Published online Aug 14, 2021. doi: 10.3748/wjg.v27.i30.5076
Peer-review started: March 31, 2021
First decision: May 28, 2021
Revised: June 7, 2021
Accepted: July 13, 2021
Article in press: July 13, 2021
Published online: August 14, 2021
Processing time: 131 Days and 18.7 Hours
Leukocytes, such as T cells and macrophages, play an important role in tumorigenesis. CC chemokine ligand (CCL) 4, which is produced by lymphocytes and macrophages, has been found to be expressed in the mucosa of the gastro
To examine CCL4 expression and its genetic polymorphism rs10491121 in patients with colorectal cancer (CRC) and evaluate their prognostic significance.
Luminex technology was used to determine CCL4 Levels in CRC tissue (n = 98), compared with paired normal tissue, and in plasma from patients with CRC (n = 103), compared with healthy controls (n = 97). Included patients had undergone surgical resection for primary colorectal adenocarcinomas between 1996 and 2019 at the Department of Surgery, Ryhov County Hospital, Jönköping, Sweden. Reverse transcription quantitative PCR was used to investigate the CCL4 gene expression in CRC tissue (n = 101). Paired normal tissue and TaqMan single nucleotide polymorphism assays were used for the CCL4 rs10491121 polymo
The CCL4 protein and messenger RNA expression levels were higher in CRC tissue than in normal paired tissue (90%, P < 0.001 and 45%, P < 0.05, respectively). CRC tissue from patients with localized disease had 2.8-fold higher protein expression levels than that from patients with disseminated disease. Low CCL4 protein expression levels in CRC tissue were associated with a 30% lower cancer-specific survival rate in patients (P < 0.01). The level of plasma CCL4 was 11% higher in CRC patients than in healthy controls (P < 0.05) and was positively correlated (r = 0.56, P < 0.01) with the CCL4 protein level in CRC tissue. The analysis of CCL4 gene polymorphism rs10491121 showed a difference (P < 0.05) between localized disease and disseminated disease in the right colon, with a dominance of allele A in localized disease. Moreover, the rate of the A allele was higher among CRC patients with mucinous cancer than among those with non-mucinous cancer.
The present study indicates that the CRC tissue levels of CCL4 and CCL4 gene polymorphism rs10491121, particularly in the right colon, are associated with clinical outcome in CRC patients.
Core Tip: Our data suggest that the CC chemokine ligand (CCL) 4 rs10491121 polymo
- Citation: Shamoun L, Landerholm K, Balboa Ramilo A, Andersson RE, Dimberg J, Wågsäter D. Association of gene and protein expression and genetic polymorphism of CC chemokine ligand 4 in colorectal cancer. World J Gastroenterol 2021; 27(30): 5076-5087
- URL: https://www.wjgnet.com/1007-9327/full/v27/i30/5076.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i30.5076
Colorectal cancer (CRC) is one of the most common malignancies and one of the leading causes of cancer death[1]. In most cases, the disease occurs sporadically through a transition from dysplastic adenomatous polyps to carcinoma. Genetic alterations and various genetic pathways cause this transition by affecting CRC induction and progression[2,3]. Details of the relationship between inflammation and CRC induction, progression, and metastasis, as well as the importance of tumor suppressor genes in inhibiting carcinogenesis, have been established by us and others[4-6]. Inflammatory factors expressed in CRC cells or in the tumor microenvironment appear to play an important role in local immune regulation and may either promote or suppress CRC pathogenesis. Communication between tumor cells and their microenvironment through secretion of factors is thought to be crucial for tumor growth. Particularly, the interaction between tumor cells and infiltrating leukocytes such as macrophages is vital for CRC progression and prognosis[4,7-9]. There is an ongoing search for molecular biomarkers to facilitate early diagnosis, determine prognosis, and guide the selection of personalized therapy for CRC patients[6,10]. The patients are identified based on pathological and clinical parameters, with CRC staging primarily based on the tumor-node-metastasis (TNM) system, as described in the American Joint Committee on Cancer classification[11].
The connection between inflammatory factors, such as cytokines and chemokines (chemotactic cytokines), and CRC is well-established[4,12]. Chemokines and their receptors have many roles in immune regulation[13] and some chemokines promote and regulate neoplastic progression, metastasis, angiogenesis, and immune cell infiltration[14-16].
The CC chemokine ligand (CCL) 4, also called macrophage inflammatory protein-1 beta, is a potent chemoattractant for monocytes/macrophages, dendritic cells, and T cells, via its cognate receptor CCR5. It is an especially effective chemoattractant for Th1 cells, which have an antitumor role[17-19]. CCL4 is mainly produced by lymphocytes and macrophages[17,18], but is also expressed in the intestinal mucosa and upregulated in gastric cancer and CRC tissue[20-22]. Genetic variations, such as single nucleotide polymorphisms (SNPs), in inflammatory genes are suggested to play a role in CRC risk and the survival of CRC patients[23,24]. The human CCL4 gene is located on chromosome 17 and recent studies have shown a relationship between the nonsynonymous SNP CCL4 rs10491121 (G > A) and oral[25], breast[26], and hepatocellular cancer[27]. To our knowledge, little is known about this SNP and CCL4 expression in patients with CRC. Therefore, the aim of the present study was to examine expression of CCL4 and its genetic polymorphism rs10491121 in patients with CRC and to evaluate their prognostic significance by identifying associations with various clinicopathological parameters and long-term survival.
Blood samples were obtained from 610 patients (344 males and 266 females) with a median age of 71 years (range: 25–94 years) who underwent surgical resection for primary colorectal adenocarcinomas between 1996 and 2019 at the Department of Surgery, Ryhov County Hospital, Jönköping, Sweden. Follow-up for the estimation of cancer-specific survival ended on the date of death or on February 23, 2021.
The study included 274 patients with rectal cancer and 336 patients with colon cancer, grouped based on the site of the primary cancer. In accordance with Liang et al[28], the colon cancer patients were divided into those with cancer localized in the right colon (cecum, ascending colon, hepatic flexure, transverse colon) (n = 194) and those with cancer localized in the left colon (splenic flexure, descending colon, sigmoid colon) (n = 142). The tumors were classified in accordance with the American Joint Committee on Cancer classification: Stage I in 105 cases, stage II in 228 cases, stage III in 196 cases, and stage IV in 81 cases. The degree of differentiation was categorized as high/medium (478 cases) or poor (132 cases) and the tumors were characterized as non-mucinous (n = 528) or mucinous (n = 82).
Healthy blood donors (n = 347) at Ryhov County Hospital, with no known CRC history and from the same geographical region as the CRC patients, were selected as the control population. This cohort comprised 216 males and 193 females, with a median age of 58 years (range: 33–68). Blood samples were collected at the start of surgery for the patients and at the time of blood donation for the controls. All blood samples were centrifuged to separate plasma and blood cells and then stored frozen, at -70°C, until analysis.
The investigation was approved by the Regional Ethical Review Board in Linköping, Sweden, and informed consent was obtained from each participant.
DNA was isolated from each blood sample using QiaAmp DNA Blood Kit (Qiagen, Hilden, Germany). The TaqMan SNP genotype assays were used for analysis of the CCL4 rs10491121 (ID C-11626804-10) genotypes (Applied Biosystems, Foster City, CA, USA). Ten nanograms of DNA were mixed with TaqMan Genotyping Master Mix (Applied Biosystems) and analyzed with the 7500 Fast Real-Time PCR System (Applied Biosystems). Amplification was performed using an initial cycle at 50°C for 2 min, followed by one cycle at 95°C for 10 min, and, lastly, 40 cycles at 95°C for 15 s and at 60°C for 1 min. The manual calling option in the allelic discrimination application ABI PRISM 7500 SDS software version 1.3.1 (Applied Biosystems) was used to assign the genotypes.
Two established human colon cancer cell lines, Caco-2 and HT-29, were purchased from American Type Culture Collection (Rockville, MD, USA). The cell lines were grown in accordance with the supplier’s instructions and the growth media were Essential Medium (Caco-2) and Mc Coy 5a (HT-29). The cell lines were stored frozen, at -78°C, until analysis.
This study utilized tumor and paired normal tissue samples from 98 of the CRC patients, of whom 52 were males and 46 females, with a median age of 68 years (range: 29–90). The tumors were located in the colon in 53 patients and in the rectum in 45 patients and were classified as stage I in 18 cases, stage II in 33 cases, stage III in 22 cases, and stage IV in 25 cases. Colorectal cancer tissue and adjacent normal colorectal mucosa (within about 5 cm from the tumor) were excised from each patient and immediately frozen at -78°C until analysis. Frozen tumor, paired normal tissue, and cell lines were thawed and homogenized in ice-cold radioimmunoprecipitation assay lysis buffer (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, United States) containing a protease inhibitor cocktail, in accordance with the manufacturer’s instructions. The lysate was placed on ice for 30 min and then centrifuged at 18000 g for 10 min. The protein content of the supernatant fluid was determined for each sample using the Bradford protein assay (Bio-Rad Laboratories, Inc., Hercules, CA, United States).
Of the CRC patients and controls, 103 and 97, respectively, were available for CCL4 protein analysis in plasma. Blood samples were drawn at the start of surgery for the patients and at the time of blood donation for the controls. The CRC patient group comprised 52 males and 51 females with a median age of 68 years (range: 29–90). Forty-six tumors were in the colon and 57 tumors in the rectum; they were classified as stage I in 18 cases, stage II in 34 cases, stage III in 25 cases, and stage IV in 26 cases. Controls for plasma samples included 51 males and 46 females with a median age of 63 years (range: 55–68).
The levels of CCL4 in plasma, colorectal tissue, and cell line lysates were measured using a Luminex bead-based technology (Bio-Rad Laboratories, Inc.) and commercially available Luminex assay for CCL4 (Bio-Rad Laboratories). The tissue and cell line levels of CCL4 protein were expressed as pg/mg of protein and the plasma CCL4 protein concentration from patients and controls as pg/mL.
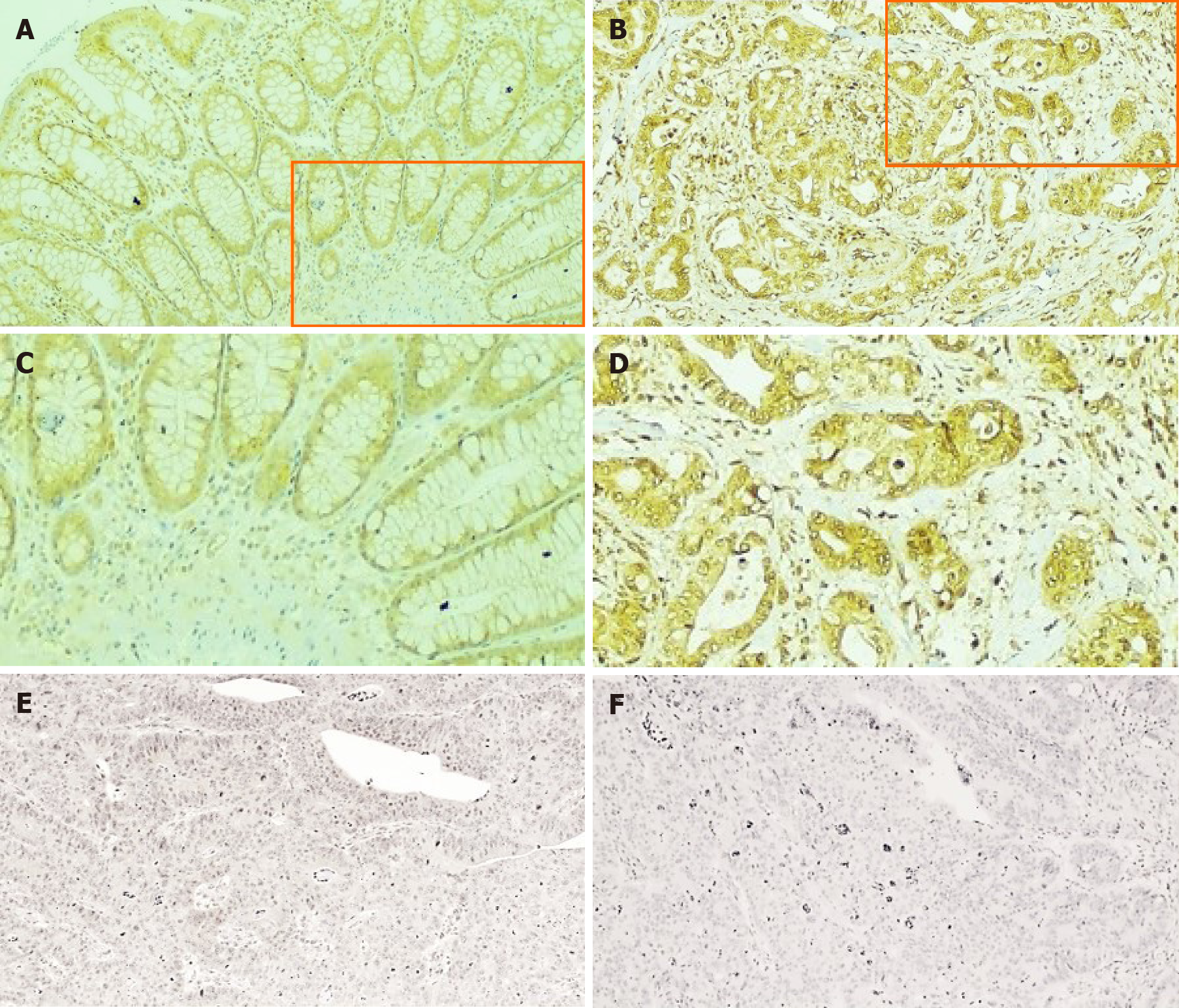
Formalin-fixed and paraffin-embedded CRC tissues from patients with CRC were obtained from the Department of Pathology of the County Hospital Ryhov, Jönköping, Sweden. Among the eight patients, six had tumors localized in the colon and two in the rectum; these were classified as stage I in two cases, stage II in two cases, stage III in two cases, and stage IV in two cases. The staining was performed on 3.5 μm sections of paraffin-embedded tissue samples to detect the CCL4 protein expression. In brief, antigen retrieval was finished by cooking at 110°C for 52 min in Diva Decloaker, 10X (Biocare Medical, Concord, CA, United States). The slides were treated with hydrogen peroxide for 5 min to prevent the occurrence of endogenous peroxidase, which might alter the interpretation of the color. Primary rabbit polyclonal CCL4 antibody against human CCL4 (Abcam, Tokyo, Japan) was used at a dilution of 1:300. The antibody was applied to the tissue sections, which were then incubated for 30 min at room temperature. The MACH 4 Universal HRP-Polymer Detection kit (Biocare Medical) was used, and the reaction was visualized using Betazoid DAB Chromogen Kit (Biocare Medical). Human kidney tissue was used as a positive control for CCL4 expression and rabbit IgG, polyclonal isotype control (Abcam) was used as a negative control and included along with each patient tissue section.
One hundred and one of the CRC patients were available for the analysis of CCL4 messenger RNA (mRNA) expression, of whom 52 were males and 49 females, with a median age of 72 years (range: 43–89). The tumors were located in the colon in 56 patients and in the rectum in 45 patients and were classified as stage I in 16 cases, stage II in 43 cases, stage III in 38 cases, and stage IV in four cases. Eighty-two available paired normal tissue samples were analyzed.
All CRC tissue samples and paired normal tissue samples were stored in RNA protect tissue reagent (Qiagen) to maintain good RNA quality. RNA was purified using the RNeasy Mini kit (Qiagen), in accordance with the manufacturer’s instructions, and eluted in nuclease-free water. The concentration and purity of the RNA and RNA integrity were determined using an Agilent 2100 Bioanalyzer with the Agilent RNA 6000 Nano Kit (Agilent Technologies, Santa Clara, CA, United States), in accordance with the manufacturer’s instructions. RNA (297 ng) was reverse transcribed in a total volume of 20 μL using Super Script III Reverse Transcription Kit with RNase Inhibitor (Thermo Fisher Scientific, Carlsbad, CA, United States), in accordance with the manufacturer’s instructions. The resulting complementary DNA and the remaining RNA were stored at -80°C.
Complementary DNA was amplified through reverse transcription quantitative PCR with TaqManTM Fast Universal PCR Master Mix (Cat.no 4366072, Applied Biosystems) and TaqMan Gene Expression Probes and Primers (RPLP0 Cat.no HS99999902 m1 and CCL4 Cat.no HS99999148 m1, Applied Biosystems) on a QuantStudio5 Real Time PCR system (Applied Biosystems), with each sample run in duplicate. Results were normalized to expression levels of human RPLP0. Relative quantification of gene expression was performed using the standard curve method and expressed in arbitrary units.
The differences in the frequencies of the CCL4 gene polymorphism between CRC patients and controls and between clinical characteristics within the CRC subgroups were analyzed using the chi-squared test. The Hardy-Weinberg equilibrium was assessed for the genotypes. Survival analysis was performed through Kaplan-Meier analysis with log-rank test. The Wilcoxon’s signed-rank test and the Mann-Whitney U-test were used for the analysis of the related and independent parameters and the Kruskal-Wallis test was used for comparing three or more classes. Correlations between parameters were analyzed using Spearman’s rank correlation test. Statistical analyses were performed using Stata Statistical Software Release 15 (Stata Corp., College Station, TX, United States) and SPSS software for Windows, version 14.0 (SPSS Inc., Chicago, IL, United States). Results were considered statistically significant at P < 0.05. The statistical methods of this study were reviewed by Roland E Andersson from Region Jönköping County, Sweden.
No statistically significant differences in the genotypic or allelic frequencies were observed between the patients and the healthy control group for CCL4 rs10491121 (Table 1). Stratification analysis of associations between this SNP and the tumor location showed no statistically significant differences between colon and rectum, or between right and left colon (data not shown). However, we noted a difference between localized (stages I + II) and disseminated (stages III + IV) cancer in the right colon, but not in the left colon, regarding the genotypic (P = 0.003) and allelic (P = 0.013) distributions (Table 2). The presence of allele A in the right colon was more common in stages I + II (40.5%) than in stages III + IV (28.2%) (Table 2) with an odds ratio (OR) = 1.73 (95% confidence interval = 1.12–2.68, P = 0.013). Analysis of the association between the genotypic or allelic distributions in non-mucinous or mucinous CRC showed statistically significant differences (Table 3). The rate of the A allele was higher (45.1%) among patients with mucinous cancer than among those with non-mucinous cancer (34.6%) with an OR = 1.56 (95% confidence interval = 1.12–2.17, P = 0.009).
| Genotype/allele | Patients | Controls |
| G/G | (250 (41.0) | 176 (43.0) |
| G/A | 281 (46.1) | 181 (44.3) |
| A/A | 79 (12.9) | 52 (12.7) |
| G | 781 (64.0) | 533 (65.2) |
| A | 439 (36.0) | 285 (34.8) |
| Variable | Cases | Genotype | Allele | |||
| Right colon | G/G | G/A | A/A | G | A | |
| Stages I + II | 116 | 38 (32.8) | 62 (53.4) | 16 (13.8) | 138 (59.5) | 94 (40.5) |
| Stages III + IV | 78 | 44 (56.4)b | 24 (30.8) | 10 (12.8) | 112 (71.8)a | 44 (28.2) |
| Left colon | ||||||
| Stages I + II | 67 | 23 (34.3) | 34 (50.8) | 10 (14.9) | 80 (59.7) | 54 (40.3) |
| Stages III + IV | 75 | 27 (36.0) | 37 (49.3) | 11 (14.7) | 91 (60.7) | 59 (39.3) |
Based on data from our cohort with up to 25 years follow-up, the Kaplan-Meier analysis revealed no differences in cancer-specific survival overall. No significant association was found between the genotypes or alleles when patients were stratified by gender, age, or degree of differentiation (data not shown). Neither the patient group nor the control group showed significant deviation from the Hardy-Weinberg equilibrium (data not shown).
The level of CCL4 protein was 90% higher in CRC tissue compared with in normal paired tissue (P < 0.001; Table 4). Moreover, the level of plasma CCL4 was 11% higher in patients than in controls (P < 0.050; Table 4) and showed a positive correlation (r = 0.56, P < 0.01) with levels in the CRC tissue.
The CRC tissue showed a gradual decrease in CCL4 protein level in relation to disease stage, and localized disease (stages I + II) showed a 2.8-fold higher CCL4 protein level than disseminated disease (stages III + IV, P = 0.001; Table 5). There was no difference in tissue levels of CCL4 protein for any of the other reported clinical features such as age, gender, location, differentiation grade, or mucinous state. Moreover, plasma levels of CCL4 were not associated with any of the investigated clinical features (data not shown). In the colon cancer cell lines, we noted that the average protein level of CCL4 was 7.8 pg/mg in HT-29 and 2.9 pg/mg in Caco-2.
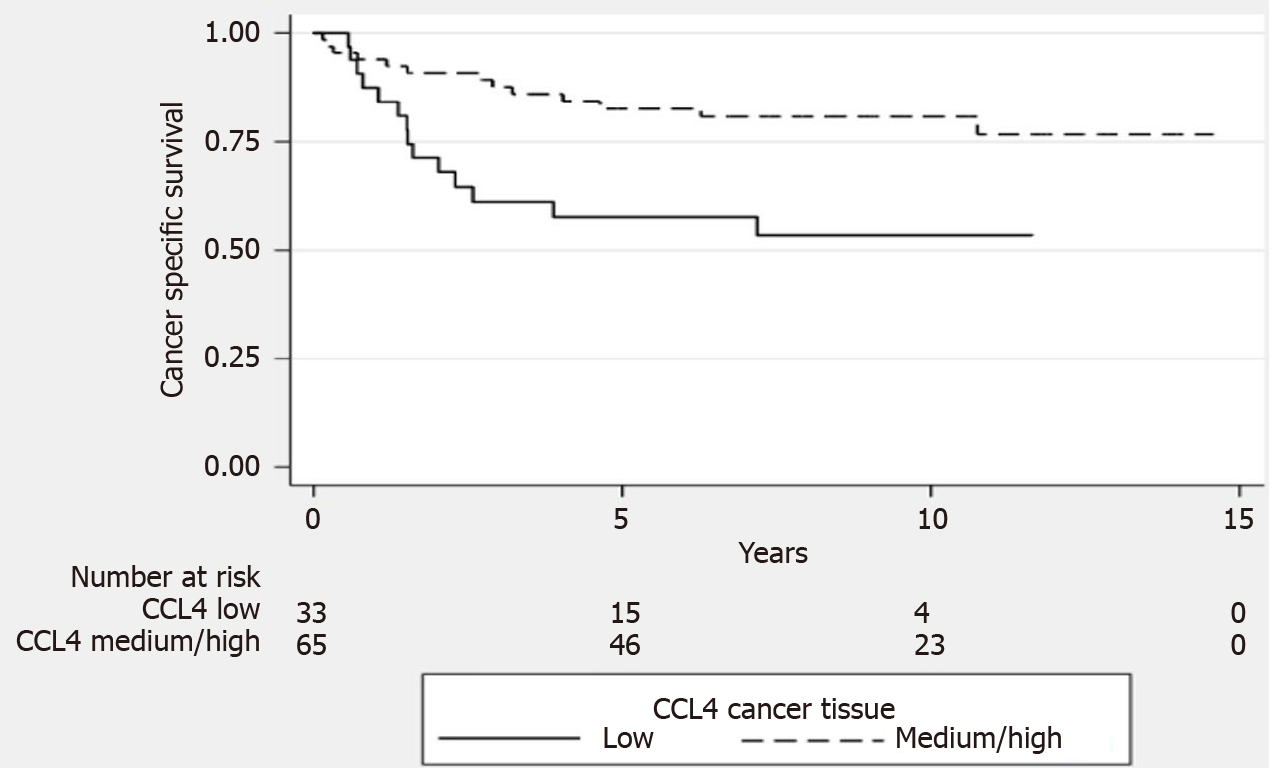
When CRC tissue levels of the CCL4 protein were used to group patients into tertiles (with low and medium/high levels), we found that the patients with low tissue levels of the CCL4 had a 30% lower cancer-specific survival rate (P = 0.005, log-rank test, Figure 1).
Immunohistochemical evaluation of CCL4 expression in CRC and normal tissue showed positive and negative to weak staining, respectively (Figure 2A-D). Cytoplasmic staining was evident predominantly in the epithelial cells. The staining of slides with isotype antibody control and with secondary antibody revealed only negative staining (Figure 2E and F).
The CCL4 mRNA expression was 45% higher in CRC tissue (median 61 AU, range: 2–494) compared with normal paired tissue (median 42 AU, range: 4-160, P = 0.044). When the relationship between the CCL4 mRNA expression and the mucinous state of the CRC tissue was analyzed, the expression level was found to be higher in mucinous CRC tissue (median 78 AU, range: 11–243) than in non-mucinous CRC tissue (median 56 AU, range: 2–494, P = 0.043). CCL4 mRNA expression levels were not associated with other reported clinical features (data not shown).
Previous studies have shown that CCL4 gene polymorphism rs10491121 (G > A) is associated with susceptibility to several cancer diseases[25-27], but little is known about this SNP in relation to CRC. The present study found no statistically significant differences between CRC patients and controls in genotype distribution or allelic frequencies of this SNP. Further, no associations were identified with clinical characteristics other than location, stage, and mucinous state. The genotypes of CCL4 gene polymorphism rs10491121 may influence gene expression, protein levels, and protein function of CCL4. However, to the best of our knowledge, there is limited data to describe the functional activity of this SNP. Regarding cancer, it has previously been shown that patients carrying the AG or GG genotype are at lower risk of developing distant metastases in breast cancer[26]. In our study, stratification analysis showed a difference in the frequency of this SNP between localized disease and disseminated disease in the right colon, but not in the left colon. Specifically, in the right colon, a dominance of the allele A was seen in localized disease as compared with in disseminated disease. Rectal, left, and right colon cancer differ with respect to histology, clinical outcomes, and expression of distinguishable genes, which has led to the hypothesis that they constitute different disease entities[28-30]. In our study, it is possible that the genotype distribution of CCL4 gene polymorphism rs10491121, together with other gene or gene-environment interactions, contributed to develo
Mucinous cancer is a subtype of CRC and is more common in proximal colon cancer than in distal colon and rectal cancer. There are conflicting results in the literature regarding the prognosis and survival rates for mucinous CRC[31]. Factors involved in the development of mucinous CRC are not yet known, but mucinous cancer has a different molecular signature than non-mucinous cancer[31]. This study demonstrated that the CCL4 gene polymorphism rs10491121 was associated with mucinous CRC, with a higher rate of the A allele than non-mucinous cancer. Moreover, we noted that higher CCL4 mRNA expression levels were associated with mucinous cancer. As far as we know, this is the first time data relating to this investigated genetic variant of CCL4 and CCL4 gene expression in mucinous CRC have been published. Further studies are needed to verify our findings and to investigate the potential function of this SNP on CCL4 expression and for prediction of the mucinous state.
Higher CCL4 mRNA expression levels in tumor tissue than in normal tissue in colon (P = 0.07) was suggested in a previous study including 20 patients[21]. In the present study, with 101 CRC patients, we confirmed that the CCL4 mRNA expression levels were higher (P = 0.044) in CRC tissue than in normal tissue. Two studies focused on CCL4 protein expression found higher levels in CRC tissue compared with in adjacent normal tissue[20,32]. These studies included only 10[20] and 25 patients[32], respectively, and found no association between CCL4 protein levels and cancer stage. In our study, CCL4 protein expression levels were examined in a large series of 98 patients with CRC. We found that CCL4 protein levels were higher in CRC tissue than in normal paired tissue. We also found that localized disease was associated with higher CCL4 protein levels than disseminated disease. The interaction between tumor cells and infiltrating leukocytes such as macrophages is a powerful relationship that affects CRC progression and prognosis[4,7-9]. CCL4 protein can be produced by lymphocytes and macrophages[17,18] and, in this study, immunostaining revealed expression of CCL4 in CRC tissue predominantly in epithelial cells. We also detected CCL4 protein in the colon cancer cell lines HT-29 and Caco-2. The lack of association between CCL4 mRNA and CCL4 protein levels in the present study could be explained by differences in transcription rate, mRNA stability, or translation efficiency within cell types such as lymphocytes, macrophages, and epithelial cancer cells in the colon and rectum.
When we grouped CRC tissue levels of the CCL4 protein as low or medium/high, we found that patients with low CCL4 protein levels in CRC tissue had lower cancer-specific survival rates. Thus, based on our data, higher CCL4 protein levels in CRC tissue are associated with less advanced disease stage and better prognosis. The properties of the tumor microenvironment and the interactions between tumor cells and infiltrating leukocytes, such as macrophages and T cells, affect CRC progression and prognosis[4,7-9]. Tumor-associated macrophages (TAMs) are commonly found in CRC tissue[7,8]. Macrophages are an important component of the tumor microenvironment and coordinate various aspects of immunity. TAMs promote tumor growth by facilitating angiogenesis, immunosuppression, and inflammation, and can also influence tumor relapse after anticancer therapies. Depending on activation status, macrophages can exert dual influences on tumorigenesis, either by antagonizing the cytotoxic activity in immune cells or by enhancing antitumor responses[7,8]. The classic view is that TAMs have one of two phenotypes, M1 or M2, which have antitumoral or protumoral activities, respectively[7,8].
The chemokine CCL4 is a potent chemoattractant for monocytes/macrophages, dendritic cells, and T cells, via its cognate receptor CCR5, and it is an especially effective chemoattractant for Th1 cells, which have an antitumor role[17-19]. TAMs adapt their phenotype in response to environmental stimuli and the presence of interferon gamma and tumor necrosis factor alpha (TNF-α) promotes polarization into phenotype M1[7,8]. In a previous study, we showed that TNF-α protein level was significantly higher in CRC tissue than in normal tissue[5]. It could be speculated that a higher level of CCL4 increases the prevalence of Th1 and TAMs, with a shift towards M1 macrophages that is stronger at lower stages than at higher stages, thus negatively affecting tumor development. However, the impact of macrophages in tumor progression remains to be fully elucidated, in part due to the contrasting roles they play depending on their polarization[7,8]. Further studies with immunohistochemical approaches will be conducted to clarify the number of TAMs, their phenotypic patterns, and the presence of T lymphocytes in our analyzed CRC tissue. From a molecular standpoint, the underlying mechanisms responsible for the differing levels of CCL4 at different disease stages remain to be clarified.
This study demonstrated that levels of plasma CCL4 were higher in patients compared with controls and were positively correlated with levels in CRC tissue. Plasma CCL4 concentration could possibly reflect levels in CRC tissues. However, we failed to detect a stage-dependent alteration of plasma CCL4 Level, which is consistent with data from another study[33]. Due to the general immunological imbalance indicated by the systemic cytokine pattern in CRC patients[33,34], we hypothesize that the production of CCL4 by lymphocytes and monocytes may be altered. Thus, the relationship between concentration of CCL4 in plasma and cancer stage may be masked.
Some limitations of our study should be mentioned. It is important to note that this study was of an exploratory nature. The patients and controls were selected from one hospital and may not represent the general populations. Additional studies with larger and more diverse populations of patients and controls are needed to validate our findings.
Our data suggested that the CCL4 rs10491121 polymorphism was associated with CRC, particularly mucinous CRC and CRC in the colon. Moreover, we observed that CCL4 protein expression was upregulated in CRC tissues compared with in normal tissue, correlated with disease stage, and that low expression was related to a lower cancer-specific survival rate. Detailed functional analysis is required to reveal the mechanisms underlying our observed associations that different levels of CCL4 in CRC tissue reflect different stages of disease. Moreover, studies with immunohistochemical approaches will be conducted to clarify the contribution of TAMs and their phenotypic patterns and the presence of T lymphocytes in our cohort of CRC tissue.
Colorectal cancer (CRC) is one of the most common malignancies and one of the leading causes of cancer death. Inflammatory factors expressed in CRC cells or in the tumor microenvironment play an important role in local immune regulation. Among these factors, chemokines like CC chemokine ligand (CCL) 4 play an important role in facilitating recruitment of leukocytes.
There is an ongoing search for molecular biomarkers to facilitate early diagnosis, and to determine the prognosis of CRC patients.
The aim of the present study was to examine expression of CCL4 and its genetic polymorphism rs10491121 in patients with CRC and to evaluate their association to clinicopathological parameters and prognostic impact.
Blood, tumor and paired normal tissue from patients with CRC and blood samples from healthy controls were subjected to an analysis of CCL4 protein using Luminex technology. Reverse transcription quantitative PCR was used to investigate the CCL4 gene expression in CRC tissue and paired normal tissue. For studies on the CCL4 rs10491121 polymorphism in CRC patients and healthy controls, TaqMan genotype assays based on polymerase chain reaction were used.
The CCL4 protein and messenger RNA expression levels were higher in CRC tissue than in normal paired tissue and the level of CCL4 protein in CRC tissue from patients with localized disease was higher than that in CRC tissue from patients with disseminated disease. Low levels of CCL4 protein in CRC tissue were associated with a 30% lower cancer-specific survival rate in patients. The level of CCL4 in plasma was higher in CRC patients than in healthy controls and was positively correlated with the CCL4 protein level in CRC tissue. There was a difference in polymorphism rs10491121 between localized disease and disseminated disease in the right colon, with a dominance of allele A in localized disease.
The results from this study indicate that CCL4 expression and gene polymorphism are good markers of prognosis. However, the findings need to be reproduced in larger cohorts.
The chemokine CCL4 deserves further attention as a clinical prognostic biomarker in CRC. Detailed functional analysis is required to reveal the mechanisms underlying the observed associations between different levels of CCL4 in CRC tissue and different stages of disease.
Manuscript source: Unsolicited manuscript
Specialty type: Biochemistry and molecular biology
Country/Territory of origin: Sweden
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Peng JY S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Liu JH
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11573] [Cited by in RCA: 13156] [Article Influence: 1879.4] [Reference Citation Analysis (4)] |
| 2. | Ahmad R, Singh JK, Wunnava A, Al-Obeed O, Abdulla M, Srivastava SK. Emerging trends in colorectal cancer: Dysregulated signaling pathways (Review). Int J Mol Med. 2021;47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 109] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 3. | Al-Sohaily S, Biankin A, Leong R, Kohonen-Corish M, Warusavitarne J. Molecular pathways in colorectal cancer. J Gastroenterol Hepatol. 2012;27:1423-1431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 174] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 4. | Mager LF, Wasmer MH, Rau TT, Krebs P. Cytokine-Induced Modulation of Colorectal Cancer. Front Oncol. 2016;6:96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 169] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 5. | Shamoun L, Kolodziej B, Andersson RE, Dimberg J. Protein Expression and Genetic Variation of IL32 and Association with Colorectal Cancer in Swedish Patients. Anticancer Res. 2018;38:321-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 6. | Yiu AJ, Yiu CY. Biomarkers in Colorectal Cancer. Anticancer Res. 2016;36:1093-1102. [PubMed] |
| 7. | Braster R, Bögels M, Beelen RH, van Egmond M. The delicate balance of macrophages in colorectal cancer; their role in tumour development and therapeutic potential. Immunobiology. 2017;222:21-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 8. | Poh AR, Ernst M. Targeting Macrophages in Cancer: From Bench to Bedside. Front Oncol. 2018;8:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 282] [Cited by in RCA: 382] [Article Influence: 54.6] [Reference Citation Analysis (0)] |
| 9. | Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423-1437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4060] [Cited by in RCA: 5752] [Article Influence: 522.9] [Reference Citation Analysis (0)] |
| 10. | Sagaert X. Prognostic biomarkers in colorectal cancer: where do we stand? Virchows Arch. 2014;464:379-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 11. | Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J Clin. 2017;67:93-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2341] [Cited by in RCA: 4396] [Article Influence: 549.5] [Reference Citation Analysis (4)] |
| 12. | Terzić J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101-2114.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1333] [Cited by in RCA: 1508] [Article Influence: 100.5] [Reference Citation Analysis (0)] |
| 13. | Raman D, Sobolik-Delmaire T, Richmond A. Chemokines in health and disease. Exp Cell Res. 2011;317:575-589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 312] [Cited by in RCA: 280] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 14. | Korbecki J, Grochans S, Gutowska I, Barczak K, Baranowska-Bosiacka I. CC Chemokines in a Tumor: A Review of Pro-Cancer and Anti-Cancer Properties of Receptors CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 Ligands. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 245] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 15. | Mollica Poeta V, Massara M, Capucetti A, Bonecchi R. Chemokines and Chemokine Receptors: New Targets for Cancer Immunotherapy. Front Immunol. 2019;10:379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 232] [Cited by in RCA: 412] [Article Influence: 68.7] [Reference Citation Analysis (0)] |
| 16. | Mukaida N, Baba T. Chemokines in tumor development and progression. Exp Cell Res. 2012;318:95-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 17. | Maurer M, von Stebut E. Macrophage inflammatory protein-1. Int J Biochem Cell Biol. 2004;36:1882-1886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 461] [Cited by in RCA: 552] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 18. | Mukaida N, Sasaki SI, Baba T. CCL4 Signaling in the Tumor Microenvironment. Adv Exp Med Biol. 2020;1231:23-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 95] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 19. | Siveke JT, Hamann A. T helper 1 and T helper 2 cells respond differentially to chemokines. J Immunol. 1998;160:550-554. [PubMed] |
| 20. | Baier PK, Eggstein S, Wolff-Vorbeck G, Baumgartner U, Hopt UT. Chemokines in human colorectal carcinoma. Anticancer Res. 2005;25:3581-3584. [PubMed] |
| 21. | Erreni M, Bianchi P, Laghi L, Mirolo M, Fabbri M, Locati M, Mantovani A, Allavena P. Expression of chemokines and chemokine receptors in human colon cancer. Methods Enzymol. 2009;460:105-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 76] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 22. | Saito S, Kitayama J, Jin ZX, Tsuno N, Kaisaki S, Seto Y, Nagawa H. Beta-chemokine, macrophage inflammatory protein-1beta (MIP-1beta), is highly expressed in diffuse type human gastric cancers. J Exp Clin Cancer Res. 2003;22:453-459. [PubMed] |
| 23. | Bondurant KL, Lundgreen A, Herrick JS, Kadlubar S, Wolff RK, Slattery ML. Interleukin genes and associations with colon and rectal cancer risk and overall survival. Int J Cancer. 2013;132:905-915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 24. | Ji H, Lu L, Huang J, Liu Y, Zhang B, Tang H, Sun D, Zhang Y, Shang H, Li Y, Lu H. IL1A polymorphisms is a risk factor for colorectal cancer in Chinese Han population: a case control study. BMC Cancer. 2019;19:181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 25. | Lien MY, Lin CW, Tsai HC, Chen YT, Tsai MH, Hua CH, Yang SF, Tang CH. Impact of CCL4 gene polymorphisms and environmental factors on oral cancer development and clinical characteristics. Oncotarget. 2017;8:31424-31434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 26. | Hu GN, Tzeng HE, Chen PC, Wang CQ, Zhao YM, Wang Y, Su CM, Tang CH. Correlation between CCL4 gene polymorphisms and clinical aspects of breast cancer. Int J Med Sci. 2018;15:1179-1186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 27. | Wang B, Chou YE, Lien MY, Su CM, Yang SF, Tang CH. Impacts of CCL4 gene polymorphisms on hepatocellular carcinoma susceptibility and development. Int J Med Sci. 2017;14:880-884. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 28. | Liang L, Zeng JH, Qin XG, Chen JQ, Luo DZ, Chen G. Distinguishable Prognostic Signatures of Left- and Right-Sided Colon Cancer: a Study Based on Sequencing Data. Cell Physiol Biochem. 2018;48:475-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 29. | Kalantzis I, Nonni A, Pavlakis K, Delicha EM, Miltiadou K, Kosmas C, Ziras N, Gkoumas K, Gakiopoulou H. Clinicopathological differences and correlations between right and left colon cancer. World J Clin Cases. 2020;8:1424-1443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 30. | Nishihara R, Glass K, Mima K, Hamada T, Nowak JA, Qian ZR, Kraft P, Giovannucci EL, Fuchs CS, Chan AT, Quackenbush J, Ogino S, Onnela JP. Biomarker correlation network in colorectal carcinoma by tumor anatomic location. BMC Bioinformatics. 2017;18:304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 31. | Penney ME, Parfrey PS, Savas S, Yilmaz YE. Associations of single nucleotide polymorphisms with mucinous colorectal cancer: genome-wide common variant and gene-based rare variant analyses. Biomark Res. 2018;6:17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 32. | De la Fuente López M, Landskron G, Parada D, Dubois-Camacho K, Simian D, Martinez M, Romero D, Roa JC, Chahuán I, Gutiérrez R, Lopez-K F, Alvarez K, Kronberg U, López S, Sanguinetti A, Moreno N, Abedrapo M, González MJ, Quera R, Hermoso-R MA. The relationship between chemokines CCL2, CCL3, and CCL4 with the tumor microenvironment and tumor-associated macrophage markers in colorectal cancer. Tumour Biol. 2018;40:1010428318810059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 33. | Kantola T, Klintrup K, Väyrynen JP, Vornanen J, Bloigu R, Karhu T, Herzig KH, Näpänkangas J, Mäkelä J, Karttunen TJ, Tuomisto A, Mäkinen MJ. Stage-dependent alterations of the serum cytokine pattern in colorectal carcinoma. Br J Cancer. 2012;107:1729-1736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 193] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 34. | Johdi NA, Mazlan L, Sagap I, Jamal R. Profiling of cytokines, chemokines and other soluble proteins as a potential biomarker in colorectal cancer and polyps. Cytokine. 2017;99:35-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |