Published online Oct 28, 2020. doi: 10.3748/wjg.v26.i40.6207
Peer-review started: June 28, 2020
First decision: July 28, 2020
Revised: August 9, 2020
Accepted: September 25, 2020
Article in press: September 25, 2020
Published online: October 28, 2020
Processing time: 122 Days and 0.9 Hours
Identifying genetic mutations in cancer patients have been increasingly important because distinctive mutational patterns can be very informative to determine the optimal therapeutic strategy. Recent studies have shown that deep learning-based molecular cancer subtyping can be performed directly from the standard hematoxylin and eosin (H&E) sections in diverse tumors including colorectal cancers (CRCs). Since H&E-stained tissue slides are ubiquitously available, mutation prediction with the pathology images from cancers can be a time- and cost-effective complementary method for personalized treatment.
To predict the frequently occurring actionable mutations from the H&E-stained CRC whole-slide images (WSIs) with deep learning-based classifiers.
A total of 629 CRC patients from The Cancer Genome Atlas (TCGA-COAD and TCGA-READ) and 142 CRC patients from Seoul St. Mary Hospital (SMH) were included. Based on the mutation frequency in TCGA and SMH datasets, we chose APC, KRAS, PIK3CA, SMAD4, and TP53 genes for the study. The classifiers were trained with 360 × 360 pixel patches of tissue images. The receiver operating characteristic (ROC) curves and area under the curves (AUCs) for all the classifiers were presented.
The AUCs for ROC curves ranged from 0.693 to 0.809 for the TCGA frozen WSIs and from 0.645 to 0.783 for the TCGA formalin-fixed paraffin-embedded WSIs. The prediction performance can be enhanced with the expansion of datasets. When the classifiers were trained with both TCGA and SMH data, the prediction performance was improved.
APC, KRAS, PIK3CA, SMAD4, and TP53 mutations can be predicted from H&E pathology images using deep learning-based classifiers, demonstrating the potential for deep learning-based mutation prediction in the CRC tissue slides.
Core Tip: Identifying genetic mutations in cancer patients have been increasingly important because distinctive mutational patterns can be very informative to determine the optimal therapy. This study aimed to investigate the feasibility of mutation prediction for the frequently occurring actionable mutations with colorectal cancer (CRC) whole-slide images. The area under the curves for receiver operating characteristic curves ranged from 0.693 to 0.809 for APC, KRAS, PIK3CA, SMAD4, and TP53, showing the potential for deep learning-based mutation prediction in the CRC pathology images. Furthermore, the prediction performance can be enhanced with the expansion of datasets.
- Citation: Jang HJ, Lee A, Kang J, Song IH, Lee SH. Prediction of clinically actionable genetic alterations from colorectal cancer histopathology images using deep learning. World J Gastroenterol 2020; 26(40): 6207-6223
- URL: https://www.wjgnet.com/1007-9327/full/v26/i40/6207.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i40.6207
Identifying genetic mutations in cancer patients has been increasingly important because mutational status can be very informative to determine the optimal therapeutic strategy[1]. However, molecular analysis is not performed routinely in every cancer patient, since it is not time and cost effective[2]. Thus, cost-effective alternatives for current molecular tests can be helpful in making appropriate treatment decisions. It has long been recognized that the histologic phenotypes reflect the genetic alterations in cancer tissues[3]. Since hematoxylin and eosin (H&E)-stained tissue slides are produced for almost every cancer patient, mutation prediction from the tissue slides can be a time- and cost-effective alternative method for individualized treatment. Thus, researchers attempted to examine the genotype–phenotype relationship in the H&E-stained tissue slides, and some gross tissue patterns related to specific molecular aberrations have been reported[4-9]. However, it remains largely unknown how specific molecular abnormalities are related to the specific histomorphologic findings, as it is not easy to capture the subtle features underlying the specific molecular alterations with the naked eye. To overcome the limitation of visual inspection of tissue structures by pathologists, various image analysis techniques have been applied for many decades to detect the subvisual characteristics of tissue patterns, not discernible to the unaided eyes[1]. Particularly, deep learning has been successfully applied to perform tasks considered too challenging for conventional image analysis techniques because it learns discriminative features directly from the large training dataset for any given task[10]. Therefore, deep learning is increasingly applied for tissue analysis tasks[11]. With the approval to use the digitized whole-slide images (WSIs) for diagnostic purposes, the digitization of tissue slides has been explosively increasing, providing huge digitized tissue data[12]. Combining the routine digitization of tissue slides with deep learning, the computer-aided analysis of WSIs could be adopted to support the evaluation of molecular alterations in H&E-stained cancer tissues in the near future. Although deep learning-based tissue analysis is still in its early phase, few promising results have been published. For example, a recent study reported that deep learning-based molecular cancer subtyping can be performed directly from the standard H&E sections obtained from patients with colorectal cancers (CRCs)[13]. Microsatellite instability can also be predicted from the tissue slides[14]. Furthermore, positive results for the mutation prediction of specific genes from histopathologic images have been reported in patients with various cancer types[3,15-17].
Motivated by these recent studies, we tried to predict the frequently occurring and clinically meaningful mutations from the H&E-stained CRC tissue WSIs with deep learning-based classifiers. Based on the frequency of mutation and prognostic values of the genes, we chose APC, KRAS, PIK3CA, SMAD4, and TP53 genes for the current study. The area under the curves (AUCs) for the receiver operating characteristic (ROC) curves ranged from 0.645 to 0.809 for The Cancer Genome Atlas (TCGA) datasets, showing the potential for deep learning-based mutation prediction in the CRC tissue slides. By combining two different datasets for training, the prediction performance can be enhanced with the expansion of datasets.
TCGA program offers the opportunity to reveal the genotype-phenotype relationship because it provides extensive archives of digital pathology slides with multi-omics test results[18]. Both frozen section tissue slides and formalin-fixed paraffin-embedded (FFPE) diagnostic slides were provided by the program. The WSIs from the TCGA-COAD (colon cancer) and TCGA-READ (rectal cancer) projects were combined in this study because colonic and rectal adenocarcinoma share similar molecular and histological features[18]. After removing the WSIs with poor quality, 629 patients were included in the present study. We chose to include the genetic alteration including frame shift insertion and deletion, missense mutations, and nonsense mutation. For APC, KRAS, PIK3CA, SMAD4, and TP53 genes, 436, 249, 133, 74, and 340 patients were confirmed to have the mutations, respectively. Deep learning did not perform optimally when there was a huge imbalance between classes[19]. In a previous study, we failed to obtain the balanced performance in tissue classification tasks unless the dataset itself was forced to have similar numbers between the classes[20]. Thus, we limited the difference in patient numbers between the mutation group and wild-type group by less than 1.4 fold through a random sampling. To match this limitation, we selected 263 patients with APC mutation as there were only 188 patients with the APC wild-type gene in the cohorts. The final patient IDs with their respective mutations are listed in Supplementary Table 1.
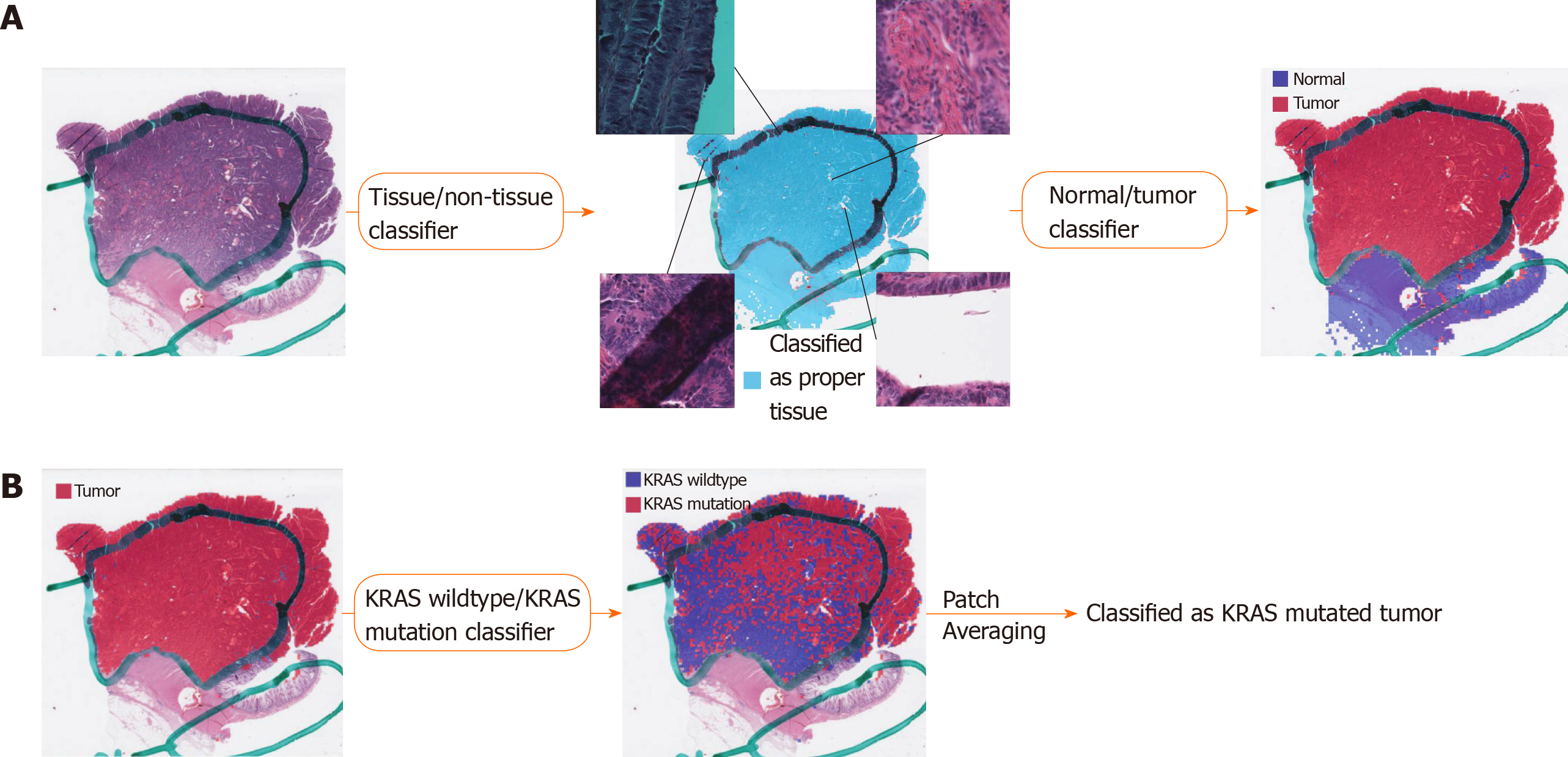
Various artifacts including air bubbles, compression artifacts, out-of-focus blur, pen markings, tissue folding, and white background are unavoidable in the WSIs. To make the prediction process fully automated, these artifacts should be automatically removed. Because it is impractical to analyze a WSI as a whole, small image patches are often sliced from a WSI and used for the analysis. Thus, we built a deep learning-based tissue/non-tissue classifier for 360 × 360 pixel image patches at 20 × magnification to remove all of these artifacts at once (Figure 1A). The classifier was a simple convolutional neural network (CNN) with 12 (5 × 5), 24 (5 × 5), and 24 (5 × 5) convolutional filters, each followed by a (2 × 2) max pooling layer. The tissue/non-tissue classifier could filter out more than 99.9% of improper patches. Next, tumor tissues should be delineated to predict the mutational status of cancer cells. Because of the freezing process for frozen tissue preparation, the frozen and FFPE tissue WSIs can differ in their morphologic features. Thus, we built separate normal/tumor classifiers for the frozen and FFPE WSIs based on the 360 × 360 pixel tissue image patches using the Inception-v3 model, a widely used CNN architecture. To train the wild-type/mutation classifiers for each gene, frozen and FFPE tissue patches with tumor probability higher than 0.9 by each tumor classifier were collected (Figure 1B). We arbitrarily chose the tumor probability as 0.9 because we decided to only include tissues with prominent tumor features. Although each slide may contain mixed regions of wild-type and mutated tissues considering the tumor heterogeneity, we assigned the same label for all tumor tissue patches in a WSI based on the mutational status of the patients. This labeling strategy was inevitable since we had no methods to delineate the wild-type and mutated regions before the classifiers could be built. The classifiers for the five genes were separately trained and validated with a patient-level ten-fold cross-validation scheme for frozen and FFPE WSIs. The slide-level mutation probability was calculated as the average of the probabilities of all the tumor patches in the WSI. For the training of the Inception-v3 models, we used a mini-batch size of 128, and the cross entropy loss function was adopted as a loss function. Deep neural networks were implemented using the TensorFlow deep learning library (http://tensorflow.org). To minimize overfitting, data augmentation techniques, including random rotations by 90°, random horizontal/vertical flipping, and random perturbation of the contrast and brightness, were applied to the tissue patches during training. In addition, 10% of the training slides were used as a validation dataset for the early stopping of the training. At least five separate classifiers were trained for each gene and tissue modality, and the classifier with the best AUC on the test dataset was included in the results.
Patient cohort: A total of 142 patients with CRC who previously underwent surgical resection in Seoul St. Mary’s hospital between 2017 and 2019 were enrolled (SMH dataset). All cases were sporadic, without any familial history of CRCs. The clinicopathological parameters including age, sex, and tumor location were retrospectively reviewed from the medical records. The study was approved by the Institutional Review Board of the College of Medicine at the Catholic University of Korea, No. KC19SESI0787.
Mutation prediction on SMH dataset: For APC, KRAS, PIK3CA, SMAD4, and TP53 genes, 66, 75, 31, 23, and 98 patients were confirmed to have the mutations, respectively. The sequencing methods are described in Supplementary Methods. Because the SMH dataset was originally collected to extra-validate the model trained on the TCGA datasets, we did not adjust the patient numbers between the classes. The normal/tumor classifier for TCGA FFPE tissues was also used to discriminate the tumor tissue patches of SMH WSIs. The normal/tumor classification accuracy was reviewed by Lee SH and Song IH and was confirmed to be valid. Again, patches with tumor probability higher than 0.9 were collected for mutational status classification. Then, the SMH data were split into ten folds, and each training fold was mixed with TCGA training fold to build new classifiers trained on both datasets. The classification results of the new classifiers on TCGA or SMH datasets were compared with the TCGA-based classifiers to investigate the effects of the expanded training dataset.
The ROC curves and their AUCs for all classifiers were presented to demonstrate the performance of each classifier. We used a permutation test with 1000 iterations to compare the differences between the two paired or unpaired ROC curves when necessary[21]. A P value of < 0.05 was considered significant.
This study aimed to investigate the feasibility of mutation prediction for the frequently occurring mutations in the CRC tissue WSIs. Since only tumor tissues would be meaningful for the prediction of the mutational status in the tissue slides, three different tissue patch classifiers were sequentially applied to discriminate between tissue/non-tissue, normal/tumor, and wild-type/mutation in order (Figure 1). Only proper tissue patches with high tumor probabilities were used to determine the mutational status (Figure 1B). Patient-level ten-fold cross validation was applied for both frozen and FFPE datasets to fully evaluate the properties of the TCGA CRC WSIs.
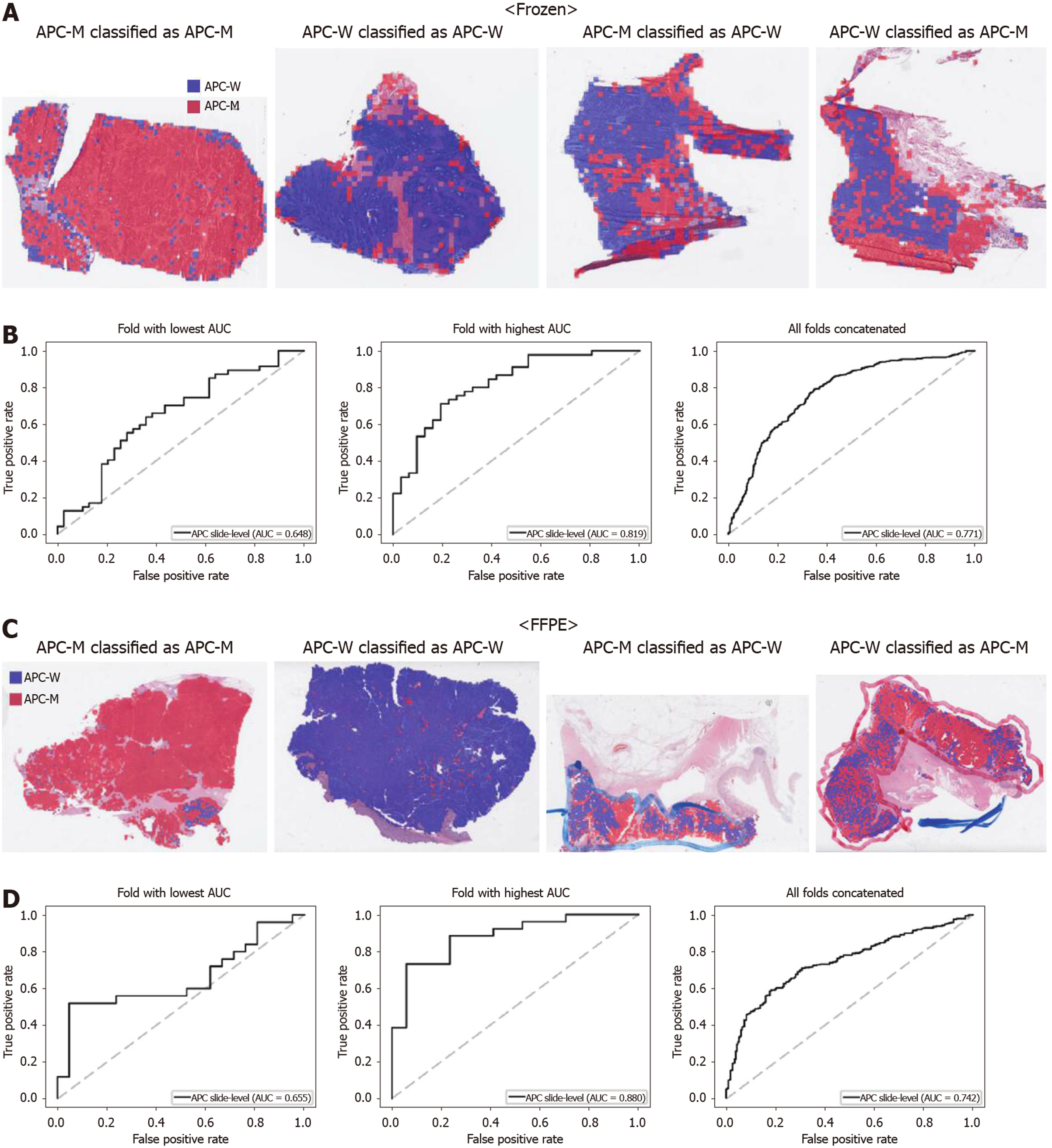
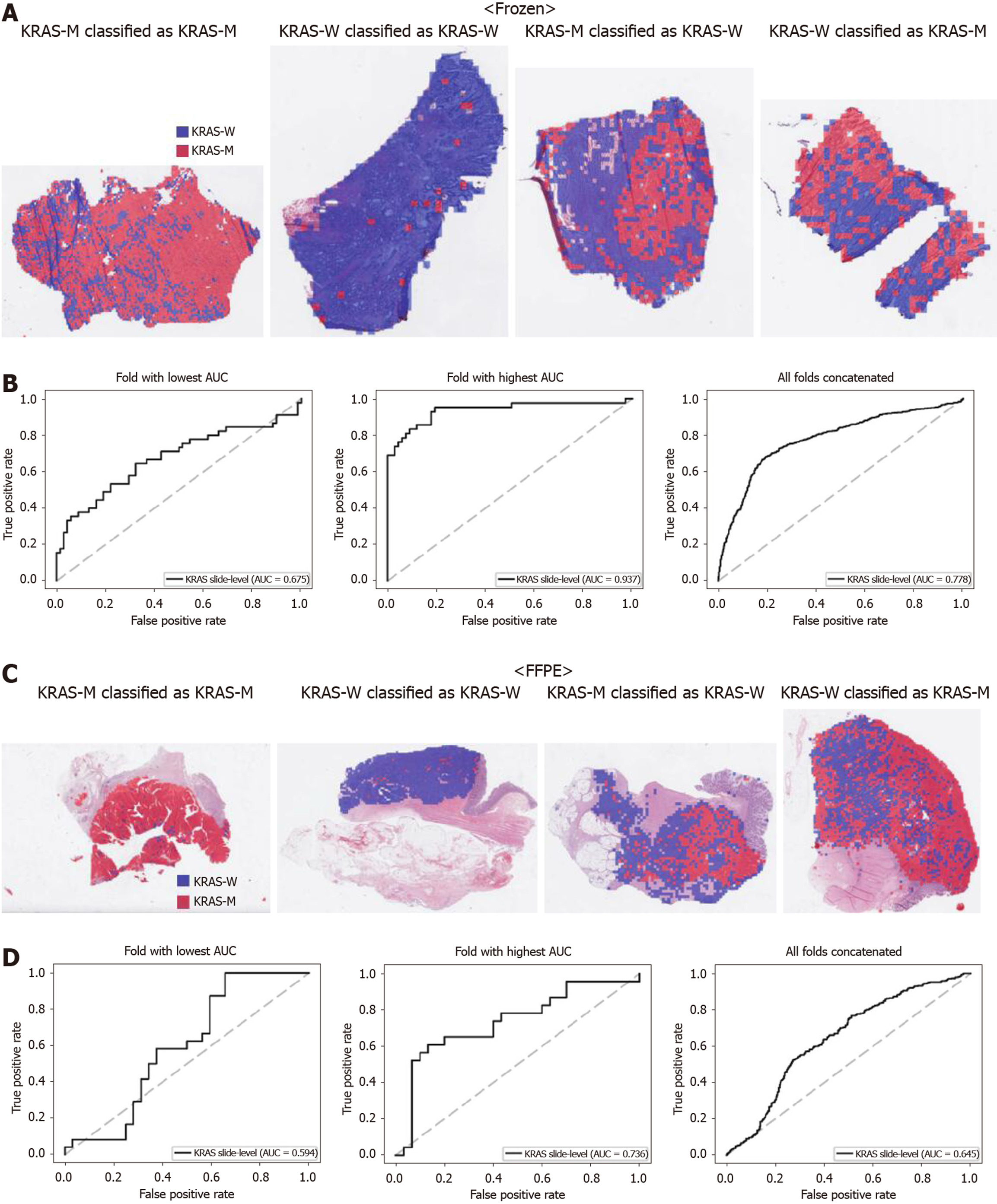
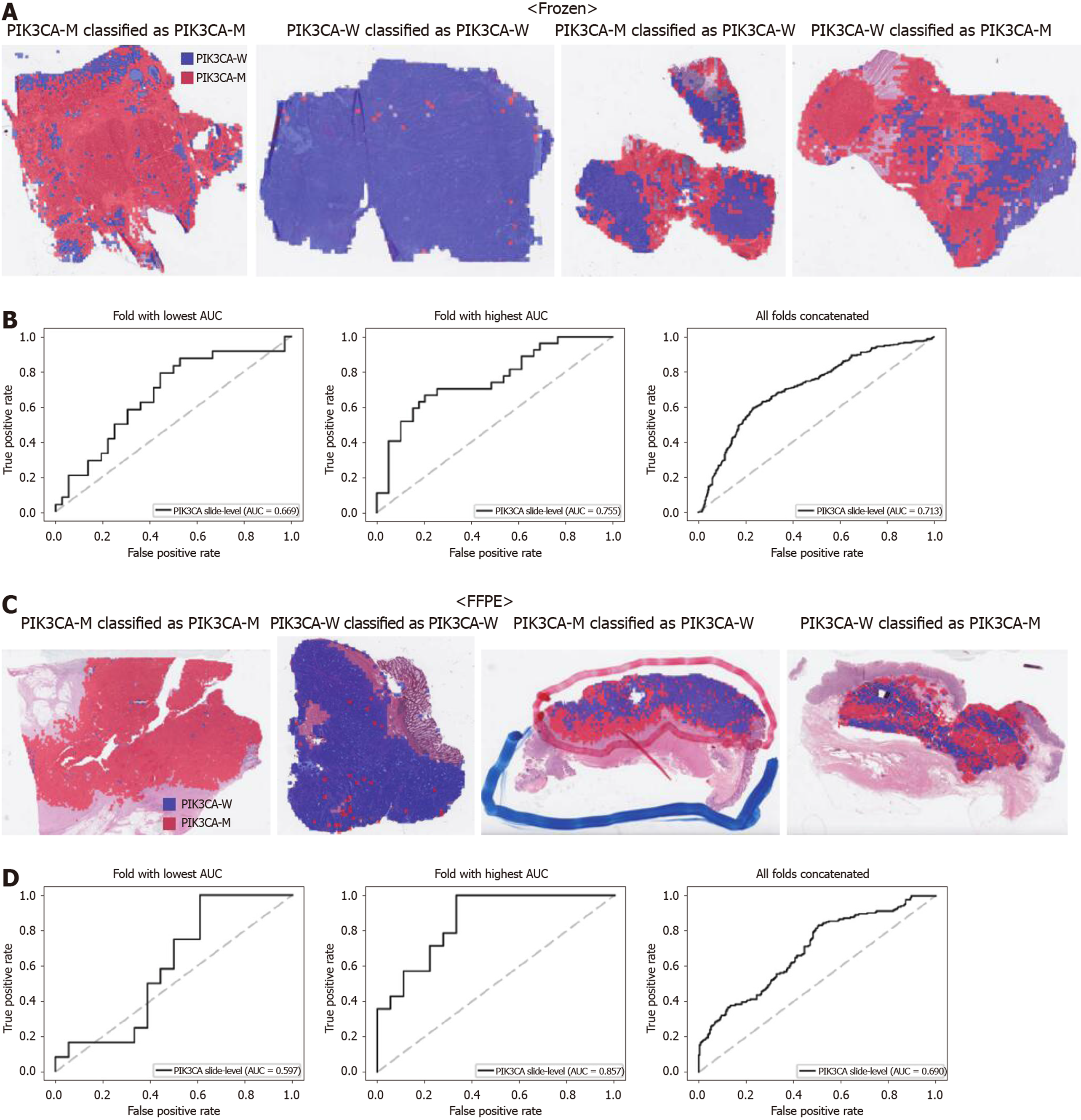
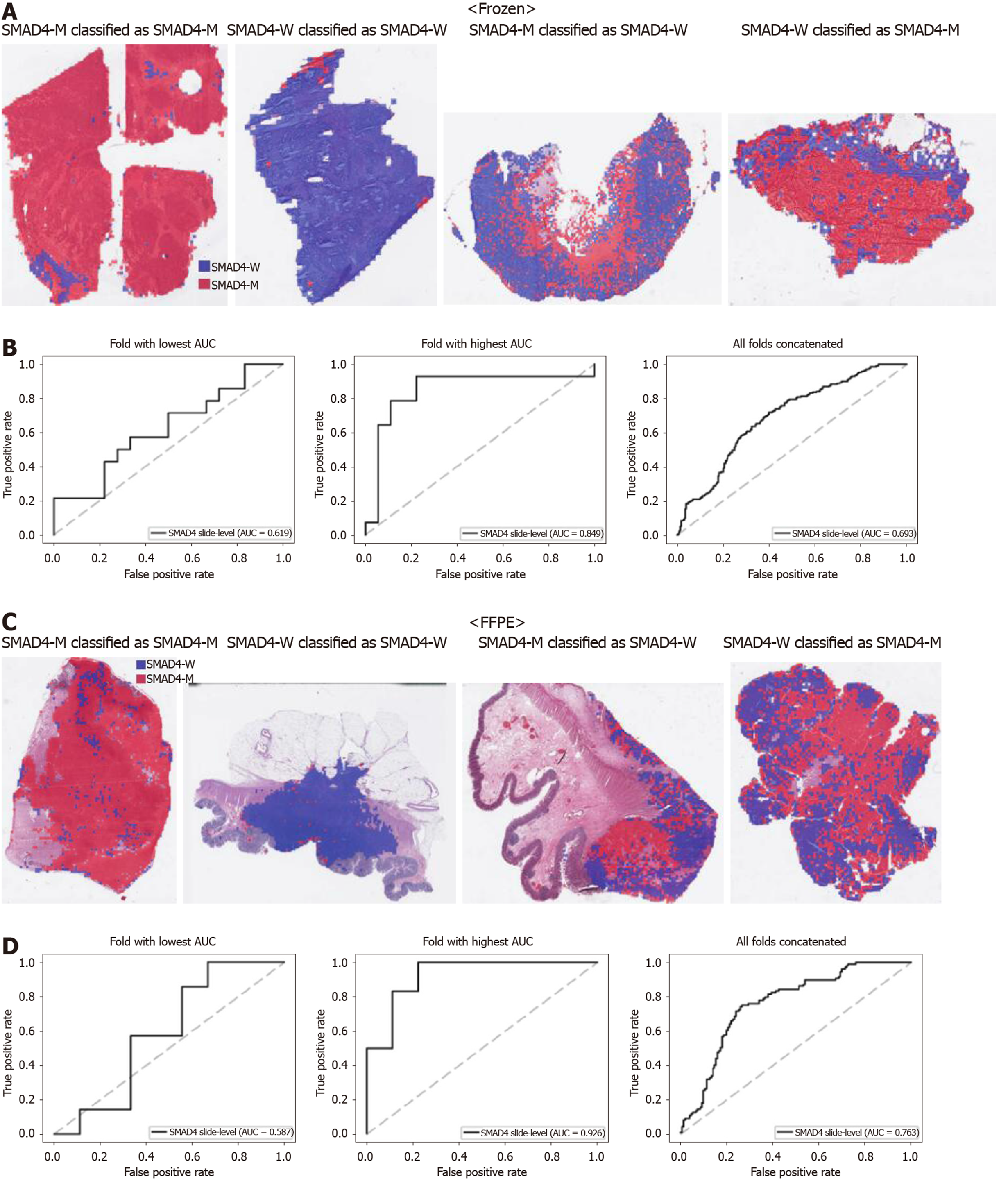
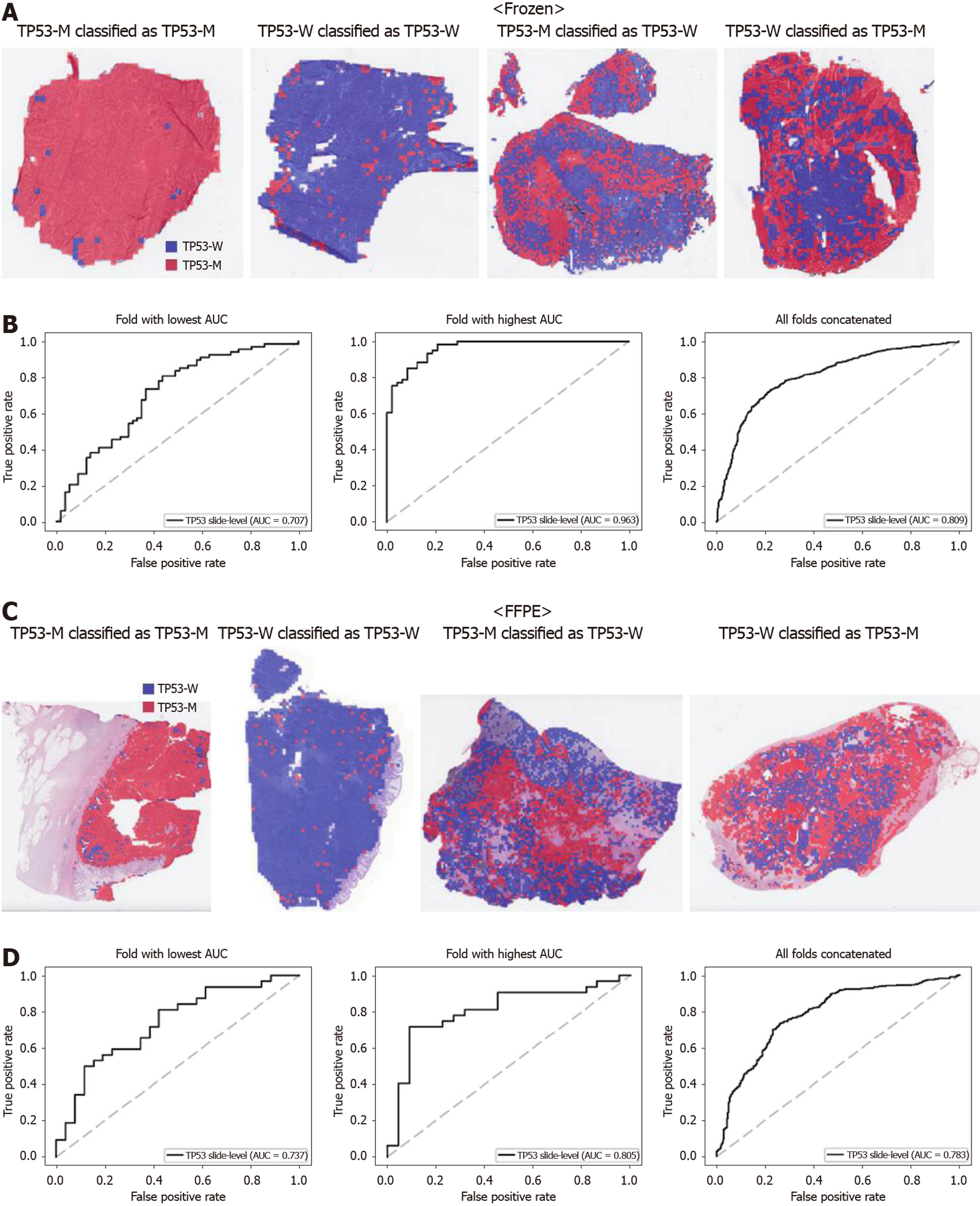
From Figures 2 to 6, the classification results for APC, KRAS, PIK3CA, SMAD4, and TP53 genes are presented for both frozen (upper panels) and FFPE (lower panels) TCGA WSIs. In A and C of every figure, the representative binary heatmaps demonstrating the distribution of tissue patches classified as wild-type or mutation are presented. From left to right, WSIs with gene mutation correctly classified as mutation, with wild-type gene correctly classified as wild type, with gene mutation falsely classified as wild-type, and with wild-type gene falsely classified as mutation are presented, which were determined by the probability threshold set to 0.5. The sensitivity and specificity of a classifier can be much improved by setting the threshold appropriately. However, we set the threshold to 0.5 in the figures for simplicity because every classifier for different folds had different optimal thresholds. To demonstrate the differences in the performance between folds, slide-level ROC curves for folds with the lowest and highest AUCs were presented (left and middle ROC curves in the figures). Finally, the overall performance was inferred based on the slide-level ROC curves drawn for the concatenated results from all ten folds (right ROC curves). For the APC gene (Figure 2), the AUCs per fold ranged from 0.648 to 0.819 for the frozen tissues and from 0.655 to 0.880 for the FFPE tissues. The concatenated AUCs were 0.771 and 0.742 for the frozen and FFPE tissues, respectively. For the KRAS gene (Figure 3), the performance was much better for the frozen tissues than for the FFPE tissues with a per fold AUC for the frozen tissues of 0.675-0.937 and a concatenated AUC of 0.778. For the FFPE tissues, the concatenated AUC was only 0.645, while the per fold AUCs ranged from 0.594 to 0.736. With regard to the PIK3CA gene (Figure 4), the lowest and highest AUCs per fold were 0.669 and 0.775 for the frozen tissues and 0.597 and 0.857 for the FFPE tissues. The concatenated AUCs were 0.713 and 0.690, respectively. For the SMAD4 gene (Figure 5), AUCs per fold ranged from 0.619 to 0.849 for the frozen tissues and from 0.587 to 0.926 for the FFPE tissues, while the concatenated AUCs were 0.693 and 0.763, respectively. With regard to the TP53 gene (Figure 6), the lowest and highest AUCs per fold were 0.707 and 0.963 for the frozen tissues and 0.737 and 0.805 for the FFPE tissues. The concatenated AUCs were 0.809 and 0.783, respectively. Overall, the wild-type/mutation classifiers for the TP53 gene yielded the highest AUCs for both frozen and FFPE tissues of the TCGA datasets. Between the ROC curves of the frozen and FFPE tissues, classifiers for the frozen tissues yielded better results for the APC and KRAS genes (P < 0.05, P < 0.001, P = 0.068, P = 0.057, and P = 0.115 between the frozen and FFPE classifiers for APC, KRAS, PIK3CA, SMAD4, and TP53 genes, respectively, by Venkatraman’s permutation test for unpaired ROC curves).
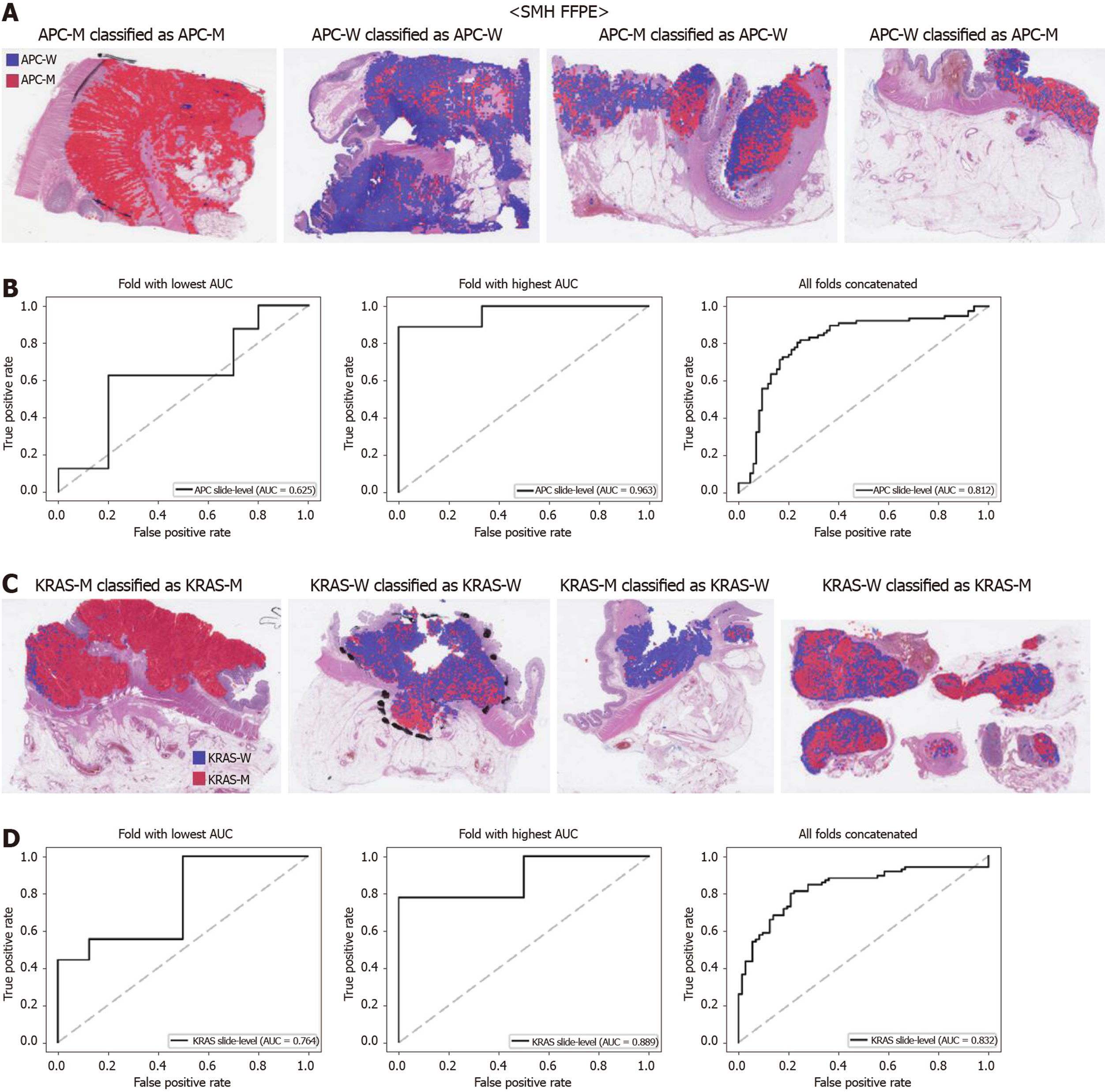
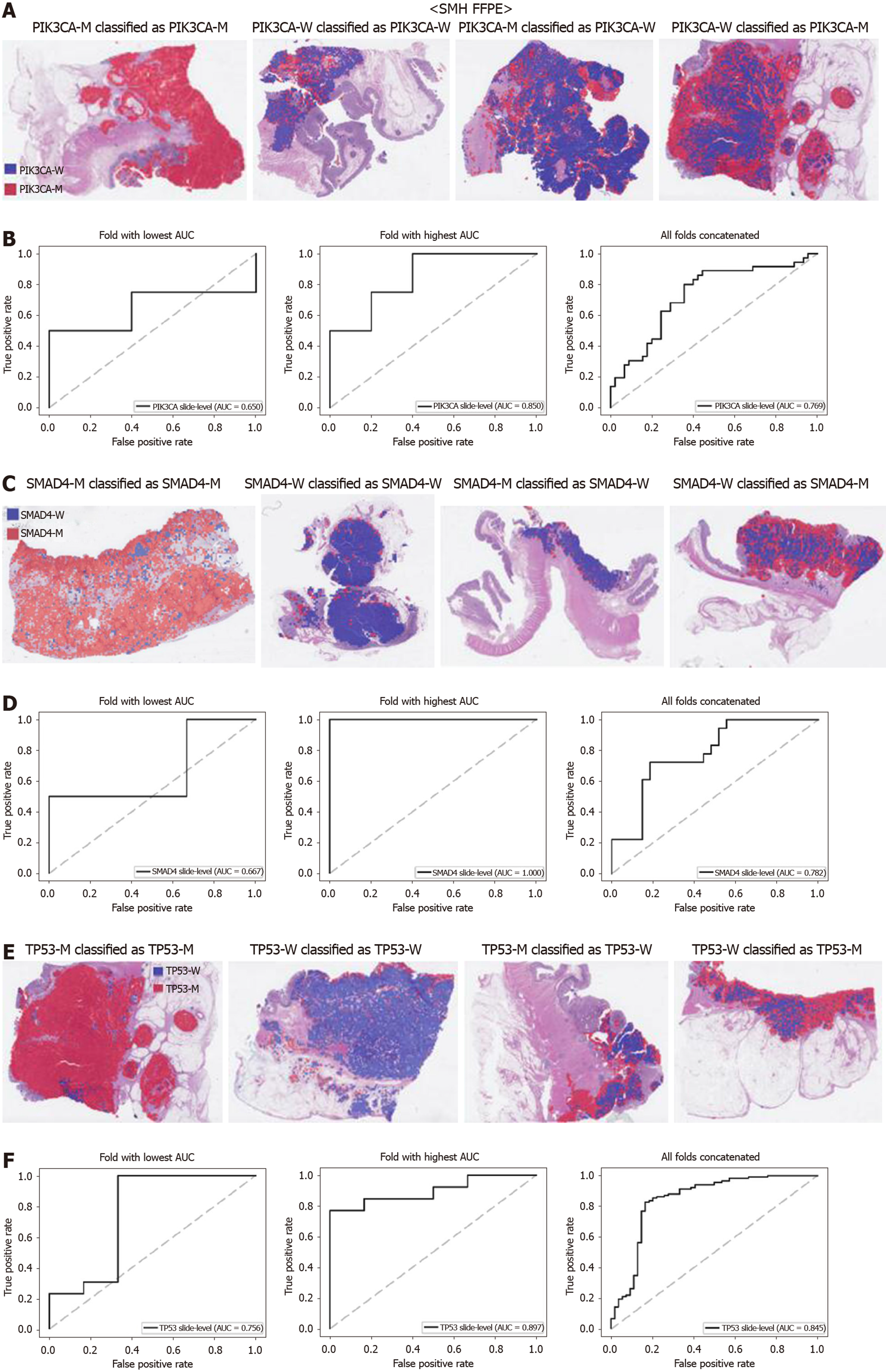
The generalizability of a deep learning model for the external dataset is an important issue to be validated. Thus, we collected our own CRC FFPE WSIs with information on genetic mutation. The normal/tumor classifier for the TCGA FFPE tissues was applied to collect the tissue patches with high tumor probabilities. Then, the mutation classifiers for each gene trained on the TCGA FFPE tissues were applied to the tumor patches. The slide-level ROC curves for the five genes are presented in Supplementary Figure 1. The AUCs were 0.654, 0.581, 0.570, 0.652, and 0.775 for APC, KRAS, PIK3CA, SMAD4, and TP53 genes, respectively. For the APC, KRAS, and PIK3CA genes, the performance of the TCGA-based mutation classifiers on the SMH dataset were worse than that on the TCGA dataset (P < 0.01, P < 0.05, P < 0.05, P = 0.107, and P = 0.263 for APC, KRAS, PIK3CA, SMAD4, and TP53 genes, respectively, by Venkatraman’s permutation test for unpaired ROC curves). These results indicated that the mutation classifiers did not have an excellent generalizability when they were trained only with the TCGA WSI datasets. It remains unclear whether the performance could be improved when more data are used for the training. Thus, we combined the TCGA and SMH datasets to train new sets of mutation classifiers. Patient-level ten-fold cross validation schemes were also used for the mixed dataset. The performance of the SMH dataset showed an obvious improvement, since the SMH data were included in the training data in this setting. The AUCs for APC and KRAS genes increased to 0.812 and 0.832 (Figure 7, P < 0.01 and P < 0.001 compared with the TCGA-trained classifiers by Venkatraman’s permutation test for paired ROC curves). Improved results were also obtained for PIK3CA, SMAD4, and TP53 with AUCs of 0.769, 0.782, and 0.845, respectively (Figure 8, P < 0.05, P < 0.01, and P < 0.05 by Venkatraman’s permutation test for paired ROC curves). More importantly, the performance of the TCGA data was also generally improved by the classifiers trained on both datasets (Supplementary Figure 2). The AUCs were 0.766, 0.694, 0.708, 0.791, and 0.822 for the APC, KRAS, PIK3CA, SMAD4, and TP53 genes, respectively (P = 0.072, P < 0.01, P = 0.091, P = 0.074, and P < 0.05 compared with the TCGA-trained classifiers). These results indicated that the deep learning-based classifiers for mutation prediction in tissue slides can yield better performance when more data are collected from various sources.
In the present study, we selected the APC, KRAS, PIK3CA, SMAD4, and TP53 genes because they were frequently occurring in both TCGA and SMH CRC datasets and had prognostic values. APC is an important tumor suppressor known to play a role in CRC development. Deactivating APC leads to the constitutive activation of the Wnt signaling pathway, which may contribute to tumor progression[22]. The frequency
In general, the APC mutation is thought to have no prognostic significance[36]. However, in a specific situation such as in a microsatellite stable proximal colon cancer, wild-type APC has been associated with poorer survival[37]. On the contrary, KRAS, PIK3CA, SMAD4, and TP53 gene mutations were associated with poorer prognosis in CRCs[34,38-40]. Thus, information on the mutational status of these genes can be useful in making therapeutic decisions for CRC patients. On occasion, a specific gene mutation can be related to a specific visual characteristic in tissue histology. For example, the PIK3CA mutation often coincides with lymphovascular invasion, tumor budding, and a high number of poorly differentiated clusters in CRC tissues[39]. However, it is not always possible to discover the visually discernible features reflecting the mutation of a specific gene. Therefore, we adopted deep learning to predict the mutational status of the five genes because the discriminative features of the mutations can be automatically learned directly from the large training data of tissue images. To our knowledge, this is the first study to evaluate the mutation prediction capabilities of deep learning models for the frequently occurring mutations in the pathologic tissue slides of CRC patients.
In all the mutation classifiers applied to the TCGA frozen and FFPE tissues, the slide-level discrimination capabilities were much better against chance performance (P < 0.001 for all five genes by permutation test). These results indicated that the Inception-v3 model learned valid features to discriminate the mutated tissue phenotypes of each gene. In the case of APC and KRAS genes, the classifiers for the frozen tissues yielded better results compared with the FFPE tissues, although the frozen sections generally showed poorer tissue quality than did the FFPE sections. It can be explained by the fact that the frozen sections provided the best representation of the tissue contents on which the genomic signatures were tested[18]. Since the FFPE sections can be taken far from the frozen tissue sections, the mutational status can be different between them, considering the heterogeneity of large tumors. When we validated the classifiers trained with the TCGA FFPE tissues on the SMH WSIs, the performance was generally poorer (Supplementary Figure 1). Deep learning operates well under a condition where both the training and test datasets come from the same distribution[41]. For the H&E-stained tissue slides, the quality may vary because they undergo multiple processes for preparation including formalin fixation, paraffin embedding, sectioning, and staining, which can be slightly different between institutes[42]. Furthermore, the ethnic difference between the TCGA and SMH datasets may also contribute to the difference in the performance. Although the difference can be negligible to human eye, deep learning can be very sensitive to the subtle difference in tissue conditions. Therefore, many researchers insisted on the necessity of using large multi-national and multi-institutional datasets to enhance the generalizability of the deep learning model[2,12]. Thus, we combined the two datasets to build new classifiers trained on both TCGA and SMH datasets. Naturally, the performance for the SMH data was greatly enhanced because the tissue features of the data were exposed to the classifiers in this setting. More importantly, the performance of the TCGA data was also enhanced by adding the WSIs from the SMH dataset for training. These results clearly demonstrated that multi-national and multi-institutional datasets can improve the performance of the mutation classifiers. However, it remains unclear how far the performance can be improved if much more data are supplied.
When we scrutinized the binary heatmaps of falsely classified WSIs, we recognized that the wild-type and mutated patches were generally aggregated rather than dispersed. The patterns implied the possibility that the tumor tissues in a tissue slide may have different mutational statuses between different regions. Large tumors can be molecularly heterogeneous, and the tumor heterogeneity can contribute to the resistance to treatment[43]. Therefore, tumor heterogeneity has been an important issue for both researchers and clinicians. To elucidate the spatial heterogeneity of a tumor, molecular methods with high spatial specificity such as multi-region sequencing and single-cell sequencing can be applied to examine a tissue sample. However, a random sampling of tissues for these molecular tests would be very inefficient. If possible regions of molecular heterogeneity in a tissue slide could be identified before the tests, molecular testing can be more specific and efficient. Furthermore, there are possibilities of false negative molecular tests because of the imprecise delineation of target regions in a tissue block[12]. Therefore, it is very important to objectively discriminate the tumor regions for the molecular evaluation of the tumor tissues. Thus, both normal/tumor and wild-type/mutation classifiers can be used to delineate the appropriate target sites for various molecular tests in cancer tissues. For example, Supplementary Figure 3 presents the heatmaps for the mutational status of all five genes in a TCGA frozen tissue slide, demonstrating how different regions of a slide can have different mutational statuses. When an overlaid probability map of mutation was drawn, areas with low and high mutational statuses can be recognized. It may not be easy to obtain this kind of information without the help of deep learning. Hence, molecular tests with high spatial specificity can be targeted to specific regions depending on the purpose of the tests. Therefore, these classifiers can make the selection of lesional regions for relevant multi-omics testing fully automated in the near future[2].
Limitations also exist for the deep learning-based tissue classifiers. One of the limitations is the sensitive nature of deep learning to minute differences in the datasets. Because of the sensitive nature, classifiers applied to very subtly different conditions should be separately built. For example, classifiers for the frozen and FFPE tissues should be separately trained for the same tasks. It requires additional data collection and training overload. In clinical practice, pathologists should take an additional step to determine the kind of classifiers that should be applied for a specific specimen. It is currently inevitable to separately build classifiers to support various real-world tasks in the pathology laboratories. Therefore, manual selection of appropriate classifiers for target tasks is a necessary step that can limit the fully automated adoption of deep learning-based classifiers in the pathology workflows.
In the current study, we used the high-throughput cancer panel to identify mutations in CRC tissues of the SMH dataset. This panel test approach makes it possible to identify diverse clinically actionable mutations in a single assay. However, it is quite expensive to prepare the equipment necessary to perform the test and to save a large number of data generated. This study demonstrated that a deep learning-based method could be a useful and effective tool for the prediction of actionable mutations from CRC WSIs. However, the interpretation of decision made by the deep learning-based classifier is unclear because of the black box nature of deep learning and should be further studied. Besides this aspect, the advantages and disadvantages between the mutation panel test (molecular test) and deep learning method were described in Table 1.
| Mutation panel test | Deep learning-based method | |
| Advantages | (1) High throughput method: Multiplex analysis of various genes; and (2) Quantitative and sensitive detection of genomic aberrations. | (1) More rapid turnaround time: Once trained, the predictions are fast (less than 5 min per gene) and fully automated; (2) Better picture of tumor heterogeneity: Heat map analysis provides insights into spatial distribution of mutations; and (3) Remote testing: It may be able to detect genetic mutation from pictures taken directly from the microscope at the remote institute. |
| Disadvantages | (1) Longer turnaround time: Run lasts from 1 to 3 d; and (2) High complexity of workflow: Requires complex sample preparation. | (1) Requires separate classifier for each gene; (2) Requires large training dataset: Neural networks work best with more data; and (3) Deep learning method is a black box: It is not straightforward to understand how the decision is made. |
Despite the limitation, with the increasing digitization of tissue slides, various computer-assisted methods will be introduced for histopathologic interpretation and clinical care. In the present study, we demonstrated the potential of deep learning-based classifiers to predict mutations in the CRC WSIs. Although the classifiers in this study are not yet enough to be used for predicting the genetic mutations in the clinic, deep learning-based methods have the potential to learn features for discriminating the wild-type tissues from the mutated tissues, which are not easily discernible to the human eye. Thus, deep learning will be increasingly adopted to discover new tissue-based biomarkers, which provide fundamental information for personalized medicine. With the accumulation of large sets of WSI data, deep learning-based tissue analyses will play important roles in the better characterization of cancer patients and will be an essential part of digital pathology in the era of precision medicine.
In the present study, we demonstrated that the APC, KRAS, PIK3CA, SMAD4 and TP53 mutation can be predicted from H&E pathology images using the deep learning-based classifiers. Furthermore, by combining the TCGA and our datasets for training, the prediction performance was enhanced. Therefore, with the accumulation of tissue image data for training, deep learning can be used to supplement current molecular testing methods in the near future.
Identifying genetic mutations in cancer patients have been increasingly important because distinctive mutational patterns can be very informative to determine the optimal therapeutic strategy. In recent years, the digitization of pathology slide images has been explosively increasing, providing huge digitized tissue data. Combining the routine digitization of pathology whole-slide images (WSIs) with deep learning, computer-aided mutation prediction with the pathology images from cancers can be a time- and cost-effective complementary method for personalized treatment.
Recent studies have reported that deep learning-based molecular cancer subtyping and microsatellite instability prediction can be performed directly from the standard hematoxylin and eosin (H&E) sections in diverse cancers. Motivated by these recent studies, we tried to predict the frequently occurring and clinically meaningful mutations from the H&E-stained colorectal cancer (CRC) tissue WSIs with deep learning-based classifiers. Cost-effective alternatives for current molecular tests can be helpful to support the decision-making process for the management of patients with CRCs.
The present study aimed to investigate the feasibility of deep learning-based mutation prediction for the frequently occurring mutations in CRCs using H&E WSIs.
We built and tested the classifiers for mutation prediction on the 629 The Cancer Genome Atlas (TCGA) CRC dataset and validated them with the 142 Seoul St. Mary Hospital (SMH) CRC dataset. Based on the frequency of mutations in both the TCGA and SMH datasets, we chose APC, KRAS, PIK3CA, SMAD4, and TP53 genes for the current study. The classifiers were trained with 360 × 360 pixel patches of tissue images. The receiver operating characteristic (ROC) curves and their area under the curves (AUCs) were presented for all the classifiers to demonstrate the performance of each classifier.
The AUCs for ROC curves ranged from 0.693 to 0.809 for the TCGA frozen WSIs and from 0.645 to 0.783 for the TCGA formalin-fixed paraffin-embedded WSIs. Moreover, the prediction performance can be enhanced with the expansion of datasets. The prediction performance was improved with the classifiers trained with both TCGA and SMH data.
The present study demonstrated that the APC, KRAS, PIK3CA, SMAD4, and TP53 mutations can be predicted from H&E pathology images using deep learning-based classifiers, showing the potential for deep learning-based mutation prediction in the CRC tissue slides.
Although the classifiers in this study were not enough to be used for predicting the genetic mutations in the clinic, we can recognize the potential of deep learning-based methods to learn features for discriminating the wild-type and mutated tissues, which are not easily discernible to the human eyes. Therefore, deep learning models can assist pathologists in the detection of cancer subtype or gene mutations.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: South Korea
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Jin Z, Sameer AS S-Editor: Huang P L-Editor: A P-Editor: Liu JH
| 1. | Hamilton PW, Bankhead P, Wang Y, Hutchinson R, Kieran D, McArt DG, James J, Salto-Tellez M. Digital pathology and image analysis in tissue biomarker research. Methods. 2014;70:59-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 136] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 2. | Djuric U, Zadeh G, Aldape K, Diamandis P. Precision histology: how deep learning is poised to revitalize histomorphology for personalized cancer care. NPJ Precis Oncol. 2017;1:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 107] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 3. | Fu Y, Jung AW, Torne RV, Gonzalez S, Vöhringer H, Jimenez-Linan M, Moore L, Gerstung M. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. bioRxiv. 2019. [DOI] [Full Text] |
| 4. | Ninomiya H, Hiramatsu M, Inamura K, Nomura K, Okui M, Miyoshi T, Okumura S, Satoh Y, Nakagawa K, Nishio M, Horai T, Miyata S, Tsuchiya E, Fukayama M, Ishikawa Y. Correlation between morphology and EGFR mutations in lung adenocarcinomas Significance of the micropapillary pattern and the hobnail cell type. Lung Cancer. 2009;63:235-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 82] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 5. | Warth A, Penzel R, Lindenmaier H, Brandt R, Stenzinger A, Herpel E, Goeppert B, Thomas M, Herth FJ, Dienemann H, Schnabel PA, Schirmacher P, Hoffmann H, Muley T, Weichert W. EGFR, KRAS, BRAF and ALK gene alterations in lung adenocarcinomas: patient outcome, interplay with morphology and immunophenotype. Eur Respir J. 2014;43:872-883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 91] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 6. | Mosquera JM, Perner S, Demichelis F, Kim R, Hofer MD, Mertz KD, Paris PL, Simko J, Collins C, Bismar TA, Chinnaiyan AM, Rubin MA. Morphological features of TMPRSS2-ERG gene fusion prostate cancer. J Pathol. 2007;212:91-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 91] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 7. | Hakimi AA, Tickoo SK, Jacobsen A, Sarungbam J, Sfakianos JP, Sato Y, Morikawa T, Kume H, Fukayama M, Homma Y, Chen YB, Sankin A, Mano R, Coleman JA, Russo P, Ogawa S, Sander C, Hsieh JJ, Reuter VE. TCEB1-mutated renal cell carcinoma: a distinct genomic and morphological subtype. Mod Pathol. 2015;28:845-853. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 117] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 8. | Weisman PS, Ng CK, Brogi E, Eisenberg RE, Won HH, Piscuoglio S, De Filippo MR, Ioris R, Akram M, Norton L, Weigelt B, Berger MF, Reis-Filho JS, Wen HY. Genetic alterations of triple negative breast cancer by targeted next-generation sequencing and correlation with tumor morphology. Mod Pathol. 2016;29:476-488. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 9. | Shia J, Schultz N, Kuk D, Vakiani E, Middha S, Segal NH, Hechtman JF, Berger MF, Stadler ZK, Weiser MR, Wolchok JD, Boland CR, Gönen M, Klimstra DS. Morphological characterization of colorectal cancers in The Cancer Genome Atlas reveals distinct morphology-molecular associations: clinical and biological implications. Mod Pathol. 2017;30:599-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 75] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 10. | LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36149] [Cited by in RCA: 20045] [Article Influence: 2004.5] [Reference Citation Analysis (0)] |
| 11. | Dimitriou N, Arandjelović O, Caie PD. Deep Learning for Whole Slide Image Analysis: An Overview. Front Med (Lausanne). 2019;6:264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 146] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 12. | Serag A, Ion-Margineanu A, Qureshi H, McMillan R, Saint Martin MJ, Diamond J, O'Reilly P, Hamilton P. Translational AI and Deep Learning in Diagnostic Pathology. Front Med (Lausanne). 2019;6:185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 163] [Cited by in RCA: 145] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 13. | Sirinukunwattana K, Domingo E, Richman S, Redmond KL, Blake A, Verrill C, Leedham SJ, Chatzipli A, Hardy C, Whalley C, Wu C, Beggs AD, McDermott U, Dunne P, Meade AA, Walker SM, Murray GI, Samuel LM, Seymour M, Tomlinson I, Quirke P, Maughan T, Rittscher J, Koelzer VH, on behalf of S:CORT consortium. Image-based consensus molecular subtype classification (imCMS) of colorectal cancer using deep learning. bioRxiv. 2019. [DOI] [Full Text] |
| 14. | Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, Marx A, Boor P, Tacke F, Neumann UP, Grabsch HI, Yoshikawa T, Brenner H, Chang-Claude J, Hoffmeister M, Trautwein C, Luedde T. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25:1054-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 992] [Cited by in RCA: 758] [Article Influence: 126.3] [Reference Citation Analysis (0)] |
| 15. | Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, Moreira AL, Razavian N, Tsirigos A. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24:1559-1567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1224] [Cited by in RCA: 1543] [Article Influence: 220.4] [Reference Citation Analysis (0)] |
| 16. | Schaumberg AJ, Rubin MA, Fuchs TJ. H&E-stained Whole Slide Image Deep Learning Predicts SPOP Mutation State in Prostate Cancer. bioRxiv. 2018. [DOI] [Full Text] |
| 17. | Kim RH, Nomikou S, Dawood Z, Jour G, Donnelly D, Moran U, Weber JS, Razavian N, Snuderl M, Shapiro R, Berman RS, Coudray N, Osman I, Tsirigos A. A Deep Learning Approach for Rapid Mutational Screening in Melanoma. bioRxiv. 2019. [DOI] [Full Text] |
| 18. | Cooper LA, Demicco EG, Saltz JH, Powell RT, Rao A, Lazar AJ. PanCancer insights from The Cancer Genome Atlas: the pathologist's perspective. J Pathol. 2018;244:512-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 124] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 19. | Kim J, Hong J, Park H. Prospects of deep learning for medical imaging. Precis Future Med. 2018;2:37-52. [RCA] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 20. | Cho KO, Lee SH, Jang HJ. Feasibility of fully automated classification of whole slide images based on deep learning. Korean J Physiol Pharmacol. 2020;24:89-99. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 21. | Venkatraman ES. A permutation test to compare receiver operating characteristic curves. Biometrics. 2000;56:1134-1138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 106] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 22. | Kwong LN, Dove WF. APC and its modifiers in colon cancer. Adv Exp Med Biol. 2009;656:85-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 184] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 23. | Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2345] [Cited by in RCA: 2382] [Article Influence: 108.3] [Reference Citation Analysis (0)] |
| 24. | Castellano E, Downward J. RAS Interaction with PI3K: More Than Just Another Effector Pathway. Genes Cancer. 2011;2:261-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 469] [Cited by in RCA: 557] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 25. | Chang YY, Lin PC, Lin HH, Lin JK, Chen WS, Jiang JK, Yang SH, Liang WY, Chang SC. Mutation spectra of RAS gene family in colorectal cancer. Am J Surg. 2016;212:537-544.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 26. | Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4616] [Cited by in RCA: 4464] [Article Influence: 120.6] [Reference Citation Analysis (0)] |
| 27. | Heinemann V, Stintzing S, Kirchner T, Boeck S, Jung A. Clinical relevance of EGFR- and KRAS-status in colorectal cancer patients treated with monoclonal antibodies directed against the EGFR. Cancer Treat Rev. 2009;35:262-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 144] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 28. | Zhao B, Wang L, Qiu H, Zhang M, Sun L, Peng P, Yu Q, Yuan X. Mechanisms of resistance to anti-EGFR therapy in colorectal cancer. Oncotarget. 2017;8:3980-4000. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 219] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 29. | Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci U S A. 2006;103:1475-1479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 352] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 30. | Stintzing S, Lenz HJ. A small cog in a big wheel: PIK3CA mutations in colorectal cancer. J Natl Cancer Inst. 2013;105:1775-1776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 31. | Cathomas G. PIK3CA in Colorectal Cancer. Front Oncol. 2014;4:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 96] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 32. | Xourafas D, Mizuno T, Cloyd JM. The impact of somatic SMAD4 mutations in colorectal liver metastases. Chin Clin Oncol. 2019;8:52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 33. | Fleming NI, Jorissen RN, Mouradov D, Christie M, Sakthianandeswaren A, Palmieri M, Day F, Li S, Tsui C, Lipton L, Desai J, Jones IT, McLaughlin S, Ward RL, Hawkins NJ, Ruszkiewicz AR, Moore J, Zhu HJ, Mariadason JM, Burgess AW, Busam D, Zhao Q, Strausberg RL, Gibbs P, Sieber OM. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 2013;73:725-735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 261] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 34. | Nakayama M, Oshima M. Mutant p53 in colon cancer. J Mol Cell Biol. 2019;11:267-276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 189] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 35. | Jauhri M, Bhatnagar A, Gupta S, Bp M, Minhas S, Shokeen Y, Aggarwal S. Prevalence and coexistence of KRAS, BRAF, PIK3CA, NRAS, TP53, and APC mutations in Indian colorectal cancer patients: Next-generation sequencing-based cohort study. Tumour Biol. 2017;39:1010428317692265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 36. | Conlin A, Smith G, Carey FA, Wolf CR, Steele RJ. The prognostic significance of K-ras, p53, and APC mutations in colorectal carcinoma. Gut. 2005;54:1283-1286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 185] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 37. | Jorissen RN, Christie M, Mouradov D, Sakthianandeswaren A, Li S, Love C, Xu ZZ, Molloy PL, Jones IT, McLaughlin S, Ward RL, Hawkins NJ, Ruszkiewicz AR, Moore J, Burgess AW, Busam D, Zhao Q, Strausberg RL, Lipton L, Desai J, Gibbs P, Sieber OM. Wild-type APC predicts poor prognosis in microsatellite-stable proximal colon cancer. Br J Cancer. 2015;113:979-988. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 38. | Deng Y, Wang L, Tan S, Kim GP, Dou R, Chen D, Cai Y, Fu X, Wang L, Zhu J, Wang J. KRAS as a predictor of poor prognosis and benefit from postoperative FOLFOX chemotherapy in patients with stage II and III colorectal cancer. Mol Oncol. 2015;9:1341-1347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 39. | Reggiani Bonetti L, Barresi V, Maiorana A, Manfredini S, Caprera C, Bettelli S. Clinical Impact and Prognostic Role of KRAS/BRAF/PIK3CA Mutations in Stage I Colorectal Cancer. Dis Markers. 2018;2018:2959801. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 40. | Mehrvarz Sarshekeh A, Advani S, Overman MJ, Manyam G, Kee BK, Fogelman DR, Dasari A, Raghav K, Vilar E, Manuel S, Shureiqi I, Wolff RA, Patel KP, Luthra R, Shaw K, Eng C, Maru DM, Routbort MJ, Meric-Bernstam F, Kopetz S. Association of SMAD4 mutation with patient demographics, tumor characteristics, and clinical outcomes in colorectal cancer. PLoS One. 2017;12:e0173345. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 41. | de Bruijne M. Machine learning approaches in medical image analysis: From detection to diagnosis. Med Image Anal. 2016;33:94-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 123] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 42. | Chang HY, Jung CK, Woo JI, Lee S, Cho J, Kim SW, Kwak TY. Artificial Intelligence in Pathology. J Pathol Transl Med. 2019;53:1-12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 102] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 43. | Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15:81-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1349] [Cited by in RCA: 2278] [Article Influence: 284.8] [Reference Citation Analysis (0)] |