Published online Jul 7, 2021. doi: 10.3748/wjg.v27.i25.3837
Peer-review started: March 11, 2021
First decision: April 5, 2021
Revised: April 14, 2021
Accepted: May 21, 2021
Article in press: May 21, 2021
Published online: July 7, 2021
Processing time: 116 Days and 19.1 Hours
Obesity is a major global health problem determined by heredity and environ
Core Tip: Obesity is closely related to the gut microbiota. The study of the gut microbiome provides a basis for the reconstruction of the gut microbiota of obese patients. Here, we discuss the characteristics of the gut microbiota in obesity, the mechanism by which the gut microbiota induces obesity, and the relationships between genetic and environmental factors and the gut microbiota in obesity.
- Citation: Liu BN, Liu XT, Liang ZH, Wang JH. Gut microbiota in obesity. World J Gastroenterol 2021; 27(25): 3837-3850
- URL: https://www.wjgnet.com/1007-9327/full/v27/i25/3837.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i25.3837
Obesity is a complex metabolic disorder caused by a variety of genetic and nongenetic factors (such as environmental factors). The World Health Organization defines obesity as having a body mass index (BMI) greater than 30, but the definition varies from country to country. In China, for example, a BMI of 28 or greater is considered obese. A comprehensive analysis shows that approximately one-third of the world’s population are overweight, and approximately 10% are obese[1]. It is predicted that by 2030, the number of obese people worldwide will reach 1.12 billion[2]. The health risk of obesity has caused widespread concern and has become an important global health problem. Obesity not only manifests as changes in appearance but is also associated with lipid and glucose metabolism disorders, chronic inflammation, oxidative stress, and an increased risk of a variety of diseases, most notably cardiovascular disease, diabetes, and cancer[3,4]. In recent years, increasing evidence has shown that an imbalance in the gut microbiota may be a factor leading to obesity[5,6].
Up to 100 trillion symbiotic microbes live in the gut, called the gut microbiota, which comprises 10 times the number of cells in the body itself[7]. The gut microbiota relies on food residues that the human body does not digest, mucus secreted by the gut, and dead cells that are shed as nutrients to maintain its high population levels[8]. The active gut microbiota will produce a large number of physiologically active substances, including short-chain fatty acids, vitamins, and health-beneficial products such as anti-inflammatory, analgesic, and antioxidant products, along with potentially harmful products such as neurotoxins, carcinogens, and immunotoxins[9,10]. These products can enter the blood, directly regulate the expression of genes, and affect human immune and metabolic processes. Therefore, a healthy gut microbiota is essential for maintaining the body’s metabolism and energy balance. An imbalance in the gut microbiota can cause metabolic disorders and increase central appetite, leading to obesity. This article reviews the research progress of the relationship between the gut microbiota and obesity.
PubMed (https://pubmed.ncbi.nlm.nih.gov/) was used for search with the following keywords: Obesity, gut microbiota, dysbiosis, energy absorption, appetite, fat storage, chronic inflammation, and circadian rhythm. More than 4000 published papers inclu
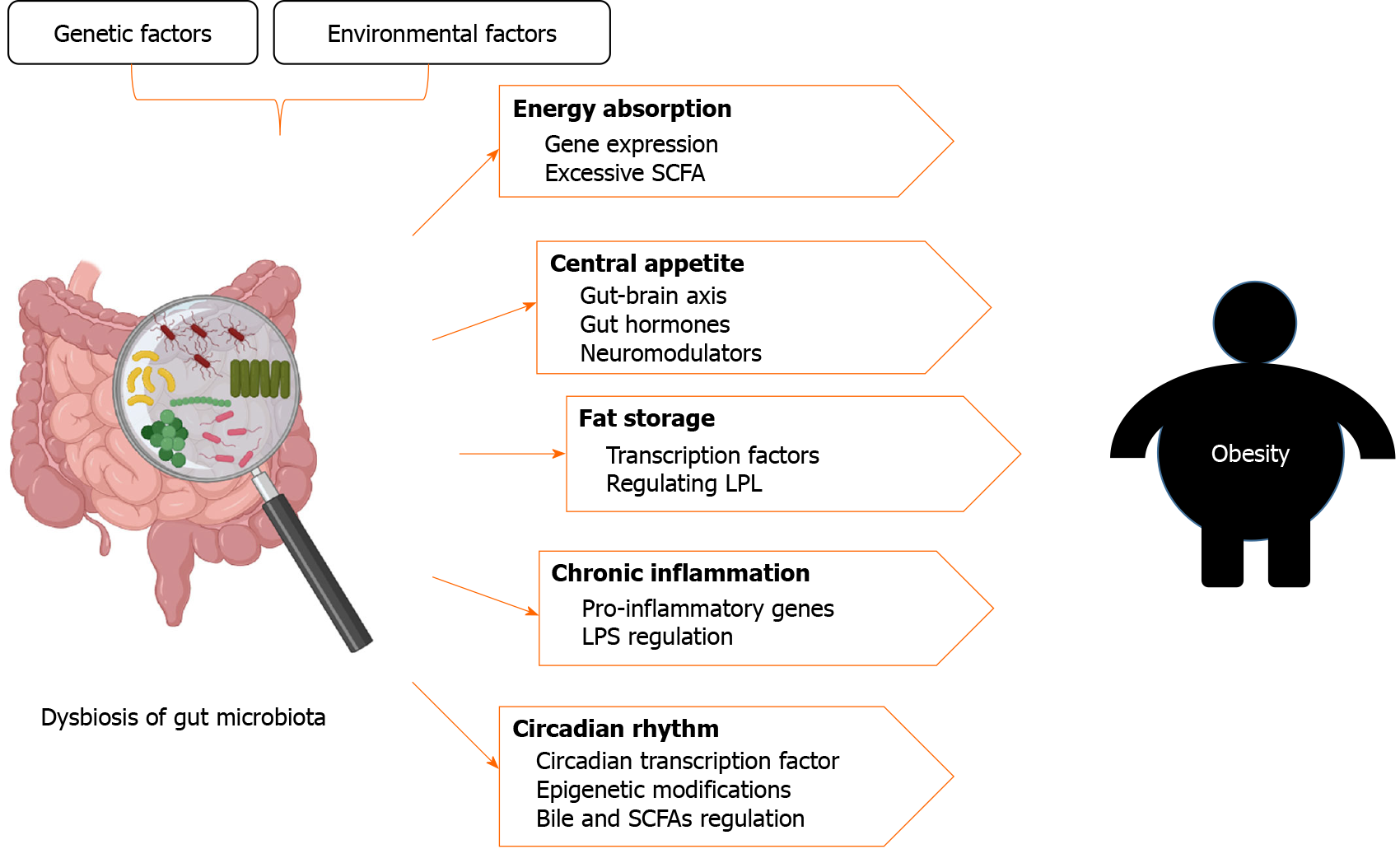
After the search, we summarize some characteristics of the gut microbiota of obese patients. Many results suggest that an increased Firmicutes/Bacteroidetes ratio at the phylum level is an important feature of the gut microbiota in obesity. The family Christensenellaceae and the genera Methanobacteriales, Lactobacillus, Bifidobacteria, and Akkermansia are usually considered as probiotics, and their relative abundance is often inversely associated with obesity. Gut microbiota regulates obesity by regulating energy absorption, central appetite, fat storage, chronic inflammation, and circadian rhythms. Finally, the effects of genetic and environmental factors on gut microbiota in obesity are discussed.
The development of the Human Microbiome Project has promoted the in-depth study of the gut microbiota[11,12]. With the application of metagenomic and 16S ribosomal RNA gene sequencing, the diversity of the gut ecosystem has been widely studied, which enables us to accurately understand the composition and function of the gut microbiota[13]. So far, the gut microbes of 6457 taxa have been collected in the gutMEGA database[14]. The normal gut microbiota of the human body is mainly composed of Firmicutes, Bacteroides, Proteus, Actinomycetes, Fusobacteria, and Verrucomi
The hypothesis that the gut microbiota may be a relevant environmental factor in obesity has led to the study of the gut microbiomes of obese individuals. The first evidence for a link between the gut microbiota and obesity was from studies of germ-free mice. Transplanting gut microbes from conventionally raised mice into germ-free mice increased the transplants’ fat content and insulin resistance levels even with reduced food intake, which proved that gut microbes can increase the accumulation of adipose tissue in the host[19]. Furthermore, 16S rRNA gene sequencing showed that obesity may be associated with two dominant bacterial phyla: Firmicutes and Bacteroidetes. The gut microbiota of obese mice showed a 50% decrease in the abundance of Bacteroidetes and a proportional increase in Firmicutes[20]. Turnbaugh et al[21] further confirmed that the Firmicutes/Bacteroidetes ratio increased signi
Other works have associated obesity with specific bacteria, such as the family Christensenellaceae and the genera Methanobacteriales, Lactobacillus, Bifidobacteria, and Akkermansia. Recently, the family Christensenellaceae was found to be associated with weight loss, and its relative abundance was inversely related to host BMI[26]. Akkermansia muciniphila is a key bacterium for weight loss. Supplementation with A. muciniphila improves metabolic parameters in overweight and obese subjects[27]. Lactobacillus and Bifidobacterium are traditional probiotics that play an important role in the balance of the human intestinal microecology. Crovesy et al[28] summarized the effect of Lactobacillus on body weight in overweight subjects and found that the beneficial effects were species specific. The abundance of Lactobacillus paracasei (L. paracasei) was negatively correlated with obesity, while the abundances of L. reuteri and L. gasseri were significantly correlated with obesity. At present, evidence of Bifidobacterium resistance to obesity has been obtained from animal experiments. Following administration of Bifidobacterium in animal models of diet-induced obesity, it also showed a strain-dependent effect on obesity[29]. However, reduced Bifidobacterium abundance in the gut is associated with obesity[30]. Million et al[31] found that M. smithii and B. animalis were associated with normal weight, while L. reuteri was associated with obesity. These findings suggest that obesity-related microorganisms are species specific and that bacteria in the same genus may have opposite effects, which may be related to the complex metabolic mechanism of obesity.
The persons with obesity can be divided into two different types: Subcutaneous obesity and visceral obesity. As mentioned above, an elevated Firmicutes/Bac
From the above analysis, the role of microorganisms in obesity is strain specific, as there are both beneficial and harmful bacteria within the same taxon. Therefore, it is difficult to classify obesity-related bacterial communities according to their taxonomic relationships. Recent studies have introduced the concept of a ‘guild’ into the study of the gut microbiota, defining a group of microorganisms that utilize similar resources or perform the same biological function as the same guild to identify potential clusters associated with specific disease phenotypes and identify candidate gut microbes that may contribute to human health and disease[37]. Using a clinical study of genetically obese children, the researchers found that the abundance of one strain of E. coli increased sharply after 30 d of eating a diet rich in nondigested carbohydrates, while the abundance levels of the other four strains decreased[37]. Therefore, guilds, as an important form of intermember organization in ecology, can better reflect the changes in specific responses of strains.
The diversity of the gut microbiota is another important factor related to obesity. Most studies have shown that the diversity and richness of the gut microbiome are reduced in obese subjects[38-40]. Wu et al[25] found that the α-diversity of obese people was significantly lower than that of healthy people, but there was no significant difference in the structure of the gut microbiota (β-diversity) between them. However, some studies believed that there was not necessarily a correlation between the diversity of the gut microbiota and diseases[41]. Strict statistical analysis showed that only in approximately 1/3 of the cases was there a significant relationship between bacterial diversity and “microbiota-related diseases”. Since bacterial ecosystems may possess a considerable degree of stability (including resistance to disease), the impression seems to be that bacterial diversity is closely related to disease occurrence and progression[41]. Obesity, as a flora-related disease without specific pathogens, may be directly related to the dysbiosis of the bacterial ecosystem. Therefore, in the face of the dilemma of diversity research, ecological theory is needed for guidance. The association between gut microbiomes and obesity has been summarized in Table 1.
| Microbiota characteristics in obesity | Preclinical or clinical | Study subjects | Ref. |
| Firmicutes/Bacteroidetes ratio increased | Preclinical | Mice | Ley et al[20], Turnbaugh et al[21] |
| Clinical | Childhood | Indiani et al[22] | |
| Clinical | Adult ukrainian population | Koliada et al[23] | |
| The relative abundance of Christensenellaceae was inversely related to host BMI | Clinical | Human | Waters et al[26] |
| Clinical | Postmenopausal women | Alemán et al[33] | |
| Clinical | Italian elderly | Tavella et al[36] | |
| Increased Akkermansia population reduced body weight | Clinical | Human | Depommier et al[27] |
| Preclinical | Mice | Anhê et al[34] | |
| Lactobacillus paracasei decreased, while Lactobacillus reuteri and Lactobacillus gasseri increased | Clinical | Human | Crovesy et al[28], Million et al[31] |
| Bifidobacteria reduced | Preclinical | Rats | Waldram et al[30] |
| Methanobacteriales smithii and Bifidobacterium were associated with normal weight | Clinical | Human | Million et al[31] |
Studies have found that genetically obese mice consume more carbohydrates and protein through the gut microbiota to provide energy for the host[42,43]. In animal experiments, it was found that in the case of no differences in diet or weight of the mice, the total body fat of the germ-free mice colonized by the ‘obese microbiota’ increased significantly compared with those colonized by the ‘lean microbiota’[21]. This finding indicates that the obese-type gut microbiota has an increased ability to absorb energy from the diet. Multiomics analysis showed increased lipid absorption in obese hosts. Colonization of Clostridia in germ-free mice downregulated the genes that control lipid absorption[44]. Therefore, the gut microbiota of obese patients can promote the absorption of energy, resulting in excessive accumulation of energy and increased weight gain.
The gut microbiota ferments hard-to-digest carbohydrates into short-chain fatty acids (SCFAs), which are either absorbed by the gut or excreted in feces. SCFAs are crucial for energy homeostasis regulation[45]. SCFAs are composed mainly of acetate, propionate, and butyrate. Acetate can produce beneficial effects on the energy metabolism of the host by secreting glucagon-like peptide-1, peptide YY, and other intestinal hormones, reduce systemic lipolysis and proinflammatory cytokine levels, and increase energy consumption and lipid oxidation[46]. Propionate promotes intestinal lipolysis and energy homeostasis in mice through the AMPK/LSD1 pathway[47]. Butyrate is the colon’s main energy source, and intestinal epithelial cells derive most of their energy from the oxidation of butyrate. The increase in butyrate-producing bacteria in the gut microbiota increases the production of butyrate, thereby improving lipid metabolism through the butyrate-SESN2/CRTC2 pathway[48]. However, excessive butyrate may reduce the proportion of probiotics and reverse the metabolic effects[48]. In recent years, SCFAs have attracted special attention due to their beneficial effects on intestinal homeostasis and energy metabolism, but their role in obesity remains controversial. Higher concentrations of fecal SCFAs are associated with gut permeability, metabolic disorder markers, obesity, and hypertension[49]. Teixeira et al[50] found that fecal SCFAs in women were positively correlated with obesity, waist circumference, and other indicators of metabolic syndrome. Overall, SCFAs seem to be a double-edged sword. Although they can protect the host from diet-induced obesity, excessive SCFAs provide extra energy for the host, thus promo
In recent years, the microbiota has emerged as one of the key regulators of gut-brain function. This gut-brain axis has received increasing attention in the study of the biological and physiological bases of obesity and its related diseases. The microbiota and the brain communicate with each other through a variety of pathways, including endocrine, immune, and neural pathways[51]. The gut microbiota affects the host’s central nervous system through the gut-brain axis. The central nervous system can also affect the composition and structure of the gut microbiota. The gut microbiota influences food intake by regulating brain function in a number of ways, such as by contributing to the production of neuromodulators, such as serotonin, which plays an important role in regulating gastrointestinal function[52]. Lactate produced by Lactobacillus and Bifidobacterium, which acts as a substrate for neuron cells, can prolong satiety after a meal[53]. The gut microbiota also participates in the gut-brain axis by regula
In 2004, researchers reported the ability of the gut microbiota to regulate fat storage[19]. The gut microbiota increases the absorption of glucose in the host’s intestine and the content of glucose in the serum, thereby increasing the expression levels of two basic transcription factors, ChREBP (carbohydrate response element binding protein) and SREBP-1 (sterol regulatory element binding protein), which induce fat synthesis in the liver. Lipoprotein lipase (LPL) helps triglycerides enter the circulatory system from the liver, where they are absorbed by fat cells. Intestinal epithelial cells can produce Fiaf, an inhibitor of LPL. Fiaf is selectively inhibited in normal mouse intestinal epithelial cells, thereby increasing the host’s energy storage. Aronsson et al[59] found that L. paracasei regulated ANGPTL4, a central player in fat storage regulation, using a mouse model with high-fat diet-induced obesity. L. paracasei could induce the expression of ANGPTL4, partly through the peroxisomal proliferator-activated receptors α and γ. ANGPTL4 inhibited LPL, leading to decreased fat storage. L. paracasei-colonized mice resisted high-fat diet-induced obesity[60]. In vitro, L. paracasei inhibits the Akt/mTOR pathway, indicating that several regulatory pathways are involved in different intracellular lipid accumulations mediated by L. paracasei.
Chronic inflammation is one of the characteristics of metabolic disorders such as obesity[61]. Evidence shows that these disorders are characterized by the gut micro
SCFAs play a key role in the interaction between the gut microbiota and the activation or inhibition of the inflammatory cascade. Butyrate is an anti-inflammatory metabolite known to inhibit pathways leading to the production of pro-inflammatory cytokines[67]. Through epigenetic interactions, butyrate stimulates adipoliolysis and mitochondrial oxidative phosphorylation, thereby achieving greater energy consump
The gut microbiota has been shown to influence host circadian rhythms in a diet-dependent manner[71]. The interruption of the circadian rhythm may lead to an increase in the incidence of obesity[72]. Microorganisms regulate lipid uptake and storage by regulating the circadian transcription factor NFIL3. The study also found that the ILC3-STAT3 signaling pathway is a key molecular pathway for the interaction between the microbiota and the circadian clock. A recent study has shown that the gut microbiota programs rhythmic histone acetylation through HDAC3 (histone deacety
Metabolites of the gut microbiota can also affect the host rhythm system. Bile metabolism is an example of the rhythmic interaction between host and gut microbes. Microbial bile salt hydrolases are related to the regulation of the circadian clock and lipid metabolism-related genes[77]. Gut bacteria such as Lachnospiraceae, Clostri
The mechanism of obesity induced by the gut microbiota is summarized in Table 2. Dysbiosis of the microbiota causes metabolic disorders and promotes the occurrence and development of obesity through the direct interaction between the microbiota and local tissues or the indirect interaction between metabolites and remote organs (Figure 1). Although we can often detect specific different microorganisms between obese and normal individuals and verify the role of the bacteria in obesity through germ-free mouse experiments, more attention should be paid to ecological theory applications in relation to the gut microbiota as a group. The gut microbiota affects appetite, energy absorption, fat storage, circadian rhythm, and chronic inflammation, leading to obesity. Therefore, targeted reconstruction of the gut microbiota structure, such as through fecal bacterial transplantation, is one of the means of treating obesity.
| Effect | Microbiota characteristics | Mechanism | Ref. |
| Increased energy absorption | Expansion of Desulfovibrio and loss of Clostridia | Elevated the expression of genes that control lipid absorption such as CD36 | Petersen et al[44] |
| Extra energy for the host | The inverse association between fecal SCFAs and gut microbiota diversity; Faecalibacterium prausnitzii, Roseburia faecis, and other Clostridiales increased; Akkermansia muciniphila, Alistipes finegoldii, Bacteroides, Christensenellaceae, Methanobrevibacter, and Oscillospira decreased | Excessive SCFAs | de la Cuesta-Zuluaga et al[49] |
| Increased appetite | A community dominated by members of the Clostridial clusters XIVa and IV | The levels of peptide YY and GLP-1 in obese patients decrease significantly | Wu et al[54], Salehi et al[55], Federico et al[56] |
| Decreased Fat storage | Germ free mice colonized with Lactobacillus paracasei | Increase the expression of ANGPTL4, and inhibit LPL, leading to decreased fat storage | Aronsson et al[59], Tazi et al[60] |
| Increased fat storage | Transplanting gut microbes from conventionally raised mice into germ-free mice | Increasing the expression of ChREBP and SREBP-1, Fiaf is inhibited, activate LPL, help triglycerides enter the circulatory system from the liver | Bäckhed et al[19] |
| Decreased chronic inflammation | Increase levels in the butyrate-producing bacteria such as Ruminococcaceae and Lachnospiraceae | Inhibit pathways leading to the production of pro-inflammatory cytokines; Stimulate adipoliolysis and mitochondrial oxidative phosphorylation, thereby achieving greater energy consumption; Reduce LPS, thereby reducing chronic low-grade inflammation | Kang et al[66], Lührs et al[67], Jia et al[68] |
| Interruption of circadian rhythm | Bile salts biotransformation bacteria such as Lachnospiraceae, Clostridiaceae, Ruminococcaceae, Lactobacillus, Bacteroides, and Bifidobacterium | Regulate transcription of key genes involved in circadian rhythm (Dbp, Per1/2) and lipid metabolism | Joyce et al[77], Parkar et al[78] |
Obesity is the result of interactions between genetic and environmental factors. It is not surprising that host genetics shapes the gut microbiota. In fact, several genetic variations explain differences in the composition and diversity of the gut microbiota in obese people. Whole-genome association was used to determine the relationship between the genetic variation of twins and different species of bacteria. More than 12 health-related gut microorganisms were identified[80]. These microorganisms are environment-acquired, but genes also have a certain impact on them, such as the associations between Bifidobacterium and the lactase gene locus[80] and AMY1-CN (CN variation of the AMY1 Locus, which encodes salivary amylase) as a genetic factor related to the composition and function of the microbiome[81]. The abundance of resistant starch-degrading microbes in the gut microbiota of high-AMY1-CN subjects increased, which resulted in a higher incidence of obesity after the microbes were transferred to germ-free mice[81]. In another study, host kinship indicated the structural similarity of the gut microbiota in wild house mice. The exon sequencing results of host genes were compared with the microbiota structure, and 20 host genes were found to be related to the diversity or abundance of the gut microbiota, including a cytokine IL12A gene with three nonconsensus mutations. Among the 20 related genes, there are a large number of homologous genes of human-microbiota interac
In addition, the gut microbiota can be transmitted from mother to child. In a passage experiment with germ-free mice, it was found that the gut microbiota of these mice was very stable. These microbiota types accounted for the majority of the gut microbiota of the mice in the normal environment, which proved that most of the gut microbiota of mice came from vertical transmission from the mother[84]. Studies have shown that the microbiota exists in the placenta, amniotic fluid, umbilical cord blood, and meconium, and maternal microorganisms may play an important role in the establishment of the child’s microbiota[85]. Therefore, maternal obesity during pregnancy is accompanied by dysbiosis of the gut microbiota and metabolic disorders, and the maternal microbiota is passed on to the child, which can lead to metabolic disorders in the child. Obesity-related bacteria are present in the placenta and amniotic fluid in obese mothers. The fetus swallows these bacteria, resulting in gut microbiota colonization in the uterus. Studies have confirmed that maternal obesity is related to dysbiosis of the gut microbiota in offspring[86]. Therefore, the increased risk of obesity in children with obese mothers can be partly explained by the spread of gut microbes from obese mothers to their offspring.
Although genes play important roles in the gut microbiota, environmental factors have a more significant impact. A study of 1000 Israelis from all over the world who have very similar eating habits and lifestyles found that their ancestry was not related to the microbiome. The overall heritability of the microbiome may be less than 2%, and more than 20% of the variation in the microbiome is related to diet, drugs, and anthropometric measurements[87].
Diet is one of the most important factors inducing obesity. The eating habits of developed countries and regions have gradually become characterized by high fat and high sugar consumption, which has contributed to the gradual increase in obesity. Gut microbes depend on the host diet to survive and harvest energy, and dietary changes have a great impact on the gut microbiota. For example, the abundance of Bacteroidetes decreased while that of Firmicutes and Proteobacteria increased in mice fed a high-fat diet. These changes were observed in mice resistant to weight gain, suggesting that dietary fat has a direct effect on the microbiome[88,89]. Sleep disturbance is another cause of obesity. Lack of sleep leads to disrupted circadian rhythms, which can affect the gut microbiome and contribute to obesity. Chronic sleep fragmentation resulted in increased food intake and reversible changes in the gut microbiota, with increases in the abundance levels of Lactobacillaceae and Ruminococcus and a decrease in the abundance of Lactobacillaceae. These factors lead to systemic and visceral white adipose tissue inflammation and changes in insulin sensitivity[90]. Stress activates genes that affect metabolism and promotes the consumption of sweet and fatty foods, thus increasing appetite and contributing to obesity[91]. Stress significantly affects the microbiota-gut-brain axis at all stages of life[51]. During stress, independent of diet, the α-diversity of the gut microbiota increased, 50% of identified genera changed, and the abundance of Bacteroides decreased, while the abundance levels of less dominant taxa increased[92]. In addition, unhealthy lifestyle choices (sedentary, lack of exercise), emotional disorders, and drugs can also contribute to obesity. In short, as a “migrant”, the gut microbiota itself is obtained from the environment; thus, environmental factors have a greater impact on it and can affect the occurrence and development of obesity.
Although there are many factors contributing to obesity, the link between gut microbiota and obesity is widely accepted. In this review, the relationship between gut microbiota and obesity and the mechanism of gut microbiota inducing obesity are provided. However, the researches on gut microbiota in obesity are limited by the underrepresentation of individuals. This drawback limits the generality of these findings. Because of the specificity of the strains, microbes from the same genus may have opposite effects on obesity, which partly explains some of the contradictory results. In addition, this review only discusses the occurrence and development of obesity from the perspective of gut microbiota, and does not make further analysis of the relevant treatment plan.
Dysbiosis of the gut microbiota has been shown to be closely linked to obesity. Many gut microorganisms have been identified to be related to obesity. They induce the occurrence and development of obesity by increasing host energy absorption, increasing central appetite, enhancing fat storage, contributing to chronic inflammation, and regulating circadian rhythms. Due to the complexity and diversity of the gut microbiota, the mechanism by which the gut microbiota induces obesity still needs to be further studied. Obesity is the result of a combination of genetic and environmental factors. Data analysis based on larger samples to clarify the mechanism of the association between the gut microbiota and obesity, functional group studies of ecological significance to identify potential pathogenic members of the gut microbiota associated with obesity, and specific microbiota management for obese individuals will be the focus of future research.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Jin W, Shimizu Y, Soto-Montenegro M S-Editor: Zhang H L-Editor: Wang TQ P-Editor: Yuan YY
| 1. | GBD 2015 Obesity Collaborators, Afshin A, Forouzanfar MH, Reitsma MB, Sur P, Estep K, Lee A, Marczak L, Mokdad AH, Moradi-Lakeh M, Naghavi M, Salama JS, Vos T, Abate KH, Abbafati C, Ahmed MB, Al-Aly Z, Alkerwi A, Al-Raddadi R, Amare AT, Amberbir A, Amegah AK, Amini E, Amrock SM, Anjana RM, Ärnlöv J, Asayesh H, Banerjee A, Barac A, Baye E, Bennett DA, Beyene AS, Biadgilign S, Biryukov S, Bjertness E, Boneya DJ, Campos-Nonato I, Carrero JJ, Cecilio P, Cercy K, Ciobanu LG, Cornaby L, Damtew SA, Dandona L, Dandona R, Dharmaratne SD, Duncan BB, Eshrati B, Esteghamati A, Feigin VL, Fernandes JC, Fürst T, Gebrehiwot TT, Gold A, Gona PN, Goto A, Habtewold TD, Hadush KT, Hafezi-Nejad N, Hay SI, Horino M, Islami F, Kamal R, Kasaeian A, Katikireddi SV, Kengne AP, Kesavachandran CN, Khader YS, Khang YH, Khubchandani J, Kim D, Kim YJ, Kinfu Y, Kosen S, Ku T, Defo BK, Kumar GA, Larson HJ, Leinsalu M, Liang X, Lim SS, Liu P, Lopez AD, Lozano R, Majeed A, Malekzadeh R, Malta DC, Mazidi M, McAlinden C, McGarvey ST, Mengistu DT, Mensah GA, Mensink GBM, Mezgebe HB, Mirrakhimov EM, Mueller UO, Noubiap JJ, Obermeyer CM, Ogbo FA, Owolabi MO, Patton GC, Pourmalek F, Qorbani M, Rafay A, Rai RK, Ranabhat CL, Reinig N, Safiri S, Salomon JA, Sanabria JR, Santos IS, Sartorius B, Sawhney M, Schmidhuber J, Schutte AE, Schmidt MI, Sepanlou SG, Shamsizadeh M, Sheikhbahaei S, Shin MJ, Shiri R, Shiue I, Roba HS, Silva DAS, Silverberg JI, Singh JA, Stranges S, Swaminathan S, Tabarés-Seisdedos R, Tadese F, Tedla BA, Tegegne BS, Terkawi AS, Thakur JS, Tonelli M, Topor-Madry R, Tyrovolas S, Ukwaja KN, Uthman OA, Vaezghasemi M, Vasankari T, Vlassov VV, Vollset SE, Weiderpass E, Werdecker A, Wesana J, Westerman R, Yano Y, Yonemoto N, Yonga G, Zaidi Z, Zenebe ZM, Zipkin B, Murray CJL. Health effects of overweight and obesity in 195 countries over 25 years. N Engl J Med. 2017;377:13-27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5669] [Cited by in RCA: 5066] [Article Influence: 633.3] [Reference Citation Analysis (2)] |
| 2. | Kelly T, Yang W, Chen CS, Reynolds K, He J. Global burden of obesity in 2005 and projections to 2030. Int J Obes (Lond). 2008;32:1431-1437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1843] [Cited by in RCA: 2072] [Article Influence: 121.9] [Reference Citation Analysis (2)] |
| 3. | Saltiel AR, Olefsky JM. Inflammatory mechanisms linking obesity and metabolic disease. J Clin Invest. 2017;127:1-4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 894] [Cited by in RCA: 1439] [Article Influence: 179.9] [Reference Citation Analysis (0)] |
| 4. | Bendor CD, Bardugo A, Pinhas-Hamiel O, Afek A, Twig G. Cardiovascular morbidity, diabetes and cancer risk among children and adolescents with severe obesity. Cardiovasc Diabetol. 2020;19:79. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 196] [Cited by in RCA: 180] [Article Influence: 36.0] [Reference Citation Analysis (1)] |
| 5. | Sonnenburg JL, Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1187] [Cited by in RCA: 1505] [Article Influence: 167.2] [Reference Citation Analysis (0)] |
| 6. | Gomes AC, Hoffmann C, Mota JF. The human gut microbiota: Metabolism and perspective in obesity. Gut Microbes. 2018;9:308-325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 304] [Article Influence: 43.4] [Reference Citation Analysis (2)] |
| 7. | Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915-1920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3394] [Cited by in RCA: 3545] [Article Influence: 177.3] [Reference Citation Analysis (5)] |
| 8. | Gentile CL, Weir TL. The gut microbiota at the intersection of diet and human health. Science. 2018;362:776-780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 457] [Cited by in RCA: 685] [Article Influence: 114.2] [Reference Citation Analysis (0)] |
| 9. | Miyamoto J, Igarashi M, Watanabe K, Karaki SI, Mukouyama H, Kishino S, Li X, Ichimura A, Irie J, Sugimoto Y, Mizutani T, Sugawara T, Miki T, Ogawa J, Drucker DJ, Arita M, Itoh H, Kimura I. Gut microbiota confers host resistance to obesity by metabolizing dietary polyunsaturated fatty acids. Nat Commun. 2019;10:4007. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 277] [Article Influence: 46.2] [Reference Citation Analysis (2)] |
| 10. | Canfora EE, Meex RCR, Venema K, Blaak EE. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat Rev Endocrinol. 2019;15:261-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 599] [Cited by in RCA: 933] [Article Influence: 155.5] [Reference Citation Analysis (1)] |
| 11. | NIH HMP Working Group; Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M. The NIH Human Microbiome Project. Genome Res. 2009;19:2317-2323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1410] [Cited by in RCA: 1388] [Article Influence: 86.8] [Reference Citation Analysis (0)] |
| 12. | Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project. Nature. 2019;569:641-648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 835] [Cited by in RCA: 776] [Article Influence: 129.3] [Reference Citation Analysis (0)] |
| 13. | Lepage P, Leclerc MC, Joossens M, Mondot S, Blottière HM, Raes J, Ehrlich D, Doré J. A metagenomic insight into our gut's microbiome. Gut. 2013;62:146-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 243] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 14. | Zhang Q, Yu K, Li S, Zhang X, Zhao Q, Zhao X, Liu Z, Cheng H, Liu ZX, Li X. gutMEGA: a database of the human gut MEtaGenome Atlas. Brief Bioinform. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 15. | Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207-214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9292] [Cited by in RCA: 8066] [Article Influence: 620.5] [Reference Citation Analysis (2)] |
| 16. | Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8:51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 865] [Cited by in RCA: 1091] [Article Influence: 121.2] [Reference Citation Analysis (0)] |
| 17. | Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480-484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6397] [Cited by in RCA: 5666] [Article Influence: 354.1] [Reference Citation Analysis (1)] |
| 18. | Bäckhed F, Fraser CM, Ringel Y, Sanders ME, Sartor RB, Sherman PM, Versalovic J, Young V, Finlay BB. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe. 2012;12:611-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 518] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 19. | Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718-15723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4530] [Cited by in RCA: 4413] [Article Influence: 210.1] [Reference Citation Analysis (4)] |
| 20. | Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070-11075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4639] [Cited by in RCA: 4466] [Article Influence: 223.3] [Reference Citation Analysis (1)] |
| 21. | Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027-1031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9796] [Cited by in RCA: 8756] [Article Influence: 460.8] [Reference Citation Analysis (1)] |
| 22. | Indiani CMDSP, Rizzardi KF, Castelo PM, Ferraz LFC, Darrieux M, Parisotto TM. Childhood obesity and Firmicutes/Bacteroidetes ratio in the gut microbiota: A Systematic Review. Child Obes. 2018;14:501-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 266] [Article Influence: 38.0] [Reference Citation Analysis (1)] |
| 23. | Koliada A, Syzenko G, Moseiko V, Budovska L, Puchkov K, Perederiy V, Gavalko Y, Dorofeyev A, Romanenko M, Tkach S, Sineok L, Lushchak O, Vaiserman A. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 2017;17:120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 469] [Cited by in RCA: 672] [Article Influence: 84.0] [Reference Citation Analysis (0)] |
| 24. | Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell MD, Wing R, Rittmann BE, Krajmalnik-Brown R. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA. 2009;106:2365-2370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1323] [Cited by in RCA: 1402] [Article Influence: 87.6] [Reference Citation Analysis (1)] |
| 25. | Wu T, Wang H-C, Lu W-W, Zhao J-X, Zhang H, Chen W. Characteristics of gut microbiota of obese people and machine learning model. Microbiol China. 2020;47:4328-4337. [DOI] [Full Text] |
| 26. | Waters JL, Ley RE. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. 2019;17:83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 427] [Cited by in RCA: 462] [Article Influence: 77.0] [Reference Citation Analysis (0)] |
| 27. | Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, Falony G, Raes J, Maiter D, Delzenne NM, de Barsy M, Loumaye A, Hermans MP, Thissen JP, de Vos WM, Cani PD. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med. 2019;25:1096-1103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 774] [Cited by in RCA: 1426] [Article Influence: 237.7] [Reference Citation Analysis (0)] |
| 28. | Crovesy L, Ostrowski M, Ferreira DMTP, Rosado EL, Soares-Mota M. Effect of Lactobacillus on body weight and body fat in overweight subjects: a systematic review of randomized controlled clinical trials. Int J Obes (Lond). 2017;41:1607-1614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 153] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 29. | Yin YN, Yu QF, Fu N, Liu XW, Lu FG. Effects of four Bifidobacteria on obesity in high-fat diet induced rats. World J Gastroenterol. 2010;16:3394-3401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 175] [Cited by in RCA: 186] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 30. | Waldram A, Holmes E, Wang Y, Rantalainen M, Wilson ID, Tuohy KM, McCartney AL, Gibson GR, Nicholson JK. Top-down systems biology modeling of host metabotype-microbiome associations in obese rodents. J Proteome Res. 2009;8:2361-2375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 192] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 31. | Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P, Valero R, Raccah D, Vialettes B, Raoult D. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int J Obes (Lond). 2012;36:817-825. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 516] [Cited by in RCA: 493] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 32. | Moreno-Navarrete JM, Serino M, Blasco-Baque V, Azalbert V, Barton RH, Cardellini M, Latorre J, Ortega F, Sabater-Masdeu M, Burcelin R, Dumas ME, Ricart W, Federici M, Fernández-Real JM. Gut microbiota interacts with markers of adipose tissue browning, insulin action and plasma acetate in morbid obesity. Mol Nutr Food Res. 2018;62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 33. | Alemán JO, Bokulich NA, Swann JR, Walker JM, De Rosa JC, Battaglia T, Costabile A, Pechlivanis A, Liang Y, Breslow JL, Blaser MJ, Holt PR. Fecal microbiota and bile acid interactions with systemic and adipose tissue metabolism in diet-induced weight loss of obese postmenopausal women. J Transl Med. 2018;16:244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 34. | Anhê FF, Roy D, Pilon G, Dudonné S, Matamoros S, Varin TV, Garofalo C, Moine Q, Desjardins Y, Levy E, Marette A. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut. 2015;64:872-883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 760] [Cited by in RCA: 849] [Article Influence: 84.9] [Reference Citation Analysis (0)] |
| 35. | Ahn SB, Jun DW, Kang BK, Lim JH, Lim S, Chung MJ. Randomized, double-blind, placebo-controlled study of a multispecies probiotic mixture in nonalcoholic fatty liver disease. Sci Rep. 2019;9:5688. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 140] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 36. | Tavella T, Rampelli S, Guidarelli G, Bazzocchi A, Gasperini C, Pujos-Guillot E, Comte B, Barone M, Biagi E, Candela M, Nicoletti C, Kadi F, Battista G, Salvioli S, O'Toole PW, Franceschi C, Brigidi P, Turroni S, Santoro A. Elevated gut microbiome abundance of Christensenellaceae, Porphyromonadaceae and Rikenellaceae is associated with reduced visceral adipose tissue and healthier metabolic profile in Italian elderly. Gut Microbes. 2021;13:1-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 175] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 37. | Wu G, Zhao N, Zhang C, Lam YY, Zhao L. Guild-based analysis for understanding gut microbiome in human health and diseases. Genome Med. 2021;13:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 101] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 38. | Denou E, Marcinko K, Surette MG, Steinberg GR, Schertzer JD. High-intensity exercise training increases the diversity and metabolic capacity of the mouse distal gut microbiota during diet-induced obesity. Am J Physiol Endocrinol Metab. 2016;310:E982-E993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 186] [Article Influence: 20.7] [Reference Citation Analysis (1)] |
| 39. | Ciobârcă D, Cătoi AF, Copăescu C, Miere D, Crișan G. Bariatric surgery in obesity: Effects on gut microbiota and micronutrient status. Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 93] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 40. | Heiss CN, Olofsson LE. Gut microbiota-dependent modulation of energy metabolism. J Innate Immun. 2018;10:163-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 193] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 41. | Ma ZS, Li L, Gotelli NJ. Diversity-disease relationships and shared species analyses for human microbiome-associated diseases. ISME J. 2019;13:1911-1919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (4)] |
| 42. | Krajmalnik-Brown R, Ilhan ZE, Kang DW, DiBaise JK. Effects of gut microbes on nutrient absorption and energy regulation. Nutr Clin Pract. 2012;27:201-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 388] [Cited by in RCA: 571] [Article Influence: 43.9] [Reference Citation Analysis (1)] |
| 43. | Festi D, Schiumerini R, Eusebi LH, Marasco G, Taddia M, Colecchia A. Gut microbiota and metabolic syndrome. World J Gastroenterol. 2014;20:16079-16094. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 342] [Cited by in RCA: 363] [Article Influence: 33.0] [Reference Citation Analysis (6)] |
| 44. | Petersen C, Bell R, Klag KA, Lee SH, Soto R, Ghazaryan A, Buhrke K, Ekiz HA, Ost KS, Boudina S, O'Connell RM, Cox JE, Villanueva CJ, Stephens WZ, Round JL. T cell-mediated regulation of the microbiota protects against obesity. Science. 2019;365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 270] [Article Influence: 54.0] [Reference Citation Analysis (1)] |
| 45. | Cani PD, Van Hul M, Lefort C, Depommier C, Rastelli M, Everard A. Microbial regulation of organismal energy homeostasis. Nat Metab. 2019;1:34-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 363] [Article Influence: 60.5] [Reference Citation Analysis (1)] |
| 46. | Hernández MAG, Canfora EE, Jocken JWE, Blaak EE. The short-chain fatty acid acetate in body weight control and insulin sensitivity. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 371] [Article Influence: 61.8] [Reference Citation Analysis (1)] |
| 47. | Wang D, Liu CD, Tian ML, Tan CQ, Shu G, Jiang QY, Zhang L, Yin Y. Propionate promotes intestinal lipolysis and metabolic benefits via AMPK/LSD1 pathway in mice. J Endocrinol. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 48. | Yu C, Liu S, Chen L, Shen J, Niu Y, Wang T, Zhang W, Fu L. Effect of exercise and butyrate supplementation on microbiota composition and lipid metabolism. J Endocrinol. 2019;243:125-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 49. | de la Cuesta-Zuluaga J, Mueller NT, Álvarez-Quintero R, Velásquez-Mejía EP, Sierra JA, Corrales-Agudelo V, Carmona JA, Abad JM, Escobar JS. Higher fecal short-chain fatty acid levels are associated with gut microbiome dysbiosis, obesity, hypertension and cardiometabolic disease risk factors. Nutrients. 2018;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 324] [Cited by in RCA: 354] [Article Influence: 50.6] [Reference Citation Analysis (0)] |
| 50. | Teixeira TF, Grześkowiak Ł, Franceschini SC, Bressan J, Ferreira CL, Peluzio MC. Higher level of faecal SCFA in women correlates with metabolic syndrome risk factors. Br J Nutr. 2013;109:914-919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 96] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 51. | Cryan JF, O'Riordan KJ, Cowan CSM, Sandhu KV, Bastiaanssen TFS, Boehme M, Codagnone MG, Cussotto S, Fulling C, Golubeva AV, Guzzetta KE, Jaggar M, Long-Smith CM, Lyte JM, Martin JA, Molinero-Perez A, Moloney G, Morelli E, Morillas E, O'Connor R, Cruz-Pereira JS, Peterson VL, Rea K, Ritz NL, Sherwin E, Spichak S, Teichman EM, van de Wouw M, Ventura-Silva AP, Wallace-Fitzsimons SE, Hyland N, Clarke G, Dinan TG. The microbiota-gut-brain axis. Physiol Rev. 2019;99:1877-2013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1156] [Cited by in RCA: 2786] [Article Influence: 464.3] [Reference Citation Analysis (2)] |
| 52. | Shajib MS, Khan WI. The role of serotonin and its receptors in activation of immune responses and inflammation. Acta Physiol (Oxf). 2015;213:561-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 263] [Article Influence: 26.3] [Reference Citation Analysis (1)] |
| 53. | Silberbauer CJ, Surina-Baumgartner DM, Arnold M, Langhans W. Prandial lactate infusion inhibits spontaneous feeding in rats. Am J Physiol Regul Integr Comp Physiol. 2000;278:R646-R653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 54. | Wu Y, He H, Cheng Z, Bai Y, Ma X. The role of neuropeptide Y and peptide YY in the development of obesity via gut-brain axis. Curr Protein Pept Sci. 2019;20:750-758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 55. | Salehi M, Purnell JQ. The role of glucagon-like peptide-1 in energy homeostasis. Metab Syndr Relat Disord. 2019;17:183-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 56. | Federico A, Dallio M, Tolone S, Gravina AG, Patrone V, Romano M, Tuccillo C, Mozzillo AL, Amoroso V, Misso G, Morelli L, Docimo L, Loguercio C. Gastrointestinal hormones, intestinal microbiota and metabolic homeostasis in obese patients: Effect of bariatric surgery. In Vivo. 2016;30:321-330. [PubMed] |
| 57. | Schéle E, Grahnemo L, Anesten F, Hallén A, Bäckhed F, Jansson JO. The gut microbiota reduces leptin sensitivity and the expression of the obesity-suppressing neuropeptides proglucagon (Gcg) and brain-derived neurotrophic factor (Bdnf) in the central nervous system. Endocrinology. 2013;154:3643-3651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 162] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 58. | Fetissov SO. Role of the gut microbiota in host appetite control: bacterial growth to animal feeding behaviour. Nat Rev Endocrinol. 2017;13:11-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 243] [Article Influence: 30.4] [Reference Citation Analysis (1)] |
| 59. | Aronsson L, Huang Y, Parini P, Korach-André M, Håkansson J, Gustafsson JÅ, Pettersson S, Arulampalam V, Rafter J. Decreased fat storage by Lactobacillus paracasei is associated with increased levels of angiopoietin-like 4 protein (ANGPTL4). PLoS One. 2010;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 201] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 60. | Tazi A, Araujo JR, Mulet C, Arena ET, Nigro G, Pédron T, Sansonetti PJ. Disentangling host-microbiota regulation of lipid secretion by enterocytes: Insights from commensals Lactobacillus paracasei and Escherichia coli. mBio. 2018;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 61. | Al Bander Z, Nitert MD, Mousa A, Naderpoor N. The gut microbiota and inflammation: An overview. Int J Environ Res Public Health. 2020;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 426] [Article Influence: 85.2] [Reference Citation Analysis (0)] |
| 62. | Tilg H, Zmora N, Adolph TE, Elinav E. The intestinal microbiota fuelling metabolic inflammation. Nat Rev Immunol. 2020;20:40-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 663] [Article Influence: 110.5] [Reference Citation Analysis (0)] |
| 63. | Burrello C, Giuffrè MR, Macandog AD, Diaz-Basabe A, Cribiù FM, Lopez G, Borgo F, Nezi L, Caprioli F, Vecchi M, Facciotti F. Fecal microbiota transplantation controls murine chronic intestinal inflammation by modulating immune cell functions and gut microbiota composition. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 64. | Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmée E, Cousin B, Sulpice T, Chamontin B, Ferrières J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761-1772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4095] [Cited by in RCA: 4563] [Article Influence: 253.5] [Reference Citation Analysis (1)] |
| 65. | Janssen AW, Kersten S. Potential mediators linking gut bacteria to metabolic health: a critical view. J Physiol. 2017;595:477-487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 66. | Kang C, Wang B, Kaliannan K, Wang X, Lang H, Hui S, Huang L, Zhang Y, Zhou M, Chen M, Mi M. Gut Microbiota Mediates the protective effects of dietary capsaicin against chronic low-grade inflammation and associated obesity induced by high-fat diet. mBio. 2017;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 164] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 67. | Lührs H, Gerke T, Schauber J, Dusel G, Melcher R, Scheppach W, Menzel T. Cytokine-activated degradation of inhibitory kappaB protein alpha is inhibited by the short-chain fatty acid butyrate. Int J Colorectal Dis. 2001;16:195-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 63] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 68. | Jia Y, Hong J, Li H, Hu Y, Jia L, Cai D, Zhao R. Butyrate stimulates adipose lipolysis and mitochondrial oxidative phosphorylation through histone hyperacetylation-associated β3 -adrenergic receptor activation in high-fat diet-induced obese mice. Exp Physiol. 2017;102:273-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 69. | Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, Petersen KF, Kibbey RG, Goodman AL, Shulman GI. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature. 2016;534:213-217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 724] [Cited by in RCA: 950] [Article Influence: 105.6] [Reference Citation Analysis (1)] |
| 70. | Frost G, Sleeth ML, Sahuri-Arisoylu M, Lizarbe B, Cerdan S, Brody L, Anastasovska J, Ghourab S, Hankir M, Zhang S, Carling D, Swann JR, Gibson G, Viardot A, Morrison D, Louise Thomas E, Bell JD. The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat Commun. 2014;5:3611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1112] [Cited by in RCA: 1183] [Article Influence: 107.5] [Reference Citation Analysis (0)] |
| 71. | Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, Pierre JF, Heneghan AF, Nadimpalli A, Hubert N, Zale E, Wang Y, Huang Y, Theriault B, Dinner AR, Musch MW, Kudsk KA, Prendergast BJ, Gilbert JA, Chang EB. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17:681-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 469] [Cited by in RCA: 619] [Article Influence: 61.9] [Reference Citation Analysis (0)] |
| 72. | Rácz B, Dušková M, Stárka L, Hainer V, Kunešová M. Links between the circadian rhythm, obesity and the microbiome. Physiol Res. 2018;67:S409-S420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 73. | Kuang Z, Wang Y, Li Y, Ye C, Ruhn KA, Behrendt CL, Olson EN, Hooper LV. The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science. 2019;365:1428-1434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 221] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 74. | Ye Y, Xu H, Xie Z, Wang L, Sun Y, Yang H, Hu D, Mao Y. Time-restricted feeding reduces the detrimental effects of a high-fat diet, possibly by modulating the circadian rhythm of hepatic lipid metabolism and gut microbiota. Front Nutr. 2020;7:596285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 75. | Zeb F, Wu X, Chen L, Fatima S, Haq IU, Chen A, Majeed F, Feng Q, Li M. Effect of time-restricted feeding on metabolic risk and circadian rhythm associated with gut microbiome in healthy males. Br J Nutr. 2020;123:1216-1226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 126] [Article Influence: 25.2] [Reference Citation Analysis (1)] |
| 76. | Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, Kuperman Y, Biton I, Gilad S, Harmelin A, Shapiro H, Halpern Z, Segal E, Elinav E. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514-529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 733] [Cited by in RCA: 935] [Article Influence: 85.0] [Reference Citation Analysis (1)] |
| 77. | Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, Hill C, Gahan CG. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci USA. 2014;111:7421-7426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 376] [Cited by in RCA: 443] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 78. | Parkar SG, Kalsbeek A, Cheeseman JF. Potential role for the gut microbiota in modulating host circadian rhythms and metabolic health. Microorganisms. 2019;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 172] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 79. | Tahara Y, Yamazaki M, Sukigara H, Motohashi H, Sasaki H, Miyakawa H, Haraguchi A, Ikeda Y, Fukuda S, Shibata S. Gut microbiota-derived short chain fatty acids induce circadian clock entrainment in mouse peripheral tissue. Sci Rep. 2018;8:1395. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 195] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 80. | Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, Spector TD, Bell JT, Clark AG, Ley RE. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe. 2016;19:731-743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 798] [Cited by in RCA: 717] [Article Influence: 79.7] [Reference Citation Analysis (0)] |
| 81. | Poole AC, Goodrich JK, Youngblut ND, Luque GG, Ruaud A, Sutter JL, Waters JL, Shi Q, El-Hadidi M, Johnson LM, Bar HY, Huson DH, Booth JG, Ley RE. Human salivary amylase gene copy number impacts oral and gut microbiomes. Cell Host Microbe. 2019;25:553-564.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 102] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 82. | Suzuki TA, Phifer-Rixey M, Mack KL, Sheehan MJ, Lin D, Bi K, Nachman MW. Host genetic determinants of the gut microbiota of wild mice. Mol Ecol. 2019;28:3197-3207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 83. | Davenport ER, Cusanovich DA, Michelini K, Barreiro LB, Ober C, Gilad Y. Genome-wide association studies of the human gut microbiota. PLoS One. 2015;10:e0140301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 190] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 84. | Moeller AH, Suzuki TA, Phifer-Rixey M, Nachman MW. Transmission modes of the mammalian gut microbiota. Science. 2018;362:453-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 187] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 85. | Koleva PT, Kim JS, Scott JA, Kozyrskyj AL. Microbial programming of health and disease starts during fetal life. Birth Defects Res C Embryo Today. 2015;105:265-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 86] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 86. | Tun HM, Bridgman SL, Chari R, Field CJ, Guttman DS, Becker AB, Mandhane PJ, Turvey SE, Subbarao P, Sears MR, Scott JA, Kozyrskyj AL; Canadian Healthy Infant Longitudinal Development (CHILD) Study Investigators. Roles of birth mode and infant gut microbiota in intergenerational transmission of overweight and obesity from mother to offspring. JAMA Pediatr. 2018;172:368-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 213] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 87. | Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, Shilo S, Lador D, Vila AV, Zmora N, Pevsner-Fischer M, Israeli D, Kosower N, Malka G, Wolf BC, Avnit-Sagi T, Lotan-Pompan M, Weinberger A, Halpern Z, Carmi S, Fu J, Wijmenga C, Zhernakova A, Elinav E, Segal E. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1342] [Cited by in RCA: 1840] [Article Influence: 262.9] [Reference Citation Analysis (0)] |
| 88. | Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, Knight R, Ahima RS, Bushman F, Wu GD. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716-24.e1-2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1234] [Cited by in RCA: 1162] [Article Influence: 72.6] [Reference Citation Analysis (0)] |
| 89. | Liu MT, Huang YJ, Zhang TY, Tan LB, Lu XF, Qin J. Lingguizhugan decoction attenuates diet-induced obesity and hepatosteatosis via gut microbiota. World J Gastroenterol. 2019;25:3590-3606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 28] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 90. | Poroyko VA, Carreras A, Khalyfa A, Khalyfa AA, Leone V, Peris E, Almendros I, Gileles-Hillel A, Qiao Z, Hubert N, Farré R, Chang EB, Gozal D. Chronic sleep disruption alters gut microbiota, induces systemic and adipose tissue inflammation and insulin resistance in mice. Sci Rep. 2016;6:35405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 333] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 91. | Kuperman Y, Issler O, Regev L, Musseri I, Navon I, Neufeld-Cohen A, Gil S, Chen A. Perifornical Urocortin-3 mediates the link between stress-induced anxiety and energy homeostasis. Proc Natl Acad Sci USA. 2010;107:8393-8398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 66] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 92. | Karl JP, Margolis LM, Madslien EH, Murphy NE, Castellani JW, Gundersen Y, Hoke AV, Levangie MW, Kumar R, Chakraborty N, Gautam A, Hammamieh R, Martini S, Montain SJ, Pasiakos SM. Changes in intestinal microbiota composition and metabolism coincide with increased intestinal permeability in young adults under prolonged physiological stress. Am J Physiol Gastrointest Liver Physiol. 2017;312:G559-G571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 252] [Article Influence: 31.5] [Reference Citation Analysis (0)] |