Published online Jun 21, 2021. doi: 10.3748/wjg.v27.i23.3182
Peer-review started: January 29, 2021
First decision: March 14, 2021
Revised: March 23, 2021
Accepted: May 7, 2021
Article in press: May 7, 2021
Published online: June 21, 2021
Processing time: 140 Days and 2.8 Hours
Global prophylactic vaccination programmes have helped to curb new hepatitis B virus (HBV) infections. However, it is estimated that nearly 300 million people are chronically infected and have a high risk of developing hepatocellular carcinoma. As such, HBV remains a serious health priority and the development of novel curative therapeutics is urgently needed. Chronic HBV infection has been attributed to the persistence of the covalently closed circular DNA (cccDNA) which establishes itself as a minichromosome in the nucleus of hepatocytes. As the viral transcription intermediate, the cccDNA is responsible for producing new virions and perpetuating infection. HBV is dependent on various host factors for cccDNA formation and the minichromosome is amenable to epigenetic modifications. Two HBV proteins, X (HBx) and core (HBc) promote viral replication by modulating the cccDNA epigenome and regulating host cell responses. This includes viral and host gene expression, chromatin remodeling, DNA methyla
Core Tip: Epigenetic regulation of the hepatitis B virus (HBV) covalently closed circular DNA (cccDNA) minichromosome is important for establishing and maintaining infection. To do this HBV manipulates several cellular pathways, resulting in an intricate and complex interplay between the virus and the host. Epigenetic silencing of the cccDNA could permanently inhibit viral transcription. Therapies such as immune modulators, small molecules, and epigenome engineering tools could silence HBV DNA to promote a functional cure.
- Citation: Singh P, Kairuz D, Arbuthnot P, Bloom K. Silencing hepatitis B virus covalently closed circular DNA: The potential of an epigenetic therapy approach. World J Gastroenterol 2021; 27(23): 3182-3207
- URL: https://www.wjgnet.com/1007-9327/full/v27/i23/3182.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i23.3182
Chronic HBV infection (CHB) remains largely incurable and is a major risk factor for the development of hepatocellular carcinoma (HCC)[1]. Regrettably, only a small percentage of chronically infected individuals are currently afforded therapy[2] and the annual rate of hepatitis-related deaths remains high. This underpins the importance of developing new therapeutics to treat the estimated 257-291 million people afflicted by this life-threatening disease[3]. Licensed direct-acting antivirals such as nucleoside/nucleotide analogues help mitigate infection by preventing viral replication but have little effect on established covalently closed circular DNA (cccDNA)[4]. Immunotherapy approaches using interferon alpha (IFN-α) have demonstrated antiviral efficacy, particularly when using pegylated formulations, and have been paired with various nucleoside/nucleotide analogues for combination therapy[5]. However cost, serious side effects, and mixed results have limited the broad clinical feasibility of IFN-based therapies in their current form. Persistence of the cccDNA as a minichromosome-like structure in the nucleus of long-lived hepatocytes may account for the inability of these antivirals to achieve cure, even after long-term treatment[6]. A sterilizing cure can only be realized following complete elimination of intrahepatic cccDNA, while permanent loss of serum hepatitis B surface antigen (HBsAg) would achieve a functional cure. The cccDNA is thought to endure for years[7] and as the template for viral mRNA synthesis[8], it drives viral replication and remains the major obstacle in the development of curative antiviral therapies.
In recent years, treatments designed to directly silence or eliminate the cccDNA minichromosome have gained interest as they may provide a means for achieving cure[9]. Gene editing tools such as designer nucleases and nickases disrupt the viral DNA via site-directed cleavage. This stimulates either targeted mutagenesis or degradation of the cccDNA[10,11]. Zinc finger (ZF) nucleases, transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR) RNA-guided nucleases have all demonstrated antiviral potential in preclinical models of infection. The simplicity of the CRISPR/Cas system has led to a surge in the field of anti-HBV gene editing, particularly as multiple studies have reported degradation of cccDNA following cleavage[12-15]. The CRISPR/Cas constructs developed by Seeger and Sohn[16] were shown to edit HBV DNA 15000 times more efficiently than IFN-α-induced APOBEC deamination. However, concerns regarding off-target cleavage, liver-specific delivery and expression, and Cas9 immunity will need to be addressed to ensure safety without compromising efficacy[17-20]. Viral vectors, including adeno-associated viruses (AAVs) and high-capacity adenoviral vectors (HCAdV) have recently been explored as hepatotropic delivery vehicles[13,21,22]. To overcome the packaging limitations of AAVs, the smaller Staphylococcus aureus endonucleases have been combined with HBV-specific guide RNAs to demonstrate cccDNA-targeting and antiviral efficacy in hNTCP-HepG2 cells[22] and transgenic mice[21]. On the other hand, the large packaging capacity of HCAdVs have been exploited to accommodate multiple HBV guide RNAs along with the larger Streptococcus pyogenes Cas9[13], which may improve the efficacy of this gene editing approach. Importantly, iterative modifications to the effector domains of traditional gene editing platforms have given rise to several new tools. CRISPR/Cas base editors comprising an APOBEC1 deaminase, Cas9 nickase, and uracil glycosylase inhibitor introduced point mutations in cccDNA as well as integrated viral DNA[23]. A number of new epigenome engineering platforms have also been developed, which could be used to generate targeted epigenetic changes to control gene expression[24]. Such an approach may be well suited to treating CHB, as epigenetic modifications regulate cccDNA minichromosome organization and can either promote or repress viral transcription[25-27]. Host and/or virus-specific epigenetic therapies could promote silencing of HBV cccDNA and achieve a functional cure. This review will explore the formation and epigenetic regulation of the cccDNA minichromosome, how host and viral factors influence transcription, and whether epigenome editing could be used to silence HBV cccDNA permanently.
cccDNA biogenesis is a multi-step process that relies on a number of host-cell DNA synthesis and repair factors[28,29]. Although the exact process is not fully understood it involves: (1) Removal of the covalently attached viral polymerase from the 5′ end of the negative DNA strand; (2) Removal of the short RNA oligomer attached to the 5′ end of the positive strand which is derived from the priming of positive-strand DNA synthesis; (3) Removal of one copy of the short terminal redundancy from the negative-strand; (4) Positive-strand elongation; and (5) Covalent ligation of the two strands[30-32]. Evidence from animal models has shown that between 1 and 50 copies of cccDNA may accumulate per infected cell[33-36]; however, more recent reports estimate up to 15 copies per cell[37,38]. Multiple rounds of infection are not necessary to establish the cccDNA pool as recycling of newly synthesized relaxed circular DNA (rcDNA) has been shown to occur, and contributes to the maintenance of cccDNA copy numbers[39-41]. Increasing evidence also indicates that the size of the pool is controlled by different host and viral mechanisms[42], but that initial de novo cccDNA formation is sufficient to maintain viral replication in the absence of rcDNA recycling[43].
Many different host factors have been implicated in cccDNA biogenesis, including host DNA polymerases[32,44], ligases[45], flap endonuclease I (FEN1)[46], topoiso
Once formed, cccDNA is organized into a nucleosome-decorated minichromosome[8,52]. It associates with histone and non-histone proteins, the latter of which are derived from both the host and the virus[29,33]. As a hepatotropic virus, the viral genome contains binding sites for ubiquitous and liver-specific transcription factors[53]. Thus, the recruitment, activity, and dynamic interplay of several host and viral factors are essential for efficient HBV gene expression[25-27]. Several histone proteins, transcription factors, chromatin modifying enzymes, as well as additional host enzymes and coactivators have been reported to bind to cccDNA (Table 1). Recrui
| Category | Epigenetic effect | Effect on HBV | Ref. | |
| Chromatin remodeling enzymes | ||||
| Histone acetyltransferase p300/CBP | Writer | Increases H3K122a | Activation | Ananthanarayanan et al[99] and Tropberger et al[221] |
| P300/CBP-associated factor (PCAF) | Writer | Activation | Levrero et al[82] and Rivière et al[93] | |
| Histone deacetylase 1 (HDAC1) | Eraser | Reduces acetylation of H3K9 and H3K27 with IFN | Inhibition | Pollicino et al[57] and Liu et al[98] |
| Sirtuin 1 and 3 (SIRT1/3) | Eraser | Reduces H3K9me3 | Inhibition | Belloni et al[105] and Ren et al[114] |
| Enhancer of zeste homolog 2 (EZH2) | Writer | Increases H3K27ac and H3K27me3 | Inhibition | Zhang et al[89] |
| Histone acetyltransferase 1 (HAT1) | Writer | Overexpression of HAT1 promotes acetylation of H3K27, H4K5 and H4K12 | Activation | Yang et al[195] |
| Mixed lineage leukemia protein 3 (MLL3) | Writer | Increases H3K4me3 | Activation | Tropberger et al[86] and Ananthanarayanan et al[99] |
| Protein arginine methyltransferase (PRMT) | Writer | PRMT5 interacts with HBc to increase H4R3me2s | Inhibition | Zhang et al[89] |
| Demethylases (KDMs) | Eraser | Increases H3K79me and function to transcriptional repression via SIRT1-mediated chromatin silencing | Inhibition | Kang et al[222] |
| Histone methyltransferase suppressor of variegation 3-9 homolog 1 (SUV39H1) | Writer | Increases H3K9me3 | Inhibition | Peng and Karpen[223,224] |
| DNA methyltransferase (DNMTs) | Writer | Inhibition | Vivekanandan et al[81] | |
| Methyl-CpG binding protein (MBPs) | Reader | Recruit chromatin remodeling and histone-modifying complexes to methylated sites resulting in histone methylation | Inhibition | Lopez-Serra and Esteller[94] |
| Cellular transcription factors | ||||
| Activating transcription factor 2 (ATF 2) | Inhibition viral transcription | Inhibition | Choi et al[225] | |
| cAMP response element binding protein (CREB) | Enhances transcription | Activation | Kim et al[226] and Song et al[227] | |
| Nuclear factor 1 (NF1) | Reader | Activation | Ori et al[228] and Shaul et al[229] | |
| Transcription factor Yin Yang 1 (YY1) | Inhibition | Belloni et al[199] and Nakanishi-Matsui et al[230] | ||
| Specificity protein 1 (SP1) | Reader | Activation | Raney and McLachlan[231], Raney et al[232] and Li and Ou[233] | |
| Nuclear Transcription Factor Y (NF-Y) | Recruit enzymes that both methylate and acetylate histone proteins | Activation | Lu and Yen[234], Maity and de Crombrugghe[235] and Nardini et al[236] | |
| Activator protein 1(AP-1) | Activation | Ren et al[111] and Choi et al[237] | ||
| TATA binding protein (TBP) | Reader | Activation | Bogomolski-Yahalom et al[238] and Chen et al[239] | |
| Prospero-related homeobox protein (Prox1) | Inhibition | Qin et al[240] | ||
| Nuclear factor kappa-B (NF-κB) | Inhibition | Lin et al[241] | ||
| Histone-lysine N-methyltransferase SETDB1 | Writer | In the absence of HBx, SETDB1 increases H3K9me2 and H3K9me3 | Inhibition | Rivière et al[93] |
| Hepatocyte factors | ||||
| Retinoid X receptors (RXRα) | Increases acetylation of histones H4 and H3 by recruiting p300 to cccDNA minichromosome | Activation | Zhang et al[89] and Nkongolo et al[242] | |
| Small heterodimer partner (SHP) | Inhibition | Oropeza et al[243] | ||
| CAAT enhancer-binding protein α and ζ (C/EBP) | C/EBP in low concentrations; C/EBP in high concentrations | Activation; Inhibition | Raney and McLachlan[231] and | |
| Hepatocyte nuclear factor 1 α and β (HNF1) | HNF1 and Oct 1 are essential co-activators of transcription. High HNF1 levels increase NF-κB expression and resulting in transcription inhibition | Activation and inhibition | Zhou and Yen[245], Zheng et al[246] and Lin et al[247] | |
| Hepatocyte nuclear factor 3 α, β, and γ (HNF3) | Functions as a chromatin remodeler | Activation | Chen et al[248] and Li et al[249] | |
| Hepatocyte nuclear factor 4 (HNF4) | Activation | Zheng et al[246], Cho et al[250] and Long et al[251] | ||
| Testicular orphan receptor 4 (TR4) | Inhibition | Lin et al[252] | ||
| Type I interferon (IFN-α) | Reduced acetylation of H3K9and H3K27 | Inhibition | Pollicino et al[57], Liu et al[98] and Yuan et al[253] | |
Epigenetic modifications are reversible heritable phenotypic changes that alter chemical signatures on chromosomes without affecting the DNA sequence. This highly regulated process has a critical impact on gene expression, cell function as well as cell behavior[54]. Epigenetic marks can be added directly to DNA or placed on histone tails. Methylation of cytosines within CpG dinucleotides is the most common DNA epigenetic mark, and is critical for controlling transcription, genomic imprinting and cell type identity maintenance[55]. The wrapping of DNA around histones creates highly condensed chromatin structures with protruding N-terminal histone tails amenable to PTMs[56]. Histone acetylation and DNA methylation are required for cccDNA formation and viral replication[57], highlighting the importance of the epigenome in HBV infection.
Numerous proteins are involved in the epigenetic regulation of genes and are generally categorized as writers, readers and erasers (Table 1)[54]. Writers are responsible for establishing epigenetic marks while readers recognize modified residues and recruit other protein complexes to regulate gene expression. Erasers are enzymes that remove epigenetic marks. Epigenetic therapies generally target one or more of these proteins, and studies using this approach for other chronic viral infections[58,59] suggest that this may be a promising treatment strategy for CHB.
In mammalian cells, methylation is a common cellular defense mechanism known to silence invading foreign DNA[60]. It involves the addition of a methyl group from the methyl donor, S-adenosyl methionine, to the fifth carbon of cytosine in DNA (5mC). DNA methylation is most commonly detected within CpG islands and is associated with transcriptional gene silencing[61]. These inheritable 5mC marks can be ‘written’ (added) de novo in unmethylated regions which are either maintained or actively ‘erased’ (removed)[62]. In mammals, methyl-CpG binding proteins (MBPs) are responsible for the identification of methylation patterns[63] and a family of DNA methyltransferases (DNMTs) is responsible for establishment and maintenance of these patterns[64]. The DNMT family consists of DNMT1, DNMT2, DNMT3A, DNMT3B and DNMT3L (DNMT3-like). DNMT1 is primarily responsible for the maintenance of the methylation patterns during cell division and shows a preference for hemimethylated DNA[65,66]. De novo methylation patterns are established by the two active DNMT3A and DNMT3B enzymes and their activity is enhanced by interaction with DNMT3L which lacks catalytic activity[67,68]. In addition, DNMT2 plays an important role in methylation of structural RNA[69]. More than 20 years ago, it was discovered that integrated HBV DNA could be methylated and this was frequently observed in CHB patients with HCC[70]. Since then methylation of non-integrated HBV DNA as well as cccDNA has been observed in patients’ liver samples[71,72]. Several studies have reported that HBV cccDNA can be methylated to various extents, resulting in repression of transcription, decreased viremia and loss of hepatitis B e antigen (HBeAg)[71,73,74]. Thus, the methylation pattern of cccDNA is a crucial component of viral replication and may affect pathogenesis.
HBV cccDNA has three predicted CpG islands which are strategically located in the regulatory elements of the viral genome[75]. The first CpG island (CpG I) overlaps the ATG start site of the sequence encoding the small HBsAg, the second (CpG II) overlaps the enhancer I and II, the core promoter, and HBx promoter sequence, and the third (CpG III) spans the SpI promoter and ATG start codon of the polymerase gene[72,76]. Among the 10 distinct HBV genotypes (A-J), CpG I is present in five genotypes (A, B, D, E, and I) whereas CpG II and III have been shown to exist in all genotypes[76]. Varying degrees of methylation have been reported to occur at the three CpG islands. In patients with CHB, methylation frequencies of 14, 0.6 and 3.7% within CpG I, II and III, respectively, were observed[77]. However, a separate computational study revealed that 50% of HBV sequences lacked CpG I, whereas CpG II and II were conserved across genotypes[76]. This suggests that methylation patterns are likely to differ between the genotypes. Vivekanandan et al[72] demonstrated that increased methylation of CpG I and II correlated with reduced viral protein production when analyzing HBV DNA samples isolated from CHB liver biopsies. In addition, the authors showed that patients with occult HBV infection had increased methylation at CpG II when compared to non-occult CHB individuals. A subsequent study compared the methylation status of CpG II in liver tissues from HBeAg+ and HBeAg- individuals[71]. Methylation was higher in HBeAg- samples (48%) compared to HBeAg+ samples (14%), indicating that an increase in methylation may reduce viral protein production. Similarly, a link between methylation of CpG III and lower HBsAg levels has been reported[74]. A cohort of cirrhosis patients failed to show an association between cccDNA methylation and HBsAg expression, but did indicate that higher methylation density was associated with lower viral load and virion productivity[73]. These results show the impact methylation can have on viral gene expression in CHB[78]. A recent comprehensive meta-analysis revealed significant hypermethylation of certain host genes depending on the geographical location of the population[79]. This included six genes in HBV-positive carcinoma tissues (p16, RASSF1A, GSTP1, APC, p15 and SFRP1), two genes in HBV-positive carcinoma sera (p16 and APC) and one gene in HBV-positive adjacent tissues (GSTP1). The study also indicated that DNA methylation could lead to the development of HBV-related HCC, an important consideration for epigenetic therapy.
The gene silencing effects of methylation have also been validated in different cell culture models of HBV replication. Transfection of HepG2 cells with methylated HBV DNA led to a reduction in viral mRNA levels, decreased HBV core antigen (HBcAg) and HBsAg expression, and reduced secretion of viral proteins[80]. In vitro infection experiments showed an increase in DNMT expression in response to HBV, which led to hypermethylation of the viral DNA, a reduction in viral mRNAs and proteins, and decreased HBV replication[81]. Furthermore, co-transfection of HBV DNA and DNMT3a was associated with decreased production of HBeAg and HBsAg[81].
HBV cccDNA is associated with host histones, whose “bead-on-a-string” organization serves to compact the viral genome and provide a means for regulating gene expression[52]. As such transcription can be controlled by epigenetic modifications to the cccDNA-bound histones, thereby regulating HBV replication[82]. Nucleosome-associated histones undergo numerous types of PTMs that are generally reversible and mainly localized at the amino-terminal histone tails. These direct modifications of the N-terminal tails most commonly include acetylation, methylation, ubiquitination, phosphorylation, SUMOylation, ADP-ribosylation, and deamination[83]. Histone modifications can also indirectly regulate chromatin structure by serving as binding sites for the recruitment of other regulatory proteins[84]. Current studies have shown that PTMs significantly impact HBV replication, maturation and infection[85,86]. Several enzymes such as histone acetyltransferases (HATs) and deacetylases (HDACs), lysine and protein arginine methyltransferases (KMTs and PRMTs), demethylases (HDMTs), kinases, phosphatases, ubiquitin ligase, and deubiquitinases, modify cccDNA-associated histones[83,87]. The epigenome is further influenced by HBx and HBc, adding to the intricate and complex interplay between host and viral proteins on the cccDNA minichromosome (discussed later). Regulation of histone modifications has been proposed as a likely method to reduce cccDNA[8,52,57]. HATs facilitate the acetylation of lysine residues, making the histone-associated DNA more accessible to transcription factors, effectors and RNA polymerase II, to promote gene expression[88,89]. Conversely, HDACs catalyze the removal of acetyl groups from lysine residues, leading to heterochromatin formation and gene repression[90,91]. Although less characterized, MBPs are reported to recruit HMTs and HDACs to promote histone methylation, which in turn recruits heterochromatin protein 1 factors (HP1) to promote DNA methylation[92-94].
HBV transcription is regulated by the acetylation status of cccDNA bound histones 3 and 4 (H3 and H4) both in cell culture models of viral replication[57,95] and in CHB patients[57]. Recruitment of HDAC1 and hypoacetylation of H3 and H4 is associated with low HBV replication and viremia in vitro and in vivo[57]. In this study, Pollicino et al[57] demonstrated that acetylation of cccDNA-bound H4 correlated with high levels of replication, an effect that was maintained when using HDAC inhibitors. Further
Genome wide studies of PTMs on the cccDNA minichromosome revealed high levels of trimethylation at lysine 4 of histone H3 (H3K4me3) and acetylation of lysine residues of histone H3 (H3K27ac and H3K122ac) at specific loci, which lead to gene activation or repression[86]. H3K4me3 modifications are crucial for active transcrip
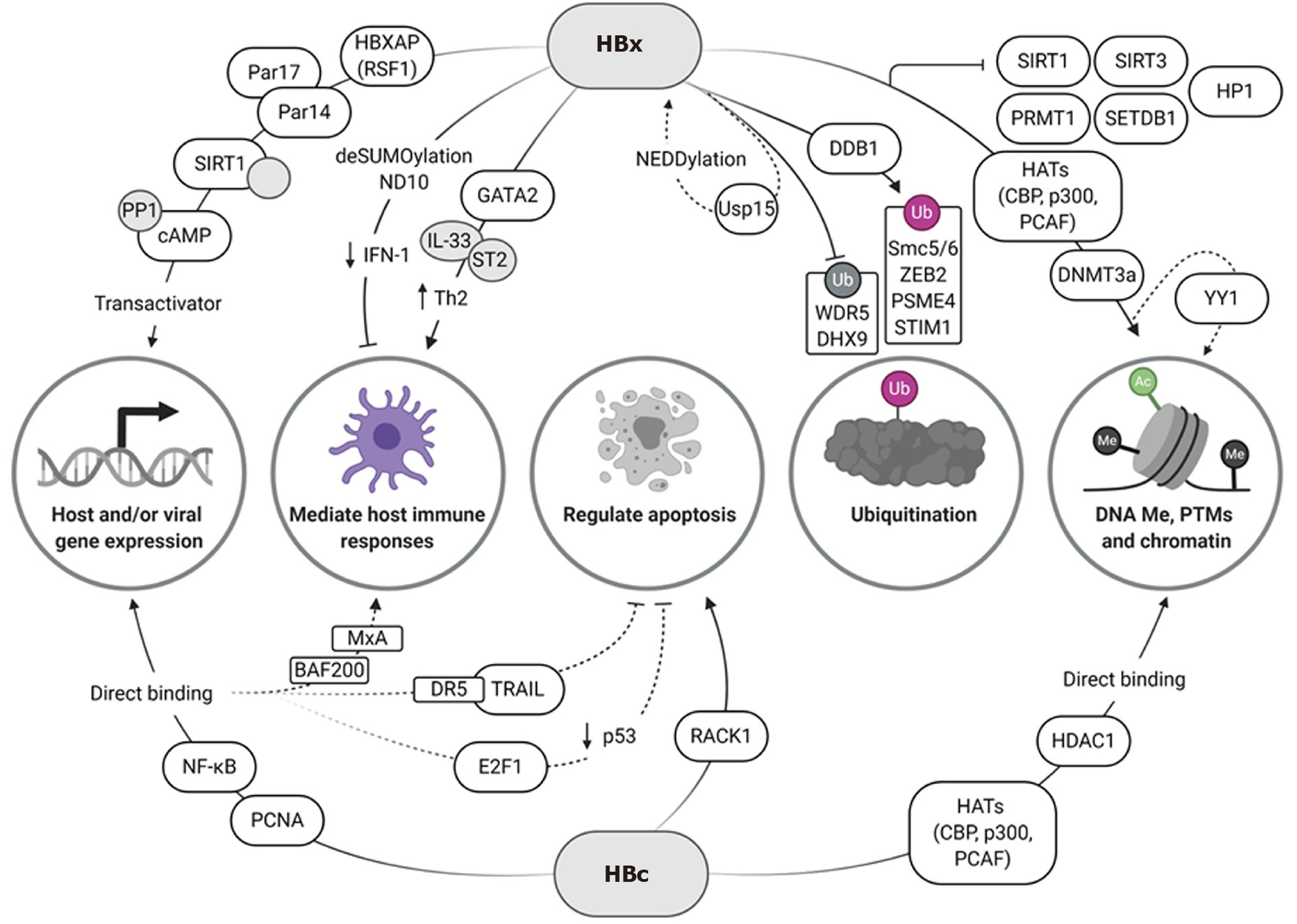
Two viral proteins, HBx and HBc, have been identified as important modulators of HBV transcription which influence the cccDNA epigenome (Figure 1). HBx is a small 17 kDa oncogenic protein that has been shown to interact with multiple host factors to manipulate both host and viral gene expression, mediate host immune responses, and control apoptosis[100]. Although HBx lacks DNA-binding motifs and does not directly bind to the cccDNA minichromosome, it is able to act as a trans-activator of viral replication by recruiting other factors to the minichromosome[101]. HBc is a 21 kDa structural polypeptide that forms the viral nucleocapsid and is essential for reverse transcription of the pgRNA[102]. As such, HBc is important for the packaging and secretion of new virions during HBV replication. Interestingly, it also forms part of the cccDNA minichromosome and is involved in its epigenetic regulation[8,57]. Unlike HBx, the HBc C-terminal domain (CTD) is capable of binding directly to cccDNA[103] as well as host gene promoter regions[104]. As such, HBc binding affects minichromosome organization as it can adjust the number and spacing of cccDNA-associated nucleosomes[57,105,106].
HBx has been shown to recruit a variety of host transcription factors and co-activators to regulate cccDNA transcription (Figure 1). These include ATF/CREB, ATF3, c/EBP, NF-κB, IL-6, Ets, Egr, SMAD4, Oct1, RXR receptor, and p53[82]. HBx-associated protein HBXAP (RSF1) has been shown to bind to HBx via its plant homology domain, and acts as a co-activator of viral transcription[107]. Recently, Saeed et al[108] demonstrated that HBx can directly bind to parvulin 14 (Par14) and 17 (Par17), which increased the stability of HBx and mediated its translocation to the nucleus and mitochondria. In addition, binding of Par14 and Par17 to the cccDNA at enhancer and promoter regions resulted in upregulation of viral transcription, an effect that was abrogated in the absence of HBx[108]. HBx has also been shown to recruit, block and promote degradation of host antiviral proteins that would normally prevent HBV replication and cccDNA persistence, as well as activate host genes to enhance viral gene expression.
Initiation of viral replication and stabilization of the cccDNA minichromosome is thought to occur through HBx-mediated PTMs of histones associated with hypomethylated CpG islands (mainly GpG I and III)[74,109]. Recruitment of HATs such as CBP, p300, and the p300/CBP-associated factor (PCAF) to the cccDNA minichromosome allows for upregulation of viral transcription[105]. In the presence of an HBx mutant, Belloni et al[105] showed increased recruitment of HDAC1 and sirtuin 1 (SIRT1) to the cccDNA, which correlated with a reduction in acetylation of cccDNA-bound H4 and viral transcription. These results suggest a role for HBx in promoting cccDNA histone acetylation to enhance viral transcription. However, the involvement of SIRT1 in the promotion or suppression of HBV replication appears to vary. Other studies found that SIRT1 positively regulated HBV replication by deacetylating PGC-1α and FXRα[110], and promoting binding of activator protein 1 to the core promoter[111]. However, these studies did not investigate the role of SIRT1 in cccDNA-dependent models of viral replication. More recently Deng et al[112] showed that in HepG2-NTCP cells, SIRT1 complexes with and stabilizes HBx, and promoted the recruitment of HBx and other co-activating factors to cccDNA. This increased viral transcription in a deacetylase-independent manner. These results suggest a dual role for SIRT1 in the regulation of HBV replication, which may be dependent on the presence or absence of HBx.
Binding of HBx to PRMT1 inhibited the methyltransferase activity of this enzyme, ultimately enhancing viral replication[113]. Recently, HBx was found to inhibit SIRT3 expression by downregulating its expression and possibly inhibiting its recruitment to the cccDNA[114]. SIRT3 promotes cccDNA-histone methylation (H3K9me3) by directly removing H3K9ac marks and recruiting histone methyltransferase SUV39H1 and SETD1A to the cccDNA[114]. However, in the presence of HBx, H3K9me3 marks were reduced and H3K4me3 and H3K9ac modifications increased to promote HBV transcription. This effect has also been observed for other transcriptional repressors. HBx reduced the chromatin structure alteration-mediated repression of histone-lysine N-methyltransferase SET domain, bifurcate 1 (SETDB1) and HP1[93]; however, the exact molecular mechanisms of this rescue remain to be elucidated. Upregulation of DNMTs by HBx and direct interaction of HBx with DNMT3A has been shown to either upregulate transcription of host genes, or block activity by facilitating host promoter methylation[115]. For example, SOCS-1, SUFU and TIRAP are downregulated, and this could be important in mediating an immune response in CHB, albeit the exact mechanisms still need to be elucidated[81,116]. HBx-mediated downregulation of tumor suppressors may be a factor leading to the development of HCC[115,117-119]. HBx-mediated deSUMOylation removes transcription factors from nuclear domain 10 (ND10), which in turn downregulates IFN-I-response pathway elements and host epigenetic modifications by p300/HDAC1, resulting in viral persistence[120]. Host gene promoters and enhancers can also be stimulated by HBx to regulate HBV replication[121-123]. Upregulation of host ST2 expression by HBx-GATA2 binding to its promoter and subsequent induction of the IL-33/ST2 axis stimulated a Th2 immune response, instead of the desired antiviral Th1 response[124], which is likely to play a role in the chronicity of infection[125].
As stated before, CREB is recruited by HBx to the cccDNA minichromosome; however, HBx also increases this activator’s longevity[126]. By binding to protein phosphatase 1, the activating phosphorylation induced by cAMP has a longer half-life, resulting in increased viral transcription[126]. Interestingly, not only does HBx mediate recruitment of factors directly to the cccDNA, but also promotes spatial localization of the minichromosome. Shen et al[127] showed that HBx and the transcription factor Yin-Yang 1 (YY1) aligned the cccDNA minichromosome with a region of the human genome rich in a highly active enhancer element, resulting in increased HBV replication.
Negative regulation by HBx has also been shown through an interaction with peroxiredoxin 1 (Prdx1), a cellular hydrogen peroxide scavenger, which recruits exosome component 5 (Excosc5) to degrade HBV RNA. Although the ability of Prdx1 to bind to HBV RNA was independent of HBx, HBx was required for degradation of the RNA[128].
Indirect epigenetic regulation can also occur through HBx-mediated proteasomal degradation of host factors, or by preventing degradation or deactivation of host factors. Damaged DNA-binding protein 1 (DDB1), an adapter protein for the cullin-RING ligase 4 (CRL4) E3 Ligase complex, is responsible for recruitment of DDB1-Cul4-associated factors (DCAFs), which allow for ubiquitination and subsequent degradation of proteins[129]. Interactions between HBx and DDB1 were implicated in early studies[130]. The interaction is mediated by a motif in HBx that mimics the DCAF proteins’ binding motif, allowing HBV to hijack the host protein ubiquitination system[131]. Years later, Decorsière et al[132] and Murphy et al[133] identified the structural maintenance of chromosomes 5/6 (Smc5/6) complex as a target for HBx-mediated proteasomal degradation. The Smc5/6 complex is a host antiviral restriction factor that enables transcriptional repression of the cccDNA minichromosome, by localizing to the ND10 without inducing an innate immune response[134]. However, the epigenetic effect is transient as it does not silence HBx mRNA expression, ultimately leading to Smc5/6 degradation and initiation of viral transcription[135]. Like Smc5/6, other HBV restriction factors are degraded by HBx-mediated polyubiquitination, to increase viral transcription and HBV replication[136]. This includes ZEB2, which inhibits HBV replication by interaction with the core promoter[137], proteasome activator subunit 4 (PSME4), which induces degradation of acetylated histones involved in upregulation of HBV[138,139], and stromal interaction molecule 1 (STIM1), known to regulate cytoplasmic calcium levels[140].
On the other hand, HBx is also able to inhibit proteasomal degradation[141]. This has recently been shown for the WD repeat domain 5 protein (WDR5), a core subunit of H3K4 methyltransferase complexes[142]. HBx stabilized WDR5 by preventing DDB1/cullin-4-induced degradation and in turn, promoted viral gene expression[142]. Shen et al[143] demonstrated how HBx could also inhibit MDM2-mediated degradation of DExH-box RNA helicase 9 (DHX9) to enhance viral DNA synthesis.
The epigenetic regulation of viral transcription is further enhanced as a result of HBx-mediated trans-activation of its own promoter[144]. Along with this, post-translational modifications of HBx through interactions with the deubiquitinating ubiquitin-specific peptidase 15 (Usp15) enzyme, increase the half-life of the viral protein[145]. This may also be a result of the close interaction of HBx with components of the proteasome machinery, such as the DDB1-CRL4 complex, which may inadvertently result in ubiquitination of HBx itself. HDM2-mediated NEDDylation of HBx was also found to occur, probably because of its close proximity to Cul4, resulting in increased stability and chromatin localization[101,146]. NEDDylation-dependent reduction of ubiquitination and subsequent degradation of HBx by E3 Ligases, such as Siah-1, may account for the increased stability of this viral protein. MLN4924 (pevonedistat), a NEDD8-activating enzyme inhibitor, has been shown to impede HBV replication by reducing cullin[147] and HBx NEDDylation[146]. In addition, MLN4924 promotes upregulation of phosphorylated extracellular signal-regulated kinases (ERKs) resulting in reduced HNF1α, HNF4α and C/EBPα transcription factor levels[147], which are known activators of HBV transcription. Overall, the maintenance of persistent HBV infection by HBx contributes to HCC, by inducing host epigenetic modifications implicated in cancer.
Direct interaction of the HBc CTD[103] with CpG islands across the cccDNA, especially CpG II, has been shown to induce hypomethylation and subsequent upregulation of viral transcription and regulation[74,148]. This hypomethylation causes increased CBP binding and resulting histone acetylation, further increasing viral gene expression[57,148] albeit at lower levels than with HBx[149,150]. The association of HBc with HDAC1 was also shown[148], as well as P300 and PCAF/CBP[151]. Increased NF-κB binding upstream of ENII induced by HBc is thought to increase pre-C promoter activity[152]. Host DNA polymerase coordinator PCNA, which is known to maintain genetic and epigenetic integrity[153], is recruited to cccDNA by HBc which upregulates viral gene expression and plays a role in the development of HCC. However, the exact mechanism of this process is still unclear[154].
HBc can additionally mediate host gene expression through the binding of HBc to endogenous promoter regions. Guo et al[104] identified nearly 3100 host promoter regions with potential HBc binding sites. Examples of these include binding to the MxA promoter to evade a host IFN-induced immune response[155] and competitive binding to BAF200, resulting in the downregulation of interferon-induced transmembrane protein 1 (IFITM1) mRNA[156]. Binding of HBc to the death receptor 5 (DR5) promoter reduced DR5 expression, thereby reducing cellular apoptosis through TRAIL which is thought to support CHB infection[157].
Transcription factor binding to mediate host expression has also been shown. The binding of HBc to E2F1 prevented its natural association with the p53 promoter, and hence decreased p53 levels[158]. This could reduce p53-related apoptosis[159] and along with DR5 downregulation further support the development of chronic infection. However, competitive binding of HBc to the receptor for activated protein kinase C 1 (RACK1) prevented phosphorylation of mitogen-activated protein kinase 7 (MKK7) which sensitized cells to apoptosis[160]. This suggests a dual role for HBc in the control of apoptosis and requires further study to determine the impact of these pathways on HBV infection. The subcellular location of HBc is dependent on cell cycle phase, with in vitro studies showing that predominantly nuclear localization occurs during G1 phase[161]. Since E2F transcription factors and p53 both have effects on cell cycling[162-164] and are differentially regulated during HBV infection, further investigation of this phenomenon and the possibility of epigenetically controlling the cell cycle is needed. HBc has also been shown to recruit APOBEC3A/B to cccDNA to promote deamination[165]. Since so many HBc binding sites are predicted to occur in endogenous promoter regions, there is the possibility that APOBEC3A/B is recruited to host genes to influence their expression, and may play a role in the development of HCC[166].
Noncoding RNAs (ncRNAs) consist of small ncRNAs (< 200 nt) and long ncRNA (lncRNA > 200 nt)[167]. Small ncRNAs, including miRNAs, are involved in post-transcriptional gene silencing[167], while lncRNA are implicated in a plethora of host functions including post-transcriptional and chromatin modifications[168]. By altering the host’s epigenetic signature, HBV dysregulates the ncRNA landscape to control viral replication and influence hepatocarcinogenesis.
miRNAs have been widely studied in HBV infection, and have been shown to facilitate or inhibit viral replication[169]. An increase in replication can occur by targeting host factors that normally restrict viral replication, for example miR-15b downregulates HNF-1α[170], or by indirectly upregulating factors to promote replication, such as miR-1 upregulation of farnesoid X receptor α (FRXα) expression[171]. Interestingly, miR-1-mediated downregulation of E2F5 and HDAC4 was implicated in G1 cell cycle arrest and upregulation of hepatocyte differentiation factors, to increase HBV replication[171]. This may be related to other viral epigenetic factors such as HBc, which showed similar associations. Indirect FRXα upregulation by miR-449a targeting of CREB5[172], HDAC4 downregulation by miR-548ah[173] and G1 arrest by miR-125b-5p-mediated inhibition of retinoblastoma protein phosphorylation[174] implicate redundant epigenetic pathways in HBV regulation.
A recent study by Moon et al[175] has suggested that miR-20a can act in an epigenetic manner and promote methylation of cccDNA. They found that AGO2-miR-20a binding to the cccDNA may recruit DNMT3 to silence gene expression; however, further studies are needed to confirm the exact mechanism. Interestingly miR-146a could play a role in cccDNA formation, through a positive feedback loop with FEN1[176], and in silencing ZEB2 expression[177].
Induction of autophagy has been associated with many miRNAs including miR-146a-5p[178], the miR-99 family[179] and miR-192-3p, which is downregulated by HBx[180]. miR-155 has also been implicated in autophagy; however, this miRNA is downregulated following HBV infection[181]. HBV upregulates expression of miR-192-5p and miR-215-5p, which results in downregulation of apoptosis[182]. Contrasting effects were observed during HBV-mediated upregulation of miR-194-5p, which resulted in the downregulation of anti-apoptotic proteins SODD and cFLIP and sensitization of liver cells to apoptosis[182]. Once again, the role of HBV in promoting and preventing apoptosis needs further clarification.
Certain cancer-related miRNAs, namely miR-15a/miR-16-1, the miR-17-92 cluster and miR-224, are also associated with decreased viral replication through modification of cccDNA promoters and histones[183]. Interestingly, miR-122, which targets the HBV polymerase ORF and core 3'UTR[184], has also been shown to inhibit HBV by targeting cyclin G1 to increase p53-mediated inhibition of replication[185]. However, HBx abrogates these antiviral effects by downregulating miR-122[183]. Host miRNAs can also indirectly inhibit viral replication by regulating various host factors[169] HBx-mediated downregulation of miR-122 alters regulation of its natural host targets, including heme oxygenase-1[186], CCNG1 and NDRG3[187]. Similarly, miR-141 was implicated in targeting both PPARA[188] and SIRT1[189]. Reduced SIRT1 inhibited autophagy, and miR-130a targets liver pyruvate kinase which is thought to reduce energy supply and hence HBV replication[190]. Induction of immune suppression may occur through the increase of miR-199a-5p, miR-221-3p and Let-7a-3p, as shown in immune tolerant HBV-infected patients[191]. HBV-miR-3, a miRNA produced by the virus, reduces replication by directly targeting viral transcripts[192] or upregulating the anti-HBV IFN immune response[193].
LncRNAs have been identified as important polyfunctional epigenetic regulators and are associated with disease progression, particularly carcinogenesis[194]. Novel lncRNAs related to development of HBV-associated HCC are continuously being identified; however, a few have also been implicated in epigenetic regulation of viral transcription. LncRNAs highly up-regulated in liver cancer (HULC), DLEU2, and lncRNA 32 have been identified as epigenetic regulators of HBV. HAT1, which is co-activated by HBx-Sp1 binding, is transported to the cccDNA by HULC in an HBc-dependent manner[195]. HULC also mediates upregulation of HBx and the subsequent HBx-STAT upregulation of miR-539, which targets APOBEC3B. This results in reduced cccDNA degradation, with resultant increased transcription of viral genes, viral persistence, and HCC progression[196]. Interestingly, the promotion of HBV transcription by the lncRNA HULC is dependent on both HBx and HBc, which forms a positive feedback loop[195]. In the case of lncRNA DLEU, HBx both upregulates its expression and is co-recruited with it to the cccDNA, where the complex may bind to the histone methyltransferase EZH2[197]. Computational analysis of this interaction suggests that binding alleviates EZH2 transcriptional repression[197]. LncRNAs have also been implicated in the host antiviral response. To increase IFN-stimulated gene (ISG) expression, lncRNA 32 binds to activating transcription factor 2 (ATF 2) and promotes gene expression. However, during HBV infection, the expression of this lncRNA is reduced[198], ultimately dampening the immune response to the virus.
A number of epigenetic strategies has been identified as having therapeutic potential for CHB[27]. Some promote the host’s natural antiviral defense mechanisms while others aim to target specific pathways involved in the epigenetic control of cccDNA.
The antiviral and epigenetic properties of IFN-α therapy are well established[199], and have been adopted as a strategy to treat CHB[5]. Type 1 IFNs interact with the IFN-α/β receptor complex to modulate transcription of ISGs, ultimately to evoke strong innate immune responses against viral infections[200]. Administration of IFN-α to HBV-infected cells and HBV-infected chimeric uPA/SCID mice resulted in the inhibition of viral replication through epigenetic regulation of cccDNA[105]. Hypoacetylation of cccDNA-bound histones was achieved following the active recruitment of histone deacetylases hSIRT1 and HDAC1, methyltransferase EZH2, and the transcriptional repressor YY1[199]. Binding of transcription factors STAT1 and STAT2 to the IFN-α sensitive response element on active cccDNA was shown to be reduced after IFN-α treatment[199]. More recently, the structural maintenance of chromosomes flexible hinge domain containing 1 (SMCHD1) and promyelocytic leukemia protein have, along with STAT1, been implicated in the stimulation of cccDNA-associated histone PTMs in response to IFN-α[201]. Furthermore, IFN-α-based treatment may regulate the SMC5/6 complex to promote transcriptional repression of cccDNA[202].
When comparing IFN-α treatment to the effects of C646 (a small molecule epigenetic modifying agent that inhibits p300/CBP), Tropberger et al[86] demonstrated that C646 reduces HBV transcription in primary human hepatocytes in a dose-dependent manner. Like IFN-α, C646 treatment reduced the levels of active PTMs but without activating the innate immune response. However, neither treatment reduced cccDNA levels[86]. When combined with lymphotoxin-β receptor activation, IFN-α treatment did eliminate cccDNA in an APOBEC-dependent manner[165]. Yet the clinical utility of currently licensed IFN-α-based treatments has proved tricky because some patients respond to therapy, but others show no benefit. High costs and adverse side effects have also limited the use of IFN-α. Alternatives are being investigated and an iterative approach has led to the identification of new IFNs with increased potency, such as the recently described IFN-α 14[203].
Epigenetic therapies and immunomodulators have shown promise as anti-HCC agents[204], of which HDAC inhibitors such as 5-Azacytidine (5-Aza) and the EZH2 inhibitor 3-Deazaneplanocin A (DZNep) have recently been explored as combination therapies[205]. Such therapies have also been proposed for the treatment of CHB to silence the cccDNA minichromosome. Two small molecule inhibitors, AGK2 and GS-5801, are currently being investigated as anti-HBV epigenetic drugs[206,207]. AGK2 is an inhibitor of the SIRT2 deacetylase, while GS-5801 prevents erasing of epigenetic marks by lysine demethylase 5 (KDM5). Both have been shown to repress viral replication; however, preliminary results from a single dose of GS-5801 suggest that H3K4me3 may not be permanently associated with cccDNA[207]. Recently Dicoumarol, an inhibitor of NAD(P)H:quinone oxidoreductase 1 (NQO1), has been shown to promote the silencing of cccDNA[208] indirectly. The epigenetic effect of Dicoumarol is thought to arise from the destabilization of the HBx/NQO1 interaction, as the upregulation of NQO1 was shown to increase the half-life of HBx[208]. Although promising, the feasibility of using epigenetic modulators to silence HBV is complex, especially as the virus employs many normal host factors to regulate gene expression. The influence on development of HBV-related HCC and viral integration are additional important considerations for advancing epigenetic therapy. For instance, treating HCC with HDAC inhibitors is likely to promote viral transcription[57], while epigenetic silencing cccDNA gene expression by increasing DNA methylation may promote hepatocarcinogenesis[209].
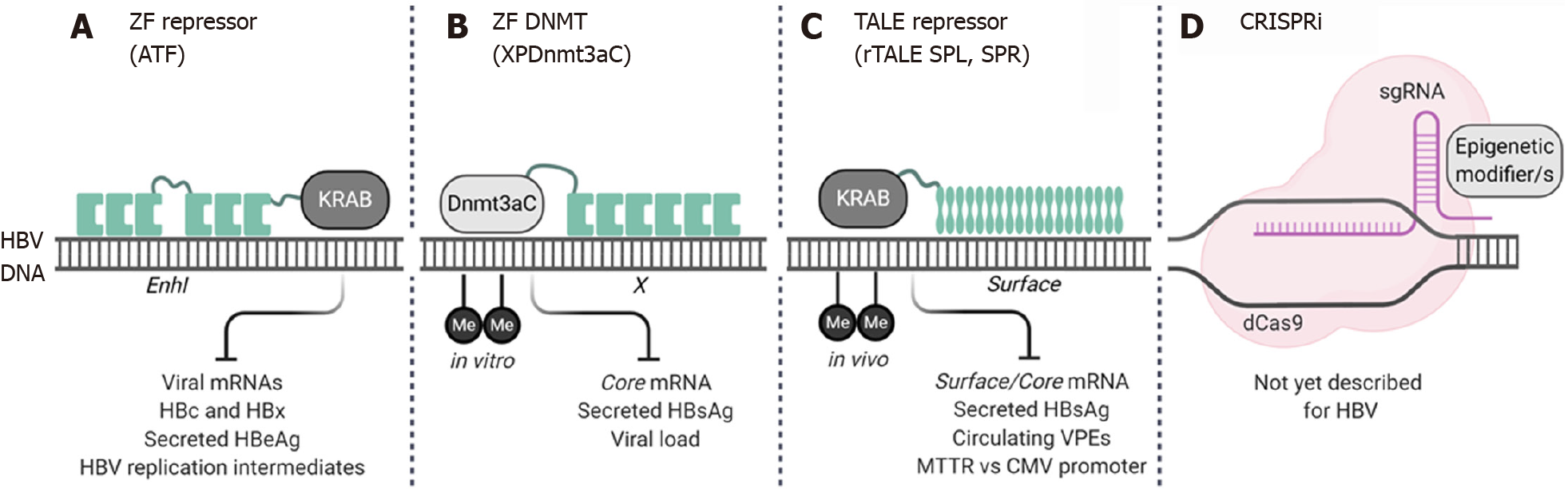
Epigenome engineering has become a popular targeted gene therapy approach to promote locus-specific epigenetic changes to treat genetic diseases[24]. Effector domains based on a variety of transcriptional activators, repressors, and chromatin remodeling enzymes including the typical writers, readers and erasers (Table 1) have been fused to DNA binding domains (DBD) to promote site-specific modifications. The targeting capabilities of epigenome engineering tools could overcome the non-specific effects epigenetic drugs may have on host gene expression. As such, cccDNA epigenome editors are currently being developed as a novel antiviral gene therapy for CHB (Figure 2).
ZF and transcription activator-like effector (TALE) DBDs have been used to direct different epigenetic effectors, primarily transcriptional repressors, to the HBV genome[210-213]. In 2013, Zhao et al[210] described the first ATF which was designed to target an 18 bp region of the HBV X or enhancer I (EnhI) sequence. The ATF was generated by joining six ZF DBDs to the Krüppelassociated box (KRAB) repression domain. Although not tested in a typical HBV replication model, the ATF achieved repression of the HBx promoter in a Hep3B reporter cell line, although accompanying cellular growth arrest was also observed[210]. Building on this study, Luo and colleagues demonstrated the antiviral activity of their EnhI-specific ATF in HepG2.2.15 cells and transgenic mice[211]. Viral replication was inhibited, in this case without affecting cellular growth, despite both models harboring integrated HBV sequences. Interestingly, the EnhI ZF-array alone achieved transcriptional repression, an effect that has previously been reported when using anti-HBV TALENs[214]. The antiviral effect of the ATF was further confirmed in the HBV transgenic mouse model where reductions in viral DNA, HBcAg and HBeAg were observed[211]. By replacing the repressor domain with a methyltransferase, ZF arrays have also been used to achieve targeted methylation of viral DNA. Xirong et al[213] fused the C-terminal region of the DNMT3A to a six-finger ZF (XPDnmt3aC) and tested the epigenome editor in HepG2 cells and HBV transgenic mice. The XPDnmt3aC was designed to bind to the HBx promoter region and achieved de novo methylation of the HBV DNA in HepG2 cells which was accompanied by a reduction in markers of viral replication. However, in transgenic mice the antiviral effect lasted for about two weeks[213], suggesting that if epigenetic modification was achieved it was not permanent.
Anti-HBV repressor TALEs (rTALEs) are engineered proteins with DBDs derived from the Xanthomonas bacteria which have been fused to the KRAB effector domain[212]. Two rTALEs, SPL and SPR, which were designed to bind to the surface ORF were tested in both in vitro and in vivo models of viral replication. Both the SPL and SPR rTALEs inhibited HBV replication, and significant reductions in HBsAg, viral mRNAs, and circulating viral particles were observed[212]. Importantly, a comparison between the liver specific mouse transthyretin (MTTR) or ubiquitously active CMV promoters was performed. The MTTR conferred liver-specific expression which may improve the safety of this epigenome engineering approach. Quantitative DNA methylation analysis using EpiTYPER™ Technology[215] was used to show that increased targeted methylation of intrahepatic HBV DNA was achieved at CpG II[212]. This confirmed the principle of this epigenome engineering approach; however, further validation of these rTALEs on cccDNA is warranted.
Interestingly, despite widespread studies using CRISPR/Cas gene editing technologies to disrupt cccDNA, the alternative epigenome editing platforms have yet to be investigated. CRISPRi and CRISPRa are RNA-guided epigenome modifiers that can repress or promote gene expression, and are generated by fusing single or multiple effector domains to a dead (nuclease deficient) Cas[216]. This means that site-directed epigenetic modifications can be achieved by designing guide RNAs to bind to the cccDNA. Although not yet reported, the field of CRISPR epigenome engineering is constantly expanding and may present a novel way of silencing cccDNA.
The epigenetic regulation of the cccDNA minichromosome to either promote or repress viral transcription involves an intricate association between host cell factors and viral proteins that is not yet fully understood. While there is currently debate around whether HBV should be considered a stealth virus[217], it is clear that it can manipulate the host cells replication, transcription and translation machinery for its own benefit. HBx appears to have a key role in viral persistence, not only as a regulator of cccDNA but also as a modulator of RNAs, ubiquitination, apoptosis, and antiviral responses. Although these intricate host-viral pathways require further clarification, the research to date has helped to identify possible feedback loops which could act as new targets for anti-HBV therapy.
There are conflicting views regarding the involvement of certain proteins, for example the epigenetic and deacetylase-independent roles of SIRT1. While some factors may have dual roles, the model systems used to evaluate epigenetic regulation of HBV need to be considered carefully. Although CHB is associated with HCC, epigenetic therapies designed to achieve a functional cure are likely to be given prior to the development of cancer. Using different liver cancer cell lines may skew the HBV epigenetic landscape as regulation of the host cells’ epigenome and gene expression is irregular. Establishing an ideal model system is however, easier said than done. Infectious primary human hepatocyte-based models require specialized facilities, as for example is required for the generation of humanized mouse models[218]. Similarly, studies on primary human hepatocytes and three-dimensional liver cultures are costly. Viruses infecting small animals, like the DHBV and woodchuck hepatitis virus, which are commonly used to test novel therapies, may also pose problems for analysis of the cccDNA epigenome. The different effects that HDAC inhibitors had on HBV and DHBV infection suggest differences between the hosts’ endogenous epigenetic pathways. Standardizing cccDNA quantification using droplet digital polymerase chain reaction and the liver biopsy-adapted chromatin immunoprecipitation (ChIP)-quantitative polymerase chain reaction technique (micro-ChIP) may help detect low levels of cccDNA as well as measure epigenetic marks[219].
Epigenetic drugs, improved interferon therapies, and epigenome editing tools are at an interesting stage of development, as new ways to silence cccDNA transcription. Accomplishing targeted epigenetic modifications and long-term viral suppression will be important, and consideration of the HCC epigenetic profile will need careful evaluation. Integrated viral sequences may also be amenable to cccDNA-specific epigenome editing, and undesirable off-target effects on the host genome need to be carefully assessed. Next-generation sequencing platforms like RNA-Seq would help to establish transcriptome profiles and identify potential off-target sites. To date only four targeted epigenome editors have been investigated, and although action on HBV DNA has been shown, a direct effect on the cccDNA minichromome is yet to be established. Furthermore, as with designer nucleases, liver-specific delivery[19] and immune responses to the foreign TALE and Cas proteins will need to be addressed[220]. Delivering epigenome editors as in vitro transcribed mRNA transcripts would avoid the packaging constraints of viral vectors[10]. Despite the current challenges associated with epigenetic therapies, there is potential for such an approach to achieve cccDNA gene silencing and perhaps cure. As this relatively new field of research continues to grow, overcoming the hurdles could lead to a promising class of new antivirals.
The authors thank the National Research Foundation, Poliomyelitis Research Foundation (PRF) and South African Medical Research Council.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: South Africa
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kayesh MEH S-Editor: Gao CC L-Editor: Webster JR P-Editor: Ma YJ
| 1. | Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol. 2016;64:S84-S101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 479] [Cited by in RCA: 710] [Article Influence: 78.9] [Reference Citation Analysis (0)] |
| 2. | Hutin Y, Nasrullah M, Easterbrook P, Nguimfack BD, Burrone E, Averhoff F, Bulterys M. Access to Treatment for Hepatitis B Virus Infection - Worldwide, 2016. MMWR Morb Mortal Wkly Rep. 2018;67:773-777. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 83] [Article Influence: 11.9] [Reference Citation Analysis (1)] |
| 3. | Polaris Observatory Collaborators. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol. 2018;3:383-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1260] [Cited by in RCA: 1215] [Article Influence: 173.6] [Reference Citation Analysis (2)] |
| 4. | Revill PA, Chisari FV, Block JM, Dandri M, Gehring AJ, Guo H, Hu J, Kramvis A, Lampertico P, Janssen HLA, Levrero M, Li W, Liang TJ, Lim SG, Lu F, Penicaud MC, Tavis JE, Thimme R; Members of the ICE-HBV Working Groups; ICE-HBV Stakeholders Group Chairs; ICE-HBV Senior Advisors; Zoulim F. A global scientific strategy to cure hepatitis B. Lancet Gastroenterol Hepatol. 2019;4:545-558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 380] [Article Influence: 63.3] [Reference Citation Analysis (1)] |
| 5. | Tan G, Song H, Xu F, Cheng G. When Hepatitis B Virus Meets Interferons. Front Microbiol. 2018;9:1611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 6. | Lythgoe KA, Lumley SF, Pellis L, McKeating JA, Matthews PC. Estimating hepatitis B virus cccDNA persistence in chronic infection. Virus Evol. 2021;7:veaa063. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 7. | Nassal M. Hepatitis B viruses: reverse transcription a different way. Virus Res. 2008;134:235-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 293] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 8. | Bock CT, Schwinn S, Locarnini S, Fyfe J, Manns MP, Trautwein C, Zentgraf H. Structural organization of the hepatitis B virus minichromosome. J Mol Biol. 2001;307:183-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 280] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 9. | Bloom K, Maepa MB, Ely A, Arbuthnot P. Gene Therapy for Chronic HBV-Can We Eliminate cccDNA? Genes (Basel). 2018;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 10. | Ely A, Singh P, Smith TS, Arbuthnot P. In vitro transcribed mRNA for expression of designer nucleases: Advantages as a novel therapeutic for the management of chronic HBV infection. Adv Drug Deliv Rev. 2021;168:134-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 11. | Wu X, Ma W, Mei C, Chen X, Yao Y, Liu Y, Qin X, Yuan Y. Description of CRISPR/Cas9 development and its prospect in hepatocellular carcinoma treatment. J Exp Clin Cancer Res. 2020;39:97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 12. | Ramanan V, Shlomai A, Cox DB, Schwartz RE, Michailidis E, Bhatta A, Scott DA, Zhang F, Rice CM, Bhatia SN. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep. 2015;5:10833. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 228] [Cited by in RCA: 234] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 13. | Schiwon M, Ehrke-Schulz E, Oswald A, Bergmann T, Michler T, Protzer U, Ehrhardt A. One-Vector System for Multiplexed CRISPR/Cas9 against Hepatitis B Virus cccDNA Utilizing High-Capacity Adenoviral Vectors. Mol Ther Nucleic Acids. 2018;12:242-253. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 14. | Liu X, Hao R, Chen S, Guo D, Chen Y. Inhibition of hepatitis B virus by the CRISPR/Cas9 system via targeting the conserved regions of the viral genome. J Gen Virol. 2015;96:2252-2261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 125] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 15. | Kostyushev D, Brezgin S, Kostyusheva A, Zarifyan D, Goptar I, Chulanov V. Orthologous CRISPR/Cas9 systems for specific and efficient degradation of covalently closed circular DNA of hepatitis B virus. Cell Mol Life Sci. 2019;76:1779-1794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 16. | Seeger C, Sohn JA. Complete Spectrum of CRISPR/Cas9-induced Mutations on HBV cccDNA. Mol Ther. 2016;24:1258-1266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 109] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 17. | Charlesworth CT, Deshpande PS, Dever DP, Camarena J, Lemgart VT, Cromer MK, Vakulskas CA, Collingwood MA, Zhang L, Bode NM, Behlke MA, Dejene B, Cieniewicz B, Romano R, Lesch BJ, Gomez-Ospina N, Mantri S, Pavel-Dinu M, Weinberg KI, Porteus MH. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med. 2019;25:249-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 598] [Cited by in RCA: 642] [Article Influence: 107.0] [Reference Citation Analysis (0)] |
| 18. | Li A, Tanner MR, Lee CM, Hurley AE, De Giorgi M, Jarrett KE, Davis TH, Doerfler AM, Bao G, Beeton C, Lagor WR. AAV-CRISPR Gene Editing Is Negated by Pre-existing Immunity to Cas9. Mol Ther. 2020;28:1432-1441. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 154] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 19. | Ates I, Rathbone T, Stuart C, Bridges PH, Cottle RN. Delivery Approaches for Therapeutic Genome Editing and Challenges. Genes (Basel). 2020;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 20. | Tong S, Moyo B, Lee CM, Leong K, Bao G. Engineered materials for in vivo delivery of genome-editing machinery. Nat Rev Mater. 2019;4:726-737. [RCA] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 153] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 21. | Li H, Sheng C, Liu H, Wang S, Zhao J, Yang L, Jia L, Li P, Wang L, Xie J, Xu D, Sun Y, Qiu S, Song H. Inhibition of HBV Expression in HBV Transgenic Mice Using AAV-Delivered CRISPR-SaCas9. Front Immunol. 2018;9:2080. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 22. | Scott T, Moyo B, Nicholson S, Maepa MB, Watashi K, Ely A, Weinberg MS, Arbuthnot P. ssAAVs containing cassettes encoding SaCas9 and guides targeting hepatitis B virus inactivate replication of the virus in cultured cells. Sci Rep. 2017;7:7401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 23. | Yang YC, Chen YH, Kao JH, Ching C, Liu IJ, Wang CC, Tsai CH, Wu FY, Liu CJ, Chen PJ, Chen DS, Yang HC. Permanent Inactivation of HBV Genomes by CRISPR/Cas9-Mediated Non-cleavage Base Editing. Mol Ther Nucleic Acids. 2020;20:480-490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 24. | Sgro A, Blancafort P. Epigenome engineering: new technologies for precision medicine. Nucleic Acids Res. 2020;48:12453-12482. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 25. | Dandri M. Epigenetic modulation in chronic hepatitis B virus infection. Semin Immunopathol. 2020;42:173-185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 26. | Hong X, Kim ES, Guo H. Epigenetic regulation of hepatitis B virus covalently closed circular DNA: Implications for epigenetic therapy against chronic hepatitis B. Hepatology. 2017;66:2066-2077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 156] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 27. | Mohd-Ismail NK, Lim Z, Gunaratne J, Tan YJ. Mapping the Interactions of HBV cccDNA with Host Factors. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 28. | Xia Y, Guo H. Hepatitis B virus cccDNA: Formation, regulation and therapeutic potential. Antiviral Res. 2020;180:104824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 131] [Article Influence: 26.2] [Reference Citation Analysis (1)] |
| 29. | Schreiner S, Nassal M. A Role for the Host DNA Damage Response in Hepatitis B Virus cccDNA Formation-and Beyond? Viruses. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 75] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 30. | Luo J, Cui X, Gao L, Hu J. Identification of an Intermediate in Hepatitis B Virus Covalently Closed Circular (CCC) DNA Formation and Sensitive and Selective CCC DNA Detection. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 31. | Königer C, Wingert I, Marsmann M, Rösler C, Beck J, Nassal M. Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses. Proc Natl Acad Sci USA. 2014;111:E4244-E4253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 186] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 32. | Qi Y, Gao Z, Xu G, Peng B, Liu C, Yan H, Yao Q, Sun G, Liu Y, Tang D, Song Z, He W, Sun Y, Guo JT, Li W. DNA Polymerase κ Is a Key Cellular Factor for the Formation of Covalently Closed Circular DNA of Hepatitis B Virus. PLoS Pathog. 2016;12:e1005893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 159] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 33. | Newbold JE, Xin H, Tencza M, Sherman G, Dean J, Bowden S, Locarnini S. The covalently closed duplex form of the hepadnavirus genome exists in situ as a heterogeneous population of viral minichromosomes. J Virol. 1995;69:3350-3357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 239] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 34. | Moraleda G, Saputelli J, Aldrich CE, Averett D, Condreay L, Mason WS. Lack of effect of antiviral therapy in nondividing hepatocyte cultures on the closed circular DNA of woodchuck hepatitis virus. J Virol. 1997;71:9392-9399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 208] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 35. | Dandri M, Burda MR, Will H, Petersen J. Increased hepatocyte turnover and inhibition of woodchuck hepatitis B virus replication by adefovir in vitro do not lead to reduction of the closed circular DNA. Hepatology. 2000;32:139-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 81] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 36. | Wong DK, Yuen MF, Yuan H, Sum SS, Hui CK, Hall J, Lai CL. Quantitation of covalently closed circular hepatitis B virus DNA in chronic hepatitis B patients. Hepatology. 2004;40:727-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 107] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 37. | Huang JT, Yang Y, Hu YM, Liu XH, Liao MY, Morgan R, Yuan EF, Li X, Liu SM. A Highly Sensitive and Robust Method for Hepatitis B Virus Covalently Closed Circular DNA Detection in Single Cells and Serum. J Mol Diagn. 2018;20:334-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 38. | Balagopal A, Hwang HS, Grudda T, Quinn J, Sterling RK, Sulkowski MS, Thio CL. Single Hepatocyte Hepatitis B Virus Transcriptional Landscape in HIV Coinfection. J Infect Dis. 2020;221:1462-1469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 39. | Wu TT, Coates L, Aldrich CE, Summers J, Mason WS. In hepatocytes infected with duck hepatitis B virus, the template for viral RNA synthesis is amplified by an intracellular pathway. Virology. 1990;175:255-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 183] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 40. | Zoulim F. New insight on hepatitis B virus persistence from the study of intrahepatic viral cccDNA. J Hepatol. 2005;42:302-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 191] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 41. | Ji M, Hu K. Recent advances in the study of hepatitis B virus covalently closed circular DNA. Virol Sin. 2017;32:454-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 42. | Köck J, Rösler C, Zhang JJ, Blum HE, Nassal M, Thoma C. Generation of covalently closed circular DNA of hepatitis B viruses via intracellular recycling is regulated in a virus specific manner. PLoS Pathog. 2010;6:e1001082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 116] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 43. | Tu T, Zehnder B, Qu B, Urban S. D e novo synthesis of hepatitis B virus nucleocapsids is dispensable for the maintenance and transcriptional regulation of cccDNA. JHEP Rep. 2021;3:100195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 44. | Tang L, Sheraz M, McGrane M, Chang J, Guo JT. DNA Polymerase alpha is essential for intracellular amplification of hepatitis B virus covalently closed circular DNA. PLoS Pathog. 2019;15:e1007742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 69] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 45. | Long Q, Yan R, Hu J, Cai D, Mitra B, Kim ES, Marchetti A, Zhang H, Wang S, Liu Y, Huang A, Guo H. The role of host DNA ligases in hepadnavirus covalently closed circular DNA formation. PLoS Pathog. 2017;13:e1006784. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 94] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 46. | Kitamura K, Que L, Shimadu M, Koura M, Ishihara Y, Wakae K, Nakamura T, Watashi K, Wakita T, Muramatsu M. Flap endonuclease 1 is involved in cccDNA formation in the hepatitis B virus. PLoS Pathog. 2018;14:e1007124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 88] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 47. | Sheraz M, Cheng J, Tang L, Chang J, Guo JT. Cellular DNA Topoisomerases Are Required for the Synthesis of Hepatitis B Virus Covalently Closed Circular DNA. J Virol. 2019;93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 48. | Wing PA, Davenne T, Wettengel J, Lai AG, Zhuang X, Chakraborty A, D'Arienzo V, Kramer C, Ko C, Harris JM, Schreiner S, Higgs M, Roessler S, Parish JL, Protzer U, Balfe P, Rehwinkel J, McKeating JA. A dual role for SAMHD1 in regulating HBV cccDNA and RT-dependent particle genesis. Life Sci Alliance. 2019;2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 49. | Wei L, Ploss A. Core components of DNA lagging strand synthesis machinery are essential for hepatitis B virus cccDNA formation. Nat Microbiol. 2020;5:715-726. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 87] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 50. | Zhao Q, Guo JT. Have the Starting Lineup of Five for Hepatitis B Virus Covalently Closed Circular DNA Synthesis Been Identified? Hepatology. 2020;72:1142-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 51. | Cai D, Mills C, Yu W, Yan R, Aldrich CE, Saputelli JR, Mason WS, Xu X, Guo JT, Block TM, Cuconati A, Guo H. Identification of disubstituted sulfonamide compounds as specific inhibitors of hepatitis B virus covalently closed circular DNA formation. Antimicrob Agents Chemother. 2012;56:4277-4288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 191] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 52. | Bock CT, Schranz P, Schröder CH, Zentgraf H. Hepatitis B virus genome is organized into nucleosomes in the nucleus of the infected cell. Virus Genes. 1994;8:215-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 153] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 53. | Quasdorff M, Protzer U. Control of hepatitis B virus at the level of transcription. J Viral Hepat. 2010;17:527-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 178] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 54. | Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1401] [Cited by in RCA: 1789] [Article Influence: 198.8] [Reference Citation Analysis (0)] |
| 55. | Jeltsch A, Jurkowska RZ. New concepts in DNA methylation. Trends Biochem Sci. 2014;39:310-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 301] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 56. | Cutter AR, Hayes JJ. A brief review of nucleosome structure. FEBS Lett. 2015;589:2914-2922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 262] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 57. | Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130:823-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 374] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 58. | Deeks SG. HIV: Shock and kill. Nature. 2012;487:439-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 441] [Cited by in RCA: 485] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 59. | Ghosh SK, Perrine SP, Williams RM, Faller DV. Histone deacetylase inhibitors are potent inducers of gene expression in latent EBV and sensitize lymphoma cells to nucleoside antiviral agents. Blood. 2012;119:1008-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 118] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 60. | Barlow DP. Methylation and imprinting: from host defense to gene regulation? Science. 1993;260:309-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 224] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 61. | Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3811] [Cited by in RCA: 4214] [Article Influence: 324.2] [Reference Citation Analysis (0)] |
| 62. | Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 567] [Cited by in RCA: 580] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 63. | Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2210] [Cited by in RCA: 2202] [Article Influence: 104.9] [Reference Citation Analysis (0)] |
| 64. | Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 556] [Cited by in RCA: 558] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 65. | Kar S, Deb M, Sengupta D, Shilpi A, Parbin S, Torrisani J, Pradhan S, Patra S. An insight into the various regulatory mechanisms modulating human DNA methyltransferase 1 stability and function. Epigenetics. 2012;7:994-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 66. | Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem. 2004;279:48350-48359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 387] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 67. | Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci USA. 2002;99:16916-16921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 378] [Cited by in RCA: 381] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 68. | Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol Chem. 2005;280:13341-13348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 210] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 69. | Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 783] [Cited by in RCA: 776] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 70. | Chen JY, Hsu HC, Lee CS, Chen DS, Zuckerman AJ, Harrison TJ. Detection of hepatitis B virus DNA in hepatocellular carcinoma: methylation of integrated viral DNA. J Virol Methods. 1988;19:257-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 71. | Guo Y, Li Y, Mu S, Zhang J, Yan Z. Evidence that methylation of hepatitis B virus covalently closed circular DNA in liver tissues of patients with chronic hepatitis B modulates HBV replication. J Med Virol. 2009;81:1177-1183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 77] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 72. | Vivekanandan P, Thomas D, Torbenson M. Hepatitis B viral DNA is methylated in liver tissues. J Viral Hepat. 2008;15:103-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 44] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 73. | Kim JW, Lee SH, Park YS, Hwang JH, Jeong SH, Kim N, Lee DH. Replicative activity of hepatitis B virus is negatively associated with methylation of covalently closed circular DNA in advanced hepatitis B virus infection. Intervirology. 2011;54:316-325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 74. | Zhang Y, Mao R, Yan R, Cai D, Zhang Y, Zhu H, Kang Y, Liu H, Wang J, Qin Y, Huang Y, Guo H, Zhang J. Transcription of hepatitis B virus covalently closed circular DNA is regulated by CpG methylation during chronic infection. PLoS One. 2014;9:e110442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 75. | Jain S, Chang TT, Chen S, Boldbaatar B, Clemens A, Lin SY, Yan R, Hu CT, Guo H, Block TM, Song W, Su YH. Comprehensive DNA methylation analysis of hepatitis B virus genome in infected liver tissues. Sci Rep. 2015;5:10478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 76. | Zhang Y, Li C, Zhang Y, Zhu H, Kang Y, Liu H, Wang J, Qin Y, Mao R, Xie Y, Huang Y, Zhang J. Comparative analysis of CpG islands among HBV genotypes. PLoS One. 2013;8:e56711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 77. | Kaur P, Paliwal A, Durantel D, Hainaut P, Scoazec JY, Zoulim F, Chemin I, Herceg Z. DNA methylation of hepatitis B virus (HBV) genome associated with the development of hepatocellular carcinoma and occult HBV infection. J Infect Dis. 2010;202:700-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 78. | Mogul D, Torbenson M, Schwarz KB. Epigenetic regulation of hepatitis B virus infection. Curr Hepatitis Rep. 2011;10:277-284. |
| 79. | Zhang C, Huang C, Sui X, Zhong X, Yang W, Hu X, Li Y. Association between gene methylation and HBV infection in hepatocellular carcinoma: A meta-analysis. J Cancer. 2019;10:6457-6465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 80. | Vivekanandan P, Thomas D, Torbenson M. Methylation regulates hepatitis B viral protein expression. J Infect Dis. 2009;199:1286-1291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 84] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 81. | Vivekanandan P, Daniel HD, Kannangai R, Martinez-Murillo F, Torbenson M. Hepatitis B virus replication induces methylation of both host and viral DNA. J Virol. 2010;84:4321-4329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 112] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 82. | Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. Control of cccDNA function in hepatitis B virus infection. J Hepatol. 2009;51:581-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 431] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 83. | Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693-705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7446] [Cited by in RCA: 7669] [Article Influence: 426.1] [Reference Citation Analysis (0)] |
| 84. | Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6156] [Cited by in RCA: 6152] [Article Influence: 246.1] [Reference Citation Analysis (0)] |
| 85. | Gong Q, Chen S, Guo J, Sun H, Zheng G, Liu Q, Ren H, He S. Chromosome remodeling related to hepatitis B virus replication in HepG2 cells. DNA Cell Biol. 2011;30:347-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 86. | Tropberger P, Mercier A, Robinson M, Zhong W, Ganem DE, Holdorf M. Mapping of histone modifications in episomal HBV cccDNA uncovers an unusual chromatin organization amenable to epigenetic manipulation. Proc Natl Acad Sci USA. 2015;112:E5715-E5724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 187] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 87. | Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 774] [Cited by in RCA: 713] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 88. | Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781-783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1183] [Cited by in RCA: 1210] [Article Influence: 75.6] [Reference Citation Analysis (0)] |
| 89. | Zhang W, Chen J, Wu M, Zhang X, Zhang M, Yue L, Li Y, Liu J, Li B, Shen F, Wang Y, Bai L, Protzer U, Levrero M, Yuan Z. PRMT5 restricts hepatitis B virus replication through epigenetic repression of covalently closed circular DNA transcription and interference with pregenomic RNA encapsidation. Hepatology. 2017;66:398-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 113] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 90. | Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6:a018713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 916] [Cited by in RCA: 1430] [Article Influence: 130.0] [Reference Citation Analysis (0)] |
| 91. | Kgatle MM. Recent Advancement in Hepatitis B Virus, Epigenetics Alterations and Related Complications. In: Kgatle MM Advances in Treatment of Hepatitis C and B. IntechOpen, 2017: 149. |
| 92. | Wang Z, Wang W, Wang L. Epigenetic regulation of covalently closed circular DNA minichromosome in hepatitis B virus infection. Biophys Rep. 2020;1-12. |
| 93. | Rivière L, Gerossier L, Ducroux A, Dion S, Deng Q, Michel ML, Buendia MA, Hantz O, Neuveut C. HBx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase. J Hepatol. 2015;63:1093-1102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 166] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 94. | Lopez-Serra L, Esteller M. Proteins that bind methylated DNA and human cancer: reading the wrong words. Br J Cancer. 2008;98:1881-1885. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 120] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 95. | Lucifora J, Durantel D, Belloni L, Barraud L, Villet S, Vincent IE, Margeridon-Thermet S, Hantz O, Kay A, Levrero M, Zoulim F. Initiation of hepatitis B virus genome replication and production of infectious virus following delivery in HepG2 cells by novel recombinant baculovirus vector. J Gen Virol. 2008;89:1819-1828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 96. | Qian G, Hu B, Zhou D, Xuan Y, Bai L, Duan C. NIRF, a Novel Ubiquitin Ligase, Inhibits Hepatitis B Virus Replication Through Effect on HBV Core Protein and H3 Histones. DNA Cell Biol. 2015;34:327-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 97. | Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1280] [Cited by in RCA: 1600] [Article Influence: 123.1] [Reference Citation Analysis (1)] |
| 98. | Liu F, Campagna M, Qi Y, Zhao X, Guo F, Xu C, Li S, Li W, Block TM, Chang J, Guo JT. Alpha-interferon suppresses hepadnavirus transcription by altering epigenetic modification of cccDNA minichromosomes. PLoS Pathog. 2013;9:e1003613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 131] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 99. | Ananthanarayanan M, Li Y, Surapureddi S, Balasubramaniyan N, Ahn J, Goldstein JA, Suchy FJ. Histone H3K4 trimethylation by MLL3 as part of ASCOM complex is critical for NR activation of bile acid transporter genes and is downregulated in cholestasis. Am J Physiol Gastrointest Liver Physiol. 2011;300:G771-G781. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 100. | Murakami S. Hepatitis B virus X protein: a multifunctional viral regulator. J Gastroenterol. 2001;36:651-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 260] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 101. | Prescott NA, Bram Y, Schwartz RE, David Y. Targeting Hepatitis B Virus Covalently Closed Circular DNA and Hepatitis B Virus X Protein: Recent Advances and New Approaches. ACS Infect Dis. 2019;5:1657-1667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 102. | Zlotnick A, Venkatakrishnan B, Tan Z, Lewellyn E, Turner W, Francis S. Core protein: A pleiotropic keystone in the HBV lifecycle. Antiviral Res. 2015;121:82-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 207] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 103. | Koumbi L, Pollicino T, Raimondo G, Stampoulis D, Khakoo S, Karayiannis P. Hepatitis B virus basal core promoter mutations show lower replication fitness associated with cccDNA acetylation status. Virus Res. 2016;220:150-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 104. | Guo Y, Kang W, Lei X, Li Y, Xiang A, Liu Y, Zhao J, Zhang J, Yan Z. Hepatitis B viral core protein disrupts human host gene expression by binding to promoter regions. BMC Genomics. 2012;13:563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 105. | Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, Raimondo G, Levrero M. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci USA. 2009;106:19975-19979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 399] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 106. | Shi L, Li S, Shen F, Li H, Qian S, Lee DH, Wu JZ, Yang W. Characterization of nucleosome positioning in hepadnaviral covalently closed circular DNA minichromosomes. J Virol. 2012;86:10059-10069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 107. | Shamay M, Barak O, Doitsh G, Ben-Dor I, Shaul Y. Hepatitis B virus pX interacts with HBXAP, a PHD finger protein to coactivate transcription. J Biol Chem. 2002;277:9982-9988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 49] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 108. | Saeed U, Kim J, Piracha ZZ, Kwon H, Jung J, Chwae YJ, Park S, Shin HJ, Kim K. Parvulin 14 and Parvulin 17 Bind to HBx and cccDNA and Upregulate Hepatitis B Virus Replication from cccDNA to Virion in an HBx-Dependent Manner. J Virol. 2019;93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 109. | Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972-1984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 694] [Article Influence: 69.4] [Reference Citation Analysis (0)] |
| 110. | Curtil C, Enache LS, Radreau P, Dron AG, Scholtès C, Deloire A, Roche D, Lotteau V, André P, Ramière C. The metabolic sensors FXRα, PGC-1α, and SIRT1 cooperatively regulate hepatitis B virus transcription. FASEB J. 2014;28:1454-1463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 111. | Ren JH, Tao Y, Zhang ZZ, Chen WX, Cai XF, Chen K, Ko BC, Song CL, Ran LK, Li WY, Huang AL, Chen J. Sirtuin 1 regulates hepatitis B virus transcription and replication by targeting transcription factor AP-1. J Virol. 2014;88:2442-2451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 100] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 112. | Deng JJ, Kong KE, Gao WW, Tang HV, Chaudhary V, Cheng Y, Zhou J, Chan CP, Wong DK, Yuen MF, Jin DY. Interplay between SIRT1 and hepatitis B virus X protein in the activation of viral transcription. Biochim Biophys Acta Gene Regul Mech. 2017;1860:491-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 113. | Benhenda S, Ducroux A, Rivière L, Sobhian B, Ward MD, Dion S, Hantz O, Protzer U, Michel ML, Benkirane M, Semmes OJ, Buendia MA, Neuveut C. Methyltransferase PRMT1 is a binding partner of HBx and a negative regulator of hepatitis B virus transcription. J Virol. 2013;87:4360-4371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 99] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 114. | Ren JH, Hu JL, Cheng ST, Yu HB, Wong VKW, Law BYK, Yang YF, Huang Y, Liu Y, Chen WX, Cai XF, Tang H, Hu Y, Zhang WL, Liu X, Long QX, Zhou L, Tao NN, Zhou HZ, Yang QX, Ren F, He L, Gong R, Huang AL, Chen J. SIRT3 restricts hepatitis B virus transcription and replication through epigenetic regulation of covalently closed circular DNA involving suppressor of variegation 3-9 homolog 1 and SET domain containing 1A histone methyltransferases. Hepatology. 2018;68:1260-1276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 115. | Zheng DL, Zhang L, Cheng N, Xu X, Deng Q, Teng XM, Wang KS, Zhang X, Huang J, Han ZG. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. J Hepatol. 2009;50:377-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 152] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 116. | Fu X, Song X, Li Y, Tan D, Liu G. Hepatitis B virus X protein upregulates DNA methyltransferase 3A/3B and enhances SOCS-1CpG island methylation. Mol Med Rep. 2016;13:301-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 117. | Park IY, Sohn BH, Yu E, Suh DJ, Chung YH, Lee JH, Surzycki SJ, Lee YI. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology. 2007;132:1476-1494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 246] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 118. | Park ES, Park YK, Shin CY, Park SH, Ahn SH, Kim DH, Lim KH, Kwon SY, Kim KP, Yang SI, Seong BL, Kim KH. Hepatitis B virus inhibits liver regeneration via epigenetic regulation of urokinase-type plasminogen activator. Hepatology. 2013;58:762-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 119. | Koumbi L, Karayiannis P. The Epigenetic Control of Hepatitis B Virus Modulates the Outcome of Infection. Front Microbiol. 2015;6:1491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 120. | Sengupta I, Das D, Singh SP, Chakravarty R, Das C. Host transcription factor Speckled 110 kDa (Sp110), a nuclear body protein, is hijacked by hepatitis B virus protein X for viral persistence. J Biol Chem. 2017;292:20379-20393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 121. | Colgrove R, Simon G, Ganem D. Transcriptional activation of homologous and heterologous genes by the hepatitis B virus X gene product in cells permissive for viral replication. J Virol. 1989;63:4019-4026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 154] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 122. | Cougot D, Wu Y, Cairo S, Caramel J, Renard CA, Lévy L, Buendia MA, Neuveut C. Correction: The hepatitis B virus X protein functionally interacts with CREB-binding protein/p300 in the regulation of CREB-mediated transcription. J Biol Chem. 2020;295:2888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 123. | Cougot D, Wu Y, Cairo S, Caramel J, Renard CA, Lévy L, Buendia MA, Neuveut C. The hepatitis B virus X protein functionally interacts with CREB-binding protein/p300 in the regulation of CREB-mediated transcription. J Biol Chem. 2007;282:4277-4287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 166] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 124. | Chen S, Wu L, Peng L, Wang X, Tang N. Hepatitis B virus X protein (HBx) promotes ST2 expression by GATA2 in liver cells. Mol Immunol. 2020;123:32-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 125. | Volarevic V, Mitrovic M, Milovanovic M, Zelen I, Nikolic I, Mitrovic S, Pejnovic N, Arsenijevic N, Lukic ML. Protective role of IL-33/ST2 axis in Con A-induced hepatitis. J Hepatol. 2012;56:26-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 121] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 126. | Cougot D, Allemand E, Rivière L, Benhenda S, Duroure K, Levillayer F, Muchardt C, Buendia MA, Neuveut C. Inhibition of PP1 phosphatase activity by HBx: a mechanism for the activation of hepatitis B virus transcription. Sci Signal. 2012;5:ra1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 127. | Shen C, Feng X, Mao T, Yang D, Zou J, Zao X, Deng Q, Chen X, Lu F. Yin-Yang 1 and HBx protein activate HBV transcription by mediating the spatial interaction of cccDNA minichromosome with cellular chromosome 19p13.11. Emerg Microbes Infect. 2020;9:2455-2464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 128. | Deng L, Gan X, Ito M, Chen M, Aly HH, Matsui C, Abe T, Watashi K, Wakita T, Suzuki T, Okamoto T, Matsuura Y, Mizokami M, Shoji I, Hotta H. Peroxiredoxin 1, a Novel HBx-Interacting Protein, Interacts with Exosome Component 5 and Negatively Regulates Hepatitis B Virus (HBV) Propagation through Degradation of HBV RNA. J Virol. 2019;93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 129. | Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34:562-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 332] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 130. | Leupin O, Bontron S, Schaeffer C, Strubin M. Hepatitis B virus X protein stimulates viral genome replication via a DDB1-dependent pathway distinct from that leading to cell death. J Virol. 2005;79:4238-4245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 143] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 131. | Li T, Robert EI, van Breugel PC, Strubin M, Zheng N. A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat Struct Mol Biol. 2010;17:105-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 177] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 132. | Decorsière A, Mueller H, van Breugel PC, Abdul F, Gerossier L, Beran RK, Livingston CM, Niu C, Fletcher SP, Hantz O, Strubin M. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature. 2016;531:386-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 414] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 133. | Murphy CM, Xu Y, Li F, Nio K, Reszka-Blanco N, Li X, Wu Y, Yu Y, Xiong Y, Su L. Hepatitis B Virus X Protein Promotes Degradation of SMC5/6 to Enhance HBV Replication. Cell Rep. 2016;16:2846-2854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 241] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 134. | Niu C, Livingston CM, Li L, Beran RK, Daffis S, Ramakrishnan D, Burdette D, Peiser L, Salas E, Ramos H, Yu M, Cheng G, Strubin M, Delaney WE IV, Fletcher SP. The Smc5/6 Complex Restricts HBV when Localized to ND10 without Inducing an Innate Immune Response and Is Counteracted by the HBV X Protein Shortly after Infection. PLoS One. 2017;12:e0169648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 119] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 135. | Mitra B, Guo H. Hepatitis B virus X protein crosses out Smc5/6 complex to maintain covalently closed circular DNA transcription. Hepatology. 2016;64:2246-2249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 136. | Minor MM, Hollinger FB, McNees AL, Jung SY, Jain A, Hyser JM, Bissig KD, Slagle BL. Hepatitis B Virus HBx Protein Mediates the Degradation of Host Restriction Factors through the Cullin 4 DDB1 E3 Ubiquitin Ligase Complex. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 137. | He Q, Li W, Ren J, Huang Y, Hu Q, Chen J, Chen W. ZEB2 inhibits HBV transcription and replication by targeting its core promoter. Oncotarget. 2016;7:16003-16011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 138. | Qian MX, Pang Y, Liu CH, Haratake K, Du BY, Ji DY, Wang GF, Zhu QQ, Song W, Yu Y, Zhang XX, Huang HT, Miao S, Chen LB, Zhang ZH, Liang YN, Liu S, Cha H, Yang D, Zhai Y, Komatsu T, Tsuruta F, Li H, Cao C, Li W, Li GH, Cheng Y, Chiba T, Wang L, Goldberg AL, Shen Y, Qiu XB. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell. 2013;153:1012-1024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 241] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 139. | Mandemaker IK, Geijer ME, Kik I, Bezstarosti K, Rijkers E, Raams A, Janssens RC, Lans H, Hoeijmakers JH, Demmers JA, Vermeulen W, Marteijn JA. DNA damage-induced replication stress results in PA200-proteasome-mediated degradation of acetylated histones. EMBO Rep. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 140. | Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE Jr, Meyer T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235-1241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1674] [Cited by in RCA: 1768] [Article Influence: 88.4] [Reference Citation Analysis (0)] |
| 141. | Hu Z, Zhang Z, Doo E, Coux O, Goldberg AL, Liang TJ. Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J Virol. 1999;73:7231-7240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 177] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 142. | Gao W, Jia Z, Tian Y, Yang P, Sun H, Wang C, Ding Y, Zhang M, Zhang Y, Yang D, Tian Z, Zhou J, Ruan Z, Wu Y, Ni B. HBx Protein Contributes to Liver Carcinogenesis by H3K4me3 Modification Through Stabilizing WD Repeat Domain 5 Protein. Hepatology. 2020;71:1678-1695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 143. | Shen B, Chen Y, Hu J, Qiao M, Ren J, Chen J, Tang N, Huang A, Hu Y. Hepatitis B virus X protein modulates upregulation of DHX9 to promote viral DNA replication. Cell Microbiol. 2020;22:e13148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 144. | Tang H, Oishi N, Kaneko S, Murakami S. Molecular functions and biological roles of hepatitis B virus x protein. Cancer Sci. 2006;97:977-983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 243] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 145. | Su ZJ, Cao JS, Wu YF, Chen WN, Lin X, Wu YL. Deubiquitylation of hepatitis B virus X protein (HBx) by ubiquitin-specific peptidase 15 (USP15) increases HBx stability and its transactivation activity. Sci Rep. 2017;7:40246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 146. | Liu N, Zhang J, Yang X, Jiao T, Zhao X, Li W, Zhu J, Yang P, Jin J, Peng J, Li Z, Ye X. HDM2 Promotes NEDDylation of Hepatitis B Virus HBx To Enhance Its Stability and Function. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 147. | Xie M, Guo H, Lou G, Yao J, Liu Y, Sun Y, Yang Z, Zheng M. Neddylation inhibitor MLN4924 has anti-HBV activity via modulating the ERK-HNF1α-C/EBPα-HNF4α axis. J Cell Mol Med. 2021;25:840-854. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 148. | Guo YH, Li YN, Zhao JR, Zhang J, Yan Z. HBc binds to the CpG islands of HBV cccDNA and promotes an epigenetic permissive state. Epigenetics. 2011;6:720-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 123] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 149. | Williams JS, Andrisani OM. The hepatitis B virus X protein targets the basic region-leucine zipper domain of CREB. Proc Natl Acad Sci USA. 1995;92:3819-3823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 126] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 150. | Andrisani OM. CREB-mediated transcriptional control. Crit Rev Eukaryot Gene Expr. 1999;9:19-32. [PubMed] |
| 151. | Chong CK, Cheng CYS, Tsoi SYJ, Huang FY, Liu F, Seto WK, Lai CL, Yuen MF, Wong DK. Role of hepatitis B core protein in HBV transcription and recruitment of histone acetyltransferases to cccDNA minichromosome. Antiviral Res. 2017;144:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 152. | Kwon JA, Rho HM. Hepatitis B viral core protein activates the hepatitis B viral enhancer II/pregenomic promoter through the nuclear factor kappaB binding site. Biochem Cell Biol. 2002;80:445-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 153. | Park SY, Jeong MS, Han CW, Yu HS, Jang SB. Structural and Functional Insight into Proliferating Cell Nuclear Antigen. J Microbiol Biotechnol. 2016;26:637-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 154. | Feng J, Yang G, Liu Y, Gao Y, Zhao M, Bu Y, Yuan H, Yuan Y, Yun H, Sun M, Gao H, Zhang S, Liu Z, Yin M, Song X, Miao Z, Lin Z, Zhang X. LncRNA PCNAP1 modulates hepatitis B virus replication and enhances tumor growth of liver cancer. Theranostics. 2019;9:5227-5245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 132] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 155. | Fernández M, Quiroga JA, Carreño V. Hepatitis B virus downregulates the human interferon-inducible MxA promoter through direct interaction of precore/core proteins. J Gen Virol. 2003;84:2073-2082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 65] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 156. | Li T, Ke Z, Liu W, Xiong Y, Zhu Y, Liu Y. Human Hepatitis B Virus Core Protein Inhibits IFNα-Induced IFITM1 Expression by Interacting with BAF200. Viruses. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 157. | Du J, Liang X, Liu Y, Qu Z, Gao L, Han L, Liu S, Cui M, Shi Y, Zhang Z, Yu L, Cao L, Ma C, Zhang L, Chen Y, Sun W. Hepatitis B virus core protein inhibits TRAIL-induced apoptosis of hepatocytes by blocking DR5 expression. Cell Death Differ. 2009;16:219-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 68] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 158. | Kwon JA, Rho HM. Transcriptional repression of the human p53 gene by hepatitis B viral core protein (HBc) in human liver cells. Biol Chem. 2003;384:203-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 159. | Fridman JS, Lowe SW. Control of apoptosis by p53. Oncogene. 2003;22:9030-9040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1017] [Cited by in RCA: 1069] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 160. | Jia B, Guo M, Li G, Yu D, Zhang X, Lan K, Deng Q. Hepatitis B virus core protein sensitizes hepatocytes to tumor necrosis factor-induced apoptosis by suppression of the phosphorylation of mitogen-activated protein kinase kinase 7. J Virol. 2015;89:2041-2051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 161. | Yeh CT, Wong SW, Fung YK, Ou JH. Cell cycle regulation of nuclear localization of hepatitis B virus core protein. Proc Natl Acad Sci USA. 1993;90:6459-6463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 76] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 162. | Chen J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb Perspect Med. 2016;6:a026104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 494] [Cited by in RCA: 810] [Article Influence: 90.0] [Reference Citation Analysis (0)] |
| 163. | Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220-5227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 766] [Cited by in RCA: 870] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 164. | Henley SA, Dick FA. The retinoblastoma family of proteins and their regulatory functions in the mammalian cell division cycle. Cell Div. 2012;7:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 199] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 165. | Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, Remouchamps C, Chou WM, Thasler WE, Hüser N, Durantel D, Liang TJ, Münk C, Heim MH, Browning JL, Dejardin E, Dandri M, Schindler M, Heikenwalder M, Protzer U. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221-1228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 640] [Cited by in RCA: 741] [Article Influence: 67.4] [Reference Citation Analysis (0)] |
| 166. | Diab A, Foca A, Zoulim F, Durantel D, Andrisani O. The diverse functions of the hepatitis B core/capsid protein (HBc) in the viral life cycle: Implications for the development of HBc-targeting antivirals. Antiviral Res. 2018;149:211-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 90] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 167. | Gomes AQ, Nolasco S, Soares H. Non-coding RNAs: multi-tasking molecules in the cell. Int J Mol Sci. 2013;14:16010-16039. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 196] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 168. | Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2868] [Cited by in RCA: 2719] [Article Influence: 302.1] [Reference Citation Analysis (0)] |
| 169. | Bandopadhyay M, Bharadwaj M. Exosomal miRNAs in hepatitis B virus related liver disease: a new hope for biomarker. Gut Pathog. 2020;12:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 170. | Dai X, Zhang W, Zhang H, Sun S, Yu H, Guo Y, Kou Z, Zhao G, Du L, Jiang S, Zhang J, Li J, Zhou Y. Modulation of HBV replication by microRNA-15b through targeting hepatocyte nuclear factor 1α. Nucleic Acids Res. 2014;42:6578-6590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 171. | Zhang X, Zhang E, Ma Z, Pei R, Jiang M, Schlaak JF, Roggendorf M, Lu M. Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA-1. Hepatology. 2011;53:1476-1485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 161] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 172. | Zhang X, Liu H, Xie Z, Deng W, Wu C, Qin B, Hou J, Lu M. Epigenetically regulated miR-449a enhances hepatitis B virus replication by targeting cAMP-responsive element binding protein 5 and modulating hepatocytes phenotype. Sci Rep. 2016;6:25389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 173. | Xing T, Zhu J, Xian J, Li A, Wang X, Wang W, Zhang Q. miRNA-548ah promotes the replication and expression of hepatitis B virus by targeting histone deacetylase 4. Life Sci. 2019;219:199-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 174. | Deng W, Zhang X, Ma Z, Lin Y, Lu M. MicroRNA-125b-5p mediates post-transcriptional regulation of hepatitis B virus replication via the LIN28B/Let-7 axis. RNA Biol. 2017;14:1389-1398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 175. | Moon IY, Choi JH, Chung JW, Jang ES, Jeong SH, Kim JW. MicroRNA20 induces methylation of hepatitis B virus covalently closed circular DNA in human hepatoma cells. Mol Med Rep. 2019;20:2285-2293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 176. | Ji M, Mei X, Jing X, Xu X, Chen X, Pan W. The cooperative complex of Argonaute-2 and microRNA-146a regulates hepatitis B virus replication through flap endonuclease 1. Life Sci. 2020;257:118089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 177. | Wang Y, Li Y. miR-146 promotes HBV replication and expression by targeting ZEB2. Biomed Pharmacother. 2018;99:576-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 178. | Fu L, Fu X, Mo J, Li X, Li R, Peng S. miR-146a-5p enhances hepatitis B virus replication through autophagy to promote aggravation of chronic hepatitis B. IUBMB Life. 2019;71:1336-1346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 179. | Lin Y, Deng W, Pang J, Kemper T, Hu J, Yin J, Zhang J, Lu M. The microRNA-99 family modulates hepatitis B virus replication by promoting IGF-1R/PI3K/Akt/mTOR/ULK1 signaling-induced autophagy. Cell Microbiol. 2017;19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 75] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 180. | Wang J, Chen J, Liu Y, Zeng X, Wei M, Wu S, Xiong Q, Song F, Yuan X, Xiao Y, Cao Y, Li C, Chen L, Guo M, Shi YB, Sun G, Guo D. Hepatitis B Virus Induces Autophagy to Promote its Replication by the Axis of miR-192-3p-XIAP Through NF kappa B Signaling. Hepatology. 2019;69:974-992. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 181. | Chen L, Ming X, Li W, Bi M, Yan B, Wang X, Yang P, Yang B. The microRNA-155 mediates hepatitis B virus replication by reinforcing SOCS1 signalling-induced autophagy. Cell Biochem Funct. 2020;38:436-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 182. | Nielsen KO, Jacobsen KS, Mirza AH, Winther TN, Størling J, Glebe D, Pociot F, Hogh B. Hepatitis B virus upregulates host microRNAs that target apoptosis-regulatory genes in an in vitro cell model. Exp Cell Res. 2018;371:92-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 183. | Zhang X, Hou J, Lu M. Regulation of hepatitis B virus replication by epigenetic mechanisms and microRNAs. Front Genet. 2013;4:202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 184. | Chen Y, Shen A, Rider PJ, Yu Y, Wu K, Mu Y, Hao Q, Liu Y, Gong H, Zhu Y, Liu F, Wu J. A liver-specific microRNA binds to a highly conserved RNA sequence of hepatitis B virus and negatively regulates viral gene expression and replication. FASEB J. 2011;25:4511-4521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 149] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 185. | Wang S, Qiu L, Yan X, Jin W, Wang Y, Chen L, Wu E, Ye X, Gao GF, Wang F, Chen Y, Duan Z, Meng S. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G(1) -modulated P53 activity. Hepatology. 2012;55:730-741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 209] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 186. | Qiu L, Fan H, Jin W, Zhao B, Wang Y, Ju Y, Chen L, Chen Y, Duan Z, Meng S. miR-122-induced down-regulation of HO-1 negatively affects miR-122-mediated suppression of HBV. Biochem Biophys Res Commun. 2010;398:771-777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 126] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 187. | Fan CG, Wang CM, Tian C, Wang Y, Li L, Sun WS, Li RF, Liu YG. miR-122 inhibits viral replication and cell proliferation in hepatitis B virus-related hepatocellular carcinoma and targets NDRG3. Oncol Rep. 2011;26:1281-1286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 188. | Hu W, Wang X, Ding X, Li Y, Zhang X, Xie P, Yang J, Wang S. MicroRNA-141 represses HBV replication by targeting PPARA. PLoS One. 2012;7:e34165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 79] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 189. | Yang Y, Liu Y, Xue J, Yang Z, Shi Y, Lou G, Wu S, Qi J, Liu W, Wang J, Chen Z. MicroRNA-141 Targets Sirt1 and Inhibits Autophagy to Reduce HBV Replication. Cell Physiol Biochem. 2017;41:310-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 190. | Duan X, Li S, Holmes JA, Tu Z, Li Y, Cai D, Liu X, Li W, Yang C, Jiao B, Schaefer EA, Fusco DN, Salloum S, Chen L, Lin W, Chung RT. MicroRNA 130a Regulates both Hepatitis C Virus and Hepatitis B Virus Replication through a Central Metabolic Pathway. J Virol. 2018;92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 191. | Singh AK, Rooge SB, Varshney A, Vasudevan M, Bhardwaj A, Venugopal SK, Trehanpati N, Kumar M, Geffers R, Kumar V, Sarin SK. Global microRNA expression profiling in the liver biopsies of hepatitis B virus-infected patients suggests specific microRNA signatures for viral persistence and hepatocellular injury. Hepatology. 2018;67:1695-1709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 192. | Yang X, Li H, Sun H, Fan H, Hu Y, Liu M, Li X, Tang H. Hepatitis B Virus-Encoded MicroRNA Controls Viral Replication. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 85] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 193. | Zhao X, Sun L, Mu T, Yi J, Ma C, Xie H, Liu M, Tang H. An HBV-encoded miRNA activates innate immunity to restrict HBV replication. J Mol Cell Biol. 2020;12:263-276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 194. | Morlando M, Fatica A. Alteration of Epigenetic Regulation by Long Noncoding RNAs in Cancer. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 134] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 195. | Yang G, Feng J, Liu Y, Zhao M, Yuan Y, Yuan H, Yun H, Sun M, Bu Y, Liu L, Liu Z, Niu JQ, Yin M, Song X, Miao Z, Lin Z, Zhang X. HAT1 signaling confers to assembly and epigenetic regulation of HBV cccDNA minichromosome. Theranostics. 2019;9:7345-7358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 196. | Liu Y, Feng J, Sun M, Yang G, Yuan H, Wang Y, Bu Y, Zhao M, Zhang S, Zhang X. Long non-coding RNA HULC activates HBV by modulating HBx/STAT3/miR-539/APOBEC3B signaling in HBV-related hepatocellular carcinoma. Cancer Lett. 2019;454:158-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 197. | Salerno D, Chiodo L, Alfano V, Floriot O, Cottone G, Paturel A, Pallocca M, Plissonnier ML, Jeddari S, Belloni L, Zeisel M, Levrero M, Guerrieri F. Hepatitis B protein HBx binds the DLEU2 LncRNA to sustain cccDNA and host cancer-related gene transcription. Gut. 2020;69:2016-2024. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 113] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 198. | Nishitsuji H, Ujino S, Yoshio S, Sugiyama M, Mizokami M, Kanto T, Shimotohno K. Long noncoding RNA #32 contributes to antiviral responses by controlling interferon-stimulated gene expression. Proc Natl Acad Sci USA. 2016;113:10388-10393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 77] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 199. | Belloni L, Allweiss L, Guerrieri F, Pediconi N, Volz T, Pollicino T, Petersen J, Raimondo G, Dandri M, Levrero M. IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J Clin Invest. 2012;122:529-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 422] [Cited by in RCA: 484] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 200. | Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519-525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 859] [Cited by in RCA: 996] [Article Influence: 76.6] [Reference Citation Analysis (0)] |
| 201. | Cheng J, Zhao Q, Zhou Y, Tang L, Sheraz M, Chang J, Guo JT. Interferon Alpha Induces Multiple Cellular Proteins That Coordinately Suppress Hepadnaviral Covalently Closed Circular DNA Transcription. J Virol. 2020;94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 202. | Allweiss L, Giersch K, Volz T, Lohse AW, Petersen J, Urban S, Luetgehmann M, Dandri M. PS-155-HBV entry inhibition after interferon alpha treatment hinders HBV rebound in hepatocytes that became negative for all HBV markers during interferon treatment. J Hepatol. 2019;70:e98. [DOI] [Full Text] |
| 203. | Chen J, Li Y, Lai F, Wang Y, Sutter K, Dittmer U, Ye J, Zai W, Liu M, Shen F, Wu M, Hu K, Li B, Lu M, Zhang X, Zhang J, Li J, Chen Q, Yuan Z. Functional Comparison of Interferon-α Subtypes Reveals Potent Hepatitis B Virus Suppression by a Concerted Action of Interferon-α and Interferon-γ Signaling. Hepatology. 2021;73:486-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 60] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 204. | Neureiter D, Stintzing S, Kiesslich T, Ocker M. Hepatocellular carcinoma: Therapeutic advances in signaling, epigenetic and immune targets. World J Gastroenterol. 2019;25:3136-3150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 47] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 205. | Hong YK, Li Y, Pandit H, Li S, Pulliam Z, Zheng Q, Yu Y, Martin RCG. Epigenetic modulation enhances immunotherapy for hepatocellular carcinoma. Cell Immunol. 2019;336:66-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 206. | Yu HB, Jiang H, Cheng ST, Hu ZW, Ren JH, Chen J. AGK2, A SIRT2 Inhibitor, Inhibits Hepatitis B Virus Replication In Vitro And In Vivo. Int J Med Sci. 2018;15:1356-1364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 207. | Gilmore S, Tam D, Dick R, Appleby T, Birkus G, Willkom M, Delaney WE, Notte GT, Feierbach B. SAT-160 - Antiviral activity of GS-5801, a liver-targeted prodrug of a lysine demethylase 5 inhibitor, in a hepatitis B virus primary human hepatocyte infection model. J Hepatol. 2017;66:S690-S691. [DOI] [Full Text] |
| 208. | Cheng ST, Hu JL, Ren JH, Yu HB, Zhong S, Wai Wong VK, Kwan Law BY, Chen WX, Xu HM, Zhang ZZ, Cai XF, Hu Y, Zhang WL, Long QX, Ren F, Zhou HZ, Huang AL, Chen J. Dicoumarol, an NQO1 inhibitor, blocks cccDNA transcription by promoting degradation of HBx. J Hepatol. 2021;74:522-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 87] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 209. | Shim YH, Yoon GS, Choi HJ, Chung YH, Yu E. p16 Hypermethylation in the early stage of hepatitis B virus-associated hepatocarcinogenesis. Cancer Lett. 2003;190:213-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 54] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 210. | Zhao X, Zhao Z, Guo J, Huang P, Zhu X, Zhou X, Yang Z, Zhao L, Xu L, Xu J, Fu L, Zhang J, Zhang X, Dong Y, Huang G, Wang Q, Li B, Song X, Yang X, Liu S, Yi S, Yu T, Yu C, Hou L, Li J, Chen W. Creation of a six-fingered artificial transcription factor that represses the hepatitis B virus HBx gene integrated into a human hepatocellular carcinoma cell line. J Biomol Screen. 2013;18:378-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 211. | Luo W, Wang J, Xu D, Bai H, Zhang Y, Li X. Engineered zinc-finger transcription factors inhibit the replication and transcription of HBV in vitro and in vivo. Int J Mol Med. 2018;41:2169-2176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 212. | Bloom K, Kaldine H, Cathomen T, Mussolino C, Ely A, Arbuthnot P. Inhibition of replication of hepatitis B virus using transcriptional repressors that target the viral DNA. BMC Infect Dis. 2019;19:802. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 213. | Xirong L, Rui L, Xiaoli Y, Qiuyan H, Bikui T, Sibo Z, Naishuo Z. Hepatitis B virus can be inhibited by DNA methyltransferase 3a via specific zinc-finger-induced methylation of the X promoter. Biochemistry (Mosc). 2014;79:111-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 214. | Bloom K, Ely A, Mussolino C, Cathomen T, Arbuthnot P. Inactivation of hepatitis B virus replication in cultured cells and in vivo with engineered transcription activator-like effector nucleases. Mol Ther. 2013;21:1889-1897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 170] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 215. | Kunze S. Quantitative Region-Specific DNA Methylation Analysis by the EpiTYPER™ Technology. Methods Mol Biol. 2018;1708:515-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 216. | Nakamura M, Gao Y, Dominguez AA, Qi LS. CRISPR technologies for precise epigenome editing. Nat Cell Biol. 2021;23:11-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 242] [Article Influence: 60.5] [Reference Citation Analysis (0)] |
| 217. | Broering R, Luo X, Liu J, Lu M. Controversial: Early Innate Responses to Hepatitis B Virus Infection, an Explanation for Viral Persistence? Virol Sin. 2021;36:163-166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 218. | Grompe M, Strom S. Mice with human livers. Gastroenterology. 2013;145:1209-1214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 99] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 219. | Lebossé F, Inchauspé A, Locatelli M, Miaglia C, Diederichs A, Fresquet J, Chapus F, Hamed K, Testoni B, Zoulim F. Quantification and epigenetic evaluation of the residual pool of hepatitis B covalently closed circular DNA in long-term nucleoside analogue-treated patients. Sci Rep. 2020;10:21097. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 220. | Crudele JM, Chamberlain JS. Cas9 immunity creates challenges for CRISPR gene editing therapies. Nat Commun. 2018;9:3497. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 190] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 221. | Tropberger P, Pott S, Keller C, Kamieniarz-Gdula K, Caron M, Richter F, Li G, Mittler G, Liu ET, Bühler M, Margueron R, Schneider R. Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell. 2013;152:859-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 184] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 222. | Kang JY, Kim JY, Kim KB, Park JW, Cho H, Hahm JY, Chae YC, Kim D, Kook H, Rhee S, Ha NC, Seo SB. KDM2B is a histone H3K79 demethylase and induces transcriptional repression via sirtuin-1-mediated chromatin silencing. FASEB J. 2018;32:5737-5750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 223. | Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007;9:25-35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 320] [Cited by in RCA: 310] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 224. | Peng JC, Karpen GH. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 2009;5:e1000435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 167] [Cited by in RCA: 161] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 225. | Choi CY, Choi BH, Park GT, Rho HM. Activating transcription factor 2 (ATF2) down-regulates hepatitis B virus X promoter activity by the competition for the activating protein 1 binding site and the formation of the ATF2-Jun heterodimer. J Biol Chem. 1997;272:16934-16939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 226. | Kim BK, Lim SO, Park YG. Requirement of the cyclic adenosine monophosphate response element-binding protein for hepatitis B virus replication. Hepatology. 2008;48:361-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 227. | Song CL, Ren JH, Ran LK, Li YG, Li XS, Chen X, Li WY, Huang AL, Chen J. Cyclin D2 plays a regulatory role in HBV replication. Virology. 2014;462-463:149-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 228. | Ori A, Atzmony D, Haviv I, Shaul Y. An NF1 motif plays a central role in hepatitis B virus enhancer. Virology. 1994;204:600-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 229. | Shaul Y, Ben-Levy R, De-Medina T. High affinity binding site for nuclear factor I next to the hepatitis B virus S gene promoter. EMBO J. 1986;5:1967-1971. [PubMed] |
| 230. | Nakanishi-Matsui M, Hayashi Y, Kitamura Y, Koike K. Integrated hepatitis B virus DNA preserves the binding sequence of transcription factor Yin and Yang 1 at the virus-cell junction. J Virol. 2000;74:5562-5568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 231. | Raney AK, McLachlan A. Characterization of the hepatitis B virus large surface antigen promoter Sp1 binding site. Virology. 1995;208:399-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 232. | Raney AK, Le HB, McLachlan A. Regulation of transcription from the hepatitis B virus major surface antigen promoter by the Sp1 transcription factor. J Virol. 1992;66:6912-6921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 47] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 233. | Li J, Ou JH. Differential regulation of hepatitis B virus gene expression by the Sp1 transcription factor. J Virol. 2001;75:8400-8406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 57] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 234. | Lu CC, Yen TS. Activation of the hepatitis B virus S promoter by transcription factor NF-Y via a CCAAT element. Virology. 1996;225:387-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 38] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 235. | Maity SN, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci. 1998;23:174-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 291] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 236. | Nardini M, Gnesutta N, Donati G, Gatta R, Forni C, Fossati A, Vonrhein C, Moras D, Romier C, Bolognesi M, Mantovani R. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell. 2013;152:132-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 223] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 237. | Choi BH, Park CJ, Rho HM. Insulin activates the hepatitis B virus X gene through the activating protein-1 binding site in HepG2 cells. DNA Cell Biol. 1998;17:951-956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 238. | Bogomolski-Yahalom V, Klein A, Greenblat I, Haviv Y, Tur-Kaspa R. The TATA-less promoter of hepatitis B virus S gene contains a TBP binding site and an active initiator. Virus Res. 1997;49:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 239. | Chen IH, Huang CJ, Ting LP. Overlapping initiator and TATA box functions in the basal core promoter of hepatitis B virus. J Virol. 1995;69:3647-3657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 240. | Qin J, Zhai J, Hong R, Shan S, Kong Y, Wen Y, Wang Y, Liu J, Xie Y. Prospero-related homeobox protein (Prox1) inhibits hepatitis B virus replication through repressing multiple cis regulatory elements. J Gen Virol. 2009;90:1246-1255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 241. | Lin YC, Hsu EC, Ting LP. Repression of hepatitis B viral gene expression by transcription factor nuclear factor-kappaB. Cell Microbiol. 2009;11:645-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 242. | Nkongolo S, Nußbaum L, Lempp FA, Wodrich H, Urban S, Ni Y. The retinoic acid receptor (RAR) α-specific agonist Am80 (tamibarotene) and other RAR agonists potently inhibit hepatitis B virus transcription from cccDNA. Antiviral Res. 2019;168:146-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 243. | Oropeza CE, Li L, McLachlan A. Differential inhibition of nuclear hormone receptor-dependent hepatitis B virus replication by the small heterodimer partner. J Virol. 2008;82:3814-3821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 244. | López-Cabrera M, Letovsky J, Hu KQ, Siddiqui A. Multiple liver-specific factors bind to the hepatitis B virus core/pregenomic promoter: trans-activation and repression by CCAAT/enhancer binding protein. Proc Natl Acad Sci USA. 1990;87:5069-5073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 123] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 245. | Zhou DX, Yen TS. The ubiquitous transcription factor Oct-1 and the liver-specific factor HNF-1 are both required to activate transcription of a hepatitis B virus promoter. Mol Cell Biol. 1991;11:1353-1359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 34] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 246. | Zheng Y, Li J, Ou JH. Regulation of hepatitis B virus core promoter by transcription factors HNF1 and HNF4 and the viral X protein. J Virol. 2004;78:6908-6914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 86] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 247. | Lin J, Gu C, Shen Z, Liu Y, Wang W, Tao S, Cui X, Liu J, Xie Y. Hepatocyte nuclear factor 1α downregulates HBV gene expression and replication by activating the NF-κB signaling pathway. PLoS One. 2017;12:e0174017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 248. | Chen M, Hieng S, Qian X, Costa R, Ou JH. Regulation of hepatitis B virus ENI enhancer activity by hepatocyte-enriched transcription factor HNF3. Virology. 1994;205:127-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 47] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 249. | Li M, Xie Y, Wu X, Kong Y, Wang Y. HNF3 binds and activates the second enhancer, ENII, of hepatitis B virus. Virology. 1995;214:371-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 55] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 250. | Cho EY, Kim HJ, Park C, So HS, Park RK, Kim HC. Impact of Nucleotide Mutations at the HNF3- and HNF4-Binding Sites in Enhancer 1 on Viral Replication in Patients with Chronic Hepatitis B Virus Infection. Gut Liver. 2013;7:569-575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 251. | Long Y, Chen E, Liu C, Huang F, Zhou T, He F, Liu L, Liu F, Tang H. The correlation of hepatocyte nuclear factor 4 alpha and 3 beta with hepatitis B virus replication in the liver of chronic hepatitis B patients. J Viral Hepat. 2009;16:537-546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 252. | Lin TJ, Yang RY, Lee HJ. Collective repression of the hepatitis B virus enhancer II by human TR4 and TR2 orphan receptors. Hepatol Res. 2008;38:79-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 253. | Yuan Y, Yuan H, Yang G, Yun H, Zhao M, Liu Z, Zhao L, Geng Y, Liu L, Wang J, Zhang H, Wang Y, Zhang XD. IFN-α confers epigenetic regulation of HBV cccDNA minichromosome by modulating GCN5-mediated succinylation of histone H3K79 to clear HBV cccDNA. Clin Epigenetics. 2020;12:135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |