Published online Sep 21, 2020. doi: 10.3748/wjg.v26.i35.5256
Peer-review started: May 25, 2020
First decision: July 29, 2020
Revised: July 29, 2020
Accepted: August 12, 2020
Article in press: August 12, 2020
Published online: September 21, 2020
Processing time: 114 Days and 14.6 Hours
Esophageal cancer poses diagnostic, therapeutic and economic burdens in high-risk regions. Artificial intelligence (AI) has been developed for diagnosis and outcome prediction using various features, including clinicopathologic, radiologic, and genetic variables, which can achieve inspiring results. One of the most recent tasks of AI is to use state-of-the-art deep learning technique to detect both early esophageal squamous cell carcinoma and esophageal adenocarcinoma in Barrett’s esophagus. In this review, we aim to provide a comprehensive overview of the ways in which AI may help physicians diagnose advanced cancer and make clinical decisions based on predicted outcomes, and combine the endoscopic images to detect precancerous lesions or early cancer. Pertinent studies conducted in recent two years have surged in numbers, with large datasets and external validation from multi-centers, and have partly achieved intriguing results of expert’s performance of AI in real time. Improved pre-trained computer-aided diagnosis algorithms in the future studies with larger training and external validation datasets, aiming at real-time video processing, are imperative to produce a diagnostic efficacy similar to or even superior to experienced endoscopists. Meanwhile, supervised randomized controlled trials in real clinical practice are highly essential for a solid conclusion, which meets patient-centered satisfaction. Notably, ethical and legal issues regarding the black-box nature of computer algorithms should be addressed, for both clinicians and regulators.
Core Tip: Deep-learning-based artificial intelligence (AI) is a breakthrough technology that has been widely explored in diagnosis, treatment and prediction of esophageal cancer. Recent studies have dealt with limitations of previous researches, including small sample size, selection bias, lack of external validation and algorithm efficiency. Favorable outcomes that are comparable to experienced endoscopists have been achieved with satisfactory robustness, indicating a real-time potential. Future randomized controlled trials are needed to further address these issues concerning AI to provide an ultimate patient-centered satisfaction, in an interpretable, ethical and legal manner.
- Citation: Zhang YH, Guo LJ, Yuan XL, Hu B. Artificial intelligence-assisted esophageal cancer management: Now and future. World J Gastroenterol 2020; 26(35): 5256-5271
- URL: https://www.wjgnet.com/1007-9327/full/v26/i35/5256.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i35.5256
Esophageal cancer (EC) is one of the top ten leading prevalent malignancies worldwide, ranking the seventh in incidence and the sixth in mortality in 2018[1]. The major histological types are squamous cell carcinoma (SCC), which is predominant worldwide, and adenocarcinoma (AC) which is more prevalent in Caucasian people[2-4]. Data collected from 12 countries have indicated that AC will possibly experience a dramatic increase in incidence up to 2030, while the incidence of SCC will continuously decrease[5]. It is estimated that EC causes absolute years of life lost reduction of 7.8 (95%CI: 2.3-12.7)[6]. Although EC is not the most common cause of admission or readmission to hospital[7], it certainly imposes economic burdens. A cohort study conducted in the United Kingdom showed that the mean net costs of care per 30 patient-days of AC were $1016, $669, and $8678 for the initial phase, continuing care phase and terminal phase, respectively[8]. The cost grows with an increase of tumor node metastasis (TNM) staging at first diagnosis[8].
Apparently, EC is a serious health threat, imposing economic burden on both high-income and low-income countries. Therefore, early diagnosis and evidence-based expert opinions on selecting the optimal treatment modality are crucial for reducing such burden. Although various diagnostic methodologies [including endoscopic ultrasonography (EUS), chromoendoscopy, optical coherence tomography (OCT), high-resolution microendoscopy (HRM), confocal laser endomicroscopy (CLE), volumetric laser endomicroscopy (VLE), and positron emission tomography (PET)], serous and genetic predictors have been developed to improve diagnostic accuracy and predict outcomes, inter-observer variabilities in interpreting images and heavy workloads limit their clinical efficiency[9-11]. A practical tool that can improve accuracy and reduce workload is in urgent need for clinical practice.
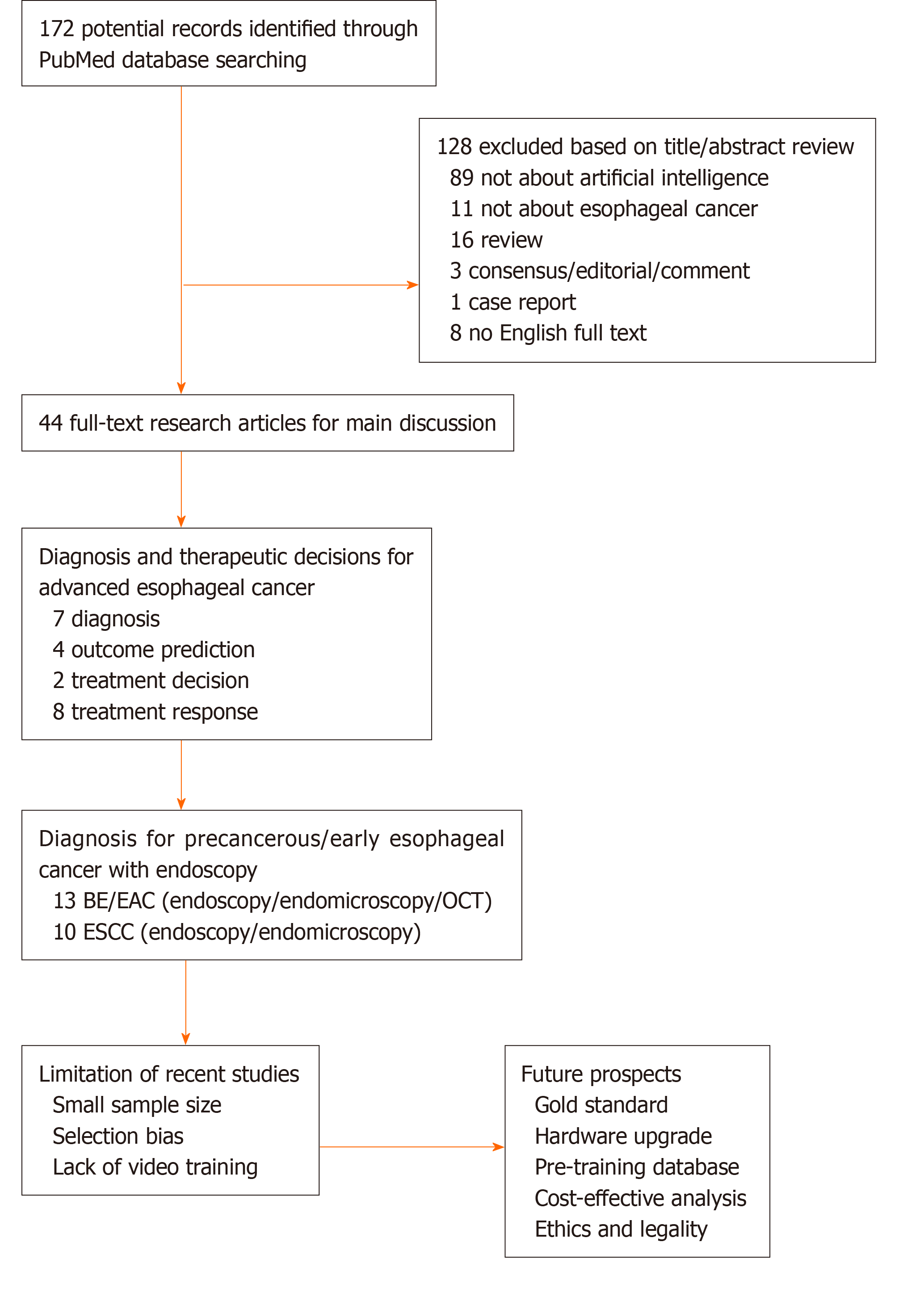
Artificial intelligence (AI), which mimics human mind’s cognitive behavior, has been an emerging hot spot globally in various disciplines. Numerous models have been attempted for machine learning (ML), and the terminologies can be referred in the previous studies[12,13]. ML models are trained by datasets to extract and transform features, thereby achieving the goal of classification and prediction by self-learning[13-15]. In gastroenterology, AI-based technologies, which are characterized by deep learning (DL) as state-of-the-art machine learning algorithms, have been mainly developed to identify dysplasia in Barrett’s esophagus (BE), SCC, gastric cancers, and Helicobacter pylori in upper gastrointestinal (UGI) tract[16], and to diagnose polyps, inflammatory bowel diseases, celiac disease, and gastrointestinal (GI) bleeding in lower GI tract[17]. Various models have been developed and studied to detect anatomical structure, discriminate dysplasia, and predict therapeutic and survival outcomes of EC. The ultimate goal of AI is to assist physicians and patients to make a superior data-based diagnosis or decision. In the following sections, we will (1) provide an overview of AI applications in diagnosis and prediction of advanced cancer; (2) specify computer-aided diagnosis (CAD) for early detection of esophageal SCC (ESCC) and esophageal adenocarcinoma (EAC) based on optical imaging; and (3) outline limitations of the existing studies and future perspectives. We searched PubMed database using terms “esophageal cancer” and “artificial intelligence” for papers published up to March 1, 2020, and initially obtained 172 studies. After exclusion of 128 items, 44 research articles that provided detailed data were included in the review and discussion (Figure 1).
EC is highly malignant, and the 5-year survival rate of late-stage EC is less than 25%[18]. Radical therapies, including surgery, chemotherapy, radiotherapy or their combination are highly essential to improve survival outcome. Accurate diagnosis, precise staging, optimal modality selection, as well as responsiveness and survival outcome prediction are necessary in making true clinical decisions. However, these decisions are made mainly based on the current guidelines and expertise in clinical practice. AI technologies have been therefore developed to enhance the reliability of those decisions in an individualized manner.
One of the important roles of AI is to detect malignant lesions. In 1996, Liu et al[19] proposed a tree-based algorithm called PREDICTOR, to classify patients with dyspeptic symptoms into EC, which achieved a discriminating accuracy of 61.3%, with sensitivity (SEN) and specificity (SPE) of 94.9% and 39.8%, respectively. In 2002, a probabilistic network-based decision-support system was developed, which could correctly predict the cancer stage of 85% of tested data in reasonable time[20]. In the same year, a robust classifier, artificial neural network (ANN), imitating neural network of the human brain, was adopted to distinguish BE from EC[21]. The ANN was trained using 160 genes selected by significance analysis of microarrays (SAM) from cDNA microarray data of esophageal lesions. This ANN outperformed cluster analysis by correctly diagnosing all the tested samples. Kan et al[22] also combined ANN with SAM-extracted 60 gene clones to accurately predict lymph node metastasis in 86% of all SCC cases, with SEN and SPE of 88% and 82%, respectively, better than clustering or predictive scoring. Kan et al[22] suggested that AI was a potential tool to detect lymph node metastasis when the SEN of coherence tomography (CT), EUS, and PET is insufficient[23,24]. Since tumor risk factors have complex nonlinear correlations, a fuzzy neural network, trained on hybridization of chaotic optimization algorithm and error back propagation (EBP), was able to correctly diagnose 87.36% of ESCC and 70.53% of dysplasia[25]. This fuzzy-logic based model outperformed traditional statistics, such as multivariate logistic regression model that was previously described by Etemadi et al[26].
While symptoms are not quite reliable and gene analysis or PET scans are expensive, a simpler noninvasive detection method may be more practical. Li et al[27] combined support vector machine (SVM), a traditional classifier, with surface-enhanced Raman spectroscopy in order to distinguish serum spectra of EC patients from healthy controls. Eventually, a combination of SVM with principle component analysis (PCA) on the basis of radial basis function (RBF), namely RBF PCA-SVM algorithm, exhibited the greatest efficacy among others with accuracy, SEN, and SPE of 85.2%, 83.3% and 86.7%, respectively.
Another significant role of AI is to predict prognosis of EC based on various demographic, clinicopathologic, hematologic, radiologic, and genetic variables. Surgery and neoadjuvant chemotherapy, radiotherapy or chemoradiotherapy are important definitive modalities for advanced EC. Selecting the optimal strategy with superior predictive outcome is of vital importance.
Traditionally, TNM staging system is used as a predictor. However, a previous study showed that it was not very accurate[28]. Hence, multiple computational algorithms were developed to assist more reliable predictions. In 2005, Sato et al[29] trained an ANN to predict survival outcome. They found that the best predictive accuracy was obtained, with 65 clinicopathologic, genetic and biologic variables for 1-year survival and 60 variables for 5-year survival. The area under ROC curve (AUC), SEN, and SPE were 0.883, 78.1%, 84.7% and 0.884, 80.7%, 86.5%, respectively. Similar results with higher SEN and SPE could be achieved in another ANN model to predict the 1- and 3-year post-operative survival of EC and esophagogastric junction cancer[30]. These two ANNs both outperformed TNM staging system[29,30].
In addition to ANN, other models were also proposed to solve certain problems. A prognostic scoring system, using serum C-reactive protein and albumin concentrations, was fused with expertise by fuzzy logic[31]. The proposed model could perform 1-year survival prediction with an AUC of 0.773. Another hierarchical forward selection (HFS), a wrapper feature selection method, was developed to solve the problem of small sample size[32]. In this SVM-validated model, clinical and PET features were learned to predict disease-free survival. The results unveiled that HFS achieved the highest accuracy of 94%, with robustness of 96%. Robustness could be further increased to 98%, if HFS was incorporated prior knowledge (pHFS).
Based on the condition and prognosis of patient, an individualized treatment strategy is needed. For instance, when chemotherapy is prescribed for a patient, what is the optimal medication with appropriate dosage and period? Generally, clinicians make decisions according to their own experience, guidelines or consensus. However, those recommendations are often fixed and human errors are sometimes inevitable. A group of Iranian experts attempted to train a multilayer neural network with particle swarm optimization and EBP algorithms, in order to determine the dosage of chemotherapy[33]. Encouraging results showed that accuracy of particle swarm optimization and EBP was both 77.3%. Zahedi et al[33] were positive about its future application as a supplementary decision-making system.
While the majority of decisions are made before treatment, is it possible to make real-time treatment decisions? The answer is YES. Maktabi et al[34] tested a relatively new hyperspectral imaging system. They found that SVM was able to detect cancerous tissue with 63% SEN and 69% SPE within 1s. It is promising that hyperspectral imaging may assist surgeons in identifying tumor borders intra-operatively in real time.
A good treatment response is crucial for consequential therapeutic decision and predicting outcome[35]. Endeavors have been made to select candidate factors that correlate with responsiveness to treatment. However, it is often extremely laborious to testify these numerous variables in clinical trials. AI technologies are potential powerful tools for this selection.
One indicator is the genetic biomarker. In 2010, Warnecke-Eberz et al[36] reported their usage of ANN to predict histopathologic responsiveness of treatment-naïve patients to neoadjuvant chemoradiotherapy by analyzing 17 genes using the TaqMan low-density arrays. Their results were promising, with 85.4% accuracy, 80% SEN, and 90.5% SPE. Radiology is another important indicator to assess tumor regression after treatment. One rationale for this exploration is that tumor heterogeneity exists within radiologic images[37]. The standardized uptake values of 18F-fluorodeoxyglucose in PET imaging were reported to have predictive potential[38]. However, this predictive power is limited[39] due to some confounding factors, such as intra-observer variations. Ypsilantis et al[40] adopted a three-slice convolutional neural network that could extract features from pre-treatment PET scans automatically to predict response to chemotherapy. It achieved a moderate accuracy of 73.4%, with SEN and SPE of 80.7% and 81.6%, respectively, which outperformed other ML algorithms trained on handcrafted PET scan features. Recently, CT radiomics three months after chemoradiotherapy were combined with dosimetric features of gross tumor volume and organs at risk to identify non-responders. Jin et al[41] found that these combinative features trained by the model of extreme gradient boosting plus PCA achieved an accuracy of 70.8%, with AUC of 0.541.
While tumor regression is an important indicator to assess responsiveness, post-treatment distant metastasis is also vital to evaluate responsiveness, which is correlated with survival outcome. In order to predict post-operative distant metastasis of ESCC, further SVM models incorporated with clinicopathological and immunohistological variables were established[42]. Finally, the SVM model with four clinicopathological features and nine immunomarkers had better performance, with accuracy, SEN, SPE, positive predictive value, and negative predictive value of 78.7%, 56.6%, 97.7%, 95.6%, and 72.3%, respectively. Another least squares SVM model was also proposed to predict post-operative lymph node metastasis in patients who received chemotherapy preoperatively, by exploiting preoperative CT radiomics[43]. Tumor length, thickness, CT value, long axis and short axis size of the largest regional lymph node were analyzed. The model reached an AUC of 0.887.
In addition to its diagnostic and predictive value, AI has learned to identify meaningful alterations in molecular and genetic level. In 2017, Lin et al[44] compared the serum chemical elements concentrations between ESCC patient and healthy controls, and found that nearly half of the elements were different between the two groups. They then trained several classifiers to perform the discrimination, with Random Forest being the best (98.38% accuracy) and SVM the second (96.56% accuracy). Later, Mourikis et al[45] developed a robust sysSVM algorithm to identify 952 genes that promoted EAC development, using 34 biological features of known cancer genes. They called these rare and highly individualized genes ”helper” genes, which function alongside known drivers.
AI may be a feasible option to help determine an optimal treatment strategy. This was previously evidenced by a study of 13 365 EACs from 33 cancer centers worldwide, which incorporated random forest algorithm, and found that the predicted survival of AI-generated therapy was superior to actual human decisions[46]. However, most of the above-mentioned ML algorithms described were developed for the sake of advanced cancer. Diagnosing EC in an early stage contributes to a far better outcome when treatment is undertaken appropriately. This is highly dependent on the development of optical imaging technologies that can directly visualize the morphology of esophageal lesions.
In recent decades, endoscopic optical imaging techniques have been rapidly advanced, which provide endoscopists a fine inspection of the morphology of esophageal mucosa, micro-vessels, and even cells. In lieu of white light imaging (WLI) and magnifying endoscopy (ME), emerging OCT, CLE, VLE, and HRM techniques have been developed to diagnose BE[47-49]. Meanwhile, the diagnosis of SCC more relies on chromoendoscopy and intra-epithelial papillary capillary loop (IPCL) observed under narrow band imaging (NBI) plus ME[50]. Although these modalities have yielded preferable diagnostic value, the interpretation of these images need expert’s experience (inter- and intra-observer variability[51]), and processing large dataset is laborious and time consuming. Researchers in medicine and information engineering have collaborated to develop different AI models for this purpose.
The current screening and surveillance recommendation for BE is endoscopic examination plus random biopsy[52] , which is limited by sampling error. AI models trained with various endoscopic modalities and pathologies aimed to overcome these shortcomings (Table 1).
| Ref. | Year | Study design | Lesions | Diagnostic method | AI technology | Dataset capacity | Validation | Outcomes | Compared to expert | Processing speed |
| Münzenmayer et al[53] | 2009 | Retrospective | BE | WLI | Color-texture analysis in a CBIR framework | 390 images with 482 ROIs | LOO (N-fold cross-validation) | Accuracy: BE/CC/EP 70%/74%/95% | NA | NA |
| van der Sommen et al[55] | 2016 | Retrospective | HGD, early EAC | WLI | SVM | 100 images | LOO | Per-image SEN/SPE: 83%/83%; Per-patient SEN/SPE: 86%/87% | Inferior | NA |
| Horie et al[56] | 2019 | Retrospective | EAC | WLI; NBI | CNN-SSD | 8 patients | Caffe DL framework | Accuracy: 90%; Per-image SEN: WLI/NBI: 69%/71%; Per-case SEN: WLI/NBI: 88%/88% | NA | 0.02 s/image |
| Ghatwary et al[57] | 2019 | Retrospective | EAC | WLI | VGG’16-based; R-CNN; Fast R-CNN; Faster R-CNN; SSD | 100 images (train 50, test 50) | 5-fold cross-validation and LOO | F-measure: 0.94 (SSD); SEN/SPE: 96%/92% (SSD) | NA | 0.1-0.2 s/image |
| Hashimoto et al[58] | 2020 | Retrospective | HGD, early EAC | WLI and NBI with both standard and near focus | CNN | 1835 images | NA | Per-image accuracy: 95.4%; Per-image SEN/SPE: 96.4%/94.2%; 98.6%/88.8% (WLI); 92.4%/99.2% (NBI) | NA | GPU gtx 1070: 0.014 s/frame; YOLO v2: 0.022 s/frame |
| Ebigbo et al[59] | 2019 | Retrospective | Early EAC | WLI; NBI | CNN-ResNet | 248 images | LOO | SEN/SPE of Augsburg database: 97%/88% (WLI); 94%/80% (NBI); SEN/SPE of MICCAI database: 92%/100% | Superior | NA |
| de Groof et al[60] | 2019 | Retrospective | Early dysplastic BE | WLI | ResNet-UNet hybrid | 1704 images (train 1544, validation 160) | 4-fold cross-validation (external validation) | Accuracy/SEN/SPE: 89%/90%/88% (dataset 4); 88%/93%/83% (dataset 5) | NA (superior to non-expert) | Classification: 0.111 s/image; Segmentation: 0.124 s/image |
| Swager et al[62] | 2017 | Retrospective | HGD, early EAC | VLE | SVM, DA, Adaboost, RF, kNN, NB, LR, LogReg | 60 images | LOO | AUC: 0.95; SEN/SPE: 90%/93% | Superior | NA |
| van der Sommen et al[63] | 2018 | Retrospective | HGD, early EAC | VLE | SVM, RF; AdaBoost; CNN, kNN; DA, LogReg | 60 frames | LOO | AUC: 0.90-0.93 | Superior | 24 ms/full dataset for clinically-inspired features |
| Struyvenberg et al[65] | 2020 | Prospective | HGD, early EAC | VLE | PCA-CAD | 3060 frames | NA | AUC of Multi-frame: 0.91; AUC of Single-frame: 0.83 | NA | 0.001 s/frame; 1.5s/full VLE scan |
| van der Putten et al[66] | 2020 | Prospective | HGD, early EAC | VLE | Multi-step PDE-CNN on an A-line basis | In-vivo: 140 images (train 111, test 29) | 4-fold cross-validation | AUC: 0.93; F1 score: 87.4% | NA | 50000 A-lines/s |
| Shin et al[67] | 2016 | Retrospective | HGD, EAC | HRM | Two-class LDA-based automated sequential classification algorithm | 230 sites (train 77, validation 153) | NA | Accuracy: 84.9%; SEN/SPE: 88%/85% | NA | 52 s/image |
| Qi et al[68] | 2006 | Retrospective | Dysplastic BE | OCT | PCA | 106 images | LOO | Accuracy: 83%; SEN/SPE: 82%/74% | NA | NA |
Endoscopy: In 2009, German experts developed a content-based image retrieval framework[53]. In this frame, novel color-texture features were combined with an interactive feedback loop. The algorithm could correctly recognize 95% of normal mucosa and 70% of BE from 390 training images, with a moderate inter-rater reliability of 0.71. The authors thought that the CAD system might be incorporated to the endoscopic system to help lesser experienced clinicians. In 2013, van der Sommen et al[54] tried an SVM algorithm, which could automatically identify and locate irregularities of esophagus on high-definition endoscopy with an accuracy of 95.9% and AUC of 0.99, taking a first step towards CAD. Later, these authors used a CAD system to automatically recognize region of interest (ROI) in dysplastic BE[55]. The SVM-based classification yielded a SEN and SPE of 83% for per-image level, and 86% and 87%, for the patient level, respectively. However, the f-score of the system, which indicates the similarity with the gold standard, was lower than experts.
In order to improve the outcome, Horie et al[56] were the first who adopted a deep CNN (Single Shot MultiBox Detector, SSD) model to detect EC from WLI and NBI images in 2018. Only 8 EACs were used in that study. The diagnostic accuracy for EAC was 90%, and SEN for WLI and NBI at patient level was both equal to 88%. The system processed one image in only 0.02 s, which is promising for a real-time job. This ability of SSD to detect EAC was assessed in another study, which outperformed their proposed regional-based CNN (R-CNN), Fast R-CNN and Faster R-CNN in both precision and speed, which achieved F-measure, SEN, and SPE of 0.94, 96% and 92%, respectively[57]. The authors stated that SSD worked faster due to its single forward pass network nature. CNN was then validated in a more recent study to detect early dysplastic BE[58]. The system was pretrained on ImageNet, and was then trained with 1853 images and tested with 458 images. The CNN accurately detected 95.4% of the dysplasia, with 96.4% SEN and 94.2% SPE. One highlight for this study is that it studied WLI and NBI images, as well as images with standard focus and near focus. Another highlight is its ability to deal with real-time videos.
Except for the above-mentioned CNNs, another CNN built upon residual net (ResNet) was introduced. Ebigbo et al[59] tested this system in two databases, Augsburg and Medical Image Computing and Computer-Assisted Intervention, with SEN both being over 90%. Later, de Groof et al[60] used a custom-made hybrid ResNet/U-Net which was pretrained on GastroNet to distinguish non-dysplastic BE from dysplasia. The system was trained using state-of-the-art ML techniques (transfer learning and ensemble learning) and validated in a sequential five datasets, with accuracy of 89% and 88% for two external validation datasets, which were slightly superior to the model pre-trained with ImageNet in its supplementary ablation experiment.
Endomicroscopy: In 2017, Hong et al[61] reported their experience in adopting CNN as a classifier to distinguish intestinal metaplasia (IM), gastric metaplasia (GM) and neoplasia (NPL) of BE using endomicroscopic images. The total accuracy was 80.77%. It performed well for IM and NPL. However, it could not identify GM in the tested samples. VLE is an advanced imaging techinique that can provide a 3-mm deep scan of the esophagus in full circumference, which is commercially available (Nvision VLETM Imaging System). In the same year, Swager et al[62] reported the first attempt of using CAD to detect NPL by adopting histology-correlated ex-vivo VLE. The authors used eight separate ML algorithms that were trained with clinically inspired features. They found that “layering and signal decay statistics” feature performed the best, with AUC, SEN, and SPE of 0.95, 90%, and 93%, respectively. Similar results were obtained by van der Sommen et al[63], with a maximum AUC of 0.93 in identifying early EAC in BE. Notably, the authors discovered that scanning depth of 0.5-1 mm was the most appropriate range for classifying tissue categories.
Since VLE produces an overwhelming number of images in a short time, a real-time CAD system is more helpful in actual clinical practice. In 2019, Trindade et al[64] reported a video case illustrating an intelligent real-time image segmentation system which employed three established features to dynamically enhance abnormal VLE images with color in endoscopic procedure. They are now undergoing a multicenter RCT (NCT03814824) to further validate this CAD system. While most studies use single frame to include ROI, a recent study tried to add neighboring VLE images to pathology-correlated ROI[65]. Hopefully, the so-called multi-frame analysis combing PCA improved the performance of single-frame analysis, from an AUC of 0.83 to 0.91. Meanwhile, the novel CAD system needs only 1.3 ms to automatically differentiate non-dysplastic BE from dysplasia in one image, and this is also a promising result for a real-time setting.
While previous studies employed ex-vivo scan images, the following study conducted by van der Putten et al[66] used in-vivo histology-correlated images. In addition, they used principle dimension encoding (PDE) to encode images into score vector. They combined this PDE with traditional ML algorithms, e.g., random forest and SVM, to classify the degree of dysplasia (high-grade dysplasia vs early EAC). They obtained an AUC of 0.93 and F1 score of 87.4%, which outperformed some traditional DL classifiers, such as Squeezenet and Inception.
Another kind of endomicroscopic technique is HRM. Shin et al[67] designed an automated imaging processing algorithm extracting epithelium morphology and BE glandular architecture features, and a classification algorithm, which distinguished NPL from dysplasia in BE with an accuracy, SEN and SPE of 84.9%, 88%, and 85% in validation dataset, respectively. This quantitative CAD is cost-effective and may be applied in clinical settings after improvement of image acquisition quality and processing speed.
OCT: OCT is also a noninvasive imaging technique that can detect BE, dysplasia and early EAC, in compensation to routine endoscopy. In 2006, Qi et al[68] attempted to extract image features using center-symmetric auto-correlation method and a PCA-based CAD algorithm was used for classification. A total of 106 pathology-paired images were included for training, which ended up with an accuracy of 83%, SEN of 82% and SPE of 74% to distinguish non-dysplastic BE from dysplasia. In general, the accuracy of OCT in identifying dysplasia is not satisfactory, which limits its application[69].
In addition to endoscopic images, pathologic morphology has also been studied. Sabo et al[70] employed an ANN-validated computerized nuclear morphometry (pseudostratification, pleomorphism, chromatin texture, symmetry and orientation) model to discriminate the degree of dysplasia in BE. The model was able to differentiate non-dysplastic BE from low-grade dysplasia with an accuracy of 89%, and low-grade dysplasia from high-grade dysplasia with an accuracy of 86%.
ESCC is the dominant histological type of esophageal cancer worldwide. Diagnosing early cancer mainly depends on endoscopic screening, which also produces a large number of images that needs special training to interpret. AI technologies have also been explored globally to address this issue (Table 2).
| Ref. | Year | Study design | Lesions | Diagnostic method | AI technology | Dataset capacity | Validation | Outcomes | Compared to expert | Processing speed |
| Liu et al[71] | 2016 | Retrospective | Early ESCC | WLI | JDPCA + CCV | 400 images | 10-fold cross-validation | Accuracy: 90.75%; AUC: 0.9471; SEN/SPE: 93.33%/89.2% | NA | NA |
| Horie et al[56] | 2019 | Retrospective | ESCC | WLI; NBI | CNN-SSD | 41 pts (train 8428 images; test 1118 images without histology distinction) | Caffe DL framework | Accuracy: 99%; Per-image SEN: 72%/86% ( WLI/NBI, respectively); Per-case SEN: 79%/89% ( WLI/NBI, respectively) | NA | 0.02 s/image |
| Cai et al[72] | 2019 | Retrospective | Early ESCC | WLI | DNN | 2615 images (train 2428, test 187) | NA | Accuracy: 91.4%; SEN/SPE: 97.8%/85.4% | Superior | NA |
| Zhao et al[74] | 2019 | Retrospective | Early ESCC | ME + NBI | Double labeling FNN | 1350 images with 1383 lesions | 3-fold cross-validation | Accuracy/SEN/SPE at lesion level: 89.2%/87%/84.1%; Accuracy at pixel level: 93% | Comparable | NA |
| Ohmori et al[73] | 2020 | Retrospective | Superficial ESCC | ME + NBI/BLI; Non-ME + WLI/NBI/BLI | CNN | 23289 images (train 22562, test 727) | Accuracy/SEN/SPE: 77%/100%/63% (Non-ME + NBI/BLI); 81%/90%76% ( Non-ME + WLI); 77%/98%/56% ( ME) | Comparable | 0.028 s/image | |
| Nakagawa et al[76] | 2019 | Retrospective | ESCC (EP-SM1/SM2+SM3) | ME; Non-ME | CNN-SSD | 15252 images (train 14338, test 914) | Caffe DL framework | Accuracy/SEN/SPE: 91%/90.1%/95.8% | Comparable | 0.033 s/image |
| Everson et al[77] | 2019 | Retrospective | ESCC IPCLs (type A/type B) | ME + NBI | CNN | 7046 images | 5-fold cross-validation+eCAM | Accuracy/SEN/SPE: 93.3%/89.3%/98% | NA | 0.026-0.037 s/image |
| Guo et al[79] | 2020 | Retrospective | Early ESCC | NBI (ME + non-ME) | CNN-SegNet | 13144 images (train 6473, validation 6671), 80 videos (47 lesions, 33 normal esophagus) | NA | Per-image SEN/SPE: 98.04%/95.03%; Per-frame SEN/SPE: 91.5%/99.9% | NA | < 0.04 s/frame; Latency <0.1 s |
| Shin et al[82] | 2015 | Retrospective | HGD, ESCC | HRM | Two-class LDA | 375 sites of images (train 104, test 104, validation 167) | NA | AUC: 0.95; SEN/SPE: 84%/95% | NA | 3.5 s/image |
| Quang et al[83] | 2016 | Retrospective | ESCC | HRM | A fully automated algorithm | 375 biopsied sites from Shin et al[82] (train 104, test 104, validation 167) | NA | AUC: 0.937; SEN/SPE: 95%/91% | NA | Average 5 s for computing |
Endoscopy: In 2016, Liu et al[71] designed an algorithm called joint diagonalization principle component analysis, which correctly detected 90.75% of EC with an AUC of 0.9471. To improve the performance of CAD system, Horie et al[56] did the first attempt to use DL to diagnose ESCC with a large number of endoscopic images. The CNN had a diagnostic accuracy of 99% for ESCC, 99% for superficial cancer, and 92% for advanced one. The SEN of CNN was 97% for per-patient level and 77% for per-image level. Later in 2019, Cai et al[72] proposed a novel CAD system called deep neural network (DNN). They used only standard WLI images to train the model. The DNN-CAD model could detect 91.4% of early ESCC, higher than senior endoscopists. By using this model, the average diagnostic performance of endoscopists improved satisfactorily, in terms of accuracy, SEN, and SPE. However, these studies excluded magnified images.
Later, Ohmori et al[73] evaluated both ME and non-ME images [including WLI and NBI/blue laser imaging (BLI)] using a CNN based on SSD to recognize SCC. The accuracy for ME, non-ME + WLI, and non-ME + NBI/BLI was 77%, 81%, and 77%, respectively, all with high SEN and moderate SPE. The result was similar to experienced endoscopists tested in this study. Zhao et al[74] conducted another study and evaluated ME + NBI images by employing a double-labeling fully convolutional network. This system used ROI-label and segmentation-label to delineate IPCLs based on the AB classification by the Japan Esophageal Society[75]. The study showed that senior observers had significant higher diagnostic accuracy than mid-level and junior ones. The model reached a diagnostic accuracy of 89.2% and 93% in lesion and pixel level, respectively, for distinguishing type A, B1 and B2 IPCLs, which was similar to that of senior group. Specifically, the model had a higher sensitivity for type A IPCL than clinicians (71.5% vs 28.2%-64.9%), which might avoid unnecessary radical treatment. Instead of identifying IPCL patterns, the study conducted by Nakagawa et al[76] aimed to predict invasion depths. The authors developed two separate SSD-based CNNs for ME and non-ME images. The ability of the system to correctly distinguish EP/submucosal (SM) 1 cancers from SM2/SM3 cancers was 91%, 92.9%, and 89.7% for the ME+ non-ME, non-ME and ME images, respectively. Regrading M and SM cancers, the differentiating accuracy was 89.7%, 90.3%, and 92.3% for the total, non-ME, and ME, respectively. The performance of this CAD model was also comparable to experts, but much faster.
A processing speed over 30 images/s is necessary for dynamic video analysis[56]. Although Horie et al[56], Ohmori et al[73] and Nakagawa et al[76] reported that their systems could process one image in 0.02, 0.027, and 0.033 s, respectively, they have not tested the systems in real-time videos. After Cai et al[72] had validated the efficacy of their DNN-CAD model, they split the video into images and then assembled them, enabling the model to delineate early cancer in real time. Everson et al[77] validated another CNN investigating IPCLs using sequential still images in real time of 0.026 to 0.037 s per image. The CNN could differentiate type A from type B IPCLs with a mean accuracy of 93.3%. Last year, Luo et al[78] reported a multicenter, comparative study, exploiting 1 036 496 endoscopic images to construct a gastrointestinal artificial intelligence diagnostic system (GRAIDS) based on the concept of DeepLab’s V3+. The GRAIDS yielded a diagnostic accuracy of UGI cancer ranging from 91.5% to 97.7% for internal, external, and prospective validation datasets, with favorable sensitivities, which were similar to experts and superior to non-experts. They also incorporated the CAD model to endoscopic videos in real time, with the highest speed of 0.008 s per image and latency less than 0.04 s. However, they did not report their outcome in distinct histology. Recently, Guo et al[79] specially developed a CNN-CAD system built on SegNet architecture, aiming at real-time application in clinical settings. In this study, 13144 NBI (ME + non-ME) images and 80 video clips were employed. In the image dataset, the SEN, SPE, and AUC were 98.04%, 95.03% and 0.989, respectively. For the video dataset, the SEN of per frame for non-ME and ME was 60.8% and 96.1%, respectively; the SEN of per lesion for non-ME and ME was both 100%. When they analyzed 33 original videos of full-range normal esophagus, they acquired a SPE of 99.95% and 90.9% for per-frame and per-case analysis, respectively. The ability of this model to process each frame with a maximum time of 0.04 s and latency less than 0.1 s set a good example for future model optimization for real-time applications[80].
Endomicroscopy: In 2007, Kodashima et al[81] used ImageJ software to label the border of nuclei under endo-cytologic images from 10 ESCC patients. They found that the computer-labelled nuclei area of ESCC was significantly different from that of normal tissues, which demonstrated the diagnostic possibility of computer. HRM is another low-cost tool that can illustrate the esophageal epithelium in cellular level, which compensates the low specificity of iodine staining and is also more cost-effective compared with CLE. In 2015, Shin et al[82] developed a 2-class linear classification algorithm using nuclei-related features to identify neoplastic squamous mucosa (HGD + cancer). It resulted in an AUC, SEN, and SPE of 0.95, 87%, 97% and 0.93, 84%, 95% for the test and validation datasets, respectively. However, the application of this system for real-time practice needs acceleration of analyzing speed. To solve this problem and reduce the cost of equipment, a smaller, tablet-interfaced HRM with real-time algorithm was developed by Quang et al[83]. The algorithm was able to automatically identify SCC with an AUC, SEN, and SPE of 0.937, 95%, and 91%, respectively, which is comparable to the result achieved by the first generation bulky laptop-interfaced HRM[82] or the combination of Lugol chromoendoscopy and HRM[84].
The exciting and promising findings of various CAD models have been summarized in detail above. Researches are ongoing worldwide because none of the studies were perfect. The limitations and problems are driving forces for evolution and innovation. We hereby discuss several major drawbacks that limit the strength of the studies.
Firstly, the most mentioned drawback is insufficient training sample size. The number of endoscopic images that the majority of studies employed ranged from 248 to about 7000 (Tables 1 and 2). The limited number of training data, lack of imaging variability, and single-center nature are likely to cause overfitting[85], which attenuates the ability of AI models to perform well in unused datasets and leads to unstable results[12,55]. To overcome this problem, various regularization methods have been developed, such as segmenting the image or using cross validation with 5 folds or even 10 folds to augment the datasets. Recently, the size of datasets has been greatly enlarged in several studies[56,73,79], the largest of which included over one million UGI images from six centers[78]. Therefore, further multicenter studies including large dataset with different kinds of images (i.e., WLI, NBI, ME and non-ME) harvested by different endoscopic systems for SCC and AC are likely to produce results with robustness and external generalizability. In addition, different AI algorithms tested in prospective external dataset need to be developed to increase the diversity of AI technology[13].
Secondly, selection bias is another contributor to limited generalizability. Most of the previous studies were retrospective and used only high-quality images. Suboptimal quality images with mucus, blur, or blood. were excluded. Additionally, unbalanced distribution of lesion types (SCC vs AC, type B1 vs B2 and B3 IPCLs), different numbers of images for each patient, and non-uniform processing method for different lesions all might cause bias in the result. Further prospective RCTs will be required in the future.
Thirdly, almost all of the studies employed still images to train AI model. Not until recently did the researchers validate the efficacy in dealing with endoscopic videos in a real-time manner. Future video-based researches are needed to narrow the gap between study and clinical practice.
Gold standard: Consensus-based ground truth for lesions is preferred over a single expert’s annotation. The committees of expert endoscopists and pathologists from different countries need to be formed to improve the precision of annotation. In addition, the AI should play a role in helping endoscopists recognize lesions and target biopsies for gold standard pathological examination, rather than replacing our “job”.
Hardware upgrade: Computers equipped with powerful GPU are needed to perform more sophisticated algorithms and process large volume of graphical data, in order to achieve the goal of real-time recognition.
Pre-training database: ImageNet and GastroNet have been introduced, which store mass datasets of manually labeled images. These databases should be constantly enriched, since CAD models with prior knowledge are prone to have better discriminative ability[60].
Cost-effect analysis: When a novel diagnostic method is introduced to clinical practice, whether it is cost-effective is an important issue. A recent multi-center add-on analysis revealed that AI is able to reduce cost of colonoscopic management of polyps[86]. Since medical cost is one of the major concerns for both patients and government, it is necessary to assess whether AI can improve diagnostic performance of EC while reducing cost of unnecessary examinations and radical therapies. Future studies concerning medical cost and reimbursement should be conducted in different countries with different healthcare and insurance systems to address this issue.
Ethics and legality: Believe it or not to believe it, it is a real question. While we have taken a giant leap of AI technology in medicine which has the potential to improve the performance of clinicians with different experience and reduce error, the black-box[87] nature of the ML algorithms truly brings doubts[88]. Can we trust the results of AI, since they lack explainability? What should we do with these computer-generated results? Are they certified to be legal evidence? Challenges for legislation, regulation, insurance and clinical practice are inevitable. Supervised RCTs and AI participation in clinical workflow are needed to provide solid evidence that AI is acceptable within the range of legal and ethical concerns[89]. Nevertheless, trends of AI are irreversible. The ultimate role of AI in medicine might be a supervised task performer[90].
In this manuscript, we provided a comprehensive review of AI technology in diagnosis, treatment decision and outcome prediction for EC. We searched only PubMed database for clinical researches and applications. Issues regarding computer science and image processing are not our topics. The CAD systems have evolved from traditional ML algorithms to neural network-based DL, and from still image analysis to real-time video processing. AI can improve non-expert’s performance while correct erroneous classification by experts[78]. Researches with larger datasets and more reliable CAD models are being conducted worldwide. It is promising that AI may facilitate early cancer screening, surveillance and treatment in high-risk regions. However, it is noteworthy that patient’s consent and satisfaction are of first priority.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Jennane R, Kravtsov V, Yoshida H S-Editor: Yan JP L-Editor: MedE-Ma JY P-Editor: Ma YJ
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55853] [Article Influence: 7979.0] [Reference Citation Analysis (132)] |
| 2. | Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1956] [Cited by in RCA: 1961] [Article Influence: 163.4] [Reference Citation Analysis (5)] |
| 3. | Simard EP, Ward EM, Siegel R, Jemal A. Cancers with increasing incidence trends in the United States: 1999 through 2008. CA Cancer J Clin. 2012;62:118-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 488] [Cited by in RCA: 520] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 4. | Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64:381-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 944] [Cited by in RCA: 1027] [Article Influence: 102.7] [Reference Citation Analysis (0)] |
| 5. | Arnold M, Laversanne M, Brown LM, Devesa SS, Bray F. Predicting the Future Burden of Esophageal Cancer by Histological Subtype: International Trends in Incidence up to 2030. Am J Gastroenterol. 2017;112:1247-1255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 304] [Article Influence: 38.0] [Reference Citation Analysis (2)] |
| 6. | Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, Dandona L, Fleming T, Forouzanfar MH, Hancock J, Hay RJ, Hunter-Merrill R, Huynh C, Hosgood HD, Johnson CO, Jonas JB, Khubchandani J, Kumar GA, Kutz M, Lan Q, Larson HJ, Liang X, Lim SS, Lopez AD, MacIntyre MF, Marczak L, Marquez N, Mokdad AH, Pinho C, Pourmalek F, Salomon JA, Sanabria JR, Sandar L, Sartorius B, Schwartz SM, Shackelford KA, Shibuya K, Stanaway J, Steiner C, Sun J, Takahashi K, Vollset SE, Vos T, Wagner JA, Wang H, Westerman R, Zeeb H, Zoeckler L, Abd-Allah F, Ahmed MB, Alabed S, Alam NK, Aldhahri SF, Alem G, Alemayohu MA, Ali R, Al-Raddadi R, Amare A, Amoako Y, Artaman A, Asayesh H, Atnafu N, Awasthi A, Saleem HB, Barac A, Bedi N, Bensenor I, Berhane A, Bernabé E, Betsu B, Binagwaho A, Boneya D, Campos-Nonato I, Castañeda-Orjuela C, Catalá-López F, Chiang P, Chibueze C, Chitheer A, Choi JY, Cowie B, Damtew S, das Neves J, Dey S, Dharmaratne S, Dhillon P, Ding E, Driscoll T, Ekwueme D, Endries AY, Farvid M, Farzadfar F, Fernandes J, Fischer F, G/Hiwot TT, Gebru A, Gopalani S, Hailu A, Horino M, Horita N, Husseini A, Huybrechts I, Inoue M, Islami F, Jakovljevic M, James S, Javanbakht M, Jee SH, Kasaeian A, Kedir MS, Khader YS, Khang YH, Kim D, Leigh J, Linn S, Lunevicius R, El Razek HMA, Malekzadeh R, Malta DC, Marcenes W, Markos D, Melaku YA, Meles KG, Mendoza W, Mengiste DT, Meretoja TJ, Miller TR, Mohammad KA, Mohammadi A, Mohammed S, Moradi-Lakeh M, Nagel G, Nand D, Le Nguyen Q, Nolte S, Ogbo FA, Oladimeji KE, Oren E, Pa M, Park EK, Pereira DM, Plass D, Qorbani M, Radfar A, Rafay A, Rahman M, Rana SM, Søreide K, Satpathy M, Sawhney M, Sepanlou SG, Shaikh MA, She J, Shiue I, Shore HR, Shrime MG, So S, Soneji S, Stathopoulou V, Stroumpoulis K, Sufiyan MB, Sykes BL, Tabarés-Seisdedos R, Tadese F, Tedla BA, Tessema GA, Thakur JS, Tran BX, Ukwaja KN, Uzochukwu BSC, Vlassov VV, Weiderpass E, Wubshet Terefe M, Yebyo HG, Yimam HH, Yonemoto N, Younis MZ, Yu C, Zaidi Z, Zaki MES, Zenebe ZM, Murray CJL, Naghavi M. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2017;3:524-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2838] [Cited by in RCA: 2962] [Article Influence: 370.3] [Reference Citation Analysis (0)] |
| 7. | Peery AF, Crockett SD, Murphy CC, Lund JL, Dellon ES, Williams JL, Jensen ET, Shaheen NJ, Barritt AS, Lieber SR, Kochar B, Barnes EL, Fan YC, Pate V, Galanko J, Baron TH, Sandler RS. Burden and Cost of Gastrointestinal, Liver, and Pancreatic Diseases in the United States: Update 2018. Gastroenterology. 2019;156:254-272.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 776] [Cited by in RCA: 1082] [Article Influence: 180.3] [Reference Citation Analysis (1)] |
| 8. | Thein HH, Jembere N, Thavorn K, Chan KKW, Coyte PC, de Oliveira C, Hur C, Earle CC. Estimates and predictors of health care costs of esophageal adenocarcinoma: a population-based cohort study. BMC Cancer. 2018;18:694. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 9. | Schreurs LM, Busz DM, Paardekooper GM, Beukema JC, Jager PL, Van der Jagt EJ, van Dam GM, Groen H, Plukker JT, Langendijk JA. Impact of 18-fluorodeoxyglucose positron emission tomography on computed tomography defined target volumes in radiation treatment planning of esophageal cancer: reduction in geographic misses with equal inter-observer variability: PET/CT improves esophageal target definition. Dis Esophagus. 2010;23:493-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 42] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 10. | Liu J, Li M, Li Z, Zuo XL, Li CQ, Dong YY, Zhou CJ, Li YQ. Learning curve and interobserver agreement of confocal laser endomicroscopy for detecting precancerous or early-stage esophageal squamous cancer. PLoS One. 2014;9:e99089. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 11. | Worrell SG, Boys JA, Chandrasoma P, Vallone JG, Dunst CM, Johnson CS, Lada MJ, Louie BE, Watson TJ, DeMeester SR. Inter-Observer Variability in the Interpretation of Endoscopic Mucosal Resection Specimens of Esophageal Adenocarcinoma: Interpretation of ER specimens. J Gastrointest Surg. 2016;20:140-4; discussion 144-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 12. | Yang YJ, Bang CS. Application of artificial intelligence in gastroenterology. World J Gastroenterol. 2019;25:1666-1683. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 211] [Cited by in RCA: 160] [Article Influence: 26.7] [Reference Citation Analysis (5)] |
| 13. | Ruffle JK, Farmer AD, Aziz Q. Artificial Intelligence-Assisted Gastroenterology- Promises and Pitfalls. Am J Gastroenterol. 2019;114:422-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 91] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 14. | Min JK, Kwak MS, Cha JM. Overview of Deep Learning in Gastrointestinal Endoscopy. Gut Liver. 2019;13:388-393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 116] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 15. | Deo RC. Machine Learning in Medicine. Circulation. 2015;132:1920-1930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1155] [Cited by in RCA: 1961] [Article Influence: 217.9] [Reference Citation Analysis (6)] |
| 16. | Mori Y, Kudo SE, Mohmed HEN, Misawa M, Ogata N, Itoh H, Oda M, Mori K. Artificial intelligence and upper gastrointestinal endoscopy: Current status and future perspective. Dig Endosc. 2019;31:378-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 87] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 17. | Le Berre C, Sandborn WJ, Aridhi S, Devignes MD, Fournier L, Smaïl-Tabbone M, Danese S, Peyrin-Biroulet L. Application of Artificial Intelligence to Gastroenterology and Hepatology. Gastroenterology. 2020;158:76-94.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 326] [Article Influence: 65.2] [Reference Citation Analysis (1)] |
| 18. | Domper Arnal MJ, Ferrández Arenas Á, Lanas Arbeloa Á. Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol. 2015;21:7933-7943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 554] [Cited by in RCA: 748] [Article Influence: 74.8] [Reference Citation Analysis (15)] |
| 19. | Liu WZ, White AP, Hallissey MT, Fielding JW. Machine learning techniques in early screening for gastric and oesophageal cancer. Artif Intell Med. 1996;8:327-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 20. | van der Gaag LC, Renooij S, Witteman CL, Aleman BM, Taal BG. Probabilities for a probabilistic network: a case study in oesophageal cancer. Artif Intell Med. 2002;25:123-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 97] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 21. | Xu Y, Selaru FM, Yin J, Zou TT, Shustova V, Mori Y, Sato F, Liu TC, Olaru A, Wang S, Kimos MC, Perry K, Desai K, Greenwald BD, Krasna MJ, Shibata D, Abraham JM, Meltzer SJ. Artificial neural networks and gene filtering distinguish between global gene expression profiles of Barrett's esophagus and esophageal cancer. Cancer Res. 2002;62:3493-3497. [PubMed] |
| 22. | Kan T, Shimada Y, Sato F, Ito T, Kondo K, Watanabe G, Maeda M, Yamasaki S, Meltzer SJ, Imamura M. Prediction of lymph node metastasis with use of artificial neural networks based on gene expression profiles in esophageal squamous cell carcinoma. Ann Surg Oncol. 2004;11:1070-1078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 23. | Fukuda M, Hirata K, Natori H. Endoscopic ultrasonography of the esophagus. World J Surg. 2000;24:216-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 24. | Himeno S, Yasuda S, Shimada H, Tajima T, Makuuchi H. Evaluation of esophageal cancer by positron emission tomography. Jpn J Clin Oncol. 2002;32:340-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 25. | Moghtadaei M, Hashemi Golpayegani MR, Malekzadeh R. A variable structure fuzzy neural network model of squamous dysplasia and esophageal squamous cell carcinoma based on a global chaotic optimization algorithm. J Theor Biol. 2013;318:164-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 26. | Etemadi A, Abnet CC, Golozar A, Malekzadeh R, Dawsey SM. Modeling the risk of esophageal squamous cell carcinoma and squamous dysplasia in a high risk area in Iran. Arch Iran Med. 2012;15:18-21. [PubMed] |
| 27. | Li SX, Zeng QY, Li LF, Zhang YJ, Wan MM, Liu ZM, Xiong HL, Guo ZY, Liu SH. Study of support vector machine and serum surface-enhanced Raman spectroscopy for noninvasive esophageal cancer detection. J Biomed Opt. 2013;18:27008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 28. | Rice TW, Apperson-Hansen C, DiPaola LM, Semple ME, Lerut TE, Orringer MB, Chen LQ, Hofstetter WL, Smithers BM, Rusch VW, Wijnhoven BP, Chen KN, Davies AR, D'Journo XB, Kesler KA, Luketich JD, Ferguson MK, Räsänen JV, van Hillegersberg R, Fang W, Durand L, Allum WH, Cecconello I, Cerfolio RJ, Pera M, Griffin SM, Burger R, Liu JF, Allen MS, Law S, Watson TJ, Darling GE, Scott WJ, Duranceau A, Denlinger CE, Schipper PH, Ishwaran H, Blackstone EH. Worldwide Esophageal Cancer Collaboration: clinical staging data. Dis Esophagus. 2016;29:707-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 107] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 29. | Sato F, Shimada Y, Selaru FM, Shibata D, Maeda M, Watanabe G, Mori Y, Stass SA, Imamura M, Meltzer SJ. Prediction of survival in patients with esophageal carcinoma using artificial neural networks. Cancer. 2005;103:1596-1605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 58] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 30. | Mofidi R, Deans C, Duff MD, de Beaux AC, Paterson Brown S. Prediction of survival from carcinoma of oesophagus and oesophago-gastric junction following surgical resection using an artificial neural network. Eur J Surg Oncol. 2006;32:533-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 31. | Wang CY, Lee TF, Fang CH, Chou JH. Fuzzy logic-based prognostic score for outcome prediction in esophageal cancer. IEEE Trans Inf Technol Biomed. 2012;16:1224-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 32. | Mi H, Petitjean C, Dubray B, Vera P, Ruan S. Robust feature selection to predict tumor treatment outcome. Artif Intell Med. 2015;64:195-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 33. | Zahedi H, Mehrshad N, Anvari K. Intelligent modelling of oesophageal cancer treatment and its use to determine the dose of chemotherapy drug. J Med Eng Technol. 2012;36:261-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 34. | Maktabi M, Köhler H, Ivanova M, Jansen-Winkeln B, Takoh J, Niebisch S, Rabe SM, Neumuth T, Gockel I, Chalopin C. Tissue classification of oncologic esophageal resectates based on hyperspectral data. Int J Comput Assist Radiol Surg. 2019;14:1651-1661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 35. | Huang FL, Yu SJ. Esophageal cancer: Risk factors, genetic association, and treatment. Asian J Surg. 2018;41:210-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 542] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 36. | Warnecke-Eberz U, Metzger R, Bollschweiler E, Baldus SE, Mueller RP, Dienes HP, Hoelscher AH, Schneider PM. TaqMan low-density arrays and analysis by artificial neuronal networks predict response to neoadjuvant chemoradiation in esophageal cancer. Pharmacogenomics. 2010;11:55-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 37. | Ganeshan B, Skogen K, Pressney I, Coutroubis D, Miles K. Tumour heterogeneity in oesophageal cancer assessed by CT texture analysis: preliminary evidence of an association with tumour metabolism, stage, and survival. Clin Radiol. 2012;67:157-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 257] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 38. | Javeri H, Xiao L, Rohren E, Komaki R, Hofstetter W, Lee JH, Maru D, Bhutani MS, Swisher SG, Wang X, Ajani JA. Influence of the baseline 18F-fluoro-2-deoxy-D-glucose positron emission tomography results on survival and pathologic response in patients with gastroesophageal cancer undergoing chemoradiation. Cancer. 2009;115:624-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 39. | Schollaert P, Crott R, Bertrand C, D'Hondt L, Borght TV, Krug B. A systematic review of the predictive value of (18)FDG-PET in esophageal and esophagogastric junction cancer after neoadjuvant chemoradiation on the survival outcome stratification. J Gastrointest Surg. 2014;18:894-905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 40. | Ypsilantis PP, Siddique M, Sohn HM, Davies A, Cook G, Goh V, Montana G. Predicting Response to Neoadjuvant Chemotherapy with PET Imaging Using Convolutional Neural Networks. PLoS One. 2015;10:e0137036. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 120] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 41. | Jin X, Zheng X, Chen D, Jin J, Zhu G, Deng X, Han C, Gong C, Zhou Y, Liu C, Xie C. Prediction of response after chemoradiation for esophageal cancer using a combination of dosimetry and CT radiomics. Eur Radiol. 2019;29:6080-6088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 42. | Yang HX, Feng W, Wei JC, Zeng TS, Li ZD, Zhang LJ, Lin P, Luo RZ, He JH, Fu JH. Support vector machine-based nomogram predicts postoperative distant metastasis for patients with oesophageal squamous cell carcinoma. Br J Cancer. 2013;109:1109-1116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 43. | Wang ZL, Zhou ZG, Chen Y, Li XT, Sun YS. Support Vector Machines Model of Computed Tomography for Assessing Lymph Node Metastasis in Esophageal Cancer with Neoadjuvant Chemotherapy. J Comput Assist Tomogr. 2017;41:455-460. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 44. | Lin T, Liu T, Lin Y, Zhang C, Yan L, Chen Z, He Z, Wang J. Serum levels of chemical elements in esophageal squamous cell carcinoma in Anyang, China: a case-control study based on machine learning methods. BMJ Open. 2017;7:e015443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 45. | Mourikis TP, Benedetti L, Foxall E, Temelkovski D, Nulsen J, Perner J, Cereda M, Lagergren J, Howell M, Yau C, Fitzgerald RC, Scaffidi P; Oesophageal Cancer Clinical and Molecular Stratification (OCCAMS) Consortium, Ciccarelli FD. Patient-specific cancer genes contribute to recurrently perturbed pathways and establish therapeutic vulnerabilities in esophageal adenocarcinoma. Nat Commun. 2019;10:3101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 46. | Rice TW, Lu M, Ishwaran H, Blackstone EH; Worldwide Esophageal Cancer Collaboration Investigators. Precision Surgical Therapy for Adenocarcinoma of the Esophagus and Esophagogastric Junction. J Thorac Oncol. 2019;14:2164-2175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 47. | Goetz M. Enhanced Imaging of the Esophagus: Confocal Laser Endomicroscopy. In: Pleskow DK, Erim T. Barrett's Esophagus. Boston: Academic Press, 2016: 123-132. |
| 48. | Gora MJ, Tearney GJ. Enhanced Imaging of the Esophagus: Optical Coherence Tomography. In: Pleskow DK, Erim T. Barrett's Esophagus. Boston: Academic Press, 2016: 105-122. |
| 49. | Falk GW, Wani S, Chandrasekhara V, Elmunzer BJ, Khashab MA, Muthusamy VR. Chandrasekhara V, Elmunzer BJ, Khashab MA, Muthusamy VR. Barrett's Esophagus: Diagnosis, Surveillance, and Medical Management. In: Chandrasekhara V, Elmunzer BJ, Khashab MA, Muthusamy VR. Clinical Gastrointestinal Endoscopy (Third Edition). Philadelphia: Content Repository Only, 2019: 279-90.e5. |
| 50. | di Pietro M, Canto MI, Fitzgerald RC. Endoscopic Management of Early Adenocarcinoma and Squamous Cell Carcinoma of the Esophagus: Screening, Diagnosis, and Therapy. Gastroenterology. 2018;154:421-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 173] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 51. | de Lange T, Halvorsen P, Riegler M. Methodology to develop machine learning algorithms to improve performance in gastrointestinal endoscopy. World J Gastroenterol. 2018;24:5057-5062. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 52. | ASGE Standards of Practice Committee, Qumseya B, Sultan S, Bain P, Jamil L, Jacobson B, Anandasabapathy S, Agrawal D, Buxbaum JL, Fishman DS, Gurudu SR, Jue TL, Kripalani S, Lee JK, Khashab MA, Naveed M, Thosani NC, Yang J, DeWitt J, Wani S; ASGE Standards of Practice Committee Chair. ASGE guideline on screening and surveillance of Barrett's esophagus. Gastrointest Endosc. 2019;90:335-359.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 262] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 53. | Münzenmayer C, Kage A, Wittenberg T, Mühldorfer S. Computer-assisted diagnosis for precancerous lesions in the esophagus. Methods Inf Med. 2009;48:324-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 54. | van der Sommen F, Aylward S, Zinger S, Schoon EJ, de With PHN. Proceedings of SPIE - The International Society for Optical Engineering; 2013 Feb 8; Proc SPIE Medical Imaging (SPIE 8670). [DOI] [Full Text] |
| 55. | van der Sommen F, Zinger S, Curvers WL, Bisschops R, Pech O, Weusten BL, Bergman JJ, de With PH, Schoon EJ. Computer-aided detection of early neoplastic lesions in Barrett's esophagus. Endoscopy. 2016;48:617-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 127] [Article Influence: 14.1] [Reference Citation Analysis (2)] |
| 56. | Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Ozawa T, Ishihara S, Kumagai Y, Fujishiro M, Maetani I, Fujisaki J, Tada T. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 273] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 57. | Ghatwary N, Zolgharni M, Ye X. Early esophageal adenocarcinoma detection using deep learning methods. Int J Comput Assist Radiol Surg. 2019;14:611-621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 58. | Hashimoto R, Requa J, Dao T, Ninh A, Tran E, Mai D, Lugo M, El-Hage Chehade N, Chang KJ, Karnes WE, Samarasena JB. Artificial intelligence using convolutional neural networks for real-time detection of early esophageal neoplasia in Barrett's esophagus (with video). Gastrointest Endosc. 2020;91:1264-1271.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 145] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 59. | Ebigbo A, Mendel R, Probst A, Manzeneder J, Souza LA, Papa JP, Palm C, Messmann H. Computer-aided diagnosis using deep learning in the evaluation of early oesophageal adenocarcinoma. Gut. 2019;68:1143-1145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 103] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 60. | de Groof AJ, Struyvenberg MR, van der Putten J, van der Sommen F, Fockens KN, Curvers WL, Zinger S, Pouw RE, Coron E, Baldaque-Silva F, Pech O, Weusten B, Meining A, Neuhaus H, Bisschops R, Dent J, Schoon EJ, de With PH, Bergman JJ. Deep-Learning System Detects Neoplasia in Patients With Barrett's Esophagus With Higher Accuracy Than Endoscopists in a Multistep Training and Validation Study With Benchmarking. Gastroenterology. 2020;158:915-929.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 222] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 61. | Hong J, Park BY, Park H. Convolutional neural network classifier for distinguishing Barrett's esophagus and neoplasia endomicroscopy images. Conf Proc IEEE Eng Med Biol Soc. 2017;2017:2892-2895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 62. | Swager AF, van der Sommen F, Klomp SR, Zinger S, Meijer SL, Schoon EJ, Bergman JJGHM, de With PH, Curvers WL. Computer-aided detection of early Barrett's neoplasia using volumetric laser endomicroscopy. Gastrointest Endosc. 2017;86:839-846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 102] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 63. | van der Sommen F, Klomp SR, Swager AF, Zinger S, Curvers WL, Bergman JJGHM, Schoon EJ, de With PHN. Predictive features for early cancer detection in Barrett's esophagus using Volumetric Laser Endomicroscopy. Comput Med Imaging Graph. 2018;67:9-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 64. | Trindade AJ, McKinley MJ, Fan C, Leggett CL, Kahn A, Pleskow DK. Endoscopic Surveillance of Barrett's Esophagus Using Volumetric Laser Endomicroscopy With Artificial Intelligence Image Enhancement. Gastroenterology. 2019;157:303-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 65. | Struyvenberg MR, van der Sommen F, Swager AF, de Groof AJ, Rikos A, Schoon EJ, Bergman JJ, de With PHN, Curvers WL. Improved Barrett's neoplasia detection using computer-assisted multiframe analysis of volumetric laser endomicroscopy. Dis Esophagus. 2020;33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 66. | van der Putten J, Struyvenberg M, de Groof J, Scheeve T, Curvers W, Schoon E, Bergman JJGHM, de With PHN, van der Sommen F. Deep principal dimension encoding for the classification of early neoplasia in Barrett's Esophagus with volumetric laser endomicroscopy. Comput Med Imaging Graph. 2020;80:101701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 67. | Shin D, Lee MH, Polydorides AD, Pierce MC, Vila PM, Parikh ND, Rosen DG, Anandasabapathy S, Richards-Kortum RR. Quantitative analysis of high-resolution microendoscopic images for diagnosis of neoplasia in patients with Barrett's esophagus. Gastrointest Endosc. 2016;83:107-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 68. | Qi X, Sivak MV, Isenberg G, Willis JE, Rollins AM. Computer-aided diagnosis of dysplasia in Barrett's esophagus using endoscopic optical coherence tomography. J Biomed Opt. 2006;11:044010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 79] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 69. | Kohli DR, Schubert ML, Zfass AM, Shah TU. Performance characteristics of optical coherence tomography in assessment of Barrett's esophagus and esophageal cancer: systematic review. Dis Esophagus. 2017;30:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 70. | Sabo E, Beck AH, Montgomery EA, Bhattacharya B, Meitner P, Wang JY, Resnick MB. Computerized morphometry as an aid in determining the grade of dysplasia and progression to adenocarcinoma in Barrett's esophagus. Lab Invest. 2006;86:1261-1271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 39] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 71. | Liu DY, Gan T, Rao NN, Xing YW, Zheng J, Li S, Luo CS, Zhou ZJ, Wan YL. Identification of lesion images from gastrointestinal endoscope based on feature extraction of combinational methods with and without learning process. Med Image Anal. 2016;32:281-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (1)] |
| 72. | Cai SL, Li B, Tan WM, Niu XJ, Yu HH, Yao LQ, Zhou PH, Yan B, Zhong YS. Using a deep learning system in endoscopy for screening of early esophageal squamous cell carcinoma (with video). Gastrointest Endosc. 2019;90:745-753.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 73. | Ohmori M, Ishihara R, Aoyama K, Nakagawa K, Iwagami H, Matsuura N, Shichijo S, Yamamoto K, Nagaike K, Nakahara M, Inoue T, Aoi K, Okada H, Tada T. Endoscopic detection and differentiation of esophageal lesions using a deep neural network. Gastrointest Endosc. 2020;91:301-309.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 74. | Zhao YY, Xue DX, Wang YL, Zhang R, Sun B, Cai YP, Feng H, Cai Y, Xu JM. Computer-assisted diagnosis of early esophageal squamous cell carcinoma using narrow-band imaging magnifying endoscopy. Endoscopy. 2019;51:333-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 75. | Oyama T, Inoue H, Arima M, Momma K, Omori T, Ishihara R, Hirasawa D, Takeuchi M, Tomori A, Goda K. Prediction of the invasion depth of superficial squamous cell carcinoma based on microvessel morphology: magnifying endoscopic classification of the Japan Esophageal Society. Esophagus. 2017;14:105-112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 224] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 76. | Nakagawa K, Ishihara R, Aoyama K, Ohmori M, Nakahira H, Matsuura N, Shichijo S, Nishida T, Yamada T, Yamaguchi S, Ogiyama H, Egawa S, Kishida O, Tada T. Classification for invasion depth of esophageal squamous cell carcinoma using a deep neural network compared with experienced endoscopists. Gastrointest Endosc. 2019;90:407-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 77. | Everson M, Herrera L, Li W, Luengo IM, Ahmad O, Banks M, Magee C, Alzoubaidi D, Hsu HM, Graham D, Vercauteren T, Lovat L, Ourselin S, Kashin S, Wang HP, Wang WL, Haidry RJ. Artificial intelligence for the real-time classification of intrapapillary capillary loop patterns in the endoscopic diagnosis of early oesophageal squamous cell carcinoma: A proof-of-concept study. United European Gastroenterol J. 2019;7:297-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 69] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 78. | Luo H, Xu G, Li C, He L, Luo L, Wang Z, Jing B, Deng Y, Jin Y, Li Y, Li B, Tan W, He C, Seeruttun SR, Wu Q, Huang J, Huang DW, Chen B, Lin SB, Chen QM, Yuan CM, Chen HX, Pu HY, Zhou F, He Y, Xu RH. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case-control, diagnostic study. Lancet Oncol. 2019;20:1645-1654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 253] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 79. | Guo L, Xiao X, Wu C, Zeng X, Zhang Y, Du J, Bai S, Xie J, Zhang Z, Li Y, Wang X, Cheung O, Sharma M, Liu J, Hu B. Real-time automated diagnosis of precancerous lesions and early esophageal squamous cell carcinoma using a deep learning model (with videos). Gastrointest Endosc. 2020;91:41-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 153] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 80. | Thakkar SJ, Kochhar GS. Artificial intelligence for real-time detection of early esophageal cancer: another set of eyes to better visualize. Gastrointest Endosc. 2020;91:52-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 81. | Kodashima S, Fujishiro M, Takubo K, Kammori M, Nomura S, Kakushima N, Muraki Y, Goto O, Ono S, Kaminishi M, Omata M. Ex vivo pilot study using computed analysis of endo-cytoscopic images to differentiate normal and malignant squamous cell epithelia in the oesophagus. Dig Liver Dis. 2007;39:762-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 82. | Shin D, Protano MA, Polydorides AD, Dawsey SM, Pierce MC, Kim MK, Schwarz RA, Quang T, Parikh N, Bhutani MS, Zhang F, Wang G, Xue L, Wang X, Xu H, Anandasabapathy S, Richards-Kortum RR. Quantitative analysis of high-resolution microendoscopic images for diagnosis of esophageal squamous cell carcinoma. Clin Gastroenterol Hepatol. 2015;13:272-279.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 66] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 83. | Quang T, Schwarz RA, Dawsey SM, Tan MC, Patel K, Yu X, Wang G, Zhang F, Xu H, Anandasabapathy S, Richards-Kortum R. A tablet-interfaced high-resolution microendoscope with automated image interpretation for real-time evaluation of esophageal squamous cell neoplasia. Gastrointest Endosc. 2016;84:834-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 61] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 84. | Protano MA, Xu H, Wang G, Polydorides AD, Dawsey SM, Cui J, Xue L, Zhang F, Quang T, Pierce MC, Shin D, Schwarz RA, Bhutani MS, Lee M, Parikh N, Hur C, Xu W, Moshier E, Godbold J, Mitcham J, Hudson C, Richards-Kortum RR, Anandasabapathy S. Low-Cost High-Resolution Microendoscopy for the Detection of Esophageal Squamous Cell Neoplasia: An International Trial. Gastroenterology. 2015;149:321-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 85. | Mutasa S, Sun S, Ha R. Understanding artificial intelligence based radiology studies: What is overfitting? Clin Imaging. 2020;65:96-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 92] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 86. | Mori Y, Kudo SE, East JE, Rastogi A, Bretthauer M, Misawa M, Sekiguchi M, Matsuda T, Saito Y, Ikematsu H, Hotta K, Ohtsuka K, Kudo T, Mori K. Cost savings in colonoscopy with artificial intelligence-aided polyp diagnosis: an add-on analysis of a clinical trial (with video). Gastrointest Endosc. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 87. | London AJ. Artificial Intelligence and Black-Box Medical Decisions: Accuracy versus Explainability. Hastings Cent Rep. 2019;49:15-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 302] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 88. | Lawrence DR, Palacios-González C, Harris J. Artificial Intelligence. Camb Q Healthc Ethics. 2016;25:250-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 89. | Yu KH, Beam AL, Kohane IS. Artificial intelligence in healthcare. Nat Biomed Eng. 2018;2:719-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 732] [Cited by in RCA: 1088] [Article Influence: 155.4] [Reference Citation Analysis (0)] |
| 90. | O'Sullivan S, Nevejans N, Allen C, Blyth A, Leonard S, Pagallo U, Holzinger K, Holzinger A, Sajid MI, Ashrafian H. Legal, regulatory, and ethical frameworks for development of standards in artificial intelligence (AI) and autonomous robotic surgery. Int J Med Robot. 2019;15:e1968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 130] [Article Influence: 21.7] [Reference Citation Analysis (0)] |