Published online Oct 14, 2021. doi: 10.3748/wjg.v27.i38.6476
Peer-review started: March 5, 2021
First decision: April 17, 2021
Revised: April 26, 2021
Accepted: September 6, 2021
Article in press: September 6, 2021
Published online: October 14, 2021
Processing time: 221 Days and 3.2 Hours
Traditional methods of developing predictive models in inflammatory bowel diseases (IBD) rely on using statistical regression approaches to deriving clinical scores such as the Crohn's disease (CD) activity index. However, traditional approaches are unable to take advantage of more complex data structures such as repeated measurements. Deep learning methods have the potential ability to automatically find and learn complex, hidden relationships between predictive markers and outcomes, but their application to clinical prediction in CD and IBD has not been explored previously.
To determine and compare the utility of deep learning with conventional algorithms in predicting response to anti-tumor necrosis factor (anti-TNF) therapy in CD.
This was a retrospective single-center cohort study of all CD patients who commenced anti-TNF therapy (either adalimumab or infliximab) from January 1, 2010 to December 31, 2015. Remission was defined as a C-reactive protein (CRP) < 5 mg/L at 12 mo after anti-TNF commencement. Three supervised learning algorithms were compared: (1) A conventional statistical learning algorithm using multivariable logistic regression on baseline data only; (2) A deep learning algorithm using a feed-forward artificial neural network on baseline data only; and (3) A deep learning algorithm using a recurrent neural network on repeated data. Predictive performance was assessed using area under the receiver operator characteristic curve (AUC) after 10× repeated 5-fold cross-validation.
A total of 146 patients were included (median age 36 years, 48% male). Concomitant therapy at anti-TNF commencement included thiopurines (68%), methotrexate (18%), corticosteroids (44%) and aminosalicylates (33%). After 12 mo, 64% had CRP < 5 mg/L. The conventional learning algorithm selected the following baseline variables for the predictive model: Complex disease behavior, albumin, monocytes, lymphocytes, mean corpuscular hemoglobin concentration and gamma-glutamyl transferase, and had a cross-validated AUC of 0.659, 95% confidence interval (CI): 0.562-0.756. A feed-forward artificial neural network using only baseline data demonstrated an AUC of 0.710 (95%CI: 0.622-0.799; P = 0.25 vs conventional). A recurrent neural network using repeated biomarker measurements demonstrated significantly higher AUC compared to the conventional algorithm (0.754, 95%CI: 0.674-0.834; P = 0.036).
Deep learning methods are feasible and have the potential for stronger predictive performance compared to conventional model building methods when applied to predicting remission after anti-TNF therapy in CD.
Core Tip: Deep learning has vast potential, but its clinical utility in predicting outcomes in Crohn’s disease (CD) has not been explored. This study showed that deep learning algorithms (a recurrent neural network) using a more complex information structure including repeated biomarker measurements had a better predictive performance compared to a conventional statistical algorithm using only baseline data. This proof-of-concept study therefore paves the way for further research in the use of deep learning methods in clinical prediction in CD.
- Citation: Con D, van Langenberg DR, Vasudevan A. Deep learning vs conventional learning algorithms for clinical prediction in Crohn's disease: A proof-of-concept study. World J Gastroenterol 2021; 27(38): 6476-6488
- URL: https://www.wjgnet.com/1007-9327/full/v27/i38/6476.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i38.6476
Crohn's disease (CD) is a heterogeneous chronic inflammatory bowel disease (IBD) that is characterized by intermittent flares, medication changes, the potential need for surgery and substantial psychological morbidity[1,2]. As with many chronic conditions, predicting disease trajectory, outcomes and response to therapies in CD are key components of clinical practice where management is tailored to the individual[3]. Precision medicine has been in part driven by the vast expansion of available electronic health data, genomic data and novel disease biomarkers[3]. However, deciphering the complex relationships between large amounts of information and multiple data types presents new analytical challenges.
Traditional approaches to constructing prediction models rely on multivariable regression approaches, typically logistic regression for classification or proportional hazards regression for longitudinal prediction[4]. The resulting predictive models are thus typically only linear combinations of the included predictors and may have limited ability to learn more complex relationships within the data. The advantage of machine learning and artificial intelligence over traditional predictive tools is the potential ability for computational algorithms to automatically find and learn complex, hidden relationships between predictive markers and outcomes[5,6]. This is especially true for deep learning or artificial neural network (ANN) methods, although their 'black box' approach has been criticized for an inability to produce a causal explanation between predictors and outcomes[6].
Despite some limitations, there is much interest in developing and testing machine learning and deep learning tools to aid decision making[5,7]. In luminal gastroenterology, machine learning is gaining traction but its use has been relatively limited to automatic image recognition in endoscopy[8-11] as well as feature selection in genomic and microbiomics data[12,13]. Although there has been great interest in predicting clinical outcomes in CD such as response to therapeutics including biologics[14-18] and immunomodulators[19,20], studies investigating the utility of machine learning models for such predictive tasks have been more limited[21-23]. In particular, the utility of deep learning or ANNs specifically in clinical prediction of CD remains unknown[7].
We aimed to evaluate the utility of deep learning algorithms compared with conventional statistical learning algorithms for clinical prediction in this proof-of-concept study. In particular, we aimed to compare these algorithms as methods of learning and prediction in a general sense, rather than to develop any specific predictive model or score.
This proof-of-concept study utilized a retrospective longitudinal cohort at a tertiary health network comprising three acute hospitals in Melbourne, Australia. The focus of the study was to compare the ability of two supervised learning algorithms (conventional statistical learning vs deep learning) to predict remission after 12 mo of treatment using clinical variables and biomarkers available at baseline. The performance of each algorithm was evaluated using cross-validation. The emphasis of the study was to compare the predictive performance of the two methods of learning rather than any specific model itself. This study was approved by the Eastern Health Office of Research & Ethics (approval number: LR 61/2015).
All adult patients > 18 years with confirmed CD according to standard criteria[24] were included if they were commenced on treatment with an anti-tumor necrosis factor (anti-TNF) agent (adalimumab or infliximab) for luminal CD and received at least one dose of the drug between January 2010 and December 2015. Patients receiving anti-TNF for perianal disease without luminal disease were excluded. Patients were followed up for 12 mo to determine rates of biochemical remission.
Response to anti-TNF was defined as having achieved biochemical remission as per serum C-reactive protein (CRP) < 5 mg/L at 12 mo. This endpoint was chosen because CRP is an accepted biomarker to reflect disease activity and predict outcomes in CD[25,26]. Additionally, normalization of CRP predicts better outcomes in CD patients in remission[27,28]. The first CRP measurement after 12 mo and before 18 mo was used. Patients who did not have a CRP measurement in this time period were excluded.
Baseline characteristics were collected via hospital and clinic records, including Montreal classification, concomitant baseline therapies, prior anti-TNF exposure and prior surgeries. Biomarker data were collected at two time points: (1) A baseline measurement defined as the most proximate measurement prior to commencing anti-TNF, up to 3 mo before commencement; and (2) A prior measurement defined as the second most proximate measurement, up to 12 mo before commencement. Only patients with complete baseline data were included, while missing prior values were imputed with the respective baseline value. The following variables were log-transformed to correct skewness: serum bilirubin, alanine aminotransferase, alkaline phosphatase and gamma-glutamyl transferase (GGT). The data underlying this article cannot be shared publicly due to privacy and ethical concerns. The data will be shared upon reasonable request to the corresponding author.
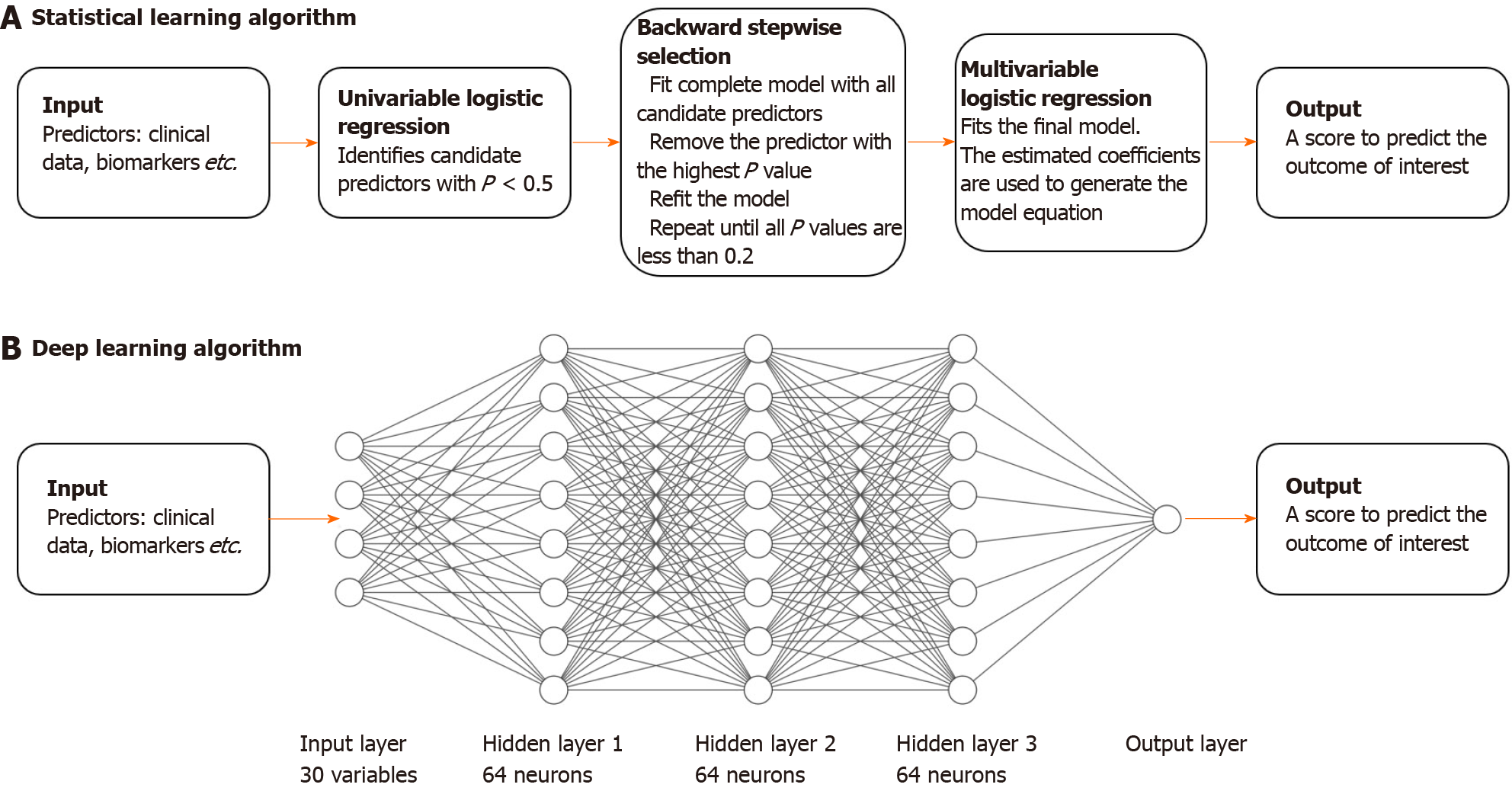
The conventional approach to developing a predictive clinical model is to run univariable and multivariable regression analysis to find useful and preferably independent predictors of the outcome of interest (see Figure 1). Criteria for variable selection usually involves significance testing (P values) or likelihood-based information criterion (such as the Akaike information criterion). In this study, logistic regression was used given the dichotomous nature of the outcome (CRP < 5 mg/L vs CRP ≥ 5 mg/L). The conventional approach typically only uses data from a single time-point, therefore we used baseline data only (the most proximate measurement for all biomarkers). For this conventional approach, we employed the following modelling algorithm: (1) Perform univariable logistic regression on each variable and retain all variables with P < 0.5; (2) Run backwards stepwise selection on all retained variables with removal criterion P > 0.2; and (3) Use the regression coefficients in the remaining multivariable model to derive the predictive score.
A basic deep learning algorithm is a feed-forward ANN[6]. An ANN is composed of layers: an input layer (consisting of all the input predictor variables), an output layer (the prediction), and a number of 'hidden' layers (see Figure 1). Nodes within a hidden layer are called 'neurons'. The hidden layers allow an ANN to learn complex, non-linear relationships between input variables and the outcome of interest. The influence of nodes in a layer on other nodes in subsequent layers is ‘trained’ or fitted using a mathematical function and ultimately determines how information is propagated through the ANN — this is analogous to fitting a regression line on data in conventional statistics. An ANN with only an input and output layer, without hidden layers, can be analogous to simple logistic regression, although they are not equivalent.
However, like the conventional statistical algorithm, a basic feed-forward ANN is still only able to model relationships between predictors at a single time-point. A recurrent neural network (RNN) is a more advanced deep learning algorithm that is able to model repeated measurements over time. Like a feed-forward ANN, information is propagated from the input layer to the output layer. However, instead of only allowing the information to pass through once, information is fed to the RNN sequentially, or 'recurrently' — that is, each set of repeated measurements is inputted once at a time allowing the RNN to update its knowledge of the relationship between the predictors and the outcome. Therefore, the algorithm is additionally able to learn and utilize the dynamics of biomarkers over time, in a way that cannot be achieved by conventional statistical learning methods.
We tested the feed-forward ANN and the RNN in three separate experiments: (1) Using all baseline clinical data in a feed-forward ANN; (2) Using only baseline biomarker data in a feed-forward ANN; and (3) Using repeated biomarker data in an RNN. In this study after hyper-parameter tuning, we used a feed-forward ANN architecture of 3 hidden layers, each with 64 neurons, and an RNN architecture of 1 hidden layer with 64 neurons.
The predictive performances of the conventional statistical algorithm and the experimental deep learning algorithm (ANN) was defined as their ability to correctly classify 12-mo CRP < 5 mg/L measured using the area under the receiver operator characteristic curve (AUC). Because the learning ability of an ANN can be arbitrarily increased, an overly powerful ANN that is trained such that it has near-perfect prediction on the original training cohort, would suffer from poor predictive ability in an external cohort (this is called ‘over-fitting’, a well-known phenomenon). Similarly, the same conventional statistical learning algorithm might result in models with different variables when applied to different cohorts. Therefore, it is important to evaluate the ability of a learning algorithm to predict outcomes in patients that are not included in the original training cohort (external validity).
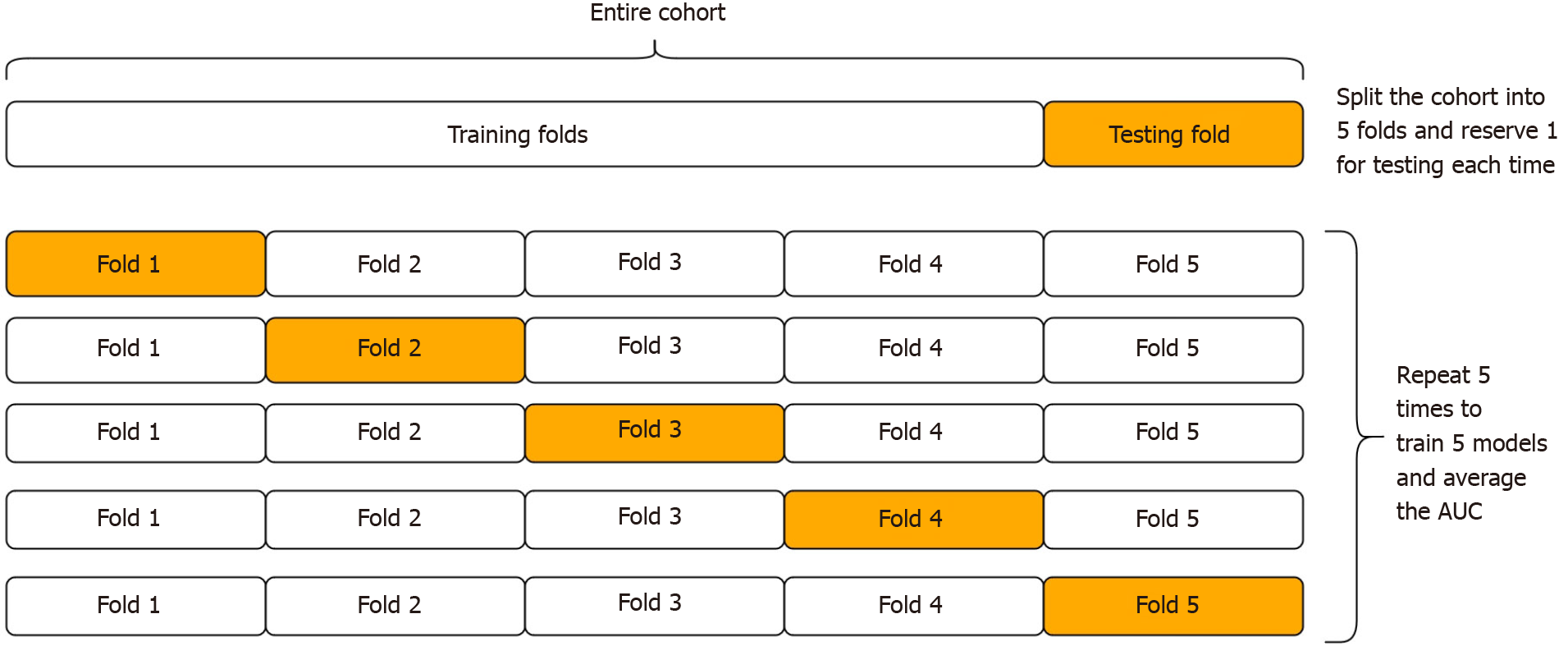
In the absence of external testing cohorts to assess external validity, cross-validation is an internal validation procedure that is suited to this purpose[4]. During cross-validation, the cohort is randomly divided into k equally sized sub-cohorts, known as ‘folds’ (where k is often 5 or 10 by convention). Then, one fold is set aside to be used to test the algorithm, after the algorithm is first trained on the remaining k-1 folds (see Figure 2). This allows the algorithms to be tested on patients that were not used during training. The process is then repeated for each fold (where each fold takes turns in being the test fold). The average AUC after repeating k times gives the cross-validated AUC. However, this procedure is not free from error, because the partitioning process may have randomly resulted in a better (or worse) than usual performance. Thus it is important to repeat the whole process a number of times, to reduce this error[29].
For this study, we used 5-fold cross-validation repeated 10 times to estimate the generalizability of each algorithm on unseen data. Statistical comparison of the cross-validated AUCs of each learning algorithm was made using the variance-corrected repeated k-fold t test instead of a conventional paired t test because of the independency violation from repeated partitioning of the same dataset[29]. For comparison, the naïve or apparent AUC of each model after training and testing on the same entire cohort was given, however this is non-informative. Sample size calculations were conducted only as a guide given the exploratory nature of the study and without prior similar studies on which to base AUC assumptions. The target sample size to detect a 10% difference in AUC with 80% power and 95% significance assuming an AUC variance of 10% was n = 157[30]. To instead detect a 15% difference in AUC under the same conditions, a sample size of n = 70 was required. The Python 3.8.4 programming language with the open-source module PyTorch was used to create the deep learning algorithm. Stata/IC 16 (Texas, United States, 2020) was used to create the statistical learning algorithm.
A total of 146 CD patients were included (see Table 1). Their median age was 36 years [inter-quartile range (IQR) 25-50], 48% were male and median disease duration since diagnosis was 5 years (IQR 1-12). The anti-TNF commenced was infliximab in 58% and adalimumab in 42%. Concomitant therapy at anti-TNF commencement included thiopurines (68%), methotrexate (18%), corticosteroids (44%) and aminosalicylates (33%). Over a quarter of patients (28%) had prior intestinal surgery, while 15% had prior exposure to anti-TNF. After 12 mo, 94 (64%) patients were in biochemical remission (CRP < 5 mg/L).
| Characteristic | n (%) |
| Age, years, median (IQR) | 36 (25-50) |
| Sex | |
| Female | 76 (52) |
| Male | 70 (48) |
| Smoker (active) | 33 (23) |
| CD behavior | |
| B1: Non-stricturing, non-penetrating | 75 (51) |
| B2: Stricturing | 56 (38) |
| B3: Penetrating/fistulizing | 15 (10) |
| CD location | |
| L1: Ileal | 41 (28) |
| L2: Colonic | 43 (29) |
| L3: Ileocolonic | 62 (42) |
| L4: Isolated UGI | 0 (0) |
| Perianal involvement | 20 (21) |
| Initial anti-TNF commenced | |
| Infliximab | 84 (58) |
| Adalimumab | 62 (42) |
| Baseline thiopurine | 99 (68) |
| Baseline methotrexate | 27 (18) |
| Baseline corticosteroids | 64 (44) |
| Baseline aminosalicylates | 48 (33) |
| Prior anti-TNF | 22 (15) |
| Prior intestinal surgery | 41 (28) |
| Disease duration, yr, median (IQR) | 5 (1-12) |
| Baseline investigations | |
| CRP, mg/L, median (IQR) | 3 (2-8) |
| Albumin, g/L, median (IQR) | 37 (36-41) |
Univariable analysis: Baseline factors associated with biochemical remission at 12 mo on univariable testing included non-complex disease behavior (B1), higher albumin and mean corpuscular hemoglobin concentration (MCHC), and lower platelets, lymphocytes and monocytes (each P < 0.05; see Table 2), while lower neutrophil count was nearly significant (P = 0.06). There was no significant association with age, sex, disease location or baseline medical therapies (see Table 2).
| Predictor | Univariable | Multivariable | ||
| OR (95%CI) | P value | Adj. OR (95%CI) | P value | |
| Age, per year | 0.98 (0.96-1.00) | 0.10 | - | - |
| Male (vs female) | 1.42 (0.72-2.82) | 0.31 | - | - |
| CD behavior | ||||
| B1 | 1.0 | Not included | ||
| B2 | 0.45 (0.22-0.94) | 0.034 | Not included | |
| B3 | 0.42 (0.13-1.29) | 0.13 | Not included | |
| CD location | ||||
| L1: ileal | 1.0 | Not included | ||
| L2: colonic | 1.33 (0.54-3.31) | 0.54 | Not included | |
| L3: ileocolonic | 0.91 (0.40-2.06) | 0.83 | Not included | |
| Ileal location (L1) | 0.94 (0.45-2.00) | 0.88 | Not included | |
| Complex disease (B2/B3) | 0.44 (0.22-0.89) | 0.021 | 0.36 (0.16-0.80) | 0.012 |
| Active smoker | 0.76 (0.40-1.47) | 0.42 | - | - |
| Perianal involvement | 1.14 (0.49-2.65) | 0.77 | Not included | |
| Anti-TNF type: infliximab (vs adalimumab) | 1.12 (0.56-2.22) | 0.75 | Not included | |
| Baseline immunomodulator | 1.24 (0.47-3.27) | 0.66 | Not included | |
| Baseline corticosteroids | 1.10 (0.56-2.18) | 0.78 | Not included | |
| Baseline aminosalicylates | 1.16 (0.56-2.40) | 0.69 | Not included | |
| Prior anti-TNF | 0.96 (0.37-2.47) | 0.94 | Not included | |
| Prior intestinal surgery | 0.71 (0.34-1.48) | 0.36 | - | - |
| Disease duration, per loge year | 0.83 (0.65-1.06) | 0.14 | - | - |
| Albumin, per g/L | 1.12 (1.03-1.22) | 0.006 | 1.08 (0.98-1.20) | 0.12 |
| Hemoglobin, per g/L | 1.01 (0.99-1.04) | 0.32 | - | - |
| HCT, per % | 0.91 (0.71-1.16) | 0.44 | - | - |
| RCC, per 109/L | 1.07 (0.84-1.36) | 0.60 | Not included | |
| MCV, per fL | 1.01 (0.96-1.07) | 0.64 | Not included | |
| MCH, per pg/cell | 1.15 (0.99-1.32) | 0.06 | - | - |
| MCHC, per mg/L | 1.05 (1.02-1.08) | 0.002 | 1.05 (1.02-1.09) | 0.004 |
| Platelets, per 100 × 109/L | 0.63 (0.43-0.93) | 0.020 | - | - |
| Neutrophils, per 109/L | 0.91 (0.82-1.00) | 0.06 | - | - |
| Lymphocytes, per 109/L | 0.66 (0.46-0.93) | 0.019 | 0.65 (0.41-1.02) | 0.06 |
| Monocytes, per 109/L | 0.23 (0.08-0.63) | 0.004 | 0.34 (0.10-1.16) | 0.09 |
| Eosinophils, per 109/L | 0.61 (0.08-4.77) | 0.64 | Not included | |
| Basophils, per 0.01 × 109/L | 0.92 (0.80-1.06) | 0.24 | - | - |
| Bilirubin, per loge µmol/L | 1.38 (0.70-2.72) | 0.36 | - | - |
| ALT, per loge IU/L | 1.04 (0.60-1.80) | 0.90 | Not included | |
| ALP, per loge IU/L | 0.55 (0.18-1.64) | 0.28 | - | - |
| GGT, per loge IU/L | 0.71 (0.46-1.09) | 0.12 | 0.69 (0.43-1.11) | 0.13 |
Multivariable analysis: After backward stepwise selection, the following variables remained in the final multivariable model: Complex disease, baseline albumin, monocytes, lymphocytes, MCHC and GGT (see Table 2). The resulting prediction model was given by the following equation (coefficients correct to two significant figures): Score = 0.079 × (albumin, g/L) + 0.050 × (MCHC, mg/L) - 1.1 × (monocytes, 109/L) - 0.43 × (lymphocytes, 109/L) - 1.0 × (complex disease, y=1|n=0) - 0.69 × loge(GGT, IU/L).
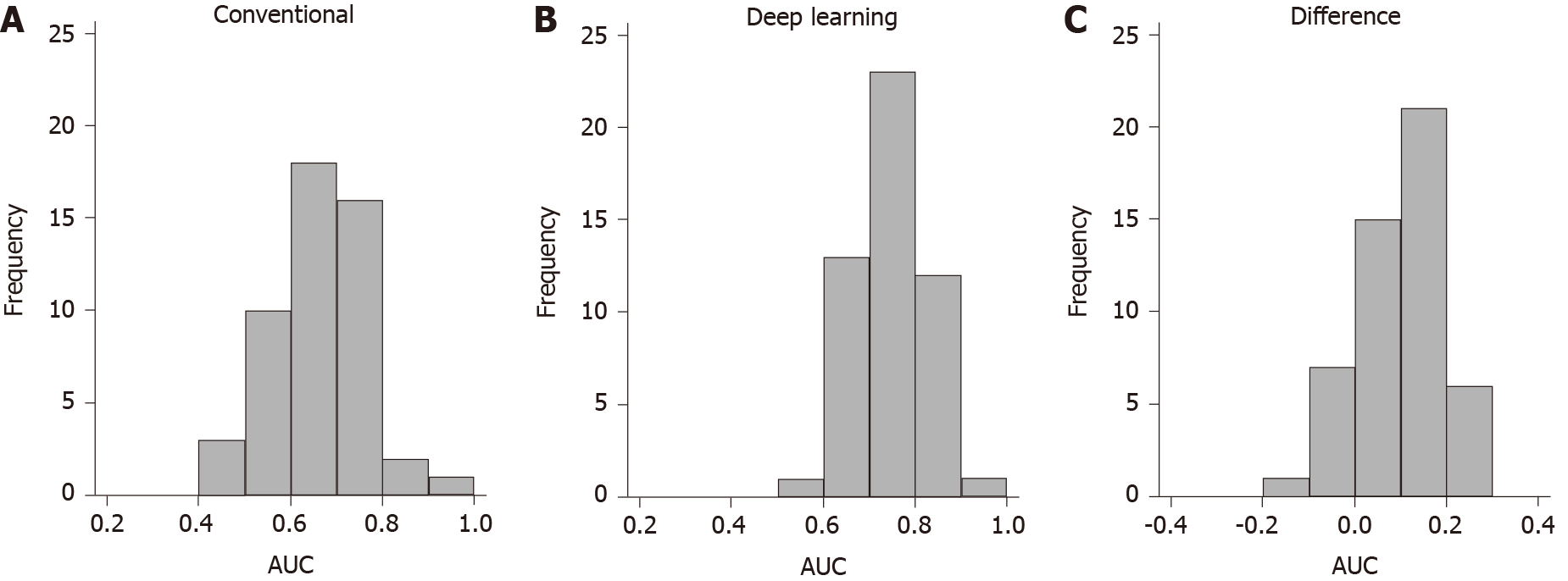
Outcome prediction: After 10× 5-fold cross validation, the average AUC of the statistical learning algorithm was 0.659 [95% confidence interval (CI): 0.562-0.756]. This suggests the statistical learning algorithm is expected to accurately classify 65.9% of patients in external cohorts who have similar characteristics to the study cohort (see Table 3). The algorithm performed better than chance (AUC > 0.5) 94% of the time and had an AUC > 0.7 in 38% of occasions (see Figure 3). The apparent naïve AUC (when trained and tested on the same data) of the model was 0.771.
Feed-forward ANN with complete baseline data: The feed-forward ANN with complete baseline data had a cross-validated AUC of 0.710 (95%CI: 0.622-0.799) (see Figure 3 and Table 3). This difference was not statistically significant using the variance corrected t test (P = 0.25). The algorithm performed better than chance 100% of the time and had good performance (AUC > 0.7) 54% of the time (see Figure 3). For comparison, the naïve AUC of the model was 0.857.
Feed-forward ANN with baseline biomarker data only: The same feed-forward ANN using only baseline biomarker data had a similar cross-validated AUC of 0.706 (95%CI: 0.621-0.791), which was again not significantly different compared to the conventional algorithm (P = 0.33) (see Table 3). The algorithm performed better than chance 100% of the time and had good performance (AUC > 0.7) 58% of the time (see Figure 3). The naïve AUC of the model was 0.776.
RNN with repeated biomarker data: The same feed-forward ANN using only baseline biomarker data had a similar cross-validated AUC of 0.754 (95%CI: 0.674-0.834), which was significantly higher than the AUC of the conventional algorithm (P = 0.036) (see Table 3). This suggests the RNN is expected to accurately classify 75.4% of patients in external cohorts who have similar characteristics to the study cohort. The RNN algorithm performed better than chance 100% of the time and had good performance (AUC > 0.7) 72% of the time (see Figure 3). For comparison, the naïve AUC of the model was 0.892.
The rapid expansion of available health data has motivated the development of machine learning and deep learning tools to predict useful outcomes in clinical medicine[5,6]. The advent of machine learning and data science techniques is especially applicable to IBD due to the heterogeneity and chronic nature of such conditions and the repeated measures of disease activity over time which provides data that may be more suitable for complex modelling techniques. For instance, those with CD typically present with a wide array of disparate disease phenotypes and underlying pathogeneses, and their response to treatment and the trajectory of their disease course varies substantially and changes based on their response[31]. This study has exhibited the potential of deep learning algorithms in predicting response to anti-TNF therapy in patients with CD. The ability to predict the likelihood of response to a given treatment is crucial for risk-benefit assessment, which in turn is crucial to facilitate shared decision making between clinicians and patients[32]. Further, although biologic therapies have revolutionized management in IBD[31], medical therapy is now the principal driver of healthcare costs[33,34] and health economic considerations will inevitably affect treatment choice. Ideally, patients should receive therapies that are both likely to work and cost-effective. Therefore, there can be no ‘one-size-fits-all’ strategy to management, and precision and personalized medicine are key objectives.
Conventional statistical learning algorithms have generated many useful clinical scores, including the CD activity index[35], the simple endoscopic score for CD[36], scores to predict response to biologic therapies[16], and scores to differentiate CD from intestinal tuberculosis[37]. The advantage of conventional scores is often their simplicity and interpretability. A simple score can be memorized and calculated at the bed side and are intuitive as they utilize important risk factors of the outcome of interest. Yet clinical scores can only apply to a rather generic subgroup of patients and are never specific to any individual, as they utilize relatively few variables. Further, conventional methods are not readily able to model more complex, non-linear or time-dependent health states. With new genomic and microbiomic profiling, as well as the rapid uptake of comprehensive electronic medical records with mass data linkage, the ability of conventional learning algorithms to select useful predictive factors may become redundant[38].
Although the advantages of deep learning for the analysis of non-numerical data types is obvious, such as image data in endoscopy[39-41] and text or speech data in natural language processing[42], the utility of deep learning for the analysis of numerical data is less clear but remains promising. A recent study has demonstrated the utility of machine learning in predicting anti-TNF response in rheumatoid arthritis, but relied on genetic markers in addition to clinical data[43]. Another recent study used machine learning to predict whether patients with ankylosing spondylitis required anti-TNF therapy, but did not evaluate whether response to therapy could be predicted[44]. It is anticipated that new data science and machine learning techniques are required to handle large amounts of data for use in clinical practice, although the optimal algorithms for this task remain unknown. Nevertheless, with the provision of comprehensive training data, machine learning tools have the potential to aid in individualized risk prediction, although no such model exists in IBD currently. In our cohort, the RNN deep learning algorithm was able to outperform the conventional algorithm after incorporating repeated biomarker measurements and thus additionally learn the non-linear temporal dynamics of the respective biomarkers — a feat that is not possible with conventional prediction models. It is expected that with enough training data, deep learning methods such as the RNN will be able to incorporate the time series data from multiple repeated health states of an individual patient over time. The clear trade-off with deep learning methods is the need for more data coordination and software to execute. However, the continued uptake of automated medical records in routine clinical practice may mitigate this limitation in future. Further, with the ever increasing breadth and volume of information from sources including comprehensive previous medical history, serum and fecal biomarkers, imaging and endoscopic data as well as genetics, the role of machine learning in prediction in chronic diseases including IBD is likely to expand.
This study has also demonstrated the importance of applying model validation techniques during model development[29]. ANNs and other powerful algorithms have the ability to learn intricate differences in data, yet poorly specified models that focus only on learning power have the propensity to learn the random variations or artefacts in the data, which are present only due to chance. This is evidenced by the RNN in this study achieving excellent AUC during training, but a reduced AUC when tested on unseen data (naïve AUC 0.892; cross-validated AUC 0.754). The same phenomenon occurred with the statistical learning algorithm but to a somewhat lesser extent (naïve AUC 0.771; cross-validated AUC 0.659). Therefore, studies developing predictive models should take care to avoid naïvely assessing predictive performance and ensure that effective cross-validation or bootstrapping methods are used for appropriate interval validation[4]. If available, external validation of predictive models in entirely new and different cohorts is the gold standard for model validation[4].
The dataset used in this study was retrospective and from a single center which subjects the results to information bias and limits their external validity. The outcome used was biochemical remission as this is a readily available as a repeated measure which allowed demonstration of more conventional and machine learning models, however it is acknowledged that clinical symptoms and/or mucosal healing are more clinically relevant end-points. Nevertheless, the goal of this study was to demonstrate the feasibility of deep learning methods in clinical prediction in this proof-of-concept study, rather than to develop a specific predictive model. Further, in practice, much larger cohorts will be required to properly train and calibrate deep learning models to maximize their utility in the real world. In future, all studies investigating specific predictive models should be subject to prospective controlled validation prior their application in clinical practice, specifically having shown that outcomes are improved after using predictive models to guide management.
In conclusion, we have demonstrated the feasibility of deep learning algorithms for clinical prediction in CD, which demonstrated an improved predictive performance compared to conventional methods. However, conventional statistical methods retain the advantage of simplicity and intuitiveness, allowing their use at the bedside. Yet with the rapid expansion of available health data, machine learning models have the potential to supersede currently conventional methods and greatly improve the development of tools for the clinical prediction of patient outcomes.
Machine learning and artificial intelligence have the potential to revolutionize precision care in inflammatory bowel diseases. The greatest area of interest has been the application of deep learning methods in automatic tumor detection during endoscopy, yet the application of such techniques in clinical outcome prediction has been lacking.
Traditional approaches to clinical prediction rely on conventional statistical algorithms such as regression, which are not suitable for more complex data such as repeated biomarker measurements.
To determine and compare the utility of deep learning with conventional algorithms in predicting response to anti-tumor necrosis factor (anti-TNF) therapy in Crohn's disease (CD).
A retrospective cohort of CD patients commenced on anti-TNF therapy was used to experimentally develop and cross-validate three supervised learning algorithms: (1) Statistical learning algorithm; (2) Feed-forward artificial neural network; and (3) Recurrent neural network with repeated data. Predictive utility was quantified using the area under the receiver operator characteristic curve (AUC).
Within our cohort of 146 patients, the conventional statistical learning algorithm had the weakest performance [AUC 0.659, 95% confidence interval (CI): 0.562-0.756], compared to the feed-forward artificial neural network (AUC 0.710, 95%CI: 0.622-0.799; P = 0.25 vs conventional) and the recurrent neural network using repeated biomarker measurements (AUC 0.754, 95%CI: 0.674-0.834; P = 0.036 vs conventional).
Deep learning methods are feasible and have the potential for stronger predictive performance compared to conventional model building methods when applied to predicting remission after anti-TNF therapy in CD.
This has been the first study to investigate the utility of deep neural networks in predicting clinical outcomes using repeated clinical data in inflammatory bowel disease. Future studies should incorporate additional data types such as genetic, imaging and endoscopic factors.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Australia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Jin B, Yu C S-Editor: Gao CC L-Editor: A P-Editor: Liu JH
| 1. | Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2693] [Cited by in RCA: 2747] [Article Influence: 119.4] [Reference Citation Analysis (2)] |
| 2. | Jackson BD, Con D, Gorelik A, Liew D, Knowles S, De Cruz P. Examination of the relationship between disease activity and patient-reported outcome measures in an inflammatory bowel disease cohort. Intern Med J. 2018;48:1234-1241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 3. | Denson LA, Curran M, McGovern DPB, Koltun WA, Duerr RH, Kim SC, Sartor RB, Sylvester FA, Abraham C, de Zoeten EF, Siegel CA, Burns RM, Dobes AM, Shtraizent N, Honig G, Heller CA, Hurtado-Lorenzo A, Cho JH. Challenges in IBD Research: Precision Medicine. Inflamm Bowel Dis. 2019;25:S31-S39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 4. | Moons KG, Altman DG, Reitsma JB, Ioannidis JP, Macaskill P, Steyerberg EW, Vickers AJ, Ransohoff DF, Collins GS. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162:W1-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2833] [Cited by in RCA: 3298] [Article Influence: 329.8] [Reference Citation Analysis (0)] |
| 5. | Chen H, Sung JJY. Potentials of AI in medical image analysis in Gastroenterology and Hepatology. J Gastroenterol Hepatol. 2021;36:31-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 6. | Le Berre C, Sandborn WJ, Aridhi S, Devignes MD, Fournier L, Smaïl-Tabbone M, Danese S, Peyrin-Biroulet L. Application of Artificial Intelligence to Gastroenterology and Hepatology. Gastroenterology. 2020;158:76-94.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 324] [Article Influence: 64.8] [Reference Citation Analysis (1)] |
| 7. | Kohli A, Holzwanger EA, Levy AN. Emerging use of artificial intelligence in inflammatory bowel disease. World J Gastroenterol. 2020;26:6923-6928. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 19] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 8. | Takenaka K, Ohtsuka K, Fujii T, Negi M, Suzuki K, Shimizu H, Oshima S, Akiyama S, Motobayashi M, Nagahori M, Saito E, Matsuoka K, Watanabe M. Development and Validation of a Deep Neural Network for Accurate Evaluation of Endoscopic Images From Patients With Ulcerative Colitis. Gastroenterology. 2020;158:2150-2157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 181] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 9. | Otani K, Nakada A, Kurose Y, Niikura R, Yamada A, Aoki T, Nakanishi H, Doyama H, Hasatani K, Sumiyoshi T, Kitsuregawa M, Harada T, Koike K. Automatic detection of different types of small-bowel lesions on capsule endoscopy images using a newly developed deep convolutional neural network. Endoscopy. 2020;52:786-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 10. | Klang E, Barash Y, Margalit RY, Soffer S, Shimon O, Albshesh A, Ben-Horin S, Amitai MM, Eliakim R, Kopylov U. Deep learning algorithms for automated detection of Crohn's disease ulcers by video capsule endoscopy. Gastrointest Endosc. 2020;91:606-613.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 152] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 11. | Sze SF, Cheung WI, Wong WC, Hui YT, Lam JTW. AmplifEYE assisted colonoscopy vs standard colonoscopy: A randomized controlled study. J Gastroenterol Hepatol. 2021;36:376-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 12. | Abbas M, Matta J, Le T, Bensmail H, Obafemi-Ajayi T, Honavar V, El-Manzalawy Y. Biomarker discovery in inflammatory bowel diseases using network-based feature selection. PLoS One. 2019;14:e0225382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 13. | Bodein A, Chapleur O, Droit A, Lê Cao KA. A Generic Multivariate Framework for the Integration of Microbiome Longitudinal Studies With Other Data Types. Front Genet. 2019;10:963. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 14. | Matsuoka K, Hamada S, Shimizu M, Nanki K, Mizuno S, Kiyohara H, Arai M, Sugimoto S, Iwao Y, Ogata H, Hisamatsu T, Naganuma M, Kanai T, Mochizuki M, Hashiguchi M. Factors predicting the therapeutic response to infliximab during maintenance therapy in Japanese patients with Crohn's disease. PLoS One. 2018;13:e0204632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 15. | Ding NS, Malietzis G, Lung PFC, Penez L, Yip WM, Gabe S, Jenkins JT, Hart A. The body composition profile is associated with response to anti-TNF therapy in Crohn's disease and may offer an alternative dosing paradigm. Aliment Pharmacol Ther. 2017;46:883-891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 16. | Barber GE, Yajnik V, Khalili H, Giallourakis C, Garber J, Xavier R, Ananthakrishnan AN. Genetic Markers Predict Primary Non-Response and Durable Response To Anti-TNF Biologic Therapies in Crohn's Disease. Am J Gastroenterol. 2016;111:1816-1822. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 81] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 17. | Ward MG, Warner B, Unsworth N, Chuah SW, Brownclarke C, Shieh S, Parkes M, Sanderson JD, Arkir Z, Reynolds J, Gibson PR, Irving PM. Infliximab and adalimumab drug levels in Crohn's disease: contrasting associations with disease activity and influencing factors. Aliment Pharmacol Ther. 2017;46:150-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 18. | Mortensen JH, van Haaften WT, Karsdal MA, Bay-Jensen AC, Olinga P, Grønbæk H, Hvas CL, Manon-Jensen T, Dijkstra G, Dige A. The Citrullinated and MMP-degraded Vimentin Biomarker (VICM) Predicts Early Response to Anti-TNFα Treatment in Crohn's Disease. J Clin Gastroenterol. 2021;55:59-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 19. | Con D, Parthasarathy N, Bishara M, Luber RP, Joshi N, Wan A, Rickard JA, Long T, Connoley DJ, Sparrow MP, Gibson PR, van Langenberg DR, Vasudevan A. Development of a Simple, Serum Biomarker-based Model Predictive of the Need for Early Biologic Therapy in Crohn's Disease. J Crohns Colitis. 2021;15:583-593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 20. | Cornish JS, Wirthgen E, Däbritz J. Biomarkers Predictive of Response to Thiopurine Therapy in Inflammatory Bowel Disease. Front Med (Lausanne). 2020;7:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 21. | Waljee AK, Wallace BI, Cohen-Mekelburg S, Liu Y, Liu B, Sauder K, Stidham RW, Zhu J, Higgins PDR. Development and Validation of Machine Learning Models in Prediction of Remission in Patients With Moderate to Severe Crohn Disease. JAMA Netw Open. 2019;2:e193721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 22. | Waljee AK, Lipson R, Wiitala WL, Zhang Y, Liu B, Zhu J, Wallace B, Govani SM, Stidham RW, Hayward R, Higgins PDR. Predicting Hospitalization and Outpatient Corticosteroid Use in Inflammatory Bowel Disease Patients Using Machine Learning. Inflamm Bowel Dis. 2017;24:45-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 23. | Noh SM, Oh EH, Park SH, Lee JB, Kim JY, Park JC, Kim J, Ham NS, Hwang SW, Yang DH, Byeon JS, Myung SJ, Yang SK, Ye BD. Association of Faecal Calprotectin Level and Combined Endoscopic and Radiological Healing in Patients With Crohn's Disease Receiving Anti-tumour Necrosis Factor Therapy. J Crohns Colitis. 2020;14:1231-1240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 24. | Maaser C, Sturm A, Vavricka SR, Kucharzik T, Fiorino G, Annese V, Calabrese E, Baumgart DC, Bettenworth D, Borralho Nunes P, Burisch J, Castiglione F, Eliakim R, Ellul P, González-Lama Y, Gordon H, Halligan S, Katsanos K, Kopylov U, Kotze PG, Krustinš E, Laghi A, Limdi JK, Rieder F, Rimola J, Taylor SA, Tolan D, van Rheenen P, Verstockt B, Stoker J; European Crohn’s and Colitis Organisation [ECCO] and the European Society of Gastrointestinal and Abdominal Radiology [ESGAR]. ECCO-ESGAR Guideline for Diagnostic Assessment in IBD Part 1: Initial diagnosis, monitoring of known IBD, detection of complications. J Crohns Colitis. 2019;13:144-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1242] [Cited by in RCA: 1161] [Article Influence: 193.5] [Reference Citation Analysis (0)] |
| 25. | Porter AC, Aubrecht J, Birch C, Braun J, Cuff C, Dasgupta S, Gale JD, Hinton R, Hoffmann SC, Honig G, Linggi B, Schito M, Casteele NV, Sauer JM. Biomarkers of Crohn's Disease to Support the Development of New Therapeutic Interventions. Inflamm Bowel Dis. 2020;26:1498-1508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 26. | Ma C, Battat R, Parker CE, Khanna R, Jairath V, Feagan BG. Update on C-reactive protein and fecal calprotectin: are they accurate measures of disease activity in Crohn's disease? Expert Rev Gastroenterol Hepatol. 2019;13:319-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 27. | Lin X, Qiu Y, Feng R, Chen B, He Y, Zeng Z, Zhang S, Chen M, Mao R. Normalization of C-Reactive Protein Predicts Better Outcome in Patients With Crohn's Disease With Mucosal Healing and Deep Remission. Clin Transl Gastroenterol. 2020;11:e00135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | Click B, Vargas EJ, Anderson AM, Proksell S, Koutroubakis IE, Ramos Rivers C, Hashash JG, Regueiro M, Watson A, Dunn MA, Schwartz M, Swoger J, Baidoo L, Barrie A 3rd, Binion DG. Silent Crohn's Disease: Asymptomatic Patients with Elevated C-reactive Protein Are at Risk for Subsequent Hospitalization. Inflamm Bowel Dis. 2015;21:2254-2261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 29. | Bouckaert RR, Frank E. Evaluating the Replicability of Significance Tests for Comparing Learning Algorithms, in Advances in Knowledge Discovery and Data Mining. In: Dai H, Srikant R, Zhang C. Lecture Notes in Computer Science. Springer: Berlin, Heidelberg, 2004. |
| 30. | Hajian-Tilaki K. Sample size estimation in diagnostic test studies of biomedical informatics. J Biomed Inform. 2014;48:193-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 647] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 31. | Torres J, Mehandru S, Colombel JF, Peyrin-Biroulet L. Crohn's disease. Lancet. 2017;389:1741-1755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1121] [Cited by in RCA: 1797] [Article Influence: 224.6] [Reference Citation Analysis (111)] |
| 32. | Con D, Jackson B, Gray K, De Cruz P. eHealth for inflammatory bowel disease self-management - the patient perspective. Scand J Gastroenterol. 2017;52:973-980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 33. | van der Valk ME, Mangen MJ, Severs M, van der Have M, Dijkstra G, van Bodegraven AA, Fidder HH, de Jong DJ, van der Woude CJ, Romberg-Camps MJ, Clemens CH, Jansen JM, van de Meeberg PC, Mahmmod N, van der Meulen-de Jong AE, Ponsioen CY, Bolwerk C, Vermeijden JR, Siersema PD, Leenders M, Oldenburg B; COIN study group and the Dutch Initiative on Crohn and Colitis. Evolution of Costs of Inflammatory Bowel Disease over Two Years of Follow-Up. PLoS One. 2016;11:e0142481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 91] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 34. | Jackson B, Con D, Ma R, Gorelik A, Liew D, De Cruz P. Health care costs associated with Australian tertiary inflammatory bowel disease care. Scand J Gastroenterol. 2017;52:851-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 35. | Best WR, Becktel JM, Singleton JW, Kern F Jr. Development of a Crohn's disease activity index. National Cooperative Crohn's Disease Study. Gastroenterology. 1976;70:439-444. [PubMed] |
| 36. | Daperno M, D'Haens G, Van Assche G, Baert F, Bulois P, Maunoury V, Sostegni R, Rocca R, Pera A, Gevers A, Mary JY, Colombel JF, Rutgeerts P. Development and validation of a new, simplified endoscopic activity score for Crohn's disease: the SES-CD. Gastrointest Endosc. 2004;60:505-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 999] [Cited by in RCA: 1318] [Article Influence: 62.8] [Reference Citation Analysis (0)] |
| 37. | Limsrivilai J, Pausawasdi N. Intestinal tuberculosis or Crohn's disease: a review of the diagnostic models designed to differentiate between these two gastrointestinal diseases. Intest Res. 2021;19:21-32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 38. | Sung JJ, Stewart CL, Freedman B. Artificial intelligence in health care: preparing for the fifth Industrial Revolution. Med J Aust. 2020;213:253-255.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 39. | Iwagami H, Ishihara R, Aoyama K, Fukuda H, Shimamoto Y, Kono M, Nakahira H, Matsuura N, Shichijo S, Kanesaka T, Kanzaki H, Ishii T, Nakatani Y, Tada T. Artificial intelligence for the detection of esophageal and esophagogastric junctional adenocarcinoma. J Gastroenterol Hepatol. 2021;36:131-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 40. | East JE, Rittscher J. Artificial intelligence for colonoscopic polyp detection: High performance vs human nature. J Gastroenterol Hepatol. 2020;35:1663-1664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 41. | Parasher G, Wong M, Rawat M. Evolving role of artificial intelligence in gastrointestinal endoscopy. World J Gastroenterol. 2020;26:7287-7298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 25] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 42. | Shung D, Tsay C, Laine L, Chang D, Li F, Thomas P, Partridge C, Simonov M, Hsiao A, Tay JK, Taylor A. Early identification of patients with acute gastrointestinal bleeding using natural language processing and decision rules. J Gastroenterol Hepatol. 2021;36:1590-1597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 43. | Guan Y, Zhang H, Quang D, Wang Z, Parker SCJ, Pappas DA, Kremer JM, Zhu F. Machine Learning to Predict Anti-Tumor Necrosis Factor Drug Responses of Rheumatoid Arthritis Patients by Integrating Clinical and Genetic Markers. Arthritis Rheumatol. 2019;71:1987-1996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 90] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 44. | Lee S, Eun Y, Kim H, Cha HS, Koh EM, Lee J. Machine learning to predict early TNF inhibitor users in patients with ankylosing spondylitis. Sci Rep. 2020;10:20299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |