Published online Jun 28, 2021. doi: 10.3748/wjg.v27.i24.3595
Peer-review started: March 2, 2021
First decision: April 5, 2021
Revised: April 13, 2021
Accepted: May 21, 2021
Article in press: May 21, 2021
Published online: June 28, 2021
Processing time: 115 Days and 1 Hours
The drug resistance rate of clinical Helicobacter pylori (H. pylori) isolates has increased. However, the mechanism of drug resistance remains unclear. In this study, drug-resistant H. pylori strains were isolated from different areas and different populations of Chinese for genomic analysis.
To investigate drug-resistant genes in H. pylori and find the genes for the early diagnosis of clarithromycin resistance.
Three drug-resistant H. pylori strains were isolated from patients with gastritis in Bama County, China. Minimal inhibitory concentrations of clarithromycin, metronidazole, and levofloxacin were determined and complete genome sequencing was performed with annotation. Hp1181 and hp1184 genes were found in these strains and then detected by reverse transcription polymerase chain reaction. The relationships between hp1181 or hp1184 and clarithromycin resistance were ascertained with gene mutant and drug-resistant strains. The homology of the strains with hp26695 was assessed through complete genome detection and identification. Differences in genome sequences, gene quantity, and gene characteristics were detected amongst the three strains. Prediction and analysis of the function of drug-resistant genes indicated that the RNA expression of hp1181 and hp1184 increased in the three strains, which was the same in the artificially induced clarithromycin-resistant bacteria. After gene knockout, the drug sensitivity of the strains was assessed.
The strains showing a high degree of homology with hp26695, hp1181, and hp1184 genes were found in these strains; the expression of the genes hp1184 and hp1181 was associated with clarithromycin resistance.
Hp1181 and hp1184 mutations may be the earliest and most persistent response to clarithromycin resistance, and they may be the potential target genes for the diagnosis, prevention, and treatment of clarithromycin resistance.
Core Tip: The World Health Organization designated clarithromycin-resistant Helicobacter pylori (H. pylori) a high priority among bacteria for antibiotic research and development, but the clarithromycin resistance mechanism remains unclear. We isolated and cultured clinical H. pylori strains, determined their minimal inhibitory concentrations, completed genome sequencing of hp1181 and hp1184 genes, analyzed their mutations, and found that the expression of the genes hp1184 and hp1181 was associated with clarithromycin resistance, which suggested that they can be used as genes for early diagnosis. This research may prove useful in the diagnosis, prevention, and treatment of clarithromycin-resistant H. pylori.
- Citation: Li XH, Huang YY, Lu LM, Zhao LJ, Luo XK, Li RJ, Dai YY, Qin C, Huang YQ, Chen H. Early genetic diagnosis of clarithromycin resistance in Helicobacter pylori. World J Gastroenterol 2021; 27(24): 3595-3608
- URL: https://www.wjgnet.com/1007-9327/full/v27/i24/3595.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i24.3595
Helicobacter pylori (H. pylori) is recognized as an important human pathogen that colonizes the gastric mucus, resulting in superficial gastritis, atrophic gastritis, and gastric cancer[1-3]. Present treatments for H. pylori infection include proton pump inhibitors, bismuth in combination with amoxicillin, metronidazole and clarithromycin[4,5]. The rate of drug resistance is increasing because of the wide use of antibiotics and high resistance rates to clarithromycin, metronidazole, and levofloxacin are associated with the failure of H. pylori eradication[6-8]. The World Health Organi
At present, the mechanism of antibiotic resistance of H. pylori is not completely understood[10,11]. It is widely accepted that the resistance to these antimicrobials is related to mutations in H. pylori genes, and clarithromycin-resistant strains present three point mutations in the region of domain V of 23S ribosomal RNA (rRNA): A2142G, A2142C, and A2143G[12,13]. In addition to the mutations, the efflux pump cluster is also involved in the development of resistance to clarithromycin[14,15]. However, there may be gene mutation sites that are not yet known, and the mechanism of drug resistance warrants further study.
We isolated and cultured H. pylori from the population in Bama County, which is a township known for the longevity of its residents in Guangxi, and randomly selected three strains of multiple drug-resistant H. pylori with resistance to clarithromycin. Complete genome sequences were analyzed to study the genomic characteristics of the strains and to elucidate the underlying mechanism of drug resistance in H. pylori.
This study had received a strict medical ethics review from Youjiang Medical University for Nationalities. Written informed consent was obtained from each patient. Gastric mucosa tissue samples were collected from the People's Hospital of Bama Yao Autonomous County in patients’ gastric body and pylorus with gastritis or gastric ulcers. Isolation and culture of H. pylori were performed at the Research Center for the Prevention and Treatment of Drug Resistant Microbial Infection, Youjiang Medical University for Nationalities. Patients investigated had not taken any antibiotics for at least 4 wk before examination. The isolation and identification of H. pylori were performed as previously described[16,17]. The bacteria were cultured on Columbia agar plates containing 5% fresh defibrinated sheep blood. The microaerophilic conditions included 5% O2, 10% CO2, and 85% N2 at 37 °C for 3 to 5 days. Suspicious colonies were confirmed by Gram staining, urease, oxidase, and catalase activity testing, and urease gene polymerase chain reaction (PCR).
The antibiotic resistance of H. pylori was measured by dilution methods with reference to the protocols of the Clinical and Laboratory Standards Institute (Wayne, PA, United States)[18]. Briefly, the density of H. pylori was adjusted to be 1 × 106 CFU/mL and incubated at 37 °C for 3 to 5 d under microaerophilic conditions. After incubation, the plates were visually examined and the minimal inhibitory concentration (MIC) was determined to be the lowest concentration that resulted in no turbidity. Metronidazole (Aladdin, d1707126), amoxicillin (Xiansheng pharmaceutical, Co., Ltd, China), levofloxacin (Shandong Lukang Pharmaceutical Group Saite Co., Ltd, China), and clarithromycin (Yangzi River Pharmaceutical Group Co., Ltd, China) were also used.
Drug-resistant strains were selected and sent to the Shenzhen Huada Gene Co., Ltd (China) for complete genome analysis. After the DNA samples were delivered, the quality of the samples was tested and then used to construct a BSlibrary. The purified genomic DNA samples including genomic DNA, bacterial artificial chromosomes, or long-length PCR products were sheared into smaller fragments by CovarisS/E210 or using a Bioruptor. The overhangs resulting from fragmentation were converted into blunt ends using T4 DNA polymerase, Klenow fragment, and T4 polynucleotide kinase. After adding an ‘A’ base to the 3' end of the blunt phosphorylated DNA fragments, adapters were ligated to the ends of the DNA fragments. The desired fragments were purified though gel-electrophoresis, selectively enriched, and amplified by PCR. The index tag was introduced into the adapter at the PCR stage as appropriate and a library quality test was conducted. Finally, the qualified BSlibrary was used for sequencing. Genomic component and gene function analyses were performed, including gene prediction, tRNA, sRNA, and gene annotation, and prediction of open reading frames by GO.
Drug-resistant genes were predicted based on the results of the complete genome sequence analysis and selected for detection by reverse transcription PCR (RT-PCR). The reaction for cDNA synthesis was held at 25 °C for 10 min, 42 °C for 60 min, and then 99 °C for 5 min. The reaction consisted of 32 cycles with each cycle composed of 1 min at 95 °C, 4 min at 56 °C, and 7 min at 70 °C. After a final extension of 15 min at 72 °C, the RT-PCR products were visualized by electrophoresis on 1% agarose gel and 15% acrylamide gel with a 200-bp ladder size marker.
Hp1181 and hp1184 knockout mutants were constructed by insertion of the KAN resistance cassette. Double-knockout mutants were made by natural transformation of the KAN resistance cassette with pBSII KS (as presented by Bi HK, Laboratory of Nanjing Medical University, China) containing an internal fragment interrupted with a cat cassette from pAV35, with selection for both KAN- and CHL-resistant colonies. Insertion of the KAN and cat resistance cassette at the desired locations in the H. pylori putative efflux genes was validated by PCR.
The MIC of clarithromycin to hp26695 was detected. Drug resistance was induced by 1/4 MIC. The culture medium was changed every 2 d and MIC was detected every 4 d. The concentration of induced drug was changed with MIC.
Three drug-resistant strains were isolated and identified by Gram staining, urease, oxidase, catalase activity testing, and urease gene PCR. The drug resistance information of these strains is summarized in Table 1.
| Strain | Metronidazole | Clarithromycin | Levofloxacin | Amoxicillin |
| Hpbs1 | 32 | 8 | 8 | 0.125 |
| Hpbs2 | 16 | 8 | 0.125 | 0.125 |
| Hpbs3 | 0.125 | 8 | 8 | 0.125 |
Based on the valid data from the previous sequencing platform, the CleanData could be assembled for each sample and the optimal assembly results were obtained after multiple adjustments. The assembly sequence was analyzed by correcting single base, circular judgment, and plasmid comparison. The results of the genome assembly statistics of each sample are displayed in Table 2. These three strains have been uploaded to the NCBI Biosample database: Hpbs1 (https://www.ncbi.nlm.nih.gov/biosample/?term=SAMN10461767). Hpbs2 (https://www.ncbi.nlm.nih.gov/biosample/?term=SAMN10663081), and Hpbs3 (https://www.ncbi.nlm.nih.gov/biosample/?term=SAMN10663175).
| Sample name | ID name | Sequence type | Sequence topology | Sequence number | Total length (bp) | GC content |
| Hpbs1 | Chromosome1 | Chromosome | Circular | 1 | 1563701 | 38.90 |
| All | All | - | 1 | 1563701 | 38.90 | |
| Hpbs2 | Chromosome1 | Chromosome | Circular | 1 | 1534481 | 38.87 |
| All | All | - | 1 | 1534481 | 38.87 | |
| Hpbs3 | Chromosome1 | Chromosome | Circular | 1 | 1534930 | 38.90 |
| All | All | - | 1 | 1534930 | 38.90 |
Gene prediction was applied to determine gene composition. The statistics are shown in Table 3.
| Sample name (#) | Genome size (#) | Total number (#) | Total length (bp) | Average length (#) | Length/genome length (%) | GC content (%) |
| Hpbs1 | 1563701 | 1571 | 1434202 | 912.92 | 91.72 | 39.49 |
| Hpbs2 | 1534481 | 1792 | 1395399 | 778.68 | 90.94 | 39.44 |
| Hpbs3 | 1534930 | 1732 | 1407495 | 812.64 | 91.70 | 39.49 |
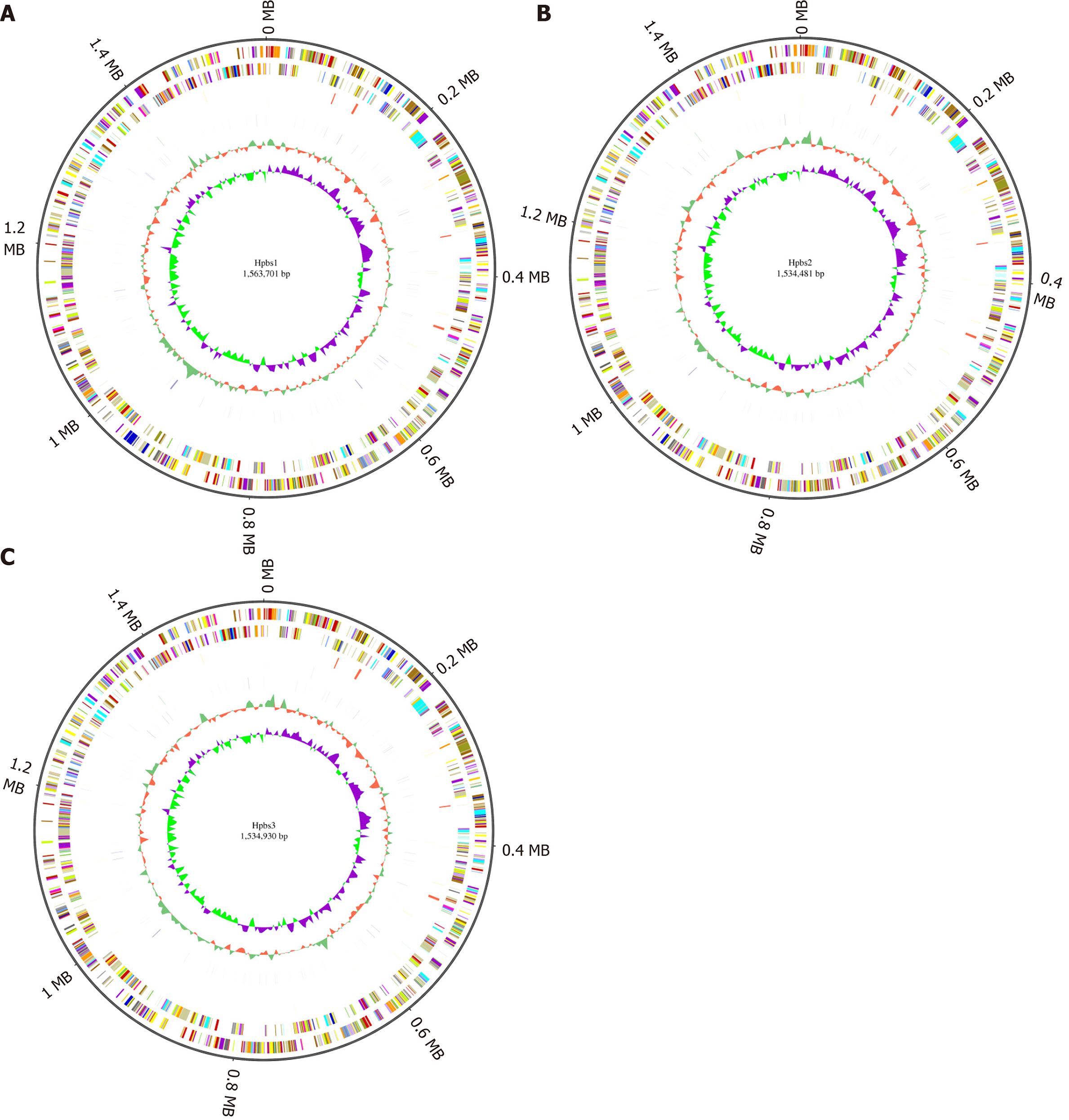
GC skew analysis was carried out using (G-C)/(G+C) calculations based on genomic sequences of strains. The results of gene distribution, ncRNA distribution, and gene annotation are demonstrated in Figure 1. Hpbs1 had 835 genes, 26 tRNAs, 6 rRNAs, and 2 sRNAs in the positive chain. It also had 736 genes, 10 tRNAs, 0 rRNAs, and 5 sRNAs in the negative chain and 157 repeats without positive or negative chain. There are 943 genes, 26 tRNAs, 6 rRNAs, 3 sRNAs, 849 genes, 10 tRNAs, 0 rRNA, 3 sRNAs, and 153 repeats in Hpbs2; there are 869 genes, 26 tRNAs, 6 rRNAs, 3 sRNAs, 863 genes, 10 tRNAs, 0 rRNA, 3 sRNAs, and 155 repeats in Hpbs3.
Functional annotation was accomplished by analysis of protein sequences. We aligned genes with databases to obtain their corresponding annotations. To demonstrate the biological meaning, the highest quality alignment result was chosen as a gene annotation. Functional annotation was completed by blast resistance genes with different databases. In this project we have finished annotations using 17 databases, including P450, VFDB, ARDB, CAZY, SWISSPROT, NOG, COG, CARD, NR, DBCAN, T3SS, TREMBL, IPR, PHI, KEGG, GO, and KOG. The annotation results are shown in Tables 4 and 5.
| Sample name (#) | Total | P450 (#) (%) | VFDB (#) (%) | ARDB (#) (%) | CAZY (#) | SWISSPROT (#) (%) | NOG (#) (%) | COG (#) (%) | CARD (#) (%) | NR (#) |
| Hpbs1 | 1571 | 22 (1.4) | 196 (12.47) | 0 (0) | 14 (0.89) | 742 (47.23) | 67 (4.26) | 1084 (69) | 14 (0.89) | 1599 (99.23) |
| Hpbs2 | 1792 | 21 (1.17) | 177 (9.87) | 0 (0) | 14 (0.78) | 751 (41.9) | 125 (6.97) | 1111 (61.99) | 13 (0.72) | 1723 (96.14) |
| Hpbs3 | 1732 | 22 (1.27) | 174 (10.04) | 0 (0) | 14 (0.75) | 750 (43.3) | 97 (5.6) | 1113 (64.26) | 15 (0.86) | 1698 (98.03) |
| Sample name (#) | DBCAN (#) (%) | T3SS (#) (%) | TREMBL (#) (%) | IPR (#) | PHI (#) (%) | KEGG (#) (%) | GO (#) (%) | KOG (#) (%) | Over all (#) (%) |
| Hpbs1 | 30 (1.9) | 175 (11.13) | 1557 (99.1) | 1234 (78.54) | 54 (3.43) | 1026 (65.3) | 957 (60.91) | 142 (9.03) | 1563 (99.49) |
| Hpbs2 | 29 (1.61) | 197 (10.99) | 1706 (95.2) | 1372 (76.56) | 52 (2.9) | 1078 (60.15) | 1056 (58.92) | 144 (8.03) | 1750 (97.65) |
| Hpbs3 | 30 (1.73) | 209 (12.06) | 1688 (97.45) | 1340 (77.36) | 51 (2.94) | 1067 (61.6) | 1030 (59.4) | 139 (8.02) | 1711 (98.78) |
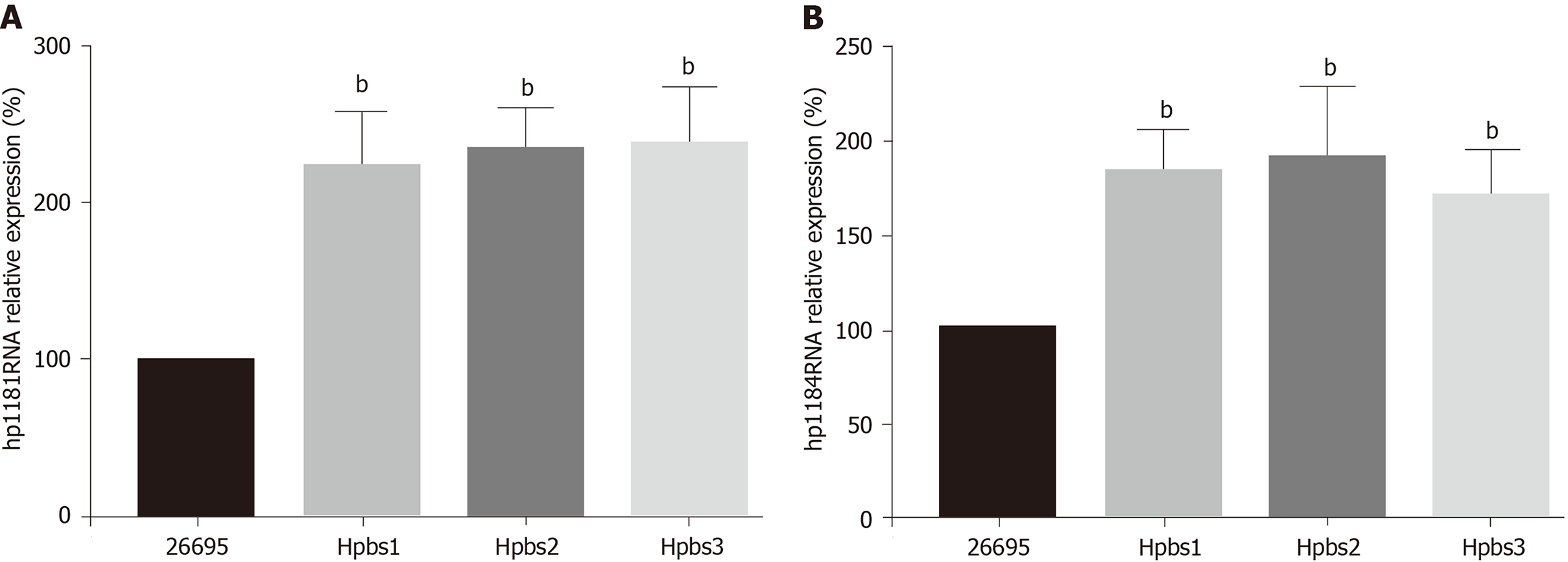
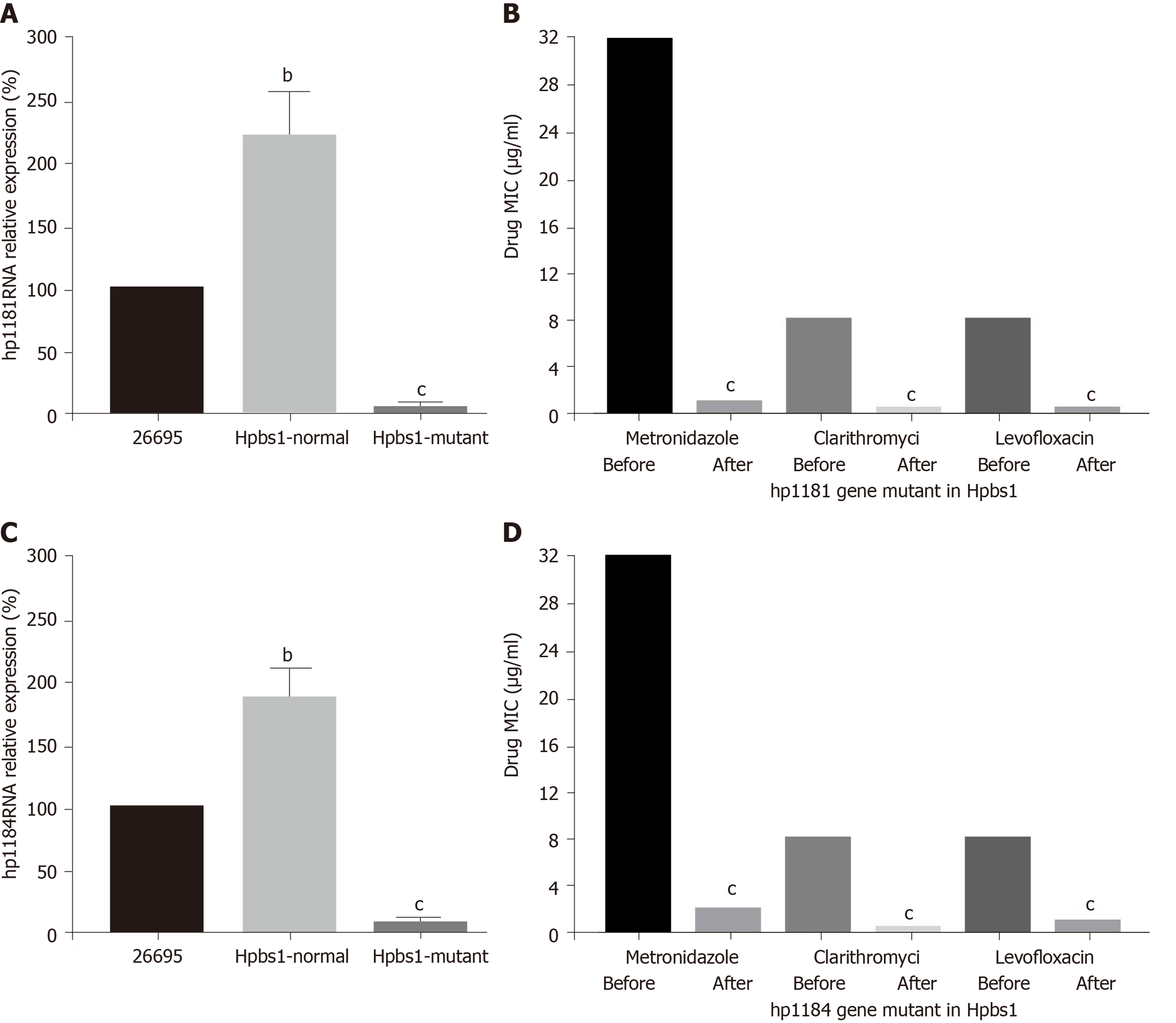
The drug resistance gene numbers of three strains were different in the CARD (Comprehensive Antibiotic Resistance Database), which are 14, 13, and 15 genes, respectively. However, after sorting, it was found that some genes were repetitive. The specific numbers and characteristics of genes are presented in the Tables 6 and 7. NP_207975.1 and NP_207972.1 were efflux pump genes of 26695 strain, i.e., hp1181 and hp1184 genes. Their drug resistance was verified by RT-PCR, as illustrated in Figure 2. After knocking out the drug-resistant genes, drug sensitivity was significantly improved, as shown in Figure 3.
| Gene ID | Subject ID | Align length | Mismatch | Gap | Gene start | Gene end | Subject start | Subject end | E value |
| GL000175 | YP_208874.1 | 97 | 39 | 0 | 2 | 98 | 4 | 100 | 6.00E-40 |
| GL000286 | YP_006374661.1 | 398 | 88 | 2 | 1 | 397 | 29 | 421 | 0 |
| GL000295 | NP_312937.1 | 1389 | 658 | 21 | 8 | 1371 | 8 | 1339 | 0 |
| GL000296 | AAK44936.1 | 124 | 35 | 0 | 1 | 124 | 1 | 124 | 4.00E-63 |
| GL000306 | NP_207975.1 | 459 | 16 | 0 | 1 | 459 | 1 | 459 | 0 |
| GL000309 | NP_207972.1 | 443 | 10 | 0 | 1 | 443 | 1 | 443 | 0 |
| GL000772 | AIL15701 | 421 | 220 | 3 | 1 | 420 | 1 | 417 | 4.00E-126 |
| GL000822 | YP_002344422.1 | 853 | 293 | 6 | 3 | 818 | 2 | 851 | 0 |
| GL000911 | NP_415611.1 | 247 | 130 | 2 | 1 | 247 | 1 | 243 | 2.00E-66 |
| GL000972 | WP_005768149.1 | 810 | 390 | 18 | 3 | 773 | 12 | 809 | 0 |
| GL001063 | AJF83452.1 | 287 | 164 | 2 | 1 | 283 | 2 | 288 | 1.00E-71 |
| GL001265 | NP_415804.1 | 262 | 141 | 1 | 1 | 261 | 1 | 262 | 2.00E-80 |
| GL001295 | YP_001332362.1 | 222 | 123 | 4 | 1 | 221 | 1 | 216 | 7.00E-51 |
| GL001455 | AJF82049.1 | 254 | 141 | 2 | 4 | 255 | 7 | 260 | 2.00E-62 |
| Subject ID | ARO number | Definition of term |
| YP_208874.1 | Neisseria gonorrhoeae FA 1090 | rpsJ is a tetracycline resistance protein identified in Neisseria gonorrhoeae. Tetracycline resistance is conferred by binding to the ribosome as a 30S ribosomal protection protein[27] |
| YP_006374661.1 | Enterococcus faecium DO | Sequence variants of Enterococcus faecium elongation factor Tu that can confer resistance to GE2270A[28] |
| NP_312937.1 | Escherichia coli O157•H7 str. Sakai | Point mutation that occurs in Escherichia coli rpoB resulting in resistance to rifampicin[29] |
| AAK44936.1 | Mycobacterium tuberculosis CDC1551 | Ribosomal protein S12 stabilizes the highly conserved pseudoknot structure formed by 16S rRNA. Amino acid substitutions in RpsL affect the higher-order structure of 16S rRNA and confer streptomycin resistance by disrupting interactions between 16S rRNA and streptomycin[30-35] |
| NP_207975.1 | Helicobacter pylori 26695 | hp1184 is a translocase that belongs to the MATE efflux pump family. It is found in H. pylori and is involved in the active efflux of antibiotics[25,26] |
| NP_207972.1 | Helicobacter pylori 26695 | hp1181 is a translocase that is part of the MFS efflux pump family. It is found in H. pylori and plays a role in the active efflux of antibiotics[25] |
| AIL15701 | Escherichia coli ATCC25922 | murA or UDP-N-acetylglucosamine enolpyruvy1 transferase catalyzes the initial step in peptidoglycan biosynthesis and is inhibited by Fosfomycin. Over-expression of murA through mutations such as Asp369Asn and Leu370I1e confers fosfomycin resistance. Extensive evidence has shown the significance of C115 mutations in conferring fosfomycin resistance since this residue represents a primary binding site for the antibiotic across many species[36-39] |
| YP_002344422.1 | Campylobacter jejuni subsp. jejuni NCTC 11168 | Campylobacter jejuni is a major bacterial infectious agent associated with gastroenteritis. Quinolone resistance is reportedly conferred by a single C-257-T nucleotide substitution in the gyrA gene[40] |
| NP_415611.1 | Escherichia coli str. K-12 substr. MG1655 | Fab G is a 3-oxoacyl-acyl carrier protein reductase involved in lipid metabolism and fatty acid biosynthesis. The bacterial biocide Triclosan blocks the final reduction step in fatty acid elongation, inhibiting biosynthesis. Point mutations in fabG can confer resistance to Triclosan[41] |
| WP_005768149.1 | Bartonella bacilliformis KC583 | Point mutation in Bartonella bacilliformis results in amino coumarin resistance[42] |
| AJF83452.1 | Acinetobacter baumannii | The LpxC gene is widely known to be involved in the biosynthesis of lipid A in Gram-negative bacteria and mutations to this gene may cause resistance to antimicrobial peptides that target the outer membrane[43,44] |
| NP_415804.1 | Escherichia coli str. K-12 substr. MG1655 | fabI is an enoyl-acy1 carrier reductase used in lipid metabolism and fatty acid biosynthesis. The bacterial biocide Triclosan blocks the final reduction step in fatty acid elongation, inhibiting biosynthesis. Point mutations in fabI can confer resistance to Triclosan and Isoniazid[41] |
| YP_001332362.1 | Staphylococcus aureus subsp. aureus str. Newman | Ar1R is a response regulator that binds to the norA promoter to activate expression. Ar1R must first be phosphorylated by Ar1S[45] |
| AJF82049.1 | Acinetobacter baumannii | The LpxA gene is widely known to be involved in the biosynthesis of lipid A in Gram-negative bacteria and mutations to this gene may cause resistance to antimicrobial peptides that target the outer membrane[43,44] |
As three strains were resistant to clarithromycin, so we analyzed and identified the sites of clarithromycin-resistant mutations. We found that three strains had mutations in A2142G, A2143G, G2144T, and some had mutations in other sites, as shown in Table 8.
| Nucleotide position | Ref | Mutation | Hpbs1 | Hpbs2 | Hpbs3 |
| 2143 | A | G | + | + | + |
| 2142 | A | G | + | + | + |
| 2144 | G | T | + | + | + |
| 2302 | A | G | - | - | + |
| 2182 | T | C | - | + | - |
| 2173 | C | T | + | + | + |
| 1513 | G | A | - | + | + |
| 2196 | C | T | + | - | - |
| 1280 | A | G | + | - | - |
| 1023 | G | A | - | - | + |
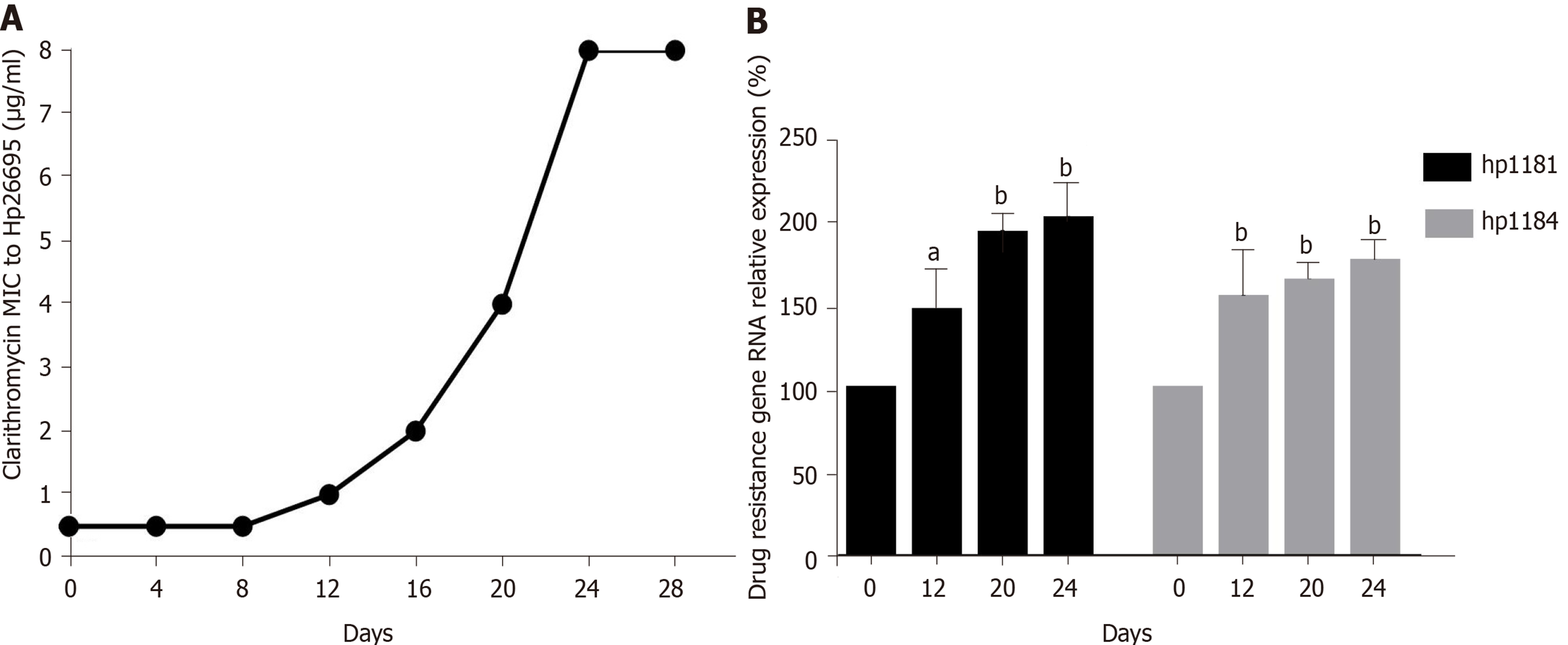
After induction with clarithromycin, hp26695 drug resistance was enhanced on the 12th day, reached the highest level on day 16, and increased to 8 μg/mL on the 24th day. The expression of hp1181 and hp1184 was also increased with increasing clarithromycin resistance, especially hp1184, as shown in Figure 4. Only A2142G and A 2143G mutations were detected in 23S RNA, with no other mutation sites being found, as shown in Table 9. These data indicated that these two genes may be involved early in the regulation of clarithromycin resistance.
| Nucleotide position | Ref | Mutation | 26695(S) | 26695(R) |
| 2142 | A | G | - | + |
| 2143 | A | G | - | + |
The treatment of H. pylori infection remains reliant on bismuth tetralogy at present. H. pylori is eradicated clinically using common antibiotics including clarithromycin, amoxicillin, metronidazole, tetracycline and levofloxacin. However, in recent years, the growing rate of antibiotic resistance has resulted in the failure of H. pylori eradication[19,20]. The most serious resistance has developed to drugs including metronidazole, clarithromycin, and levofloxacin star. The common mechanisms of bacterial resistance involve the production of inactivated enzymes, change in the target position of antibacterial drugs, change in the permeability of bacterial outer membrane, effects on the active outflow system, and formation of bacterial biofilm and cross resistance[21-23]. There are some differences in the mechanisms of drug resistance of each kind of bacteria; however, the same kind of bacteria still have different resistances to the same antibiotic in different areas[24]. The mechanism of drug resistance of H. pylori remains unclear and needs further study.
We selected drug-resistant strains using metronidazole, clarithromycin, and levofloxacin for genome sequencing analysis. We found that there were no significant differences in the number of drug-resistant genes in the CARD database. This may be because two kinds of antibiotic resistance can develop and the drug-resistant genes in H. pylori are mainly hp1181 and hp1184. Hp1181 encodes a putative NDA translocase that is related to the major facilitator superfamily and is an integral membrane protein; hp1184 encodes another translocase that belongs to the MATE family, resulting in the aforementioned susceptibility. These can contribute to resistance via a multidrug-resistant efflux protein, active-efflux of antibiotics, and other efflux pump genes, such as HefA. After knockout of these two genes, the MICs of the drugs were significantly decreased and the sensitivity was increased. It is noteworthy that in addition to these two genes, the GE2270A gene of Enterococcus and MurA gene of Escherichia coli also show a correlation. It is likely that the drug-resistant plasmids of other strains invade H. pylori through transformation or other mechanisms. Bacteria other than H. pylori in the gastric mucosa of patients can indirectly confirm this view. The main reason for this may be long-term acid resistant treatment, gastric erosion, or intestinal bacterial reflux. This will lead to drug resistance becoming more difficult to prevent and control. In addition, all three strains have clarithromycin resistance. The mechanism of resistance to clarithromycin is mainly reflected in the mutations A2142G, A2143G, and G2144T. In addition, it is common that there are several mutations in the same strain.
Hp1181 and hp1184 are related to multidrug resistance and to clarithromycin resistance, which has been previously reported in the literature[25,26]. The RNA expression of hp1181 and hp1184 were increased with the emergence of clarithromycin resistance, with hp1184 showing the fastest increase. Therefore, these genes are also involved in the regulation of drug resistance and may be one of the mechanisms of H. pylori resistance to clarithromycin. Compared with the clinical isolates, 23S RNA mutation sites of H. pylori were less frequent in artificially induced strains that had only A2142G and A2143G mutations. These may be attributed to the single factor of artificial induction that is not as complex as human stomach environment. More importantly, hp1181 and hp1184 mutations may be the earliest and most persistent response to clarithromycin resistance, and they may be the main target genes for the diagnosis, prevention, and treatment of clarithromycin resistance.
The genetic characteristics of multidrug-resistant strains in this area were preliminarily identified: The relationship between hp1181 or hp1184 and clarithromycin resistance was ascertained through genome sequencing analysis and gene function identification of drug-resistant H. pylori from Bama County, Guangxi Province. Our study further provided an improved experimental basis for the prevention and treatment of drug resistance of H. pylori.
Hp1181 and hp1184 mutations may be the earliest and most persistent response to clarithromycin resistance, and they may be the main target genes for the diagnosis, prevention, and treatment of clarithromycin resistance.
Helicobacter pylori (H. pylori) is recognized as an important human pathogen associated with superficial gastritis, atrophic gastritis, gastric cancer, etc., each of which has become a serious threat to human health and survival. The rate of drug resistance is increasing due to the wide use of antibiotics and high rates of resistance to clarithromycin, metronidazole, and levofloxacin are associated with the failure of H. pylori eradication. At present, the mechanism of antibiotic resistance of H. pylori is not completely understood. It is very difficult to prevent drug resistance and improve the rate of eradication of the target, thus warranting exploration of the mechanism of drug resistance to H. pylori, and provision of an experimental basis for the prevention and treatment of drug resistance.
Clarithromycin-resistant H. pylori urgently needs new antibiotics; however, antibiotic research and development are very difficult. If we can detect drug resistance by detecting drug-resistant genes in a timeous manner, this may help to alleviate the problem of clarithromycin resistance.
The objectives of this study were to investigate drug-resistant genes in H. pylori, and find a gene suited to early diagnosis of clarithromycin resistance, thereby rationalizing the rate of use of the drug.
H. pylori strains were isolated and cultured, minimal inhibitory concentrations were measured, and complete genome sequence was determined. Prediction and analysis of the function of drug-resistant genes indicated that the RNA expression of hp1181 and hp1184 increased in the H. pylori strains, which was the same in the artificially induced clarithromycin-resistant bacteria. The relationships between hp1181 or hp1184 and clarithromycin resistance were confirmed with gene mutant and drug-resistant strains.
Hp1181 and hp1184 genes were found in these H. pylori strains. Their expression was associated with clarithromycin resistance.
Hp1181 and hp1184 mutations may be the earliest and most persistent response to clarithromycin resistance, and they may be the main target genes for the diagnosis, prevention, and treatment of clarithromycin resistance.
The relationship between hp1181 or hp1184 and clarithromycin resistance was demonstrated, providing an improved experimental basis for early diagnosis of clarithromycin resistance in H. pylori.
The authors thank Huang YN and Huang MY working in Guangxi Bama Yao Autonomous County People Hospital who helped to collect gastric mucosal samples from clinical patients.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Araujo RLC, Fujiwara N, Scorsetti M S-Editor: Gao CC L-Editor: Wang TQ P-Editor: Wang LL
| 1. | Yoon K, Kim N, Lee JW, Yoon H, Shin CM, Park YS, Lee DH. Annual eradication rate of bismuth-containing quadruple therapy as second-line treatment for Helicobacter pylori infection: A 15-year prospective study at a tertiary hospital in Korea. Helicobacter. 2020;25:e12685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 2. | Fischbach W, Malfertheiner P. Helicobacter Pylori Infection. Dtsch Arztebl Int. 2018;115:429-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (1)] |
| 3. | Diaconu S, Predescu A, Moldoveanu A, Pop CS, Fierbințeanu-Braticevici C. Helicobacter pylori infection: old and new. J Med Life. 2017;10:112-117. [PubMed] |
| 4. | Lahner E, Carabotti M, Annibale B. Treatment of Helicobacter pylori infection in atrophic gastritis. World J Gastroenterol. 2018;24:2373-2380. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 75] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (1)] |
| 5. | Suzuki S, Esaki M, Kusano C, Ikehara H, Gotoda T. Development of Helicobacter pylori treatment: How do we manage antimicrobial resistance? World J Gastroenterol. 2019;25:1907-1912. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 53] [Cited by in RCA: 79] [Article Influence: 13.2] [Reference Citation Analysis (2)] |
| 6. | Malfertheiner P, Megraud F, O'Morain CA, Gisbert JP, Kuipers EJ, Axon AT, Bazzoli F, Gasbarrini A, Atherton J, Graham DY, Hunt R, Moayyedi P, Rokkas T, Rugge M, Selgrad M, Suerbaum S, Sugano K, El-Omar EM; European Helicobacter and Microbiota Study Group and Consensus panel. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut. 2017;66:6-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2220] [Cited by in RCA: 1984] [Article Influence: 248.0] [Reference Citation Analysis (1)] |
| 7. | Savoldi A, Carrara E, Graham DY, Conti M, Tacconelli E. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Systematic Review and Meta-analysis in World Health Organization Regions. Gastroenterology 2018; 155: 1372-1382. e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 617] [Cited by in RCA: 823] [Article Influence: 117.6] [Reference Citation Analysis (0)] |
| 8. | Liu DS, Wang YH, Zeng ZR, Zhang ZY, Lu H, Xu JM, Du YQ, Li Y, Wang JB, Xu SP, Chen Y, Lan CH, Cheng H, Jiang MD, Zhang LX, Huo LJ, Chen SY, Zhang GX, Wu KC, Zhu X, Chen YX, Zhu Y, Shu X, Xie Y, Lu NH. Primary antibiotic resistance of Helicobacter pylori in Chinese patients: a multiregion prospective 7-year study. Clin Microbiol Infect 2018; 24: 780.e5-780. e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 9. | Ge X, Cai Y, Chen Z, Gao S, Geng X, Li Y, Jia J, Sun Y. Bifunctional Enzyme SpoT Is Involved in Biofilm Formation of Helicobacter pylori with Multidrug Resistance by Upregulating Efflux Pump Hp1174 (gluP). Antimicrob Agents Chemother. 2018;62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 10. | Bińkowska A, Biernat MM, Łaczmański Ł, Gościniak G. Molecular Patterns of Resistance Among Helicobacter pylori Strains in South-Western Poland. Front Microbiol. 2018;9:3154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 11. | Zhang XY, Shen WX, Chen CF, Sheng HH, Cheng H, Li J, Hu F, Lu DR, Gao HJ. Detection of the clarithromycin resistance of Helicobacter pylori in gastric mucosa by the amplification refractory mutation system combined with quantitative real-time PCR. Cancer Med. 2019;8:1633-1640. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 12. | Matta AJ, Zambrano DC, Pazos AJ. Punctual mutations in 23S rRNA gene of clarithromycin-resistant Helicobacter pylori in Colombian populations. World J Gastroenterol. 2018;24:1531-1539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 17] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (1)] |
| 13. | Alarcón-Millán J, Fernández-Tilapa G, Cortés-Malagón EM, Castañón-Sánchez CA, De Sampedro-Reyes J, Cruz-Del Carmen I, Betancourt-Linares R, Román-Román A. Clarithromycin resistance and prevalence of Helicobacter pylori virulent genotypes in patients from Southern México with chronic gastritis. Infect Genet Evol. 2016;44:190-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 14. | Kouitcheu Mabeku LB, Eyoum Bille B, Tepap Zemnou C, Tali Nguefack LD, Leundji H. Broad spectrum resistance in Helicobacter pylori isolated from gastric biopsies of patients with dyspepsia in Cameroon and efflux-mediated multiresistance detection in MDR isolates. BMC Infect Dis. 2019;19:880. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 15. | Attaran B, Falsafi T, Ghorbanmehr N. Effect of biofilm formation by clinical isolates of Helicobacter pylori on the efflux-mediated resistance to commonly used antibiotics. World J Gastroenterol. 2017;23:1163-1170. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 40] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 16. | Su P, Li Y, Li H, Zhang J, Lin L, Wang Q, Guo F, Ji Z, Mao J, Tang W, Shi Z, Shao W, Zhu X, Zhang X, Tong Y, Tu H, Jiang M, Wang Z, Jin F, Yang N. Antibiotic resistance of Helicobacter pylori isolated in the Southeast Coastal Region of China. Helicobacter. 2013;18:274-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 136] [Article Influence: 11.3] [Reference Citation Analysis (1)] |
| 17. | Ranjbar R, Farsani FY, Dehkordi FS. Phenotypic analysis of antibiotic resistance and genotypic study of the vacA, cagA, iceA, oipA and babA genotypes of the Helicobacter pylori strains isolated from raw milk. Antimicrob Resist Infect Control. 2018;7:115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 18. | Humphries RM, Kircher S, Ferrell A, Krause KM, Malherbe R, Hsiung A, Burnham CA. The Continued Value of Disk Diffusion for Assessing Antimicrobial Susceptibility in Clinical Laboratories: Report from the Clinical and Laboratory Standards Institute Methods Development and Standardization Working Group. J Clin Microbiol. 2018;56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 19. | Goderska K, Agudo Pena S, Alarcon T. Helicobacter pylori treatment: antibiotics or probiotics. Appl Microbiol Biotechnol. 2018;102:1-7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 159] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 20. | Thung I, Aramin H, Vavinskaya V, Gupta S, Park JY, Crowe SE, Valasek MA. Review article: the global emergence of Helicobacter pylori antibiotic resistance. Aliment Pharmacol Ther. 2016;43:514-533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 410] [Cited by in RCA: 553] [Article Influence: 61.4] [Reference Citation Analysis (2)] |
| 21. | Munita JM, Arias CA. Mechanisms of Antibiotic Resistance. Microbiol Spectr. 2016;4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 927] [Cited by in RCA: 1346] [Article Influence: 168.3] [Reference Citation Analysis (0)] |
| 22. | Lee CR, Lee JH, Park M, Park KS, Bae IK, Kim YB, Cha CJ, Jeong BC, Lee SH. Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front Cell Infect Microbiol. 2017;7:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 333] [Cited by in RCA: 591] [Article Influence: 73.9] [Reference Citation Analysis (0)] |
| 23. | Pang Z, Raudonis R, Glick BR, Lin TJ, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. 2019;37:177-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 562] [Cited by in RCA: 1215] [Article Influence: 173.6] [Reference Citation Analysis (0)] |
| 24. | Chung The H, Baker S. Out of Asia: the independent rise and global spread of fluoroquinolone-resistant Shigella. Microb Genom. 2018;4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 25. | Falsafi T, Ehsani A, Attaran B, Niknam V. Association of hp1181 and hp1184 Genes With the Active Efflux Phenotype in Multidrug-Resistant Isolates of Helicobacter pylori. Jundishapur J Microbiol. 2016;9:e30726. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 26. | van Amsterdam K, Bart A, van der Ende A. A Helicobacter pylori TolC efflux pump confers resistance to metronidazole. Antimicrob Agents Chemother. 2005;49:1477-1482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 79] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 27. | Kubanov A, Vorobyev D, Chestkov A, Leinsoo A, Shaskolskiy B, Dementieva E, Solomka V, Plakhova X, Gryadunov D, Deryabin D. Molecular epidemiology of drug-resistant Neisseria gonorrhoeae in Russia (Current Status, 2015). BMC Infect Dis. 2016;16:389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 28. | Miele A, Goldstein BP, Bandera M, Jarvis C, Resconi A, Williams RJ. Differential susceptibilities of enterococcal species to elfamycin antibiotics. J Clin Microbiol. 1994;32:2016-2018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 29. | Jin DJ, Gross CA. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 519] [Cited by in RCA: 517] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 30. | Ballif M, Harino P, Ley S, Coscolla M, Niemann S, Carter R, Coulter C, Borrell S, Siba P, Phuanukoonnon S, Gagneux S, Beck HP. Drug resistance-conferring mutations in Mycobacterium tuberculosis from Madang, Papua New Guinea. BMC Microbiol. 2012;12:191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 31. | Finken M, Kirschner P, Meier A, Wrede A, Böttger EC. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol Microbiol. 1993;9:1239-1246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 231] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 32. | Okamoto S, Tamaru A, Nakajima C, Nishimura K, Tanaka Y, Tokuyama S, Suzuki Y, Ochi K. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol Microbiol. 2007;63:1096-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 203] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 33. | Sreevatsan S, Pan X, Stockbauer KE, Williams DL, Kreiswirth BN, Musser JM. Characterization of rpsL and rrs mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from diverse geographic localities. Antimicrob Agents Chemother. 1996;40:1024-1026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 126] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 34. | Nair J, Rouse DA, Bai GH, Morris SL. The rpsL gene and streptomycin resistance in single and multiple drug-resistant strains of Mycobacterium tuberculosis. Mol Microbiol. 1993;10:521-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 105] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 35. | Brzostek A, Sajduda A, Sliwiński T, Augustynowicz-Kopeć E, Jaworski A, Zwolska Z, Dziadek J. Molecular characterisation of streptomycin-resistant Mycobacterium tuberculosis strains isolated in Poland. Int J Tuberc Lung Dis. 2004;8:1032-1035. [PubMed] |
| 36. | Wanke C, Amrhein N. Evidence that the reaction of the UDP-N-acetylglucosamine 1-carboxyvinyltransferase proceeds through the O-phosphothioketal of pyruvic acid bound to Cys115 of the enzyme. Eur J Biochem. 1993;218:861-870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 53] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 37. | Kim DH, Lees WJ, Kempsell KE, Lane WS, Duncan K, Walsh CT. Characterization of a Cys115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry. 1996;35:4923-4928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 147] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 38. | Cheng G, Hu Y, Lu N, Li J, Wang Z, Chen Q, Zhu B. Identification of a novel fosfomycin-resistant UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) from a soil metagenome. Biotechnol Lett. 2013;35:273-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 39. | Takahata S, Ida T, Hiraishi T, Sakakibara S, Maebashi K, Terada S, Muratani T, Matsumoto T, Nakahama C, Tomono K. Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int J Antimicrob Agents. 2010;35:333-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 143] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 40. | Hormeño L, Palomo G, Ugarte-Ruiz M, Porrero MC, Borge C, Vadillo S, Píriz S, Domínguez L, Campos MJ, Quesada A. Identification of the main quinolone resistance determinant in Campylobacter jejuni and Campylobacter coli by MAMA-DEG PCR. Diagn Microbiol Infect Dis. 2016;84:236-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 41. | Khan R, Kong HG, Jung YH, Choi J, Baek KY, Hwang EC, Lee SW. Triclosan Resistome from Metagenome Reveals Diverse Enoyl Acyl Carrier Protein Reductases and Selective Enrichment of Triclosan Resistance Genes. Sci Rep. 2016;6:32322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 42. | Battisti JM, Smitherman LS, Samuels DS, Minnick MF. Mutations in Bartonella bacilliformis gyrB confer resistance to coumermycin A1. Antimicrob Agents Chemother. 1998;42:2906-2913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 43. | Moffatt JH, Harper M, Harrison P, Hale JD, Vinogradov E, Seemann T, Henry R, Crane B, St Michael F, Cox AD, Adler B, Nation RL, Li J, Boyce JD. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother. 2010;54:4971-4977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 515] [Cited by in RCA: 617] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 44. | Beceiro A, Moreno A, Fernández N, Vallejo JA, Aranda J, Adler B, Harper M, Boyce JD, Bou G. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob Agents Chemother. 2014;58:518-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 182] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 45. | Fournier B, Aras R, Hooper DC. Expression of the multidrug resistance transporter NorA from Staphylococcus aureus is modified by a two-component regulatory system. J Bacteriol. 2000;182:664-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 92] [Article Influence: 3.7] [Reference Citation Analysis (0)] |