Published online Sep 21, 2020. doi: 10.3748/wjg.v26.i35.5287
Peer-review started: May 10, 2020
First decision: July 29, 2020
Revised: August 3, 2020
Accepted: August 13, 2020
Article in press: August 13, 2020
Published online: September 21, 2020
Processing time: 129 Days and 17.5 Hours
Serum amyloid A1 (SAA1) is an acute-phase protein involved in acute or chronic hepatitis. Its function is still controversial. In addition, the effect of the expression of SAA1 and its molecular function on the progression in hepatocellular carcinoma (HCC) is still unclear.
To demonstrate the expression of SAA1 and its effect on the prognosis in HCC and explain further the correlation of SAA1 and immunity pathways.
SAA1 expression in HCC was conducted with The Cancer Genome Atlas-Liver Hepatocellular Carcinoma (TCGA-LIHC) in GEPIA tool, and the survival analysis based on the SAA1 expression level was achieved in the Kaplan-Meier portal. The high or low expression group was then drawn based on the median level of SAA1 expression. The correlation of SAA1 and the clinical features were conducted in the UALCAN web-based portal with TCGA-LIHC, including tumor grade, patient disease stage, and the TP53 mutation. The correlation analysis between SAA1 expression and TP53 mutation was subjected to the TCGA portal. The tumor purity score and the immune score were analyzed with CIBERSORT. The correlation of SAA1 expression and tumor-infiltrating lymphocytes was achieved in TISIDB web-based integrated repository portal for tumor-immune system interactions. GSE125336 dataset was used to test the SAA1 expression in the responsive or resistant group with anti-PD1 therapy. Gene set enrichment analysis was applied to evaluate the gene enrichment signaling pathway in HCC. The similar genes of SAA1 in HCC were identified in GEPIA, and the protein-protein interaction of SAA1 was conducted in the Metascape tool. The expression of C-X-C motif chemokine ligand 2, C-C motif chemokine ligand 23, and complement C5a receptor 1 was studied and overall survival analysis in HCC was conducted in GEPIA and Kaplan-Meier portal, respectively.
SAA1 expression was decreased in HCC, and lower SAA1 expression predicted poorer overall survival, progression-free survival, and disease-specific survival. Furthermore, SAA1 expression was further decreased with increased tumor grade and patient disease stage. Also, SAA1 expression was further downregulated in patients with TP53 mutation compared with patients with wild type TP53. SAA1 expression was negatively correlated with the TP53 mutation. Lower SAA1 predicted poorer survival rate, especially in the patients with no hepatitis virus infection, other than those with hepatitis virus infection. Moreover, the SAA1 expression was negatively correlated with tumor purity. In contrast, SAA1 expression was positively correlated with the immune score in HCC, and the correlation analysis between SAA1 expression and tumor-infiltrating lymphocytes also showed a positive correlation in HCC. Decreased SAA1 was closely associated with the immune tolerance of HCC. C-X-C motif chemokine ligand 2 and C-C motif chemokine ligand 23 genes were identified as the hub genes associated with SAA1, which could also serve as favorable prognosis markers for HCC.
SAA1 is downregulated in the liver tumor, and it is closely involved in the progression of HCC. Lower SAA1 expression indicates lower survival rate, especially for those patients without hepatitis virus infection. Lower SAA1 expression also suggests lower immune infiltrating cells, especially for those with immune cells exerting anti-tumor immune function. SAA1 expression is closely associated with the anti-tumor immune pathways.
Core Tip: In this study, we identified the downregulation of serum amyloid A1 (SAA1) in hepatocellular carcinoma (HCC). SAA1 expression could predict the favorable prognosis for HCC patients, especially for those patients without hepatitis virus infection. SAA1 expression was closely associated with anti-tumor immune signaling pathways. We also identified two signature genes associated with SAA1, suggesting a favorable prognosis function of SAA1 for HCC patients.
- Citation: Zhang W, Kong HF, Gao XD, Dong Z, Lu Y, Huang JG, Li H, Yang YP. Immune infiltration-associated serum amyloid A1 predicts favorable prognosis for hepatocellular carcinoma. World J Gastroenterol 2020; 26(35): 5287-5301
- URL: https://www.wjgnet.com/1007-9327/full/v26/i35/5287.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i35.5287
Serum amyloid A1 (SAA1) is a kind of acute-phase protein that is also a member of the coding gene of serum amyloid A (SAA) protein, which consists of SAA2, SAA3, and SAA4[1,2]. SAA1, SAA2, and SAA3 contribute to the acute phase response, and SAA4 expression is constitutively expressed[3]. At present, SAA protein is regarded as the most important biomarker in acute inflammation and tissue injury, and it could serve as a marker for viral or bacterial infection. The sensitivity of SAA1 for inflammatory infection diagnosis is superior to C-reaction protein, which was confirmed in the diagnosis of several diseases, such as tuberculosis, lepriasis, Crohn’s disease, and rheumatoid arthritis[4-6]. In recent years, the importance of SAA in the malignant tumor has attracted much more attention. Some studies reported that serum SAA is a biomarker for some solid tumors, such as stomach, colon, pancreas, breast, and lung cancers, and that elevated SAA is also correlated with tumor stage[7-10]. SAA protein is mainly synthesized by the liver, and some literature has revealed its expression changes in liver diseases, such as liver injury and virus infection[11]. However, the expression and clinical correlation of SAA in hepatocellular carcinoma (HCC) are still unclear. SAA1, which is an important preproprotein of SAA, should also be examined. Thus, this study mainly focused on the significance of SAA1 in HCC.
HCC is a major malignant tumor of the digestive system with a high lethality rate, especially in Asians[12]. Clinical data have shown that hepatitis virus infection could be an important reason for the high frequency of HCC in Asians[13,14]. The clinical treatment for HCC includes chemotherapy, radiotherapy, immunotherapy, and interventional treatment[15]. The representative chemotherapy drug is sorafenib, from which some patients benefit in prolonging survival. However, clinical practice also revealed the true frequency of drug resistance, which contributed to the poor outcome of HCC patients[16]. The complex tumor microenvironment and side effects also interfered with the effect of radiotherapy or interventional therapy[17]. In recent years, immune-based treatments have attracted much attention. Immune checkpoint blockade therapy has been widely conducted in clinical trials (e.g., the PD-1 blockade antibody[18]) with unsatisfactory outcomes. Considering the significance of the anti-PD1 antibody in other tumors, some researchers hypothesized that immune tolerance might contribute to the poor outcome. Researchers had been trying to explore solutions to overcome immune tolerance. Driving immune cytotoxic cells to kill tumor cells could be an encouraging strategy.
In this study, SAA1 expression was decreased in HCC, and its lower expression represented poorer prognosis, especially in those without hepatitis virus infection, suggesting that immune signaling pathways might be involved in SAA1-mediated HCC progression. The molecular mechanism exploration also confirmed the close association between SAA1 and immune tolerance. In summary, downregulation of SAA1 in HCC may be a candidate target for HCC therapy, especially in the practice of anti-tumor immunity.
The expression of SAA1, C-X-C motif chemokine ligand 2 (CXCL2), C-C motif chemokine ligand 23 (CCL23), and complement C5a receptor 1 (C5AR1) in the HCC tumor tissues and normal liver tissues was conducted in the GEPIA portal (http://gepia2.cancer-pku.cn/#index)[19]. Normal liver tissues were comprised of surrounding non-tumor and genotype-tissue expression liver tissues. The correlation of SAA1 with the clinical features (e.g., tumor grade, patient disease stage, and TP53 mutation) was subjected to UALCAN (http://ualcan.path.uab.edu/index.html)[20]. The correlation of SAA1 expression with the frequency of TP53 mutation was conducted in Liver Hepatocellular Carcinoma (LIHC) of The Cancer Genome Atlas (TCGA) portal (http://www.tcgaportal.org).
The overall survival (OS), progression-free survival (PFS), recurrence-free survival (RFS), and disease-specific survival (DSS) were subjected to Kaplan Meier plotter (http://kmplot.com/analysis/)[21]. The portal can assess the effect of SAA1 on the survival rate in HCC. The TCGA samples and gene expression omnibus (GEO) series samples were included in the liver tumor of the Kaplan Meier plotter portal. The standard level of high or low expression group was achieved by the best cutoff, which was between the lower and upper quartiles, and the best performing threshold was used as a cutoff.
The correlation between SAA1 expression and the tumor purity of HCC was measured in TIMER (http://timer.cistrome.org/)[22], a comprehensive resource for systematic analysis of immune infiltrates. The interaction between SAA1 and the immune system was assessed in TISIDB[23], an integrated repository portal for tumor-immune system interactions. In this study, the transcriptomics data of SAA1 and clinical data of LIHC from TCGA were included to evaluate the correlation, elucidating the potential interaction between SAA1 and the immune system.
The enrichment analysis of the Kyoto encyclopedia of genes and genomes pathways (KEGG) was conducted with gene set enrichment analysis (GSEA)[24]. The standard of sample clustering was based on the median level of SAA1 expression. The number of permutations was set as 1000, and the enrichment statistic was weighted. The Signal2Nosie was used as a metric for ranking genes. The minimum sets were above 15 genes. The normalized enrichment score (NES) was used to evaluate the enrichment intensity of SAA1 in indicated pathways. A P value < 0.05 and a false discovery rate < 0.05 for an enrichment gene sets were considered as the statistically significant.
GSE125336 data set was from GEO, a public functional genomics data repository portal. The sequencing data were downloaded via the SangerBox tool (https://shengxin.ren/). This dataset was collected from the GPL21103 platform (Illumina Hiseq 4000).
Similar genes that have a similar expression pattern with SAA1 in HCC were obtained from GEPIA. The top 300 similar genes were subjected to protein-protein interaction (PPI) analysis in Metascape[25].
The data were shown as mean ± standard deviation, and the difference between the two groups was determined by student’s t-test. The correlation analysis was assessed by the Spearman method. The log-rank method was used to perform survival analysis. P value < 0.05 was considered a significant statistical difference.
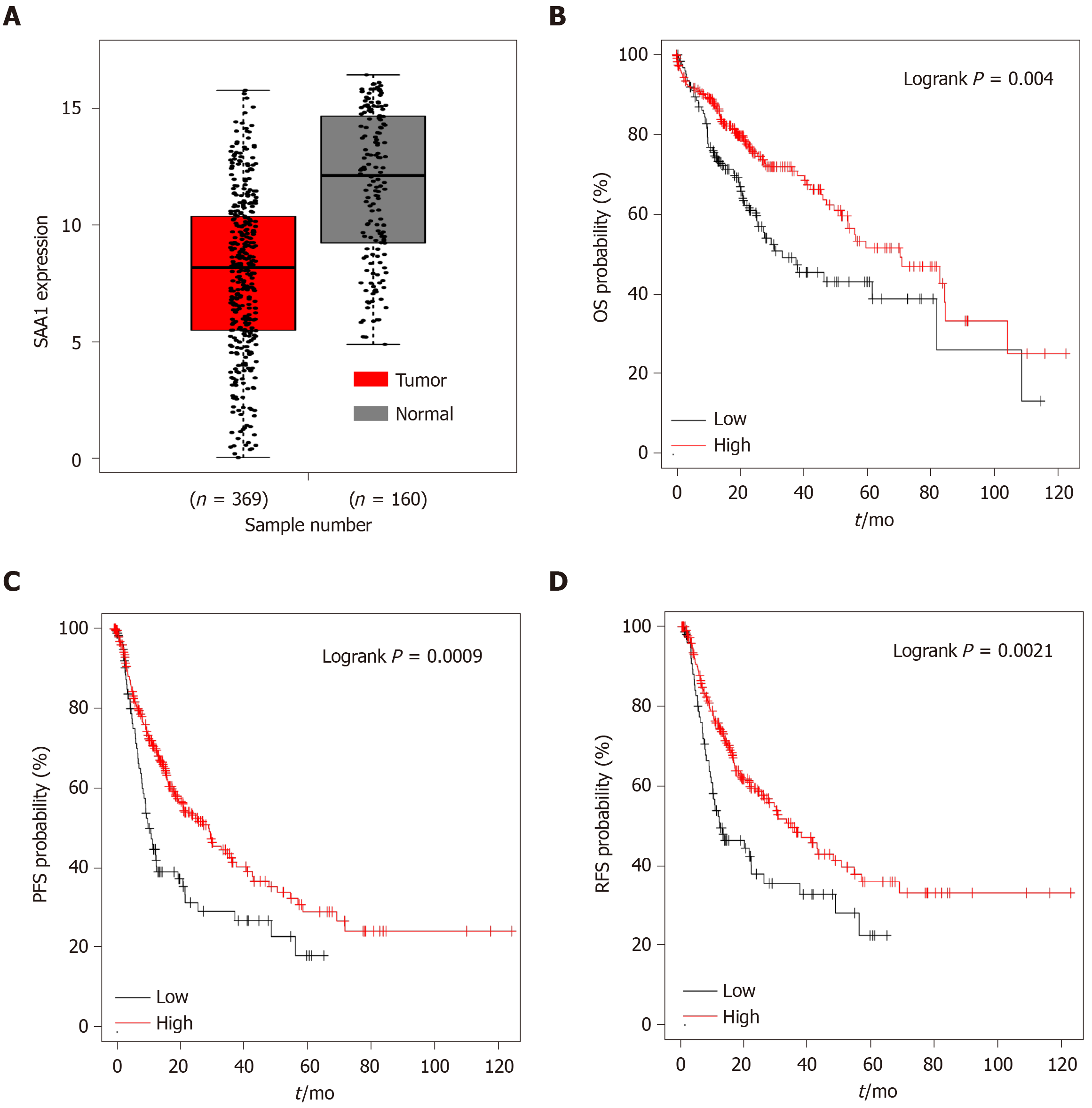
SAA1 as an acute-phase protein, which was reported to be involved in the regulation of pro-inflammation or anti-inflammation signaling, is controversial[4,26]. Furthermore, the expression of SAA1 in HCC was also unclear. Firstly, we evaluated the expression of SAA1 in HCC and found that SAA1 expression was decreased in liver tumor tissues compared with normal liver tissues (Figure 1A). Then, to determine the clinical significance of the downregulated SAA1 in HCC, we conducted survival analysis. As shown in Figure 1B-D, the patients with lower SAA1 showed poorer survival rates from OS, PFS, and RFS analysis. In detail, the median survival time of HCC patients with low SAA1 expression was significantly shorter than those with high SAA1 expression (OS: 33.5 mo vs 70.5 mo; PFS: 11.33 mo vs 29.3 mo; DSS: 81.87 mo vs 84.83 mo; RFS: 11.97 mo vs 34.4 mo). The Cox proportional hazards model also showed favorable prognosis of low SAA1 expression, and the hazard ratio value with 95% confidence interval (CI) of low SAA1 expression in OS, PFS, DSS, and RFS was 0.6 (0.43-0.85), 0.59 (0.43-0.81), 0.59 (0.37-0.91), 0.58 (0.41-0.83) respectively (Table 1). These results demonstrated that the decreased level of SAA1 in HCC could serve as a good prognostic biomarker for HCC patients.
| Survival analysis | Median time in mo | HR (95%CI) | P value | |
| SAA1 high | SAA1 low | |||
| OS | 70.5 | 33.5 | 0.6 (0.43-0.85) | 0.004 |
| PFS | 29.3 | 11.33 | 0.59 (0.43-0.81) | 0.00091 |
| DSS | 84.73 | 81.87 | 0.59 (0.37-0.91 | 0.017 |
| RFS | 34.4 | 11.97 | 0.58 (0.41-0.83) | 0.0021 |
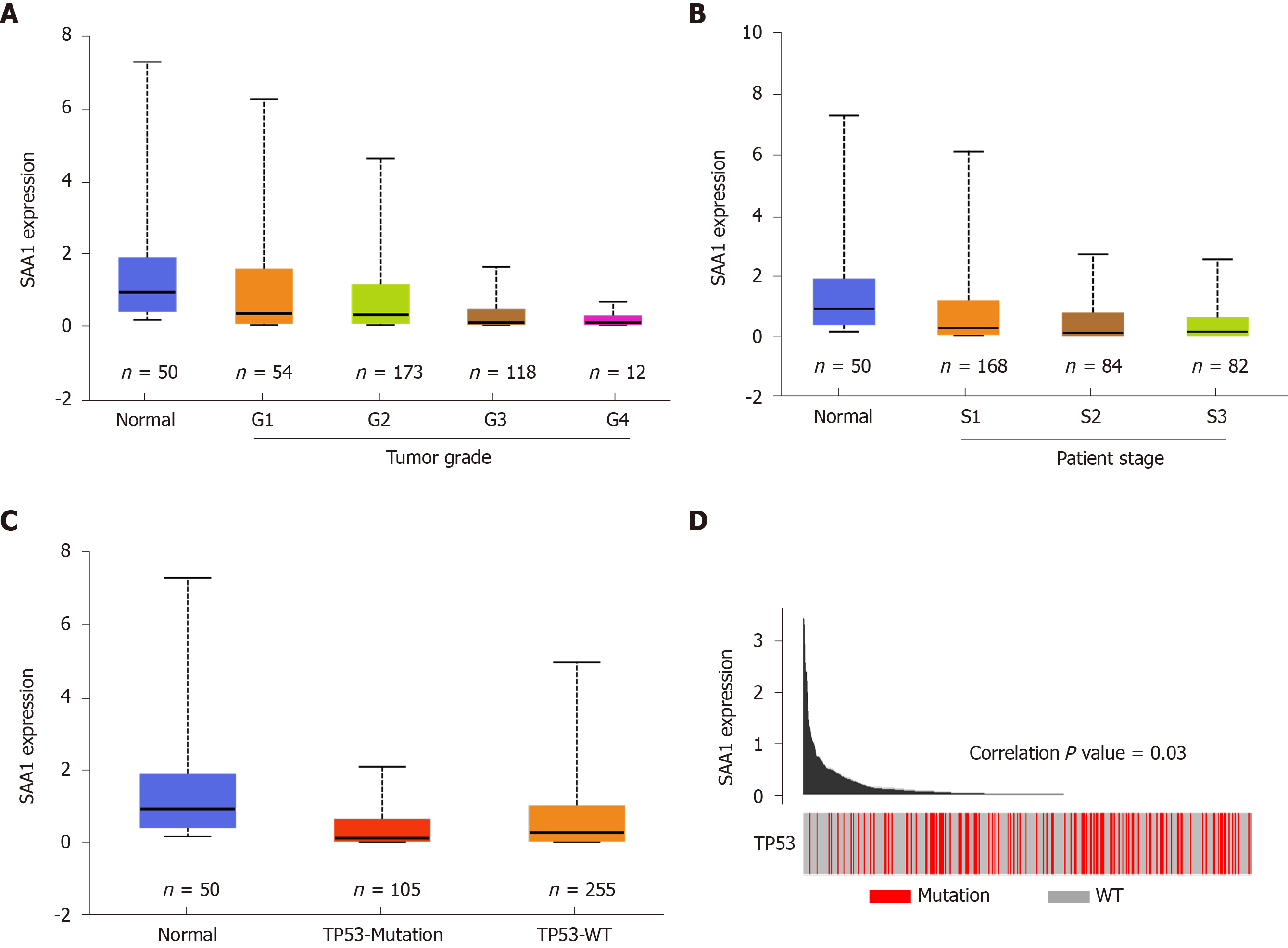
SAA1 expression is downregulated in the liver tumor tissues, as above demonstrated, and the clinical significance of SAA1 in survival time was also confirmed, as shown in Figure 1. To understand further the expression of SAA1 in the development of HCC, we analyzed SAA1 expression in some clinical features, such as tumor grade, patient disease stage, and the TP53 mutation or not. As shown in Figure 2A, SAA expression was further decreased with the increase of tumor grade. Similarly, SAA1 expression was also decreased with increased patient disease stage (Figure 2B), suggesting that SAA1 was closely involved in the progression of HCC. Moreover, TP53 mutation is an important risk factor contributing to the poor prognosis in HCC[27]. Thus, this study included the TP53 mutation in the analysis of SAA1 in HCC, suggesting interestingly that SAA1 expression was further decreased in tumor tissues with TP53 mutation than those without TP53 mutation (Figure 2C). The correlation analysis between SAA1 expression and the frequency of TP53 mutation also showed that lower SAA1 expression was accompanied by the high incidence of TP53 mutation (P < 0.05, Figure 2D).
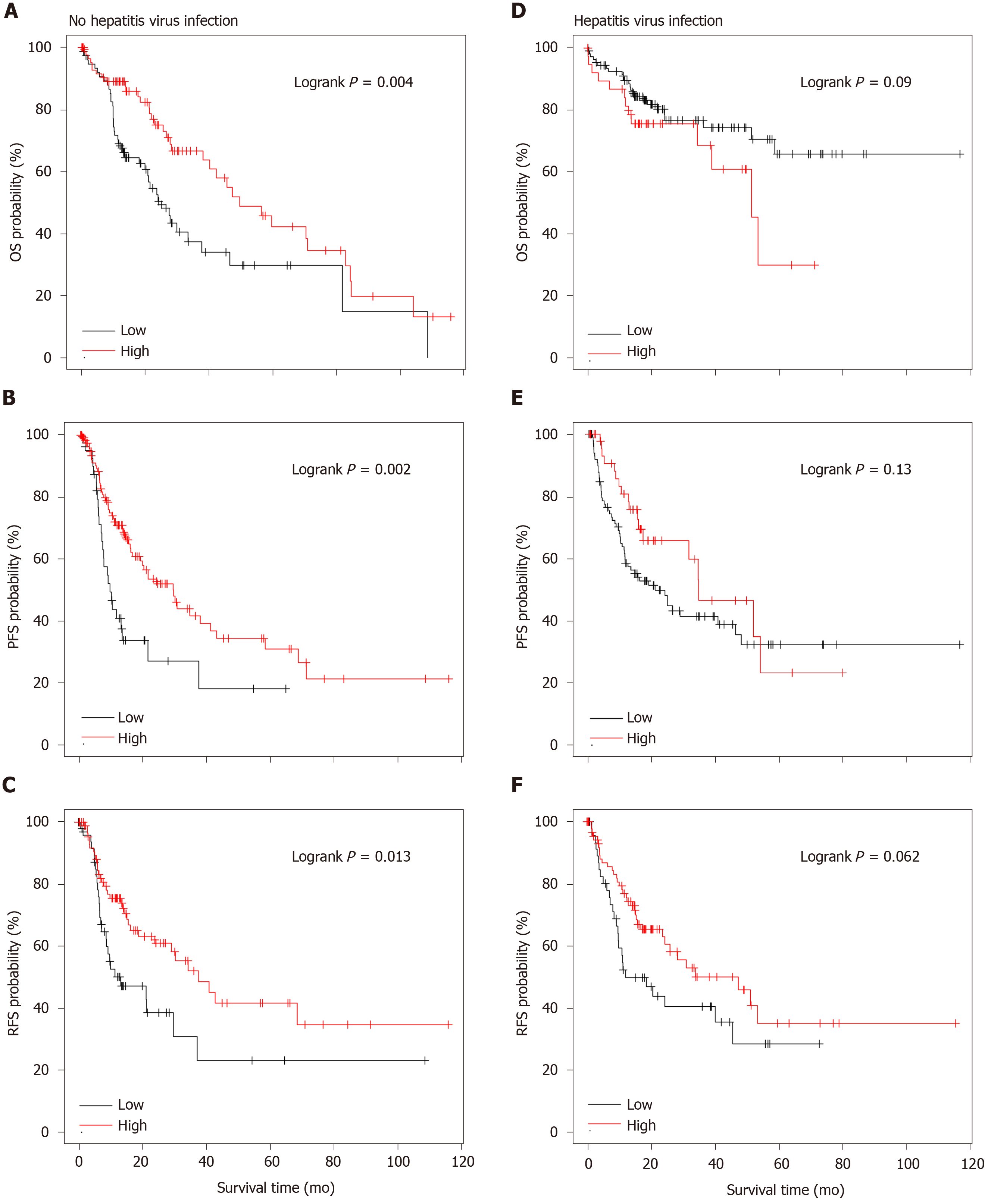
As shown in Figure 1, SAA1 could be a favorable prognostic biomarker for HCC. Considering the importance of SAA1 in the regulation of inflammation and the interaction between hepatitis virus infection and tumor in HCC, we evaluated the potential application of SAA1 as a prognostic biomarker in HCC patients with and without hepatitis virus infection. The patients with high SAA1 presented with a good possibility to survive in OS, PFS, and RFS analysis (Figure 3). On the contrary, there was no significant difference in the HCC patients with hepatitis virus infection (Figure 3D-F), suggesting that hepatitis virus infection could affect the value of SAA1 in the prediction of HCC prognosis.
SAA1 was downregulated in HCC, and the decreased SAA1 expression could predict the poor prognosis of HCC patients, especially in patients without hepatitis virus infection (Figure 3). However, there was no statistical significance in those patients with hepatitis virus infection. As an inflammation-responsive gene, some reports confirmed the expression difference of SAA1 in the physiological response, and the hepatitis virus infection was the most important driving factor in the development of HCC. Thus, it was easy to understand the difference of SAA1 as a prognosis factor in HCC patients with and without hepatitis virus infection.
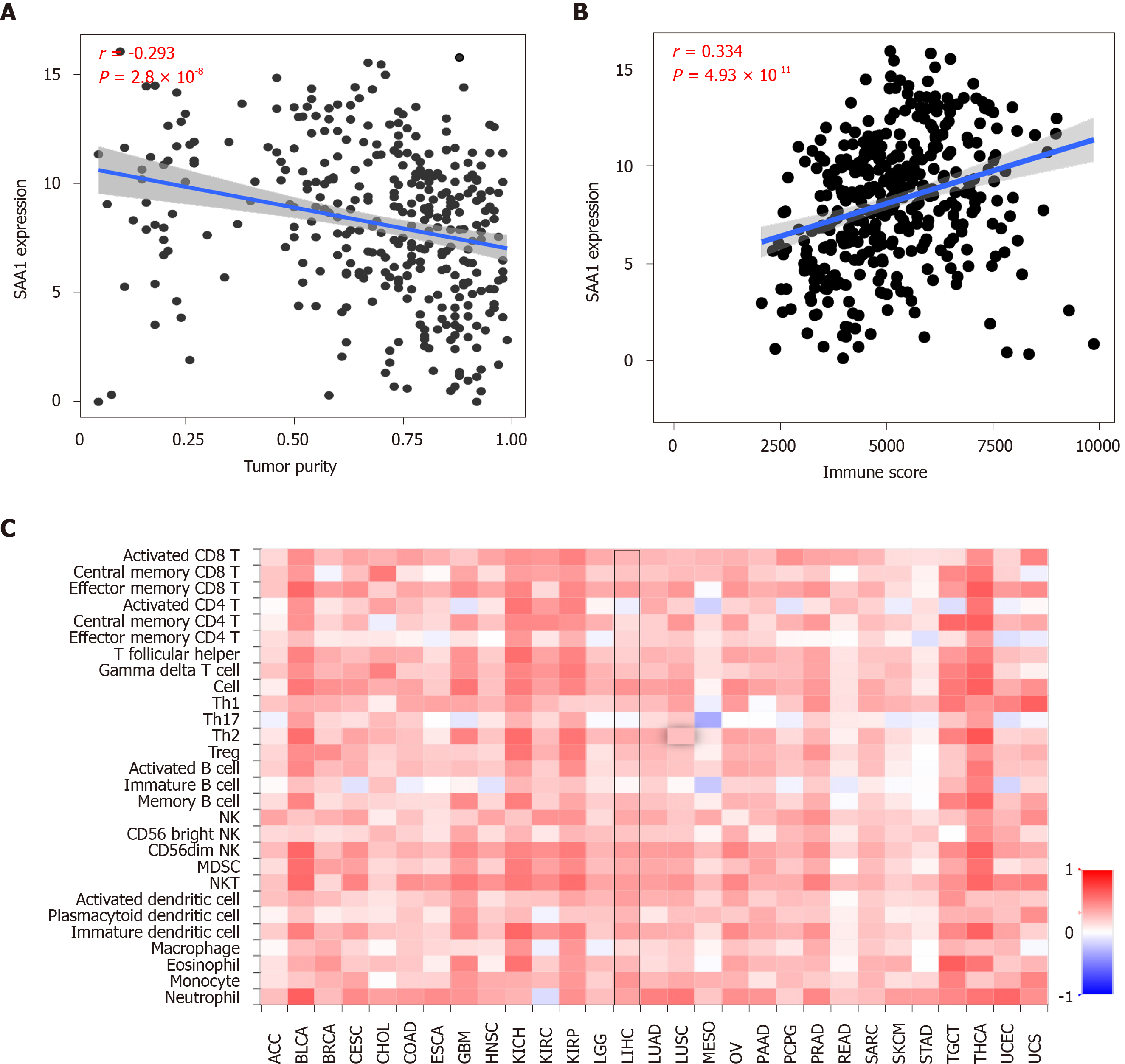
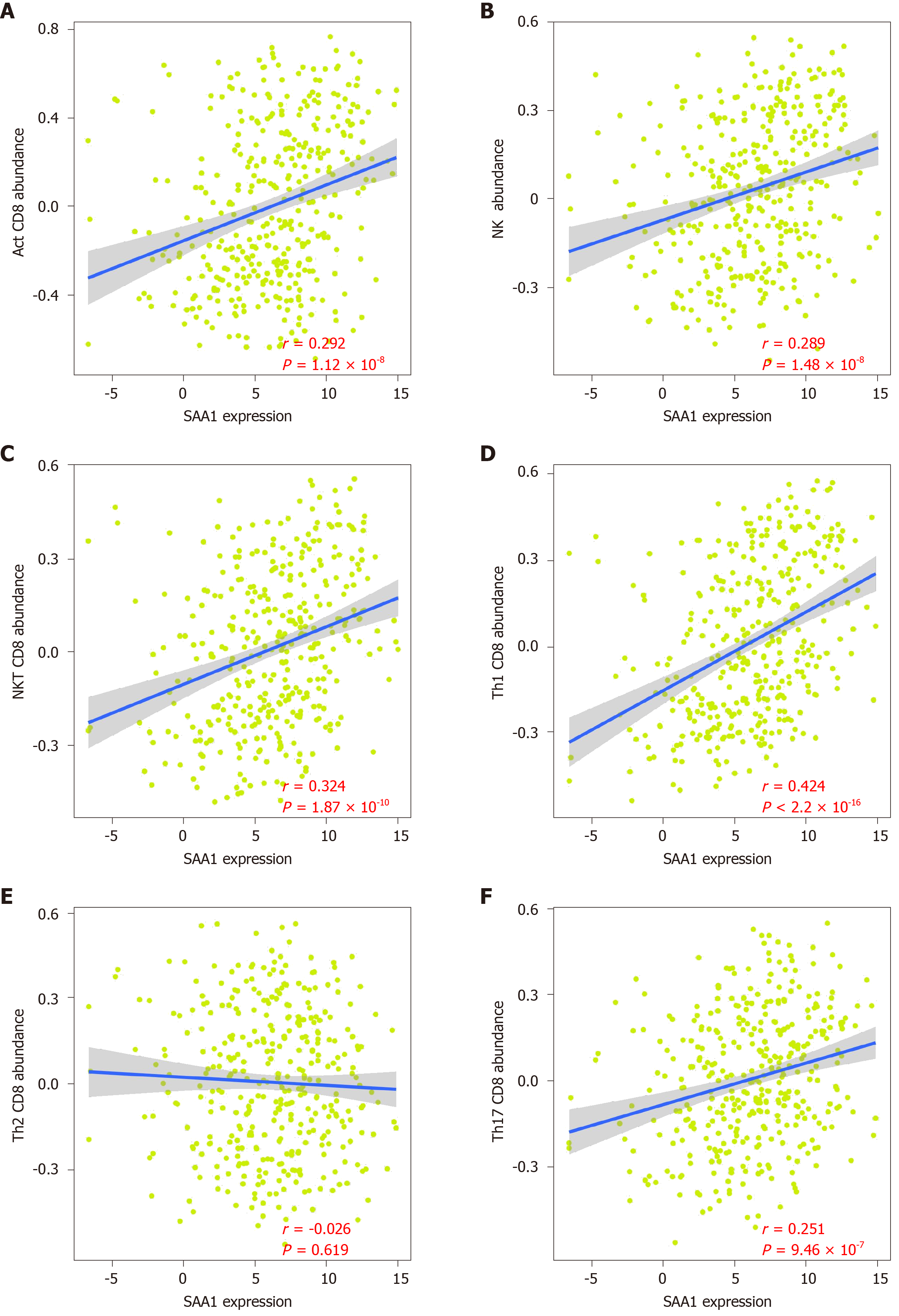
As attention increases regarding immune regulation in the development of HCC, we further evaluated the importance of SAA1 in the tumor immunomodulation. Firstly, the correlation of SAA1 expression and tumor purity was analyzed, and the negative correlation between SAA1 and tumor purity was confirmed (Figure 4A), which was also consistent with the downregulated SAA1 expression in HCC (as mentioned in Figure 1). Moreover, the CIBERSORT method was applied to evaluate the correlation between SAA1 and immune score. The results showed a significant positive correlation (P < 4.93 × 10-11, Figure 4B), suggesting that the lower expression of SAA1 in HCC was followed with a low immune score. Next, this study analyzed the correlation between SAA1 and 27 kinds of tumor-infiltrating lymphocytes (TILs) across human pan-cancers. As shown in Figure 4C, SAA1 expression was widely positively correlated with TILs in many human cancer types, especially in bladder urothelial carcinoma, glioblastoma multiforme, kidney chromophobe, kidney renal papillary cell carcinoma, testicular germ cell tumor, and thyroid carcinoma. The positive correlation between SAA1 expression and TILs was also observed in LIHC. SAA1 was remarkably correlated with the abundance of activated CD8 T cells (r = 0.292, P = 1.12 × 10-8, Figure 5A), natural killer (NK) cells (r = 0.289, P = 1.48 × 10-8, Figure 5B), natural killer T (NKT) cells (r = 0.324, P = 1.87 × 10-10, Figure 5C), T helper (Th)1 cells (r = 0.424, P = 2.2 × 10-16, Figure 5D), and Th17 cells (r = 0.251, P = 9.46 × 10-7, Figure 5F), but there was no significant correlation between SAA1 and the abundance of Th2 cells (P = 0.619, Figure 5E).
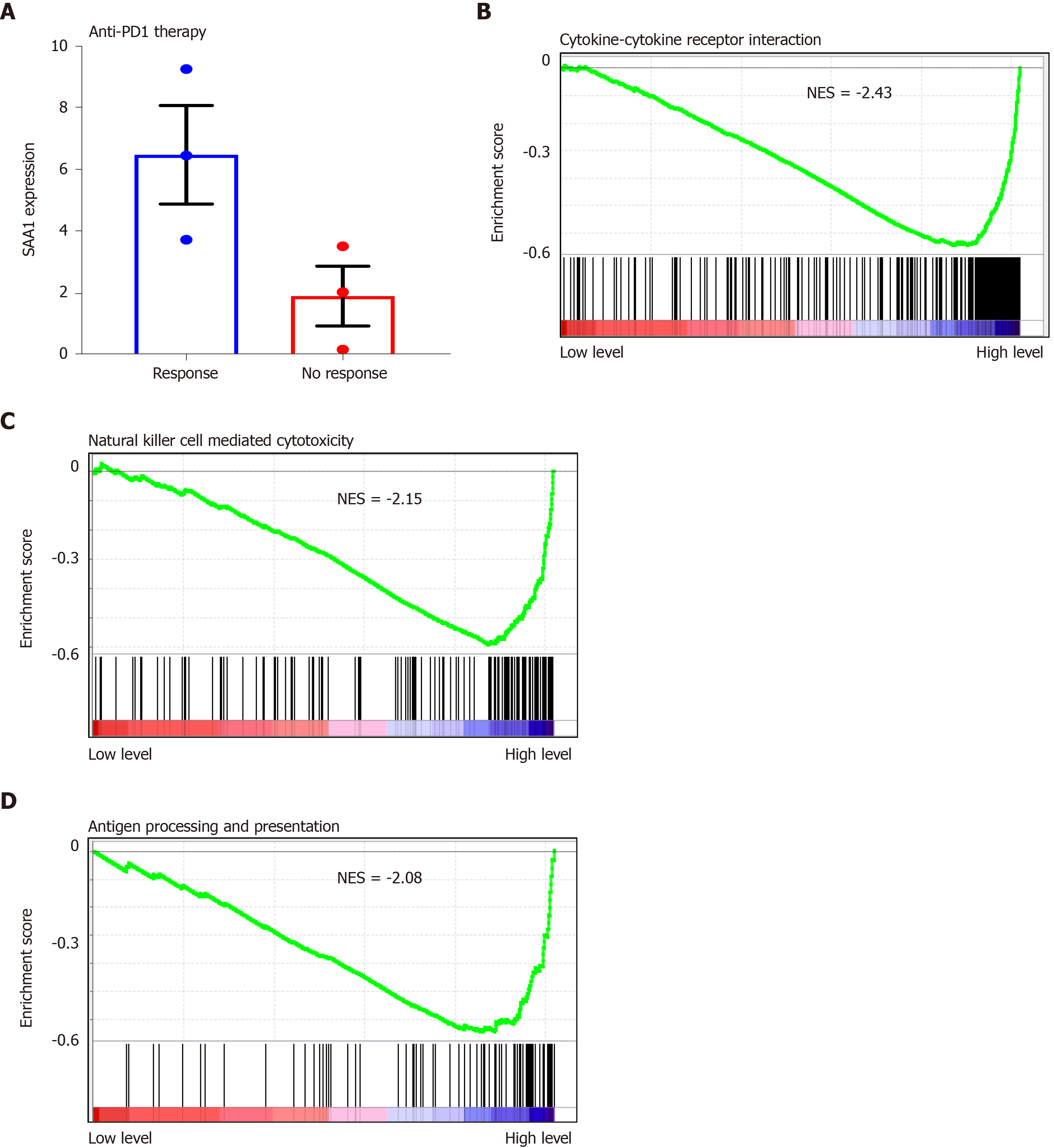
The decreased SAA1 expression was followed by lower immune score, especially with the lower cytotoxic T cell infiltration in the HCC, suggesting that SAA1 might contribute to the immune tolerance of HCC. To explain the hypothesis, the GSE125336 dataset was included, and the data showed that SAA1 expression was decreased in the patient group resistant to anti-PD1 therapy than those responsive to anti-PD1 therapy (Figure 6A). The GSEA was included to evaluate the significant pathways of SAA1 in LIHC. The results showed that the most enriched pathways related to decreased SAA1 included cytokine-cytokine receptor interaction (NES = -2.43, Figure 6B), NK cell-mediated cytotoxicity (NES = -2.15, Figure 6C), and antigen processing and presentation (NES = -2.08, Figure 6D).
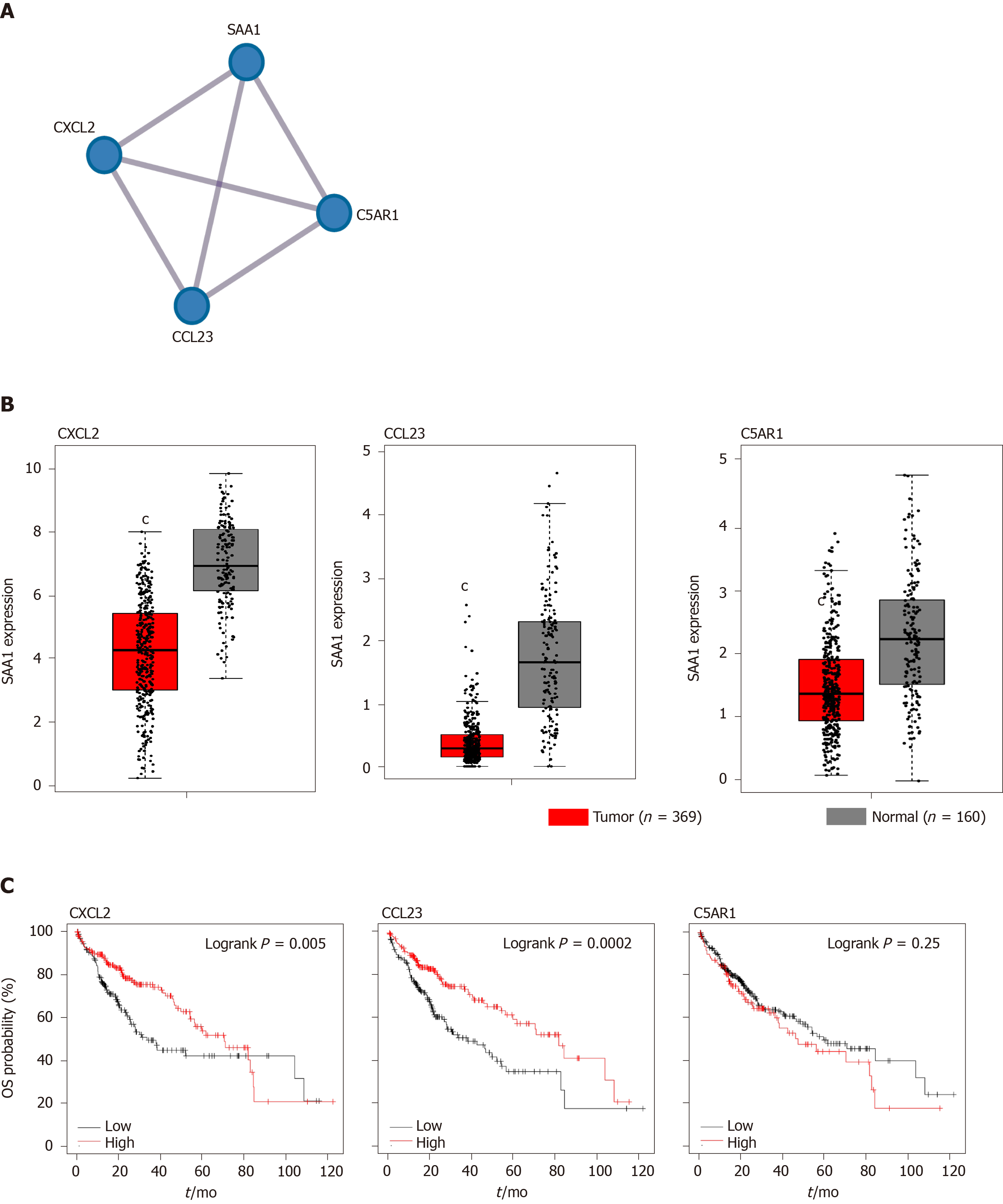
The gene enrichment analysis showed that higher SAA1 was enriched in the anti-tumor immunity pathways, including cytokine-cytokine receptor interaction, NK cell-mediated cytotoxicity, and antigen processing and presentation (Figure 6). High SAA1 predicted the favorable prognosis for HCC patients. To evaluate further the importance of SAA1 in the prognosis of HCC, we first conceived the similar genes of SAA1, which were analyzed with PPI analysis. As Figure 7A shows, SAA1 closely interacted with CXCL2, CCL23, and C5AR1. To determine the role of the three associated genes in HCC, their expression levels were assessed (Figure 7B). CXCL2, CCL23, and C5AR1 were all decreased in HCC, which was similar to SAA1. Furthermore, the results of the overall survival analysis of the three genes are presented in Figure 7C and show that higher CXCL2 or CCL23 predicted better survival time. However, there was no clinical value of C5AR1 in the prediction of prognosis for HCC. Taken together, these results indicated that CXCL2 and CCL23, as an interacted protein of SAA1, could be complementary prognostic biomarkers for HCC patients.
SAA1, as the most important preproprotein of SAA, is an acute-phase protein involved in viral and bacterial infection, autoimmune disease, and some tumor pathogenesis[28]. To date, most SAA1 research focuses on its application as a disease marker. As an inflammation-related gene, SAA1 has been reported as a biomarker for the detection of stroke, ankylosing spondylitis, and acute aortic and acute hepatic injury[29-31]. However, whether SAA1 exerts * pro-inflammatory or anti-inflammatory role is still controversial. Lee et al[4] reported that SAA1 protein could promote inflammatory intestinal disease by inducing pro-inflammatory Th17 cell differentiation[4]. On the contrary, Cheng et al[26] identified that SAA1 could decrease lipopolysaccharide (LPS)-induced intestinal inflammation by directly binding to LPS to form a complex and induce LPS clearance by macrophage.
Similar to the inflammatory disease, the biological functions of SAA1 in tumors are different. SAA1 was reported to be overexpressed in ovarian and renal cell carcinoma[32,33]; and serum SAA1 expression is positively correlated with the development of melanoma. Besides, SAA1 induced interleukin-10 production in neutrophils from melanoma patients, suggesting that SAA1 is a negative prognostic marker in melanoma[34]. In HCC, SAA1 was identified as the hub gene in the PPI analysis of differentially expressed genes[35], with unclear expression.
Hence, this study aimed to evaluate the expression of SAA1 and its clinical significance in HCC. Firstly, we confirmed the downregulated expression of SAA1 in HCC tumor tissues (Figure 1A). The correlation analysis between SAA1 expression and tumor purity further confirmed its decreased expression in HCC tumor tissues (Figure 4A). Moreover, SAA1 expression decreased with increased tumor grade and disease stage (Figure 2A and B); and the lower SAA1 expression was accompanied with the higher frequency of TP53 mutation (Figure 2C and D), a marker of poor prognosis in HCC. The above results suggested that decreased SAA1 expression was closely involved in the progression of HCC. In the further study of clinical significance, the effect of SAA1 on the survival rate revealed that the lower SAA1 expression in HCC predicted worse survival time (Figure 1B-D), especially in HCC patients without hepatitis virus infection (Figure 3).
Considering the induction of SAA1 in hepatitis by virus infection or tissue injury, the hepatitis virus infection-induced SAA1 expression could interfere with the prognostic value of SAA1 in HCC. This data also indicated that the immune signaling pathways might be closely associated with SAA1-mediated HCC progression. Interestingly, the immune infiltrating analysis showed that lower SAA1 expression represented lower immune score and immune cells infiltration (Figure 4B and C), especially in the cytotoxic T cells and anti-tumor associated immune cells, including activated CD8 T, NK, NKT, Th1, and Th17 cells (Figure 5). In the exploration of potential molecular mechanisms, the lower SAA1 might contribute to immune tolerance (Figure 6), which could be a potential therapeutic target for enhanced anti-tumor immunity. More significantly, this study also identified two signature genes that interacted with SAA1 as compensative prognostic biomarkers (Figure 7), which could enhance the prognostic value of SAA1 in HCC.
In summary, this study identified the downregulated expression of SAA1 as a potential prognostic biomarker for HCC, and decreased SAA1 was closely associated with immune tolerance signaling pathways. SAA1 could be an encouraging drug target for anti-tumor immunity. More experiments should be conceived to confirm the significance of SAA1 in HCC immunotherapy.
Serum amyloid A1 (SAA1) is regarded as an important regulator in the immune network. Recently, SAA1 was reported to regulate the development of some cancers, and it may function as a biomarker for some cancers.
SAA1 is a potential biomarker in some cancers, but its expression and function in hepatocellular carcinoma (HCC) are still unclear.
The project was designed to determine the expression level of SAA1 in HCC and to analyze the association between SAA1 expression and prognosis of HCC patient and its potential regulation on the immune network.
GEPIA web-based analytical tool was subjected to evaluate the expression of SAA1 in HCC. The patients from The Cancer Genome Atlas-Liver Hepatocellular Carcinoma (TCGA-LIHC) were sub-grouped according to the median expression level of SAA1. Then, the Kaplan-Meier portal was used to analyze the survival curve of the high or low SAA1 expression groups. UALCAN tool was used to evaluate the expression of SAA1 in different tumor grades, stages, and TP53 mutation or not. The CIBERSORT method was subjected to test the correlation between SAA1 expression and immune infiltration score in HCC. TISIDB integrated portal was conducted to reveal the association between SAA1 level and the tumor-infiltrating lymphocytes. GSE125336 dataset was subjected to analyze the SAA1 level according to the anti-PD1 response. Gene set enrichment analysis method was subjected to analyze the enriched signaling pathways based on SAA1 in HCC. The co-expression genes of SAA1 was subjected to Metascape to evaluate the hub genes. These hub genes were subjected to GEPIA and Kaplan-Meier to analyze the expression and overall survival.
SAA1 level was downregulated in the liver tumor, and the lower expression could function as a prognostic biomarker in overall survival, progression-free survival, and disease-specific survival of HCC. Besides, the SAA1 expression level was closely associated with tumor grades and patient stages. More interestingly, the HCC patients with TP53 mutation showed a lower expression of SAA1. SAA1 could act as a good prognostic marker in HCC patients without hepatitis infection. SAA1 expression was positively correlated with the immune infiltration score and tumor-infiltrating lymphocytes. Low SAA1 expression was negatively correlated with anti-immune signaling, including cytokine-cytokine receptor interaction, natural killer cell-mediated cytotoxicity, and antigen processing and presentation. CXCL2 and CCL23 were identified as the hub genes that interacted with SAA1 and acted as prognostic markers for HCC.
SAA1 expression is low in HCC, and its expression is closely associated with the progression of HCC. Besides, SAA1 can act as a poor prognostic biomarker for HCC patients. More interestingly, SAA1 is closely involved in the regulation of the immune infiltrating process.
In this study, SAA1 was identified as a negative regulator for HCC, and its expression might be a poor prognostic biomarker for HCC patients. Interestingly, the SAA1 expression was closely related to the tumor-infiltrating immune cells network. However, these findings were based on the expression levels. SAA1 is a secreted protein from the liver, and the secreted levels could be much more practical clinically, especially in the prognosis analysis.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sitkin S S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Li JH
| 1. | Kuret T, Sodin-Šemrl S, Mrak-Poljšak K, Čučnik S, Lakota K, Erman A. Interleukin-1β Induces Intracellular Serum Amyloid A1 Expression in Human Coronary Artery Endothelial Cells and Promotes its Intercellular Exchange. Inflammation. 2019;42:1413-1425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 2. | Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, Lee JY, Ziel JW, Miraldi ER, Domingos AI, Bonneau R, Littman DR. An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell. 2015;163:381-393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 423] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 3. | Kovacevic A, Hammer A, Stadelmeyer E, Windischhofer W, Sundl M, Ray A, Schweighofer N, Friedl G, Windhager R, Sattler W, Malle E. Expression of serum amyloid A transcripts in human bone tissues, differentiated osteoblast-like stem cells and human osteosarcoma cell lines. J Cell Biochem. 2008;103:994-1004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 4. | Lee JY, Hall JA, Kroehling L, Wu L, Najar T, Nguyen HH, Lin WY, Yeung ST, Silva HM, Li D, Hine A, Loke P, Hudesman D, Martin JC, Kenigsberg E, Merad M, Khanna KM, Littman DR. Serum Amyloid A Proteins Induce Pathogenic Th17 Cells and Promote Inflammatory Disease. Cell. 2020;180:79-91.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 283] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 5. | D'Haens G, Kelly O, Battat R, Silverberg MS, Laharie D, Louis E, Savarino E, Bodini G, Yarur A, Boland BS, Afif W, Li XJ, Hale M, Ho J, Kondragunta V, Huang B, Kuy C, Okada L, Hester KD, Bray KR, Mimms L, Jain A, Singh S, Collins A, Valasek MA, Sandborn WJ, Vermeire S, Dulai PS. Development and Validation of a Test to Monitor Endoscopic Activity in Patients With Crohn's Disease Based on Serum Levels of Proteins. Gastroenterology. 2020;158:515-526.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 73] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 6. | Yamada T, Okuda Y, Takasugi K, Itoh K, Igari J. Relative serum amyloid A (SAA) values: the influence of SAA1 genotypes and corticosteroid treatment in Japanese patients with rheumatoid arthritis. Ann Rheum Dis. 2001;60:124-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 7. | Wu DC, Wang KY, Wang SSW, Huang CM, Lee YW, Chen MI, Chuang SA, Chen SH, Lu YW, Lin CC, Lee KW, Hsu WH, Wu KP, Chen YJ. Exploring the expression bar code of SAA variants for gastric cancer detection. Proteomics. 2017;17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 8. | Liang B, Li C, Zhao J. Identification of key pathways and genes in colorectal cancer using bioinformatics analysis. Med Oncol. 2016;33:111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 107] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 9. | Djurec M, Graña O, Lee A, Troulé K, Espinet E, Cabras L, Navas C, Blasco MT, Martín-Díaz L, Burdiel M, Li J, Liu Z, Vallespinós M, Sanchez-Bueno F, Sprick MR, Trumpp A, Sainz B, Al-Shahrour F, Rabadan R, Guerra C, Barbacid M. Saa3 is a key mediator of the protumorigenic properties of cancer-associated fibroblasts in pancreatic tumors. Proc Natl Acad Sci USA. 2018;115:E1147-E1156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 132] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 10. | Ignacio RMC, Gibbs CR, Kim S, Lee ES, Adunyah SE, Son DS. Serum amyloid A predisposes inflammatory tumor microenvironment in triple negative breast cancer. Oncotarget. 2019;10:511-526. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 11. | Kluve-Beckerman B, Dwulet FE, Benson MD. Human serum amyloid A. Three hepatic mRNAs and the corresponding proteins in one person. J Clin Invest. 1988;82:1670-1675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 58] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 12. | Gu J, Zhang X, Miao R, Ma X, Xiang X, Fu Y, Liu C, Niu W, Qu K. A three-long non-coding RNA-expression-based risk score system can better predict both overall and recurrence-free survival in patients with small hepatocellular carcinoma. Aging (Albany NY). 2018;10:1627-1639. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 13. | Nahon P, Layese R, Bourcier V, Cagnot C, Marcellin P, Guyader D, Pol S, Larrey D, De Lédinghen V, Ouzan D, Zoulim F, Roulot D, Tran A, Bronowicki JP, Zarski JP, Riachi G, Calès P, Péron JM, Alric L, Bourlière M, Mathurin P, Blanc JF, Abergel A, Serfaty L, Mallat A, Grangé JD, Attali P, Bacq Y, Wartelle C, Dao T, Thabut D, Pilette C, Silvain C, Christidis C, Nguyen-Khac E, Bernard-Chabert B, Zucman D, Di Martino V, Sutton A, Roudot-Thoraval F, Audureau E; ANRS CO12 CirVir Group. Incidence of Hepatocellular Carcinoma After Direct Antiviral Therapy for HCV in Patients With Cirrhosis Included in Surveillance Programs. Gastroenterology. 2018;155:1436-1450.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 181] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 14. | Gil-García AI, Madejón A, Francisco-Recuero I, López-López A, Villafranca E, Romero M, García A, Olveira A, Mena R, Larrubia JR, García-Samaniego J. Prevalence of hepatocarcinoma-related hepatitis B virus mutants in patients in grey zone of treatment. World J Gastroenterol. 2019;25:5883-5896. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 5] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 15. | Baldan Ferrari G, Coelho França Quintanilha J, Berlofa Visacri M, Oliveira Vaz C, Cursino MA, Sampaio Amaral L, Brito Bastos, Pereira TT, de Oliveira Guarnieri JP, de Godoy Torso N, Passos Lima CS, Moriel P. Outcomes in hepatocellular carcinoma patients undergoing sorafenib treatment: toxicities, cellular oxidative stress, treatment adherence, and quality of life. Anticancer Drugs. 2020;31:523-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 16. | De Mattia E, Cecchin E, Guardascione M, Foltran L, Di Raimo T, Angelini F, D'Andrea M, Toffoli G. Pharmacogenetics of the systemic treatment in advanced hepatocellular carcinoma. World J Gastroenterol. 2019;25:3870-3896. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 55] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (1)] |
| 17. | Wang ZX, Li J, Wang EX, Xia DD, Bai W, Wang QH, Yuan J, Li XM, Niu J, Yin ZX, Xia JL, Fan DM, Han GH. Validation of the six-and-twelve criteria among patients with hepatocellular carcinoma and performance score 1 receiving transarterial chemoembolization. World J Gastroenterol. 2020;26:1805-1819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 4] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 18. | Voutsadakis IA. PD-1 inhibitors monotherapy in hepatocellular carcinoma: Meta-analysis and systematic review. Hepatobiliary Pancreat Dis Int. 2019;18:505-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 19. | Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5550] [Cited by in RCA: 7089] [Article Influence: 886.1] [Reference Citation Analysis (0)] |
| 20. | Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649-658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2365] [Cited by in RCA: 4228] [Article Influence: 528.5] [Reference Citation Analysis (0)] |
| 21. | Menyhárt O, Nagy Á, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci. 2018;5:181006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 343] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 22. | Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77:e108-e110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2728] [Cited by in RCA: 4093] [Article Influence: 511.6] [Reference Citation Analysis (0)] |
| 23. | Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, Chu KC, Wong CY, Lau CY, Chen I, Chan NW, Zhang J. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35:4200-4202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 561] [Cited by in RCA: 1437] [Article Influence: 287.4] [Reference Citation Analysis (0)] |
| 24. | Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, Rostamianfar A, Wadi L, Meyer M, Wong J, Xu C, Merico D, Bader GD. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc. 2019;14:482-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1112] [Cited by in RCA: 1203] [Article Influence: 200.5] [Reference Citation Analysis (0)] |
| 25. | Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3766] [Cited by in RCA: 8793] [Article Influence: 1465.5] [Reference Citation Analysis (0)] |
| 26. | Cheng N, Liang Y, Du X, Ye RD. Serum amyloid A promotes LPS clearance and suppresses LPS-induced inflammation and tissue injury. EMBO Rep. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 103] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 27. | Ge PL, Li SF, Wang WW, Li CB, Fu YB, Feng ZK, Li L, Zhang G, Gao ZQ, Dang XW, Wu Y. Prognostic values of immune scores and immune microenvironment-related genes for hepatocellular carcinoma. Aging (Albany NY). 2020;12:5479-5499. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 28. | Kramer HB, Lavender KJ, Qin L, Stacey AR, Liu MK, di Gleria K, Simmons A, Gasper-Smith N, Haynes BF, McMichael AJ, Borrow P, Kessler BM. Elevation of intact and proteolytic fragments of acute phase proteins constitutes the earliest systemic antiviral response in HIV-1 infection. PLoS Pathog. 2010;6:e1000893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 69] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 29. | Shridas P, De Beer MC, Webb NR. High-density lipoprotein inhibits serum amyloid A-mediated reactive oxygen species generation and NLRP3 inflammasome activation. J Biol Chem. 2018;293:13257-13269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 66] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 30. | Zapata-Arriaza E, Mancha F, Bustamante A, Moniche F, Pardo-Galiana B, Serrano-Gotarredona P, Navarro-Herrero S, Pallisa E, Faura J, Vega-Salvatierra Á, Penalba A, Escudero-Martínez I, Ramos-Herrero VD, Azurmendi L, Charles Sanchez J, Montaner J. Biomarkers predictive value for early diagnosis of Stroke-Associated Pneumonia. Ann Clin Transl Neurol. 2019;6:1882-1887. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 31. | Yuan ZY, Zhang XX, Wu YJ, Zeng ZP, She WM, Chen SY, Zhang YQ, Guo JS. Serum amyloid A levels in patients with liver diseases. World J Gastroenterol. 2019;25:6440-6450. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 15] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 32. | Urieli-Shoval S, Finci-Yeheskel Z, Dishon S, Galinsky D, Linke RP, Ariel I, Levin M, Ben-Shachar I, Prus D. Expression of serum amyloid a in human ovarian epithelial tumors: implication for a role in ovarian tumorigenesis. J Histochem Cytochem. 2010;58:1015-1023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 33. | Wood SL, Rogers M, Cairns DA, Paul A, Thompson D, Vasudev NS, Selby PJ, Banks RE. Association of serum amyloid A protein and peptide fragments with prognosis in renal cancer. Br J Cancer. 2010;103:101-111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 34. | De Santo C, Arscott R, Booth S, Karydis I, Jones M, Asher R, Salio M, Middleton M, Cerundolo V. Invariant NKT cells modulate the suppressive activity of IL-10-secreting neutrophils differentiated with serum amyloid A. Nat Immunol. 2010;11:1039-1046. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 253] [Cited by in RCA: 242] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 35. | Zhang Y, Wang S, Xiao J, Zhou H. Bioinformatics analysis to identify the key genes affecting the progression and prognosis of hepatocellular carcinoma. Biosci Rep. 2019;39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |