Published online Sep 21, 2021. doi: 10.3748/wjg.v27.i35.5908
Peer-review started: February 27, 2021
First decision: April 18, 2021
Revised: April 29, 2021
Accepted: August 24, 2021
Article in press: August 24, 2021
Published online: September 21, 2021
Processing time: 199 Days and 13.1 Hours
Colonoscopy remains the gold standard investigation for colorectal cancer screening as it offers the opportunity to both detect and resect pre-malignant and neoplastic polyps. Although technologies for image-enhanced endoscopy are widely available, optical diagnosis has not been incorporated into routine clinical practice, mainly due to significant inter-operator variability. In recent years, there has been a growing number of studies demonstrating the potential of convolutional neural networks (CNN) to enhance optical diagnosis of polyps. Data suggest that the use of CNNs might mitigate the inter-operator variability amongst endoscopists, potentially enabling a “resect and discard“ or ”leave in“ strategy to be adopted in real-time. This would have significant financial benefits for healthcare systems, avoid unnecessary polypectomies of non-neoplastic polyps and improve the efficiency of colonoscopy. Here, we review advances in CNN for the optical diagnosis of colorectal polyps, current limitations and future directions.
Core Tip: A convolutional neural network (CNN) is a specific type of artificial intelligence deep learning. These networks may play an important role in the coming years in assisting endoscopists to optically diagnose colorectal polyps. CNNs can mitigate the inter-operator variability amongst endoscopists, potentially enabling a “resect and discard” or “leave in” strategy to be adopted. This would improve the efficiency of colonoscopy, reduce healthcare costs and reduce adverse events for patients by avoiding unnecessary resections of non-neoplastic polyps. In this article, we expand on the most relevant studies in this field and discuss limitations and future directions that will determine fulfilment of the potential of CNN in the optical diagnosis of colorectal polyps.
- Citation: Kader R, Hadjinicolaou AV, Georgiades F, Stoyanov D, Lovat LB. Optical diagnosis of colorectal polyps using convolutional neural networks. World J Gastroenterol 2021; 27(35): 5908-5918
- URL: https://www.wjgnet.com/1007-9327/full/v27/i35/5908.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i35.5908
Colorectal cancer (CRC) is the third most commonly diagnosed cancer worldwide[1] and thus, a significant burden on global healthcare systems. Most CRCs develop in a relatively predictable, stepwise sequence from mutation-accumulating neoplastic polyps, such as adenomas and sessile serrated lesions (SSL)[2]. Current evidence-based societal guidelines unequivocally accept colonoscopy to be the gold standard tool for screening of CRC[3]. Colonoscopy offers the opportunity to both detect and resect neoplastic polyps[4] and its implementation, especially as part of bowel cancer screening programs, has been linked to a significant reduction in the incidence of the CRC and CRC-related mortality[5].
Over 90% of polyps detected at colonoscopy are either small (6-9 mm) or diminutive (≤ 5 mm), entities that are thought to harbour a very low risk for developing into CRC[6]. Furthermore, almost half of these polyps are non-neoplastic in nature; and frequently hyperplastic[7]. Accurate differentiation of neoplastic from non-neoplastic polyps can prevent the unnecessary resection of the latter, avoiding an intervention which is not cost-effective and which carries risks of significant morbidity[8].
Recent years have seen significant research activity in the use of artificial intelligence (AI), particularly convolutional neural networks (CNN), to optically diagnose colorectal polyps. The field is gaining increasing momentum. The aim of this review article is to summarise and critically appraise the available medical literature related to advances in CNN for optical diagnosis of colorectal polyps and highlight the field’s current limitations and future directions.
The term “optical diagnosis” refers to the use of advanced imaging techniques for real-time, in-vivo polyp characterisation and evaluation to guide therapeutic decisions[9]. Accurate optical diagnosis of diminutive polyps would enable identification of hyperplastic polyps in the rectosigmoid region, where they are commonly found, and allow the endoscopist to confidently take a “diagnose and leave” approach instead of resecting the lesion. Equally, for diminutive adenomas, accurate optical diagnosis would prompt the endoscopist to remove the lesion on the spot and discard the specimen without the need for histological assessment (“resect and discard”strategy)[9].
The American Society of Gastrointestinal Endoscopy established the Preservation and Incorporation of Valuable endoscopic Innovations (PIVI) to provide thresholds that are required of endoscopic technology in order to implement a “resect and discard”(PIVI 1) and “diagnose and leave” (PIVI 2) strategy[9]. PIVI 1 requires ≥ 90% concordance in post-polypectomy surveillance intervals when comparing the combination of optical diagnosis for diminutive adenomas with histopathology assessment of all other polyps against decisions based solely on histopathology evaluation of all identified polyps[10]. PIVI 2 requires a technology to achieve a negative predictive value (NPV) of ≥ 90% for diminutive adenomatous polyps in the rectosigmoid region[9].
There has been extensive research in image enhanced endoscopy (IEE), such as narrow band imaging (NBI), to assist endoscopists in optical diagnosis to characterise diminutive polyps[11-13]. Using IEE, expert endoscopists in academic centres have consistently demonstrated an optical diagnosis accuracy that exceeds PIVI thresholds[14-16], however, studies have often found community and non-expert endoscopists to fall short of these minimal thresholds[17]. An example is the multi-centre DISCARD-2 study which evaluated the optical diagnosis accuracy of 28 community endoscopists using NBI. Disappointingly, the endoscopists’ optical diagnosis derived colonoscopy surveillance intervals only matched 68% of the histopathology derived intervals[18]. Although widely available, technologies for optical diagnosis has not been incorporated into routine clinical practice with one of the main barriers being the inter-operator variability amongst endoscopists[19].
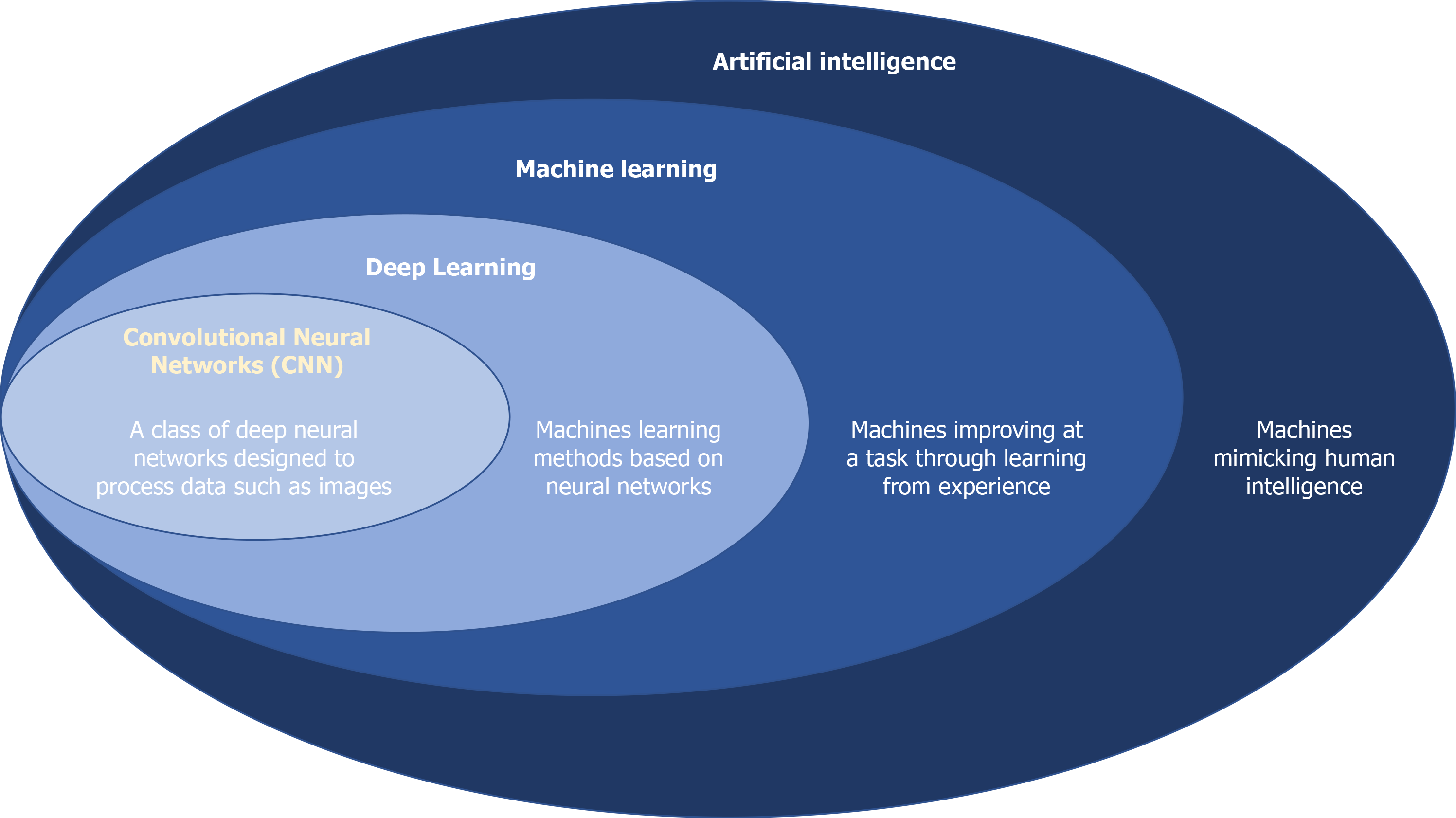
AI is the ability of computers to perform tasks that traditionally require human intelligence (Figure 1)[20]. Machine learning (ML) is a subset of AI, whereby computers continuously learn from data without explicit human programming[21]. This can be used to predicate a polyp’s histology. ML models can be trained using unsupervised or supervised techniques. Unsupervised learning is when the input and output data are not paired. Supervised ML is more labour intensive as it requires paired input and output data for training. An example of a supervised ML model for optical diagnosis is to annotate a bounding box around a polyp (input data), commonly referred to as a region of interest, and label it with the histology of the polyp (output data). The model automatically learns to extract features that allow it to differentiate polyp subtypes and output a diagnosis based on the histology classification system it was trained with but the annotation process is time consuming for the clinician.
Deep learning is a subset of ML, whereby algorithms use multiple layers within a neural network[22], mimicking the human brain, to extract high level features from input data. CNNs are the most commonly used network in the application of deep learning to optically diagnose polyps. They provide an objective output, bypassing the human inter and intra-operator variability, and can develop classification algorithms without exhaustive effort as they do not require human-crafted feature extraction or extensive pre-processing of data[23].
Building a CNN model typically involves three separate datasets; a training set, a validation set and a test set[24]. The training set is used to develop the model so that it predicts a label (e.g., adenomatous or hyperplastic polyp for polyp characterisation) based on features extracted from the endoscopic image by the algorithm itself. The validation set is used to avoid over-fitting into the training dataset through fine tuning of the hyperparameters of the model. Finally, the testing set is used as an independent dataset to evaluate the generalisability of the CNN. With smaller datasets, cross-validation can be used to assess the model’s robustness. In cross-validation, the data is split into equal parts (e.g., 4 parts), with one part held out as a validation dataset. This process is repeated multiple times, with the results of each split eventually pooled together to decide how robust the model is[24]. CNNs evaluated using cross-validation should still be assessed against an independent test set to examine their generalisability[24].
It is only in the last few years that the use of CNNs in optical diagnosis of colorectal polyps has been extensively investigated, with various studies emerging (Table 1). Many of these studies have in fact demonstrated the capability of CNNs to surpass the PIVI 2 threshold in order to support a “leave in” strategy for rectosigmoid hyperplastic polyps (Table 2). This was first demonstrated by Chen et al[25], who used a single centre, retrospective, still image dataset of 2157 polyps to train a CNN and reported a sensitivity for identifying adenomas of 96.3% , specificity 78.1%, and NPV of 91.5% when evaluating a test set of 284 colonic and rectal diminutive adenomatous and hyperplastic polyps. Using colonic diminutive polyps is a common strategy to assess against PIVI 2 due to difficulties in obtaining large datasets of diminutive rectosigmoid polyps. An important limitation of this study is that it used magnified narrow-band imaging (NBI) data. This recently developed modality is not yet readily available in most endoscopy departments, although it will become more widely used with time.
| Ref. | Study design (training/testing) | Multi-centre study | Dataset | Image quality | Classification system | Lesion number (training/testing) | SSL excluded | Endoscopic processor | Image modality (training) | Real-time capability |
| Komeda et al[37] | Retrospective | Single | Video | Not specified | Adenoma/non-adenoma | Not specified/10 | No | Not specified | WLI, NBI, chromoendoscopy | Not specified |
| Chen et al[25] | Retrospective/prospective | Single | Still | HQ | Hyperplastic/neoplastic | 2157/284 | Yes | Olympus 260 + 290 | Magnified NBI | Real-time (approximately 450 ms) |
| Byrne et al[23] | Retrospective/prospective | Single | Video | All images | NICE Type 1/NICE Type 2 | 220/125 | Yes | Olympus 190 | NBI-NF | Real-time ( approximately 50 ms) |
| Zachariah et al[26] | Prospective | Two | Still | Adequate and HQ | Adenomatous/serrated polyp | 5278/634 | No | Olympus 190 (90%), 180 (7%), Pentax i10(3%) | WLI, NBI, i-SCAN | Real-time ( approximately 13 ms) |
| Ozawa et al[38] | Retrospective/prospective | Single | Still | HQ | Hyperplastic/adenomatous/SSL/CRC/other | WLI: 17566/783 | No | Olympus 260 + 290 | WLI, NBI | Real-time (approximately 20 ms) |
| Jin et al[31] | Retrospective/prospective | Single | Still | HQ | Hyperplastic/adenomatous | 2150/300 | Yes | Olympus 290 | NBI-NF | Real-time (approximately 10 ms) |
| Song et al[39] | Retrospective/prospective | Single | Still | HQ | Serrated polyp/benign adenoma/MSM/DSMC | 624/545 | No | Olympus 290 | NBI-NF | Real-time ( approximately 20-40 ms) |
| Rodriguez-Diaz et al[28] | Retrospective/prospective | Two | Still | Not specified | Neoplastic (adenomas, CRC)/non-neoplastic (hyperplastic, normal) | 607/280 | Training: Yes | Olympus 190 | NBI-NF, NBI (digital magnification) | Real-time (approximately 100 ms) |
| van der Zander et al[27] | Retrospective/prospective | Not specified | Still | HQ | Benign (hyperplastic)/pre-malignant (adenomatous, SSL, T1 CRC) | 398/60 | No | Fujifilm, Pentax | WLI, BLI, i-SCAN | Real-time (approximately 14.8 ms) |
| Ref. | Image Modality (testing) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy for neoplasia (%) | PIVI 1 achieved (%) | PIVI 2 achieved (%) |
| Komeda et al[37] | Not specified | - | - | - | - | 70 | - | - |
| Chen et al[25] | Magnified NBI | 96.3 | 78.1 | 89.6 | 91.5 | 90.1 | - | Yes (91.5) |
| Byrne et al[23] | NBI-NF | 98 | 83 | 90 | 97 | 94 | - | Yes (97) |
| Zachariah et al[26] | NBI | - | - | - | 96.5 | 93.1 | Yes (98.3) | Yes (96.5) |
| WLI | - | - | - | 88.9 | 92.8 | Yes (90.8) | No (88.9) | |
| Ozawa et al[38]1 | NBI | 97 | - | 84 | 88 | - | - | - |
| WLI | 98 | - | 85 | 88 | - | - | - | |
| Jin et al | NBI-NF | 83.3 | 91.7 | 93.3 | 78.6 | 86.7 | - | - |
| Song et al[39] | NBI-NF (test set 1) | 84.1 | 74 | 88.3 | 67.7 | - | - | - |
| NBI-NF (test set 2) | 88.5 | 72.1 | 88.6 | 84.7 | - | - | - | |
| Rodriguez-Diaz et al[28] | NBI-NF (90%) + NBI (10%) | 95 | 88 | - | 93 | - | Yes (94 (20/90 LC)) | Yes (98 (6/68 LC)) |
| van der Zander et al[27] | WLI + BLI | 95.6 | 93.3 | 97.7 | 87.5 | 95.0 | - | No (87.5) |
Byrne et al[23] further advanced the field by training a CNN with NBI-near focus (NBI-NF) which is more commonly used in Europe and North America. It was trained with 220 polyp positive videos and when tested against 125 diminutive polyps which were collected prospectively, the model diagnosed 106 polyps with high confidence, achieving a sensitivity for identifying NBI International Colorectal Endoscopic (NICE) type 1 polyps of 98%, specificity 83% and NPV of 97%. A novelty worth highlighting in this study was the use of images derived from videos, an approach that reduces selection bias compared to retrospective still images as endoscopists usually capture high quality polyp views that are free from motion blur and surface artifact. An additional advantage of this CNN is that it simplified the clinical workflow as it automatically diagnoses polyps without requiring a still image of the polyp to be captured. Limitations of the study are that SSLs, normal tissue and lymphoid aggregates were excluded from the final analysis and the videos used to train and test the CNN were captured from colonoscopies performed by a single expert endoscopist and hence, potentially less generalisable to novice users.
The most commonly used imaging modalities amongst community endoscopists are white light imaging (WLI) and NBI without magnification. Using a large retrospective still image training set of 5278 polyps and tested against 634 polyps, Zachariah et al[26]’s CNN fell short of PIVI 2 in WLI (NPV of 88.9% and accuracy 92.8%) but achieved the threshold in NBI without magnification (NPV of 90.8% and accuracy 93.1%). This study advanced the field as it demonstrated the capabilities of CNNs to optically diagnose polyps in standard NBI modality and also to differentiate adenomas from serrated polyps through the inclusion of SSLs in its dataset.
Whilst the majority of CNNs have been trained and tested using Olympus data, studies are emerging using data from other manufacturers. van der Zander et al[27] recently developed a CNN using Fujifilm data in high definition white light (HDWL) and blue light imaging (BLI). The CNN was more efficacious when it used a unique multimodal imaging approach where it combined both HDWL and BLI images of the same polyp in its decision process compared to a single imaging modality. When evaluated against 60 prospectively collected diminutive polyps, it did not reach the PIVI 2 threshold with a NPV of 87.5% but did achieve an optical diagnosis accuracy of 95% (sensitivity for identifying pre-malignant polyps 95.6% and specificity 93.3%) and demonstrated superiority to both expert and novice endoscopists in human benchmark testing.
In comparison to PIVI 2, there are fewer studies evaluating the performance of CNNs against PIVI 1. The CNN presented in Zachariah et al[26] reached PIVI 1 thresholds in both WLI and NBI with normal magnification, achieving concordance with histology-based colonoscopy surveillance intervals in 90.9% and 98.3% of patients, for each respective modality. Rodrigues-Diaz et al[28] used a single centre retrospective still image dataset to train a CNN with 607 polyps and tested against 90 diminutive polyps where it achieved a high confidence diagnosis in 78% of cases, with a 94% agreement with histology-based colonoscopy surveillance intervals. Tested against 68 rectosigmoid polyps, the model diagnosed 88% of polyps with high confidence, achieving PIVI 2 thresholds with a NPV of 97%.
There is also potential to expand the use of optical diagnosis CNNs outside of the ”resect and discard” and “leave in strategy”. A dilemma that can complicate issuing post-polypectomy surveillance intervals is discrepancies between endoscopic and histological diagnosis and classification of polyps with tissue fragmentation in the specimen retrieval process playing an important role. Shahidi et al[29]’s proof of concept study used a CNN to resolve discrepancies in polyps ≤ 3 mm in size. Tested against 900 polyps that were ≤ 3 mm and optically diagnosed as adenomatous by an expert endoscopist, the CNN diagnosed the adenomas with high confidence in 644 polyps, with 256 polyps deemed to be of sub-optimal imaging quality. However, of these high confidence diagnoses, the pathologists diagnosed 15.4% as normal mucosa, 13.2% as hyperplastic polyp and 0.3% as SSL. In this context, a CNN could help to mitigate against the risk of under-surveillance.
Whilst CNN’s diagnostic accuracy excels in many studies, without real-time capabilities, they would have no clinical utility. Prior to the era of deep learning, computer aided diagnosis algorithms lacked real-time capability, but most CNNs do not share this problem and often process data at a rate that exceeds the 25 frames per second that is generated in a video recording of a colonoscopy procedure. Given the excellent performance in ex-vivo studies and the real-time capabilities displayed by CNNs, the future appears promising for their integration in colonoscopy.
The complexity of CNN models’ decision process is often referred to as a “black box” and represents an important barrier to its acceptance by both clinicians and patients[30]. Opening the ‘’black box’’ to display the raw features which informed the CNN’s decision is important for transparency especially from a safety standpoint[28]. Transparency can help identify biases within the neural network and aid root-cause analyses in cases of patient harm, for example, if a neoplastic polyp that subsequently develops into a CRC is originally misdiagnosed as non-neoplastic by the CNN model.
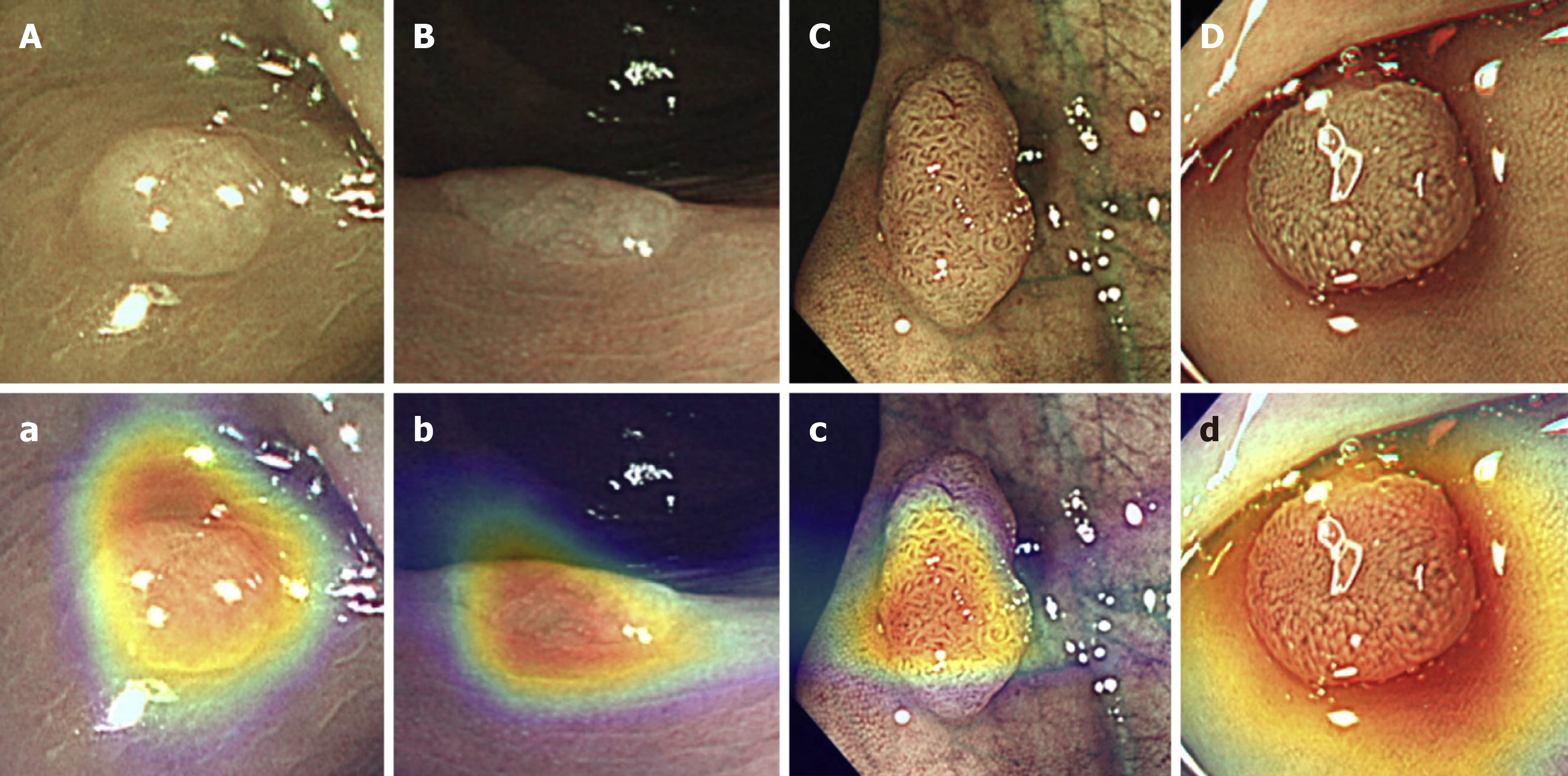
For polyp characterisation, important steps have been taken to open the black box. Jin et al[31] developed a CNN that generated a coloured heat map, overlaid to the polyp, to help the endoscopist comprehend the specific aspects of the image that contributed to the CNN’s prediction (Figure 2). This could help the endoscopist to decide which information is relevant and which decisions are truly based on appropriate image analysis. If, for example, the heatmap is overlaid to normal mucosa, then the endoscopist would quickly be able to appreciate this and disregard the CNN’s diagnosis.
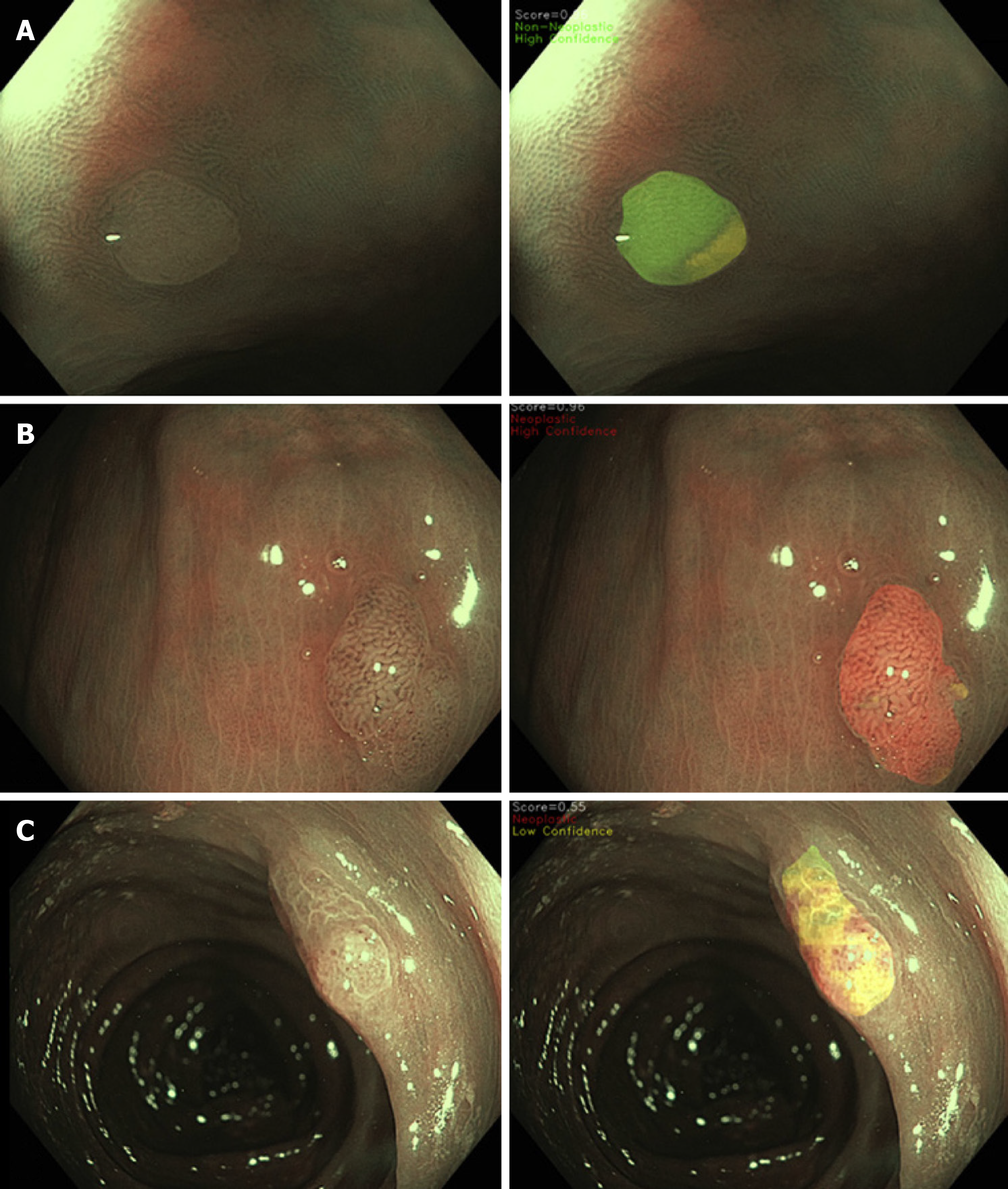
More recently, in order to further enhance CNN transparency, Rodriguez-Diaz et al[28] developed a colour coded segmentation model (Figure 3). In this model, the CNN divides the polyp into distinct segments to allow the endoscopist to identify the specific regions within the image that is informing the CNN’s decision. The CNN predicts the histology of each subregion of the segmented polyp, with high confidence neoplastic diagnoses coloured in red, high confidence non-neoplastic in green, and low confidence/indeterminate diagnoses in yellow, with the final predication resulting from an aggregate of all the analysed regions. The end result is a detailed spatial colour coded histology map of the polyp surface, which the endoscopist can visualise and incorporate into their decision process[28], enhancing the interpretability of this CNN model in comparison to others. However, an important limitation to this advanced CNN is that it currently lacks the ability to operate at a video rate.
Further research in the interpretability of CNN models is required to improve its acceptance[32] and accelerate its translation to clinical practise.
Despite the promise shown by CNNs this far, it is crucial to recognise that there are various limitations that need to be overcome before they can become part of the endoscopic clinical workflow. The most significant limitations are the reliance on retrospective datasets[33], which are inherently subject to selection bias, and the lack of prospective studies and randomised controlled trials[34]. Most studies train and test CNNs using high quality images of polyps, free from “noise” such as motion blur and polyp surface artifact (e.g., mucus, stool or blood). The extent to which CNNs pre-clinical results are reproducible in the real-world setting, where ‘noise’ is frequently encountered, remains to be seen.
To the best of our knowledge, there have been no prospective randomised controlled clinical trials evaluating optical diagnosis CNN in-vivo. This is partly due to clinical trials being time consuming and expensive, and an alternative pragmatic approach could be the use of a benchmark test in the form a publicly available external dataset to compare different CNN models[35]. No such datasets currently exist for polyp characterisation and therefore the generalisability of CNN models remains poorly understood. Generalisability refers to the CNN performance with different endoscope models and clinical settings from the site that the data was generated to train the CNN. To date, only one study[36] has evaluated generalisability, and this was limited to a small testing set of 69 polyp images from two population cohorts (Australian and Japanese) using two separate endoscope manufactures (Olympus and Fujifilm). Despite the small test-set, this study highlighted the concerns of generalisability as the operator area under the curve fell from 94.3% for the internal set, to 84.5% and 90.3% for the external testing sets (NBI and BLI respectively).
Another important limitation is that studies often exclude polyps that are not adenomas or hyperplastic polyps, restricting the possible classification outputs of CNNs. This, in turn, limits their clinical utility as polyps such as SSL and inflammatory polyps would be misclassified due to limitations in the initial training phase of the CNNs when the categorisation system is established.
Research in this field is likely to continue to expand and future directions to consider include: (1) Guidelines to identify the role of CNNs in the clinical workflow, specifically, whether it is a second reader, a concurrent reader or a provider of an independent diagnosis[30]; (2) Prospective multi-centre randomised clinical trials; (3) Publicly available external datasets for benchmark testing and evaluation of the generalisability of CNN models in different clinical settings and population cohorts; and (4) Acquiring datasets inclusive of all polyp sub-types to advance CNN classification systems.
In summary, this is an exciting time for the endoscopy community. CNNs diagnostic performance has excelled in ex-vivo studies and in human benchmarking testing. CNNs are likely to be a key adjunct in optically diagnosing polyps and have renewed optimism that implementation of a “resect and discard” and “leave in” strategy is feasible due to the potential to alleviate the inter-operator variability amongst endoscopists. This would bring significant financial benefits to healthcare systems, avoid unnecessary polypectomies of non-neoplastic polyps and improve the efficiency of colonoscopy. However, prospective multi-centre randomised controlled trials and publicly available datasets for benchmark testing are required to further evaluate the efficacy and generalisability of CNNs. Furthermore, with these models now emerging in endoscopy units, it’s imperative that guidelines are developed to establish their role in the clinical workflow.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United Kingdom
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cavdar SC S-Editor: Ma YJ L-Editor: A P-Editor: Li JH
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55843] [Article Influence: 7977.6] [Reference Citation Analysis (132)] |
| 2. | Song M, Emilsson L, Bozorg SR, Nguyen LH, Joshi AD, Staller K, Nayor J, Chan AT, Ludvigsson JF. Risk of colorectal cancer incidence and mortality after polypectomy: a Swedish record-linkage study. Lancet Gastroenterol Hepatol. 2020;5:537-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 77] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 3. | US Preventive Services Task Force. Bibbins-Domingo K, Grossman DC, Curry SJ, Davidson KW, Epling JW Jr, García FAR, Gillman MW, Harper DM, Kemper AR, Krist AH, Kurth AE, Landefeld CS, Mangione CM, Owens DK, Phillips WR, Phipps MG, Pignone MP, Siu AL. Screening for Colorectal Cancer: US Preventive Services Task Force Recommendation Statement. JAMA. 2016;315:2564-2575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1249] [Cited by in RCA: 1387] [Article Influence: 154.1] [Reference Citation Analysis (1)] |
| 4. | Rutter MD, East J, Rees CJ, Cripps N, Docherty J, Dolwani S, Kaye PV, Monahan KJ, Novelli MR, Plumb A, Saunders BP, Thomas-Gibson S, Tolan DJM, Whyte S, Bonnington S, Scope A, Wong R, Hibbert B, Marsh J, Moores B, Cross A, Sharp L. British Society of Gastroenterology/Association of Coloproctology of Great Britain and Ireland/Public Health England post-polypectomy and post-colorectal cancer resection surveillance guidelines. Gut. 2020;69:201-223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 260] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 5. | Zauber AG, Winawer SJ, O'Brien MJ, Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, Shi W, Bond JH, Schapiro M, Panish JF, Stewart ET, Waye JD. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. 2012;366:687-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1952] [Cited by in RCA: 2287] [Article Influence: 175.9] [Reference Citation Analysis (2)] |
| 6. | Lieberman D, Moravec M, Holub J, Michaels L, Eisen G. Polyp size and advanced histology in patients undergoing colonoscopy screening: implications for CT colonography. Gastroenterology. 2008;135:1100-1105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 355] [Cited by in RCA: 329] [Article Influence: 19.4] [Reference Citation Analysis (1)] |
| 7. | Rex DK, Overhiser AJ, Chen SC, Cummings OW, Ulbright TM. Estimation of impact of American College of Radiology recommendations on CT colonography reporting for resection of high-risk adenoma findings. Am J Gastroenterol. 2009;104:149-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 100] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 8. | El Hajjar A, Rey JF. Artificial intelligence in gastrointestinal endoscopy: general overview. Chin Med J (Engl). 2020;133:326-334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 9. | ASGE Technology Committee. Abu Dayyeh BK, Thosani N, Konda V, Wallace MB, Rex DK, Chauhan SS, Hwang JH, Komanduri S, Manfredi M, Maple JT, Murad FM, Siddiqui UD, Banerjee S. ASGE Technology Committee systematic review and meta-analysis assessing the ASGE PIVI thresholds for adopting real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest Endosc. 2015;81:502.e1-502.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 249] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 10. | Rex DK. Can we do resect and discard with AI-assisted colon polyp ‘optical biopsy’? Tech Gastrointest Endosc. 2019;150638. [RCA] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 11. | Ignjatovic A, East JE, Suzuki N, Vance M, Guenther T, Saunders BP. Optical diagnosis of small colorectal polyps at routine colonoscopy (Detect InSpect ChAracterise Resect and Discard; DISCARD trial): a prospective cohort study. Lancet Oncol. 2009;10:1171-1178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 287] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 12. | Ignjatovic A, Thomas-Gibson S, East JE, Haycock A, Bassett P, Bhandari P, Man R, Suzuki N, Saunders BP. Development and validation of a training module on the use of narrow-band imaging in differentiation of small adenomas from hyperplastic colorectal polyps. Gastrointest Endosc. 2011;73:128-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 13. | Rastogi A, Keighley J, Singh V, Callahan P, Bansal A, Wani S, Sharma P. High accuracy of narrow band imaging without magnification for the real-time characterization of polyp histology and its comparison with high-definition white light colonoscopy: a prospective study. Am J Gastroenterol. 2009;104:2422-2430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 139] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 14. | Repici A, Hassan C, Radaelli F, Occhipinti P, De Angelis C, Romeo F, Paggi S, Saettone S, Cisarò F, Spaander M, Sharma P, Kuipers EJ. Accuracy of narrow-band imaging in predicting colonoscopy surveillance intervals and histology of distal diminutive polyps: results from a multicenter, prospective trial. Gastrointest Endosc. 2013;78:106-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 15. | Wallace MB, Crook JE, Coe S, Ussui V, Staggs E, Almansa C, Patel MK, Bouras E, Cangemi J, Keaveny A, Picco M, Riegert-Johnson D. Accuracy of in vivo colorectal polyp discrimination by using dual-focus high-definition narrow-band imaging colonoscopy. Gastrointest Endosc. 2014;80:1072-1087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 16. | Singh R, Jayanna M, Navadgi S, Ruszkiewicz A, Saito Y, Uedo N. Narrow-band imaging with dual focus magnification in differentiating colorectal neoplasia. Dig Endosc. 2013;25 Suppl 2:16-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 17. | Rees CJ, Rajasekhar PT, Wilson A, Close H, Rutter MD, Saunders BP, East JE, Maier R, Moorghen M, Muhammad U, Hancock H, Jayaprakash A, MacDonald C, Ramadas A, Dhar A, Mason JM. Narrow band imaging optical diagnosis of small colorectal polyps in routine clinical practice: the Detect Inspect Characterise Resect and Discard 2 (DISCARD 2) study. Gut. 2017;66:887-895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 151] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 18. | Kuiper T, Marsman WA, Jansen JM, van Soest EJ, Haan YC, Bakker GJ, Fockens P, Dekker E. Accuracy for optical diagnosis of small colorectal polyps in nonacademic settings. Clin Gastroenterol Hepatol. 2012;10:1016-20; quiz e79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 19. | Ladabaum U, Fioritto A, Mitani A, Desai M, Kim JP, Rex DK, Imperiale T, Gunaratnam N. Real-time optical biopsy of colon polyps with narrow band imaging in community practice does not yet meet key thresholds for clinical decisions. Gastroenterology. 2013;144:81-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 186] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 20. | Sharma P, Pante A, Gross SA. Artificial intelligence in endoscopy. Gastrointest Endosc. 2020;91:925-931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 21. | Alagappan M, Brown JRG, Mori Y, Berzin TM. Artificial intelligence in gastrointestinal endoscopy: The future is almost here. World J Gastrointest Endosc. 2018;10:239-249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 106] [Cited by in RCA: 103] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 22. | Vakli P, Deák-Meszlényi RJ, Hermann P, Vidnyánszky Z. Transfer learning improves resting-state functional connectivity pattern analysis using convolutional neural networks. Gigascience. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 23. | Byrne MF, Chapados N, Soudan F, Oertel C, Linares Pérez M, Kelly R, Iqbal N, Chandelier F, Rex DK. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94-100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 412] [Article Influence: 68.7] [Reference Citation Analysis (0)] |
| 24. | van der Sommen F, de Groof J, Struyvenberg M, van der Putten J, Boers T, Fockens K, Schoon EJ, Curvers W, de With P, Mori Y, Byrne M, Bergman JJGHM. Machine learning in GI endoscopy: practical guidance in how to interpret a novel field. Gut. 2020;69:2035-2045. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 86] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 25. | Chen PJ, Lin MC, Lai MJ, Lin JC, Lu HH, Tseng VS. Accurate Classification of Diminutive Colorectal Polyps Using Computer-Aided Analysis. Gastroenterology. 2018;154:568-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 276] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 26. | Zachariah R, Samarasena J, Luba D, Duh E, Dao T, Requa J, Ninh A, Karnes W. Prediction of Polyp Pathology Using Convolutional Neural Networks Achieves "Resect and Discard" Thresholds. Am J Gastroenterol. 2020;115:138-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 84] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 27. | van der Zander QEW, Schreuder RM, Fonollà R, Scheeve T, van der Sommen F, Winkens B, Aepli P, Hayee B, Pischel AB, Stefanovic M, Subramaniam S, Bhandari P, de With PHN, Masclee AAM, Schoon EJ. Optical diagnosis of colorectal polyp images using a newly developed computer-aided diagnosis system (CADx) compared with intuitive optical diagnosis. Endoscopy. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 28. | Rodriguez-Diaz E, Baffy G, Lo WK, Mashimo H, Vidyarthi G, Mohapatra SS, Singh SK. Real-time artificial intelligence-based histologic classification of colorectal polyps with augmented visualization. Gastrointest Endosc. 2021;93:662-670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 42] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 29. | Shahidi N, Rex DK, Kaltenbach T, Rastogi A, Ghalehjegh SH, Byrne MF. Use of Endoscopic Impression, Artificial Intelligence, and Pathologist Interpretation to Resolve Discrepancies Between Endoscopy and Pathology Analyses of Diminutive Colorectal Polyps. Gastroenterology. 2020;158:783-785.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 30. | Ahmad OF, Mori Y, Misawa M, Kudo SE, Anderson JT, Bernal J, Berzin TM, Bisschops R, Byrne MF, Chen PJ, East JE, Eelbode T, Elson DS, Gurudu SR, Histace A, Karnes WE, Repici A, Singh R, Valdastri P, Wallace MB, Wang P, Stoyanov D, Lovat LB. Establishing key research questions for the implementation of artificial intelligence in colonoscopy: a modified Delphi method. Endoscopy. 2021;53:893-901. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 31. | Jin EH, Lee D, Bae JH, Kang HY, Kwak MS, Seo JY, Yang JI, Yang SY, Lim SH, Yim JY, Lim JH, Chung GE, Chung SJ, Choi JM, Han YM, Kang SJ, Lee J, Chan Kim H, Kim JS. Improved Accuracy in Optical Diagnosis of Colorectal Polyps Using Convolutional Neural Networks with Visual Explanations. Gastroenterology. 2020;158:2169-2179.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 91] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 32. | Yang YJ, Bang CS. Application of artificial intelligence in gastroenterology. World J Gastroenterol. 2019;25:1666-1683. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 211] [Cited by in RCA: 160] [Article Influence: 26.7] [Reference Citation Analysis (5)] |
| 33. | Namikawa K, Hirasawa T, Yoshio T, Fujisaki J, Ozawa T, Ishihara S, Aoki T, Yamada A, Koike K, Suzuki H, Tada T. Utilizing artificial intelligence in endoscopy: a clinician's guide. Expert Rev Gastroenterol Hepatol. 2020;14:689-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 34. | Pannala R, Krishnan K, Melson J, Parsi MA, Schulman AR, Sullivan S, Trikudanathan G, Trindade AJ, Watson RR, Maple JT, Lichtenstein DR. Artificial intelligence in gastrointestinal endoscopy. VideoGIE. 2020;5:598-613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 35. | Misawa M, Kudo SE, Mori Y, Hotta K, Ohtsuka K, Matsuda T, Saito S, Kudo T, Baba T, Ishida F, Itoh H, Oda M, Mori K. Development of a computer-aided detection system for colonoscopy and a publicly accessible large colonoscopy video database (with video). Gastrointest Endosc. 2021;93:960-967.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 105] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 36. | Zorron Cheng Tao Pu L, Maicas G, Tian Y, Yamamura T, Nakamura M, Suzuki H, Singh G, Rana K, Hirooka Y, Burt AD, Fujishiro M, Carneiro G, Singh R. Computer-aided diagnosis for characterization of colorectal lesions: comprehensive software that includes differentiation of serrated lesions. Gastrointest Endosc. 2020;92:891-899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 37. | Komeda Y, Handa H, Watanabe T, Nomura T, Kitahashi M, Sakurai T, Okamoto A, Minami T, Kono M, Arizumi T, Takenaka M, Hagiwara S, Matsui S, Nishida N, Kashida H, Kudo M. Computer-Aided Diagnosis Based on Convolutional Neural Network System for Colorectal Polyp Classification: Preliminary Experience. Oncology. 2017;93 Suppl 1:30-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 129] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 38. | Ozawa T, Ishihara S, Fujishiro M, Kumagai Y, Shichijo S, Tada T. Automated endoscopic detection and classification of colorectal polyps using convolutional neural networks. Therap Adv Gastroenterol. 2020;13:1756284820910659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 76] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 39. | Song EM, Park B, Ha CA, Hwang SW, Park SH, Yang DH, Ye BD, Myung SJ, Yang SK, Kim N, Byeon JS. Endoscopic diagnosis and treatment planning for colorectal polyps using a deep-learning model. Sci Rep. 2020;10:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |