Published online Aug 28, 2020. doi: 10.3748/wjg.v26.i32.4786
Revised: June 4, 2020
Accepted: August 4, 2020
Published online: August 28, 2020
Processing time: 113 Days and 12.1 Hours
Hepatocellular carcinoma (HCC), often diagnosed at advanced stages without curative therapies, is the fifth most common malignant cancer and the second leading cause of cancer-related mortality. Polo-like kinase 1 (PLK1) is activated in the late G2 phase of the cell cycle and is required for entry to mitosis. Interestingly, PLK1 is overexpressed in many HCC patients and is highly associated with poor clinical outcome. Baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5) is also highly overexpressed in HCC and plays key roles in this malignancy.
To determine the expression patterns of PLK1 and BIRC5, as well as their correlation with p53 mutation status and patient clinical outcome.
The expression patterns of PLK1 and BIRC5, and their correlation with p53 mutation status or patient clinical outcome were analyzed using a TCGA HCC dataset. Cell viability, cell apoptosis, and cell cycle arrest assays were conducted to investigate the efficacy of the PLK1 inhibitors volasertib and GSK461364 and the BIRC5 inhibitor YM155, alone or in combination. The in vivo efficacy of volasertib and YM155, alone or in combination, was assessed in p53-mutated Huh7-derived xenograft models in immune-deficient NSIG mice.
Our bioinformatics analysis using a TCGA HCC dataset revealed that PLK1 and BIRC5 were overexpressed in the same patient subset and their expression was highly correlated. The overexpression of both PLK1 and BIRC5 was more frequently detected in HCC with p53 mutations. High PLK1 or BIRC5 expression significantly correlated with poor clinical outcome. PLK1 inhibitors (volasertib and GSK461364) or a BIRC5 inhibitor (YM155) selectively targeted Huh7 cells with mutated p53, but not HepG2 cells with wild-type p53. The combination treatment of volasertib and YM155 synergistically inhibited the viability of Huh7 cells via apoptotic pathway. The efficacy of volasertib and YM155, alone or in combination, was validated in vivo in a Huh7-derived xenograft model.
PLK1 and BIRC5 are highly co-expressed in p53-mutated HCC and inhibition of both PLK1 and BIRC5 synergistically compromises the viability of p53-mutated HCC cells in vitro and in vivo.
Core tip: A bioinformatics analysis using a TCGA hepatocellular carcinoma (HCC) dataset revealed that Polo-like kinase 1 (PLK1) and baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5) were overexpressed in the same patient subset and their expression was highly correlated. Overexpression of both PLK1 and BIRC5 was more frequently detected in HCC with p53 mutations. High PLK1 or BIRC5 expression significantly correlated with poor clinical outcome. The PLK1 inhibitors volasertib and GSK461364 or BIRC5 inhibitor YM155 selectively targeted Huh7 cells with mutated p53, but not HepG2 cells with wild-type p53. Combination treatment with volasertib and YM155 synergistically inhibited the viability of Huh7 cells by inducing apoptosis. The efficacy of volasertib and YM155, alone or in combination, was validated in vivo in a Huh7-derived xenograft model in immuno-deficient NSIG mice.
- Citation: Li Y, Zhao ZG, Luo Y, Cui H, Wang HY, Jia YF, Gao YT. Dual targeting of Polo-like kinase 1 and baculoviral inhibitor of apoptosis repeat-containing 5 in TP53-mutated hepatocellular carcinoma. World J Gastroenterol 2020; 26(32): 4786-4801
- URL: https://www.wjgnet.com/1007-9327/full/v26/i32/4786.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i32.4786
According to the World Health Organization: GLOBOCAN, hepatocellular carcinoma (HCC) is the fifth most common cancer worldwide. More than 800000 new cases are diagnosed each year, and China alone accounts for more than half (55%) of all cases[1]. Most HCC patients (80%) are diagnosed at advanced stages, with a median survival of only 6-8 mo due to the lack of effective therapies. Overall, HCC has an extremely high mortality rate (95%) and is the second leading cause of cancer mortality. The most common risk factors for HCC are chronic infection with two liver tropic viruses [hepatitis B virus (HBV) and hepatitis C virus (HCV)], exposure to dietary aflatoxin B1, or alcohol consumption, all of which can lead to liver cirrhosis[2].
In recent years, extensive efforts have focused on the molecular carcinogenesis of HCC. The tumor suppressor p53 is one of the most frequently mutated genes in all cancers, including HCC[3-7]. Mutations in p53 resulting in p53 inactivation or gain of function significantly contribute to disease malignancies; chemo-resistance to cisplatin (CDDP), doxorubicin, and 5-fluorouracil (5-FU); and drug resistance to targeted therapies including histone deacetylase inhibitors[8,9].
Cell cycle dysregulation and apoptosis are essential cellular processes required for cancer cells to survive, proliferate, and evade immune elimination and therapeutic treatments. Polo-like kinase 1 (PLK1) is a master cell cycle regulator in mitosis that mediates centrosome disjunction and movement, CDK1/cyclin B activation, spindle assembly, and cytokinesis[10,11]. PLK1 has been shown to be an oncogene in many cancer types including HCC[12]. PLK1 expression is tightly regulated in a cell cycle-dependent manner in normal cells and is upregulated in various cancers[13-15], suggesting that it is a promising therapeutic target. Indeed, it has been shown that PLK1 inhibitors have potent activity in targeting various cancer types, including HCC.
Baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5), also known as Survivin, is the smallest family member of the inhibitor of apoptosis (IAP) proteins. BIRC5 expression is tightly regulated and only expressed during G2-M phase in normal cells[16]. Transcriptomic analysis identified BIRC5 as the fourth most highly transcribed gene in cancer cells compared to normal cells[17]. BIRC5 is a multifunctional protein that regulates chromosome segregation and cytokinesis during the cell cycle and promotes cell survival by inhibiting both intrinsic and extrinsic apoptosis pathways[16]. Furthermore, suppression of BIRC5 expression in cancer cells induces cell apoptosis and growth inhibition[16-18].
However, how p53, PLK1, and BIRC5 interact in HCC has not been well defined. In this study, we revealed that PLK1 and BIRC5 are significantly co-expressed in HCC together with a number of other important cell cycle factors. More interestingly, PLK1 and BIRC5 expression were observed to be highly upregulated in HCC cells with mutated p53 compared to those wild-type p53. Both PLK1 inhibitors and a BIRC5 inhibitor selectively targeted p53-mutated HCC cells. Furthermore, dual targeting of PLK1 and BIRC5 showed synergistic effects both in vitro and in vivo in p53-mutated cells and their derived xenografts.
HepG2 and Huh7 cell lines were obtained from our laboratory for long-term storage. The cells were cultured in DMEM (high glucose) containing 10% fetal bovine serum and 1% penicillin/streptomycin. GSK461364 (S2193), volasertib (S2235), and YM155 (S1130) were purchased from Selleck. A CellTiter-Glo 2.0 cell viability assay kit (G9241) was purchased from Promega. Annexin V-FITC (Cat#: 1001) and propidium iodide (Cat#: 1056) were purchased from BioVision.
The Liver Hepatocellular Carcinoma (TCGA, PanCancer Atlas) dataset was downloaded from the Cancer Genome Atlas website. The data were normalized using the RNA-Seq with the Expectation Maximization (RSEM) algorithm[19]. Co-expression data were downloaded using cBioPortal and the co-expression heatmap was generated using GraphPad Prism. Individual pairs of co-expressed gene plots, differential expression of PLK1 and BIRC5 in p53/RB1-wild-type and -mutated HCC patients, and the PLK1 and BIRC5 associated pathway networks were generated using cBioPortal. The Wilcoxon rank-sum test was used to assess the differences in PLK1 and BIRC5 expression between groups. For both overall clinical survival and disease/progression-free survival analysis, we generated Kaplan-Meier survival plots using the cBioPortal to compare patients with high PLK1 or BIRC5 expression levels and those with low PLK1 or BIRC5 levels. The log-rank test was used to compare the survival distribution between patients with differential expression of PLK1 or BIRC5.
The cell viability assay was performed following the manufacturer’s instructions. Briefly, 2000 HepG2 or Huh7 cells were seeded into each well of white 96-well plates overnight. The cells were then treated with a 2-fold serial dilution of GSK461364 (0-50 nM), volasertib (0-50 nmol/L), or YM155 (0-20 M). At 72 h post treatment, the cells were lysed using CellTiter-Glo 2.0 Reagent, and cell viability was then measured using a VICTOR Nivo Multimode Microplate Reader from PerkinElmer.
The cell apoptosis assay was performed following the manufacturer’s instructions. Briefly, 100000 HepG2 or Huh7 cells were seeded into each well of 6-well plates overnight. The cells were then treated with GSK461364, volasertib, or YM155. At 24 h post treatment, all the cells, including attached and detached cells, were harvested by trypsin treatment. Subsequently, the cells were stained with annexin V and propidium iodide, and analyzed by flow cytometry. Then, the percentage of annexin V-positive cells for each sample was calculated and plotted.
To assess cell cycle arrest, 100000 HepG2 or Huh7 cells were seeded into each well of 6-well plates overnight. The cells were then treated with GSK461364, volasertib, or YM155. At 24 h post treatment, all the cells, including attached and detached cells, were harvested by trypsin treatment. Subsequently, the cells were fixed with chilled 100% ethanol, stained with propidium iodide, and analyzed by flow cytometry. Then, the percentage of each cell cycle phase for each sample was calculated and plotted.
All animal experiments and procedures were conducted under the protocol approved by the Animal Care and Use Committee at Nankai University. Eight-week-old male NOD-PrkdcscidIL2rgtm1-HFK (NSIG) mice were purchased from Beijing HFK Bioscience Co. Ltd. The mice were fed in a controlled environment (22 ± 2 °C, 50% ± 10% humidity, 12-h light/dark cycle, free access to water and standard diet). After 1 week of adaptive feeding, the mice were subcutaneously injected with 2 × 106 of Huh7 cells that had been harvested from cell culture and suspended in a 2:1 mixture of appropriate medium and Matrigel. Following cell inoculation, the mice were monitored daily for tumor engraftment and health conditions. When the tumor size became palpable, the mice were treated with vehicle or volasertib (10 mg/kg) weekly for 4 wk via the intraperitoneal route or received a continuous infusion of YM155 (3 mg/kg) for 7 d and off for the last 3 wk using a Alzet Osmotic pumps (Model 1007D), alone or in combination. The tumor growth and mouse health conditions were monitored daily and body weight was measured every week. At the end of the 4-wk treatment, the mice were euthanized using CO2, and the tumor mass was dissected, weighed, and imaged.
Statistical analyses were performed with SPSS 22.0. The results are presented as the means ± SD from at least three independent experiments. Differences among multiple groups were analyzed using one-way ANOVA, while comparisons between two groups were conducted using Student’s t-test. A P value of < 0.05 was considered statistically significant.
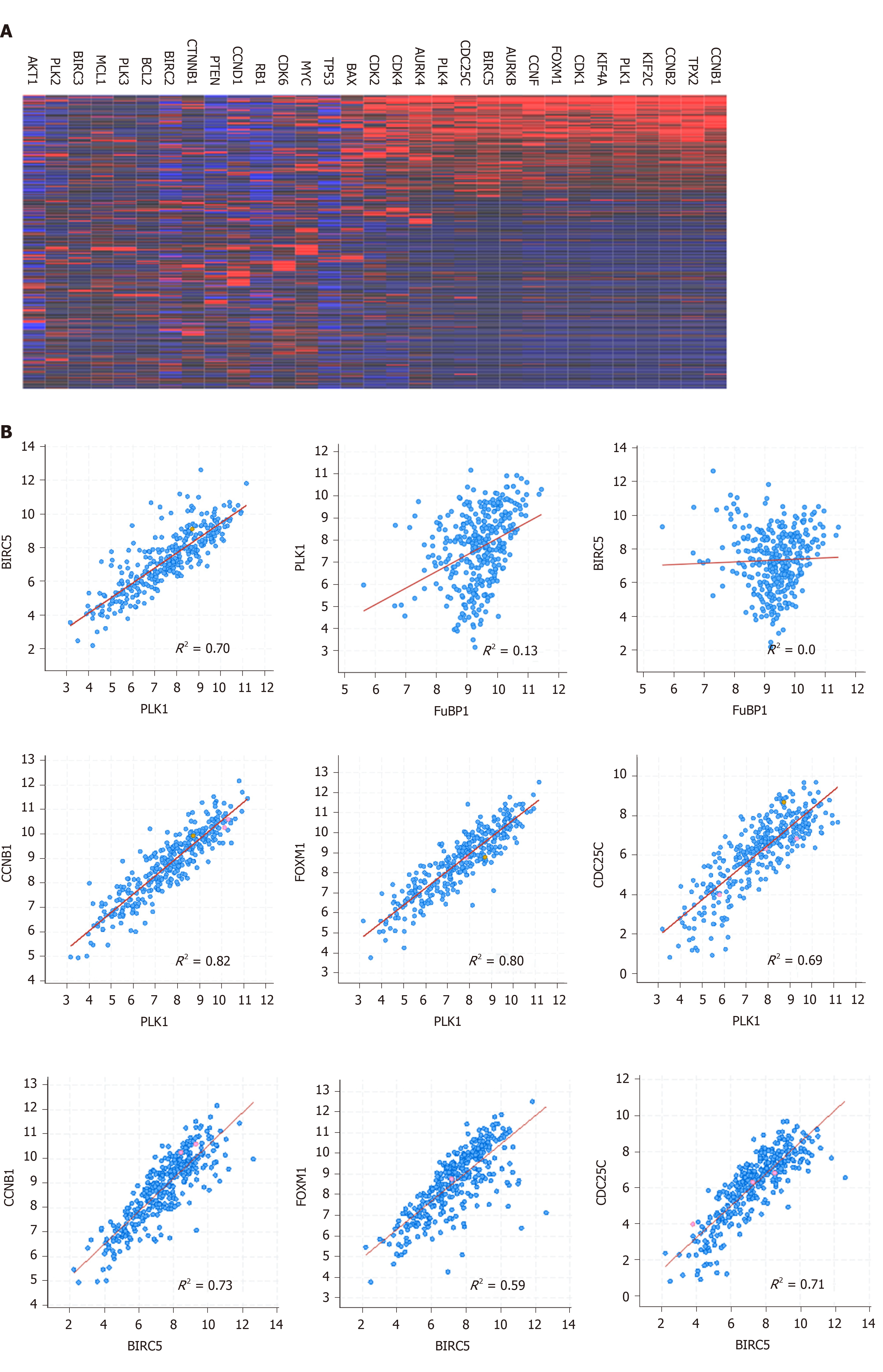
Both PLK1 and BIRC5 are expressed at the G2/M phase and play key roles in regulating the cell cycle in normal cells[10,11,16]. The expression of PLK1 and BIRC5 is frequently upregulated in cancer cells[12,17,18]. To assess the expression of PLK1 and BIRC5 in HCC tumor cells, we analyzed a transcriptome dataset (n = 374) available at TCGA and observed that PLK1 and BIRC5 were highly expressed in a subset of HCC patients. More interestingly, the expression of PLK1 and BIRC5 was significantly correlated in the same subset together with a number of other important cell cycle factors, including CCNB1, CCNB2, CDK1, FOXM1, AURKA, AURKB, and CDC25C (Figure 1A and B). Notably, PLK1 and PLK4, but not other family members PLK2 or PLK3, showed higher co-expression in the same subset (Figure 1A). Similarly, BIRC5 was not co-expressed with other family members BIRC2 or BIRC3, or other apoptotic genes, such as BCL-2 and MCL-1 (Figure 1A). In addition, we did not detect any significant correlation between FUBP1 expression and that of either PLK1 or BIRC5, although FuBP1 is overexpressed in 80% of HCC tumors with chronic hepatitis C (CHC)[20]. Altogether, these results indicated that PLK1 and BIRC5 may coordinate to mediate HCC malignancy.
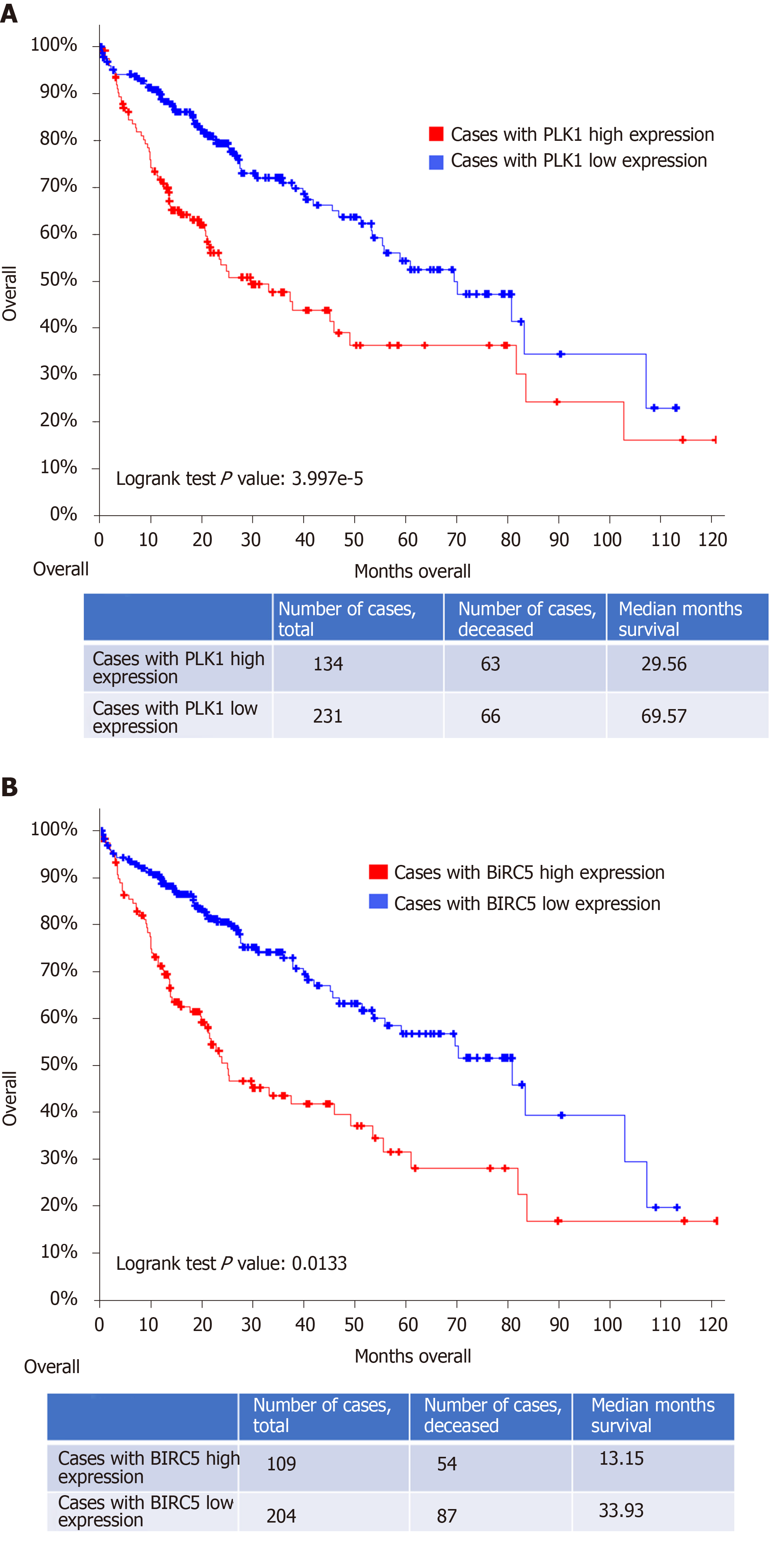
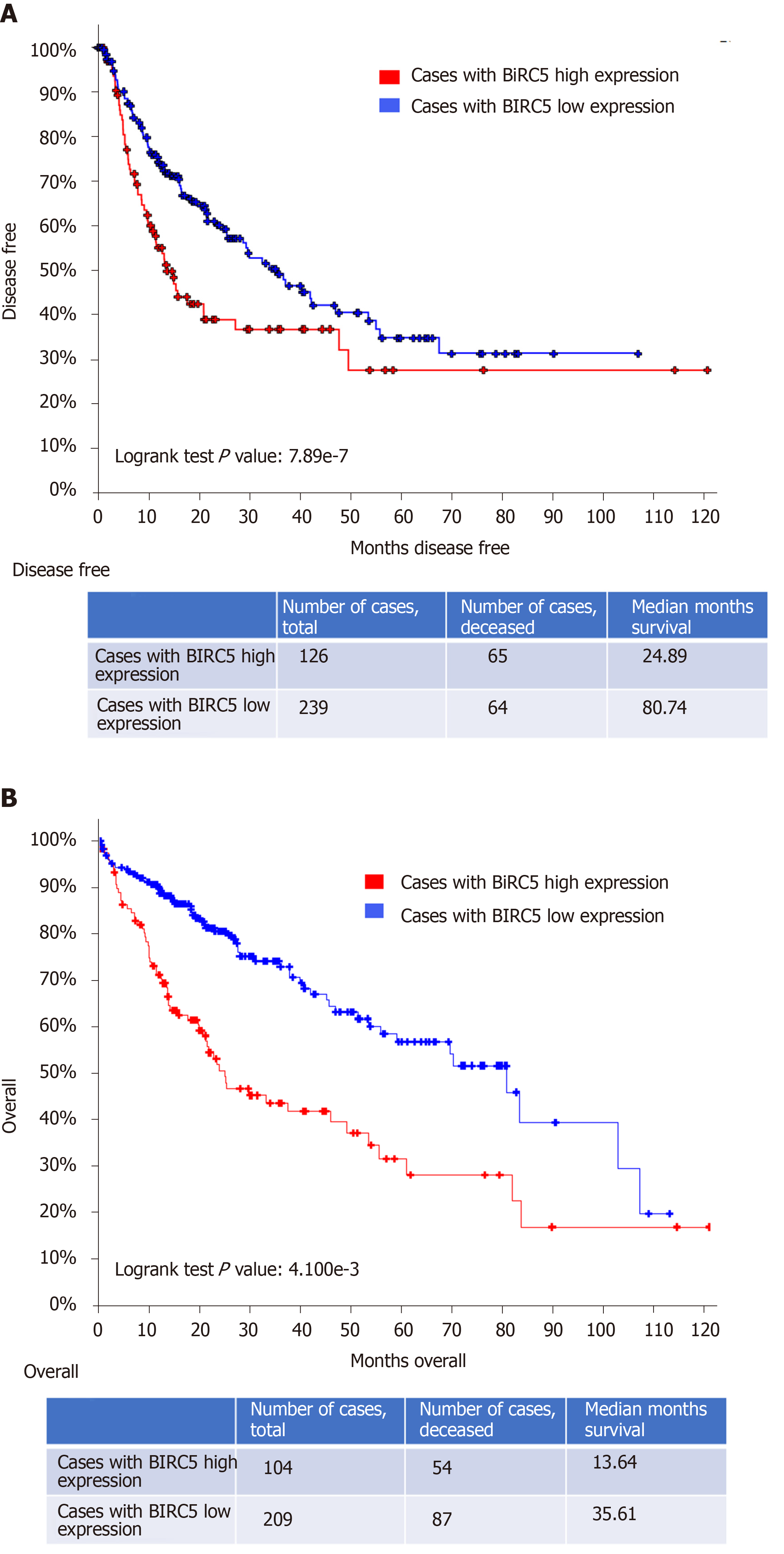
In addition, poor overall clinical survival and disease/progression-free survival after initial treatment were observed to be significantly correlated with high PLK1 expression in patients with HCC (n = 134 and 109, respectively) compared to HCC patients with low PLK1 expression (n = 231 and 204, respectively) (Figure 2A and B). Similarly, poor overall clinical survival and disease/progression-free survival after initial treatment were observed to be significantly correlated high BIRC5 expression in patients with HCC (n = 126 and 104, respectively) compared to HCC patients with low BIRC5 expression (n = 239 and 209, respectively) (Figure 3A and B). These data suggested that dual targeting of PLK1 and BIRC5 in HCC patients with high expression of both PLK1 and BIRC5 may be a promising therapeutic strategy for anti-tumor treatments.
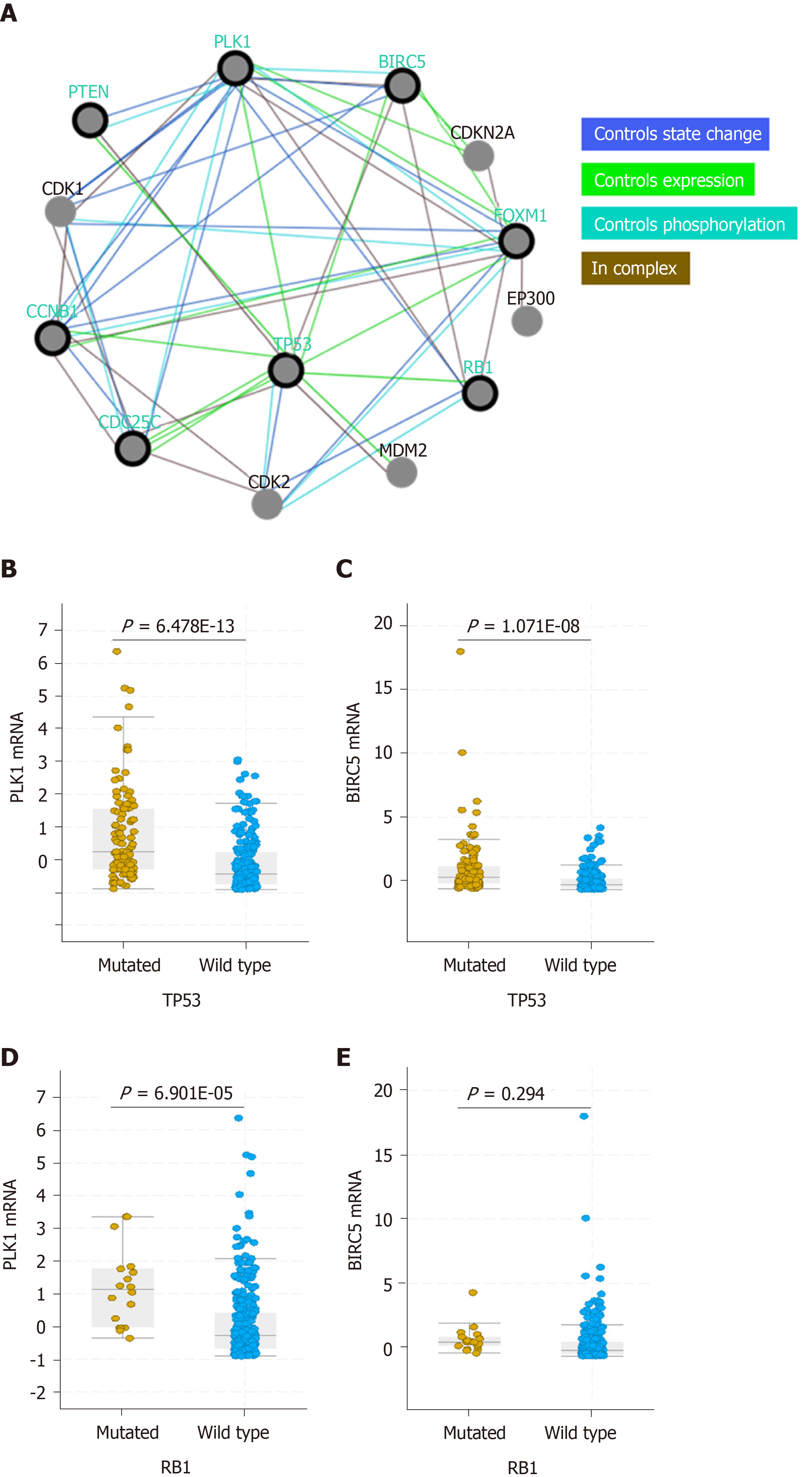
Previous studies have shown that p53 regulates the expression of PLK1 and BIRC5[21,22] and that PLK1, in turn, regulates p53 turnover (Figure 4A)[23]. In this study, we observed that both PLK1 and BIRC5 were upregulated in patients with p53-mutated HCC compared to those with wild-type p53 HCC (Figure 4B and C). These results suggest that mutations in or the functional loss of p53 in HCC may contribute to upregulation of PLK1 and BIRC5 expression, resulting in poor clinical outcome (Figures 2 and 3). In contrast, only high expression of PLK1, but not BIRC5, was observed in patients with RB1-mutated HCC compared to those with wild-type RB1 HCC (Figure 4D and E). These results suggest that the mutation status of the tumor suppressor p53 is more relevant than that of the tumor suppressor RB1 in mediating PLK1 and BIRC5 co-expression in HCC patients.
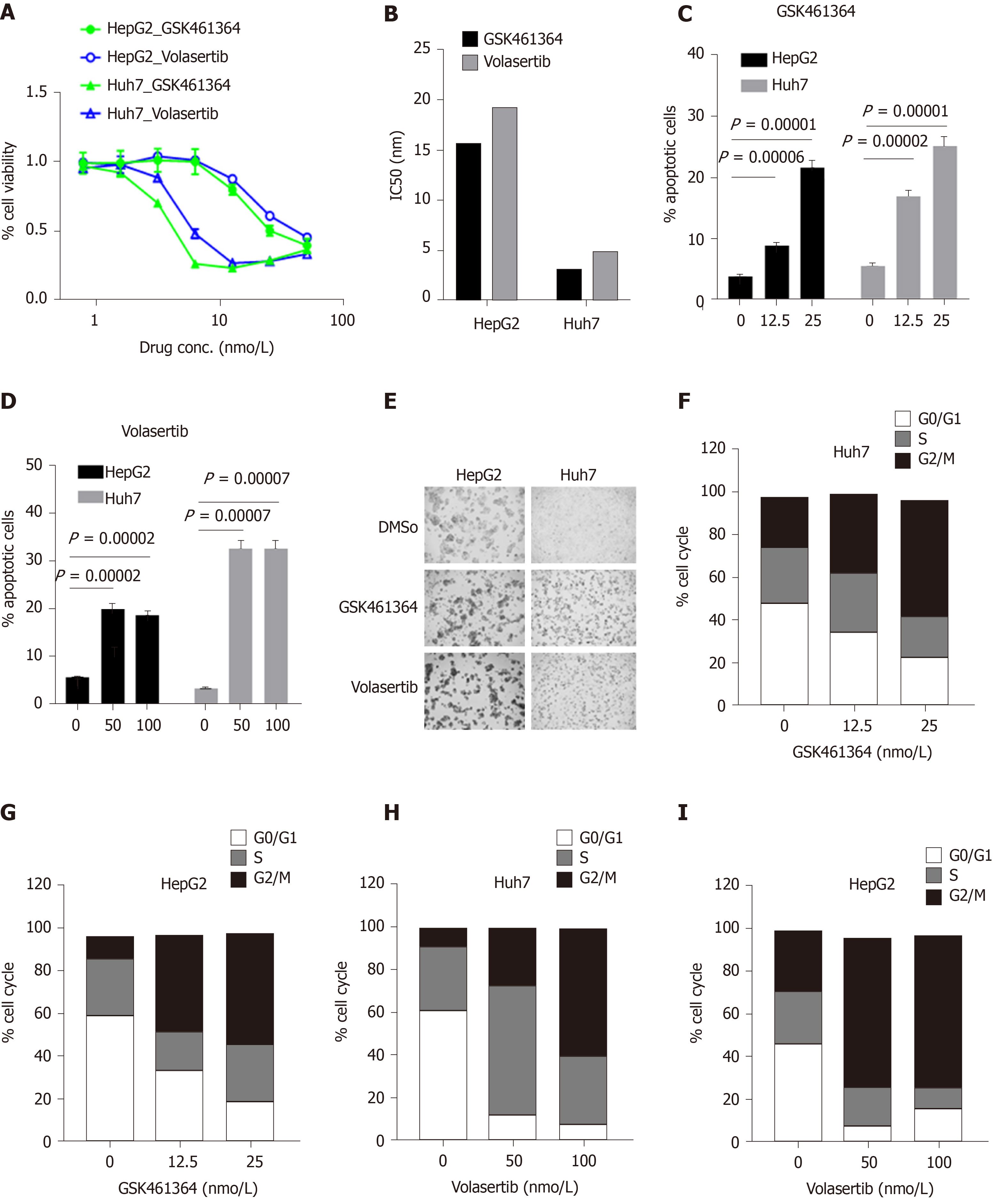
In this study, we investigated the in vitro efficacy of two potent and selective PLK1 inhibitors, GSK462364 and volasertib, against the p53-mutated HCC cell line Huh7 and the p53-wild-type HCC cell line HepG2. These two independently developed PLK1 inhibitors showed potent anti-tumor activity against both cell lines 72 h post treatment, but significantly lower IC50 values were observed in p53-mutated Huh7 cells (IC50 = 3.2 and 4.9 nmol/L, respectively) compared to those measured for p53-wild-type HepG2 cells (IC50 = 15.7 and 19.2 nmol/L, respectively) (Figure 5A and B). Treatment with both inhibitors induced robust apoptosis in both cell lines in a dose-dependent manner. In line with the cell viability assay results (Figure 5A and B), these PLK1 inhibitors more potently induced cell apoptosis in Huh7 cells than in HepG2 cells in a dose-dependent manner (12.5 and 25 nmol/L for GSK361464; 50 and 100 nmol/L for volasertib, respectively) 24 h post treatment (Figure 5C and D). Treatment with both PLK1 inhibitors for 24 h induced greater cellular morphological changes in Huh7 cells than in HepG2 cells (Figure 5E). Moreover, both inhibitors induced substantial cell cycle arrest at G2/M phase in Huh7 and HepG2 cells in a dose-dependent manner 24 h post treatment (Figure 5F-I).
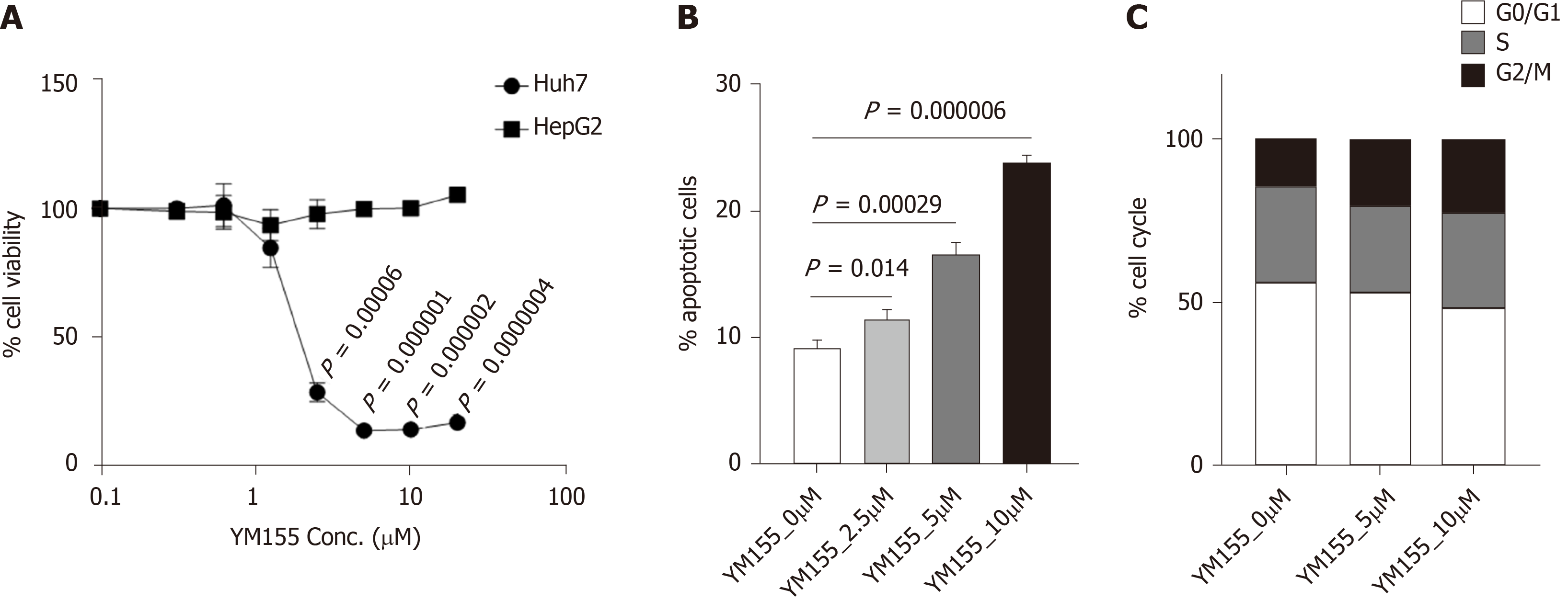
YM155 is a selective BIRC5 suppressor that inhibits BIRC5 expression at the transcriptional level. In this study, treatment with YM155 (dose range of 0-20 μmol/L) induced growth inhibition in p53-mutated Huh7 cells but not in p53-wild-type HepG2 cells 72 h post treatment (Figure 6A). These data suggest that the BIRC5 inhibitor YM155 is highly selective in targeting p53-mutated Huh7 cells over p53-wild-type HepG2 cells. YM155 treatment for 24 h induced robust cell apoptosis in a dose-dependent manner in Huh7 cells (Figure 6B). Similar to the assayed PLK1 inhibitors, YM155 treatment for 24 h in Huh7 cells induced cell cycle arrest at G2/M phase in a dose-dependent manner (Figure 6C), but to a much less extent than that observed for the PLK1 inhibitors.
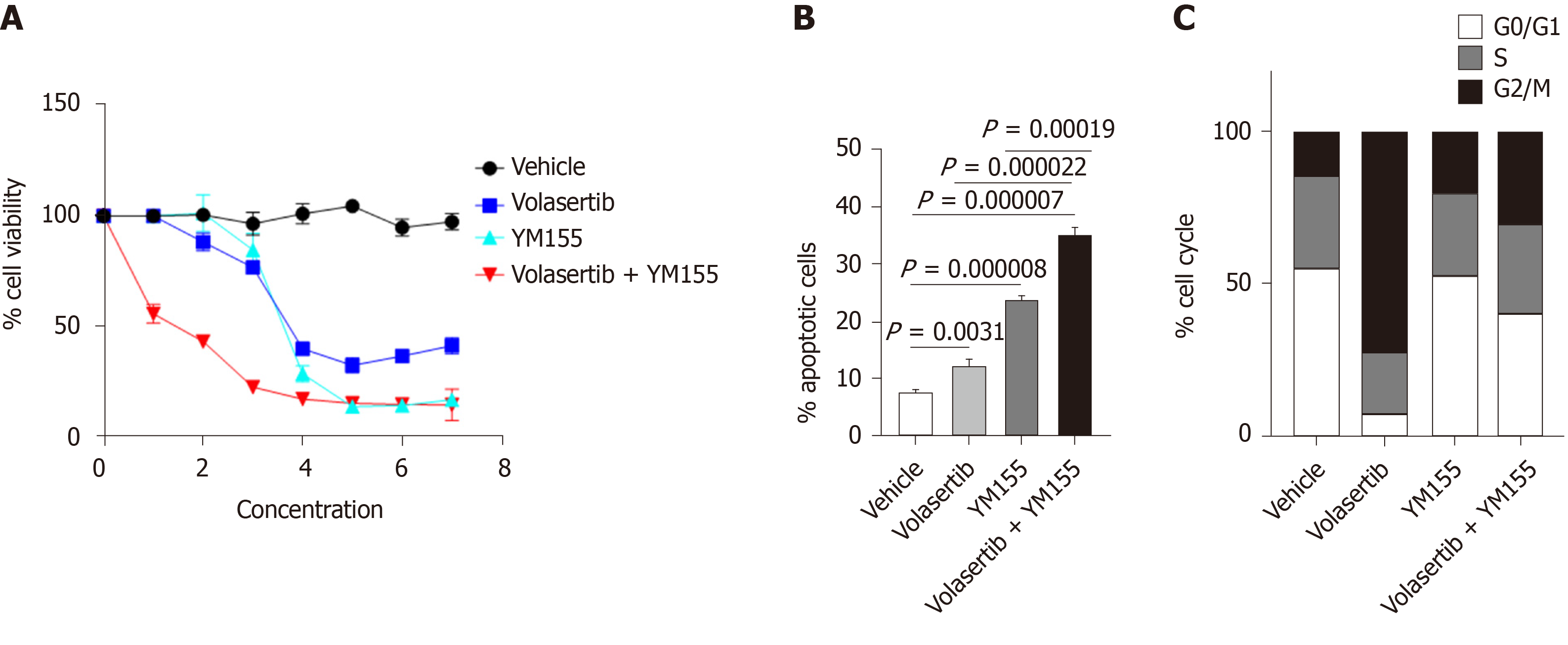
Co-expression of PLK1 and BIRC5 in the same subset of HCC patients suggested that dual targeting of PLK1 and BIRC5 may be a promising therapeutic strategy for these patients. Therefore, we investigated the in vitro efficacy of combining the PLK1 inhibitor volasertib, which reached a phase III clinical trial, and the BIRC5 inhibitor YM155, which reached a phase II clinical trial, in p53-mutated Huh7 cells. The combination treatment of volasertib (dose range: 0-50 nmol/L) and YM155 (dose range: 0-20 μmol/L) exhibited synergistic anti-tumor activity in the in vitro cell viability assay 72 h post treatment (Figure 7A). Consistent with these results, this combination treatment also showed synergistic induction of cell apoptosis 24 h post treatment (Figure 7B). In line with the data presented in Figures 5H and 6C, treatment with volasertib or YM155 alone markedly induced cell cycle arrest at G2/M phase, while the combination treatment of volasertib and YM155 showed less G2/M arrest than volasertib alone but more than that using YM155 alone (Figure 7C).
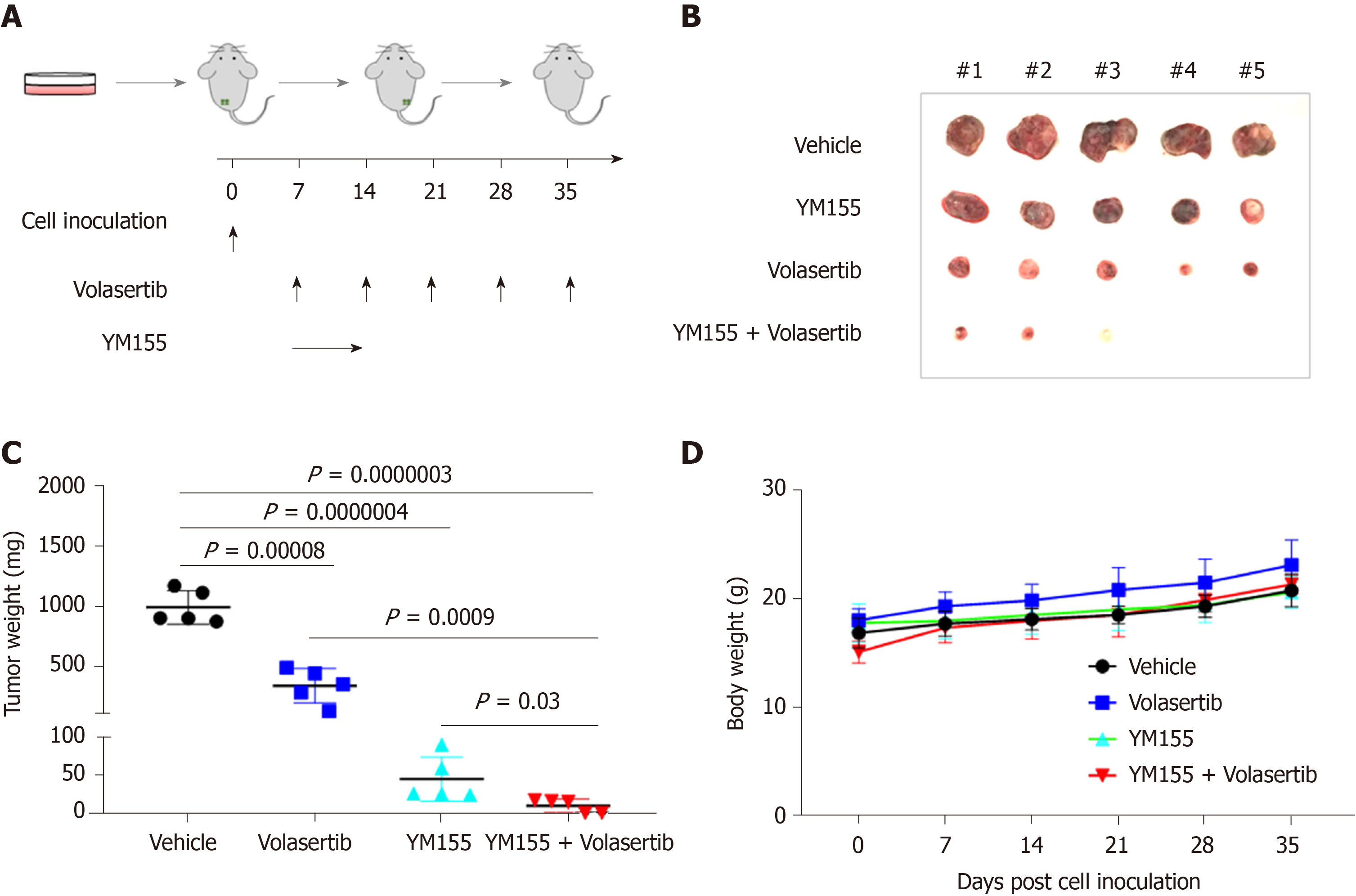
To validate the synergistic effect of the volasertib and YM155 combination treatment, we subcutaneously injected p53-mutated Huh7 cells into immunodeficient NSIG mice to establish mouse xenograft models. When the tumor became palpable, the mice were treated with volasertib (10 mg/kg, IP, weekly) for 4 wk and YM155 (3 mg/kg, continuous infusion for the first week and off for the following 3 wk), alone or in combination (Figure 8A). Single-agent treatment with volasertib or YM155 significantly inhibited tumor growth in vivo, and their combined use resulted in even smaller tumor masses in 3 out of 5 mice and no tumor masses in the last two mice in the group (Figure 8B and C). Furthermore, no toxicity was observed under the combination treatment schedule (Figure 8D).
In this study, we performed bioinformatic analysis for a TCGA RNA sequencing dataset from a large cohort of HCC patients (n = 374). Our analysis showed that two potential therapeutic targets, PLK1 and BIRC5, were highly expressed in a subset of HCC patients and that their expression correlated well in all patients. More interestingly, upregulation of PLK1/BIRC5 expression was frequently observed in HCC patients with mutations in p53. This observation led use to hypothesize that dual targeting PLK1 and BIRC5 in these patients would have superior therapeutic efficacy and could allow for the development of precision medicine to treat HCC patients with high PLK1 and BIRC5 expression, especially when p53 is also mutated.
In HCC cells with high co-expression of PLK1 and BIRC5, other cell cycle factors, including CCNB1, CCNB2, CDK1, FOXM1, AURKA, AURKB, and CDC25C were also observed to be highly expressed. In contrast, other PLK family members (PLK2 and PLK3), IAP family members (BIRC2 and BIRC3), and BCL-2 family members (BCL-2 and MCL-1) were not co-expressed with PLK1 and BIRC5. These results may provide a gene signature panel to identify HCC patients who will likely respond to PLK1 inhibition and BIRC5 treatment, although this possibility requires further validation.
For the first time, we provide a link between PLK1/BIRC5 co-expression and upregulation to the mutation status of p53, but not the p53 expression status, in HCC patients. The tumor suppressor p53 plays key roles in cell cycle arrest, DNA repair, and apoptosis. PLK1 is a direct target for the p53 transcription factor, which binds to the PLK1 promoter to suppress its expression[21]. In response to DNA damage, wild-type p53 but not mutant p53 suppresses PLK1 expression in an E2F1-dependent manner[24]. Therefore, PLK1 expression is suppressed in cells with wild-type p53, and PLK1 is upregulated in cells with mutated p53 that lead to the loss or inactivation of p53. Functioning within a negative regulatory feedback loop, PLK1 negatively regulates p53 transcriptional activation via physical interaction and through the phosphorylation of PLK1[25]. A similar scenario may also apply to the p53-BIRC5 axis, since wild-type but not mutant p53 also transcriptionally represses BIRC5 expression to promote p53-dependent cell apoptosis[22,26,27]. BIRC5 overexpression negatively regulates the expression of wild-type p53[28]. BIRC5 and PLK1 interact during mitosis, and PLK1 phosphorylates BIRC5 at serine 20[29]. In turn, BIRC5 regulates PLK1 localization to the kinetochore for the recruitment and dynamic localization of the BIRC5-containing chromosomal passenger complex (CPC) during cell division throughout mitosis[30,31]. However, no direct interaction between BIRC5 and p53 has been reported yet. Altogether, these results may explain why HCC cells with mutated p53 express higher levels of PLK1 and BIRC5 and suggest that PLK1 and BIRC5 work coordinately in contributing to cancer malignancies in cancer cells with p53 mutations. Based on this, one would expect that HCC cells with mutated p53 would be more sensitive to PLK1 and BIRC5 inhibition. Inhibition of PLK1 using small molecules such as GSK461364A and volasertib was previously shown to have differential anti-tumor activity and cell apoptosis based on p53 mutation status[32-35]. Inhibition of BIRC5 by YM155 leads to decreased BIRC5 expression and increased expression of PUMA, a direct target of p53, which results in cell apoptosis[36]. Indeed, this phenomenon is observed in Huh7 cells with mutated p53 and in HepG2 cells with wild-type p53. It has been shown that HuH7 cells harbor a homozygous p53 mutation (Y220C), which is a destabilizing mutation that results in partial DNA-binding activity compared to wild-type p53[37]. This type of mutant p53 can be pharmaceutically reactivated and functionally rescued by p53 activators such as ARP-246[38,39]. FuBP1 is overexpressed in 80% of HCC tumors with chronic hepatitis C[20] and has been shown to inhibit p53 function[37]. However, we did not detect any significant correlation between FUBP1 expression and that of either PLK1 or BIRC5, indicating that FUBP1 is not functionally correlated with PLK1 and BIRC5 in contributing to HCC malignancy.
Our data also showed that expression of PLK1 was upregulated in HCC patients with mutations in the tumor suppressor RB1 compared to those with wild-type RB1. It has also been shown that RB1 loss is associated with a higher sensitivity to PLK1 inhibitors in triple-negative breast cancer[40]. In contrast to PLK1, BIRC5 did not show differential expression in HCC patients with/without RB1 mutations. This result indicated that high co-expression of PLK1/BIRC5 is selective for HCC patients with p53 mutations and therefore provides strong evidence for the potential value of single or dual targeting of PLK1 and BIRC5 in p53-mutated HCC cells.
Moreover, the results of our study showed that dual targeting of PLK1 and BIRC5 showed synergistic antitumor activity in vitro and in vivo in p53-mutated Huh7 cells. Therefore, our study provides valuable insights for therapeutic development targeting PLK1 and BIRC5 for the subset of HCC patient population with the PLK1/BIRC5 co-expression signature in p53-mutated HCC patients as well as other cancer models.
Hepatocellular carcinoma (HCC) is the fifth most common malignant cancer and the second leading cause of cancer-related mortality. HCC is often diagnosed at advanced stages without curative therapies. Therefore, there is an unmet need of preclinical studies to develop novel therapeutic strategies to treat HCC, especially at advanced stages. Polo-like kinase 1 (PLK1) is activated at the late G2 phase of the cell cycle and is required for entry to mitosis. Interestingly, PLK1 is overexpressed in many HCC patients and is highly associated with poor clinical outcome. Baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5) is also highly overexpressed in HCC and plays key roles in HCC cell survival, cell proliferation, and disease progression of HCC.
More biomarkers are required for the diagnosis and treatment of HCC. However, how p53, PLK1, and BIRC5 interact in HCC has not been well defined.
To determine the expression pattern of PLK1 and BIRC5, as well as their correlation with mutation status of p53 and patient clinical outcome.
The expression of PLK1 and BIRC5 and their correlation with the mutation status p53 were analyzed using a TCGA HCC dataset. Cell-based studies were conducted to investigate the efficacy of PLK1 and BIRC5 inhibitors, alone or in combination, the results of which were further validated in p53-mutated Huh7-derived xenografts in immune-deficient NSIG mice.
Our bioinformatic analysis using an HCC dataset from TCGA revealed that PLK1 and BIRC5 were overexpressed in the same subset of HCC patients and that their expression was highly correlated in all HCC patients. Both PLK1 and BIRC5 overexpression was more frequently detected in HCC with p53 mutations, compared to that observed in HCC with wild-type p53. High PLK1 or BIRC5 expression was significantly correlated with poor clinical outcome. Both PLK1 inhibitors volasertib and GSK461364 or the BIRC5 inhibitor YM155 selectively targeted Huh7 cells, which express Y220C-mutated p53 that is aberrantly stable and transcriptionally inactive, but not HepG2 cells, which express wild-type p53. Combination treatment with volasertib and YM155 synergistically inhibited the cell viability of Huh7 cells by promoting cell apoptosis. The efficacy of volasertib and YM155, alone or in combination, was further validated in vivo in a Huh7-derived xenograft model in immuno-deficient NSIG mice.
PLK1 and BIRC5 are highly co-expressed in p53-mutated HCC and dual targeting of PLK1 and BIRC5 synergistically inhibits the cell viability of p53-mutated HCC cells in vitro through the induction of cell apoptosis as well as the tumor growth of p53-mutated HCC cells in vivo.
The results of this study provides valuable insights for therapeutic development for the subset of the HCC patient population with the PLK1/BIRC5 co-expression signature in p53-mutated HCC patients as well as for other cancer models in the future.
The authors would like to thank Li C from the Animal Laboratory of Tianjin Third Central Hospital for her help in animal preparation and Professor Li SJ from Nankai University for aid in biostatistics during the preparation of this manuscript.
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
| 1. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20108] [Cited by in RCA: 20516] [Article Influence: 2051.6] [Reference Citation Analysis (20)] |
| 2. | Janevska D, Chaloska-Ivanova V, Janevski V. Hepatocellular Carcinoma: Risk Factors, Diagnosis and Treatment. Open Access Maced J Med Sci. 2015;3:732-736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 126] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 3. | Zhou X, Hao Q, Lu H. Mutant p53 in cancer therapy-the barrier or the path. J Mol Cell Biol. 2019;11:293-305. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 173] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 4. | Hussain SP, Schwank J, Staib F, Wang XW, Harris CC. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene. 2007;26:2166-2176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 411] [Cited by in RCA: 439] [Article Influence: 24.4] [Reference Citation Analysis (1)] |
| 5. | Tornesello ML, Buonaguro L, Tatangelo F, Botti G, Izzo F, Buonaguro FM. Mutations in TP53, CTNNB1 and PIK3CA genes in hepatocellular carcinoma associated with hepatitis B and hepatitis C virus infections. Genomics. 2013;102:74-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 131] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 6. | Shirabe K, Toshima T, Taketomi A, Taguchi K, Yoshizumi T, Uchiyama H, Harimoto N, Kajiyama K, Egashira A, Maehara Y. Hepatic aflatoxin B1-DNA adducts and TP53 mutations in patients with hepatocellular carcinoma despite low exposure to aflatoxin B1 in southern Japan. Liver Int. 2011;31:1366-1372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 7. | Parrales A, Iwakuma T. Targeting Oncogenic Mutant p53 for Cancer Therapy. Front Oncol. 2015;5:288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 252] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 8. | Li S, Gao M, Li Z, Song L, Gao X, Han J, Wang F, Chen Y, Li W, Yang J. p53 and P-glycoprotein influence chemoresistance in hepatocellular carcinoma. Front Biosci (Elite Ed). 2018;10:461-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 9. | Hientz K, Mohr A, Bhakta-Guha D, Efferth T. The role of p53 in cancer drug resistance and targeted chemotherapy. Oncotarget. 2017;8:8921-8946. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 298] [Cited by in RCA: 419] [Article Influence: 59.9] [Reference Citation Analysis (0)] |
| 10. | Combes G, Alharbi I, Braga LG, Elowe S. Playing polo during mitosis: PLK1 takes the lead. Oncogene. 2017;36:4819-4827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 122] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 11. | Colicino EG, Hehnly H. Regulating a key mitotic regulator, polo-like kinase 1 (PLK1). Cytoskeleton (Hoboken). 2018;75:481-494. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 12. | Jeong SB, Im JH, Yoon JH, Bui QT, Lim SC, Song JM, Shim Y, Yun J, Hong J, Kang KW. Essential Role of Polo-like Kinase 1 (Plk1) Oncogene in Tumor Growth and Metastasis of Tamoxifen-Resistant Breast Cancer. Mol Cancer Ther. 2018;17:825-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 13. | Ramani P, Nash R, Sowa-Avugrah E, Rogers C. High levels of polo-like kinase 1 and phosphorylated translationally controlled tumor protein indicate poor prognosis in neuroblastomas. J Neurooncol. 2015;125:103-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 14. | Tut TG, Lim SH, Dissanayake IU, Descallar J, Chua W, Ng W, de Souza P, Shin JS, Lee CS. Upregulated Polo-Like Kinase 1 Expression Correlates with Inferior Survival Outcomes in Rectal Cancer. PLoS One. 2015;10:e0129313. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 15. | Zhang R, Shi H, Ren F, Liu H, Zhang M, Deng Y, Li X. Misregulation of polo-like protein kinase 1, P53 and P21WAF1 in epithelial ovarian cancer suggests poor prognosis. Oncol Rep. 2015;33:1235-1242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 16. | Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res. 2008;14:5000-5005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 526] [Cited by in RCA: 582] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 17. | Velculescu VE, Madden SL, Zhang L, Lash AE, Yu J, Rago C, Lal A, Wang CJ, Beaudry GA, Ciriello KM, Cook BP, Dufault MR, Ferguson AT, Gao Y, He TC, Hermeking H, Hiraldo SK, Hwang PM, Lopez MA, Luderer HF, Mathews B, Petroziello JM, Polyak K, Zawel L, Kinzler KW. Analysis of human transcriptomes. Nat Genet. 1999;23:387-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 531] [Cited by in RCA: 521] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 18. | Garg H, Suri P, Gupta JC, Talwar GP, Dubey S. Survivin: a unique target for tumor therapy. Cancer Cell Int. 2016;16:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 327] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 19. | Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10832] [Cited by in RCA: 14510] [Article Influence: 1036.4] [Reference Citation Analysis (0)] |
| 20. | Rabenhorst U, Beinoraviciute-Kellner R, Brezniceanu ML, Joos S, Devens F, Lichter P, Rieker RJ, Trojan J, Chung HJ, Levens DL, Zörnig M. Overexpression of the far upstream element binding protein 1 in hepatocellular carcinoma is required for tumor growth. Hepatology. 2009;50:1121-1129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 75] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 21. | McKenzie L, King S, Marcar L, Nicol S, Dias SS, Schumm K, Robertson P, Bourdon JC, Perkins N, Fuller-Pace F, Meek DW. p53-dependent repression of polo-like kinase-1 (PLK1). Cell Cycle. 2010;9:4200-4212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 100] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 22. | Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S, Wen SF, Wang L, Kirschmeier P, Bishop WR, Nielsen LL, Pickett CB, Liu S. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21:2613-2622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 417] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 23. | Dias SS, Hogan C, Ochocka AM, Meek DW. Polo-like kinase-1 phosphorylates MDM2 at Ser260 and stimulates MDM2-mediated p53 turnover. FEBS Lett. 2009;583:3543-3548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Zhou Z, Cao JX, Li SY, An GS, Ni JH, Jia HT. p53 Suppresses E2F1-dependent PLK1 expression upon DNA damage by forming p53-E2F1-DNA complex. Exp Cell Res. 2013;319:3104-3115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 25. | Ando K, Ozaki T, Yamamoto H, Furuya K, Hosoda M, Hayashi S, Fukuzawa M, Nakagawara A. Polo-like kinase 1 (Plk1) inhibits p53 function by physical interaction and phosphorylation. J Biol Chem. 2004;279:25549-25561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 210] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 26. | Zhou M, Gu L, Li F, Zhu Y, Woods WG, Findley HW. DNA damage induces a novel p53-survivin signaling pathway regulating cell cycle and apoptosis in acute lymphoblastic leukemia cells. J Pharmacol Exp Ther. 2002;303:124-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 103] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 27. | Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem. 2002;277:3247-3257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 576] [Cited by in RCA: 583] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 28. | Wang Z, Fukuda S, Pelus LM. Survivin regulates the p53 tumor suppressor gene family. Oncogene. 2004;23:8146-8153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 64] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 29. | Colnaghi R, Wheatley SP. Liaisons between survivin and Plk1 during cell division and cell death. J Biol Chem. 2010;285:22592-22604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 30. | Sun SC, Liu HL, Sun QY. Survivin regulates Plk1 localization to kinetochore in mouse oocyte meiosis. Biochem Biophys Res Commun. 2012;421:797-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 31. | Chu Y, Yao PY, Wang W, Wang D, Wang Z, Zhang L, Huang Y, Ke Y, Ding X, Yao X. Aurora B kinase activation requires survivin priming phosphorylation by PLK1. J Mol Cell Biol. 2011;3:260-267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 81] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 32. | Tyagi S, Bhui K, Singh R, Singh M, Raisuddin S, Shukla Y. Polo-like kinase1 (Plk1) knockdown enhances cisplatin chemosensitivity via up-regulation of p73α in p53 mutant human epidermoid squamous carcinoma cells. Biochem Pharmacol. 2010;80:1326-1334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 33. | Degenhardt Y, Greshock J, Laquerre S, Gilmartin AG, Jing J, Richter M, Zhang X, Bleam M, Halsey W, Hughes A, Moy C, Liu-Sullivan N, Powers S, Bachman K, Jackson J, Weber B, Wooster R. Sensitivity of cancer cells to Plk1 inhibitor GSK461364A is associated with loss of p53 function and chromosome instability. Mol Cancer Ther. 2010;9:2079-2089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 74] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 34. | Van den Bossche J, Domen A, Peeters M, Deben C, De Pauw I, Jacobs J, De Bruycker S, Specenier P, Pauwels P, Vermorken JB, Lardon F, Wouters A. Radiosensitization of Non-Small Cell Lung Cancer Cells by the Plk1 Inhibitor Volasertib Is Dependent on the p53 Status. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 35. | Van den Bossche J, Deben C, De Pauw I, Lambrechts H, Hermans C, Deschoolmeester V, Jacobs J, Specenier P, Pauwels P, Vermorken JB, Peeters M, Lardon F, Wouters A. In vitro study of the Polo-like kinase 1 inhibitor volasertib in non-small-cell lung cancer reveals a role for the tumor suppressor p53. Mol Oncol. 2019;13:1196-1213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 36. | Yan X, Su H. YM155 Down-Regulates Survivin and Induces P53 Up-Regulated Modulator of Apoptosis (PUMA)-Dependent in Oral Squamous Cell Carcinoma Cells. Med Sci Monit. 2017;23:1963-1972. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 37. | Dixit U, Pandey AK, Liu Z, Kumar S, Neiditch MB, Klein KM, Pandey VN. FUSE Binding Protein 1 Facilitates Persistent Hepatitis C Virus Replication in Hepatoma Cells by Regulating Tumor Suppressor p53. J Virol. 2015;89:7905-7921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 38. | Zhang Q, Bykov VJN, Wiman KG, Zawacka-Pankau J. APR-246 reactivates mutant p53 by targeting cysteines 124 and 277. Cell Death Dis. 2018;9:439. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 189] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 39. | Lehmann S, Bykov VJ, Ali D, Andrén O, Cherif H, Tidefelt U, Uggla B, Yachnin J, Juliusson G, Moshfegh A, Paul C, Wiman KG, Andersson PO. Targeting p53 in vivo: a first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J Clin Oncol. 2012;30:3633-3639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 306] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 40. | Witkiewicz AK, Chung S, Brough R, Vail P, Franco J, Lord CJ, Knudsen ES. Targeting the Vulnerability of RB Tumor Suppressor Loss in Triple-Negative Breast Cancer. Cell Rep. 2018;22:1185-1199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 83] [Article Influence: 11.9] [Reference Citation Analysis (0)] |