Published online Apr 21, 2021. doi: 10.3748/wjg.v27.i15.1595
Peer-review started: January 21, 2021
First decision: February 9, 2021
Revised: February 17, 2021
Accepted: March 13, 2021
Article in press: March 13, 2021
Published online: April 21, 2021
Processing time: 82 Days and 21.1 Hours
Expression of the full-length isoform of Abelson interactor 1 (ABI1), ABI1-p65, is increased in colorectal carcinoma (CRC) and is thought to be involved in one or more steps leading to tumor progression or metastasis. The ABI1 splice isoform-L (ABI1-SiL) has conserved WAVE2-binding and SH3 domains, lacks the homeo-domain homologous region, and is missing the majority of PxxP- and Pro-rich domains found in full-length ABI1-p65. Thus, ABI1-SiL domain structure suggests that the protein may regulate CRC cell morphology, adhesion, migration, and metastasis via interactions with the WAVE2 complex pathway.
To investigate the potential role and underlying mechanisms associated with ABI1-SiL-mediated regulation of CRC.
ABI1-SiL mRNA expression in CC tissue and cell lines was measured using both qualitative reverse transcriptase-polymerase chain reaction (RT-PCR) and real-time quantitative RT-PCR. A stably ABI1-SiL overexpressing SW480 cell model was constructed using Lipofectamine 2000, and cells selected with G418. Image J software, CCK8, and transwell assays were used to investigate SW480 cell surface area, proliferation, migration, and invasion. Immunoprecipitation, Western blot, and co-localization assays were performed to explore intermolecular interactions between ABI1-SiL, WAVE2, and ABI1-p65 proteins.
ABI1-SiL was expressed in normal colon tissue and was significantly decreased in CRC cell lines and tissues. Overexpression of ABI1-SiL in SW480 cells significantly increased the cell surface area and inhibited the adhesive and migration properties of the cells, but did not alter their invasive capacity. Similar to ABI1-p65, ABI1-SiL still binds WAVE2, and the ABI1-p65 isoform in SW480 cells. Furthermore, co-localization assays confirmed these intermolecular interactions.
These results support a model in which ABI1-SiL plays an anti-oncogenic role by competitively binding to WAVE2 and directly interacting with phosphorylated and non-phosphorylated ABI1-p65, functioning as a dominant-negative form of ABI1-p65.
Core Tip: The purpose of this study was to investigate the role and mechanism of Abelson interactor 1 splice isoform-L (ABI1-SiL) in the metastatic behavior of colorectal carcinoma cells. Our results showed that ABI1-SiL played a key role in cell surface area, adhesion, and migration, but not in proliferation and apoptosis in SW480 cells. ABI1-SiL may have anti-oncogenic roles by competitively binding to WAVE2, and directly interacting with phosphorylated and non-phosphorylated ABI1-p65, functioning as a dominant-negative molecule towards ABI1-p65.
- Citation: Li K, Peng YF, Guo JZ, Li M, Zhang Y, Chen JY, Lin TR, Yu X, Yu WD. Abelson interactor 1 splice isoform-L plays an anti-oncogenic role in colorectal carcinoma through interactions with WAVE2 and full-length Abelson interactor 1. World J Gastroenterol 2021; 27(15): 1595-1615
- URL: https://www.wjgnet.com/1007-9327/full/v27/i15/1595.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i15.1595
Globally, colorectal carcinoma (CRC) is the third most commonly diagnosed cancer and the fourth most common cause of cancer death[1]. Despite treatment advances for CRC over the past two decades, novel molecular therapeutic strategies are required to generate informative biomarkers and identify new targets[2,3].
Abelson interactor protein-1 (ABI1) is increased and/or phosphorylated in many malignant tumors and has been proposed as a potential tumor promoter[4-11]. By forming complexes with Eps8-Sos1[6], NWasp[12], and WAVE2[13,14], ABI1 has been shown to play critical roles in the regulation of cell proliferation, apoptosis, adhesion, and migration in breast cancer, Bcr-Abl-induced leukemia, and melanoma, both in vitro and in vivo[4-15]. Recent findings have indicated that ABI1-p65 is increased and involved in colonic tumorigenesis and that it mediates the invasive properties of CRC[7,8]. Recently, using a CRC tissue chip, we showed that increased expression of ABI1-p65 is significantly correlated with infiltration and associated with shorter overall survival[16]. Taken together, this evidence strongly suggests that ABI1 is involved in one or more of the steps leading to tumor progression or metastasis.
Alternative splicing is one of the main engines that drive proteome diversity, and up to 94% of genes are estimated to be alternatively spliced in humans[17,18]. There are many abnormal mRNA splice isoforms specifically associated with cancer progression and metastasis, some of which exert antagonistic functions[2,19,20]. Numerous structurally distinct isoforms of ABI1 exist in mammalian cells[12,21]. The ABI1-p65 isoform is increased in CRC and is thought to be involved in one or more of the steps leading to tumor progression and metastasis. However, the specific role, if any, that other splice isoforms of ABI1, such as ABI1 splice isoform-L (ABI1-SiL), play in CRC development remains unknown. As Figure 1 shows, ABI1-SiL is a splice isoform of ABI1 resulting from the alternative splicing of exons III, IV, and VIII through X. ABI1-SIL has conserved WAVE2-binding and SH3 domains, as well as several phosphorylated sites, such as pY213, pY421/pY435, and pY506, but lacks the homeo-domain homologous region (HHR) and the majority of the PxxP- and Pro-rich domain[16,21]. The domain structure of ABI1-SiL suggests that it might regulate cell morphology, adhesion, migration, and even metastasis of CRC cells, similar to the ABI1-p65 isoform, by interacting with WAVE2 or other proteins.
In this study, we generated polymerase chain reaction (PCR) primers capable of specific detection of the ABI1-SiL isoform. We demonstrated that, compared with ABI1-p65, the expression of ABI1-SiL was significantly decreased in CRC tissues and cell lines, and overexpression of ABI1-SiL repressed migration and adhesion in SW480 colon cancer cells. We showed that both ABI1-p65 and ABI1-SiL were able to interact and co-localize with WAVE2 and ABI1-p65 in SW480 cells. Taken together, this evidence is the first to demonstrate that ABI1-SiL is an antagonistic splice isoform of ABI1-p65 that functions as a potential anti-oncogene.
The CRC cell lines HCT-116, HT-29, LoVo, LS174T, SW480, and SW620 (the Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China.), as well as the normal colon cell line CRL-1541 (ATCC, Rockville, MD, United States), were cultured in DMEM (HyClone, Beijing, China) supplemented with 10% fetal bovine serum (FBS) (Sigma, St. Louis, MO, United States). HEK-293T cells (ClonTech, Mountain View, CA, United States) were grown in DMEM (HyClone, Beijing, China) supplemented with 10% FBS. Plasmid-mediated transfection of SW480 cells with pCMV6 and pCMV6-ABI1-GFP (green fluorescence protein) transcript variant 12 (ABI1-Tv12) was performed as previously described using Lipofectamine (Invitrogen, CA, United States)[4,22]. Both pCMV6 and the vector containing human ABI1 cDNA (NM_001178125.1, ABI1-Tv12, encoding ABI1-SiL) were purchased from OriGene Technologies, Inc. (Rockville, MD, United States). G418 (500 µg/mL) was used to select stable cell lines overexpressing ABI1-SiL.
CRC and matched adjacent tissue samples were obtained from 24 patients. All CRC cases were diagnosed using enteroscopy, and primary carcinoma of the colon-rectum was histologically identified. All samples were anonymized prior to this study. The local ethics committee of the Peking University, People’s Hospital approved the study.
Tissue samples, as well as the cell lines described above, were analyzed for the expression of transcript variant-12 (ABl1-SiL)[21]. Total RNA was isolated from patient samples, CRC cell lines, and transfected SW480 cells using an RNeasy mini kit (QIAGEN, Valencia, CA, United States) as previously described[23]. cDNA was subsequently generated using oligo-(dT) primers and the SuperScript III First-strand Synthesis System (Invitrogen, Carlsbad, CA, United States). PCR was performed to confirm the expression of ABI1-SiL[21] using the following primers: Forward, 5’-CCTCTCAGCTTCGGAGAATGG-3’ and reverse, 5’-AGGAGGATTTGT TCTCGACAGTGT-3’. A 101 bp-amplified fragment in the PCR products was shown to be ABI1-SiL (confirmed by DNA sequencing). Quantitative PCR (qPCR) was performed using either human ABI1 universal 1 (ABI1-U1) (transcript variant 1-12) primers (forward: 5’-ACACTGGGACGGAATACTCCTTAT-3’; reverse: 5’-CTACCACTGTTTTCTCGACTTCCA-3’) or human ABI1 universal 2 (ABI1-U2) (transcript variant 1-11) primers (forward: 5’-AGTGGCACGAAGAGAGATTGG-3’; reverse: 5’-CTGTGTAATCGATAGGTTTCCGAAT-3’) and SYBR Green Master Mix (Applied Biosystems, Foster City, CA, United States). PCR reactions began with a 10 min incubation at 95 °C for AmpliTaq Gold activation, followed by 40 cycles at 95 °C for 15 s for the denaturation step and 60 °C for 1 min for the primer annealing and extension step. qPCR was performed using the DNA Engine Option 2 System (Bio-Rad, CA, United States). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous housekeeping control for relative quantification of transcription (forward primer: 5’-AACGACCCCTTCATTGAC-3’, reverse primer: 5’-TCCACGACATACTCAGCAC-3’). The percentage of ABI1-SiL among all variants was calculated as (1-2-[Ct-ABI1-U2-Ct-ABI1-U1]) × 100%.
To examine the potential roles of ABI1-SiL overexpression on differentiation and invasion of SW480 cells, a series of additional genetic markers (including POU, CD44, KRT20, MMP1-4, and WAVE2) were also analyzed by qPCR (primers sequences are not shown).
Lipofectamine-mediated transfection of SW480 cells was performed as previously described[4]. Cells were plated in six-well plates 24 h prior to transfection, and 2 µg of plasmid DNA was used for each transfection. To overexpress ABI1-SiL, a pCMV6 vector specifically coding for ABI1-SiL or a control pCMV6 empty vector was used for stable transfection of SW480 cells or transient transfection of 293T cells. Forty-eight hours after transfection, transfected cells were treated with G418 (500 µg/mL). Individual G418-resistant clones were picked after 3-4 wk. Clones were analyzed by qPCR and Western blot for ABI1-SiL expression, and clones with the highest ABI1-SiL expression were chosen for further studies. Clones 3 and 6 overexpressing ABI1-SiL were named SW480-ABI1-SiL3 and SW480-ABI1-SiL6, respectively. SW480 cells transfected with the empty pCMV6 vector were named SW480-V.
To analyze the subcellular localization of ABI1-SiL in SW480 cells and to test for co-localization with ABI1-p65 and/or WAVE2, a pCMV6 vector encoding a GFP-ABI1-SiL fusion protein was used for both transient and stable transfections. In transient experiments, 48 h after transfection, the 293T cells underwent subcellular localization studies by fixing the cells in 4% paraformaldehyde in phosphate buffer saline (PBS) for 10 min, followed by fluorescence microscopy analysis.
Western blot analyses were performed as previously described[4]. We used the following antibodies: GAPDH (1:2000 dilution; #CB100999, Proteintech, IL, United States), ABI1-p65 (1:1000 dilution; D147-3, BML, MA, United States), WAVE2 (1:500 dilution; 07-410, Upstate, NY, United States), GFP (1:200 dilution; 1G6; Pregene Co. Beijing, China), and NAP-1 (1:1000 dilution; #12140-1-AP, Proteintech, IL, United States) protein A/G (Sc-2003, Santa Cruz, CA, United States). The protease inhibitor cocktail was purchased from Sigma (St. Louis, MO, United States). Cells were lysed in lysis buffer (20 mmol/L HEPES, pH 7.2; 150 mmol/L NaCl; 1% Triton X-100; and 10% glycerol). Total protein lysates (150 µg each) from all tested cell lines were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, transferred to a nitrocellulose membrane, and immunoblotted with appropriate antibodies. Immunoprecipitation analysis was performed as described previously[4]. Briefly, SW480-V and/or SW480-ABI1-SiL6 cells were lysed as described above and incubated with appropriate antibodies bound to sepharose beads. Immunoprecipitates were resolved by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with appropriate antibodies. Membranes were then washed with TBST buffer and incubated with secondary antibodies conjugated to HRP (horseradish peroxidase) (Jackson ImmunoResearch Laboratories Beijing, China). Bound antibodies were detected using the SuperSignal West Pico Trial Kit (Thermo Scientific, Rockford, IL, United States), and images were acquired using an ImageQuant 350 system (GE Healthcare, Piscataway, NJ, United States) and analyzed using ImageJ software (NIH, Bethesda, MD, United States).
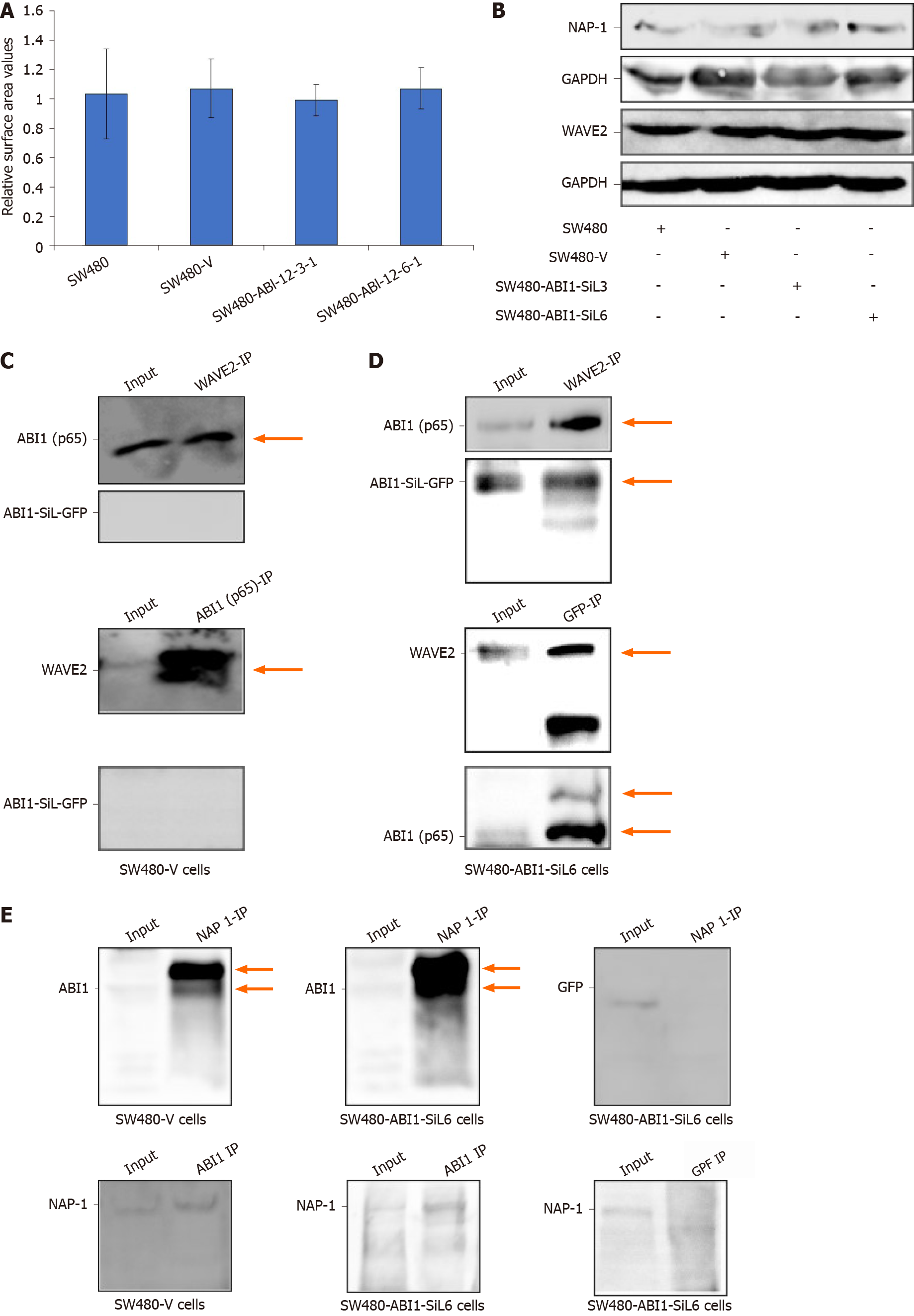
To measure the surface area, all four tested cell lines were routinely cultured and images were captured using a Cannon EOS600D camera. ImageJ was used to measure the cell surface area of 100 cells per condition. The measured values were normalized to the mean value of the SW480 parental cells.
For adhesion assays, 2.5 × 105 cells from each cell line were plated in a six-well plate (2.5 mL/well) coated with or without either 0.2% BSA or 0.1% gelatin, or fibronectin (5 μg/mL) (BD Biosciences, Bedford, MA, United States) and incubated at 37 °C/5% CO2 for 6 h. Non-adherent cells were removed, and adherent cells were washed three times with 1 mL pre-warmed DMEM medium. To determine the number of adherent cells, the number of adherent cells within five randomly selected microscopic fields was counted in three independent experiments.
Cell migration assays were performed using a 24-well transwell chamber (Corning, New York, NY, United States) with 8.0-μm pore polycarbonate filter inserts, and cell invasion assays were performed using inserts of Transwell plates (8.0-μm pores, Corning Costar Corp., Cambridge, MA, United States) coated with Matrigel (Costar, Cambridge, MA, United States)[22]. Cells (2.5 × 104 cells/well) suspended in serum-free DMEM containing 0.2% BSA were placed in the upper chamber of each transwell. In each lower chamber, 600 μL of DMEM supplemented with 10% FBS was added. The inserts were then incubated at 37 °C in a humidified atmosphere containing 5% CO2 for 24 h (migration) and 48 h (invasion), respectively. Cells that had not penetrated the filters were removed using cotton swabs. Cells that migrated to the bottom of the insert were fixed in 4% paraformaldehyde for 10 min and stained with 0.1% crystal violet for 30 min, followed by rinsing with PBS and examination under a bright-field microscope equipped with a 100 × objective. Migratory or invasive activity was expressed as the average number of migrated cells per microscopic field over five fields in each assay from three independent experiments.
Cell proliferation assays were performed using Cell Counting Kit-8 (CCK-8; Dojindo, Rockville, MD, United States) according to the manufacturer’s instructions. All tested cell lines were seeded at 2.5 × 104 cells per well of a 96-well plate and cultured in 100 μL of DMEM supplemented with 10% FBS. At the indicated time points, culture medium was exchanged for 110 μL of DMEM supplemented with CCK-8 reagent (10 μL CCK-8 and 100 μL DMEM), and cells were incubated for an additional 2 h. Absorbance was measured for each well at a wavelength of 450 nm, with the reference wavelength set at 600 nm. An increase or decrease in absorbance values at 450 nm in the experimental wells relative to the initial value was indicative of cell growth or death, respectively. Cell growth was monitored at 24 h, 48 h, 72 h, and 96 h, and was repeated in replicates of three wells per time point.
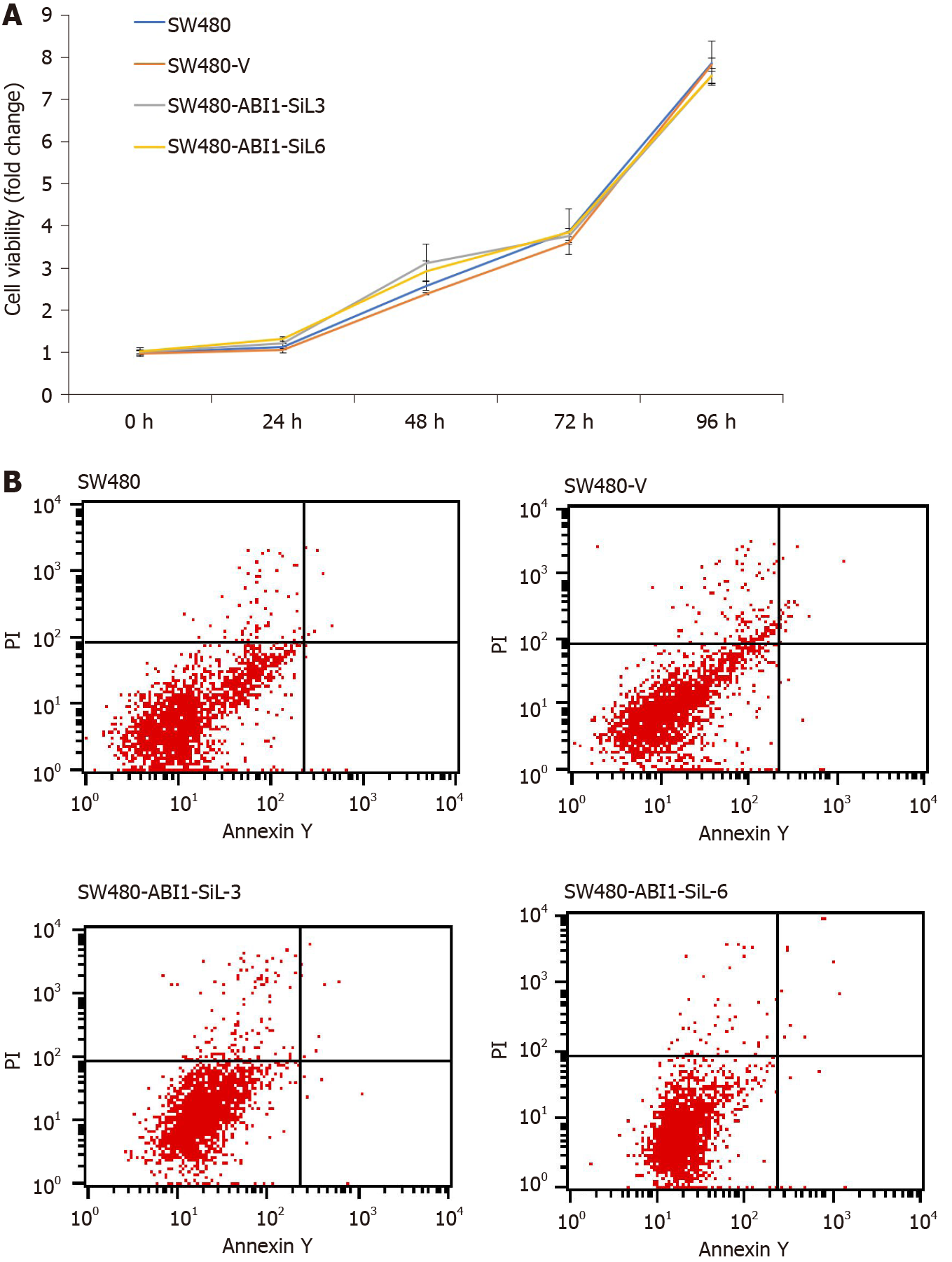
Apoptosis assays were performed immediately after selection with G418. Cells were washed twice and seeded at a density of 2 × 105 cells/mL in DMEM and 10% FBS in the presence of G418 (for SW480-V, SW480-ABI1-SiL3, and SW480-ABI1-SiL6). After 48 h of culture, apoptosis was monitored using the Annexin V-FITC Apoptosis Detection Kit (KeyGen Biotech, #KGA107, Shanghai, China). For FACS analysis, 1 × 105 cells were co-stained with Annexin V-EGFP and with the fluorescent dye propidium iodide (PI). Cells that stained negative for both PI and annexin V were considered viable cells; PI-negative and annexin V-positive stained cells were considered early apoptotic cells; PI-positive and annexin V-positive stained cells were considered to be in the later stages of apoptosis. All assays were carried out independently, in triplicate.
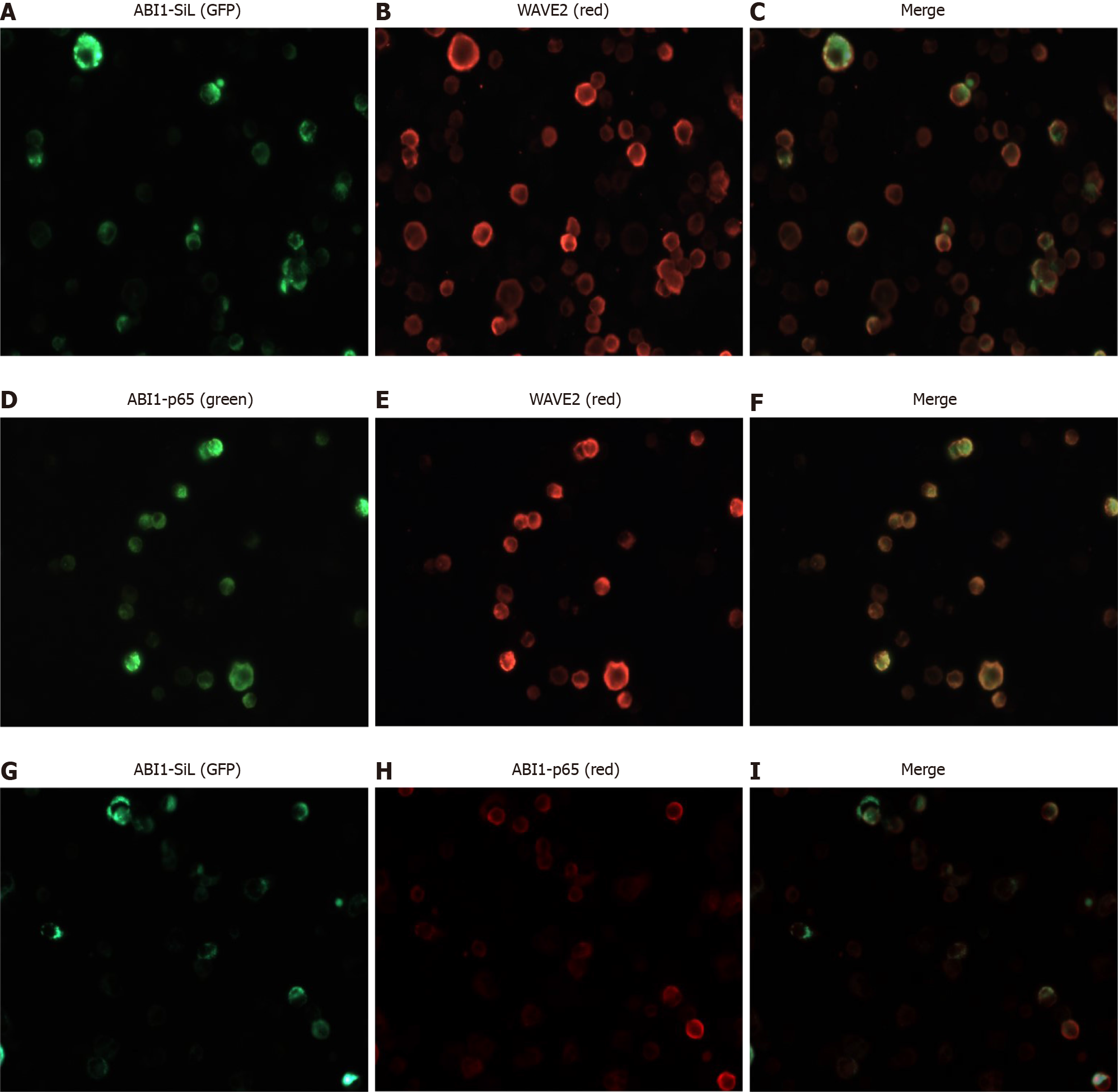
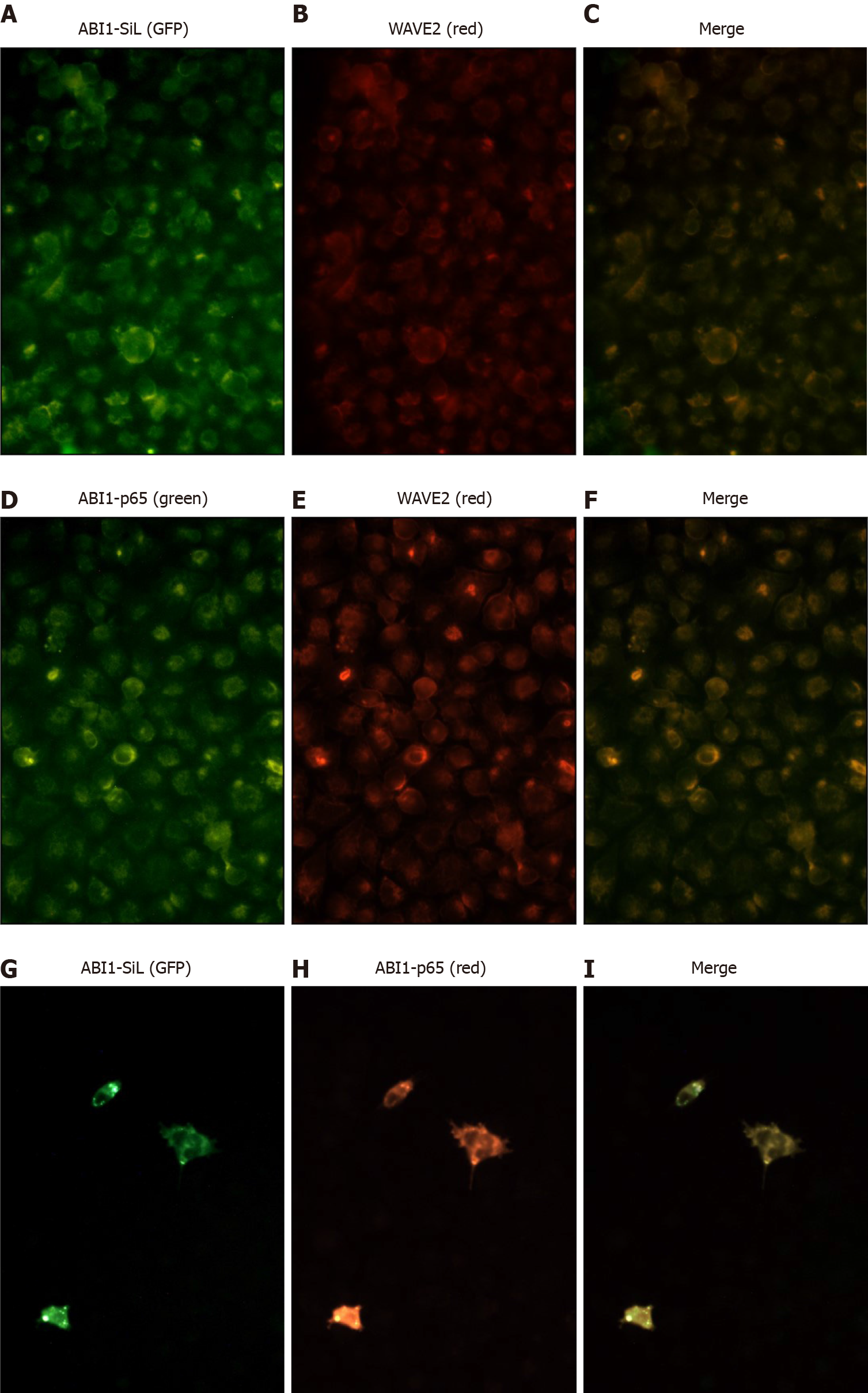
Samples of 293T cells were cultured on fibronectin-coated coverslips and transiently transfected with either the ABI1-SiL-GFP plasmid or a control plasmid, as well as SW480-ABI1-SiL6. The cells were fixed in 4% paraformaldehyde in PBS for 10 min and permeabilized with 0.2% Triton X-100 in PBS for 8 min. Non-specific binding was blocked by incubation of the coverslips with 3% bovine serum albumin in PBS, followed by incubation with primary antibodies (anti-ABI1-p65 and or anti-WAVE2) in 1% bovine serum albumin/PBS. After extensive washing with PBS, cells were incubated with appropriate Alexa-conjugated secondary antibodies. Cytoskeleton was detected by Phalloidin staining. The slides were examined under a Nikon Eclipse TE2000-U fluorescence microscope, and images were captured and analyzed using Nikon NIS Elements imaging software.
SPSS version 16.0 software (SPSS Inc., Chicago, IL, United States) was used for statistical analyses. Data from all quantitative assays are expressed as the mean ± SD and were analyzed statistically using one-way analysis of variance (ANOVA) followed by Dunnett test and general linear model analysis (univariate). P values less than 0.05 were considered statistically significant.
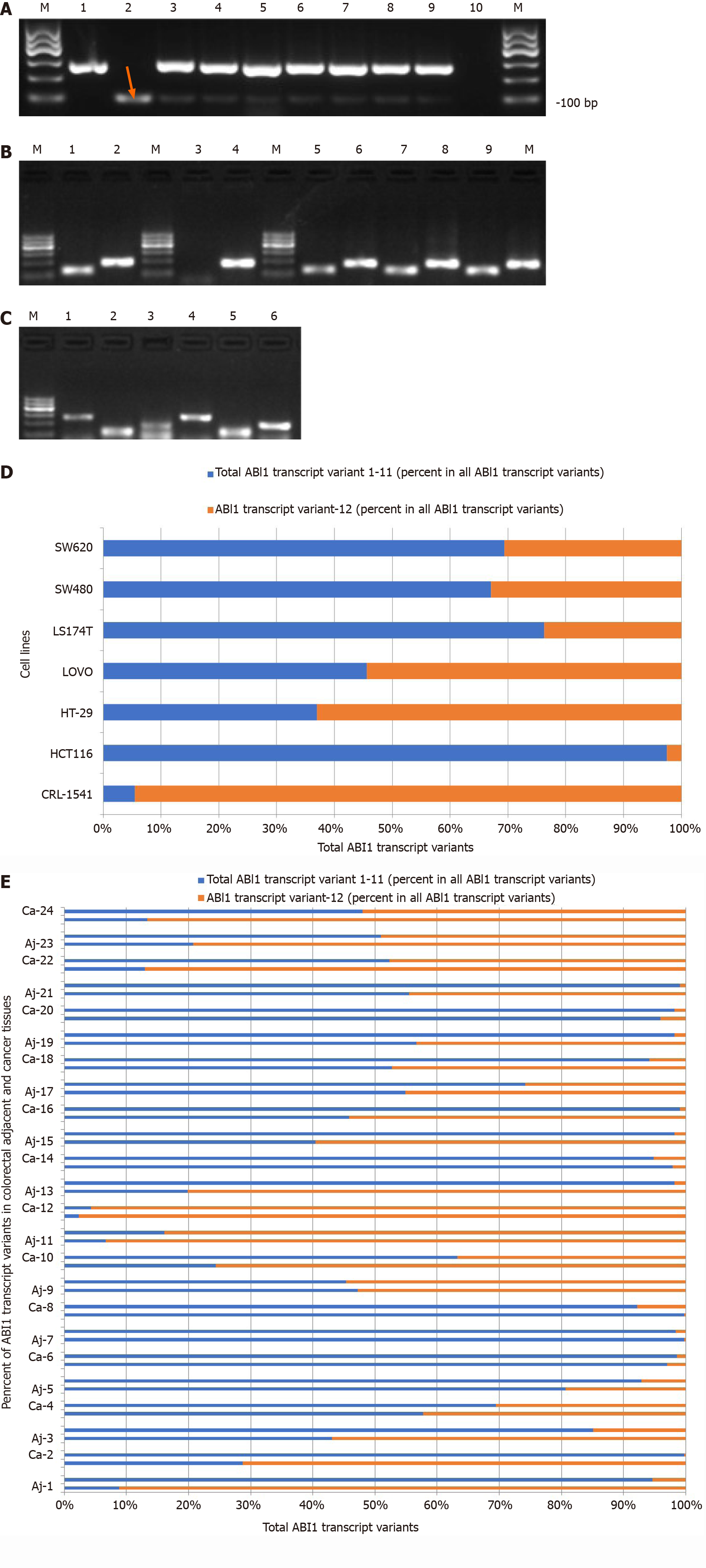
The ABI1 gene encodes a member of the Abelson interactor family of adaptor proteins. At least 12 alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene[21]. There are significant differences in the exons and functional domains contained in the different isoforms of ABI1, which may result in structural differences in the proteins that lead to differing functions between ABI1-SiL and other transcript variants (Figure 1A). To detect the expression of ABI1-SiL, we designed a set of qualitative primers and two sets of universal quantitative primers that detect this isoform specifically, as shown in Figure 1B. Using qualitative PCR, two visible bands were amplified, and we verified through sequencing that the 101-bp band represents the transcript variant-12 specific PCR product (Figure 2A). We demonstrated that ABI1-SiL was expressed in all tested CRC cells and control CRL-1541 cells.
Next, we tested the reliability of the primers designed for quantitative analysis of ABI1-SiL expression. ABI1-SiL and 12 plasmids (single and/or mixtures), as well as the cDNA from CRL-1541 cells and HCT116 cells, were used as templates to perform qPCR. PCR amplified products were examined by 1.5% agarose gel electrophoresis (Figure 2B). Taken with the analysis of the amplification curve, the melting curve, and the sequencing data for the PCR products (data not shown), these observations indicated that we have successfully established a quantitative assay specific for detection of ABI1-SiL.
In previous studies, we and other groups have shown that the full-length isoform of ABI1 (ABI1-p65) is increased in colon cancer cells and tissues and involved in the progression and development of colon cancer[7,8,22]. To analyze the potential role of ABI1-SiL in colon cancer, we examined the expression level of ABI1-SiL in CRC cells and tissues using qPCR. As Figure 2D and E shows, the ratio of the ABI1-SiL isoform to other ABI1 splice isoforms was significantly decreased in all CRC cells compared with control CRL-1541 cells and was also decreased in 16 of 24 CRC tissues compared with adjacent tissues. These observations indicated that ABI1-SiL may play an anti-oncogenic role in colorectal tissues.
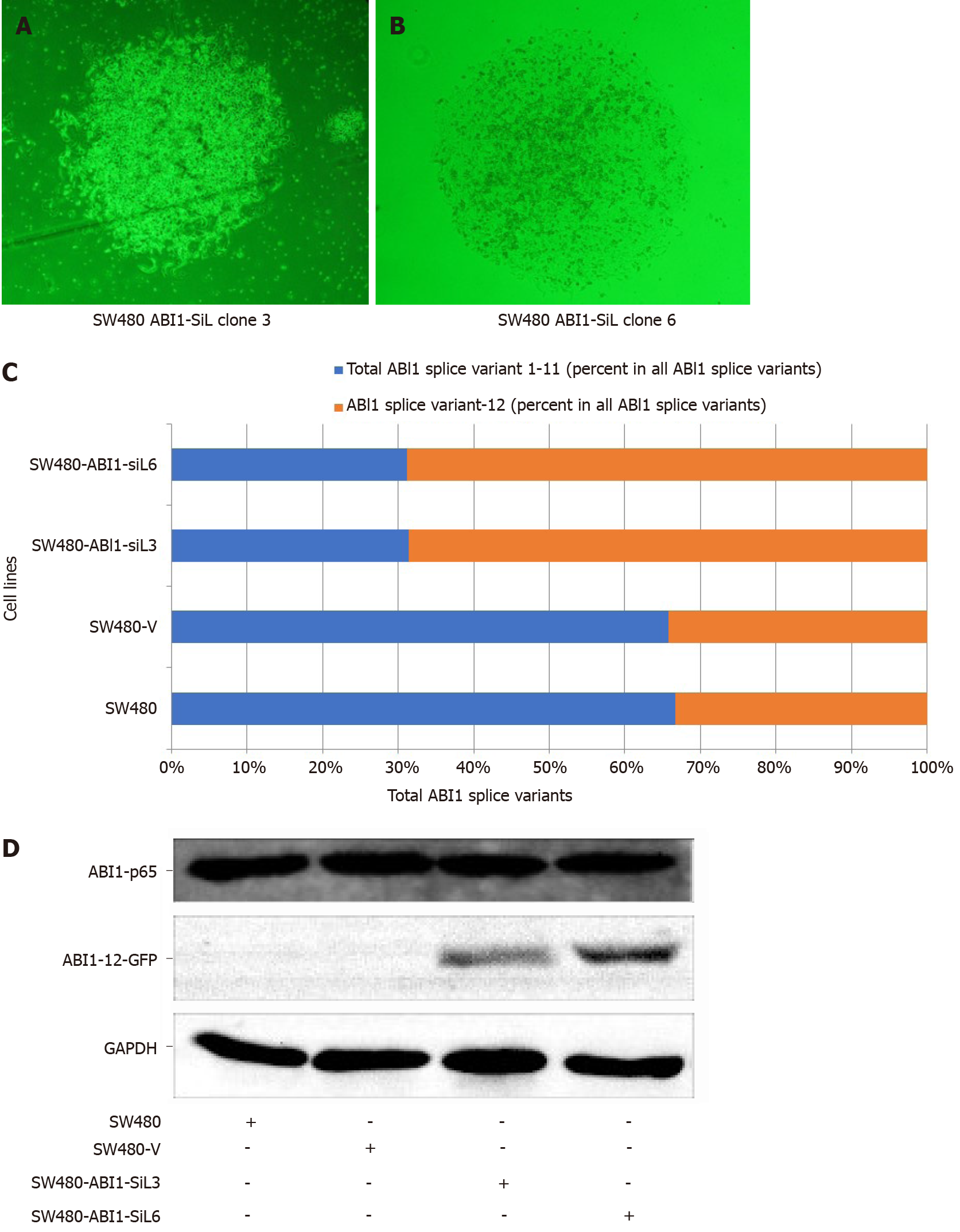
To test the function of ABI1-SiL in cellular phenotypes of colon cancer, we overexpressed ABI1-SiL in SW480 colon cancer cells. The full-length cDNA of ABI1-SiL, or an empty vector control plasmid, was introduced into SW480 by Lipofectamine-mediated transfection. We confirmed ABI1-SiL expression in two stable cell lines, SW480-ABI1-SiL3 and SW480-ABI1-SiL6 (Figure 3A and B). As shown in Figure 3C, the expression of the cDNA in SW480 cells resulted in a significant increase in the proportion of the ABI1-SiL isoform relative to other isoforms by about 30%, as compared to the parental SW480 and SW480-V cell lines. We also demonstrated through Western blot that the ABI1-SiL-GFP fusion protein was expressed in SW480-ABI1-SiL3 and SW480-ABI1-SiL6 cells, but not in parental SW480 and SW480-V cells (Figure 3D). These data confirm that we successfully constructed a SW480 colon cancer cell model overexpressing ABI1-SiL.
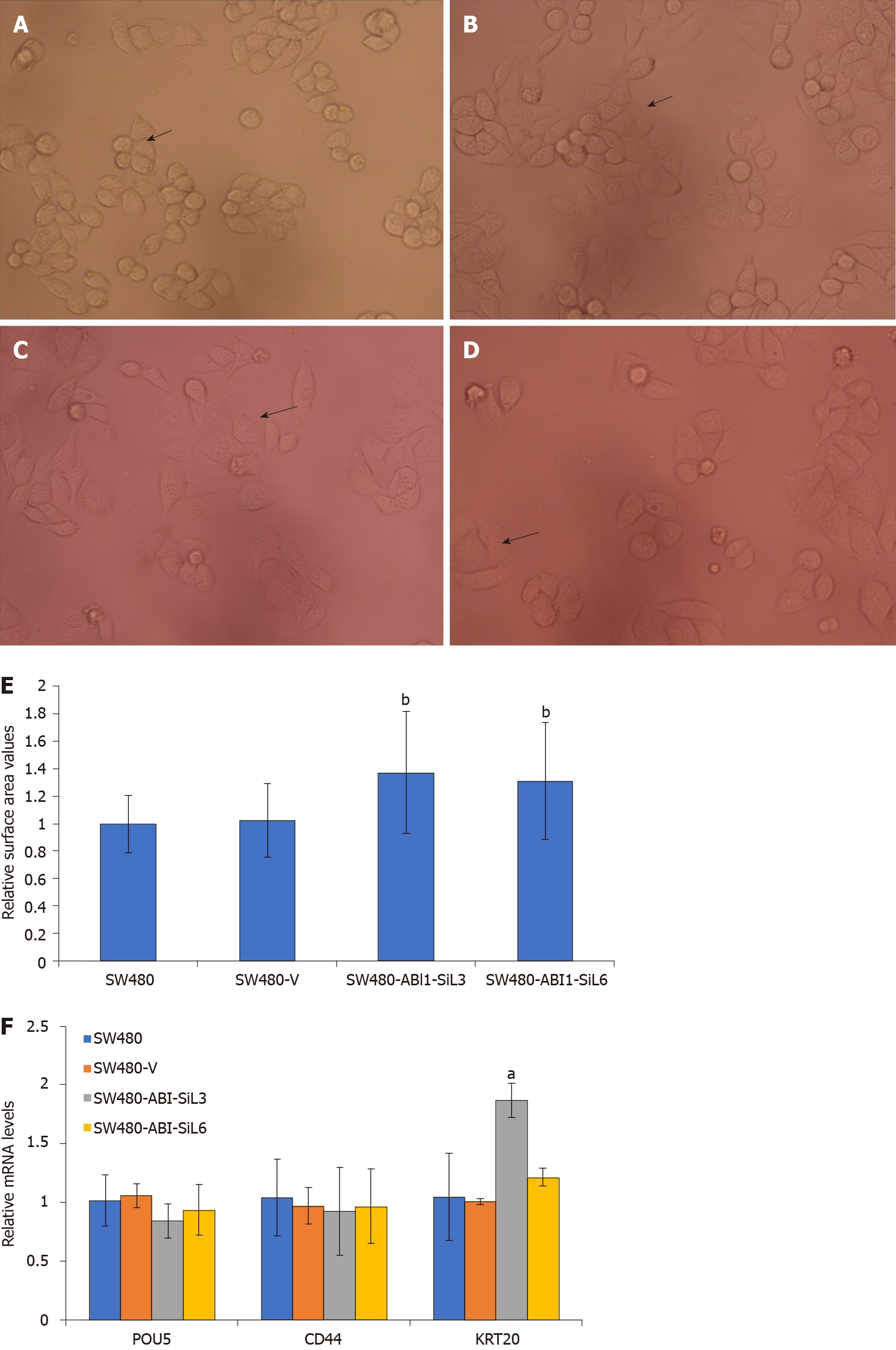
We observed that the SW480-ABI1-SiL3 and SW480-ABI1-SiL6 stable cell lines were substantially larger and were more similar in morphology to epithelial cells than the parental and empty vector transfected cells (Figure 4A-D, see arrows). To quan-titatively assess this difference in cellular morphology, Image J and SPSS software were used to examine the cell surface area of the transfected and control cell lines. As Figure 4E shows, the cellular surface area was significantly increased by about 40% in SW480-ABI1-SiL3 and SW480-ABI1-SIL6 cells compared with the parental and control SW480 cells (P < 0.01). To determine whether ABI1-SiL-induced changes in cellular morphology are due to effects on differentiation of SW480 cells, we used qPCR to analyze the expression of POUS, CD44, and KRT20, but we observed no difference in the expression of these genes, except for KRT20 in SW480-SiL3 (Figure 4E).
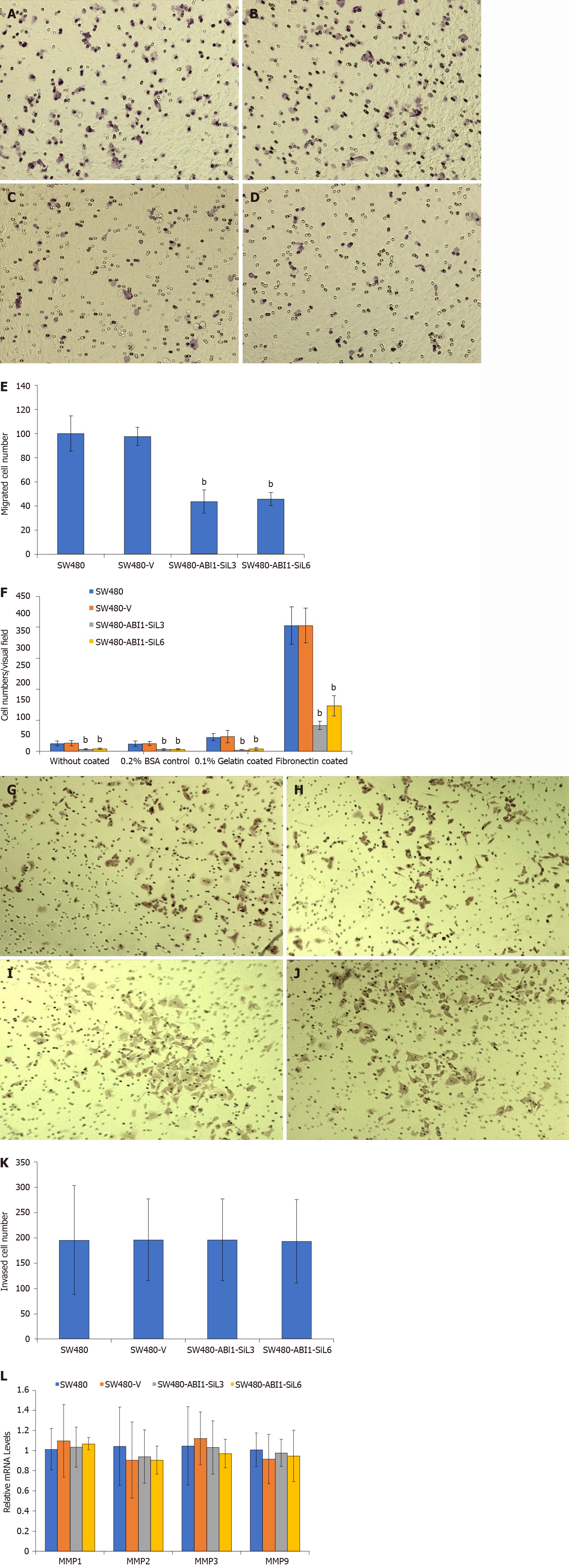
We performed adhesion, migration, and invasion assays to test whether ABI1-SiL overexpression activated these cellular mechanisms. In the adhesion assay, we discovered that ABI1-SiL overexpression significantly impaired the adhesive capability in all tested conditions (0.2% BSA, gelatin, fibronectin, and no coating) (P < 0.01, Figure 5A). Among the tested coating conditions, the adhesion assay showed that ABI1-SiL had the strongest inhibitory effect on adhesion in the presence of fibronectin. In the migration assay, ABI1-SiL overexpression also greatly inhibited the migration of SW480 cells in vitro (P < 0.01) (Figure 5B-F). Interestingly, ABI1-SiL overexpression showed no inhibitory effect on invasion of SW480 cells (P > 0.05; Figures 5G-K). Furthermore, expression analysis using qPCR revealed that ABI1-SiL overexpression had no effect on expression of matrix metalloproteases 1, 2, 3, or 4 (Figure 5L).
It has been reported that the full-length isoform of ABI1 (ABI1-p65) promotes proliferation and inhibits apoptosis of colon cancer cells[24-26], but the action of ABI1-SiL in colon cancer cells is unknown. To further determine the effect of ABI1-SiL overexpression on the proliferation and apoptosis of SW480 cells, we performed CCK-8 cell viability and apoptosis assays. As Figure 6A and B shows, overexpression of ABI1-SiL had no effects on proliferation or apoptosis of SW480 cells. These observations confirmed that changes in migration and invasion did not result from alterations in cell proliferation and apoptosis.
It has been widely accepted that ABI1 regulates cancer cell morphology, adhesion, and migration by directly interacting with WAVE2 and forming the ABI1-WAVE2-NAP-1 complex[27-29]. We confirmed the expression of ABI1-p65, WAVE2 and NAP-1 in all tested cell lines. As Figures 3D, 7A, and 7B demonstrate, ABI1-SiL overexpression did not alter the expression of ABI-p65 and WAVE2 at either the mRNA or protein level, as well as NAP-1 at the protein level. The SW480-Vector and SW480-ABI1-SiL6 cell lines were used to analyze potential mechanisms underlying the effect of ABI1-SiL expression on cell morphology, adhesion, and migration of SW480 cells. Western blot and co-immunoprecipitation (co-IP) revealed that both ABI1-p65 and ABI1-SiL-GFP were able to interact with WAVE2 in SW480-ABI1-SiL6 cells, as determined by successful bidirectional co-IP of ABI1-p65, ABI1-SiL-GFP, and WAVE2 (Figure 7C and D). Interestingly, we also observed evidence of a direct interaction between ABI1-p65, phosphorylated ABI1-p72, and ABI1-SiL-GFP (Figure 7D). Additionally, we showed that ABI1-SiL does not bind to NAP-1, but ABI1-p65 and phosphorylated ABI1-p72 do in SW480-ABI1-SiL6 cells (Figure 7E). Finally, 293T cells, transfected with ABI1-SiL-GFP and stable SW480-ABI1-SiL6, were used to examine the co-localization of ABI1-SiL-GFP and WAVE2 or ABI1-p65. As shown in Figures 8 and 9, ABI1-SiL co-localizes with both WAVE2 and ABI1-p65. These observations suggested that the ABI1-SiL isoform inhibits the adhesion and migration of SW480 cells in vitro by competitively interacting with WAVE2 and directly binding to ABI1-p65. These results provide a novel molecular mechanism explaining the functions of distinct ABI1 isoforms in CRC cells.
In this study, we have identified a novel function for ABl1-SiL in adhesion and migration of colon cancer SW480 cells and propose a mechanism underlying this function. Our model is based on a combination of gene expression and mechanistic studies of ABI1-SiL in colon cancer cells and tissues. We and other groups have previously shown that overexpressed non-phosphorylated ABI1-p65 functions as a pro-oncogene in development and progression of CRC[7,8,22]. Now, we wanted to address the question of whether there are other ABI1 isoforms that exist in colon tissues and cells that play additional roles in colon cancer. Identification of novel alternatively spliced isoforms with dysregulated expression in cancer could lead to the development of tumor-specific molecular targets for prognosis and therapy[30].
At present, there is no commercial antibody available that can detect the ABI1-SiL protein. Here, we report a set of PCR-based methods that can specifically measure ABI1-SiL mRNA expression. Unlike the full-length ABI1-p65 isoform, ABI1-SiL was significantly decreased in colorectal tissues and cells compared with the adjacent tissues and CRL-1541 normal colon cells (Figure 2). This observation, coupled with the structural differences between ABI1-SiL and other isoforms, especially ABI1-p65 (Figure 1A), led us to postulate that ABI1-SiL might function as an anti-oncogene.
ABI1-SiL mRNA expression was significantly decreased in SW480 cells, so we selected this cell line to investigate the effects of ABI1-SiL expression on cellular phenotypes in colon cancer cells. We successfully constructed SW480 cell lines that stably overexpress ABI1-SiL. We observed that ABI1-SiL overexpression in SW480 cells resulted in enlarged cell size and impaired adhesion and migration, but had no effect on invasion, proliferation, apoptosis, or transcriptional regulation (Figures 3-6). ABI1-SiL overexpression did not alter the mRNA expression levels of ABI1-p65, WAVE2, MMPs, POU, CD44, or KRT20. In combination with its decresion in CRC tissues and cells, these observations suggest that ABI1-SiL could function as an anti-oncogene and that the underlying mechanism needs to be explored.
It is widely accepted that an ABI1-WAVE2 complex is involved in epithelial morphogenesis[31], adhesion, migration, and invasion of cancer cells[13,28,32]. As shown in Figure 1A, the conserved WAVE2-binding and SH3 domains of ABI1-SiL provide the same sites as ABI1-p65 to competitively bind to WAVE2 and/or the proline-rich domain (PRD) of other molecules, including the PRD of ABI1-p65, but the lack of the HHR and the majority of the PxxP- and Pro-rich domain makes it likely to function as a native dominant negative that suppresses the pro-oncogenic functions of ABI1-p65. Bidirectional co-IP experiments confirmed interactions between ABI1-SiL, WAVE2, and ABI1-p65. In control SW480 cells, ABI1-p65 interacts with WAVE2, while in SW480 ABI1-SiL6 cells, both ABI1-p65 and ABI1-SiL are able to bind to WAVE2 (Figure 7C and D). These observations suggested a possible mechanism by which ABI1-SiL competitively binds to WAVE2. Increased ABI-SIL expression could inhibit ABI1-p65-WAVE2 complex formation, resulting in reduced adhesion and migration. Competitive binding of WAVE2 to ABI1 isoforms suggests the possibility of fine regulation of WAVE2 complex function in an ABI1 isoform-dependent manner. Interestingly, we also found that ABI1-SiL might interact directly with ABI1-p65, thus indirectly decreasing the formation of the ABI1-p65-WAVE2 complex. These intermolecular interactions were further confirmed by the co-localization of ABI1-SiL-GFP with WAVE2 and ABI1-p65 (Figures 8 and 9). WAVE2, ABI1, and NAP-1 are required for the WAVE regulatory complex (WRC) which activates the Arp2/3 complex to control branched actin polymerization in response to Rac activation[33-35]. As Figure 7E shows, the fact that ABI1-SiL (lacks a HHR domain required for binding to Nap1) does not bind to NAP-1, but ABI1-p65 and phosphorylated ABI1-p72 do in SW480-ABI1-SiL6 cells, suggests that ABI1-SiL overexpression disturbed WRC assembly and further impaired SW480 adhesion and migration. Additionally, all of the above observations were also observed in leukemia K562 and gastric cancer NCI-N87 cells (data not shown).
The present results indicate possible functions of ABI1-SiL in the regulation of SW480 morphogenesis, adhesion, and migration through interactions with WAVE2 and ABI1-p65, respectively. Although the overexpression of ABI1-SiL results in increased cell surface area and a more epithelial-like morphology, we did not find alterations in expression levels of any mRNA involved in differentiation of SW480 cells. Future studies will explore correlations between ABI1 splicing-isoform expression and KRAS or BRAF mutations or MSI in patients with colon cancer. Similarly, relationships and underlying mechanisms will be investigated to connect ABI1-SiL overexpression with mesenchymal-epithelial transition, and determine the roles of other ABI1 isoforms in the cellular and molecular phenotypes underlying colon cancer. Furthermore, ABI1 is present in several intrinsic protein complexes that regulate actin cytoskeletal remodeling, and the existence of several ABI1 isoforms raises the possibility of isoform-specific roles in other ABI1-specific functions[36]. In conclusion, we have shown for the first time herein that ABI1-SiL regulates adhesion and migration in SW480 cells via a mechanism involving fine regulation of WAVE2 complex activity, providing a putative biomarker and novel therapeutic target for colon cancer.
In addition, there are still some other limitations in the current study. First, we need more specific and accurate quantitative methods[37,38] to investigate the expression pattern of ABI1-SiL in a large-scale population in the future, and evaluate its potential application value as a clinical molecular diagnostic marker. Second, we need to systematically analyze the exon sequences of all ABI1 transcriptional variants, and use splice switching oligonucleotides[39] and/or gene modification techniques[40] to explore clinical interventions targeting ABI1-SiL. Finally, we also need to rely on animal models to further verify the underlying mechanism of the anti-oncogenic role of ABI1-SiL.
These results support a model in which ABI1-SiL plays an anti-oncogenic role by competitively binding to WAVE2 and directly interacting with phosphorylated and non-phosphorylated ABI1-p65, functioning as a dominant-negative form of ABI1-p65.
Abnormally expressed and/or phosphorylated Abelson interactor 1 (ABI1) participates in the metastasis and progression of colorectal carcinoma (CRC), but the roles of ABI1 splice isoforms (ABI1-SIs) are unknown. Exploring the roles and underlying mechanism of ABI1-SIs has important theoretical and practical significance for molecular diagnosis and targeted therapy of CRC.
ABI1-SIs-L (ABI1-SiL) plays an anti-oncogenic role in CRC, and it may be a potential molecular marker and therapeutic target in CRC.
To exploring the roles and underlying mechanism of ABI1-SiL in CRC.
We report a set of polymerase chain reaction-based methods that can specifically measure ABI1-SiL mRNA expression and constructed an ABI-SiL overexpressing SW480 cell model. CCK8, transwell, immunoprecipitation, Western blot, and co-localization assays were used to identify the anti-oncogenic role of ABI-SiL and the underlying mechanism in CRC.
ABI-SiL had a potential anti-tumor effect in CRC. Overexpression of ABI-SiL did not affect the proliferation and invasion of SW480 cells, but can increase the surface area and migration of SW480 cells by competitively binding WAVE2 and ABI1-p65.
ABI1-SiL plays an antioncogenic role in CRC cells by competitively binding WAVE2 and ABI1-p65.
Targeting ABI1-SIL expression is a potential treatment for colorectal cancer metastasis.
We are very grateful to Dr. Dai ZH for his reasonable suggestions and comments on the revision of the manuscript.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Schlevogt B, Schulte B, Tamori A S-Editor: Gao CC L-Editor: Wang TQ P-Editor: Liu JH
| 1. | Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490-1502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1965] [Cited by in RCA: 2284] [Article Influence: 207.6] [Reference Citation Analysis (1)] |
| 2. | Miura K, Fujibuchi W, Unno M. Splice isoforms as therapeutic targets for colorectal cancer. Carcinogenesis. 2012;33:2311-2319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 3. | Yi Q, Tang L. Alternative spliced variants as biomarkers of colorectal cancer. Curr Drug Metab. 2011;12:966-974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 4. | Yu W, Sun X, Clough N, Cobos E, Tao Y, Dai Z. Abi1 gene silencing by short hairpin RNA impairs Bcr-Abl-induced cell adhesion and migration in vitro and leukemogenesis in vivo. Carcinogenesis. 2008;29:1717-1724. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | Maia V, Ortiz-Rivero S, Sanz M, Gutierrez-Berzal J, Alvarez-Fernández I, Gutierrez-Herrero S, de Pereda JM, Porras A, Guerrero C. C3G forms complexes with Bcr-Abl and p38α MAPK at the focal adhesions in chronic myeloid leukemia cells: implication in the regulation of leukemic cell adhesion. Cell Commun Signal. 2013;11:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 6. | Taki T, Shibuya N, Taniwaki M, Hanada R, Morishita K, Bessho F, Yanagisawa M, Hayashi Y. ABI-1, a human homolog to mouse Abl-interactor 1, fuses the MLL gene in acute myeloid leukemia with t(10;11)(p11.2;q23). Blood. 1998;92:1125-1130. [PubMed] |
| 7. | Steinestel K, Brüderlein S, Lennerz JK, Steinestel J, Kraft K, Pröpper C, Meineke V, Möller P. Expression and Y435-phosphorylation of Abelson interactor 1 (Abi1) promotes tumour cell adhesion, extracellular matrix degradation and invasion by colorectal carcinoma cells. Mol Cancer. 2014;13:145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 8. | Steinestel K, Brüderlein S, Steinestel J, Märkl B, Schwerer MJ, Arndt A, Kraft K, Pröpper C, Möller P. Expression of Abelson interactor 1 (Abi1) correlates with inflammation, KRAS mutation and adenomatous change during colonic carcinogenesis. PLoS One. 2012;7:e40671. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 9. | Wang C, Tran-Thanh D, Moreno JC, Cawthorn TR, Jacks LM, Wang DY, McCready DR, Done SJ. Expression of Abl interactor 1 and its prognostic significance in breast cancer: a tissue-array-based investigation. Breast Cancer Res Treat. 2011;129:373-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 10. | Sun X, Li C, Zhuang C, Gilmore WC, Cobos E, Tao Y, Dai Z. Abl interactor 1 regulates Src-Id1-matrix metalloproteinase 9 axis and is required for invadopodia formation, extracellular matrix degradation and tumor growth of human breast cancer cells. Carcinogenesis. 2009;30:2109-2116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 11. | Chen H, Wu X, Pan ZK, Huang S. Integrity of SOS1/EPS8/ABI1 tri-complex determines ovarian cancer metastasis. Cancer Res. 2010;70:9979-9990. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 79] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 12. | Hu J, Mukhopadhyay A, Craig AW. Transducer of Cdc42-dependent actin assembly promotes epidermal growth factor-induced cell motility and invasiveness. J Biol Chem. 2011;286:2261-2272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | Li Y, Clough N, Sun X, Yu W, Abbott BL, Hogan CJ, Dai Z. Bcr-Abl induces abnormal cytoskeleton remodeling, beta1 integrin clustering and increased cell adhesion to fibronectin through the Abl interactor 1 pathway. J Cell Sci. 2007;120:1436-1446. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 41] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 14. | Leng Y, Zhang J, Badour K, Arpaia E, Freeman S, Cheung P, Siu M, Siminovitch K. Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphorylation required for WAVE2 activation. Proc Natl Acad Sci USA. 2005;102:1098-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 171] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 15. | Watahiki A, Waki K, Hayatsu N, Shiraki T, Kondo S, Nakamura M, Sasaki D, Arakawa T, Kawai J, Harbers M, Hayashizaki Y, Carninci P. Libraries enriched for alternatively spliced exons reveal splicing patterns in melanocytes and melanomas. Nat Methods. 2004;1:233-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 16. | Li K, Li M, Liu Y, Ma S, Yu W. [The relationship between ABI1 upregulation and the clinicopathologic features and prognosis of colon adenocarcinoma]. Zhonghua Putong Waike Zazhi. 2015;30:643-646. |
| 17. | Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2521] [Cited by in RCA: 2814] [Article Influence: 165.5] [Reference Citation Analysis (0)] |
| 18. | Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470-476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4232] [Cited by in RCA: 3864] [Article Influence: 227.3] [Reference Citation Analysis (0)] |
| 19. | Lapuk AV, Volik SV, Wang Y, Collins CC. The role of mRNA splicing in prostate cancer. Asian J Androl. 2014;16:515-521. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 20. | Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33:5311-5318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 493] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 21. | National Center for Biotechnology Information. ABI1: abl interactor 1 [Homo sapiens (human)]. [cited Jan 15, 2021]. Database: NCBI Gene [Internet]. Available from: http://www.ncbi.nlm.nih.gov/gene/10006. |
| 22. | Yu W, Ma S, Wang L, Zuo B, Li M, Qiao Z, Pan X, Liu Y, Wang J. Upregulation of GPR34 expression affects the progression and prognosis of human gastric adenocarcinoma by PI3K/PDK1/AKT pathway. Histol Histopathol. 2013;28:1629-1638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 23. | Ikeguchi A, Yang HY, Gao G, Goff SP. Inhibition of v-Abl transformation in 3T3 cells overexpressing different forms of the Abelson interactor protein Abi-1. Oncogene. 2001;20:4926-4934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 24. | Wang C, Navab R, Iakovlev V, Leng Y, Zhang J, Tsao MS, Siminovitch K, McCready DR, Done SJ. Abelson interactor protein-1 positively regulates breast cancer cell proliferation, migration, and invasion. Mol Cancer Res. 2007;5:1031-1039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | Cui MH, Yu WD, Dong JQ, Liu YL. [Effect of Abl-interacting protein 1 overexpression upon human gastric cancer cell proliferation in vitro]. Zhonghua Yixue Zazhi. 2009;89:3111-3115. [PubMed] |
| 26. | Kotula L. Abi1, a critical molecule coordinating actin cytoskeleton reorganization with PI-3 kinase and growth signaling. FEBS Lett. 2012;586:2790-2794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 27. | Dubielecka PM, Ladwein KI, Xiong X, Migeotte I, Chorzalska A, Anderson KV, Sawicki JA, Rottner K, Stradal TE, Kotula L. Essential role for Abi1 in embryonic survival and WAVE2 complex integrity. Proc Natl Acad Sci USA. 2011;108:7022-7027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 60] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 28. | Mendoza MC, Er EE, Zhang W, Ballif BA, Elliott HL, Danuser G, Blenis J. ERK-MAPK drives lamellipodia protrusion by activating the WAVE2 regulatory complex. Mol Cell. 2011;41:661-671. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 144] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 29. | Kheir WA, Gevrey JC, Yamaguchi H, Isaac B, Cox D. A WAVE2-Abi1 complex mediates CSF-1-induced F-actin-rich membrane protrusions and migration in macrophages. J Cell Sci. 2005;118:5369-5379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 64] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 30. | Sebestyén E, Zawisza M, Eyras E. Detection of recurrent alternative splicing switches in tumor samples reveals novel signatures of cancer. Nucleic Acids Res. 2015;43:1345-1356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 138] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 31. | Bryce NS, Reynolds AB, Koleske AJ, Weaver AM. WAVE2 regulates epithelial morphology and cadherin isoform switching through regulation of Twist and Abl. PLoS One. 2013;8:e64533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 32. | Takahashi K. WAVE2 Protein Complex Coupled to Membrane and Microtubules. J Oncol. 2012;2012:590531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 33. | Wu S, Ma L, Wu Y, Zeng R, Zhu X. Nudel is crucial for the WAVE complex assembly in vivo by selectively promoting subcomplex stability and formation through direct interactions. Cell Res. 2012;22:1270-1284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 34. | Burianek LE, Soderling SH. Under lock and key: spatiotemporal regulation of WASP family proteins coordinates separate dynamic cellular processes. Semin Cell Dev Biol. 2013;24:258-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 60] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 35. | Ryu JR, Echarri A, Li R, Pendergast AM. Regulation of cell-cell adhesion by Abi/Diaphanous complexes. Mol Cell Biol. 2009;29:1735-1748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 83] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 36. | Dubielecka PM, Cui P, Xiong X, Hossain S, Heck S, Angelov L, Kotula L. Differential regulation of macropinocytosis by Abi1/Hssh3bp1 isoforms. PLoS One. 2010;5:e10430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 37. | Sun B, Zheng YL. Simultaneous Quantification of Multiple Alternatively Spliced mRNA Transcripts Using Droplet Digital PCR. Methods Mol Biol. 2018;1768:387-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 38. | Wang H, Wang H, Duan X, Sun Y, Wang X, Li Z. Highly sensitive and multiplexed quantification of mRNA splice variants by the direct ligation of DNA probes at the exon junction and universal PCR amplification. Chem Sci. 2017;8:3635-3640. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 39. | Wan J. Antisense-mediated exon skipping to shift alternative splicing to treat cancer. Methods Mol Biol. 2012;867:201-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 40. | Maruyama R, Yokota T. Creation of DMD Muscle Cell Model Using CRISPR-Cas9 Genome Editing to Test the Efficacy of Antisense-Mediated Exon Skipping. Methods Mol Biol. 2018;1828:165-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |