Published online Aug 28, 2020. doi: 10.3748/wjg.v26.i32.4817
Peer-review started: March 12, 2020
First decision: May 15, 2020
Revised: July 2, 2020
Accepted: August 20, 2020
Article in press: August 20, 2020
Published online: August 28, 2020
Processing time: 168 Days and 17.5 Hours
Polymorphisms of human leukocyte antigen (HLA) genes are suggested to increase the risk of gastric cancer (GC).
To investigate the HLA allele frequencies of patients with GC relative to a control group in terms of CagA+ multiple (≥ 2) EPIYA-C repeats.
The patient group comprised 94 patients [44 GC and 50 duodenal ulcer (DU) patients], and the control group comprised 86 individuals [(50 non-ulcer dyspepsia patients and 36 people with asymptomatic Helicobacter pylori (H. pylori)]. Polymerase chain reaction was performed for the amplification of the H. pylori cagA gene and typing of EPIYA motifs. HLA sequence-specific oligonucleotide (SSO) typing was performed using Lifecodes SSO typing kits (HLA-A, HLA-B HLA-C, HLA-DRB1, and HLA-DQA1-B1 kits).
The comparison of GC cases in terms of CagA+ multiple (≥ 2) EPIYA-C repeats showed that only the HLA-DQB1*06 allele [odds ratio (OR): 0.37, P = 0.036] was significantly lower, but significance was lost after correction (Pc = 0.1845). The HLA-DQA1*01 allele had a high ratio in GC cases with multiple EPIYA-C repeats, but this was not significant in the univariate analysis. We compared allele frequencies in the DU cases alone and in GC and DU cases together using the same criterion, and none of the HLA alleles were significantly associated with GC or DU. Also, none of the alleles were detected as independent risk factors after the multivariate analysis. On the other hand, in a multivariate logistic regression with no discriminative criterion, HLA-DQA1*01 (OR = 1.848), HLA-DQB1*06 (OR = 1.821) and HLA-A*02 (OR = 1.579) alleles were detected as independent risk factors for GC and DU.
None of the HLA alleles were detected as independent risk factors in terms of CagA+ multiple EPIYA-C repeats. However, HLA-DQA1*01, HLA-DQB1*0601, and HLA-A*2 were independent risk factors with no criterion in the multivariate analysis. We suggest that the association of these alleles with gastric malignancies is not specifically related to cagA and multiple EPIYA C repeats.
Core tip: The development of gastric cancer (GC) is suggested to be related to the interactions of bacterial virulence and host genetic factors with the immune response of the host. The effects of polymorphisms in the human leukocyte antigen (HLA) gene may regulate the degree of the inflammatory response of the host leading to the gastric malignancies. We could not detect any prominent HLA alleles between the patient and control groups in terms of CagA+ multiple (≥ 2) EPIYA-C repeat numbers. HLA-DQA1*01, HLA-DQB1*06, and HLA-A*02 were detected as independent risk factors for the risk of GC and duodenal ulcer with no criterion in multivariate analyses. We suggest that the association of these alleles with gastric malignancies is not specifically related to cagA and multiple EPIYA C repeats.
- Citation: Saribas S, Demiryas S, Yilmaz E, Uysal O, Kepil N, Demirci M, Caliskan R, Dinc HO, Akkus S, Gareayaghi N, Kirmusaoglu S, Ozbey D, Tokman HB, Koksal SS, Tasci I, Kocazeybek B. Association between human leukocyte antigen gene polymorphisms and multiple EPIYA-C repeats in gastrointestinal disorders. World J Gastroenterol 2020; 26(32): 4817-4832
- URL: https://www.wjgnet.com/1007-9327/full/v26/i32/4817.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i32.4817
Gastric cancer (GC) is the third most common cause of death among all cancer types (8.2% of all cancers, 2018, World Health Organization)[1]. GC has been closely associated with Helicobacter pylori (H. pylori) infections, which generally cause mild gastrointestinal symptoms. However, in a few infected patients, they may progress to peptic ulcer (PU) (10%-15%) or GC (1%-3%)[2].
Generally, H. pylori infections cause gastric inflammation and a chronic inflammatory response that result in progressive mucosal damage. Eventually, the gastric mucosa transforms into metaplastic and dysplastic epithelia, which leads to gastric adenocarcinomas[3]. The development of GC is suggested to be related to the interactions between bacterial virulence factors, host genetic factors such as the human leukocyte antigen (HLA) gene, the immune response of the host, and environmental factors such as diet and smoking[4]. Some of the host factors may be related to polymorphisms genes such as HLA genes, which regulate the strength of the inflammatory response and influence the probability of specific clinical results[5].
CagA is among the most important virulence factors specific to H. pylori and is delivered into the gastric epithelium cells by the type IV secretion system (T4SS) of the bacterium. It is involved with intracellular signal transduction pathways and leads to the malignant transformation of gastric epithelial cells[6]. CagA subtypes are defined by the EPIYA variants (Glu-Pro-Ile-Tyr-Ala) in their C-terminal region, and there are four types of EPIYA motifs: EPIYA-A, -B, -C, and -D[7]. H. pylori isolates from East Asia are associated with a higher incidence of GC and contain EPIYA A-B-D motifs. However, in Western countries, GC cases have EPIYA A-B-C motifs. H. pylori strains with multiple EPIYA-C repeats have higher phosphorylation capacity and SHP-2 binding affinity and are significantly associated with GC[8].
HLA class I and II proteins bind to bacterial antigen peptides or tumor proteins and present these peptides and proteins to T cells, which leads to the differentiation of T cells into cytotoxic or helper T cells. In HLA gene polymorphism, a variety of alleles may occupy the same locus. A limited number of studies have focused on the relation between gastrointestinal pathologies such as the development of GC risk and HLA polymorphisms in different geographic regions and populations[9,10,11].
In addition to HLA gene polymorphisms, the virulence factors of H. pylori strains may also be related to the intensity of the inflammatory response[3]. The limited studies on HLA polymorphisms have focused on MHC class II and specifically the relation of HLA polymorphisms and the development of GC risk[10,12,13]. In these studies, the following HLA class II polymorphisms were suggested to be associated with the development of GC risk: DQB1*03, HLA-DRB1*04, and HLA-DQA1*01/*03[14,15].
Reported HLA allele frequencies related to GC pathologies with H. pylori positivity have been contradictory among different ethnic groups. The regional and ethnic differences are very important for the association of HLA gene polymorphism and GC risk. These contradictory results may be attributed to factors such as differences in populations, research designs, environmental factors, H. pylori virulence factors, and host genetic factors, such as polymorphisms of HLA alleles. For example, the HLA-DQB1*0301 allele was positively associated with GC in Caucasian populations but negatively associated with it in Taiwanese populations[13,16]. This HLA polymorphism had no effect in Japanese populations[17]. HLA-DQB1*0401 82 and *0602 alleles increase the GC risk in European and Indonesian populations[9,18].
To the best of our knowledge, no studies have compared GC and duodenal ulcer (DU) cases with controls in regard to HLA allele frequencies in terms of differentiation by the CagA+ multiple (≥ 2) EPIYA-C repeat numbers. Therefore, we aimed to investigate the allele frequencies of HLA class I and II in a patient group [H. pylori (+) GC and DU patients] and compared the results to those of a control group [H. pylori (+) non-ulcer dyspepsia (NUD) and asymptomatic H. pylori] in terms of CagA+ multiple EPIYA-C repeats for the first time in a Turkish population.
This case–control study was conducted between July 10, 2014 and November 9, 2017. The patient group comprised 94 patients, including 44 (46.8%) GC and 50 (53.2%) DU patients with 58 (61.7%) males, 36 (38.3%) females, a mean age of 49.6 years, and age range of 19–79 years. The control group comprised 86 individuals including 50 (58.1%) NUD patients and 36 (41.9%) people with asymptomatic H. pylori. This group had 30 (34.9%) males, 56 (65.1%) females, a mean age of 47.3 years, and age range of 18–86 years. All of the GC + DU patients and the NUD + asymptomatic H. pylori members of the control group members had H. pylori.
The NUD + Asymptomatic H. pylori control group was matched with the GC + DU patient group according to the age and gender distribution of the patient group (P > 0.05). Blood samples for the genotyping of HLA alleles were collected when obtaining biopsy samples (from the corpus and antrum) on the same day. The antrum and corpus biopsy specimens were stored and used in molecular studies.
We excluded patient and control group individuals with autoimmune diseases and who were under 18 years old, had previous gastric surgery and H. pylori eradication treatment with antibiotics, antisecretory drugs and bismuth salts, in the month prior to sampling. The study was reviewed and approved by the Clinical Research Ethics Board of Istanbul University-Cerrahpasa, Cerrahpasa Faculty of Medicine (No. 83045809/32-38/A-15/2014). The study was also conducted according to the standards of the Declaration of Helsinki. All study participants or their legal guardians provided informed written consent prior to the study.
H. pylori DNA extractions were performed using the antrum and corpus biopsy specimens. Genomic DNA extraction (Real Genomics Quality Nucleic Acid/ Purification system; RBC Bioscience Laboratories, Taipei, Taiwan) and QIAamp DNA mini prep kits (Qiagen, Hilden Germany) were used.
An H. pylori-QLS 1.0 kit (Fluorion, Iontek, Istanbul, Turkey) was used for the detection of 156 bp of the ureC gene in H. pylori DNA extractions[19].
Primers reported in related studies were used for the detection of the H. pylori cagA gene (349 bp) (Table 1)[20,21]. The polymerase chain reaction (PCR) cycles were as follows: Denaturation at 95 ºC for 2 min, followed by 45 cycles of 95 ºC for 30 s, 45 s at 53 ºC, and 45 s at 72 ºC. The final elongation was done for 5 min at 72 ºC.
| Primer | Primer sequence (59R39) | Ref. |
| cagA-F | GATAACAGGCAAGCTTTTGAGG1 | [19,20] |
| cagA-R | CTGCAAAAGATTGTTTGGCAGA1 | |
| cagA28F | TTCTCAAAGGAGCAATTGGC2 | [21] |
| cagA-P1C | GTCCTGCTTTCTTTTTATTAACTTKAGC2 | |
| cagA-P2CG | TTTAGCAACTTGAGCGTAAATGGG2 | |
| cagA-P2TA | TTTAGCAACTTGAGTATAAATGGG2 | |
| cagA-P3E | ATCAATTGTAGCGTAAATGGG2 | |
| “cag empty PCR” | GCTTGCTTGTATTGGCCTTG / GCATGCACATTCCCTAAAGT3 | [22] |
The amplification of H. pylori DNA for EPIYA motifs was done using the forward primer cagA28F and reverse primers cagA-P1C, cagA-P2CG, cagA-P2TA, and cagA-P3E (Table 1)[22]. In the PCR assay, the protocol steps were as follows: Initial denaturation step, one cycle at 95 ºC for 2 min, 50 cycles at 95 ºC for 30 s, 57 ºC for 45 s, and 72 ºC for 35 s; and a final extension at 72 ºC for 5 min. After the PCR amplification, PCR products were sequenced bidirectionally using a Sequence Reagent Mix kit with an ABI Prism (310) analyzer (Applied Biosystems, United States).
An empty-site-positive PCR assay was used to confirm the EPIYA-negative H. pylori strains[23]. To confirm cagPAI in all of the strains, amplification was performed with two primers (forward 468 HP519 and reverse 496 HP549 primers of the reference HP519 and HP549 H. pylori strains; Table 1, cag empty PCR). In the PCR assay, the protocol steps were as follows: Initial denaturation at 95 ºC for 2 min, 40 cycles at 95 ºC for 30 s, 57 ºC for 30 s, and 72 ºC for 20 s; followed by a final extension at 72 ºC for 5 min.
Whole blood samples (10 mL) were collected from the patient and control group cases during a biopsy procedure. An EZ1 DNA extraction kit (Qiagen, Germany) was used in a DNA isolation device (Bio Robot EZ1; Qiagen, Germany) for the DNA isolation procedure from 3-mL blood samples collected in tubes containing ethylenediaminetetraacetic acid. Isolated DNA samples were stored at -70 °C until laboratory studies. HLA typing at low levels (2 digits) of HLA-A, HLA-B, HLA-C, and HLA-DQ alleles were done with a Luminex 100/200 instrument with sequence-specific oligonucleotide (SSO) probes bound to color-coded microbeads. LIFECODES SSO HLA typing kits (Lifecodes, Immucor, Germany) were used for the typing of HLA-A HLA-B, HLA-DRB1, and HLA-DQA1/B1. This typing test is a reverse sequence-specific oligonucleotide (rSSO) DNA typing assay using SSO probes and color-coded microspheres.
The PCR mixture was composed of 15 μL of lifecodes Master Mix, 200 ng of genomic DNA, and 2.5 U Taq polymerase in a final volume of 50 μL. In the PCR assay, the steps were as follows: Initial denaturation step at 95 °C for 5 min; 40 cycles including 8 cycles at 95 °C for 30 s, 60 °C for 45 s, and 72 °C for 45 s, followed by 32 cycles at 95 °C for 30 s, 63 °C for 45 s, and 72 °C for 45 s, and a final extension step at
Hardy-Weinberg (H-W) equilibrium and Linkage Disequilibrium (LD) were examined for HLA-A, HLA-B, HLA-C, HLA-DR1, HLA-DQA1 and HLA-DQB1 allele polymorphisms[24]. Genepop software version 4.7 was used to calculate the Hardy–Weinberg equilibrium and LD. H–W equilibrium was present for P-values > 0.05. The allele frequencies of the GC + DU patient and the NUD + Asymptomatic H. pylori control groups and subgroups were compared by the chi-squared (χ2) test and Fisher’s exact test (Tables 2-5). Corrected P values (Pc) were calculated by multiplying with the allele numbers (A = 15, B = 26, DRB1 = 12, DQA1 = 6, and DQB1 = 5) in each locus by Bonferroni correction. Multivariate logistic regression (enter method) was used to assess the relation of HLA alleles and the risk of GC and DU development in terms of CagA+ multiple EPIYA-C repeat numbers (Table 6). P < 0.05 was used to determine significance. SPSS 25.0 (IBM Corporation, Armonk, NY, United States) was used for the analyses.
| HLA alleles | Patient groupGC + DU, H. pylori (+) (n = 94, alleles = 198) | Control groupNUD + Asymptomatic, H. pylori (+) (n = 86, alleles = 172) | OR | 95%CI | P value | |
| Minimum | Maximum | |||||
| HLAs increasing susceptibility to GC/DU | ||||||
| HLA-A*02 | 52 (27.6) | 38 (22) | 1.34 | 0.833 | 2.182 | 0.2239 |
| HLA-B*35 | 40 (21.3) | 26 (15.1) | 1.51 | 0.880 | 2.615 | 0.1329 |
| HLA-DRB1*13 | 36 (19) | 24 (14) | 1.46 | 0.831 | 2.567 | 0.1880 |
| HLA-DQA1*01 | 76 (40) | 30 (17.4) | 3.21 | 1.968 | 5.242 | 0.0001 |
| HLA-DQB1*06 | 52 (27.6) | 20 (11.6) | 2.90 | 1.652 | 5.139 | 0.0002 |
| HLAs making resistant to GC/DU | ||||||
| HLA-A*03 | 14 (7.4) | 20 (11.6) | 0.61 | 0.298 | 1.252 | 0.1787 |
| HLA-B*50 | 0 (0) | 10 (5.8) | 0.08 | 0.010 | 0.680 | 0.0201 |
| HLA-DRB1*04 | 16 (8.5) | 24 (14) | 0.57 | 0.293 | 1.120 | 0.1038 |
| HLA-DQA1*05 | 28 (14.9) | 54 (31.4) | 0.38 | 0.228 | 0.639 | 0.0003 |
| HLA-DQB1*03 | 42 (23) | 64 (37.2) | 0.48 | 0.305 | 0.770 | 0.0022 |
| GC (≥ 2) EPIYA-C (n = 26, alleles = 52) | GC (< 2) EPIYA-C (n = 18, alleles = 36) | OR | 95%CI | P value | |||
| Minimum | Maximum | ||||||
| HLA-A*02 | 12 (23) | 10 (27) | 0.78 | 0.29 | 2.06 | 0.6170 | |
| HLA-B*35 | 12 (23) | 10 (27) | 0.78 | 0.29 | 2.06 | 0.6170 | |
| HLA-DRB1*13 | 12 (23) | 12 (33) | 0.60 | 0.23 | 1.54 | 0.2903 | |
| HLA-DQA1*01 | 24 (46) | 16 (44) | 1.07 | 0.45 | 2.51 | 0.8742 | |
| HLA-DQB1*06 | 12 (23) | 16 (44) | 0.37 | 1.14 | 2.90 | 0.0369 | |
| DU (≥ 2) EPIYA-C (n = 14, alleles = 28) | DU (< 2) EPIYA-C (n = 36, alleles = 72) | ORc | 95%CI | P value | |||
| Minimum | Maximum | ||||||
| HLA-A*02 | 10 (36) | 20 (27) | 1.44 | 0.53 | 3.62 | 0.4380 | |
| HLA-B*35 | 4 (14) | 14 (19) | 0.80 | 0.24 | 0.680 | 0.0201 | |
| HLA-DRB1*13 | 6 (21) | 6 (8) | 3 | 0.87 | 10.26 | 0.0801 | |
| HLA-DQA1*01 | 10 (36) | 26 (36) | 0.9 | 0.39 | 2.44 | 0.9704 | |
| HLA-DQB1*06 | 8 (28) | 16 (22) | 1.4 | 0.52 | 3.75 | 0.5340 | |
| GC(≥ 2) EPIYA-C (n = 26, alleles=52) | DU(≥ 2) EPIYA-C (n = 14, alleles=28) | OR | 95%CI | P value | |||
| Minimum | Maximum | ||||||
| HLA-A*02 | 12 (23) | 10 (36) | 0.54 | 0.19 | 1.47 | 0.2302 | |
| HLA-B*35 | 12 (23) | 4 (14) | 1.80 | 0.52 | 6.21 | 0.3527 | |
| HLA-DRB1*13 | 12 (23) | 6 (21) | 1.10 | 0.36 | 3.83 | 0.8663 | |
| HLA-DQA1*01 | 24 (46) | 10 (36) | 1.54 | 0.59 | 3.97 | 0.3689 | |
| HLA-DRB1*06 | 12 (23) | 8 (28) | 0.75 | 0.26 | 2.12 | 0.5889 | |
| P value | OR | 95%CI | ||
| Lower | Upper | |||
| HLA-DQA1*01 | 0.004 | 1.848 | 1.215 | 2.811 |
| HLA-DQA1*05 | 0.050 | |||
| HLA-DQB1*03 | 0.061 | |||
| HLA-DQB1*06 | 0.009 | 1.821 | 1.163 | 2.850 |
| HLA-A*02 | 0.040 | 1.579 | 1.021 | 2.442 |
| HLA-A*25 | 0.999 | |||
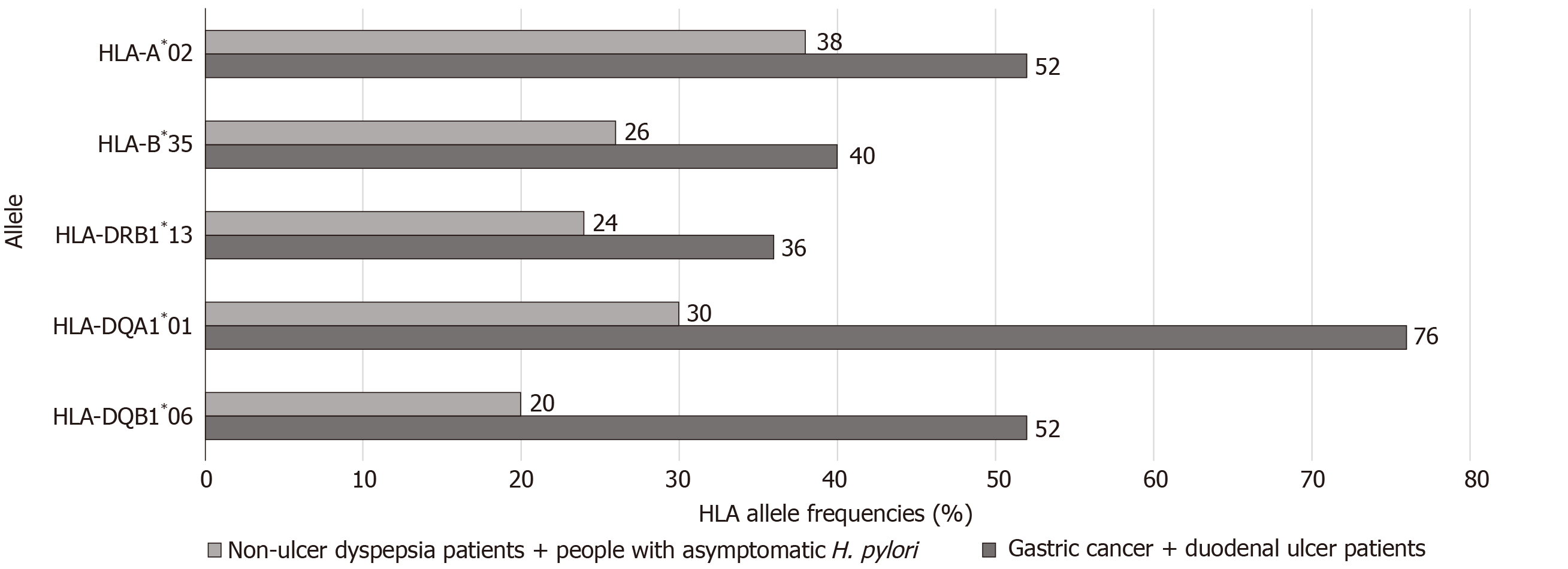
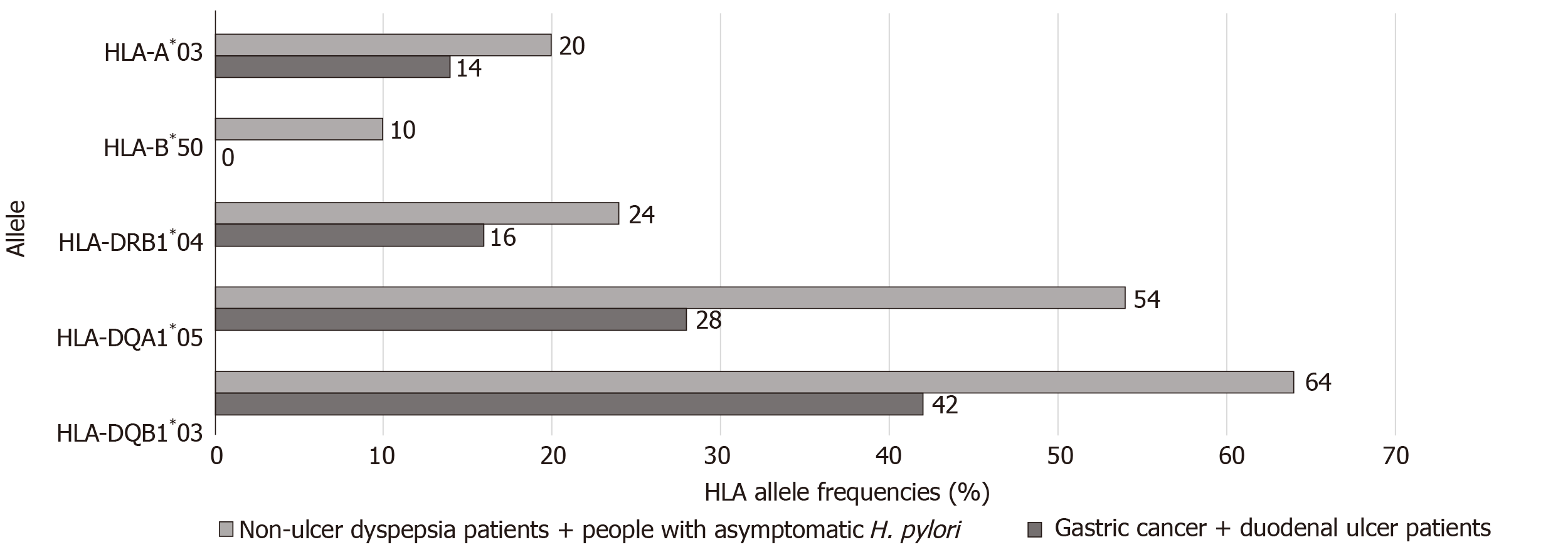
Sixty-four HLA alleles (41 for class I and 23 for class II) were found between the 94 HLA alleles tested in all of the groups (Table 7). The maximum numbers of alleles in the GC + DU patient group were 52 for HLA-A*02 (27.6%), 40 for HLA-B*35 (21.3%), 36 for HLA-DRB1*13 (19%), 76 for HLA-DQA1*01 (40%), and 52 for HLA-DQB1*06 (27.6%) (Figure 1). The maximum numbers of alleles in the NUD + Asymptomatic H. pylori control group were 20 for HLA-A*03 (11.6%), 10 for HLA-B*50 (5.81%), 24 for HLA-DRB1*04 (13.95%), 54 for HLA-DQA1*05 (31.4%), and 64 for HLA-DQB1*03 (37.2%) (Figure 2). Among GC cases, the genotype frequencies of HLA- B, DQA1 and DQB1 loci were in H-W equilibrium. Among DU cases, all of the genotype frequencies were in H-W equilibrium. Moreover, Among NUD cases, all of the genotype frequencies in H-W equilibrium except HLA-DQB1 cases and in asymptomatic H. pylori control group, all of the genotype frequencies were in H-W equilibrium except HLA-A cases. H-W equilibrium is only valid with a very large sample size and therefore, we suggest that some of our genotype frequencies are not in H-W equilibrium. The deviations from H-W equilibrium is larger at small sample sizes like this study and smaller at large sample sizes. HLA-DR1*13-HLA-DQA1*01-HLA-DQB1*06 haplotype frequency (10/44, 22% for GC; 2/50, 4% for DU, 2/50, 4% for NUD and 0 for NGIS) showed LD and had an odds ratio (OR) value as 6.143 (95%CI: 1.33-28.31, P = 0.0027) in the comparison of patient and control group cases. Moreover, HLA-DR1*13-HLA-DQA1*01-HLA-DQB1*06 haplotype frequency compared between GC and DU cases and OR was detected as 10.58 (95%CI: 2.27-49.34).
| HLA-A | Allele frequency | HLA-B | Allele frequency | HLA-DRB1 | Allele frequency | HLA-DQA1 | Allele frequency | HLA-DQB1 | Allele frequency | |||||
| Alleles | Cases (n = 94) | Control (n = 86) | Alleles | Cases (n = 94) | Control (n = 86) | Alleles | Cases (n = 94) | Control (n = 86) | Alleles | Cases (n = 94) | Control (n = 86) | Alleles | Cases (n = 94) | Control (n = 86) |
| 01 | 22 (11.4) | 24 (13.9) | 07 | 18 (9.5) | 22 (12.8) | 01 | 14 (7.4) | 8 (4.6) | 01 | 76 (40) | 30 (17.4) | 02 | 20 (10.6) | 26 (15.1) |
| 02 | 52 (27.6) | 38 (22) | 08 | 6 (3.2) | 6 (3.5) | 03 | 10 (5.3) | 10 (5.8) | 02 | 20 (10.6) | 20 (11.6) | 03 | 42 (23) | 64 (37.2) |
| 03 | 14 (7.4) | 20 (11.6) | 13 | 6 (3.2) | 4 (2.3) | 04 | 16 (8.5) | 22 (12.7) | 03 | 12 (6.4) | 22 (12.8) | 04 | 26 (13.8) | 28 (16.2) |
| 11 | 18 (9.5) | 18 (10.4) | 14 | 6 (3.2) | 4 (2.3) | 07 | 10 (5.3) | 20 (11.6) | 04 | 16 (8.5) | 20 (11.6) | 05 | 48 (25.5) | 34 (19.8) |
| 23 | 10 (5.3) | 4 (2.3) | 15 | 2 (1) | 6 (3.5) | 08 | 4 (2.1) | 6 (3.5) | 05 | 28 (14.9) | 54 (31.4) | 06 | 52 (27.6) | 20 (11.6) |
| 24 | 26 (13.8) | 24 (13.9) | 18 | 10 (5.3) | 10 (5.8) | 10 | 6 (3.2) | 4 (2.3) | 06 | 36 (19.1) | 26 (15.1) | |||
| 25 | 0 (0) | 2 (1.1) | 27 | 6 (3.2) | 4 (2.3) | 11 | 34 (18) | 42 (24.4) | ||||||
| 26 | 4 (2.1) | 4 (2.3) | 35 | 40 (21.3) | 26 (15.1) | 12 | 6 (3.2) | 2 (1.2) | ||||||
| 29 | 8 (4.2) | 8 (4.6) | 37 | 4 (2.1) | 0 (0) | 13 | 36 (19) | 24 (14) | ||||||
| 30 | 4 (2.1) | 6 (3.4) | 38 | 8 (4.2) | 2 (1.2) | 14 | 18 (9.5) | 8 (4.6) | ||||||
| 31 | 2 (1) | 4 (2.3) | 39 | 0 (0) | 4 (2.3) | 15 | 28 (14.9) | 22 (12.8) | ||||||
| 32 | 16 (8.5) | 10 (5.8) | 40 | 6 (3.2) | 6 (3.5) | 16 | 6 (3.2) | 4 (2.3) | ||||||
| 33 | 6 (3.1) | 4 (2.3) | 41 | 4 (2.1) | 2 (1.2) | |||||||||
| 68 | 4 (2.1) | 6 (3.4) | 44 | 8 (4.2) | 14 (8.1) | |||||||||
| 69 | 2 (1) | 0 (0) | 45 | 0 (0) | 2 (1.2) | |||||||||
| 48 | 2 (1) | 2 (1.2) | ||||||||||||
| 49 | 6 (3.2) | 4 (2.3) | ||||||||||||
| 50 | 0 (0) | 10 (5.8) | ||||||||||||
| 51 | 24 (12) | 30 (17.4) | ||||||||||||
| 52 | 14 (7.4) | 2 (1.2) | ||||||||||||
| 53 | 0 (0) | 2 (1.2) | ||||||||||||
| 54 | 2 (1) | 0 (0) | ||||||||||||
| 55 | 6 (3.2) | 8 (4.6) | ||||||||||||
| 57 | 2 (1) | 2 (1.2) | ||||||||||||
| 58 | 8 (4.2) | 0 (0) | ||||||||||||
| 78 | 0 (0) | 0 (0) | ||||||||||||
When comparing the prominent alleles detected, only HLA-DQA1*01 (OR: 3.211, P = 0.0001) and HLA-DQB1*06 (OR: 2.906, P = 0.0002) were significantly higher in the GC + DU patient group. The values also stayed significant after Bonferroni correction (DQB1*06: Pc = 0.001; DQA1*01: Pc = 0.0006). The prominent alleles in the NUD + Asymptomatic H. pylori control group were HLA-B*50 (OR: 0.086, P = 0.02), HLA-DQA1*05 (OR: 0.384, P = 0.0003), and HLA-DQB1*03 (OR: 0.485, P = 0.0022), but after Bonferroni correction for multiple comparisons, the changes were statistically significant for only HLA-DQA1*05 (Pc = 0.0018) and HLA-DQB1*03 (Pc = 0.011) and not HLA-B*50, (Pc = 0.52) (Table 2).
Multiple EPIYA-C repeat numbers and CagA positivity were found in 40 (42.5%) subjects in the GC + DU patient group. Multiple EPIYA-C repeats were observed in 26 (59%) GC subgroup cases and 14 (28%) DU subgroup cases in the GC + DU patient group, 2 (2.3%) NUD cases, and no asymptomatic H. pylori cases. Other EPIYA motifs and their numbers are shown in Table 8. It was not possible to perform statistical analysis due to small number of positive cases with CagA+ multiple EPIYA-C repeats in the NUD+ Asymptomatic H. pylori control group (n = 2). Instead, we compared two groups without using any criteria with regard to HLA alleles.
| EPIYA-C repeat patterns | Patient group | Control group | Total (n = 140) | ||||||
| Gastric cancer | Duodenal ulcer | Non-ulcer dyspepsia | Individuals with normal gastrointestinal system | ||||||
| ABC | 12 | (27.2) | 34 | (68) | 28 | (93.3) | 16 | (100) | 90 |
| AC | 2 | (4.5) | 2 | (4) | - | - | 4 | ||
| BC | 2 | (4.5) | - | - | - | 2 | |||
| ABCC | 16 | (36.4) | 8 | (16) | - | - | 24 | ||
| BCC | 2 | (4.6) | 2 | (4) | 2 | (6.7) | - | 6 | |
| ACCC | - | - | - | - | - | ||||
| ABCC | 8 | (18.2) | 4 | (8) | - | - | 12 | ||
| AB | 2 | (4.6) | - | - | - | 2 | |||
| Total | 44 | (100) | 50 | (100) | 30 | (100) | 16 | (100) | 140 |
Thus, we used the HLA-DQB1*06, HLA-DRB1*13, HLA-B*35, HLA-DQA1 and HLA-A*02 alleles (i.e., the alleles with the maximum numbers in the GC + DU patient group) to investigate their effects on GC/DU in terms of the CagA+ multiple EPIYA-C repeats, which is suitable for our purposes. First, we compared alleles in GC cases in terms of CagA+ multiple EPIYA-C repeat numbers. Only the HLA-DQB1*06 allele (OR: 0.37, 95%CI: 1.149-0.942, P = 0.0369) was significantly associated with GC, but after Bonferroni correction, there was no significant association between DQB1*06 and GC (DQB1*06, Pc = 0.1845).
The HLA-DQA1*01 allele had a high ratio in the multiple EPIYA-C repeat group, but the univariate analysis did not show any significant association. None of the selected alleles were significantly higher in the GC cases in terms of CagA+ multiple EPIYA-C repeats (Table 3). Using the same criterion, we also compared allele frequencies in the DU cases, but none of the HLA alleles were significantly higher (Table 4). When we compared selected allele frequencies in the GC and DU cases together using this criterion, again, none of the HLA alleles were significantly higher (Table 5). HLA-DR1*13-HLA-DQA1*01-HLA-DQB1*06 haplotype frequency was 4/24, 16% and 2/12, 16% for GC and DU cases with multiple EPIYA-C repeat, respectively. No difference was detected between GC and DU cases in terms of multiple EPIYA-C repeat.
Multivariate logistic regression analyses were carried out for the risk of GC and DU development alone and both subcases of the GC + DU patient group combined involving HLA-DQB1*06, HLA-DRB1*13, HLA-A*02, HLA-DQA1*01, and HLA-B*35 alleles, and none of the alleles were detected as independent risk factors for the risk of GC and DU development in terms of CagA+ multiple EPIYA-C repeats. However, a multivariate logistic regression analysis was done without any specific criteria using only the significantly different detected alleles between GC + DU patient and the NUD+Asymptomatic H. pylori control groups. The results showed that HLA-DQA1*01 (P = 0.004, OR = 1.848, 95%CI, 1.215-2.811), HLA-DQB1*06 (P = 0.009, OR = 1.821, 95%CI, 1.163-2.850), and HLA-A*02 (P = 0.04, OR = 1.579, 95%CI, 1.021-2.442) were risk factors for the development of GC and DU (Table 6).
The development of GC and PU or DU is influenced by the virulence factors of H. pylori along with the host’s genetic, epigenetic, and environmental factors. A variety of clinical consequences of H. pylori infection may arise depending on the variability of host response to the specific virulence factors of H. pylori[7]. For example, genes coding HLA class II molecules (HLA-DP/DQ/DR) may have genetically variable coding loci that lead to HLA gene polymorphisms. Therefore, specific HLA class II alleles were hypothesized to be related to the risks of some gastroduodenal malignancies such as GC and DU or PU development in patients with H. pylori infection.
In the literature, there are only traditional comparison studies using only H. pylori positivity for the association between HLA gene polymorphisms and the diseases caused by H. pylori infections. However, we focused on CagA+ multiple EPIYA-C repeats for the comparison of our study groups for HLA alleles for the first time in Turkey. The incidence of GC in Turkey is higher than in Eastern countries and lower than in Western countries at 5.7 and 9.6 cases per 100000 people for women and men, respectively. The mean age of occurrence is 56 years. It is the second and third leading cause of cancer-related deaths in men and women in Turkey, respectively[25].
The higher incidence in Turkey is mainly associated with dietary factors, and the differences in incidence are especially significant in the central, northeastern, and eastern regions of the country. Salt is commonly used for food preservation, and wood charcoal with dried cow dung is commonly used for cooking in these regions, which are known to have carcinogenic effects on food. Another important factor for the development of GC is H. pylori infections[25].
Studies have shown an association between HLA gene polymorphisms and autoimmune diseases, and in genetically susceptible individuals, persistent bacterial infections can lead to autoimmune responses (e.g., HLA-DR4-restricted autoimmune chronic synovitis following Lyme disease)[26]. While evaluating the effects of H. pylori infection on the pathogenesis of GC, the relationship between bacteria and the host should be considered because H. pylori infections may cause strong immune responses by causing the secretion of cytokines from the epithelial cells and gastric mucosa infiltration with neutrophils, macrophages, and lymphocytes. After the interaction of H. pylori with dendritic cells in luminal and subepithelial regions, dendritic cells may transform naive T cells into immunosuppressive Treg cells, and consequently, developed Th1 and Th17 cells may cause atrophic gastritis, epithelial hyperplasia, and intestinal metaplasia[27]. H. pylori infections tend to cause chronic inflammation, which can increase an individual’s risk for the development of GC.
A commonly seen (90%) type of GC is adenocarcinoma, which is known to originate from the epithelial cells in chronic inflammation states[28]. Moreover, cytokines, which are the effector cells of inflammatory responses, may regulate a variety of immunologic events, including the inflammation, proliferation, and differentiation of epithelial cells. In the progression of gastric carcinogenesis, initially, H. pylori strongly induces specific cytokines, but the immune response generally is not sufficient to clear the H. pylori infection completely from the human epithelial cells. As a result, chronic inflammation may occur[29].
Consequently, the tissue damage increases along with parietal cell atrophy and may progress to dysplasia and GC through the combined effects of the various factors of the host and the environment. A subtype of T cells, Th17 cells, and their associated inflammatory cytokines, interleukin (IL)-17A, IL-23, and IL-1β, have important roles in the development of GC, colorectal cancer, ovarian cancer, and hepatocellular carcinoma. Cytokine IL-23 plays a major role in the primary activation of IL-17A. Th17 responses are reported to be increased during H pylori infections. The IL-17/IL-23 axis is believed to have an important role in the progression of chronic inflammation and related pathologies like gastric neoplasms[30].
Other than their main roles in chronic inflammation in gastric epithelial cells, cytokines also have specific polymorphisms in their genes. Polymorphisms in cytokine genes may modify the effect of gene-environment interaction and increase the degree of cytokine expression in the promoter regions of the genes. Polymorphisms in genes coding various cytokines such as IL-1β, IL-1Ra, IL-8, IL-10, and tumor necrosis factor-α are also suggested to be associated with the risk of GC. The IL1RN2 allele polymorphism is related to the risk of GC[31]. Wu et al[32] found a relation between IL-17F, A7488G and GC, and Felipe et al[33] also reported a relation between IL-8 (rs4073)–251A/T gene polymorphism and GC development.
Some virulent factors of H. pylori seem to be associated with GC risk, including vacuolating cytotoxin A, cytotoxin associated antigen A, DU promoting gene protein A, and outer inflammatory protein with blood group antigen binding adhesins. Moreover, the cagA gene of H. pylori strains with multiple EPIYA-C repeats and EPIYA-D motif in its cagA gene are suggested to increase the risk of GC development. However, the role of host polymorphisms and the virulence factors of H. pylori in the risk of GC development varies among regions and ethnicities[31].
Several studies suggest that atrophic gastritis and GC risk are increased by CagA-positive H. pylori strains. An association has been reported between multiple EPIYA-C phosphorylation sites and GC. In a recent meta-analysis including 23 studies, Li et al[8] evaluated the association of EPIYA motifs and gastroduodenal pathologies. They concluded that the EPIYA-D motif was significantly related to GC risk, and multiple EPIYA-C motifs were related to PU and DU in Asia countries.
Conversely, in the United States and Europe, multiple EPIYA-C motifs were commonly associated with GC risk. Multiple EPIYA-C repeats cause stronger binding of CagA to SHP-2 than a single EPIYA-C. Multiple EPIYA C repeats are associated with a higher risk of GC[34,35]. The functionality of CagA is increased with multiple EPIYA-C phosphorylation sites and is involved in cellular phenotypic changes. Therefore, H. pylori strains with multiple EPIYA-C sites are related to the risk of GC.
We could not find any studies specifically evaluating the interaction between EPIYA-C repeats, HLA alleles, and gastric pathologies. In our study, there were only two cases in the control group with CagA+ multiple EPIYA-C repeats, and it was not possible to make a comparison as the number was too low and our criteria were not met. HLA-DQA1, HLA-DRB1*13, HLA-A*02, HLA-B*35, and HLA-DQB1*06 alleles were shown to contribute to the susceptibility to GC and DU and were used for a comparison between GC and DU subgroups of patients. However, we did not compare the higher HLA alleles detected in the control group as it was not possible to determine an HLA allele with a protective effect without including a control group.
In the comparison within the GC subgroup, due to our criterion, only the HLA-DQB1*06 allele was significantly low in the GC subgroup without EPIYA C repeats, but the difference was not statistically significant after Bonferroni correction. The higher frequency of the HLA-DQB1*06 allele suggests that this allele remains influential even in GC cases without multiple EPIYA-C repeats. With the presence of the HLA-DQB1*06 allele, we can suggest that mechanisms other than multiple EPIYA-C repeats may contribute to the development of GC. On the other hand, in the GC subgroup, the number of HLA-DQA1*01 alleles was high in the multiple EPIYA-C repeats group, but the result was not significant after the univariate analysis.
In the DU subgroup, none of the alleles were found to be significantly predominant in terms of frequencies. In addition, when the analyses were repeated while including GC and DU subgroups together, none of the HLA alleles were shown to either effective or protective. A multivariate logistic regression analysis was performed for GC and DU subgroups alone and together, and none of the alleles were detected as independent risk factors. We then performed a multivariate logistic regression without including any discriminative criterion and only using the significantly different detected alleles between patient and control groups. As a result, HLA-DQA1*01, HLA-DQB1*0601, and HLA-A*2 alleles were found to be independent risk factors for the risk of GC and DU.
There are no previous studies to compare our results based on the discrimination criteria for the selection of HLA alleles. Our results partly coincided with those of a meta-analysis of Asian populations by Wang et al[36]. In that study, the susceptibility genes for H. pylori infection were reported as DQB1*0401, DQA1*0103, and DQA1*0301. Garza-González et al[37] reported that the HLA-DQA1*0503 allele served as an independent protective factor for GC. In our study, we also detected HLA-DQA1*05 as a protective allele.
Similar to our results, Quintero et al[9] reported that the DQB1*0602 allele increased the risk of GC risk in a Southern European population infected with H. pylori. On the other hand, the results of several case–control studies conducted with a Japanese population were contradictory. In contrast to our results, Azuma et al[38] found that the number of DQA1*0102 alleles was significantly lower in their study group of H. pylori-infected patients with GC than in control subjects. However, our data did not support their findings for the DQA1* alleles.
In a study by Herrera-Goepfert et al[39], patients with GC displayed a high frequency of HLA-DQA1*0601, similar to our results. After our univariate analysis, DQA1*01 and HLA-DQB1*06 alleles were found to be positively associated with GC and DU, and DQB1*03 was found to be negatively associated with DU. These results are consistent with those of Herrera-Goepfert et al[39], Wu et al[13], and Quintero et al[9] .
The reason for the conflicting results may be the different methods used in HLA typing, ethnicities, and the fact that GC is a heterogeneous disease. Specific HLA alleles may have the capability to modulate the presentation of peptides derived from H. pylori infection to T cells. As a result of this presentation, the type or severity of T cell response may affect the proliferation of a lineage-specific malignant T cell clones[5].
In the study by Li et al[10], the HLA-CW*03 ratio was significantly higher in cases with increased risk of GC and H. pylori-infected patients. A variety of exogenous stimulations such as toxins of H. pylori with gastric and bile juices always affect the gastric mucosa[40]. These stimulations cause epithelial cells to secrete IL-12 and induce natural killer (NK) cells. These NK cells secrete IFN-g, which initiates the Th1 immune response from naive T cells and causes phenotypic changes in epithelial cells, resulting in the upregulation of HLA-DR and HLA-B27 genes[41].
Different mechanisms may be responsible for the synergistic effect of H. pylori infection and specific HLA genotypes. Specific HLA genotypes may influence the immune response and lead to carcinogenesis after the initial infection[17]. However, the common consensus is that genetically different host responses against the virulence factors of H. pylori may cause inflammation with varying intensities, as well as gastric epithelial erosions with different stages.
Genetic and epigenetic factors may also influence the severity of inflammation and thereby contribute to different clinical outcomes. Upregulation of significant MHC class II type genes in epithelial cells may be related to the activation of T cells in the lamina propria and macrophages, as well as the consequent release of cytokines such as interferon-γ. Confirming this hypothesis, upregulated expression of MHC class II genes in gastric epithelial cells had a positive correlation with the T cells in the lamina propria in children with H. pylori infections according to Lopes et al[42].
We could not detect any HLA allele as an independent risk factor for GC under our criterion, but without any criterion, some HLA alleles may regulate host susceptibility to H. pylori infection in the presence of its virulence factors. It can be suggested that some immunogenetic factors of the host may have important effects for H. pylori-initiated inflammation leading to carcinogenesis. Although various host or H. pylori virulence factors may have a role in the development of GC, specific host factors like HLAs could also modulate GC susceptibility or the resistance of individuals. Individuals who are at risk due to the susceptibility of HLA alleles to GC should be monitored prior to the initiation of carcinogenesis.
We only focused on the association between HLA gene polymorphisms and multiple EPIYA-C repeats of H. pylori DNAs isolated from the gastric biopsy specimens. Only two of our control group cases had multiple EPIYA-C repeats, so we compared two subgroups of the study group. We could not find a significant difference between GC and DU subgroups in terms of multiple EPIYA-C repeats. On the other hand, in a simple comparison between study and control groups, HLA-DQA1*01 (OR = 1.848), HLA-DQB1*06 (OR = 1.821), and HLA-A*2 (OR = 1.579) alleles were detected as independent risk factors for GC and DU.
We believe that there is an association between HLA allele polymorphisms and gastric pathologies, but this is not a definite reality because of differences between regions and ethnicities and the virulence factors of H. pylori. The same HLA alleles sometimes show a positive correlation and sometimes show a negative association in different regions and ethnicities. We suggest that the role of host polymorphisms of the host and H. pylori virulence factors in the risk of GC development varies among countries of different regions and different ethnicities.
This study has some limitations. The resolution of the HLA kits was low, which prevented an exact HLA comparison with other studies. We also did not evaluate some HLA alleles, such as HLA-C. Other important result of our study was that HLA-DR1*13-HLA-DQA1*01-HLA-DQB1*06 haplotype frequency was detected significantly higher in our GC subgroup cases more than both DU and control group cases. This results is similar for the locus type but different for HLA allele types from the study of Ando et al[43] They reported DRB1*04:05-DQA1*03:03-DQB1*04:01 haplotype frequency with 10%–30% in Japanese population and the risk of GC development.
This is the first study to evaluate the association between HLA alleles with GC and DU and asymptomatic H. pylori cases. Even though no HLA alleles were detected in multivariate analysis, HLA-DQB1*06 was significantly less frequent in the GC subgroup with multiple EPIYA repeats in the univariate analysis. However, this HLA allele was not detected as an independent risk factor in the multivariate analysis. In the multivariate logistic regression analysis using significantly different alleles and no discriminative criteria, HLA-DQA1*01 (OR = 1.848), HLA-DQB1*06 (OR = 1.821), and HLA-A*02 (OR = 1.579) alleles were found to be risk factors for GC and DU. However, we suggest that these HLA alleles make individuals prone to the development of GC without cagA and multiple EPIYA C repeats of the host. To clarify the effects of HLA alleles on the pathogenesis of gastric malignancies, more comprehensive, prospective, large-scale studies with high HLA resolution detection should be performed in the future.
The development of gastric cancer (GC) is suggested to be related to the interactions between bacterial virulence factors, host genetic factors such as the human leukocyte antigen (HLA) gene, the immune response of the host, and environmental factors. Some of the host factors may be polymorphisms in the host genes such as HLA genes, which regulate the strength of the inflammatory response and influence the probability of a specific clinical outcome.
We seek to determine which HLA class I and II alleles differ in gastrointestinal pathologies such as GC and duodenal ulcer (DU) in Turkey.
We investigated the allele frequencies of HLA class I and II in a patient group [Helicobacter pylori (H. pylori)-positive GC and DU patients] and compared the results to a control group (H. pylori-positive non-ulcer dyspepsia patients and asymptomatic individuals with H. pylori) in terms of CagA+ multiple (≥ 2) EPIYA-C repeats for the first time in a Turkish population.
In this case-control study, amplification of the H. pylori cagA gene and typing of EPIYA motifs were performed by PCR. HLA allele types were identified by sequence-specific oligonucleotide typing kits (HLA-A, HLA-B HLA-C, HLA-DRB1, and HLA-DQA1/B1 kits).
None of the alleles were detected as independent risk factors after multivariate analysis in terms of CagA+ multiple (≥ 2) EPIYA-C repeats. On the other hand, in a multivariate logistic regression with no discriminative criterion, HLA-DQA1*01 [odds ratio (OR) = 1.848], HLA-DQB1*06 (OR = 1.821), and HLA-A*02 (OR = 1.579) alleles were detected as independent risk factors for GC and DU.
We suggest that the association of these alleles with gastric malignancies is not specifically related to cagA and multiple EPIYA C repeats.
Specific HLA alleles maybe related to the gastric malignancies and could be for the indication of GCs in order to scan populations before the development of GC. The alleles may also be cost-effective to find individuals with a higher risk for GC development.
We thank Assoc. Prof. Dr. Murat Telli from the Department of Biology in Bolu Abant Izzet Baysal University, Turkey for the calculation of H-W equilirum and LD.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Turkey
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: de Melo FF, Du Y, Sun X S-Editor: Liu M L-Editor: A P-Editor: Li JH
| 1. | Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941-1953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3585] [Cited by in RCA: 4902] [Article Influence: 700.3] [Reference Citation Analysis (1)] |
| 2. | Malvehy Rovira J, Barranco Peña F, Terradas Mercader P. [Serratia marcescens and neonatal sepsis]. An Esp Pediatr. 1988;29:23-25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 60] [Reference Citation Analysis (1)] |
| 3. | Koulis A, Buckle A, Boussioutas A. Premalignant lesions and gastric cancer: Current understanding. World J Gastrointest Oncol. 2019;11:665-678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 4. | Burkitt MD, Duckworth CA, Williams JM, Pritchard DM. Helicobacter pylori-induced gastric pathology: insights from in vivo and ex vivo models. Dis Model Mech. 2017;10:89-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 83] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 5. | Crux NB, Elahi S. Human Leukocyte Antigen (HLA) and Immune Regulation: How Do Classical and Non-Classical HLA Alleles Modulate Immune Response to Human Immunodeficiency Virus and Hepatitis C Virus Infections? Front Immunol. 2017;8:832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 143] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 6. | Ansari S, Yamaoka Y. Helicobacter pylori Virulence Factors Exploiting Gastric Colonization and its Pathogenicity. Toxins (Basel). 2019;11:677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 167] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 7. | Chang WL, Yeh YC, Sheu BS. The impacts of H. pylori virulence factors on the development of gastroduodenal diseases. J Biomed Sci. 2018;25:68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 127] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 8. | Li Q, Liu J, Gong Y, Yuan Y. Association of CagA EPIYA-D or EPIYA-C phosphorylation sites with peptic ulcer and gastric cancer risks: A meta-analysis. Medicine (Baltimore). 2017;96:e6620. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 9. | Quintero E, Pizarro MA, Rodrigo L, Piqué JM, Lanas A, Ponce J, Miño G, Gisbert J, Jurado A, Herrero MJ, Jiménez A, Torrado J, Ponte A, Díaz-de-Rojas F, Salido E. Association of Helicobacter pylori-related distal gastric cancer with the HLA class II gene DQB10602 and cagA strains in a southern European population. Helicobacter. 2005;10:12-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 10. | Li Z, Chen D, Zhang C, Li Y, Cao B, Ning T, Zhao Y, You W, Ke Y. HLA polymorphisms are associated with Helicobacter pylori infected gastric cancer in a high risk population, China. Immunogenetics. 2005;56:781-787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 11. | Watanabe Y, Aoyama N, Sakai T, Shirasaka D, Maekawa S, Kuroda K, Wambura C, Tamura T, Nose Y, Kasuga M. HLA-DQB1 locus and gastric cancer in Helicobacter pylori infection. J Gastroenterol Hepatol. 2006;21:420-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 12. | Magnusson PKE, Enroth H, Eriksson I, Held M, Nyrén O, Engstrand L, Hansson LE, Gyllensten UB. Gastric cancer and human leukocyte antigen: distinct DQ and DR alleles are associated with development of gastric cancer and infection by Helicobacter pylori. Cancer Res. 2001;61:2684-2689. [PubMed] |
| 13. | Wu MS, Hsieh RP, Huang SP, Chang YT, Lin MT, Chang MC, Shun CT, Sheu JC, Lin JT. Association of HLA-DQB1*0301 and HLA-DQB1*0602 with different subtypes of gastric cancer in Taiwan. Jpn J Cancer Res. 2002;93:404-410. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 14. | Yoshitake S, Okada M, Kimura A, Sasazuki T. Contribution of major histocompatibility complex genes to susceptibility and resistance in Helicobacter pylori related diseases. Eur J Gastroenterol Hepatol. 1999;11:875-880. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 15. | Ohtani M, Azuma T, Yamazaki S, Yamakawa A, Ito Y, Muramatsu A, Dojo M, Yamazaki Y, Kuriyama M. Association of the HLA-DRB1 gene locus with gastric adenocarcinoma in Japan. Dig Liver Dis. 2003;35:468-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 16. | Lee JE, Lowy AM, Thompson WA, Lu M, Loflin PT, Skibber JM, Evans DB, Curley SA, Mansfield PF, Reveille JD. Association of gastric adenocarcinoma with the HLA class II gene DQB10301. Gastroenterology. 1996;111:426-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 58] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 17. | Ohmori M, Yasunaga S, Maehara Y, Sugimachi K, Sasazuki T. DNA typing of HLA class I (HLA-A) and class II genes (HLA-DR, -DQ and -DP) in Japanese patients with gastric cancer. Tissue Antigens. 1997;50:277-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 18. | Zhao Y, Wang J, Tanaka T, Hosono A, Ando R, Soeripto S, Ediati Triningsih FX, Triono T, Sumoharjo S, Astuti EY, Gunawan S, Tokudome S. Association between HLA-DQ genotypes and haplotypes vs Helicobacter pylori infection in an Indonesian population. Asian Pac J Cancer Prev. 2012;13:1247-1251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 19. | He Q, Wang JP, Osato M, Lachman LB. Real-time quantitative PCR for detection of Helicobacter pylori. J Clin Microbiol. 2002;40:3720-3728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 88] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 20. | van Doorn LJ, Figueiredo C, Rossau R, Jannes G, van Asbroek M, Sousa JC, Carneiro F, Quint WG. Typing of Helicobacter pylori vacA gene and detection of cagA gene by PCR and reverse hybridization. J Clin Microbiol. 1998;36:1271-1276. [PubMed] |
| 21. | Erzin Y, Koksal V, Altun S, Dobrucali A, Aslan M, Erdamar S, Dirican A, Kocazeybek B. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA, babA2 genotypes and correlation with clinical outcome in Turkish patients with dyspepsia. Helicobacter. 2006;11:574-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 90] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 22. | Argent RH, Zhang Y, Atherton JC. Simple method for determination of the number of Helicobacter pylori CagA variable-region EPIYA tyrosine phosphorylation motifs by PCR. J Clin Microbiol. 2005;43:791-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 86] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 23. | Occhialini A, Marais A, Urdaci M, Sierra R, Muñoz N, Covacci A, Mégraud F. Composition and gene expression of the cag pathogenicity island in Helicobacter pylori strains isolated from gastric carcinoma and gastritis patients in Costa Rica. Infect Immun. 2001;69:1902-1908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 49] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Rousset F. genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour. 2008;8:103-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6512] [Cited by in RCA: 5330] [Article Influence: 444.2] [Reference Citation Analysis (0)] |
| 25. | Tural D, Selçukbiricik F, Akar E, Serdengeçti S, Büyükünal E. Gastric cancer: a case study in Turkey. J Cancer Res Ther. 2013;9:644-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 26. | Arvikar SL, Crowley JT, Sulka KB, Steere AC. Autoimmune Arthritides, Rheumatoid Arthritis, Psoriatic Arthritis, or Peripheral Spondyloarthritis Following Lyme Disease. Arthritis Rheumatol. 2017;69:194-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 27. | Rezalotfi A, Ahmadian E, Aazami H, Solgi G, Ebrahimi M. Gastric Cancer Stem Cells Effect on Th17/Treg Balance; A Bench to Beside Perspective. Front Oncol. 2019;9:226. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 28. | Bockerstett KA, DiPaolo RJ. Regulation of Gastric Carcinogenesis by Inflammatory Cytokines. Cell Mol Gastroenterol Hepatol. 2017;4:47-53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 88] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 29. | Blogowski W, Madej-Michniewicz A, Marczuk N, Dolegowska B, Starzyńska T. Interleukins 17 and 23 in patients with gastric neoplasms. Sci Rep. 2016;6:37451. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 30. | Dixon BREA, Hossain R, Patel RV, Algood HMS. Th17 Cells in Helicobacter pylori Infection: a Dichotomy of Help and Harm. Infect Immun. 2019;87:e00363-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 31. | de Brito BB, da Silva FAF, de Melo FF. Role of polymorphisms in genes that encode cytokines and Helicobacter pylori virulence factors in gastric carcinogenesis. World J Clin Oncol. 2018;9:83-89. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 12] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 32. | Wu X, Zeng Z, Chen B, Yu J, Xue L, Hao Y, Chen M, Sung JJ, Hu P. Association between polymorphisms in interleukin-17A and interleukin-17F genes and risks of gastric cancer. Int J Cancer. 2010;127:86-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 100] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 33. | Felipe AV, Silva TD, Pimenta CA, Kassab P, Forones NM. lnterleukin-8 gene polymorphism and susceptibility to gastric cancer in a Brazilian population. Biol Res. 2012;45:369-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 34. | Batista SA, Rocha GA, Rocha AM, Saraiva IE, Cabral MM, Oliveira RC, Queiroz DM. Higher number of Helicobacter pylori CagA EPIYA C phosphorylation sites increases the risk of gastric cancer, but not duodenal ulcer. BMC Microbiol. 2011;11:61. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 35. | El Khadir M, Alaoui Boukhris S, Benajah DA, Ibrahimi SA, Chbani L, Bouguenouch L, El Rhazi K, El Abkari M, Nejjari C, Mahmoud M, Bennani B. Helicobacter pylori CagA EPIYA-C motifs and gastric diseases in Moroccan patients. Infect Genet Evol. 2018;66:120-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 36. | Wang J, Zhang Q, Liu Y, Han J, Ma X, Luo Y, Liang Y, Zhang L, Hu Y. Association between HLA-â…¡gene polymorphism and Helicobacter pylori infection in Asian and European population: A meta-analysis. Microb Pathog. 2015;82:15-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 37. | Garza-González E, Bosques-Padilla FJ, Pérez-Pérez GI, Flores-Gutiérrez JP, Tijerina-Menchaca R. Association of gastric cancer, HLA-DQA1, and infection with Helicobacter pylori CagA+ and VacA+ in a Mexican population. J Gastroenterol. 2004;39:1138-1142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 38. | Azuma T, Ito S, Sato F, Yamazaki Y, Miyaji H, Ito Y, Suto H, Kuriyama M, Kato T, Kohli Y. The role of the HLA-DQA1 gene in resistance to atrophic gastritis and gastric adenocarcinoma induced by Helicobacter pylori infection. Cancer. 1998;82:1013-1018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 39. | Herrera-Goepfert R, Zúñiga J, Hernández-Guerrero A, Rodríguez-Reyna T, Osnalla N, Ruíz-Morales J, Vargas-Alarcón G, Yamamoto-Furusho JK, Mohar-Betancourt A, Hernández-Pando R, Granados J. [Association of the HLA-DQB*0501, allele of the major histocompatibility complex with gastric cancer in Mexico]. Gac Med Mex. 2004;140:299-303. [PubMed] |
| 40. | Kitamura H, Honma I, Torigoe T, Asanuma H, Sato N, Tsukamoto T. Down-regulation of HLA class I antigen is an independent prognostic factor for clear cell renal cell carcinoma. J Urol. 2007;177:1269-72; discussion 1272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 41. | Engstrand L, Scheynius A, Påhlson C, Grimelius L, Schwan A, Gustavsson S. Association of Campylobacter pylori with induced expression of class II transplantation antigens on gastric epithelial cells. Infect Immun. 1989;57:827-832. [PubMed] |
| 42. | Lopes AI, Victorino RM, Palha AM, Ruivo J, Fernandes A. Mucosal lymphocyte subsets and HLA-DR antigen expression in paediatric Helicobacter pylori-associated gastritis. Clin Exp Immunol. 2006;145:13-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 43. | Ando T, Ishikawa T, Kato H, Yoshida N, Naito Y, Kokura S, Yagi N, Takagi T, Handa O, Kitawaki J, Nakamura N, Hasegawa G, Fukui M, Imamoto E, Nakamura C, Oyamada H, Isozaki Y, Matsumoto N, Nagao Y, Okita M, Nakajima Y, Kurokawa M, Nukina M, Ohta M, Mizuno S, Ogata M, Obayashi H, Park H, Kitagawa Y, Nakano K, Yoshikawa T. Synergistic effect of HLA class II loci and cytokine gene polymorphisms on the risk of gastric cancer in Japanese patients with Helicobacter pylori infection. Int J Cancer. 2009;125:2595-2602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |