapy for at least 48 wk; and (3) Available serum samples and liver specimens at baseline and 48 wk. The exclusion criteria were: (1) A history of hepatitis C virus or hepatitis D virus infection; (2) Human immunodeficiency virus; (3) Inflammatory diseases such as rheumatoid arthritis, diabetes, auto
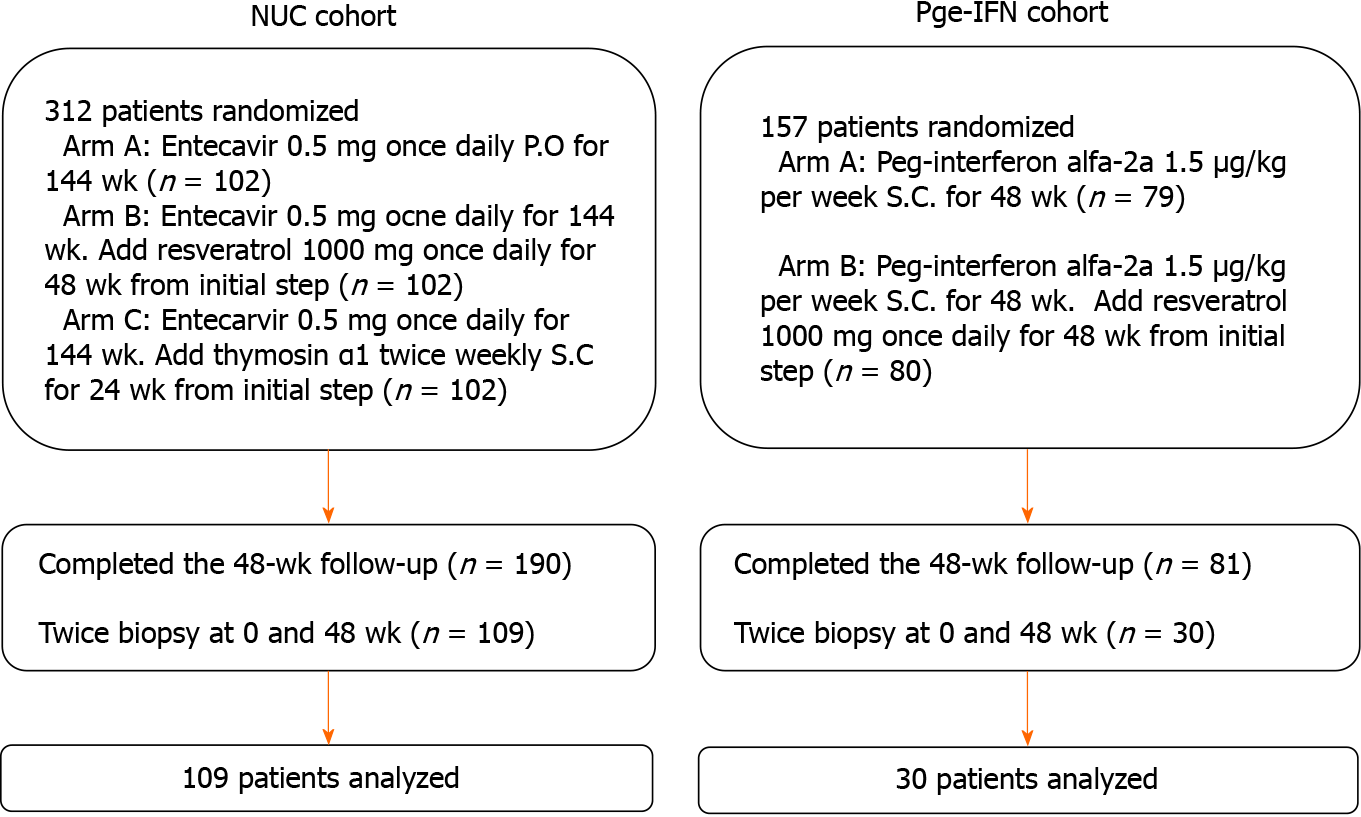
The treatment regimens followed the clinical practice guidelines OF the Asian-Pacific Association for the Study of the Liver on the management of hepatitis B[21]. In one of the original trials, the patients received entecavir (ETV) (Cosunter Pharmaceutical, China) 0.5 mg once daily po for 144 wk with/without resveratrol 1000 mg once daily for 48 wk or thymosin α1 twice-weekly sc for 24 wk. In the other trial, the patients received interferon (IFN, Kawin Technology, China) 1.5 μg/kg per week sc for 48 wk with/without resveratrol 1000 mg once daily for 48 wk. The patients who underwent liver biopsy before and after 48 wk of treatment were included in the present study. In the two trials, 139 patients completed 48 wk treatment with both biopsies (Figure 1). In each original trial[22-24], the patients were randomly assigned to the trial drugs.
The quantification of HBcrAg was performed using a fully automated Lumipulse chemiluminescence enzyme immunoassay analyzer (Fujirebio Inc., Tokyo, Japan) according to the manufacturer’s instructions. Serum was pretreated with sodium dodecyl sulfate and incubated with monoclonal antibodies against denatured HBcAg and HBeAg. After washing and incubation with secondary antibodies, the concentrations of HBcrAg were determined by relative chemiluminescence intensity and compared with a standard curve. Because the general analytic measurement range of the assay was between 1000 U/mL (3 log10 U/mL) and 10000000 U/mL (7 log10 U/mL), serial dilutions of the serum sample were needed when the serum qHBcrAg level was above the detection limit of the assay.
Fasting venous blood was centrifuged at 4000 rpm for 10 min to obtain serum. All laboratory assessments were performed at baseline and weeks 4, 12, 24, 48, and 96. HBV DNA was detected by quantitative polymerase chain reaction (PCR) using the Roche COBAS AmpliPrep/COBAS TaqMan system (Roche Diagnostics, Basel, Switzerland). The lowest detection limit was 20 IU/mL. Hepatitis B surface antigen (HBsAg), anti-HBs, HBeAg, anti-HBe, and anti-HBc were detected by chemiluminescence microparticle immunoassays using the Architect i2000SR platform and Abbott Architect reagents (Abbott Laboratories, Abbott Park, IL, United States). Serum HBsAg levels were measured with a dynamic range of 0-250 IU/mL. If qHBsAg levels were > 250 IU/mL, the samples were retested with a stepwise dilution of 1:10,000. ALT and aspartate aminotransferase were measured at each participating medical site. ALT, HBsAg, HBeAg, and HBV DNA were directly detected immediately at each time point. The HBV genotype was determined at screening. HBV pgRNA was measured at Peking University Health Science Center (Beijing, China), as previously described[25]. HBV genotypes were determined by real-time PCR with Taqman probe technology (Shanghai ZJ Bio-Tech, Shanghai, China). A FibroScan system was used to measure liver stiffness (Echosens, Paris, France).
All patients in this study underwent liver biopsy before treatment and after 48 wk of treatment. The remaining liver tissue was stored in liquid nitrogen. Quantitative intrahepatic cccDNA was detected by PCR-fluorescent probing (SUPBIO Biotechnology, Beijing, China) following the manufacturer’s instructions. Intrahepatic HBV DNA was detected by quantitative PCR using the Roche COBAS AmpliPrep/COBAS TaqMan system (Roche Diagnostics, Basel, Switzerland).
Continuous data were expressed as means ± SD and analyzed using Student’s t-test or Mann-Whitney U-test, as appropriate based on the results of the Kolmogorov-Smirnov test. Intragroup analyses were performed using the paired ˆ-test or repeated-measure analysis of variance. Categorical variables were reported as numbers and percentages (%) and analyzed using Fisher’s exact test. The correlation between two continuous variables was analyzed using Spearman’s bivariate correlation, with a two-tailed significance level of P < 0.01. Receiver operating characteristic (ROC) curves were generated to compare the relative sensitivity and specificity of HBcrAg as a predictor of HBeAg seroconversion. A multivariable logistic regression model was used to determine whether the level of HBcrAg was a risk factor of HBeAg seroconversion. Otherwise, two-tailed P values of < 0.05 were considered statistically significant in all analyses. The statistical analysis were performed with SPSS 18.0 (IBM, Armonk, NY, United States).
From the two original trials, 139 patients were eligible (Table 1). Among them, 69.1% were men, and 30.9% were women. There were more patients with HBV genotype C (79.9%) than B (20.1%). At baseline, the mean levels of intrahepatic HBV cccDNA were 26.65 ± 11.03 copies/cell. The mean levels of serum HBV DNA and HBsAg were 7.59 ± 1.05 log10 IU/mL and 3.81 ± 0.69 log10 IU/mL, respectively. The detailed baseline characteristics of the patients in the ETV and Peg-IFN cohorts are shown in Supple
| Characteristic | n = 139 |
| Age, yr, median (range) | 30.4 (19-62) |
| Sex (male/female, %) | 96/43 (69.1/30.9) |
| HBV genotype, n (%) | |
| B | 28 (20.1) |
| C | 111 (79.9) |
| ALT, U/mL, mean ± SD | 174.07 ± 119.82 |
| Serum HBV DNA, log10 IU/mL, mean ± SD | 7.59 ± 1.05 |
| Serum pgRNA, log10 copies/mL, mean ± SD | 7.82 ± 1.17 |
| ihHBV cccDNA, copies/cell | 26.65 ± 11.03 |
| ihHBV DNA, copies/cell | 385.13 ± 86.09 |
| Serum HBsAg, log10 IU/mL, mean ± SD | 3.81 ± 0.69 |
| Serum HBeAg, log10 S/CO, mean ± SD | 2.49 ± 0.82 |
| HBcrAg, log10 U/mL, mean ± SD | 5.32 ± 1.18 |
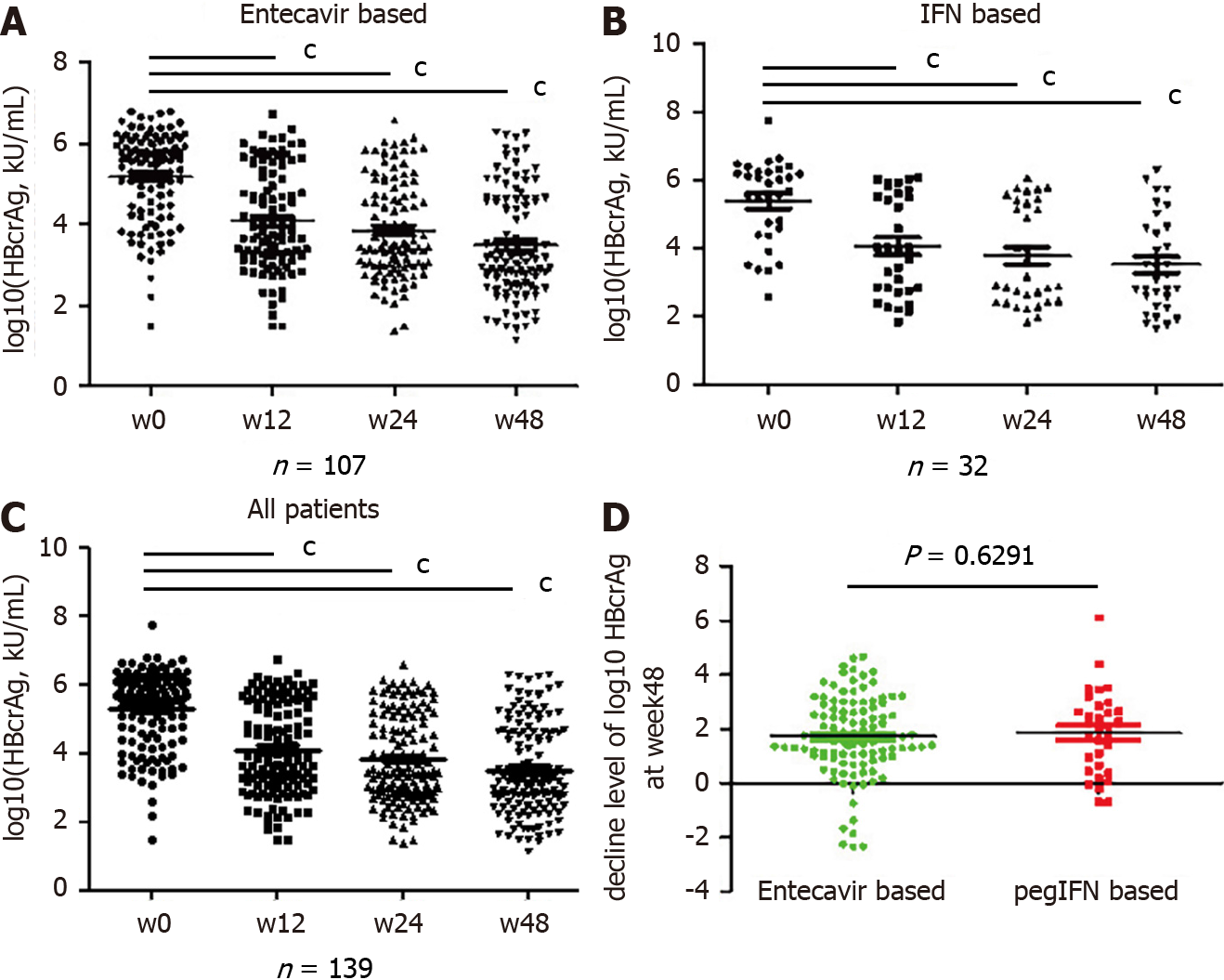
The serum qHBcrAg levels were different among the 139 patients with different phases of HBV infection and treatment. Indeed, the levels of qHBcrAg were 2.30-7.80 log10 U/mL. The mean levels were 5.24 ± 1.07 log10 U/mL in the ETV group (Figure 2A) and 5.38 ± 1.22 in the Peg-IFN group (Figure 2B) at baseline. The mean levels of qHBcrAg were 5.32 ± 1.18 log10 U/mL at baseline and 3.50 ± 1.31 log10 U/mL at week 48, showing decreases at each time point after treatment initiation (all P < 0.0001. Figure 2A-C), and there were no differences between the two groups (P = 0.6291, Figure 2D).
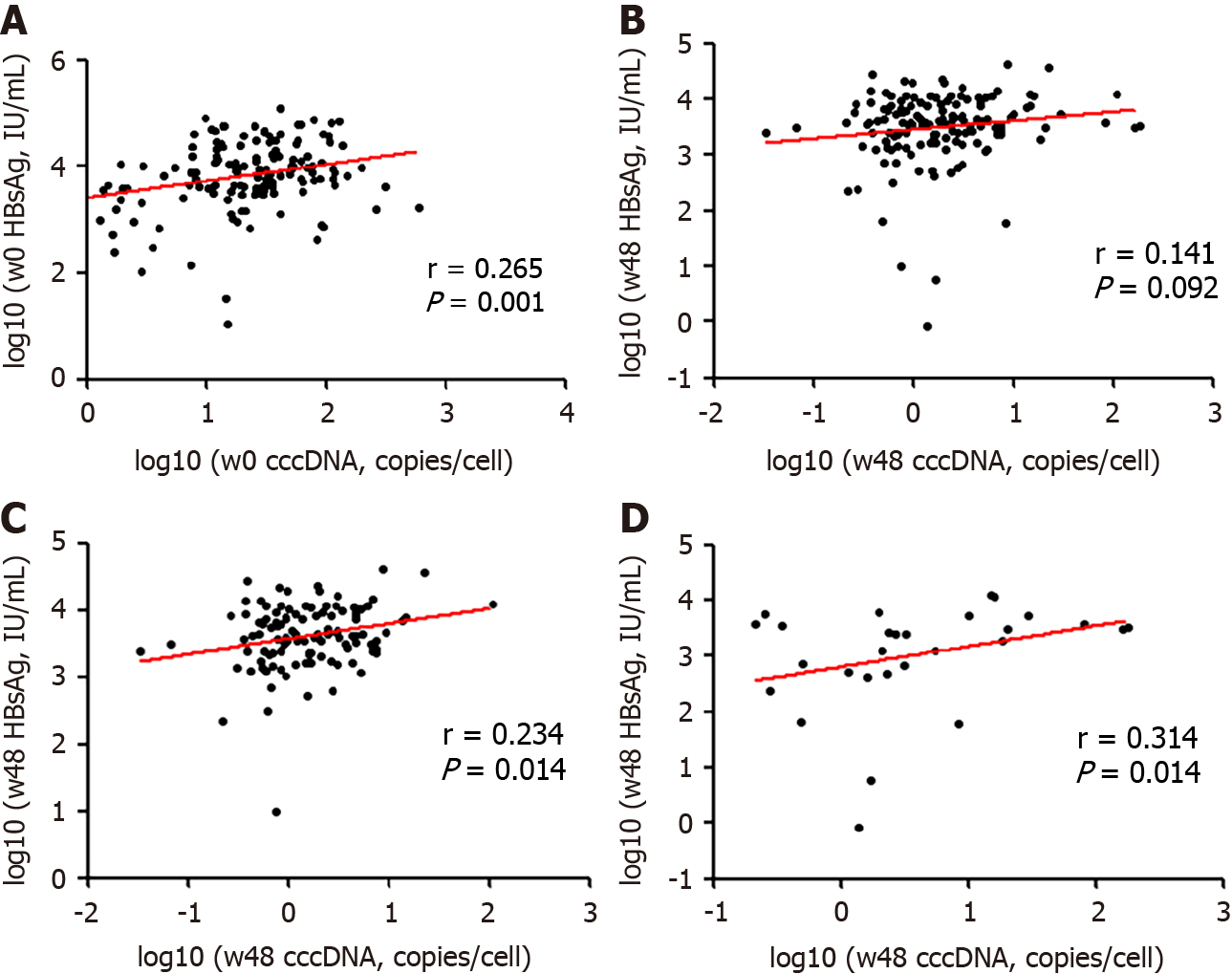
Among the 139 patients, the serum qHBcrAg levels were positively correlated with cccDNA (r = 0.527, P < 0.0001; r = 0.323, P = 0.0001) and pgRNA (r = 0.597, P < 0.0001; r = 0.592, P = 0.0001) levels before and after treatment (Figure 3A and B). The correlation of serum qHBcrAg levels and intrahepatic HBV DNA levels was statistically sig
Serum HBsAg was weakly correlated with cccDNA levels at baseline (r = 0.265, P < 0.001) and week 48 (r = 0.141, P = 0.092; Figure 4A and B). The correlation between HBsAg and cccDNA levels at week 48 was weak in both treatment cohorts (Figure 4C and D).
The levels of serum HBcrAg were significantly lower in patients with HBeAg conversion compared with those without (Figure 5A). The area under the ROC curve of HBcrAg levels for the prediction of HBeAg seroconversion was 0.643 [95% con
qHBcrAg better represents intrahepatic levels of HBV cccDNA than HBV DNA or HBeAg[19,20,26], but data are still lacking in support of its wide clinical application. Therefore, this study aimed to investigate serum qHBcrAg levels in patients with CHB and assess the correlation of serum qHBcrAg with pgRNA and cccDNA. As expected, serum qHBcrAg was significantly and positively associated with the intrahepatic levels of cccDNA in CHB, and the association was stronger than the correlation between serum qHBsAg and intrahepatic cccDNA. Furthermore, antiviral therapy reduced the serum levels of HBcrAg and HBV DNA and the intrahepatic levels of cccDNA. The results suggest that serum HBcrAg levels correlate with HBV virological markers and could be used to predict CHB treatment outcomes, especially HBeAg seroconversion.
Serum levels of HBV DNA have been considered for many decades as a marker of the intrahepatic levels of cccDNA in untreated patients with CHB, but not when they are under treatment with NUCs. Indeed, under treatment, the decrease of the in
Of importance, decreases of the serum HBcrAg level were positively associated with decreases of the intrahepatic levels of cccDNA, even under therapy. Hence, the serum levels of HBcrAg could be a biomarker for the intrahepatic levels of cccDNA. In addition, because the changes in HBcrAg parallel those of intrahepatic cccDNA during treatment, it might be a better long-term prognostic indicator of CHB outcomes than other biomarkers (i.e. serum HBV DNA and HBeAg). HBcrAg levels are determined by the transcription level of cccDNA. Therefore, decline of the HBcrAg levels with NUCs is slower than the decline of the serum levels of HBV DNA in patients. The slower decline might also explain why the serum levels of HBcrAg were positively correlated with the intrahepatic levels of cccDNA, either before or after ETV treatment.
In addition, the levels of serum qHBcrAg were higher in this study than in a previous study[31], which might have been the result of the high proportion of HBeAg-positive patients in this study. Considering that viral replication and host immune responses can be influenced by the HBV genotype, the distribution of the serum levels of HBcrAg was examined between patients carrying the HBV genotypes B and C, but the difference was not statistically significant. HBcrAg is a pre-core protein encoded by the pre-core/core regions of the HBV genome. Hence, the production of HBcrAg is not affected by the promoters found in the S region[18], possibly explaining the similar distribution of HBcrAg between the genotypes.
The serum levels of qHBsAg reflect the intrahepatic levels of cccDNA[19,20]. Still, in this study, the correlation between qHBsAg and intrahepatic cccDNA was weaker than the correlation between serum qHBcrAg and intrahepatic cccDNA, both without and under treatment. Most patients with HBsAg loss or seroconversion still had detectable levels of intrahepatic cccDNA and HBV DNA. Thus, according to the currently available evidence, serum levels of qHBcrAg could be more appropriate than HBsAg as a biomarker of the intrahepatic levels of cccDNA, but that needs to be confirmed in larger studies. The predictive value of qHBcrAg for HBeAg seroconversion was the best at 24 and 48 wk. That could be because baseline qHBcrAg is not predictive of the response to treatment, and that 12 wk is too early to observe a proper response. Additional studies are necessary to determine the best timing of qHBcrAg measurement for the prognosis of CHB. Nevertheless, qHBcrAg levels at the other time points were still associated with HBeAg seroconversion, as observed in previous studies[32-35]. HBsAg and HBeAg each represent only a part of the process of HBV production and assembly, and that could explain why they have a lower prognostic value, especially HBsAg at 48 wk[30].
The study has limitations. It was a secondary analysis of patients from two different trials with two different antiviral treatments and different regimens. Not all patients underwent two biopsies, leading to a selection bias and a small sample size. Im
Serum qHBcrAg levels were correlated with the intrahepatic levels of cccDNA in patients with CHB and might be an acceptable surrogate marker for cccDNA. HBcrAg levels correlated with HBV virological markers and could be used to predict CHB treatment outcomes, especially HBeAg seroconversion. Hence, serum qHBcrAg might be used in the clinical setting for monitoring intrahepatic HBV status and determining the long-term prognosis of patients with CHB.
Quantitative hepatitis B core-related antigen (qHBcrAg) had a better correlation with intrahepatic hepatitis B virus (HBV) covalently closed circular DNA (cccDNA) than either HBV DNA or hepatitis B e antigen (HBeAg).
Data are still lacking for the widespread clinical application of qHBcrAg.
This study aimed to investigate the serum qHBcrAg levels in patients with chronic hepatitis B (CHB) and to assess the correlation of serum qHBcrAg with pregenomic RNA (pgRNA), cccDNA, and HBeAg seroconversion.
This was a secondary analysis of patients who underwent percutaneous liver biopsy in two multicenter, randomized, controlled clinical trials. Serum qHBcrAg, pgRNA, HBV DNA, hepatitis B core antigen, and HBeAg and liver cccDNA, and HBV DNA were measured. The correlations of serum qHBcrAg with other biomarkers were tested.
Serum qHBcrAg levels were positively associated with pgRNA (r = 0.597, P < 0.0001) and cccDNA (r = 0.527, P < 0.0001) levels. HBcrAg predicted HBeAg seroconversion, with an area under the receiver operating characteristics curve of 0.788 at 24 wk and 0.825 at 48 wk.
Serum HBcrAg levels correlated with HBV virological markers and could be used to predict HBeAg seroconversion.
Serum qHBcrAg might be used in the clinical setting to monitor intrahepatic HBV status and determine the long-term prognosis of patients with CHB.
We are very grateful to the following professors for their selfless help in the process of collecting and treating clinical cases: Yu L, Chen YP, Shang J, Liu LG, Zhang SQ, Jiang YF, Zhang MX, Tong QX, Zhang LL, Tan YW, Ma AL, Dang SS, Xu B, Jin ZJ, Li J and Li XB.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: El-Nakeep S, Simsek H S-Editor: Gao CC L-Editor: Filipodia P-Editor: Yuan YY
| 1. | Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386:1546-1555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1806] [Cited by in RCA: 1998] [Article Influence: 199.8] [Reference Citation Analysis (4)] |
| 2. | Terrault NA, Lok ASF, McMahon BJ, Chang KM, Hwang JP, Jonas MM, Brown RS Jr, Bzowej NH, Wong JB. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology. 2018;67:1560-1599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2290] [Cited by in RCA: 2843] [Article Influence: 406.1] [Reference Citation Analysis (0)] |
| 3. | Trépo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet. 2014;384:2053-2063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1004] [Cited by in RCA: 1157] [Article Influence: 105.2] [Reference Citation Analysis (0)] |
| 4. | Wang H, Men P, Xiao Y, Gao P, Lv M, Yuan Q, Chen W, Bai S, Wu J. Hepatitis B infection in the general population of China: a systematic review and meta-analysis. BMC Infect Dis. 2019;19:811. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 163] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 5. | Liu J, Zhang S, Wang Q, Shen H, Zhang M, Zhang Y, Yan D, Liu M. Seroepidemiology of hepatitis B virus infection in 2 million men aged 21-49 years in rural China: a population-based, cross-sectional study. Lancet Infect Dis. 2016;16:80-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 159] [Article Influence: 17.7] [Reference Citation Analysis (1)] |
| 6. | Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology. 2009;50:661-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2125] [Cited by in RCA: 2171] [Article Influence: 135.7] [Reference Citation Analysis (0)] |
| 7. | Tang H, Banks KE, Anderson AL, McLachlan A. Hepatitis B virus transcription and replication. Drug News Perspect. 2001;14:325-334. [PubMed] |
| 8. | Tang CM, Yau TO, Yu J. Management of chronic hepatitis B infection: current treatment guidelines, challenges, and new developments. World J Gastroenterol. 2014;20:6262-6278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 111] [Cited by in RCA: 102] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 9. | Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972-1984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 694] [Article Influence: 69.4] [Reference Citation Analysis (0)] |
| 10. | Yang HC, Kao JH. Persistence of hepatitis B virus covalently closed circular DNA in hepatocytes: molecular mechanisms and clinical significance. Emerg Microbes Infect. 2014;3:e64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 11. | Shi M, Sun WL, Hua YY, Han B, Shi L. Effects of entecavir on hepatitis B virus covalently closed circular DNA in hepatitis B e antigen-positive patients with hepatitis B. PLoS One. 2015;10:e0117741. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 12. | Zheng Q, Zhu YY, Chen J, Liu YR, You J, Dong J, Zeng DW, Gao LY, Chen LH, Jiang JJ. Decline in intrahepatic cccDNA and increase in immune cell reactivity after 12 weeks of antiviral treatment were associated with HBeAg loss. J Viral Hepat. 2014;21:909-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Thampanitchawong P, Piratvisuth T. Liver biopsy:complications and risk factors. World J Gastroenterol. 1999;5:301-304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 190] [Cited by in RCA: 208] [Article Influence: 8.0] [Reference Citation Analysis (3)] |
| 14. | Tanaka E, Matsumoto A, Yoshizawa K, Maki N. Hepatitis B core-related antigen assay is useful for monitoring the antiviral effects of nucleoside analogue therapy. Intervirology. 2008;51 Suppl 1:3-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 24] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 15. | Liu YY, Liang XS. Progression and status of antiviral monitoring in patients with chronic hepatitis B: From HBsAg to HBV RNA. World J Hepatol. 2018;10:603-611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 16. | Mak LY, Wong DK, Cheung KS, Seto WK, Lai CL, Yuen MF. Review article: hepatitis B core-related antigen (HBcrAg): an emerging marker for chronic hepatitis B virus infection. Aliment Pharmacol Ther. 2018;47:43-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 167] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 17. | Shimakawa Y, Ndow G, Njie R, Njai HF, Takahashi K, Akbar SMF, Cohen D, Nayagam S, Jeng A, Ceesay A, Sanneh B, Baldeh I, Imaizumi M, Moriyama K, Aoyagi K, D'Alessandro U, Mishiro S, Chemin I, Mendy M, Thursz MR, Lemoine M. Hepatitis B Core-related Antigen: An Alternative to Hepatitis B Virus DNA to Assess Treatment Eligibility in Africa. Clin Infect Dis. 2020;70:1442-1452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 18. | Kimura T, Rokuhara A, Sakamoto Y, Yagi S, Tanaka E, Kiyosawa K, Maki N. Sensitive enzyme immunoassay for hepatitis B virus core-related antigens and their correlation to virus load. J Clin Microbiol. 2002;40:439-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 191] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 19. | Suzuki F, Miyakoshi H, Kobayashi M, Kumada H. Correlation between serum hepatitis B virus core-related antigen and intrahepatic covalently closed circular DNA in chronic hepatitis B patients. J Med Virol. 2009;81:27-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 159] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 20. | Matsuzaki T, Tatsuki I, Otani M, Akiyama M, Ozawa E, Miuma S, Miyaaki H, Taura N, Hayashi T, Okudaira S, Takatsuki M, Isomoto H, Takeshima F, Eguchi S, Nakao K. Significance of hepatitis B virus core-related antigen and covalently closed circular DNA levels as markers of hepatitis B virus re-infection after liver transplantation. J Gastroenterol Hepatol. 2013;28:1217-1222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 21. | Sarin SK, Kumar M, Lau GK, Abbas Z, Chan HL, Chen CJ, Chen DS, Chen HL, Chen PJ, Chien RN, Dokmeci AK, Gane E, Hou JL, Jafri W, Jia J, Kim JH, Lai CL, Lee HC, Lim SG, Liu CJ, Locarnini S, Al Mahtab M, Mohamed R, Omata M, Park J, Piratvisuth T, Sharma BC, Sollano J, Wang FS, Wei L, Yuen MF, Zheng SS, Kao JH. Asian-Pacific clinical practice guidelines on the management of hepatitis B: a 2015 update. Hepatol Int. 2016;10:1-98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1985] [Cited by in RCA: 1959] [Article Influence: 217.7] [Reference Citation Analysis (0)] |
| 22. | Sonneveld MJ, van Oord GW, van Campenhout MJ, De Man RA, Janssen HLA, de Knegt RJ, Boonstra A, van der Eijk AA. Relationship between hepatitis B core-related antigen levels and sustained HBeAg seroconversion in patients treated with nucleo(s)tide analogues. J Viral Hepat. 2019;26:828-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 23. | Song G, Rao H, Feng B, Wei L. Prediction of spontaneous HBeAg seroconversion in HBeAg-positive chronic hepatitis B patients during the immune clearance phase. J Med Virol. 2014;86:1838-1844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 24. | Song G, Yang R, Rao H, Feng B, Ma H, Jin Q, Wei L. Serum HBV core-related antigen is a good predictor for spontaneous HBeAg seroconversion in chronic hepatitis B patients. J Med Virol. 2017;89:463-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 25. | Maasoumy B, Wiegand SB, Jaroszewicz J, Bremer B, Lehmann P, Deterding K, Taranta A, Manns MP, Wedemeyer H, Glebe D, Cornberg M. Hepatitis B core-related antigen (HBcrAg) levels in the natural history of hepatitis B virus infection in a large European cohort predominantly infected with genotypes A and D. Clin Microbiol Infect. 2015;21:606.e1-606.10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 115] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 26. | Wang L, Cao X, Wang Z, Gao Y, Deng J, Liu X, Zhuang H. Correlation of HBcrAg with Intrahepatic Hepatitis B Virus Total DNA and Covalently Closed Circular DNA in HBeAg-Positive Chronic Hepatitis B Patients. J Clin Microbiol. 2019;57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 27. | Wong DK, Seto WK, Cheung KS, Chong CK, Huang FY, Fung J, Lai CL, Yuen MF. Hepatitis B virus core-related antigen as a surrogate marker for covalently closed circular DNA. Liver Int. 2017;37:995-1001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 139] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 28. | Chen EQ, Feng S, Wang ML, Liang LB, Zhou LY, Du LY, Yan LB, Tao CM, Tang H. Serum hepatitis B core-related antigen is a satisfactory surrogate marker of intrahepatic covalently closed circular DNA in chronic hepatitis B. Sci Rep. 2017;7:173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 83] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 29. | van Campenhout MJ, Brouwer WP, van Oord GW, Xie Q, Zhang Q, Zhang N, Guo S, Tabak F, Streinu-Cercel A, Wang J, Pas SD, Sonneveld MJ, de Knegt RJ, Boonstra A, Hansen BE, Janssen HL. Hepatitis B core-related antigen levels are associated with response to entecavir and peginterferon add-on therapy in hepatitis B e antigen-positive chronic hepatitis B patients. Clin Microbiol Infect. 2016;22:571.e5-571.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 30. | Inoue T, Tanaka Y. The Role of Hepatitis B Core-Related Antigen. Genes (Basel). 2019;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 31. | Wong DK, Tanaka Y, Lai CL, Mizokami M, Fung J, Yuen MF. Hepatitis B virus core-related antigens as markers for monitoring chronic hepatitis B infection. J Clin Microbiol. 2007;45:3942-3947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 141] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 32. | Ma H, Yang RF, Li XH, Jin Q, Wei L. HBcrAg Identifies Patients Failing to Achieve HBeAg Seroconversion Treated with Pegylated Interferon Alfa-2b. Chin Med J (Engl). 2016;129:2212-2219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 33. | Chuaypen N, Posuwan N, Payungporn S, Tanaka Y, Shinkai N, Poovorawan Y, Tangkijvanich P. Serum hepatitis B core-related antigen as a treatment predictor of pegylated interferon in patients with HBeAg-positive chronic hepatitis B. Liver Int. 2016;36:827-836. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 63] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 34. | Matsumoto A, Yatsuhashi H, Nagaoka S, Suzuki Y, Hosaka T, Tsuge M, Chayama K, Kanda T, Yokosuka O, Nishiguchi S, Saito M, Miyase S, Kang JH, Shinkai N, Tanaka Y, Umemura T, Tanaka E. Factors associated with the effect of interferon-α sequential therapy in order to discontinue nucleoside/nucleotide analog treatment in patients with chronic hepatitis B. Hepatol Res. 2015;45:1195-1202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 35. | Martinot-Peignoux M, Lapalus M, Maylin S, Boyer N, Castelnau C, Giuily N, Pouteau M, Moucari R, Asselah T, Marcellin P. Baseline HBsAg and HBcrAg titres allow peginterferon-based 'precision medicine' in HBeAg-negative chronic hepatitis B patients. J Viral Hepat. 2016;23:905-911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |