Published online Jun 14, 2020. doi: 10.3748/wjg.v26.i22.3034
Peer-review started: February 11, 2020
First decision: March 26, 2020
Revised: April 10, 2020
Accepted: April 24, 2020
Article in press: April 24, 2020
Published online: June 14, 2020
Processing time: 124 Days and 10.6 Hours
The incidence of inflammatory bowel disease, a chronic intestinal inflammatory disorder that includes Crohn’s disease (CD) and ulcerative colitis, is rising. Circular RNAs are considered valuable diagnostic biomarkers for CD. Current evidence supports the views that epithelial-mesenchymal transition (EMT) plays an important role in CD pathogenesis, and that hsa-miR-130a-3p can inhibit transforming growth factor-β1 (TGF-β1)-induced EMT. Our previous study revealed that hsa_circRNA_102610 was upregulated in CD patients. Moreover, we predicted an interaction between hsa_circRNA_102610 and hsa-miR-130a-3p. Thus, we hypothesized that hsa_circRNA_102610 may play roles in the proliferation and EMT of intestinal epithelial cells by sponging hsa-miR-130a-3p to participate in the pathogenesis of CD.
To explore the mechanism of hsa_circRNA_102610 in the pathogenesis of CD.
The relative expression levels of hsa_circRNA_102610 and hsa-miR-130a-3p in patients were detected by quantitative reverse transcription-polymerase chain reaction. The proliferation of human intestinal epithelial cells (HIECs) and normal-derived colon mucosa cell line 460 (NCM460) cells was detected by cell counting kit-8, 5-ethynyl-2’-deoxyuridine staining and cell cycle assays following overexpression or downregulation of hsa_circRNA_102610. Cell proliferation assays were performed as described above in a rescue experiment with hsa-miR-130a-3p mimics. The interaction of hsa_circRNA_102610 and hsa-miR-130a-3p was verified by fluorescence in situ hybridization and dual luciferase reporter assays. The relative expression levels of CyclinD1, mothers against decapentaplegic homolog 4 (SMAD4), E-cadherin, N-cadherin and Vimentin were detected by western blotting following hsa_circRNA_102610 overexpression, TGF-β1-induced EMT or hsa-miR-130a-3p mimic transfection (in rescue experiments).
Upregulation of hsa_circRNA_102610 was determined to be positively correlated with elevated fecal calprotectin levels in CD (r = 0.359, P = 0.007) by Pearson correlation analysis. Hsa_circRNA_102610 promoted the proliferation of HIECs and NCM460 cells, while hsa-miR-130a-3p reversed the cell proliferation-promoting effects of hsa_circRNA_102610. Fluorescence in situ hybridization and dual luciferase reporter assays showed that hsa_circRNA_102610 directly bound hsa-miR-130a-3p in NCM460 and 293T cells. An inverse correlation between downregulation of hsa-miR-130a-3p and upregulation of hsa_circRNA_102610 in CD patients was observed (r = -0.290, P = 0.024) by Pearson correlation analysis. Moreover, overexpression of hsa_circRNA_102610 promoted SMAD4 and CyclinD1 protein expression validated by western-blotting. Furthermore, over-expression of hsa_circRNA_102610 promoted TGF-β1 induced EMT in HIECs and NCM460 cells via targeting of hsa-miR-130a-3p, with increased expression of Vimentin and N-cadherin and decreased expression of E-cadherin.
Hsa_circRNA_102610 upregulation in CD patients could promote the proliferation and EMT of intestinal epithelial cells via sponging of hsa-miR-130a-3p.
Core tip: The incidence of inflammatory bowel disease, a chronic intestinal inflammatory disorder that includes Crohn’s disease (CD) and ulcerative colitis is rising. Our previous study revealed that hsa_circRNA_102610 is upregulated in CD patients. However, further investigation is required to explore how hsa_circRNA_102610 participates in the pathogenesis of CD. The results of this study indicate that hsa_circRNA_102610 overexpression in CD patients could accelerate the proliferation and epithelial-mesenchymal transition of intestinal epithelial cells via sponging of hsa-miR-130a-3p. Thus, hsa_circRNA_102610 may promote CD progression. Hsa_circRNA_102610 may serve as a potential target for CD therapy and novel drug research. Exogenously delivered hsa-miR-130a-3p could possibly act as a sponge of hsa_circRNA_102610.
- Citation: Yin J, Ye YL, Hu T, Xu LJ, Zhang LP, Ji RN, Li P, Chen Q, Zhu JY, Pang Z. Hsa_circRNA_102610 upregulation in Crohn’s disease promotes transforming growth factor-β1-induced epithelial-mesenchymal transition via sponging of hsa-miR-130a-3p. World J Gastroenterol 2020; 26(22): 3034-3055
- URL: https://www.wjgnet.com/1007-9327/full/v26/i22/3034.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i22.3034
Circular RNAs (circRNAs) are endogenous covalently closed circular biomolecules generated by back-splicing. Due to their unique structure without a 5’cap or 3’poly A tail, circRNAs are more stable than linear RNAs, such as microRNAs (miRNAs) and lncRNAs. They are expressed in eukaryotes with tissue-specific and disease-specific characteristics[1]. Thus, circRNAs are potential biomarkers for disease prediction, diagnosis and prognostic analysis.
To date, circRNAs have been confirmed to participate in various diseases, including colorectal cancer, hepatic carcinoma and rheumatoid arthritis[2-5]. CircRNAs are also considered valuable diagnostic biomarkers for Crohn’s disease (CD)[6-8]. Among the known biological functions of circRNAs, the miRNA-sponging function is one of the most extensively studied. By this mechanism, circRNAs can act as competing endogenous RNAs due to the presence of similar miRNA-binding site sequences on the mRNA targets of the corresponding miRNAs[9,10]. Our previous study demonstrated that hsa_circRNA_102610 was upregulated in CD patients[7]. Furthermore, miRNA response element (MRE) analysis suggested the existence of a potential interaction between hsa_circRNA_102610 and hsa-miR-130a-3p.
Hsa-miR-130a-3p is generally considered a tumor suppressor because it is downregulated in multiple types of cancers[11]. In addition, it participates in various biological processes related to tumorigenesis, such as epithelial mesenchymal transition (EMT), cell viability-related processes, invasion and apoptosis[12-16]. Overexpression of hsa-miR-130a-3p markedly inhibits GC cell EMT and tumorigenesis, by targeting TBL1XR1 to induce E-cadherin expression and reduce N-cadherin, Twist, and MMP2 expression[11]. Moreover, a mothers against dec-apentaplegic homolog 4 (SMAD4)-dependent mechanism was recently discovered to inhibit transforming growth factor-β1 (TGF-β1)-induced EMT via hsa-miR-130a-3p in EC-1 cells, resulting in upregulation of E-cadherin and downregulation of N-cadherin and Vimentin[13].
Current evidence supports the view that EMT plays an important role in CD pathogenesis. Intestinal fibrosis accompanying CD is triggered by multiple factors. EMT induced by TGF-β or IL-13 makes an important contribution to fibrosis by inducing the generation of new mesenchymal cells from the epithelium[17,18]. Moreover, miRNAs have been confirmed to participate in the regulation of the pathologic processes of inflammatory bowel disease (IBD). Downregulation of the miR-200 family (miR-141, miR-200a, miR-200c and miR-429) in the epithelium of fibrotic CD intestinal tissue accompanied by significantly elevated rates of cytokeratin-18 or Vimentin-positive epithelial staining in CD strictures is associated with EMT[19]. Functional studies have demonstrated that miR-200b can inhibit TGF-β1-induced EMT in IECs[20].
With regard to hsa-miR-130a-3p, there are no research reports on its role in CD. Thus, according to the MRE analysis results, we hypothesized that the expression of hsa-miR-130a-3p might be reduced in CD patients and that hsa_circRNA_102610 might participate in the regulation of hsa-miR-130a-3p and its downstream pathway proteins. In this study, a correlation analysis between hsa_circRNA_102610 and hsa-miR-130a-3p expression was carried out in CD patients. Further studies were performed to explore the mechanism by which hsa_circRNA_102610 regulates hsa-miR-130a-3p and its downstream proteins related to EMT and cell proliferation.
Recently diagnosed CD patients and healthy controls were recruited at Suzhou Affiliated Hospital of Nanjing Medical University (Suzhou, Jiangsu Province, China) from 2018 to 2019. The sample collection methods were described in our previous study[7]. Informed consent was obtained from all the participants. Ethical approval was obtained from the Ethics Committee of Nanjing Medical University. The methods of RNA extraction and reverse transcription quantitative polymerase chain reaction (RT-qPCR) were described in our previous study[7]. Primers for RT-qPCR are listed in Table 1.
| Primer | Forward (5’-3’) | Reverse (5’-3’) |
| miR130a-3p | GATGCTCTCAGTGCAATGTTA | TATGGTTGTTCTGCTCTCTGTCTC |
| U6 | ATTGGAACGATACAGAGAAGATT | GGAACGCTTCACGAATTTG |
| hsa_circRNA_102610 | ACAGTGTGCCAAGTTACTCG | AGTGCAAGATAAAGGCCCAA |
| β-actin | GTGGCCGAGGACTTTGATTG | CCTGTAACAACGCATCTCATATT |
A hsa_circRNA_102610 overexpression plasmid was constructed in pLC5-ciR by Geneseed (Guangzhou, Guangdong, China). Small interfering RNA (siRNA) targeting the splice junction region of hsa_circRNA_102610 was synthesized by GenePharma (Shanghai, China).
Hsa-miR-130a-3p mimics and hsa-miR-130a-3p inhibitor were synthesized by GenePharma. The sequences are listed in Table 2.
| RNA | Sequence (5'-3') |
| hsa_circRNA_102610 si-1 | GCUGAUUUUUCAGGGUCAATTUUGACCCUGAAAAAUCAGCTT |
| hsa_circRNA_102610 si-2 | UUCAGGGUCAACUUUGUAGTTCUACAAAGUUGACCCUGAATT |
| hsa-miR-130a-3p mimics | CAGUGCAAUGUUAAAAGGGCAUGCCCUUUUAACAUUGCACUGUU |
| hsa-miR-130a-3p inhibitor | AUGCCCUUUUAACAUUGCACUG |
| hsa_circRNA_102610 probe | FAM-GCACACTACAAAGTTGTTGACCCTGAAAAATCAGCAGAA |
| hsa-miR-130a-3p probe | CY3-ATGCCCTTTTAACATTGCACTG |
Human intestinal epithelial cells (HIECs) and normal-derived colon mucosa cell line 460 (NCM460) cells were cultured in Roswell Park Memorial Institute 1640 Medium or Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Cells at 60%-70% confluence were transfected with the hsa_circRNA_102610 overexpression plasmid hsa_ circRNA_102610-pLC5-ciR, the hsa-miR-130a-3p mimics, the hsa-miR-130a-3p inhibitor, or hsa_circRNA_102610 siRNA fragments with Lipofectamine 3000 (Life Technologies, Carlsbad, CA, United States) according to the manufacturer’s instructions for 48 h.
HIECs and NCM460 cells were seeded in 60 mm cell culture dishes. Cells at 60-70% confluence were transfected according to the above protocol. Subsequently, the cells were fixed with precooled 70% ethanol overnight at -20°C and labeled with propidium iodide (PI, Beyotime, Shanghai, China). The cell cycle distribution was detected by flow cytometry (FACSCalibur, BD, San Jose, CA, United States).
HIECs and NCM460 cells were seeded in 24-well plates at a density of 2 × 104 cells/mL. Cells at 60-70% confluence were transfected according to the above protocol the next day. On the third day, cells were harvested and reseeded in 96-well plates at a density of 2 × 104 cells/mL. A cell counting kit-8 (CCK-8) (Dojindo, Shanghai, China) was used to detect cell viability. A Cell-Light™ 5-ethynyl-2’-deoxyuridine (EdU) Apollo®567 In Vitro Imaging Kit (RiboBio, Guangzhou, Guangdong, China) was used to measure the cellular proliferation rate.
MiRNAs that may bind to hsa_circRNA_102610 were predicted by TargetScan and miRanda. The top 5 miRNAs potentially binding to microRNA response elements in hsa_circRNA_102610 were predicted by Arraystar’s custom miRNA target prediction software (Arraystar, Rockville, MD, United States).
Target mRNAs of miRNAs were predicted by TargetScan7.1 and miRDB_V5.0. Pathway analysis of the upregulated circRNAs in CD was performed with the Database for Annotation, Visualization and Integrated Discovery. A network containing hsa_circRNA_102610, the top 5 potentially binding miRNAs, and their target genes was generated by Cytoscape 3.7.0.
CircRNAs determined to be upregulated in CD patients in our previous study related to EMT were chosen according to their predicted binding miRNAs. A hierarchical clustering analysis of target circRNAs was performed by MeV based on expression values.
The expression level and location of hsa_circRNA_102610 or hsa-miR-130a-3p were detected by fluorescence in situ hybridization (FISH) using FAM- and CY3-labeled specific probes, respectively (GenePharma, Shanghai, China). 4’, 6-diamidino-2-phenylindole was used to stain cell nuclei. Cellular fluorescence images were collected by a confocal laser scanning microscope (Carl Zeiss, Göschwitzer Strasse, Jena, Germany). The sequences of the probes are listed in Table 2.
A fragment containing a total of 273 bp of hsa_circRNA_102610 including the hsa-miR-130a-3p response element was synthesized (GenScript, Nanjing, Jiangsu Province, China) and cloned into the plasmid psiCHECK-2 (Promega, Madison, WI, United States). In addition, a counterpart containing mutations in the hsa-miR-130a-3p response element was constructed. Then, 293T and NCM460 cells were transfected with 500 ng of plasmids with Lipofectamine 3000. The relative luciferase activity was assayed by a Dual-Luciferase®@ Reporter Assay System (Promega, Madison, WI, United States) 48 h after transfection.
HIECs and NCM460 cells were treated with TGF-β1 at a concentration of 10 ng/mL for 48 h. Phase microscopy was used to observe morphological changes. The expression levels of EMT markers such as Vimentin, N-cadherin, and E-cadherin were detected by western blotting.
Western blotting was performed as follows: Briefly, cellular proteins of NCM460 cells and HIECs were extracted with RIPA Lysis Buffer (Beyotime, Shanghai, China) containing 1% phenylmethanesulfonyl fluoride. Proteins (30 μg/well) were separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis gels and transferred to polyvinylidene difluoride membranes by a wet blotting system (Mini-PROTEAN® Tetra, Bio-Rad, Hercules, CA, United States). The membranes were then blocked in 5% nonfat milk at room temperature for 1.5 h and incubated with primary antibodies overnight at 4°C. Primary antibodies against CyclinD1 (CST-2878S, Cell Signaling Technology, Danvers, MA, United States), SMAD4 (CST-38454S, Cell Signaling Technology, Danvers, MA, United States), N-cadherin (CST-13116S, Cell Signaling Technology, Danvers, MA, United States), E-cadherin (CST-3195S, Cell Signaling Technology, Danvers, MA, United States) and Vimentin (CST-5741S, Cell Signaling Technology, Danvers, MA, United States) were used in this study. After washing with TBST three times, the membranes were incubated with HRP-linked secondary antibodies at room temperature for 1.5 h. Tanon High-sig ECL Western Blotting Substrate (Tanon, Shanghai, China) was used to detect the signals with GAPDH as a loading control.
Statistical significance was analyzed by GraphPad Prism 5 (GraphPad, La Jolla, CA, United States). Comparisons between two groups were performed by Student’s t-test. One-way ANOVA test was used to determine differences among groups. Correlations between parameters were assessed by Pearson correlation coefficients. Statistical significance was considered at P < 0.05.
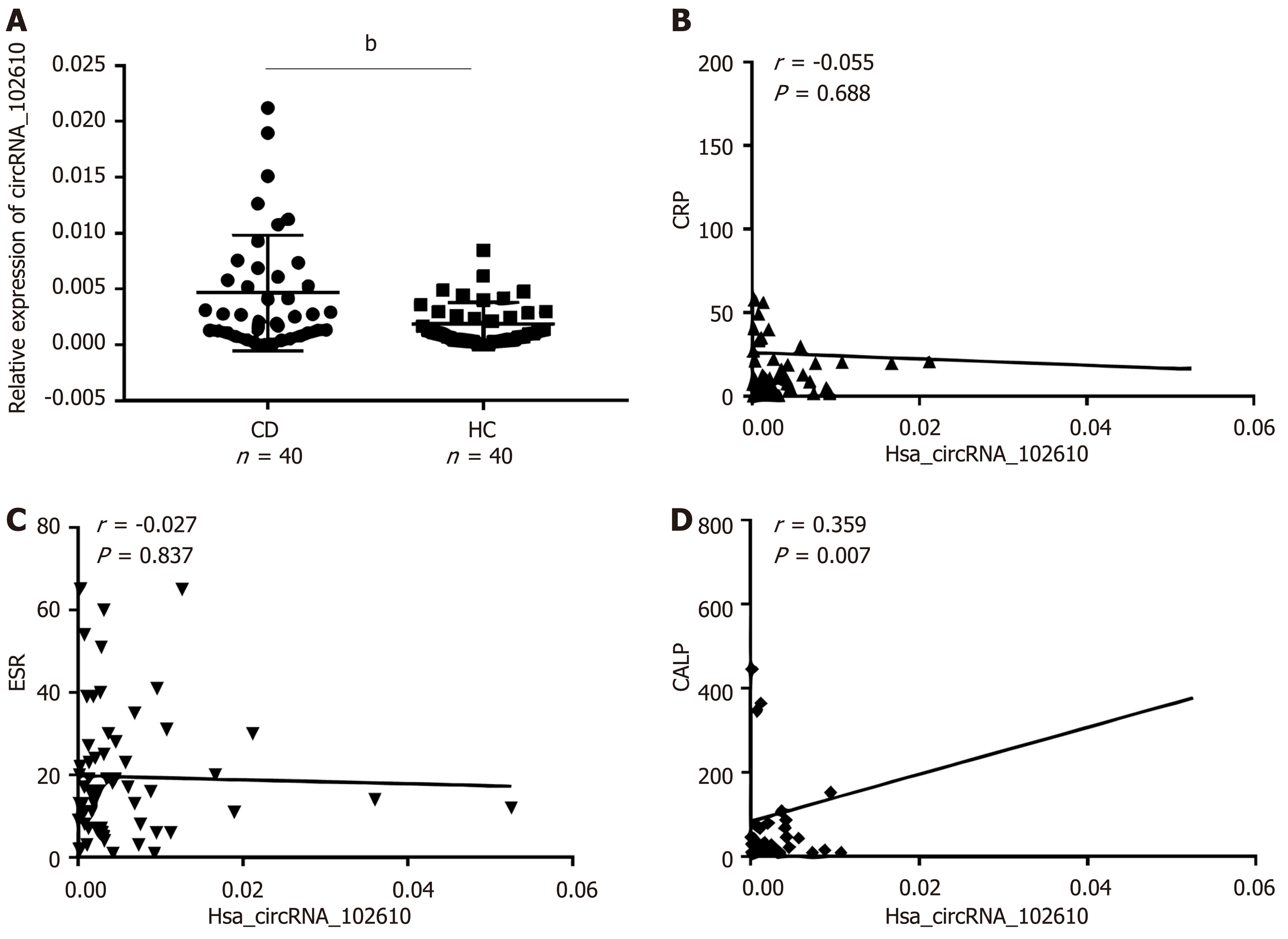
Our previous study showed that hsa_circRNA_102610 is upregulated in CD patients. To investigate whether the upregulation of hsa_circRNA_102610 correlates with the degree of inflammation, the calprotectin (CALP) level, C-reactive protein level and erythrocyte sedimentation rate (ESR) were included in the correlation analysis. The results are shown in Figure 1. There was no correlation between the upregulation of hsa_circRNA_102610 (Figure 1A) and C-reactive protein (Figure 1B) or ESR level (Figure 1C). However, it was found that hsa_circRNA_102610 expression was positively correlated with the CALP level (Figure 1D).
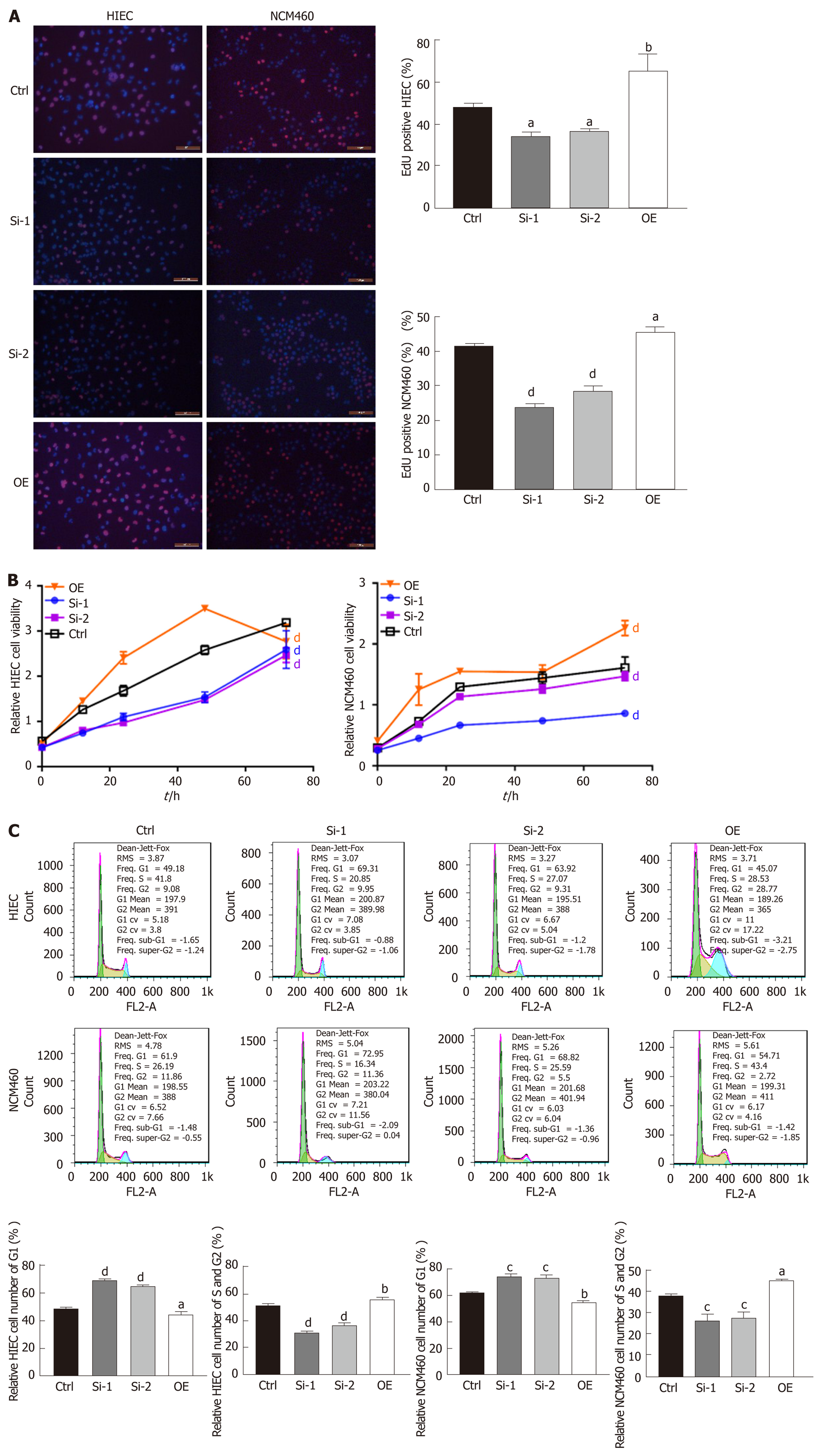
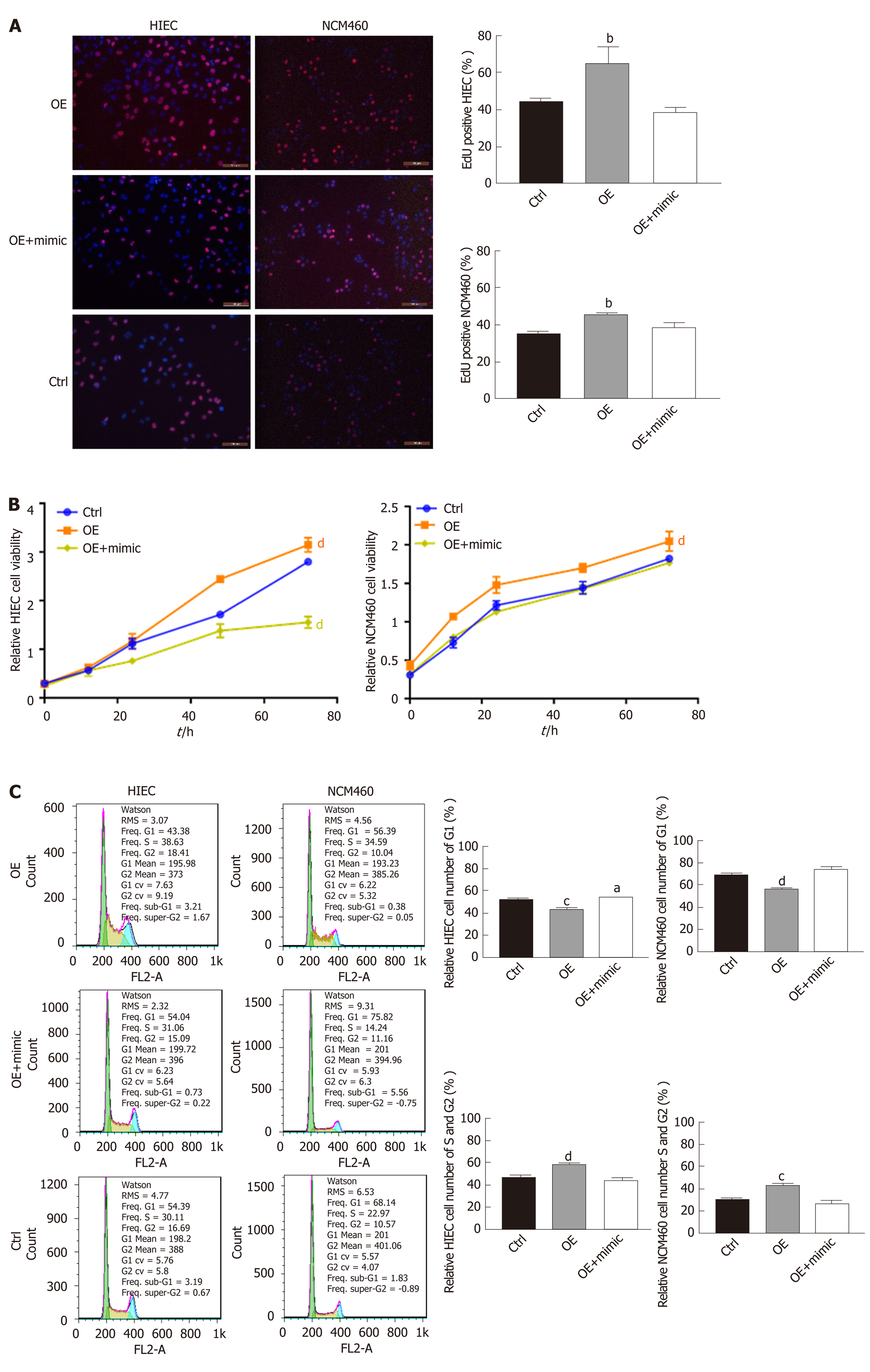
EdU staining suggested that proliferation rates were lower among HIECs and NCM460 cells with hsa_circRNA_102610 downregulation, and higher among cells with hsa_circRNA_102610 overexpression (Figure 2A). In line with the EdU staining results, the CCK-8 assay showed that downregulation of hsa_circRNA_102610 decreased the viability of HIECs and NCM460 cells, while overexpression of hsa_circRNA_102610 promoted viability (Figure 2B). Moreover, cell cycle detection by flow cytometry demonstrated that hsa_circRNA_102610 downregulation caused cell cycle arrest in HIECs and NCM460 cells, while hsa_circRNA_102610 overexpression promoted cell cycle progression. The relative HIEC and NCM460 cell numbers in G1 phase were decreased following hsa_circRNA_102610 overexpression, while the relative numbers of cells in S phase and G2 phase were increased. The opposite results were observed with hsa_circRNA_102610 downregulation (Figure 2C).
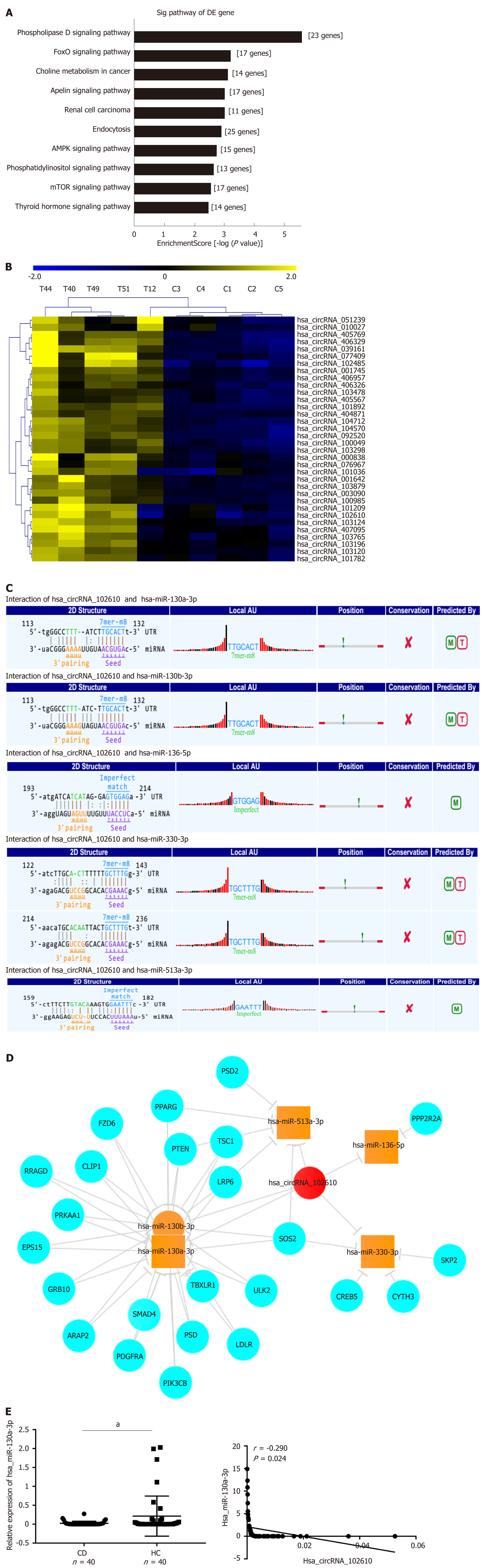
Gene Ontology and pathway analyses were performed to study the upregulated circRNAs (092520, 102610, 004662, and 103124) in CD, which we validated in a recently published article[7]. The results suggested that the AMPK and mTOR signaling pathways participate in CD pathology (Figure 3A). Further hierarchical clustering analysis indicated that hsa_circRNA_102610 (Figure 3B) was among the circRNAs associated with EMT according to MRE analysis. MRE analysis of hsa_circRNA_102610 showed the top 5 possible binding miRNAs, and hsa-miR-130a-3p was the most likely miRNA. The predicted binding sites of hsa_circRNA_102610 and these 5 miRNAs are listed in Figure 3C. Furthermore, a network consisting of hsa_circRNA_102610, the miRNAs and the corresponding proteins was generated by Cytoscape (Figure 3D). SMAD4 was one of the proteins predicted to bind to hsa-miR-130a-3p. It is generally thought that the AMPK and mTOR signaling pathways are related to cell proliferation, and hsa-miR-130a-3p could regulate EMT by controlling SMAD4 expression[13]. Moreover, an inverse correlation between downregulation of hsa-miR-130a-3p and upregulation of hsa_circRNA_102610 in CD patients was observed (Figure 3E). Thus, we infer that hsa_circRNA_102610 may participate in cell proliferation and SMAD4-related EMT by sponging hsa-miR-130a-3p.
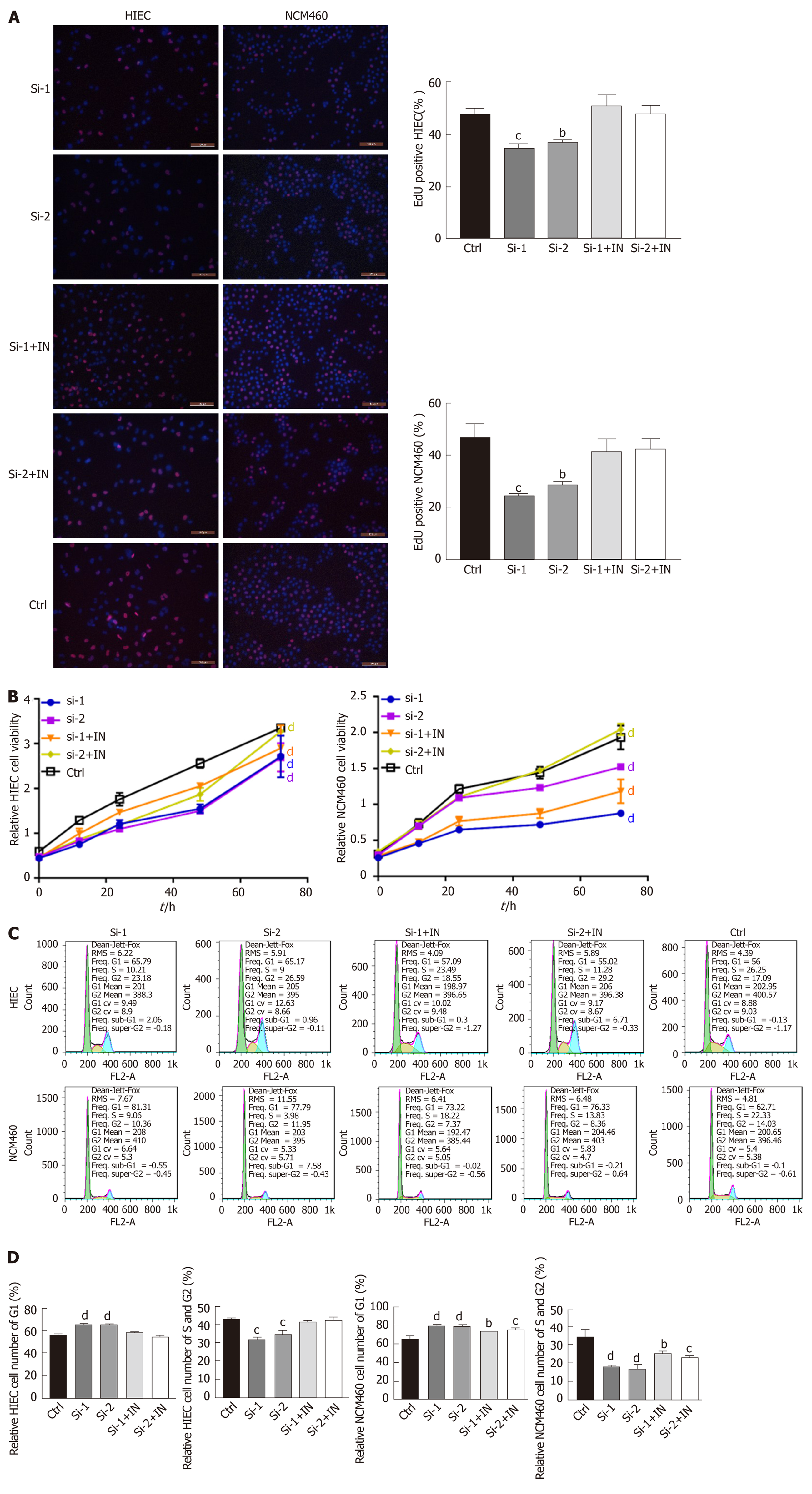
To explore whether hsa-miR-130a-3p can reverse the effects of hsa_circRNA_102610 in intestinal epithelial cells, CCK-8 assays, EdU staining and cell cycle detection were performed. The results showed that the hsa-miR-130a-3p inhibitor could reverse the decrease in cell proliferation caused by hsa_circRNA_102610 downregulation (Figure 4). Conversely, the hsa-miR-130a-3p mimics partially attenuated the increase in NCM460 and HIEC proliferation induced by hsa_circRNA_102610 overexpression (Figure 5).
MRE analysis predicted the existence of binding sites between hsa_circRNA_102610 and hsa-miR-130a-3p. A previous study demonstrated that hsa-miR-130a-3p can reduce cell viability and suppress proliferation, invasion, and TGF-β1-induced EMT[13].
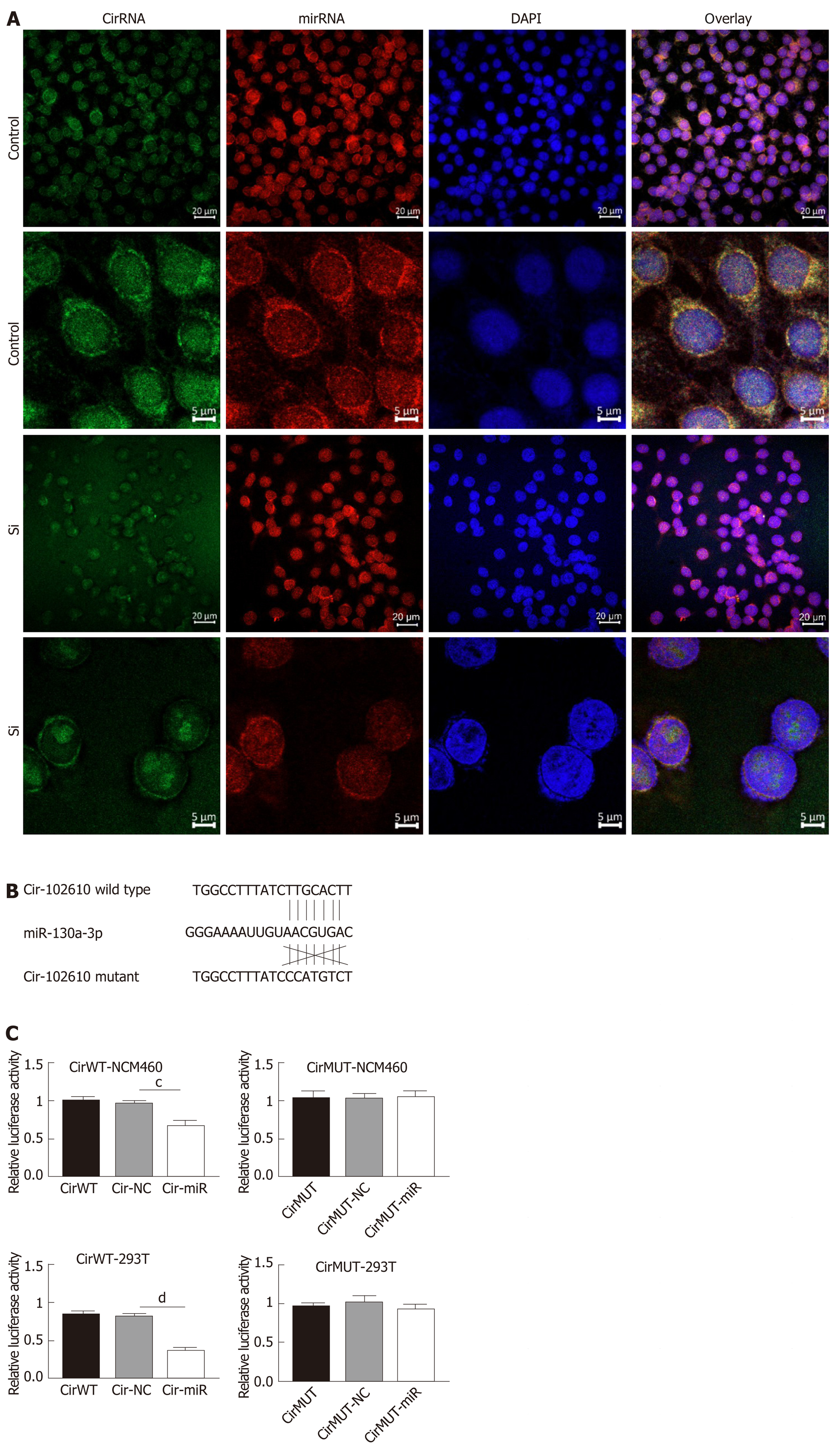
The above results suggest that hsa_circRNA_102610 and hsa-miR-130a-3p exhibit opposing functions. Hence, we hypothesized that hsa-circRNA_102610 might regulate cell proliferation and EMT by sponging hsa-miR-130a-3p. To verify this hypothesis, FISH and luciferase reporter assays were performed. The results (Figure 6A) suggested that hsa_circRNA_102610 (in green) and hsa-miR-130a-3p (in red) colocalized in both the cytoplasm and nucleus in intestinal epithelial cells. There were stronger signals near the nuclear membrane and cellular tentacles. Downregulation of hsa_circRNA_102610 attenuated these specific colocalizations in the cytoplasm (Figure 6A).
The results of the luciferase reporter assay (Figure 6B and C) showed that the cells cotransfected with hsa-miR-130a-3p mimics and psiCHECK-2 plasmids containing the wild-type binding sites of hsa-miR-130a-3p in hsa_circRNA_102610 exhibited lower relative luciferase activity than the cells transfected with the miR-control construct. Cells cotransfected with hsa-miR-130a-3p mimics and psiCHECK-2 containing mutated binding sites displayed no significant change in luciferase activity. These trends were consistent in 293T and NCM460 cells. These results indicate that hsa_circRNA_102610 directly binds to hsa-miR-130a-3p.
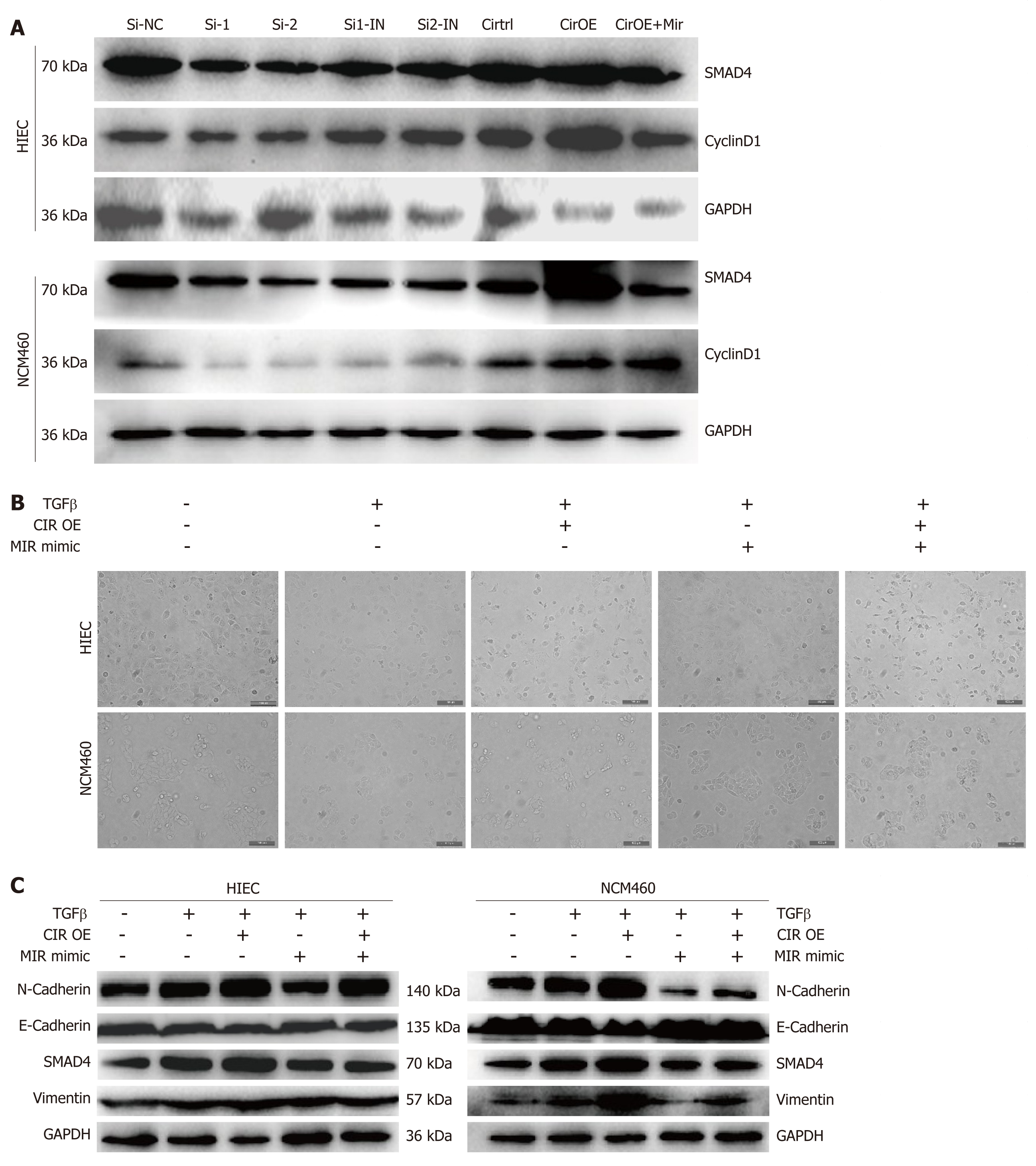
HIEC and NCM460 cell cycle progression was promoted by overexpression of hsa_circRNA_102610, and bioinformatics analysis indicated that hsa_circRNA_102610 might participate in EMT through hsa-miR-130a-3p and SMAD4 regulation. Therefore, we detected SMAD4 and CyclinD1 protein expression following hsa_circRNA_102610 upregulation or downregulation. The results in Figure 7A show that hsa_circRNA_102610 overexpression was accompanied by elevated expression of SMAD4 and CyclinD1. Reduced expression of SMAD4 and CyclinD1 was observed when hsa_circRNA_102610 was downregulated.
Previous studies have demonstrated that hsa-miR-130a-3p could inhibit TGF-β1-induced EMT by targeting SMAD4[13,20]. To explore whether hsa_circRNA_102610 participates in EMT regulation, cell-cell junctions were observed by phase contrast microscopy. Moreover, the expression levels of SMAD4 and EMT-related proteins (E-cadherin, N-cadherin and Vimentin) were detected by western blotting.
The results (Figure 7B) showed a loss of cell-cell junctions in response to TGF-β1 stimulation, while hsa-miR-130a-3p protected cells from morphological changes. Overexpression of hsa_circRNA_102610 promoted the TGF-β1-induced loss of cell-cell junctions, and hsa-miR-130a-3p mimics reversed the effects of hsa_circRNA_102610.
Regarding EMT related proteins (Figure 7C), TGF-β1 stimulation increased the expression of SMAD4, Vimentin and N-cadherin, but decreased the expression of E-cadherin expression. Overexpression of hsa_circRNA_102610 promoted this effect, while hsa-miR-130a-3p mimics transfection reversed it to some extent. Therefore, our studies indicate that hsa_circRNA_102610 promotes TGF-β1-induced EMT in intestinal epithelial cells by targeting hsa-miR-130a-3p.
IBD is a chronic intestinal inflammatory disorder with rising incidence that includes CD and ulcerative colitis[21,22]. Currently, the etiology of IBD is still not clear. The sensitivity and specificity of biomarkers for IBD diagnosis are limited[23,24]. CircRNAs have recently been discovered as potential biomarkers for multiple diseases, such as colorectal cancer, hepatic carcinoma and rheumatoid arthritis[2-5].
Our previous studies have suggested that hsa_circRNA_102610 was valuable for the diagnosis of CD exhibiting an AUC of 0.78, a sensitivity of 60.53% and a specificity of 78.85% in ROC curve analysis[7]. In this study, our further analysis showed that hsa_circRNA_102610 levels were positively correlated with CALP levels. CALP is an increasingly accepted sensitive biomarker for the assessment of CD[25,26]. Thus, although the positive correlation between hsa_circRNA_102610 and CALP in CD patients is weak (r = 0.359, P = 0.007), this finding indicates a possible role of hsa_circRNA_102610 in CD. However, the exact mechanism of hsa_circRNA_102610 in the development of CD is still unclear.
Given this result, we next overexpressed or knocked down hsa_circRNA_102610 in intestinal epithelial cells. The CCK8 and EdU staining results demonstrated that cell growth was promoted by hsa_circRNA_102610 overexpression. Cell cycle progression was promoted by hsa_circRNA_102610, as indicated by increased proportions of S and G2 phase cells, while knockdown of hsa_circRNA_102610 had the opposite effect. Excessive cell growth is strongly associated with inflammation[27]. We infer that cell growth is probably induced by overexpression of hsa_circRNA_102610 in CD patients and we hypothesize that chronic intestinal inflammation may be related to upregulation of hsa_circRNA_102610. However, the above hypothesis should be confirmed in vivo in animal models. This will be the focus of our future studies.
It has been widely reported that circRNAs act as sponges of miRNAs[4]. The MRE analysis in this study suggested that interactions occur between hsa_circRNA_102610 and 5 miRNAs (Figure 3C). Among them, an interaction with hsa-miR-130a-3p was predicted to be the most likely one. It has been reported that hsa-miR-130a-3p participates in EMT inhibition and cell proliferation in various cell types in a SMAD4-dependent manner[12-14]. Thus, hsa_circRNA_102610 is likely involved in regulating the downstream pathways of hsa-miR-130a-3p. The RT-qPCR analysis results obtained with clinical samples showed that downregulation of hsa-miR-130a-3p was inversely correlated with upregulation of hsa_circRNA_102610 in CD. In vitro studies demonstrated that hsa-miR-130a-3p mimics could reverse the growth promoting effect of hsa_circRNA_102610 overexpression on intestinal epithelial cells. Conversely, knockdown of hsa_circRNA_102610 blocked cell growth, and a hsa-miR-130a-3p inhibitor reversed these effects. In addition, FISH and luciferase reporter assays indicated a direct interaction of hsa_circRNA_102610 with hsa-miR-130a-3p. Thus, the research results in this study suggest that hsa_circRNA_102610 play roles in the regulation of intestinal epithelial cell proliferation by sponging hsa-miR-130a-3p.
Intestinal strictures and fistulas are common in CD. The mechanism of intestinal fibrosis is complex, and includes epigenetic and genetic modulations, microbiotic regulation, and extracellular matrix-related processes, among other factors[28]. Among these factors, EMT is an important contributor to the production of new mesenchymal cells from epithelial tissue[17,29]. Research by Shameer and colleagues demonstrated that epithelial expression of the EMT marker Vimentin was significantly elevated in a strictured CD group[19]. It is becoming apparent that EMT participates in CD-associated intestinal fibrosis and fistulas[17,30]. In addition, it is universally accepted that inflammation can induce cell proliferation[31]. In our study, the results of CCK-8 assays, EdU staining, and cell cycle detection by flow cytometry suggested that hsa_circRNA_102610 overexpression could promote HIEC and NCM460 proliferation and upregulate cyclinD1 expression, as verified by western blotting. The expression of the EMT related protein E-cadherin was reduced in response to hsa_circRNA_102610 overexpression, while the expression of SMAD4, Vimentin, N-cadherin and the cell cycle related protein CyclinD1 was upregulated. These results further suggested that hsa_circRNA_102610 regulates specific signaling pathways downstream of hsa-miR-130a-3p in a SMAD4-dependent manner.
In conclusion, we studied, for the first time, the mechanism by which hsa_circRNA_102610 participates in the regulation of intestinal epithelial cell EMT and proliferation, based on our previous discovery that hsa_circRNA_102610 is upregulated in CD patients[7]. The results of this study imply that hsa_ circRNA_102610 can promote the growth and TGF-β1-induced EMT of intestinal epithelial cells by sponging hsa-miR-130a-3p (Figure 8)[32,33]. It has been reported that synthetic circRNAs can act as sponges to competitively inhibit miRNAs of interest. Accordingly, hsa_circRNA_102610 may serve as a potential target for CD therapy and novel drug research. Externally delivered hsa-miR-130a-3p could possibly act as a sponge of hsa_circRNA_102610.
However, there were limitations in our study. First, this research was conducted only on CD patients and intestinal epithelial cells. In vivo mechanistic studies should be carried out on animal models to further confirm the role of hsa_circRNA_102610 in CD development. Second, how hsa_circRNA_102610-mediated regulation of intestinal epithelial cell EMT and proliferation plays a role in CD pathological processes, such as the formation of intestinal strictures, requires extensive investigation.
CircRNAs are considered valuable diagnostic biomarkers for Crohn's disease (CD). Our previous study demonstrated that hsa_circRNA_102610 was upregulated in CD patients. Furthermore, miRNA response element (MRE) analysis suggested the existence of a potential interaction between hsa_circRNA_102610 and hsa-miR-130a-3p. Current evidence supports the views that epithelial-mesenchymal transition (EMT) plays an important role in CD pathogenesis, and that hsa-miR-130a-3p can inhibit transforming growth factor-β1 (TGF-β1)-induced EMT.
Further investigation is required to explore the mechanism of hsa_circRNA_102610 in the pathogenesis of CD.
This study was designed to investigate whether the upregulation of hsa_circRNA_102610 correlates with the degree of inflammation in Crohn's disease. The CALP level, C-reactive protein (CRP) level and erythrocyte sedimentation rate (ESR) were included in the correlation analysis. Furthermore, the roles that hsa_circRNA_102610 may play in the proliferation and EMT of intestinal epithelial cells were studied in normal-derived colon mucosa cell line 460 (NCM460) and human intestinal epithelial cells (HIECs).
The relative expression levels of hsa_circRNA_102610 and hsa-miR-130a-3p in CD patients were detected by quantitative reverse transcription-polymerase chain reaction. The proliferation of HIECs and NCM460 cells was detected by cell counting kit-8, EdU staining and cell cycle assays following overexpression or downregulation of hsa_circRNA_102610. Cell proliferation assays were performed as described above in a rescue experiment with hsa-miR-130a-3p mimics. The interaction of hsa_circRNA_102610 and hsa-miR-130a-3p was verified by fluorescence in situ hybridization and dual luciferase reporter assays. The relative expression levels of CyclinD1, SMAD4, E-cadherin, N-cadherin and Vimentin were detected by western blotting following hsa_circRNA_102610 overexpression, TGF-β1-induced EMT or hsa-miR-130a-3p mimic transfection (in rescue experiments).
Upregulation of hsa_circRNA_102610 was determined to be positively correlated with elevated fecal calprotectin levels in CD (r = 0.359, P = 0.007) by Pearson correlation analysis. Hsa_circRNA_102610 promoted the proliferation of HIECs and NCM460 cells, while hsa-miR-130a-3p reversed the cell proliferation-promoting effects of hsa-circRNA_102610. Fluorescence in situ hybridization and dual luciferase reporter assays showed that hsa_circRNA_102610 directly bound hsa-miR-130a-3p in NCM460 and 293T cells. An inverse correlation between downregulation of hsa-miR-130a-3p and upregulation of hsa_circRNA_102610 in CD patients was observed (r = -0.290, P = 0.024) by Pearson correlation analysis. Moreover, overexpression of hsa_circRNA_102610 promoted SMAD4 and CyclinD1 protein expression, as validated by western blotting. Furthermore, overexpression of hsa_circRNA_102610 promoted TGF-β1-induced EMT in HIECs and NCM460 cells via targeting of hsa-miR-130a-3p, with increased expression of Vimentin and N-cadherin and decreased expression of E-cadherin.
Hsa_circRNA_102610 upregulation in CD patients could promote the proliferation and EMT of intestinal epithelial cells via sponging of hsa-miR-130a-3p.
Hsa_circRNA_102610 may serve as a potential target for CD therapy and novel drug research. Externally delivered hsa-miR-130a-3p could possibly act as a sponge of hsa_circRNA_102610.
Mei-Fen Li and Guang-Hua Zhai contributed to the laboratory management. Colleagues in the Department of Gastroenterology and the Department of Clinical Laboratory helped greatly in sample collection. We sincerely thank them for their contribution.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Christodoulou DK, Tanabe S S-Editor: Wang J L-Editor: Webster JR E-Editor: Zhang YL
| 1. | Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3273] [Cited by in RCA: 3138] [Article Influence: 523.0] [Reference Citation Analysis (0)] |
| 2. | Chen S, Zhang L, Su Y, Zhang X. Screening potential biomarkers for colorectal cancer based on circular RNA chips. Oncol Rep. 2018;39:2499-2512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 3. | Zhu M, Xu Y, Chen Y, Yan F. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed Pharmacother. 2017;88:138-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 142] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 4. | Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, Li N, Zhou W, Yu Y, Cao X. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151-1164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 756] [Cited by in RCA: 916] [Article Influence: 114.5] [Reference Citation Analysis (0)] |
| 5. | Zheng F, Yu X, Huang J, Dai Y. Circular RNA expression profiles of peripheral blood mononuclear cells in rheumatoid arthritis patients, based on microarray chip technology. Mol Med Rep. 2017;16:8029-8036. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 98] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 6. | Ye YL, Yin J, Hu T, Zhang LP, Wu LY, Pang Z. Increased circulating circular RNA_103516 is a novel biomarker for inflammatory bowel disease in adult patients. World J Gastroenterol. 2019;25:6273-6288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 28] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 7. | Yin J, Hu T, Xu L, Li P, Li M, Ye Y, Pang Z. Circular RNA expression profile in peripheral blood mononuclear cells from Crohn disease patients. Medicine (Baltimore). 2019;98:e16072. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 8. | Qiao YQ, Cai CW, Shen J, Zheng Q, Ran ZH. Circular RNA expression alterations in colon tissues of Crohn's disease patients. Mol Med Rep. 2019;19:4500-4506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 9. | Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1116] [Cited by in RCA: 1589] [Article Influence: 176.6] [Reference Citation Analysis (0)] |
| 10. | Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell. 2018;71:428-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 988] [Cited by in RCA: 1481] [Article Influence: 211.6] [Reference Citation Analysis (0)] |
| 11. | Wang S, Han H, Hu Y, Yang W, Lv Y, Wang L, Zhang L, Ji J. MicroRNA-130a-3p suppresses cell migration and invasion by inhibition of TBL1XR1-mediated EMT in human gastric carcinoma. Mol Carcinog. 2018;57:383-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 12. | Wang Y, Du J, Niu X, Fu N, Wang R, Zhang Y, Zhao S, Sun D, Nan Y. MiR-130a-3p attenuates activation and induces apoptosis of hepatic stellate cells in nonalcoholic fibrosing steatohepatitis by directly targeting TGFBR1 and TGFBR2. Cell Death Dis. 2017;8:e2792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 13. | Tian X, Fei Q, Du M, Zhu H, Ye J, Qian L, Lu Z, Zhang W, Wang Y, Peng F, Chen J, Liu B, Li Q, He X, Yin L. miR-130a-3p regulated TGF-β1-induced epithelial-mesenchymal transition depends on SMAD4 in EC-1 cells. Cancer Med. 2019;8:1197-1208. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 14. | Liu Y, Li Y, Wang R, Qin S, Liu J, Su F, Yang Y, Zhao F, Wang Z, Wu Q. MiR-130a-3p regulates cell migration and invasion via inhibition of Smad4 in gemcitabine resistant hepatoma cells. J Exp Clin Cancer Res. 2016;35:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 72] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 15. | Chen X, Yue B, Zhang C, Qi M, Qiu J, Wang Y, Chen J. MiR-130a-3p inhibits the viability, proliferation, invasion, and cell cycle, and promotes apoptosis of nasopharyngeal carcinoma cells by suppressing BACH2 expression. Biosci Rep. 2017;37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 16. | Qu R, Sun Y, Li Y, Hu C, Shi G, Tang Y, Guo D. MicroRNA-130a-3p suppresses cell viability, proliferation and invasion in nasopharyngeal carcinoma by inhibiting CXCL12. Am J Transl Res. 2017;9:3586-3598. [PubMed] |
| 17. | Jiang H, Shen J, Ran Z. Epithelial-mesenchymal transition in Crohn's disease. Mucosal Immunol. 2018;11:294-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 18. | Chen Y, Ge W, Xu L, Qu C, Zhu M, Zhang W, Xiao Y. miR-200b is involved in intestinal fibrosis of Crohn's disease. Int J Mol Med. 2012;29:601-606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 75] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 19. | Mehta SJ, Lewis A, Nijhuis A, Jeffery R, Biancheri P, Di Sabatino A, Feakins R, Silver A, Lindsay JO. Epithelial down-regulation of the miR-200 family in fibrostenosing Crohn's disease is associated with features of epithelial to mesenchymal transition. J Cell Mol Med. 2018;22:5617-5628. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 20. | Chen Y, Xiao Y, Ge W, Zhou K, Wen J, Yan W, Wang Y, Wang B, Qu C, Wu J, Xu L, Cai W. miR-200b inhibits TGF-β1-induced epithelial-mesenchymal transition and promotes growth of intestinal epithelial cells. Cell Death Dis. 2013;4:e541. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 108] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 21. | Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, Sung JJY, Kaplan GG. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2018;390:2769-2778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2677] [Cited by in RCA: 4091] [Article Influence: 511.4] [Reference Citation Analysis (110)] |
| 22. | Li Y, Chen B, Gao X, Hu N, Huang M, Ran Z, Liu Z, Zhong J, Zou D, Wu X, Ren J, Sheng J, Zheng P, Wang H, Chen M, Chen J, Xi P, Lu J, Handel M, Liu Y, Fan H, Qian J. Current diagnosis and management of Crohn's disease in China: results from a multicenter prospective disease registry. BMC Gastroenterol. 2019;19:145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 23. | Dulai PS, Peyrin-Biroulet L, Danese S, Sands BE, Dignass A, Turner D, Mantzaris G, Schölmerich J, Mary JY, Reinisch W, Sandborn WJ. Approaches to Integrating Biomarkers Into Clinical Trials and Care Pathways as Targets for the Treatment of Inflammatory Bowel Diseases. Gastroenterology. 2019;157:1032-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 24. | Sommer F, Rühlemann MC, Bang C, Höppner M, Rehman A, Kaleta C, Schmitt-Kopplin P, Dempfle A, Weidinger S, Ellinghaus E, Krauss-Etschmann S, Schmidt-Arras D, Aden K, Schulte D, Ellinghaus D, Schreiber S, Tholey A, Rupp J, Laudes M, Baines JF, Rosenstiel P, Franke A. Microbiomarkers in inflammatory bowel diseases: caveats come with caviar. Gut. 2017;66:1734-1738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (1)] |
| 25. | Kennedy NA, Jones GR, Plevris N, Patenden R, Arnott ID, Lees CW. Association Between Level of Fecal Calprotectin and Progression of Crohn's Disease. Clin Gastroenterol Hepatol. 2019;17:2269-2276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 26. | Mowat C, Digby J, Strachan JA, Wilson R, Carey FA, Fraser CG, Steele RJ. Faecal haemoglobin and faecal calprotectin as indicators of bowel disease in patients presenting to primary care with bowel symptoms. Gut. 2016;65:1463-1469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 135] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 27. | Schwiebs A, Herrero San Juan M, Schmidt KG, Wiercinska E, Anlauf M, Ottenlinger F, Thomas D, Elwakeel E, Weigert A, Farin HF, Bonig H, Scholich K, Geisslinger G, Pfeilschifter JM, Radeke HH. Cancer-induced inflammation and inflammation-induced cancer in colon: a role for S1P lyase. Oncogene. 2019;38:4788-4803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 28. | Rieder F, Fiocchi C. Intestinal fibrosis in IBD--a dynamic, multifactorial process. Nat Rev Gastroenterol Hepatol. 2009;6:228-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 272] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 29. | Flier SN, Tanjore H, Kokkotou EG, Sugimoto H, Zeisberg M, Kalluri R. Identification of epithelial to mesenchymal transition as a novel source of fibroblasts in intestinal fibrosis. J Biol Chem. 2010;285:20202-20212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 235] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 30. | Lovisa S, Genovese G, Danese S. Role of Epithelial-to-Mesenchymal Transition in Inflammatory Bowel Disease. J Crohns Colitis. 2019;13:659-668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 92] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 31. | Kiraly O, Gong G, Olipitz W, Muthupalani S, Engelward BP. Inflammation-induced cell proliferation potentiates DNA damage-induced mutations in vivo. PLoS Genet. 2015;11:e1004901. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 121] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 32. | Liu X, Abraham JM, Cheng Y, Wang Z, Wang Z, Zhang G, Ashktorab H, Smoot DT, Cole RN, Boronina TN, DeVine LR, Talbot CC, Liu Z, Meltzer SJ. Synthetic Circular RNA Functions as a miR-21 Sponge to Suppress Gastric Carcinoma Cell Proliferation. Mol Ther Nucleic Acids. 2018;13:312-321. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 164] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 33. | Wang Z, Ma K, Cheng Y, Abraham JM, Liu X, Ke X, Wang Z, Meltzer SJ. Synthetic circular multi-miR sponge simultaneously inhibits miR-21 and miR-93 in esophageal carcinoma. Lab Invest. 2019;99:1442-1453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |