Published online Jan 21, 2020. doi: 10.3748/wjg.v26.i3.335
Peer-review started: October 15, 2019
First decision: November 13, 2019
Revised: December 11, 2019
Accepted: December 21, 2019
Article in press: December 21, 2019
Published online: January 21, 2020
Processing time: 85 Days and 15.2 Hours
Obesity is a risk factor for colorectal cancer, yet metabolic distinctions between healthy right and left colon tissue, before cancer is diagnosed, remains largely unknown. This study compared right-ascending and left-descending colon tissue metabolomes to identify differences from the stool metabolome in normal weight, overweight, and obese adults.
To examine right and left colon tissue metabolites according to body mass index that may serve as mechanistic targets for interventions and biomarkers for colon cancer risk.
Global, non-targeted metabolomics was applied to assess right-ascending and left-descending colon tissue collected from healthy adults undergoing screening colonoscopies to test the hypothesis that BMI differentially impacts colon tissue metabolite profiles. The colon tissue and stool metabolome of healthy adults (n = 24) was analyzed for metabolite signatures and metabolic pathway networks implicated in progression of colorectal cancer.
Ascending and descending colon contained 504 host, food, and microbiota-derived metabolites from normal weight, overweight and obese adults grouped according to body mass index. Amino acids, lipids, and nucleotides were among the chemical types that further differentiated from the stool metabolite profiles. Normal weight adults had 46 significantly different metabolites between ascending and descending colon tissue locations, whereas there were 37 metabolite differences in overweight and 28 metabolite differences for obese adults (P < 0.05). Obese adults had trimethylamine N-oxide, endocannabinoids and monoacylglycerols with different relative abundances identified between ascending and descending colon. Primary and secondary bile acids, vitamins, and fatty acids also showed marked relative abundance differences in colon tissue from overweight/obese adults.
There were metabolite profile differences between right-ascending and left-descending colon tissue in healthy adults. Colon lipids and other metabolites in obese and overweight adults were distinguished from normal weight participants and associated with gut inflammation, nutrient absorption, and products of microbiota metabolism.
Core tip: This study identified metabolite profile differences between right-ascending and left-descending colon from normal, overweight or obese adults. We also show that stool metabolite composition does not accurately reflect the right-ascending colon. There is limited knowledge of human colon small molecules and metabolite signatures that may impact colon cancer risk. Colon cancer of the right-ascending colon has a poorer prognosis and reduced survival outcome when compared to colon cancer on the left-descending colon. Diet and lifestyle are additional factors of overweight and obesity that may influence colon tissue metabolite composition with respect to inflammation. Right and left colon metabolite profiles may be helpful to evaluate after interventions that seek to prevent or mitigate cancer risk.
- Citation: Baxter BA, Parker KD, Nosler MJ, Rao S, Craig R, Seiler C, Ryan EP. Metabolite profile comparisons between ascending and descending colon tissue in healthy adults. World J Gastroenterol 2020; 26(3): 335-352
- URL: https://www.wjgnet.com/1007-9327/full/v26/i3/335.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i3.335
Body mass index (BMI) of 30% or greater is an established risk factor for colon cancer in men and women[1,2]. Obesity is a complex lipid-storage disease with metabolic aberrations locally in the gut and systemically in the host that increase risk for multiple chronic diseases[3]. Similar relationships occur for obesity and the incidence of larger (vs smaller) colon adenomas[3,4]. Weight gain from early to middle adulthood increases risk[5], whereby middle-aged obese adults had a 60% increase risk of right-side colon cancer compared to the left-side[6]. We and others have previously shown that stool reveals changes in microbial communities[7], and modulation by diet[8], yet this may not accurately reflect metabolic differences between the right and left side colon tissue[9].
Right-cancer patients have a worse prognosis with a median survival of 76.6 mo while left-sided have median survival of 101 mo[10] and right-sided tumors are significantly larger in size with a higher tumor grade when compared to left side colon cancer[11,12]. African American and non-Hispanic blacks have 24% greater odds of right-sided colon cancer[13]. Physical inactivity, excess body weight, alcohol, smoking, and a central deposition of adiposity are consistent risk factors for colorectal cancer. High consumption of red meat more than 3 times/wk has been associated with 2-fold increased risk for colon cancer and these food components merit attention in the tissue of healthy adults[14].
Metabolomics is a high- throughput screening methodology that is sensitive for detection of exogenous and endogenous (microbial, host and food) products of metabolism[15] and can aid in identification of disease risk biomarkers[16]. Metabolite profiling analysis of ascending and descending colon tissue was conducted herein to assess metabolic differences between colon locations that differ from stool. This study utilized normal weight, overweight and obese adults for investigation of colonic compounds that may impact colon cancer risk[17]. Metataxonomics of colon tissue by location has varied results[18,19] and provided rationale for using metabolomics. The major objective of this study was to identify metabolic pathways that distinguished ascending and descending colon tissue and to reveal metabolites altered by overweight and obesity that may pose elevated risks for developing cancer. We hypothesized that lipids (e.g., fatty acids, bile acids, phospholipids, monoacylglycerol, and endocannabinoids) are distinct in type and abundance between the ascending and descending colon, and that colon tissue metabolomes will differ according to BMI when compared to stool in overweight and obese adults.
Ninety-three healthy adults were contacted prior to a scheduled colonoscopy in Fort Collins CO. Forty adult males and females provided written informed consent to collect a stool sample and an ascending and descending colon tissue biopsy. Twenty-four individuals (colon and stool) were assessed for non-targeted metabolomics. Eligible participants were provided a stool kit and study instructions. The gastroenterology clinical nurses and research staff confirmed study code number assignments and ensured the completed de-identification at the site of colonoscopy procedure. Colorado State University study personnel were contacted by clinic staff for sample retrieval immediately following procedure. Three study groups were BMI 20-24.9 for normal weight (n = 9), BMI 25-29.9 for overweight (n = 9) and BMI 30+ for obese (n = 6) adults. One normal weight female (BMI 24) had the right and left colon and stool sample applied for metataxonomic analysis (16S rRNA gene sequencing). Participant’s inclusion criteria for this study were at least 18 years of age, a scheduled routine colonoscopy, no prior history of colorectal cancer diagnosis, non-smoker, and not having taken antibiotics for at least one month prior to the standard of care, routine screening colonoscopy.
The colon tissue collected for this study was visually determined by the gastroenterologist performing the procedure to be normal, healthy tissue without polyps. Each participant had an about 5 mm biopsy of ascending (right) and descending (left) colon tissue and a self-collected stool sample prior to bowel preparation. Colonoscopy was completed by Centers for Gastroenterology-Fort Collins and University of Colorado-Health North Gastroenterology Clinic (Fort Collins, CO, United States). Samples were de-identified for personal information and study ID coded before storage and metabolite processing at Colorado State University. The number of polyps removed by the doctor with the respective location was provided following the procedure. This study received IRB approval, and include protocol number; Colorado State University IRB No. 15-6051, and University of Colorado Health IRB No. 0010144. Participants in this study had no history of diseases related to the liver or biliary tract and they did not have previous procedures such as cholecystectomy or ileal resections. This study did not collect information regarding the family history of colorectal cancer and did not perform hereditary genetic or epi-genetic screening history on the patients. Table 1 shows the study participant characteristics.
| Characteristics | Total (n = 24) | Normal (n = 9) | Overweight (n = 9) | Obese (n = 6) |
| Sex | ||||
| Males | 5 | 0 | 4 | 1 |
| Females | 19 | 9 | 5 | 5 |
| BMI (mean ± SD, kg/m2) | 27.6 ± 5.8 | 22 ± 1 | 26.7 ± 1.3 | 35 ± 5 |
| Total number of people with polyps removes | 14 | 6 | 4 | 4 |
| Total number polyps removed | - | 14 | 13 | 22 |
| Polyp Location | ||||
| Cecum | - | 1 | 0 | 3 |
| Ascending | - | 6 | 2 | 10 |
| Transverse | 2 | 1 | 1 | |
| Sigmoid | - | 2 | 4 | 6 |
| Rectum | - | 0 | 6 | 0 |
| Descending | - | 3 | 0 | 2 |
| History of hypertension | ||||
| Yes | 6 | 2 | 3 | 2 |
| No | 18 | 7 | 6 | 4 |
| History of type 2 diabetes | ||||
| Yes | 3 | 0 | 2 | 1 |
| No | 21 | 9 | 7 | 5 |
| Taking dietary supplements | ||||
| Yes | 14 | 4 | 8 | 2 |
| No | 10 | 5 | 1 | 4 |
| Smoking history | ||||
| Past smoker | 9 | 2 | 6 | 1 |
| Never | 15 | 6 | 4 | 5 |
Stool samples were self-collected by participants in a pre-labeled study coded container and frozen at -80 °C. Approximately 5 mm of normal healthy colon tissue were stored immediately at -80 °C following collection. Samples were shipped on dry ice to Metabolon, Inc. (Durham, NC, United States) and a single participant sample underwent DNA extraction for metataxonomics.
Tissue and stool metabolite extraction was completed using 80% methanol as previously described[7], prior to ultrahigh performance liquid chromatography-tandem mass spectroscopy (UPLC-MS/MS) as completed by Metabolon, Inc. Positive and negative ion modes were chosen to provide broad, non-targeted detection of metabolites.
Samples were extracted using the automated MicroLab STAR® system from Hamilton Company. A set of recovery standards were added prior to the first step in the extraction process for quality control purposes. To remove protein, dissociate small molecules bound to protein or trapped in the precipitated protein matrix, and to recover chemically diverse metabolites, proteins were precipitated with methanol under vigorous shaking for 2 min (Glen Mills GenoGrinder 2000) followed by centrifugation. The resulting extract was divided into five fractions: two for analysis by two separate reverse phase (RP)/UPLC-MS/MS methods with positive ion mode electrospray ionization (ESI), one for analysis by RP/UPLC-MS/MS with negative ion mode ESI, one for analysis by HILIC/UPLC-MS/MS with negative ion mode ESI, and one sample was reserved for backup. Samples were placed briefly on a TurboVap® (Zymark) to remove the organic solvent. The sample extracts were stored overnight under nitrogen before preparation for analysis.
The UPLC-MS/MS portion of the platform was based on a Waters ACQUITY ultra-performance liquid chromatography (UPLC) and a Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer operated at 35000 mass resolution. The protocol has been previously described by our lab[7].
Raw data were extracted, peak-identified, and processed using Metabolon’s hardware and software. Compounds were identified by comparison to library entries of purified standards or recurrent unknown entities as previously described[7].
Healthy colon tissue or stool metabolite profiles were semi-quantified in terms of relative abundance and median scaled to 1. Fold differences were calculated for normal weight, overweight and obese (colon tissue and stool) and for colon tissue between ascending and descending sites. A matched-pairs 2-way ANOVA was completed using the scaled relative abundance of each metabolite, experimental groups in ArrayStudio on log transformed data, were used for normal weight, overweight and obese. Metabolite profile distinctions between ascending and descending colon tissue were evaluated using P < 0.05 for statistical significance with matched pair t-test. An estimate of the false discovery rate (Q value) was calculated to account the multiple comparisons across metabolites that are typical of metabolomics-based studies with a Q value ≥ 0.01. A linear regression analyses for colon metabolites were preformed to compare the groups with polyp removal to no polyp removal, after adjusting for the effect of weight category of the subjects.
Principal component analysis and hierarchical clustering were applied to understand the similarities and differences between samples and/or groups of samples in a complex dataset. Unsupervised clustering was performed using the ward D2 method[20]. Random forest (RF) analysis, a supervised classification technique, was applied for identifying candidate biomarkers. To determine which variables (biochemicals) make the largest contribution to the classification of BMI, a “variable importance” measure was computed. We used the “Mean Decrease Accuracy” as this metric prediction accuracy[21].
DNA was extracted from colon tissue and stool with the MoBio PowerSoil Kit according to manufacturer protocols. Amplification of the V4 region of the 16S rRNA gene and amplicon sequencing followed the standards outlined by the Earth Microbiome Project. Raw FASTQ-formatted forward reads were imported into the Quantitative Insights Into Microbial Ecology 2 (QIIME 2) platform[22]. A feature table comprised of amplicon sequence variants (ASVs) was inferred from reads using the DADA2 algorithm[23]. Taxonomy was assigned to each representative ASV sequence using Naïve Bayes classifiers trained against 99% OTU reference collections from Greengenes 13_8 or SILVA 132. The raw feature table, representative sequences, and taxonomy tables were exported from QIIME 2 for further processing using R[24]. Following import, a master table comprised of ASV IDs with corresponding representative sequences, full and truncated Green-genes and SILVA taxonomic lineages, and absolute abundances for all ASVs within each sample was constructed. This master table served as the entry point for all downstream processing and analysis. Comparisons of microbiota composition proceeded from the compositional data analysis paradigm with count zero multiplicative replacement prior to applying the centred log-ratio (clr) transformation[25]. Taxon abundance are depicted as proportions (i.e., relative abundances). Supplemental Methods for additional details regarding amplification conditions, library preparation, sequencing, and a comprehensive account of analytical approaches.
Metataxonomics sequence data supporting the conclusions of this manuscript are available via NCBI SRA BioProject Accession PRJNA594611 and on this project’s GitHub repository located at github.com/kdprkr/ConjurersBrew, along with each of the materials needed to reproduce the analysis.
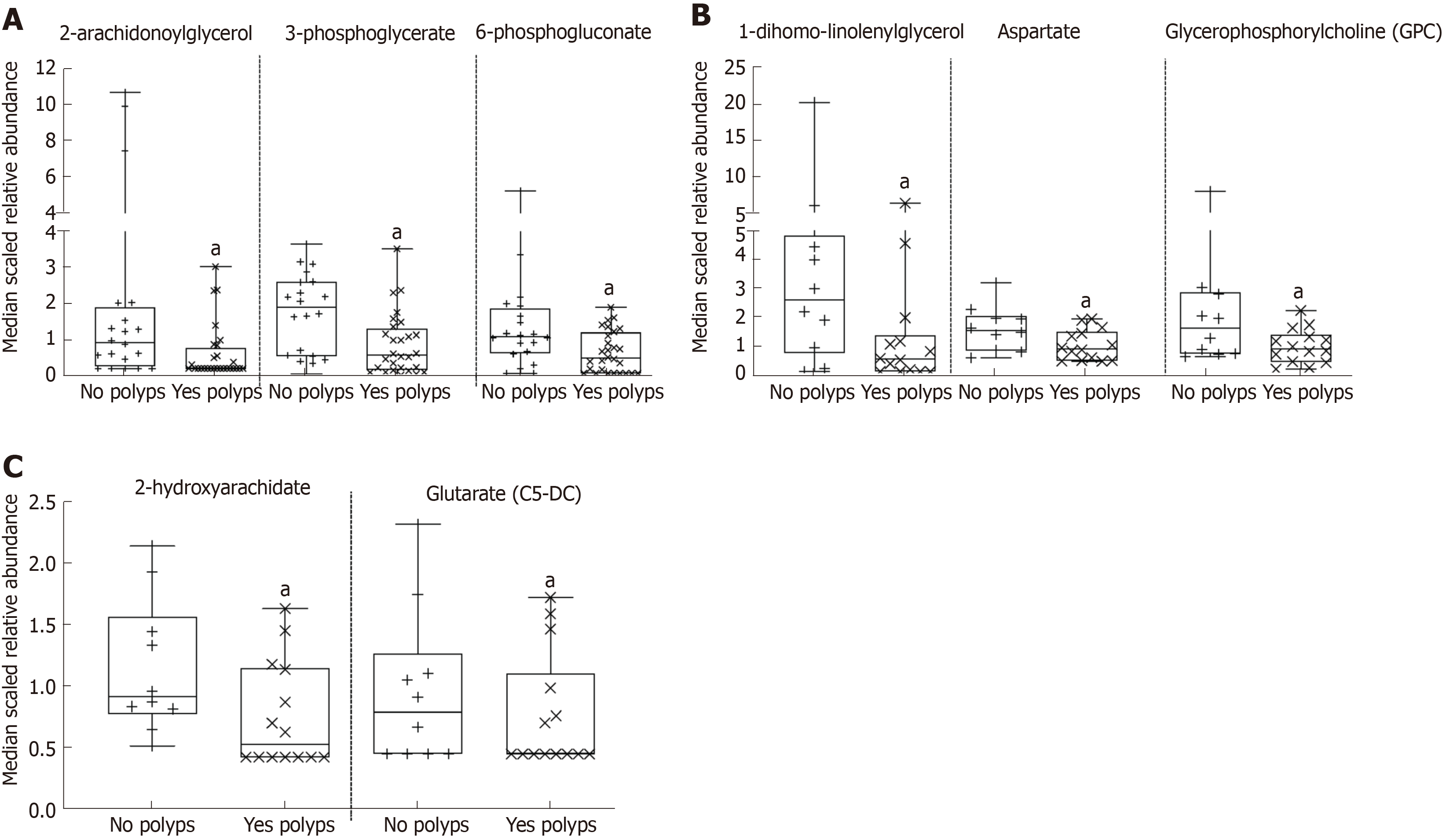
The stool metabolome of healthy adults classified according to BMI as normal weight, overweight or obese had a total of 842 named compounds (Supplemental Table 1). The 842 stool metabolites consisted of 175 amino acids, 26 peptides, 33 carbohydrates, 11 energy, 345 lipids, 46 nucleotides, 47 cofactors and vitamins, and 159 were classified as exogenous and referred to as xenobiotics. There were 98 stool metabolites that significantly differ according to BMI were 22 amino acids, 1 peptide, 8 nucleotides, 2 cofactors and vitamins, 9 xenobiotics and 56 lipids. Principal component analyses (PCA) for the stool metabolome (Supplemental Figure 1A) and hierarchal clustering (Supplemental Figure 1B) did not clearly separate participants by BMI groups. The 504 colon metabolites with known identity and 44 unnamed/unknown compounds from 24 male and female participants are provided in Supplemental Table 2. The colon metabolome contained 93 amino acids, 13 peptides, 35 carbohydrates, 10 metabolites were classified under TCA cycle and oxidative phosphorylation and there were 20 cofactors and vitamins. The largest portion of the colon tissue metabolome were lipids (about 50%, 262 lipid metabolites) that span 40 sub-metabolic pathways. Other notable small molecules from colon were 38 nucleotides and 32 exogenous, xenobiotic metabolites. Fourteen out of twenty-four healthy participants had 1-14 polyps removed during screening colonoscopy. A regression analysis for colon metabolites adjusting for the effect of weight was done, revealing 17 colon metabolites that had lower expression correlated with polyps removed (Supplemental Table 3). Figure 1A, shows 2-arachidonoylglycerol, 3-phosphoglycerate, and 6-phosphogluconate had a lower relative abundance in ascending and descending tissue from participants with polyps removed. Figure 1B-C, shows 1-dihomo-linolenylglycerol, aspartate, and glycerophosphorycholine (GPC) with lower expression in ascending colon tissue, while glutarate (C5-DC), and 2-hydroxyarachidate had lower expression in descending tissue from participants with polyps removed (Figures 1B and C).
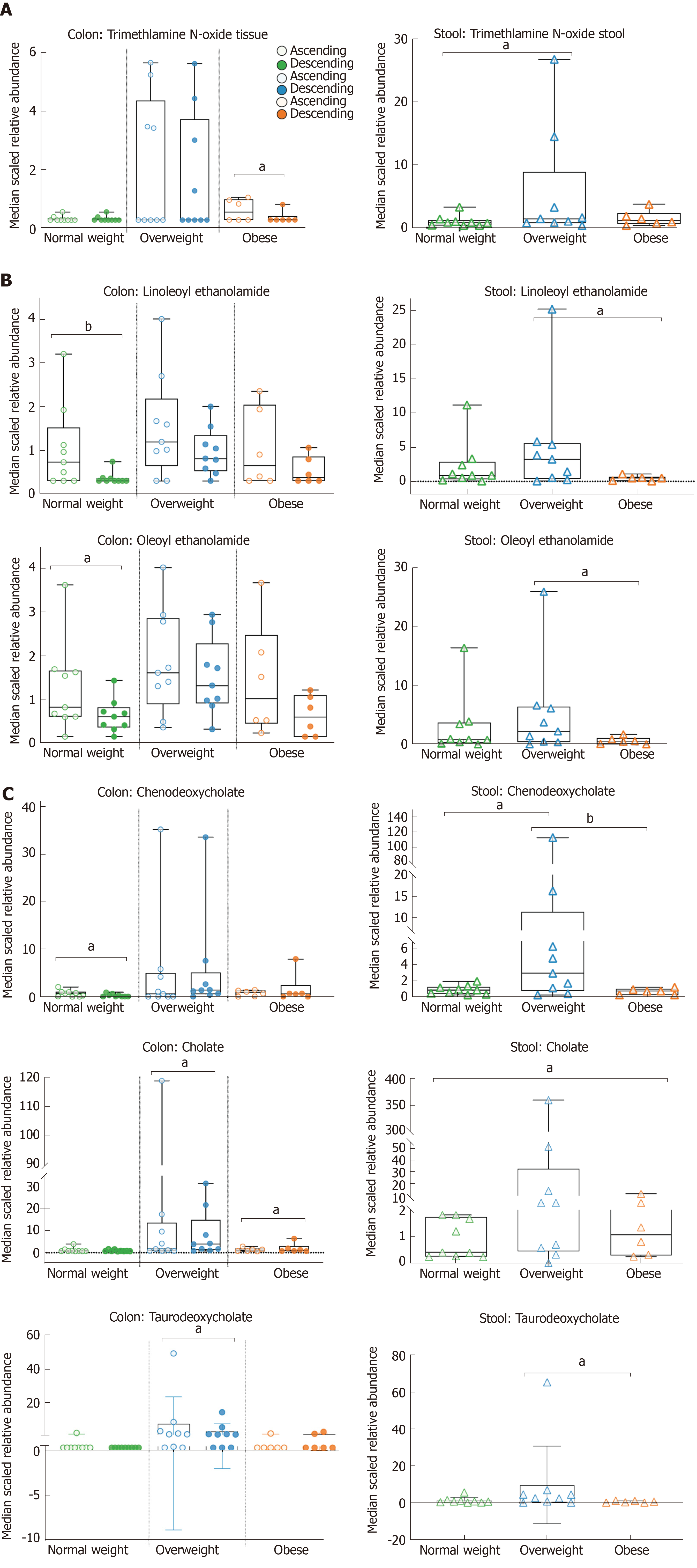
Random forest analysis of colon tissue comparing normal weight vs overweight/obese yielded 30 metabolites with a predictive accuracy of 56% for the overweight/obese phenotype (Supplemental Figure 2A). BMI associated colon metabolites were bile acids and cofactors/vitamins (e.g., biliverdin, alpha-tocopherol, and pyridoxate). The predictive accuracy for metabolites in ascending versus descending colon was 73%, and 24 of the top 30 metabolites were classified as lipids (Supplemental Figure 2B). Table 2 further shows the 12 metabolites with statistically significant fold difference identified in right/left colon tissue and in stool by BMI comparisons. Figure 2 shows lipids that are significantly different between weight groups for colon tissue and stool (P < 0.05). The phospholipid, trimethylamine N-oxide (TMAO), was 2.80-fold difference from ascending in obese adults, and 6.23-fold difference from stool in overweight adults when compared to normal weight adults (Figure 2A). Endocannabinoids, linoleoylethanolamide (2.11-fold) and oleoylethanolamide (1.60-fold) difference from ascending in normal weight adults and decrease in stool of obese adults (Figure 2B). Furthermore, the median scaled relative abundance of primary and secondary bile acids (chenodeoxycholate 2.89-fold and 0.41-fold, cholate 0.55-fold, and taurodeoxycholate 1.49-fold) had significant differences by colon location and stool (Figure 2C). The primary bile acid, chenodeoxycholate is 2.89-fold difference from ascending in normal weight and 0.41-fold difference from descending in overweight adults, while 21.80-fold difference in stool of overweight adults when compared to normal weight. Cholate is 0.55-fold difference from obese descending colon and 61.06-fold difference from overweight stool when compared to normal weight (Figure 2C). The secondary bile acid, taurodeoxycholate, had 1.49-fold difference from ascending colon in overweight adults and 9.32-fold difference in stool of overweight adults compared to normal weight (Table 2 and Figure 2C).
| Sub pathway | Biochemical name | Fold difference colon ascending/descending | Fold difference stool BMI comparisons | ||||||||||
| NW | P value | OW | P value | OB | P value | Ow NW | P value | OB NW | P value | OB OW | P value | ||
| Methione, Cysteine, SAM and taurine metabolism | N-acetylglut-amine | 1.47 | 0.027 | 1.15 | 0.412 | 1.13 | 0.534 | 1.20 | 0.711 | 3.44 | 0.029 | 0.35 | 0.096 |
| Endocannabinoid | Oleoyl ethanolamide | 1.6 | 0.038 | 1.06 | 0.790 | 1.55 | 0.109 | 0.23 | 0.261 | 1.94 | 0.174 | 0.12 | 0.026 |
| Linoleoyl ethanolamide | 2.11 | 0.005 | 1.26 | 0.351 | 1.52 | 0.171 | 0.21 | 0.172 | 2.49 | 0.146 | 0.08 | 0.012 | |
| Phospholipid metabolism | Trimethylamine N-oxide | 0.92 | 0.832 | 1.02 | 0.969 | 2.8 | 0.049 | 1.47 | 0.494 | 6.23 | 0.025 | 0.24 | 0.149 |
| Phosphatidylethanolamine | 1-palmitoyl-2-oleoyl-GPE (16:0/18:1) | 0.77 | 0.145 | 0.66 | 0.029 | 0.7 | 0.113 | 2.63 | 0.044 | 2.57 | 0.244 | 1.02 | 0.322 |
| Lysophospholipid | 2-palmitoyl-GPC (16:0)1 | 1.52 | 0.019 | 1.18 | 0.317 | 0.93 | 0.709 | 0.84 | 0.667 | 2.81 | 0.061 | 0.30 | 0.039 |
| 1-linoleoyl-GPG (18:2)1 | 1.55 | 0.030 | 1.26 | 0.239 | 1.05 | 0.819 | 1.25 | 0.335 | 3.45 | 0.001 | 0.36 | 0.010 | |
| Primary bile acid metabolism | Cholate | 1.12 | 0.612 | 0.84 | 0.447 | 0.55 | 0.041 | 3.13 | 0.512 | 61.06 | 0.019 | 0.05 | 0.114 |
| Chenodeoxycholate | 2.89 | 0.049 | 0.41 | 0.094 | 0.9 | 0.872 | 0.76 | 0.697 | 21.80 | 0.005 | 0.03 | 0.004 | |
| Secondary bile acid metabolism | Taurodeoxycholate | 1.13 | 0.482 | 1.49 | 0.027 | 0.78 | 0.249 | 0.43 | 0.434 | 9.32 | 0.080 | 0.05 | 0.024 |
| Pyrimidine metabolism, cytidine containing | Cytidine | 1.93 | 0.036 | 1.46 | 0.216 | 1.12 | 0.748 | 1.25 | 0.469 | 2.06 | 0.047 | 0.61 | 0.249 |
| Sub pathway | Biochemical name | Fold difference ascending/descending | |||||
| NW | P value | OW | P value | OB | P value | ||
| Long chain fatty acids | Palmitate (16:0) | 1.3 | 0.02 | 1.0 | 0.75 | 0.7 | 0.04 |
| 5 | 1 | 4 | 4 | 3 | 4 | ||
| Margarate (17:0) | 1.2 | 0.13 | 0.8 | 0.33 | 0.7 | 0.03 | |
| 1 | 1 | 8 | 1 | 7 | |||
| Stearate (18:0) | 1.3 | 0.01 | 0.9 | 0.61 | 0.6 | 0.01 | |
| 9 | 3 | 4 | 5 | 8 | 6 | ||
| Oleate/vaccenate (18:1) | 1.5 | 0.04 | 1.2 | 0.36 | 0.9 | 0.80 | |
| 8 | 5 | 4 | 0 | ||||
| Nonadecanoate (19:0) | 1.1 | 0.29 | 0.8 | 0.26 | 0.6 | 0.01 | |
| 2 | 8 | 5 | 9 | 1 | |||
| Arachidate (20:0) | 1.3 | 0.02 | 0.9 | 0.56 | 0.6 | 0.02 | |
| 5 | 5 | 3 | 4 | 9 | 5 | ||
| Eicosenoate (20:1) | 1.4 | 0.04 | 1.1 | 0.56 | 1.0 | 0.88 | |
| 8 | 1 | 1 | 2 | 3 | 3 | ||
| Erucate (22:1n9) | 1.5 | 0.03 | 1.0 | 0.81 | 1.1 | 0.47 | |
| 2 | 1 | 4 | 1 | 8 | 2 | ||
| Polyunsaturated fatty acid | Dihomo-linolenate (20:3n3 or n6) | 1.5 | 0.04 | 1.2 | 0.28 | 1.1 | 0.56 |
| 9 | 1 | 6 | 5 | 6 | 7 | ||
| Dihomo-linoleate (20:2n6) | 1.6 | 0.01 | 1.1 | 0.41 | 1.2 | 0.46 | |
| 8 | 7 | 8 | 7 | 3 | |||
| Fatty acid, dicarboxylate | Dodecadienoate (12:2)1 | 0.9 | 0.64 | 0.8 | 0.26 | 0.7 | 0.03 |
| 5 | 5 | 7 | 1 | 1 | 2 | ||
| Ketone bodies | 3-hydroxybutyrate (BHBA) | 0.4 | 0.00 | 0.9 | 0.90 | 1.0 | 0.82 |
| 8 | 5 | 7 | 4 | 7 | 5 | ||
| Fatty acid, metabolism (Acyl choline) | Linoleoylcholine1 | 1.2 | 0.34 | 1.1 | 0.62 | 2.0 | 0.02 |
| 6 | 4 | 3 | 6 | 1 | |||
| Fatty acid, monohydroxy | 2-hydroxystearate | 1.3 | 0.04 | 0.9 | 0.67 | 0.7 | 0.13 |
| 2 | 5 | 7 | 9 | 5 | |||
| Eicosanoid | 15-HETE | 1.6 | 0.04 | 1.1 | 0.51 | 1.9 | 0.03 |
| 9 | 5 | 8 | 5 | 3 | 9 | ||
| Endocannabinoid | Oleoyl ethanolamide | 1.6 | 0.03 | 2.2 | 0.79 | 1.5 | 0.10 |
| 8 | 3 | 5 | 9 | ||||
| Palmitoyl ethanolamide | 1.3 | 0.02 | 1.0 | 0.56 | 1.1 | 0.42 | |
| 7 | 4 | 8 | 1 | 4 | 1 | ||
| Linoleoyl ethanolamide | 2.1 | 0.00 | 1.2 | 0.35 | 1.5 | 0.17 | |
| 1 | 5 | 6 | 1 | 2 | 1 | ||
| Phosphatidylethanolamine | 1-stearoyl-2-oleoyl-GPE (18:0/18:1) | 0.8 | 0.28 | 0.6 | 0.01 | 0.7 | 0.15 |
| 1 | 4 | 6 | 6 | ||||
| 1-stearoyl-2-linoleoyl-GPE (18:0/18:2)1 | 0.9 | 0.87 | 0.6 | 0.04 | 0.6 | 0.09 | |
| 7 | 5 | 5 | 3 | 5 | 7 | ||
| 1,2-dioleoyl-GPE (18:1/18:1) | 0.6 | 0.07 | 0.6 | 0.03 | 0.7 | 0.19 | |
| 9 | 5 | 8 | 3 | ||||
| 1-oleoyl-2-docosahexaenoyl-GPE (18:1/22:6)1 | 0.6 | 0.01 | 0.7 | 0.08 | 0.4 | 0.00 | |
| 2 | 3 | 1 | 3 | 1 | |||
| Lysophospholipid | 1-palmitoleoyl-GPC (16:1)1 | 1.5 | 0.03 | 1.3 | 0.20 | 1.2 | 0.33 |
| 9 | 6 | 1 | 2 | 8 | 3 | ||
| 1-linoleoyl-GPC (18:2) | 1.6 | 0.00 | 1.5 | 0.01 | 1.4 | 0.08 | |
| 8 | 7 | 1 | 3 | 5 | |||
| 1-arachidonoyl-GPC (20:4n6)1 | 1.74 | 0.035 | 1.55 | 0.092 | 1.39 | 0.285 | |
| 1-linoleoyl-GPS (18:2)1 | 1.8 | 0.00 | 1.4 | 0.07 | 1.2 | 0.31 | |
| 9 | 2 | 1 | 7 | 6 | 4 | ||
| 1-palmitoyl-GPG (16:0)1 | 2.1 | 0.00 | 1.3 | 0.18 | 0.8 | 0.51 | |
| 1 | 5 | 8 | 3 | 3 | 5 | ||
| 1-stearoyl-GPG (18:0) | 2.1 | 0.00 | 1.3 | 0.16 | 0.8 | 0.66 | |
| 6 | 2 | 6 | 9 | 5 | |||
| 1-oleoyl-GPG (18:1)1 | 1.6 | 0.01 | 1.0 | 0.66 | 1.1 | 0.47 | |
| 7 | 8 | 6 | 6 | ||||
| 1-palmitoyl-GPI (16:0) | 1.5 | 0.04 | 1.1 | 0.55 | 0.7 | 0.31 | |
| 8 | 1 | 3 | 4 | 7 | 8 | ||
| 1-stearoyl-GPI (18:0) | 1.4 | 0.03 | 1.0 | 0.66 | 0.8 | 0.50 | |
| 9 | 4 | 8 | 3 | 7 | 7 | ||
| 1-oleoyl-GPI (18:1) | 1.6 | 0.02 | 1.2 | 0.24 | 0.9 | 0.81 | |
| 4 | 7 | 8 | 7 | 4 | 8 | ||
| 1-linoleoyl-GPI (18:2)1 | 1.4 | 0.01 | 1.1 | 0.48 | 1.2 | 0.22 | |
| 8 | 7 | 1 | 8 | 6 | 5 | ||
| Plasmalogen | 1-(1-enyl-palmitoyl)-2-oleoyl-GPE (P-16:0/18:1)1 | 0.7 | 0.08 | 0.6 | 0.02 | 0.6 | 0.13 |
| 1 | 9 | 2 | 2 | 9 | 8 | ||
| 1-(1-enyl-palmitoyl)-2-oleoyl-GPC (P-16:0/18:1)1 | 0.7 | 0.10 | 0.6 | 0.01 | 0.7 | 0.16 | |
| 4 | 5 | 3 | 9 | 3 | |||
| 1-(1-enyl-stearoyl)-2-oleoyl-GPE (P-18:0/18:1) | 0.7 | 0.23 | 0.6 | 0.03 | 0.7 | 0.31 | |
| 9 | 4 | 4 | 3 | 8 | 1 | ||
| Monoacylglycerol | 1-palmitoylglycerol (16:0) | 1.3 | 0.25 | 2.6 | 0.00 | 1.2 | 0.53 |
| 5 | 3 | 1 | 1 | 2 | 7 | ||
| 1-palmitoleoylglycerol (16:1)1 | 1.6 | 0.09 | 2.2 | 0.00 | 1.1 | 0.62 | |
| 2 | 3 | 7 | 8 | 3 | |||
| 1-oleoylglycerol (18:1) | 1.6 | 0.20 | 2.2 | 0.03 | 0.7 | 0.50 | |
| 5 | 5 | 6 | 4 | 7 | |||
| 1-linolenoylglycerol (18:3) | 1.5 | 0.07 | 2.0 | 0.00 | 1.0 | 0.82 | |
| 4 | 4 | 7 | 5 | 7 | 3 | ||
| 1-dihomo-linolenylglycerol (20:3) | 1.5 | 0.25 | 2.2 | 0.04 | 0.7 | 0.57 | |
| 5 | 1 | 6 | 7 | 4 | |||
| 1-arachidonylglycerol (20:4) | 1.5 | 0.21 | 2.3 | 0.01 | 1.1 | 0.78 | |
| 1 | 8 | 4 | 7 | 2 | 3 | ||
| 2-palmitoleoylglycerol (16:1)1 | 1.3 | 0.25 | 1.7 | 0.03 | 0.9 | 0.89 | |
| 4 | 7 | 7 | 4 | 6 | 8 | ||
| 2-arachidonoylglycerol (20:4) | 1.3 | 0.25 | 1.6 | 0.03 | 0.9 | 0.75 | |
| 1 | 3 | 8 | 5 | 1 | 2 | ||
| Diacylglycerol | Linoleoyl-linolenoyl-glycerol (18:2/18:3) (2)1 | 1.3 | 0.51 | 1.6 | 0.29 | 3.3 | 0.03 |
| 4 | 7 | 1 | 6 | 2 | 9 | ||
| Ceramides | N-palmitoyl-sphingosine (d18:1/16:0) | 0.9 | 0.73 | 0.6 | 0.00 | 0.7 | 0.04 |
| 6 | 1 | 9 | 8 | 3 | 9 | ||
| N-stearoyl-sphingosine (d18:1/18:0)1 | 1.0 | 0.57 | 0.7 | 0.03 | 0.8 | 0.33 | |
| 9 | 8 | 1 | 3 | 9 | |||
| N-palmitoyl-heptadecasphingosine (d17:1/16:0)1 | 0.9 | 0.95 | 0.7 | 0.04 | 0.8 | 0.20 | |
| 9 | 7 | 4 | 2 | 0 | |||
| Hexosylceramides (HCER) | Glycosyl-N-palmitoyl-sphingosine (d18:1/16:0) | 0.7 | 0.04 | 0.6 | 0.00 | 0.6 | 0.02 |
| 4 | 8 | 2 | 3 | 7 | 9 | ||
| Primary bile acid | Glycochenodeoxycholate | 0.9 | 0.75 | 1.8 | 0.00 | 0.8 | 0.54 |
| 8 | 6 | 3 | 7 | 0 | |||
| Taurochenodeoxycholate | 1.0 | 0.98 | 1.6 | 0.45 | 0.8 | 1 | |
| 4 | 7 | 2 | 6 | 6 | |||
| Secondary bile acid | Glycolithocholate sulfate | 0.9 | 0.70 | 1.4 | 0.01 | 1 | 1 |
| 5 | 3 | 2 | |||||
| Deoxycholate | 0.9 | 0.79 | 0.6 | 0.12 | 0.7 | 0.00 | |
| 7 | 8 | 3 | 3 | 9 | |||
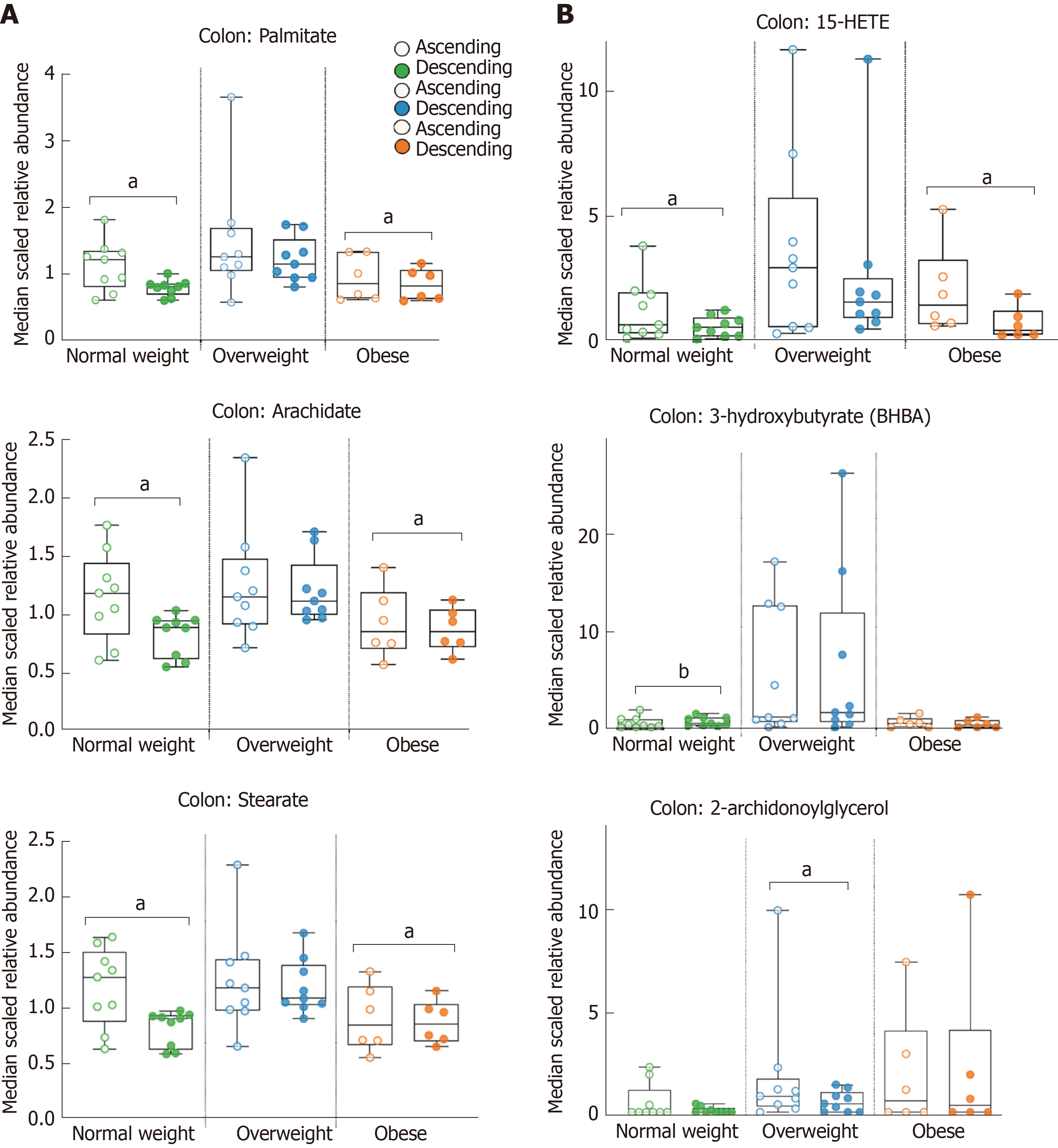
We next utilized metabolomics to distinguish ascending and descending colon tissue. There were 87 metabolites with statistically significant fold differences between ascending vs descending colon by BMI. These included, 46 metabolites in normal weight, 37 metabolites in overweight, and 28 metabolites in obese. Notably, the number of metabolites distinguishing ascending and descending colon decreased as BMI increased. We found that 62% of the metabolites distinguishing ascending and descending colon were lipids. Table 3 shows 54 colon tissue lipids with statistically significant fold difference between ascending and descending colon. In normal weight adults there were 29 colon lipids, 24 of which were fatty acids and lysophospholipid (1.48-fold – 2.16-fold difference) from ascending, and 5 that were from descending colon. Overweight adults had 24 significantly different colon lipids; 11 metabolites from ascending, (8 derived from monoacylglycerols 1.67-fold – 2.61-fold), and 13 metabolites with higher abundance from descending tissue (Table 3). Obese adults had the fewest significant differences in colon lipids between ascending and descending tissue (15 identified metabolites). Table 3 shows 4 metabolites increased in ascending tissue and 11 increased in descending tissue that were primarily fatty acids. Figure 3 shows median scaled relative abundance of right and left colon tissue lipids, including those that are food derived long chain fatty acids and microbiome-products. The long chain fatty acids; palmitate 1.35-fold, arachidate 1.35-fold, and stearate 1.39-fold from ascending tissue in normal weight, and palmitate 0.73-fold, arachidate 0.69-fold, and stearate 0.68-fold from descending tissue in obese adults (Figure 3A). Figure 3B shows microbiome-influenced metabolites that show significant fold difference in colon; 15-HETE from ascending tissue is 1.69-fold in normal weight and 1.93-fold in obese while, 3-hydroxybutyrate is 0.48-fold from descending in normal weight, and 2-archidonoylglycerol is 1.68-fold from ascending tissue in overweight adults (Figure 3B). This study suggests as BMI increases lipid diversity decrease in the colon and primarily in the ascending colon.
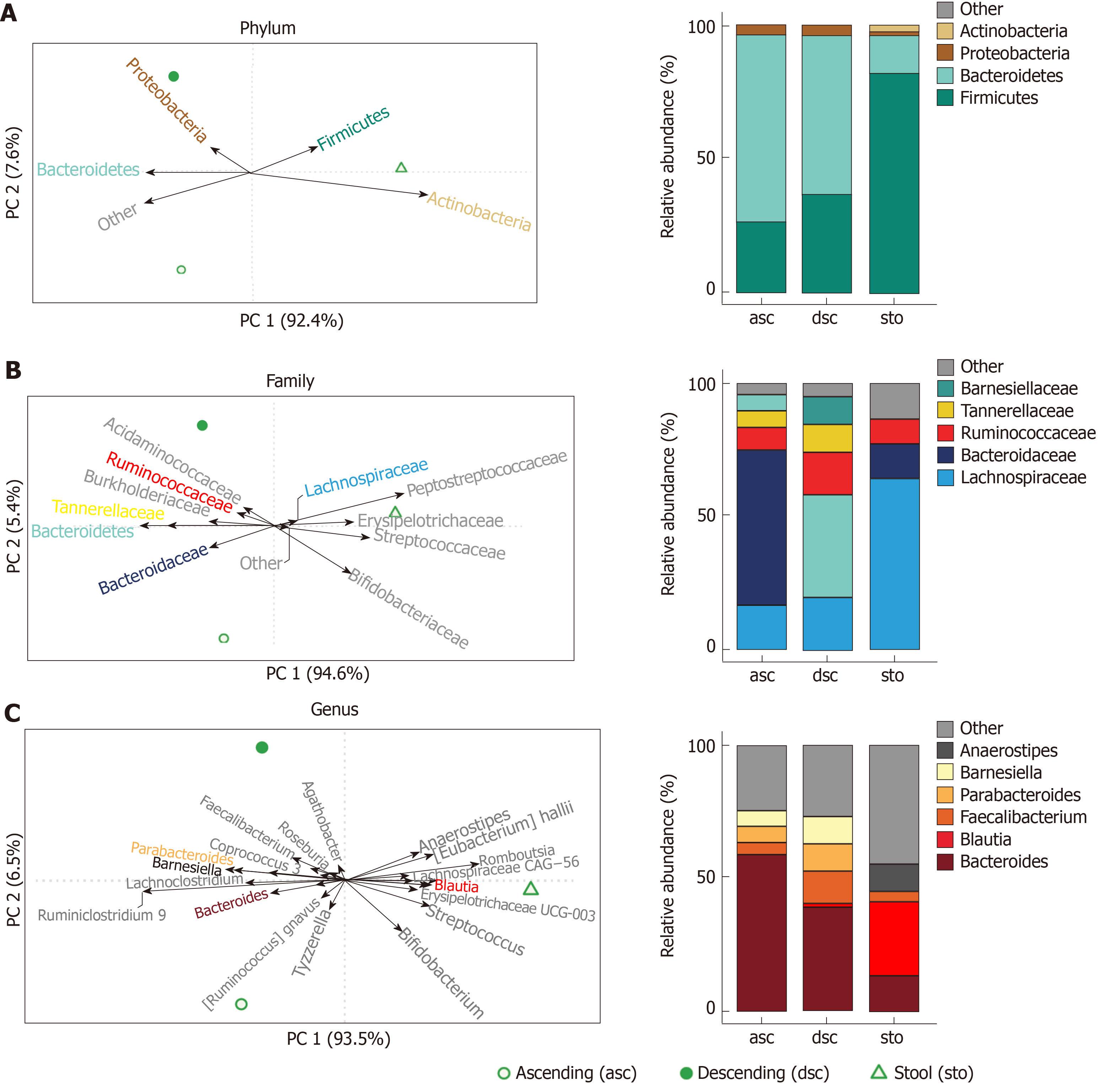
To explore associations of taxonomic groups with ascending colon, descending colon, and stool in a healthy weight adult female, we constructed a compositional PCA biplot from centred log-ratio transformed relative abundances. We observed marked separation across all three sample types at the phylum-, family-, and genus-levels. (Figure 4, Supplementary Table 4). Differences between ascending and descending colon were driven by increased abundance for several taxa in the Firmicutes phylum, including Anaerostipes, Blautia, Dorea, and Fusicatenibacter (all members of the Lachnospiraceae family), as well as Streptococcus and Romboutsia (members of the Streptococcaceae and Peptostreptococcaceae families, respectively). Stool samples were also differentiated by the genus Bifidobacterium (a member of the phylum Actinobacteria). Comparisons of the composition between colon samples (ascending versus descending) indicated enrichments for Bacteroides, Ruminiclostridium 9, Ruminococcus gnavus, and Tyzzerella in the ascending colon, while the descending colon harbored more Barnesiella, Faecalibacterium, Parabacteroides, Parasutterella, and Roseburia (Figure 4C and Supplemental Table 4).
This study demonstrated colon tissue metabolite profile differences between normal weight, overweight and obese adults, and metabolic distinctions between ascending and descending colon within each of the BMI groups. A healthy human colon tissue metabolome had not previously been established across multiple metabolic pathways and revealed 504 known metabolites in both ascending and descending colon locations. Metabolomics has been widely employed for understanding changes that may result from colon metabolism, but the actual metabolite measurements for association with gut health have been from plasma[26], urine[27], stool[7,9], and from cancerous tissue [28] or other digestive disease conditions[29].
Our findings support that a healthy normal weight colon tissue metabolome involves complex lipid metabolism and that differences in lipid metabolite abundance between the right and left colon is associated with regulation of body weight. Differences were identified for right and left colon metabolites from the endocannabinoid pathway that may signify control over energy metabolism, which regulates appetite, lipolysis, and energy expenditure. The endocannabinoid pathway is implicated in both homeostatic and hedonic food intakes that result in increased hunger[30]. Specific endocannabinoids, such as the monounsaturated oleoylethanolamide, saturated palmitoylethanolamide and polyunsaturated linoleoylethanolamide showed higher relative abundance in normal weight adults from ascending colon compared to descending colon, and relative higher abundance from stool in overweight adult compared to normal weight. These lipids are also important for regulating metabolism in immune and neuronal cells[31]. Oleoylethanolamide levels in the mucosal layer of the proximal small intestine was shown to increase with nutrient availability and may be another factor in the regulation of satiety[32]. Palmitoylethanolamide has supportive roles for reducing inflammation and eliciting neuroprotective effects[33], while linoleoylethanolamide has reported anti-inflammatory functions[34].
Bile acids are important signaling molecules which contribute to regulation of whole-body glucose, lipid metabolism, and body weight[35]. Primary and secondary bile acids were also identified for distinctions in abundance by colon location and from stool between overweight/obese and normal weight adults. Primary bile acids produced in the liver and increased bile acids in colon tissue may indicate altered reabsorption of bile acid by the liver and that results in subsequent alteration to metabolism by intestinal microbiota[36]. Primary bile acids, cholic acid and chenodeoxycholic acid, are derived from cholesterol by an enzymatic reaction occurring mainly in the liver[37]. Chenodeoxycholate has shown to increase colonic transit and improves bowel function[38]. Dietary cholic acid supplementation in rats caused a significant increase in colon tumors[39]. Interestingly, this study showed that primary bile acids; glycocholate, glycochenodeoxycholate, and taurocheno-deoxycholate had 1.26-fold – 1.86-fold difference in overweight colon and merit attention as a mechanism with alongside other lipid classes to increase cancer risk in people. Concentrations of bile salts was shown to be higher in the proximal colon and bile-acid profiles were hypothesized to increase the risk of proximal cancer[40].
Population- based studies have shown that individuals who consume high-fat and high-beef foods display elevated levels of fecal secondary bile acids, as do patients diagnosed with colonic carcinomas[3,41]. Secondary bile acid, taurodeoxycholate are generated from primary bile acids and were shown to be increased in obese children plasma with insulin resistance when compared with their non-insulin resistant counterparts, unveiling the influence of the gut microbiota on the host metabolism[42]. Glycochenodeoxycholate a secondary bile acid produced by microbial flora in the large intestine was associated with colorectal cancer in women[43], and high levels of deoxycholate in blood, bile feces, and mucosa were increased in colorectal cancer[7,37]. This study showed elevated glycochenodeoxycholate from descending colon in overweight adults, and elevated deoxycholate from descending colon in obese adults. Impaired bile acid signaling and dysbiosis may contribute to type 2 diabetes and other metabolic disease associated with obesity and colorectal cancer risk[44]. This study had limitations in the total sample size for each BMI group and did not control for different dietary intake patterns. The lack of gender balance in each BMI subgroup was also a potential source of bias for sex-based differences that may exist in colon tissue metabolite profiles. Future studies that control these variables merit attention because the colon tissue metabolite signatures that emerged herein did demonstrate metabolic relevance to the high risk of overweight and obesity in the progression of proximal and distal colon cancers.
Colonic TMAO abundance in obesity was a major finding from this study with respect to risk for cancer and supports a role for phospholipids from choline metabolism. TMAO was identified herein for increased abundance in ascending colon of obese adults and in stool of overweight adults. Deng et al[26] showed increased plasma levels of TMAO in patients with right sided colon cancer when compared to left sided colon cancer patients. High urine concentration of TMAO also directly correlated to the consumption of a high meat containing diet[45] and higher total milk and dairy consumption in plasma[46]. The increased levels in serum and urine were also shown to be associated with predisposition to impaired glucose homeostasis in high fat diet-fed mice[47]. Links between colorectal cancer and TMAO was detected in a genome-wide systems analysis[48] and in the development of colorectal cancer[26]. Our findings also revealed microbiome-influenced metabolites in the colon tissue that were not in the stool, such as the ketone body, 3-hydroxybutyrate, an eicosanoid; 15-Hydroxyeicostetraenoic acid (15-HETE), and the monoacylglycerol; 2-arachidonoylglycerol. Ketone bodies are strongly affected by obesity-related metabolic disorders and are utilized in the body as an energy source[49]. In visceral adipose tissue from obese subjects, 15-HETE was higher than in healthy subjects[50]. These aforementioned metabolic changes support the differences in microbiota between stool and colon and between colon locations (ascending vs descending). Right and left colon microbiota analysis for differences in healthy adults has been limited[19]. We observed an increased abundance of Bacteroides in the ascending colon and Proteobacteria in the descending colon that were consistent with Flynn et al[19]. Given that stool did not recapitulate the composition of the colonic mucosa-associated microbiota and metabolites, additional investigations with larger cohorts of each BMI group is warranted that will assess impacts of intervention strategies to reduce disease risk.
Fatty acids were also statistically supported in abundance by right and left location, and alongside BMI. Food-derived long chain fatty acids are found in dairy fat, coconut oil, palm kernel oil, peanut oil and vegetable oils. Monounsaturated long-chain fats such as oleic acid, and palmitoleic acid are found in animal fats, olive, canola and safflower oil. Oleic acid enhances insulin action and inhibits glucose production[51], but also demonstrates cardiovascular benefits when it replaces heart-damaging saturated fat[52]. Palmitic acid is the first fatty acid produced during fatty acid synthesis, and is the precursor to longer fatty acids, while excess carbohydrates in the body may also be converted to palmitic acid. This analysis revealed major differences in long chain fatty acids as relevant to normal weight ascending colon and were also significantly different in the opposite direction of obese adults, namely increased in descending colon. Polyunsaturated long chain fats include linoleic acid, alpha-linolenic acid (seeds and nuts), arachidonic acid (meat, eggs, and algae) and eicosapentaenoic acid (fish oil)[53]. Margarate, also known as heptadecanoic acid is a biomarker of long-term milk fat intake[54], and was elevated in the obese adults for descending colon. Stearate fed to mice showed 70% reduction of visceral fat[55], and reduced metastasis tumor burden in a breast cancer mouse model[56]. Arachidate is necessary for the function of all cells, especially in the nervous system, skeletal muscle and immune systems[57].
In conclusion, our study identified important metabolic differences between the right and left colon tissue of healthy adults and highlighted a wide range of lipids from normal weight, overweight and obese adults. The magnitude and abundance of a metabolite difference between ascending and descending colon tissue has not been previously evaluated and warrants further investigation for screening risk of proximal versus distal colon cancers.
Obesity is a risk factor for colorectal cancer, yet metabolic distinctions between healthy right and left colon tissue, before cancer is diagnosed, remains largely unknown.
Colon cancer of the ascending colon has a poorer prognosis and survival when compared to colon cancer on the descending colon. Stool metabolite composition does not accurately reflect proximal/ascending/right sided colon. Development of healthy colon tissue small molecule signatures for ascending and descending colon will aid in our understanding of how to improve gut metabolism and may help prevent or mitigate colorectal cancer risk.
This study compared right-ascending and left-descending colon tissue metabolomes and sought to identify differences from the stool metabolome in normal weight, overweight, and obese adults.
Global, non-targeted metabolomics was applied to assess right-ascending and left-descending colon tissue collected from healthy adults undergoing screening colonoscopies to test the hypothesis that body mass index (BMI) differentially impacts colon tissue metabolite profiles. The colon tissue and stool metabolome of healthy adults was analyzed for metabolite signatures and metabolic pathway networks implicated in progression and prevention of colorectal cancer.
This is the first report using metabolomics to compare the right-ascending vs left--descending colon tissue of healthy adults. Our findings show that BMI was associated with metabolite profile differences between the ascending and descending colon. Disturbances in multiple metabolic pathways of the right and left colon from being overweight/obese may have important implications for increasing colorectal cancer risk.
There were metabolite profile differences between right-ascending and left-descending colon tissue in healthy adults receiving routine, screening colonoscopies. BMI impacts the number, type and magnitude of metabolite differences between the ascending and descending colon. Colon lipids and other metabolites in obese and overweight adults were distinguished from normal weight participants and associated with gut inflammation, nutrient absorption, and products of microbiota metabolism.
Right and left colon tissue metabolites that differ in relative abundance between normal weight, overweight, obese adults may be sensitive biomarkers for colon cancer risk. Diet and lifestyle influence right and left sided colon tissue metabolite composition that shape inflammation and cancer risk in overweight and obese adults. Development of healthy colon tissue small molecule signatures for ascending and descending colon will aid in our understanding of how to improve gut metabolism and may help prevent or mitigate colorectal cancer risk.
We would like to thank the study participants, Erica Borresen, Renee Oppel and Luke Schwerdtfeger for assisting with recruitment, consent, and sample procurement from participating gastroenterology/endoscopy clinics and transporting samples to Colorado State University. We also thank Dr. Daniel Hampton, Dr. Rebecca Dunphy and Dr. Nicole Kershner for tissue biopsy collections from screening colonoscopies.
Manuscript source: Unsolicited Manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: United States
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Hidaka E, Manenti A, Zheng YW S-Editor: Ma YJ L-Editor: A E-Editor: Ma YJ
| 1. | Zeng H, Lazarova DL. Obesity-related colon cancer: dietary factors and their mechanisms of anticancer action. Clin Exp Pharmacol Physiol. 2012;39:161-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 2. | Tarasiuk A, Mosińska P, Fichna J. The mechanisms linking obesity to colon cancer: An overview. Obes Res Clin Pract. 2018;12:251-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 3. | Lee GH, Malietzis G, Askari A, Bernardo D, Al-Hassi HO, Clark SK. Is right-sided colon cancer different to left-sided colorectal cancer? - a systematic review. Eur J Surg Oncol. 2015;41:300-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 314] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 4. | Giovannucci E, Colditz GA, Stampfer MJ, Willett WC. Physical activity, obesity, and risk of colorectal adenoma in women (United States). Cancer Causes Control. 1996;7:253-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 191] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 5. | Song M, Hu FB, Spiegelman D, Chan AT, Wu K, Ogino S, Fuchs CS, Willett WC, Giovannucci EL. Adulthood Weight Change and Risk of Colorectal Cancer in the Nurses' Health Study and Health Professionals Follow-up Study. Cancer Prev Res (Phila). 2015;8:620-627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 6. | Moore LL, Bradlee ML, Singer MR, Splansky GL, Proctor MH, Ellison RC, Kreger BE. BMI and waist circumference as predictors of lifetime colon cancer risk in Framingham Study adults. Int J Obes Relat Metab Disord. 2004;28:559-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 210] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 7. | Brown DG, Rao S, Weir TL, O'Malia J, Bazan M, Brown RJ, Ryan EP. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016;4:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 191] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 8. | Hullar MA, Fu BC. Diet, the gut microbiome, and epigenetics. Cancer J. 2014;20:170-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 122] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 9. | Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013;8:e70803. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 414] [Cited by in RCA: 494] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 10. | Wang CB, Shahjehan F, Merchea A, Li Z, Bekaii-Saab TS, Grothey A, Colibaseanu DT, Kasi PM. Impact of Tumor Location and Variables Associated With Overall Survival in Patients With Colorectal Cancer: A Mayo Clinic Colon and Rectal Cancer Registry Study. Front Oncol. 2019;9:76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 11. | Meguid RA, Slidell MB, Wolfgang CL, Chang DC, Ahuja N. Is there a difference in survival between right- versus left-sided colon cancers? Ann Surg Oncol. 2008;15:2388-2394. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 370] [Cited by in RCA: 387] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 12. | Lim DR, Kuk JK, Kim T, Shin EJ. Comparison of oncological outcomes of right-sided colon cancer versus left-sided colon cancer after curative resection: Which side is better outcome? Medicine (Baltimore). 2017;96:e8241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 82] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 13. | Gonzalez EC, Roetzheim RG, Ferrante JM, Campbell R. Predictors of proximal vs. distal colorectal cancers. Dis Colon Rectum. 2001;44:251-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 97] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 14. | Bernstein AM, Song M, Zhang X, Pan A, Wang M, Fuchs CS, Le N, Chan AT, Willett WC, Ogino S, Giovannucci EL, Wu K. Processed and Unprocessed Red Meat and Risk of Colorectal Cancer: Analysis by Tumor Location and Modification by Time. PLoS One. 2015;10:e0135959. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 98] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 15. | Kim S, Kim J, Yun EJ, Kim KH. Food metabolomics: from farm to human. Curr Opin Biotechnol. 2016;37:16-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 81] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 16. | Hale VL, Jeraldo P, Mundy M, Yao J, Keeney G, Scott N, Cheek EH, Davidson J, Greene M, Martinez C, Lehman J, Pettry C, Reed E, Lyke K, White BA, Diener C, Resendis-Antonio O, Gransee J, Dutta T, Petterson XM, Boardman L, Larson D, Nelson H, Chia N. Synthesis of multi-omic data and community metabolic models reveals insights into the role of hydrogen sulfide in colon cancer. Methods. 2018;149:59-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 17. | Montrose DC, Zhou XK, Kopelovich L, Yantiss RK, Karoly ED, Subbaramaiah K, Dannenberg AJ. Metabolic profiling, a noninvasive approach for the detection of experimental colorectal neoplasia. Cancer Prev Res (Phila). 2012;5:1358-1367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14:20-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1265] [Cited by in RCA: 1612] [Article Influence: 161.2] [Reference Citation Analysis (0)] |
| 19. | Flynn KJ, Ruffin MT 4th, Turgeon DK, Schloss PD. Spatial Variation of the Native Colon Microbiota in Healthy Adults. Cancer Prev Res (Phila). 2018;11:393-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 20. | Ward JH. Hierarchical Grouping to Optimize an Objective Function. J Am Stat Assoc. 1963;58:236-244. [RCA] [DOI] [Full Text] [Cited by in Crossref: 9993] [Cited by in RCA: 5398] [Article Influence: 87.1] [Reference Citation Analysis (0)] |
| 21. | Breiman L. Random Forests. Mach Learn. 2001;45:5-32. [RCA] [DOI] [Full Text] [Cited by in Crossref: 56052] [Cited by in RCA: 34150] [Article Influence: 2845.8] [Reference Citation Analysis (0)] |
| 22. | Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS 2nd, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12449] [Cited by in RCA: 11613] [Article Influence: 1935.5] [Reference Citation Analysis (0)] |
| 23. | Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581-583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18515] [Cited by in RCA: 17764] [Article Influence: 1973.8] [Reference Citation Analysis (0)] |
| 24. | R-Core-Team. R: A language and environment for statistical computing. 3.5.2 ed. Vienna: R Foundation for Statistical Computing, 2019. . |
| 25. | Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ. Microbiome Datasets Are Compositional: And This Is Not Optional. Front Microbiol. 2017;8:2224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1061] [Cited by in RCA: 1488] [Article Influence: 186.0] [Reference Citation Analysis (0)] |
| 26. | Deng K, Han P, Song W, Wang Z, Zhang F, Xie H, Zhao W, Xu H, Cai Y, Rong Z, Yu X, Cui BB, Li K. Plasma metabolomic profiling distinguishes right-sided from left-sided colon cancer. Clin Chim Acta. 2018;487:357-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 27. | Wang H, Tso V, Wong C, Sadowski D, Fedorak RN. Development and validation of a highly sensitive urine-based test to identify patients with colonic adenomatous polyps. Clin Transl Gastroenterol. 2014;5:e54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 28. | Griffin JL, Shockcor JP. Metabolic profiles of cancer cells. Nat Rev Cancer. 2004;4:551-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 565] [Cited by in RCA: 520] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 29. | Scoville EA, Allaman MM, Brown CT, Motley AK, Horst SN, Williams CS, Koyama T, Zhao Z, Adams DW, Beaulieu DB, Schwartz DA, Wilson KT, Coburn LA. Alterations in Lipid, Amino Acid, and Energy Metabolism Distinguish Crohn's Disease from Ulcerative Colitis and Control Subjects by Serum Metabolomic Profiling. Metabolomics. 2018;14:17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 145] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 30. | Naughton SS, Mathai ML, Hryciw DH, McAinch AJ. Fatty Acid modulation of the endocannabinoid system and the effect on food intake and metabolism. Int J Endocrinol. 2013;2013:361895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 31. | Matias I, Bisogno T, Di Marzo V. Endogenous cannabinoids in the brain and peripheral tissues: regulation of their levels and control of food intake. Int J Obes (Lond). 2006;30 Suppl 1:S7-S12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 103] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 32. | Fu J, Astarita G, Gaetani S, Kim J, Cravatt BF, Mackie K, Piomelli D. Food intake regulates oleoylethanolamide formation and degradation in the proximal small intestine. J Biol Chem. 2007;282:1518-1528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 203] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 33. | Andresen SR, Bing J, Hansen RM, Biering-Sørensen F, Johannesen IL, Hagen EM, Rice AS, Nielsen JF, Bach FW, Finnerup NB. Ultramicronized palmitoylethanolamide in spinal cord injury neuropathic pain: a randomized, double-blind, placebo-controlled trial. Pain. 2016;157:2097-2103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 34. | Ishida T, Nishiumi S, Tanahashi T, Yamasaki A, Yamazaki A, Akashi T, Miki I, Kondo Y, Inoue J, Kawauchi S, Azuma T, Yoshida M, Mizuno S. Linoleoyl ethanolamide reduces lipopolysaccharide-induced inflammation in macrophages and ameliorates 2,4-dinitrofluorobenzene-induced contact dermatitis in mice. Eur J Pharmacol. 2013;699:6-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 35. | Hasenour CM, Ridley DE, Hughey CC, James FD, Donahue EP, Shearer J, Viollet B, Foretz M, Wasserman DH. 5-Aminoimidazole-4-carboxamide-1-β-D-ribofuranoside (AICAR) effect on glucose production, but not energy metabolism, is independent of hepatic AMPK in vivo. J Biol Chem. 2014;289:5950-5959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 36. | Li T, Chiang JY. Bile acids as metabolic regulators. Curr Opin Gastroenterol. 2015;31:159-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 202] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 37. | Ajouz H, Mukherji D, Shamseddine A. Secondary bile acids: an underrecognized cause of colon cancer. World J Surg Oncol. 2014;12:164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 268] [Cited by in RCA: 283] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 38. | Rao AS, Wong BS, Camilleri M, Odunsi-Shiyanbade ST, McKinzie S, Ryks M, Burton D, Carlson P, Lamsam J, Singh R, Zinsmeister AR. Chenodeoxycholate in females with irritable bowel syndrome-constipation: a pharmacodynamic and pharmacogenetic analysis. Gastroenterology. 2010;139:1549-1558, 1558.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 137] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 39. | McSherry CK, Cohen BI, Bokkenheuser VD, Mosbach EH, Winter J, Matoba N, Scholes J. Effects of calcium and bile acid feeding on colon tumors in the rat. Cancer Res. 1989;49:6039-6043. [PubMed] |
| 40. | Gervaz P, Bucher P, Morel P. Two colons-two cancers: paradigm shift and clinical implications. J Surg Oncol. 2004;88:261-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 234] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 41. | Nagengast FM, Grubben MJ, van Munster IP. Role of bile acids in colorectal carcinogenesis. Eur J Cancer. 1995;31A:1067-1070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 240] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 42. | Mastrangelo A, Martos-Moreno GÁ, García A, Barrios V, Rupérez FJ, Chowen JA, Barbas C, Argente J. Insulin resistance in prepubertal obese children correlates with sex-dependent early onset metabolomic alterations. Int J Obes (Lond). 2016;40:1494-1502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 43. | Cross AJ, Moore SC, Boca S, Huang WY, Xiong X, Stolzenberg-Solomon R, Sinha R, Sampson JN. A prospective study of serum metabolites and colorectal cancer risk. Cancer. 2014;120:3049-3057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 44. | Shapiro H, Kolodziejczyk AA, Halstuch D, Elinav E. Bile acids in glucose metabolism in health and disease. J Exp Med. 2018;215:383-396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 328] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 45. | Stella C, Beckwith-Hall B, Cloarec O, Holmes E, Lindon JC, Powell J, van der Ouderaa F, Bingham S, Cross AJ, Nicholson JK. Susceptibility of human metabolic phenotypes to dietary modulation. J Proteome Res. 2006;5:2780-2788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 279] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 46. | Rohrmann S, Linseisen J, Allenspach M, von Eckardstein A, Müller D. Plasma Concentrations of Trimethylamine-N-oxide Are Directly Associated with Dairy Food Consumption and Low-Grade Inflammation in a German Adult Population. J Nutr. 2016;146:283-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 142] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 47. | Gao X, Liu X, Xu J, Xue C, Xue Y, Wang Y. Dietary trimethylamine N-oxide exacerbates impaired glucose tolerance in mice fed a high fat diet. J Biosci Bioeng. 2014;118:476-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 270] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 48. | Xu R, Wang Q, Li L. A genome-wide systems analysis reveals strong link between colorectal cancer and trimethylamine N-oxide (TMAO), a gut microbial metabolite of dietary meat and fat. BMC Genomics. 2015;16 Suppl 7:S4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 138] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 49. | Giesbertz P, Padberg I, Rein D, Ecker J, Höfle AS, Spanier B, Daniel H. Metabolite profiling in plasma and tissues of ob/ob and db/db mice identifies novel markers of obesity and type 2 diabetes. Diabetologia. 2015;58:2133-2143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 117] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 50. | Candi E, Tesauro M, Cardillo C, Lena AM, Schinzari F, Rodia G, Sica G, Gentileschi P, Rovella V, Annicchiarico-Petruzzelli M, Di Daniele N, Melino G. Metabolic profiling of visceral adipose tissue from obese subjects with or without metabolic syndrome. Biochem J. 2018;475:1019-1035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 64] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 51. | Obici S, Feng Z, Morgan K, Stein D, Karkanias G, Rossetti L. Central administration of oleic acid inhibits glucose production and food intake. Diabetes. 2002;51:271-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 493] [Cited by in RCA: 447] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 52. | Voelker R. Oleic Acid Can Make Heart Claim Without Hard Evidence. JAMA. 2019;321:23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 53. | Kingsbury KJ, Paul S, Crossley A, Morgan DM. The fatty acid composition of human depot fat. Biochem J. 1961;78:541-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 134] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 54. | Wolk A, Vessby B, Ljung H, Barrefors P. Evaluation of a biological marker of dairy fat intake. Am J Clin Nutr. 1998;68:291-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 161] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 55. | Shen MC, Zhao X, Siegal GP, Desmond R, Hardy RW. Dietary stearic acid leads to a reduction of visceral adipose tissue in athymic nude mice. PLoS One. 2014;9:e104083. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 56. | Evans LM, Toline EC, Desmond R, Siegal GP, Hashim AI, Hardy RW. Dietary stearate reduces human breast cancer metastasis burden in athymic nude mice. Clin Exp Metastasis. 2009;26:415-424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 57. | Tallima H, El Ridi R. Arachidonic acid: Physiological roles and potential health benefits - A review. J Adv Res. 2018;11:33-41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 398] [Article Influence: 49.8] [Reference Citation Analysis (0)] |