Published online Jul 21, 2021. doi: 10.3748/wjg.v27.i27.4358
Peer-review started: January 31, 2021
First decision: May 13, 2021
Revised: May 15, 2021
Accepted: June 22, 2021
Article in press: June 22, 2021
Published online: July 21, 2021
Processing time: 169 Days and 2.4 Hours
Since it was first reported in December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has spread rapidly around the world to cause the ongoing pandemic. Although the clinical manifestations of SARS-CoV-2 infection are predominantly in the respiratory system, liver enzyme abnormalities exist in around half of the cases, which indicate liver injury, and raise clinical concern. At present, there is no consensus whether the liver injury is directly caused by viral replication in the liver tissue or indirectly by the systemic inflammatory response. This review aims to summarize the clinical manifestations and to explore the underlying mechanisms of liver dysfunction in patients with SARS-CoV-2 infection.
Core Tip: The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is one of the most influential emerging infectious diseases worldwide. Accumulating evidence suggests that liver injury is common in COVID-19 patients, and many severe cases tend to be associated with dysregulated liver functions. In this review, we summarize the currently available data of liver enzyme abnormalities in patients confirmed to have COVID-19 and analyze multiple risk factors for liver injury. However, the mechanism of liver impairment seems to be multifactorial. The evidence of direct liver injury triggered by SARS-CoV-2 infection or indirect liver injury induced by overwhelmed cytokine storm will also be discussed.
- Citation: Huang YK, Li YJ, Li B, Wang P, Wang QH. Dysregulated liver function in SARS-CoV-2 infection: Current understanding and perspectives. World J Gastroenterol 2021; 27(27): 4358-4370
- URL: https://www.wjgnet.com/1007-9327/full/v27/i27/4358.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i27.4358
Coronavirus disease 2019 (COVID-19) has broken out worldwide and was declared as a global pandemic by the World Health Organization (WHO) on March 11, 2020 after it was first reported in December 2019. The virus was later isolated[1] and was named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee for Taxonomy of Viruses. To date, this virus has spread to 220 countries, infecting 179601602 people with 3891974 deaths (as of June 24, 2021, source: Johns Hopkins University; https://coronavirus.jhu.edu/map.html), which constitutes a public health emergency of international concern.
The severity of symptoms in patients with COVID-19 ranges from mild self-limited respiratory disease to severe progressive pneumonia and respiratory failure, with an overall mortality rate of 2.3%[2]. The typical clinical manifestations of COVID-19 include elevated body temperature (> 37.3 °C), dry cough, dyspnea, leukocytosis, lung infiltration, and no significant improvement after antibiotic treatment for 3 d[3]. In addition to the respiratory system, various organs are reported to be involved in the process of SARS-CoV-2 infection, which caused complications that aggravate the infection[4]. Up to 50% of COVID-19 patients have been reported to have abnormal liver biochemical indicators, including elevated expression of aminotransferases, glutamyl transferase, and alkaline phosphatase (ALP)[3,5,6], and the proportion of infected patients with abnormal liver functions is greater in highly epidemic areas than other regions[7]. In addition, 58%-78% of patients with severe COVID-19 have apparent liver damage. Liver injury in turn increases the risk of other outcomes, such as cardiac arrest, requirement for intubation, acute respiratory distress syndrome, arrhythmia and shock[8]. Therefore, it is crucial to understand the mechanisms of the aberrant liver functions caused by this novel respiratory virus[9]. It is currently believed that three mechanisms may account for the liver damage in COVID-19 patients: (1) Hepatic bile duct endothelial cells and hepatocytes are directly infected by SARS-CoV-2 mediated by the cellular receptors; (2) A cytokine storm is induced by overactivated immune responses and causes inflammatory liver damage; and (3) Multiple risk factors such as underlying liver diseases may cause secondary liver damage following SARS-CoV-2 infection. In this review, we summarize the clinical manifestations and explore the potential underlying mechanisms of liver dysfunctions in patients with SARS-CoV-2 infection.
Coronaviruses (CoVs) are a group of enveloped positive single-stranded RNA viruses that belong to the Coronaviridae family of order Nidovirales[10]. CoVs are further grouped into α-, β-, γ-, and δ-coronaviruses. Among them, α- and β-CoVs can infect mammals and γ-coronaviruses infect birds and δ-coronaviruses infect both[11]. SARS-CoV-2, initially isolated from human airway epithelial cells, was identified as the seventh member of the CoVs capable of infecting humans. It genetically belongs to the β-CoV genus in the same clade with SARS-CoV and middle-east respiratory syndrome coronavirus (MERS-CoV)[1,12].
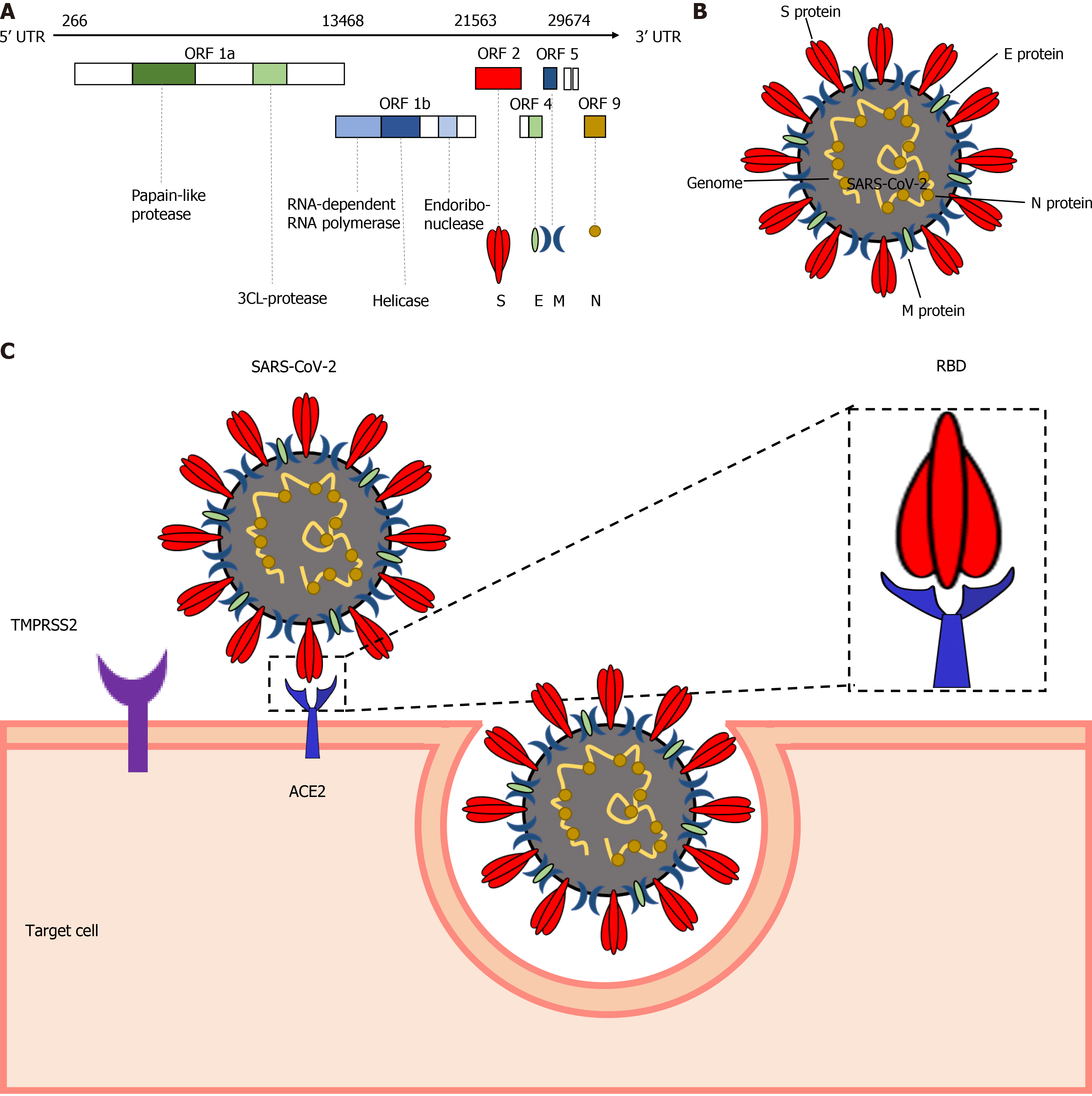
The SARS-CoV-2 genome shares around 76.5% amino acid sequence identity with SARS-CoV[13] , which might be responsible for the shared transmission mode and pathogenesis of these two viruses. The SARS-CoV-2 genome contains 14 open reading frames, encoding 27 proteins (Figure 1A). Four major structural proteins are encoded by the 3’-terminus of the genome, including the spike surface glycoprotein (S), matrix protein (M), small envelope protein (E), and nucleocapsid protein (N)[11] (Figure 1B). The S protein initiates the infection by interacting with the cellular receptor through its receptor binding domain (RBD). SARS-CoV-2 infects the target cells by S protein on the surface of the virion binding to cellular angiotensin-converting enzyme 2 (ACE2) receptors followed by fusion of the cellular and viral membranes (Figure 1C). In this process, transmembrane protease serine 2 (TMPRSS2), together with the endosomal cysteine proteases cathepsin B and L (CatB/L), are used for S protein priming. The S protein of SARS-CoV-2 is efficiently cleaved at the junction of S1 and S2 proteins to produce an S2 subunit to facilitate SARS-CoV-2 entry[14]. Considering the major role played in the pathogenesis, S protein is considered to be a potential target for vaccine design and drug development[9]. E protein is involved in the assembly and release of virions. As the most abundant viral protein, M protein plays an important role in RNA packaging[15] and N protein mediates the transcription and replication of viral RNA[16].
The variability of the SARS-CoV-2 genome is lower when compared to other human CoVs. There were 149 mutations in 103 sequenced SARS-CoV-2 genomes in the early stage of the pandemic[17]. The virus was classified into two major variants (L and S) through population genetic analysis. The L type, derived from the S type of SARS-CoV-2, was reported to be evolutionarily more aggressive[17]. Recently, a more transmissible phylogenetic cluster (named lineage B.1.1.7), which has 23 specific genetic changes in S protein of SARS-CoV-2, was reported in the UK, and rapidly became the dominant strain in London. Mutation N501Y affects the structure of the RBD and P681H is adjacent to the furin-cleavage site, which increases the binding affinity of the virus to ACE2 receptors by 1000-fold. Moreover, the deletion 69-70del on the S protein facilitates viral evasion from immune surveillance. Collectively, these variations have raised concerns about potential antigenic drift of SARS-CoV-2 similar to that of influenza virus[18].
SARS-CoV-2 is a zoonotic virus that spreads in humans through respiratory droplets[19]. Comparative sequence analysis of the SARS-CoV-2 genome has revealed a striking similarity with bat CoVs, indicating that bats may be the origin of this virus[20,21]. Although many COVID-19 cases initially reported in China were traced to have connection with Huanan Seafood Wholesale Market in Wuhan[6,22,23], several reports clearly indicate that the global spread of the SARS-CoV-2 might have preceded the discovery of the first case in Wuhan. The epidemic likely started between October 6 and December 11, 2019 in the USA, Italy and France[24,25];however, debate still exists about the source of SARS-CoV-2. Since the transmission of SARS-CoV-2 needs intermediate hosts, pangolins have been suggested as a probable intermediate host[26]. In February 2020, evidence of human-to-human transmission of SARS-CoV-2 was first reported[27]. Thereafter, a case of SARS-CoV-2 provided proof that human-to-human transmission occurred during the incubation period[28]. With the appearance of the second-generation patients and many medical staff in the hospital being infected, the strong infectious ability of SARS-CoV-2 was confirmed[29]. With the detection of SARS-CoV-2 RNA in fecal sample from a COVID-19 patient in the USA, SARS-CoV-2 could also be transmitted through the fecal–oral route[30]. Since September 2020, the route of transmission from contaminated objects to people has been reported by tracing the origin of local confirmed cases in Qingdao, Tianjin, Dalian, Chengdu and other cities in China, further complicating epidemic prevention and control.
The common clinical manifestations of patients with COVID-19 are cough, fever, shortness of breath, and other respiratory symptoms[31]. Gastrointestinal symptoms including nausea, vomiting, and diarrhea are also observed in some patients. The abnormal values of liver enzymes have frequently been noted in patients with COVID-19. Existing data suggest that, in most cases, liver enzyme elevations are mild to moderate and rarely severe. Limited pathophysiological studies have indicated that SARS-CoV-2 infection can cause liver injury, although SARS-CoV-2-induced liver injury is not the leading cause of death.
Liver injury in hospitalized patients is mainly manifested by changes in biochemical liver markers. Many studies have reported the presence of liver dysfunction in COVID-19 patients (Table 1). Respiratory failure is a common characteristic in severe cases, characterized by an imbalance of oxygen supply leading to an increase in transaminases[32]. As reported, 14%-53% of patients have elevated aspartate aminotransferase (AST), alanine aminotransferase (ALT), g-glutamyltransferase (GGT), hypoalbuminemia, and slightly elevated bilirubin[33]. The elevation of AST (39.4%) is more significant than that of ALT (28.1%) in severe cases. Among these markers affected by viral infections, elevated AST is related to a higher risk of death, while elevated GGT[33-38], ALP[34-36], together with bilirubin levels[6,31,33-35], are associated with biliary tract damage. Some patients have hypoalbuminemia, an independent predictor of mortality, which may be related to inflammation and poor prognosis[39,40]. Some studies have also reported C-reactive protein (CRP) elevation in many severe COVID-19 cases[6,31,35,41-44], indicating that overactivation of the immune system might account for disease severity, which can be used to predict worse outcomes in COVID-19 patients. In the elderly population with underlying liver disease, liver enzymes, especially transaminases, are elevated due to lobular necrosis[45].
| Region | No. of patients | Rate of abnormal liver test | Hepatocyte injury markers | Bile duct injury or cholestasis markers | Hepatic clearance markers | Infection-related biomarkers | Refs. | ||
| ALT | AST | ALP | GGT | Total bilirubin | CRP | ||||
| Shenzhen, China | 417 | 76.3% | 23.4% | 14.80% | N/A | 24.4% | 11.5% | N/A | [33] |
| Wuhan, China | 115 | N/A | 9.57% | 14.78% | 5.21% | 13.4% | 6.96% | N/A | [34] |
| Wuhan, China | 99 | 43% | 28% | 35% | N/A | N/A | 18% | 86% | [6] |
| Wuhan, China | 1099 | N/A | 21.3% | 22.20% | N/A | N/A | 10.5% | 60.7% | [31] |
| Shanghai, China | 148 | 37.20% | 18.2% | 21.6% | 4.10% | 17.60% | 6% | 8.7-32.3% | [35] |
| Wuhan, China | 69 | N/A | 33% | 28% | N/A | N/A | N/A | 67% | [41] |
| Fuyang, China | 125 | N/A | 20.8% | 21.60% | N/A | N/A | N/A | 70.4% | [42] |
| Japan | 22 | 68.20% | 54.5% | N/A | N/A | 54.50% | N/A | N/A | [36] |
| Turkey | 554 | N/A | 27.6% | 4% | N/A | N/A | [37] | ||
| Zaragoza, Spain | 531 | 64.3% | 28.6% | 40.90% | N/A | 47.30% | N/A | N/A | [38] |
| Wuhan, China | 81 | N/A | 29.5% | 17.90% | N/A | N/A | 3.6% | 41.8% | [43] |
| New York, United States | 5700 | N/A | 39% | 58.40% | N/A | N/A | N/A | 6.4-26.9% | [44] |
Mild portal vein inflammation, moderate microvesicular steatosis, and necrotizing inflammation have been observed in liver autopsy tissue of COVID-19[10]. This is likely due to increase of monocyte chemoattractant protein 1 (MCP-1) in patients infected with SARS-CoV-2, which aggravates fatty hepatitis[46]. Autopsy results from COVID-19 patients in Wuhan have shown infiltration of monocytes and lymphocytes in the portal vein area, accompanied by hepatic sinus congestion and microthrombus formation[45]. Cytologically, hepatocyte degeneration with focal lobular necrosis and neutrophil infiltration are observed together with swollen mitochondria, endoplasm reticulum expansion, and decreased glycogen particles in the hepatocytes. The histological manifestations are apoptosis and binuclear hepatocytes. In addition, immunohistochemical results have shown a lack of CD4+ cells and CD8+ T cells in liver tissue, suggesting that immunopathological insult may not be the main factor leading to liver injury.
SARS-CoV-2 initiates membrane fusion and cytoplasmic invasion by entering respiratory endothelial cells expressing ACE2 and TMPRSS2, triggering an initial immune response characterized by inflammatory cytokine production. Afterwards, the downstream proinflammatory immune responses of pathogenic Th1 cells and monocyte signaling pathways are activated[47]. Two hypotheses have been proposed to explain the mechanisms of liver dysfunctions in COVID-19 patients: direct injury by SARS-CoV-2[48] and indirect injury from a cytokine storm.
It has been reported that SARS-CoV-2 infection of various types of liver cells causes liver injury through direct cytopathic effects[48]. As a functional receptor for SARS-CoV-2 S protein binding, ACE2 is widely expressed on the surface of various human cells. Due to the low expression level of ACE2 in liver tissue (approximately 0.31%), SARS-CoV-2 was once considered unlikely to infect the liver. However, Li et al[49] systematically evaluated the expression of host receptor genes interacting with SARS-CoV-2 in liver tissue and their distribution patterns in different cell types using single-cell transcriptomics. ACE2 protein is expressed in various cell types in the liver, especially in bile duct cells[50], likely leading to direct injury to the liver (Figure 2). Boettler et al[51] proved that SARS-CoV-2 has a stronger affinity for bile duct cells than hepatocytes because of higher ACE2 receptor expression.
It has been proposed that SARS-CoV-2 infection of bile duct cells is the source of viral nucleic acid detection in feces[51]. Even in patients with negative throat swabs for viral RNA, 48% of feces were positive for SARS-CoV-2 RNA, which may be derived from portal vein viremia. Although SARS-CoV-2 can infect bile duct cells, the histological characteristics of bile duct injury have not been observed. Wang et al[48] observed many SARS-CoV-2 virions in the hepatocyte cytoplasm of two cases with COVID-19. Most of the virosomes were found to have complete envelopes with crown protrusions, providing evidence that SARS-CoV-2 can enter and replicate in hepatocytes. However, the low expression level of ACE2 on hepatocytes cannot fully explain the hepatotropic nature of SARS-CoV-2. One possibility is that there are alternative receptors or co-receptors other than ACE2. This hypothesis has gradually been confirmed by the discovery of more co-receptors that facilitate SARS-CoV-2 entry. For example, tyrosine-protein kinase receptor UFO (AXL), heparan sulfate, scavenger receptor B type 1 (SR-B1), neuropilin-1 (NRP1) and CD147 have been identified as attachment factors for SARS-CoV-2 infection[52-57], although the abundance of these co-receptors in liver tissue remains to be determined. The other possibility is that the expression of ACE2 in hepatocytes is temporarily upregulated when the virus enters the cells, assisting infection. Again, more evidence is needed to support this hypothesis.
The variable symptoms in severe and asymptomatic patients with COVID-19 suggest that host immune responses contribute to the pathogenesis. SARS-CoV-2 infection leads to excessive production of a series of cytokines by macrophages and neutrophils known as a cytokine storm[47], which is one of the main features in COVID-19 patients and may be a key factor affecting disease severity and mortality. Cytokines are self-antigens that induce autoimmunity, and could account for the multisystem inflammatory syndrome often seen in COVID-19 patients. CD4+ and CD8+ T cells in the peripheral blood of COVID-19 patients are highly activated via Toll-like receptors. Elevated proinflammatory CD4+ T cells and cytotoxic granular CD8+ T cells suggests an antiviral immune response and excessive activation of T cells in the peripheral blood[10]. The host cells are attacked by activated T cells leading to necrosis and apoptosis, and many damage-related pattern molecules are involved to amplify the inflammatory signals. Lymphopenia is also observed in the disordered cytokine storm, but it is still unknown whether it is caused by destruction of lymphocytes or tissue infiltration. It has been demonstrated that the proinflammatory cytokines are elevated significantly in patients with severe COVID-19, leading to severe pneumonia. The inflammatory cytokines not only protect the host cells from injury but also cause blood clotting, thereby blocking the vessels and resulting in decrease of oxygen saturation and sepsis, which is considered to be one of the causes of insufficient lung ventilation. Therefore, hypoxic hepatitis is common in patients with severe COVID-19, especially elderly patients with right congestive heart failure who are prone to have hypoxic–ischemic liver injury[48].
More specifically, the cytokine storm induced by SARS-CoV-2 is characterized by high expression of interleukin (IL)-6 and tumor necrosis factor-α (TNF-α). Activated pathogenic Th1 cells produce proinflammatory cytokines, such as granulocyte– macrophage colony-stimulating factor (GM-CSF) and IL-6. GM-CSF in turn stimulates CD4+ T cells to secrete large quantities of cytokines, such as TNF-α, IL-10, IL-6 and type Ⅰ interferons[58] (Figure 2). The inflammatory response is obvious in COVID-19 patients as indicated by increased expression of inflammatory biomarkers such as lactate dehydrogenase (LDH), IL-6, IL-2, CRP, serum ferritin, and D-dimer[59-61]. It has been reported that 28% of patients who died of multiple organ damage, including liver failure, was due to an excessive inflammatory response. In the case of liver injury, ACE2 expression is upregulated due to the compensated proliferation of hepatic parenchymal cells derived from bile duct epithelial cells, which increases the opportunities of viral infection in liver tissue.
Neutralizing cytokine release syndrome (CRS) that blocks the signal transduction pathway of these cytokines contributes to the development of effective drugs for the treatment of patients with severe COVID-19, and targeting CRS has significant implications for reducing mortality. One such example is tocilizumab, an IL-6 inhibitor, which improves the condition of patients within 15 d and reduces mortality considerably[62]. Although the cytokine storm is involved in liver injury, these cytokines also function in viral suppression. Therefore, application of treatment regimens to block cytokine signal transduction must be careful. Clinically, the application of synthetic corticosteroid dexamethasone worsens outcomes in milder COVID-19 patients, but reduces mortality in severe cases[63]. Likewise, other studies have supported that immunosuppression has a beneficial effect if given at a late stage, whereas immunostimulation enhances antiviral activity in the early stage[64], indicating that the timing of treatment and the specific patient population need to be identified to personalize the therapeutic schemes.
Current evidence is insufficient to conclude that liver injury in COVID-19 patients results entirely from SARS-CoV-2 infection. Considering the multisystem involvement in severe COVID-19 patients, liver injury may be caused by multiple factors, including drug toxicity, systemic inflammation, liver congestion, microvascular thrombosis, or damage induced by cytotoxic T cells and innate immune responses following SARS-CoV-2 infection.
Drug toxicity: Currently there is no specific treatment for COVID-19, although initial clinical guidelines recommended using antiviral drugs and monoclonal antibodies such as remdesivir, chloroquine, tocilizumab, lopinavir, ritonavir, and traditional Chinese medicine. However, it has been reported that both lopinavir and ritonavir have little clinical effect on COVID-19 patients[65]. According to WHO and National Health Commission of China guidelines, patients with moderate symptoms should receive timely treatment with Coriolus versicolor, abidor, chloroquine phosphate, and recombinant human interferon α-2b. For severe/critical patients, respiratory support, appropriate hormone therapy, and traditional Chinese medicine may be beneficial[66]. Chloroquine phosphate has been proved to cause significant liver damage[67]. A more recent study reported that ACE inhibitors and angiotensin Ⅱ receptor blockers might contribute to liver impairment in COVID-19 patients, although more studies are needed to confirm these findings[33]. In addition, fever is one of the main symptoms for COVID-19 patients. Paracetamol, which is a commonly used antipyretic drug, has been proven to cause fulminant liver failure, so the possibility of excessive use of paracetamol leading to elevated ALT cannot be ruled out[59,68].
Liver congestion: Forty percent of patients are at high risk of venous thromboembolism (VTE) when admitted to hospital. High-risk VTE patients have elevated expression of ALT, AST and CRP[69]. It is well known that viral infection leads to a hypercoagulable state, increasing the risk of thromboembolism[70,71]. The incidence of VTE is 20% on day 7 and 42% on day 21, even among hospitalized patients taking anticoagulants[72]. Recent studies have found that in some younger patients, microvascular thrombosis causes end-stage organ damage and contributes to liver injury. Anticoagulant drugs can be used to prevent the development of VTE. A few studies have concluded that 80% of COVID-19 patients with cirrhosis and portal hypertension received anticoagulant therapy, such as low-molecular-weight heparin, and those patients do not have serious bleeding complications[73,74]. However, Wang et al[69] have reported that 11% of COVID-19 patients with high risk of VTE also have a high risk of bleeding. Therefore, evaluating the risk of thromboembolism and rational use of anticoagulants for the treatment of COVID-19 patients is critical.
Underlying liver diseases: Chronic liver diseases, including chronic viral hepatitis and nonalcoholic fatty liver disease (NAFLD), affect approximately 300 million people in China and constitute a major global burden of disease. Although chronic liver disease does not increase the infection risk of patients with SARS-CoV-2 in general, patients with liver cirrhosis or hepatocellular carcinoma tend to be more susceptible to SARS-CoV-2 infection due to systemic immunodeficiency. Furthermore, the underlying liver diseases may worsen disease progression in patients with COVID-19. A study consisting of > 17 million people in the United States found that although < 1% of the confirmed COVID-19 cases had chronic liver diseases, chronic liver disease was an independent risk factor for death from SARS-CoV-2 infection[75].
Hepatitis B virus (HBV) does not predispose COVID-19 patients to more severe outcomes, although HBV patients infected with SARS-CoV-2 normally present with aggravated liver damage together with more severe thrombocytopenia and monocytopenia, coagulation dysfunction, and more disturbed hepatic function in relation to albumin production and lipid metabolism[76], and these co-infected patients have a tendency to HBV reactivation[67]. Astrilizumab and baritinib, which are used clinically to antagonize adverse immune reactions, tend to induce HBV reactivation, leading to impaired liver function in HBV patients[77]. However, it is currently unclear whether viral infection aggravates cholestasis in patients with the cholestatic disease. Further in-depth research should focus on persistent liver damage and active viral replication in HBV patients after co-infection with SARS-CoV-2. Recent studies have demonstrated the possible impact of SARS-CoV-2 infection on NAFLD. It has been shown that SARS-CoV-2 infection increases the possibility of NAFLD progressing to nonalcoholic hepatitis. ACE inhibitors are commonly used in the anti-inflammatory and anti-obesity treatment of NAFLD. Although there have been no reports on the correlation between the use of ACE inhibitors and mortality, one study has shown that ACE inhibitors upregulate expression of ACE2 receptor, thereby increasing the viral entry in patients taking such drugs[78].
How the underlying liver diseases affect liver function in patients with COVID-19 remains elusive. Whatever mechanisms are involved in liver damage, we need to be more cautious in managing COVID-19 patients with underlying liver diseases. For example, the British Liver Foundation recommends that patients taking steroids or immunosuppressive drugs maintain strict social distance[79]. The European Association for Liver Research and the European Association for Clinical Microbiology and Infectious Diseases recommend priority outpatient visits, prehospital assessment of risk factors, reduction of exposure through modification of waiting areas, reduction of waiting times, and use of online system to order medication.
Although liver dysfunction is common in patients with COVID-19, the detailed mechanisms remain incompletely understood. Available evidence suggests that the severity of liver damage is partly due to direct infection by SARS-CoV-2 to the liver and partly due to the cytokine storm produced by an overactive immune response. In addition, multiple risk factors, such as drug toxicity and liver congestion, need to be taken into account. For COVID-19 patients with underlying liver diseases, the causes of liver damage are more complex and more personalized management should be considered. Future directions should focus on deciphering the virus–host interactions to better understand the detailed molecular mechanisms of the liver impairment following SARS-CoV-2 infection. In the meantime, rapid development of effective preventive vaccines and specific antiviral drugs is critical to control this ongoing pandemic.
Manuscript source: Invited manuscript
Specialty type: Microbiology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Mohammadi M, Şehirli AÖ S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Xing YX
| 1. | Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W; China Novel Coronavirus Investigating and Research Team. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382:727-733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18987] [Cited by in RCA: 17646] [Article Influence: 3529.2] [Reference Citation Analysis (0)] |
| 2. | Epidemiology Working Group for NCIP Epidemic Response; Chinese Center for Disease Control and Prevention. [The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China]. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41:145-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1341] [Reference Citation Analysis (0)] |
| 3. | Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35178] [Cited by in RCA: 30121] [Article Influence: 6024.2] [Reference Citation Analysis (3)] |
| 4. | Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631-637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3643] [Cited by in RCA: 4149] [Article Influence: 197.6] [Reference Citation Analysis (0)] |
| 5. | Xu XW, Wu XX, Jiang XG, Xu KJ, Ying LJ, Ma CL, Li SB, Wang HY, Zhang S, Gao HN, Sheng JF, Cai HL, Qiu YQ, Li LJ. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-CoV-2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368:m606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1298] [Cited by in RCA: 1263] [Article Influence: 252.6] [Reference Citation Analysis (0)] |
| 6. | Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507-513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14869] [Cited by in RCA: 12976] [Article Influence: 2595.2] [Reference Citation Analysis (1)] |
| 7. | Feng G, Zheng KI, Yan QQ, Rios RS, Targher G, Byrne CD, Poucke SV, Liu WY, Zheng MH. COVID-19 and Liver Dysfunction: Current Insights and Emergent Therapeutic Strategies. J Clin Transl Hepatol. 2020;8:18-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 289] [Cited by in RCA: 278] [Article Influence: 55.6] [Reference Citation Analysis (0)] |
| 8. | Desai J, Patel U, Arjun S, Farraj K, Yeroushalmi K, Paz SG, Im J, Castillo A, Rammohan R, Mustacchia P. Impact of Liver Injury in COVID-19 Patients: Single-center Retrospective Cohort Analysis. J Clin Transl Hepatol. 2020;8:476-478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 9. | Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1-23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1889] [Cited by in RCA: 2096] [Article Influence: 209.6] [Reference Citation Analysis (0)] |
| 10. | Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420-422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5228] [Cited by in RCA: 5785] [Article Influence: 1157.0] [Reference Citation Analysis (2)] |
| 11. | Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, Sheng J, Quan L, Xia Z, Tan W, Cheng G, Jiang T. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe. 2020;27:325-328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1413] [Cited by in RCA: 1502] [Article Influence: 300.4] [Reference Citation Analysis (0)] |
| 12. | Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8473] [Cited by in RCA: 7602] [Article Influence: 1520.4] [Reference Citation Analysis (0)] |
| 13. | Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, Zhong W, Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457-460. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1252] [Cited by in RCA: 1316] [Article Influence: 263.2] [Reference Citation Analysis (0)] |
| 14. | Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020; 181: 271-280. e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11946] [Cited by in RCA: 14273] [Article Influence: 2854.6] [Reference Citation Analysis (0)] |
| 15. | Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178:104792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 610] [Cited by in RCA: 547] [Article Influence: 109.4] [Reference Citation Analysis (0)] |
| 16. | Neuman BW, Buchmeier MJ. Supramolecular Architecture of the Coronavirus Particle. Adv Virus Res. 2016;96:1-27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 85] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 17. | Tang XL, Wu CC, Li X, Song YH, Yao XM, Wu XK, Duan YG, Zhang H, Wang YR, Qian ZH, Cui J, Lu J. On the origin and continuing evolution of SARS-CoV-2. Guojia Kexue Pinglun. 2020;6:1012-1023. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1089] [Cited by in RCA: 867] [Article Influence: 173.4] [Reference Citation Analysis (0)] |
| 18. | Rambaut A, Loman N, Pybus O, Barclay W, Barrett J, Carabelli A, Connor T, Peacock T, Robertson DL, Volz E. on behalf of COVID-19 Genomics Consortium UK (CoG-UK) Preliminary genomic characterisation of an emergent SARS-CoV-2 Lineage in the UK defined by a novel set of spike mutations. Genom Epidemiol. 2020. |
| 19. | Chu DK, Akl EA, Duda S, Solo K, Yaacoub S, Schünemann HJ; COVID-19 Systematic Urgent Review Group Effort (SURGE) study authors. Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis. Lancet. 2020;395:1973-1987. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2542] [Cited by in RCA: 2387] [Article Influence: 477.4] [Reference Citation Analysis (0)] |
| 20. | Zhang YZ, Holmes EC. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell. 2020;181:223-227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 612] [Cited by in RCA: 511] [Article Influence: 102.2] [Reference Citation Analysis (0)] |
| 21. | Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6893] [Cited by in RCA: 7499] [Article Influence: 1499.8] [Reference Citation Analysis (0)] |
| 22. | Li X, Zai J, Wang X, Li Y. Potential of large "first generation" human-to-human transmission of 2019-nCoV. J Med Virol. 2020;92:448-454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 94] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 23. | Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020;382:1199-1207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11224] [Cited by in RCA: 9319] [Article Influence: 1863.8] [Reference Citation Analysis (0)] |
| 24. | Deslandes A, Berti V, Tandjaoui-Lambotte Y, Alloui C, Carbonnelle E, Zahar JR, Brichler S, Cohen Y. SARS-CoV-2 was already spreading in France in late December 2019. Int J Antimicrob Agents. 2020;55:106006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 146] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 25. | Amendola A, Bianchi S, Gori M, Colzani D, Canuti M, Borghi E, Raviglione MC, Zuccotti GV, Tanzi E. Evidence of SARS-CoV-2 RNA in an Oropharyngeal Swab Specimen, Milan, Italy, Early December 2019. Emerg Infect Dis. 2021;27:648-650. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 26. | Zhang T, Wu Q, Zhang Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr Biol. 2020;30:1578. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 167] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 27. | Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514-523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6483] [Cited by in RCA: 5423] [Article Influence: 1084.6] [Reference Citation Analysis (0)] |
| 28. | Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, Zimmer T, Thiel V, Janke C, Guggemos W, Seilmaier M, Drosten C, Vollmar P, Zwirglmaier K, Zange S, Wölfel R, Hoelscher M. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Engl J Med. 2020;382:970-971. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2799] [Cited by in RCA: 2491] [Article Influence: 498.2] [Reference Citation Analysis (0)] |
| 29. | Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323:1061-1069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14113] [Cited by in RCA: 14767] [Article Influence: 2953.4] [Reference Citation Analysis (0)] |
| 30. | Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, Diaz G, Cohn A, Fox L, Patel A, Gerber SI, Kim L, Tong S, Lu X, Lindstrom S, Pallansch MA, Weldon WC, Biggs HM, Uyeki TM, Pillai SK; Washington State 2019-nCoV Case Investigation Team. First Case of 2019 Novel Coronavirus in the United States. N Engl J Med. 2020;382:929-936. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4155] [Cited by in RCA: 3822] [Article Influence: 764.4] [Reference Citation Analysis (1)] |
| 31. | Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS; China Medical Treatment Expert Group for Covid-19. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382:1708-1720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19202] [Cited by in RCA: 18877] [Article Influence: 3775.4] [Reference Citation Analysis (7)] |
| 32. | Ebert EC. Hypoxic liver injury. Mayo Clin Proc. 2006;81:1232-1236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 92] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 33. | Cai Q, Huang D, Yu H, Zhu Z, Xia Z, Su Y, Li Z, Zhou G, Gou J, Qu J, Sun Y, Liu Y, He Q, Chen J, Liu L, Xu L. COVID-19: Abnormal liver function tests. J Hepatol. 2020;73:566-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 623] [Cited by in RCA: 661] [Article Influence: 132.2] [Reference Citation Analysis (0)] |
| 34. | Zhang Y, Zheng L, Liu L, Zhao M, Xiao J, Zhao Q. Liver impairment in COVID-19 patients: A retrospective analysis of 115 cases from a single centre in Wuhan city, China. Liver Int. 2020;40:2095-2103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 310] [Article Influence: 62.0] [Reference Citation Analysis (0)] |
| 35. | Fan Z, Chen L, Li J, Cheng X, Yang J, Tian C, Zhang Y, Huang S, Liu Z, Cheng J. Clinical Features of COVID-19-Related Liver Functional Abnormality. Clin Gastroenterol Hepatol. 2020;18:1561-1566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 559] [Cited by in RCA: 556] [Article Influence: 111.2] [Reference Citation Analysis (0)] |
| 36. | Abe K, Yamamoto T, Matsumoto K, Kikuchi K, Miura R, Tachizawa N, Asaoka Y, Takezawa T, Matsunaga N, Obi S, Tanaka A. Clinical Features and Liver Injury in Patients with COVID-19 in the Japanese Population. Intern Med. 2020;59:2353-2358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 37. | Medetalibeyoglu A, Catma Y, Senkal N, Ormeci A, Cavus B, Kose M, Bayramlar OF, Yildiz G, Akyuz F, Kaymakoglu S, Tukek T. The effect of liver test abnormalities on the prognosis of COVID-19. Ann Hepatol. 2020;19:614-621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 38. | Bernal-Monterde V, Casas-Deza D, Letona-Giménez L, de la Llama-Celis N, Calmarza P, Sierra-Gabarda O, Betoré-Glaria E, Martínez-de Lagos M, Martínez-Barredo L, Espinosa-Pérez M, M Arbones-Mainar J. SARS-CoV-2 Infection Induces a Dual Response in Liver Function Tests: Association with Mortality during Hospitalization. Biomedicines. 2020;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 39. | Aziz M, Fatima R, Lee-Smith W, Assaly R. The association of low serum albumin level with severe COVID-19: a systematic review and meta-analysis. Crit Care. 2020;24:255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 130] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 40. | Huang J, Cheng A, Kumar R, Fang Y, Chen G, Zhu Y, Lin S. Hypoalbuminemia predicts the outcome of COVID-19 independent of age and co-morbidity. J Med Virol. 2020;92:2152-2158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 210] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 41. | Wang Z, Yang B, Li Q, Wen L, Zhang R. Clinical Features of 69 Cases With Coronavirus Disease 2019 in Wuhan, China. Clin Infect Dis. 2020;71:769-777. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 557] [Cited by in RCA: 702] [Article Influence: 140.4] [Reference Citation Analysis (0)] |
| 42. | Wang R, Pan M, Zhang X, Han M, Fan X, Zhao F, Miao M, Xu J, Guan M, Deng X, Chen X, Shen L. Epidemiological and clinical features of 125 Hospitalized Patients with COVID-19 in Fuyang, Anhui, China. Int J Infect Dis. 2020;95:421-428. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 148] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 43. | Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J, Fan Y, Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20:425-434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2493] [Cited by in RCA: 2310] [Article Influence: 462.0] [Reference Citation Analysis (0)] |
| 44. | Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW; the Northwell COVID-19 Research Consortium; Barnaby DP, Becker LB, Chelico JD, Cohen SL, Cookingham J, Coppa K, Diefenbach MA, Dominello AJ, Duer-Hefele J, Falzon L, Gitlin J, Hajizadeh N, Harvin TG, Hirschwerk DA, Kim EJ, Kozel ZM, Marrast LM, Mogavero JN, Osorio GA, Qiu M, Zanos TP. Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA. 2020;323:2052-2059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6024] [Cited by in RCA: 6518] [Article Influence: 1303.6] [Reference Citation Analysis (0)] |
| 45. | Li J, Fan JG. Characteristics and Mechanism of Liver Injury in 2019 Coronavirus Disease. J Clin Transl Hepatol. 2020;8:13-17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 199] [Cited by in RCA: 206] [Article Influence: 41.2] [Reference Citation Analysis (2)] |
| 46. | Boeckmans J, Rodrigues RM, Demuyser T, Piérard D, Vanhaecke T, Rogiers V. COVID-19 and drug-induced liver injury: a problem of plenty or a petty point? Arch Toxicol. 2020;94:1367-1369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 47. | Hussman JP. Cellular and Molecular Pathways of COVID-19 and Potential Points of Therapeutic Intervention. Front Pharmacol. 2020;11:1169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 106] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 48. | Wang Y, Liu S, Liu H, Li W, Lin F, Jiang L, Li X, Xu P, Zhang L, Zhao L, Cao Y, Kang J, Yang J, Li L, Liu X, Li Y, Nie R, Mu J, Lu F, Zhao S, Lu J, Zhao J. SARS-CoV-2 infection of the liver directly contributes to hepatic impairment in patients with COVID-19. J Hepatol. 2020;73:807-816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 353] [Cited by in RCA: 457] [Article Influence: 91.4] [Reference Citation Analysis (0)] |
| 49. | Li MY, Li L, Zhang Y, Wang XS. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty. 2020;9:45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1028] [Cited by in RCA: 1068] [Article Influence: 213.6] [Reference Citation Analysis (0)] |
| 50. | Chai XQ, Hu LF, Zhang Y, Han WY, Lu Z, Ke AW, Zhou J, Shi GM, Fang N, Fan J, Cai JB, Lan F. Specific ACE2 Expression in Cholangiocytes May Cause Liver Damage After 2019-nCoV Infection. 2020 Preprint. Available from: bioRxiv. [DOI] [Full Text] |
| 51. | Boettler T, Newsome PN, Mondelli MU, Maticic M, Cordero E, Cornberg M, Berg T. Care of patients with liver disease during the COVID-19 pandemic: EASL-ESCMID position paper. JHEP Rep. 2020;2:100113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 312] [Cited by in RCA: 334] [Article Influence: 66.8] [Reference Citation Analysis (1)] |
| 52. | Chu H, Hu B, Huang X, Chai Y, Zhou D, Wang Y, Shuai H, Yang D, Hou Y, Zhang X, Yuen TT, Cai JP, Zhang AJ, Zhou J, Yuan S, To KK, Chan IH, Sit KY, Foo DC, Wong IY, Ng AT, Cheung TT, Law SY, Au WK, Brindley MA, Chen Z, Kok KH, Chan JF, Yuen KY. Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat Commun. 2021;12:134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 123] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 53. | Wang S, Qiu Z, Hou Y, Deng X, Xu W, Zheng T, Wu P, Xie S, Bian W, Zhang C, Sun Z, Liu K, Shan C, Lin A, Jiang S, Xie Y, Zhou Q, Lu L, Huang J, Li X. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31:126-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 341] [Cited by in RCA: 368] [Article Influence: 92.0] [Reference Citation Analysis (0)] |
| 54. | Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, van der Meer F, Kallio K, Kaya T, Anastasina M, Smura T, Levanov L, Szirovicza L, Tobi A, Kallio-Kokko H, Österlund P, Joensuu M, Meunier FA, Butcher SJ, Winkler MS, Mollenhauer B, Helenius A, Gokce O, Teesalu T, Hepojoki J, Vapalahti O, Stadelmann C, Balistreri G, Simons M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856-860. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1507] [Cited by in RCA: 1380] [Article Influence: 276.0] [Reference Citation Analysis (0)] |
| 55. | Daly JL, Simonetti B, Klein K, Chen KE, Williamson MK, Antón-Plágaro C, Shoemark DK, Simón-Gracia L, Bauer M, Hollandi R, Greber UF, Horvath P, Sessions RB, Helenius A, Hiscox JA, Teesalu T, Matthews DA, Davidson AD, Collins BM, Cullen PJ, Yamauchi Y. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861-865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 669] [Cited by in RCA: 937] [Article Influence: 187.4] [Reference Citation Analysis (0)] |
| 56. | Clausen TM, Sandoval DR, Spliid CB, Pihl J, Perrett HR, Painter CD, Narayanan A, Majowicz SA, Kwong EM, McVicar RN, Thacker BE, Glass CA, Yang Z, Torres JL, Golden GJ, Bartels PL, Porell RN, Garretson AF, Laubach L, Feldman J, Yin X, Pu Y, Hauser BM, Caradonna TM, Kellman BP, Martino C, Gordts PLSM, Chanda SK, Schmidt AG, Godula K, Leibel SL, Jose J, Corbett KD, Ward AB, Carlin AF, Esko JD. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020; 183: 1043-1057. e15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 877] [Cited by in RCA: 879] [Article Influence: 175.8] [Reference Citation Analysis (0)] |
| 57. | Ulrich H, Pillat MM. CD147 as a Target for COVID-19 Treatment: Suggested Effects of Azithromycin and Stem Cell Engagement. Stem Cell Rev Rep. 2020;16:434-440. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 259] [Cited by in RCA: 290] [Article Influence: 58.0] [Reference Citation Analysis (0)] |
| 58. | Zhou YG, Fu BQ, Zheng XH, Wang DS, Zhao CC, Qi YJ, Sun R, Tian ZG, Xu XL, Wei HM. Aberrant pathogenic GM-CSF+ T cells and inflammatory CD14+CD16+ monocytes in severe pulmonary syndrome patients of a new coronavirus. 2020 Preprint. Available from: bioRxiv. [DOI] [Full Text] |
| 59. | Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054-1062. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17476] [Cited by in RCA: 18200] [Article Influence: 3640.0] [Reference Citation Analysis (0)] |
| 60. | Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, Wang T, Zhang X, Chen H, Yu H, Zhang M, Wu S, Song J, Chen T, Han M, Li S, Luo X, Zhao J, Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620-2629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2835] [Cited by in RCA: 3418] [Article Influence: 683.6] [Reference Citation Analysis (0)] |
| 61. | Herold T, Jurinovic V, Arnreich C, Lipworth BJ, Hellmuth JC, von Bergwelt-Baildon M, Klein M, Weinberger T. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J Allergy Clin Immunol. 2020;146:128-136.e4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 696] [Cited by in RCA: 713] [Article Influence: 142.6] [Reference Citation Analysis (0)] |
| 62. | Luo P, Liu Y, Qiu L, Liu X, Liu D, Li J. Tocilizumab treatment in COVID-19: A single center experience. J Med Virol. 2020;92:814-818. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 834] [Cited by in RCA: 876] [Article Influence: 175.2] [Reference Citation Analysis (0)] |
| 63. | RECOVERY Collaborative Group, Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, Staplin N, Brightling C, Ustianowski A, Elmahi E, Prudon B, Green C, Felton T, Chadwick D, Rege K, Fegan C, Chappell LC, Faust SN, Jaki T, Jeffery K, Montgomery A, Rowan K, Juszczak E, Baillie JK, Haynes R, Landray MJ. Horby Dexamethasone in Hospitalized Patients with Covid-19. N Engl J Med. 2021;384:693-704. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6762] [Cited by in RCA: 7389] [Article Influence: 1847.3] [Reference Citation Analysis (1)] |
| 64. | Fajgenbaum DC, June CH. Cytokine Storm. N Engl J Med. 2020;383:2255-2273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1324] [Cited by in RCA: 2103] [Article Influence: 420.6] [Reference Citation Analysis (0)] |
| 65. | Li YP, Xie ZW, Lin WY, Cai WP, Wen CY, Guan YJ, Mo XN, Wang J, Wang YP, Peng P, Chen XD, Hong WX, Xiao GM, Liu JX, Zhang LG, Hu FY, Li F, Zhang FC, Deng XL, Li LH. An exploratory randomized controlled study on the efficacy and safety of lopinavir/ritonavir or arbidol treating adult patients hospitalized with mild/moderate COVID-19 (ELACOI). 2020 Preprint. Available from: medRxiv. [DOI] [Full Text] |
| 66. | World Health Organization. Infection prevention and control during health care when novel coronavirus (nCoV) infection is suspected: interim guidance. [cited 25 January 2020]. Available from: https://apps.who.int/iris/handle/10665/330674. |
| 67. | Wu J, Yu J, Shi X, Li W, Song S, Zhao L, Zhao X, Liu J, Wang D, Liu C, Huang B, Meng Y, Jiang B, Deng Y, Cao H, Li L. Epidemiological and clinical characteristics of 70 cases of coronavirus disease and concomitant hepatitis B virus infection: A multicentre descriptive study. J Viral Hepat. 2021;28:80-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 68. | de la Rica R, Borges M, Aranda M, Del Castillo A, Socias A, Payeras A, Rialp G, Socias L, Masmiquel L, Gonzalez-Freire M. Low Albumin Levels Are Associated with Poorer Outcomes in a Case Series of COVID-19 Patients in Spain: A Retrospective Cohort Study. Microorganisms. 2020;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 69. | Wang T, Chen R, Liu C, Liang W, Guan W, Tang R, Tang C, Zhang N, Zhong N, Li S. Attention should be paid to venous thromboembolism prophylaxis in the management of COVID-19. Lancet Haematol. 2020;7:e362-e363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 258] [Cited by in RCA: 273] [Article Influence: 54.6] [Reference Citation Analysis (0)] |
| 70. | Ranucci M, Ballotta A, Di Dedda U, Bayshnikova E, Dei Poli M, Resta M, Falco M, Albano G, Menicanti L. The procoagulant pattern of patients with COVID-19 acute respiratory distress syndrome. J Thromb Haemost. 2020;18:1747-1751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 679] [Cited by in RCA: 680] [Article Influence: 136.0] [Reference Citation Analysis (0)] |
| 71. | Spiezia L, Boscolo A, Poletto F, Cerruti L, Tiberio I, Campello E, Navalesi P, Simioni P. COVID-19-Related Severe Hypercoagulability in Patients Admitted to Intensive Care Unit for Acute Respiratory Failure. Thromb Haemost. 2020;120:998-1000. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 343] [Cited by in RCA: 433] [Article Influence: 86.6] [Reference Citation Analysis (1)] |
| 72. | Middeldorp S, Coppens M, van Haaps TF, Foppen M, Vlaar AP, Müller MCA, Bouman CCS, Beenen LFM, Kootte RS, Heijmans J, Smits LP, Bonta PI, van Es N. Incidence of venous thromboembolism in hospitalized patients with COVID-19. J Thromb Haemost. 2020;18:1995-2002. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1017] [Cited by in RCA: 1093] [Article Influence: 218.6] [Reference Citation Analysis (0)] |
| 73. | Iavarone M, D'Ambrosio R, Soria A, Triolo M, Pugliese N, Del Poggio P, Perricone G, Massironi S, Spinetti A, Buscarini E, Viganò M, Carriero C, Fagiuoli S, Aghemo A, Belli LS, Lucà M, Pedaci M, Rimondi A, Rumi MG, Invernizzi P, Bonfanti P, Lampertico P. High rates of 30-day mortality in patients with cirrhosis and COVID-19. J Hepatol. 2020;73:1063-1071. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 276] [Article Influence: 55.2] [Reference Citation Analysis (2)] |
| 74. | Loffredo L, Pastori D, Farcomeni A, Violi F. Effects of Anticoagulants in Patients With Cirrhosis and Portal Vein Thrombosis: A Systematic Review and Meta-analysis. Gastroenterology 2017; 153: 480-487. e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 298] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 75. | Williamson EJ, Walker AJ, Bhaskaran K, Bacon S, Bates C, Morton CE, Curtis HJ, Mehrkar A, Evans D, Inglesby P, Cockburn J, McDonald HI, MacKenna B, Tomlinson L, Douglas IJ, Rentsch CT, Mathur R, Wong AYS, Grieve R, Harrison D, Forbes H, Schultze A, Croker R, Parry J, Hester F, Harper S, Perera R, Evans SJW, Smeeth L, Goldacre B. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430-436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4343] [Cited by in RCA: 4209] [Article Influence: 841.8] [Reference Citation Analysis (0)] |
| 76. | Liu R, Zhao L, Cheng X, Han H, Li C, Li D, Liu A, Gao G, Zhou F, Liu F, Jiang Y, Zhu C, Xia Y. Clinical characteristics of COVID-19 patients with hepatitis B virus infection - a retrospective study. Liver Int. 2021;41:720-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 77. | Varona Pérez J, Rodriguez Chinesta JM. [Risk of hepatitis B reactivation associated with treatment against SARS-CoV-2 (COVID-19) with corticosteroids]. Rev Clin Esp (Barc). 2020;220:534-536. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 78. | Prins GH, Olinga P. Potential implications of COVID-19 in non-alcoholic fatty liver disease. Liver Int. 2020;40:2568. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 79. | Sivaraj V, Pittrof R, Davies O, Kulasegaram R. Homelessness among an inpatient HIV-positive cohort at a tertiary care hospital in central London. Int J STD AIDS. 2020;31:705-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |