Published online May 7, 2021. doi: 10.3748/wjg.v27.i17.1993
Peer-review started: December 14, 2020
First decision: February 11, 2021
Revised: February 24, 2021
Accepted: March 24, 2021
Article in press: March 24, 2021
Published online: May 7, 2021
Processing time: 135 Days and 14.7 Hours
Previous studies have suggested that long non-coding RNAs (lncRNA) TP73-AS1 is significantly upregulated in several cancers. However, the biological role and clinical significance of TP73-AS1 in pancreatic cancer (PC) remain unclear.
To investigate the role of TP73-AS1 in the growth and metastasis of PC.
The expression of lncRNA TP73-AS1, miR-128-3p, and GOLM1 in PC tissues and cells was detected by quantitative real-time polymerase chain reaction. The bioinformatics prediction software ENCORI was used to predict the putative binding sites of miR-128-3p. The regulatory roles of TP73-AS1 and miR-128-3p in cell proliferation, migration, and invasion abilities were verified by Cell Counting Kit-8, wound-healing, and transwell assays, as well as flow cytometry and Western blot analysis. The interactions among TP73-AS1, miR-128-3p, and GOLM1 were explored by bioinformatics prediction, luciferase assay, and Western blot.
The expression of TP73-AS1 and miRNA-128-3p was dysregulated in PC tissues and cells. High TP73-AS1 expression was correlated with a poor prognosis. TP73-AS1 silencing inhibited PC cell proliferation, migration, and invasion in vitro as well as suppressed tumor growth in vivo. Mechanistically, TP73-AS1 was validated to promote PC progression through GOLM1 upregulation by competitively binding to miR-128-3p.
Our results demonstrated that TP73-AS1 promotes PC progression by regulating the miR-128-3p/GOLM1 axis, which might provide a potential treatment strategy for patients with PC.
Core Tip: In this study, the expression level of TP73-AS1 in pancreatic cancer (PC) was measured and its clinical significance was assessed. In vitro and in vivo experiments were performed to determine the roles of TP73-AS1 in the progression and development of PC. Moreover, the underlying molecular mechanisms were also illustrated, which could provide a novel therapeutic target for patients with PC.
- Citation: Wang B, Sun X, Huang KJ, Zhou LS, Qiu ZJ. Long non-coding RNA TP73-AS1 promotes pancreatic cancer growth and metastasis through miRNA-128-3p/GOLM1 axis. World J Gastroenterol 2021; 27(17): 1993-2014
- URL: https://www.wjgnet.com/1007-9327/full/v27/i17/1993.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i17.1993
Pancreatic cancer (PC) is the fourth most frequent cause of cancer-related deaths with an extremely poor prognosis, especially in patients with advanced-stage PC [1,2]. Although standard treatments have been improved in recent years, the effectiveness of these treatments was still limited and surgical resection was the only chance to obtain curative treatment[2,3]. Hence, it is necessary to seek for new treatment to optimize therapeutic approaches.
Long non-coding RNAs (lncRNAs) are small endogenous non-coding RNAs whose lengths are larger than 200 nucleotides. LncRNAs have the capacity to regulate various biological processes such as tumor initiation, growth, metastasis, chemoresistance, and radioresistance by directly binding to partially complimentary sequences in their target genes[4-7]. Moreover, emerging evidence has revealed that lncRNAs could play crucial roles in the progression of PC[8,9]. LncRNA-BX111 was upregulated in pancreatic cancer and high BX111 expression was correlated with advance tumor-node-metastasis (TNM) stage, lymphatic invasion, and distant metastasis, as well as poor clinical prognosis in patients with PC[10]. Further investigation revealed that BX111 contributed to metastasis and progression of PC by regulating expression of ZEB1 and its downstream proteins E-cadherin and MMP2[10]. PVT1 was identified as a regulator of gemcitabine sensitivity with a genome-wide and piggyBac transposon-based genetic screening platform[11]. Therefore, lncRNAs may be new biological markers for disease diagnosis and could be taken as new drug targets, which would provide a new strategy for PC.
Dysregulation of TP73-AS1 has been identified in several human cancer types, including glioma, hepatocellular carcinoma, and non-small cell lung cancer[12-14]. However, little is known about the expression pattern and biological roles of TP73-AS1 in PC. In this study, the expression level of TP73-AS1 in PC was measured and its clinical significance was assessed. In vitro and in vivo experiments were performed to determine the roles of TP73-AS1 in the progression and development of PC. Further investigation indicated that the 3’ untranslated region (UTR) of GOLM1 harbors a functional response element for miR-128-3p. Besides, miR-128-3p-3p could abrogate TP73-AS1-mediated expression of GOLM1, which suggested that TP73-AS1 could act as a molecular sponge to decrease miR-128-3p expression, thereby resulting in partial abolition of the translational repression of its target gene GOLM1 in PC cells. Therefore, we hope that the underlying molecular mechanisms of TP73-AS1 could provide a novel therapeutic target for patients with PC.
A total of 116 clinical PC tissues and corresponding normal tissues from surgical resection were collected at Shanghai General Hospital of Shanghai Jiao Tong University between April 2007 and July 2010. PC was diagnosed by pathological examinations. Patients were excluded if they received any treatments such as chemotherapy, radiotherapy, or molecular targeted therapy prior to surgery. All patients provided informed written consent prior to the use of these clinical materials for research purpose. This study was conducted in accordance with the ethical guidelines of the Declaration of Helsinki and approved by the Ethics Committee of Shanghai First People's Hospital (No. 2014-07DF), School of Medicine, Shanghai Jiaotong University (Shanghai, China). All human tissues were immediately frozen in liquid nitrogen until being used.
Human PC cell lines (SW1990, PANC-1, BXPC-3, AsPc-1, and Capan-1) and human pancreatic duct epithelial cell line (H6C7) were obtained from the Shanghai Cell Bank of Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Invitrogen, Carlsbad, CA, United States) supplemented with 10% fatal bovine serum (FBS, Invitrogen), 100 U/mL penicillin, and 100 μg/mL streptomycin (Hyclone, South Logan, UT, United States) in a humidified incubator (5% CO2) at 37 °C. Small interfering RNAs (siRNAs) targeting TP73-AS1 (si-TP73-AS1#1, si-TP73-AS1#2, and si-TP73-AS1#3) and negative control (si-control) were purchased from GenePharma (Shanghai, China). MiR-128-3p, miR-NC, anti-miR-128-3p, and anti-miR-NC were obtained from Thermofisher. Cell transfection was performed using FuGENE HD Transfection Reagent (Roche, United States) according to the manufacturer’s instructions.
Total RNA was isolated using TRIzol reagent (Invitrogen) from PC tissues and cell lines. RNA was reversely transcribed into cDNA using the PrimeScript™RT reagent Kit with gDNA Eraser (TakaRa, Dalian, China). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using FastStart Universal SYBR Green Master (Roche, Basel, Switzerland) on a Bio-Rad RT-PCR cycler (Bio-Rad, Hercules, United States). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and small RNA RNU6B (U6) were used as the internal controls for lncRNA/mRNA and miRNAs, respectively. Relative expression values of genes were analyzed by the 2−ΔΔCt method.
For cell proliferation assay, cells were plated into 96-well plates with four replicate wells per group and then incubated with 10 μL of CCK-8 reagent (Dojindo, Kumamoto, Japan). The absorbance was measured at 450 nm with a microplate reader (Bio-Tek, Winooski, United States) 2 h later. For colony formation assay, approximately 600 cells were plated into 6-well plates with three replicates. Cells were fixed with 10% formaldehyde and stained with 0.5% crystal violet 14 d later. Cell apoptosis was detected using Annexin-V-fluorescein isothiocyanate apoptosis detection kit (BD, Franklin Lakes, United States) according to the manufacturer's instructions. The apoptosis rate of cells was measured on a BD FACSAria™ II flow cytometer (BD).
Cell migration and invasion were evaluated using the Boyden chambers (Millipore; Merck KGaA, Germany) with an 8 mm pore size. Briefly, a total of 2 × 104 cells in 100 μL serum-free medium were transferred into the upper chamber, and the lower chamber was filled with medium containing 10% FBS. After 24 h incubation, cells were fixed with 10% formaldehyde for 15 min and stained with 0.5% crystal violet for 20 min at room temperate. The migrated cells were counted under an X71 inverted microscope in six randomly selected fields and captured using a microscope (Nikon). The invasion assay was performed in the same way as the migration assay did except that the inserts were pre-coated with Matrigel (BD).
The putative miR-128-3p binding sites in TP73-AS1 and GOLM1 3′UTR were synthesized and inserted into pMIR-REPORT™ miRNA Expression Reporter Vector (Thermofisher). Their corresponding mutants were generated using MutanBEST Kit (TaKaRa). AsPc-1 and Capan-1 cells were co-transfected with these reporter plasmids and pMIR-REPORT β-gal, miR-128-3p mimics, or miR-128-3p inhibitors using Lipofectamine 3000 (Invitrogen). Luciferase activity was measured using Dual-Luciferase Reporter Assay System (Promega, WI, United States) 48 h after the transfection.
PANC-1 and ASPC-1 cells transfected with miR-NC or miR-128-3p were harvested with revised importance-performance analysis lysis buffer (Cell Signaling Technology, Danvers, MA, United States) containing a proteinase inhibitor cocktail (Roche, IN, United States). The lysates were incubated with magnetic beads conjugated with human anti-Ago2 antibody and normal rabbit immunoglobulin G. Then RNA was isolated from the mixture with TRIzol reagent for qRT-PCR analysis.
Western blot was performed as we previously described[15]. The primary antibodies used in this study are listed as following: Anti-GOLM1 (H00051280-PW1, Abnova, Taiwan), anti-GAPDH (#10494-1-AP, Proteintech, IL, United States), anti-E-cadherin (14-3249-82, CST, United States), anti-N-cadherin (MA1-91128, CST, United States), anti-Vimentin (PA5-27231, CST, United States), anti-Caspase-3 (700182, CST, United States), and anti-Bcl-2 (MA5-11757, CST, United States).
All animal experiments were performed in compliance to institutional guidelines approved by the Use Committee for Animal Care and this study was approved by the Ethics Committees of Shanghai First People's Hospital of Shanghai Jiao Tong University (approval No. 201804SF). Female BALB/c-nude mice (4–6 wk of age) were purchased from Shanghai SJA Laboratory Animal Company (Shanghai, China) and maintained under specific pathogen free conditions. Capan-1 cells (1 × 107; transfected with si-control or si-TP73-AS1#1) mingled with 100 μL serum-free medium were injected subcutaneously into to the flanks of the nude mice. All mice were sacrificed 4 wk after injection and then tumors were isolated and photographed. Tumor volumes were calculated using the formula length × width2/2 and tumor weights were measured. For tail vein injection, 1 × 106 cells in serum-free medium were injected into 6 wk-old BALB/c-nude mice via the tail vein. Five weeks after injection, all mice were sacrificed and lung tissues were finally embedded with paraffin and subjected to hematoxylin and eosin (H&E) staining.
Data are shown as the mean ± SD. The differences between groups were analyzed by Student’s t-test or Chi-square test. The cumulative overall survival was calculated using the Kaplan-Meier method, and the log-rank test was used to analyze differences in the survival times. Data were analyzed using GraphPad software 7.0. P < 0.05 was considered significant.
To explore the roles of TP73-AS1 in PC, we first detected the expression of TP73-AS1 in the human pancreatic duct epithelial cell line (H6C7) and five PC cell lines by qRT-PCR. Our data indicated that TP73-AS1 expression was higher in all PC cell lines than in H6C7, especially in AsPc-1 and Capan-1 cells (Figure 1A). Then the expression of TP73-AS1 in 116 pairs of PC tissues and adjacent non-cancerous tissues was measured, and the results revealed that TP73-AS1 expression was significantly increased in PC tissues compared to the corresponding non-cancerous tissues (Figure 1B and C). Furthermore, it was shown that in the tissue samples of stages I and II PC patients, the expression levels of TP73-AS1 were lower than those in stage III PC patients (Figure 1D). Moreover, the associations between TP73-AS1 expression and the clinicopathological characteristics in PC patients were analyzed. The results suggested that increased TP73-AS1 expression was significantly correlated with tumor size, vessel infiltration, and TNM stage. Besides, no correlation was found between TP73-AS1 expression and other clinical pathological features (Table 1). Kaplan-Meier survival results suggested that patients with higher TP73-AS1 expression had a shorter overall survival than those with lower TP73-AS1 expression (Figure 1E). These data indicated that TP73-AS1 might play a vital role in the progression of PC.
| Characteristic | (n) | TP73-AS1 expression | P value | |
| n = 116 | Low expression | High expression | ||
| Age | 0.7101 | |||
| < 60 | 65 | 32 | 33 | |
| ≥ 60 | 51 | 23 | 28 | |
| Gender | 0.7026 | |||
| Male | 74 | 34 | 40 | |
| Female | 42 | 21 | 21 | |
| Tumor differentiation | 0.5723 | |||
| Poor | 69 | 31 | 38 | |
| Middle and well | 47 | 24 | 23 | |
| Tumor size | 0.008 | |||
| ≤ 2 cm | 48 | 31 | 17 | |
| > 2 cm | 68 | 26 | 42 | |
| Tumor site | 0.4555 | |||
| Head | 64 | 28 | 36 | |
| Body | 52 | 27 | 25 | |
| Vessel infiltration | 0.001 | |||
| Negative | 82 | 47 | 35 | |
| Positive | 34 | 8 | 26 | |
| Lymph node metastasis | 0.5526 | |||
| No | 78 | 35 | 43 | |
| Yes | 38 | 20 | 18 | |
| TNM stage | 0.0008 | |||
| I-II | 84 | 48 | 36 | |
| III | 32 | 7 | 25 | |
In order to assess the biological functions of TP73-AS1, we knocked down TP73-AS1 by transfecting specific siRNAs in AsPc-1 and Capan-1 cells, which have higher endogenous TP73-AS1 expression. The knockdown efficacy was confirmed by qRT-PCR analysis. The expression of TP73-AS1 was markedly decreased in AsPc-1 and Capan-1 cells after transfecting with siRNAs targeting TP73-AS1 (Figure 2A). CCK-8 (Figure 2B) and colony formation assay (Figure 2C) showed that knockdown of TP73-AS1 in PC cells markedly restrained cell proliferation. Furthermore, cell apoptosis was highly promoted by depletion of TP73-AS1 in AsPc-1 and Capan-1 cells (Figure 2D). In addition, in the transwell assay, TP73-AS1 silencing could effectively impede the invasive ability of PC cells (Figure 2E and F). These data revealed that TP73-AS1 acts as an oncogene and depletion of TP73-AS1 inhibits PC cell growth and invasion in vitro.
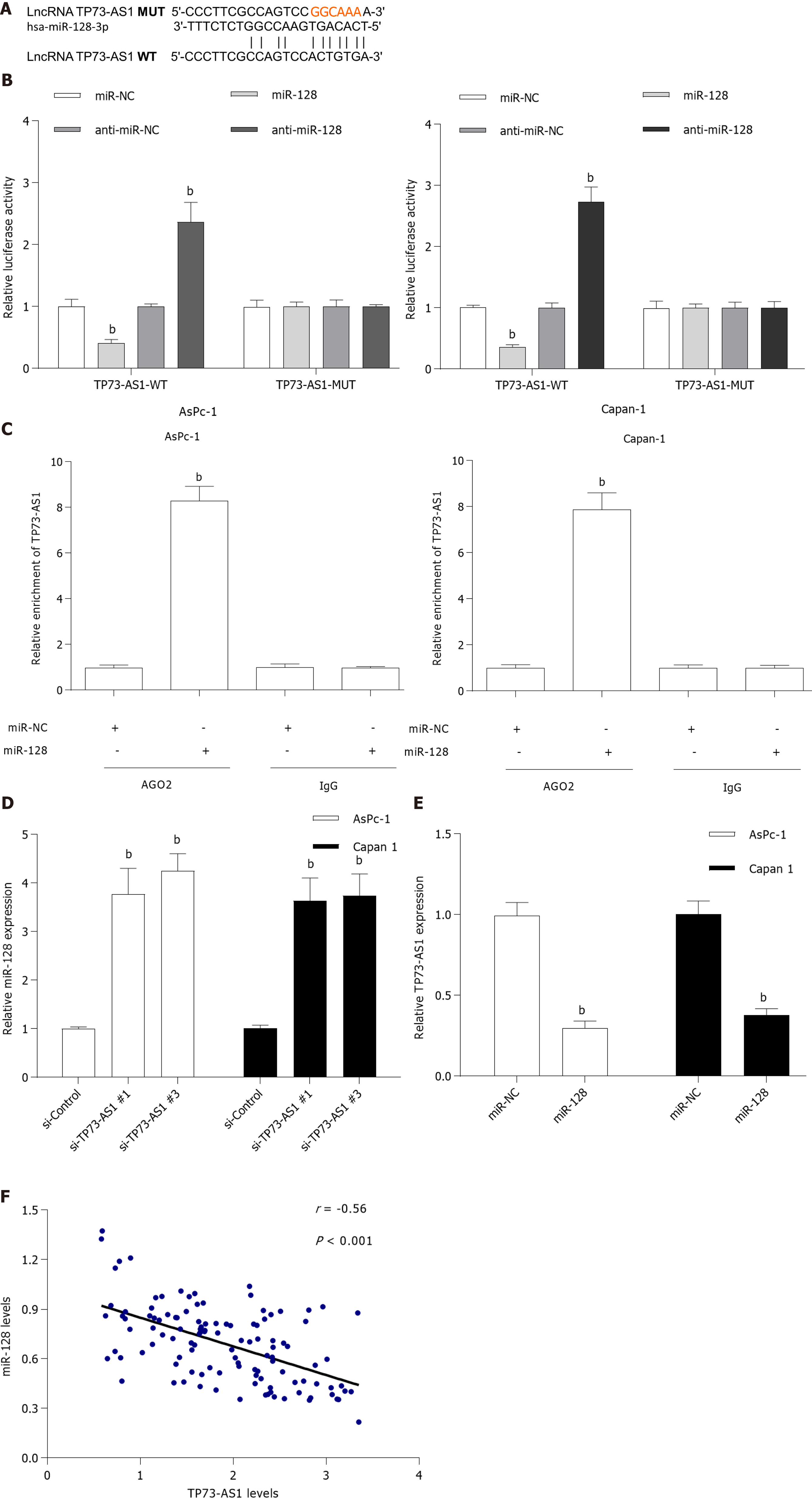
Accumulating evidence suggests that lncRNAs could bind to miRNAs and function as a molecular sponge in the tumorigenesis of various cancers[4]. To elucidate the underlying mechanism of TP73-AS1 involved in PC progression, the potential target miRNAs of TP73-AS1 were forecasted with bioinformatics analysis software (http://starbase.sysu.edu.cn). Among these potential targets, miR-128-3p was chosen for further study because it had been validated as a tumor suppressor in PC (Figure 3A)[16]. Dual-luciferase reporter assay was used to validate the relationship between TP73-AS1 and miR-128-3p. The activity of the wild-type luciferase reporter gene was significantly reduced following transfection with miR-128-3p mimics, whereas the activity of the reporter gene containing the mutant sequence showed no significant change, which indicated that TP73-AS1 could bind to the specific sites of miR-128-3p (Figure 3B). Moreover, anti-Ago2 RNA immunoprecipitation in AsPc-1 and Capan-1 cells transiently overexpressing miR-128-3p could significantly increase the amount of TP73-AS1 (Figure 3C), which could further validate their binding potential. The expression of miR-128-3p was significantly increased in PC cells transfected with si-TP73-AS1#1 and si-TP73-AS1#3 (Figure 3D). Then, we measured miR-128-3p expression level and the relationship between TP73-AS1 and miR-128-3p expression in PC tissues. Interestingly, qRT-PCR assay showed that the miR-128-3p level was remarkably reduced in PC tissues (Figure 3E) and the endogenous miR-128-3p level was negatively correlated with TP73-AS1 in PC tissues (Figure 3F). These results suggested that TP73-AS1 might function as a competing endogenous RNA (ceRNA) for miR-128-3p.
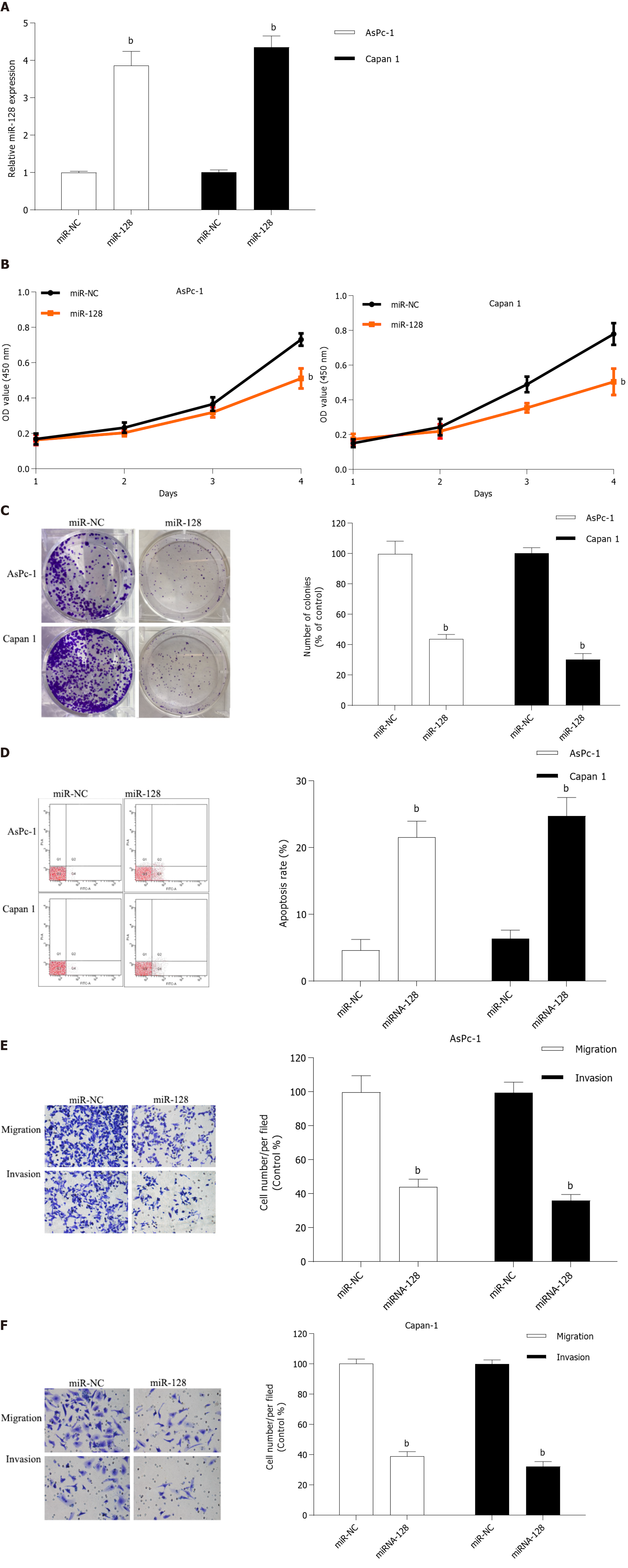
MiR-128-3p was reported to be a tumor suppressor in several cancers, including PC[16,17]. But its effects in the progression of PC are largely unknown. To explore the roles of miR-128-3p in PC cell growth and mobility, miR-128-3p mimics were transfected in AsPc-1 and Capan-1 cells (Figure 4A). CCK-8 (Figure 4B) and colony formation assays (Figure 4C) revealed that miR-128-3p had significant negative regulation effects on the ability of cell proliferation in AsPc-1 and Capan-1 cells. Flow cytometry assay indicated that overexpression of miR-128-3p increased the number of apoptotic cells both in AsPc-1 and Capan-1 cells (Figure 4D). Meanwhile, transwell assays demonstrated that miR-128-3p overexpression significantly restrained PC cell migration and invasion (Figure 4E and F). Considering the downregulation of miR-128-3p in PC tissues, our results manifested that miR-128-3p could act as a tumor suppressor by regulating PC cell proliferation and invasion.
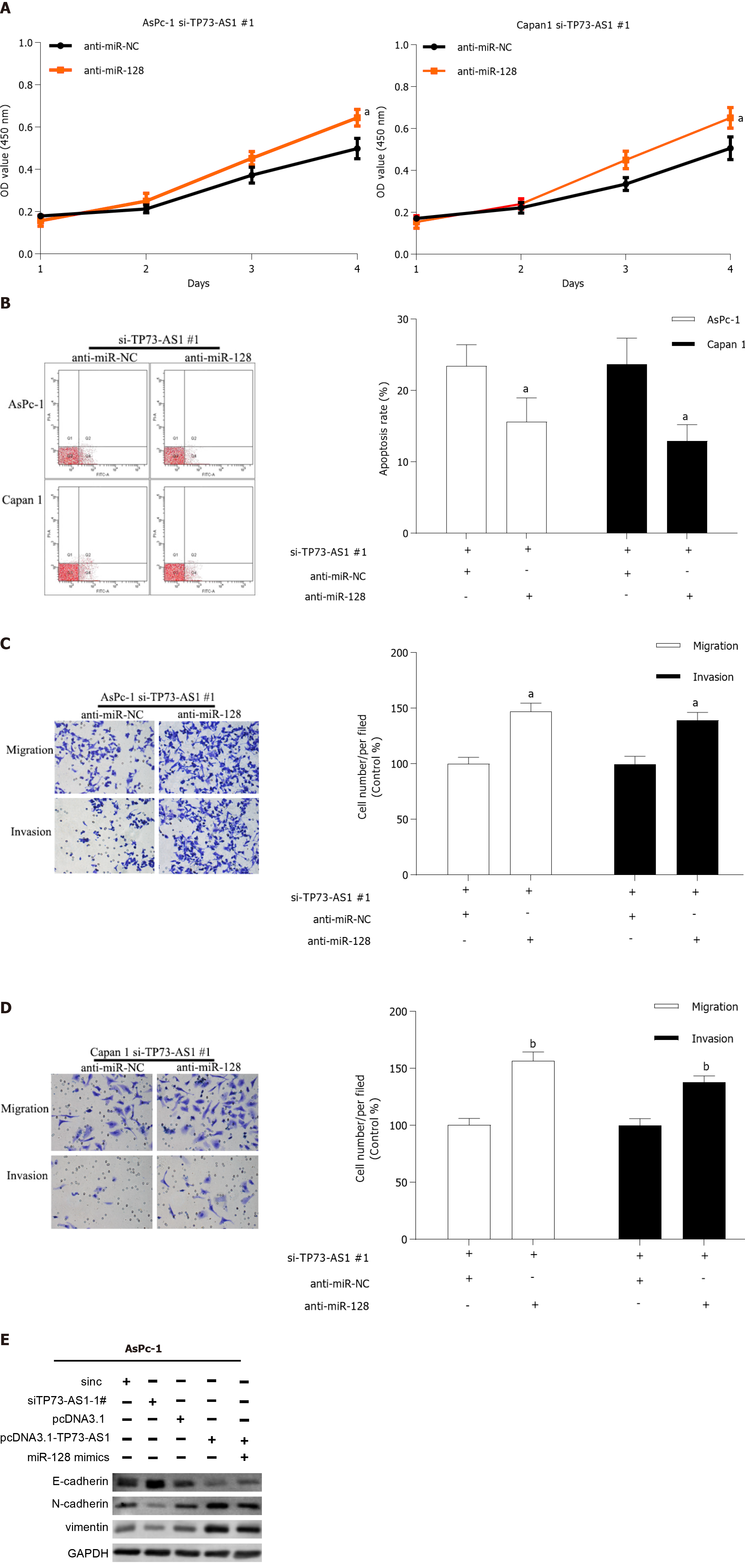
To determine if TP73-AS1 knockdown could exert anti-proliferation and anti-metastasis function by mediating miR-128-3p, anti-miR-128-3p was transfected into AsPc-1 and Capan-1 cells after TP73-AS1 silencing. Functional experiments demonstrated that the TP73-AS1-mediated pro-proliferation (Figure 5A) and anti-apoptosis (Figure 5B) effect was dramatically abrogated by anti-miR-128-3p transfection in TP73-AS1 silencing PC cells. In addition, the inhibitory effects of TP73-AS1 silencing on cell metastasis were rescued by anti-miR-128-3p transfection (Figure 5C and D). Moreover, epithelial-mesenchymal transition (EMT)-related proteins were detected by Western blot. Consistent with the functional assays above, the results showed that TP73-AS1 could regulate EMT-related proteins by regulating miR-128-3p (Figure 5E). All these data indicated that TP73-AS1 is involved in PC progression, at least partly through miR-128-3p.
Since TP73-AS1 was demonstrated to bind to miR-128-3p, we assessed whether TP73-AS1 could indirectly affect the target gene of miR-128-3p by serving as a ceRNA. Based on online bioinformatics analysis, GOLM1 3’UTR was found to possess a putative recognition site for miR-128-3p (Figure 6A). The luciferase reporter assay was carried out and the results showed that the luciferase activity of plasmid carrying GOLM1 3’UTR-WT was significantly decreased by transfecting miR-128-3p mimics both in AsPc-1 and Capan-1 cells (Figure 6B). However, these effects were abolished when the binding sequences were mutated. Transfecting miR-128-3p mimics led to a significant decrease of GOLM1 mRNA and protein expression in AsPc-1 and Capan-1 cells (Figure 6C). To further explore the correlation between TP73-AS1 and GOLM1, the mRNA and protein expression of GOLM1 was detected after TP73-AS1 silencing. As expected, the mRNA and protein expression of GOLM1 was remarkably decreased after silencing TP73-AS1 both in AsPc-1 and Capan-1 cells (Figure 6D). Moreover, the level of GOLM1 mRNA was significantly down-regulated in PC tissues compared to the corresponding non-cancerous tissues (Figure 6E). Interestingly, an inverse correlation was identified between miR-128-3p and GOLM1 mRNA levels in PC tissues (Figure 6F). In contrast, the positive relationship between TP73-AS1 and GOLM1 mRNA levels was observed in PC tissues (Figure 6G). The protein expression of GOLM1 was increased after anti-miR-128-3p transfection in TP73-AS1 silencing PC cells (Figure 6H), which suggested that TP73-AS1 could regulate the expression of GOLM1 by acting as a sponge for miR-128-3p in vitro.
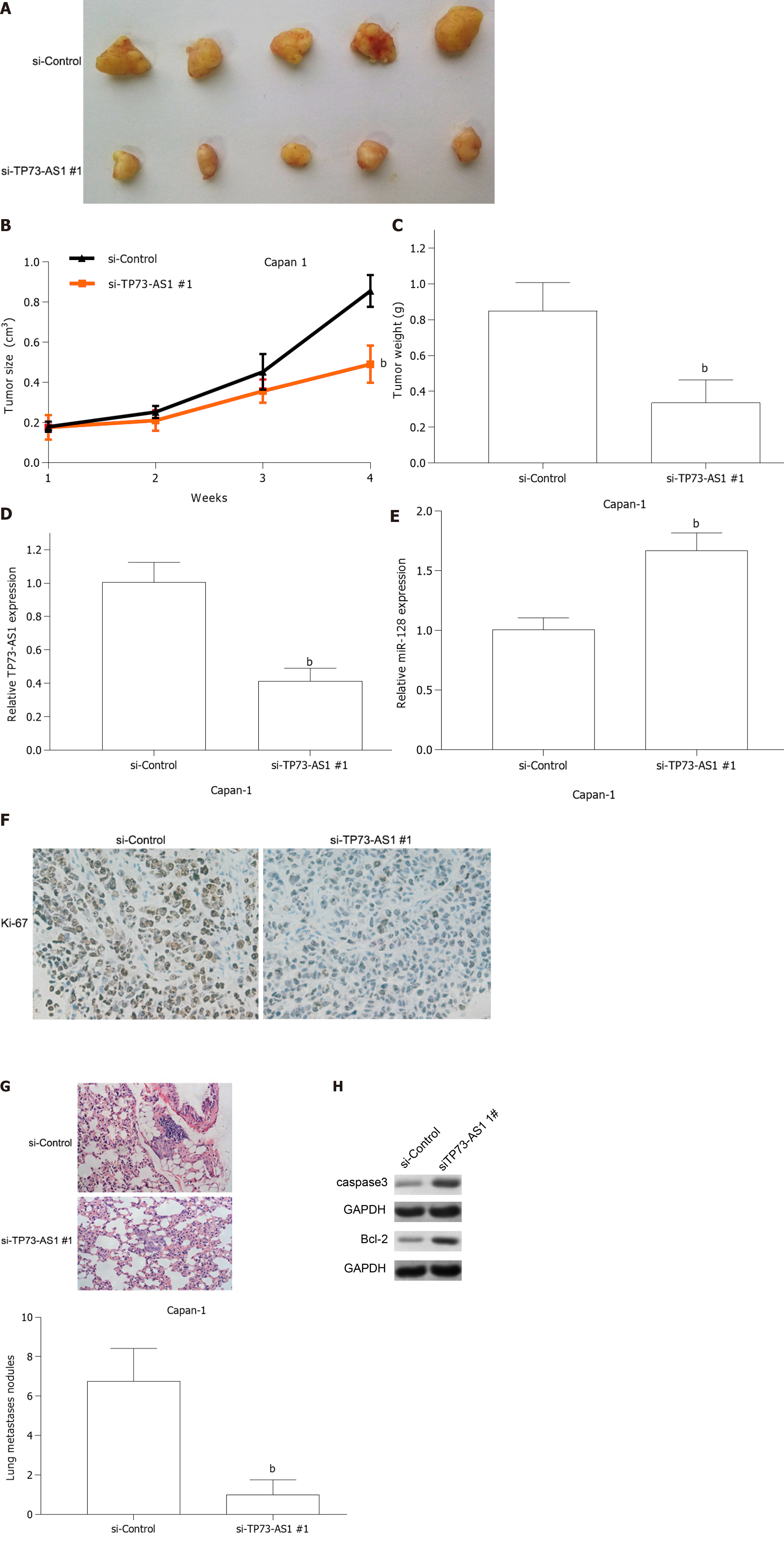
To further elucidate the biological roles of TP73-AS1 in PC tumorigenesis in vivo, Capan-1 cells transfected with si-TP73-AS1#1 or si-Control were implanted into nude mice via subcutaneous injection. Four weeks later, the subcutaneous tumors were collected. Tumor growth curve and tumor weight from the si-TP73-AS1#1 group showed lower size and lighter tumor weight (Figure 7A-C). Moreover, qRT-PCR analysis suggested that the expression of TP73-AS1 was decreased (Figure 7D) and the expression of miR-128-3p (Figure 7E) was increased in the si-TP73-AS1#1 group. In addition, Ki-67 immunostaining indicated that the subcutaneous tumors formed by TP73-AS1 silencing Capan-1 cells showed fewer Ki-67 positive cells compared to the control group (Figure 7F). Together, the in vitro and in vivo results suggested that TP73-AS1 might function as an oncogene in the progression of PC. To investigate the metastatic potential of TP73-AS1 in vivo, Capan-1 cells transfected with si-TP73-AS1#1 or si-Control were injected into the mice via the tail vein. As shown in Figure 7G, silencing TP73-AS1 remarkably decreased the number and size of lung metastatic lesions as detected by H&E staining. Moreover, we detected apoptotic markers in the tumors from the two groups and the results showed that apoptosis-related genes were significantly altered (Figure 7H).
Increasing numbers of studies have shown that lncRNAs are involved in both normal development and pathological processes of human diseases by chromatin modification, genomic imprinting, RNA decay, and sponge-like miRNAs[5,18,19]. Dysregulation of lncRNAs might influence cell proliferation, metastasis, angiogenesis, and drug resistance[18,20,21]. It has been previously reported that increased expression of TP73-AS1 is associated with a poorer prognosis and shorter survival in patients with hepatocellular carcinoma[13]. High TP73-AS1 expression was also observed and associated with poor overall survival of patients with osteosarcoma[22]. TP73-AS1 was up-regulated in both colorectal cancer tissues and colorectal cancer cells and high TP73-AS1 expression was associated with metastasis and advanced clinical stages in patients with colorectal cancer[23]. Above studies suggested that TP73-AS1 might act as an oncogene in tumor progression, which encouraged us to explore the expression and biological function of TP73-AS1 in PC. In agreement with these studies, we found that TP73-AS1 was significantly increased and associated with tumor size, vessel infiltration, and poor prognosis in PC patients. Furthermore, our results showed that knockdown of TP73-AS1 suppressed the proliferation and invasion of PC cells in vitro and the tumor growth in vivo.
Emerging studies demonstrated that lncRNAs could function as ceRNAs to regulate gene expression through competitively binding to miRNAs[4,5]. To further investigate the mechanism of the TP73-AS1 in PC, bioinformatics analysis predicted that TP73-AS1 is a target of miR-128-3p. Numerous studies have indicated that miR-128-3p could act as a tumor suppressive role in many tumors, including glioma, breast cancer, and non-small cell lung cancer[24-26]. In our present study, we found that miR-128-3p could significantly suppress PC cell growth and invasion. Luciferase reporter assay confirmed the relationship between TP73-AS1 and miR-128-3p. Mechanistical study showed that TP73-AS1 could mediate PC cell proliferation, migration, and invasion by sponging miR-128-3p and a negative correlation between TP73-AS1 and miR-128-3p expression was observed in PC tissues. Further investigation indicated that the 3’UTR of GOLM1 harbors a functional response element for miR-128-3p. GOLM1, a type II transmembrane protein, has been reported to be induced by virus infection[27,28]. Recent studies have shown that GOLM1 commonly expressed in epithelial cells of normal tissues was significantly upregulated in tumor tissues, which suggested a possible oncogenic role of GOLM1 in tumor progression[29,30]. Moreover, clinicopathological features showed that GOLM1 was correlated with Edmondson grade, vascular invasion, TNM stage, overall survival, as well as Vimentin expression[31]. GOLM1 was also reported to promote prostate cancer cell growth, migration, and invasion, and inhibited cell apoptosis via the PI3K/AKT/mTOR signaling axis[32]. The role of GOLM1 was unclear in PC and our data showed that the mRNA expression of GOLM1 was increased in PC tissues. Moreover, ectopic expression of miR-128-3p significantly inhibited the expression of GOLM1 at both the mRNA and protein level. Most interestingly, miR-128-3p-3p could abrogate TP73-AS1-mediated expression of GOLM1, which suggested that TP73-AS1 could act as a molecular sponge to decrease miR-128-3p expression, thereby resulting in partial abolition of the translational repression of its target gene GOLM1 in PC cells.
KRAS gene, the most common genetic driver in PC, is mutated in about 93% of PC s[33,34]. The KRAS protein is a small GTPase, which is responsible for interacting with cell membrane growth factor receptors and controlling the switch of multiple signaling pathways and cellular processes. Oncogenic KRAS mutations have been found in 95% of pancreatic ductal adenocarcinoma tissues[35,36]. Decades of research have discovered and clarified the complex picture of KRAS-regulated biological processes, including cell metabolism, tumor cell signaling, the tumor microenvironment, micropinocytosis, apoptosis, and redox homeostasis[37,38]. In our research, ASPC-1 and Capan-1 cells were the two PC cell lines that we selected, both of which contained mutations in the KRAS gene. As our results show, the regulatory roles of TP73-AS1 in cell proliferation, migration, and invasion ability were verified by Cell Counting Kit-8, wound-healing, and transwell assays in ASPC-1 and Capan-1 cells. Due to the vital role that KRAS could play in PC, we are also curious about the role of TP73-AS1 in KRAS wild cells. Therefore, in our further research, we would select BXPC-3 cell line, which contains wild KRAS gene, for in vitro and in vivo functional assays of TP73-AS1 to detect whether KRAS gene could modulate the function of TP73-AS1 in PC.
In summary, our data suggested that TP73-AS1 could function as an oncogenic lncRNA in PC progression. Moreover, TP73-AS1 could promote tumor growth and invasion by acting as a ceRNA to promote GOLM1 expression by sponging miR-128-3p in PC.
Pancreatic cancer (PC) is the fourth most frequent cause of cancer-related deaths in the world. Emerging evidence has revealed that long non-coding RNAs (lncRNAs) could play crucial roles in the progression of PC. However, the biological role and clinical significance of TP73-AS1 in PC remain unclear.
Treatments for PC are still limited, and surgical resection could be the only chance to obtain curative treatment. We hope to provide a novel therapeutic target for patients with PC.
The present study aimed to investigate the role of TP73-AS1 in the growth and metastasis of PC.
Quantitative reverse transcription-polymerase chain reaction was used to detect the expression of lncRNA TP73-AS1, miR-128-3p, and GOLM1 in PC tissues and cells. The regulatory roles of TP73-AS1 in cell proliferation, migration, and invasion ability were verified by Cell Counting Kit-8, wound-healing, and transwell assays. The bioinformatics prediction software ENCORI was used to predict the putative binding sites of miR-128-3p. The interactions among TP73-AS1, miR-128-3p, and GOLM1 were explored by bioinformatics prediction, luciferase assay, and Western blot.
Our data suggested that TP73-AS1 and miRNA-128-3p were dysregulated in PC tissues and cells. TP73-AS1 silencing inhibited PC cell proliferation, migration, and invasion in vitro as well as suppressed tumor growth in vivo. Moreover, TP73-AS1 could promote tumor growth and invasion by acting as a competing endogenous RNA to promote GOLM1 expression by sponging miR-128-3p in PC.
TP73-AS1 could promote PC cell proliferation and metastasis by modulating the miR-128-3p/GOLM1 axis.
TP73-AS1 could promote PC progression, which might provide a potential treatment strategy for patients with PC.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Koustas E, Mikulic D, Shimada S, Tantau AI S-Editor: Fan JR L-Editor: Wang TQ P-Editor: Wang LL
| 1. | Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8789] [Cited by in RCA: 9562] [Article Influence: 869.3] [Reference Citation Analysis (0)] |
| 2. | Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1397] [Cited by in RCA: 1717] [Article Influence: 190.8] [Reference Citation Analysis (1)] |
| 3. | Tsai S, Evans DB. Therapeutic Advances in Localized Pancreatic Cancer. JAMA Surg. 2016;151:862-868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 4. | Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4631] [Cited by in RCA: 6021] [Article Influence: 501.8] [Reference Citation Analysis (0)] |
| 5. | Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2963] [Cited by in RCA: 3493] [Article Influence: 249.5] [Reference Citation Analysis (0)] |
| 6. | Ebahimzadeh K, Shoorei H, Mousavinejad SA, Anamag FT, Dinger ME, Taheri M, Ghafouri-Fard S. Emerging role of non-coding RNAs in response of cancer cells to radiotherapy. Pathol Res Pract. 2021;218:153327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 7. | Wang C, Chen Y, Chen K, Zhang L. Long Noncoding RNA LINC01134 Promotes Hepatocellular Carcinoma Metastasis via Activating AKT1S1 and NF-κB Signaling. Front Cell Dev Biol. 2020;8:429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 8. | Duguang L, Jin H, Xiaowei Q, Peng X, Xiaodong W, Zhennan L, Jianjun Q, Jie Y. The involvement of lncRNAs in the development and progression of pancreatic cancer. Cancer Biol Ther. 2017;18:927-936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 9. | Chen X, Wang J, Xie F, Mou T, Zhong P, Hua H, Liu P, Yang Q. Long noncoding RNA LINC01559 promotes pancreatic cancer progression by acting as a competing endogenous RNA of miR-1343-3p to upregulate RAF1 expression. Aging (Albany NY). 2020;12:14452-14466. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 10. | Deng SJ, Chen HY, Ye Z, Deng SC, Zhu S, Zeng Z, He C, Liu ML, Huang K, Zhong JX, Xu FY, Li Q, Liu Y, Wang CY, Zhao G. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene. 2018;37:5811-5828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 159] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 11. | You L, Chang D, Du HZ, Zhao YP. Genome-wide screen identifies PVT1 as a regulator of Gemcitabine sensitivity in human pancreatic cancer cells. Biochem Biophys Res Commun. 2011;407:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 101] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 12. | Zhang R, Jin H, Lou F. The Long Non-Coding RNA TP73-AS1 Interacted With miR-142 to Modulate Brain Glioma Growth Through HMGB1/RAGE Pathway. J Cell Biochem. 2018;119:3007-3016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 13. | Li S, Huang Y, Fu Y, Tang D, Kang R, Zhou R, Fan XG. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. J Exp Clin Cancer Res. 2017;36:51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 125] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 14. | Zhang L, Fang F, He X. Long noncoding RNA TP73-AS1 promotes non-small cell lung cancer progression by competitively sponging miR-449a/EZH2. Biomed Pharmacother. 2018;104:705-711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 15. | Deng B, Wang B, Fang J, Zhu X, Cao Z, Lin Q, Zhou L, Sun X. MiRNA-203 suppresses cell proliferation, migration and invasion in colorectal cancer via targeting of EIF5A2. Sci Rep. 2016;6:28301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 16. | Jiang J, Feng X, Zhou W, Wu Y, Yang Y. MiR-128 reverses the gefitinib resistance of the lung cancer stem cells by inhibiting the c-met/PI3K/AKT pathway. Oncotarget. 2016;7:73188-73199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 17. | Liu T, Zhang X, Du L, Wang Y, Liu X, Tian H, Wang L, Li P, Zhao Y, Duan W, Xie Y, Sun Z, Wang C. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer. 2019;18:43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 233] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 18. | Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298-1307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1761] [Cited by in RCA: 2110] [Article Influence: 175.8] [Reference Citation Analysis (0)] |
| 19. | Li N, Yang G, Luo L, Ling L, Wang X, Shi L, Lan J, Jia X, Zhang Q, Long Z, Liu J, Hu W, He Z, Liu H, Liu W, Zheng G. lncRNA THAP9-AS1 Promotes Pancreatic Ductal Adenocarcinoma Growth and Leads to a Poor Clinical Outcome via Sponging miR-484 and Interacting with YAP. Clin Cancer Res. 2020;26:1736-1748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 20. | Pan S, Shen M, Zhou M, Shi X, He R, Yin T, Wang M, Guo X, Qin R. Long noncoding RNA LINC01111 suppresses pancreatic cancer aggressiveness by regulating DUSP1 expression via microRNA-3924. Cell Death Dis. 2019;10:883. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 21. | Kong X, Liu CX, Wang GD, Yang H, Yao XM, Hua Q, Li XY, Zhang HM, Ma MZ, Su Q, Lv K. LncRNA LEGLTBC Functions as a ceRNA to Antagonize the Effects of miR-34a on the Downregulation of SIRT1 in Glucolipotoxicity-Induced INS-1 Beta Cell Oxidative Stress and Apoptosis. Oxid Med Cell Longev. 2019;2019:4010764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 22. | Yang G, Song R, Wang L, Wu X. Knockdown of long non-coding RNA TP73-AS1 inhibits osteosarcoma cell proliferation and invasion through sponging miR-142. Biomed Pharmacother. 2018;103:1238-1245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Cai Y, Yan P, Zhang G, Yang W, Wang H, Cheng X. Long non-coding RNA TP73-AS1 sponges miR-194 to promote colorectal cancer cell proliferation, migration and invasion via up-regulating TGFα. Cancer Biomark. 2018;23:145-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 24. | Papagiannakopoulos T, Friedmann-Morvinski D, Neveu P, Dugas JC, Gill RM, Huillard E, Liu C, Zong H, Rowitch DH, Barres BA, Verma IM, Kosik KS. Pro-neural miR-128 is a glioma tumor suppressor that targets mitogenic kinases. Oncogene. 2012;31:1884-1895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 149] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 25. | Zhu Y, Yu F, Jiao Y, Feng J, Tang W, Yao H, Gong C, Chen J, Su F, Zhang Y, Song E. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin Cancer Res. 2011;17:7105-7115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 197] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 26. | Cai J, Fang L, Huang Y, Li R, Xu X, Hu Z, Zhang L, Yang Y, Zhu X, Zhang H, Wu J, Li J, Zeng M, Song E, He Y, Li M. Simultaneous overactivation of Wnt/β-catenin and TGFβ signalling by miR-128-3p confers chemoresistance-associated metastasis in NSCLC. Nat Commun. 2017;8:15870. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 140] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 27. | Wei H, Hao X, Li B, Li X, Hou J, Qiao Y, Zhang R. GP73 is a potential marker for evaluating AIDS progression and antiretroviral therapy efficacy. Mol Biol Rep. 2013;40:6397-6405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 28. | Kladney RD, Cui X, Bulla GA, Brunt EM, Fimmel CJ. Expression of GP73, a resident Golgi membrane protein, in viral and nonviral liver disease. Hepatology. 2002;35:1431-1440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 176] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 29. | Mao Y, Yang H, Xu H, Lu X, Sang X, Du S, Zhao H, Chen W, Xu Y, Chi T, Yang Z, Cai J, Li H, Chen J, Zhong S, Mohanti SR, Lopez-Soler R, Millis JM, Huang J, Zhang H. Golgi protein 73 (GOLPH2) is a valuable serum marker for hepatocellular carcinoma. Gut. 2010;59:1687-1693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 194] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 30. | Ye QH, Zhu WW, Zhang JB, Qin Y, Lu M, Lin GL, Guo L, Zhang B, Lin ZH, Roessler S, Forgues M, Jia HL, Lu L, Zhang XF, Lian BF, Xie L, Dong QZ, Tang ZY, Wang XW, Qin LX. GOLM1 Modulates EGFR/RTK Cell-Surface Recycling to Drive Hepatocellular Carcinoma Metastasis. Cancer Cell. 2016;30:444-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 175] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 31. | Bao YX, Cao Q, Yang Y, Mao R, Xiao L, Zhang H, Zhao HR, Wen H. Expression and prognostic significance of golgiglycoprotein73 (GP73) with epithelial-mesenchymal transition (EMT) related molecules in hepatocellular carcinoma (HCC). Diagn Pathol. 2013;8:197. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 32. | Yan G, Ru Y, Wu K, Yan F, Wang Q, Wang J, Pan T, Zhang M, Han H, Li X, Zou L. GOLM1 promotes prostate cancer progression through activating PI3K-AKT-mTOR signaling. Prostate. 2018;78:166-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 33. | Haigis KM. KRAS Alleles: The Devil Is in the Detail. Trends Cancer. 2017;3:686-697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 265] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 34. | di Magliano MP, Logsdon CD. Roles for KRAS in pancreatic tumor development and progression. Gastroenterology. 2013;144:1220-1229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 315] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 35. | Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801-1806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3216] [Cited by in RCA: 3019] [Article Influence: 177.6] [Reference Citation Analysis (0)] |
| 36. | Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:2140-2141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 253] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 37. | Tape CJ, Ling S, Dimitriadi M, McMahon KM, Worboys JD, Leong HS, Norrie IC, Miller CJ, Poulogiannis G, Lauffenburger DA, Jørgensen C. Oncogenic KRAS Regulates Tumor Cell Signaling via Stromal Reciprocation. Cell. 2016;165:1818. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 68] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 38. | Liang C, Qin Y, Zhang B, Ji S, Shi S, Xu W, Liu J, Xiang J, Liang D, Hu Q, Ni Q, Xu J, Yu X. Metabolic plasticity in heterogeneous pancreatic ductal adenocarcinoma. Biochim Biophys Acta. 2016;1866:177-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |